Abstract
Family B2 or adhesion G protein–coupled receptors (AGPCRs) are distinguished by variable extracellular regions that contain a modular protease, termed the GPCR autoproteolysis-inducing domain that self-cleaves the receptor into an N-terminal fragment (NTF) and a C-terminal fragment (CTF), or seven transmembrane domain (7TM). The NTF and CTF remain bound after cleavage through noncovalent interactions. NTF binding to a ligand(s) presented by nearby cells, or the extracellular matrix anchors the NTF, such that cell movement generates force to induce NTF/CTF dissociation and expose the AGPCR tethered peptide agonist. The released tethered agonist (TA) binds rapidly to the 7TM orthosteric site to activate signaling. The orphan AGPCR, GPR114 was reported to be uncleaved, yet paradoxically capable of activation by its TA. GPR114 has an identical cleavage site and TA to efficiently cleave GPR56. Here, we used immunoblotting and biochemical assays to demonstrate that GPR114 is a cleaved receptor, and the self-cleavage is required for GPR114 TA-activation of Gs and no other classes of G proteins. Mutagenesis studies defined features of the GPR114 and GPR56 GAINA subdomains that influenced self-cleavage efficiency. Thrombin treatment of protease-activated receptor 1 leader/AGPCR fusion proteins demonstrated that acute decryption of the GPR114/56 TAs activated signaling. GPR114 was found to be expressed in an eosinophilic-like cancer cell line (EoL-1 cells) and endogenous GPR114 was efficiently self-cleaved. Application of GPR114 TA peptidomimetics to EoL-1 cells stimulated cAMP production. Our findings may aid future delineation of GPR114 function in eosinophil cAMP signaling related to migration, chemotaxis, or degranulation.
Keywords: adhesion G protein–coupled receptor (AGPCR), tethered peptide agonist, ADGRG5, GAIN domain, autoproteolysis, receptor activation, cyclic AMP (cAMP), orthosteric agonist
G protein–coupled receptors (GPCRs) are the largest class of membrane receptors. They are investigated heavily as pharmaceutical targets and over 700 approved drugs act on them (1). Adhesion GPCRs (AGPCRs) or class B2 GPCRs are the second largest GPCR subfamily and are involved in a variety of physiological processes and diseases. Most of the 33 human AGPCRs are orphan receptors that are distinguished for containing of a variety of extracellular adhesive domains and a GPCR autoproteolysis-inducing (GAIN) domain (2, 3). The GAIN domain is found at the C-terminal end of the extracellular regions. The GAIN domain has two subdomains, a variable length α-helical-rich GAINA and the conserved β-sheet GAINB. AGPCRs self-cleave at the GPCR proteolytic site (GPS) located between β-strand 12 and β-strand 13 of GAINB to generate a two-fragment receptor consisting of the N-terminal fragment (NTF) and the C-terminal fragment (CTF) or the 7TM domain (4). The NTF and CTF remain noncovalently bound at the plasma membrane preceding receptor activation. The GPS of most AGPCRs including those of GPR114 and GPR56 consist of the conserved residues HL/T such that the T is the first residue of β-strand 13 of GAINB and becomes the first residue of the CTF after constitutive self-cleavage (4, 5, 6). In the holoreceptor form, β-strand 13 is highly hydrophobic and embedded within the hydrophobic core of the GAINB subdomain (4).
The current hypothesis of adhesion GPCR activation by orthosteric agonism begins with ligand binding to the NTF adhesive modules to affix the NTF (2). Cellular movement opposed to the anchored NTF creates shear force sufficient to dissociate the NTF from the CTF. This releases β-strand 13 from the core of the GAINB subdomain, after which it undergoes a rapid transition into a tethered agonist (TA) and occupation of the 7TM orthosteric site (7, 8). This stabilizes an active conformation of the 7TM to promote G protein signaling. This model was recently bolstered by the solution of seven active-state AGPCR structures (9, 10, 11, 12). The structures were derived from CTF-only constructs and revealed a common hook-like, partial α-helical conformation that the TAs had adopted when they occupied the orthosteric sites of the G protein-complexed 7TM domains (3). Low resolution structures of two AGPCR holoreceptors were solved and although the precise position of the β-strand-13 within the GAIN domains could not be assigned, the GAIN domains exhibited a range of positions that were ≥3 nm away from the entrance to the 7TM orthosteric site (9). Therefore, the TA must undergo a great conformational and positional change when dissociated from the NTF to gain access to the orthosteric site.
Despite the clarity added to the activation mechanism from these new structures, alternative models of AGPCR tethered agonism persist. The TA was hypothesized to transition to the orthosteric binding pocket while being encrypted within the GAIN domain (10). Similarly, AGPCR TAs were proposed to gain access to the orthosteric site after partial, transient exposure from flexible GAIN domains (13). Alternatively, the TA was hypothesized to be prebound in the orthosteric site and activated by isomerization when the receptor is stimulated by a ligand (10). Some of the support for these models was derived from the study that indicated that GPR114 was not a cleaved AGPCR, even though the overexpressed receptor could raise cAMP levels when treated with synthetic peptidomimetics of its TA, a finding that is seemingly at odds with the theory that a preisomerized tethered peptide agonist occupies the orthosteric site (14). Based on the prospect that GPR114 was not cleaved, it was not reconciled why a CTF-only GPR114 construct was chosen for the newer structural work (10).
Here, we investigated the paradox of GPR114 tethered agonism regarding its purported noncleavage. Our results showed that GPR114 is cleaved in endogenous- and over-expression contexts while defining elements of the GAIN domain that affect cleavage efficiency. Cleavage was requisite to TA-stimulated GPR114 signaling that occurs exclusively through Gs. A GPR114 fusion protein engineered to reveal its TA upon exogenous protease cleavage acutely activated signaling after fusion protein proteolysis. Cell lines were profiled to identify those that express endogenous GPR114 and produce the protein. An eosinophilic-like cancer cell line (EoL-1 cells) produced efficiently cleaved GPR114 and activated cAMP production in response to synthetic GPR114 TA peptidomimetics (15).
Results
GPR114 is a cleaved adhesion GPCR with a dissociable NTF
The majority of adhesion GPCRs contains a functional extracellular self-cleaving protease termed the GAIN domain (4). The AGPCR GPS consensus cleavage site is His, Leu/Thr. GPR114/ADGRG5 and GPR56/ADGRG1 contain consensus GPSs, have identical seven-residue TAs, and exhibit the ability to be activated by synthetic TA peptidomimetics based on the GPR114 TA and stalk sequence (16). However, an overexpression study concluded that GPR114/ADGRG5 was a noncleaved AGPCR (14). This prompted confounding models to explain the simultaneous presence of the GPR114 TA within the GAIN domain and its observed occupation in the orthosteric site of a CTF-only active-state GPR114 7TM receptor (10).
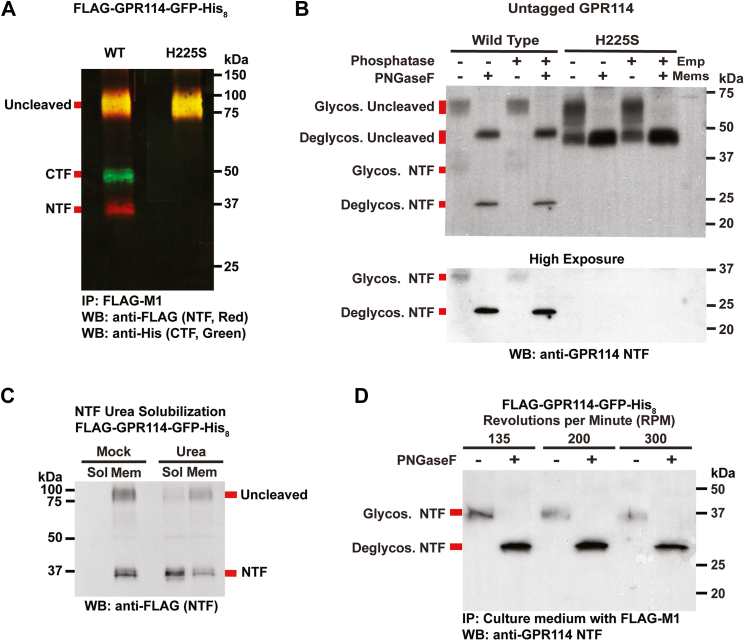
We investigated whether GPR114 has autoproteolytic activity by examining dually tagged WT and GPS-cleavage deficient (H225S) mutant human receptors that had a FLAG tag at the N-termini and a GFP-His8 module at the C termini. Detergent extracts from Sf9 cells expressing the receptors were subjected to anti-FLAG immunoprecipitation followed by dual color Western blotting with FLAG- and His-tag antibodies (Fig. 1A). WT GPR114 was partially cleaved as observed as a single color (red) NTF band at ∼36 kDa and single color (green) CTF band observed at ∼48 kDa. The predicted molecular weight of the GPR114 CTF is 33.3 kDa; however, AGPCR CTFs are known to run aberrantly fast by SDS-PAGE to ∼22 to 25 kDa. The added mass of the GFP-His8 module accounts for the observed ∼48 kDa CTF band. Cleavage of overexpressed GPR114 was partially efficient as an uncleaved band of ∼90 kDa was observed (yellow, merged). Cleavage-deficient GPR114-H225S only had the ∼90 kDa protein, supporting our finding that WT GPR114 is cleaved.
Figure 1.
GPR114 is a self-cleaved adhesion GPCR.A, WT and H225S FLAG-GPR114-GFP-His8 were immunoprecipitated from Sf9 cell membrane detergent extracts and immunoblotted with FLAG and Penta-His antibodies to detect the NTF and CTF, respectively. B, untagged WT and H225S GPR114 cell membranes were untreated or treated with PNGaseF and/or alkaline phosphatase and immunoblotted with a GPR114 NTF-specific antibody. C, FLAG-GPR114-GFP-His8 cell membranes were treated with buffer (Mock) or 7M urea and centrifuged to recover the membranes (Mem) and solubilized material (Sol). The fractions were immunoblotted with the Invitrogen GPR114 Polyclonal Antibodies (Cat# PA5-21701). D, FLAG-GPR114-GFP-His8Sf9 suspension cultures were shaken at different speeds (135 rpm is standard) and clarified media supernatants from the cultures were subjected to FLAG-M1 immunoprecipitation. On bead treatment ± PNGase F was performed prior to elution with SDS-sample buffer and immunoblotting with the GPR114 NTF-specific antibody. All Western blot images are representative of multiple trials. CTF, C-terminal fragment; GPCR, G protein–coupled receptor; NTF, N-terminal fragment.
We next examined the cleavage efficiency, glycosylation, and phosphorylation of untagged GPR114. WT and H225S GPR114 membranes were prepared from Sf9 cells and treated with PNGaseF and/or alkaline phosphatase prior to immunoblotting with a commercial GPR114 antibody that detects the NTF (Fig. 1B). Two broad, fuzzy bands were observed for nontreated WT GPR114 at ∼65 kDa and ∼35 kDa. When these receptor membranes were treated with PNGaseF, the bands became sharp and had collapsed to ∼45 kDa and ∼23 kDa, in line with the predicted MWs of uncleaved, mature holoreceptor and the isolated NTF. These results matched the observation of GPR114 partial cleavage obtained with the epitope-tagged receptor. Untagged GPR114 H225S displayed a smear of bands predominantly at the ∼65 kDa position, consistent as the uncleaved receptor with minor hypoglycosylated species. PNGaseF treatment collapsed these species to a sharp band of ∼45 kDa. No molecular weight shifts were observed upon alkaline phosphatase treatment suggesting that phosphorylation events do not account for any of the observed differences in molecular weights of GPR114 bands.
Epitope-tagged GPR114 membrane homogenates were next treated with 7M urea or a mock buffer to assess NTF solubilization. We previously used urea to release multiple AGPCR NTFs from the membrane as a proxy to force-induced NTF release and TA-mediated signaling (16). The soluble (Sol) and membrane (Mem) fractions were visualized by Western blotting with the FLAG M1 antibody to detect the GPR114 NTF. The majority of the uncleaved ∼90 kDa full-length receptor remained in the membrane fraction upon urea treatment, whereas the majority of the ∼36 kDa NTF was solubilized, demonstrating that the NTF was peripherally associated with the membrane (Fig. 1C). There are many documented instances of AGPCR NTF shedding from the cell surface to the extracellular space and a few accounts in which experimental shaking or vibrating cells was purported to induce NTF release (10, 14, 17, 18). Figure 1D shows that under standard Sf9 suspension cell culture conditions (135 rpm rotation), the FLAG-tagged GPR114 NTF was recovered from culture medium supernatant by anti-FLAG immunoprecipitation. PNGaseF treatment was included during immunoprecipitation to aid Western blot transfer of the deglycosylated NTF and to help authenticate the NTF band. With our culture conditions, increased Sf9 cell culture shaking did not induce additional GPR114 NTF shedding.
Comparative analysis of GPR114 and GPR56 GAIN domain cleavage efficiency
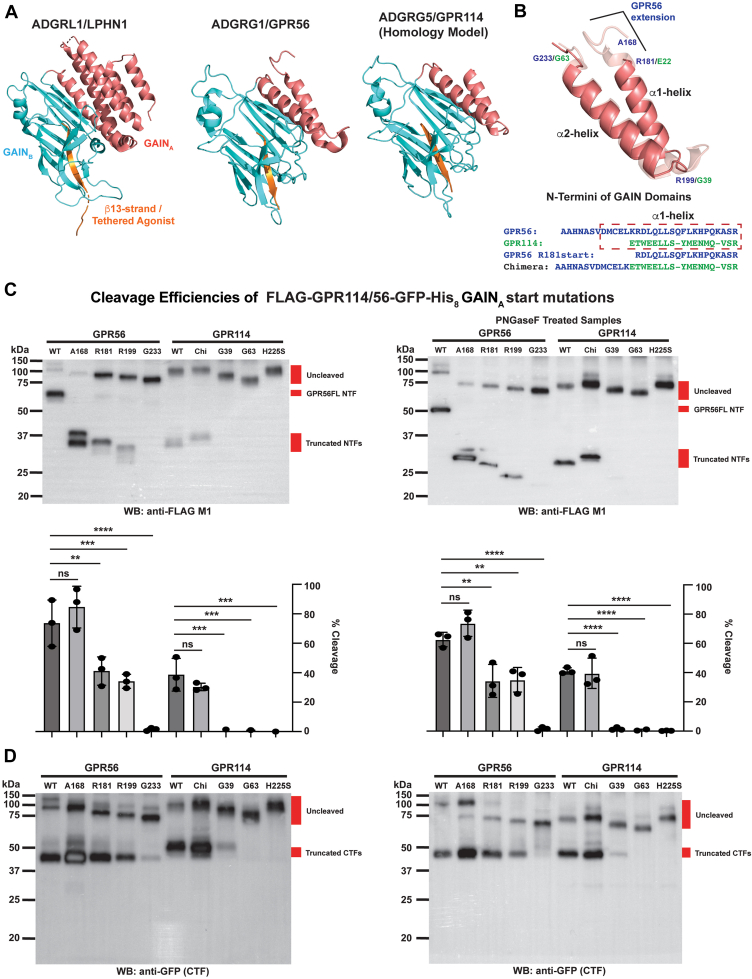
GPR56 has an N-terminal Pentraxin/Laminin/neurexin/sex-hormone-binding-globulin-Like (PLL) domain followed by a short GAINA subdomain that consists of two antiparallel α-helices followed by a typical GAINB subdomain that is comprised of 13 β-strands (Fig. 2A) (19). In comparison, the latrophilin-1 GAIN domain structure shows commonality of the GAINB subdomain and its more complex 5 α-helical GAINA subdomain (4). A structure of the GPR114 GAIN domain has not been solved, so we built a GPR114 homology model using the Iterative Threading ASSEmbly Refinement program (20). The GPR114 GAIN domain was predicted to be highly similar to GPR56 with both GAINA subdomains possessing two antiparallel α-helices. However, the GPR56 α1-helix is extended by ∼13 residues (Fig. 2B).
Figure 2.
Requirements of the GPR114 and GPR56 GAINBsubdomain and GAINAsubdomain α1 and α2 helices for self-cleavage efficiency.A, LPHN1 (PDB 4DLQ) and GPR56 (PDB 5KVM) have GAINA subdomains that consist of a five α-helix bundle or two antiparallel α-helices, respectively. A GPR114 GAIN domain homology model was built using ITASSER and predicts that the GPR114 GAINA subdomain resembles that of GPR56, but it has a shortened α1-helix. B, zoomed overlay view of the GPR114 and GPR56 GAINA subdomains with the amino acid positions denoting the start sites of truncated and chimeric receptors (blue text, GPR56 and green text, GPR114). C and D, comparison of self-cleavage efficiencies of WT, truncated, and chimeric (Chi) GPR56 and GPR114 receptors in Sf9 cell membranes by quantitative immunoblotting with C, the FLAG M1 antibody (NTF) and D, the GFP antibody (CTF). The analyses were conducted ± PNGase F due to the propensity of some glycosylated receptor species to have altered efficiencies of immunoblot electrophoretic transfer. Percent receptor cleavage was determined by pixel densitometry ratioing the relative band intensities of the free NTFs to the uncleaved receptor band intensities. Representative immunoblots are shown for experiments performed in triplicate. Error bars are the mean ± S.D. One-way ANOVAs were used for statistical analyses. ns, not significant, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001. CTF, C-terminal fragment; GAIN, GPCR autoproteolysis-inducing domain; GPCR, G protein–coupled receptor; ITASSER, Iterative Threading ASSEmbly Refinement; LPHN1, latrophilin-1; NTF, N-terminal fragment; PDB, Protein Data Bank.
We used the GPR56 GAIN structure and the GPR114 GAIN homology model to design receptor truncation mutants to examine the influence of GAINA elements toward GAINB self-cleavage efficiency. Relative quantitative immunoblotting with the FLAG antibody was performed by ratioing the pixel densitometry values of uncleaved to cleaved products as a means of estimating cleavage efficiency (Fig. 2C). Measurements were made from samples treated ± PNGaseF to account for instances of inefficient electrophoretic protein transfer to polyvinylidene fluoride that we occasionally observed for glycosylated species. The PNGaseF treatment also permitted us to assign a new glycosylation site(s) within the GPR114 CTF, but not the GPR56 CTF (Fig. 2C). Anti-GFP detection of the epitope-tagged CTFs is shown below the quantified NTF blots as a secondary means to qualitatively evaluate cleavage efficiency.
WT GPR56 (65–75% cleaved) was more efficiently cleaved than WT GPR114 (∼40% cleaved). GPR56-A168-start is truncated to ablate the PLL domain, and the resultant receptor was cleaved as efficiently as full length GPR56. The GPR56-R181-start construct is equivalent to maturated GPR114 with a shortened GAINA α1-helix. Interestingly, the cleavage efficiency of GPR56-R181-start was reduced in comparison to GPR56-A168-start to a level equivalent to GPR114 (∼40%). We created a chimeric receptor in which the GPR114 α1-helix was extended with GPR56 sequence to test whether the full α1-helix could improve GPR114 cleavage efficiency; however, no improvement was observed and we could not conclude whether this was due to our choice of residues added into the chimeric construct or indicates that there is an inherent difference in the catalytic properties of the GPR56 and GPR114 GAINB subdomains. Further truncations of the full α1 helix and the α1 and α2 helices together of the GPR56 and GPR114 GAINA subdomains indicate that the latter is likely the case. The GPR56-R199-start and GPR114-G39-start constructs remove the respective α1-helices. The GPR56 truncation retained substantial self-cleavage ability (∼37%), whereas the GPR114 truncation lost the ability to self-cleave altogether. Removal of the full GAINA subdomains in the GPR56-G223-start and GPR114-G63-start constructs fully abrogated self-cleavage of both receptors, which aligns with a previous report showing that the GAINB subdomain alone is not sufficient for cleavage (4).
GPR114 exclusively activates Gs via cleavage-dependent tethered agonism
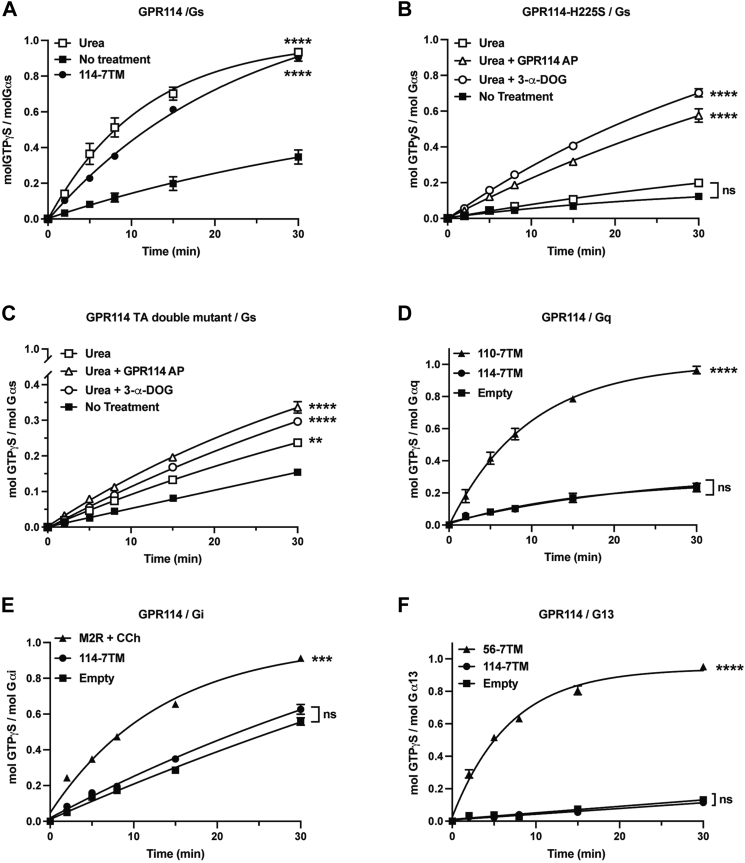
GPR114 potentiated cAMP accumulation in response to TA peptidomimetics in HEK293 cells and its active state structure was solved in complex with Gs (10, 14). We investigated the full coupling profile of GPR114 using direct G protein activation assays while evaluating the requirement of NTF dissociation for tethered agonism. GPR114 membrane homogenates were treated ± 7M urea to dissociate the NTF (Fig. 1C) and then reconstituted with purified Gs heterotrimer. The kinetics of Gs GTPγS binding was measured to assess receptor activation (Fig. 3A). The untreated receptor displayed slow Gs activation kinetics that was markedly potentiated by urea-mediated NTF dissociation (i.e. TA release from the GAIN domain). The improvement in Gs activation kinetics nearly matched the kinetics observed when a CTF-only GPR114 was used to stimulate the G protein.
Figure 3.
GPR114 exclusively couples to Gs and is activated by its tethered peptide agonist following NTF/CTF dissociation.A–C, untagged GPR114 holoreceptor and GPR114 7TM membranes were prepared from Sf9 cells and mock treated (solid symbols) or 7M urea treated (open symbols). Receptors were reconstituted with Gs and GPR114-AP or 3-α-DOG as indicated prior to measurement of [35S]-GTPγS binding kinetics. D–F, GPR114 7TM membrane stimulation of Gq, Gi, G13 GTPγS binding kinetics were compared to control GPCR membranes including GPR110 7TM, M2 muscarinic receptor plus carbachol, and GPR56 7TM respectively. Biological triplicate reactions with triplicate technical replicates are presented. Error bars are the means ± S.D. AUCs were calculated using GraphPad Prism and Tukey’s Multiple Comparison tests were used to evaluate statistical significances to the no treatment or empty (no receptor) membrane conditions; ns, not significant, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001. AUC, Area under the Curve; CTF, C-terminal fragment; GPCR, G protein–coupled receptor; NTF, N-terminal fragment.
This analysis was repeated for the cleavage-deficient GPR114-H225S receptor (Fig. 3B). Overall, Gs activation kinetics were diminished and there was no urea-dependent activation, which is consistent with the observation that urea dissociated the NTF from cleaved GPR114, but not the uncleavable mutant receptor. GPR114-H225S was found to be a functional receptor as its ability to activate Gs was potentiated by the GPR56/GPR114 partial agonist, 3-α-DOG, and the GPR114 activating peptide/TA peptidomimetic (Fig. 3B) (14, 16). We further assessed the importance of receptor fragment dissociation-dependent tethered agonism. Full-length GPR114 with a dual TA mutant in which the P6 Leu and P7 Met of the TA were mutated to alanines was tested for the ability to activate Gs in a urea-dependent manner (Fig. 3C). This TA dual mutant was first shown for other AGPCRs such as latrophilin 3 and later for GPR56 to maintain cleavage efficiency while abolishing TA-mediated receptor activation (21, 22). For GPR114, this dual TA mutant had decreased basal activity that was modestly improved by urea pretreatment. Receptor activity was further potentiated by GPR114-AP or 3-α-DOG, demonstrating that the mutant receptor retained functionality.
The G protein coupling specificity of GPR114 has not been assessed; therefore, we tested the ability of TA-activated GPR114-7TM to stimulate representative members of the other three families of G proteins (Gq, Gi, and G13) (Fig. 3, D–F). Positive receptor controls were evaluated alongside GPR114-7TM for each G protein including, GPR110-7TM (Gq), the M2 muscarinic acetylcholine receptor with carbachol (Gi), and the GPR56-7TM (G13). GPR114-7TM showed no ability to activate any of these G proteins, leading us to conclude that GPR114 is an exclusive Gs coupler.
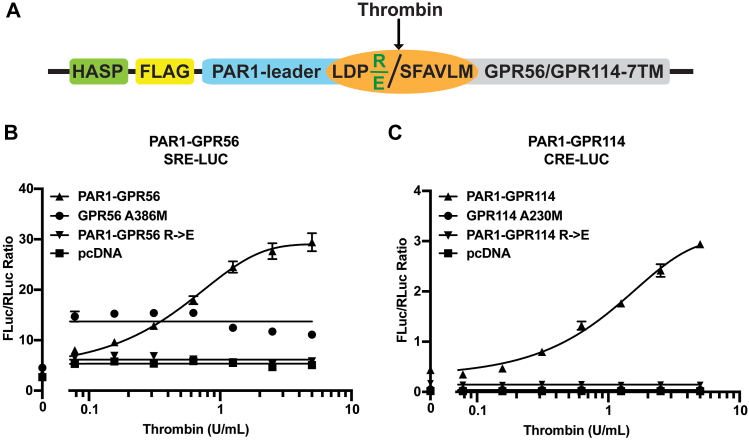
PAR1-GPR114 and PAR1-GPR56 chimeric receptors are activated by acute thrombin treatment that decrypts the TA
The previous experiments with urea-mediated dissociation of AGPCR NTFs are inferred to be a proxy of the physiological ligand- and force-based means for releasing the TA from the GAIN domain so that it may acquire access to the 7TM domain orthosteric site. To further test GPR114 activation by acute decryption of the tethered agonist, we engineered chimeric AGPCRs consisting of the protease-activated receptor (PAR)—1 leader sequence fused to the GPR56 and GPR114 CTFs (Fig. 4A). Cleavage by treatment with the exogenous protease, thrombin, will acutely release the TA permitting measurements of activated downstream signaling as shown previously for latrophilin-3 (7). The PAR1-GPR56 and PAR1-GPR114 fusions were coexpressed with the serum response element (SRE)- or cyclic AMP response element (CRE)-luciferase reporters, respectively in HEK293T cells. Serum-starved cells were treated with thrombin to measure thrombin-dependent luminescence (Fig. 4, B and C). Thrombin stimulated both WT PAR-1/AGPCR fusions, but not versions in which the thrombin cut site was mutated (R→E). GPR56-A386M and GPR114-A230M are CTF-only receptors that have compromised TAs due to removal of the first three critical TA residues (T, Y, and F) (16). GPR56A386M had a modest basal SRE-LUC luminescence signal that was not stimulated by thrombin, although all concentrations of thrombin had a slightly elevated signal, which is consistent with an endogenous PAR response signature in HEK293T cells. GPR114-A230M had negligible CRE luciferase (CRE-LUC) activity that was not stimulated by thrombin.
Figure 4.
PAR1-GPR114 and PAR1-GPR56 chimeric receptors are activated acutely by thrombin-mediated cleavage and tethered agonist exposure.A, schematic of the PAR1-GPR56 and -GPR114 chimeric receptor sequence features. HASP is the HA signal peptide, followed by the 1X FLAG tag, the PAR1-leader sequence is residues 1 to 41 of the PAR1 receptor with thrombin cleavage site fused to a modified, partially functional tethered agonist of GPR56 or GPR114 (SFAVLM versus the authentic TYFAVLM) followed by the remaining CTFs. The thrombin cleavage site is LDPR with point mutant E in the P1′ position used to obviate cleavage. B, HEK293T cell-based GPR56 SRE-luciferase or (C) GPR114 CRE-luciferase assays were used to measure thrombin-dependent reporter activation. GPR56 A386M 7TM and GPR114 A230M 7TM receptors were used as partially active controls. Data are presented as the ratios of the FLuc reporter signal to RLuc balancer signal and are biological triplicates. Error bars depict the mean ± S.D. In some cases, error bars were smaller than the plotted symbols. CRE-luciferase, cyclic AMP response element luciferase; CTF, C-terminal fragment; HASP, hemagglutinin signal peptide; PAR, protease-activated receptor; SRE, serum response element.
We attempted to measure the PAR1-AGPCR activities in orthogonal G protein reconstitution assays in receptor-expressed Sf9 membranes. The PAR1-GPR56 fusion exhibited strong thrombin-dependent activation of purified G13, but the PAR1-GPR114 fusion had no ability to activate Gs (Fig. S1, A and B). Immunoblotting of the Sf9 receptor membranes showed proper processing of the PAR-1/GPR56 fusion, whereas the PAR-1/GPR114 fusion appeared to be improperly processed and was thus not functionally produced in insect cells (Fig. S1C).
Endogenous GPR114 is properly cleaved in an eosinophilic-like cell line and can be activated by a TA peptidomimetic
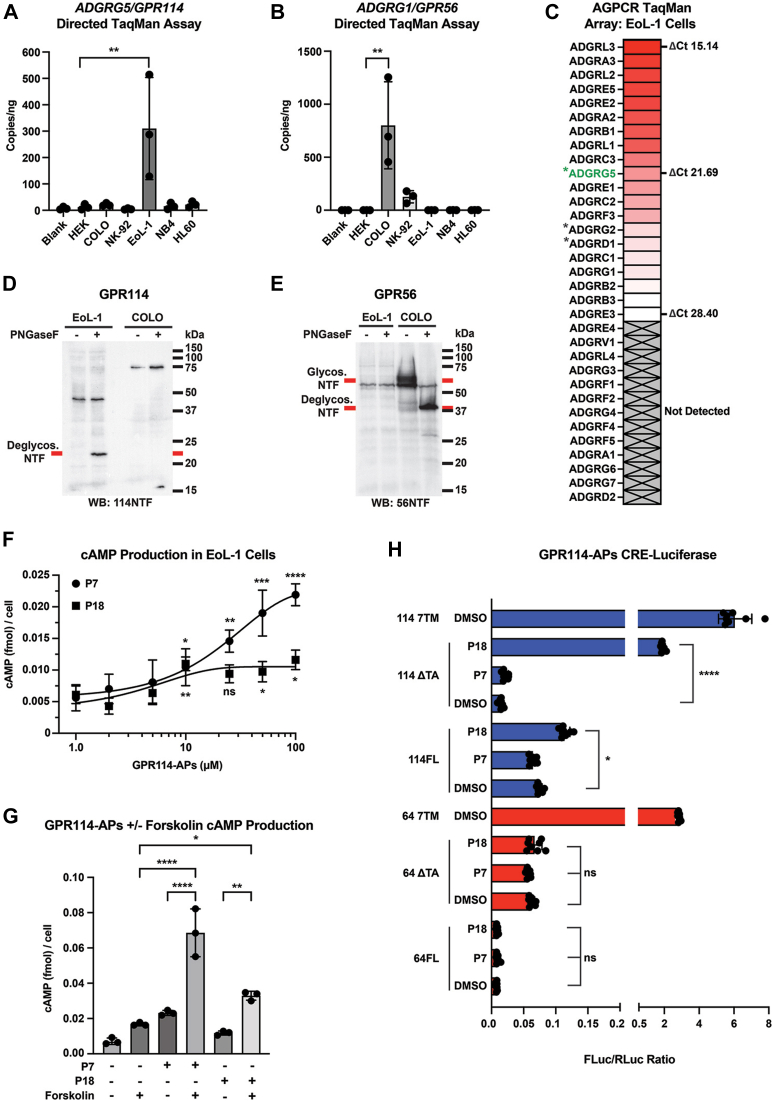
GPR114 expression data from tissues and cells have been enigmatic, with the Protein Atlas database and literature reporting GPR114 expression in eosinophils, testes, spleen, and the lungs (23, 24). Using these clues of potential endogenous GPR114 expression, we sought human cell line(s) that express GPR114 with the intention to use them to evaluate endogenous GPR114 self-cleavage and signaling. Total RNA was harvested from human embryonic kidney 293T (HEK293T), colorectal carcinoma 205 (COLO205), natural killer-92 (NK-92), human acute promyelocytic NB4, human eosinophilic leukemia (EoL-1), and human promyelocytic leukemia (HL-60) cell lines, converted to complementary DNA (cDNA) and introduced into directed TaqMan quantitative PCR assays to measure GPR114 and GPR56 expression. Standard curves with known amounts of cDNA plasmid standards were developed for both assays to permit absolute quantification (Fig. S2). GPR114 was expressed in the human eosinophil-like leukemia cell line EoL-1, while GPR56 was expressed in the human colon carcinoma cell line, COLO205 (Fig. 5, A and B). These data are consistent with the reports of GPR114 eosinophil expression and the emerging understanding that GPR56 may have a role colorectal cancer progression (24, 25, 26, 27). We used the cDNA generated from the EoL-1 cell line to probe expression of nonolfactory GPCRs using a human TaqMan GPCR array card (Table S2). Multiple adhesion GPCRs were expressed with appreciable GPR114/ADGRG5 expression that was the highest among the ADGRG subfamily in which low levels of GPR56/ADGRG1 and GPR64/ADGRG2 were detected (Fig. 5C).
Figure 5.
GPR114 is expressed, self-cleaved, and stimulates cAMP production in EoL-1 cells. (A) GPR114 and (B) GPR56 TaqMan gene expression assays were performed to measure cDNA copy numbers from HEK293T, COLO205, NK-92, EoL-1, NB4, and HL60 cell lines. TaqMan assays were calibrated for absolute quantification in Fig. S2. C, TaqMan GPCR Array measurement of relative AGPCR expression levels in EoL-1 cells. ∗Denotes known Gs-coupled AGPCRs. Immunoblotting of (D) GPR114 and (E) GPR56 in EoL-1 and COLO205 cell membrane samples that were treated ± PNGaseF to aid visualization of the respective NTF bands. F, GPR114-AP peptidomimetics P7 and P18 stimulated cAMP production EoL-1 cells. G, forskolin-potentiation of GPR114-AP P7 and P18 stimulated cAMP production in EoL-1 cells. H, GPR114-AP peptidomimetics P7 and P18 stimulation of CRE-Luc activity in HEK293T cells overexpressing full length (FL) or compromised tethered agonist (ΔTA) 7TM versions of GPR114 or GPR64. WT 7TM receptors have intact tethered agonists. Error bars are the mean ± S.D. One-way ANOVAs were used for statistical analyses. ns, not significant, ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001. AGPCR, adhesion G protein–coupled receptors; CRE-Luc, cyclic AMP response element luciferase; cDNA, complementary DNA; GPCR, G protein–coupled receptor.
The relative protein levels of GPR114 and GPR56 were measured in membranes prepared from the EoL-1 and COLO205 cells. EoL-1 and COLO205 membranes were treated ± PNGaseF and immunoblotted with NTF-specific antibodies for GPR56 and GPR114 to visualize the respective NTFs and potential uncleaved receptor species (Fig. 5, D and E). Glycosylated GPR114 species could not be detected, possibly due to the difficulty in electrophoretic transfer (Fig. 1B), but a clear 23 kDa band emerged when the EoL-1 membranes were treated with PNGaseF. This endogenous GPR114 band coincides with the 23 kDa deglycosylated NTF band shown in Figure 1B for overexpressed, untagged GPR114. Authenticity of the 23 kDa GPR114 NTF band was verified using two different GPR114 NTF-specific commercial antibodies (Fig. S3). Interestingly, no uncleaved GPR114 could be detected in the EoL-1 cells even upon PNGaseF treatment. This finding of efficient self-cleavage of endogenous GPR114 parallels our finding that endogenous, platelet GPR56 was more efficiently cleaved than it was in overexpression systems (28). Figure 5E shows the clear presence of GPR56, but not GPR114 in membranes prepared from COLO205 cells. A broad ∼62 kDa GPR56 NTF band was found in the COLO205 membrane sample that collapsed to ∼41 kDa upon PNGaseF treatment. No detectable uncleaved, endogenous GPR56 was observed in the COLO205 membranes.
Next, we explored GPR114 signaling in EoL-1 cells using a calibrated ELISA-based cAMP production assay (Fig. S4). EoL-1 cells were stimulated with two GPR114-APs (i.e. TA peptidomimetics) and/or forskolin. Figure 5F shows GPR114-AP concentration-dependent stimulation of cAMP production in EoL-1 cells. Both GPR114-APs induced cAMP production, although the P18 GPR114-AP exhibited a plateau effect at ≥10 μM, suggesting that it may have peaked solubility. We took advantage of the fact that Gαs-specific stimulation of adenylyl cyclase, but not many other inputs can be synergistically stimulated with forskolin. Figure 5G shows significant increases in cAMP accumulation when EoL-1 cells were treated with forskolin and the GPR114-APs. These combined data provide probable evidence that endogenous Gs-coupled GPR114 stimulates cAMP production in EoL-1 cells in response to its synthetic TA peptidomimetics, although we could not exclude the possibility that these peptide agonists may stimulate alternative AGPCR(s). A few AGPCRs known to couple to Gs are expressed in EoL-1 cells (Asterisked, Fig. 5C). In particular, a low level of ADGRG2/GPR64 expression was detected and its TA displays moderate homology to GPR114 (2, 12, 29, 30). As EoL-1 cells are refractory to transfection, we used HEK293 T cell-based overexpression CRE-Luc assays to directly compare GPR114-AP stimulation of GPR114 and GPR64. Engineered, TA-activated versions of GPR114 and GPR64 (7TM) provided robust CRE-Luc activity that was independent of exogenous GPR114-AP application (Fig. 5H). The P18 GPR114 AP stimulated cAMP production of full length and a TA-compromised version of GPR114 but did not do so for equivalent versions of GPR64. The P7 GPR114-AP did not stimulate cAMP production in any condition. From these combined data in Figure 5H with overexpressed receptors, we conclude that the P18 GPR114-AP most likely provides on-target stimulation of endogenous GPR114 in EoL-1 cells (Fig. 5F), whereas the shorter P7 GPR114-AP promiscuously stimulates cAMP accumulation via a yet to be determined GPCR.
Discussion
We demonstrate that GPR114 is a self-cleaved adhesion GPCR and cleavage is required for the receptor to be activated by its tethered peptide agonist. GPR114 was previously reported to be noncleaved despite possessing a consensus GPS cleavage site and TA that are identical to the efficiently cleaved AGPCR, GPR56 (14). Our results help to rectify alternative models of AGPCR TA activation that were based in part, on the prior assumption that GPR114 was not cleaved (10, 14). It is our position that AGPCR self-cleavage and dissociation of the NTF are prerequisites for TA intramolecular binding to the 7TM domain, the orthosteric agonistic mode of AGPCR activation.
Using overexpressed human GPR114 and GPR56, we made comparative measurements of GAIN domain elements that influenced receptor cleavage efficiencies. Both receptors possess minimal GAINA subdomains consisting of two antiparallel α-helices (Fig. 2, A and B, the GPR114 GAINA was derived from a homology model (4, 19). The GAINA α1-helix is shorter in GPR114 than in GPR56 and it is not contiguous with the GPR56 N-terminal PLL domain. Excising the GPR56 PLL domain had no influence on cleavage efficiency but trimming down the GPR56 α1-helix to approximate its length in GPR114 reduced GPR56 cleavage efficiency to that of GPR114. Successively removing the GPR114 and GPR56 α1-helices or the α1-helices and α2-helices reduced and eliminated receptor cleavage altogether. Combined, we concluded that both GAINA α-helices of GPR56 and GPR114 support GAINB domain proteolytic efficiency and that the GPR114 GAINB domain is inherently less efficient than that of GPR56. A recent study found that residues Ser876 and Asn893 of the BAI2 GAINB domain β-sheet network lack important conformational support of the GPS catalytic His,Leu/Thr residues resulting in lost cleavage efficiency of BAI2. In autoproteolysis-competent AGPCRs these residues are replaced with a Phe and Gly, respectively, and support cleavage (31). The β-strand 10 Phe and β-strand 11 Gly support residues are conserved in GPR114 and GPR56 thereby affirming our observations that both receptors are autoproteolysis-competent. Further molecular analysis will be required to explain the inherent cleavage efficiency difference between GPR56 and GPR114.
GPR114 was demonstrated to be a Gs coupler as it raised cAMP levels in overexpression systems and formed an active-state complex with Gs and nanobody 35 (10, 14). In our hands, GPR114 activated Gs and provided no ability to activate representative members of the other 3 G protein classes (i/q/13). Our reconstitution setup also permits direct comparison of the activities of AGPCR holoreceptors to the freed CTF/7TMs. NTF dissociation provided strong Gs activation demonstrating that GPR114 behaves like other cleaved adhesion GPCRs; its TA is concealed within the GAIN domain and requires full release from the NTF to activate the 7TM. Both of these findings will help inform future studies directed at understanding the biological function of GPR114, which is currently unknown.
Clues of its function are emerging based on an understanding of its tissue and cell distribution profile. GPR114 is likely expressed in eosinophils due to detection of its mRNA in circulating the eosinophils, lung, spleen, and colon (23, 24, 32). Each of these tissues is involved in immune responses where eosinophils become distributed (33). We tested various immune cell and eosinophil-like cell lines for GPR114 RNA expression using quantitative real-time reverse-transcription PCR assays. Moderate levels of GPR114 expression were found in the EoL-1 eosinophilic-like cancer cell line. A GPCR expression array was conducted for EoL-1 cells and GPR114 was detected as a midlevel expressed adhesion GPCR, but the highest expressed member of the ADGRG subfamily. Immunoblotting prepared EoL-1 cell membranes with a GPR114 NTF-directed antibody showed the maturated, deglycosylated NTF at its predicted molecular weight of 23 kDa. No uncleaved GPR114 product was detected, which is in line with our previous findings for other AGPCRs; the receptors may be misprocessed in heterologous overexpression systems, but the endogenously produced proteins are processed efficiently in native tissues/cells (28).
Our future work will assess the physiological function of GPR114 in eosinophil biology. A primary function of eosinophils is to release cytotoxic factors that are stored in granules to increase inflammatory responses in peripheral tissues (33). Degranulation is stimulated by inositol trisphosphate-dependent calcium mobilization yet decreased by cAMP signaling (34, 35). It remains uncertain how the inhibitory and stimulatory eosinophil degranulation signals are regulated. Our study demonstrated P18 GPR114-AP dependent rises of cAMP production in EoL-1 cells (Fig. 5F). We suspect that endogenous EoL-1 GPR114 is responsible for the observed cAMP accumulation, as the CRE-Luc results (Fig. 5H) showed that the P18 TA peptidomimetic exhibited on-target specificity for GPR114 and not the other Gs-coupled ADGR”G” subfamily member expressed in EoL-1 cells, ADGRG2/GPR64. Due to the conserved homology, AGPCR TA peptidomimetics may cross-activate multiple receptors (9, 36). This appears to be the case for the GPR114/GPR56-AP peptidomimetic P7, which stimulated cAMP production in EoL-1 cells, but did not activate GPR114 nor GPR64 in the CRE-Luc assay. The peptidomimetic P7 sequence, TYFAVLM is identical to the TAs of GPR114 and GPR56 and contains the three most critical residues found in the TAs of most AGPCRs, the P3 position Phe, the P6 Leu, and the P7 Met (2, 9). We suspect that peptidomimetic P7 is capable of activating another AGPCR present in EoL-1 cells and illustrate this example to caution the use of TA peptidomimetics as tools to provide absolute proof of AGPCR signaling specificity in endogenous cells and tissues.
No protein ligand that interacts with the GPR114 NTF has been found. Its identity is of particular interest for providing clues of GPR114 function and for understanding the potential specificity of ligand binding to those AGPCR NTFs that consist of minimal GAIN domains like GPR114. One speculation for the potential location of the GPR114 ligand is on eosinophils themselves. Eosinophils play a critical role in peripheral tissue inflammatory responses (33). Eosinophils make up ∼5% of white blood cells at ∼30 to 350 cell per ml of blood. However, an inflammatory response can increase localized peripheral eosinophil counts dramatically to 500 to 5000 cells/ml (37). The accumulated eosinophils may crowd to a point where trans-eosinophil ligand/GPR114 NTF interaction pairs form. Transiently adhered eosinophils may move in relation to the adhesive linkage thereby shearing the GPR114 NTF off and activating cAMP signaling, perhaps for cell migratory purposes. Alternatively, we envision that GPR114-dependent cAMP signaling may be activated during migration along and across the endothelium as eosinophils move toward a proinflammatory tissue site. Upon interaction with a potential ligand on the endothelial surface, the NTF will shed and drive cAMP production. cAMP alterations can be promigratory or antimigratory depending on context (38, 39). Alternatively, this form of GPR114-dependent cAMP signaling may serve to keep migrating eosinophils quiescent until they reach a peripheral destination where degranulation is favored.
Experimental procedures
Molecular cloning
Plasmids created or used in this study are described in Table S1. In brief, human GPR56 and GPR114 sequences were used as templates for PCR-based subcloning of full length, truncated, and mutagenized cDNAs into pFastBac-1 and pcDNA3.1 expression vectors. Baculoviruses were generated following the Bac-to-Bac system manufacturer’s instructions (Invitrogen). All constructs examining cleavage efficiencies included an N-terminal hemagglutinin signal peptide followed by a FLAG epitope and a tobacco etch virus proteolytic site. A human rhinovirus 3C protease cleavage site followed by a GFP and His8 module was added to the C-termini to create N- and C-terminal epitope-tagged GPR114 and GPR56 constructs. The GPR56/114 chimeric construct consisted of codons 168 to 181 of GPR56 in frame with codons 22 to 528 of full-length, maturated GPR114.
Constructs to express N-terminal fusions of the human FPR2 (PAR1) leader sequence with thrombin sites encoding residues LDPRS (Leu, Asp, Pro, Arg, Ser) appended to the C terminally His8-tagged 7TM domains of ADGRG5/GPR114 and ADGRG1/GPR56 were purchased as gene blocks with human codon optimization from Integrated DNA Technologies (7). Site-directed mutagenesis of the thrombin sites to encode LDPES was performed to create versions of the chimeras not susceptible to thrombin cleavage. The gene blocks were digested with EcoRI and NotI and subcloned into pFastbac-1 and pcDNA3.1 (Invitrogen).
Sf9 culture, baculovirus, and recombinant AGPCR membrane production
Spodoptera frugiperda 9 (Sf9) cells were cultured in shake flasks at 27 °C in ESF921 medium (Expression Systems). Recombinant bacmids were produced using the Bac-to-Bac system (Invitrogen) and transfected into S. frugiperda 9 (Sf9) cells using Fugene HD (Promega). Baculoviral supernatants were harvested 5 days after transfection. For viral amplification, Sf9 cells grown to 2.0 to 3.0 × 106 cells/ml were infected with a 1/100 dilution of virus. Amplified viruses were harvested 72 h postinfection.
For AGPCR membrane homogenate preparation, Sf9 cells growing at 2.0 to 3.0 × 106 cells/ml were infected with 1/100th volume of amplified baculovirus and harvested 48 h postinfection. Sf9 AGPCR cell pellets were resuspended in lysis buffer (20 mM Hepes pH 7.4, 1 mM EDTA, 1 mM EGTA, and protease inhibitor cocktail (phenylmethylsulfonyl fluoride, Nα-p-tosyl-L-lysine-chloromethyl ketone, L-1-p-tosylamino-2-phenylethyl-chloro ketone, leupeptin, and lima bean trypsin inhibitor). Cells were lysed using a nitrogen cavitation device (Parr Industries). The lysates were centrifuged at 600g for 10 min and the resultant supernatant was centrifuged at 100,000g for 40 min at 4 °C. Membrane pellets were Dounce homogenized in lysis buffer containing 10% w/v sucrose and overlaid on a 25% and 40% w/v sucrose cushion in a Beckman SW28 tube. The membranes were centrifuged to equilibrium at 104,000g at 4 °C. Enriched plasma membranes were collected from the 25/40% sucrose interface, diluted in lysis buffer and centrifuged at 100,000g for 40 min. The membrane pellet was Dounce homogenized into 20 mM Hepes pH 7.4, 1 mM EGTA, and 11% w/v sucrose and cryopreserved at −80 °C. The concentration of total protein in the membrane homogenates was determined by Bradford assay.
Enrichment of FLAG-tagged AGPCRs
Prepared FLAG-AGPCR membrane homogenates (400 mg) were solubilized in 1 ml of 20 mM Hepes pH 7.4, 150 mM NaCl, 1% w/v dodecyl maltoside (DDM), 0.2% w/v cholesterol hemisuccinate (CHS), and protease inhibitor cocktail for 1 h at 4 °C. The detergent insoluble material was precipitated by centrifugation at 100,000g and 5 mM CaCl2 and a 25 μl bed volume of FLAG-M1 antibody resin (Sigma-Aldrich) were added to the soluble detergent extract for an 1 h incubation with gentle rotation. The resin was washed thrice with 20 mM Hepes pH 7.4, 150 mM NaCl, 5 mM CaCl2, 1% w/v DDM, 0.2% w/v CHS, protease inhibitor cocktail. FLAG-tagged AGPCRs were eluted with 150 μl of 20 mM Hepes pH 7.4, 150 mM NaCl, 1% w/v DDM, 0.2% w/v CHS, protease inhibitor cocktail, 5 mM EDTA, and 0.1 mg/ml 3X FLAG peptide (Sigma-Aldrich).
Detection of AGPCR NTFs in cell culture media
Sf9 cultures (200 ml) infected with FLAG-tagged AGPCR viruses were harvested 48 h postinfection by centrifugation at 10,000g. The 10,000g supernatant was centrifuged at 200,000g to precipitate AGPCR-containing exosomes. The 200,000g clarified media supernatants were adjusted to contain 20 mM Hepes pH 7.4, 150 mM NaCl, 5 mM CaCl2, and 1% w/v DDM, 0.2% w/v CHS. A 50 μl bed volume of FLAG-M1 antibody resin was added and allowed to batch bind overnight at 4 °C. The resin was pelleted, washed with 20 mM Hepes pH 7.4, 150 mM NaCl, 5 mM CaCl2, 1% w/v DDM, 0.2% CHS, washed with the same buffer lacking CaCl2 and eluted with 150 μl of 20 mM Hepes pH 7.4, 150 mM NaCl, 1% w/v DDM, 0.2% CHS, protease inhibitor cocktail, 5 mM EDTA, and 0.1 mg/ml 3X FLAG peptide (Sigma-Aldrich).
SDS-PAGE and immunoblotting
AGPCR cell membranes, purified receptors, or eluants from FLAG M1 resin purifications were mixed with reducing SDS-sample buffer and resolved by 12% SDS-PAGE followed by transfer to polyvinylidene fluoride membrane for immunoblotting. Samples were not heated prior to gel loading. The FLAG M1 monoclonal antibody (1:1000, Sigma-Aldrich) or FLAG polyclonal antibody (1:5000, Sigma-Aldrich) were used for detection of N-terminal FLAG-tagged receptors. The penta-His monoclonal antibody (1:1000, Qiagen) was used to detect C terminally His8-tagged receptors. Secondary antibodies, purchased from Li-Cor, were IRDye 800 CW Donkey anti-rabbit (Cat# 926-32213), IRDye 800 CW Donkey anti-mouse (Cat# 926-32212), IRDye 680 CW Donkey Anti-rabbit (Cat# 926-68023), IRDye 680 CW Donkey anti-mouse (Cat# 926-68022) (1:5000).
Chemiluminescent Western blotting was used to detect purified WT GPR114 and uncleaved mutant GPR114-H225S receptors. Invitrogen GPR114 Polyclonal Antibodies (Cat# PA5-21701) and (Cat# PA5-52390) were used to detect the N-terminal fragment of GPR114. The secondary antibody used was anti-Rabbit IgG peroxidase-linked species-specific whole antibody from donkey (1:5000, Thermo Fisher Scientific). Gel images were acquired using iBright FL1000 (Invitrogen).
Luciferase reporter assays
Low-passage HEK293T cells were plated at 5.5 × 106 cells per 10 cm dish in Dulbecco’s modified Eagle’s medium (DMEM) + 10% (v/v) fetal bovine serum (FBS) 24 h prior to polyethylenimine-mediated transfection with 6.86 μg of AGPCR in pcDNA3.1, 2.74 μg SRE Luciferase reporter (pGL4.33 (Promega) for GPR56 or rat insulin II promoter CRE-Luciferase for GPR114), and 27.5 ng of phRLuc-N1 (PerkinElmer), or the reporters alone. Each transfection was balanced to 9.6 μg total DNA with pcDNA3.1 as necessary. The transfected cells were lifted 6 h later with 0.05% w/v trypsin in Puck’s Saline G containing 1 mM EGTA, pooled, and used to seed white, sterile 96-well Nunclon Delta Surface plates (Thermo Fisher Scientific) at 80,000 cells per well in fresh DMEM plus 10% w/v FBS. The medium was aspirated at 36 h and replaced with 100 μl of serum-free DMEM. After 4 h of serum starvation, synthetic peptide GPR56-AP in dimethyl sulfoxide or thrombin (Chrono-log) in 20 mM Tris–HCl pH 6.5, 150 mM NaCl, 0.1% PEG-8000 was added to appropriate wells and incubated for an additional 4 h. The final dimethyl sulfoxide content in each well did not exceed 1% v/v. The plates were centrifuged for 5 min at 500g and the top 50 μl of medium was removed from each well. Cells were lysed by addition of 50 μl Steady-Glo reagent (Promega) and luminescence was measured using a TriStar2 plate reader (Berthold Technologies) as described (16). Hundred microliters of Renilla luciferase quench buffer containing 3 μM coelenterazine h was added to each well to measure the RLuc signal (40). Raw firefly luciferase (Fluc) units were presented as ratioed data with respect to Renilla luciferase (RLuc) units.
AGPCR/G protein signaling reconstitution assays
Mock (5 μg/assay) and urea-treated (equiv. vol.) GPR114 and GPR56 membranes were reconstituted with purified Gαs, i, q, or 13 (100 nM) and Gβ1Gγ2 (500 nM) in preincubation buffer containing 50 mM Hepes, pH 7.4, 1 mM DTT, 1 mM EDTA, 20 μM GDP, 3 μg/ml bovine serum albumin. The G protein-reconstituted AGPCR membranes were incubated on ice and then at 25 °C, 2 min prior to initiation of biological triplicate reactions by addition of an equal volume of buffer containing 50 mM Hepes, pH 7.4, 1 mM DTT, 1 mM EDTA, 20 μM GDP, 3 μg/ml bovine serum albumin, 10 mM MgCl2, 50 mM NaCl, 2 μM [35S]-GTPγS (20,000 cpm/pmol). Samples were removed as technical triplicates at times 2, 5, 8, 15, and 30 min and quenched in buffer containing 20 mM Tris (pH 7.7), 100 mM NaCl, 10 mM MgCl2, 1 mM GTP, and 0.08% (m/v) deionized polyoxyethylene 10 lauryl ether C12E10. The samples were then filtered through Whatman GF/C glass microfiber filters (Cytiva), dried, and subjected to liquid scintillation counting.
RNA isolation and cDNA synthesis
Cells (1.0 × 106) were solubilized in 1 ml TRIzol reagent (Invitrogen) and incubated at 22 °C for 5 min. A total of 0.2 ml of chloroform was added and inverted to mix. Aqueous and organic layers were separated by centrifugation at 12,000g for 15 min at 4 °C. The RNA-containing aqueous layer was isolated and added to an equal volume of chloroform. Centrifugation was repeated and the aqueous layer was isolated and added to an equal volume of isopropanol and mixed prior to centrifugation at 21,000g for 5 min. The RNA pellet was washed with 70% ethanol, dried, and solubilized in Tris-EDTA buffer (10 mM Tris, pH 7.7 and 1 mM EDTA). RNA concentration was determined via Nanodrop. RNA (16 μg) was converted to cDNA in 160 μl using the Transcriptor First Strand cDNA Synthesis Kit according to the manufacture’s protocol (Roche Catalog No.04897030001).
TaqMan human GPCR array
A total of 160 μl of synthesized cDNA was added to 290 μl of PCR-grade water. 450 μl of TaqMan Gene Expression Master Mix (Applied Biosystems Catalog No. 4369016) was added to the reaction prior to loading 100 μl to each funnel of the TaqMan Human GPCR Array (Applied Biosystems Part No. 4365295). The array was centrifuged twice at 1000g for 1 min. Quantitative PCR was performed on a QuantStudio7 (Applied Biosystems) using the following cycle parameters: 1 μl reaction volume, with one cycle of 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min.
TaqMan gene expression assay
Gene expression assays were performed following the product manual (Thermo Fisher Scientific TaqMan Gene Expression Assay Protocols). TaqMan Gene Expression Assays were purchased from Thermo Fisher Scientific for ADGRG5–Hs00917307_m1 (Catalog No. 4351372), ADGRG1–Hs00938474_m1 (Catalog No. 4351372), and Ribosomal 18S–Hs03928985_g1 (Catalog No. 4331182). Quantitative PCR was performed using a QuantStudio7 (Applied Biosystems) using the following cycle parameters: 20 μl reaction volume, with one cycle at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Eight-point standard curves for the ADGRG1 and ADGRG5 expression assays were generated using plasmid cDNA (Fig. S2). An absolute quantification relationship was developed using a linear curve fit model.
Cell lines used for ADGRG1 and ADGRG5 expression identification
Human embryonic kidney 293T (HEK293T) cells were cultured in DMEM + 10% FBS. Colorectal carcinoma 205 (COLO205) cells were gifted from the Narla Lab at the University of Michigan and cultured in RPMI-1640 medium with 10% FBS. Natural killer-92 (NK-92) cells were purchased from American Type Culture Collection and cultured in RPMI-1640 + 10% FBS, 100 U/ml interleukin-2. Human acute promyelocytic (NB4), human eosinophilic leukemia (EoL-1), human leukemia promyeloblast (HL-60) cells were purchased from American Type Culture Collection and cultured in RPMI-1640 + 10% FBS.
Measurement of cAMP accumulation
The cAMP ELISA assay kit from Cayman Chemical (Item No. 581001) was used to measure cAMP accumulation. Cells were collected and resuspended at a concentration of 1.0 × 106 cells/ml in Tyrode’s buffer pH 7.4 (119 mM NaCl, 5 mM KCl, 25 mM Hepes, 2 mM CaCl2, 2 mM MgCl2, 6 g/L glucose, and 100 μM IBMX). A total of 1.0 × 106 cells were added to an Eppendorf tube and were incubated with peptide (2 μM, 5 μM, 10 μM, 25 μM, 50 μM, and 100 μM) for 30 min. Forskolin was used at 200 μM for synergy experiments. Cells were centrifuged at 100g for 30 s to pellet cells. The cell pellet was then resuspended in 75 μl of 0.1 N HCl for 20 min to lyse the cells. The lysed cells were centrifuged at 21,000g for 5 min at 4 °C. Clarified supernatant was collected and diluted 1:2 with ELISA buffer from the kit. The remainder of the assay was carried out according to the manufacturer’s protocol.
Statistical analysis
All data are presented as the mean ± SD of three or more independent experiments (biological replicates). GraphPad Prism (https://www.graphpad.com/) was used for all statistical analyses. The threshold for significance was p < 0.05.
Data availability
All the data are contained within the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank Kelly Bernadyn for her help with statistical analyses and Georgios Skiniotis and Moran Shelav-Benami for the kind gift of FLAG- and -GFP-His6 dually tagged GPR114 and GPR56 expression constructs.
Author contributions
T. F. B., A. V., R. A., F. K., and G. G. T. investigation; T. F. B., A. V., R. A., F. K., and G. G. T. methodology; T. F. B., A. V., R. A., F. K., and G. G. T. writing–original draft; T. F. B., A. V., R. A., F. K., and G. G. T. conceptualization; T. F. B., A. V., R. A., F. K., and G. G. T. data curation; G. G. T. supervision; T. F. B. and G. G. T. writing–reviewing and editing.
Funding and additional information
This work was funded by NIH Grant GM149539 to G. G. T. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Kirill Martemyanov
Supporting information
References
- 1.Rask-Andersen M., Masuram S., Schiöth H.B. The druggable genome: evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu. Rev. Pharmacol. Toxicol. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]
- 2.Vizurraga A., Adhikari R., Yeung J., Yu M., Tall G.G. Mechanisms of adhesion G protein-coupled receptor activation. J. Biol. Chem. 2020;295:14065–14083. doi: 10.1074/jbc.REV120.007423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta C., Bernadyn T.F., Tall G.G. Structural clarity is brought to adhesion G protein-coupled receptor tethered agonism. Basic Clin. Pharmacol. Toxicol. 2022 doi: 10.1111/bcpt.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araç D., Boucard A.A., Bolliger M.F., Nguyen J., Soltis S.M., Südhof T.C., et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krasnoperov V., Lu Y., Buryanovsky L., Neubert T.A., Ichtchenko K., Petrenko A.G. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J. Biol. Chem. 2002;277:46518–46526. doi: 10.1074/jbc.M206415200. [DOI] [PubMed] [Google Scholar]
- 6.Lin H.H., Chang G.W., Davies J.Q., Stacey M., Harris J., Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 7.Mathiasen S., Palmisano T., Perry N.A., Stoveken H.M., Vizurraga A., McEwen D.P., et al. G12/13 is activated by acute tethered agonist exposure in the adhesion GPCR Adgrl3. Nat. Chem. Biol. 2020;16:1343–1350. doi: 10.1038/s41589-020-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liebscher I., Schön J., Petersen S.C., Fischer L., Auerbach N., Demberg L.M., et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014;9:2018–2026. doi: 10.1016/j.celrep.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barros-Alvarez X., Nwokonko R.M., Vizurraga A., Matzov D., He F., Papasergi-Scott M.M., et al. The tethered peptide activation mechanism of adhesion GPCRs. Nature. 2022;604:757–762. doi: 10.1038/s41586-022-04575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ping Y.Q., Xiao P., Yang F., Zhao R.J., Guo S.C., Yan X., et al. Structural basis for the tethered peptide activation of adhesion GPCRs. Nature. 2022;604:763–770. doi: 10.1038/s41586-022-04619-y. [DOI] [PubMed] [Google Scholar]
- 11.Qu X., Qiu N., Wang M., Zhang B., Du J., Zhong Z., et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature. 2022;604:779–785. doi: 10.1038/s41586-022-04580-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao P., Guo S., Wen X., He Q.T., Lin H., Huang S.M., et al. Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4. Nature. 2022;604:771–778. doi: 10.1038/s41586-022-04590-8. [DOI] [PubMed] [Google Scholar]
- 13.Beliu G., Altrichter S., Guixà-González R., Hemberger M., Brauer I., Dahse A.K., et al. Tethered agonist exposure in intact adhesion/class B2 GPCRs through intrinsic structural flexibility of the GAIN domain. Mol. Cell. 2021;81:905–921. doi: 10.1016/j.molcel.2020.12.042. [DOI] [PubMed] [Google Scholar]
- 14.Wilde C., Fischer L., Lede V., Kirchberger J., Rothemund S., Schöneberg T., et al. The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 2016;30:666–673. doi: 10.1096/fj.15-276220. [DOI] [PubMed] [Google Scholar]
- 15.Mayumi M. EoL-1, a human eosinophilic cell line. Leuk. Lymphoma. 1992;7:243–250. doi: 10.3109/10428199209053629. [DOI] [PubMed] [Google Scholar]
- 16.Stoveken H.M., Hajduczok A.G., Xu L., Tall G.G. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6194–6199. doi: 10.1073/pnas.1421785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paavola K.J., Sidik H., Zuchero J.B., Eckart M., Talbot W.S. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal. 2014;7:ra76. doi: 10.1126/scisignal.2005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen S.C., Luo R., Liebscher I., Giera S., Jeong S.J., Mogha A., et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in schwann cell development mediated by interaction with laminin-211. Neuron. 2015;85:755–769. doi: 10.1016/j.neuron.2014.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman G.S., Ackerman S.D., Ding C., Koide A., Leon K., Luo R., et al. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron. 2016;91:1292–1304. doi: 10.1016/j.neuron.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng W., Zhang C., Li Y., Pearce R., Bell E.W., Zhang Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods. 2021;1 doi: 10.1016/j.crmeth.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry-Hauser N.A., VanDyck M.W., Lee K.H., Shi L., Javitch J.A. Disentangling autoproteolytic cleavage from tethered agonist-dependent activation of the adhesion receptor ADGRL3. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vizurraga A.L., Robertson M.J., Yu M., Skiniotis G., Tall G.G. Hexahydroquinoline derivatives are selective agonists for the adhesion G protein-coupled receptor ADGRG1/GPR56. Mol. Pharmacol. 2023;104:28–41. doi: 10.1124/molpharm.123.000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjarnadóttir T.K., Geirardsdóttir K., Ingemansson M., Mirza M.A., Fredriksson R., Schiöth H.B. Identification of novel splice variants of adhesion G protein-coupled receptors. Gene. 2007;387:38–48. doi: 10.1016/j.gene.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y.M., van de Garde M.D., Cheng K.F., Baars P.A., Remmerswaal E.B., van Lier R.A., et al. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011;90:735–740. doi: 10.1189/jlb.0211092. [DOI] [PubMed] [Google Scholar]
- 25.Ji B., Feng Y., Sun Y., Ji D., Qian W., Zhang Z., et al. GPR56 promotes proliferation of colorectal cancer cells and enhances metastasis via epithelial-mesenchymal transition through PI3K/AKT signaling activation. Oncol. Rep. 2018;40:1885–1896. doi: 10.3892/or.2018.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S., Chatterjee T., Godoy C., Wu L., Liu Q.J., Carmon K.S. GPR56 drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-mediated mechanism. Mol. Cancer Res. 2019;17:2196–2207. doi: 10.1158/1541-7786.MCR-19-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ke N., Sundaram R., Liu G., Chionis J., Fan W., Rogers C., et al. Orphan G protein-coupled receptor GPR56 plays a role in cell transformation and tumorigenesis involving the cell adhesion pathway. Mol. Cancer Ther. 2007;6:1840–1850. doi: 10.1158/1535-7163.MCT-07-0066. [DOI] [PubMed] [Google Scholar]
- 28.Yeung J., Adili R., Stringham E.N., Luo R., Vizurraga A., Rosselli-Murai L.K., et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28275–28286. doi: 10.1073/pnas.2008921117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D.L., Sun Y.J., Ma M.L., Wang Y.J., Lin H., Li R.R., et al. Gq activity- and β-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. Elife. 2018;7 doi: 10.7554/eLife.33432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demberg L.M., Rothemund S., Schöneberg T., Liebscher I. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/Adgrg2. Biochem. Biophys. Res. Commun. 2015;464:743–747. doi: 10.1016/j.bbrc.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Pohl F., Seufert F., Chung Y.K., Volke D., Hoffmann R., Schöneberg T., et al. Structural basis of GAIN domain autoproteolysis and cleavage-resistance in the adhesion G-protein coupled receptors. bioRxiv. 2023 doi: 10.1101/2023.03.12.532270. [preprint] [DOI] [Google Scholar]
- 32.Hsiao C.C., Chu T.Y., Wu C.J., van den Biggelaar M., Pabst C., Hébert J., et al. The adhesion G protein-coupled receptor GPR97/ADGRG3 is expressed in human granulocytes and triggers antimicrobial effector functions. Front. Immunol. 2018;9:2830. doi: 10.3389/fimmu.2018.02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diny N.L., Rose N.R., Čiháková D. Eosinophils in autoimmune diseases. Front. Immunol. 2017;8:484. doi: 10.3389/fimmu.2017.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroegel C., Chilvers E.R., Giembycz M.A., Challiss R.A., Barnes P.J. Platelet-activating factor stimulates a rapid accumulation of inositol (1,4,5)trisphosphate in guinea pig eosinophils: relationship to calcium mobilization and degranulation. J. Allergy Clin. Immunol. 1991;88:114–124. doi: 10.1016/0091-6749(91)90308-b. [DOI] [PubMed] [Google Scholar]
- 35.Kita H., Abu-Ghazaleh R.I., Gleich G.J., Abraham R.T. Regulation of Ig-induced eosinophil degranulation by adenosine 3',5'-cyclic monophosphate. J. Immunol. 1991;146:2712–2718. [PubMed] [Google Scholar]
- 36.Demberg L.M., Winkler J., Wilde C., Simon K.U., Schön J., Rothemund S., et al. Activation of adhesion G protein-coupled receptors: agonist specificity of stachel sequence-derived peptides. J. Biol. Chem. 2017;292:4383–4394. doi: 10.1074/jbc.M116.763656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Sullivan J.A., Bochner B.S. Eosinophils and eosinophil-associated diseases: an update. J. Allergy Clin. Immunol. 2018;141:505–517. doi: 10.1016/j.jaci.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M.O., Ryu J.M., Suh H.N., Park S.H., Oh Y.M., Lee S.H., et al. cAMP promotes cell migration through cell junctional complex dynamics and actin cytoskeleton remodeling: implications in skin wound healing. stem cells Dev. 2015;24:2513–2524. doi: 10.1089/scd.2015.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ydrenius L., Molony L., Ng-Sikorski J., Andersson T. Dual action of cAMP-dependent protein kinase on granulocyte movement. Biochem. Biophys. Res. Commun. 1997;235:445–450. doi: 10.1006/bbrc.1997.6822. [DOI] [PubMed] [Google Scholar]
- 40.Dyer B.W., Ferrer F.A., Klinedinst D.K., Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are contained within the manuscript.