Significance
Low organ- and cell-specific mRNA (Messenger RNA) transfection in vivo represents one of the most critical challenges in the field of mRNA medicine. Herein, this study reported the design of a new generation of dual-targeted mRNA nanoparticles (NPs), which can be inhaled and home to lung tumor cells and inflammatory macrophages, for highly efficient expression of any proteins of interest in vivo. Considering the vital roles of cancer cells and macrophages in lung-related diseases (e.g., tumors, infections, and inflammations), this platform technology may potentially pave the way for the development of new inhaled mRNA medicine for treating lung cancers and pneumonia, as well as inhaled mRNA vaccines against infectious diseases.
Keywords: inhalation delivery, dual-targeted mRNA nanoparticles, lung tissue transfection, lung cancer, pneumonia
Abstract
Messenger RNA (mRNA)–based therapeutics are transforming the landscapes of medicine, yet targeted delivery of mRNA to specific cell types while minimizing off-target accumulation remains challenging for mRNA-mediated therapy. In this study, we report an innovative design of a cationic lipid- and hyaluronic acid–based, dual-targeted mRNA nanoformulation that can display the desirable stability and efficiently transfect the targeted proteins into lung tissues. More importantly, the optimized dual-targeted mRNA nanoparticles (NPs) can not only accumulate primarily in lung tumor cells and inflammatory macrophages after inhalation delivery but also efficiently express any desirable proteins (e.g., p53 tumor suppressor for therapy, as well as luciferase and green fluorescence protein for imaging as examples in this study) and achieve efficacious lung tissue transfection in vivo. Overall, our findings provide proof-of-principle evidence for the design and use of dual-targeted mRNA NPs in homing to specific cell types to up-regulate target proteins in lung tissues, which may hold great potential for the future development of mRNA-based inhaled medicines or vaccines in treating various lung-related diseases.
With the capability to produce any proteins of interest, messenger RNA (mRNA)–based therapeutics are transforming the therapeutics and vaccination landscapes (1–4). Considering the great success of mRNA nanoparticle (NP) vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV 2) and their ongoing applications in eliciting cross-neutralizing antibodies for COVID-19 variants in the clinic (5–7), future extensive use of NP-mediated mRNA medicine in treating many other major human diseases besides infectious diseases can be well predicted (1, 2, 8, 9). However, safe and efficacious delivery of mRNA to specific organs and cells remains a great challenge in the development of mRNA-mediated therapies. Many studies showed that the widely used mRNA delivery platform of lipid NPs presents an affinity to the liver when they were systemically administered, which was thought to be triggered by the natural physiological advantages of the liver such as a slow blood flow (10, 11) and discontinuous vasculature feature in hepatic sinusoids (12). In addition, there are many diseases in nonliver tissues, among which lung-related diseases have attracted great attentions.
Different from other major organs, the lung has the physiological characteristics of direct contact with the air in the external environment (13, 14). Therefore, the lung tissue cells are susceptible to the invasion and infection of various pathogenic microorganisms (15) or toxic substances (16) and gradually lead to the production, development, and deterioration of lung-related diseases. The prevalence of respiratory diseases is due to the deterioration of the environment and the emergence of multiple pathogens, which draws aggressive attention to developing therapeutic drugs and introducing more treatment methods, especially during the current pandemic.
Importantly, the delivery route will substantially affect the biodistribution (17–21) and therapeutic efficacy of the mRNA nanotherapeutics. The inhalation of formulating drugs is regarded as a highly potential way to lung because lung inhalation does not undergo first-pass metabolism (22). Patients and physicians could prefer inhalation delivery of medications for its convenience, and the inhaled mRNA NPs may also enable local transfection in specific lung cells that are not easily targeted via systemic administration (organ- and cell-specific targeting). Moreover, the rational chemical design of mRNA NPs such as those modified with targeting ligands can further improve their accumulation in specific types of cells in lung. For example, targeting tumor cells in lung for effective lung cancer treatment and homing to inflammatory macrophages for robust pulmonary inflammation therapy. Despite the significant promise, such a therapeutic strategy and mRNA nanoformulations that may provide high efficacy and specificity for treating lung cancer and/or pulmonary inflammation remain largely unexplored.
Herein, we developed a new class of polymer-lipid hybrid mRNA NPs (Fig. 1 A and B), composed of I) FDA-approved polymer hyaluronic acid (HA) to both i) load/protect the mRNA-cationic lipid and to ii) target CD44 proteins (23, 24) overexpressed on the surface of cancer cells and/or macrophages (25), II) 1, 2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(ethylene glycol) (DSPE-PEG) to stabilize the NP system, and III) targeting moiety of mannose (Man) conjugated to DSPE-PEG on mRNA NP surface to further dual-target glucose transporters (26, 27) overexpressed on the surface of cancer cells and/or macrophages (28). Three different types of mRNA encoding different proteins including luciferase (Luc), enhanced green fluorescence protein (EGFP), and wide-type p53 were used as model mRNAs in this study (i.e., Luc-mRNA, EGFP-mRNA, and p53-mRNA, respectively) for efficient delivery to the lung. Our results demonstrated that the optimized HDP-Man (HDPM) NPs could not only selectively deliver different types of mRNA to the target cells (e.g., H1299 lung cancer cells and proinflammation macrophages) with extremely high efficacy both in vitro and in vivo but also effectively produce the target proteins of interest for different applications. To the best of our knowledge, this is the first-of-its-kind dual-targeted NP delivery platform for mRNA delivery, which may address the critical challenges of mRNA delivery to specific cell types in the lung tissues. Considering the significant role of cancer cells and macrophages in many human diseases, this proof-of-concept work could also benefit the future development of inhaled mRNA-based medicines or vaccines for treating various diseases (e.g., lung cancers, pneumonia relief, infectious diseases, and other respiratory diseases).
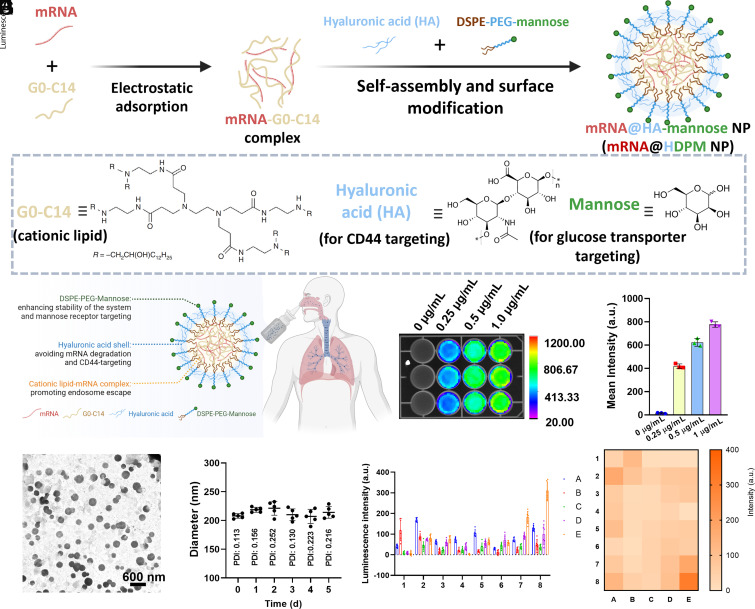
Fig. 1.
Characterization and optimization of dual-targeted mRNA HDPM NPs from 40 nanoformulations. Schematic illustration of the (A) preparation and (B) inhalation delivery of dual-targeted mRNA HDPM NPs. (C) TEM image and (D) time-dependent DLS size of mRNA NPs in PBS for a period of 5 d. (E) Screening of the in vitro transfection efficacy of Luc-mRNA delivered by 40 different nanoformulations. Corresponding heat map results summarized in (F). NPs composed of different ratios of HA, G0-C14, DSPE-PEG, and DSPE-PEG-Man. In detail, NP core materials include Conditions [A: 2.5 μL G0-C14 (0.5 mg/mL) without HA], [B: 2.5 μL G0-C14 (0.5 mg/mL) with 0.63 μL HA (0.5 mg/mL), [C: 2.5 μL G0-C14 (0.5 mg/mL) with 0.83 μL HA (0.5 mg/mL)], [D: 2.5 μL G0-C14 (0.5 mg/mL) with 1.25 μL HA (0.5 mg/mL)], or [E: 2.5 μL G0-C14 (0.5 mg/mL) with 2.5 μL HA (0.5 mg/mL)]; and NP surface modification materials include Conditions [1: No PEG], [2: 1.00 μL DSPE-PEG (5 mg/mL), [3: 0.75 μL DSPE-PEG (5 mg/mL) and 0.25 μL DSPE-PEG-Man (5 mg/mL)], [4: 0.67 μL DSPE-PEG (5 mg/mL) and 0.33 μL DSPE-PEG-Man (5 mg/mL)], [5: 0.50 μL DSPE-PEG (5 mg/mL) and 0.50 μL DSPE-PEG-Man (5 mg/mL)], [6: 0.33 μL DSPE-PEG (5 mg/mL) and 0.67 μL DSPE-PEG-Man (5 mg/mL)], [7: 0.25 μL DSPE-PEG (5 mg/mL) and 0.75 μL DSPE-PEG-Man (5 mg/mL)], or [8: 1 μL DSPE-PEG-Man (5 mg/mL)]. (G) In vitro bioluminescence images and (H) the mean bioluminescence intensity of H1299 cells at 24 h post-treatment of the dual-targeted HDPM NPs at different concentrations of Luc-mRNA (expressing Luc).
Results
Synthesis and Characterization of Dual-Targeted mRNA NPs.
The inhaled, dual-targeted mRNA HDPM NPs were fabricated via the self-assembly of G0-C14 cationic lipids (29) and negatively charged mRNA followed by encapsulation in HA and DSPE-PEG-Man (Fig. 1 A and B). To optimize the stable dual-targeted NPs that can further encapsulate mRNA, we first used different concentrations of G0-C14 and HA to synthesize the NPs. The dynamic light scattering (DLS) results showed that the use of a low concentration of G0-C14 and a high volume of HA was beneficial for synthesizing the NPs with a size of about 200 nm (SI Appendix, Fig. S1); we selected the concentration of 0.5 mg/mL for both G0-C14 and HA for NP synthesis in the following experiments. The transmission electron microscope (TEM) image (Fig. 1C) and DLS results (Fig. 1D) further confirmed the average size of dual-targeted NPs was approximately 200 nm with a uniform sphere morphology. According to the standard line (SI Appendix, Fig. S2), the encapsulation efficiency of Cy5-labeled mRNA in dual-targeted HDPM NPs was calculated to be 50%. Additionally, the dual-targeted mRNA NPs had excellent stability in phosphate-buffered solution (PBS) as their size did not change obviously within 5 d (Fig. 1D).
We then compared the in vitro mRNA transfection efficiency of the dual-targeted mRNA HDPM NPs with other control mRNA NPs such as i) nontargeted PLGA-DSPE-PEG (PDP) NPs synthesized in our previous study (8) and ii) single-targeted HA-DSPE-PEG (HDP) NPs. We utilized the Luc-expressing mRNA (Luc-mRNA) as the model mRNA loaded in these NPs and monitored the bioluminescence expression in both H1299 cells (model human nonsmall cell lung carcinoma cell line with the overexpression of CD44 proteins and glucose transporters) and HeLa cells (another cancer cell line also with the overexpression of CD44 proteins and glucose transporters). SI Appendix, Fig. S3A shows that the bioluminescence generated by the mRNA NP-mediated Luc expression was higher in groups treated by the Luc-mRNA HDP NPs than those treated by Luc-mRNA PDP NPs at the same mRNA dosage (approximately 2 times higher in H1299 cells and 2.5 times higher in HeLa cells). The results could be ascribed to the CD44 binding ability of HA in the mRNA NPs as we designed. Moreover, the modification of Man targeting ligands on the mRNA NP surface further improved the Luc expression in both cell lines as the bioluminescence expression was 2 times higher in H1299 cells treated by Luc-mRNA HDPM NPs than those treated by Luc-mRNA HDP NPs at the same mRNA dosage (v.s. 3 times higher in HeLa cells) (SI Appendix, Fig. S3B). The result was credited to the additional glucose transporter uptake of these HDPM NPs (besides the CD44-targeting ability possessed by both HDPM NPs and HDP NPs).
Optimization and In Vitro Transfection of Dual-Targeted mRNA NPs.
To deeply investigate the essential roles of Man- and HA-mediated dual targeting effects, we synthesized a NP library including different ratios of DSPE-PEG/DSPE-PEG-Man and G0-C14/HA, and compared their bioluminescence in H1299 cells. As shown in Fig. 1 E and F, the introduction of both DSPE-PEG-Man and HA enhanced the in vitro Luc-mRNA transfection ability in our system, through which we optimized the ratio of these components; we used the largest amounts of DSPE-PEG-Man and HA (8E NPs) to obtain the dual-targeted mRNA NPs for the subsequent studies. Next, we used an imaging system that can detect bioluminescence to study the mRNA expression efficacy using HDPM NPs at different concentrations of Luc-mRNA (0.25, 0.5, and 1 μg/mL). As observed from Fig. 1 G and H, the HDPM NPs presented a good bioluminescence expression with a concentration-dependent tendency.
We then moved forward to initially evaluating the targeting ability of our NPs by the cell uptake experiment at different time points after the incubation with H1299 cells. To access the cell uptake ability, we replaced the Luc-mRNA with red fluorescent Cy 5-labeled mRNA in our synthetic NPs. Compared with our previous nontargeted PDP nanoformulation and single-targeted HDP nanoformulation groups, the dual-targeted HDPM NPs were easily engulfed into the H1299 cells (SI Appendix, Fig. S4). Remarkably, obvious red fluorescence of dual-targeted HDPM NPs was observed in the cytoplasm at 0.5 h postincubation with H1299 cells, whereas no naked-eye-detectable fluorescence was present in the cells from the PDP NPs group and single-targeted HDP NPs group (at the tested conditions). Moreover, while the uptake of NPs increased in all groups with the increasing incubation time, the dual-targeted HDPM NPs group showed the highest signals of mRNA in the cytoplasm of H1299 cells among all the groups. In addition, we also tested the targeting ability of our HDPM NPs in lipopolysaccharide (LPS)-induced proinflammatory macrophages. When compared with nontargeted PDP NPs and single-targeted HDP NPs, the dual-targeted HDPM NPs showed the best uptake in LPS-induced proinflammatory macrophages which at different tested time points (SI Appendix, Figs. S5 and S6). These results were also attributed to the HA and mannose targeting capability of the NPs to recognize the proinflammation macrophages. Taken all together, these results preliminarily supported the potential targeting ability of HDPM NPs, highlighting the potential superiority of the dual-targeted design of HDPM NPs for mRNA delivery selectively in the targeted cells compared with the use of nontargeted NPs and single-targeted NPs.
After verifying the active dual-targeting ability of our designed NPs, we then tested the transfection efficiency of the Luc-mRNA HDPM NPs after incubation with H1299 cancer cells for different periods of time. Specifically, H1299 cells were seeded in 24-well plates with the amount of about 5,000 cells per well. After overnight incubation, the various concentrations (0.25 μg/mL, 0.5 μg/mL and 1 μg/mL) of the synthesized NPs were placed into the cells for further coincubation. Subsequently, we studied the transfection process by recording the bioluminescence of cells in corresponding groups after coincubation for different periods of time (3, 6, 12, 24, and 48 h). At the tested timepoints, the bioluminescence intensity gradually increased and reached the maximum at 24 h after coincubation, allowing us to select 24 h as the incubation time for evaluating Luc expression after the Luc-mRNA NP transfection (SI Appendix, Fig. S7). Meanwhile, we also tested the in vitro biocompatibility of these mRNA HDPM NPs in different cells including both the cancer cells and the normal cells (myoblast cell line C2C12 and fibroblast cell line NIH/3T3 were used as model normal cell lines). As revealed in SI Appendix, Fig. S8, no obvious cytotoxicity was observed at the tested concentrations of Luc-mRNA HDPM NPs in all these tested cell lines, indicating that our mRNA NPs presented good biocompatibility.
Versatile HDPM NP Platform in Delivering Different Types of Functional mRNA In Vitro.
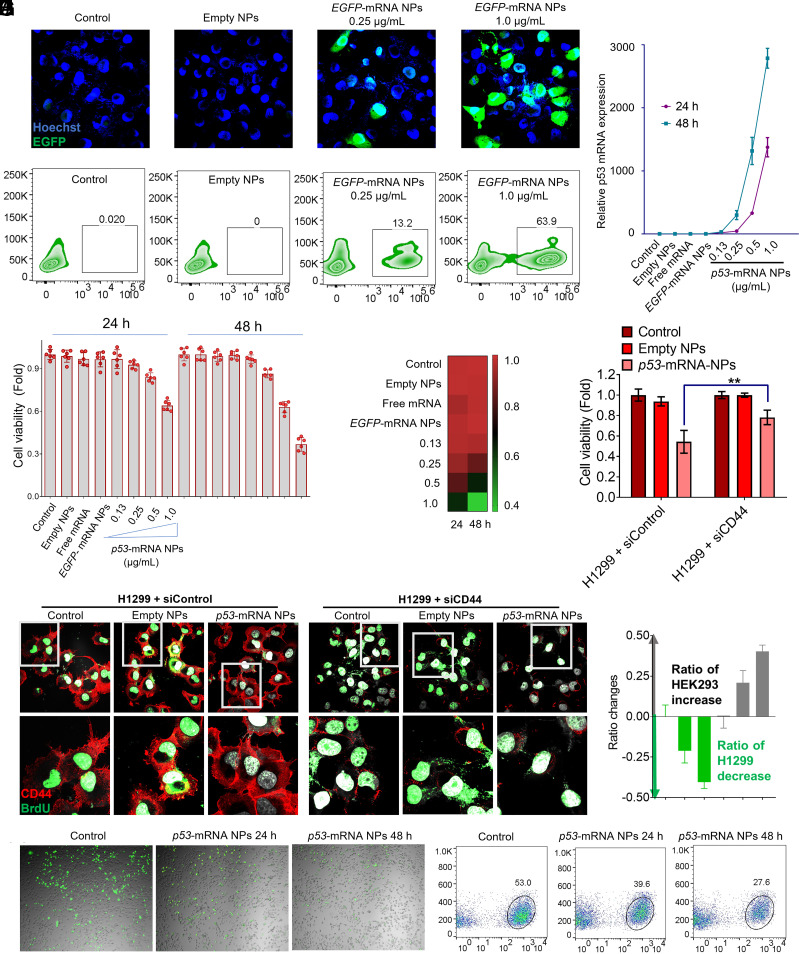
After confirming the effective and safe transfection of the Luc-mRNA by our dual-targeted NPs, we further tested whether this unique HDPM NP platform could also serve as a versatile platform to protect other types of mRNA at the cellular level. Functional mRNA such as EGFP-mRNA (encoding fluorescent proteins EGFP) and p53-mRNA (encoding tumor suppressor proteins p53) were utilized in the following studies. As shown in the confocal images and flow cytometry results (Fig. 2 A and B), the EGFP-mRNA encapsulated HDPM NPs could successfully induce the EGFP expression with a high efficiency in H1299 cells, while control group and empty NPs group did not show any expression of EGFP.
Fig. 2.
Versatile HDPM NP platform in delivering different types of functional mRNA in vitro, and its ability in selectively delivering mRNA to target cells. (A) Confocal laser scanning microscope (CLSM) images of H1299 cells after 24 h-treatment of empty HDPM NPs, or EGFP-mRNA HDPM NPs at the mRNA concentrations of 0.25 and 1.0 μg/mL (X40 magnification; Green, EGFP expression; Blue, Hoechst). (B) Flow cytometry analysis of H1299 cells after 24 h-treatment of empty HDPM NPs, or EGFP-mRNA HDPM NPs at the mRNA concentrations of 0.2 and 1.0 μg/mL. (C) Quantitative analysis of p53-mRNA expression in H1299 cells was determined by RT-qPCR after 24 h- or 48 h-treatment of empty HDPM NPs, free p53-mRNA, EGFP-mRNA HDPM NPs, or p53-mRNA HDPM NPs at the mRNA concentrations of 0.13, 0.25, 0.5 and 1.0 μg/mL. (D) Viability of H1299 cells after 24 h- or 48 h-treatment of empty HDPM NPs, free p53-mRNA, EGFP-mRNA HDPM NPs, or p53-mRNA HDPM NPs at the mRNA concentrations of 0.13, 0.25, 0.5, and 1.0 μg/mL. (E) Heat-map summary on the viability of H1299 cells after different treatments in figure (D). (F) CLSM images of control-siRNA or CD44-siRNA pretransfected H1299 cells after the treatments of empty HDPM NPs or p53-mRNA HDPM NPs at the mRNA concentrations of 1.0 μg/mL (60× magnification; Red, CD44 protein; Green, thymidine analog BrdU). (G) Viability of control-siRNA or CD44-siRNA pretransfected H1299 cells after the treatment of empty HDPM NPs or p53-mRNA HDPM NPs. Statistical significance was calculated using two-tailed student’s t test (**P < 0.01; n = 6, Mean ± SEM). (H) CLSM images of the cocultured, GFP-labeled H1299 and nonlabeled HEK293 cells at an initial ratio of 1:1. The mixed cells were treated with p53-mRNA HDPM NPs for 24 h or 48 h (Green dots, H1299-GFP cells; Gray dots, HEK293 cells). (I) Flow cytometry analysis of the cell population in figure (H) via the green fluorescence signals (green fluorescence positive-group, H1299-GFP cells; green fluorescence negative-group, HEK293 cells). (J) Analysis of the ratio changes between H1299-GFP and HEK293 cells. Data are presented as mean ± SEM (n = 3).
To further confirm whether our dual-targeted HDPM NPs could also effectively deliver p53-mRNA for therapeutic functions, we moved on to carry out more studies in H1299 cells with p53-mRNA HDPM NP-treatments. The RT-qPCR experiments revealed the successful delivery of p53-mRNA by HDPM NPs to the cytoplasm of H1299 cells, and the p53-mRNA expression increased along with increasing concentration and incubation time (Fig. 2C). As shown in Fig. 2D, the viability of H1299 cells decreased to about 50% and 30% after coincubation with p53-mRNA HDPM NPs (at the mRNA dosage of 1.0 μg/mL) for 24 h and 48 h, respectively, as well as displayed a concentration-dependent tendency. The therapeutic efficacy of p53-mRNA HDPM NPs on H1299 cells were also summarized with a heat-map form (Fig. 2E). These results primarily supported that our HDPM NPs could be used as a versatile platform to protect/deliver p53-mRNA and generate therapeutic effects specifically on cancer cells.
Enhanced and Selective Delivery of p53-mRNA via HDPM NPs to Lung Cancer Cells.
As cell proliferation results could be monitored by the thymidine analog BrdU (5-bromo-2′-deoxyuridine) (30) after its integration into newly generated DNA followed by the detection using the corresponding anti-BrdU antibody, we then used this assay to further verify cell proliferation inhibition ability of p53-mRNA and the CD44 targeting ability of the HDPM NPs. In the control groups (H1299 cells pretransfected control siRNA without any functions), incubation with p53-mRNA HDPM NPs could effectively decrease the signals of thymidine analog BrdU (green) compared with those without NP treatment and those with empty NP treatment (Fig. 2 F, Left), indicating the treatment of p53-mRNA HDPM NPs effectively inhibited the proliferation of cancer cells. However, in the groups pretreated with siCD44 (H1299 cells pretransfected functional siRNA to knockdown CD44; reflected by the significantly reduced red signals), p53-mRNA HDPM NPs’ effect on reducing the signals of thymidine analog BrdU (green) was somehow impaired (Fig. 2 F, Right) compared with the same treatment in the siControl pretransfected cells, indicating CD44-targeting design of mRNA HDPM NPs could promote the delivery of loaded mRNA to target cells with CD44 overexpression and improve the transfection efficacy. Moreover, the therapeutic effects of p53-mRNA HDPM NPs were also reduced in the siCD44 pretransfected H1299 cells compared with the same treatment in siControl pre-transfected H1299 cells (Fig. 2G), further indicating the CD44-targeting design of mRNA HDPM NPs could enhance the delivery of therapeutic mRNA to target cells with CD44 overexpression and potentially generate enhanced therapeutic efficacy.
Subsequently, we also conducted a competitive cell growth assay to further confirm the p53-mRNA HDPM NPs could selectively deliver therapeutic mRNA to glucose transporter/CD44 overexpressed H1299 cells for effective killing of cancer cells, while being safe and biocompatible for normal cells. HEK293 cells (normal human cells as example) and GFP-labeled H1299 cells were used in this part of studies and assessed through confocal and flow cytometry. In brief, the same number of HEK293 cells and GFP-labeled H1299 cells were coincubated with p53-mRNA-encapsulated NPs for 24 h and 48 h. As can be observed in our results (Fig. 2H), with the treatment of p53-mRNA HDPM NPs, the ratios of GFP-labeled H1299 cells (green) significantly decreased with the increase of incubation time. At 48 h, HEK293 cells (gray) almost dominated the total number of the cell population. The quantitative analysis of flow cytometry results also revealed that, compared with the control group of 53% GFP-labeled H1299 cells detected, the ratios of GFP-labeled H1299 cells decreased to 39.6% and 27.6% after 24 h- and 48 h-incubation with p53-mRNA HDPM NPs, respectively (Fig. 2I). Quantitative ratio changes in HEK293 and H1299 cells were also presented in Fig. 2J, showing the significantly increased number of HEK293 normal cells and decreased number of H1299 cancer cells. These data indicated our dual-targeted HDPM NPs could selectively deliver mRNA to glucose transporter/CD44 overexpressed cancer cells for therapeutic applications, but not normal cells without the over-expression of glucose transporter/CD44.
p53-mRNA HDPM NPs Effectively Expressed p53 Proteins and Activated Apoptosis Pathways in Cancer Cells.
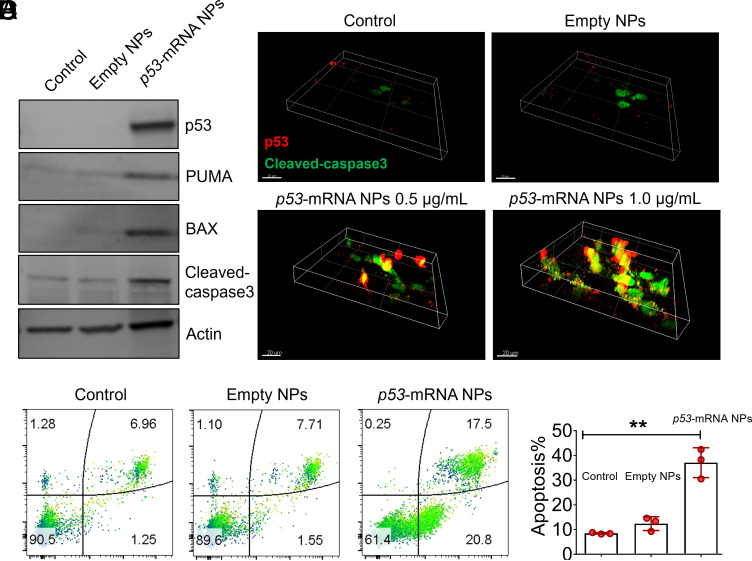
After primarily confirming the selective delivery of p53-mRNA via HDPM NPs to lung cancer cells and their effective killing effects on cancer cells, we then investigated the anticancer mechanism of our p53-mRNA HDPM NPs at the cellular level. As shown in the western blotting (WB) results (Fig. 3A), the treatment of p53-mRNA HDPM NPs effectively up-regulated the expression of p53 proteins (one kind of important tumor suppressors), followed by the remarkable upregulation of PUMA, BAX, and cleaved-caspase 3 proteins which would result in the apoptosis of H1299 cells. Such kind of p53-induced apoptosis was further confirmed by the flow cytometry assay (Fig. 3 B and C), as the treatment of p53-mRNA HDPM NPs significantly increased the ratios of apoptotic cells in H1299 compared with the control group and the empty NPs group. The cleaved-caspase 3-related pathways were also confirmed by immunofluorescence (IF) staining images shown in Fig. 4D. After the treatment of p53-mRNA HDPM NPs, the expression of p53 proteins along with cleaved-caspase 3 proteins could be prominently up-regulated in H1299 compared with the control group and the empty NPs group.
Fig. 3.
p53-mRNA HDPM NPs effectively expressed p53 proteins and activated apoptosis pathways in cancer cells. (A) WB analysis of the cell apoptosis–associated signaling pathway in p53-unexpressed H1299 cells after the treatment with empty HDPM NPs or p53-mRNA NPs. p53, PUMA, BAX, and cleaved-caspase 3 proteins were monitored, respectively. Actin served as the loading control. (B) Flow cytometry analysis of the apoptosis of H1299 cells after the treatment with empty HDPM NPs or p53-mRNA NPs. (C) Quantitative analysis of the cell apoptosis using the FlowJo software. Data were presented as mean ± SEM (n = 3), and statistical significance was calculated using two-tailed student’s t test (**P < 0.01). (D) Volume view of CLSM images of H1299 cells after the treatment with empty HDPM NPs or p53-mRNA NPs at the mRNA concentrations of 0.5 and 1.0 μg/mL [p53, red staining; Cleaved-caspase 3, green staining; (Scale bar, 20 μm)].
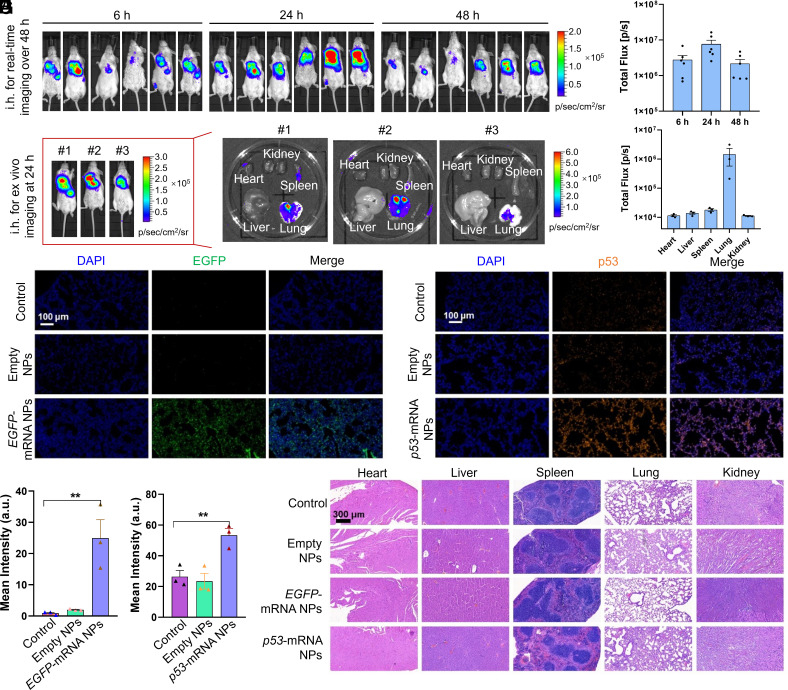
Fig. 4.
Biocompatible HDPM NPs effectively delivered different types of mRNA in vivo and expressed different proteins of interest in the lung. (A) In vivo bioluminescence images of BALB/c mice at different time points after receiving Luc-mRNA NPs via inhalation delivery. (B) Quantitative analysis of total luminescence flux of lung at different time points of Fig. 4A. Data were presented as mean ± SEM (n = 6) (C) Ex vivo bioluminescence images of different main organs at 24 h after inhalation from another group of 3 mice. (D) Quantitative analysis of luminescence flux from different organs in Fig. 4C. Data were presented as mean ± SEM (n = 3) (E) Fluorescence images of lung tissue section from mice after the treatment of empty HDPM NPs or EGFP-mRNA HDPM NPs (Nucleus, blue; EGFP, green). (F) Quantitative analysis of EGFP signals from the images in Fig. 4E. Statistical significance was calculated using two-tailed student’s t test (**P < 0.01). (G) Fluorescence images of lung tissue section from mice after the treatment of empty HDPM NPs or p53-mRNA HDPM NPs (Nucleus, blue; p53 protein, orange). (H) Quantitative analysis of p53 signals from the images in Fig. 4G. Statistical significance was calculated using two-tailed student’s t test (**P < 0.01). (I) H&E staining images of the sections from main organs, collected from the mice receiving different treatment.
Biocompatible HDPM NPs Effectively Delivered Different Types of mRNA In Vivo and Expressed Different Proteins of Interest in the Lung.
After confirming the excellent in vitro performance of our mRNA HDPM NPs, we next tested whether these NPs could effectively transfect the lung tissues in vivo. The mice were anesthetized and then administrated mRNA HDPM NPs through the spray. We first used the Cy 5-labeled Luc-mRNA as a model mRNA to investigate the biodistribution of mRNA-loaded NPs through the comparation between inhalation (i.h.) administration and intravenous (i.v.) injection. As shown in SI Appendix, Fig. S9, all the fluorescent signals from the Cy 5-labeled Luc-mRNA HDPM NPs were located in the lung of the i.h. administration group, while in the i.v. injection group, the fluorescent signals were majorly located in the liver and kidneys (not much in the lung). Moreover, the total fluorescent signals from the Cy 5-labeled Luc-mRNA HDPM NPs seemed to be reduced (cleared) quite a lot after passing the blood circulation to different organs. These results indicated that, different from i.v. injection method, inhalation delivery could increase the local concentrations of mRNA in lung, which could reduce the dosage of mRNA NPs used for treatment and avoid the potential systemic toxicity caused by i.v. injected high-dosage of mRNA NPs (in order to achieve the same amount of mRNA NPs in lung as i.h. administration), thereby highlighting the importance of inhalation delivery manner for the highly efficient delivery to lung tissues.
In the subsequent studies, we went on to check the Luc expression over the time after i.h. administration of our Luc-mRNA HDPM NPs in mice (Fig. 4 A and B). As shown in the in vivo bioluminescence images, after 6 h postinhalation, nearly all the bioluminescence was observed in lung as anticipated, confirming the superior mRNA transfection-induced protein expression of our HDPM NPs in lung via i.h. delivery. At 24 h postinhalation, the bioluminescence became stronger in the lung, indicating the sustained expression of target proteins by our NP delivery platform. Moreover, obvious bioluminescence could still be observed in lung 48 h after i.h., suggesting that our HDPM NPs could maintain the level of desired protein expression at least over 48 h in vivo. Additionally, in another group of studies, we sacrificed the mice at 24 h after i.h. of the mRNA NPs and harvested main organs to evaluate the Luc expression by bioluminescence imaging. As shown in Fig. 4 C and D, significantly strong bioluminescence signals were observed in lung, whereas negligible signals in spleen, kidney, heart, and liver were monitored.
Next, we used EGFP-mRNA- and p53-mRNA-loaded NPs to further confirm that our HDPM NPs could effectively deliver different types of mRNA in vivo and expressed different proteins of interest in the lung. At 24 h post-inhalation of PBS, empty HDPM NPs or mRNA-loaded HDPM NPs, the mice were sacrificed and lungs were collected for IF staining to detect the expression of EGFP or p53 proteins. As depicted in Fig. 4 E and F, compared with control group and empty HDPM NPs group, the EGFP protein level in the EGFP-mRNA HDPM NPs group was significantly higher, which was credited to the efficient EGFP-mRNA translation in lung tissues by our inhaled NPs. Similarly, after inhalation of p53-mRNA HDPM NPs, significant p53 protein expression was also observed at 24 h postinhalation (Fig. 4 G and H).
Moreover, the main organs were collected and stained with Hematoxylin and Eosin (H&E) for in vivo biocompatibility evaluation. As shown in Fig. 4I, no obvious signs of organ injury were observed in the main organs including heart, liver, spleen, lung, and kidney, indicating the potential good biocompatibility of the mRNA HDPM NPs. Specifically, as the mRNA HDPM NPs also showed signals in the spleen (although negligible) in Fig. 4C, we photographed the spleen organ isolated from Balb/c mice after inhalation of PBS or p53-mRNA HDPM NPs for 7 d. As also shown in SI Appendix, Fig. S10, there was no significant size or other difference between the two groups, indicating that our mRNA HDPM NPs did not cause any significant pathological alterations in the spleen. Meanwhile, based on the H&E staining images from Fig. 4I, we observed no histopathological abnormalities in the spleen sections of the mRNA HDPM NP-treated group compared to the control group. The H&E staining revealed a normal and nontoxic tissue architecture in the spleen, with distinct white pulp and red pulp regions, intact lymphoid follicles, and a well-preserved network of sinuses and splenic cords. These results suggested that our mRNA HDPM NPs did not induce any detectable signs of toxicity or pathological changes in the spleen.
Additionally, the blood of mice in different groups was also analyzed to evaluate whether there were any inflammation induced by the NPs via detecting the representative proinflammatory cytokines of IL-12, IL-6, TNF-α, and IFN-γ. According to the blood results in SI Appendix, Fig. S11, no obvious differences were been monitored between control group and different NP groups (empty HDPM NPs, EGFP-mRNA HDPM NPs, or p53-mRNA HDPM NPs group), further indicating the good biocompatibility and application potential of our NPs. We also investigated the blood biocompatibility by conducting hemolysis quantification of RBC (red blood cells) incubated with p53-mRNA HDPM NPs and empty HDPM NPs at different mRNA concentrations for 0.5 h. SI Appendix, Fig. S12 showed that no significant hemolysis was found in the presence of mRNA HDPM NPs or empty HDPM NPs, and all the hemolysis rates are less than 2%, which indicated excellent blood biocompatibility of our mRNA NPs. Meanwhile, we evaluated the blood biochemical parameters and blood hematology after inhalation of the HDPM NPs for 7 d to further confirm the biocompatibility of the HDPM NPs. As shown in SI Appendix, Fig. S13 A–D, there was no significant difference between the control group and the p53-mRNA HDPM NPs group in the blood parameters of alanine aminotransferase, blood urea nitrogen, aspartate aminotransferase and lactate dehydrogenase. As for blood hematology analysis, macrophages, white blood cells, RBC, hemoglobin, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, and platelet counts were measured. Compared with the control group, no statistically significant difference was shown in the mRNA HDPM NPs group in all the parameters (SI Appendix, Fig. S13 E–L).
Collectively, all these data provided solid evidence that our HDPM NPs are safe/biocompatible and promising for effective mRNA transfection and generating target proteins of interest in lung tissues. It is also worth briefly mentioning that although this NP platform was designed for inhaled delivery of mRNA, we also tried the intramuscular injection of our mRNA HDPM NPs as it is the most common vaccination way just like the currently FDA-approved mRNA vaccines. As shown in SI Appendix, Fig. S14, bright bioluminescence could be observed over at least 48 h after the injection of Luc-mRNA HDPM NPs, confirming that our HDPM NPs could also be potentially used in the application of mRNA vaccines.
Inhaled Delivery of mRNA via Dual-Targeted HDPM NPs to Cancer Cells or Proinflammation Macrophages in Lung Diseases Models (Mice with Orthotopic Lung Cancer or LPS-Induced Pneumonia).
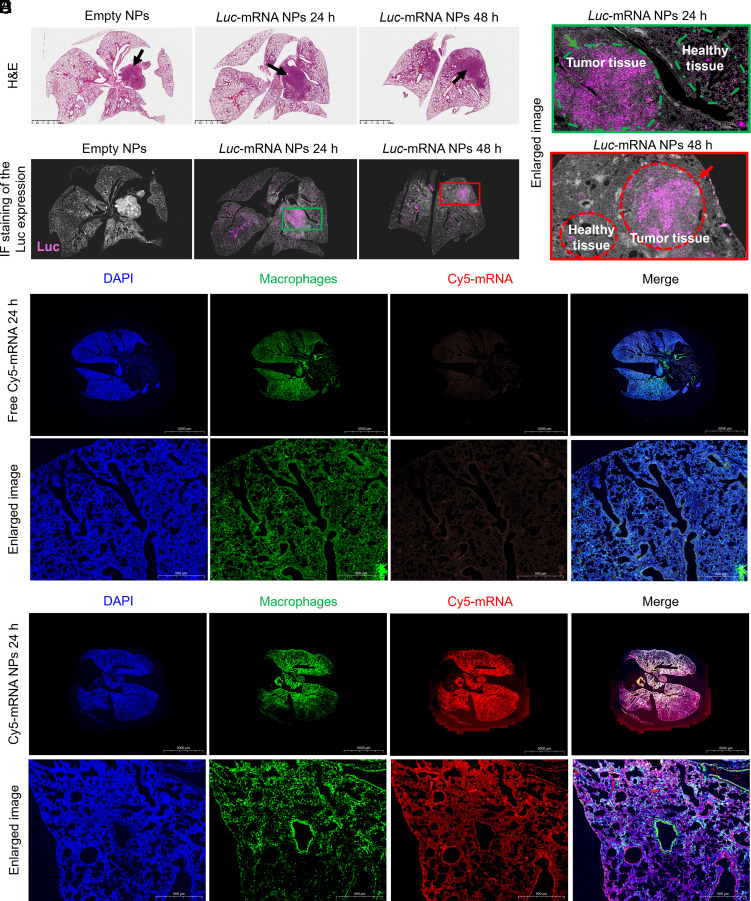
Based on the potent mRNA transfection ability in healthy lung tissue, we further used the mice models of orthotopic nonsmall cell lung cancer and LPS-induced pneumonia to evaluate the targeted delivery of mRNA to cancer cells or proinflammation macrophages in lung via our inhaled HDPM NPs in vivo. For the orthotopic nonsmall cell lung cancer mice model, after receiving different treatments (i.h. administration of empty HDPM NPs or Luc-mRNA HDPM NPs at 24 h and 48 h), the whole lung tissues were carefully collected for further H&E staining (Fig. 5A) and IF staining (Fig. 5B). When comparing the H&E staining results with the IF staining results, we could easily observe that the expression of Luc proteins was almost detected in the lung tumor area, while negligible signals in the healthy lung tissue area were observed (Fig. 5C), supporting the lung tumor targeting ability of our mRNA HDPM NPs. These results could be attributed to the dual-targeting ability of the HDPM NPs at the same time, i.e., HA part of the NPs for CD44-targeting and Mannose part on the surface or the NPs for glucose transporter targeting, respectively.
Fig. 5.
Targeted delivery of mRNA via HDPM NPs to cancer cells or proinflammation macrophages in lung diseases models (mice with orthotopic lung cancer or LPS-induced pneumonia) in vivo. (A) Histological examination (H&E staining) of lung tissues from mice with orthotopic nonsmall cell lung cancer after i.h. administration of empty HDPM NPs or Luc-mRNA HDPM NPs at 24 h and 48 h. (Scale bars, 2.5 mm); Black arrows, orthotopic tumor positions. (B) IF staining of lung tissues in Fig. 5A evaluating the location of target protein expression. Pink, Luc protein. (C) Enlarged images from Fig. 5B. (D) IF staining images and enlarged views of lung tissue sections from mice with LPS-induced pneumonia after i.h. administration of naked Cy5-mRNA at 24 h. Blue, nucleus; red, Cy5-mRNA; green, proinflammatory macrophages. (E) IF staining images and enlarged views of lung tissue sections from mice with LPS-induced pneumonia after i.h. administration of Cy5-mRNA HDPM NPs at 24 h. Blue, nucleus; Red, Cy 5-mRNA; Green, proinflammatory macrophages.
For the LPS-induced pneumonia mice model, after receiving different treatments (i.h. administration of free Cy5-mRNA or Cy5-mRNA HDPM NPs at 24 h), the whole lung tissues were carefully collected for further IF staining (Fig. 5 D and E). Consistent with the in vitro results, the in vivo studies also revealed the excellent targeting ability of the HDPM NPs for mRNA delivery to proinflammatory macrophages, as reflected the excellent colocalization between the macrophages (green) and Cy5-mRNA HDPM NPs (red). At the same time, because naked mRNA cannot be taken up by cells, most of the signals from Cy5-mRNA disappeared at 24 h-post treatment (at the tested conditions). These results strongly supported the good targeting efficacy and effective delivery of mRNA by our HDPM NPs to the proinflammatory lung tissues of pneumonia.
In summary, all the above results not only supported the biosafety of our dual-targeted mRNA HDPM NPs via inhalation delivery manner but also presented the satisfactory mRNA delivery in lung tissues, especially in target disease cells including lung cancer cells and proinflammatory macrophages, which highlight the promising application of our HDPM in developing mRNA-based therapeutics or vaccines for lung-related diseases.
Discussion
Inspired by the previous accomplishment especially in combating SARS-CoV-2, mRNA delivery has drawn much attention, and plenty of nanoplatforms and strategies were developed for tackling different diseases (1–4). The safe and efficacious delivery of mRNA to specific organs and cells remains a great challenge but extremely promising in the development of mRNA-mediated therapies, which needs new targeting strategies. Organ-specific targeting strategy could enable the mRNA therapeutics to arrive at the targeted organ tissues, but it is still not enough as it is hard for these mRNA therapeutics only take actions in the disease or target cells. Toxicity issues or unwanted protein expressions may still happen in the healthy tissues of the targeted organ. Therefore, the cell targeting of mRNA therapeutics, which directly contributes to precision medicine with more accurate therapies at the cellular level, is especially attractive and meaningful. However, realizing such kinds of specific cell-targeting is very difficult. Current common problems include i) lacking effective and specific targeting ligands for different cell types, ii) complexity of the chemical modification, iii) blocking of the targeting modification by the formation of protein corona (if systemic delivery via blood circulation is used as the administration route), and iv) difficulties in high-throughput production of targeted NPs with controlled quality from lab-scale synthesis to the clinical scale production (31–36). Targeted NP delivery platforms that can simultaneously address the difficulties in both organ-specific targeting and cell-specific targeting are thus urgently needed.
Among the main organs, the lung tissues are more susceptible to the external pathogenic microorganisms or toxic substances and lastly developed into lung-related diseases, so targeting the diseased lung tissues and specific cells could be valuable but challenging. In our work, we have demonstrated the perfect match between our dual-targeted NPs and inhalation way for efficient mRNA delivery to lung tumor cells and inflammatory macrophages. In particular, the NPs consisted of HA and DSPE-PEG-Mannose for CD44 proteins and glucose transporters targeting respectively, which lastly guaranteed the dual-targeting effects of lung tumor cells and inflammatory macrophages. Three types of mRNA including Luc-mRNA, EGFP-mRNA, and p53-mRNA were introduced to evaluate in vitro and in vivo lung tumor cells and inflammatory macrophages targeting, and all presented excellent performance, showing the promising potential of our dual-targeted mRNA NP strategy. In the representative lung tumor model and LPS-induced pneumonia model, our dual-targeted NPs showed excellent targeting ability and then endowed effective transfection, indicating that our NPs could be a powerful platform to deliver various therapeutic mRNAs to diseased lung cells for corresponding treatments in future applications. Besides the potential in lung cancer and lung inflammation treatment, our dual-targeted mRNA NPs may also be utilized to treat pulmonary cancer metastasis. Because the lung is a common metastatic site for many cancers and such cancer metastasis is among the major reasons for treatment failure in various cancers, the developed dual-targeted mRNA NPs might also be useful in tackling these tough lung metastasis problems.
In summary, this dual-targeting strategy in our study can also inspire the future design of a panel of dual-targeted mRNA NPs used for various disease conditions and different disease cells, which can address the current critical challenges in low cell-specificity of mRNA transfection. Moreover, our results not only highlighted the promise of the inhaled mRNA therapeutics to treat pulmonary diseases (such as cancers and viral infection) but also revealed the broad applications of the inhaled mRNA NPs for other lung disease management and vaccine development. For example, in treating lung inflammation and infections which are both common diseases at present, our strategy might be further optimized to i) deliver specific mRNAs for long-term expression of target proteins to relieve pneumonia, ii) deliver mRNA vaccine via inhalation to prevent viral infection, or iii) target immune cells to efficiently trigger the immune responses to combat lung inflammation and infections.
As our work is still in the infancy stage, there are still some aspects that need to be further investigated before future translation. For example, we only utilized one molecular weight of the DSPE-PEG-Mannose for glucose transporter targeting, and more representative DSPE-PEG-Mannose with different molecular weights can be introduced for optimized cell targeting ability. In addition, the ratios between mRNA and total other components could be further modified, which might affect the in vitro and in vivo mRNA delivery/targeting performance. Another potential problem is that the size of our NPs was about 200 nm which may not be advantageous for in vivo targeting, so the size should be optimized and commercial microfluidic technology (37, 38) could be introduced. Based on our promising in vitro results, other diseased lung cells could also be used for NPs targeting evaluation in the future studies, which could expand the potential of our dual-targeted NPs. More detailed biocompatibility evaluation in large animals could also be conducted for our NPs which is highly important for the future applications. Encouraged by the current market of mRNA medicine, ongoing clinical trials, and the irreplaceable role of the delivery platforms, we firmly believe that our dual-targeting NP strategy holds tremendous potential in delivering various mRNAs to wanted sites for corresponding disease treatment with high translational potential.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Materials and Methods, including all materials; synthesis of mRNA NPs; characterization of the targeted mRNA NPs; cell lines; in vitro transfection efficiency of targeted Luc-mRNA NPs; in vitro transfection efficacy of targeted EGFP-mRNA NPs; anti-cancer efficacy of targeted p53-mRNA NPs; in vitro and in vivo targeting efficacy of HA-Man-PEG NPs to inflammatory macrophages; animals; in vivo transfection of targeted Luc-mRNA NPs; in vivo transfection efficacy of targeted EGFP-mRNA or p53-mRNA NPs; in vivo mRNA targeted delivery to the orthotopic lung tumor site; and in vivo biocompatibility evaluation. Animal protocol was approved by the Institutional Animal Care and Use Committee at Harvard Medical School/Brigham and Women’s Hospital and Zhejiang University.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the American Lung Association (ALA) Cancer Discovery Award (LCD1034625; W.T.), ALA Courtney Cox Cole Lung Cancer Research Award (2022A017206; W.T.), Harvard Medical School/Brigham and Women’s Hospital Health & Technology Innovation Fund (2023A004452; W.T.), Nanotechnology Foundation (2022A002721; W.T.), and Farokhzad Family Distinguished Chair Foundation (018129, W.T.). W.T. also acknowledges the support from Novo Nordisk ValidatioNN (2023A009607), Khoury Innovation Award (2020A003219), American Heart Association (AHA) Transformational Project Award (23TPA1072337), and AHA’s Second Century Early Faculty Independence Award (23SCEFIA1151841).
Author contributions
Z.T., Y.X., N.K., and W.T. designed research; Z.T., X.Y., Y.X., X.H., H.L., F.X., C.L., and N.K. performed research; W.C., Y.L., S.K., N.K., and W.T. contributed new reagents/analytic tools; Z.T., X.Y., and Y.X. analyzed data; X.Y., W.C., Y.L., S.K., and W.T. wrote the paper; and N.K. and W.T. supervised the project.
Competing interests
Z.T., Y.X., W.C., N.K., and W.T. are inventors on a U.S. patent application (BWH2023-022) entitled ‘Inhalation Delivery of mRNA via Targeted NPs for Lung Tissue Transfection’ and filed by Brigham and Women’s Hospital related to the technology disclosed herein.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are available within this manuscript and the associated SI Appendix.
Supporting Information
References
- 1.Huang X. G., et al. , The landscape of mRNA nanomedicine. Nat. Med. 28, 2273–2287 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Xiao Y. F., et al. , Emerging mRNA technologies: Delivery strategies and biomedical applications. Chem. Soc. Rev. 51, 3828–3845 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Liu C., et al. , mRNA-based cancer therapeutics. Nat. Rev. Cancer 23, 526–543 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Chen S., et al. , Nanotechnology-based mRNA vaccines. Nat. Rev. Methods Primers 3, 63 (2023). [Google Scholar]
- 5.Tang Z. M., et al. , A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 5, 847–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X. G., et al. , Nanotechnology-based strategies against SARS-CoV-2 variants. Nat. Nanotechnol. 17, 1027–1037 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Tang Z. M., et al. , Insights from nanotechnology in COVID-19 treatment. Nano Today 36, 101019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong N., et al. , Intravesical delivery of KDM6A-mRNA via mucoadhesive nanoparticles inhibits the metastasis of bladder cancer. Proc. Natl. Acad. Sci. U.S.A. 119, e2112696119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao W., Peppas N. A., Robotic pills for gastrointestinal-tract-targeted oral mRNA delivery. Matter 5, 775–777 (2022). [Google Scholar]
- 10.Tsoi K. M., et al. , Mechanism of hard-nanomaterial clearance by the liver. Nat. Mater. 15, 1212–1221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y.-N., et al. , Nanoparticle–liver interactions: Cellular uptake and hepatobiliary elimination. J. Control Release 240, 332–348 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Augustin H. G., Koh G. Y., Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 357, eaal2379 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Cotes J. E., Chinn D. J., Miller M. R., Lung Function: Physiology, Measurement and Application in Medicine (John Wiley & Sons, 2009). [Google Scholar]
- 14.Lopez-Vidriero M., Mucus as a natural barrier. Respiration 55, 28–32 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Leiva-Juarez M. M., Kolls J. K., Evans S. E., Lung epithelial cells: Therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. 11, 21–34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright J. R., Pulmonary surfactant: A front line of lung host defense. J. Clin. Invest. 111, 1453–1455 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao P., Le Z., Liu L., Chen Y., Therapeutic delivery to the brain via the lymphatic vasculature. Nano Lett. 20, 5415–5420 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Lokugamage M. P., et al. , Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 5, 1059–1068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel A. K., et al. , Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 31, e1805116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., et al. , Exosomes decorated with a recombinant SARS-CoV-2 receptor-binding domain as an inhalable COVID-19 vaccine. Nat. Biomed. Eng. 6, 791–805 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melamed J. R., et al. , Ionizable lipid nanoparticles deliver mRNA to pancreatic beta cells via macrophage-mediated gene transfer. Sci. Adv. 9, eade1444 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patton J. S., Byron P. R., Inhaling medicines: Delivering drugs to the body through the lungs. Nat. Rev. Drug Discov. 6, 67–74 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Zhang S., et al. , Targeting chronic lymphocytic leukemia cells with a humanized monoclonal antibody specific for CD44. Proc. Natl. Acad. Sci. U.S.A. 110, 6127–6132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattheolabakis G., Milane L., Singh A., Amiji M. M., Hyaluronic acid targeting of CD44 for cancer therapy: From receptor biology to nanomedicine. J. Drug Target. 23, 605–618 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Penno M. B., et al. , Expression of CD44 in human lung tumors. Cancer Res. 54, 1381–1387 (1994). [PubMed] [Google Scholar]
- 26.Gonzalez P. S., et al. , Mannose impairs tumour growth and enhances chemotherapy. Nature 563, 719–723 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Panneerselvam K., Freeze H. H., Mannose enters mammalian cells using a specific transporter that is insensitive to glucose. J. Biol. Chem. 271, 9417–9421 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Higashi K., et al. , Correlation of Glut-1 glucose transporter expression with [18 F] FDG uptake in non-small cell lung cancer. Eur. J. Nucl. Med. 27, 1778–1785 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Zhu X., et al. , Long-circulating siRNA nanoparticles for validating prohibitin1-targeted non-small cell lung cancer treatment. Proc. Natl. Acad. Sci. U.S.A. 112, 7779–7784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gratzner H. G., Leif R. C., Ingram D., Castro A., The use of antibody specific for bromodeoxyuridine for the immunofluorescent determination of DNA replication in single cells and chromosomes. Exp. Cell Res. 95, 88–94 (1975). [DOI] [PubMed] [Google Scholar]
- 31.Zhang P., et al. , Cancer nanomedicine toward clinical translation: Obstacles, opportunities, and future prospects. Med 4, 147–167 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Rosenblum D., Joshi N., Tao W., Karp J. M., Peer D., Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 9, 1410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., et al. , Nano-bio interactions in cancer: From therapeutics delivery to early detection. Acc. Chem. Res. 54, 291–301 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Chen W., et al. , Macrophage-targeted nanomedicine for the diagnosis and treatment of atherosclerosis. Nat. Rev. Cardiol. 19, 228–249 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao W., et al. , siRNA nanoparticles targeting CaMKIIγ in lesional macrophages improve atherosclerotic plaque stability in mice. Sci. Transl. Med. 12, eaay1063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X., et al. , Synthesis of siRNA nanoparticles to silence plaque-destabilizing gene in atherosclerotic lesional macrophages. Nat. Protoc. 17, 748–780 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shepherd S. J., et al. , Scalable mRNA and siRNA lipid nanoparticle production using a parallelized microfluidic device. Nano Lett. 21, 5671–5680 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeki M., Uno S., Niwa A., Okada Y., Tokeshi M., Microfluidic technologies and devices for lipid nanoparticle-based RNA delivery. J. Control Release 344, 80–96 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are available within this manuscript and the associated SI Appendix.