Significance
This paper describes the characterization of endogenous retroviruses in chronic lymphocytic leukemia (CLL), the most common leukemia in the Western world. In this study, we identified a signature of dysregulated human endogenous retroviruses (HERVs) in CLL. In addition, our analysis suggested the potential contribution of HERVs in CLL pathogenesis.
Keywords: CLL, HERVs, RNA-Seq
Abstract
Chronic lymphocytic leukemia (CLL) is one of the most diagnosed forms of leukemia worldwide and it is usually classified into two forms: indolent and aggressive. These two forms are characterized by distinct molecular features that drive different responses to treatment and clinical outcomes. In this context, a better understanding of the molecular landscape of the CLL forms may potentially lead to the development of new drugs or the identification of novel biomarkers. Human endogenous retroviruses (HERVs) are a class of transposable elements that have been associated with the development of different human cancers, including different forms of leukemias. However, no studies about HERVs in CLL have ever been reported so far. Here, we present the first locus-specific profiling of HERV expression in both the aggressive and indolent forms of CLL. Our analyses revealed several dysregulations in HERV expression occurring in CLL and some of them were specific for either the aggressive or indolent form of CLL. Such results were also validated by analyzing an external cohort of CLL patients and by RT-qPCR. Moreover, in silico analyses have shown relevant signaling pathways associated with them suggesting a potential involvement of the dysregulated HERVs in these pathways and consequently in CLL development.
Chronic lymphocytic leukemia (CLL) is the most common human leukemia, accounting for about 10,000 new cases annually in the United States and representing approximately 30% of all leukemia cases (1). This disease occurs in two forms named “aggressive” and “indolent,” which are both characterized by the clonal expansion of CD5-positive B-lymphocytes (1–3). In more detail, the aggressive form is characterized by high ZAP-70 expression and unmutated IgH VH (IGHV), while the indolent shows low ZAP-70 expression and mutated IGHV (1–3). In addition to the expression levels of ZAP-70 and mutational status of IGHV, several other studies have revealed numerous genetic alterations in CLL, including single-nucleotide polymorphisms (SNPs), chromosomal alterations, and dysregulation in noncoding RNA expression, such as microRNA (miRNA) and tRNA-derived ncRNAs (tsRNAs), some of which can be used to determine prognosis and to guide management strategies (4, 5). However, alterations in protein-coding genes and noncoding RNAs are not solely responsible for cancer development. Recent studies point out the biological relevance of human endogenous retroviruses (HERVs), a class of transposable elements, in cancer biogenesis and also their potential clinical applications as novel diagnostic, prognostic, and treatment response biomarkers for several tumor types (6–19). HERVs are remnants of exogenous retroviruses integrated into the human genome during evolution, accounting for 8% of the human genome (20–22). Indeed, HERVs share genomic similarities with other exogenous retroviruses. However, due to accumulated mutations, HERVs have preserved the features of their original provirus to a highly variable extent, ranging from the retention of a complete set of long terminal repeats (LTRs) and retroviral genes to the retention of only fragments of the parental viral genomes (20, 21, 23). Based on their sequences, HERVs are usually classified into groups (20, 21). Precisely, their nomenclature refers to the single-letter amino acid code of the tRNA complementary to the primer binding site formerly used to prime reverse transcription (20, 21). Among them, the HERV-K family, which harbors the lysine tRNA binding site, includes some of the most recent HERVs acquired by humans around three million years ago, and it is commonly subdivided into 11 subgroups (HML-1 through HML-11) (24–28). Due to their relatively recent integration into the human genome, members of the HERV-K subgroup, called HML-2, still contain genes with intact open reading frame (ORF) that can encode retroviral proteins (24–28). Although the expression of several HERV groups has been seen to be deregulated in several human cancers, their exact roles in cancer development and progression remain unknown. Interestingly, some recent studies highlighted the importance of HERVs in different forms of leukemias (29–32), but no studies about HERVs in CLL have been conducted so far.
Because of their relevance in cancer development, in this study, we decided to perform locus-specific profiling of HERVs in CLL to identify the signatures of dysregulated HERVs that characterize the indolent and aggressive forms of CLL. Moreover, in silico analyses have been performed to infer their potential biological functions and their potential oncogenic effect.
Results
HERV Characterization in Indolent and Aggressive CLL.
To investigate HERV expression in CLL, we performed RNA sequencing (RNA-Seq) experiments in B cells taken from ten patients with the aggressive form of CLL carrying unmutated IGHV, ten patients with the indolent form of CLL carrying mutated IGHV, and four healthy donors (SI Appendix, Table S1). HERV characterization at the locus-specific level was conducted by developing an in-house workflow that relied on Telescope (33), which leverages a Bayesian statistical model to quantify HERVs addressing uncertainty in fragment assignment by reassigning ambiguously mapped fragments to the most probable source transcript.
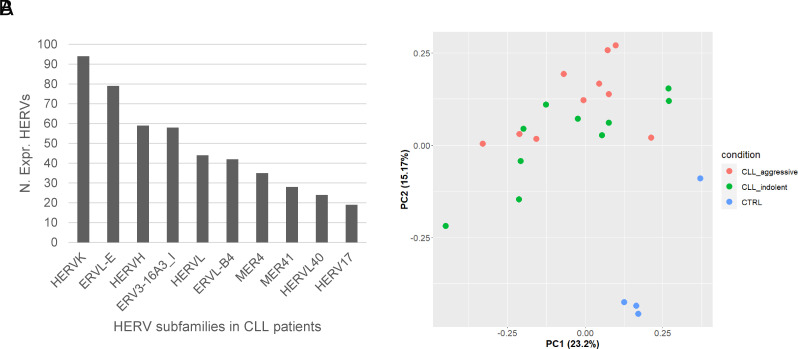
Among the 14,968 HERV genomic loci evaluated in this study, 765 (5,1 %) were found expressed [geometric mean Read Per Million (RPM) > 1] across all our samples and, therefore, considered for the downstream analyses. Among them, several HERV groups were detected, with the HERV-K, ERVL-E, and HERVH being the top three most represented groups (Fig. 1A). A complete list of the expressed HERV groups can be found in Dataset S1.
Fig. 1.
Expressed HERVs in CLL. (A) Bar plot showing the most expressed HERV groups identified in CD5+ B cells taken from CLL patients; (B) PCA plot showing the distribution of CLL aggressive, CLL indolent, and normal B cell samples in a two-dimensional space using the expression of the top 200 most variable HERVs based on their MAD values.
Afterward, we performed a principal component analysis (PCA) of the top 200 most variable HERVs based on their median absolute deviation (MAD) values. The results showed that CLL and control samples form two well-separated clusters indicating that CD5+ B cells of CLL patients have remarkably different HERV expression patterns compared to normal B cells (Fig. 1B). On the other hand, the aggressive and indolent samples of CLL do not seem to form different clusters (Fig. 1B). Noteworthily, considering the PC1 and PC2 (Fig. 1B), one of the healthy samples showed a certain difference compared to the other healthy ones. To further investigate this point, we explored the PC3 component as well, and, as shown in SI Appendix, Fig. S1, such a difference is reduced.
Dysregulation of HERVs in Indolent and Aggressive CLLs.
To identify dysregulated HERVs in CLL, we performed differential expression analyses comparing the CD5+ B cells from CLL patients with either the aggressive or the indolent form against the control samples from healthy donors. Specifically, a total of 130 and 82 HERVs were found dysregulated (|Log2FC| > 0.58 and FDR < 0.05) in the aggressive and indolent forms, respectively (Fig. 2A and Dataset S2). On the other hand, a direct comparison of the aggressive form against the indolent one did not show strong significant differences in HERV expression (Dataset S2). Therefore, we used the results obtained from the first two comparisons for further analyses. In more detail, we observed a bigger number of dysregulated HERVs in the aggressive form of CLL compared to the indolent one and a predominance of down-regulated HERVs (aggressive = 91; indolent = 59) over the up-regulated ones (aggressive = 39; indolent = 23) in both CLL forms. Notably, around 90% and 83% of the HERVs found down- and up-regulated, respectively, in the indolent form of CLL were also dysregulated with the same trend in aggressive CLL (Fig. 2B) (no HERVs were found dysregulated with the opposite trend between the two forms of CLL).
Fig. 2.
Differentially expressed HERVs in CLL. (A) Volcano plots showing the differentially expressed HERVs found in either the aggressive or indolent form of CLL compared to normal B cells; (B) Venn diagrams showing the down-regulated and up-regulated HERVs found in common between the aggressive and indolent forms of CLL; (C) Bipartite graphs reporting the log2FC values of the differentially expressed HERVs found in common in the aggressive and indolent forms of CLL. P-values were calculated by applying one-tailed Wilcoxon signed rank test.
Noteworthily, several of the HERVs that were found dysregulated in common between the indolent and the aggressive form of CLL presented a statistically significant tendency to have an increased magnitude of change in aggressive CLL while in others, the level of dysregulation was similar between the two forms (Fig. 2C).
These results indicated a more dramatic effect on HERV expression occurring in the aggressive form of CLL compared to the indolent one. Furthermore, we built two heatmaps, one for the aggressive form of CLL and one for the indolent form, using the normalized counts of the identified differentially expressed HERVs. The results for both cohorts revealed two well-defined clusters, one for tumors and one for normal samples confirming major dysregulations in HERV expression occurring in both forms of CLL (Fig. 3A). From a genomic standpoint, the differentially expressed HERVs were found dispersed throughout the genome, although some chromosomes such as the 17, the 16, the 9, and the 20 had higher percentages of differentially expressed HERVs calculated over the total number of HERV loci per chromosome (Fig. 3 B and C and Dataset S3). Moreover, chromosomes 10 and 18 did not present any dysregulated HERV in both CLL forms while chromosome 13 did not show any dysregulated HERVs in the indolent form only (Fig. 3 B and C and Dataset S3). Overall, we observed significant differences in the percentages of differentially expressed HERVs across chromosomes between the aggressive and indolent forms of CLL (Wilcoxon rank sum test P-value = 0.00035).
Fig. 3.
Distribution of differentially expressed HERVs in CLL. (A) Heatmaps with sample clustering of the differentially expressed HERVs identified in either the aggressive or indolent forms of CLL compared to normal B cells; (B) Circos plots showing the genomic distribution of the HERV loci found up (red dots) or down-regulated (blue dots) in the aggressive and indolent forms of CLL; (C) Bar plot showing the percentage of differentially expressed HERVs across the human chromosomes normalized by the number of HERV loci in the corresponding chromosome; (D) Bar plot reporting the top 10 most dysregulated HERV groups (% of dysregulated HERVs per group) identified in the aggressive (dark gray bars) and indolent (light gray bars) forms of CLL.
Among these dysregulated HERVs, several groups were found affected. In more detail, in terms of the absolute number of dysregulated HERVs per group, we observed that the HERVH, MER4, ERVL-B4, HERVL, and HERV-K groups were the top 5 most dysregulated in the aggressive form of CLL, while the HERVL, HERVH, ERV3-16A3_I, MER4, and PABL_A were the most affected in the indolent form. Notably, by normalizing the number of dysregulated HERVs per group, we identified that HERV groups such as HERVIP10HF, MER4, HERVL40, and HERVK had remarkable differences in terms of the percentage of dysregulated HERVs identified in either the aggressive or indolent form of CLL suggesting a potential involvement of these groups in CLL progression (Fig. 3D). Overall, we observed significant differences in the percentages of differentially expressed HERVs across the top 10 most represented HERV groups between the aggressive and indolent forms of CLL (Wilcoxon rank sum test P-value = 0.0091). The complete lists of the differentially expressed HERV groups found in either the aggressive or indolent form of CLL are reported in Dataset S4.
Validation of Dysregulated HERVs in CLL.
In order to validate the results obtained from our RNA-Seq experiments, we analyzed a publicly available external cohort of CLL (34). Precisely, the RNA-Seq data generated from the external cohort were analyzed in order to identify the differentially expressed HERVs in both the aggressive and indolent forms of CLL and see how they overlap with those ones identified from our cohort. In this external CLL cohort, we identified 83 and 97 differentially expressed HERVs in the aggressive and indolent forms, respectively. Among them, only 21 and 20 dysregulated HERVs in the aggressive and indolent forms, respectively, overlapped with those identified in our internal cohort suggesting that sample selection and experimental condition (different library preparation, Illumina RNA-seq machine, and read length, as specified in the Material and Methods for our internal cohort and in NCBI BioProject ID: PRJNA488107 for the external cohort) play a role in the observed differentially expressed HERVs (Dataset S5). Nonetheless, such analysis revealed a subset of HERVs which might be dysregulated in a significant portion of CLL patients. Moreover, only 13 of the dysregulated HERVs identified from the aggressive and indolent forms of CLL were in common between these two CLL forms (Fig. 4A) indicating that some of the remaining dysregulated HERVs may be specific for either the aggressive or the indolent form of CLL, respectively. Noteworthily, several of these differentially expressed HERVs were internal to host genes (mRNAs and/or lncRNAs), and some of them (four in indolent and six in aggressive forms of CLL) were significantly and positively correlated with the expression of the host genes (Dataset S6). In addition, we checked whether alterations in the karyotype and other common chromosomal abnormalities in CLL (e.g., 11q deletion, and 13q deletion) were correlated with the detected dysregulation in HERV expression, but no direct links were observed (SI Appendix, Fig. S2).
Fig. 4.
Validated differentially expressed HERVs in CLL. (A) Venn diagram reporting the number of HERVs identified as differentially expressed in the aggressive and indolent forms of CLL, in common between the two cohorts (internal and external); (B) Box plots showing the 2−ΔCt values of HERVL_17p11.2b and HUERSP3_11p15.1 in our internal cohort of CLL samples across the three conditions (CLL aggressive, CLL indolent, and CTRL). P-values were calculated by applying a one-tailed Wilcoxon rank sum test.
To further validate the obtained results from the RNA-Seq, we picked the most up-regulated (HERVL_17p11.2b) and the most down-regulated (HUERSP3_11p15.1) HERVs to confirm their expression by real-time RT-PCR. These HERVs were abundantly expressed in our samples (HERVL_17p11.2b average TPM CLL aggressive = 11217.6; CLL indolent = 20769.4; CTRL = 399.8; HUERSP3_11p15.1 average TPM CLL aggressive = 600.4; CLL indolent = 918.2; CTRL = 3253.4) compared to standard housekeeping genes (ACTB average TPM CLL aggressive = 1197.8; CLL indolent = 1269.5; CTRL = 1611.7; GAPDH average TPM CLL aggressive = 467.2; CLL indolent = 454.7; CTRL = 424.7) and were also located inside annotated genes giving us the technical advantage to design and use real-time RT-PCR assays that were specific for these two HERVs (as Real-Time PCR assays are much more accurate if primers are designed inside different exons of annotated genes). Precisely, HERVL_17p11.2b (Chr17:20998063-20998876), belonging to the HERVL group, was located within the last exon of a predicted protein-coding gene called LOC124900389 (Gene ID: 124900389) (SI Appendix, Fig. S3), while HUERSP3_11p15.1 (Chr11:17370374-17378687), belonging to the ERV1 group, was located within the last intron and the last exon of its host gene NCR3LG1 (SI Appendix, Fig. S3). The real-time RT-PCR confirmed significant overexpression of HERVL_17p11.2b in both aggressive and indolent forms of CLL against normal B cells (Fig. 4B and SI Appendix, Figs. S4 and S5). At the same time, HUERSP3_11p15.1 was confirmed to be down-regulated in both CLL forms compared to normal control samples (Fig. 4B and SI Appendix, Fig. S4). In conclusion, these results confirm our RNA-Seq data.
HERV-specific Pathway Analysis and Correlation Networks with Hallmarks in CLL.
In order to identify the potential biological functions of the 21 and 20 validated differentially expressed HERVs in the aggressive and indolent forms of CLL, respectively, we performed a mechanistic network-based topological pathway analysis using MITHrIL (35). First, we used a Pearson correlation formula to determine the correlation of the differentially expressed HERVs in either the aggressive or indolent forms of CLL with the differentially expressed genes (DEGs) identified in the respective form (Dataset S7) (Materials and Methods). The list of DEGs whose expression statistically correlates with a given HERV was then used as input for MITHrIL to identify the signaling pathways potentially connected with a specific HERV, as described in ref. 20. The results of the HERV-specific pathways analysis showed that several cancer-related signaling pathways were enriched in both forms of CLL (Fig. 5 A and B). In more detail, we observed that each HERV had different biological pathways associated with it which barely overlapped with the others associated with the other HERVs. Moreover, even in the case of HERVs dysregulated in common between the aggressive and the indolent forms of CLL, the pathways related to them seemed to be CLL form specific (Dataset S8). A relevant example is HERVL_17p11.2b. Such HERV was the most up-regulated HERV in both CLL forms; however, it showed a greater number of significant signaling pathways potentially associated with it in the aggressive form of CLL only compared with the other HERVs. Examples of such pathways include PI3K-AKT, MAPK, ErbB, TNF, and NOTCH signaling pathways. Collectively, these results seem to suggest that each HERV may be transcriptionally regulated by different biological pathways in a tumor-specific manner.
Fig. 5.
HERV-specific pathway analysis. Heatmaps that show the results of the HERV-specific pathway analysis for the aggressive (A) and indolent (B) forms of CLL. In detail, the columns are the enriched pathways while the rows are the differentially expressed HERVs. The values plotted in the heatmap are the “corrected accumulators” generated by MITHrIL as an indicator of the magnitude of pathway dysregulation. Corrected accumulator values with an associated not statistically significant P-value have been assigned the value 0 and plotted with the color white in the heatmaps.
To look more into the detail of potential functional connections, we made CLL form–specific correlation networks using as nodes the HERVs found differentially expressed in either the aggressive or indolent forms of CLL and a selected list of genes that are considered “hallmarks” of CLL (the distribution of the TPM values for the differentially expressed HERVs and hallmarks genes for the aggressive and indolent forms of CLL is reported in SI Appendix, Fig. S6). Such analyses revealed several statistically significant correlations between our identified differentially expressed HERVs in either the aggressive or the indolent forms of CLL and the hallmark genes involved in the development of CLL (Fig. 6). Among them, HERVL_17p11.2b has been observed to be significantly correlated with the expression of BTK in both CLL forms suggesting a possible transcriptional regulation of such HERV through the PI-3 K pathways (Fig. 6). Notably, from the previously described HERV-specific pathway analysis, the PI-3 K signaling pathway was the most up-regulated pathway among the enriched ones for HERVL_17p11.2b. To determine whether BTK regulates HERVL_17p11.2b expression, as suggested by the correlation and pathway analyses, we treated CLL samples with ibrutinib (a BTK inhibitor). We observed that in three out of three treated CLL samples with viability >50% at 48 h, ibrutinib treatment inhibited HERVL_17p11.2b expression (Fig. 7). This suggests that BTK at least in part regulates HERVL_17p11.2b expression.
Fig. 6.
Correlation networks between deregulated HERVs and hallmarks genes in CLL. Correlation networks for the aggressive (A) and indolent (B) forms of CLL where the blue nodes are the differentially expressed HERVs, the red nodes are the hallmark genes for CLL and the edge are either the positive (red) or negative (blue) correlation between them. Red and blue arrows by the HERV nodes indicate the up- or downregulation of the HERV, respectively, compared with normal B cell samples. HERVs with the highlighted borders are dysregulated in both CLL forms. Correlations between the differentially expressed HERVs and the hallmark genes that were not statistically significant were not plotted in the correlation networks.
Fig. 7.
Real-Time RT-PCR results in CLL samples treated with ibrutinib. Bar plot that shows the expression of HERVL_17p11.2b as 2−ΔCt in three CLL samples untreated or treated with 10 μm of ibrutinib at either 24 or 48 h (two biological replicates per condition).
Discussion
The expression signatures of HERV group members and their functional role in solid cancers and hematological malignancies have been an emerging topic in the last several years. Several recent studies found dysregulation of HERV expression in several types of cancer (36). On the other hand, the exact roles of these elements in cancer development and progression remain unknown. Since no studies describing HERV expression in CLL have been carried out, and because of the relevance of HERVs in cancer development, we performed a locus-specific profiling of HERV expression in CLL and identified the signatures of dysregulated HERVs in indolent and aggressive forms of CLL. Although our analyses allow an accurate estimation of HERV expression with a locus-specific resolution, it was not possible to determine whether the transcription of the individual HERVs was starting either from their LTRs or from the promoter region of the host genes due to the technical limitations of short-read sequencing platforms. Nevertheless, our data demonstrated that among the ~15 K HERVs considered in our analysis, 765 (5,1 % relative to the whole set of analyzed HERV loci) were found expressed in our samples. HERV-K, ERVL-E, and HERV-H were the three most represented expressed groups in CLL, and HERV-H, and MER4 groups were the most dysregulated. Interestingly, in our previous study, HERV-K and HERV-H were most expressed in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), while HERV-K, HERV-H, and MER4 were the most dysregulated (20). Although the higher representation of the HERV-K, ERVL-E, and HERV-H groups in our results was probably due to the larger number of elements included in these groups, the MER4 group still presented a considerable number of expressed and dysregulated HERVs considering other larger HERV groups (Datasets S1 and S3). Nonetheless, this suggests the importance of these 3 HERV groups in cancer in general.
We also showed that among the top 10 most differentially expressed HERV groups in CLL, 9 showed a lower number of members dysregulated in indolent CLL compared to the aggressive form (Fig. 3D), and generally, we found that the HERVs dysregulated in common between the aggressive and indolent forms of CLL when compared with healthy controls had a statistically significant tendency to increase their magnitude of dysregulation when they go from the indolent to the aggressive form of CLL (Fig. 2C), indicating a potential role of HERVs in CLL progression. However, this tendency may be also due to higher contamination of normal B cells in the indolent CLL samples compared to the aggressive ones, potentially representing a limitation for this type of analysis. Moreover, we are also aware of the limited number of samples used in this report, and further studies may be necessary to confirm the role of the identified dysregulated HERVs in CLL.
Worth mentioning, the comparison of CLL samples (aggressive and indolent samples pooled together) vs. the control samples showed differences that were similar to those identified in the individual forms of CLL when compared with control samples while a direct comparison of the aggressive vs. indolent forms of CLL did not show significant results in terms of the adjusted P-value. This is likely due to the large variability in HERV and gene expression in primary CLL samples collected from different patients. However, the relatively large fold-change values, significant P-values (Dataset S2), and the tendency to increase the magnitude of change in HERV expression in the aggressive form of CLL compared to the indolent one (Fig. 2C) seem to suggest a differential HERV expression between the two CLL forms that is not entirely captured by this cohort size.
Because of the repetitive nature of most HERVs, most of the previous reports only analyzed their expression by sequencing and did not show confirmation of the expression of individual HERVs (not at the group level) by other methods. Interestingly, in our study, both the most up-regulated and the most down-regulated in CLL HERVs (HERVL_17p11.2b and HUERSP3_11p15.1, respectively) overlapped with annotated genes. This enabled us to design a real-time RT-PCR assay for HERVL_17p11.2b (we designated it Retro 1) and use an existing real-time RT-PCR assay for HUERSP3_ 11p15.1 confirming our profiling data by real-time RT-PCR. Interestingly, Retro 1 (containing HERVL_17p11.2b in its 3′ UTR) is an uncharacterized protein coding gene (LOC124900389, Gene ID: 124900389) that showed drastic overexpression in our CLL samples (SI Appendix, Fig. S4). Its expression is also positively correlated with BTK expression in aggressive and indolent CLL (Fig. 6) and treatment with a BTK inhibitor (ibrutinib) suppressed Retro 1 expression (Fig. 7) in three of three CLL samples with viability over 50%. Moreover, HERVL_17p11.2b showed several significant signaling pathways potentially associated with it only in the aggressive form of CLL (Fig. 5), suggesting that this element may be regulated by different biological pathways in a tumor-specific manner. It remains to be determined whether Retro 1 is a novel CLL marker or a gene involved in the pathogenesis of CLL. Overall, our study reveals the potentially important role of human endogenous retroviruses in the biology of CLL.
Materials and Methods
Patient and Healthy Controls Sample Selection.
All human samples (both from CLL patients and healthy donors) utilized in this study were collected and analyzed under the protocol approved by the Institutional Review Board of The Ohio State University. The CLL Research Consortium provided us with a total of 20 samples collected from CLL patients enrolled upon written informed consent. According to IgVH mutational gene status (VH%), ZAP‐70 expression, and karyotype analysis (SI Appendix, Table S1), 10 of these samples were classified as indolent CLL and 10 as aggressive CLL. VH% indicates the percentage of homology between the observed sequence of IGHV in CLL samples and the reference IGVH sequence and only the samples with more than 98% homology were considered unmutated according to standard CLL classification (3). CLL samples were mostly (>95%) composed of CD5+/CD19+ B cells. Normal CD19+ B cell samples used for real-time RT-PCR and RNA sequencing were from frozen peripheral blood mononuclear cells (PBMCs) of four healthy donors described in ref. 37. RNA was extracted using TRIzol reagent (Thermo Fisher) following the standard protocol.
Real-time RT-PCR.
The expression of up-regulated HERVL_17p11.2b and down-regulated HUERSP3_11p15.1 was analyzed by real-time RT-PCR. The assay for HERVL_17p11.2b (Retro 1) was designed by custom TaqMan RNA assays (Thermo Fisher) as follows: Retro1_ex1_DIR2 (18-mer) GCCCCGGAGGACAACGTG; Retro1_ex2_REV2 (19-mer); TGCGTGGTCACCTGGAGAT.
Retro1_Probe (17-mer) GGACAGGGGCACTGCAG. Assay ID: APYMUFA.
Assay name: Retro1. Cat number: 4331348. For HUERSP3_11p15.1, we used the following assay:
Gene: NCR3LG; Gene ID: 374383; TaqMan Probe#: Hs02340611_m1. The data were normalized
using TaqMan assay TBP, catalog # Hs00427620 (Thermo Fisher). All real-time RT-PCR experiments were carried out using standard Thermo Fisher protocols.
RNA Sequencing.
Samples with acceptable RNA quality (RIN > 7; assessed with Agilent 6000 RNA Nano chip) and quantity (200 ng or more; assessed with Invitrogen Qubit High Sensitivity RNA assay) were used in library preparation. NEBNext® Ultra™ II Directional (stranded) RNA Library Prep Kit for Illumina (NEB #E7760L) was used as the core kit and coding genes were enriched using the NEBNext Poly (A) mRNA Magnetic Isolation Module (NEB #E7490). RNA fragmentation time: 10 min. PCR cycles: 12. NEBNext Multiplex Oligos for Illumina Unique Dual Index Primer Pairs (NEB #6446/L) were used to index each sample for multiplexing purposes. Final libraries were quantified using Qubit High Sensitivity DNA assay and pooled to yield ~20 million clusters/sample. mRNA-seq libraries were sequenced using Illumina NovaSeq 6000 flow cell with Paired-End 100 bp format to ~20 million clusters/sample.
Preprocessing of the RNA-Seq Data for HERV Analysis.
Raw sequencing reads in FASTQ format were quality trimmed, and adapters were removed using Trim Galore (v.0.6.6) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were then mapped to the human genome (HG38 assembly) using Bowtie2 (v. 2.4.5) (38), allowing up to 100 alignments per read (-k 100). Afterward, the mapped reads in SAM format were converted into BAM format, sorted (for coordinates), and indexed using Samtools (v. 1.6) (39). Finally, HERV quantification was performed using Telescope (v. 1.0.3) (33) with its GTF annotation file containing ~15,000 largely intact HERVs (small HERV fragments are not reported). The generated raw read count matrices were then used as input for all the downstream analyses presented in this study (see the following sections). The same methods as previously described have been used to preprocess the RNA-Seq data of the external publicly available CLL cohort (NCBI BioProject ID: PRJNA488107) (34).
Differential HERV Expression Analysis.
To perform the differential HERV expression analysis, we first scaled the raw read counts via RPM and filtered out low-expressed HERVs, whose geometric mean value was less than one across all samples. Afterward, the raw read counts of the retained HERVs were log2-transformed leveraging Voom and then used for the differential expression analysis, leveraging the Limma R package (v3.52.4) (40). HERVs with a |Log2FC| > 0.58 (|Linear FC| > 1.5) and an adjusted P-value (Benjamini-Hochberg correction) < 0.05 were considered differentially expressed. The plots generated for showing the results of the differential HERV expression analysis, such as volcano plots, heatmaps, Venn diagrams, and circos plots, have been drawn using EnhancedVolcano (v.1.16.0) (41), pheatmap (v.1.0.12) (https://cran.r-project.org/web/packages/pheatmap/index.html), InteractiVenn (42), and circlize (v.0.4.15) (43), respectively. Regarding the heatmaps, they were built by using the scaled expression (RPM) of HERVs. The same methods as previously described have been used to identify the differentially expressed HERVs from the RNA-Seq data of the external publicly available CLL cohort (NCBI BioProject ID: PRJNA488107) (34).
Preprocessing of the RNA-Seq Data for Gene Analysis.
Raw sequencing reads in FASTQ format were quality trimmed, and adapters were removed using Trim Galore (v0.6.6) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were then mapped to the human genome (HG38 assembly) using HISAT2 (v. 2.1.0) (44). Afterward, the mapped reads in SAM format were converted into BAM format, sorted for coordinates, and indexed using samtools (v.1.6) (39). Sorted BAM files were finally used as input for featureCounts (v.2.0.0) (45) in order to count the mapped reads to the gene coordinates reported in the GTF annotation file downloaded from GENCODE (v.41).
Differential Gene Expression Analysis.
To perform the differential gene expression analysis, we first scaled the raw read counts via RPM and filtered out low-expressed genes, whose geometric mean value was less than one across all samples. Afterward, the raw read counts of the retained genes were log2-transformed leveraging the Voom function and then used for the differential expression analysis, leveraging the Limma R package (v3.52.4) (40). Genes with a |Log2FC| > 0.58 (|Linear FC| > 1.5) and an adjusted P-value (Benjamini-Hochberg correction) < 0.05 were considered differentially expressed.
HERV-Specific Mechanistic Network-Based Pathway Analysis.
To predict the effects of the dysregulated HERVs identified in the aggressive and indolent forms of CLL on signaling pathways, we performed a mechanistic network-based topological pathway analysis using MITHrIL (35). For each differentially expressed HERV, we used the list of its correlated differentially expressed genes (identified as described in the previous paragraph) with their Entrez IDs and respective Log2FC values as input for MITHrIL (35). Only the differentially expressed genes with a P-value < 0.05 (corresponding to an average of |R | =0.73 for both aggressive and indolent forms) were considered correlated for that specific HERV and used for the HERV-specific pathway analysis. The heatmaps showing the pathways analysis results were generated by the ComplexHeatmap R package (46) using the Corrected Accumulator values generated by MITHrIL (35).
Correlation Network Analysis for Hallmark Genes in CLL.
The differentially expressed HERVs identified from the aggressive and indolent forms of CLL were correlated with the hallmark gene of CLL. HERV and gene raw read count matrices were normalized using the RPM formula to scale the raw library sizes. Subsequently, we correlated the HERV expression (Pearson correlation) with the hallmark gene of CLL. Only the genes correlated with that specific HERV with a P-value < 0.05 (corresponding to an average of |R | =0.74 and |R | =0.73 for aggressive and indolent forms, respectively) were used for the correlation networks. Finally, correlation networks between HERVs and selected genes have been generated by Cytoscape (v3.9.1) (47) for both CLL forms.
Statistical Analysis.
The Wilcoxon signed-rank test was employed in Figs. 2C and 3 C and D, while the Wilcoxon rank-sum test was used in Fig. 4B. The analyses were performed by the R stats package (v.4.2.2).
CLL Sample Treatments with Ibrutinib.
Frozen CLL samples were thawed, plated into RPMI media with 20% FBS, and treated with 10um ibrutinib for 24 h and 48 h. Total RNA was extracted using TRIzol reagent (Thermo Fisher) following the standard protocol, and real-time RT-PCR was carried out as described above.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Acknowledgments
This work was supported by National Cancer Institute Grants R35-CA197706 (to C.M.C.), by The Ohio State University Comprehensive Cancer Center, and by the NIH under grant number P30 CA016058, and a grant from Chronic Lymphocytic Leukemia (CLL) Global Research Foundation (to C.M.C.). We thank the Genomic Shared Resource at The Ohio State University Comprehensive Cancer Center, Columbus, OH for the RNA-Seq. We thank Dr. Emanuela Ghia (University of California, San Diego) for providing CLL sample information.
Author contributions
A.L.F., G.N., Y.P., and C.M.C. designed research; A.L.F., G.N., L.T., A. Palamarchuk, V.B., L.R., P.N.T., T.K., and Y.P. performed research; A.L.F., G.N., S.A., and A. Pulvirenti analyzed data; and A.L.F., G.N., Y.P., and C.M.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: J.M.C., Tufts University; and I.R., Thomas Jefferson University.
Contributor Information
Yuri Pekarsky, Email: pekarsky.yuri@osumc.edu.
Carlo M. Croce, Email: carlo.croce@osumc.edu.
Data, Materials, and Software Availability
RNA-Seq data have been deposited in NCBI GEO (GSE233157). Previously published data were used for this work (34).
Supporting Information
References
- 1.Pekarsky Y., Zanesi N., Croce C. M., Molecular basis of CLL. Semin. Cancer Biol. 20, 370–376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullrich F., Croce C. M., Molecular biology of chronic lymphocytic leukemia. Basic and Clinical Oncology 2, 9–32 (2001). [Google Scholar]
- 3.Cheson B. D., Chronic Lymphocytic Leukemia: Epidemiological, Familial, and Genetic Aspects. CLL 2, 1–30 (2001). [Google Scholar]
- 4.Kipps T. J., et al. , Chronic lymphocytic leukaemia. Nat. Rev. Dis. Primers 3, 16096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pekarsky Y., Croce C. M., Noncoding RNA genes in cancer pathogenesis. Adv. Biol. Regul. 71, 219–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang-Johanning F., et al. , Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 68, 5869–5877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhardwaj N., Coffin J. M., Endogenous retroviruses and human cancer: is there anything to the rumors? Cell Host. Microbe 15, 255–259 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Tavakolian S., Goudarzi H., Faghihloo E., Evaluating the expression level of HERV-K env, np9, rec and gag in breast tissue. Infect. Agent. Cancer 14, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin X., et al. , The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci. Adv. 5, eaat9820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curty G., et al. , Human endogenous retrovirus K in cancer: A potential biomarker and immunotherapeutic target. Viruses 12, 726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golkaram M., et al. , HERVs establish a distinct molecular subtype in stage II/III colorectal cancer with poor outcome. npj Genom. Med. 6, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecuelle J., et al. , MER4 endogenous retrovirus correlated with better efficacy of anti-PD1/PD-L1 therapy in non-small cell lung cancer. J. Immunother. Cancer 10, e004241 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y., et al. , Screening and identification of human endogenous retrovirus-K mRNAs for breast cancer through integrative analysis of multiple datasets. Front. Oncol. 12, 820883 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manca M. A., et al. , HERV-K and HERV-H Env proteins induce a humoral response in prostate cancer patients. Pathogens 11, 95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., et al. , CDK2 inhibition enhances antitumor immunity by increasing IFN response to endogenous retroviruses. Cancer Immunol. Res. 10, 525–539 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Pan D., et al. , SETDB1 restrains endogenous retrovirus expression and antitumor immunity during radiotherapy. Cancer Res. 82, 2748–2760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko E.-J., et al. , Effect of human endogenous retrovirus-K env gene knockout on proliferation of ovarian cancer cells. Genes Genomics 44, 1091–1097 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Mao J., et al. , TERT activates endogenous retroviruses to promote an immunosuppressive tumour microenvironment. EMBO Rep. 23, e52984 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng K. W., et al. , Antibodies against endogenous retroviruses promote lung cancer immunotherapy. Nature 616, 563–573 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Ferlita A., et al. , Transcriptome analysis of human endogenous retroviruses at locus-specific resolution in non-small cell lung cancer. Cancers 14, 4433 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandi N., Tramontano E., Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front. Immunol. 9, 2039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jern P., Coffin J. M., Effects of retroviruses on host genome function. Annu. Rev. Genet. 42, 709–732 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Wildschutte J. H., et al. , Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. U.S.A. 113, E2326–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burn A., Roy F., Freeman M., Coffin J. M., Widespread expression of the ancient HERV-K (HML-2) provirus group in normal human tissues. PLoS Biol. 20, e3001826 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holloway J. R., Williams Z. H., Freeman M. M., Bulow U., Coffin J. M., Gorillas have been infected with the HERV-K (HML-2) endogenous retrovirus much more recently than humans and chimpanzees. Proc. Natl. Acad. Sci. U.S.A. 116, 1337–1346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montesion M., Bhardwaj N., Williams Z. H., Kuperwasser C., Coffin J. M., Mechanisms of HERV-K (HML-2) transcription during human mammary epithelial cell transformation. J. Virol. 92, e01258-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhardwaj N., Montesion M., Roy F., Coffin J. M., Differential expression of HERV-K (HML-2) proviruses in cells and virions of the teratocarcinoma cell line Tera-1. Viruses 7, 939–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes J. F., Coffin J. M., Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. U.S.A. 101, 1668–1672 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triviai I., et al. , Endogenous retrovirus induces leukemia in a xenograft mouse model for primary myelofibrosis. Proc. Natl. Acad. Sci. U.S.A. 111, 8595–8600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deniz Ö., et al. , Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat. Commun. 11, 3506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel K., et al. , Identification of differentially expressed human endogenous retrovirus families in human leukemia and lymphoma cell lines and stem cells. Front. Oncol. 11, 637981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monne M., et al. , Expression profiles of human endogenous retrovirus in chronic myeloid leukemia at diagnosis and after TKI therapy. Blood 140, 12175–12177 (2022). [Google Scholar]
- 33.Bendall M. L., et al. , Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput. Biol. 15, e1006453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastore A., et al. , Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL. Nat. Commun. 10, 1874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alaimo S., et al. , Post-transcriptional knowledge in pathway analysis increases the accuracy of phenotypes classification. Oncotarget 7, 54572–54582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansz N., Faulkner G. J., Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 22, 147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pekarsky Y., et al. , Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 5071–5076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., et al. , 1000 Genome Project Data Processing Subgroup, The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie M. E., et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blighe K., Rana S., Lewis M., EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version (2018). https://github.com/kevinblighe/EnhancedVolcano.
- 42.Heberle H., Meirelles G. V., da Silva F. R., Telles G. P., Minghim R., InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 16, 169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu Z., Gu L., Eils R., Schlesner M., Brors B., Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Data Availability Statement
RNA-Seq data have been deposited in NCBI GEO (GSE233157). Previously published data were used for this work (34).