Abstract
Background: The nck-associated protein 1 (NCKAP1) of the disulfidptosis-related gene is essential in programmed cell death. However, a comprehensive analysis of the biological significance of NCKAP1 in pan-cancer is lacking.
Methods: Gene expression matrices and clinical expression information of cancers were obtained from The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEX) databases. A comprehensive analysis of NCKAP1 expression, biological function, gene mutation, immune cell infiltration, DNA methylation, and drug sensitivity profiles in pan-cancer was performed using the Timer2.0, HPA, GEPIA, STRING, cBioPortal, UALCAN and CellMiner databases. The prognostic value of NCKAP1 was investigated based on COX regression analysis and the Kaplan-Meier(K-M) curves. A nomogram was established to verify the clinical value of NCKAP1 for LUAD. The correlation between NCKAP1 and immune cells and signaling pathways were investigated by single-sample gene set enrichment analysis(ssGSEA). Validation was performed using PCR, Western Blot (WB), and Transwell assays.
Result: Significant differences in expression levels, mutation levels, and methylation levels of NCKAP1 between tumor and normal samples. NCKAP1 affects the prognosis of various cancers. NCKAP1 is strongly associated with microsatellite instability (MSI) and tumor mutational burden (TMB). The Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses indicate that NCKAP1 is strongly associated with cell death and tumor immunity. The expression of NCKAP1 affects the sensitivity to various drugs. Moreover, NCKAP1 is an independent predictor of prognosis in LUAD patients. The results of ssGSEA showed that elevated NCKAP1 expression was positively correlated with multiple immune-related signaling pathways. PCR analysis showed that the expression of NCKAP1 was increased in LUAD cells. Transwell invasion assay showed that overexpression of NCKAP1 resulted in enhanced invasion of LUAD cells.
Conclusions: We comprehensively analyzed the relationship between NCKAP1 and pan-cancer and its potential clinical value. NCKAP1 could be a potential immune marker for various cancers (especially LUAD), providing new insights and insights for cancer therapy.
Keywords: Pan-cancer, immune, Lung adenocarcinoma, prognosis, biomarkers, Transwell.
Introduction
In most countries, cancer is the leading or second leading cause of death before age 70. Cancer incidence and mortality rates have continued to increase over the past few decades, representing a significant public health challenge worldwide 1, 2. Comprehensive research into common hallmarks of cancer has developed rapidly in recent years, and pan-cancer studies are widely used to explore mechanisms of cancer development and potential biomarkers, which can significantly impact cancer therapy and improve its prognosis 3-6.
Lung cancer ranks second among all cancers and first in mortality, with approximately 2.227 million new lung cancer cases and nearly 1.796 million deaths related to lung cancer in 2020 1. An important reason for this high mortality rate is that more than half of patients have intermediate to advanced lung cancer at the time of initial diagnosis when treatment options are limited and the prognosis is poor 7, 8. Thus, searching for potential immune checkpoints is crucial for treating and diagnosing lung cancer.
In recent years, cell death-related genes have been used to find cures for cancer. For example, the concept of ferroptosis has been widely studied since its introduction in 2012 9. Ferroptosis is an iron-dependent form of regulated cell death progression characterized by excessive lipid peroxidation and subsequent plasma membrane rupture. The association between ferroptosis and cancer has been thought to exist since its inception and has been studied for many years. This observation has been progressively applied to tumor immunotherapy and other areas 9, 10. A recent study has proposed a new mode of cell death, disulfidosis, which refers to cell death in cells with high SLC7A11 expression due to glucose starvation, resulting in the collapse of disulfide bonds of actin cytoskeletal proteins and actin. This mode of death differs from apoptosis and ferroptosis 11, 12. Preclinical results show that metabolic treatment with glucose transporter inhibitors can cause sulfation and inhibit cancer growth 13. In addition, previous studies have demonstrated that cellular disulfide metabolism is disordered following oxidative stress, which may impact cancer cell survival and proliferation 14, 15. Therefore, it is reasonable to believe disulfidptosis is also closely linked to cancer.
In addition to SLC7A11, which plays a vital role in disulfidosis, NCKAP1 is also a central gene in disulfidosis. The deletion of NCKAP1 inhibits cell death like SLC7A11 and attenuates cellular sulfide metabolism 11.
Therefore, we comprehensively analyzed the differential expression of NCKAP1 in pan-cancer and its impact on the prognosis of pan-cancer. It also explores how NCKAP1 affects the prognosis of pan-cancer from the gene and immune levels. In addition, we performed drug sensitivity analysis based on NCKAP1 to guide the clinical application of drugs. We found that LUAD had positive results in several studies of NCKAP1, so we focused on analyzing the association between NCKAP1 and LUAD and demonstrated that NCKAP1 has the potential to be a therapeutic target for LUAD. Overall, NCKAP1 is expected to open up new pathways for the treatment of a variety of cancers.
Materials and Methods
Data Download
Clinical information and sequencing data for 33 common tumors were obtained from the TCGA database. We also downloaded the TCGA-GTEx data for these tumors to complete the study from UCSC XENA (https://xenabrowser.net/datapages/).
Expression Analysis of NCKAP1
The expression profiles of NCKAP1 in tumor and normal samples were compared by TIMER2.0 (http://timer.cistrome.org/) 16. Because some tumors did not have normal samples for comparison, we processed and visualized the TCGA-GTEx database sequencing data using the "ggplot2" package within the "R" software package. Protein expression levels of NCKAP1 in normal tissue and corresponding tumor tissue were obtained from the Human Protein Atlas database (https://www.proteinatlas.org). We also downloaded NCKAP1 Immunohistochemistry (IHC) images of tumor cells and the corresponding normal cells.
Prognostic and Survival Analysis
The significance of NCKAP1 in predicting pan-cancer progression-free interval (PFI) was explored using batch univariate COX regression analysis. Visualize the results with the "forestploter" package in the "R" software. With the help of "Survival" and "SurvMiner" packages in "R" software, we divided the samples into two groups based on the median NCKAP1 expression: a high-expression group and a low-expression group. Survival curves (based on Overall survival [OS]) for various tumors were plotted using Kaplan-Meier curves.
Correlation of the infiltration of immune cells with NCKAP1
Using the TIMER 2.0 tool, we investigated the correlation between NCKAP1 expression and immune cell infiltration in different tumor tissues using computational methods such as Timer, EPIC, MCPcounter, CIBERSORT, QUANTISEQ, xCELL, and TIDE. It is worth noting that these algorithms have counted differences in results due to their different working principles. The analysis of the tumor microenvironment receives the influence of multiple factors (different databases, different tumors, differences in detection methods, etc.). These algorithms are applied in different scenarios. TIMER is suitable for estimating the infiltration of multiple immune cell types in tumor tissues and their relationship with tumor development 17. The EPIC algorithm mainly estimates tumor samples' purity and impurity content, providing a basis for subsequent immune cell analysis 18. The MCPcounter algorithm is suitable for estimating the number of different immune and non-immune cell types in tumor tissues and exploring their relationship with tumor progression 19. CIBERSORT is used to analyze the relative abundance of multiple immune cell types in tumor tissues 20. The QUANTISEQ algorithm estimates the abundance of multiple immune cell types in tumor tissues with some flexibility 21. The xCell is suitable for estimating the relative number of multiple immune and non-immune cell types in tumor tissue 22. The TIDE algorithm is mainly used to predict the response to tumor immunotherapy 23.
Functional Enrichment Analysis
To validate the biological function of NCKAP1, we obtained the 100 genes most closely related to NCKAP1 (in cancer) from the GEPIA database (http://gepia2.cancer-pku.cn). The 100 genes were subsequently studied by GO and KEGG analysis. P-values < 0.05 were considered statistically significant. The "ggplot" package of "R" software visualized the results. We also obtained the top 20 proteins with the highest correlation with NCKAP1 from the String database (https://cn.string-db.org), and the results were constructed into a network using Cytoscape software. We aim to elucidate the function of NCKAP1 through these genes and proteins. The functional analysis results of these two gene collections can also be validated against each other to make the findings more accurate.
The mutation level of the NCKAP1 and promoter methylation analysis
A comprehensive analysis of NCKAP1 mutations in 10,967 samples from the TCGA database was performed using the cBioPortal database (https://www.cbioportal.org/). We compared the values of NCKAP1 methylation in normal and tumor samples using the UALCAN tool (https://ualcan.path.uab.edu/index.html).
Relationship between NCKAP1 expression and TMB, MSI
We calculated the TMB of each tumor using the "tmb" function of the "maftools" package and obtained the MSI scores of the tumors from the study of Russell Bonneville et al. 24. Correlations between NCKAP1 and TMB, MSI were also analyzed separately. The results are listed in Supplementary Table S1. We then visualized the results with the "fmsb" package of "R" software.
Drug sensitivity analysis of NCKAP1
"NCI-60 compound activity data" and "RNA-seq expression profiles" were downloaded from the CellMiner database to analyze the drug sensitivity of NCKAP1 in pan-cancer. The data were processed using the "impute" and "limma" packages in the "R" package, and drug sensitivity was calculated using Pearson correlation analysis. And |cor|>0.3 and P<0.05 were screening conditions. Finally, the results were visualized by "ggplot2" and "ggpubr".
Correlation analysis of NCKAP1 and immune cells and signaling pathways in LUAD
The TCGA-LUAD cohort contained a total of 598 samples. Of these, 539 were tumor samples. 59 were normal samples. To investigate the role of NCKAP1 in immunity to LUAD. We divided 539 tumor samples into two groups (high and low expression groups) based on the median NCKAP1 expression values of the 539 samples. Then, we downloaded the immune-related CELLMAKER and signaling pathways from the GSEA website. And ssGSEA analysis was performed on the two groups with the help of the "GSEABase" and "GSVA" packages in the "R" package. We then calculated the correlation between NCKAP1 expression and several immune checkpoint inhibitors (PDCD1, PDCD2, CD47, CD86, CD276, and CTLA4) in the TCGA-LUAD cohort and visualized the results using the "circlize" package.
Cell cultuCorrelation analysis of NCKAP1 with clinical factors
We used univariate and multivariate COX regression analyses to analyze the effects of various clinical factors and NCKAP1 expression on the prognosis of LUAD patients. The nomogram and calibration curves were then constructed using the "rms" and "survival" packages.
Cell culture
We purchased Lung epithelial cells (BEAS-2B) and three lung adenocarcinoma cell lines (A549, CALU-3, HCC78), all from Hunan Fenghui Biotechnology Co., cells were cultured on DMEM medium (Gibco, USA) supplemented with 10ml of fetal bovine serum (FBS) (Gibco, USA). The medium was placed in a 37°C, 5% CO2 incubator for stationary culture to reach 80-90% density.
Reverse Transcription-Polymerase Chain Reaction
RNA was extracted from these four cells using the Redzol reagent (Sebasun, Beijing, China). Synthesis of cDNA using a reverse transcription kit (Toyobo Biotech Co., Ltd., Shanghai, China), and then the mRNA expression level was detected by iQ5 Real-Time PCR (Applied Biosystems, USA). The PCR reaction program was (95°C for 60 s, then 95°C for 15 s, 60°C for 60 s, 40 cycles). All PCR experiments were repeated three times. Finally, all results were normalized by GAPDH. Relative expression levels for these genes were calculated using 2 -ΔΔCt. The primer sequences were: GAPDH, F-5′-TCAGCAATGCCTCCTGCAC-3′, R-5′-TCTGGGTGGCAGTGATGGC-3′. NCKAP1, F-5′-TGCTGTAGAAACCCGCAACA-3′, R-5′-TCTGGGTGGCAGTGATGGC-3′.
Plasmid Transfection Experiment and Western Blot Analysis
The human NCKAP1 cDNA-containing plasmid was sourced from Shanghai Genechem Co., Ltd. (Shanghai, China). The plasmid was prepared using the pUC ori replicon. Cells were seeded in 6-well plates when the cell density was greater than 80%. Liposome 2000 was used for transfection and subsequently incubated in a 5% CO2 incubator. Transfection efficiency was assessed by Western Blot assay after 48 hours. Cellular proteins were extracted with RIPA lysate. The extracted proteins and markers were separated by electrophoresis with the help of SDS-PAGE gel (Western Protein Marker I: G2086, Servicebio, China). The protein blot was transferred onto a PVDF membrane (Microporous, USA). It was then blocked with 5% skimmed milk. Primary antibodies were incubated at 4 °C for 12 hours (NCKAP1 antibody: ab126061, GAPDH antibody: ab9485, Abcam, UK). The membranes were washed four times with TBST and incubated with a secondary antibody for 1.5 hours at room temperature (Goat Anti-Rabbit IgG H&L (HRP), Abcam, UK). The cell membranes were placed in Western LightningTM Chemiluminescent Reagent Chromogen (PerkinElmer, USA) for 30 seconds and then immediately placed in an exposure cassette, and finally scanned and imaged with an Epson Perfection V39 scanner, and then analyzed for the expression of each group of protein bands by Image-J software.
Transwell Invasion experiment
The BD matrix gel was thawed at 4 °C overnight. It was mixed with serum-free medium at 1:8, and 60 μL of diluted matrix gel was aspirated and added to the upper chamber of the Transwell (24-well plate cell chamber of Corning Incorporated, USA). Then, they were incubated in an incubator (37°C, 5% CO2) for 3 hours. The excess liquid in the upper chamber was then aspirated. It was then placed in the incubator for 30 min to hydrate the basement membrane. 500 μL of medium containing 10% fetal bovine serum was added to the lower chamber of the 24-well plate. Then, 200 μL of cell suspension was inoculated into the upper chamber. After 48 h of incubation in the incubator, 4% paraformaldehyde was added to the 24-well plate, and the bottom surface of the chambers was immersed in the solution for 15 min to complete fixation. Excess liquid was subsequently aspirated, and the bottom surface of the chambers was immersed in 0.1% crystal violet for 5 minutes to stain. After cleaning the chamber, the cells were observed under a microscope, counting the top, bottom, right, left and center of the membrane and taking the average.
Statistical Analysis
The "R" version of the software used in the paper is version 4.3.0. The statistical result P<0.05 was considered statistically significant. (* for p < 0.05, ** for p < 0.01, *** for p < 0.001).
Result
Tumor abbreviations and corresponding full names are shown in Figure 1.
Figure 1.
Abbreviations and full names of tumors.
Expression of NCKAP1
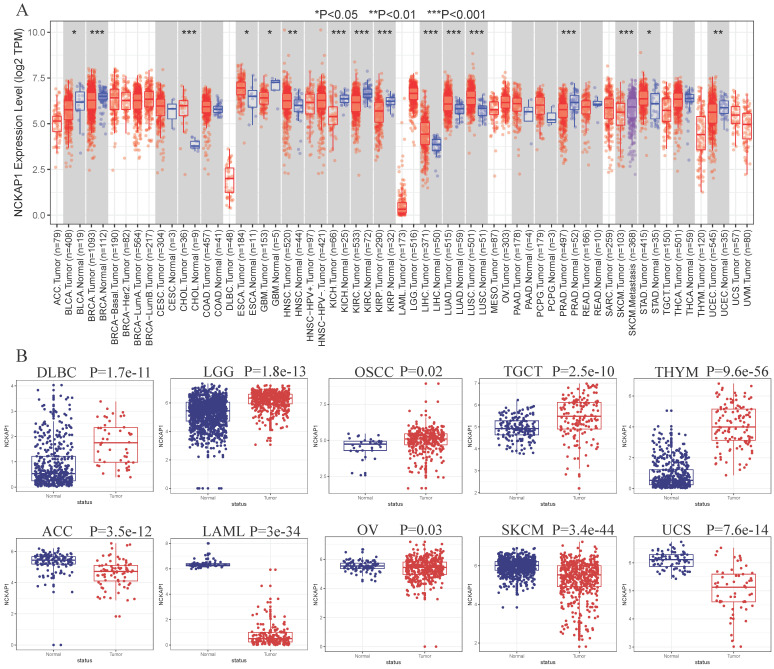
NCKAP1 expression in tumor and normal samples is shown in Figure 2A-B. Based on the results, we know that NCKAP1 expression levels were higher in these tumors than in the normal samples, including CHOL, ESCA, HNSC, LIHC, LUAD, LUSC, STAD, DLBC, TGCT, LGG, OSCC, and THYM. In contrast, NCKAP1 was expressed at lower levels in these tumors than in normal samples such as BLCA, BRCA, GBM, KICH, KIRC, KIRP, PRAD, UCEC, ACC, SKCM, LAML, OV, UCS.
Figure 2.
(A) Expression levels of NCKAP1 in pan-cancerous tissues. (B) Differential expression of NCKAP1 in TCGA-GTEX database in multiple cancers (DLBC, LGG, OSCC, TGCTT, THYM, ACC, LAML, OV, SKCM, UCS).
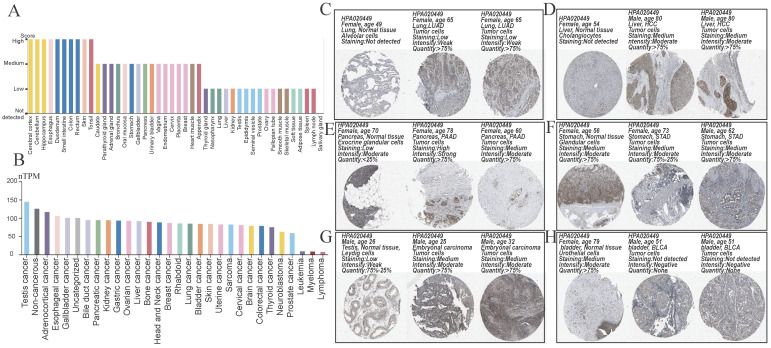
We obtained a gradient map of NCKAP1 expression levels in normal tissues and its expression levels in various cancers from the HPA database. NCKAP1 is highly expressed in the brain, esophagus, and intestine (Figure 3A). Among the cancers, the highest levels of NCKAP1 were in the testicular, gallbladder, and esophageal cancers (Figure 3B). IHC images were obtained from the HPA database. Lung adenocarcinoma, hepatocellular carcinoma, pancreatic carcinoma, and testicular carcinoma showed higher staining than the corresponding normal samples (Figure 3C, D, E, G). The staining degree of urothelial carcinoma was lower than that of normal tissues (Figure 3H). In contrast, staining in gastric cancer did not differ significantly from normal tissue (Figure 3F). This result is consistent with our analysis above.
Figure 3.
(A) Expression of NCKAP1 in normal tissues. (B) Expression of NCKAP1 in cancer. (C-H) IHC images in multiple cancers and their adjacent tissues.
Prognostic analysis
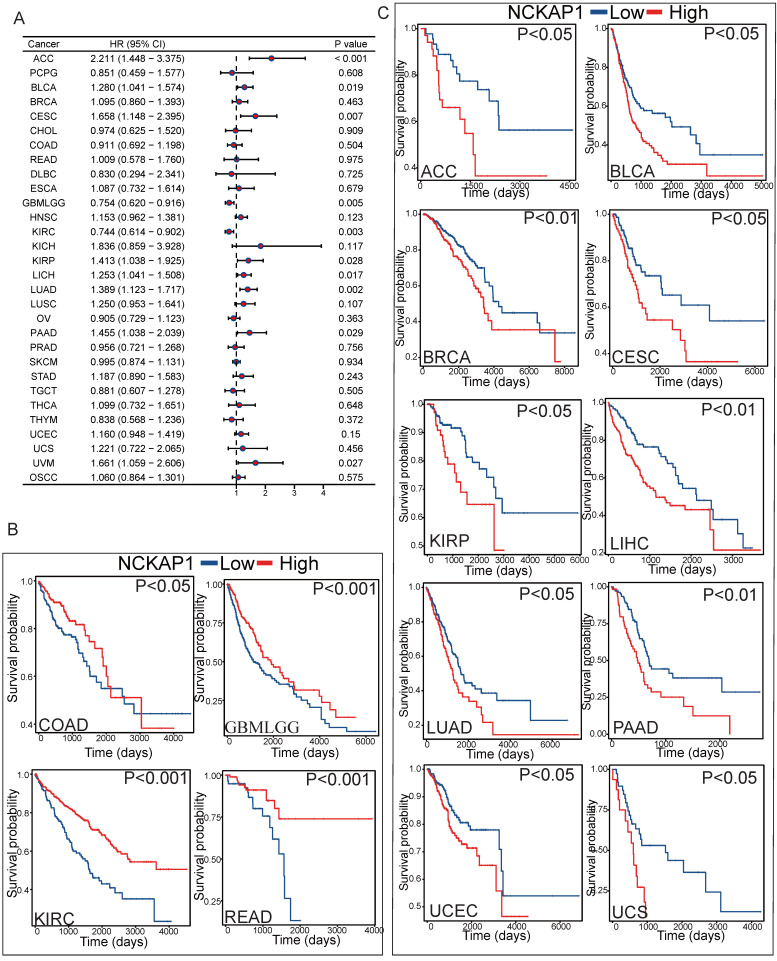
To further investigate the relationship between NCKAP1 and tumor prognosis. The relationship between NCKAP1 expression and PFI in pan-cancer patients was investigated by COX regression analysis. NCKAP1 was associated with a good prognosis in GBMLGG and KIRC. In contrast, NCKAP1 was a poor prognostic factor for ACC, BLCA, CESC, KIRP, LICH, LUAD, PAAD, and UVM (Figure 4A). We then plotted the difference in OS between the NCKAP1 high- and low-expression groups and showed that the low-expression group had a better prognosis in COAD, KIRC, BMLGG, and READ patients (Figure 4B). The high expression of NCKAP1 resulted in lower OS in ACC, BLCA, CESC, KIRP, LIHC, LUAD, BRCA, PAAD, UCEC, and UCS samples (Figure 4C). These findings suggest that NCKAP1 does indeed have potential as a prognostic marker. However, in some tumors, there was no clear correlation between the expression level of NCKAP1 and the survival time (Supplementary Figure S1).
Figure 4.
(A) Forest plot showing the results of univariate Cox regression of NCKAP1 on PFI in TCGA pan-cancer. (B-C) Kaplan-Meier curves for NCKAP1 high/low-risk groups in COAD, GBMLGG, KIRC, READ, ACC, BLCA, BRCA, CESC, KIRP, LIHC, LUAD, PAAD, UCEC, UCS.
Correlation between immune cells and NCKAP1
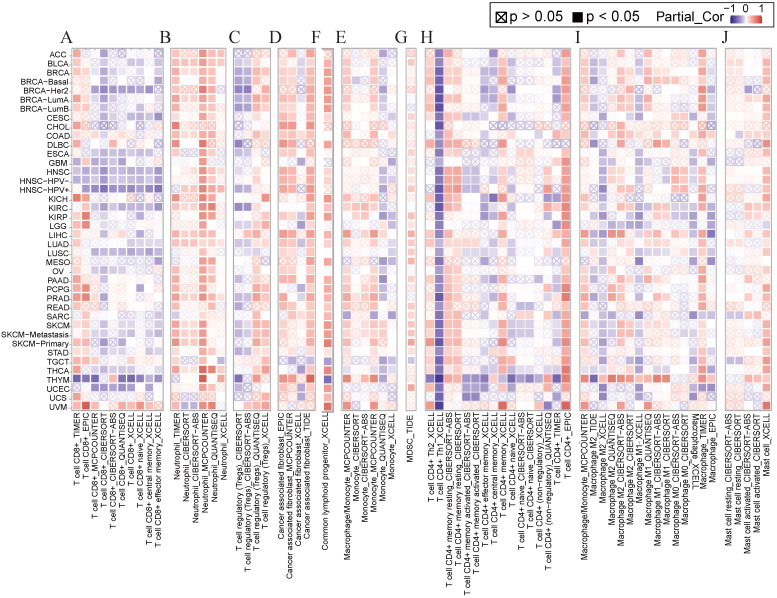
Several studies have shown that immune cell infiltration into the tumor immune microenvironment is strongly associated with tumorigenesis initiation, progression, and prognosis 25, 26. We applied various algorithms such as EPIC, CiberSort, timer, and XCELL to investigate the relationship between NCKAP1 and immune cells including CD8+ T cells, neutrophils, T cell regulatory cells (Tregs), tumor-associated fibroblasts (CAF), common lymphoid progenitor cells (CLP), monocytes, myeloid suppressor cells (MDSC), CD4+ T cells, macrophages, and mast cells. In general, NCKAP1 was positively correlated with the infiltration of a variety of immune cells, such as neutrophils, CAF, CLP, monocytes, MDSC, CD4+ T cells, and giant cells. NCKAP1 was also found to be negatively correlated with CD8+ T cells (Figure 5). However, the results of different algorithms also varied, and this result needs to be verified in a larger sample.
Figure 5.
(A-J) Relationship of NCKAP1 with CD8+ T cells, neutrophils, Treg, CAF, CLP, monocytes, MDSC, CD4+ T cells, macrophages, and mast cells in pan-cancer analyzed by multiple algorithms obtained from TIMER2.0.
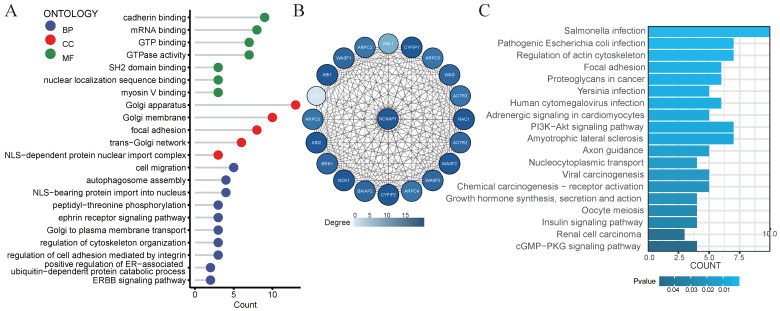
GO analysis and the KEGG analysis
Consider that NCKAP1 is a crucial gene for disulfidptosis. We decided to explore the biological functions of the NCKAP1 gene. We obtained the 100 genes with the most significant correlation with NCAKP1 from GEPIA2 (Supplementary Table S2). We performed GO and KEGG analyses on these 100 genes. The results of GO analysis suggested that NCKAP1 was primarily enriched in calcineurin binding, myosin V binding, regulation of cytoskeletal organization, ERBB signaling pathway, ephrin receptor signaling pathway, and so on (Figure 6A). In contrast, KEGG analysis showed that NCKAP1 was mainly enriched in the regulation of actin cytoskeleton, focal adhesion, and proteoglycans in cancer, PI3K-Akt signaling pathway, viral carcinogenesis, chemical carcinogenesis - receptor activation, renal cell carcinoma, cGMP-PKG signaling pathway and other aspects (Figure 6C). In light of this result, NCKAP1 may be known to play an essential role in the cytoskeleton in regulating disulfidptosis. In addition, it is closely linked to various signaling pathways, inflammation, and cancer progression. In addition, we obtained the PPI networks of the top 20 proteins relevant to NCKAP1 from the STRING database. We visualized the results using Cytoscape software (Figure 6B) and performed GO and KEGG analysis on the 20 genes, and the results matched our functional analysis of NCKAP1 above (Supplementary Table S3). These observations suggest that the level of NCKAP1 expression may indeed influence cell death and cancer progression.
Figure 6.
(A) Results of GO analysis of the 100 genes most associated with NCKAP1 expression in pan-cancer. (B) PPI network of the 20 most relevant proteins of NCKAP1. (C) Results of KEGG analysis of the 100 genes most associated with NCKAP1 expression in pan-cancer.
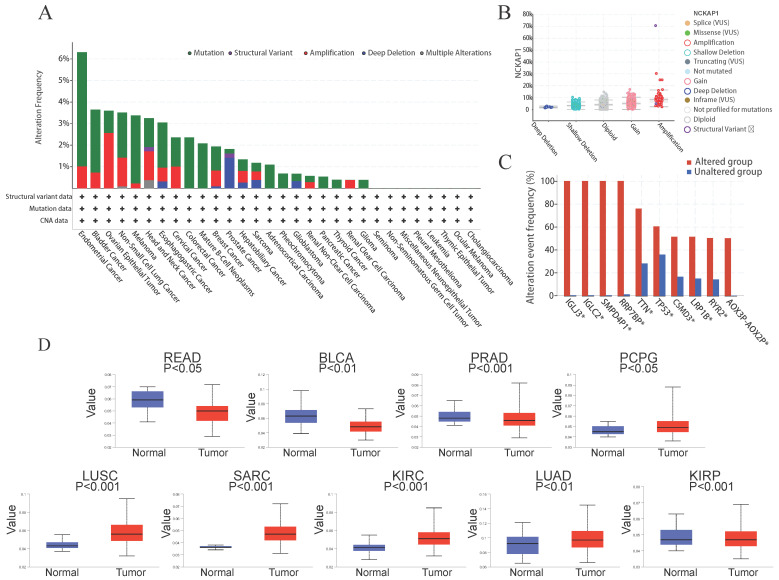
The mutation level of the NCKAP1
Many studies have found that the progression of many cancers is caused by genetic mutations 27. So, we explored the relationship between NCKAP1 and pan-cancer at the gene level. Using the cBioPortal software, we analyzed mutations in 10 967 samples from the TCGA database and found that mutations in NCKAP1 are indeed found in various cancers. In endometrial cancer, the mutation rate of NCKAP1 is even more than 6%. Mutations in this gene were found in over 3% of bladder cancer, ovarian epithelial tumors, non-small-cell lung cancer, melanoma, head and neck cancer, and esophagogastric cancer (Figure 7A). Notably, the primary forms of mutations in NCKAP1 were amplification and gain (Figure 7B). Alterations in genes such as IGLJ3, IGLC2, SMPD4P1, RRP7BP, TTN, TP53, CSMD3, LRP1B, RYR2, AOX3P-AOX2P were more common in the altered group than in the unaltered group (Figure 7C). This situation suggests that NCKAP1 may act with other genes in tumor progression, which we will analyze specifically in the follow-up.
Figure 7.
(A) Summary of NCKAP1 mutations in TCGA pan-cancer data. (B) Mutation types of NCKAP1 in pan-cancer. (C) Comparison of the frequency of associated gene alterations in the NCKAP1 altered and unaltered groups. (D) Promoter methylation levels of NCKAP1 in READ, BLCA, PRAD, PCPG, LUSC, SARC, KIRC, LUAD, and KIRP.
Several studies have demonstrated that methylation of the DNA promoter can affect transcriptional processes and be involved in tumor development 28. Therefore, we compared the promoter methylation levels of NCKAP1 in tumor and normal tissues. Consistent with the results shown in Figure 7D, NCKAP1 promoter methylation levels in READ, BLCA, and PRAD were lower than in the normal samples. Moreover, the levels of NCKAP1 promoter methylation were higher in the PCPG, SARC, LUSC, KIRC, LUAD, and KIRP samples than in the normal samples, and these differences were statistically significant (P < 0.05). These results suggest that the expression of NCKAP1 in cancer may be receiving the influence of promoter methylation. While comparing some tumors and corresponding normal samples, the methylation levels of NCKAP1 were also not significantly different (Supplementary Figure S2).
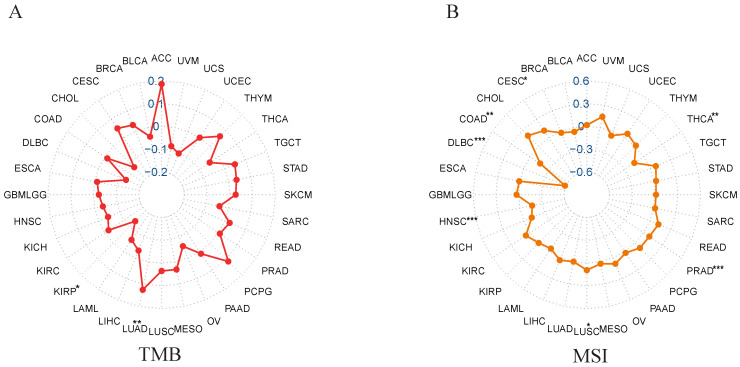
Correlation of NCKAP1 with TMB and MSI
We analyzed the correlation between NCKAP1 and MSI/TMB in pan-cancer. From the correlation analysis between NCKAP1 and TMB, we learned that the expression of NCKAP1 had a positive correlation with LUAD (P<0.01), While NCKAP1 was found to have a negative correlation with KIRP (P<0.05) (Figure 8A). Based on Figure 8B, we observed a positive correlation between NCKAP1 expression and CESC (P<0.05) and LUSC (P<0.05). In contrast, NCKAP1 expression was negatively correlated with the following seven tumors: COAD (P<0.01), PRAD (P<0.001), HNSC (P<0.001), THCA (P<0.01), DLBC (P<0.001). Specific results are in Supplementary Table S1. These results suggest that NCKAP1 may influence cancer progression at the mutational level and potentially provide ideas for immunotherapy of tumors (specifically discussed later).
Figure 8.
(A) Correlation between NCKAP1 and TMB in pan-cancer. (B) Correlation between NCKAP1 and MSI in pan-cancer.
Drug sensitivity of NCKAP1
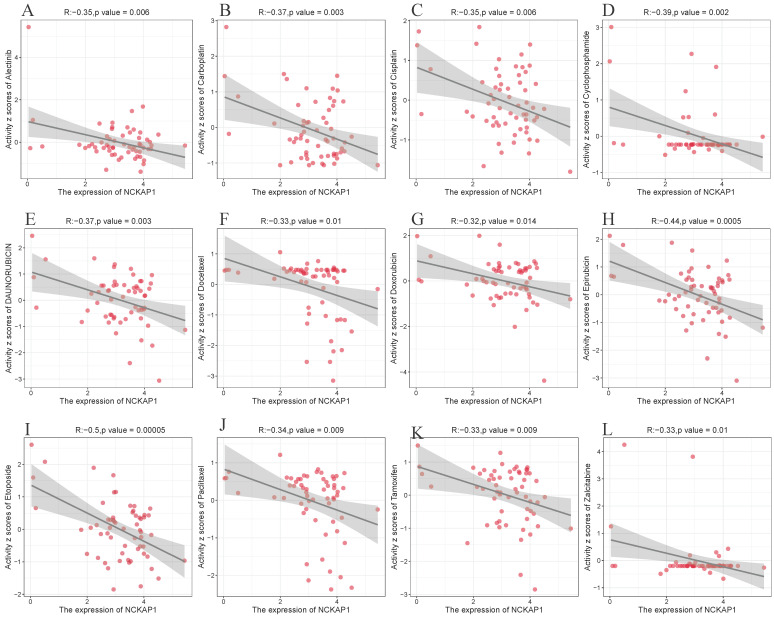
We explored its clinical value after determining that NCKAP1 is closely associated with cancer. We analyzed NCKAP1 with multiple drug sensitivities based on the CellMiner database. After using |cor|>0.3 and P<0.05 as screening criteria, we found 67 drugs with a high degree of association with NCKAP1. Of these 67 drugs, 64 were negatively correlated with NCKAP1, and three were positively correlated (Irofulven, Kahalide F, Trametinib). Specific results are in the Supplementary Table S4. We chose to visualize the results of several drugs commonly used for chemotherapy in the clinic. NCKAP1 was negatively correlated with Alectinib, Carboplatin, Cisplatin, Cyclophosphamide, Daunorubicin, Docetaxel, Doxorubicin, Epirubicin, Etoposide, Paclitaxel, Tamoxifen, and Zalcitabine (Figure 9). These findings suggest that NCKAP1 expression profiling can guide physicians in selecting chemotherapeutics in the clinic.
Figure 9.
(A-L) NCKAP1 in pan-cancer with drug sensitivity (Alectinib, Carboplatin, Cisplatin, Cyclophosphamide, Daunorubicin, Docetaxel, Doxorubicin, Epirubicin, Etoposide, Paclitaxel, Tamoxifen, and Zalcitabine) analysis.
Detailed analysis of NCKAP1 and LUAD
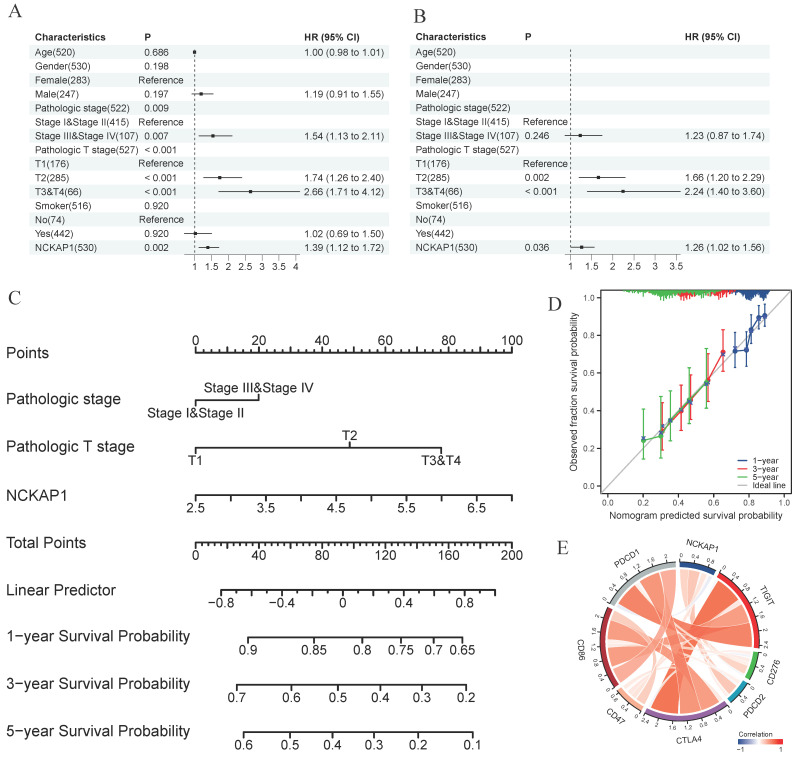
In our pan-cancer study, NCKAP1 was strongly linked to LUAD. NCKAP1 is highly expressed in LUAD, leading to a poorer prognosis. To determine whether NCKAP1 could be a therapeutic target for LUAD. We performed univariate and multivariate COX regression analyses of age, gender, pathological stage, T-stage, smoking or not, and NCKAP1 expression levels in LUAD patients. The results showed that NCKAP1 was indeed a factor that independently influenced the prognosis of NCKAP1 (P<0.05) (Figure 10A, B). To further investigate the role of NCKAP1 in clinical evaluation, we constructed a nomogram based on NCKAP1, pathological staging, and T-staging (Figure 10C). We validated the nomogram by calibration curves. The ideal line of the calibration curves graph was our predicted survival of LUAD patients; we found that the 1-, 3-, and 5-year survival curves of the patients almost overlapped with our predicted curves (Figure 10D), which indicated that the performance of the column line graph we built based on NCKAP1 was good. So, we considered that NCKAP1 could be a potential therapeutic target for LUAD. We first analyzed the relationship between NCKAP1 and some important therapeutic targets (PDCD1, PDCD2, CD47, CD86, CD276, and CTLA4) and found that these genes were mainly positively correlated in LUAD (Figure 10E).
Figure 10.
Results of the univariate COX regression analysis (A) and the multivariate COX regression analysis (B) were performed for the TCGA-LUAD cohort. (C) A Nomogram was constructed based on the results of multivariate COX regression analysis. (D)Calibration chart to verify the accuracy of the Nomogram. (E) Correlation between NCKAP1 and suppressive immune checkpoints.
The results of ssGSEA
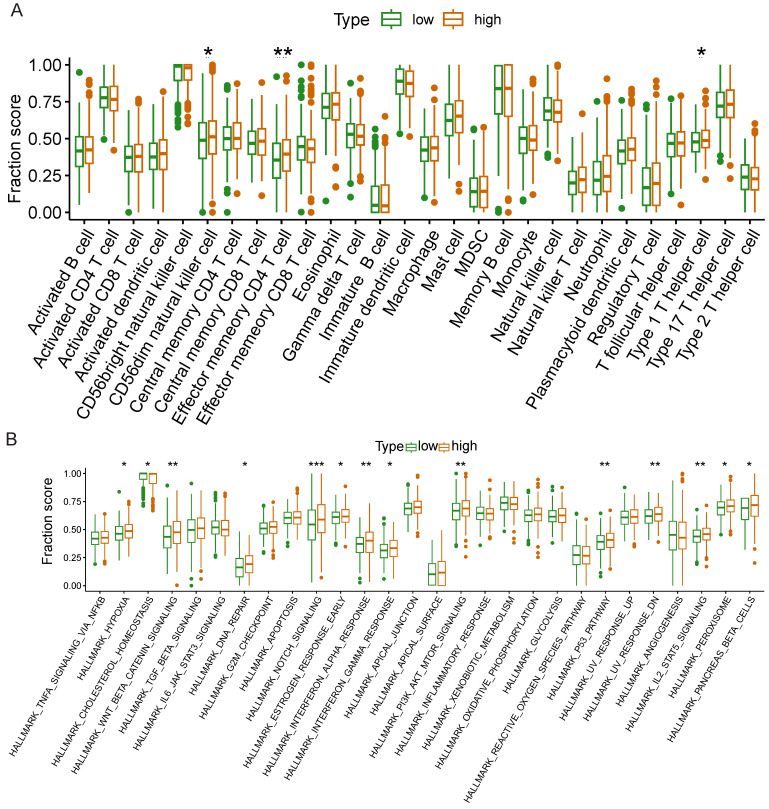
We further investigated whether NCKAP1 could be used as a therapeutic target of NCKAP1 by the relationship of NCKAP1 in immune cells and signaling pathways in LUAD. Based on median NCKAP1 expression values, we divided 539 TCGA-LUAD patients into high-expression and low-expression groups. We performed ssGSEA analysis to study the correlation between NCKAP1 and immune cells and signaling pathways. From Figure 11A, we found that the high-expression group had higher enrichment scores in cd56dim natural killer cells, effector memory CD4+ T cells, and type 1 T helper cells. These immune cells play an essential role in the anti-tumor effects of LUAD. This result suggests that high NCKAP1 expression and tumor immune escape may be related. The results of immune-related signaling pathways showed that the high expression group had higher enrichment scores in these signaling pathways (hallmark_hypoxia, hallmark_wnt_beta_catenin_signaling, hallmark_dna_repair, hallmark_notch_signaling, hallmark_estrogen_signaling, and hallmark_notch_signaling, hallmark_estrogen_response_early, hallmark_interferon_alpha_response, hallmark_interferon_gamma_response, hallmark_pi3k_akt_mtor_signaling, hallmark_p53_pathway, hallmark_uv_response_dn, hallmark_il2_ stat5_signaling, hallmark_peroxisome, hallmark_pancreas_beta_cell) (Figure 11B). These findings suggest that NCKAP1 can influence LUAD progression via multiple signaling pathways.
Figure 11.
Differences in immune cells(A) and immune-related signaling pathways(B) between high and low expression groups of NCKAP1.
Results of the Experimental Validation
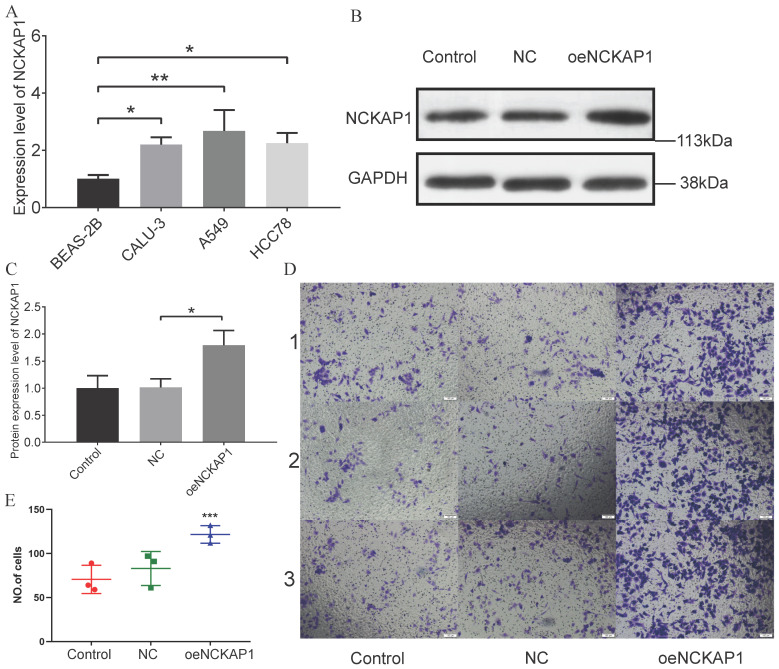
Based on the results of PCR experiments, we found that the NCKAP1 did have a high mRNA expression level in lung adenocarcinoma cells (A549, CALU-3, and HCC78) (Figure 12A). The PCR data are shown in Supplementary Table S5. Then, we randomly divided the A549 cells into three groups. We did not perform any treatment on A549 cells in the first group, named the “Control group.” A blank plasmid was added to the second group of A549 cells, termed the “NC group.” In the third group, we transfected A549 cells with the plasmid that caused NCKAP1 overexpression and named it “oeNCKAP1 group”. WB experiments verified our transfection efficiency, and the expression of NCKAP1 in the oeNCKAP1 group was higher than in the other two groups (Figure 12B, C). A transwell invasion assay was then carried out on these three groups of cells. The images of the experimental results were also organized and placed in Figure 12D (magnified 100 under the microscope). The statistical results of the experiment are shown in Figure 12E. As can be seen, the invasion ability of A549 cells in the oeNCKAP1 group was significantly increased. The results of these experiments are consistent with our findings above.
Figure 12.
(A) PCR validation of NCKAP1 expression in four cells (BEAS-2B, A549 cells, CALU-3, and HCC78). (B-C) WB results for Control, NC, and oeNCKAP1. Experimental (D) and statistical (E) results of the Transwell invasion test.
Discussion
Altered metabolism is an essential hallmark of cancer progression, which often leads to a high dependence of cancer cells on specific nutrients or metabolic pathways that are often breakthroughs in cancer therapy. For example, cuproptosis, a copper-dependent regulation of cell death, is a mode of cell death induced by the toxic stress of tricarboxylic acid (TCA) cycle proteins in mitochondria, which has been progressively used in the treatment and prognosis of many cancers 29. Disulfidptosis is a novel form of cell death in which the signals leading to cell death are recognized by immune cells within the tumor to activate specific immune processes. It alters cellular and humoral immunity and affects the treatment and prognosis of cancer patients. Previous studies have shown that disulfide metabolism in cancer cells affects the biological processes of tumor metastasis and drug resistance 30, 31. And a number of studies have demonstrated the correlation between DRGs and cancers, for example, DRGs can be used in the clinical diagnosis of HCC to predict the prognosis and therapeutic goals 32. DRGs are also closely related to the treatment and prognosis of breast cancer 33.
NCKAP1 is a critical gene in disulfidptosis and regulates various processes, such as apoptosis, migration, and invasion, and plays a vital role in pathogenesis 34. Therefore, we focused on the expression of the NCKAP1 gene in various cancers to explore its potential as a potential therapeutic target.
We first investigated the expression of NCKAP1 in different tumors and normal tissues. The results showed that NCKAP1 was highly expressed in CHOL, ESCA, HNSC, LIHC, LUAD, LUSC, STAD, SLYC, DLBC, TGCT, LGG, OSCC and THYM. However, the expression of NCKAP1 was lower in the BLCA, BRCA, GBM, KICH, KIRC, KIRP, PRAD, UCEC, ACC, SKCM, LAML, OV, and UCS. Increased expression of NCKAP1 in ACC, BLCA, CESC, KIRP, LICH, LUAD, PAAD, and UVM abnormally predicted poor tumor prognosis. These results suggest that NCKAP1 may be essential in tumorigenesis and prognosis. Therefore, we need further investigate the function of NCKAP1 in various tumors.
The critical impact of the type and number of immune cells in the tumor immune microenvironment on cancer progression and prognosis is well recognized. In the current study, NCKAP1 was found to correlate with immune cells, and in particular, the negative correlation with CD8+ T cells sparked our interest. CD8+ T cells play an essential role in the immune response of tumors, and their massive infiltrates are usually considered to portend an improved prognosis 35, 36. Our results suggest that high expression of NCKAP1 may be associated with the suppression of CD8+ T cells. This point could also explain that aberrant expression of NCKAP1 leads to poorer prognosis in various cancers. Although our study provides important insights, we would also like to acknowledge the differences in immune infiltration correlations obtained by different algorithms, which may be influenced by the working mechanism of each algorithm and the purpose of the analysis. In addition, immune cell infiltration analysis is also influenced by different databases. However, it is noteworthy that our study observed a consistent trend in some immune cell types (CD8+ T cell, neutrophils, CAF, etc.), and the scope of application of different algorithms varied, which emphasizes the necessity of a multifaceted analysis. In summary, our study provides valuable clues for further investigation of the potential role of NCKAP1 in tumor immunoregulation. Despite some complexities and discrepancies, our findings remain scientifically essential and provide a solid foundation for more in-depth studies and clinical applications in the future.
We first performed GO and KEGG analysis on NCKAP1. The analysis showed that NCKAP1 is not only a critical gene for disulfidptosis, but it is also closely related to various signaling pathways (cGMP-PKG, ERBB signaling pathway, PI3K-Akt signaling pathway, etc.), inflammation, and cancer progression (viral oncogenesis, chemical oncogene receptor activation, renal cell carcinoma). For example, the PI3K-Akt signaling pathway can promote tumor cell proliferation and metastasis 37. It can also affect the activation and function of T cells, B cells, and macrophages. The abnormally activated PI3K-Akt signaling pathway may inhibit the activity of immune cells, leading to immune escape 38, 39. cGMP-PKG signaling pathway affects tumor immunity by influencing the activation status of immune cells 40. Thus, the multifunctionality of NCKAP1 in cancer and immunity suggests that it is expected to provide breakthroughs in cancer treatment and immunotherapy development.
We decided to study the relationship between NCKAP1 and cancer at the genetic level. Gene mutation and DNA methylation have become the focus of attention in recent years through in-depth cancer studies. Genetic mutations are closely linked to cancer's occurrence, development, and metastasis. For example, DNA replication should be considered a hallmark of cancer because it drives cancer progression 41; abnormal upregulation or downregulation of DNA methylation is also believed to contribute to cancer formation and progression or may serve as a marker of cancer development. Hypermethylated DNA is also prevalent in breast cancer 42. Our study found that promoter methylation levels of NCKAP1 differed in multiple cancers, combined with NCKAP1 differences in cancers. We suggest that the promoter methylation level influences the expression level of NCKAP1. It has been shown that DNA methylation can affect tumor development and biological characteristics by influencing gene transcription 43. The mutation rate of NCKAP1 in endometrial, bladder, ovarian epithelial, non-small cell lung, melanoma, head and neck, and esophageal cancers was more than 3%. In addition, we found that the mutation rates of IGLJ3, IGLC2, SMPD4P1, RRP7BP, TTN, TP53, CSMD3, LRP1B, RYR2, and AOX3P-AOX2P genes were also higher than those of the unaltered group after mutation of the NCKAP1 gene. This suggests that NCKAP1 mutations also affect the stability of other genes, thereby influencing tumor progression. Mutations in multiple genes may act synergistically in tumor progression, leading to more complex tumor characteristics. When the mutation rate of multiple genes is increased simultaneously, it indicates that these genes may be involved in common signaling pathways or biological processes. These results will help us gain insight into the effects of NCKAP1 mutations on tumorigenesis and growth. This finding prompted our decision to explore the association of NCKAP1 with TMB and MSI in pan-cancer.
TMB is usually defined as the number of genes mutated in the tumor genome. TMB is highly correlated with the efficacy of PD-1/PD-L1 inhibitors, and TMB can be used to predict the prognosis of patients 44. MSI is a feature reflecting microsatellite repeat instability in tumor DNA. As the relationship between TMB, MSI and tumors is complex, we conducted a correlation analysis of NCKAP1 with TMB and MSI to gain further insight into the immunotherapeutic potential of NCKAP1 in cancer. Correlation analysis of NCKAP1 expression with TMB revealed that NCKAP1 expression positively correlated with LUAD expression (P<0.01). This suggests that in LUAD, NCKAP1 may contribute to poor prognosis by promoting tumor growth and metastasis. So, NCKAP1 has potential as a therapeutic target for LUAD. However, in KIRP, NCKAP1 was negatively correlated with TMB (p<0.05), but high expression of NCKAP1 also led to poor prognosis in KIRP. This suggests a possible immune escape mechanism involved by NCKAP1 in KIRP, and we can target NCKAP1 to reverse immune tolerance and thus enhance tumor response to immunotherapy. In the correlation analysis between MSI and NCKAP1, we found that the expression of NCKAP1 was positively correlated with the expression of both CESC and LUSC. The expression of NCKAP1 was negatively correlated with COAD, PRAD, HNSC, THCA, and DLBC. These results suggest that NCKAP1 may differentially regulate immune features in different tumor types. Further studies will help to reveal the exact role and potential mechanisms of NCKAP1 in these tumors. These results provide helpful information for individualized immunotherapy and help optimize treatment strategies to improve the efficacy of immunotherapy. Therefore, we will focus on the relationship between NCKAP1 and the response to specific drug therapy.
Based on the CellMiner database, we sought to determine the drug sensitivity of NCKAP1 in cancer. Sixty-seven clinically used drugs significantly correlated with NCKAP1 (Supplementary Table S4). Three drugs showed positive correlations (Irofulven, Kahalide F, Trametinib), which may present a potential therapeutic opportunity. The other 64 drugs showed a significant pairwise negative correlation, including common anti-tumor drugs such as carboplatin, cisplatin, cyclophosphamide, and paclitaxel. Therefore, increased NCKAP1 expression leads to increased resistance to chemotherapeutic drugs in tumor patients. This study's results will help guide us in using clinical chemotherapeutic agents.
In these study species above, LUAD always had positive results. Therefore, we decided to investigate the relationship between NCKAP1 and LUAD further. After univariate and multivariate COX regression analyses, we determined that NCKAP1 was an independent prognostic predictor of LUAD. And based on NCKAP1, we established a nomogram to predict the 1-,3-, and 5- survival of LUAD patients. Moreover, NCKAP1 was positively correlated with common immunotherapy targets. These results suggest the feasibility of NCKAP1 as a therapeutic target for LUAD. We analyzed the relationship between NCKAP1 expression and immune cells and immune signaling pathways to explore this potential further. What intrigued our interest was that the NCKAP1 high-expression group had higher enrichment scores than the low-expression group in multiple signaling pathways. The enrichment of the high expression group in signaling pathways related to immune cell activation (e.g., PI3K-Akt-mTOR, IL-2-STAT5, etc.) suggests that it plays a role in promoting immune cell activation 45-47. Its enrichment in the Notch signaling pathway suggests that NCKAP1 has an essential role in regulating the immune pathway and may be involved in the differentiation and function of immune cells to influence the host immune response to tumors 48, 49. While hallmark_wnt_beta_catenin_ signaling, hallmark_dna_repair, and hallmark_hypoxia pathways also play essential roles in tumor progression species 50. For example, the Wnt/β-catenin signaling pathway is associated with cell proliferation, differentiation, and stem cell properties, and its aberrant activation has been linked to the development of various cancers 51. NCKAP1 may also play a role in tumor development and prognosis through these pathways. Enriching these signaling pathways may suggest that NCKAP1 may be an immunotherapeutic target for LUAD. By interfering with the function of NCKAP1, the activation status of immune cells could be adjusted, and the host immune response to tumors could be enhanced.
In conclusion, we investigated the differential expression and prognosis of NCKAP1 in pan-cancer. By comprehensively analyzing NCKAP1 in pan-cancer with respect to gene mutation, promoter methylation, and immune microenvironment, we can better understand the function of NCKAP1 in cancer. We also analyzed NCKAP1 drug sensitivity in tumors to guide clinical therapy. Finally, we demonstrate that NCKAP1 plays a role in tumor development and prognosis, with the potential to be a therapeutic target for various cancers. We also found NCKAP1 as an independent prognostic factor for LUAD and provided a new avenue for immunotherapy in LUAD.
Supplementary Material
Supplementary tables.
Acknowledgments
Funding
This study was funded by Henan Provincial Health Commission (LHGJ20190422), Henan Provincial Health Commission (LHGJ20210486) and the Key Scientific Item of Henan Province Education Department (21A320037).
Availability of data and materials
All data used in this work can be acquired from The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEX) databases.
Ethics approval and consent to participate
The patient data in this work is from publicly available patients consent to a complete data set without ethical approval.
Author contributions
Ankang Zhu and Yan Zong designed this work. Ankang Zhu performed the validation of the experiments, Yinuo Li and Yan Zong integrated and analyzed the data. Yan Fan, Shaodong Liu, Ankang Zhu wrote this manuscript. Xingcai Gao edited and revised the manuscript. All authors approved this manuscript.
Abbreviations
- TCGA
The Cancer Genome Atlas
- TMB
Tumor mutational burden
- NCKAP1
Nck-associated protein 1
- MSI
Microsatellite instability
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
- ssGSEA
Single-sample GSEA
- K-M
Kaplan-Meier
- WB
Western Blot
- OS
Overall survival
- IHC
Immunohistochemistry
- DRGs
Disulfidptosis-related genes
- PFI
Progression-free interval
- OS
Overall Survival
- Tregs
T cell regulatory cells
- CAF
Tumor-associated fibroblasts
- CLP
Common lymphoid progenitor cells
- MDSC
Myeloid suppressor cells
- LUAD
Lung adenocarcinoma
- TCA
Tricarboxylic acid
References
- 1.Sung H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):p.. doi: 10.3322/caac.21660. 209-249. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL. et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):p.. doi: 10.3322/caac.21708. 7-33. [DOI] [PubMed] [Google Scholar]
- 3.Saidak Z. et al. A pan-cancer analysis of the human tumor coagulome and its link to the tumor immune microenvironment. Cancer Immunol Immunother. 2021;70(4):p.. doi: 10.1007/s00262-020-02739-w. 923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z. et al. The Basic Characteristics of the Pentraxin Family and Their Functions in Tumor Progression. Front Immunol. 2020;11:p.. doi: 10.3389/fimmu.2020.01757. 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He J. et al. Intra-Tumoral Expression of SLC7A11 Is Associated with Immune Microenvironment, Drug Resistance, and Prognosis in Cancers: A Pan-Cancer Analysis. Front Genet. 2021;12:p.. doi: 10.3389/fgene.2021.770857. 770857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J. et al. A Comprehensive Prognostic and Immune Analysis of SLC41A3 in Pan-Cancer. Front Oncol. 2020;10:p.. doi: 10.3389/fonc.2020.586414. 586414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Canales J. et al. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat Res. 2016;170:p.. doi: 10.1007/978-3-319-40389-2_2. 25-46. [DOI] [PubMed] [Google Scholar]
- 8.Bade BC, Dela CC. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med. 2020;41(1):p.. doi: 10.1016/j.ccm.2019.10.001. 1-24. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X. et al. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):p.. doi: 10.1038/s41580-020-00324-8. 266-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X. et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):p.. doi: 10.1038/s41571-020-00462-0. 280-296. [DOI] [PubMed] [Google Scholar]
- 11.Liu X. et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25(3):p.. doi: 10.1038/s41556-023-01091-2. 404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S. et al. Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework. Front Endocrinol (Lausanne) 2023;14:p.. doi: 10.3389/fendo.2023.1180404. 1180404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng P. et al. Disulfidptosis: a new target for metabolic cancer therapy. J Exp Clin Cancer Res. 2023;42(1):p.. doi: 10.1186/s13046-023-02675-4. 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogg PJ, Biological regulation through protein disulfide bond cleavage. Redox Rep. 2002; 7(2): p. 71-7. [DOI] [PubMed]
- 15.Daly EB. et al. Secretion of phosphoglycerate kinase from tumour cells is controlled by oxygen-sensing hydroxylases. Biochim Biophys Acta. 2004;1691(1):p.. doi: 10.1016/j.bbamcr.2003.11.004. 17-22. [DOI] [PubMed] [Google Scholar]
- 16.Li T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):p.. doi: 10.1093/nar/gkaa407. W509-W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):p.. doi: 10.1158/0008-5472.CAN-17-0307. e108-e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Racle J, Gfeller D. EPIC: A Tool to Estimate the Proportions of Different Cell Types from Bulk Gene Expression Data. Methods Mol Biol. 2020;2120:p.. doi: 10.1007/978-1-0716-0327-7_17. 233-248. [DOI] [PubMed] [Google Scholar]
- 19.Becht E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):p.. doi: 10.1186/s13059-016-1113-y. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):p.. doi: 10.1038/nmeth.3337. 453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29(8):p.. doi: 10.1093/bioinformatics/btt090. 1083-5. [DOI] [PubMed] [Google Scholar]
- 22.Aran D. et al. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):p.. doi: 10.1186/s13059-017-1349-1. 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):p.. doi: 10.1038/s41591-018-0136-1. 1550-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonneville R, Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol. 2017. 2017. [DOI] [PMC free article] [PubMed]
- 25.Stenstrom J. et al. Regulatory T lymphocyte infiltration in metastatic breast cancer-an independent prognostic factor that changes with tumor progression. Breast Cancer Res. 2021;23(1):p.. doi: 10.1186/s13058-021-01403-0. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren L. et al. Systematic pan-cancer analysis identifies APOC1 as an immunological biomarker which regulates macrophage polarization and promotes tumor metastasis. Pharmacol Res. 2022;183:p.. doi: 10.1016/j.phrs.2022.106376. 106376. [DOI] [PubMed] [Google Scholar]
- 27.Boca SM. et al. Patient-oriented gene set analysis for cancer mutation data. Genome Biol. 2010;11(11):p.. doi: 10.1186/gb-2010-11-11-r112. R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q. et al. Gene body methylation in cancer: molecular mechanisms and clinical applications. Clin Epigenetics. 2022;14(1):p.. doi: 10.1186/s13148-022-01382-9. 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z. et al. Cuproptosis-Related Risk Score Predicts Prognosis and Characterizes the Tumor Microenvironment in Hepatocellular Carcinoma. Front Immunol. 2022;13:p.. doi: 10.3389/fimmu.2022.925618. 925618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong Y. et al. Nck-associated protein 1 associates with HSP90 to drive metastasis in human non-small-cell lung cancer. J Exp Clin Cancer Res. 2019;38(1):p.. doi: 10.1186/s13046-019-1124-0. 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon MR. et al. NCK-associated protein 1 regulates metastasis and is a novel prognostic marker for colorectal cancer. Cell Death Discov. 2023;9(1):p.. doi: 10.1038/s41420-023-01303-6. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T. et al. Disulfidptosis classification of hepatocellular carcinoma reveals correlation with clinical prognosis and immune profile. Int Immunopharmacol. 2023;120:p.. doi: 10.1016/j.intimp.2023.110368. 110368. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y. et al. Single-cell sequencing and bulk RNA data reveal the tumor microenvironment infiltration characteristics of disulfidptosis related genes in breast cancer. J Cancer Res Clin Oncol. 2023;149(13):p.. doi: 10.1007/s00432-023-05109-y. 12145-12164. [DOI] [PubMed] [Google Scholar]
- 34.Chen J. et al. NCKAP1 is a Prognostic Biomarker for Inhibition of Cell Growth in Clear Cell Renal Cell Carcinoma. Front Genet. 2022;13:p.. doi: 10.3389/fgene.2022.764957. 764957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:p.. doi: 10.1186/s40425-016-0165-6. 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M. et al. CD8(+) T Cell-Associated Gene Signature Correlates with Prognosis Risk and Immunotherapy Response in Patients with Lung Adenocarcinoma. Front Immunol. 2022;13:p.. doi: 10.3389/fimmu.2022.806877. 806877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng Y. et al. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front Oncol. 2022;12:p.. doi: 10.3389/fonc.2022.819128. 819128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J. et al. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18(1):p.. doi: 10.1186/s12943-019-0954-x. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:p.. doi: 10.1146/annurev-immunol-032712-095946. 675-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raingeaud J. et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270(13):p.. doi: 10.1074/jbc.270.13.7420. 7420-6. [DOI] [PubMed] [Google Scholar]
- 41.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:p.. doi: 10.1146/annurev-pathol-012414-040424. 425-48. [DOI] [PubMed] [Google Scholar]
- 42.Jackson K. et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol Ther. 2004;3(12):p.. doi: 10.4161/cbt.3.12.1222. 1225-31. [DOI] [PubMed] [Google Scholar]
- 43.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):p.. doi: 10.1016/j.cell.2013.03.008. 38-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samstein RM. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):p.. doi: 10.1038/s41588-018-0312-8. 202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vergadi E. et al. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J Immunol. 2017;198(3):p.. doi: 10.4049/jimmunol.1601515. 1006-1014. [DOI] [PubMed] [Google Scholar]
- 46.Solomon I. et al. CD25-T(reg)-depleting antibodies preserving IL-2 signaling on effector T cells enhance effector activation and antitumor immunity. Nat Cancer. 2020;1(12):p.. doi: 10.1038/s43018-020-00133-0. 1153-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones DM. et al. Dynamic Roles for IL-2-STAT5 Signaling in Effector and Regulatory CD4(+) T Cell Populations. J Immunol. 2020;205(7):p.. doi: 10.4049/jimmunol.2000612. 1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garis M, Garrett-Sinha LA. Notch Signaling in B Cell Immune Responses. Front Immunol. 2020;11:p.. doi: 10.3389/fimmu.2020.609324. 609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hopkins JL. et al. DNA repair defects in cancer and therapeutic opportunities. Genes Dev. 2022;36(5-6):p.. doi: 10.1101/gad.349431.122. 278-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jing X. et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18(1):p.. doi: 10.1186/s12943-019-1089-9. 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13(1):p.. doi: 10.1186/s13045-020-00990-3. 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.
Data Availability Statement
All data used in this work can be acquired from The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEX) databases.