Significance
Rett syndrome (RTT) is an X-linked neurodevelopmental disorder that predominantly affects girls. RTT is caused by loss of function variants in a single gene, MECP2. Girls with RTT develop normally during their first year of life, but then experience neurological abnormalities, including breathing and movement difficulties, loss of speech, and seizures. This study investigates the function of the MeCP2 protein in the brain and how MeCP2 activity is modulated by sensory experience in early life. Evidence is presented that sensory experience affects MeCP2 function and that this is required for synaptic pruning in the brain. These findings provide insight into MeCP2 function and clues as to what goes awry in the brain when the function of MeCP2 is disrupted.
Keywords: Rett syndrome, MeCP2, synapse refinement, LGN, phosphorylation
Abstract
Mutations in MECP2 give rise to Rett syndrome (RTT), an X-linked neurodevelopmental disorder that results in broad cognitive impairments in females. While the exact etiology of RTT symptoms remains unknown, one possible explanation for its clinical presentation is that loss of MECP2 causes miswiring of neural circuits due to defects in the brain’s capacity to respond to changes in neuronal activity and sensory experience. Here, we show that MeCP2 is phosphorylated at four residues in the mouse brain (S86, S274, T308, and S421) in response to neuronal activity, and we generate a quadruple knock-in (QKI) mouse line in which all four activity-dependent sites are mutated to alanines to prevent phosphorylation. QKI mice do not display overt RTT phenotypes or detectable gene expression changes in two brain regions. However, electrophysiological recordings from the retinogeniculate synapse of QKI mice reveal that while synapse elimination is initially normal at P14, it is significantly compromised at P20. Notably, this phenotype is distinct from the synapse refinement defect previously reported for Mecp2 null mice, where synapses initially refine but then regress after the third postnatal week. We thus propose a model in which activity-induced phosphorylation of MeCP2 is critical for the proper timing of retinogeniculate synapse maturation specifically during the early postnatal period.
Rett Syndrome (RTT) is an X-linked neurodevelopmental disorder caused by loss-of-function variants in methyl-CpG-binding protein 2 (MeCP2), a transcriptional repressor that binds to methylated cytosines across the genome (1, 2). Girls with RTT develop normally during the first year of life, then exhibit developmental regression during early childhood, characterized by breathing abnormalities, hand wringing, movement difficulties, seizures, loss of speech, and decelerated head growth (3). While the full etiology underlying the abnormal cognitive and social skills seen in RTT remains unclear, several lines of evidence suggest that synaptic and circuit defects contribute to these symptoms. For example, in mouse models of RTT, abnormalities in various aspects of synaptic development and transmission were observed, including defects in the maturation of excitatory synapses (4), disruption of long-term synaptic plasticity in the hippocampus (5, 6), and altered synaptic scaling (7, 8). Notably, the emergence of RTT symptoms, in both mice and humans, coincides with periods when spontaneous neural activity and sensory experience profoundly influence nervous system development (9, 10). Moreover, mouse models of RTT or MeCP2 duplication syndrome harboring Mecp2 loss- or gain-of-function mutations, respectively, exhibit a myriad of defects in activity-dependent developmental processes, including dendritic arborization, the maturation of dendritic spines, and synapse elimination (4, 11–13).
Numerous studies in neurons have shown that the MeCP2 protein becomes newly phosphorylated in response to neuronal activity (14–16). Our laboratory and others have identified multiple sites of activity-induced MeCP2 phosphorylation and dephosphorylation, spanning the methyl-DNA-binding domain (MBD), transcriptional repressor domain (TRD), and C-terminal domain (CTD) of MeCP2 (17–22). A variety of the effects of the association of MeCP2 with methylated DNA and the NCoR corepressor complex have been ascribed to these post-translational modifications. However, while various Mecp2 knock-in mouse lines abolishing individual phosphorylation events have been reported to exhibit behavioral and cellular phenotypes (18, 21, 23), a clear understanding of the contribution of these phosphorylation events to the functions of MeCP2 remains elusive.
Here, we sought to further assess the contribution of activity-induced phosphorylation to MeCP2 function. By mass spectrometry, we confirmed the sites of activity-dependent MeCP2 phosphorylation and then generated a quadruple knock-in (QKI) mouse line in which the four major sites of inducible MeCP2 phosphorylation are mutated to alanines so that the MeCP2 protein is refractory to activity-induced phosphorylation. QKI mice are viable, grossly normal, and exhibit none of the well-characterized RTT phenotypes, making it possible to assess the function of activity-dependent MeCP2 phosphorylation without the confounding effects observed in Mecp2 null mice. We hypothesized that QKI mice might be specifically defective in steps of activity-dependent brain development, as these would likely be stages when MeCP2 would be subject to activity-induced phosphorylation. To this end, we focused our assessment on the refinement of synapses between retinal ganglion cells (RGCs) and thalamocortical (TC) relay neurons in the dorsal lateral geniculate nucleus (dLGN), during a developmental window when many convergent retinal inputs are eliminated, while the remaining inputs are strengthened. Previous studies have shown that this synapse elimination and strengthening is both activity- and MeCP2-dependent (13, 24, 25). We find that, as with Mecp2 null mice (13), developmental refinement of the synapses between RGCs and TC neurons in the dLGN is impaired in QKI mice, with more RGC inputs to each relay neuron in QKI mice. Notably, this disruption is observed earlier in QKI mice, between the ages of postnatal day (P)14 and P20, compared to Mecp2 null mice, which show aberrant refinement after P20. Thus, the timing of the synaptic defect in QKI mice suggests that activity-dependent phosphorylation of MeCP2 is critical during the spontaneous activity-dependent phase of retinogeniculate synaptic refinement.
Results
Characterization of Activity-Induced Sites of MeCP2 Phosphorylation.
We initially sought to assess activity-responsive MeCP2 post-translational modifications, both in vitro and in vivo, to confirm and extend previous investigations using cultured neurons (18) and epileptic rodent brains (21). To this end, we injected wild-type (WT) mice with either a saline control or kainic acid (KA) to induce seizures and then harvested hippocampal tissue after 1 h. In parallel, cultured WT mouse cortical neurons were silenced with the voltage-gated sodium channel antagonist tetrodotoxin and the NMDA receptor antagonist AP5 overnight, then either left untreated as a control or harvested 30 min following treatment with either brain-derived neurotrophic factor (BDNF, 50 ng/mL) or elevated levels of extracellular potassium chloride (KCl, 55 mM) to induce membrane depolarization. In each case, MeCP2 protein was immunoprecipitated and subjected to quantitative mass spectrometry to evaluate changes in post-translational modifications.
Consistent with the results of a previous study, which used phosphotryptic mapping of MeCP2 from 32P labeled neurons to identify sites of MeCP2 phosphorylation (18), this analysis showed induction of MeCP2 phosphorylation at serine (S) 86, S274, and S421 in response to the various stimuli (Fig. 1A). Similarly, stimulus-induced phosphorylation of the MeCP2 peptide containing residues threonine (T) 308 and T311 was also observed. Although quantitative mass-spectrometry is unable to distinguish between phosphorylation at these two nearby residues, activity-induced MeCP2 T308 phosphorylation has been previously reported and validated with an MeCP2 phospho-T308-specific antiserum (18), strongly suggesting that the detected phospho-peptide species corresponds to phospho-T308. In addition to these four sites, MeCP2 S424 phosphorylation has been previously reported in epileptic mouse brain extracts (21). While we detected S424 phosphorylation in cultured cortical neurons following KCl-mediated depolarization, this modification was not consistently induced in the hippocampus in response to KA. Likewise, phosphorylation at MeCP2 S80, which has been previously reported to undergo rapid dephosphorylation in response to activity (21), was detected in our dataset but did not show alterations upon stimulation either in vitro or in vivo. Finally, our analysis did not reveal evidence of stimulus-induced changes in other types of MeCP2 post-translational modifications, such as lysine acetylation or arginine methylation. We note that some variability was observed in MeCP2 phosphorylation in our hippocampal samples that might be explained by animal-to-animal variability in activity-dependent responses to KA (26).
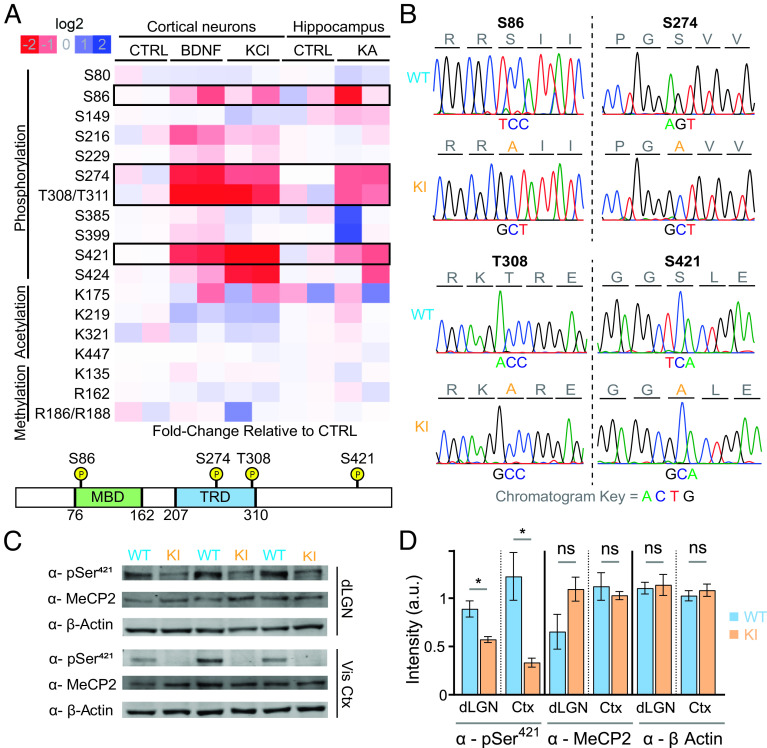
Fig. 1.
Activity-dependent modifications of MeCP2 and generation of MeCP2 quadruple knock-in (QKI) mice. (A) Top: Quantitative assessment of stimulus-responsive MeCP2 post-translational modifications. Neuronal activity was induced in vivo with injection of KA for 1 h or saline (control) in mice, after which hippocampi were dissected. Cultured mouse cortical neurons were either left untreated (control) or stimulated at in vitro day 7 with BDNF (50 ng/mL) or KCl (55 mM) for 30 min to induce depolarization. After induction, MeCP2 protein was isolated using immunoprecipitation, after which samples underwent quantitative mass spectrometry to assess post-translational modifications. Phosphorylation at residues S86, S274, T308/T311, and S421 of MeCP2 were found to be induced by neuronal activity. Bottom: Schematic depiction of the MeCP2 protein showing the characterized methyl-DNA-binding domain (MBD) and transcriptional repression domain (TRD) as well as the location of the four sites of activity-responsive phosphorylation targeted for mutation. (B) Sanger sequencing confirms the expected base substitutions in QKI mice. (C) Western blotting of dLGN and visual cortex from P27-P32 mice with the indicated antisera confirms the loss of MeCP2 S421 phosphorylation in QKI mice without affecting the total levels of MeCP2 protein (n = 3 mice). Equal amounts of protein lysates from WT and QKI brain tissue were loaded on a denaturing protein gel, followed by western blotting with an MeCP2-S421 phospho-specific antiserum, an antibody against total MeCP2, and an antibody specific for β-actin as a loading control. (D) Quantification of western blots shown in (C). Levels of MeCP2 phosphorylated-S421 are significantly reduced in QKI dLGN and visual cortex compared to WT animals (Left). In contrast, levels of total MeCP2 protein are unaffected in QKI mice in both dLGN and visual cortex (Middle) (n = 3 mice). Protein levels in each lane were normalized to the corresponding β-actin control (Right). *P < 0.05, unpaired, two-tailed Student’s t test.
While we cannot exclude the presence of other modifications not detected by these methods, our mass spectrometry findings indicate that neuronal stimulation drives the acute phosphorylation of MeCP2 predominantly at four previously reported sites. Intriguingly, three of these sites lie in well-characterized domains important for MeCP2 function—S86 lies in the MBD of MeCP2 and could thus influence MeCP2–chromatin interactions, whereas S274 and T308 both lie in the TRD, where phosphorylation of T308 has been previously shown to disrupt MeCP2 association with the NCoR complex in in vitro peptide pulldown experiments (18). While single knock-in mice preventing phosphorylation of S421 or T308 have been generated previously and been shown to have phenotypic differences compared to WT mice (18, 23), the apparent coordinate regulation of these four sites suggests that they may act in concert to modulate neuronal MeCP2 function. We therefore sought to evaluate the effect of mutating these four phosphorylation sites together.
Generation of MeCP2 QKI Mice.
To coordinately abolish activity-induced MeCP2 phosphorylation at S86, S274, T308, and S421, we generated mice in which all four of these sites are converted to alanines (MeCP2 S86A, S274A, T308A, S421A quadruple knock-in, or QKI, mice) using homologous recombination techniques (23). Sanger sequencing of genomic tail DNA from the resulting animals was used to confirm the presence of the four targeted sequence changes (Fig. 1B).
Because relatively subtle changes in the level of MeCP2 protein can give rise to significant abnormalities in nervous system development (27), we performed quantitative western blotting in two distinct brain regions using a total MeCP2 antibody to confirm that MeCP2 protein levels are not significantly altered in QKI mice. Additionally, we used an MeCP2 phospho-S421-specific antiserum to confirm that S421 phosphorylation is significantly decreased in both dLGN and visual cortex lysates. We detected a more pronounced loss of phospho-S421 in the visual cortex than in the dLGN, perhaps reflecting a higher level of MeCP2 protein expression in the dLGN and partial recognition of unphosphorylated MeCP2 by our phospho-S421 antibody (Fig. 1 C and D). We also found that the absence of MeCP2 phosphorylation in QKI mice did not significantly affect MeCP2 methyl-DNA binding in vivo, as assayed by chromatin immunoprecipitation (ChIP) sequencing (SI Appendix, Fig. S1A). Consistent with the finding that total MeCP2 protein levels and binding are unchanged, QKI mice are viable, grossly normal, and have normal body weights (SI Appendix, Fig. S1B). QKI mice exhibited none of the overt RTT behavioral phenotypes typically seen with decreased MeCP2 protein levels, as judged by an established phenotypic scoring method that measures mobility, gait, hindlimb clasping, tremor, breathing, and general body condition (28) (SI Appendix, Fig. S1C). Additionally, Mecp2 null mice exhibited premature lethality at postnatal day 54 (29), whereas QKI mice show no significant difference in longevity from WT mice for up to 1 y (n = 10 WT, 12 QKI) (SI Appendix, Fig. S1D). Thus, the lack of these four phosphorylation events is not sufficient to give rise to pronounced RTT phenotypes, alter MeCP2 protein expression, or affect MeCP2 binding to methyl-DNA.
Developmental Synaptic Refinement Is Aberrant in the dLGN of QKI Mice.
While prior studies have implicated MeCP2 in synaptic regulation, the pleiotropic nature of RTT phenotypes has made it difficult to determine whether MeCP2 acts as a direct regulator of activity-driven circuit refinement or whether the synaptic and cognitive deficits observed in RTT mouse models reflect secondary effects of a broader form of neuronal dysfunction. Given the absence of characteristic RTT phenotypes in QKI mice, we went on to assess potential defects in postnatal activity-dependent circuit development in these animals. For these studies, we focused on the retinogeniculate synapses formed between RGCs and TC relay neurons of the dLGN, which undergo a well-characterized, sequential process of vision-insensitive (P0 to P20) and vision-sensitive (P20 to P30) refinement over the course of early postnatal development (10, 24, 30–33). We note that while mice eyes first open at P14, the lens does not become fully transparent until around P18, which may explain why the vision-sensitive phase of retinogeniculate synapse refinement does not begin until P20 (34, 35). Importantly, previous work has shown that the process of vision-sensitive synaptic refinement is disrupted in Mecp2 null mice (13).
To directly investigate the role of activity-induced MeCP2 phosphorylation in this refinement process, we performed whole-cell recordings from TC neurons in acute dLGN slices of QKI and WT littermates at P27-P32 (hereafter referred to as P30 for simplicity), an age after the bulk of synaptic remodeling has occurred, and when there are clear defects in retinogeniculate connectivity in Mecp2 null mice (13, 24, 25, 36) (Fig. 2). Using established methods, inward excitatory postsynaptic currents (EPSCs) at −70 mV and outward EPSCs at +40 mV were recorded in a voltage clamp configuration in response to extracellular electrical stimulation of the RGC axons that make up the optic tract. We held each recorded cell at these two potentials to isolate inward AMPA receptor (AMPAR)-mediated EPSCs at −70 mV and outward EPSCs at +40 mV, comprising a fast AMPAR-mediated current and a slow NMDA receptor (NMDAR)-mediated current (30, 37). Recordings of currents at minimal, intermediate, and maximal stimulus intensities were made at each holding potential. Particular effort was made to determine the single fiber (SF) response: the amplitude of the EPSC that is first seen following incremental increases in stimulus intensity after failures of synaptic transmission at weaker intensities. The average SF response provides a measure of the strength of a given RGC input. We also use the single fiber response measurements to estimate the number of retinogeniculate inputs that converge onto each TC neuron, using a metric called the fiber fraction (24). The fiber fraction (FF) is the amplitude of the SF EPSC divided by the amplitude of the EPSC in response to maximal stimulus intensity, and thus quantifies the average contribution of each RGC input relative to the total retinal drive to each TC neuron. As such, a lower FF is indicative of more numerous synaptic inputs to each TC neuron, as seen early in postnatal development. Previous studies have shown that in WT mice, the FF increases as spontaneous neuronal activity drives refinement of the retinogeniculate synapse from birth until P20, with further increases as the retinogeniculate synapse enters the vision-sensitive period of refinement, indicative of a shift to fewer RGC inputs (24).
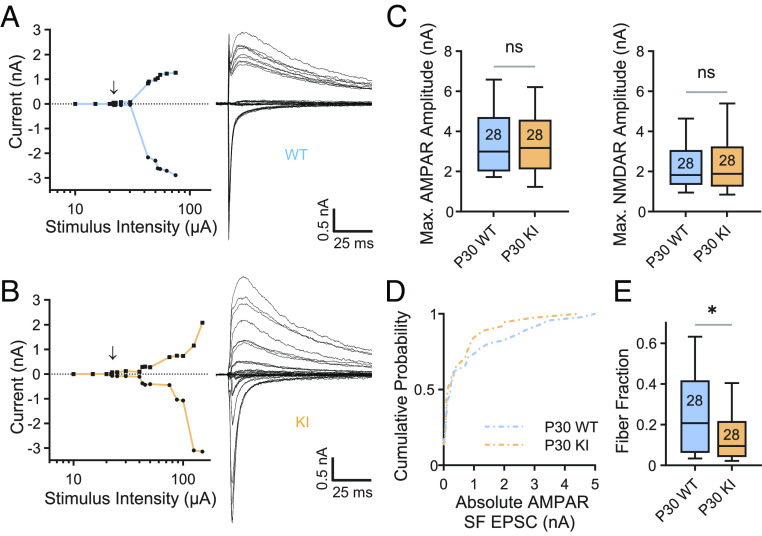
Fig. 2.
Retinogeniculate synaptic refinement is aberrant in QKI mice. (A and B) Representative example recordings from P30 WT (A) and QKI (B) relay neurons demonstrating abnormal synaptic refinement in QKI mice. Left: Plot of peak AMPAR EPSC (circles) and NMDAR (squares) plotted against stimulus intensity. The arrow indicates the emergence of the first single fiber (SF) following failures of synaptic transmission at lower stimulus intensities. Incremental increases in stimulus intensity were used to sequentially activate additional RGC axons. Right: Superimposed representative EPSCs evoked by optic tract stimulation over a range of intensities. At each stimulus intensity, recordings were performed at both −70 mV and +40 mV. Inward currents at −70 mV predominantly represent AMPAR-mediated conductances, whereas outward currents at +40 mV represent AMPAR- and NMDAR-mediated conductances. Stimulus artifacts are blanked for clarity. (C) Maximal EPSC amplitudes at −70 mV (AMPAR, Left) and +40 mV (NMDAR, Right) are not significantly different between WT and QKI mice at P30. Numbers in boxplots represent numbers of cells recorded. (D) Cumulative probability distributions of AMPAR-mediated SF amplitudes recorded at −70 mV from P30 WT and QKI mice. WT: n = 46 from six mice; KI: n = 37 from seven mice. (E) The fiber fraction (FF = SF EPSC amplitude/maximal EPSC amplitude) is significantly reduced in P30 QKI mice. *P < 0.05, two-tailed Mann–Whitney U test. Box, 25 to 75% interquartile range; line within box, median; whiskers, 10 to 90% range. WT: n = 28 from six mice; KI: n = 28 from seven mice.
During our electrophysiological recordings, when we incrementally increased the stimulus intensity, we observed that the peak amplitudes of both the AMPAR and NMDAR EPSCs increased in WT and QKI TC neurons, indicating that more of the inputs that innervate the recorded TC cell had been activated (Fig. 2 A and B). These recordings revealed a difference between the inputs that converge onto TC neurons in QKI mice vs. WT mice. Like the Mecp2 null mouse, maximal AMPAR- and NMDAR-mediated amplitudes are normal in QKI mice at P30 (Fig. 2C). However, unlike Mecp2 null mice where SF inputs are significantly weaker than those of their WT littermates (13), the cumulative distribution of SF AMPAR amplitudes is not significantly different in QKI mice when compared to WT mice (Fig. 2D). Nevertheless, examination of the cumulative distributions reveals a clear trend in which a subset of inputs from QKI mice are shifted to the left of WT mice, suggesting that fewer inputs strengthen in the dLGN of QKI mice. Notably, QKI mice exhibited fewer SF inputs that are greater than 600 pA, a threshold strength that was previously defined as a strong input capable of driving TC neuron firing (31, 38). Since the analysis of maximal currents is not significantly different when WT and QKI mice are compared (Fig. 2C), the presence of a subset of weaker inputs in QKI mice significantly reduces the median FF in QKI mice, indicative of an increase in the number of inputs onto TC neurons in these mice (Fig. 2E). This increase in the number of inputs is consistent with phenotypes previously described in Mecp2 null mice (13). However, these data suggest that while a subset of TC neurons in QKI mice receives more convergent retinal inputs than TC neurons in WT mice, the overall phenotype of QKI mice is milder than that previously reported of Mecp2 null mice.
Retinogeniculate Synapse Refinement Is Delayed in QKI Mice.
We next investigated when the defects in QKI mice emerge during retinogeniculate synapse development. The initial phase of retinogeniculate synaptic refinement, extending from prenatal ages through the third postnatal week (P20), is mediated by spontaneous activity in the form of correlated cholinergic and glutamatergic bursts of action potentials, or waves, from the retina (10, 24, 32, 33, 39). In Mecp2 null mice, we previously showed that retinogeniculate refinement proceeds relatively normally until the beginning of the vision-sensitive phase of synaptic refinement (approximately P20), after which connectivity regresses (13). Between P20 and P30, the number of inputs to TC neurons increases, while the average strength of the inputs decreases in Mecp2 null mice compared to WT mice. We thus asked whether a similar time course of disrupted refinement is seen in QKI mice. Given previous findings that MeCP2 S421 phosphorylation is present in the visual cortex during the third postnatal week (23), we first investigated whether early-stage, vision-insensitive refinement at this time is disrupted in QKI mice. In this regard, whole-cell recordings from TC neurons in acute dLGN slices prepared from P14-15 WT and QKI littermates revealed no differences in single fiber AMPAR- and NMDAR-mediated EPSC amplitudes, maximal EPSC amplitudes, or FF between the two genotypes (Fig. 3 A, Left, 3C). Thus, mimicking trends previously observed in Mecp2 null mice (13), the early spontaneous activity-driven period of refinement is unaffected by the loss of activity-induced MeCP2 phosphorylation, with both WT and QKI mice exhibiting a predominance of weak RGC synapses onto each TC neuron around P14.
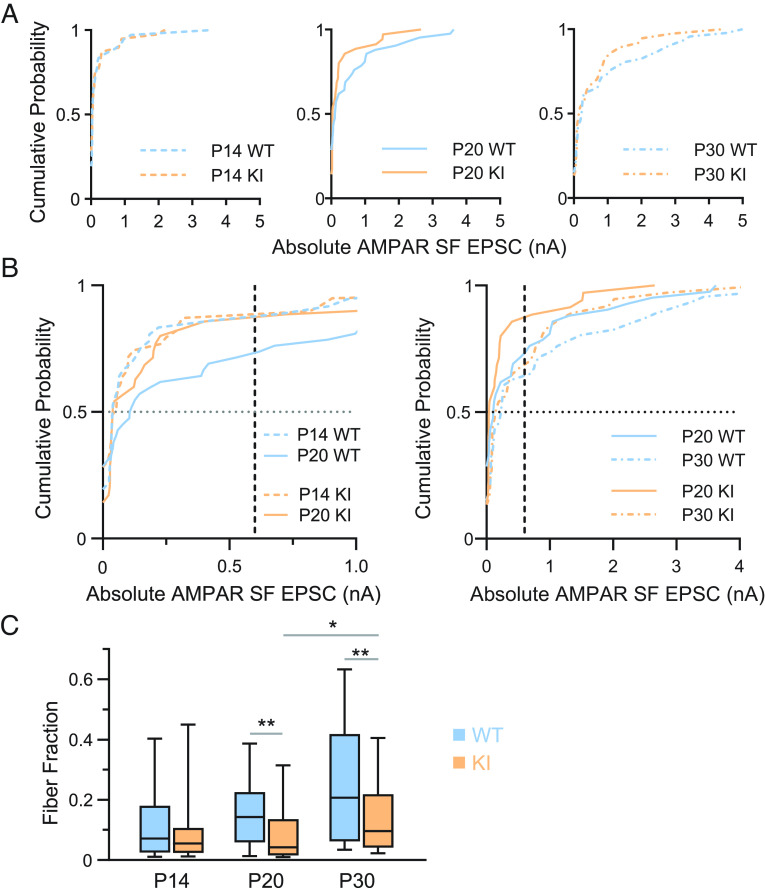
Fig. 3.
Delayed retinogeniculate refinement in QKI mice. (A) Cumulative probability distributions of AMPAR-mediated single fiber (SF) amplitudes at −70 mV for P14 (Left), P20 (Middle), and P30 (Right) QKI and WT mice. SF AMPAR EPSC amplitude distributions of WT and QKI mice overlap at P14. By P20, a subset of inputs in WT mice have strengthened, shifting the distribution to the right compared to that of QKI (WT vs. QKI: third and fourth quartiles P < 0.02; full distribution P = 0.5). By P30, the QKI distribution shifts toward that of the WT, indicating some strengthening of SF inputs in the mutant (WT vs. QKI: third and fourth quartiles P = 0.16; full distribution P = 0.64). (B) Comparison of cumulative probability distributions of AMPAR-mediated SF amplitudes (from panel A) between P14 and P20 (Left), and P20 and P30 (Right), from WT and QKI mice. The median (dotted line at y = 0.5) WT SF strengthens by 187% (0.039 at P14 to 0.11 at P20), whereas the median QKI SF slightly weakens by −27% (0.054 at P14 to 0.039 at P20). AMPAR-mediated SFs begin to strengthen from P20 to P30 in the QKI (Right), visualized by the median values at y = 0.5 in the WT (0.11 at P20 to 0.23 at P30, a 109% increase) vs. in the QKI (0.039 at P20 to 0.15 at P30, a 285% increase). Dashed lines at x = 0.6 nA highlight the greater proportion of weak RGC inputs less than 0.6 nA in the QKI relative to WT at P20. By P30, the proportion of weak RGC inputs less than 0.6 nA is relatively closer in value between QKI and WT mice. (C) The FF is not significantly different between WT and QKI mice at P14. By P20, the FF is significantly lower, indicating the presence of more RGC inputs to each relay neuron in QKI mice. While the FF of QKI mice increases significantly between P20 and P30, it does not recover to WT levels. All statistics were performed using a two-tailed Mann–Whitney U test, Kolmogorov–Smirnov, or Kruskal–Wallis when comparing more than two groups. ns, P > 0.05; *P < 0.05; **P < 0.01. P14 WT: n = 26 cells from six mice; KI: n = 26 cells from six mice. P20 WT: n = 25 cells from four mice; KI: n = 26 cells from four mice. P30 WT: n = 28 from six mice; KI: n = 28 from seven mice.
We next performed recordings from P19-P21 WT and QKI mice (hereafter referred to as P20 for simplicity) and found that the FF is abnormal in QKI mice at P20. First, comparison of the cumulative distribution of SF peak amplitudes suggests that while WT mice exhibit a significant increase in SF peak amplitudes between P14 and P20, very few RGC inputs strengthen in QKI mice over the same developmental window (Fig. 3 A and B, Left). Notably, the median SF amplitude at −70 mV strengthens by 182% in WT mice from P14 to P20 (0.039 to 0.11), whereas the median SF amplitude weakens by 28% in QKI mice over this period (0.054 to 0.039). Quantification of the third and fourth quartiles of the cumulative distributions, which represent the subset of inputs that have strengthened, reveals a significant difference between WT and QKI mice at P20 (P < 0.02, Mann–Whitney U test). This is in contrast to Mecp2 null mice, in which the disruption of refinement does not occur until after P20 (13). Strikingly, between P20 and P30, SF inputs of QKI mice significantly strengthen (P20 vs. P30, P = 0.035, Kruskal–Wallis, n = 35 to 37), so that they are no longer significantly different in strength than the SF inputs of WT mice (WT vs. QKI mice, third and fourth quartiles, P = 0.16) (Fig. 3 B, Right).
We then compared maximal currents and the FF across development of WT and QKI mice. Maximal peak AMPAR and NMDAR EPSC currents were not significantly different at any point during development; however, the FF shows distinct developmental trends between the two genotypes (Fig. 3C). The median FF in WT mice doubles between P14 and P20 (0.07 to 0.14), consistent with the pruning of retinal inputs onto TC neurons, while the median FF in QKI mice does not significantly change over this same period (0.06 to 0.04). Instead, the FF in QKI mice only increases between P20 vs. P30 (P = 0.0050, Kruskal–Wallis, n = 26 to 28), although not to the same level as in WT mice (Fig. 3C). Taken together, retinogeniculate refinement appears to be delayed in QKI mice at P20, but partially recovers by P30.
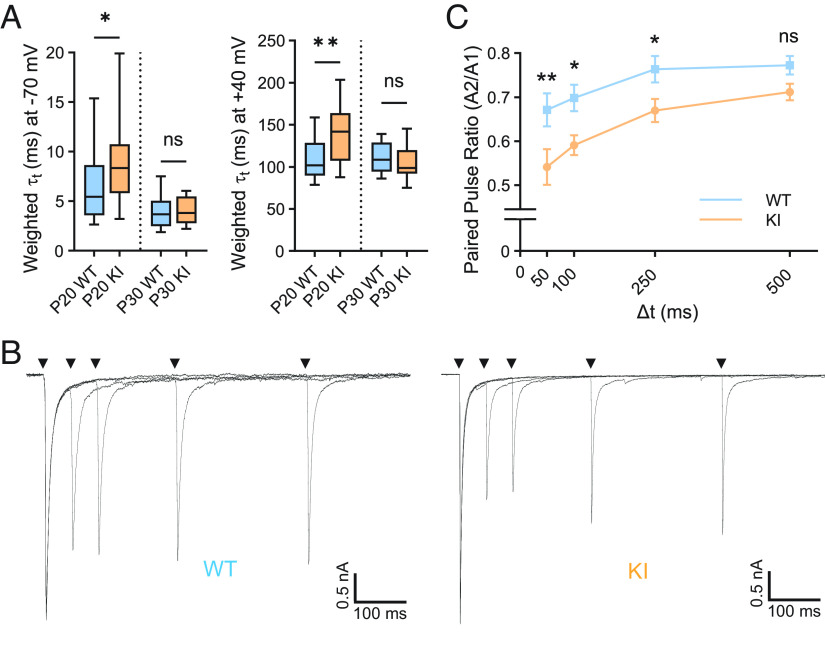
We next examined other synaptic features that are known to change during postnatal retinogeniculate development (24, 30). We found no significant differences in the AMPAR/NMDAR ratio (calculated as the ratio of the maximal AMPAR-mediated EPSC at −70 mV divided by the maximal NMDAR-mediated EPSC at +40 mV) when WT and QKI mice are compared. However, there are notable differences in the EPSC waveform when WT and QKI mice are compared; EPSCs recorded from P20 QKI mice at both holding potentials exhibit significantly slower decay kinetics (τ) than those of WT mice at the same age. By P30, EPSC decay kinetics have accelerated in QKI mice so that AMPAR and NMDAR decay kinetics are similar to those seen in WT mice (Fig. 4A).
Fig. 4.
Retinogeniculate synaptic properties are altered in QKI mice. (A) Decay kinetics of EPSCs at −70 mV (Left) and +40 mV (Right) are significantly slower in QKI at P20, but not at P30, as compared to WT mice. (B) Superimposed pairs of EPSCs recorded at a holding potential of −70 mV with varying ISIs (50, 100, 250, and 500 ms) from P29-32 WT (Left) and QKI (Right) mice. Stimulus artifacts were blanked for clarity. (C) Mean paired pulse ratio (PPR) ± SEM, calculated by dividing the amplitude of the second EPSC (A2) by the amplitude of the first EPSC (A1), shows greater short-term depression in QKI mice as compared to WT mice at three interstimulus intervals (ISIs) tested. WT: n = 17 from three mice; KI: n = 14 cells from four mice. Statistics in (A) were performed using a two-tailed Mann–Whitney U test, and those in (B) using a one-way ANOVA and Fisher’s LSD of multiple comparisons. ns, P > 0.05; *P < 0.05; **P < 0.01.
Retinogeniculate Synapse Refinement Defects in QKI Mice Are Presynaptic.
One possible explanation for the change in retinogeniculate connectivity in QKI mice is that probability of presynaptic vesicle release from RGCs is altered in QKI animals. Toward this end, we stimulated the optic tract twice in rapid succession with varying interstimulus intervals (ISIs) to determine paired-pulse ratios (PPRs) in P29-32 WT and QKI mice. PPRs were found to be significantly decreased in QKI mice compared to their WT littermates across a range of ISIs (Fig. 4 B and C), indicating that vesicles in QKI mice are depleted faster and/or recover more slowly than in WT mice. Importantly, Mecp2 null mice have been previously reported to not exhibit alterations in PPR at P30, and prior studies have shown that deficits in synaptic refinement do not necessarily lead to changes in PPRs (13, 37). Our findings thus suggest that activity-induced MeCP2 phosphorylation contributes to retinogeniculate synapse refinement at least in part by modulating presynaptic vesicle release in RGCs.
RGC Axon Targeting to dLGN Is Not Disrupted in QKI Mice.
The deficits observed in synaptic refinement in QKI mice at P20 led us to hypothesize that these effects might be due to defects in the structural development of the retinogeniculate synapse. To investigate this possibility, we examined eye-specific RGC axon segregation in QKI mice. While the segregation of retinal axons into eye-specific domains is largely complete by P8-P10 in WT mice (13, 40), a failure to maintain this organization has been previously observed in Mecp2 null animals when assessed later in development (P46-P51) (13). To determine whether eye-specific segregation is altered in QKI mice, we injected cholera toxin subunit B conjugated to different fluorescent dyes into each eye of littermate WT and QKI mice at P58-P64 to visualize retinal projections to the dLGN (SI Appendix, Fig. S2 A and B). Segregation was quantified by calculating the logarithm of the ratio of ipsilateral to contralateral intensity for each fluorophore at every pixel, known as the R-value. The mean variance of R from each section was then used to assess segregation, where a high value indicates a high degree of eye-specific segregation and a low value suggests a low degree of segregation (41). Surprisingly, in contrast to the significant defect in eye-specific segregation observed in Mecp2 null mice, this analysis revealed no significant differences in eye-specific segregation between WT and QKI mice (SI Appendix, Fig. S2C). These results are consistent with the idea that, in contrast to Mecp2 null mice, retinogeniculate circuits in QKI mice do not regress with age.
No Detectable Changes in Gene Expression in Visual Circuits of QKI Mice.
Given the well-characterized role of MeCP2 as a transcriptional repressor, we next sought to characterize transcriptional changes in QKI mice that might give rise to the observed electrophysiological abnormalities. As three of the activity-dependent phosphorylation sites of MeCP2 occur within its MBD and TRD, one possibility is that activity-dependent MeCP2 phosphorylation relieves the repressor function of MeCP2 by inhibiting the binding of MeCP2 to methylated DNA and/or inhibiting the interaction of MeCP2 with NCoR, thus allowing for or facilitating the induction of activity-dependent genes. In the absence of activity-dependent phosphorylation of MeCP2 in QKI mice, the failure to relieve MeCP2 repression might thus result in a defect in the induction of activity-dependent genes. To investigate this possibility, we used bulk RNA-seq to profile gene expression in the dLGN and visual cortex of WT and QKI mice at P16 (n = 10 per genotype) and P15 (n = 6 per genotype), around the time of eye-opening, but before the onset of vision-sensitive synaptic refinement, as we hypothesized that gene expression changes would precede the observed synaptic refinement defects observed at P20. However, differential gene expression analysis identified no significant gene expression changes between WT and QKI mice at these early developmental stages, or later during the vision-sensitive period when we assessed gene expression at P30 (n = 6 per genotype) (SI Appendix, Fig. S3). The absence of MeCP2 phosphorylation in QKI mice might have been predicted to affect gene expression in this circuit, given previous reports that MeCP2 phosphorylation disrupts the association of MeCP2 with the NCoR corepressor complex in vitro and attenuated visual experience-dependent gene responses have been previously reported in the visual cortex of MeCP2 T308A knock-in mice (18). However, these effects were not seen after substantial backcrossing of the mice (SI Appendix, Fig. S4).
We next dark-reared mice from P20-P27 [known as late dark rearing (LDR)] and then acutely re-exposed the mice to light for either 0 or 4 h to assess possible differences in vision-dependent gene expression at this later time. However, no significant gene expression differences were observed between WT and QKI samples from either the dLGN or visual cortex at both 0 and 4 h of light stimulation after LDR (SI Appendix, Figs. S5 and S6). One possible explanation for these findings is that alterations in gene expression in QKI mice are subtle, especially given that overall gene expression fold-changes in Mecp2 null mice, while encompassing many genes, are quite modest and thus may be below the level of detection in QKI mice. Subtle differences in gene expression between LDR WT and QKI mice may also have been missed, given that the synaptic phenotype of QKI mice is recovering between P20-P30.
Reasoning that bulk RNA-seq might not be sensitive enough to detect cell type-specific differences in gene expression between WT and QKI mice, we also performed single-nucleus RNA-seq of the dLGN from P20 WT and QKI mice. We chose P20 mice for this analysis because the retinogeniculate synapse refinement defect in QKI mice is most apparent at this time during development. Cell types were assigned on the basis of known marker genes, including Gad1 for inhibitory neurons, Aldoc for astrocytes, and Slc17a7 for excitatory neurons (42). Clusters expressing the same marker genes were then combined, after which differential gene expression analysis between WT and QKI mice was performed within each cluster and across all cells (a total of 34,693 nuclei were included in final analysis) (SI Appendix, Fig. S7). Across all cells, this analysis identified no differentially expressed genes between WT and QKI mice. Furthermore, we saw no differentially expressed genes between WT and QKI mice within excitatory neurons, inhibitory neurons, astrocytes, oligodendrocytes and oligodendrocyte precursors, or endothelial cells. Within the microglia cluster, we detected only one differentially expressed gene, Atg16l2. While the lack of differentially expressed genes might seem surprising, it is possible that any differentially expressed genes between WT and QKI mice are below the level of current detection, that the detected synaptic changes are due to gene expression alterations that occur presynaptically within RGCs, or that the synaptic refinement defects are being driven by a function of MeCP2 separate from its role as a transcriptional repressor.
Discussion
Previous studies have suggested a role for MeCP2 in both spontaneous activity-dependent and experience-driven synaptic regulation. For example, in addition to deficits in experience-dependent retinogeniculate synapse remodeling in Mecp2 null mice (13), MeCP2 loss has also been reported to accelerate the critical period of ocular dominance plasticity in the visual cortex and to disrupt vision- and activity-dependent homeostatic synaptic scaling responses (7, 8, 43). However, it has been difficult to distinguish the direct contribution of MeCP2 to activity-driven processes from the secondary effects stemming from the generalized RTT-associated neural dysfunction. To begin to address this issue, we generated a mouse line in which MeCP2 is no longer susceptible to activity-induced phosphorylation. While these animals appear grossly normal and fail to exhibit hallmark symptoms of RTT, they have deficits in retinogeniculate circuit refinement.
QKI mice do not phenocopy the significant deficits in retinogeniculate refinement reported previously in Mecp2 null mice (13). Rather than presenting with a milder form or a subset of the characteristics of Mecp2 null mice, the QKI mice exhibit a refinement defect that is distinct from that of Mecp2 null mice. While the two mutants exhibit similarly aberrant retinogeniculate connectivity at P30, the phenotype of each of the mutants is a consequence of different developmental trajectories. In Mecp2 null mice, refinement is not significantly different from that of WT littermates during the vision-independent phase of development (from birth to P20), after which the retinogeniculate circuit in Mecp2 null mice regresses. By contrast, synaptic connectivity in QKI mice appears normal at P14, but after P14 synapse elimination and strengthening is disrupted (Fig. 3 B, Left, 3C). Thus, QKI mice exhibit disrupted synaptic refinement after P14, whereas this disruption occurs later, after P20, in Mecp2 null mice. Importantly, the timing of developmental retinogeniculate refinement of WT littermates of QKI mice is remarkably similar to that reported previously for WT littermates of Mecp2 null mice (13), justifying the validity of this comparison between these two studies.
By P20, when compared to WT littermates, the strength of individual RGC axons is weaker, decay kinetics of EPSCs are slower, and RGC inputs fail to eliminate correctly in QKI mice. Notably, in QKI mice, it is only after P20 that there is further refinement, with significant strengthening of SF inputs to TC neurons, a decrease in retinal inputs to TC neurons, and acceleration of EPSC decay kinetics. However, synaptic refinement fails to fully normalize by P30, as the number of inputs onto TC neurons is still significantly greater in QKI mice than in their WT littermates. It is possible that the functional difference between WT and QKI mice diminishes further with age; however, recordings at later time points are difficult to obtain due to technical limitations. The fact that eye-specific segregation is indistinguishable between WT and QKI adult mice at a later age further suggests that, while initially delayed in QKI mice, the degree of refinement of the retinogeniculate synapse eventually begins to recover toward the level observed in WT mice. Together, these findings provide strong support for a direct role for activity-regulated MeCP2 phosphorylation in activity-dependent synaptic remodeling prior to the vision-sensitive period of synaptic refinement.
Our findings complement a previous study that examined the function of phosphorylation of S421 in mice in which serine 421 of MeCP2 was converted to an alanine (MeCP2 S421A mice) (23). In MeCP2 S421A mice, there were no detectable activity-induced transcriptional differences, and chromatin immunoprecipitation sequencing (ChIP-seq) revealed normal binding of MeCP2 across the neuronal genome, similar to the lack of gene expression and MeCP2 binding differences observed in QKI mice compared to WT mice. Acute slice recordings from the visual cortex of P16-17 S421A mice revealed that spontaneous miniature inhibitory postsynaptic currents (mIPSCs), but not spontaneous excitatory postsynaptic currents, were disrupted. Taken together with our findings in QKI mice, the electrophysiological defects in MeCP2 S421A mice suggest that activity-induced MeCP2 phosphorylation is likely critical for proper synaptic development in multiple brain regions during the early postnatal period.
The differences in synaptic refinement observed in QKI mice compared to Mecp2 null animals are notable, although the precise mechanisms underlying this finding remain unclear. One possible explanation for our data is that activity-dependent phosphorylation of MeCP2 is highest in the dLGN and retina during the vision-insensitive phase and decreases in the later experience-dependent phase of retinogeniculate refinement. Thus, in QKI mice, the inability to phosphorylate MeCP2 would lead to constitutive MeCP2 repressor activity between P14 and P20 and a defect in retinogeniculate development. In contrast, in Mecp2 null mice, the absence of MeCP2 before P20 may have no effect on synaptic refinement because the normal developmental process that leads to MeCP2 phosphorylation in WT mice would have inactivated MeCP2 during this developmental window. Instead, a synaptic defect emerges at a later timepoint in Mecp2 null mice, perhaps due to a loss of the unphosphorylated form of MeCP2 and its typical repressor activity during the vision-sensitive period of development. Because the phospho-mutant form of MeCP2 would be expected to mimic the unphosphorylated WT form of MeCP2, this could also explain why the QKI mice show relative, but not complete, rescue of the phenotype after P20. Future experiments to determine the time course of MeCP2 phosphorylation in the dLGN and retina, as well as to determine how MeCP2 phosphorylation affects corepressor recruitment and direct binding to heterochromatin will be important (44, 45).
Despite the known role of MeCP2 as a transcriptional repressor, we were surprised to find that loss of activity-dependent phosphorylation of MeCP2 does not have a detectable impact on gene expression in the dLGN or visual cortex at the time points assessed. This raises the possibility that the differences seen in synaptic refinement between WT and QKI mice reflect a function of MeCP2 distinct from the ability of MeCP2 to repress gene transcription. However, given the well-established function of MeCP2 as a repressor of neuronal gene expression, it seems more likely that alterations in gene expression do underlie the synaptic defects in QKI mice but that these differences are too subtle to detect with current methods. This possibility is consistent with the relatively small fold-changes in gene expression detected when comparing WT and Mecp2 null mice (46). It is noteworthy that our ChIP-seq data show that binding of MeCP2 to DNA is also not altered in QKI mice (SI Appendix, Fig. S1A), although these findings do not rule out the possibility that the dynamics of MeCP2-DNA binding are changed in QKI mice. Such a change seems possible, since one site of MeCP2 phosphorylation, S86, resides within the methyl binding domain of MeCP2 (Fig. 1B). In this regard, single-molecule tracking of WT and QKI MeCP2 to examine DNA binding dynamics at different timepoints following membrane depolarization might be a worthwhile future direction (47).
Further analysis to detect subtle differences in gene expression across additional developmental timepoints between WT and QKI mice might also provide insight into how activity-induced phosphorylation of MeCP2 affects gene expression in the dLGN. It is also possible that presynaptic mechanisms could account for the failed refinement in QKI mice, such as disrupted feedforward excitation from RGCs, in line with our PPR studies showing that vesicle release probability is altered in RGCs in QKI mice. Thus, gene expression changes in RGCs, rather than in the dLGN or visual cortex, could be responsible for the synaptic defects that we have observed in QKI mice. Future studies will be needed to look at potential changes in gene expression of the many different types of RGCs that innervate the dLGN (48, 49).
Beyond the retinogeniculate synapse, further study of the QKI mice will be needed to address the broader contribution of MeCP2 phosphorylation to postnatal experience-dependent circuit remodeling. In this regard, MeCP2 has been implicated in the control of ocular dominance plasticity as well as the timing of critical period plasticity (43, 50). Likewise, it will be of interest to examine potential deficits in neuroplasticity in adult QKI mice. It will also be important to dissect the contribution of individual MeCP2 phosphorylation sites to these phenotypes through the assessment of synaptic refinement in single knock-in MeCP2 phospho-mutant mouse strains, as well as to elucidate the mechanisms through which activity-dependent phosphorylation of MeCP2 affects synaptic refinement.
Materials and Methods
Generation of Mecp2 Quadruple Knock-In Mice.
All animal experiments were approved by the NIH and the Harvard Medical School Institutional Animal Care and Use Committee and were conducted in compliance with the relevant ethical regulations. MeCP2 quadruple knock-in mice were generated by homologous recombination in embryonic stem (ES) cells as described previously (18, 23) with minor modifications. The Mecp2 targeting construct generated previously (23) contains a floxed neomycin-resistance cassette (NEO) in Mecp2 intron 3 and 4.1 kb of the Mecp2 locus upstream (including exon 3, which contains S86) and 3.9 kb downstream (including Mecp2 exon 4, which contains S274, T308, and S421). MeCP2 S86A, S274A, T308A, and S421A mutations were added to this targeting construct, and the sequence of the entire targeting construct was verified by Sanger sequencing prior to use in gene targeting. Gene targeting was performed at the IDDRC Mouse Gene Manipulation Core at Boston Children’s Hospital. The construct was linearized by digesting with NotI and electroporated into JM8 Agouti C57BL/6N mouse ES cells. Genomic DNA was isolated from G418-resistant ES cell clones and was screened by PCR for each arm of the targeting construct. One primer recognized a sequence located outside the targeted region, while the other primer recognized a sequence located in the other arm of the targeting construct. A 1.8-kb increase in the size of the product indicated successful insertion of the NEO cassette into the Mecp2 locus. Two confirmed MeCP2 S86A; S274A; T308A; S421A ES cell clones were transfected using a Cre recombinase-expressing plasmid. Appropriate excision of the loxP-flanked NEO cassette was subsequently confirmed by PCR genotyping, and the presence of the four mutations was verified by PCR and Sanger sequencing. The MeCP2 QKI ES cell clones were injected into C57BL/6 blastocysts and subsequently implanted into pseudopregnant females. The resulting chimeric offspring were mated with C57BL/6J mice, and the offspring were screened by PCR genotyping of tail genomic DNA for the presence of the residual loxP site to confirm germline transmission of the mutant allele. The sequence of all four mutations was verified by Sanger sequencing of tail genomic DNA. Mice were housed under a standard 12-h light cycle unless otherwise indicated (see late dark-rear methods).
Quantitative Mass Spectrometry for MeCP2 Modifications.
Induction of neuronal activity.
MeCP2 immunoprecipitation for quantitative mass spectrometry was performed from both cultured embryonic mouse cortical neurons and adult mouse hippocampus. For cultured cortical neurons, embryonic day 16.5 (E16.5) cortices from WT C57BL/6 mice were dissociated with Papain (Sigma). Neurons were plated at 30 million cells in 15-cm dishes precoated overnight with 40 μg/mL poly-D-lysine and 4 μg/mL laminin. Neurons were grown in Neurobasal media (Gibco) containing 1X B-27 Supplement, 1X GlutaMAX supplement, and 1X penicillin-streptomycin. After 6 d in vitro (DIV6), neurons were silenced for 16 h with 1 μM TTX and 100 μM AP5 to prevent spurious activity. On DIV7, neurons were either left untreated (control) or stimulated for 30 min by depolarizing with 55 mM potassium chloride (KCl) or by adding 50 ng/mL of brain-derived neurotrophic factor (BDNF). For each condition (untreated, KCl, BDNF), 180 million neurons were used, and two biological replicates were performed. After 30 min, cells were harvested by washing once with cold phosphate-buffered saline (PBS), then scraped in 4 mL cold NE1 buffer per 30 million cells [NE1: 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.9, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol (DTT), 1X Protease Inhibitor Cocktail (Roche), 1% Phosphatase Inhibitor Cocktail 2 and 3 (Sigma), and 10 mM sodium butyrate]. For adult mouse hippocampus, 8-wk-old male WT C57BL/6 mice were injected intraperitoneally with 15 mg/kg kainic acid or a saline control. Mice were killed 1 h after injection, and the hippocampus was dissected. Hippocampi from six mice were combined for each control and kainic acid condition, and two biological replicates were performed. Hippocampi from a single brain were homogenized with a pellet pestle homogenizer in 50 μL NE1 buffer; then, the volume was increased to 4 mL NE1 for cell lysis.
MeCP2 immunoprecipitation.
For both cultured cortical neurons and hippocampal tissue, cell lysis was ensured by pipetting up and down 10 times and rotating for 10 min at 4 °C. Nuclei were pelleted by spinning at 500 × g for 10 min at 4 °C. Nuclei were resuspended in one pellet volume of NE1 buffer, and 0.9 μL Benzonase (Sigma) per 90 million cells or hippocampi from three brains was added, and rotated for 30 min at 4 °C. Benzonase activity was quenched by adding 5.67 volumes of NE1 250 (NE1 buffer with 250 mM NaCl and 0.5% Triton X-100) and rotating for 20 min at 4 °C. Insoluble material was removed by spinning at 16,000 × g for 20 min at 4 °C. The nuclear supernatant was precleared by adding 30 μL Protein A Dynabeads (Invitrogen) per 90 million neurons or hippocampi from three brains and rotating for 30 min at 4 °C. MeCP2 antibody (14) was conjugated to Protein A Dynabeads in NE1 250 by rotating at 4 °C for 1.5 h. Antibody-coupled beads were then added to precleared nuclear supernatant, and immunoprecipitation was performed by rotating tubes for 1.5 h at 4 °C (for 90 million cells or hippocampi from three brains, 90 μL of MeCP2 antibody with 270 μL beads were used). After immunoprecipitation, beads were washed four times for 5 min each in NE1 250 with rotation at 4 °C. Proteins were eluted from beads by adding 1× sample buffer [1× lithium dodecyl sulfate (LDS) (Life Technologies) in PBS with 5% Beta-Mercaptoethanol] and boiling for 15 min at 95 °C. Also, 10% of eluted sample was used to verify successful MeCP2 IP by western blot, and 90% of supernatant was electrophoresed on a 10% Bis-Tris gel (Invitrogen) and stained with QC Colloidal Coomassie (Bio-Rad). The MeCP2 bands were excised using clean razor blades and submitted for mass spectrometry.
TMT quantitative mass spectrometry and analysis.
Mass spectrometry was performed at the Thermo Fisher Center for Multiplexed Proteomics at Harvard Medical School. Bands were destained, reduced with DTT, and alkylated before digesting overnight with Trypsin. Peptides were extracted from gels, dried, and desalted. Cleaned peptides were labeled with Tandem Mass Tag (TMT)10 reagents. Labeling reactions were combined, cleaned, and dried down. Peptides were resuspended in 5% acetonitrile and 5% formic acid, and the entire sample was analyzed on an Orbitrap Fusion Mass spectrometer. Peptides were separated using a gradient of 6 to 28% acetonitrile in 0.125% formic acid over 180 min. Peptides were detected (MS1) and quantified (MS3) in the Orbitrap. Peptides were sequenced (MS2) in the ion trap. MS2 spectra were searched using the SEQUEST algorithm against a Uniprot composite database derived from the mouse proteome containing its reverse complements and known contaminants. MeCP2 was confirmed as the top protein in the sample, and then, raw files were searched with a composite database including only the MeCP2 sequence, contaminants, and reverse complements to identify modifications of interest. Peptide spectral matches were filtered to a 1% false discovery rate (FDR) using the target-decoy strategy combined with linear discriminant analysis. Proteins were quantified only from peptides with a summed signal to noise threshold of ≥200 and MS2 isolation specificity of 0.5. For each modification, the total summed signal to noise was multiplied by the MeCP2 normalization factor for each sample. The log2 fold-change for each treatment condition compared to its control was calculated and displayed in the heatmap.
MeCP2 Chromatin Immunoprecipitation Sequencing (ChIP-Seq).
MeCP2 ChIP-seq.
MeCP2 ChIP-seq in MeCP2 QKI and WT mice was performed as described in ref. 46, with minor modifications. Forebrain (cortex + hippocampus) was dissected from 8-to-11-wk-old male MeCP2 QKI and WT mice and flash frozen. Tissue was homogenized in 1% formaldehyde and cross-linked by rotating for 10 min at room temperature. Cross-linking was quenched with 125 mM glycine for 5 min at room temperature. Tissue was pelleted by spinning at 500 × g for 5 min at 4 °C, washed once with PBS, and spun again at 500 × g for 5 min at 4 °C. Cells were lysed by resuspending in Buffer L1 (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) pH 8, 1 mM EGTA pH 8, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and 1X protease inhibitors) and rotating for 10 min at 4 °C. Nuclei were pelleted by spinning at 500 × g for 5 min at 4 °C, and then washed by rotating for 10 min at 4 °C in Buffer L2 (10 mM Tris pH 8, 200 mM NaCl, and 1X protease inhibitors). Nuclei were pelleted by spinning at 500 × g for 5 min, and then resuspended in Buffer LB3 (10 mM Tris pH 8, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and 1X protease inhibitors). Chromatin was sonicated in a Diagenode Bioruptor (High power, 65 cycles, 30 s on, and 45 s off). Insoluble material was removed by spinning at 16,000 × g for 10 min at 4 °C, and Triton X-100 was added to soluble chromatin to a final concentration of 1%. Chromatin was precleared for 2 h with Protein A Dynabeads, then incubated with Protein A Dynabeads conjugated to MeCP2 antibody (14) overnight at 4 °C. Beads were washed at 4 °C twice with low salt buffer (20 mM Tris pH 8, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS), twice with high salt buffer (20 mM Tris pH 8, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS), twice with LiCl wash buffer (10 mM Tris pH 8, 1 mM EDTA, 1% NP-40, 250 mM LiCl, and 1% sodium deoxycholate), and once with TE Buffer (50 mM Tris pH 8 and 10 mM EDTA). Chromatin was eluted off beads in TE buffer with 1% SDS at 65 °C for 1 h, and crosslinking was reversed overnight at 65 °C. Chromatin was treated with RNase A for 30 min at 37 °C and Proteinase K for 2 h at 55 °C. DNA was phenol-chloroform extracted and purified with the QIAGEN PCR purification kit. MeCP2 ChIP and input libraries were generated using the NuGEN Ovation Ultralow System V2 following manufacturer instructions. Libraries were sequenced on an Illumina NextSeq 500 with 75-bp single-end reads.
MeCP2 ChIP-seq analysis.
MeCP2 ChIP-seq was analyzed as in ref. 46, with minor modifications. Sequencing reads were trimmed with Trimmomatic (v0.36) to remove adapters and low-quality sequence (settings: LEADING:5 TRAILING:5 SLIDINGWINDOW:4:20 MINLEN:50). Trimmed reads were mapped to the mm10 genome with Bowtie2 (v2.2.9) with default parameters. PCR duplicate reads were removed with SAMtools (v0.1.19) rmdup. Reads were extended to 250 bp (to approximate fragment length) with UCSC tools (v363) bedextendranges. Reads for each sample were randomly down-sampled to equal numbers using the GNU shuf utility. MeCP2 ChIP-seq and input reads were quantified in mm10 RefSeq gene bodies (excluding the first 3 kb, which is relatively depleted for MeCP2 binding and DNA methylation, and thus excluding genes less than 4.5 kb in length) using Bedtools map (v2.26). For DNA methylation, bisulfite sequencing data from the 10-wk-old mouse cortex from ref. 51 was used to calculate the DNA methylation density as the number of cytosines/(number of cytosines + thymines) in either the CA or CG dinucleotide context in each gene body. Genes with fewer than five total CA, CG, MeCP2 ChIP, or input reads in their gene bodies were excluded from analysis. To generate smooth-line correlation plots, genes were sorted by CA or CG DNA methylation density, and a sliding window was defined by 400-gene bins with 40-gene steps. The mean log2 fold-change MeCP2 ChIP/input for each bin was plotted for each MeCP2 QKI or WT sample.
dLGN Acute Slice Preparation for Electrophysiology.
Acute sections of brain tissue containing the dLGN as well as the optic tract comprising RGC axons were prepared as previously described (30, 37). Anesthesia of mice was performed through inhalation of isoflurane followed by immediate decapitation. The mouse head was immersed into an ice-cold, oxygenated solution containing (in mM): 130 K-gluconate, 15 KCl, 0.05 EGTA, 20 HEPES, and 25 glucose (pH 7.4 with NaOH; Sigma). A single 250-μm-thick parasagittal slice was cut for each mouse in this ice-cold solution using a sapphire blade (Delaware Diamond Knives) on a vibratome (VT1000S; Leica). Slices were then recovered at 32 °C for 20 to 30 min in oxygenated artificial cerebrospinal fluid (ACSF) in mM: 125 NaCl, 26 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 1.0 MgCl2, 2.0 CaCl2, and 25 glucose (Sigma), adjusted to 310 to 312 mOsm with water.
Electrophysiology.
Whole-cell voltage clamp recordings of TC neurons in dLGN were performed as previously described (30) with borosilicate glass pipettes (1-2.5 MOhms, Sutter Instrument) filled with an internal solution (in mM): 35 CsF, 100 CsCl, 10 EGTA, 10 HEPES, and an L-type Ca2+ antagonist 0.1 methoxyverapamil (290 to 300 mOsm, pH 7.3; Sigma). Experiments were performed at room temperature in oxygenated ACSF containing 50 μM of the GABAA receptor antagonist picrotoxin (Tocris). Recordings from synapses that were silent (failure of transmission at −70 mV, but EPSC evident at +40 mV) were averaged for at least three to five trials. Single RGC fiber EPSCs were obtained using a threshold method as previously described (13, 24). The fiber fraction (FF = single fiber current amplitude ÷ maximal current amplitude) was calculated to approximate RGC convergence onto each TC neuron in the dLGN. Further details are in SI Appendix.
For Paired Pulse Ratio (PPR) experiments, the optic tract was stimulated twice with randomized interstimulus intervals (ISIs) of 50, 100, 250, and 500 ms while holding the cell at −70 mV. PPR was calculated by dividing A2/A1. A2 is the peak amplitude of the second EPSC, and A1 is the peak amplitude of the first EPSC. For ISIs of 50, 100, and 250 ms, the average waveform of the first EPSC at an ISI of 500 ms was subtracted from the second EPSC to obtain a more accurate measure of A2. Averages of three to six trials of each ISI were used to calculate PPR. To inhibit modulatory and postsynaptic contributions to short-term synaptic plasticity at the retinogeniculate synapse (52–54), recordings were performed in the presence of (in μM): 5 CGP-55845 (GABAB-receptor antagonist), 10 cyanopindolol (5-HT1 receptor antagonist), 50 cyclothiazide (AMPAR desensitization inhibitor), 10 DPCPX (A1 adenosine receptor antagonist), and 50 LY-341495 (mGluR antagonist).
Western Blotting.
Tissue for western blotting was dissected as described for electrophysiology in ice-cold potassium gluconate. Following anesthesia and decapitation, the most anterior portion of the brain containing the olfactory bulb was cut and discarded using a razor blade, creating a flat surface for the remainder of the brain that was superglued onto a Leica VT1000S vibratome. Three 250 to 300 μm coronal sections of the most posterior visual cortex were cut and placed into a 24-well plate with ice-cold potassium gluconate. Sections continued to be collected, from posterior to anterior, until dLGN was visible. The dLGN and the most dorsal portions of the visual cortex were microdissected with a scalpel under a dissecting microscope, placed into separate tubes, and flash-frozen with liquid nitrogen.
Flash-frozen tissue was homogenized in ice-cold RIPA buffer [50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, protease inhibitors, and 1% Phosphatase Inhibitor Cocktail 2 and 3 (Sigma)] and incubated on ice for 15 min. Sonication of lysates was performed using a Diagenode Bioruptor on high power for 10 cycles (30 s on then 30 s off), followed by centrifugation (16,000 × g, 10 min, 4 °C) to pellet the insoluble fraction. The supernatant was carefully transferred to a new tube; then, NuPage lithium dodecyl sulfate (LDS) 4× sample buffer was added to a final concentration of 1×, as well as 2-mercaptoethanol (5% final concentration). Samples were boiled at 95 °C for 15 min, electrophoresed on 4 to 12% Bis-Tris gels (Life Technologies), and transferred to nitrocellulose membranes, which were blocked with 5% dry milk in TBS-T for 1 h at room temperature. Membranes were incubated with primary antibodies overnight at 4 °C, washed three times with TBS-T, incubated with IR dye secondary antibodies for 1 h, and again washed with TBS-T three times. Western blots were imaged using the Li-Cor Odyssey platform and quantified using ImageJ/Fiji. The following primary antibodies and dilutions were used: custom anti-pS421 MeCP2 antibody (1:1,000, rabbit, generated in the Greenberg lab in ref. 22), anti-total MeCP2 N-terminal antibody (1:2,000, mouse, clone Men-8 from Sigma-Aldrich), and β-actin (1:2,000, mouse, Abcam ab8226).
LDR for Bulk RNA Sequencing.
Mice were housed under standard conditions in a room set to a 12-h light/dark cycle, with food and water ad libitum. LDR experiments were performed as previously described (24, 37), with cages of mice reared in the aforementioned conditions until P20, and then placed in a custom-designed, ventilated, light-proof cabinet located in a dark room using night vision goggles (Pulsar). Mice were then re-exposed to light at P27 by moving their cages to a brightly lit separate chamber in the cabinet for 4 uninterrupted hours. Unstimulated control mice remained in the dark chamber and were anesthetized by isoflurane and decapitated in a dark room to limit the induction of experience-dependent gene expression upon exposure to light. Lights were turned on immediately after the decapitated head was submerged in an ice-cold potassium gluconate solution. An acute slice preparation as described above was performed to dissect out the dLGN and visual cortex for qRT-PCR or bulk RNA sequencing.
RNA Isolation for qRT-PCR.
Tissue samples were triturated with a needle and syringe with TRIzol (Life Technologies, 15596026) to subsequently harvest RNA. The RNeasy Micro Kit (Qiagen, 74004) was then used to purify RNA. RNA (120 ng) across all samples were converted to cDNA using the High-Capacity cDNA Kit (Thermo Fisher, 4368813) according to the manufacturer’s instructions. cDNA was diluted by 20-fold and standard qRT-PCR was performed using SYBR Green Master Mix (Thermo Fisher, A25743). qPCR was performed with technical triplicates using a QuantStudio 3 qPCR machine (Applied Biosystems). Expression for each qPCR target gene was normalized to the housekeeping gene Gapdh. The primer sets used to amplify the genes of interest are listed in SI Appendix.
Single-Nucleus RNA-Seq (SnRNA-Seq).
To isolate dLGN nuclei, we combined flash-frozen dLGN tissue dissected from P20 mice in ACSF (in mM): 125 NaCl, 26 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 1.0 MgCl2, 2.0 CaCl2, and 25 glucose. Three male mice per genotype (MeCP2-WT/Y and MeCP2-QKI/Y) were dissected for each replicate, for a total of 12 mice dissected over two replicates. To the tissue, we added 1 mL of buffer HB (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM Tricine-KOH, pH 7.8, supplemented with RNase inhibitors, 1 mM DTT, 0.15 mM spermine, 0.5 mM spermidine) and homogenized with a Dounce 5× with a loose pestle and 10× with a tight pestle. Then, 5% IGEPAL CA-630 (32 μL) was added prior to homogenization with a tight pestle five more times and filtering through a 40-μm strainer into a 50 mL conical collection tube. Then, 1 mL working solution (50% iodixanol, 25 mM KCl, 5 mM MgCl2, and 20 mM Tricine-KOH, pH 7.8, supplemented with RNase inhibitors, DTT, spermine and spermidine) was added. Homogenized tissue was gently layered on top of 100 μL of 30% iodixanol on top of a layer of 100 μL of 40% iodixanol (diluted from working solution). Samples were centrifuged at 1,500 × g for 15 min. Nuclei (50 μL) were collected from the 30/40% iodixanol interface. An aliquot of each sample was mixed with trypan blue, and nuclei were counted using a standard hemocytomter, for a total of ~10,000 nuclei per pooled sample.
SnRNA-seq was performed using the 10X Genomics Chromium Next GEM Single Cell 3′ v3.1 (Dual Index) kit. Each reaction lane was loaded with up to 10,000 nuclei from each pooled dLGN sample. Subsequent steps for cDNA amplification and library preparation were conducted according to the manufacturer’s protocol (10X Genomics). Samples were sequenced using Illumina NextSeq500 with 28 bp (R1), 90 bp (R2), and 10 bp (index). Our final dataset includes 34,693 total nuclei collected from WT and QKI samples. Methods for analysis of SnRNA-seq data are in SI Appendix.
RNA Sequencing (RNA-Seq).
RNA-seq.
Tissue was dissected from WT and QKI male littermates (MeCP2-WT/Y and MeCP2-QKI/Y) in either ACSF (P16, P20 datasets) or the K-gluconate solution used for electrophysiology (P15, P30, and LDR datasets). Tissue was homogenized in TRIzol reagent, and RNA was chloroform extracted and then purified using the Qiagen RNeasy Micro Kit with on-column DNase treatment according to the manufacturer’s instructions. Total RNA (150 to 200 ng for dLGN libraries, 500 ng for visual cortex libraries) was used to generate libraries following rRNA depletion (NEBNext, E6310X) according to the manufacturer’s instructions (NEBNext, E7420). Libraries were sequenced on an Illumina NextSeq 500 with 85-bp single-end reads. All libraries that were compared as a set for differential gene expression analysis were prepared and sequenced at the same time to reduce batch effects, before being subsequently analyzed with our standardized RNA-seq data analysis pipeline (below).
RNA-seq analysis.
Analysis was performed as described previously (46). Briefly, reads were trimmed with Trimmomatic (v0.36) (55) to remove Illumina adaptors and low-quality sequence (settings: LEADING:5 TRAILING:5 SLIDINGWINDOW:4:20 MINLEN:50). Trimmed reads were mapped to the mm10 RefSeq transcriptome and genome using STAR (v2.5.2b) (56). Reads were counted in the full gene body (TSS to TTS of longest isoform, in order to include intronic reads) using Subread featureCounts. Differential expression was performed with the R package edgeR (v3.34.1) (57) or DESeq2 (v1.34.0) (58). Genes with low counts were filtered by keeping only genes with rowSums(cpm(y)>1) >= (number of samples per group). Differentially expressed genes were defined by an FDR < 0.05 with no fold-change cut-off. For DESeq2 analyses, shrinkage of effect sizes was performed using lfcshrink and the apeglm method following differential expression analysis (59).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Drs. Hume Stroud, Elizabeth Pollina, Lucas Cheadle, Christopher Davis, David Harmin, Thomas Schwarz, Bruce Bean, Pascal Kaeser, David Ginty, Gord Fishell, Lisa Goodrich, and all members of the Greenberg and Chen labs for discussions and input on the project. This work was supported by the Rett Syndrome Research Trust, NIH R01NS048276 (M.E.G.), R01EY013613 (C.C.), K99NS112415 (L.D.B.), the Stuart H.Q. & Victoria Quan Fellowship (T.W.), and the Harvard Department of Neurobiology Graduate Fellowship (T.W.).
Author contributions
C.P.T., T.W., L.D.B., S.T., W.R., C.C., and M.E.G. designed research; C.P.T., T.W., L.D.B., E.L., A.S., S.T., K.M., C.L., and W.R. performed research; L.D.B., E.L., A.S., R.S., and W.R. contributed new reagents/analytic tools; C.P.T., T.W., L.D.B., S.T., K.M., C.L., W.R., and C.C. analyzed data; and C.P.T., T.W., L.D.B., E.C.G., C.C., and M.E.G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: G.M., Oregon Health & Science University; and A.E.W., Duke University.
Preprint Servers: This manuscript was deposited as a preprint on July 3, 2023: bioRxiv 2023.07.03.547549; https://doi.org/10.1101/2023.07.03.547549.
Contributor Information
Chinfei Chen, Email: Chinfei.Chen@childrens.harvard.edu.
Michael E. Greenberg, Email: Michael_Greenberg@hms.harvard.edu.
Data, Materials, and Software Availability
All genomic data have been deposited in the NCBI Gene Expression Omnibus under accession number: GSE245026 (60). All other data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Amir R. E., et al. , Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Lewis J. D., et al. , Purification, sequence, and cellular localization of a novel chromosomal protein that binds to Methylated DNA. Cell 69, 905–914 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Chahrour M., Zoghbi H. Y., The story of Rett syndrome: From clinic to neurobiology. Neuron 56, 422–437 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Chao H. T., Zoghbi H. Y., Rosenmund C., MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron 56, 58–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asaka Y., Jugloff D. G. M., Zhang L., Eubanks J. H., Fitzsimonds R. M., Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol. Dis. 21, 217–227 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Na E. S., et al. , A mouse model for MeCP2 duplication syndrome: MeCP2 overexpression impairs learning and memory and synaptic transmission. J. Neurosci. 32, 3109–3117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu Z., et al. , The Rett syndrome protein MeCP2 regulates synaptic scaling. J. Neurosci. 32, 989–994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackman M. P., Djukic B., Nelson S. B., Turrigiano G. G., A critical and cell-autonomous role for MeCP2 in synaptic scaling up. J. Neurosci. 32, 13529–13536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crair M. C., Neuronal activity during development: Permissive or instructive? Curr. Opin. Neurobiol. 9, 88–93 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Huberman A. D., Feller M. B., Chapman B., Mechanisms underlying development of visual maps and receptive fields. Annu. Rev. Neurosci. 31, 479–509 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballas N., Lioy D. T., Grunseich C., Mandel G., Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat. Neurosci. 12, 311–317 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapleau C. A., et al. , Dendritic spine pathologies in hippocampal pyramidal neurons from Rett syndrome brain and after expression of Rett-associated MECP2 mutations. Neurobiol. Dis. 35, 219–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noutel J., Hong Y. K., Leu B., Kang E., Chen C., Experience-dependent retinogeniculate synapse remodeling is abnormal in MeCP2-deficient mice. Neuron 70, 35–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W. G., et al. , Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science (80-.) 302, 885–889 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Damen D., Heumann R., MeCP2 phosphorylation in the brain: From transcription to behavior. Biol. Chem. 394, 1595–1605 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Bellini E., et al. , MeCP2 post-translational modifications: A mechanism to control its involvement in synaptic plasticity and homeostasis? Front. Cell. Neurosci. 8, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L., et al. , MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E2982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert D. H., et al. , Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499, 341–345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzales M. L., Adams S., Dunaway K. W., LaSalle J. M., Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol. Cell. Biol. 32, 2894–2903 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefanelli G., et al. , Brain phosphorylation of MeCP2 at serine 164 is developmentally regulated and globally alters its chromatin association. Sci. Rep. 6, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao J., et al. , Phosphorylation of MeCP2 at serine 80 regulates its chromatin association and neurological function. Proc. Natl. Acad. Sci. U.S.A. 106, 4882–4887 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z., et al. , Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen S., et al. , Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72, 72–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooks B. M., Chen C., Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron 52, 281–291 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Hooks B. M., Chen C., Vision triggers an experience-dependent sensitive period at the retinogeniculate synapse. J. Neurosci. 28, 4807–4817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusina E., Bernard C., Williamson A., The kainic acid models of temporal lobe epilepsy. eNeuro 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy J., Cheval H., Selfridge J., Bird A., The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 27, 631–652 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Guy J., Gan J., Selfridge J., Cobb S., Bird A., Reversal of neurological defects in a mouse model of Rett syndrome. Science (80-) 315, 1143–1147 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guy J., Hendrich B., Holmes M., Martin J. E., Bird A., A mouse Mecp2-null mutation causes neurological symptoms that mimic rett syndrome. Nat. Genet. 27, 322–326 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Chen C., Regehr W. G., Developmental remodeling of the retinogeniculate synapse. Neuron 28, 955–966 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Litvina E. Y., Chen C., Functional convergence at the retinogeniculate synapse. Neuron 96, 330–338.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demas J., Eglen S. J., Wong R. O. L., Developmental loss of synchronous spontaneous activity in the mouse retina is independent of visual experience. J. Neurosci. 23, 2851–2860 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feller M. B., Wellis D. P., Stellwagen D., Werblin F. S., Shatz C. J., Requirement for cholinergic synaptic transmission in the propagation of spontaneous retinal waves. Science (80-.) 272, 1182–1187 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Piatigorsky J., Lens differentiation in vertebrates: A review of cellular and molecular features. Differentiation 19, 134–153 (1981). [DOI] [PubMed] [Google Scholar]
- 35.Greiling T. M. S., Clark J. I., The transparent lens and cornea in the mouse and zebra fish eye. Semin. Cell Dev. Biol. 19, 94–99 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong Y. K., Chen C., Wiring and rewiring of the retinogeniculate synapse. Curr. Opin. Neurobiol. 21, 228–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheadle L., et al. , Visual experience-dependent expression of Fn14 is required for retinogeniculate refinement. Neuron 99, 525–539.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X., Chen C., Different roles for AMPA and NMDA receptors in transmission at the immature retinogeniculate synapse. J. Neurophysiol. 99, 629–643 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Meister M., Wong R. L., Baylor D. A., Shatz C. J., Synchronous bursts of action potentials in ganglion cells of the developing mammalian retina. Science (80-.) 252, 939–943 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Jaubert-Miazza L., et al. , Structural and functional composition of the developing retinogeniculate pathway in the mouse. Vis. Neurosci. 22, 661–676 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Torborg C. L., Feller M. B., Unbiased analysis of bulk axonal segregation patterns. J. Neurosci. Methods 135, 17–26 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Kalish B. T., et al. , Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc. Natl. Acad. Sci. U.S.A. 115, E1051–E1060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan K., et al. , MeCP2 regulates the timing of critical period plasticity that shapes functional connectivity in primary visual cortex. Proc. Natl. Acad. Sci. U.S.A. 112, E4782–E4791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyst M. J., et al. , Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 16, 898–902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guy J., et al. , A mutation-led search for novel functional domains in MeCP2. Hum. Mol. Genet. 27, 2531–2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boxer L. D., et al. , MeCP2 represses the rate of transcriptional initiation of highly methylated long genes. Mol. Cell 77, 294–309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piccolo F. M., et al. , MeCP2 nuclear dynamics in live neurons results from low and high affinity chromatin interactions. Elife 8, 1–22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran N. M., et al. , Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron 104, 1039–1055.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rheaume B. A., et al. , Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun. 9, 2759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Picard N., Fagiolini M., MeCP2: An epigenetic regulator of critical periods. Curr. Opin. Neurobiol. 59, 95–101 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Lister R., et al. , Global epigenomic reconfiguration during mammalian brain development. Science (80-.) 341, 1237905-1–1237905–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C., Blitz D. M., Regehr W. G., Contributions of receptor desensitization and saturation to plasticity at the retinogeniculate synapse. Neuron 33, 779–788 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Hauser J. L., Liu X., Litvina E. Y., Chen C., Prolonged synaptic currents increase relay neuron firing at the developing retinogeniculate synapse. J. Neurophysiol. 112, 1714–1728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reggiani J. D. S., et al. , Brainstem serotonin neurons selectively gate retinal information flow to thalamus. Neuron 111, 711–726.e11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu A., Ibrahim J. G., Love M. I., Heavy-Tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzeng C. P., Whitwam T., Boxer L. D., Greenberg M. E., Activity-induced MeCP2 phosphorylation regulates retinogeniculate synapse refinement. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245026. Deposited 10 October 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All genomic data have been deposited in the NCBI Gene Expression Omnibus under accession number: GSE245026 (60). All other data are included in the article and/or SI Appendix.