Abstract
Neuroglobin is a hemoprotein expressed in several nervous system cell lineages with yet unknown physiological functions. Neuroglobin presents a very similar structure to that of the related globins hemoglobin and myoglobin, but shows an hexacoordinate heme as compared to the pentacoordinated heme of myoglobin and hemoglobin. While several reactions of neuroglobin have been characterized in vitro, the relative importance of most of those reactions in vivo is yet undefined. Neuroglobin, like other heme proteins, can reduce nitrite to nitric oxide, providing a possible route to generate nitric oxide in vivo in low oxygen conditions. The reaction kinetics are highly dependent on the nature of the distal residue, and replacement of the distal histidine His64(E7) can increase the reaction rate constants by several orders of magnitude. However, mutation of other distal pocket positions such as Phe28(B10) or Val68(E11) has more limited impact on the rates. Computational analysis using myoglobin as template, guided by the structure of dedicated nitrite reductases like cytochrome cd1 nitrite reductase, has pointed out that combined mutations of the residues B10 and CD1 could increase the nitrite reductase activity of myoglobin, by mimicking the environment of the distal heme pocket in cytochrome cd1 nitrite reductase. As neuroglobin shows high sequence and structural homology with myoglobin, we hypothesized that such mutations (F28H and F42Y in neuroglobin) could also modify the nitrite reductase activity of neuroglobin. Here we study the effect of these mutations. Unfortunately, we do not observe in any case an increase in the nitrite reduction rates. Our results provide some further indications of nitrite reductase regulation in neuroglobin and highlight the minor but critical differences between the structure of penta- and hexacoordinate globins.
Keywords: Neuroglobin, Nitrite reduction, Nitric oxide, Protein engineering
Highlights
-
•
Previous work pinpoint a role of B10 and CD1 amino acids in globin nitrite reduction.
-
•
Neuroglobin F28H and F42Y mutations failed to increase nitrite reduction rates.
-
•
Neuroglobin F42Y mutation alters nitrite reduction mechanism.
-
•
Differences between pentacoordinated and hexacoordinated globins are highlighted.
Abbreviations
- Cygb
cytoglobin
- cyt cd1NR
cytochrome cd1 Nitrite Reductase
- Hb
hemoglobin
- KD
dissociation constant
- Mb
myoglobin
- Ngb
neuroglobin
- WT
wild-type
1. Introduction
Neuroglobin (Ngb) is a heme protein highly conserved in vertebrates. Discovered in 2000 [1], its physiological role is still unknown. In mammals, Ngb is expressed mainly in nervous system and endocrine tissues [2,3]. Ngb evolved from a common ancestor to vertebrate myoglobin (Mb) and hemoglobin (Hb), diverging before the appearance of the ancestral Mbs and Hbs. Despite their close relationship, Ngb presents a singular heme iron coordination, with both proximal and distal histidine residues coordinating the iron in the ferrous and ferric states; in Mb and Hb only the proximal histidine coordinates the heme, and the distal side of the heme is available for ligand binding. In contrast to cytochromes, which also bear hexacoordinate heme, but do not readily bind exogenous ligands, the distal histidine of hexacoordinate globins is in a dynamic binding and dissociation equilibrium, allowing for the binding of ligands in the sixth heme coordination site [4]. Unlike Mb and Hb, the ferrous dioxygen complex (FeII-O2) of Ngb is unstable towards oxidation and therefore is not suitable for the traditional oxygen transport and delivery roles of most globins [4]. These observations have led to the exploration of other roles for Ngb [2,3].
Ngb can catalyze a variety of reactions through its heme iron, including detoxification of reactive oxygen species [[4], [5], [6]] and reduction of pro apoptotic cytochrome c [[7], [8], [9]]. Ngb may also be involved in the regulation of guanosine diphosphate exchange for the Gα protein [10,11]. Although a role in general oxidative stress response is very possible, the nature of the actual physiological role(s) of Ngb is yet undefined [2,3].
Another potential physiological mechanism of Ngb involves the regulation of nitric oxide (NO) metabolism [12]. When Ngb is bound to oxygen, it can scavenge NO by reaction of the ferrous dioxygen heme species with NO producing nitrate and ferric heme. Alternatively, deoxy (ferrous) Ngb can reduce nitrite and generate NO [13,14]. This is a reaction that can be regulated by the redox state of the Ngb thiols, and is faster when the intramolecular disulfide bond between Cys46 and Cys55 is formed [15]. The physiological relevance of this NO-generating reaction for Ngb is still unclear, but does not require oxygen, unlike the reaction catalyzed by NO synthases [16], and thus could provide a source of NO in tissues when oxygen supply is low [14,17].
The reaction of Ngb with nitrite has been previously studied [15,[18], [19], [20]]. Some features of the reaction, such as the importance of the distal histidine, have been also explored in an evolutionarily related globin, cytoglobin (Cygb) [18,[20], [21], [22], [23], [24]]. In Ngb and Cygb, the kinetics of the reaction are highly dependent on the nature of the distal residue. Mutation of this amino acid -His64(E7)- can increase the reaction rates by several orders of magnitude, with reaction rate constants up to 1120 M−1s−1 (Ngb H64A), as compared to the value for the WT protein (0.52 M−1s−1) [15,19]. The mutation of other distal pocket positions such as Phe28(B10) or Val68(E11) has a more limited impact on the rates [19].
Computational analysis have tried to mimic the heme environment of dedicated nitrite reductases like cytochrome cd1 Nitrite Reductase (cyt cd1NR) using the Mb scaffold. These studies have pointed out that mutations of residues B10 and CD1 (L29H and F43Y in Mb) could increase the nitrite reductase activity of Mb by providing an electronic environment optimal for the stabilization of heme-bound nitrite [25]. As Ngb shows high sequence and structural homology with myoglobin (Fig. 1), we hypothesized that the mutations proposed for Mb could also increase the nitrite reductase activity of Ngb. Here we study the effect of the Ngb mutations F28H and F42Y, structurally equivalent to Mb L29H and F43Y mutations. Unfortunately, we do not observe in any case an increase in the reduction rate constants in the Ngb mutants. Our results provide further indications of nitrite reductase regulation in Ngb and highlight the minor but critical differences between the structure of penta- and hexa-coordinate globins.
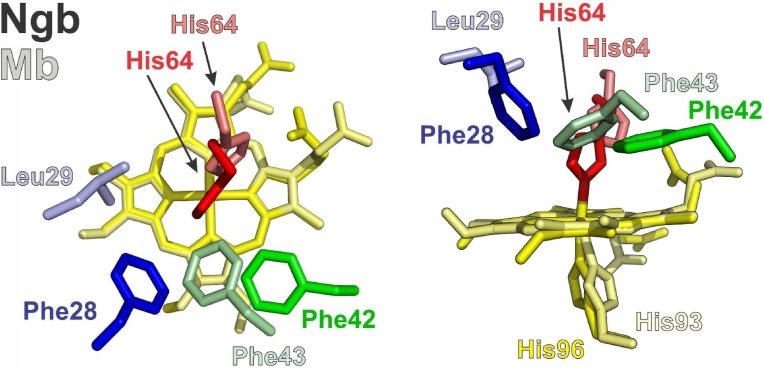
Fig. 1.
Location of selected heme pocket residues in Ngb and Mb. The relative location of neuroglobin residues studied in this work (Phe28 and Phe 42) is shown. The left panel presents a top view and the right panel a side view, relative to the heme plane. Heme moieties and side chains are shown as sticks: Ngb Phe 28, blue; Ngb Phe 42, green, Ngb His64, red, Ngb His 96 and heme, yellow; Mb Leu 29, light blue; Mb Phe43, light green; Mb His64, light red, Mb His93 and Ngb heme, pale yellow. The proximal histidines are omitted in the top view for clarity. Protein data Bank entries 1OJ6 (Ngb) and 2W6W (Mb) were used. Figure was generated with PyMOL 0.99rc6 [26].
2. Materials and methods
Reagents and protein preparation – Reagents were obtained from Sigma unless stated otherwise.
Protein expression and purification – Plasmids for the expression of Ngb mutants were generated using standard molecular biology techniques. The wild-type (WT) human Ngb protein was expressed using the pET28-Ngb plasmid as previously described [19]. The mutations F28H, F42Y and F28H/F42Y were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using adequate primers. Purification of the proteins from SoluBL21 cells (Genlantis, San Diego, CA) expressing the corresponding plasmids was accomplished by DEAE anion exchange chromatography followed by centrifugation through an Amicon Ultra centrifugal filter (Millipore) with a 50 kDa cutoff to remove high-molecular weight contaminants. The flow-through was concentrated using a 10 kDa cutoff Amicon Ultra centrifugal filter (Millipore) and buffer exchanged to 100 mM phosphate buffer (pH 7.4) as previously described [19].
Determination of melting temperatures – Thermal denaturation of WT and mutant Ngbs in their ferric heme form was monitored by UV–Visible spectroscopy as previously reported [27] with minor modifications. The experiments were conducted in a Cary100 Spectrophotometer (Agilent Technologies, Santa Clara, CA). Reactions were carried out in 10 mM phosphate-buffered saline (pH 7.4) with stirring. The temperature was increased from 20 to 100 °C; for each step, the temperature was maintained for ∼5 min and the spectra were then recorded. Melting temperatures (Tm) were determined by fitting the absorbance changes in the Soret peak to the Santoro-Bolen equation [28].
Determination of autoxidation rates – The autoxidation of the ferrous dioxygen complexes was studied at 37 °C in sodium phosphate buffer 100 mM, pH 7.4 as previously described [19]. Briefly, the ferrous dioxygen complexes were prepared by reduction of the globins with excess sodium dithionite (Acros Organics) to form the reduced, ferrous deoxy form (FeII) of the protein, followed by gel filtration through a Sephadex G25-column (PD10, GE Healthcare) equilibrated with aerobic buffer. As the protein is eluted in the column and dithionite is removed from the sample, the ferrous deoxy globin binds dissolved oxygen from the buffer to yield a final elution sample containing mostly the ferrous dioxygen species. These concentrated protein samples were mixed with prewarmed buffer at 37 °C and the spectral changes were monitored for 30–60 min in a Cary-50 spectrophotometer (Agilent Technologies, Santa Clara, CA). In the case of the F28H and F28H/F42Y mutants, the fast autoxidation yields an almost completely oxidized, ferric protein (FeIII) after the G25-column. In these cases, a modified procedure was used [19]. The ferrous deoxy Ngb species was prepared by titration of the ferric protein with dithionite in a closed cuvette. The reaction was then started by addition of aerobic buffer in a 1:4 (v:v) ratio. All the reaction rates were determined at 37 °C using a Cary 50 spectrophotometer with a thermostated cell holder. Spectral changes were monitored between 450 and 700 nm for slower reactions, or between 380 and 480 nm (Soret peak) or 500 and 600 nm (Q bands) for fast reactions. Scans were taken every 12 or 6 s at a scan rate of 2400 nm/min. The wavelengths showing maximal absorbance changes between ferrous dioxygen and ferric Ngb species were used to determine the autoxidation rates. Spectral changes were fit to a single-exponential equation using Origin 8.0 software (OriginLab Corp., Northampton, MA).
Nitrite reduction experiments – Nitrite reduction experiments were performed in 100 mM sodium phosphate buffer, pH 7.4, at 37 °C. The reactions were followed in the presence of 2.5 mM sodium dithionite as described previously [15,19,20]. The presence of excess dithionite generates quantitatively the ferrous deoxy heme species, reduces the ferric species formed during the reaction back to the ferrous deoxy form, and consumes oxygen avoiding side reactions between nitrite and ferrous dioxygen heme [29]. In these conditions, ferrous deoxy heme reacts with nitrite producing NO and ferric heme. The ferric species is reduced to ferrous deoxy heme by dithionite, and the NO binds to ferrous deoxy heme until all the heme is bound to NO, with the ferrous nitrosyl species (FeII-NO) being the final product. Samples of Ngbs (5–10 μM) were mixed with different concentrations of nitrite (typically between 2 and 20 mM) and the spectral changes were monitored in the 350–700 nm range. Absorbance changes were fit to a single or double exponential equation as described [15,19,20].
Statistical Analysis – Data were analyzed using Origin 8.0 (OriginLab Corporation, Northampton, MA) and expressed as mean ± standard deviation of the mean.
3. Results
Mutant selection – We selected the mutations F28H, F42Y and the double mutant F28H/F42Y by analogy with the mutants selected for Mb in the computational study by Lin et al. [25]. In Ngb, F28 and F42 are the structural equivalent of myoglobin residues L29 and F43, respectively (Fig. 1). Heterologous expression of the WT protein and mutants were performed as described [19]. There were no obvious differences in yield or stability for the mutants studied.
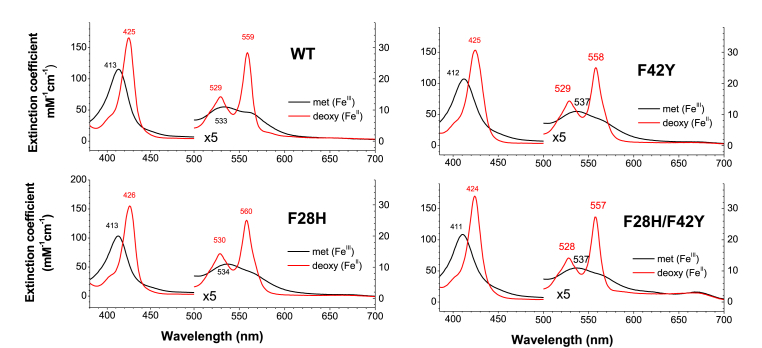
Spectral properties - The three mutant Ngbs (F28H, F42Y, F28H/F42Y) did not present major changes in the spectra of their ferrous deoxy or ferric species (Fig. 2). The spectrum of the ferrous deoxy heme is similar for all the Ngb mutants, showing a double peak with maxima around 530 nm and 560 nm. These spectra are consistent with distal histidine coordination to yield a hexacoordinate, ferrous deoxy heme species similar to those observed for WT Ngb and Cygb. The F28H mutant spectra were the most similar to the WT spectra, with shifts of 1 nm or less for the peaks of both ferrous and ferric species. Soret and Q-band maxima for ferrous deoxy and ferric F42Y and F28H/F42Y species showed hypsochromic (blue) shifts of 1–2 nm for both ferrous deoxy and ferric species, except for the ferric Q band (537 nm), which presented a bathochromic (red) shift of 4 nm vs the WT spectra (Fig. 2). These changes in the Soret peak are smaller, but comparable to those observed in F43Y and L29H/F43Y Mb [30].
Fig. 2.
Spectral properties of wild type Neuroglobin and mutants. UV–Vis spectra for the ferric (FeIII) and ferrous deoxy (FeII) species are shown in black and red respectively. Q-band region (500–700 nm) is expanded for clarity.
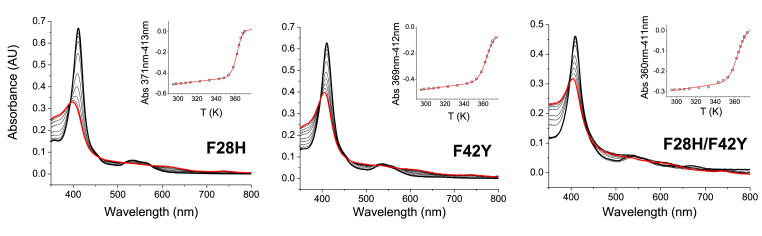
Thermal stability of Ngb mutants - In the study of Mb mutant L29H/F43Y, unfolding data revealed that the substituted tyrosine residue can form a covalent cross-link with a vinyl group of the heme porphyrin ring [30]. These changes were apparent during chemical denaturation studies, as denaturation of the native-like Mb species results in absorbance decrease in the protein-bound Soret peak at 406 nm with concomitant increase in a Soret feature at around 370 nm, consistent with heme release. In contrast, chemical unfolding of cross-linked L29H/F43Y Mb resulted in a decrease in Soret intensity and blue shift to 410 nm without absorbance increase at 370 nm [30]. To determine if such a cross-link forms in the analogous Ngb mutations, we performed thermal denaturation studies of the F28H, F42Y and F28H/F42Y mutants. Spectral changes during the thermal denaturation of F28H, F42Y and F28H/F42Y are shown in Fig. 3. The three mutants show a very high thermal stability (Tm > 90 °C, Fig. 3, Table 1), that is only slightly lower than that observed for the WT protein [31]. Indeed, the denaturation is not completed in any of the mutants by the experimental upper limit of the instrument (around 98 °C), likely giving rise to higher than usual experimental error in the Tm values as indicated in Table 1. We do not observe spectral differences in the denaturation process of the F42Y or F28H/F42Y Ngb mutants compared to those observed for F28H or WT, suggesting that in the Ngb mutants the tyrosine does not form a covalent cross-link with the heme, as reported for the Mb L29F/F43Y mutant [30].
Fig. 3.
Thermal denaturation of WT Ngb and mutants. The spectra for the ferric heme species determined at increasing temperatures are shown. The initial spectrum (20 °C) is denoted by a thick black line, and the final spectrum (approx. 98 °C) is denoted by a thick red line. Intermediate spectra are shown as thin black lines. The insets show the changes in the Soret peak absorbance with the temperature and the fit of the absorbance changes to the Santoro−Bolen equation (red lines).
Table 1.
Melting temperatures of wild-type and mutant neuroglobins.
| Neuroglobin | Tm (°C) |
|---|---|
| Wild-type | 100a |
| F28H | 90.3 ± 1.6 |
| F42Y | 93.6 ± 4.2 |
| F28H/F42Y | 93.4 ± 5.0 |
Values determined in 10 mM phosphate buffered saline, pH 7.4. a WT value from Ref. [31] in 10 mM phosphate buffer, pH 7.0.
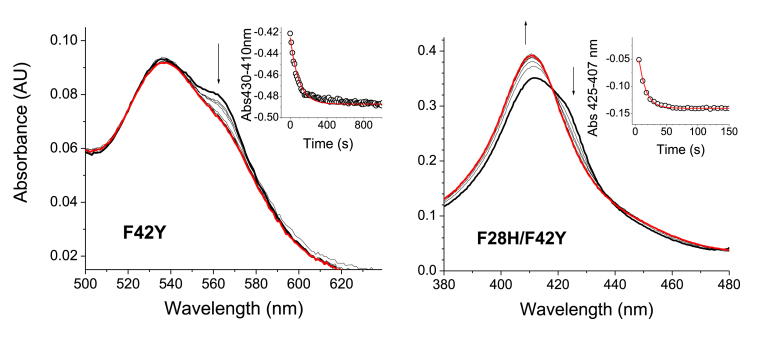
Autoxidation of neuroglobin mutants – Like other globins, ferrous deoxy WT Ngb can bind molecular oxygen to form a ferrous dioxygen complex. However, unlike bona fide oxygen carriers like Mb or Hb, the ferrous dioxygen complex of WT Ngb decays quickly with formation of superoxide and ferric Ngb (k = 0.23 min−1, corresponding to a half-life of 3 min for WT Ngb [19]). We determined the autoxidation rates for all the Ngb mutants generated in this work. Our previous work indicates that for hexacoordinated globins, these rates are very sensitive to mutations around the heme pocket [19] but usually not altered by changes far from the heme moiety [8,22]. In addition, we have observed that the autoxidation rates in hexacoordinate globins depends mainly on the heme redox potential, with more negative potentials corresponding to faster autoxidation rates. Therefore we can also use the autoxidation rates as a surrogate for changes in redox potential [19]. The spectral changes for the reaction of the Ngb mutants are shown in Fig. 4 and our calculated rates are shown in Table 2.
Fig. 4.
Autoxidation of Ngb mutants. The plots show selected spectra during the course of the autoxidation reaction for each mutant. As changes for Ngb F28H/F42Y in the Q-band were very small, the Soret band is shown instead. The insets show the fitting of the decay to a single exponential equation. The initial spectrum is denoted by a thick black line, and the final spectrum is denoted by a thick red line Arrows indicate the direction of the spectral change.
Table 2.
Autoxidation rates of wild-type and mutant neuroglobins.
| Neuroglobin | Rate (min−1) |
|---|---|
| Wild-type | 0.23 ± 0.03a |
| F28H | 11.3 ± 0.7a |
| F28H/F42Y | 5.03 ± 0.47 |
| F42Y | 0.82 ± 0.24 |
Values determined at 37 °C in 100 mM sodium phosphate, pH 7.4. a values from Ref. [19].
The F42Y mutant shows a faster autoxidation than for the WT enzyme, with a 3.5-fold increase in the autoxidation rate. Notably, there is no significant formation of a ferrous dioxygen species, and the reaction seems to involve the direct outer sphere oxidation of the ferrous deoxy heme, without transient formation of a ferrous dioxygen species. This behavior is unusual in Ngb, which usually favors inner sphere oxidation of the ferrous dioxygen complex, but has been observed for the Ngb F28H and V68A mutants [19]. Both autoxidation mechanisms, involving ferrous dioxygen or ferrous deoxy species usually coexist in heme proteins, albeit the outer sphere mechanisms is, in most cases, only apparent at low oxygen concentrations [32].
The effect of the F28H mutation on autoxidation rates is more dramatic, as already observed [19] and consistent with similar trends reported for myoglobin B10 substitutions [32,33]. The replacement of the phenylalanine in position B10 causes 50-fold (F28H) and 22-fold (F28H/F42Y) increases in the autoxidation rates. Similar to the changes observed during the reaction of F28H [19], and F42Y, the reaction for F28H/F42Y mutant does not show significant buildup of a ferrous dioxygen complex, and also appears to proceed via direct outer sphere oxidation of the ferrous deoxy heme.
Nitrite reduction by Ngb mutants – The ferrous deoxy heme reacts with nitrite yielding ferric heme and ferrous nitrosyl heme as final products [13,14,34]. The reaction can be described according to the equations:
| FeII + NO2− + H+ → FeIII + NO + OH− | (equation 1) |
| FeII + NO → FeIINO | (equation 2) |
Where reaction 1 is rate limiting. We monitored nitrite reduction using excess dithionite as described previously [15,19,20]. Under these conditions, the ferric heme species is reduced to ferrous deoxy heme, with the overall reaction yielding 100% ferrous-nitrosyl species as final product. The spectral changes during the reaction and the observed rate constants at pH 7.4 are shown in Fig. 5 and Table 3.
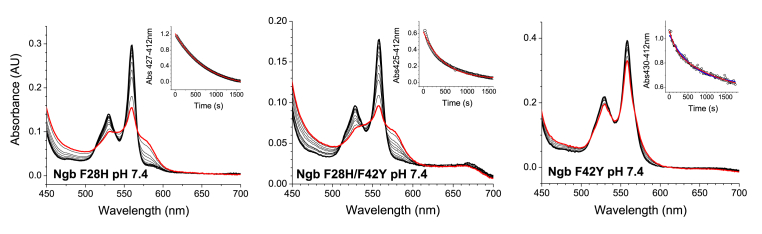
Fig. 5.
Absorbance decay for the reaction of neuroglobin mutants with nitrite at pH 7.4. Traces correspond to the reaction of F28H (10 μM) with 5 mM nitrite; F28H/F42Y (6 μM) with 10 mM nitrite and F42Y (9 μM) with 10 mM nitrite. The fit of the observed data to a single exponential equation is indicated by a solid red line for the F28H and F28H/F42Y mutants. In the case of the F42Y mutant, the fit to a double exponential equation is denoted by solid red line and the fit to a single exponential equation is indicated by a solid blue line.
Table 3.
Nitrite reductase rates of selected wild-type globins and mutants.
The Ngb F28H mutant has a slightly slower nitrite reduction rate than WT, as shown previously [19] (Table 3). The F42Y and F28H/F42Y mutants show even slower bimolecular rate constants, with a 3-fold decrease for F28H/F42Y and a 7-fold decrease for F42Y. It is also apparent in Fig. 5 that a very small fraction of the protein has reacted for F42Y, whereas the reaction of F28H and F28H/F42Y, although incomplete, has progressed substantially. The decay of the ferrous deoxy species for F28H and F28H/F42Y follows kinetics consistent with a pseudo-first order process, as generally observed for this reaction [15,19,20]. However, the F42Y mutant consistently shows a more complex pattern. We fitted the decay for F42Y to a double exponential fit, which provided a better fit of the observed data (Fig. 5, Fig. 6, Fig. 7).
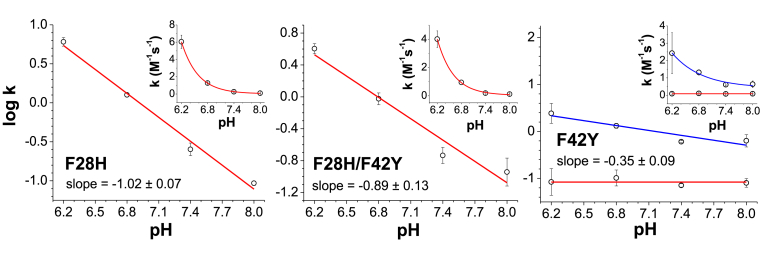
Fig. 6.
Dependence of the reaction rate constants versus the pH for the neuroglobin mutants. The main panels show the logarithm of the reaction rate constants versus the pH; the insets show the reaction rate constants versus the pH. The linear fit of the logarithm of the rate constants is denoted by a solid red line. In the case of the F42Y mutant, red lines indicate the fit of the slow phase, and blue lines denote the fit of the fast phase rate constants. The slope of the linear fit is indicated in each panel.
Fig. 7.
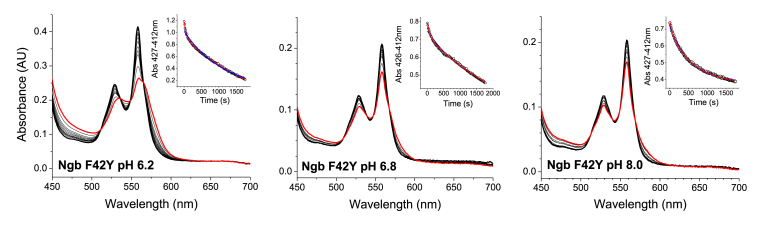
Absorbance decay for the reaction of neuroglobin F42Y mutant at varying pH. Traces correspond to the reaction of F42Y (9 μM) with 10 mM nitrite (pH 6.2) 10 mM nitrite (pH 6.8) and 20 mM nitrite pH (8.0). Fit to a double exponential equation is denoted by red lines. Fit to a single exponential equation is indicated by blue lines.
pH dependence of the nitrite reduction reaction - As noted in equation (1), the reduction is dependent upon the concentration of protons, as the reacting species is usually HNO2 [34,36]. To determine if the mutants show a similar pattern, we studied the reaction for the three Ngb mutants in the 6.2–8.0 pH range. The observed values are shown in Fig. 6. For F28H and F28H/F42Y mutants, the observed bimolecular reaction rate constants vary linearly with [H+] and the calculated slope is within experimental error of the expected value of −1 according to equation (1). This is similar to the behavior observed for the nitrite reduction process in most heme proteins [15,20,34,37]. As noted above for the F42Y mutant, the traces do not fit well to a single exponential equation and are better fit to a double exponential equation. The fast phase represents around 25% of the absorbance change at pH 6.2 and its contribution decreases as the pH increases; a single exponential fit describes the decay accurately at pH 8.0 (Fig. 7). Also, the observed rate for the fast phase decreases at increasing pH, whereas the slow phase is not sensitive to the pH changes. The dependence of the fast phase does not yield a slope of −1 as expected from equation (1) (Fig. 6), but that may be due to the larger experimental error given the smaller contribution of the fast phase to the absorbance change observed. Altogether, it appears that the reaction of the F42Y mutant with nitrite involves mainly the reaction with the anion NO2− species, and only a small population of the protein, lost at high pH, can react via the nitrous acid species.
4. Discussion
Nitrite reduction is a notable process in biology. In bacteria, this step is a critical element of the denitrification process [38]. Plants and cyanobacteria may use non symbiotic hemoglobins to produce NO from nitrite in hypoxic conditions [39,40]. Finally, substantial evidence indicates that nitrite reduction supports vasodilation in mammalian circulation [41,42], a reaction with important physiological and clinical ramifications [14,17].
As nitrite reduction is one of the possible roles of Ngb in vivo, we have conducted several studies trying to elucidate the role of the heme pocket on the regulation of nitrite reduction rates [15,19,20]. In this context, we wanted to determine if the modifications proposed by Lin et al. to modify the heme pocket of myoglobin to create a distal pocket environment similar to that of cyt cd1NR [25] would have the same effect in Ngb. These changes involve the replacement of the B10 residue (Leu29 in Mb, Phe28 in Ngb) with a histidine and the amino acid in position CD1 (Phe43 in Mb, Phe42 in Ngb) with a tyrosine residue. The introduction of the H-bond donor side chains would, in theory, improve the stability of the intermediate heme-bound nitrite species.
The replacement of these amino acids did yield a functional protein, with minor spectral and stability changes. However, we did not observe an increase in the nitrite reductase rates for the F28H/F42Y mutant. Of note, the same group encountered problems with the function of these mutations in Mb, as the F43Y mutation caused the formation of a covalent bond between the tyrosine side chain and the vinyl group of the heme [30]. We did not observe this phenomenon in Ngb, even though the orientation of Ngb F42Y could place the tyrosine in the proximity of one of the heme vinyl groups (Fig. 8). Studies by the same group noted that further modifications were required to actually increase Mb nitrite reductase activity, and only the mutations L29H, F43H and H64A in Mb increased nitrite reductase rates by 8-fold [35] (Table 3).
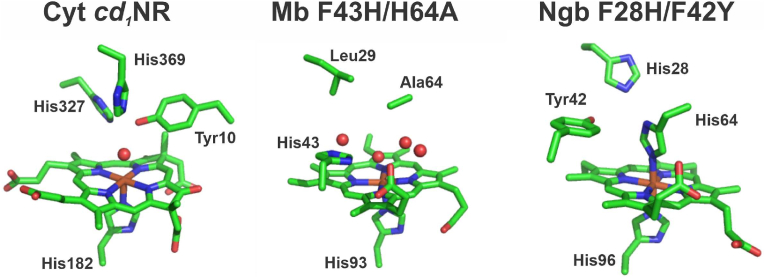
Fig. 8.
Comparison of the distal heme environment in cytochrome cd1 Nitrite Reductase, Mb F43H/H64A mutant and neuroglobin F28H/F42Y mutant. Left, Pseudomonas aeruginosa cyt cd1NR; Middle, sperm whale F43H/H64 A Mb; Right, putative structure of the human Ngb F28H/F42Y mutant. Heme and selected amino acids are shown as sticks. Water molecules (oxygen atom) in the proximity of the heme iron are shown as red spheres. Ngb structure was generated with PyMOL 0.99rc6 using the mutagenesis option and selecting for the rotamer with less steric clashes. Figure was generated with PyMOL 0.99rc6 [26]. Protein Data Bank entries 1NIR (cyt cd1NR), 5HLQ (Mb), and 1OJ6 (Ngb) were used.
The comparison of the cyt cd1NR, Mb and Ngb structures indicates that the distal pocket -where oxygen or other ligands like nitrite/nitrous acid will bind-is more constricted in Ngb (Fig. 8). In fact, the dissociation equilibrium of Ngb His64 is the main limitation to ligand binding in Ngb [4,6,15,19,43]. We and others have studied the His64(E7) role in Mb nitrite reductase activity, and observed that Mb H64A, H64L and H64V mutations decrease nitrite reduction in Mb, whereas the H64L and H64A replacements vastly increase the rate constants for Ngb [15,44]. This fact is likely related to the more crowded distal pocket in Ngb, whereas in WT myoglobin the distal heme pocket is already large enough to accommodate the nitrite molecule (Fig. 8). In fact, in Mb His64 side chain is too far to coordinate the heme directly, but optimally placed to stabilize polar ligands [45,46]. As noted above, the H64A mutation in Mb can lead to an increase in nitrite reduction rate constants when combined with L29H and F43H mutations [35]. It is possible that the similar combination F28H/F42H/H64A mutant could increase reactivity in Ngb, but this effect is probably modest compared to the effect of H64A alone [19]. Notably, the Mb F43H/H64A mutant introduces a large network of water molecules in the Mb pocket that could be displaced by the nitrite without disturbing the distal pocket side chains [35], which seems unlikely to happen in the more closed Ngb pocket, even for the F28H/F42H/H64A mutant (Fig. 8).
In globins, the residue in position CD1 has an important role in heme binding and stability. It has been shown that even conservative mutations to hydrophobic, non-aromatic amino acids cause fast heme loss [47]. Also, as noted above, F43Y Mb can undergo a reaction with one of the heme vinyl groups forming a covalent adduct [30]. We do not observe this effect in our Ngb mutants, but there is an interesting effect on nitrite reduction. In this case, the pH dependence of the reaction is largely diminished, indicating a reaction mainly through the nitrite species and less dependent on nitrous acid. The deviation of the reaction rates from the expected linear dependence has been noted for Ngb at high pH, and it is more noticeable in Cygbs [20], but is notably exacerbated for the F42Y mutant (Fig. 6, Fig. 7). Given the slow reaction rates for the F42Y mutant, it seems plausible that this state is reached due to a much slower reaction with nitrous acid, rather than due to an increase in the rate of reaction for nitrite. In the absence of structural data, we speculate that this can be due to a further stabilization of the His64 binding to the heme iron, perhaps through a hydrogen bond interaction of the tyrosine hydroxyl with the histidine, directly or via a water molecule. The presence of a histidine in position B10 avoids this effect in the F28H/F42Y mutant, probably by providing some electrostatic repulsion with His64 that leads to a faster dissociation rate of the distal histidine. In support of this hypothesis, Smagghe et al. have shown that different mutations of the native Phe(B10) residue in rice hexacoordinate hemoglobin cause an increase of the distal histidine dissociation rate, and overall dissociation constants for the distal histidine [48].
The effects of B10 mutations in globins have been widely studied. This side chain is critical in regulating binding to the distal heme side, and in particular autoxidation rates in both penta- [32,33,49,50] and hexacoordinated globins [19,48,51,52]. We observe a mildly destabilizing effect, with a decrease of 10 °C in the Tm values, but other than the large increase in autoxidation rates, the effect on the nitrite reduction reaction is small. It is possible that in Ngb, the contribution of the B10 residue is mainly transduced by a gating effect due to the steric bulk of the native Phe, and that this gating regulates the binding of gaseous ligands [51]. Moreover, the F28H Ngb mutant apparently does not form stabilizing interactions with the bound nitrite molecule, directly or through H-bonding (Fig. 8).
Another possible limitation in the redesign of nitrite reductase ability of Ngb is the starting heme redox potential. A higher redox potential of the heme seems to favor the reaction, at least in the bacterial nitrite reductases. The cd1 heme of Thiosphaera pantotropha cyt cd1NR has a redox potential of around +250 mV [38], compatible with the potentials of its physiological redox partners [53]. The cyt cd1NR redox potential may be also optimal for a fast nitrite reduction process. The value reported for cyt cd1NR is still considerably more positive than the potential of Mb (+59 mV) [54] and higher than Ngb (−118/−129 mV) [4,19]. It is unclear, however, if this factor is a main driver of the nitrite reduction process for globins. In the case of Ngb, large changes in the reaction rate constant are achieved by mutation of the His64 residue, whereas the redox potential of the heme was almost unchanged for the His64 mutants studied, indicating that other aspects can also influence the reaction [19].
Altogether, even though we did not generate Ngb mutants with increased nitrite reductase activity, we present important observations about Ngb reactivity in the context of related globin proteins. We found that the Ngb F42Y mutation can further stabilize the distal histidine binding, rendering the protein almost inactive towards nitrite while keeping other properties such as thermal stability and autoxidation relatively intact. Our results showcase the specific features and differences between the long studied, pentacoordinate globins and the growing family of hexacoordinated globins.
Funding
This work was supported by funding from Vitalant and the Hemophilia Center of Western Pennsylvania (to Stephen Y. Chan, University of Pittsburgh), National Institutes of Health Grants number F32 HL162381 and K99 HL168224 (to M.R.D) and National Institutes of Health Grant number R01 HL125886 to Mark T. Gladwin and J.T.
Declaration of competing interest
The authors declare the following competing financial interest(s): J.T. is a shareholder and officer of Globin Solutions, Inc. M.R.D. and J.T. are co-inventors of provisional and pending patents for the use of recombinant Cygb, Ngb, and other heme-based molecules as antidotes for carbon monoxide poisoning. Globin Solutions, Inc., has licensed this technology.
Acknowledgements
M.W. was part of the Summer Undergraduate Research Program at the University of Pittsburgh.
Data availability
Data will be made available on request.
References
- 1.Burmester T., Weich B., Reinhardt S., Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407(6803):520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 2.Burmester T., Hankeln T. Function and evolution of vertebrate globins. Acta Physiol. 2014;211(3):501–514. doi: 10.1111/apha.12312. [DOI] [PubMed] [Google Scholar]
- 3.Keppner A., Maric D., Correia M., Koay T.W., Orlando I.M.C., Vinogradov S.N., Hoogewijs D. Lessons from the post-genomic era: globin diversity beyond oxygen binding and transport. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewilde S., Kiger L., Burmester T., Hankeln T., Baudin-Creuza V., Aerts T., Marden M.C., Caubergs R., Moens L. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J. Biol. Chem. 2001;276(42):38949–38955. doi: 10.1074/jbc.M106438200. [DOI] [PubMed] [Google Scholar]
- 5.Brunori M., Giuffre A., Nienhaus K., Nienhaus G.U., Scandurra F.M., Vallone B. Neuroglobin, nitric oxide, and oxygen: functional pathways and conformational changes. Proc. Natl. Acad. Sci. U.S.A. 2005;102(24):8483–8488. doi: 10.1073/pnas.0408766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trent J.T., 3rd, Watts R.A., Hargrove M.S. Human neuroglobin, a hexacoordinate hemoglobin that reversibly binds oxygen. J. Biol. Chem. 2001;276(32):30106–30110. doi: 10.1074/jbc.C100300200. [DOI] [PubMed] [Google Scholar]
- 7.Raychaudhuri S., Skommer J., Henty K., Birch N., Brittain T. Neuroglobin protects nerve cells from apoptosis by inhibiting the intrinsic pathway of cell death. Apoptosis. 2010;15(4):401–411. doi: 10.1007/s10495-009-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tejero J. Negative surface charges in neuroglobin modulate the interaction with cytochrome c. Biochem. Biophys. Res. Commun. 2020;523(3):567–572. doi: 10.1016/j.bbrc.2019.12.089. [DOI] [PubMed] [Google Scholar]
- 9.Tiwari P.B., Chapagain P.P., Uren A. Investigating molecular interactions between oxidized neuroglobin and cytochrome c. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-28836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitatsuji C., Kurogochi M., Nishimura S., Ishimori K., Wakasugi K. Molecular basis of guanine nucleotide dissociation inhibitor activity of human neuroglobin by chemical cross-linking and mass spectrometry. J. Mol. Biol. 2007;368(1):150–160. doi: 10.1016/j.jmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Wakasugi K., Nakano T., Morishima I. Oxidized human neuroglobin acts as a heterotrimeric Galpha protein guanine nucleotide dissociation inhibitor. J. Biol. Chem. 2003;278(38):36505–36512. doi: 10.1074/jbc.M305519200. [DOI] [PubMed] [Google Scholar]
- 12.Tejero J., Shiva S., Gladwin M.T. Sources of vascular nitric oxide and reactive oxygen species and their regulation. Physiol. Rev. 2019;99(1):311–379. doi: 10.1152/physrev.00036.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tejero J., Gladwin M.T. The globin superfamily: functions in nitric oxide formation and decay. Biol. Chem. 2014;395(6):631–639. doi: 10.1515/hsz-2013-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dent M.R., DeMartino A.W., Tejero J., Gladwin M.T. Endogenous hemoprotein-dependent signaling pathways of nitric oxide and nitrite. Inorg. Chem. 2021;60(21):15918–15940. doi: 10.1021/acs.inorgchem.1c01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiso M., Tejero J., Basu S., Azarov I., Wang X., Simplaceanu V., Frizzell S., Jayaraman T., Geary L., Shapiro C., Ho C., Shiva S., Kim-Shapiro D.B., Gladwin M.T. Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 2011;286(20):18277–18289. doi: 10.1074/jbc.M110.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuehr D.J., Haque M.M. Nitric oxide synthase enzymology in the 20 years after the Nobel Prize. Br. J. Pharmacol. 2019;176(2):177–188. doi: 10.1111/bph.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundberg J.O., Weitzberg E., Gladwin M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008;7(2):156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 18.Petersen M.G., Dewilde S., Fago A. Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J. Inorg. Biochem. 2008;102(9):1777–1782. doi: 10.1016/j.jinorgbio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Tejero J., Sparacino-Watkins C.E., Ragireddy V., Frizzell S., Gladwin M.T. Exploring the mechanisms of the reductase activity of neuroglobin by site-directed mutagenesis of the heme distal pocket. Biochemistry. 2015;54(3):722–733. doi: 10.1021/bi501196k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corti P., Ieraci M., Tejero J. Characterization of zebrafish neuroglobin and cytoglobins 1 and 2: zebrafish cytoglobins provide insights into the transition from six-coordinate to five-coordinate globins. Nitric Oxide. 2016;53:22–34. doi: 10.1016/j.niox.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Reeder B.J., Ukeri J. Strong modulation of nitrite reductase activity of cytoglobin by disulfide bond oxidation: implications for nitric oxide homeostasis. Nitric Oxide. 2018;72:16–23. doi: 10.1016/j.niox.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Kaliszuk S.J., Morgan N.I., Ayers T.N., Sparacino-Watkins C.E., DeMartino A.W., Bocian K., Ragireddy V., Tong Q., Tejero J. Regulation of nitrite reductase and lipid binding properties of cytoglobin by surface and distal histidine mutations. Nitric Oxide. 2022;125:12–22. doi: 10.1016/j.niox.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X., Tong J., Zweier J.R., Follmer D., Hemann C., Ismail R.S., Zweier J.L. Differences in oxygen-dependent nitric oxide metabolism by cytoglobin and myoglobin account for their differing functional roles. FEBS J. 2013;280(15):3621–3631. doi: 10.1111/febs.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Hemann C., Abdelghany T.M., El-Mahdy M.A., Zweier J.L. Characterization of the mechanism and magnitude of cytoglobin-mediated nitrite reduction and nitric oxide generation under anaerobic conditions. J. Biol. Chem. 2012;287(43):36623–36633. doi: 10.1074/jbc.M112.342378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y.-W., Nie C.-M., Liao L.-F. Rational design of a nitrite reductase based on myoglobin: a molecular modeling and dynamics simulation study. J. Mol. Model. 2012;18:4409–4415. doi: 10.1007/s00894-012-1451-y. [DOI] [PubMed] [Google Scholar]
- 26.DeLano W.L. 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 27.Amdahl M.B., Petersen E.E., Bocian K., Kaliszuk S.J., DeMartino A.W., Tiwari S., Sparacino-Watkins C.E., Corti P., Rose J.J., Gladwin M.T., Fago A., Tejero J. The zebrafish cytochrome b5/cytochrome b5 reductase/NADH system efficiently reduces cytoglobins 1 and 2: conserved activity of cytochrome b5/cytochrome b5 reductases during vertebrate evolution. Biochemistry. 2019;58(29):3212–3223. doi: 10.1021/acs.biochem.9b00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoro M.M., Bolen D.W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27(21):8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 29.Keszler A., Piknova B., Schechter A.N., Hogg N. The reaction between nitrite and oxyhemoglobin: a mechanistic study. J. Biol. Chem. 2008;283(15):9615–9622. doi: 10.1074/jbc.M705630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan D.-J., Yuan H., Li W., Xiang Y., He B., Nie C.-M., Wen G.-B., Lin Y.-W., Tan X. How a novel tyrosine–heme cross-link fine-tunes the structure and functions of heme proteins: a direct comparitive study of L29H/F43Y myoglobin. Dalton Trans. 2015;44(43):18815–18822. doi: 10.1039/c5dt03040d. [DOI] [PubMed] [Google Scholar]
- 31.Hamdane D., Kiger L., Dewilde S., Uzan J., Burmester T., Hankeln T., Moens L., Marden M.C. Hyperthermal stability of neuroglobin and cytoglobin. FEBS J. 2005;272(8):2076–2084. doi: 10.1111/j.1742-4658.2005.04635.x. [DOI] [PubMed] [Google Scholar]
- 32.Brantley R.E., Jr., Smerdon S.J., Wilkinson A.J., Singleton E.W., Olson J.S. The mechanism of autooxidation of myoglobin. J. Biol. Chem. 1993;268(10):6995–7010. [PubMed] [Google Scholar]
- 33.Carver T.E., Brantley R., Singleton E., Arduini R., Quillin M.L., Phillips G., Olson J. A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autooxidation rate. J. Biol. Chem. 1992;267(20):14443–14450. [PubMed] [Google Scholar]
- 34.Doyle M.P., Pickering R.A., DeWeert T.M., Hoekstra J.W., Pater D. Kinetics and mechanism of the oxidation of human deoxyhemoglobin by nitrites. J. Biol. Chem. 1981;256(23):12393–12398. [PubMed] [Google Scholar]
- 35.Wu L.B., Yuan H., Gao S.Q., You Y., Nie C.M., Wen G.B., Lin Y.W., Tan X. Regulating the nitrite reductase activity of myoglobin by redesigning the heme active center. Nitric Oxide. 2016;57:21–29. doi: 10.1016/j.niox.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Doyle M.P., LePoire D.M., Pickering R.A. Oxidation of hemoglobin and myoglobin by alkyl nitrites inhibition by oxygen. J. Biol. Chem. 1981;256(23):12399–12404. [PubMed] [Google Scholar]
- 37.Shiva S., Huang Z., Grubina R., Sun J., Ringwood L.A., MacArthur P.H., Xu X., Murphy E., Darley-Usmar V.M., Gladwin M.T. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ. Res. 2007;100(5):654–661. doi: 10.1161/01.RES.0000260171.52224.6b. [DOI] [PubMed] [Google Scholar]
- 38.Baker S.C., Saunders N.F., Willis A.C., Ferguson S.J., Hajdu J., Fülöp V. Cytochrome cd1 structure: unusual haem environments in a nitrite reductase and analysis of factors contributing to β-propeller folds. J. Mol. Biol. 1997;269(3):440–455. doi: 10.1006/jmbi.1997.1070. [DOI] [PubMed] [Google Scholar]
- 39.Sturms R., DiSpirito A.A., Hargrove M.S. Plant and cyanobacterial hemoglobins reduce nitrite to nitric oxide under anoxic conditions. Biochemistry. 2011;50(19):3873–3878. doi: 10.1021/bi2004312. [DOI] [PubMed] [Google Scholar]
- 40.Tiso M., Tejero J., Kenney C., Frizzell S., Gladwin M.T. Nitrite reductase activity of nonsymbiotic hemoglobins from Arabidopsis thaliana. Biochemistry. 2012;51(26):5285–5292. doi: 10.1021/bi300570v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosby K., Partovi K.S., Crawford J.H., Patel R.P., Reiter C.D., Martyr S., Yang B.K., Waclawiw M.A., Zalos G., Xu X., Huang K.T., Shields H., Kim-Shapiro D.B., Schechter A.N., Cannon R.O., 3rd, Gladwin M.T. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat. Med. 2003;9(12):1498–1505. doi: 10.1038/nm954. [DOI] [PubMed] [Google Scholar]
- 42.van Faassen E.E., Bahrami S., Feelisch M., Hogg N., Kelm M., Kim-Shapiro D.B., Kozlov A.V., Li H., Lundberg J.O., Mason R., Nohl H., Rassaf T., Samouilov A., Slama-Schwok A., Shiva S., Vanin A.F., Weitzberg E., Zweier J., Gladwin M.T. Nitrite as regulator of hypoxic signaling in mammalian physiology. Med. Res. Rev. 2009;29:683–741. doi: 10.1002/med.20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fago A., Hundahl C., Dewilde S., Gilany K., Moens L., Weber R.E. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J. Biol. Chem. 2004;279(43):44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- 44.Yi J., Heinecke J., Tan H., Ford P.C., Richter-Addo G.B. The distal pocket histidine residue in horse heart myoglobin directs the O-binding mode of nitrite to the heme iron. J. Am. Chem. Soc. 2009;131(50):18119–18128. doi: 10.1021/ja904726q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dou Y., Olson J.S., Wilkinson A.J., Ikeda-Saito M. Mechanism of hydrogen cyanide binding to myoglobin. Biochemistry. 1996;35(22):7107–7113. doi: 10.1021/bi9600299. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda-Saito M., Hori H., Andersson L., Prince R., Pickering I., George G., Sanders C., 2nd, Lutz R., McKelvey E., Mattera R. Coordination structure of the ferric heme iron in engineered distal histidine myoglobin mutants. J. Biol. Chem. 1992;267(32):22843–22852. [PubMed] [Google Scholar]
- 47.Hargrove M.S., Krzywda S., Wilkinson A.J., Dou Y., Ikeda-Saito M., Olson J.S. Stability of myoglobin: a model for the folding of heme proteins. Biochemistry. 1994;33(39):11767–11775. doi: 10.1021/bi00205a012. [DOI] [PubMed] [Google Scholar]
- 48.Smagghe B.J., Kundu S., Hoy J.A., Halder P., Weiland T.R., Savage A., Venugopal A., Goodman M., Premer S., Hargrove M.S. Role of phenylalanine B10 in plant nonsymbiotic hemoglobins. Biochemistry. 2006;45(32):9735–9745. doi: 10.1021/bi060716s. [DOI] [PubMed] [Google Scholar]
- 49.Draghi F., Miele A.E., Travaglini-Allocatelli C., Vallone B., Brunori M., Gibson Q.H., Olson J.S. Controlling ligand binding in myoglobin by mutagenesis. J. Biol. Chem. 2002;277(9):7509–7519. doi: 10.1074/jbc.M109206200. [DOI] [PubMed] [Google Scholar]
- 50.Travaglini Allocatelli C., Cutruzzola F., Brancaccio A., Vallone B., Brunori M. Engineering Ascaris hemoglobin oxygen affinity in sperm whale myoglobin: role of tyrosine B10. FEBS Lett. 1994;352(1):63–66. doi: 10.1016/0014-5793(94)00918-x. [DOI] [PubMed] [Google Scholar]
- 51.Nienhaus K., Nienhaus G.U. Influence of distal residue B10 on CO dynamics in myoglobin and neuroglobin. J. Biol. Phys. 2007;33(5–6):357–370. doi: 10.1007/s10867-008-9059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ezhevskaya M., Trandafir F., Moens L., Dewilde S., Van Doorslaer S. EPR investigation of the role of B10 phenylalanine in neuroglobin—evidence that B10Phe mediates structural changes in the heme region upon disulfide-bridge formation. J. Inorg. Biochem. 2011;105(9):1131–1137. doi: 10.1016/j.jinorgbio.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 53.Lojou E., Cutruzzola F., Tegoni M., Bianco P. Electrochemical study of the intermolecular electron transfer to Pseudomonas aeruginosa cytochrome cd1 nitrite reductase. Electrochim. Acta. 2003;48(8):1055–1064. [Google Scholar]
- 54.Varadarajan R., Zewert T.E., Gray H.B., Boxer S.G. Effects of buried ionizable amino acids on the reduction potential of recombinant myoglobin. Science. 1989;243(4887):69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.