Abstract
Pasture-grazed dairy cows, deer, and sheep were tested for the presence of ammonia-hyperproducing (HAP) bacteria in roll tubes containing a medium in which tryptone and Casamino Acids were the sole nitrogen and energy sources. Colonies able to grow on this medium represented 5.2, 1.3, and 11.6% of the total bacterial counts of dairy cows, deer, and sheep, respectively. A total of 14 morphologically distinct colonies were purified and studied further. Restriction fragment length polymorphisms of 16S rRNA genes indicated that all isolates differed from the previously described HAP bacteria, Clostridium aminophilum, Clostridium sticklandii, and Peptostreptococcus anaerobius. Carbon source utilization experiments showed that five isolates (C2, D1, D4, D5, and S1) were unable to use any, or very few, of the carbon sources tested. Biochemical tests and phylogenetic analyses of 16S ribosomal DNA sequences indicated that all isolates were monensin sensitive; that D1 and S1 belonged to the genus Peptostreptococcus, that D4 and D5 belonged to the family Bacteroidaceae, where D4 was similar to Fusobacterium necrophorum; and that C2 was most similar to an unidentified species from the genus Eubacterium. Growth on liquid medium containing tryptone and Casamino Acids as the sole nitrogen and energy source showed that D1, D4, and S1 grew rapidly (specific growth rates of 0.40, 0.35, and 0.29 h−1, respectively), while C2 and D5 were slow growers (0.25 and 0.10 h−1, respectively). Ammonia production rates were highest in D1 and D4, which produced 945.5 and 748.3 nmol/min per mg of protein, respectively. Tests of individual nitrogen sources indicated that D1 and D4 grew best on tryptone, S1 grew equally well on Casamino Acids or tryptone, and C2 and D5 grew poorly on all nitrogen sources. The intact proteins casein and gelatin did not support significant growth of any of the isolates. These isolates extend the diversity of known HAP rumen bacteria and indicate the presence of significant HAP bacterial populations in pasture-grazed New Zealand ruminants.
Protein degradation in the rumen often proceeds at a rate which exceeds the ability of the microbial population to utilize the resulting breakdown products. This can lead to excessive absorption of ammonia from the rumen to the liver, where it is converted to urea and excreted in the urine (22). This results in an inefficient use of nitrogen by the animal, and ruminant nutrition research has therefore been directed at improving nitrogen retention in the rumen. Bacteria are the most active proteolytic organisms in the rumen (5, 27), and many species of rumen bacteria are known to be proteolytic (1, 3, 13, 32, 37). The commonly isolated proteolytic bacteria are also able to break down peptides and amino acids, and it was assumed that they were responsible for the ruminal degradation of intact protein through to ammonia. However, studies comparing the specific activities of ammonia production between mixed ruminal bacteria and the well-known proteolytic bacteria noted that no individual bacterium had an activity which could explain the activity of the mixed ruminal culture (33). Subsequently, three gram-positive, monensin-sensitive, ammonia hyperproducing (HAP) bacteria were isolated from the rumen. These included two previously described bacteria, Clostridium sticklandii and Peptostreptococcus anaerobius, and a new organism, Clostridium aminophilum (9, 10, 28). These organisms seemed to occupy the niche in the rumen of peptide and amino acid fermentation, because they were unable to hydrolyze intact protein and fermented few, if any, carbohydrates. Moreover, the monensin-sensitive nature of these organisms appeared to explain previous observations of large decreases in ruminal deamination upon monensin treatment and pointed to their importance in the ruminal fermentation of peptides and amino acids.
Ruminants in New Zealand are predominantly forage grazed, and they are especially vulnerable to nitrogen loss via ruminal ammonia overflow. New Zealand forages are typically rich in protein but low in soluble carbohydrates (17), and studies with sheep receiving fresh forage diets have shown that much of the available protein can be lost via this process (22). A recent study of the proteolytic bacteria present in cattle fed fresh forage in New Zealand has identified species of the genera Streptococcus, Eubacterium, and Butyrivibrio as being important members of the proteolytic flora (2), but HAP bacteria in New Zealand ruminants have not been investigated. The present study reports the first screening of dairy cows, sheep, and deer fed fresh forage for the presence of HAP bacteria and the isolation and preliminary characterization of five HAP rumen bacteria.
MATERIALS AND METHODS
Bacterial strains and media.
C. aminophilum F, C. sticklandii SR, Peptostreptococcus anaerobius C, and Streptococcus bovis JB1 were supplied by Jim Russell at Cornell University; Butyrivibrio fibrisolvens H17c, Clostridium clostridiiforme, Eubacterium cellulosolvens 5494, Eubacterium limosum ATCC 8486 Eubacterium ruminantium, Fibrobacter succinogenes AC3, Lachnospira multiparus ATCC 19307, Megasphaera elsdenii, Peptostreptococcus productus SF50, Prevotella ruminicola 118b, Ruminococcus albus 8, Ruminococcus flavefaciens FD1, and Selenomonas ruminantium subsp. lactilytica GA31 were obtained from Rod Mackie at the University of Illinois at Urbana-Champaign; and Enterococcus faecalis CG110 was supplied by Don Clewell at the University of Michigan. All other bacterial strains were obtained from the Rumen Microbiology Culture Collection at the AgResearch Grasslands Research Centre, Palmerston North, New Zealand. CC medium was prepared as previously described (21), except that the rumen fluid was not preincubated to remove fermentable carbohydrates. HAP medium was the same as the basal medium of Chen and Russell (9), except that cysteine-HCl (0.6 g/liter) replaced Na2S · 9H2O and tryptone and Casamino Acids (Difco, Detroit, Mich.) were added at 15 g/liter each to replace Trypticase. Purified agar (2% [wt/vol]; Difco) or high-purity agarose (1% agarose MP; Boehringer Mannheim N.Z., Ltd., Auckland, New Zealand) was used to solidify media.
Rumen sampling and enumeration of HAP bacteria.
Rumen samples were collected from four fistulated animals of each ruminant species sampled. Samples were taken from lactating Freisian dairy cows grazing on a mixed ryegrass-clover pasture at the Dairying Research Corporation in Hamilton, New Zealand. Samples were also taken from castrated Red deer and Romney wethers grazing on mixed ryegrass-clover pastures at the Department of Animal Sciences at Massey University and AgResearch Grasslands, respectively, both in Palmerston North, New Zealand.
Grab samples were taken from both the raft and the liquid phase of cow and deer rumen contents, while samples from sheep were taken directly from the rumen contents adjacent to the fistula. Samples were placed in plastic screw-cap containers, where their pH was recorded with a portable pH meter. A further sample was taken from each animal and squeezed through two layers of cheese cloth, and 50 ml of the fluid was collected and acidified for NH3 determinations. The samples were taken immediately to the laboratory, where 100 g of each was mixed with 100 ml of anaerobic mineral salts buffer (2) and blended for 1 min in a Waring blender under a CO2 atmosphere. One milliliter of the treated sample was diluted in 9 ml of anaerobic diluent (6) and then serially diluted in both HAP and CC broths. Appropriate dilutions were mixed with corresponding molten agar medium, mixed, and rolled into roll tubes. Roll tubes were incubated at 39°C and counted after colonies had finished appearing (usually 3 to 4 days). Dilution broths were incubated overnight and used as enrichments for subsequent isolation of HAP bacteria.
HAP isolates were purified from dilution broths and roll tubes by being streaked onto HAP medium plates and incubated at 38°C in an anaerobic chamber with a 92% CO2–8% H2 atmosphere (Coy Laboratory Products, Inc., Grass Lake, Mich.). Individual colonies of distinct morphology were picked, restreaked, and grown repeatedly on HAP medium plates until microscopic examination indicated colony purity.
Growth and biochemical tests.
Routine growth of HAP bacteria was carried out in HAP liquid medium. Carbon source utilization tests used HAP liquid medium in which tryptone and Casamino Acids were replaced by (NH4)2SO4 (6 g/liter), and the substrate to be tested was added at a final concentration of 10 g/liter, except for arabinose, melezitose, melibiose, and trehalose (which were added at 5 g/liter) and lactate (which was added at 7.7 g/liter). Nitrogen source tests used HAP liquid medium in which cysteine-HCl was replaced with Na2S · 9H2O (0.25 g/liter) and which lacked tryptone and Casamino Acids. To this medium was added the nitrogen sources to be tested: NH4Cl, 6.0 g/liter; tryptone, 15.0 g/liter; Casamino Acids, 15.0 g/liter; casein Hammarsten, 15.0 g/liter; and gelatin, 15.0 g/liter. Further growth and biochemical tests were carried out as previously described (15). End products of fermentation were analyzed by gas-liquid chromatography. One-milliliter samples of culture were centrifuged at 12,000 × g for 10 min at 4°C, and 1 μl of the supernatant was analyzed with a nitroterephthalic acid-modified polyethylene glycol column (DB-FFAP; 30 m by 0.53 mm by 1.0-μm film thickness; J & W Scientific, Folsom, Calif.) attached to a Hewlett-Packard 6890 series gas chromatography system. Helium was the carrier gas at a flow rate of 5 ml/min. The oven temperature started at 85°C, ramped to 200°C at 10°C/min, was held at 200°C for 10 min, and then decreased to 50°C and held for 5 min before the next sample was injected. Peaks were detected with a flame ionization detector, identified by comparison with standards, and integrated with Hewlett-Packard ChemStation software (version 4.02). Gas chromatography was carried out as previously described (38). Protein concentrations were estimated with the Bradford assay (4). Ammonia was determined by the colorimetric method of Chaney and Marbach (8), and absorbance readings were measured in a Spectramax 250 plate reader (Molecular Devices, Sunnyvale, Calif.). Specific activities of ammonia production were calculated from measurements of ammonia production and bacterial protein concentrations taken throughout the growth of each isolate on HAP liquid medium.
DNA extractions.
DNA was extracted from rumen samples by a bead-beating technique. Briefly, 1 ml of blended rumen sample was added to 1.2 g of 0.1-mm-diameter zirconia-silica beads and centrifuged at 10,000 × g for 5 min at 4°C, and the supernatant was removed. One milliliter of DNAzol (Gibco BRL, Life Technologies, Auckland, New Zealand) was added to the pellet, and the mixture was homogenized twice for 2 min with a Mini-beadbeater (Biospec Products, Ltd., Bartlesville, Okla.) with a 2-min period on ice between each treatment. The homogenate was centrifuged at 10,000 × g for 15 min at 4°C, and the supernatant was precipitated with ethanol (23). The DNA pellet was redissolved in Tris-EDTA (TE) buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), phenol-chloroform extracted, precipitated with ethanol, and finally dissolved and stored in TE buffer (23). DNA from individual cultures of HAP bacteria and isolates was extracted by the method of Saito and Miura (34).
16S rDNA RFLPs and sequence analysis.
Restriction fragment length polymorphisms (RFLPs) were performed with PCR-amplified 16S rRNA genes of HAP bacteria and isolates. The universal ribosomal DNA (rDNA) primers fD1* (5′ GAGTTTGATCCTGGCTCAG 3′) and rD1* (5′ AAGGAGGTGATCCAGCC 3′) were used to PCR amplify 16S rRNA genes by using purified DNAs from HAP bacteria and isolates as templates. PCR mixtures contained 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 2.5 mM MgCl2; 0.2 mM (each) dATP, dCTP, dGTP, and dTTP; 1 μM (each) primer; and 0.5 U of Taq DNA polymerase (GIBCO BRL). PCRs were carried out in heat-sealed capillaries in a model FTS-1 capillary thermal sequencer (Corbett Research, Sydney, Australia). The amplification conditions were denaturation at 95°C for 3 min, followed by 6 cycles of 95°C for 30 s, 55°C for 15 s, and 72°C for 30 s; 25 cycles of 95°C for 15 s, 55°C for 5 s, and 72°C for 30 s; and a final cycle of 72°C for 3 min. PCR products were precipitated with ethanol and digested with the restriction endonucleases MspI, CfoI, and HaeIII (Boehringer Mannheim N.Z. Ltd.) according to the manufacturer’s instructions. Restriction fragments were separated in 1.2% (wt/vol) agarose gels, stained with ethidium bromide, and photographed under UV illumination (23). The 16S rRNA genes of isolates C2, D1, D4, and S1 were sequenced (36) directly from PCR-amplified products with internal primers to conserved regions of the 16S rRNA gene (20). Sequencing reactions were carried out with an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Corp., Norwalk, Conn.), and the sequence was determined with a model 373A automated sequencer (Applied Biosystems, Foster City, Calif.). Closely related sequences obtained from GenBank and the Ribosomal Database Project were aligned with the HAP bacterial sequences, and approximately 1,100 unambiguous bases were used to construct similarity matrices (18) and phylogenetic trees (35).
Dot blots and 16S rDNA probing.
Detection of bacterial DNAs with rDNA probes was carried out with a dot blot format. DNAs isolated from deer, sheep, and dairy cow rumen samples and control DNAs from C. aminophilum, C. sticklandii, and P. anaerobius were diluted in denaturation buffer (0.4 M NaOH, 10 mM EDTA) and boiled for 10 min. The denatured DNAs were applied to Zeta-probe nylon membrane with a Bio-dot microfiltration apparatus (Bio-Rad Laboratories, Hercules, Calif.) attached to a vacuum line and were washed three times with 0.4 M NaOH. The membranes were neutralized in 2× SSC (0.3 M NaCl, 0.03 M sodium citrate) and air dried. Membrane prehybridizations, hybridizations, and stringency washes were performed as previously described (19), except that membranes probed with the F966 and SR836 probes were washed at 50°C, those probed with the C72 probe were washed at 52°C, and those probed with the universal fD1* probe were washed at 48°C. Probes were 3′ labelled with digoxigenin-11-ddUTP with terminal transferase (Boehringer Mannheim N.Z., Ltd.) and added to hybridization buffer at 10 ng/ml. Probe hybridization was detected with a chemiluminescent system which uses alkaline phosphatase-labeled antidigoxigenin antibody and the chemiluminescent substrate CSPD (Boehringer Mannheim N.Z., Ltd.). Chemiluminescence on membranes was recorded by autoradiography.
Nucleotide sequence accession number.
The 16S rDNA sequences of C2, D1, D4, and S1 have been deposited with GenBank under accession no. AF044945, AF044947, AF044948, and AF044946, respectively.
RESULTS
Medium formulation and verification.
In order to screen for HAP rumen bacteria, it was first necessary to formulate a medium to select for these organisms. The defined medium of Chen and Russell (9) was modified to include Casamino Acids, because C. aminophilum and P. anaerobius utilize this better than Trypticase (9, 10). The ability of this modified (HAP) medium to support the growth of HAP bacteria was verified by streaking overnight cultures of C. aminophilum, C. sticklandii, and P. anaerobius onto HAP medium plates. Also, the rumen bacteria P. ruminicola 23, S. bovis NCFB 2476, S. ruminantium ATCC 12561, Clostridium proteoclasticum ATCC 51982, and B. fibrisolvens H17c were treated in a similar manner. All three HAP bacteria produced colonies after overnight incubation and could be successfully transferred onto fresh HAP medium plates. None of the common rumen bacteria grew under these conditions, even after prolonged incubation.
Rumen samplings and HAP enumerations.
Samples taken from fistulated animals grazing on fresh forage were inoculated into both HAP and CC medium roll tubes, and after 4 days, the incubation colonies appearing on each medium were counted (Table 1). Colonies on HAP roll tubes represented from 1.3 to 11.6% of the total bacterial count on CC roll tubes in the ruminants sampled. Counts on HAP roll tubes were most numerous in samples from dairy cows; these also had the most numerous total bacterial populations. There appeared to be no direct relationship between rumen pH or ammonia concentration and either the total number or percentage of HAP organisms present in the ruminants sampled.
TABLE 1.
Bacterial counts on HAP and CC roll tubes from New Zealand ruminants
| Sample source | pH | NH3 concn (mM) | No. of bacteria ml of rumen fluid−1 ona:
|
% of HAP bacteria | |
|---|---|---|---|---|---|
| HAP medium | CC medium | ||||
| Cows | 6.0 | 8.5 | (4.99 ± 0.64) × 109 | (9.63 ± 2.06) × 1010 | 5.2 |
| Deer | 6.5 | 16.0 | (7.09 ± 3.68) × 108 | (5.51 ± 2.62) × 1010 | 1.3 |
| Sheep | 5.8 | 15.7 | (1.56 ± 0.21) × 109 | (1.34 ± 0.61) × 1010 | 11.6 |
Values are means (± standard errors) of duplicate samples taken from four animals per species.
Isolation of obligate peptide- and amino acid-fermenting bacteria.
Initial samplings indicated a relatively high level of putative HAP bacteria in New Zealand ruminants. Fourteen colonies of distinct morphology, color, and size were isolated from broth and roll tubes from the HAP enumeration experiments described above. Isolates fell into two distinct categories: isolates C2, D1, D4, D5, and S1, which grew well on HAP medium plates and broths; and the remaining isolates, which grew poorly on HAP medium plates and could not be successfully transferred to the broth form of the medium. The ability of the latter group of isolates to grow weakly on HAP medium plates was not due to impurities within the agar, because when purified agarose was used to solidify the medium, the isolates continued to grow slowly (result not shown). To examine substrate utilization more closely, the growth of isolates was tested against a range of carbohydrates (Table 2). Isolates C2, D1, D4, D5, and S1 fermented few, if any, of the commonly utilized carbon sources. D1 and D5 failed to grow on any of the carbon sources tested, while C2, D4, and S1 grew only on lactate or weakly on pyruvate. The remaining isolates used a variety of carbon sources.
TABLE 2.
Carbon source utilization by the isolates in this study
| Carbon source | Growtha of isolateb:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C2 | D1 | D4 | D5 | S1 | C3 | C4 | C7 | C8 | C10 | D3 | S1b | S3a | S3b | |
| Glucose | − | − | − | − | − | + | w | + | − | − | + | + | + | + |
| Fructose | − | − | − | − | − | + | − | + | + | − | + | + | + | + |
| Galactose | − | − | − | − | − | + | w | − | − | − | + | − | w | − |
| Mannose | − | − | − | − | − | + | − | − | − | w | + | + | + | + |
| Rhamnose | − | − | − | − | − | − | w | − | − | − | w | − | − | − |
| Ribose | − | − | − | − | − | + | − | − | − | − | w | − | w | − |
| Xylose | − | − | − | − | − | + | w | − | − | − | w | − | − | − |
| Arabinose | − | − | − | − | − | − | − | − | − | − | w | − | − | − |
| Maltose | − | − | − | − | − | + | − | + | + | − | + | − | w | w |
| Cellobiose | − | − | − | − | − | + | w | − | − | − | + | − | + | + |
| Trehalose | − | − | − | − | − | − | − | − | − | − | + | − | w | w |
| Sucrose | − | − | − | − | − | + | w | + | − | w | + | − | + | + |
| Lactose | − | − | − | − | − | + | − | w | − | − | + | − | w | − |
| Melibiose | − | − | − | − | − | + | w | − | − | − | + | − | + | w |
| Raffinose | − | − | − | − | − | + | w | + | − | w | + | − | + | − |
| Melezitose | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Starch | − | − | − | − | − | − | − | + | w | − | + | w | − | − |
| Cellulose | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Lactate | − | − | + | − | + | − | − | − | − | − | − | − | − | − |
| Pyruvate | w | − | w | − | w | w | w | − | − | − | w | − | − | − |
| Mannitol | − | − | − | − | − | + | − | − | − | − | w | − | − | − |
| Sorbitol | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| Dulcitol | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Salicin | − | − | − | − | − | + | w | + | − | w | + | − | + | + |
| Glycerol | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| HAP broth | + | + | + | + | + | − | − | − | − | − | − | − | − | − |
| HAP agar | + | + | + | + | + | w | w | w | w | w | w | w | w | w |
+, change of greater than 0.1 in OD600; w, change of between 0.05 and 0.1 in OD600; −, change of less than 0.05 in OD600. Results represent determinations from duplicate cultures.
C, D, and S signify that isolates were obtained from cows, deer, and sheep, respectively.
16S RFLPs and probing for known HAP bacteria.
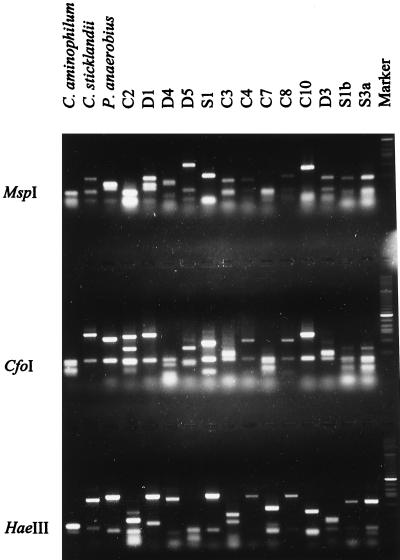
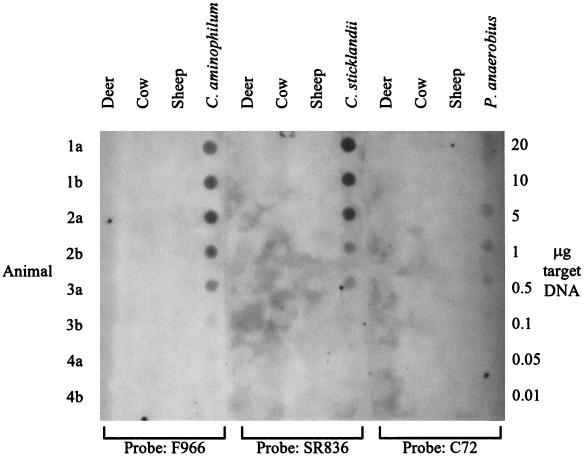
As a rapid and convenient method for examining genetic similarity among HAP isolates and their relationship to C. aminophilum, C. sticklandii, and P. anaerobius, 16S rRNA genes from each organism were amplified by PCR and subjected to RFLP analysis (Fig. 1). The RFLP patterns of all of the HAP isolates were different from those of C. aminophilum, C. sticklandii, and P. anaerobius. The patterns of the HAP isolates were also different from each other, with the exception of isolates S1b and S3a, which were identical. To determine whether C. aminophilum, C. sticklandii, and P. anaerobius were present within rumen samples taken from New Zealand ruminants, we probed DNA extracted from these samples with rDNA probes (19) specific for each of these organisms (Fig. 2). We found it was necessary to raise the temperature of the final probe stringency wash to 50°C for C. aminophilum and C. sticklandii and to 52°C for P. anaerobius to eliminate nonspecific hybridization (result not shown). Under these conditions, the probes detected 0.05 μg of purified DNA from each bacterium (Fig. 2), which represents 5.4 × 105, 1.3 × 106, and 2.1 × 106 C. aminophilum, C. sticklandii, and P. anaerobius cells ml−1, respectively. However, we observed no hybridization of the C. aminophilum, C. sticklandii, or P. anaerobius probes to DNA extracted from rumen samples from any of the animals sampled (Fig. 2). The universal probe, fD1*, hybridized to each of the sample spots, confirming the presence of DNA in these samples (result not shown).
FIG. 1.
16S rDNA RFLPs of HAP bacteria and isolates from New Zealand ruminants. PCR-amplified 16S ribosomal genes were digested with the restriction endonucleases MspI, CfoI, and HaeIII and separated in a 1.2% (wt/vol) agarose gel. The marker lane contains a 1-kb ladder (GIBCO BRL).
FIG. 2.
Dot blots of DNA extracted from deer, cow, and sheep rumen samples hybridized with oligonucleotide probes F966, SR836, and C72, which are specific for C. aminophilum, C. sticklandii, and P. anaerobius, respectively. All rumen DNA samples were applied at 1 μg per dot. Each membrane also included control target DNA spotted at the amounts shown to the right of the figure, except for the P. anaerobius membrane, in which control DNA was applied only up to 5 μg.
Growth and biochemical tests.
Carbohydrate utilization patterns indicated that isolates C2, D1, D4, D5, and S1 were typical of HAP bacteria, while 16S RFLPs showed that they were different from previously described HAP bacteria. To characterize the HAP isolates in more detail, they were subjected to a number of growth and biochemical tests alongside cultures of C. aminophilum, C. sticklandii, and P. anaerobius (Table 3). Isolates C2 and D1 stained gram positive and were a thick, slightly curved rod and a coccus, respectively. Isolates D4, D5, and S1 stained gram negative and were a thin rod, a medium rod and a coccus, respectively. All isolates were sensitive to 5 μM monensin, although D4 showed weak growth after 24 h of incubation.
TABLE 3.
Characteristics of HAP isolates in this study
| Test | Result for HAP group
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Isolates
|
Bacteria
|
|||||||
| C2 | D1 | D4 | D5 | S1 | C. aminophilum | C. sticklandii | P. anaerobius | |
| Gram stain | + | + | − | − | − | + | + | + |
| Morphology | Rod | Coccus | Rod | Rod | Coccus | Rod | Rod | Coccus |
| Growth | ||||||||
| Aerobic | − | − | − | − | − | − | − | − |
| 25°C | − | − | − | − | + | w | + | w |
| 45°C | + | + | + | + | + | + | + | w |
| 5 μM monensin | − | − | w | − | − | − | − | − |
| Indole | − | − | + | − | + | + | − | − |
| Catalase | − | + | − | − | + | + | − | + |
| Urease | − | − | − | − | − | − | − | − |
| Heat | − | − | − | − | − | − | − | − |
| H2S production | − | + | w | − | w | + | − | + |
| Lecithinase | − | − | + | − | − | − | − | − |
| Lipase | − | − | − | − | − | − | − | − |
| Fermentation products (mM)b | ||||||||
| Acetate | 3.7 | 12.3 | 17.6 | 14.7 | 18.6 | 18.7 | 31.7 | 12.0 |
| Propionate | 4.6 | 1.6 | 4.4 | 1.4 | ||||
| n-Butyrate | 4.8 | 20.8 | 6.4 | 8.9 | 12.8 | 11.1 | ||
| iso-Butyrate | 3.7 | 3.4 | 6.3 | |||||
| n-Valerate | ||||||||
| iso-Valerate | 4.1 | 8.1 | 4.1 | |||||
| n-Caproate | ||||||||
| iso-Caproate | 10.4 | 9.6 | ||||||
Results are from duplicate determinations. Growth was recorded as follows: +, change of greater than 0.1 in OD600; w, change of between 0.05 and 0.1 in OD600; −, change of less than 0.05 in OD600. Other reactions were recorded as follows: +, positive reaction; −, negative reaction; w, weak reaction.
Values are end products from overnight growth in HAP medium. Lactate, succinate, formate, and ethanol were not resolved. Note that no hydrogen was detected in the culture headspaces of any of the HAP isolates or control bacteria.
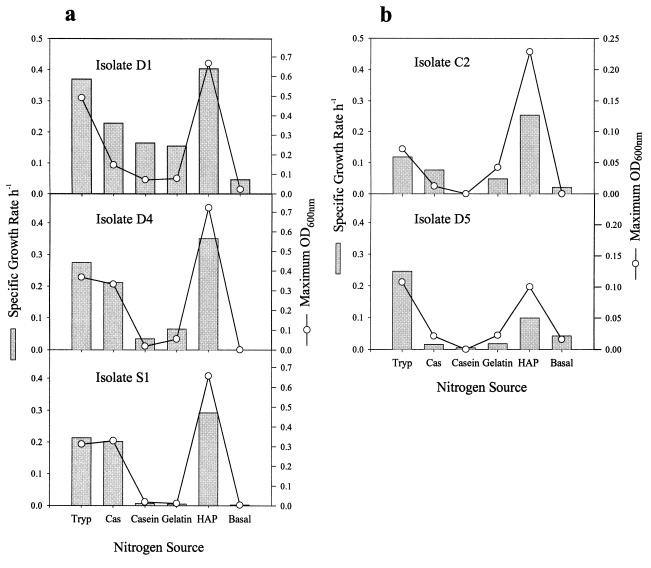
Growth rates and nitrogen source preference.
Results from growth in HAP liquid medium indicated that the HAP isolates fell into two categories: isolates D1, D4, and S1, which grew rapidly, attaining a maximum optical density at 600 nm (OD600) of around 0.7, with specific growth rates of 0.29 to 0.40 h−1; and isolates C2 and D5, which grew slowly, reaching a maximum OD600 of 0.1 to 0.2 with specific growth rates of 0.10 to 0.25 h−1 (Fig. 3). Because HAP liquid medium contains both tryptone and Casamino Acids, we decided to test these sources of nitrogen separately to investigate their preferred form of nitrogen. Liquid media containing casein or gelatin as the sole nitrogen source were also tested to investigate whether the isolates could utilize intact protein, while basal medium containing NH4Cl served as a control for residual growth from carryover of inoculum. Measurements of the specific growth rate and maximum OD600 of the fast-growing isolates (Fig. 3a) showed a preference by D1 and D4 for tryptone as the nitrogen source, while isolate S1 grew equally well either on Casamino Acids or tryptone. There was poor growth of these three isolates when either casein or gelatin was supplied as the sole nitrogen source. Of the slow-growing isolates (Fig. 3b), C2 had poor growth on all of the individual nitrogen sources, with significant growth only on HAP medium. Isolate D5 grew poorly on all nitrogen sources tested, including HAP medium, but its highest specific growth rate and maximum OD600 were attained on medium containing tryptone. With the exception of isolate D5, all organisms grew better on HAP medium, in which both tryptone and Casamino Acids are supplied.
FIG. 3.
Nitrogen source utilization by HAP isolates. Fast (a)- and slow (b)-growing isolates were grown on media containing either tryptone (Tryp), Casamino Acids (Cas), casein, gelatin, tryptone plus Casamino Acids (HAP), or NH4Cl (Basal) as the sole nitrogen source. OD600 readings were taken throughout the growth of each isolate and were used to calculate the maximum specific growth rate and maximum OD600 for each nitrogen source. Results are the means of duplicate cultures.
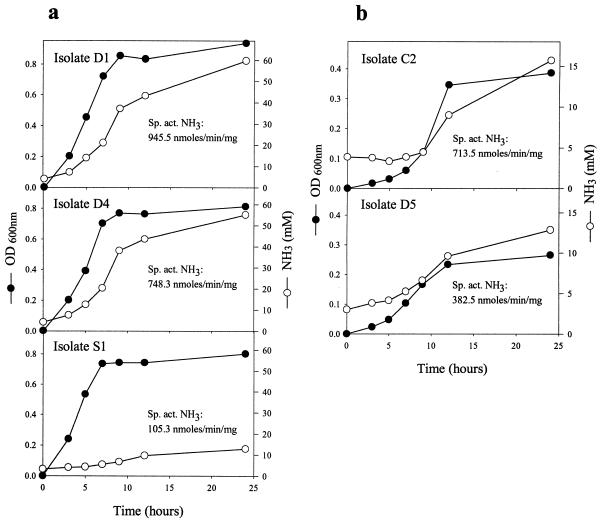
Ammonia production.
The production of high levels of ammonia from the fermentation of amino acids and peptides is a distinguishing characteristic of HAP bacteria. Ammonia production by each of the isolates during growth in HAP liquid medium was examined (Fig. 4). Ammonia accumulation in the fast-growing cultures, D1 and D4 (Fig. 4a), followed growth, and ammonia was produced at the greatest rate at the mid- to late log phase. Isolates D1 and D4 produced the greatest amount of ammonia of the isolates tested and had specific activities of ammonia production of 945.5 and 748.3 nmol/min/mg, respectively. Although isolate S1 grew quickly, it produced relatively small amounts of ammonia (105.3 nmol/min/mg), which appeared mainly in the stationary phase of growth. The slower-growing isolates, C2 and D5 (Fig. 4b), produced comparatively small amounts of ammonia which accumulated mainly in the mid- to late log phase of growth. However, due to their lower cell growth and therefore cell mass, they had relatively high specific activities of ammonia production. Under the same conditions, C. aminophilum, C. sticklandii, and P. anaerobius had specific activities of ammonia production of 676.2, 551.7, and 640.5 nmol/min/mg, respectively.
FIG. 4.
Ammonia production by HAP isolates. Cultures of HAP isolates grown on HAP medium were sampled and assayed for ammonia (8). Results are the means of duplicate cultures.
Phylogenetic analysis.
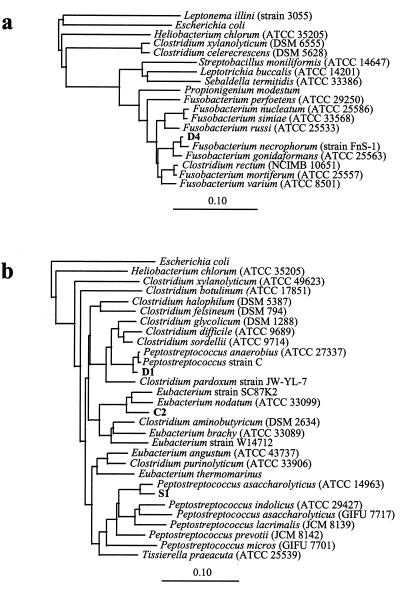
The sequences of the 16S rRNA genes of isolates C2, D1, D4, and S1 were determined and were compared to sequences from closely related species (Fig. 5). D4 was similar to Fusobacterium necrophorum (Fig. 5a [98.8%]), while D1 was closely related to P. anaerobius (Fig. 5b [99.0%]). However, in the case of D1, examination of sequence alignments with P. anaerobius revealed an additional 155 bp in the D1 sequence from position 50 to position 204. The remaining two isolates showed lower sequence homology with published data. Isolate C2 was most similar to Eubacterium sp. strain SC87K (94.4%), while isolate S1 showed the greatest similarity to Peptostreptococcus asaccharolyticus (96.9%).
FIG. 5.
Unrooted phylogenetic trees showing the relationship of isolate D4 (a) and isolates C2, D1, and S1 (b) to closely related bacteria. The trees were derived from similarity matrices (18) by the method of Fitch and Margoliash with the PHYLIP package (12). The scale bar represents a 10% difference in nucleotide sequences.
DISCUSSION
The isolation of HAP bacteria from the rumen has raised questions about their prevalence within ruminants and their contribution to ruminal peptide and amino acid degradation. Decreases in ruminal deamination and accumulation of peptides after treatment with ionophores such as monensin (39) and high rates of ammonia production have been cited as evidence for a significant role of HAP bacteria in ruminal nitrogen metabolism (33). Alternatively, the effects of ionophores have been partially explained by decreases in deaminase activity of ionophore-adapted bacteria (25) and by an alteration of membrane permeability within the prominent peptidolytic organism P. ruminicola, such that large peptides are unable to be metabolized (26). In the present study, enumerations from forage-grazed sheep, deer, and dairy cows indicate high levels of HAP bacteria in all of the commonly farmed New Zealand ruminants. The medium used to isolate HAP bacteria was not completely selective for obligate amino acid and peptide fermenters, because 9 of the 14 putative HAP isolates were also able to ferment a variety of carbohydrates. The reason for this is not clear, but the growth of these nine isolates only on the solid form of the medium indicates that they may degrade the agar sufficiently to provide trace amounts of usable substrates. Therefore, the roll tube enumerations are likely to overestimate obligate HAP bacterial numbers. In comparison, the isolation of C. aminophilum, C. sticklandii, and P. anaerobius involved several days of enrichment in a basal medium containing 15 g of Trypticase per liter (10, 33), followed by plating and picking of individual colonies. These procedures are likely to select only those organisms capable of rapid growth on Trypticase and may have overlooked slow-growing peptide fermenters. Estimations of C. aminophilum, C. sticklandii, and P. anaerobius populations by most-probable-number methods have placed their populations at 107 to 108 ml−1 (10, 33), while ribosomal probes have estimated their numbers at approximately 1% each of the total 16S rRNA (19). Oligonucleotide probing in the present study failed to detect C. aminophilum, C. sticklandii, or P. anaerobius DNA in any of the ruminants sampled, supporting the conclusion that these organisms do not form a significant part of the ruminal flora in pasture-grazed New Zealand ruminants. More-sensitive probing techniques are required before ruling out the presence of small populations of C. aminophilum, C. sticklandii, or P. anaerobius.
Like the previously described HAP bacteria, the isolates from the present study are monensin sensitive, despite the fact that three of the five isolates are gram negative. However, growth, biochemical, and phylogenetic data indicate that all of these HAP isolates are distinct from the previously described rumen HAP bacteria. Isolate D1 belongs in the genus Peptostreptococcus (16), where it shares many of the same characteristics as P. anaerobius. On the other hand, 16S rDNA RFLP analysis demonstrated a clear difference between these organisms, and 16S rRNA sequencing showed that, although closely related to P. anaerobius (99.0% similarity), D1 has an extra 155 bp in its helix 6 region and therefore probably represents a new Peptostreptococcus species. Similar elongated helix 6 structures have been observed in some thermophilic bacteria (29, 30) and plant mitochondria (24), but their significance remains unclear. It is interesting to note that the P. anaerobius-specific oligonucleotide probe was designed in this region (19), and this explains the failure to detect D1 sequences during the DNA-probing experiments. Isolate S1 is a gram-negative, non-spore-forming, anaerobic coccus which grows on lactate and, weakly, on pyruvate, but no other carbon sources supported growth. Morphological and growth characteristics place S1 in the family Veillonellaceae. Production of acetate and n-butyrate in a 2:1 molar ratio (18.6:8.9 mM), lack of H2 production, and preference for amino acids for growth support the inclusion of S1 in the genus Acidaminococcus (31). However, 16S rDNA sequence analysis places S1 closest to the gram-positive species P. asaccharolyticus. Further tests are required before a final assignment of S1 is possible. Isolates D4 and D5 are gram-negative, nonmotile, non-spore-forming rods. Their growth and biochemical characteristics place them in the family Bacteroidaceae, where they most closely resemble the description of the genus Fusobacterium (14). Phylogenetic analysis of D4 confirms this placement, showing 99.2% similarity to F. necrophorum. Isolate C2 is a nonmotile, non-spore-forming, gram-positive, slightly curved rod which produces acetate, n-butyrate, and ammonia as end products of fermentation. Growth on HAP medium was slow, and only pyruvate was fermented weakly. The 16S rDNA sequence indicates that its closest relative is Eubacterium sp. strain SC87K, but the percentage similarity (94.4%) indicates a distant relationship, and therefore C2 is best described as Eubacterium sp.
The preferred form of nitrogen for growth of D1, D4, D5, and C2 was tryptone (a pancreatic digest of casein), suggesting that these organisms utilize peptides more efficiently than the corresponding amino acids. In this respect, these isolates are similar to C. sticklandii, which grew faster on Trypticase (also an enzymatic digest of casein) than it did on Casamino Acids (10). Isolate S1, on the other hand, utilized Casamino Acids equally well as tryptone and is similar in this respect to C. aminophilum and P. anaerobius (9, 10). The inability of the HAP isolates to use intact proteins (casein and gelatin) as nitrogen sources may indicate a lack of extracellular proteinases. Previous studies of C. aminophilum and C. sticklandii grown in the presence of proteolytic rumen bacteria have shown that more ammonia was produced in cocultures with either gelatin hydrolysate or Trypticase as the substrate than from the same cultures grown individually (10). The reliance of HAP bacteria on peptides and amino acids for growth, coupled with their general lack of carbohydrate fermentation, places these organisms firmly in the ruminal niche of obligate peptide and amino acid fermentation. Presumably HAP bacteria depend on proteolytic organisms to carry out primary hydrolysis of protein and larger peptides and use the smaller peptides and amino acids liberated as fermentation substrates. Therefore, one might expect HAP bacteria to be closely associated with proteolytic populations in the rumen to enable access to their substrates. In the rumen, free peptides and amino acids do not accumulate to a significant degree (27), and it is likely that HAP bacteria are at least partly responsible for this rapid turnover. Most of the obligate amino acid- and peptide-fermenting isolates described in this study had high specific activities of ammonia production, comparable with those reported from C. aminophilum, C. sticklandii, and P. anaerobius (9, 10). In particular, isolates D1 and D4 grew rapidly and accumulated ammonia up to a concentration of 60 mM in batch culture. C2 and D5 produced much less ammonia, although their specific activities of ammonia production are still higher than those reported for common rumen bacteria (33). Isolate S1 appears anomalous, because it grew rapidly in HAP medium, yet produced only 13 mM ammonia after 24 h of growth. The rumen bacterium Acidaminococcus fermentans metabolizes glutamate via the hydroxyglutarate pathway (7) and produces ammonia in equimolar proportions to acetate. A similar pathway in S1 may explain the production of ammonia at similar levels to acetate.
The bacterial isolates described in this study extend the known diversity of organisms involved in ruminal ammonia production. The absence of significant populations of C. aminophilum, C. sticklandii, and P. anaerobius is most likely due to the fresh forage diet encountered by New Zealand ruminants. It is likely that the presence of high protein and low soluble carbohydrate in New Zealand pastures, combined with the semicontinuous nature of grazing, has produced conditions which favor the proliferation of a distinct group of amino acid- and peptide-fermenting bacteria. The diversity and numbers of these new HAP isolates suggest that these bacteria are probably responsible for considerable losses of amino acid or peptide nitrogen via deamination and subsequent excretion as urea. This is particularly severe in New Zealand ruminants, and previous measurements with sheep fed five different fresh forages have shown that from 61.5 to 81.1% of plant protein nitrogen ingested is excreted as urea (11). In the absence of completely selective media for HAP bacteria, population estimates of isolates C2, D1, D4, D5, and S1 are difficult to obtain. Therefore, we have begun designing specific rDNA probes for these organisms which will allow quantitation of individual HAP populations in New Zealand pasture-grazed ruminants and help provide estimates of their relative contributions to ruminal ammonia production.
ACKNOWLEDGMENTS
This work was funded by Public Good Science Funding from the Foundation for Research in Science and Technology, a Lotteries Science Research Grant from the New Zealand Lotteries Grants Board, and an OECD Fellowship (Co-operative Research Programme, Biological Resource Management for Sustainable Agricultural Systems) awarded to A.V.K.
The assistance of Vicki Carruthers of the Dairying Research Corporation in sampling dairy cows and constructive criticism of the manuscript by Keith Joblin are gratefully acknowledged.
REFERENCES
- 1.Abou Akkada A R, Blackburn T H. Some observations on the nitrogen metabolism of rumen proteolytic bacteria. J Gen Microbiol. 1963;31:461–469. doi: 10.1099/00221287-31-3-461. [DOI] [PubMed] [Google Scholar]
- 2.Attwood G T, Reilly K. Identification of proteolytic rumen bacteria isolated from New Zealand cattle. J Appl Bacteriol. 1995;79:22–29. doi: 10.1111/j.1365-2672.1995.tb03119.x. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn T H, Hobson P N. Further studies on the isolation of proteolytic bacteria from the sheep rumen. J Gen Microbiol. 1962;29:69–81. doi: 10.1099/00221287-29-1-69. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brock F M, Forsberg C W, Buchanan-Smith J G. Proteolytic activity of rumen microorganisms and effects of proteinase inhibitors. Appl Environ Microbiol. 1982;44:561–569. doi: 10.1128/aem.44.3.561-569.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant M P, Burkey L A. Cultural methods and some characteristics of the more numerous groups of bacteria in the bovine rumen. J Dairy Sci. 1953;36:205–217. [Google Scholar]
- 7.Buckel W, Barker H A. Two pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol. 1974;117:1248–1260. doi: 10.1128/jb.117.3.1248-1260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaney A L, Marbach E P. Modified reagents for determination of urea and ammonia. Clin Chem. 1962;8:130–132. [PubMed] [Google Scholar]
- 9.Chen G, Russell J B. Fermentation of peptides and amino acids by a monensin-sensitive ruminal peptostreptococcus. Appl Environ Microbiol. 1988;54:2742–2749. doi: 10.1128/aem.54.11.2742-2749.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Russell J B. More monensin-sensitive, ammonia-producing bacteria from the rumen. Appl Environ Microbiol. 1989;55:1052–1057. doi: 10.1128/aem.55.5.1052-1057.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan R, Ulyatt M J. Quantitative digestion of fresh herbage by sheep. VI. Utilization of nitrogen in five herbages. J Agric Sci Camb. 1980;94:47–56. [Google Scholar]
- 12.Felsenstein J. PHYLIP (Phylogeny Inference Package), 3.5c ed. Seattle, Wash: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 13.Fulghum R S, Moore W E C. Isolation, enumeration, and characteristics of proteolytic ruminal bacteria. J Bacteriol. 1963;85:808–815. doi: 10.1128/jb.85.4.808-815.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holdeman L V, Kelley R W, Moore W E C. Bacteroides. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins Co.; 1984. pp. 602–662. [Google Scholar]
- 15.Holdeman L V, Moore W E C. Anaerobe laboratory manual. Blacksburg: Virginia Polytechnic Institute and State University; 1972. [Google Scholar]
- 16.Holdeman-Moore L V, Johnson J L, Moore W E C. Genus Peptostreptococcus. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 1082–1092. [Google Scholar]
- 17.Johns A T. Pasture quality and ruminant digestion. 1. Seasonal change in botanical and chemical composition of pasture. N Z J Sci Technol. 1955;A37:301–311. [Google Scholar]
- 18.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 19.Krause D O, Russell J B. An rRNA approach for assessing the role of obligate amino acid-fermenting bacteria in ruminal amino acid deamination. Appl Environ Microbiol. 1996;62:815–821. doi: 10.1128/aem.62.3.815-821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. pp. 115–147. [Google Scholar]
- 21.Leedle J A Z, Hespell R B. Differential carbohydrate media and anaerobic replica plating techniques in delineating carbohydrate-utilizing subgroups in rumen bacterial populations. Appl Environ Microbiol. 1980;39:709–719. doi: 10.1128/aem.39.4.709-719.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacRae J C, Ulyatt M J. Quantitative digestion of fresh herbage by sheep. II. The sites of digestion of some nitrogenous constituents. J Agric Sci Camb. 1974;82:309–319. [Google Scholar]
- 23.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 24.Neefs J-M, de Peer Y V, De Rijk P, Goris A, De Wachter R. Compilation of small subunit RNA sequences. Nucleic Acids Res. 1991;19:1987–2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newbold C J, Wallace R J, McKain N. Effects of the ionophore tetronasin on nitrogen metabolism by ruminal microorganisms in vitro. J Anim Sci. 1990;68:1103–1109. doi: 10.2527/1990.6841103x. [DOI] [PubMed] [Google Scholar]
- 26.Newbold C J, Wallace R J, Watt N D. Properties of ionophore-resistant Bacteroides ruminicola enriched by cultivation in the presence of tetronasin. J Appl Microbiol. 1992;72:65–70. doi: 10.1111/j.1365-2672.1992.tb04883.x. [DOI] [PubMed] [Google Scholar]
- 27.Nugent J H A, Mangan J L. Characteristics of the rumen proteolysis of fraction I (18S) leaf protein from lucerne (Medicago sativa L) Br J Nutr. 1981;46:39–58. doi: 10.1079/bjn19810007. [DOI] [PubMed] [Google Scholar]
- 28.Paster B J, Russell J B, Yang C M J, Chow J M, Woese C R, Tanner R. Phylogeny of the ammonia-producing ruminal bacteria Peptostreptococcus anaerobius, Clostridium sticklandii, and Clostridium aminophilum sp. nov. Int J Syst Bacteriol. 1993;43:107–110. doi: 10.1099/00207713-43-1-107. [DOI] [PubMed] [Google Scholar]
- 29.Patel B K C, Love C A, Stackebrandt E. Helix 6 of the 16S rRNA of the bacterium Desulfotomaculum australicum exhibits an unusual structural idiosyncrasy. Nucleic Acids Res. 1992;20:5483. doi: 10.1093/nar/20.20.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redburn A C, Patel B K C. Phylogenetic analysis of Desulfotomaculum thermobenzoicum using polymerase chain reaction-amplified 16S rRNA-specific DNA. FEMS Microbiol Lett. 1993;113:81–86. doi: 10.1111/j.1574-6968.1993.tb06492.x. [DOI] [PubMed] [Google Scholar]
- 31.Rogosa M. Veillonellaceae. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 680–685. [Google Scholar]
- 32.Russell J B, Bottje W G, Cotta M A. Degradation of protein by mixed cultures of rumen bacteria: identification of Streptococcus bovis as an actively proteolytic rumen bacterium. J Anim Sci. 1981;53:242–252. doi: 10.2527/jas1981.531242x. [DOI] [PubMed] [Google Scholar]
- 33.Russell J B, Strobel H J, Chen G. Enrichment and isolation of a ruminal bacterium with a very high specific activity of ammonia production. Appl Environ Microbiol. 1988;54:872–877. doi: 10.1128/aem.54.4.872-877.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito H, Miura K I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace R J, Brammall M L. The role of different species of bacteria in the hydrolysis of protein in the rumen. J Gen Microbiol. 1985;131:821–832. [Google Scholar]
- 38.Wallace R J, Joblin K N. Proteolytic activity of a rumen anaerobic fungus. FEMS Microbiol Lett. 1985;29:19–25. [Google Scholar]
- 39.Whetstone H D, Davis C L, Bryant M P. Effects of monensin on breakdown of protein by ruminal microorganisms in vitro. J Anim Sci. 1981;53:803–809. doi: 10.2527/jas1981.533803x. [DOI] [PubMed] [Google Scholar]