Abstract
The goal of this work was to construct Escherichia coli strains capable of enhanced arginine production. The arginine biosynthetic capacity of previously engineered E. coli strains with a derepressed arginine regulon was limited by the availability of endogenous ornithine (M. Tuchman, B. S. Rajagopal, M. T. McCann, and M. H. Malamy, Appl. Environ. Microbiol. 63:33–38, 1997). Ornithine biosynthesis is limited due to feedback inhibition by arginine of N-acetylglutamate synthetase (NAGS), the product of the argA gene and the first enzyme in the pathway of arginine biosynthesis in E. coli. To circumvent this inhibition, the argA genes from E. coli mutants with feedback-resistant (fbr) NAGS were cloned into plasmids that contain “arg boxes,” which titrate the ArgR repressor protein, with or without the E. coli carAB genes encoding carbamyl phosphate synthetase and the argI gene for ornithine transcarbamylase. The free arginine production rates of “arg-derepressed” E. coli cells overexpressing plasmid-encoded carAB, argI, and fbr argA genes were 3- to 15-fold higher than that of an equivalent system overexpressing feedback-sensitive wild-type (wt) argA. The expression system with fbr argA produced 7- to 35-fold more arginine than a system overexpressing carAB and argI genes on a plasmid in a strain with a wt argA gene on the chromosome. The arginine biosynthetic capacity of arg-derepressed DH5α strains with plasmids containing only the fbr argA gene was similar to that of cells with plasmids also containing the carAB and argI genes. Plasmids containing wt or fbr argA were stably maintained under normal growth conditions for at least 18 generations. DNA sequencing identified different point mutations in each of the fbr argA mutants, specifically H15Y, Y19C, S54N, R58H, G287S, and Q432R.
The mammalian urea cycle is the main chemical pathway for the “detoxification” of ammonia by conversion to urea which is efficiently eliminated in the urine. Hyperammonemia, a clinical problem with severe consequences for the central nervous system, is usually caused by liver disease or inherited metabolic disorders. This work was undertaken to engineer Escherichia coli strains for enhanced incorporation of ammonia into arginine. The availability of such a biological system could then be used for development of therapy for the removal of ammonia in hyperammonemic patients. For example, these bacteria can be used to colonize the intestine for the purpose of incorporating free intestinal ammonia into arginine. As arginine contains three nitrogen atoms compared to the one atom in glutamate, the “flux” through this biosynthetic pathway, if enhanced, would result in incorporation of a large number of nitrogen atoms into organic compounds.
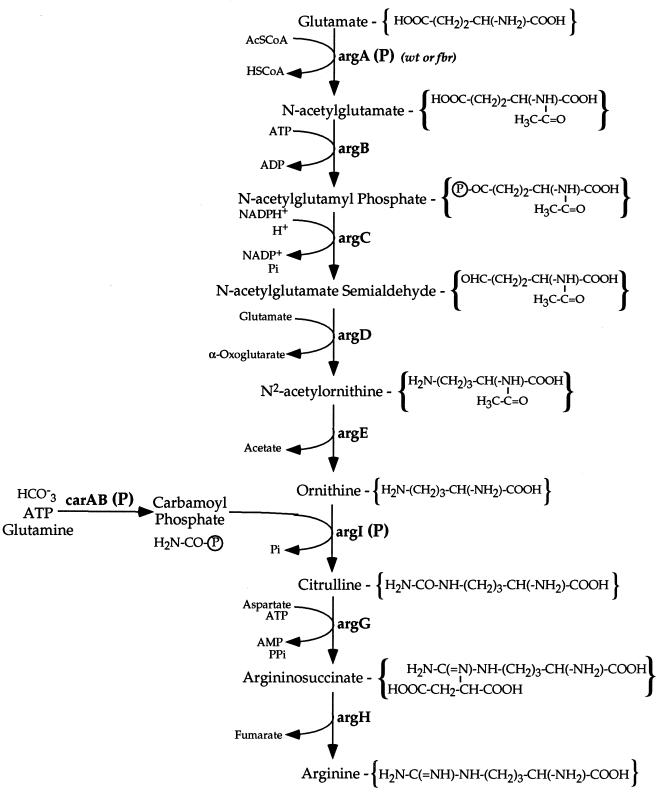
In E. coli, biosynthesis of arginine from glutamate is carried out by a series of reactions initiated by the acetylation of glutamate by N-acetylglutamate synthetase (NAGS) encoded by argA (2) (Fig. 1). Arginine biosynthesis is regulated via transcriptional repression of the arg regulon and by feedback inhibition of NAGS by arginine (2, 6, 10, 19). l-Arginine represses argA expression with a ratio greater than 250 and inhibits NAGS activity (Ki = 0.02 mM) (9). In order to develop a system for enhanced biosynthesis of arginine by E. coli, effective transcriptional derepression of arg biosynthetic genes and feedback-resistant (fbr) NAGS enzymes are required. Previous attempts to overproduce arginine in Serratia marcescens by using this strategy were unsuccessful, since the bacteria carrying the chromosomal fbr argA mutations were unstable, giving rise to argA mutants with reduced activity or with altered affinity for glutamate (8, 15, 16). Recently, we have reported the use of a plasmid system in E. coli for enhanced biosynthesis of arginine by means of derepression of the arginine regulon and simultaneous overexpression of the E. coli carAB genes encoding carbamyl phosphate synthetase and the argI gene for ornithine transcarbamylase on a plasmid (18). Arginine production in these bacteria was 6- to 16-fold higher than controls, but only if exogenous ornithine was added to the incubation medium, since ornithine production was limited due to feedback inhibition of NAGS by arginine. In order to circumvent the requirement of exogenous ornithine, fbr NAGS activity was needed. The argA genes from the three fbr NAGS E. coli strains of Eckhardt and Leisinger (5), as well as from two newly isolated fbr argA strains, were incorporated into the previously engineered arginine-producing systems, and the modified expression systems were investigated with respect to arginine production and strain stability. Different single-base substitutions in argA genes were found in each of the fbr NAGS strains.
FIG. 1.
Pathway of arginine biosynthesis in E. coli. (P), overexpression of the genes on engineered plasmids; wt, feedback sensitive; fbr, feedback resistant; AcSCoA, acetyl coenzyme A; HSCoA, coenzyme A.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. E. coli strains used in this study were the K-12 derivatives, DH5α (Gibco-BRL, Bethesda, Md.) and MG1655R, an argR strain incapable of producing the arginine repressor (obtained from Werner Maas, New York University). The argA mutant strains (EE11, EE17, and EE51) and the parent strain A1Rthy (5) were obtained from Dieter Haas, Mikrobiologisches Institut ETH, Zurich, Switzerland. The E. coli PT2 strain (11) used for the isolation of new fbr NAGS mutants and the pAI1 plasmid, a pBR322 derivative containing the E. coli argI gene (12), were obtained from Nicolas Glansdorff, University of Brussels, Brussels, Belgium.
TABLE 1.
E. coli strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 7 |
| MG1655R | argR::Tn7 | W. Maas |
| A1Rthy | argD argR his proAB thyA nalA | 5 |
| EE11 | argD argR argA213(11) his proAB thyA nalA | 5 |
| EE17 | argD argR argA214(17) his proAB thyA nalA | 5 |
| EE51 | argD argR argA215(51∗) his proAB thyA nalA | 5 |
| PT2 | F−argD proAB his ilvA metB rpsL | 11 |
| PT2M216 | F−argD proAB his ilvA metB rpsL argA216 | This study |
| PT2M217 | F−argD proAB his ilvA metB rpsL argA217 | This study |
The argA allele numbers given in this study were assigned by the E. coli Genetic Stock Center and replace the designations within parentheses given in reference 5.
TABLE 2.
Expression plasmids used in this study
| Plasmida | Expressed plasmid gene(s) |
|---|---|
| pABI | carAB argI |
| pABIA | carAB argI argA |
| pABIM213 | carAB argI argA213 |
| pABIM214 | carAB argI argA214 |
| pABIM215 | carAB argI argA215 |
| pABIM216 | carAB argI argA216 |
| pABIM217 | carAB argI argA217 |
| pABA | carAB argA |
| pABM213 | carAB argA213 |
| pABM215 | carAB argA215 |
| pA | argA |
| pM213 | argA213 |
| pM214 | argA214 |
| pM215 | argA215 |
| pM216 | argA216 |
| pM217 | argA217 |
| pM218 | argA218 |
| pM219 | argA219 |
All plasmids except pABI (18) were constructed for this study and are derivatives of pUC19 containing the trc promoter and argR titrating “arg boxes” from the argI gene and LacIq.
Plasmid nomenclature.
All expression plasmids engineered and used in this study are derivatives of pUC19 (Table 2). They contain the genes to be overexpressed under the transcription control of the lactose-regulated trc (trp-lac hybrid) promoter and also contain argR titrating (operator) boxes from the argI gene (12, 18). The plasmid names include single-letter abbreviations for each overexpressed gene (AB for carAB, I for argI, A for wild-type [wt] argA, and M for fbr mutant argA). The argA genes in the strains of Eckhardt and Leisinger (5) have been assigned new allele numbers, as described in Table 1, footnote a, and these new designations are used for naming the plasmids (for example, the argA gene from strain EE11 is designated M213). Thus, plasmid pABIM213 expresses carAB, argI, and the fbr mutant argA213 genes, while pM213 expresses only the fbr mutant argA213 gene. Plasmids obtained from other laboratories may not conform to this naming scheme and have their original designation and are referenced.
Media and growth conditions.
Luria broth (LB) was used routinely for liquid cultures. Bacto Agar (Difco Laboratories, Detroit, Mich.) was added at 1.5% for solid media. Ampicillin (obtained from Sigma Chemical Co., St. Louis, Mo.) was used at 50 to 100 μg/ml.
Isolation of new argA fbr mutants.
The general scheme of Eckhardt and Leisinger (5) was followed with some modifications. Rather than starting with an argR strain, the arginine regulon of strain PT2 was derepressed by introducing plasmid pAI1 (from N. Glansdorff [12]) which contains an operator with “arg boxes” from the argI gene. Five milliliters of PT2 cells carrying the pAI1 plasmid that had been grown in LB to 108 CFU/ml was treated with 100 μl of the mutagen ethyl methanesulfonic acid (100%) (Sigma Chemical Co.) at 37°C for 1 h. The cells were washed with saline and grown overnight in LB with 100 μg of ampicillin per ml. The resulting cultures were washed with saline and then plated on minimal medium with required amino acids, arginine, and ampicillin, but without proline. After 4 days of incubation, colonies that showed satellite effects (able to synthesize and excrete proline to feed the surrounding PT2 cells) were picked and repurified. The isolates were compared for proline excretion with the known argA fbr strains as described before (5). Isolates PT2M216 and PT2M217 were chosen as new argA fbr strains.
Construction of an inducible system for enhanced arginine production.
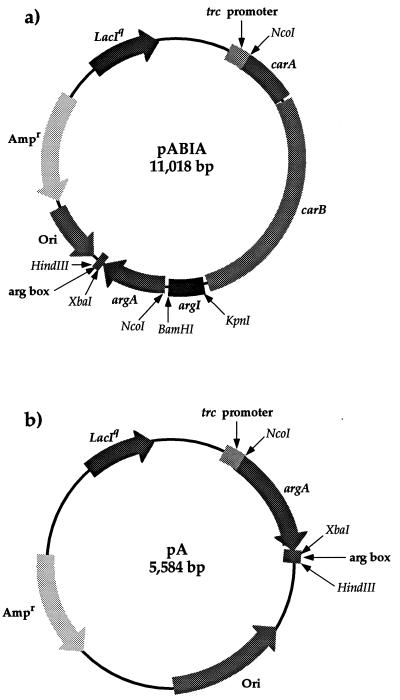
Recombinant DNA techniques were employed by routine protocols (13). The plasmids overexpressing carAB and argI (pABI) have been described before (18). The argA genes, without promoter sequence, from the feedback-sensitive wt (A1Rthy and PT2) and fbr (EE11, EE17, EE51, PT2M216, and PT2M217) strains were amplified by PCR and cloned into pABI as follows. The genes were engineered by PCR to include an intercistronic region with a ribosome binding site and an NcoI site near the initiation codon. Forward and reverse primers containing flanking BamHI and XbaI/HindIII sites, respectively (5′-CCCGGATCCTCAGGAGTAAAAGAGCCATGGTAAAGGAACGTAAAACC-3′ and 5′-CCCAAGCTTTCTAGATTACCCTAAATCCGCCATCAA-3′), were used to amplify the entire gene from chromosomal DNA. The resulting fragment (1,373 bp) was cut with BamHI and XbaI and cloned into pABI to obtain plasmids containing argR titrating boxes, carAB, argI, and either a feedback-sensitive wt (pABIA) or fbr (pABIM) argA gene (Fig. 2a). The wt or fbr argA genes were also cloned into plasmids containing arg boxes without carAB or argI producing pA (wt) and pM (mutant) plasmid derivatives. They were produced by linearizing the pABIA and pABIM derivatives with NcoI to remove carAB and argI followed by religation (Fig. 2b). As the fbr argA gene from PT2M217 contained two separate mutations, the isolated gene was restricted with NcoI and PstI and PstI and XbaI (to separate the mutations), and the fragments were cloned into pA to obtain pM218 and pM219, respectively, each harboring a single mutation. The inserts were sequenced for verification. To construct plasmids containing carAB and wt or fbr argA, but not argI, the pABIA and pABIM plasmids were digested with KpnI and BamHI to remove the argI fragment, filled in using the Klenow enzyme, and ligated.
FIG. 2.
pABIA (linked as an operon) (a) and pA vectors (b) engineered for this study. The vectors contained the gene(s) downstream from a control region which includes the trc promoter and a lac operator. The constructs also contained an arg box cloned from the argI gene for binding and titration of the arginine repressor. Ori, ColE1 replication origin.
We verified the overexpression of plasmid genes by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and specific enzyme assays as described before (18; data not shown).
Arginine biosynthesis experiments.
A single colony of cells transformed with the engineered plasmid or parent vector was inoculated into 10 ml of LB with the appropriate antibiotic and grown to saturation at 37°C. The saturated culture was diluted 15-fold and grown to log phase (A600 = 0.5 to 0.6) and then induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C for 2 h. Three 45-ml aliquots were centrifuged for 10 min at 1,500 × g in a Beckman TJ-6 centrifuge, and the bacterial pellets were washed once with 10 ml of M9 minimal medium without a nitrogen source. One pellet was resuspended in 1 ml of M9 minimal medium and used to determine free arginine at time zero. The other two pellets were resuspended in 1 ml of M9 minimal medium containing 20 mM l-glutamine or 20 mM l-glutamine plus 5 mM l-ornithine. After incubation for 3 h, the cells were sonicated and centrifuged to remove membranes and cell debris. After precipitation of the soluble proteins with 50% trichloroacetic acid, the free arginine concentration was determined colorimetrically by the Sakaguchi procedure (17). Arginine levels at 3 h were normalized to the dry weight of bacteria after subtraction of the time zero value; free arginine production rates are reported as nanomoles per milligram (dry weight) per hour. In a separate experiment to determine the linearity of arginine production, DH5α cells with plasmids carrying wt or fbr argA genes were grown and induced as described above, washed, resuspended in M9 minimal medium with glucose and 20 mM glutamine, and then incubated at 37°C; samples were removed every hour for 3 h to determine arginine concentration.
Stability of arginine production by pA and pM plasmids.
Single colonies of DH5α containing plasmid pA (wt), pM214, or pM215 were inoculated into 25 ml of LB containing 100 μg of ampicillin per ml (LB-Amp) and grown for 12 h at 37°C. Cells were diluted in fresh LB-Amp medium to an A600 of 0.05 and grown for 3 to 4 h to an A600 of 0.8 at 37°C. The cultures were then passaged as described above two more times and grown for four generations after each passage. At the end of the third passage, the cultures were subcultured in fresh medium and grown for 12 h. Before each passage, 50 ml of culture was treated with 0.5 mM IPTG for 2 h and total free arginine in the cells and culture media was determined as described above.
Sequencing.
Plasmid DNA containing a wt (pA) or mutant fbr argA gene (pM213, pM214, pM215, pM216, or pM217) was restricted with XbaI and BamHI and subcloned into pUC19. Sequencing of both strands was performed by the dideoxynucleotide chain-termination method (14), using the Sequenase DNA sequencing kit (U.S. Biochemical Corp.) and [α-35S]dATP (NEN).
Nucleotide sequence accession numbers.
The nucleotide sequences of fbr NAGS mutants of E. coli argA213, argA214, argA215, argA216, and argA218 and argA219 have been deposited in GenBank under the accession numbers AF008115, AF008116, AF008117, AF008118, and AF008119, respectively.
RESULTS AND DISCUSSION
Arginine biosynthesis.
The free arginine synthesis rates of arg-derepressed MG1655R and DH5α strains containing the engineered plasmids are shown in Table 3. The free arginine production rate in arg-derepressed cells containing a pABIM plasmid expressing carAB, argI, and fbr argA (pABIM213, pABIM214, pABIM215, pABIM216, or pABIM217) incubated in minimal medium with glutamine as the nitrogen source ranged from 39.3 to 105.9 nmol/mg (dry wt)/h compared with 6.6 to 15.2 nmol/mg (dry wt)/h in cells containing pABIA expressing carAB, argI, and feedback-sensitive argA. The rate of synthesis of free arginine in cells containing pABI expressing carAB and argI and chromosomal (wt) argA was only 2.6 to 5.3 nmol/mg (dry wt)/h. The addition of exogenous ornithine to cells containing pABIM plasmids (pABIM213, pABIM214, pABIM215, pABIM216, and pABIM217) resulted in only small increases in the synthesis of arginine (Table 3). On the other hand, the addition of ornithine to cells containing pABIA increased arginine production 2.5- to 3-fold. This result indicates that overexpression of fbr argA, unlike feedback-sensitive wt argA, allows endogenous ornithine production to almost saturate the arginine synthetic capacity of the cells.
TABLE 3.
Arginine production by E. coli K-12 strains transformed with different plasmidsa
| Plasmid | Arginine concnb
|
|||
|---|---|---|---|---|
| MG1655R
|
DH5α
|
|||
| Gln | Gln + Orn | Gln | Gln + Orn | |
| pUC19 | 3.8 (2.9–4.5) | 12.2 (11.2–12.8) | 1.1 (0.8–1.6) | 2.5 (1.6–3.2) |
| pABI | 5.3 (4.5–5.9) | 13.3 (11.5–14.8) | 2.6 (1.5–3.5) | 8.9 (7.3–10.5) |
| pABIA | 15.2 (12.8–16.9) | 48.2 (42.5–55.6) | 6.6 (4.8–8.1) | 27.9 (23.4–30.8) |
| pABIM213 | 105.9 (98.5–110.5) | 107.8 (95.6–114.4) | 67.2 (60.2–75.4) | 76.5 (70.3–81.2) |
| pABIM214 | 89.7 (85.4–95.5) | 112.7 (98.3–120.4) | 101.9 (98.7–105.6) | 110.6 (102.4–115.5) |
| pABIM215 | 103.7 (95.8–110.8) | 126.7 (115.4–134.3) | 100.6 (95.3–106.4) | 105.6 (100.3–112.2) |
| pABIM216 | 99.8 (95.5–104.8) | 122.5 (115.3–132.5) | 82.2 (76.6–90.8) | 90.6 (85.2–98.9) |
| pABIM217 | 50.7 (45.5–55.2) | 64.7 (60.5–72.4) | 39.3 (32.3–45.4) | 75.3 (70.2–81.3) |
| pABA | 28.2 (21.9–37.5) | 54.9 (50.3–63.3) | 9.6 (6.8–11.8) | 30.9 (25.8–35.4) |
| pABM213 | 109.0 (98.4–115.7) | 111.3 (105.4–120.8) | 74.2 (68.3–80.4) | 97.9 (90.4–102.3) |
| pABM215 | 98.2 (91.9–107.5) | 111.9 (104.3–119.3) | 79.1 (70.8–86.2) | 106.1 (95.8–112.3) |
The bacterial strains were transformed with the parent vector or with plasmid expressing carbamyl phosphate synthetase, ornithine transcarbamylase, and wt or fbr NAGS.
Mean values and range of data from three experiments. E. coli K-12 strains MG1655R and DH5α were used. Cultures were incubated with glutamine (Gln) (20 mM) or glutamine (20 mM) plus ornithine (Orn) (5 mM) for 3 h, and total arginine production was determined and reported as nanomoles of arginine per milligram (dry weight) per hour.
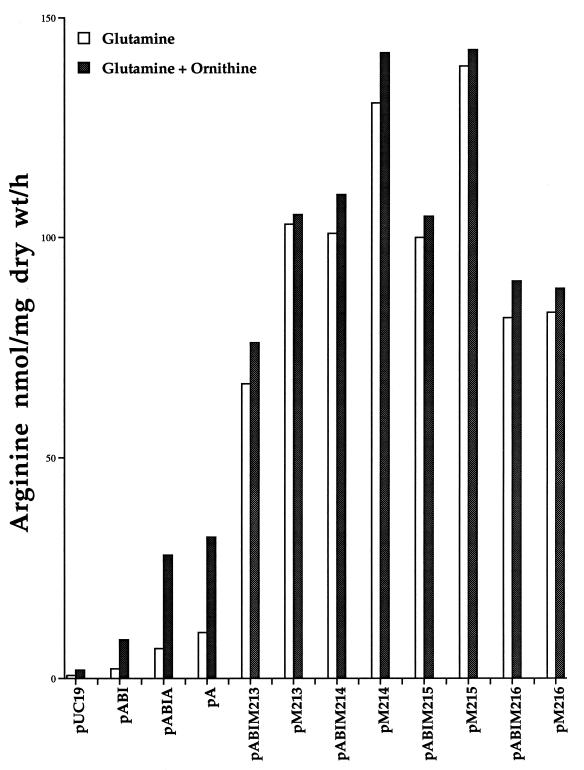
In subsequent experiments, free arginine synthesis rates were measured in arg-derepressed DH5α strains containing plasmid derivatives expressing only wt argA (pA) or fbr argA (pM213, pM214, pM215, and pM216) without carAB or argI. Arginine production in these cells was linear for at least 3 h under the experimental conditions used (data not shown). The free arginine production rate of cells containing a plasmid expressing fbr argA (pM213, pM214, pM215, and pM216) was 66.8 to 139 nmol/mg (dry wt)/h compared to 10.4 nmol/mg (dry wt)/h in cells containing pA expressing wt argA (Fig. 3). Again, the addition of exogenous ornithine to these strains only slightly increased the production of arginine. The argA217 gene contains two separate mutations (S54N and Q432R), and the two mutations were separated to yield the plasmids pM218 and pM219 expressing fbr mutant argA218 (S54N) and argA219 (Q432R), respectively. Arginine production in DH5α containing pM217, pM218, and pM219 was similar in all three (91 to 108 nmol/mg [dry wt]/h), suggesting that either of the two mutations can independently produce a fbr argA strain.
FIG. 3.
Arginine biosynthesis in E. coli DH5α containing the engineered plasmids or the parent vectors. The relevant expressed genes of vectors are shown in Table 2. The induced cultures were incubated with glutamine (20 mM) or with glutamine (20 mM) plus ornithine (5 mM) for 3 h, and total arginine production was determined and reported as nanomoles of arginine per milligram (dry weight) per hour.
The free arginine biosynthesis rate of DH5α cells containing a plasmid expressing fbr argA (pM213, pM214, pM215, pM216, or pM217) was 6- to 10-fold higher than that of cells containing pA expressing wt argA and 16- to 26-fold higher than that of cells containing pABI expressing carAB and argI. Free arginine biosynthesis in the DH5α strain was higher in cells containing a plasmid expressing fbr argA (pM213, pM214, pM215, pM216, or pM217) than in cells containing a pABIM plasmid (pABIM213, pABIM214, pABIM215, pABIM216, or pABIM217) which also expresses the carAB and argI genes.
The arginine biosynthetic capacity of DH5α containing pA or pABIA in the absence of exogenous ornithine was two- to fourfold higher than in cells containing pABI. This result indicates that despite the feedback inhibition of NAGS by arginine, some formation of endogenous ornithine from glutamate is occurring when wt NAGS is present in excess. In vitro assays have revealed that the inhibition of NAGS activity by arginine is not complete (9). Thus, it is likely that in cells overexpressing plasmid wt argA, some residual enzyme activity is available for the first step of arginine biosynthesis.
Stability of arginine production by pA and pM plasmids.
Uninduced DH5α cells containing a plasmid expressing fbr argA (pM214 or pM215) or wt argA (pA) subcultured in fresh media and grown for a total of 18 generations were stable. Their free arginine production capacities were similar before each passage, yielding 90 to 107 nmol/mg (dry wt)/h in cells containing pM214 or pM215 and 6 to 16 nmol/mg (dry wt)/h in cells containing pA. Continuously induced cultures (treated with IPTG) were unstable. Thus, uninduced plasmids carrying fbr argA appear to be stable under normal growth conditions and retain their capacity to produce arginine as long as the argA gene is not expressed during growth.
These results offer a possible explanation for the observation that the fbr NAGS mutants of S. marcescens overproducing arginine were unstable and lost their capacity to produce arginine during subculturing (8, 15, 16). The instability of the strains was attributed to poor growth and high mutability of the argA allele. This would be expected, since the arginine genes were continuously expressed, “forcing” enhanced arginine production during growth and thus allowing the selection of mutants with attenuated arginine production. The plasmid-derived inducible arginine expression system described in this report offers an advantage over chromosomal expression, as the plasmid fbr argA can be expressed only when needed without affecting the growth of bacteria. We also demonstrate that E. coli can be engineered for increased production of arginine without the need for endogenous ornithine when fbr NAGS is produced.
Identification of argA mutations.
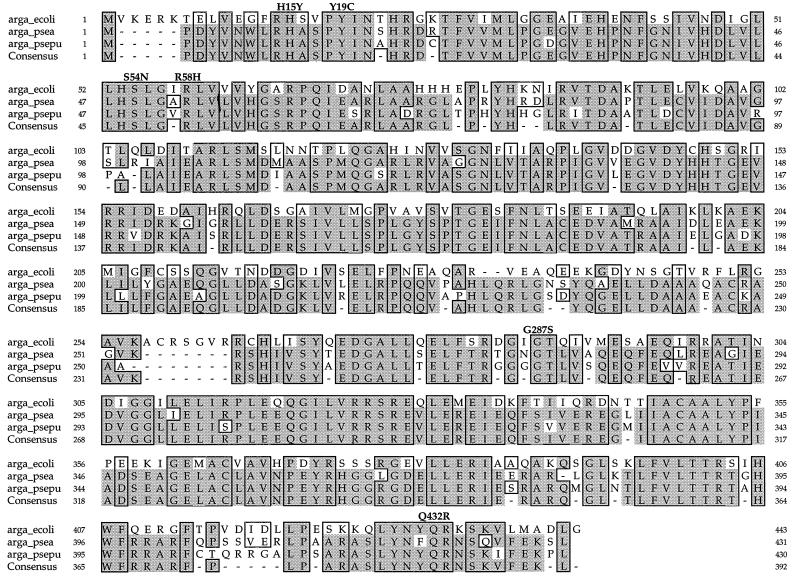
Sequence analysis of the four mutant (EE11, EE17, EE51, and PT2M216) and wt (A1Rthy) argA genes revealed different single-base substitutions in each of the four mutant alleles. The argA213 and argA216 mutations were G-to-A transitions at nucleotides 173 and 859, respectively, replacing Arg-58 with His (R58H) and Gly-287 with Ser (G287S). In argA214, a C-to-T transition at nucleotide 43 replaced His-15 with Tyr (H15Y), whereas in argA215, an A-to-G transition at nucleotide 56 resulted in substitution of Tyr-19 with Cys (Y19C). The argA217 gene (in PT2M217) contained two separate single-base substitutions: a G-to-A transition at nucleotide 161 replaced Ser-54 with Asn (S54N), while an A-to-G transition at nucleotide 1295 replaced Gln-432 with Arg (Q432R). Both of the mutations in argA217 were found to independently produce a fbr argA phenotype. All six amino acids affected by the mutations (H15Y, Y19C, S54N, R58H, G287S, and Q432R) were at sites conserved in the three prokaryotes (Fig. 4). In all of the mutant and wt argA genes sequenced, we found the nucleotide at position 1167 to be G, rather than the previously reported T (1), and in argA213, the nucleotide at 207 was T instead of C. However, these base changes did not result in a change in the amino acid residues.
FIG. 4.
Multiple-sequence alignment of argA homologs. The sequences used were argA homologs from E. coli (1) (arga_ecoli), Pseudomonas aeruginosa (4) (arga_psea), and Pseudomonas putida (4) (arga_psepu). The argA homologs were aligned by using the PILEUP and PRETTY programs of the Genetics Computer Group. The three prokaryotic NAGS sequences showed 45% identity and an additional 12% similarity. Identical residues are shaded, and homologous residues detected by the PAM250 matrix of amino acid similarity (3) (determined by using the SEQVU program and manual editing) are boxed (functionally similar amino acids follow: D and E; F and Y; G and W; N and D; K and R; Q and E; L and M; I and V; and A, S, and T). The mutations H15Y (argA214), Y19C (argA215), S54N (argA218), R58H (argA213), G287S (argA216), and Q432R (argA219) are indicated above the sequence. Gaps introduced to optimize alignment are indicated by hyphens.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant 1PO1-HD32652 from the National Institute of Child Health and Human Development.
We thank Werner Maas, Nicolas Glansdorff, and Dieter Haas for their help in providing E. coli strains and plasmids and Ruoqiong Chen for technical assistance.
REFERENCES
- 1.Brown K, Finch P W, Hickson I D, Emmerson P F. Complete nucleotide sequence of the Escherichia coli argA gene. Nucleic Acids Res. 1987;15:10586. doi: 10.1093/nar/15.24.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dayhoff M O, Schwarz R M, Orcutt B C. A model of evolutionary change in proteins. Matrices for detecting distant relationships. In: Dayhoff M O, editor. Atlas of protein sequence and structure, vol. 5, suppl. 3. Washington, D.C: National Biomedical Research Foundation; 1978. pp. 345–358. [Google Scholar]
- 4.Dharmsthiti S, Krishnapillai V. DNA sequence conservation at the gene level in a conserved chromosomal segment in two Pseudomonas species. J Genet. 1993;72:1–14. [Google Scholar]
- 5.Eckhardt T, Leisinger T. Isolation and characterization of mutants with a feedback resistant N-acetylglutamate synthase in Escherichia coli K 12. Mol Gen Genet. 1975;138:225–232. doi: 10.1007/BF00269349. [DOI] [PubMed] [Google Scholar]
- 6.Haas D, Leisinger T. In vitro assay and some properties of N-acetylglutamate synthetase from Escherichia coli. Pathol Microbiol. 1974;40:140–141. [PubMed] [Google Scholar]
- 7.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 8.Kisumi M, Takagi T, Chibata I. Construction of an L-arginine-producing mutant in Serratia marcescens. Use of the wide substrate specificity of acetylornithinase. J Biochem. 1978;84:881–890. doi: 10.1093/oxfordjournals.jbchem.a132200. [DOI] [PubMed] [Google Scholar]
- 9.Leisinger T, Haas D. N-Acetylglutamate synthase of Escherichia coli: regulation of synthesis and activity by arginine. J Biol Chem. 1975;250:1690–1693. [PubMed] [Google Scholar]
- 10.Maas W. The arginine repressor of Escherichia coli. Microbiol Rev. 1994;58:631–640. doi: 10.1128/mr.58.4.631-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mountain A, Mannton N H, Munton R N, Baumberg S. Cloning of a Bacillus subtilis restriction fragment complementing auxotrophic mutants of eight Escherichia coli genes of arginine biosynthesis. Mol Gen Genet. 1984;197:82–89. doi: 10.1007/BF00327926. [DOI] [PubMed] [Google Scholar]
- 12.Piette J, Cunin R, Van Vilet F, Charlier D, Crabeel M, Ota Y, Glansdorff N. Homologous control sites and DNA transcription starts in the related argG and argI genes of Escherichia coli K12. EMBO J. 1982;1:853–857. doi: 10.1002/j.1460-2075.1982.tb01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takagi T, Sugiura M, Kisumi M. Instability of an arginine-overproducing mutant of Serratia marcescens and its stabilization. J Biochem. 1986;99:357–364. doi: 10.1093/oxfordjournals.jbchem.a135490. [DOI] [PubMed] [Google Scholar]
- 16.Takagi T, Kisumi M, Chibata I. Construction of an arginine-producing strain of Serratia marcescens. Appl Microbiol Biotechnol. 1985;21:378–382. [Google Scholar]
- 17.Tomlinson G, Viswanatha T. Determination of the arginine content of proteins by the Sakaguchi procedure. Anal Biochem. 1974;60:15–24. doi: 10.1016/0003-2697(74)90127-4. [DOI] [PubMed] [Google Scholar]
- 18.Tuchman M, Rajagopal B S, McCann M T, Malamy M H. Enhanced production of arginine and urea by genetically engineered Escherichia coli K-12 strains. Appl Environ Microbiol. 1997;63:33–38. doi: 10.1128/aem.63.1.33-38.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vyas S, Maas W K. Feedback inhibition of acetylglutamate synthase by arginine in Escherichia coli. Arch Biochem Biophys. 1963;100:452–456. doi: 10.1016/0003-9861(63)90124-3. [DOI] [PubMed] [Google Scholar]