Abstract
Convolutional neural networks (CNNs) have shown promising performance in various 2D computer vision tasks due to availability of large amounts of 2D training data. Contrarily, medical imaging deals with 3D data and usually lacks the equivalent extent and diversity of data, for developing AI models. Transfer learning provides the means to use models trained for one application as a starting point to another application. In this work, we leverage 2D pre-trained models as a starting point in 3D medical applications by exploring the concept of Axial-Coronal-Sagittal (ACS) convolutions. We have incorporated ACS as an alternative of native 3D convolutions in the Generally Nuanced Deep Learning Framework (GaNDLF), providing various well-established and state-of-the-art network architectures with the availability of pre-trained encoders from 2D data. Results of our experimental evaluation on 3D MRI data of brain tumor patients for i) tumor segmentation and ii) radiogenomic classification, show model size reduction by ~22% and improvement in validation accuracy by ~33%. Our findings support the advantage of ACS convolutions in pre-trained 2D CNNs over 3D CNN without pre-training, for 3D segmentation and classification tasks, democratizing existing models trained in datasets of unprecedented size and showing promise in the field of healthcare.
Keywords: Deep learning, ImageNet, Transfer learning, MRI, segmentation, classification
1. Introduction
Deep learning (DL) based approaches are continuously being developed for various medical imaging tasks, including segmentation, classification, and detection, for a wide range of modalities (i.e., MRI, CT, X-Ray), regularly outperforming earlier approaches [1,2]. However, DL is computationally expensive and requires large amounts of annotated data for model training limiting their applicability in problems where large amounts of annotated datasets are unavailable [3]. Transfer learning (TL) is a popular approach to overcome this issue by initializing a DL model with pre-trained weights, thereby reducing convergence time and concluding at a superior state, while utilizing otherwise insufficient data [4,5]. The basic idea of TL involves re-using model weights trained for a problem with a large available dataset as the initialization point for a completely different task. The foundation behind this idea is that convolutional layers extract general, lower-level features (such as edges, patterns, and gradients) that are applicable across a wide variety of images [6]. The latter layers of a convolutional neural network (CNN) learn features more specific to the image of the particular task by combining the previous lower-level features. Leveraging weights of trained models has proven to be a better initialisation point for DL model training, when compared to random initialization [4,7–10].
There are numerous pre-trained models available for applications on 2D imaging data, such as ImageNet [11], YOLO [12], and MS-COCO [13], however, universally applicable pre-trained models are not available for utilization on 3D data like medical images due to the lack of associated large and diverse data. Current application of pre-trained CNN for 3D medical image segmentation and classification can be divided in three categories depending on the dimensionality of the input data:
-
2D Approaches
Here, a 3D input volume is considered as a stack of 2D slices, and a multi-slice planar (2D) network is applied on each 2D slice independently [14,15]. Some earlier approaches considered 3D medical images as tri-planar representation where axial, coronal, and sagittal views are considered as 3 channels of the input data. But such 2D representation learning is fundamentally weak in capturing 3D contexts. Some DL based approaches for classification of brain cancer MRI images use representative 2D slices as the input data rather than utilizing full 3D volume [16,17].
-
3D Approaches
In this case, a 3D network is trained using native 3D convolution layers that are useful in capturing spatial correlations present along the 3rd dimension, in order to capture 3D contextual information [18–21]. Significant improvement in classification accuracy was observed in [22] with the use of native 3D convolutions compared to 2D convolutions. Although data in adjacent slices, across each of the three axes, are correlated and can be potentially used to yield a better model, this suffers from two weaknesses: a) reduced model stability due to random weight initialization (since there are no available pre-trained models) and b) unnecessarily high memory consumption.
-
Hybrid Approaches
There are few studies using a hybrid of the two aforementioned approaches, i.e. 2D & 3D. An ensemble-based learning framework built upon a group of 2D and 3D base learners was designed in [23]. Another strategy is to train multiple 2D networks on different viewpoints and then generate final segmentation results by 3D volumetric fusion net [24]. A similar approach was proposed in [25], which consists of a 2D DenseUNet for intra-slice feature extraction and its 3D counterpart for aggregating volumetric contexts. Finally, Ni et al. trained a 2D deep network for 3D medical image segmentation by introducing the concept of elastic boundary projection [26].
Current literature shows inadequate exploration on the application of 2D pre-trained models in native 3D applications. As medical datasets are limited when compared with those from the computer vision domain, TL of models trained in the latter can be beneficial in medical applications.
In this paper, we explore the concept of Axial-Coronal-Sagittal (ACS) convolution to utilize pre-trained weights of models trained on 2D datasets to perform natively 3D operations. This is achieved by splitting the 2D kernels into 3 parts by channels and convolving separately across Axial-Coronal-Sagittal views to enable development of native 3D CNNs for both classification and segmentation workloads. This way, we can take advantage of the 3D spatial context, as well as the available pre-trained 2D models to pave the way towards building better models for medical imaging applications. Multiple options of pre-trained models for use in 3D datasets have been made publicly available through the Generally Nuanced Deep Learning Framework (GaNDLF) [27]6.
2. Methods
In this work, we leverage the concept of Axial-Coronal-Sagittal (ACS) proposed in [28] and incorporate it into the Generally Nuanced Deep Learning Framework (GaNDLF) [27]7 which supports a wide variety of model architectures, loss functions, pre-processing, and training strategies.
2.1. ACS Convolutions
Convolution operations in CNNs can be classified as either 2D or 3D. The 2D convolutional layers use 2D filter kernels (K × K) and capture 2D spatial correlation, whereas 3D convolutional kernels (K × K × K) are used in native 3D convolutional layers capturing 3D context (Fig. 1). As mentioned earlier, each of these approaches have their own advantages and disadvantages.
Fig. 1:
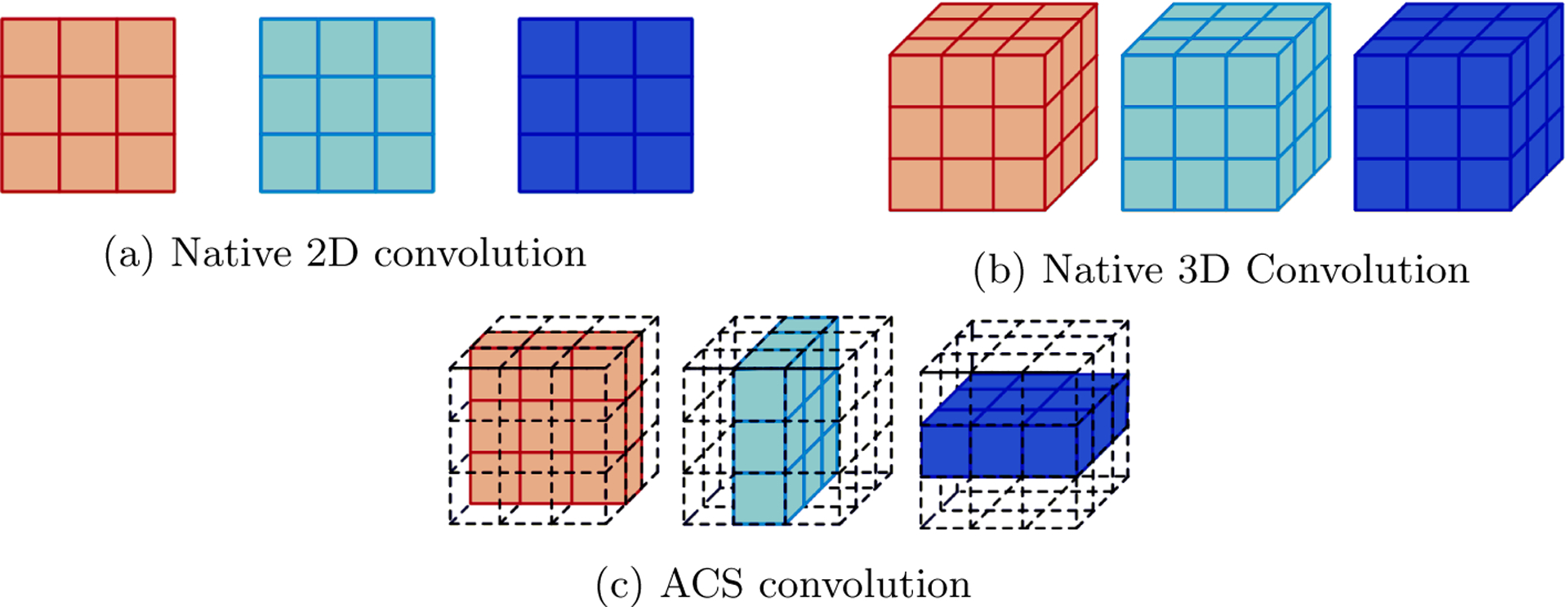
Comparison of various types of convolution for 3D medical data (Figure adopted from [28].
Yang et al. [28] introduced the concept of Axial-Coronal-Sagittal (ACS) convolutions to learn the spatial representation of three dimensions from the combination of each of the three (A-C-S) views (Fig. 1(c)). The basic concept of the ACS convolutions is to split the kernel into three parts (K ×K ×1), (K ×1×K) and (1 × K × K) and run multiple 2D convolution filters across the three views (axial, coronal, and sagittal). For any convolution layer, let us consider the number of input channels as Ci and number of output channels as Co. The number of output channels in ACS convolution are then set as:
| (1) |
Thus 2D convolutions are transformed into 3 dimensions by simultaneously performing computations across axial, coronal, and sagittal axes. The final output is then obtained by the concatenation of three convolved feature maps without any additional fusion layer.
The concept of ACS convolutions can be used as a generic plug-and-play replacement of 3D convolution enabling development of native 3D CNNs using 2D pre-trained weights as illustrated in Fig. 2.
Fig. 2:
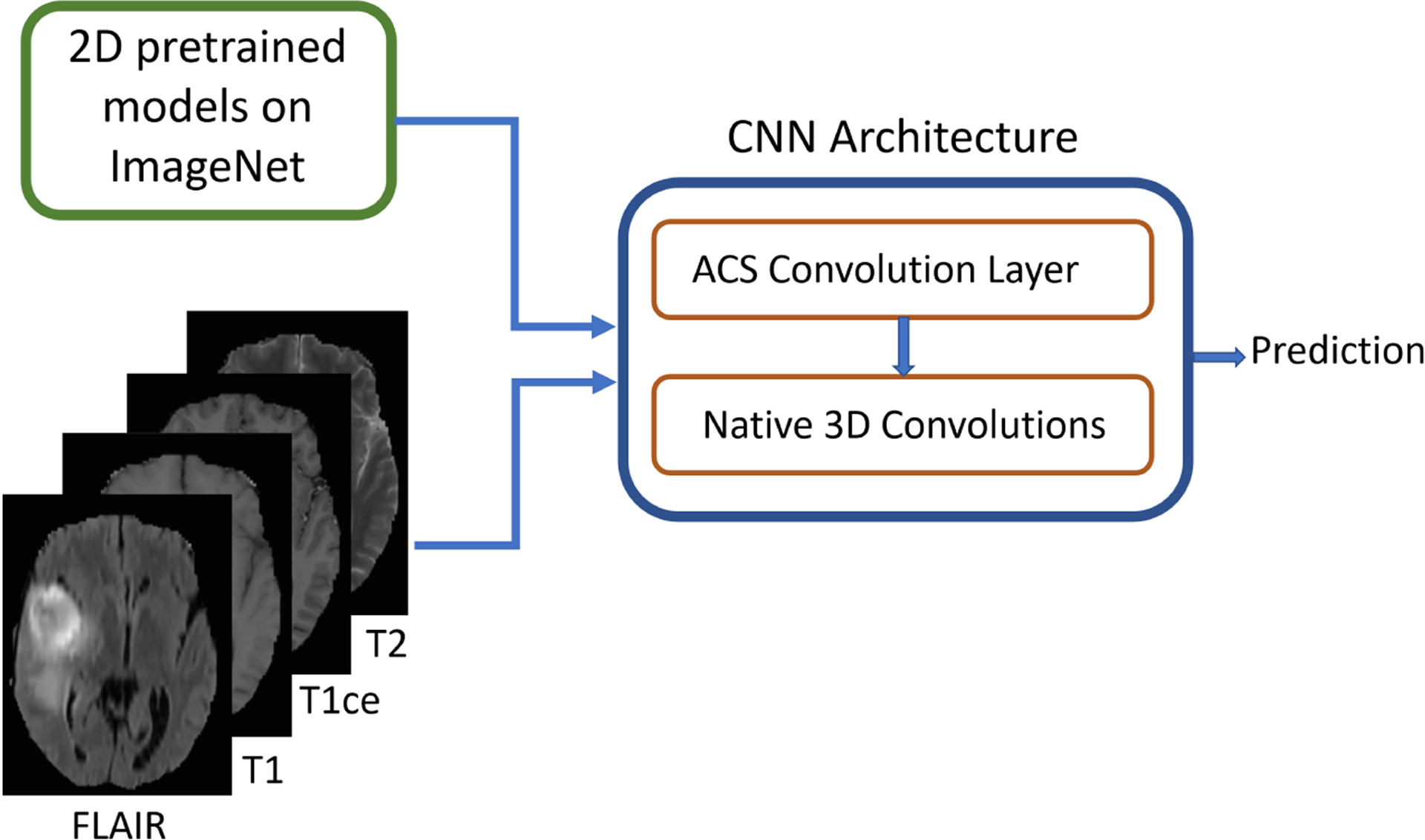
Idea of integrating ACS convolutions with pre-trained 2D model weights to enable native 3D convolutions on 3D medical data (e.g., MRI)
2.2. Architecture design
We have incorporated the concept of ACS convolutions in GaNDLF, which is a framework for training models for segmentation, classification, and regression in a reproducible and deployable manner [27]. GaNDLF has several architectures for segmentation and classification, as well as a wide range of data pre-processing and augmentation options along with the choice of several training hyper parameters and loss functions. We integrated several encoders from [29], pre-trained on ImageNet [11] into this framework, including variants of VGG [30], ResNet [31], DenseNet [32], and EfficientNet [33]. We have created a mechanism to combine the outputs of these encoders with either a segmentation or a classification head depending on the task, as shown in Fig. 3. The segmentation head consists of a set of upsampling layers similar to the decoder mechanism of the UNet network topology/architecture [34], where the user has the flexibility to choose the number of upsampling layers and the number of feature maps in each layer. The classification head consists of a average pooling layer applied on the top of feature maps obtained from the encoder. Dropout can be set between range 0 to 1 to reduce overfitting to the training data before the final classification layer.
Fig. 3:
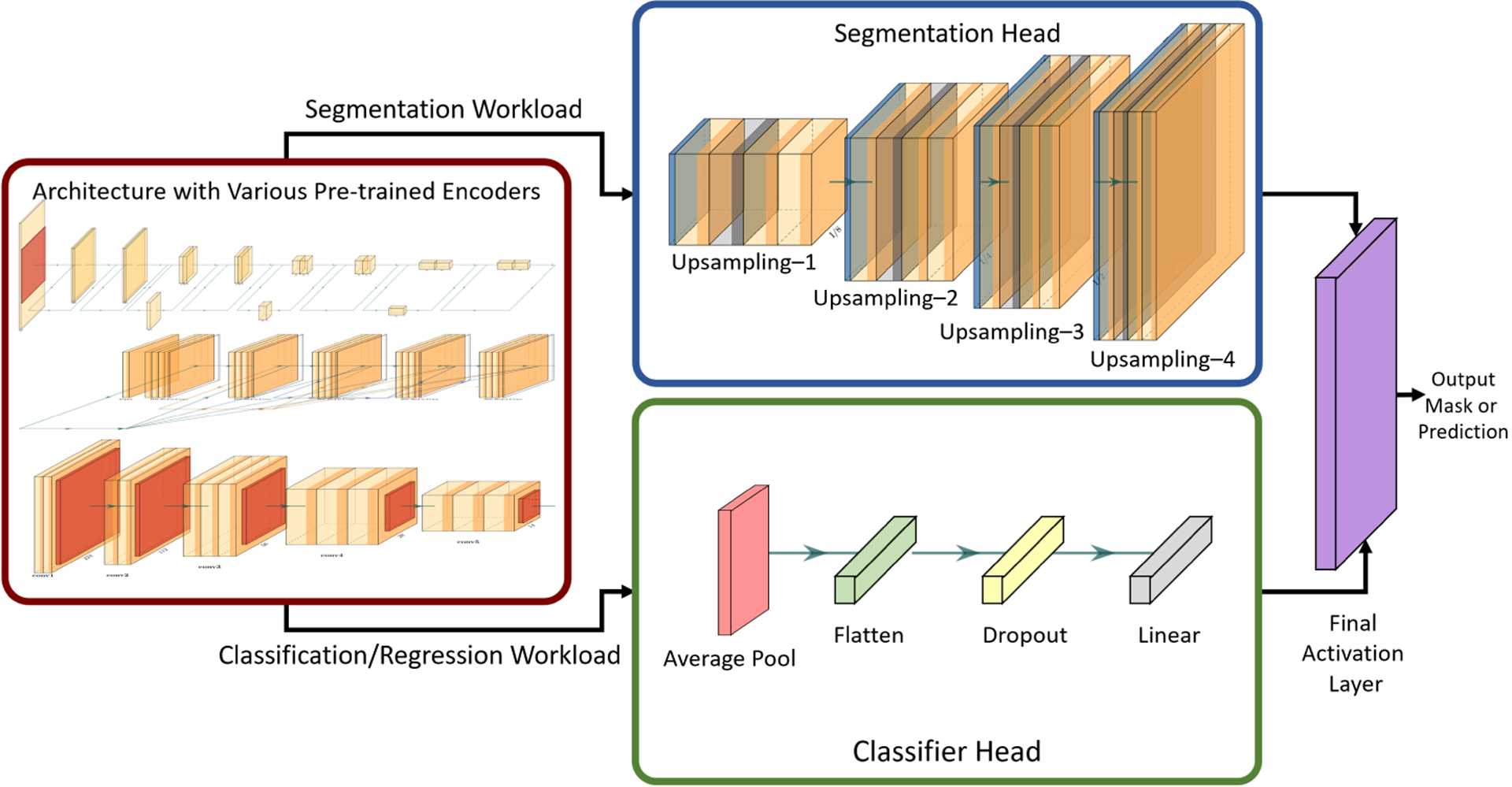
The architecture that allows using different pre-trained encoders with either a segmentation or classifier head for specific workloads.
While the 2D pre-trained weights could be directly loaded for applications on 2D data, we have replaced the usual convolution layer with an ACS convolution layer in GaNDLF, enabling their use for training on 3D medical data in a native manner, regardless of the number of input modalities. In comparison with the 2D models, ACS convolution layers do not introduce any additional computation cost, memory footprint, or model size.
2.2.1. Design for segmentation
Gliomas are among the most common and aggressive brain tumors and accurate delineation of the tumor sub-regions is important in clinical diagnosis. We trained two different architectures for segmentation through GaNDLF. UNet [34] with residual connections (ResUNet) is one of the famous architectures for 2D and 3D medical segmentation. It consists of encoder and decoder modules and feature concatenation pathways. The encoder is a stack of convolutional and downsampling layers for feature extraction from the input images, and the decoder consists of a set of upsampling layers (applying transpose convolutions) to generate the fine-grained segmentation output.
We trained two different models using the publicly available multi-parametric magnetic resonance imaging (mpMRI) data of 369 cases from training set of the International Brain Tumor Segmentation [35–37] (BraTS2020) challenge. This dataset consists of four multi-parametric magnetic resonance imaging (mpMRI) scans per subject/case, with the exact modalities being: a) native (T1) and b) post-contrast T1-weighted (T1-Gd), c) T2-weighted (T2), and d) T2 fluid attenuated inversion recovery (T2-FLAIR). These models are evaluated on 125 unseen cases from the BraTS2020 validation dataset. We first trained a standard ResUNet architecture of depth = 4 and base filters = 32 such that weights of all the layers were randomly initialized. We then built another architecture by using pre-trained ResNet50 as an encoder with depth = 4 and the standard UNet decoder. For each of these experiments, 40 patches of 64×64×64 were extracted from each subject. Various training parameters were also kept constant, like the choice of optimizer (we used SGD), scheduler (modified triangular) with learning rate of 0.001, and loss function based on the Dice similarity coefficient (DSC) [38]. Maximum number of epochs was set to 250 with patience of 30 for early stopping. The performance is evaluated on clinically-relevant tumor regions, i.e., whole tumor (considered for radiotherapy), tumor core (considered for surgical resection) as well as enhancing tumor.
2.2.2. Design for classification
Glioblastoma (GBM) is the most aggressive and common adult primary malignant brain tumor and epidermal growth factor receptor variant III (EGFRvIII) mutation is considered a driver mutation and therapeutic target in GBM [39–41]. Usually, the presence of EGFRvIII is determined by the analysis of actual tissue specimens and is stated as positive or negative. We focus on non-invasive prediction of EGFRvIII status by analysis of these pre-operative and pre-processed MRI data. Residual Networks (ResNets) [31] introduced the idea of skip connections which enabled design of much deeper CNNs. GaNDLF supports variants of ResNet, including ResNet18, ResNet34, ResNet50, ResNet101, and ResNet152, each having different number of layers.
We use an internal private cohort of 146 patients containing four structural mpMRI modalities (T1, T2, T1-Gd and T2-FLAIR) such that the positive and negative classes were equally distributed. These 146 cases were distributed in Training (80%) and Validation (20%) sets for experimentation. We used cross entropy loss function, adam optimiser and cosine annealing scheduler with learning rate of 0.0001. As the dataset is smaller, we set the maximum number epochs to 100 and patience of 30 epochs for early stopping.
3. Results
In this section we present the quantitative results of the segmentation and classification workloads described above, to showcase the feasibility and performance of ACS convolutions on 3D medical imaging data. Specifically, we compare the 2D pre-training approach with the random initialization to evaluate the superiority of the ACS convolutions over usual 3D convolution operations.
3.1. Brain Tumor Segmentation workload
The segmentation model is trained on the publicly available training data of BraTS2020 challenge. We then quantitatively evaluate the performance of the final models on the unseen BraTS2020 validation data by submitting results to the online evaluation platform (CBICA Image Processing Portal). Table 1 lists the number of parameters of each model, as well as the comparative performance, in terms of Dice Similarity Coefficient (DSC) and the 95th percentile of the Hausdorff distance between the predicted ground truth labels.
Table 1:
Results on Brain Tumor Segmentation (BraTS2020) validation dataset
| Metric | Region | Standard ResUNet | ResNet50+UNet (Random init.) | ResNet50+UNet (Pre-trained) |
|---|---|---|---|---|
| DSC | Whole Tumor | 0.8771 | 0.8775 | 0.8736 |
| Tumor Core | 0.7735 | 0.7458 | 0.7719 | |
| Enhancing Tumor | 0.7138 | 0.69508 | 0.7017 | |
| Hausdorff95 | Whole Tumor | 13.2425 | 7.6747 | 9.5384 |
| Tumor Core | 14.7492 | 8.6579 | 15.4840 | |
| Enhancing Tumor | 34.8858 | 41.00332 | 40.2053 | |
| #Parameters | - | 33.377 Million | 25.821 Million | 25.821 Million |
| #Epochs for convergence | - | 104 epochs | 250 epochs | 95 epochs |
3.2. Binary Classification of Brain Tumor Molecular Status
For the performance evaluation of the classification workload, we have used the structural mpMRI scans in-tandem as input (i.e., passing all the scans together at once as separate channels) similar to the segmentation workload. The classification model performance on training and validation data is summarized in Table 2, illustrating the effectiveness of pre-trained weights.
Table 2:
Results on EGFR Classification
| ResNet50(Random initialization) | ResNet50(Pre-trained on ImageNet) | |
|---|---|---|
| Training Accuracy | 0.7203 | 0.9915 |
| Training Loss | 0.5736 | 0.3292 |
| Val Accuracy | 0.5357 | 0.7142 |
| Val Loss | 0.6912 | 0.5758 |
4. Discussion
In this work, we have assessed the functionality of transfer learning for 3D medical data based on the available 2D models pre-trained on ImageNet for segmentation and classification. The framework that this functionality is evaluated is designed such that deep learning network architecture’s first and last layers are flexible to be able to process input images of any size with varying number of channels or modalities, and provide the final prediction based on the relevant number of classes for the specified task. The rest of the layers are initialized with pre-trained weights from the ImageNet models and are further fine-tuned.
The results of brain tumor segmentation using i) 3D U-Net with residual connections, ii) randomly initialized ResNet50 encoder & UNet decoder, and iii) pre-trained ResNet50 encoder & UNet decoder are shown in Table 1. In these architectures, the obvious difference was in the encoders being randomly initialised in the former i) 3DUNet and ii) ResNet50 and pre-trained in the latter ResNet50-UNet (iii). As the pre-trained decoders are not available from ImageNet, the decoder was initialised with random weights in all the three architectures. We hypothesize that this might be the reason for comparable segmentation performance in terms of dice and hausdorff95 scores, while the difference in number of parameters is significant. It should be observed that the ResNet50-UNet (ii and iii) has only 25.821 Million parameters, which is around 22% less compared to 33.377 Million parameters of the standard ResUNet (i), with the same encoder-decoder depth. The randomly initialised ResNet50-UNet model oscillates around the same performance and did not converge in the specified maximum number of epochs (250). On the other hand, the same model initialised with pre-trained weights converged within 95 epochs. Thus models initialized with pre-trained weights have advantage of better convergence speed as well as smaller model size. Importantly, smaller models are more preferable in the clinical setting due to their higher feasibility for deployment in low-resource environments.
Baseline results of binary classification for the determination of the EGFRvIII mutational status are reported in Table 2, with ResNet50 architecture. We did not use any additional data augmentation techniques. As the data in this task were limited, the effect of pre-trained weights are clearly observed resulting in better accuracy. Fig. 4 shows the plots of cross entropy loss and accuracy in training with respect to epochs. Similar performance is observed for validation set as well. The weights of the model with lowest validation loss are stored for reproducibility and the accuracy and loss values reported in Table 2 are for the saved model with lowest validation loss.
Fig. 4:
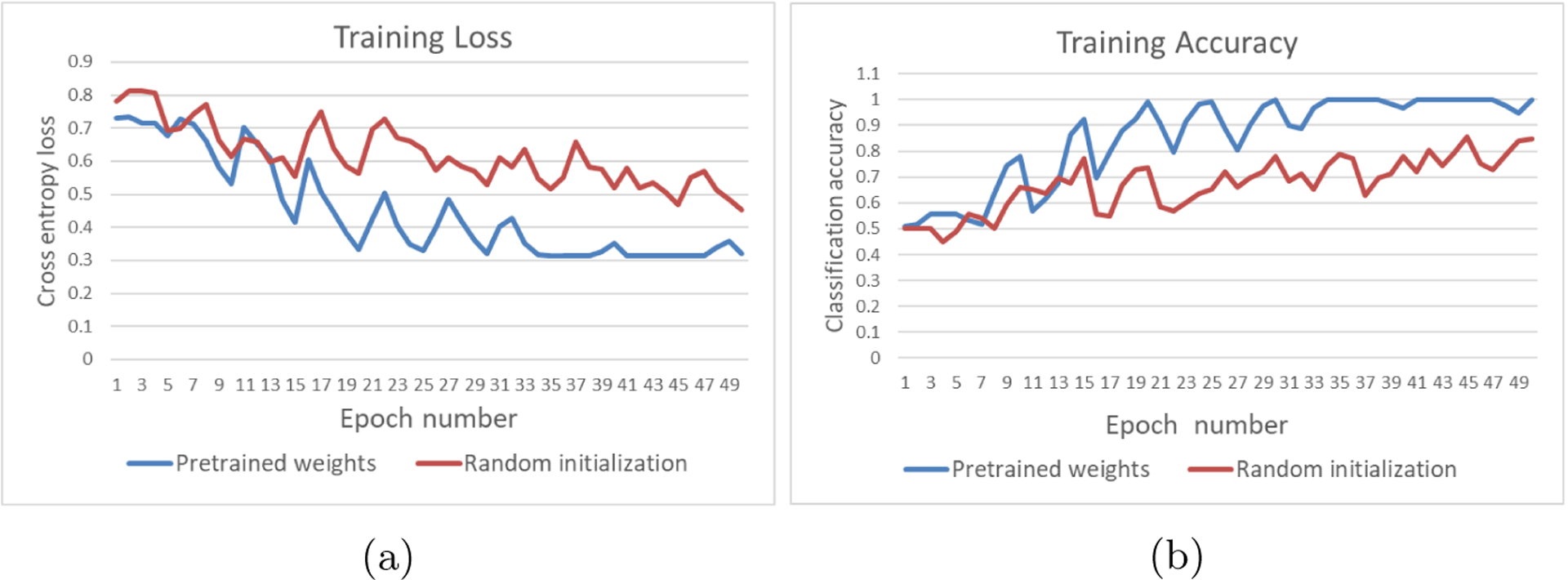
Comparison plots of training loss and accuracy for binary classification of EGFRvIII mutation status. These plots are for ResNet50 architecture with and without use of 2D pre-trained weights from ImageNet
Our findings support the incorporation of 2D pre-trained models towards improving the performance on 3D medical image segmentation and classification workloads, with demonstrably smaller model size (Table 1). Large increase in accuracy is specially observed in those applications where sufficient labelled data are not available. Incorporating this functionality in GaNDLF provides a readily available solution to researchers towards an end-to-end solution for several computational tasks, along with support for pre-trained encoders, making it a robust application framework for deployment and integration in clinical workflows. Future studies can explore this mechanism by applying it to compare randomly initialized and pre-trained models for convergence speed (in both centralized and federated learning settings [8,9,42,43]), performance gains in applications requiring 3D datasets, model optimization allowing deployment in low-resource environments, and privacy analysis.
Acknowledgments
Research reported in this publication was partly supported by the National Institutes of Health (NIH) under award numbers NIH/NCI:U01CA242871 and NIH/NINDS:R01NS042645. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH.
Footnotes
References
- 1.Zhou SK, Greenspan H, Davatzikos C, Duncan JS, Van Ginneken B, Madabhushi A, Prince JL, Rueckert D, and Summers RM, “A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises,” Proceedings of the IEEE, vol. 109, no. 5, pp. 820–838, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Wang X, Zhang K, Fung K-M, Thai TC, Moore K, Mannel RS, Liu H, Zheng B, and Qiu Y, “Recent advances and clinical applications of deep learning in medical image analysis,” Medical Image Analysis, p. 102444, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varoquaux G and Cheplygina V, “Machine learning for medical imaging: methodological failures and recommendations for the future,” NPJ digital medicine, vol. 5, no. 1, pp. 1–8, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yosinski J, Clune J, Bengio Y, and Lipson H, “How transferable are features in deep neural networks?,” Advances in neural information processing systems, vol. 27, 2014. [Google Scholar]
- 5.Goodfellow I, Bengio Y, and Courville A, Deep learning. MIT press, 2016. [Google Scholar]
- 6.Aloysius N and Geetha M, “A review on deep convolutional neural networks,” in 2017 international conference on communication and signal processing (ICCSP), pp. 0588–0592, IEEE, 2017. [Google Scholar]
- 7.Pan SJ and Yang Q, “A survey on transfer learning,” IEEE Transactions on Knowledge and Data Engineering, vol. 22, no. 10, pp. 1345–1359, 2010. [Google Scholar]
- 8.Pati S, Baid U, Edwards B, Sheller M, Wang S-H, Reina GA, Foley P, Gruzdev A, Karkada D, Davatzikos C, et al. , “Federated learning enables big data for rare cancer boundary detection,” arXiv preprint arXiv:2204.10836, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A, Milchenko M, Xu W, Marcus D, Colen RR, et al. , “Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data,” Scientific reports, vol. 10, no. 1, pp. 1–12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baid U, Pati S, Thakur S, Edwards B, Sheller M, Martin J, and Bakas S, “NIMG-32. THE FEDERATED TUMOR SEGMENTATION (FETS) INITIATIVE: THE FIRST REAL-WORLD LARGE-SCALE DATA-PRIVATE COLLABORATION FOCUSING ON NEURO-ONCOLOGY,” Neuro-Oncology, vol. 23, pp. vi135–vi136, 11 2021. [Google Scholar]
- 11.Deng J, Dong W, Socher R, Li L-J, Li K, and Fei-Fei L, “Imagenet: A large-scale hierarchical image database,” in 2009 IEEE conference on computer vision and pattern recognition, pp. 248–255, Ieee, 2009. [Google Scholar]
- 12.Redmon J and Farhadi A, “Yolov3: An incremental improvement,” arXiv preprint arXiv:1804.02767, 2018. [Google Scholar]
- 13.Lin T-Y, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, and Zitnick CL, “Microsoft coco: Common objects in context,” in European conference on computer vision, pp. 740–755, Springer, 2014. [Google Scholar]
- 14.Chen J, Yang L, Zhang Y, Alber M, and Chen DZ, “Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation,” Advances in neural information processing systems, vol. 29, 2016. [Google Scholar]
- 15.Yu Q, Xie L, Wang Y, Zhou Y, Fishman EK, and Yuille AL, “Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8280–8289, 2018. [Google Scholar]
- 16.Díaz-Pernas FJ, Martínez-Zarzuela M, Antón-Rodríguez M, and González-Ortega D, “A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network,” in Healthcare, vol. 9, p. 153, MDPI, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ismael SAA, Mohammed A, and Hefny H, “An enhanced deep learning approach for brain cancer mri images classification using residual networks,” Artificial intelligence in medicine, vol. 102, p. 101779, 2020. [DOI] [PubMed] [Google Scholar]
- 18.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, and Ronneberger O, “3d u-net: learning dense volumetric segmentation from sparse annotation,” in International conference on medical image computing and computer-assisted intervention, pp. 424–432, Springer, 2016. [Google Scholar]
- 19.Zhao W, Yang J, Sun Y, Li C, Wu W, Jin L, Yang Z, Ni B, Gao P, Wang P, et al. , “3d deep learning from ct scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas,” Cancer research, vol. 78, no. 24, pp. 6881–6889, 2018. [DOI] [PubMed] [Google Scholar]
- 20.Milletari F, Navab N, and Ahmadi S-A, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” in 2016 fourth international conference on 3D vision (3DV), pp. 565–571, IEEE, 2016. [Google Scholar]
- 21.Baid U, Talbar S, Rane S, Gupta S, Thakur MH, Moiyadi A, Sable N, Akolkar M, and Mahajan A, “A novel approach for fully automatic intra-tumor segmentation with 3d u-net architecture for gliomas,” Frontiers in computational neuroscience, p. 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trivizakis E, Manikis GC, Nikiforaki K, Drevelegas K, Constantinides M, Drevelegas A, and Marias K, “Extending 2-d convolutional neural networks to 3-d for advancing deep learning cancer classification with application to mri liver tumor differentiation,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 3, pp. 923–930, 2019. [DOI] [PubMed] [Google Scholar]
- 23.Zheng H, Zhang Y, Yang L, Liang P, Zhao Z, Wang C, and Chen DZ, “A new ensemble learning framework for 3d biomedical image segmentation,” in Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 5909–5916, 2019. [Google Scholar]
- 24.Xia Y, Xie L, Liu F, Zhu Z, Fishman EK, and Yuille AL, “Bridging the gap between 2d and 3d organ segmentation with volumetric fusion net,” in MICCAI, 2018. [Google Scholar]
- 25.Li X, Chen H, Qi X, Dou Q, Fu C-W, and Heng P-A, “H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes,” IEEE transactions on medical imaging, vol. 37, no. 12, pp. 2663–2674, 2018. [DOI] [PubMed] [Google Scholar]
- 26.Ni T, Xie L, Zheng H, Fishman EK, and Yuille AL, “Elastic boundary projection for 3d medical image segmentation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2109–2118, 2019. [Google Scholar]
- 27.Pati S, Thakur SP, Bhalerao M, Baid U, Grenko C, Edwards B, Sheller M, Agraz J, Baheti B, Bashyam V, et al. , “Gandlf: A generally nuanced deep learning framework for scalable end-to-end clinical workflows in medical imaging,” arXiv preprint arXiv:2103.01006, 2021. [Google Scholar]
- 28.Yang J, Huang X, He Y, Xu J, Yang C, Xu G, and Ni B, “Reinventing 2d convolutions for 3d images,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 8, pp. 3009–3018, 2021. [DOI] [PubMed] [Google Scholar]
- 29.Yakubovskiy P, “Segmentation models pytorch.” https://github.com/qubvel/segmentation_models.pytorch, 2020.
- 30.Simonyan K and Zisserman A, “Very deep convolutional networks for large-scale image recognition,” arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
- 31.He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778, 2016. [Google Scholar]
- 32.Huang G, Liu Z, Van Der Maaten L, and Weinberger KQ, “Densely connected convolutional networks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4700–4708, 2017. [Google Scholar]
- 33.Tan M and Le Q, “Efficientnet: Rethinking model scaling for convolutional neural networks,” in International conference on machine learning, pp. 6105–6114, PMLR, 2019. [Google Scholar]
- 34.Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, pp. 234–241, Springer, 2015. [Google Scholar]
- 35.Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, et al. , “The multimodal brain tumor image segmentation benchmark (brats),” IEEE transactions on medical imaging, vol. 34, no. 10, pp. 1993–2024, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, and Davatzikos C, “Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features,” Scientific data, vol. 4, no. 1, pp. 1–13, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M, et al. , “Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge,” arXiv preprint arXiv:1811.02629, 2018. [Google Scholar]
- 38.Zijdenbos AP, Dawant BM, Margolin RA, and Palmer AC, “Morphometric analysis of white matter lesions in mr images: method and validation,” IEEE transactions on medical imaging, vol. 13, no. 4, pp. 716–724, 1994. [DOI] [PubMed] [Google Scholar]
- 39.Binder ZA, Thorne AH, Bakas S, Wileyto EP, Bilello M, Akbari H, Rathore S, Ha SM, Zhang L, Ferguson CJ, et al. , “Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development,” Cancer cell, vol. 34, no. 1, pp. 163–177, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakas S, Akbari H, Pisapia J, Martinez-Lage M, Rozycki M, Rathore S, Dahmane N, O’Rourke DM, and Davatzikos C, “In vivo detection of egfrviii in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: The ϕ-indexin vivo egfrviii detection in glioblastoma via mri signature,” Clinical Cancer Research, vol. 23, no. 16, pp. 4724–4734, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbari H, Bakas S, Pisapia JM, Nasrallah MP, Rozycki M, Martinez-Lage M, Morrissette JJ, Dahmane N, O’Rourke DM, and Davatzikos C, “In vivo evaluation of egfrviii mutation in primary glioblastoma patients via complex multiparametric mri signature,” Neuro-oncology, vol. 20, no. 8, pp. 1068–1079, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K, et al. , “The future of digital health with federated learning,” NPJ digital medicine, vol. 3, no. 1, pp. 1–7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baid U, Pati S, Kurc TM, Gupta R, Bremer E, Abousamra S, Thakur SP, Saltz JH, and Bakas S, “Federated learning for the classification of tumor infiltrating lymphocytes,” arXiv preprint arXiv:2203.16622, 2022. [Google Scholar]


