Abstract
Sulfation widely exists in the eukaryotic proteome. However, understanding of the biological functions of sulfation in peptides and proteins has been hampered by the lack of methods to control its spatial or temporal distribution in the proteome. Herein, we report that fluorosulfate can serve as a latent precursor of sulfate in peptides and proteins, which can be efficiently converted into sulfate by hydroxamic acid reagents under physiologically relevant conditions. Photocaging the hydroxamic acid reagents further allowed for light-controlled activation of functional sulfopeptides. This work provides a valuable tool for probing functional roles of sulfation in peptides and proteins.
Graphical Abstract
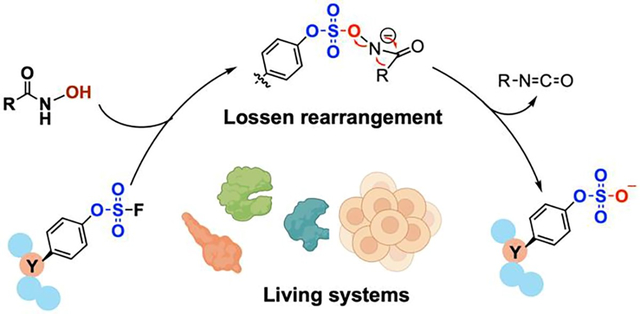
O-Sulfation of the tyrosine residue is a post-translational modification (PTM) that widely exists in eukaryotic peptides and proteins (Figure 1a), and has been implicated to regulate a variety of biological functions such as immune response, hemostasis, and pathogen evasion.1–2 However, only a small fraction of the sulfoproteome has been annotated.3–4 A long-standing challenge for studying the sulfoproteome is that sulfation is highly heterogeneous, with various sulfopeptides and sulfoproteins exist in different sulfoforms.5 The seminal works of Schultz,6 Liu,7 Chatterjee,8 Niu,9 and Xiao10 that incorporate sulfotyrosine (sY) into proteins as a non-canonical amino acid (ncAA) represent notable examples to address this challenge. Expanding upon these advances, methods that allow researchers to spatiotemporally control sulfation in the proteomic context would be highly valuable for studying their functional roles in biology.11
Figure 1. Background and our approach.
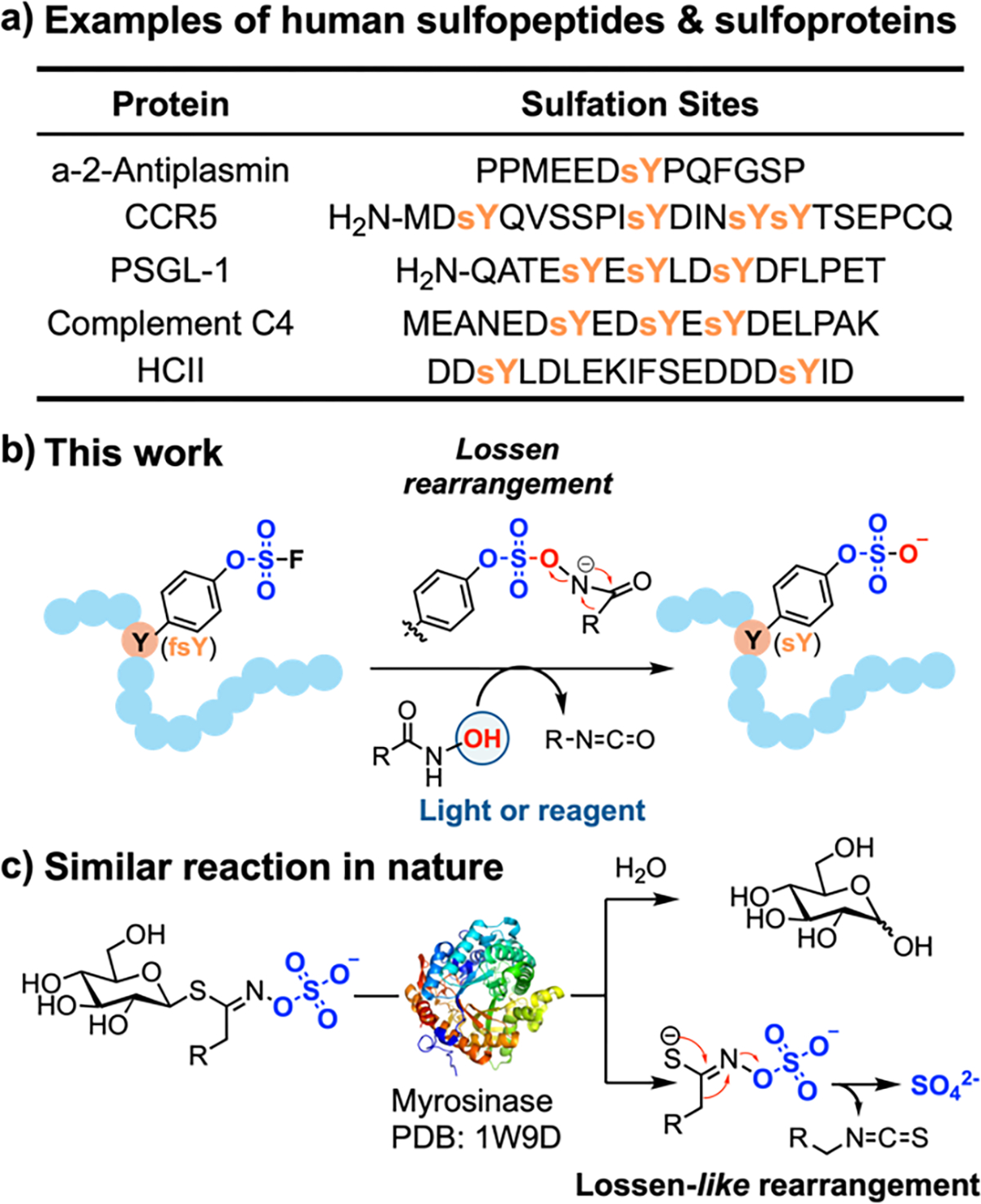
a) Sulfation widely exists in diverse bioactive peptides and proteins. b) In this work, fluorosulfate is incorporated in peptides and proteins as a latent sulfate and can be efficiently converted into sulfate by hydroxamic acid reagents under physiologically relevant conditions. c) Our approach mirrors the myrosinase-catalyzed Lossen-like rearrangement of glucosinolates in nature.
Caging strategies have been developed for various protein PTMs to probe how these PTMs regulate dynamic cellular events. Although a broad collection of caging groups are available for a variety of PTMs, a caging group that stably protects sulfotyrosine (sY) residues in peptides and proteins and can be efficiently removed under physiological conditions remains elusive.12 The reasons for such a knowledge gap includes the high energy barrier for chemically activating the sulfate group for coupling chemistries, the lability of sY to acid, heat, and high-energy ionization, and the strong electron-withdrawing propensity of sulfate that renders commonly used benzylic ester caging groups unstable.13–15 On the other hand, while multiple alkyl and aryl esters have been successfully used as protecting groups of sY in solid-phase peptide synthesis,17–18 such as 2,2,2-trichloroethyl (TCE),18–19 2,2-dichlorovinyl (DCV),20–21 2,2,2-trifluoroethyl (TFE),22 neopentyl,23–24 and phenyl25 sulfate diesters, their deprotection conditions (e.g., hydrogenolysis,18–21, 25 strong base,22 heating,23 high salt concentraton,24 etc.) are incompatible with living systems.
In 2014, Sharpless et al. reported the reactivity of fluorosulfate in Sulfur(VI) Fluoride exchange (SuFEx) reaction.26–29 Compared to other halogen-substituted sulfate derivatives, fluorosulfate not only has a size closest to that of sulfate, but is also far less electrophilic due to the π-donation from fluorine to sulfur.30 As a result, fluorosulfate has demonstrated excellent metabolic stability in vivo.31–32 The chemical inertness of fluorosulfate has allowed its tyrosine derivative, L-fluorosulfotyrosine (fsY), to be incorporated into peptides and proteins via solid-phase peptide synthesis12, 33 and ncAA mutagenesis.13, 32 Herein, we demonstrate that fluorosulfate can serve as a latent sulfate in sulfopeptides and sulfoproteins and can be efficiently converted into sulfate (hereafter denoted as “decaging”) by hydroxamic acid (HA) reagents under physiologically relevant conditions. Mechanistic studies revealed an unusual Lossen rearrangement pathway of fluorosulfate activation and decaging (Figure 1b) that is analogous to the myrosinase-mediated Lossen-like rearrangement of glucosinolate in nature (Figure 1c).34
Our initial investigation confirmed that fluorosulfate is stable in various aqueous physiologically relevant conditions, such as buffer solution, cell lysate, and serum at neutral pH. Specifically, negligible (<5%) hydrolysis of fluorosulfate could be detected in aqueous buffer at neutral pH after 24 hours (Table S1, entries 1–2). Fluorosulfate also remained mostly intact after 12 hours in serum and after 48 hours in cell lysate (Table S2). Even tetramethylguanidine, a reagent previously reported to promote SuFEx reaction in aqueous solution,35 showed no reactivity against fluorosulfate alone (Table S1). Interestingly, we found that the hydrolysis product of N-hydroxylsuccimide (Table S3 and Figure S2), N-hydroxylsuccinic acid monoamide (3), converted a fsY-containing hexapeptide 1 into the corresponding sulfopepitde 2 in 57% yield in one hour (Figure 2a). Encouraged by this finding, we examined other HA derivatives (Figure 2a, S4). Acetohydroxamic acid (4) promoted the reaction to 78% over one hour. Good yield (95%) of 2 was obtained using aromatic benzohydroxamic acid (5) under the same condition. The highest efficiency was observed when the cationic HA 6 and heteroaromatic HA 7 were used, achieving quantitative conversion in 30 minutes. Other non-HA α-nucleophile reagents such as oxime 8,36 2-aminoxime 9, and 1-hydroxybenzotriazole (10)37 resulted in lower reaction efficiency. In contrast, triisopropylsilyl ether (TIPS)-masked HA 11 showed no reactivity until potassium fluoride (KF) was added to remove the TIPS protecting group (Table S4 and Figure S5), confirming that HA is the reactive center for fluorosulfate activation. It is also noteworthy that the decaging reaction mediated by 7 proceeded with no detectable side reaction in the presence of 20 equivalents of amino acids including lysine, histidine, tyrosine, and cysteine (Table S5).
Figure 2. Reagent screen and mechanistic investigation.
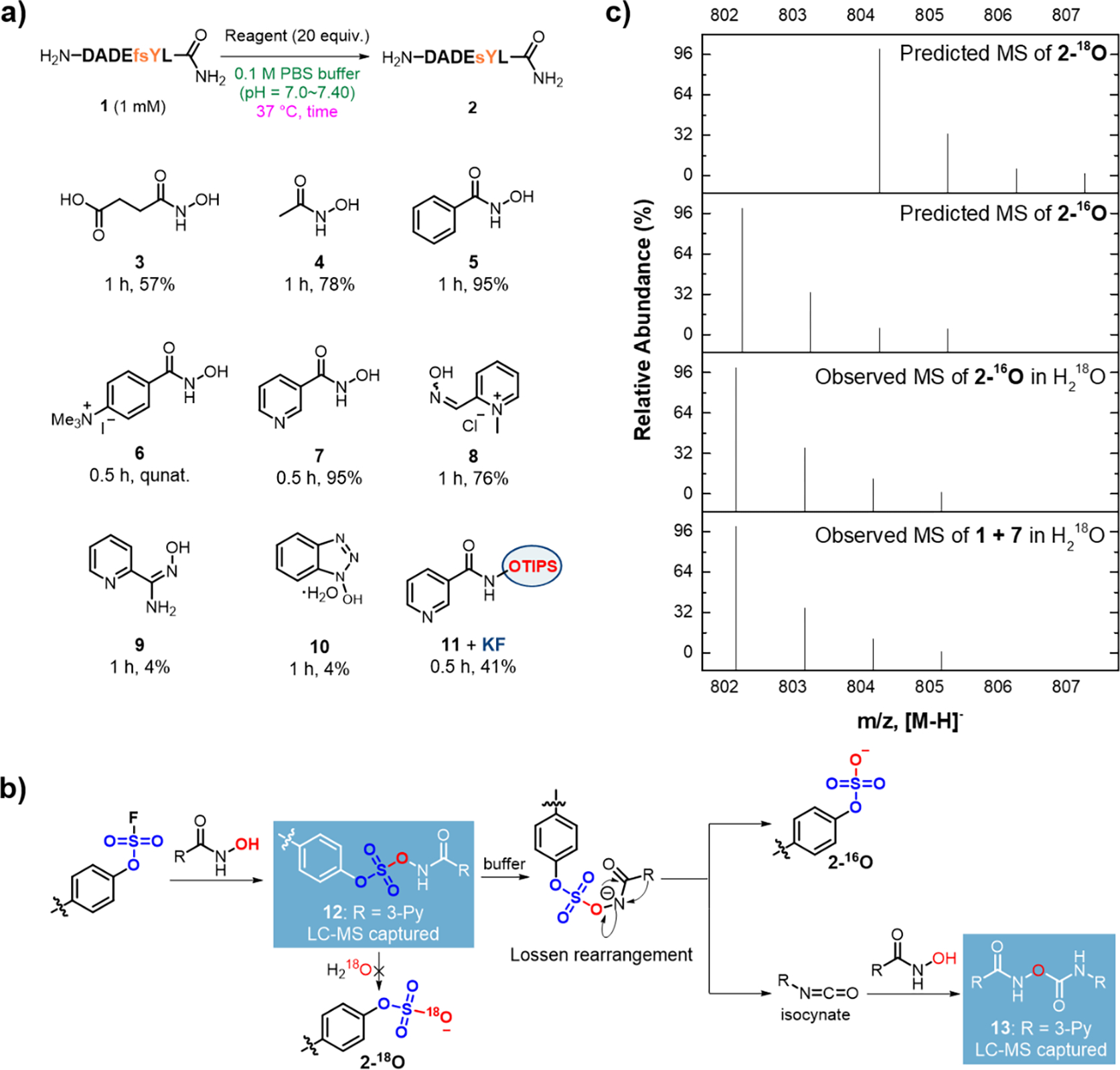
a) A variety of HA reagents were investigated for their ability to activate fluorosulfate in model peptide 1. Yields were determined by HPLC. b) Real-time LC-MS reaction monitoring identified two adducts of 7, 12 and 13, suggesting a Lossen rearrangement mechanism. c) No 18O-labeled products were found from the reaction in H218O buffer, suggesting that the sulfate product was not generated from direct hydrolysis.
To gain insight into the reaction mechanism, the reaction with 1 as the substrate and reagent 7 was monitored using liquid chromatography mass spectrometry (LC-MS) to capture the reaction intermediates (Figure 2b and Figure S6). An adduct (12) of 7 and 1 was detected, confirming the nucleophilic coupling between the HA reagent and the substrate. Surprisingly, an isocyanate adduct 13 was also detected within 10 minutes at 37 °C, suggesting an uncommon intramolecular Lossen rearrangement pathway. To further probe this possibility, we performed the decaging reaction of 1 by 7 in the buffer prepared exclusively using H218O. This reaction yielded 2 that contained no 18O isotope (Figure 2c and Figure S7), suggesting that the conversion of fluorosulfate into sulfate is not through direct hydrolysis. These results further support a Lossen rearrangement mechanism.38–40 Such a pathway is similar to the myrosinase-catalyzed Lossen-like rearrangement of glucosinolate in Brassia plants, in which an inorganic sulfate and isothiocyante are generated from a thiohydroximate-O-sulfate intermediate (Figure 1c).34
We then examined the decaging of various fluorosulfate-containing peptides mediated by 7 under physiological pH. Notably, peptides that contains multiple fsY residues, or nucleophilic residues (e.g., lysine or cysteine) were successfully decaged in high yields (Figure 3a). In addition, decaging can be achieved in a light-mediated fashion. The fsY residues in 16 was efficiently decaged after a 2-nitrobenzyl-caged reagent 20 was exposed to 370 nm UV light irradiation (Figure 3b).41 No conversion was observed in dark or without 20 (Figure S31).
Figure 3. Decaging of fluorosulfate-containing peptides and proteins under physiologically relevant conditions.
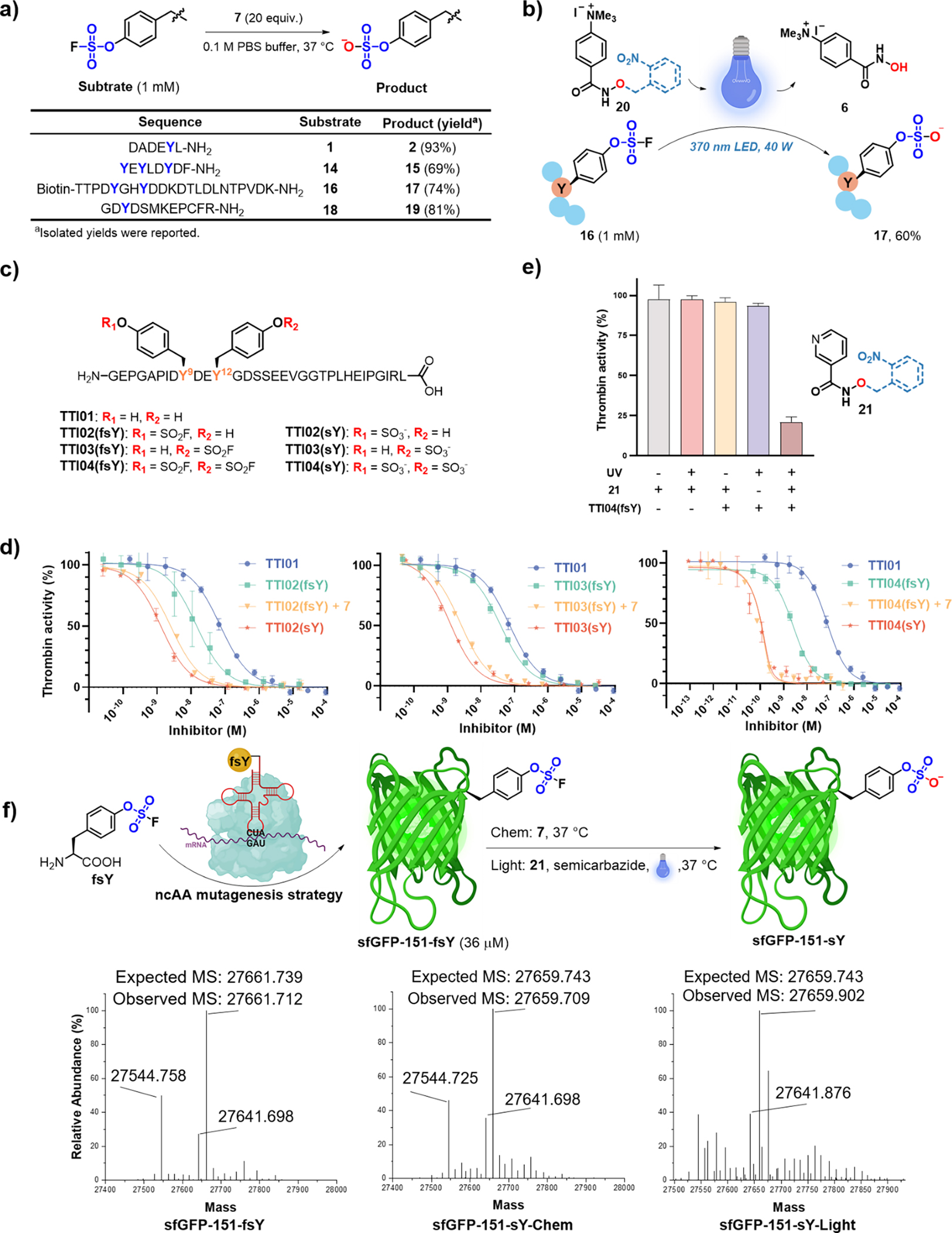
a) Fluorosulfate decaging in fsY-containing synthetic peptides. b) Light-mediated fluorosulfate decaging in C5aR1 22mer peptide using photocaged reagent 20. c) TTI peptide sequences and sulfation patterns. d) Thrombin inhibition assay of TTI peptides. Data were fitted to the Morrison inhibition model and error bars represent the standard deviation of three independent measurements. e) Light-mediated activation and decaging of fluorosulfate-containing TTI peptide TTI04(fsY) regulates its sulfation-dependent thrombin inhibitory activity. f) Fluorosulfate decaging in fsY-containing protein sfGFP-151-fsY and its corresponding high-resolution mass spectrometry.50
We used tsetse thrombin inhibitor (TTI)42 peptides as a model system to probe the utility of the HA reagents in controlling the bioactivities associated with sulfation under physiologically relevant conditions. We used a standard human α-thrombin activity assay with Chromozym TH substrate to determine the inhibitory effects of the TTI peptides (latent) consisting of fsY residues at position 9 and 12: TTI02(fsY), TTI03(fsY), and TTI04(fsY), and the corresponding sY-containing TTI peptides (active): TTI02(sY), TTI03(sY), and TTI04(sY) (Figure 3c).43–44 Although the latent TTI peptides still exhibited minor inhibitory effects compared to the non-sulfated control TTI01, the active TTI peptides demonstrated significantly higher potencies (Figure 3d and Figure S9).45 The latent TTI peptides that were decaged in situ by reagent 7 all showed similar inhibitory effects as the purified active TTI peptides (Figure 3d). These results confirmed that fluorosulfate can serve as an effective latent sulfate in peptides, and can be facilely decaged in aqueous solution at neutral pH. Light-controlled decaging is also possible. For example, while the latent TTI04 (fsY) remains inactive for thrombin inhibition at 3.7 nM in the presence of 2-nitrobenzyl protected reagent 21 in dark, after irradiation thrombin activity was reduced to 21% (Figure 3e).
The small size of fluorine atom allows fsY to be facilely incorporated into proteins as a ncAA.32, 46 Following the procedure established by Wang et al.,32 we cloned the fsY-specific aminoacyl tRNA synthetase FsTyrRS and an optimal pyrrolysyl tRNA into plasmids for fsY incorporation into proteins. A sfGFP gene containing a TAG codon at position 151 was co-transformed along with the genes containing the FsTyrRA/tRNA pair into B95 E. coli cells.47 The targeted sfGFP-151-fsY was successfully expressed in a 12 mg/L yield. Tandem MS results verified the incorporation of fsY at the TAG-specified position-151 (Figure S10).32, 48 Next, to confirm the conversion from fluorosulfate to sulfate in sfGFP-151-fsY by 7, as well as the integrity of the resulting sulfoprotein, we performed whole protein intact mass analyses of sfGFP-151-fsY before and after decaging using high-resolution Orbitrap mass spectrometry, which is capable of achieving sub-5 ppm mass accuracy49 and can confidently resolve the 1.996 Da mass shift after decaging (Figure 3f and Figure S11).50 Similarly, incorporation of fsY at position 3 of sfGFP and the subsequent decaging by 7 were also confirmed (Figure S12–S13). Furthermore, we showed the light-mediated decaging of sfGFP-151-fsY by the photocaged reagent 19 (Figure 3f), highlighting the potential of our approach for the spatiotemporal release of caged sulfoproteins.
Last, we tested the cytocompatibility of the fluorosuflate decaging reagents. Previously, cesium carbonate (Cs2CO3)/ethylene glycol33 or 2 M ammonium acetate (NH4OAc) aqueous solution24, 43 was used to remove the protecting groups for sulfate in peptides and small molecules. However, these conditions were found to be strongly denaturing to proteins (Figure S15) and highly toxic to live cells (Figure S16 and S17). In contrast, our reagents caused no protein denaturation and has low toxicity to cells at various concentrations. To mimic the cell membrane-bound sulfoproteins,51 we examined in situ fluorosulfate decaging on the surface of live Staphylococus aureus (S. aureus) cells (Figure 4a–d). S. aureus cells were chosen because there are no known endogenous sulfopeptides expressed on their surface, and the endogenous sortase A on their surface can be used to ligate peptides.52–53 A fluorescently labeled peptide 22 consisting of a Tobacco Etch Virus (TEV) protease cleavage sequence54 and a LPETG sortase A-recognition motif was ligated to the cell surface of S. aureus (Figure 4a, S18 and S19). The cell surface-ligated peptide was then decaged by reagent 7. Compared to the phosphate-buffered saline (PBS) buffer control, neither the cell surface ligation nor the fsY decaging experiments caused significant reduction of cell viability (Figure 4b). LC-MS analysis of the peptide residues cleaved after the decaging reaction (24) confirmed that the fsY were successfully converted into sY on live cell surface (Figure 4c). Finally, reagents 5-7 also exhibited low toxicity to mammalian cells even at millimolar concentrations based on the MTT assay (Figure 4d).55
Figure 4. Cytocompatibility of the reagents.
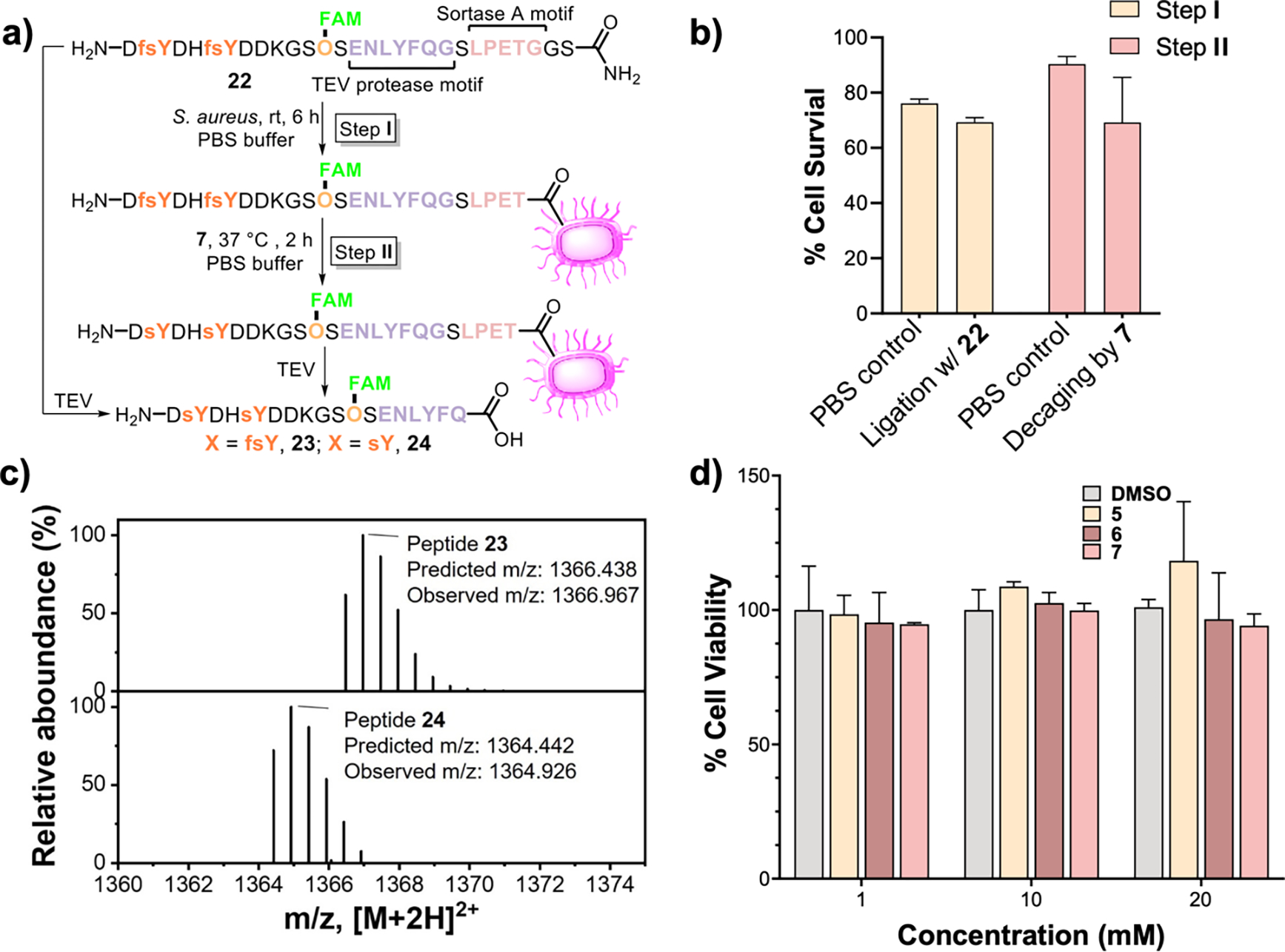
a) Sortase A-mediated ligation of peptide 22 onto the S. aureus cell surface and its decaging followed by the TEV protease cleavage. b) Percent of S. aureus cell survived after sortase A-mediated ligation of 22 (Step I) and after fluorosulfate decaging by 7 (Step II) compared to the cells treated with PBS. The average data of two trials were plotted. c) LC-MS analysis of samples after the TEV cleavage identified the decaged peptide (24, bottom) compared to the cleaved peptide before decaging (23, top). d) MTT assay of the mammalian HEK-293T cells after incubation with various concentrations of reagents 5, 6, and 7. The average data of three trials were plotted.
In conclusion, we demonstrated that fluorosulfate is a physiologically compatible latent sulfate in peptides and proteins. Fluorosulfate is stable in neutral aqueous buffers, cell lysates, and serum, and can be efficiently converted into sulfate by easily modified and readily accessible HA reagents under physiologically relevant conditions via Lossen rearrangement. Leveraging the facile incorporation of fluorosulfate-containing amino acid fsY via solid-phase peptide synthesis and ncAA mutagenesis, our reported approach can be applied to studying a wide range of sulfopeptides and sulfoproteins in their physiological states. The excellent compatibility of our reagents with both bacterial and mammalian cells suggest that they are promising candidates for decaging fluorosulfate-containing peptides and proteins in experiments involving live systems.
Supplementary Material
ACKNOWLEDGMENT
The research is supported by a startup fund from Boston College to J.N., the NIH Director’s New Innovator Award (1DP2HG011027-01) to J.N., and NIH R35GM136437 to A.C. The NSF MRI award CHE-2117246, and the NIH HEI-S10 award 1S10OD026910-01A1 are also acknowledged.
ABBREVIATIONS
- CCR5
C-C chemokine receptor type 5
- PGSL-1
P-selectin glycoprotein ligand-1
- HCII
heparin cofactor II
- C5aR1
complement component 5a receptor 1
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Mehta AY; Heimburg-Molinaro J; Cummings RD; Goth CK Emerging patterns of tyrosine sulfation and O-glycosylation cross-talk and co-localization. Curr. Opin. Struct. Biol. 2020, 62, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Huttner WB Sulphation of tyrosine residues—a widespread modification of proteins. Nature 1982, 299, 273–276. [DOI] [PubMed] [Google Scholar]
- (3).Moore KL The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 2003, 278, 24243–24246. [DOI] [PubMed] [Google Scholar]
- (4).Baeuerle PA; Huttner WB Tyrosine sulfation of yolk proteins 1, 2, and 3 in Drosophila melanogaster. J. Biol. Chem. 1985, 260, 6434–6439. [PubMed] [Google Scholar]
- (5).Ludeman JP; Stone MJ The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 2014, 171, 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Liu CC; Schultz PG Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat. Biotechnol. 2006, 24, 1436–1440. [DOI] [PubMed] [Google Scholar]
- (7).Li X; Liu CC Site-Specific Incorporation of Sulfotyrosine Using an Expanded Genetic Code. Methods Mol. Biol. 2018, 1728, 191–200 [DOI] [PubMed] [Google Scholar]
- (8).Italia JS; Peeler JC; Hillenbrand CM; Latour C; Weerapana E; Chatterjee A Genetically encoded protein sulfation in mammalian cells. Nat. Chem. Biol. 2020, 16, 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).He X; Chen Y; Beltran DG; Kelly M; Ma B; Lawrie J; Wang F; Dodds E; Zhang L; Guo J; Niu W Functional genetic encoding of sulfotyrosine in mammalian cells. Nat. Commun. 2020, 11, 4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen Y; Jin S; Zhang M; Hu Y; Wu K-L; Chung A; Wang S; Tian Z; Wang Y; Wolynes PG; Xiao H Unleashing the potential of noncanonical amino acid biosynthesis to create cells with precision tyrosine sulfation. Nat. Commun. 2022, 13, 5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kehoe JW; Bertozzi CR Tyrosine sulfation: a modulator of extracellular protein–protein interactions. Chem. Biol. 2000, 7, R57–R61. [DOI] [PubMed] [Google Scholar]
- (12).So WH; Wong CTT; Xia J Peptide photocaging: A brief account of the chemistry and biological applications. Chin. Chem. Lett. 2018, 29, 1058–1062. [Google Scholar]
- (13).Yang B; Wang N; Schnier PD; Zheng F; Zhu H; Polizzi NF; Ittuveetil A; Saikam V; DeGrado WF; Wang Q; Wang PG; Wang L Genetically introducing biochemically reactive amino acids dehydroalanine and dehydrobutyrine in proteins. J. Am. Chem. Soc. 2019, 141, 7698–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yang W; Eken Y; Zhang J; Cole LE; Ramadan S; Xu Y; Zhang Z; Liu J; Wilson AK; Huang X Chemical synthesis of human syndecan-4 glycopeptide bearing O-, N-sulfation and multiple aspartic acids for probing impacts of the glycan chain and the core peptide on biological functions. Chem. Sci. 2020, 11, 6393–6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tiruchinapally G; Yin Z; El-Dakdouki M; Wang Z; Huang X Divergent heparin oligosaccharide synthesis with preinstalled sulfate esters. Chem. Eur. J. 2011, 17, 10106–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Al-Horani RA; Desai UR Chemical sulfation of small molecules–advances and challenges. Tetrahedron 2010, 66, 2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Stone MJ; Payne RJ Homogeneous sulfopeptides and sulfoproteins: synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Acc. Chem. Res. 2015, 48, 2251–61. [DOI] [PubMed] [Google Scholar]
- (18).Ingram LJ; Taylor SD Introduction of 2, 2, 2-Trichloroethyl-Protected Sulfates into Monosaccharides with a Sulfuryl Imidazolium Salt and Application to the Synthesis of Sulfated Carbohydrates. Angew. Chem. Int. Ed. 2006, 45, 3503–3506. [DOI] [PubMed] [Google Scholar]
- (19).Bunschoten A; Kruijtzer JA; Ippel JH; de Haas CJ; van Strijp JA; Kemmink J; Liskamp RM A general sequence independent solid phase method for the site specific synthesis of multiple sulfated-tyrosine containing peptides. Chem. Commun. 2009, 2999–3001. [DOI] [PubMed] [Google Scholar]
- (20).Ali AM; Taylor SD Efficient solid-phase synthesis of sulfotyrosine peptides using a sulfate protecting-group strategy. Angew. Chem. Int. Ed. 2009, 48, 2024–2026. [DOI] [PubMed] [Google Scholar]
- (21).Ali AM; Taylor SD Synthesis of disulfated peptides corresponding to the N-terminus of chemokines receptors CXCR6 (CXCR6 1–20) and DARC (DARC 8–42) using a sulfate-protecting group strategy. J. Pept. Sci. 2010, 16, 190–9. [DOI] [PubMed] [Google Scholar]
- (22).Desoky AY; Hendel J; Ingram L; Taylor SD Preparation of trifluoroethyl-and phenyl-protected sulfates using sulfuryl imidazolium salts. Tetrahedron 2011, 67, 1281–1287. [Google Scholar]
- (23).Simpson LS; Widlanski TS A comprehensive approach to the synthesis of sulfate esters. J. Am. Chem. Soc. 2006, 128, 1605–1610. [DOI] [PubMed] [Google Scholar]
- (24).Simpson LS; Zhu JZ; Widlanski TS; Stone MJ Regulation of chemokine recognition by site-specific tyrosine sulfation of receptor peptides. Chem. Biol. 2009, 16, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liu C; Yang C; Hwang S; Ferraro SL; Flynn JP; Niu J A General Approach to O-Sulfation by a Sulfur (VI) Fluoride Exchange Reaction. Angew. Chem. Int. Ed. 2020, 132, 18593–18599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dong J; Krasnova L; Finn M; Sharpless KB Sulfur (VI) fluoride exchange (SuFEx): another good reaction for click chemistry. Angew. Chem. Int. Ed. 2014, 53, 9430–9448. [DOI] [PubMed] [Google Scholar]
- (27).Barrow AS; Smedley CJ; Zheng Q; Li S; Dong J; Moses JE The growing applications of SuFEx click chemistry. Chem. Soc. Rev. 2019, 48, 4731–4758. [DOI] [PubMed] [Google Scholar]
- (28).Li S; Li G; Gao B; Pujari SP; Chen X; Kim H; Zhou F; Klivansky LM; Liu Y; Driss H; Liang D-D; Lu J; Wu P; Zuilhof H; Moses J; Sharpless KB SuFExable polymers with helical structures derived from thionyl tetrafluoride. Nat. Chem. 2021, 13, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Miloserdov F; Zuilhof H Binding S (VI) to alkynes. Nat. Synth. 2022, 1, 415–416. [Google Scholar]
- (30).Lee C; Cook AJ; Elisabeth JE; Friede NC; Sammis GM; Ball ND The Emerging Applications of Sulfur(VI) Fluorides in Catalysis. ACS Catal. 2021, 6578–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Mukherjee H; Debreczeni J; Breed J; Tentarelli S; Aquila B; Dowling JE; Whitty A; Grimster NP A study of the reactivity of S (VI)–F containing warheads with nucleophilic amino-acid side chains under physiological conditions. Org. Biomol. Chem. 2017, 15, 9685–9695. [DOI] [PubMed] [Google Scholar]
- (32).a) While reactivities of fluorosulfate with cellular nucleophiles have been reported, these examples all require the close spatial proximity through ligand-receptor binding. For more detail, please see references: Wang N; Yang B; Fu C; Zhu H; Zheng F; Kobayashi T; Liu J; Li S; Ma C; Wang PG; Wang Q; Wang L Genetically Encoding Fluorosulfate-l-tyrosine To React with Lysine, Histidine, and Tyrosine via SuFEx in Proteins in Vivo. J. Am. Chem. Soc. 2018, 140, 4995–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen W; Dong J; Plate L; Mortenson DE; Brighty GJ; Li S; Liu Y; Galmozzi A; Lee PS; Hulce JJ; Cravatt BF; Saez E; Powers ET; Wilson IA; Sharpless KB; Kelly JW Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J. Am. Chem. Soc. 2016, 138, 7353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Han Y; Yang Z; Hu H; Zhang H; Chen L; Li K; Kong L; Wang Q; Liu B; Wang M; Lin J; Chen PR Covalently engineered protein minibinders with enhanced neutralization efficacy against escaping SARS-CoV-2 variants. J. Am. Chem. Soc. 2022, 144, 5702–5707. [DOI] [PubMed] [Google Scholar]
- (33).Chen W; Dong J; Li S; Liu Y; Wang Y; Yoon L; Wu P; Sharpless KB; Kelly JW Synthesis of Sulfotyrosine-Containing Peptides by Incorporating Fluorosulfated Tyrosine Using an Fmoc-Based Solid-Phase Strategy. Angew. Chem. Int. Ed. 2016, 55, 1835–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bones AM; Rossiter JT The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol. Plant. 1996, 97, 194–208. [Google Scholar]
- (35).Choi EJ; Jung D; Kim JS; Lee Y; Kim BM Chemoselective Tyrosine Bioconjugation through Sulfate Click Reaction. Chem. Eur. J. 2018, 24, 10948–10952. [DOI] [PubMed] [Google Scholar]
- (36).Eddleston M; Buckley NA; Eyer P; Dawson AH Management of acute organophosphorus pesticide poisoning. Lancet 2008, 371, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wei M; Liang D; Cao X; Luo W; Ma G; Liu Z; Li L A Broad-Spectrum Catalytic Amidation of Sulfonyl Fluorides and Fluorosulfates. Angew. Chem. Int. Ed. 2021, 60, 7397–7404. [DOI] [PubMed] [Google Scholar]
- (38).Hurd CD; Bauer L A novel rearrangement of hydroxamic acids using sulfonyl chlorides. J. Am. Chem. Soc. 1954, 76, 2791–2792. [Google Scholar]
- (39).Hackley BE Jr.; Plapinger R; Stolberg M; Wagner-Jauregg T Acceleration of the hydrolysis of organic fluorophosphates and fluorophosphonates with hydroxamic acids. J. Am. Chem. Soc. 1955, 77, 3651–3653. [Google Scholar]
- (40).Zhang G; Cui Y; Zhao Y; Cui Y; Bao S; Ding C A Practical Approach to Ureas and Thiocarbamates: SO2F2-Promoted Lossen Rearrangement of Hydroxamic Acid. ChemistrySelect 2020, 5, 7817–7821. [Google Scholar]
- (41).Semicarbazide was added to scavenge the nitrosobenzaldehyde produced in the light-mediated reaction (Figure S8). For more detailed information, please see reference: Hansen MJ; Velema WA; Lerch MM; Szymanski W; Feringa BL Wavelength-selective cleavage of photoprotecting groups: strategies and applications in dynamic systems. Chem. Soc. Rev. 2015, 44, 3358–3377. [DOI] [PubMed] [Google Scholar]
- (42).Cappello M; Bergum PW; Vlasuk GP; Furmidge BA; Pritchard DI; Aksoy S Isolation and characterization of the tsetse thrombin inhibitor: a potent antithrombotic peptide from the saliva of Glossina morsitans morsitans. Am. J. Trop. Med. Hyg. 1996, 54, 475–480. [DOI] [PubMed] [Google Scholar]
- (43).Calisto BM; Ripoll-Rozada J; Dowman LJ; Franck C; Agten SM; Parker BL; Veloso RC; Vale N; Gomes P; de Sanctis D; Payne RJ; Pereira PJB Sulfotyrosine-mediated recognition of human thrombin by a tsetse fly anticoagulant mimics physiological substrates. Cell Chem. Biol. 2021, 28, 26–33. [DOI] [PubMed] [Google Scholar]
- (44).Spannagl M; Bichler J; Birg A; Lill H; Schramm W Development of a chromogenic substrate assay for the determination of hirudin in plasma. Blood Coagul. Fibrinolysis 1991, 2, 121–127. [DOI] [PubMed] [Google Scholar]
- (45).Williams JW; Morrison JF The kinetics of reversible tight-binding inhibition. Methods Enzymol. 1979, 63, 437–467. [DOI] [PubMed] [Google Scholar]
- (46).Chatterjee A; Sun SB; Furman JL; Xiao H; Schultz PG A versatile platform for single-and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 2013, 52, 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Xiao H; Nasertorabi F; Choi S.-h.; Han GW; Reed SA; Stevens RC; Schultz PG Exploring the potential impact of an expanded genetic code on protein function. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 6961–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Weerapana E; Wang C; Simon GM; Richter F; Khare S; Dillon MB; Bachovchin DA; Mowen K; Baker D; Cravatt BF Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Scheffler K; Viner R; Damoc E High resolution top-down experimental strategies on the Orbitrap platform. J. Proteomics 2018, 175, 42–55. [DOI] [PubMed] [Google Scholar]
- (50).a) The 27544.758 Da and 27544.725 Da mass peaks correspond to the misincorporation of glutamine into sfGFP at position of 151 (expected exact mass: 27544.804 Da), a known byproduct of the ncAA mutagenesis method.37–38 The observed 27641.698 Da and 27641.876 Da mass peak corresponds to an intramolecular reaction of fluorosulfotyrosine with nucleophilic residue (expected exact mass: 27641.732 Da). For more information, please see references: Italia JS; Addy PS; Wrobel CJ; Crawford LA; Lajoie MJ; Zheng Y; Chatterjee A An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat. Chem. Biol. 2017, 13, 446–450. [DOI] [PubMed] [Google Scholar]; b) Italia JS; Latour C; Wrobel CJJ; Chatterjee A Resurrecting the bacterial tyrosyl-tRNA Synthetase/tRNA pair for expanding the genetic code of both E. coli and eukaryotes. Cell Chem. Biol. 2018, 25, 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Hille A; Huttner WB Occurrence of tyrosine sulfate in proteins–a balance sheet: 2. Membrane proteins. Eur. J. Biochem. 1990, 188, 587–596. [DOI] [PubMed] [Google Scholar]
- (52).Reja RM; Wang W; Lyu Y; Haeffner F; Gao J Lysine-targeting reversible covalent inhibitors with long residence time. J. Am. Chem. Soc. 2022, 144, 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Rentero Rebollo I; McCallin S; Bertoldo D; Entenza JM; Moreillon P; Heinis C Development of potent and selective S. aureus sortase A inhibitors based on peptide macrocycles. ACS Med. Chem. Lett. 2016, 7, 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kapust RB; Tözsér J; Copeland TD; Waugh DS The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 2002, 294, 949–955. [DOI] [PubMed] [Google Scholar]
- (55).Mosmann T Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


