Abstract
The food chain, especially raw minced meat, is thought to be responsible for an increase in the incidence of vancomycin-resistant enterococci (VRE) in human nosocomial infections. Therefore, 555 samples from 115 batches of minced beef and pork from a European Union-licensed meat-processing plant were screened for the occurrence of VRE. The processed meat came from 45 different slaughterhouses in Germany. Enterococci were isolated directly from Enterococcosel selective agar plates and also from Enterococcosel selective agar plates supplemented with 32 mg of vancomycin per liter. In addition, peptone broth was used in a preenrichment procedure, and samples were subsequently plated onto Enterococcosel agar containing vancomycin. To determine resistance, 209 isolates from 275 samples were tested with the glycopeptides vancomycin, teicoplanin, and avoparcin and 19 other antimicrobial substances by using a broth microdilution test. When the direct method was used, VRE were found in 3 of 555 samples (0.5%) at a concentration of 1.0 log CFU/g of minced meat. When the preenrichment procedure was used, 8% of the samples were VRE positive. Our findings indicate that there is a low incidence of VRE in minced meat in Germany. In addition, the resistance patterns of the VRE isolates obtained were different from the resistance patterns of clinical isolates. A connection between the occurrence of VRE in minced meat and nosocomial infections could not be demonstrated on the basis of our findings.
In the last decade, enterococci became the second most frequently reported cause of surgical wound infections and nosocomial urinary tract infections and the third most frequently reported cause of bacteremia (23, 33). Ampicillin and aminoglycosides have been considered the drugs of choice for treatment of serious enterococcal infections (5). The number of enterococci that are resistant to ampicillin and aminoglycosides has increased (4, 14, 15, 31, 32). The glycopeptide antibiotics vancomycin and teicoplanin are important substances for treatment of severe hospital infections. Diseases caused by enterococci which are resistant to the β-lactam antimicrobial agent ampicillin and aminoglycoside antibiotics can be treated only with glycopeptides (5, 24). Unfortunately, resistance to vancomycin and teicoplanin (both of which are glycopeptides) has also been reported. In the United States, the Centers for Disease Control and Prevention reported that there was a 20-fold increase in the occurrence of vancomycin-resistant enterococci (VRE) associated with nosocomial infections from 1989 to 1990 (6). Therefore, the therapeutic use of glycopeptides should be restricted; otherwise, infections caused by such multiresistant enterococci will become untreatable (3, 7, 17, 35). The high level of resistance to glycopeptides (MIC, >512 mg/liter) is inducible and is encoded by the vanA gene cluster, which is carried on transposons similar to or related to Tn1546 (34). Transfer of resistance can occur via conjugative plasmids. Conjugation experiments have shown that the rates of transfer between vanA donors and vanA-negative recipient strains of enterococci range from 2 × 10−7 to >2 × 10−4 (20).
The source of glycopeptide-resistant enterococci is not known. One possibility is that these organisms are spread via the food chain. Some data have indicated that raw poultry and raw minced meat may harbor VRE (2, 18, 37). In this context, adding the glycopeptide avoparcin, a mycelial product of Streptomyces candidus, to animal feed was thought to be responsible for the development of glycopeptide resistance in enterococci in animals (2, 18). Because this possibility could not be ruled out, the use of avoparcin as a feed additive was banned in Germany in January 1996 and in the whole European Union (EU) later. Furthermore, the possible use of resistant Enterococcus faecium strains as human probiotic strains and as starter cultures for a number of cheese products has been discussed as a possible source of VRE (37). The increased use of glycopeptide antibiotics in hospitals and the widespread use of glycopeptide antibiotics for treatment of patients may be another source of VRE (35).
In this study the incidence of VRE in fresh minced beef and pork was investigated to determine the importance of raw minced meat as a possible vector for the transfer of VRE from animals to humans. Fresh minced pork and beef samples from an EU-admitted meat-processing plant with a broad catchment area in Germany were tested to obtain representative data. In order to assess the potential human risk through the food chain, we also investigated the occurrence and resistance of enterococci to a broad range of antimicrobial substances that are used either as therapeutic agents or as feed additives.
MATERIALS AND METHODS
Pork and beef skeletal muscles (but no cheek meat or sticking or diaphragm muscles) were comminuted with a mincer and packaged for retail markets. The minced meat investigated was produced in an EU-admitted meat-processing plant in Berlin, Germany. The meat originated from different abattoirs throughout Germany. Enterococci were isolated from 275 meat samples collected on 55 days (5 days per week) between May and November 1996; all of the enterococci isolated, including VRE, were identified to the species level. In the period between November 1996 and April 1997 the occurrence of enterococci and VRE was determined, but the organisms were not identified to the species level. The antibiotic susceptibility patterns of all isolates were determined.
Hygiene control.
To minimize the risk of contaminating the meat with enterococci from the environment, processing hygiene and disinfection were performed strictly according to the EU guidelines (12). The results of these measures were checked with a wet swab-dry swab technique (9, 25). Briefly, a wet swab and a dry swab were moved over a defined area of each surface in the meat-processing plant. Subsequently, the swabs were spread onto Enterococcosel agar (ECSA) (Becton Dickinson) and ECSA containing 32 mg of vancomycin (Sigma Chemical Co., St. Louis, Mo.) per liter (ECSA-VA) for quantitative detection. After enrichment by incubation in dilution broth for 24 h at 37 ± 0.5°C, a qualitative procedure to detect enterococci on ECSA and ECSA-VA was performed.
Total aerobic mesophilic colony counts for the minced meat samples.
The minced meat samples were stored at 4 ± 0.5°C immediately after they were produced, and they were analyzed 3 to 6 h after processing. The number of aerobic mesophilic CFU per gram in each meat sample was determined by the surface plating method (drop plating) as described in the German national regulations (8). The resulting values were transformed into log values, and statistical parameters were calculated with a computer program (BIAS 5.0; Ackermann, Frankfurt am Main, Germany).
Isolation of enterococci.
A 25-g portion of minced meat from each sample (the total weight of each sample was 125 g) was placed in 225 ml of buffered peptone water (BPW) and homogenized with a stomacher for 1 min. Then 1 ml of the diluted sample was plated onto ECSA. The agar plates were incubated for 24 h at 37 ± 0.5°C aerobically. From each ECSA plate, which represented one meat sample, two typical black colonies on the underlying black agar with colony diameters of ca. 1 mm were isolated randomly and used for further investigation. Up to 10 colonies per production day (batch) were isolated (five samples per day). The total enterococcal colony count was also determined.
Isolation of VRE.
A direct culturing method was used for quantitative detection of VRE in the meat samples. The procedure used was the procedure used to determine the total enterococcal counts with ECSA-VA instead of ECSA (see above).
In addition, an enrichment procedure for qualitative isolation of VRE was performed. After overnight incubation at 37 ± 0.5°C, 0.1 ml of each BPW enrichment broth was plated onto ECSA-VA. The agar plates were incubated at 37 ± 0.5°C aerobically overnight. If growth was observed, one representative colony was isolated from each plate and investigated further.
Species identification.
The Enterococcus species were identified with a Rapid ID 32 Strep identification kit (bioMérieux, Marcy l’Etoile, France). As recommended by Nusser (28) and Reuter (30), growth at 10 ± 0.5, 45 ± 0.5, and 50 ± 0.5°C and growth in the presence of 6.5% NaCl were examined, and the potassium tellurite reaction was performed to confirm the results. The catalase reaction test and an experiment to determine the Lancefield serotype (Slidex StreptoD; bioMérieux) were also performed (26). To distinguish Enterococcus gallinarum and Enterococcus casseliflavus from the E. faecium group and from Enterococcus faecalis, acidification of methyl-α-d-glucopyranoside (Sigma) was examined for all isolates (10).
Determination of the MICs.
The MICs of 19 antibiotics and three feed additives were determined by using the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS) (27) and the broth microdilution method. Microtiter plates containing the test substances in cation-adjusted Mueller-Hinton broth supplemented with 3% lysed horse blood (PML Microbiologicals, Portland, Oreg.) were used. The microtiter plates were allowed to reach room temperature before inoculation. A 0.5-McFarland unit suspension, prepared as recommended by the NCCLS (27), was diluted 1:30 and homogenized, and 10 μl was inoculated into each well, which resulted in a final inoculum of 5 × 105 CFU/ml. The test results were determined visually after incubation for 16 to 20 h at 37 ± 0.5°C by using the instructions given by the NCCLS (27). The MIC ranges described by the NCCLS (27) were suitable for enterococci in most cases. When no specific ranges for enterococci were mentioned, the ranges for gram-positive bacteria were used. To interpret the results obtained with the three feed additives tested and the new antibiotic substance LY 333328, the interpretive guidelines for related substances were used (i.e., the vancomycin guidelines were used for avoparcin and LY 333328, and the erythromycin guidelines were used for the macrolides tylosin and virginiamycin).
Each Enterococcus isolate was tested once. The enterococci were tested with the following 22 substances: amoxicillin-clavulanic acid (2:1), ampicillin, ceftriaxone, erythromycin, penicillin G, ciprofloxacin, clindamycin, imipenem, teicoplanin, rifampin, gentamicin, vancomycin, chloramphenicol, avoparcin, tylosin, trimethoprim-sulfamethoxazole (1:19), methicillin, LY 333328, virginiamycin, cephalothin, streptomycin, and tetracyline. LY 333328 is a newly developed glycopeptide which is currently available only for research purposes. Avoparcin, virginiamycin, and tylosin are used as feed additives. Pseudomonas aeruginosa ATCC 27853, E. faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 25922 were used as reference strains.
β-Lactamase test.
Production of β-lactamase by isolated enterococci was investigated with the BR66 identification stick test, which is a nitrocefin-based test (Oxoid). S. aureus ATCC 29213 was used as a positive control, and E. faecalis ATCC 29212 was used as a negative control.
vanA gene.
All VRE strains were grown for 24 h at 37 ± 0.5°C in BPW. Isolation of bacterial DNA, amplification of a vanA gene fragment by PCR, and agarose gel electrophoresis were performed as described by Ausubel et al. (1) and Klare et al. (18). After the cells were lysed with 0.5% sodium dodecyl sulfate, proteins were removed by digestion with proteinase K (Boehringer, Mannheim, Germany). A cetyltrimethylammonium bromide solution (Sigma) was used to remove cell wall debris, polysaccharides, and the remaining proteins. To extract the bacterial DNA for amplification of the vanA gene, the DNA was precipitated with isopropanol and transferred to a fresh tube containing 70% ethanol. The DNA concentration was determined with a model TKO 100 DNA fluorometer (Hoefer). The following oligonucleotides were used as primers for amplification of the 377-bp fragment of the vanA gene: vanA 1 (5′-TCT GCA ATA GAG ATA GCC GC-3′; vanA sequence positions 443 to 462 [11]) and vanA 2 (5′-GG AGT AGC TAT CCC AGC ATT-3′; vanA sequence positions 819 to 800) (primers were obtained from TIB MOL BIOL, Berlin, Germany). Each PCR mixture contained 10 μl of Mg2+-free buffer, 3 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, 0.4 μl of primer vanA 1 (50 pmol/100 μl), 0.44 μl of primer vanA 2 (50 pmol/100 μl), 1.25 μl of polymerase (2.5 U), 50 μl of mineral oil, 20 ng of template DNA, and enough water to bring the volume to 100 μl. The solutions were obtained from a PrimeZyme polymerase kit (catalog no. 100-652; Biometra, Göttingen, Germany). A thermal cycler (model TRIO Thermoblock; Biometra) was programmed for 30 cycles; cycle 1 consisted of 94°C for 3 min, 55°C for 1 min, and 72°C for 1 min, cycles 2 through 29 each consisted of 94°C for 1 min, 55°C for 30 s, and 72°C for 30 s, and cycle 30 consisted of 94°C for 1 min, 55°C for 1 min, and 72°C for 4 min. Gel electrophoresis was performed for 90 min in a 1.4% agarose gel (Low EEO; Appligene) at 100 V. E. faecium 64/3, a vanA-negative strain, and vanA-positive strain E. faecium 70/90 were used as reference strains.
RESULTS
Hygiene and disinfection in the meat-processing plant.
The surfaces in the meat-processing plant were examined to determine if they were contaminated with enterococci immediately before the minced meat was processed. Enterococci were detected in 7 of 48 surface samples only when the enrichment procedure was used. No VRE were detected on the premises after the disinfection procedure was performed. The disinfection measures were performed as recommended in the EU guidelines (25) (48 surfaces were tested).
Microecological results.
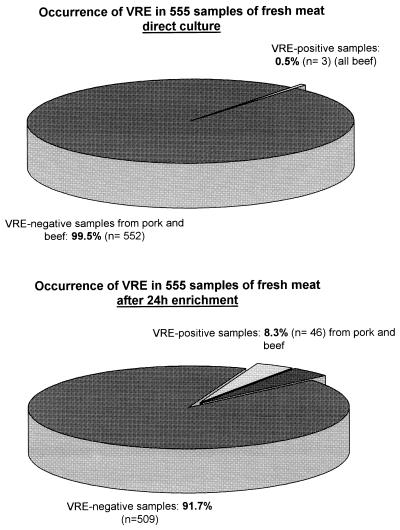
The total bacterial counts in the minced meat samples were between 2.7 × 103 and 9.3 × 106 CFU/g. Enterococci were present at concentrations between 0.5 × 101 and 7.1 × 102 CFU/g. VRE at a concentration of 10 CFU per g of minced meat were found in 3 of 555 samples (0.5%) when no enrichment procedure was performed. In 46 of 555 samples (8.3%) VRE at concentrations ranging from 1 to 9 CFU per g of minced meat were found after overnight enrichment in BPW. A total of 509 meat samples (91.7%) were VRE negative (Fig. 1). The occurrence of VRE was distributed nearly equally over the investigation period.
FIG. 1.
Occurrence and distribution of VRE in fresh minced meat samples after direct plating and after a 24-h enrichment procedure.
Most of the enterococci isolated from non-VRE-selective ECSA plates were identified as E. faecalis isolates (182 of 209 isolates [87%]), and 8 of the 209 enterococci (4%) were identified as E. faecium isolates. In addition, isolates were identified as E. casseliflavus (six isolates [3%]), E. gallinarum (five isolates [2%]), Enterococcus durans (four isolates [2%]), Enterococcus hirae (three isolates [1%]), and Enterococcus avium (one isolate [<1%]). There was no noticeable difference between the beef and pork samples with respect to the dominance of E. faecalis. (Table 1).
TABLE 1.
Identification of Enterococcus isolates (n = 209) to the species levela
| Species | No. of isolates obtained from:
|
Total no. of isolates | |
|---|---|---|---|
| Minced beef | Minced pork | ||
| E. faecalis | 97 | 85 | 182 |
| E. faecium | 2 | 6 | 8 |
| E. casseliflavus | 0 | 6 | 6 |
| E. gallinarum | 3 | 2 | 5 |
| E. durans | 3 | 1 | 4 |
| E. hirae | 3 | 0 | 3 |
| E. avium | 0 | 1 | 1 |
| Total | 108 | 101 | 209 |
There was no noticeable difference between minced pork and minced beef.
A total of 34 VRE strains isolated during the first investigation period were identified to the species level. Thirteen of these 34 VRE isolates were identified as E. faecium, 3 were identified as E. durans, and 1 was identified as E. hirae; thus, 17 of the 34 isolates (50%) were members of the E. faecium group, which includes E. faecium, E. durans, E. hirae, and Enterococcus mundtii, four species that are phylogenetically and phenotypically related. Twelve of the 34 VRE isolates (35%) were identified as E. faecalis, and 5 (15%) were identified as E. gallinarum. There was no noticeable difference between the beef and pork samples. The VRE strains were nearly evenly distributed between the beef and pork samples (data not shown).
MIC determination.
The MIC data obtained in vitro with 22 antibiotic substances and the enterococci isolated from non-VRE-selective ECSA plates (n = 209) revealed that all of the isolates were susceptible to ampicillin and only one Enterococcus strain was susceptible to methicillin. The combination of amoxicillin and clavulanic acid was very effective; no strain showed resistance. A total of 134 of the 209 isolates were resistant to or exhibited intermediate reactions with ceftriaxone, and 192 of the 209 isolates were resistant to or exhibited intermediate reactions with cephalothin. The carbapenem substance imipenem was effective against 172 enterococci; 9 strains were resistant to imipenem, and 28 exhibited intermediate reactions.
None of the enterococci isolated exhibited resistance to the glycopeptides vancomycin, teicoplanin, and LY 333328 and the aminoglycoside gentamicin. All of the isolates except two enterococci were susceptible to streptomycin. A total of 144 isolates exhibited intermediate resistance or were resistant to the macrolide erythromycin; 42, 75, 6, 197, and 51 strains exhibited intermediate resistance or were resistant to tetracycline, ciprofloxacin, chloramphenicol, clindamycin, and rifampin, respectively. The MIC test performed with the three feed additives showed that none of the 209 isolates exhibited resistance to avoparcin, 6 isolates were resistant to virginiamycin, and 15 isolates were resistant to tylosin (Table 2). The strains isolated from beef exhibited resistance patterns similar to the patterns exhibited by the strains isolated from pork (data not shown).
TABLE 2.
Antibiotic resistance patterns of enterococci (n = 209) isolated from minced beef and pork samplesa
| Group | Antibiotic(s) or chemothera- peutic agent | % of enteroccal strains
|
||
|---|---|---|---|---|
| Susceptible | Intermediate resistance | Resistant | ||
| β-Lactams | Ampicillin | 100 | 0 | 0 |
| Penicillin G | 99.5 | 0 | 0.5 | |
| Methicillin | 0.5 | 0 | 99.5 | |
| β-Lactams in combination with β-lactamase inhibitor | Amoxicillinclavulanic acid | 100 | 0 | 0 |
| Cephalosporins | Ceftriaxone | 35.9 | 28.7 | 35.4 |
| Cephalothin | 8.1 | 23 | 68.9 | |
| Carbapenems | Imipenem | 82.3 | 13.4 | 4.3 |
| Glycopeptides | Vancomycin | 100 | 0 | 0 |
| Teicoplanin | 100 | 0 | 0 | |
| LY 333328 | 100 | 0 | 0 | |
| Aminoglycosides | Gentamicin | 100 | 0 | 0 |
| Streptomycin | 99 | 0 | 1 | |
| Macrolides | Erythromycin | 31.1 | 51.9 | 17 |
| Tetracyclines | Tetracycline | 79.9 | 0 | 20.1 |
| Quinolones | Ciprofloxacin | 64.1 | 32.1 | 3.8 |
| Amphenicols | Chloramphenicol | 97.1 | 1.9 | 1 |
| Other antibiotics | Clindamycin | 5.7 | 4.8 | 89.5 |
| Rifampin | 75.6 | 15.3 | 9.1 | |
| Trimethoprim-sulfamethoxazole | 100 | 0 | 0 | |
| Feed additives | Avoparcin | 100 | 0 | 0 |
| Virginiamycin | 97.1 | 0 | 2.9 | |
| Tylosin | 93.3 | 2.9 | 3.8 | |
Results were interpreted by using the recommendations of the NCCLS (27).
All VRE strains (n = 38) exhibited intermediate resistance or were resistant to methicillin, cephalothin, vancomycin, erythromycin, clindamycin, avoparcin, virginiamycin, and tylosin. They were susceptible to ampicillin, amoxicillin-clavulanic acid, gentamicin, and the new glycopeptide LY 333328. A total of 10 of the 38 strains isolated were resistant or exhibited intermediate resistance to penicillin G; 23, 19, 30, 13, 37, 10, 11, 8, and 7 of the strains exhibited intermediate resistance or were resistant to ceftriaxone, imipenem, teicoplanin, streptomycin, tetracycline, ciprofloxacin, chloramphenicol, trimethoprim-sulfamethoxazole (1:19), and rifampin, respectively (Table 3). These organisms differed from the enterococci isolated from nonselective plates. None of the enterococci isolated in this study showed β-lactamase activity.
TABLE 3.
Antibiotic resistance patterns of VRE (n = 38) isolated from minced beef and pork samplesa
| Group | Antibiotic(s) or chemothera- eutic agent | % of VRE strains
|
||
|---|---|---|---|---|
| Susceptible | Intermediate resistance | Resistant | ||
| β-Lactams | Ampicillin | 100 | 0 | 0 |
| Penicillin G | 73.7 | 0 | 26.3 | |
| Methicillin | 0 | 0 | 100 | |
| β-Lactams in combination with β-lactamase inhibitor | Amoxicillinclavulanic acid | 100 | 0 | 0 |
| Cephalosporins | Ceftriaxone | 39.5 | 21 | 39.5 |
| Cephalothin | 0 | 10.5 | 89.5 | |
| Carbapenems | Imipenem | 50 | 15.8 | 34.2 |
| Glycopeptides | Vancomycin | 0 | 0 | 100 |
| Teicoplanin | 21 | 10.5 | 68.4 | |
| LY 333328 | 100 | 0 | 0 | |
| Aminoglycosides | Gentamicin | 100 | 0 | 0 |
| Streptomycin | 65.8 | 0 | 34.2 | |
| Macrolides | Erythromycin | 0 | 0 | 100 |
| Tetracyclines | Tetracycline | 2.6 | 0 | 97.4 |
| Quinolones | Ciprofloxacin | 73.7 | 26.3 | 0 |
| Amphenicols | Chloramphenicol | 71.1 | 0 | 28.9 |
| Other antibotics | Clindamycin | 0 | 0 | 100 |
| Rifampin | 81.6 | 2.6 | 15.8 | |
| Trimethoprim-sulfamethoxazole | 78.9 | 0 | 21.1 | |
| Feed additives | Avoparcin | 0 | 0 | 100 |
| Virginiamycin | 0 | 15.8 | 84.2 | |
| Tylosin | 0 | 0 | 100 | |
Results were interpreted by using the recommendations of the NCCLS (27).
vanA gene.
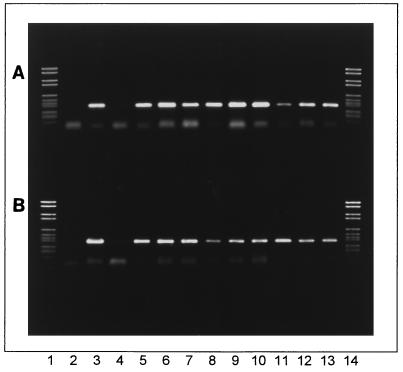
The vanA-specific 377-bp fragment was amplified from the genomic DNA of all VRE. The results obtained for some strains are shown in Fig. 2. The vanA gene fragments of E. faecium, E. faecalis, E. durans, E. gallinarum, and E. hirae were amplified from both beef and pork isolates. The positive control strain E. faecium 70/90 contained the typical 377-bp fragment, and the negative control strain E. faecium 64/3 did not.
FIG. 2.
Detection of the vanA gene in VRE from minced beef and pork. (A) Lanes 1 and 14, DNA molecular weight marker VI (catalog no. 1062590; Boehringer Mannheim) with bands at 2,176, 1,766, 1,230, 1,033, 653, 517, 453, 394, 298, 234-220, and 154 bp; lane 2, blank; lane 3, E. faecium 70/90 (positive control); lane 4, E. faecium 64/3 (negative control); lane 5, E. faecium VRE 3; lane 6, E. faecium VRE 4; lane 7, E. faecium VRE 5; lane 8, E. faecium VRE 6; lane 9, E. faecium VRE 7; lane 10, E. faecalis VRE 9; lane 11, E. durans VRE 10; lane 12, E. faecalis VRE 11; lane 13, E. faecium VRE 12. (B) Lanes 1 and 14, DNA molecular weight marker VI; lane 2, blank; lane 3, E. faecium 70/90 (positive control); lane 4, E. faecium 64/3 (negative control); lane 5, E. faecalis VRE 13; lane 6, E. gallinarum VRE 14; lane 7, E. gallinarum VRE 15; lane 8, E. faecium VRE 16; lane 9, E. gallinarum VRE 17; lane 10, E. faecium VRE 18; lane 11, E. hirae VRE 19; lane 12, E. faecalis VRE 20; lane 13, E. faecalis VRE 21. The main band of the positive control strain and the VRE strains represents the vanA-specific 377-bp fragment.
DISCUSSION
Minced meat was used in this investigation because the production process creates a risk that microbiological contamination will occur. The incidence of VRE in minced meat should therefore be greater than the incidence of VRE in other meat products. As the meat investigated originated from different counties in Germany, we assumed that our conclusions were representative. The disinfection of the meat plant was assessed strictly according to the EU guidelines. The possibility that the meat was contaminated with enterococci, especially VRE, due to insufficient hygiene practices could be excluded. The use of plastic gloves by workers avoided contamination of the meat with enterococci and VRE from humans. Other investigations of meat obtained from retail outlets (38) did not differentiate between human and environmental origins of the VRE detected.
The use of ECSA and BPW resulted in greater recovery of enterococci than the use of other media (29); therefore, these media were considered suitable for isolation and enrichment of enterococci and VRE.
Enterococci were detected in nearly all samples of minced meat at levels ranging from 0.5 × 101 to 7.1 × 102 CFU/g. VRE were detected directly in only 3 of the 555 samples investigated (0.5%). VRE were found after overnight enrichment in 48 of the 555 samples (8.3%). These findings suggest that VRE occurred in very low numbers and only sporadically in minced meat. The use of an enrichment procedure is essential for detection of VRE. Using pork samples obtained from retail outlets, Wegener et al. (38) found that up to 27% of the samples were VRE positive after enrichment. The considerable difference between these results and our results may be explained by possible human contamination of the meat in the retail outlets. However, the difference may be due to less effective disinfection in a meat-processing plant. The enterococci may have persisted in the factory, and they could be of environmental or animal origin. Therefore, the difference between the results of Wegener and our results cannot be satisfactorily explained.
Enterococci isolated from non-VRE-selective ECSA plates were identified to the species level. The results showed that E. faecalis was the dominant species in minced meat (87% of the isolates). The percentage of E. faecium was about 4%, and the percentages of the other species identified were less than 4%. The VRE isolates identified to the species level belonged mainly to E. faecium or the E. faecium species group. Wegener et al. (38) found that food contaminated with vancomycin-resistant E. faecium strains was an important possible source of infections in the community, based on their study in Denmark and a study performed in Belgium (13). Thus, the low incidence of E. faecium in minced meat in our study may indicate that food contamination is not an important source of VRE in Germany.
The susceptibility of enterococci isolated from non-VRE-selective ECSA to the glycopeptides vancomycin, teicoplanin, and avoparcin is consistent with the findings of Knudtson and Hartmann (21). In 50 pork samples Knudtson and Hartmann did not find any enterococcal isolates which exhibited multiple resistance to these antibiotics. However, Klare et al. (18) isolated five vancomycin-resistant E. faecium strains with vanA resistance from meat samples obtained from 13 different butcher shops. In view to our findings, the possibility that microbiological contamination from the environment occurred could not be ruled out. These findings (18) cannot represent the microbiological situation in fresh meat in general. In an earlier study Klare et al. (19) isolated E. faecium strains from stools of patients in intensive care units in a hospital in Berlin, Germany. They found that 49 of 52 VRE strains exhibited intermediate resistance or were resistant to ampicillin. This indicates that VRE from meat have different antibiotic resistance patterns than VRE responsible for untreatable infections in hospitals. These findings are supported by the results of other authors who detected 11% vancomycin-resistant E. faecium in stools of healthy volunteers (37) and found about 3.5% VRE-positive patients in a hospital in which no VRE had been isolated previously (13). This indicates a natural reservoir for VRE.
All Enterococcus strains, including VRE isolated from raw minced meat in this study, exhibited susceptibility to the antibiotic ampicillin, which is currently used to treat human enterococcal infections. In the case of infections caused by enterococci from meat, patients can be treated with ampicillin as the antibiotic of choice. The use of glycopeptides is not necessary. For the future, a new antibiotic, LY 333328, seems to be a substance that could be used to treat infections caused by enterococci resistant to ampicillin, aminoglycosides, and the “old” glycopeptides vancomycin and teicoplanin. The results of other investigations support our finding that enterococci isolated from clinical samples exhibit different resistance patterns than enterococci isolated from meat (16). Furthermore, a recent study of the genetic similarity of clinical and food enterococci isolated in Germany demonstrated with pulsed-field gel electrophoresis that these strains are not genetically related (22). van den Braak et al. (36) found VRE in fecal samples from vegetarians more often than they found VRE in fecal samples from nonvegetarians (9.7 versus 4.7%). These authors indicated that there may be no association between meat consumption and intestinal colonization with VRE. We concluded, therefore, that enterococci isolated from raw minced meat cannot be considered the main source of untreatable nosocomial infections of humans. However, these strains can acquire additional resistance in hospitals and can have the same antibiotic resistance patterns as the clinical strains, even when they are not genetically related. In addition, there are a lot of other virulence factors sensu stricto, including hemolysin, adhesin, aggregation substances, and proteases, that should be considered. In light of the observations mentioned above, we concluded that it is very likely that clinical strains in Germany originate mainly from sources other than meat.
The results of the direct culturing method and the enrichment procedure indicate that low levels of contamination of minced meat with enterococci and with VRE occur when minced meat is processed under prescribed hygienic rules according to EU regulations. The enterococcal flora of raw minced meat is dominated by E. faecalis. E. faecium, which harbors the vanA resistance gene in most cases, was detected only in a small percentage of samples. The level of VRE detected by direct culturing (0.5%) is very low compared with the levels of other food-borne pathogens, such as S. aureus. The level of VRE isolates after enrichment (8.3%) was the worst case observed in this study. In the enrichment procedure even one colony in 25 g of minced meat would have resulted in a positive test.
This study started a few months after avoparcin was banned in Germany. Our data could represent the baseline incidence of VRE occurring in the absence of avoparcin. However, in a previous study performed with a smaller number of samples we found VRE in 1 of 45 samples from nine batches and no VRE in 180 samples from 36 batches (22). The study was performed in 1995 before avoparcin was banned in Germany (22). Therefore, the results of this study performed within 1 year after the German avoparcin ban suggest that the use of avoparcin may not have been responsible for the development of vancomycin resistance in animals. Hence, it seems that glycopeptide resistance is ubiquitous and may not be connected with the use of avoparcin.
ACKNOWLEDGMENTS
We thank A. Busch (Hoffmann-LaRoche, Grenzach-Wyhlen, Germany) for intense discussions and for technical support. We thank L. Bräutigam, D. Jaeger, L. Jäger, and R. Ludewig for excellent technical assistance. We also thank U. Köpke and J. Louwers for support and for commenting on the manuscript. We thank W. Witte and I. Klare (Robert Koch Institut, Wernigerode, Germany) for kindly providing strains.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 2.Bates J, Jordens J Z, Griffith D T. Farm animals as putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–517. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 3.Bodnar R U R, Noskin G A, Suriano T, Cooper I, Reisberg B, Peterson L R. Use of in-house studies of molecular epidemiology and full species identification for controlling spread of vancomycin-resistant Enterococcus faecalis isolates. J Clin Microbiol. 1996;34:2129–2132. doi: 10.1128/jcm.34.9.2129-2132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyce J M, Opal S M, Potter-Bynoe G, LaForge R G, Zervos M J, Furtado G, Victor G, Meideros A A. Emergence and nosocomial transmission of ampicillin-resistant enterococci. Antimicrob Agents Chemother. 1992;36:1032–1039. doi: 10.1128/aac.36.5.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calia F M. Glycopeptides, polypeptides, tertiary amines, furans, nitroimidazoles, and other organic compounds. In: Andriole V T, editor. Current infectious disease drugs. Philadelphia, Pa: Current Medicine; 1996. pp. 117–138. [Google Scholar]
- 6.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 7.Chow J W, Kuritza A, Shlaes D M, Green M, Sahm D F, Zervos M J. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–1611. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutsches Institut für Normung. DIN 10161-2. Mikrobiologische Untersuchung von Fleisch und Fleischerzeugnissen. Bestimmung der aeroben Keimzahl bei 30°C—Tropfplattenverfahren. Berlin, Germany: Beuth Verlag; 1984. [Google Scholar]
- 9.Deutsches Institut für Normung. DIN 10113-1. Bestimmung des Oberflächenkeimgehaltes auf Einrichtungs- und Bedarfsgegenständen im Lebensmittelbereich - Teil 1: Quantitatives Tupferverfahren. Berlin, Germany: Beuth Verlag; 1997. [Google Scholar]
- 10.Devriese L A, Pot B, Kersters K, Lauwers S, Haesebrouck F. Acidification of methyl-d-glycopyranoside: a useful test to differentiate Enterococcus casseliflavus and Enterococcus gallinarum from Enterococcus faecium species group and from Enterococcus faecalis. J Clin Microbiol. 1996;34:2607–2608. doi: 10.1128/jcm.34.10.2607-2608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutka-Malen S, Molinas C, Arthur C, Courvalin P. The VanA glycopeptide resistance protein is related to d-alanyl-d-alanine ligase cell wall biosynthesis enzymes. Mol Gen Genet. 1990;224:364–372. doi: 10.1007/BF00262430. [DOI] [PubMed] [Google Scholar]
- 12.European Community. Richtlinie 94/65/EG des Rates vom 14. Dezember 1994 zur Festlegung von Vorschriften für die Herstellung und das Inverkehrbringen von Hackfleisch/Faschiertem und Fleischzubereitungen. Amtsblatt der Europäischen Gemeinschaften, Nr. L 368. Luxembourg, Luxembourg: Amt für amtliche Veröffentlichungen; 1994. [Google Scholar]
- 13.Gordts B, van Landuyt H, Ieven M, Vandamme P, Goosens H. Vancomycin-resistant enterococci colonizing the intestinal tracts of hospitalized patients. J Clin Microbiol. 1995;33:2842–2846. doi: 10.1128/jcm.33.11.2842-2846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grayson M L, Elliopoulus G M, Wennersten C B, Ruoff K L, De Girolami P C, Ferrera M, Moellering R C. Increasing resistance to β-lactam antibiotics among clinical isolates of Enterococcus faecium: a 22-year review at one institution. Antimicrob Agents Chemother. 1991;35:2180–2184. doi: 10.1128/aac.35.11.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman D J, Gerding D N. Antimicrobial resistance among enterococci. Antimicrob Agents Chemother. 1991;35:1–4. doi: 10.1128/aac.35.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klare I, Witte W. Glykopeptidresistente Enterokokken: zur Situation in Deutschland. Hyg Mikrobiol. 1997;1:31–38. [Google Scholar]
- 17.Klare I, Heier H, Claus H, Witte W. Environmental strains of Enterococcus faecium strains with inducible high-level resistance to glycopeptides. FEMS Microbiol Lett. 1993;106:23–29. doi: 10.1111/j.1574-6968.1993.tb05930.x. [DOI] [PubMed] [Google Scholar]
- 18.Klare I, Heier H, Claus H, Reissbrodt R, Witte W. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol Lett. 1995;125:165–172. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 19.Klare I, Collatz E, Al-Obeid S, Wagner J, Rodloff A C, Witte W. Glykopeptidresistenz bei Enterococcus faecium aus Besiedlungen und Infektionen von Patienten aus Intensivstationen Berliner Kliniken und einem Transplantationszentrum. Z Antimikrob Antineopl Chemother. 1992;10:45–53. [Google Scholar]
- 20.Klein, G., A. Pack, and G. Reuter. 1995. Glykopeptidresistenz, Resistenzübertragung und -muster bei Probiotikastämmen aus dem Genus Enterococcus. Immun. Infekt. 23(Suppl. 1):52.
- 21.Knudtson L M, Hartmann P A. Enterococci in pork processing. J Food Prot. 1993;56:6–9. doi: 10.4315/0362-028X-56.1.6. [DOI] [PubMed] [Google Scholar]
- 22.Lemcke R, Bülte M. 38. Arbeitstagung des Arbeitsgebietes Lebensmittelhygiene der Deutschen Veterinärmedzinischen Gesellschaft in Garmisch-Partenkirchen. Gießen, Germany: DVG; 1997. Feintypisierung Vancomycin-resistenter Enterokokken (VRE) aus Geflügel- und Schweinefleisch; pp. 162–169. [Google Scholar]
- 23.Lemmen R, Daschner F. Veränderung des Erregerspektrums bei schweren nosokomialen Infektionen. Chemother J. 1996;5:2–4. [Google Scholar]
- 24.Lerner S A. Aminoglycosides. In: Andriole V T, editor. Current infectious disease drugs. Philadelphia, Pa: Current Medicine; 1996. pp. 1–12. [Google Scholar]
- 25.Louwers J, Klein G. Eignung von Probenahmemethoden zur Umgebungsuntersuchung in fleischgewinnenden und -verarbeitenden Betrieben mit EU-Zulassung. Berl Muench Tieraerztl Wochenschr. 1994;107:367–373. [PubMed] [Google Scholar]
- 26.Murray B E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 3rd ed. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 28.Nusser E. Charakterisierung klinischer Isolate und einiger Kultursammlungs- und Probiotika-Produktionsstämme der Spezies Enterococcus faecium und Enterococcus faecalis durch extrazelluläre Enzyme, Chemotherapeutika-Resistenz und Plasmid-Darstellung. Vet. Med. dissertation. Berlin, Germany: Freie Universität Berlin; 1991. [Google Scholar]
- 29.Pack, A., and G. Klein. 1997. Unpublished data.
- 30.Reuter G. Culture media for enterococci and group D streptococci. In: Corry J E L, Curtis G D W, Baird R M, editors. Culture media for food microbiology. Amsterdam, The Netherlands: Elsevier Science B. V.; 1995. pp. 51–61. [Google Scholar]
- 31.Rhinehart E, Smith N E, Wennersten C, Gorss E, Freeman J, Eliopoulos G M, Moellering R C, Jr, Goldmann D A. Rapid dissemination of beta-lactamase-producing, aminoglycoside-resistant Enterococcus faecalis among patients and staff on an infant-toddler surgical ward. N Engl J Med. 1990;323:1814–1818. doi: 10.1056/NEJM199012273232606. [DOI] [PubMed] [Google Scholar]
- 32.Sapico F L, Canawati H N, Ginunas V J, Gilmore D S, Montgomerie J Z, Tuddenham W J, Facklam R R. Enterococci highly resistant to penicillin and ampicillin: an emerging clinical problem? J Clin Microbiol. 1989;27:2091–2095. doi: 10.1128/jcm.27.9.2091-2095.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaberg D R, Culver D H, Gaynes R P. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91:72–75. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 34.Shlaes D M, Binczewski B. Enterococcal resistance to vancomycin and related cyclic glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1990;9:106–110. doi: 10.1007/BF01963634. [DOI] [PubMed] [Google Scholar]
- 35.Uttley A H C, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;335:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 36.van den Braak N, Kreft D, van Belkum A, Verbrugh H, Endtz H. Vancomycin-resistant enterococci in vegetarians. Lancet. 1997;350:146–147. doi: 10.1016/S0140-6736(05)61856-9. [DOI] [PubMed] [Google Scholar]
- 37.van der Auwera P, Pensart N, Korten V, Murray B E, Leclerq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–1136. doi: 10.1093/infdis/173.5.1129. [DOI] [PubMed] [Google Scholar]
- 38.Wegener H C, Madsen M, Nielsen N, Aarestrup F M. Isolation of vancomycin resistant Enterococcus faecium from food. Int J Food Microbiol. 1997;35:57–66. doi: 10.1016/s0168-1605(96)01221-4. [DOI] [PubMed] [Google Scholar]