Abstract
Background
Powdery mildew is a major disease that causes great losses in soybean yield and seed quality. Disease-resistant varieties, which are generated by reducing the impact of susceptibility genes through mutation in host plants, would be an effective approach to protect crops from this disease. The Mildew Locus O (MLO) genes are well-known susceptibility genes for powdery mildew in plant. In this study, we utilized the CRISPR/Cas9 system to induce targeted mutations in the soybean GmMLO genes to improve powdery mildew resistance.
Results
A dual-sgRNA CRISPR/Cas9 construct was designed and successfully transferred into the Vietnamese soybean cultivar DT26 through Agrobacterium tumefaciens-mediated transformation. Various mutant forms of the GmMLO genes including biallelic, chimeric and homozygous were found at the T0 generation. The inheritance and segregation of CRISPR/Cas9-induced mutations were confirmed and validated at the T1 and T2 generations. Out of six GmMLO genes in the soybean genome, we obtained the Gmmlo02/Gmmlo19/Gmmlo23 triple and Gmmlo02/Gmmlo19/Gmmlo20/Gmmlo23 quadruple knockout mutants at the T2 generation. When challenged with Erysiphe diffusa, a fungus that causes soybean powdery mildew, all mutant plants showed enhanced resistance to the pathogen, especially the quadruple mutant. The powdery mildew severity in the mutant soybeans was reduced by up to 36.4% compared to wild-type plants. In addition, no pleiotropic effect on soybean growth and development under net-house conditions was observed in the CRISPR/Cas9 mutants.
Conclusions
Our results indicate the involvement of GmMLO02, GmMLO19, GmMLO20 and GmMLO23 genes in powdery mildew susceptibility in soybean. Further research should be conducted to investigate the roles of individual tested genes and the involvement of other GmMLO genes in this disease infection mechanism. Importantly, utilizing the CRISPR/Cas9 system successfully created the Gmmlo transgene-free homozygous mutant lines with enhanced resistance to powdery mildew, which could be potential materials for soybean breeding programs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-023-04549-5.
Keywords: CRISPR/Cas9, GmMLO genes, Powdery mildew resistance, Soybean, Targeted mutation
Background
Soybean [Glycine max (L.) Merrill] is one of the most important economic legume crops that is grown and consumed all over the world as a source of protein and oil for animal feed and human food [1]. However, global soybean production is seriously threatened by many diseases, one of which is powdery mildew caused by an obligate biotrophic fungus Erysiphe diffusa (Cooke & Peck) (syn. Microsphaera diffusa (Cooke & Peck) [2–4]. Powdery mildew is considered a major soybean disease [5] that negatively impacts yield in the largest soybean production countries such as the United States, Canada, Brazil, China and Germany. The average, annual yield loss was estimated at 13% because of this disease [6]. However, when environmental conditions are optimal for fungal growth, the yield loss of some susceptible varieties can approach 35%-60% [7, 8].
The use of resistant varieties would be an effective method to protect plant from powdery mildew. Although resistant genes (R-genes) against powdery mildew have been identified in some plant species. Up to now, only one R-gene named the resistance-to-M. diffusa 1 (Rmd1) was cloned and characterized in soybean [9], however, it is still a challenge to introduce the R-gene to the local cultivar. Reducing the impact of susceptibility genes (S-genes) through mutation in host plants is now considered as an effective alternative approach for disease resistant breeding. Recessive mutations of S-genes can limit pathogen infection of host plants and provide durable and broad-spectrum resistance [10]. The Mildew Locus O (MLO) genes are well-known S-genes for powdery mildew in different plant species. The MLO genes encode seven-transmembrane domain proteins [11], which are conserved throughout monocots and dicots [12]. These proteins are located in the plasma membrane and contain a 20 amino acid-long calmodulin (CaM)-binding domain, which is required for susceptibility to powdery mildew infection [13, 14]. Previous reports demonstrated the important role of MLO genes in the susceptibility to powdery mildew in barley [15, 16], Arabidopsis [17], tomato [18], pea [19, 20], pepper [21], bread wheat [22, 23], rose [24], apple [25], and grapevine [26, 27]. In addition, loss-of-function mutant alleles of MLO genes generated by different mutagenic approaches, such as chemical, RNAi, TALLEN and TILLING provided complete or enhanced resistance to powdery mildew in various plant species [17, 22, 28]. Therefore, inducing loss-of-function mutations in MLO is a potential strategy to improve powdery mildew resistance in important crops.
Recently, CRISPR/Cas9 was mentioned as the most effective and precise approach for trait improvement in crop plants [29]. This system has been successfully utilized to induce targeted mutations of MLO genes for enhancing powdery mildew resistance in wheat, grapevine, and tomato [27, 30–32]. In soybean, 39 GmMLO genes were previously predicted using comparative phylogenetic analysis from soybean and Arabidopsis genomes but their respective functions have not been defined [33, 34]. The aims of this study were to utilize the CRISPR/Cas9 system for inducing targeted mutations of selected GmMLO genes in a Vietnamese elite soybean cultivar DT26, and investigate their functions in powdery mildew susceptibility. The CRISPR/Cas9-induced Gmmlo mutant lines were generated and the inheritance of GmMLO mutations was assessed through generations. The homozygous mutant lines were subsequently identified for powdery mildew challenges. Moreover, pleiotropic effects of GmMLO mutations on soybean growth and development were analyzed under the net-house condition. The results here would provide a potential system to generate local soybean cultivars with enhanced powdery mildew resistance.
Results
Target selection and CRISPR/Cas9 vector validation
MLO which encodes a membrane-associated protein with seven transmembrane domains is conserved throughout monocots and dicots. Loss-of-function mlo mutations confer durable and broad-spectrum resistance to powdery mildew in various crop species [12, 28]. In soybean, there are 39 MLO genes, of which GmMLO2, GmMLO10, GmMLO18, GmMLO19, GmMLO20, and GmMLO23 are closely related to AtMLO2, AtMLO6 and AtMLO12, which were shown to play a role in powdery mildew susceptibility in Arabidopsis thaliana [34]. Publicly-available soybean RNA-Seq Atlas indicated that the above mentioned GmMLO genes are expressed in various soybean tissues at low levels (Fig. S1). To investigate the roles of these soybean MLO genes in powdery mildew resistance, we employed the CRISPR/Cas9 system to induce knock-out mutations in all six GmMLO genes.
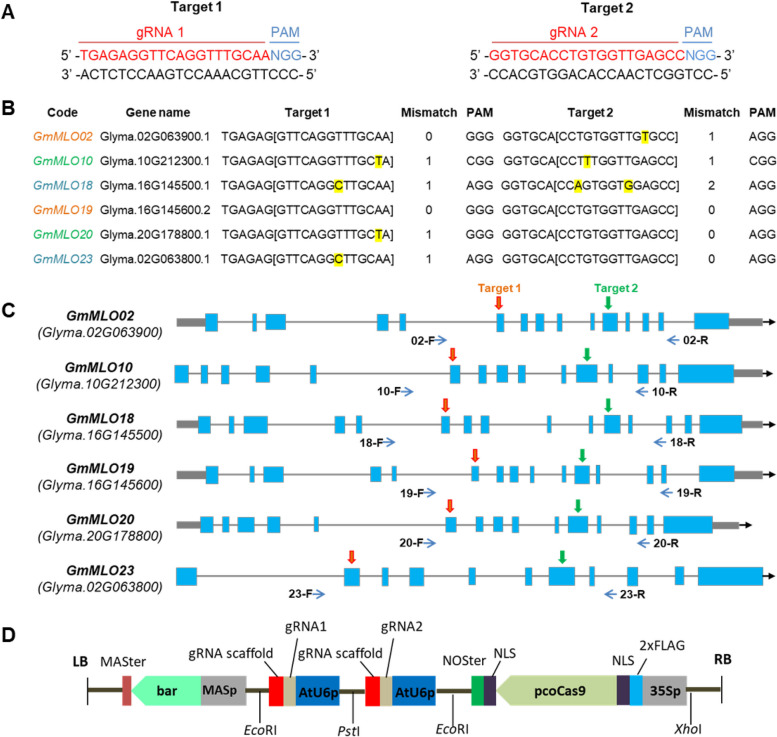
Based on the Williams 82 (W82) reference genome, we identified two potential target sites which were highly conserved and located within the exons of these six MLO genes (Fig. 1A-C). The sgRNA target 1 contained a single-mismatch in the GmMLO20, GmMLO23, GmMLO18, GmMLO10 while the sgRNA target 2 contained a single-mismatch in the GmMLO02, GmMLO10 and double-mismatch in the GmMLO18 (Fig. 1B). Sequencing results of these MLO genes in the Vietnamese elite soybean (DT26) showed that the target sites in GmMLO02, GmMLO18, GmMLO19, GmMLO20, GmMLO23 were identical to the reference Williams 82 genome. Whereas a single nucleotide polymorphism was found in the sgRNA target 1 of GmMLO10. Despite these mismatches, these were the most conserved target sites available for all six genes; thus, we selected these two target sites to generate a dual-sgRNA CRISPR/Cas9 vector reagent (Fig. 1D).
Fig. 1.
GmMLO gene maps, gRNA sequences, target locations and CRISPR/Cas9 vector. A gRNA and PAM sequences. B GmMLO genes and target sequences. Letters in yellow indicate mismatches that are within the gRNA core (PAM-1 to PAM -14). Different colors in the code column indicate different pairs of homologous genes. C GmMLO gene structures and target locations, primers for genotyping are indicated by arrows. D T-DNA region for soybean transformation. bar, herbicide resistant gene as selection marker; pcoCas9, Cas9 codon-optimized gene, driven by 35SPPDK promoter (35Sp) and two sgRNAs driven by Arabidopsis U6 promoter (AtU6p); MASp, Manopine Synthase promoter; MASter, Manopine Synthase terminator; NOSter, Nopaline synthase terminator; LB/RB, Left and Right Border
To validate the efficacy of the dual-sgRNA-CRISPR/Cas9 construct, the CRISPR/Cas9 vector was mobilized into Agrobacterium rhizogenes K599 strain for soybean hairy root transformation. Ten independent in vitro hairy root lines were used for mutagenesis analysis by PCR- agarose gel electrophoresis. Low mobility DNA bands were observed in 30% of the root samples indicating that large deletion(s) was induced in GmMLO20 (Fig. S2A). Sequencing of PCR amplicons derived from one hairy root line (HR1) indeed showed a 1214-bp deletion in GmMLO20 (Fig. S2B). Therefore, these results indicated that the CRISPR/Cas9 construct was sufficiently efficent for stable soybean transformation.
Generation of transgenic soybean and characterization of GmMLO induced mutations
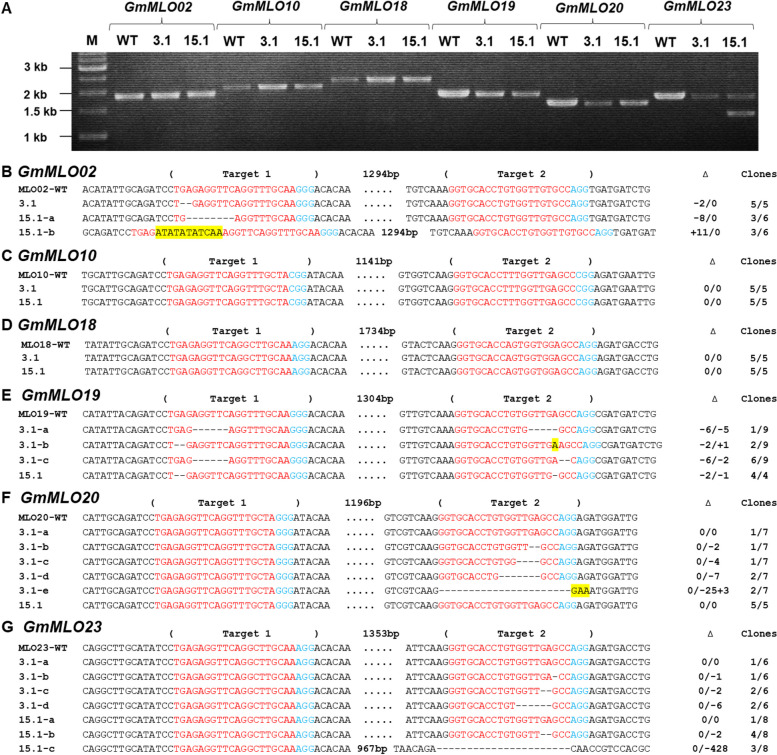
Two transgenic lines 3.1 and 15.1 were generated using Agrobacterium-mediated transformation. The presence of transgenes was confirmed by herbicide leaf painting (Fig. S3F, G) and PCR amplification using primers for the bar gene and the 35S promoter spanning sequences (Table S3). CRISPR/Cas-induced mutations in the six GmMLO genes were first evaluated by PCR using gene specific primers followed by agarose gel electrophoresis (Fig. 2A). However, low mobility PCR amplicons, indicative of large deletions, were found only for the GmMLO23 gene in the 15.1 line. Sanger sequencing of wild-type and low mobility amplicons indicated biallelic mutations of -428 bp and -2 bp alleles in the GmMLO23 gene of the 15.1 line. We further used Sanger sequencing of PCR amplicons with wild-type mobility to determine if small deletions were induced that were undetectable by gel electrophoresis. Indeed, various small insertions (from + 1 to + 11 bp) and deletions (from -1 bp to -25 bp) were observed at the target sites of the GmMLO genes in the two transgenic lines (Fig. 2B-G). We also observed chimeric mutations (more than two different alleles) in certain GmMLO genes, which suggests that CRISRP/Cas9 activity may occur late during shoot development. However, we found no mutation in the GmMLO10 and GmMLO18 genes of both two transgenic lines. In summary, the 3.1 line carried a homozygous mutation of the GmMLO02 gene and chimeric forms for the other three tested GmMLO genes including GmMLO19, GmMLO20 and GmMLO23. For the 15.1 line, homozygous, biallelic and chimeric mutant forms were found in the GmMLO19, GmMLO02 and GmMLO23 genes, respectively, but no indel was observed in the GmMLO20 gene (Table S2).
Fig. 2.
Identification and characterization of induced MLO mutations in T0 transgenic soybean plants. A Gel electrophoresis of PCR-amplicons of GmMLO target expanding regions. WT, non-transgenic wild-type plant; 3.1, 15.1, transgenic lines; M, 1 kb DNA ladder. B-G Sequence alignment of targeted regions in GmMLO genes (GmMLO02, GmMLO10, GmMLO18, GmMLO19, GmMLO20 and GmMLO23) of T0 transgenic lines. Target sequences and PAMs are indicated in red and blue color, respectively. Inserted nucleotides are shown in yellow. a/b/c/d/e indicates different alleles for each T0 line; ∆ indicates targeted sequence changes: 0 for no change, -for deletion, + for insertion. Clones indicate number of colonies with the respective alleles out of total of clones sequenced
Assessment of the inheritance of GmMLO mutations
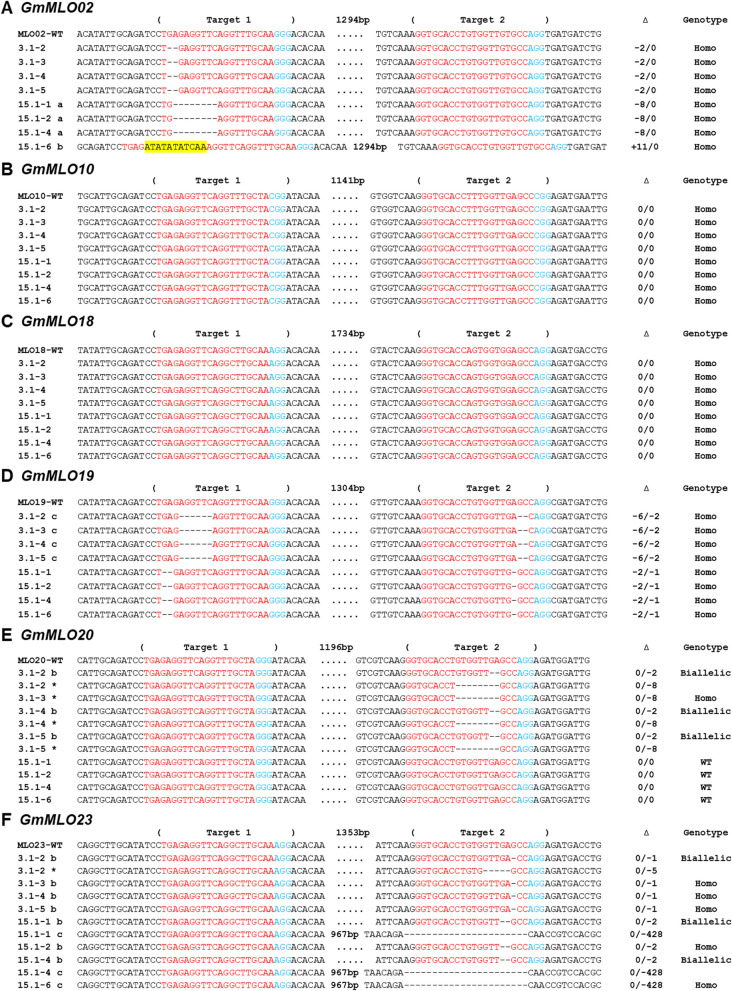
Gel electrophoresis and sequencing were conducted to assess the inheritance of CRISPR/Cas9-induced mutations at the T1 generation (Figs. S4, 3A-F). The large indel (-428 bp) in the GmMLO23 gene was passed to T1 progeny of the 15.1 line and visually observed by DNA band shifts on the agarose gel (Fig. S4). The inheritance of certain induced mutations including -2/0 (for line 3.1), -8/0 and + 11/0 (for line 15.1) of GmMLO02; -6/2 (for line 3.1) and -2/1 (for line 15.1) of GmMLO19; 0/-2 (for line 3.1) of GmMLO20; and 0/-1 (for line 3.1), 0/-2 and 0/-428 (for line 15.1) of GmMLO23 genes was confirmed by Sanger sequencing (Fig. 3). The absence of other indels indicated that they were chimeric mutations at the T0 generation that did not pass through the germline. No mutation was found in the GmMLO10 and GmMLO18 genes at the T1 generation (Fig. 3C, D). In addition, we also detected new indels in the T1 plants suggesting that late CRISPR/Cas activity occurring in the T0 plants that were subsequently passed through the germline (Fig. 3E, F). Particularly, 0/-8 allele was observed in the GmMLO20 gene of the 3.1 offspring, while 0/-5 mutations occurred in the GmMLO23 gene from some T1 plants of this T0 line.
Fig. 3.
Inheritance and segregation of targeted induced mutations at T1 generation. Target sequences and PAMs are indicated in red and blue color, respectively. Inserted nucleotides are shown in yellow. a/b/c indicates different alleles inherited from T0 for each T1 line; * indicates new alleles appeared at T1 generation; ∆ indicates targeted sequence changes: 0 for no change,—for deletion, + for insertion
Four T1 plants (3.1–3, 3.1–5, 15.1–2 and 15.1–6) harboring homozygous mutations in GmMLO genes were used for offspring analysis. Of these, 3.1–3 carried homozygous mutations in four GmMLO genes (GmMLO02, GmMLO19, GmMLO20 and GmMLO23), while 3.1–5 contained biallelic mutations in GmMLO20 and homozygous mutant alleles in three GmMLO genes (GmMLO02, GmMLO19 and GmMLO23) (Fig. 3). Meanwhile, both 15.1–2 and 15.1–6 were homozygous for induced mutations in GmMLO02, GmMLO19 and GmMLO23 genes, but carried no mutation in the GmMLO10, GmMLO18 and GmMLO20 genes. Sequencing data of selected T2 plants showed all CRIRSPR/Cas9-induced mutations from the four T1 lines, which demonstrated stable inheritance of these indels (Fig. S5). Progenies derived from four (4) T2 lines (3.1–3-41, 3.1–5-44, 15.1–2-2, and 15.1–6-4), representing various combinations of GmMLO mutations, were selected for fungal challenges (Table 1).
Table 1.
Mutant characterization of T2 CRISPR/Cas9-edited soybeans
| Events | Induced indels at Target 1/Target 2 | |||||
|---|---|---|---|---|---|---|
| GmMLO02 | GmMLO10 | GmMLO18 | GmMLO19 | GmMLO20 | GmMLO23 | |
| 3.1–3-41 | -2/0 | WT | WT | -6/-2 | 0/-8 | 0/-1 |
| 3.1–5-44 | -2/0 | WT | WT | -6/-2 | 0/-2 | 0/-1 |
| 15.1–2-2 | -8/0 | WT | WT | -2/-1 | WT | 0/-2 |
| 15.1–6-4 | + 11/0 | WT | WT | -2/-1 | WT | 0/-248 |
Evaluation of powdery mildew resistance of GmMLO mutant soybeans
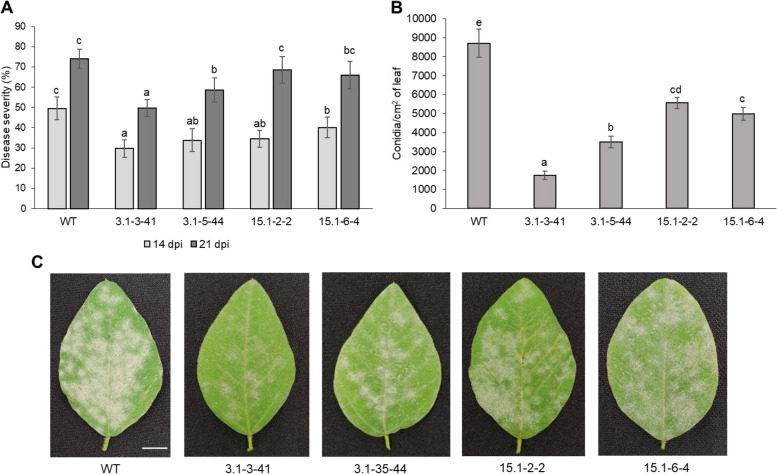
E. diffusa conidia from powdery mildew infected soybean leaves was isolated and confirmed by morphological characterization and 16S sequence analysis (Fig. S6), then used for the artificial infection of wild-type and T3 mutant plants. At 14 days post inoculation (dpi), all mutant plants showed reduced disease severity (19.1% to 40% reduction) compared to wild-type control plants (Fig. 4A; Table 2), with plants derived from 3.1–3-41 line showing the highest resistance to E. diffusa infection. At 21 dpi, a significant decrease in disease severity was only observed in 3.1–3-41 and 3.1–5-44 plants. In addition to decreased disease severity, the conidial density in the infected leaf surfaces was dramatically reduced in all mutant soybean lines (1.5 to 5 times), which indicated the inhibition of fungal development (Fig. 4B, C).
Fig. 4.
Soybean powdery mildew resistant assessment. A Disease severity assessment at 14 dpi and 21 dpi. Error bars indicate standard deviations, n = 5–6. Each time point was analyzed independently using one-way ANOVA followed by a post hoc Turkey’s test (P < 0.05). B Quantification of conidia per cm2 leaf surface at 21 dpi. Error bars indicate standard deviations, n = 6. Statistical analysis was done using one-way ANOVA followed by a post hoc Turkey’s test. Significant difference was considered at P < 0.05. C The representative leaves were collected 21 days after inoculation. Scale bar = 1 cm. WT, control line (DT26 cultivar); 3.1–3-41, 3.1–5-44, 15.1–2-2, 15.1–6-4, T2 offspring Gmmlo soybean mutant lines
Table 2.
Powdery mildew symptom reduction of mutant lines compared to WT
| Lines | Disease reduction (%) | Average reduction (%) | |
|---|---|---|---|
| 14 dpi | 21 dpi | ||
| 3.1–3-41 | 40.0 | 32.8 | 36.4 |
| 3.1–5-44 | 31.7 | 20.8 | 26.3 |
| 15.1–2-2 | 30.5 | 7.5 | 19.0 |
| 15.1–6-4 | 19.1 | 10.9 | 15.0 |
Note: Disease reduction was calculated as WT disease severity subtracted mutant disease severity divided by WT disease severity and × 100. 3.1–3-41, 3.1–5-44, 15.1–2-2, 15.1–6-4, T2 offspring Gmmlo soybean mutant lines
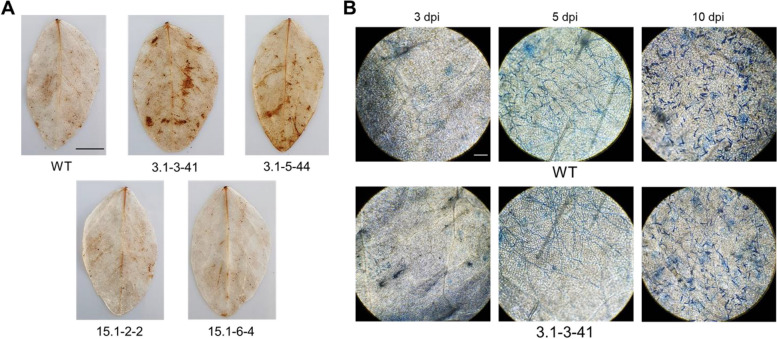
3, 3'-diaminobenzidine (DAB) staining showed stronger H2O2 accumulation with more brown spots in infected leaves of lines 3.1–3-41 and 3.1–5-44 as compared to wild-type plants, indicating a stronger reactive oxygen species (ROS) response to fungal infection. In contrast, there was no visible difference in DAB staining between the infected leaves of two lines 15.1–2-2, 15.1–6-4 and wild-type leaves (Fig. 5A). Histological analysis of the 3.1–3-41 line, which showed the lowest infection levels of soybean powdery mildew, also exhibited a delay of E. diffusa hyphae development (Fig. 5B). Particularly, at 3 dpi, hyphae were found on the infected leaf surfaces of wild-type plants, but not in the 3.1–3-41 line. At 5 dpi, both developed hyphae and conidiophores were observed on the leaf surfaces of wild-type plants, while only hyphae were found on the leaves of the 3.1–3-41 line. At 10 dpi, the conidial density was much higher in wild-type leaves as compared to the 3.1–3-41 line. We further assessed the enhanced powdery mildew resistance of the mutant soybean lines under net-house farming conditions with high pathogenic pressure of E. diffusa. The disease severity was measured and recorded at 2.6 and 3.5 for the 3.1–3-41 and 3.1–5-44 lines, respectively. However, severity increased up to 4.7 in infected plants from the 15.1–2-2 and 15.1–6-4 mutant lines, as well as the wild-type (Fig. S7). Altogether, our results indicated that the two mutant lines 3.1–3-41 and 3.1–5-44, which carry homozygous mutations of GmMLO02, GmMLO19, GmMLO20 and GmMLO23 genes, showed the least susceptibility to powdery mildew infection.
Fig. 5.
Powdery mildew fungal development and damages to soybean leaves. A The accumulation of hydrogen peroxide on powdery mildew infected leaves at 24 hpi using DAB staining method. WT, control line (DT26 cultivar); 3.1–3-41, 3.1–5-44, 15.1–2-2, 15.1–6-4, T2 offspring Gmmlo soybean mutant lines. Scale bar = 1 cm. B E. diffusa hyphae development and conidia formation in the wild-type leaves and in the T2 offspring targeted mutant line 3.1–3-41 at 3, 5 and 10 dpi. Scale bar = 100 µm
Growth and development of soybean homozygous mutant lines
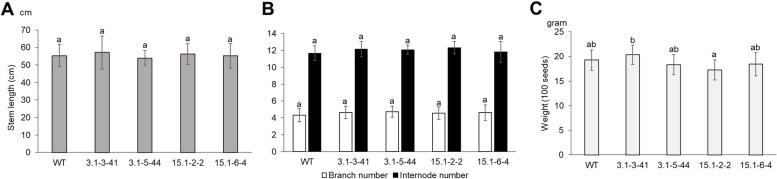
The morphology and agronomic traits of T3 plants carrying homozygous mutations of GmMLO02, GmMLO19, GmMLO20 and GmMLO23 were assessed under net-house conditions (Fig. 6). No significant differences in plant height, branch and internode number were observed between mutants and wild-type plants (Fig. 6A, B). For agronomical traits, the seed weights varied slightly between the wild-type and the mutant lines, but we found no statistically significant difference (Fig. 6C). In addition, mutant soybean plants showed no change in total pods per plant as compared to the wild-type (around 30 pods per plant), except line 15.1–6-4, which had about 22 pods per plant (Fig. S8A). However, this mutant line also exhibited the highest rate of 3-seeded pods (Fig. S8B). Altogether, targeted mutations in GmMLO genes had no obvious negative effects on soybean growth and development under net-house conditions.
Fig. 6.
Growth and development of T2 offspring Gmmlo mutant soybeans under the net-house conditions. A Stem length. B Branch and internode number. C Weight of 100 seeds. WT, control line (DT26 cultivar); 3.1–3-41, 3.1–5-44, 15.1–2-2, 15.1–6-4, T2 offspring Gmmlo soybean mutant lines. Error bars indicate standard deviations, n = 9–14. Statistical analysis was done using one-way ANOVA followed by a post hoc Turkey’s test. Significant difference was considered at P < 0.05
Identification of transgene-free homozygous mutants
Transgene-free mutant soybeans were screened at the T1 and T2 generations using herbicide leaf-painting (Fig. S9A) and PCR with specific primers for bar gene and for transgene region spanning the pFGC vector and 35SPPDK promoter of pcoCas9 (Fig. S9B; Table S1). All tested T1 plants were resistant to herbicide. In addition, PCR results also confirmed the presence of transgenes in these plants (Fig. S9B; Table S3). At the T2 generation, we identified 4 plants from the T0 line 3.1 were negative for the presence of transgenes and susceptible to herbicide (Fig. S9B; Table S3). Of which, two lines 3.1–3-41 and 3.1–5-44 were then confirmed to carry quadruple null mutations in four GmMLO genes. In line with previous reports, our results indicate that transgene-free mutant soybean could be obtained as early as the T2 generation using the CRISPR/Cas9 system.
Discussion
Inducing loss-of-function mutations in MLO genes using the CRISPR/Cas9 system is a promising approach to generate powdery mildew resistant cultivars in important crops, such as wheat [22], grapevine [27, 35], and tomato [30, 31]. In this study, we utilized the dual-gRNA CRISPR/Cas9 system to simultaneously knock-out the four soybean MLO homologs—GmMLO02, GmMLO19, GmMLO20, and GmMLO23—in the Vietnamese soybean elite cultivar DT26, and resulted in elevated resistance to powdery mildew.
In soybean, six out of 39 putative GmMLO genes—GmMLO02, GmMLO10, GmMLO18, GmMLO19, GmMLO20, and GmMLO23 were found to be orthologous to three AtMLO genes (AtMLO2, AtMLO6, AtMLO12), which are required for resistance to powdery mildew in Arabidopsis [17]. Thus, we designed a plasmid vector for simultaneous targeting of all six GmMLO genes. The use of a dual-gRNA CRISPR/Cas9 system increases editing frequency either single or multiple genes in soybean [36–38]. We found the dual-gRNA CRISPR/Cas9 system induced mutations in GmMLO02, GmMLO19, GmMLO20 and GmMLO23 in the two soybean events, however, no mutation was found in the GmMLO10 and GmMLO18 in these events (Fig. 2; Table S2). The lack of mutations in these latter two genes may be due to mismatches in the ‘seed’ sequence (10–12 bp proximal to the PAM), which could significantly reduce Cas9's ability to bind and cleave its target [39]. Indeed, we found one to two mismatches located in the seed region of the targets of the GmMLO10 and GmMLO18 in the soybean cultivar DT26 (Fig. 1B). In addition, we also found a mismatch in target 1 of GmMLO20 and GmMLO23 or in target 2 of GmMLO02 inhibited the cleavage activity of Cas9 in soybean plants.
Functional MLO genes associated with powdery mildew resistance are exclusively grouped in clade V for dicots [12]. In this study, we showed that at least four members of GmMLO in this clade are involved in powdery mildew resistance in soybean. We found that not only the quadruple mutants (Gmmlo02/Gmmlo19/Gmmlo20/Gmmlo23) but also the triple mutant (Gmmlo02/Gmmlo19/Gmmlo23) exhibited significantly enhanced E. diffusa resistance compared to the wild-type at 14 dpi (Fig. 4A). However, the quadruple mutants showed more resistant to the pathogen compared to the wild-type and the triple mutants at 21dpi, as exemplified by fewer conidia formation and hyphae development of E. diffusa (Figs. 4B, C, and 5B). In addition, H2O2 production and accumulation, visualized as brownish precipitates upon DAB staining, were more prominent in mildew-infected leaf of the quadruple GmMLO mutant lines at 21 hpi (Fig. 5A). The correlation between the production of H2O2 and resistance to powdery mildew in our soybean mutants is similar to reports in barley [40], cucumber [41] and grapevine [27] where H2O2 accumulation, and subsequent host cell death, is one of defense mechanisms in mlo plants to powdery mildew [17, 28, 40]. Hence, our results showed these four GmMLO genes are functionally conserved and contribute to powdery mildew resistance in soybean. Functional redundancy, especially unequal genetic redundancy, of MLO members in contribution to powdery mildew resistance was reported for Arabidopsis, grape and tomato [17, 26, 42]. In Arabidopsis, Atmlo2 single mutant plants displayed partial powdery mildew resistance, whereas Atmlo2/Atmlo6/Atmlo12 triple mutant plants were fully resistant. A similar scenario was observed in tomato, where the SlMLO1 is the major powdery mildew susceptibility factor, and SlMLO5 and SlMLO8, have minor function [42]. In grape, knock-down of at least three MLO genes including VvMLO7, VvMLO11 and VvMLO6 significantly reduced powdery mildew severity [26]. Taken together, our data suggest that GmMLO02, GmMLO19, and GmMLO23 are functionally redundant with GmMLO20 as powdery mildew susceptibility factors in soybean. Complete resistance to powdery mildew was obtained in the knock-out and knock-down of three MLO genes in Arabidopsis and tomato, respectively [17, 42]. However, our mutant plants did not exhibit complete resistance to powdery mildew. This may indicate that additional GmMLO genes, other than the four mutated in our study, also function to some extent in the susceptibility of soybean to powdery mildew infection. Further researches need to be performed to clarify the contribution of single tested GmMOL genes as well as the interaction of these genes in the mechanism of powdery mildew resistance in soybean.
In plants, MLO genes have been implicated in various physiological processes [12]. Disruption of these genes were accompanied by undesired pleiotropic effects such as leaf chlorosis and reduced grain yield in barley and wheat [14, 21, 31], reduced growth in A. thaliana [17], smaller plant size in pepper [21], as well as senescence-like chlorosis and necrosis in grapevine [27]. In our study, no pleiotropic phenotype was observed in the Gmmlo mutants under net-house conditions. The resistant lines carrying loss-of-function mutations in four GmMLO genes, i.e., GmMLO02, GmMLO19, GmMLO20 and GmMLO23, showed no obvious difference in morphology, development and seed production compared to wild-type plants. The transgene-free Gmmlo mutant soybean lines with the highest resistance to powdery mildew created in this study should be useful genetic materials for breeding programs for increased disease resistance.
Conclusions
In summary, we were successful in using a dual-gRNA CRISPR/Cas9 system to simultaneously knock-out the four soybean MLO homologs—GmMLO02, GmMLO19, GmMLO20, and GmMLO23—in the Vietnamese soybean elite cultivar DT26. The CRISPR/Cas9-induced Gmmlo mutant lines exhibited enhanced resistance to soybean powdery mildew. Moreover, the Gmmlo transgene-free mutant lines showed no obvious difference in morphology, development and productivity compared to wild-type plants. Our results indicate the involvement of four GmMLO genes in soybean powdery mildew susceptibility and provide a potential strategy for improving disease resistance of local soybean cultivars.
Methods
CRISPR/Cas9 vector construction
One pFGC-CRISPR/Cas9 vector carrying dual sgRNAs, each independently driven by a Arabidopsis thaliana AtU6 promoter, was constructed for simultaneously inducing targeted mutations of multiple selected GmMLO genes in soybean. Particularly, AtU6 promoter and gRNA scaffold were derived from the pBlu/gRNA vector, a gift from Robert Stupar's laboratory (RRID: Addgene_59188). Double-stranded DNA oligonucleotides of sgRNA were cloned into the pBlu/gRNA vector by BpiI sites. The expected fragments (AtU6-gRNA- scaffold) were excised by EcoRI sites and used for gel purification. The cassette of a plant-codon-optimized Cas9 driven by 35S promoter was generated from the HBT-pcoCas9 vector, a gift from Jen Sheen's laboratory (RRID: Addgene_52254) using EcoRI and XhoI sites. All designed cassettes were assembled in the pFGC5941 backbone by EcoRI sites to generate the final construct pFGC5941-gRNA1-gRNA2-Cas9. The designed construct was validated and confirmed by Sanger sequencing.
Soybean hairy root transformation
Soybean in vitro hairy root transformation was performed following a previously reported method [43] to evaluate the targeted editing activity of the designed CRISPR/Cas9 vector. Briefly, cotyledons from 4-day-old seedlings of Vietnamese elite cultivar DT26 obtained from Legumes Research and Development Center, Field Crops Research Institute, Vietnam Academy of Agricultural Sciences were used as explants for infection with A. rhizogenes K599 strain harboring the pFGC5941-gRNA1-gRNA2-Cas9 construct. Seven days after co-cultivation, induced soybean hairy roots were transferred to selection medium (MS medium with 3 mg/L glufosinate). Genomic DNA was extracted from herbicide-resistant hairy roots using the CTAB method [44] and used for induced mutant identification and characterization.
Stable soybean transformation and transgene confirmation
Soybean transformation was performed using Agrobacterium tumefaciens via cotyledon node infection as previously described [37, 45] (Fig. S3A-E). Regenerated plants on selection medium were transferred to perlite and vermiculite mixture (1:3 v/v) for acclimatization, then cultured in plastic pots containing TRiBAT® compost (Green Saigon Biotechnology Limited Company, Vietnam) under greenhouse conditions. Glufosinate solution (200 mg/L) was painted onto three trifoliate leaves of each plant for herbicide resistant tests. Genomic DNA of herbicide-resistant plants was extracted by the CTAB method [44] and used for transgene confirmation with specific primers (Table S2).
Induced mutant identification and characterization
The target spanning regions on the GmMLO genes were amplified using specific primers (Table S2) and analysed by 1% agarose gel electrophoresis to detect DNA band shifts. The PCR amplicons were then purified and ligated to the pJET1.2 cloning vector (Thermo Fisher Scientific, USA) for Sanger sequencing by ABI PRISM® 3100 Avant Genetic Analyzer system (Applied Biosystems, USA). The sequencing data were analysed by the FinchTV chromatogram viewer program (Geospiza) and MEGA X [46].
Plant cultivation and morphological characterization
Mature soybean seeds were imbibed on moist paper for 48 h at 26oC, then sown in plant pots (26 cm depth, 21 cm diameter top) containing the TRiBAT® compost mixture (Green Saigon Biotechnology Limited Company, Vietnam) and organic soil (Minh Hiep Thanh Cooperative, Vietnam) (1:3 v/v). Soybean plants were grown under net-house conditions and fertilized with NPK (15:5:20) at the V3 stage, NPK (16:16:16) at 40 and 65 days-old stages. Plant morphological parameters and soybean yield traits including stem length, branch and internode number, total pods, seeds per pod and seed weight were collected and analysed at the R8 stage. Seeds were harvested, dried and stored in seed room at 40% humidity and 4°C for further experiments.
E. diffusa susceptibility assessment
Soybean leaves with symptoms of powdery mildew infection were provided by Legumes Research and Development Center, Field Crops Research Institute, Vietnam Academy of Agricultural Sciences. Conidiophore and conidia of powdery mildew fungus were visually confirmed and isolated from the infected leaves. Fungal genomic DNA was extracted using a EZ-10 Spin Colum Fungal Genomic DNA Mini-Preps Kit (Bio Basic, Canada) and used for species confirmation by PCR with ITS1/PM6 specific primers [47, 48]. The E. diffusa isolate was subsequently maintained and propagated on a susceptible soybean under growth chamber condition. The fungus was infected into selected T3 Gmmlo mutant plants (n ≥ 5 for each mutant line) at the V2 stage using the leaf brushing method as described by Kang and Mian [49]. Infected plants were kept in a growth chamber (22°C ± 1°C and 100% RH) for 12 h to promote fungal germination, penetration, and development. The disease symptom and severity were recorded and analysed at 14- and 21-days post inoculation (dpi) as previous description by Pessina et al. [25].
For the net-house test, mature seeds of selected T3 mutant lines were directly sown beside the susceptible cultivar showing powdery mildew symptoms. The disease severity was observed and scored at V3, V5 and V7 stages based on the symptom scales (0 to 5 grades) proposed by Tran et al. [50].
Histological analysis
E. diffusa infected leaves were collected at 3, 5 and 10 dpi and submerged in ethanol-acetic acid solution (3:1 v/v) to remove chlorophyll [25]. The treated leaves were stained with 250 µg/mL trypan blue in lactoglycerol solution [lactic acid:glycerol:water 1:1:1 (v/v/v)] for 15 min, then rinsed in the same solution at room temperature as described by Vogel and Somerville [51]. The samples were then mounted and captured under 100X magnification for visualization of hyphae development and conidia germination.
DAB staining was conducted to assess hydrogen peroxide (H2O2) accumulation in infected leaves at 24 hpi followed the method by Yu et al. [41]. The stained leaves were boiled in ethanol-lactic acid-glycerol (3:1:1 v/v/v) for 20 min and then transferred to pre-chilled 95% ethanol before being photographed.
Data analysis
Agronomic traits and disease severity data were analyzed with SPSS Statistics software (version 20.0, IBM, Armonk, NY). The mean values and standard deviation of the mean (SD) were calculated and presented reflecting three replicates. Statistical significance was conducted using one-way ANOVA followed by a post-hoc Turkey’s test at P < 0.05.
Supplementary Information
Additional file 1: Fig. S1. Transcriptomics analysis of GmMLO genes in different tissues of soybean plant. Data were obtained from RNA-Seq Atlas of Glycine max. Fig. S2. Induced mutation analysis of hairy roots. A Gel electrophoresis (agarose 1%) of GmMLO20 edited region in wild-type (WT) and hairy root samples (HR1, HR2, HR3) with large deletions. M: 1 kb DNA marker. Shifted bands in lines HR1, HR2 and HR3 indicated the induced mutations of targeted genes. B Sequencing result of the HR1 line for the edited regions of GmMLO20 compared to wild-type allele. Target sequences and PAMs are indicated in red and blue, respectively. Δ indicates targeted sequence changes: - for deletion. Clones indicate number of colonies with the respective alleles out of total of clones sequenced. Fig. S3. Soybean transformation procedure. A Cotyledons at 5 days on the co-cultivation medium. B, C Shoot induction at 14 and 28 days on the selection medium. D Shoot elongation. E Rooted plants on the rooting medium. F, G Leaf painting using 200 mg/L glufosinate. Fig. S4. Gel electrophoresis of PCR-amplicons of GmMLO target expanding regions at T1 generation. WT: Non transgenic wild-type plant; 3.1-2 to 3.1-5: T1 plants from 3.1 line; 15.1-1 to 15.1-6: T1 plants from 15.1 line; M: 1 kb DNA ladder. PCR products amplified by specific primers for extended regions of GmMLO02, GmMLO10, GmMLO18, GmMLO19, GmMLO20 and GmMLO23 genes. Fig. S5. Inheritance of induced mutations in GmMLO02 (A), GmMLO19 (B), GmMLO20 (C) and GmMLO23 (D) genes in T2 plants. Target sequences and PAMs are indicated in red and blue color, respectively. Inserted nucleotides are shown in yellow. Δ indicates targeted sequence changes: 0 for no change, - for deletion, + for insertion. Fig. S6. E. diffusa isolation and characterization. A Conidiophore and conidia of E. diffusa isolated from infected leaves. Scale bar = 10 μm. B Internal transcribed spacer (ITS) sequences of the collected E. diffusa. C Nucleotide BLAST result of the E. diffusa ITS sequence on GenBank, NCBI. Fig. S7. Powdery mildew resistant assessment of T2 offspring Gmmlo mutant lines under the net- house conditions. Infection levels were recorded using a 0 to 5 scale (described by Tran et al., 2015), which according to strong resistance to severe infection. Infection levels were calculated as the average of 20-30 biological replicates and three experiments. Statistical analysis was done using one-way ANOVA followed by a post hoc Turkey’s test. Significant difference was considered at P < 0.05. Fig. S8. Soybean seed production under the net-house conditions. A The total number of pods per plant. B The frequency of pods with 3 seeds. WT: Control line (DT26 cultivar); 3.1-3-41, 3.1-5-87 44, 15.1-2-2, 15.1-6-4: T2 offspring Gmmlo soybean mutant lines. Error bars indicate standard deviations, n = 9-14. Statistical analysis was done using one-way ANOVA followed by a post hoc Turkey’s test. Significant difference was considered at P < 0.05. Fig. S9. Transgene inheritance and segregation at different generations. A Representative results of herbicide leaf painting with glufosinate solution (200 mg/L) on wild-type (WT), herbicide resistant line (3.1) and herbicide susceptible line (3.1-3-41). B Gel electrophoresis of PCR amplicons of transgenes at T0, T1 and T2 generations. M: 1 kb DNA ladder; WT: non transgenic wild-type plant; (+): positive control (CRISPR/Cas9 vector); bar: herbicide resistance gene; 35S:pFGC: transgene region spanning pFGC vector and 35SPPDK promoter of pcoCas9. Table S1. Sequences of oligonucleotides and primer sets used in this study. Table S2. Genotypes of T0 mutant lines. Table S3. Inheritance and segregation of transgenes at different transgenic soybean generations.
Acknowledgements
We would like to thank Dr. Van Tuong Nguyen for her suggestion on the manuscript, and Quyen Phan for taking care of mutant plants in the greenhouse.
Authors’ contributions
TPB, HL and PTD designed the experiments. TPB, HL, DTT, CXN and NTL performed the experiments. TTT performed E. diffusa susceptibility assessment at the net-house condition. TPB and HL analyzed the data and wrote the manuscript. CXN, PVN, GS, MGS, NBP, HHC and PTD revised and proofread the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Foundation for Science and Technology Development of Vietnam (106.03–2019.11).
Availability of data and materials
All the data generated or analyzed during this study are included in this published article and its supplementary information files. The ITS sequence of E. diffusa was deposited into GenBank with accession number OQ933656. The partial nucleotide sequences of GmMLO genes were deposited into GenBank with accession numbers OQ945362-OQ945367. The materials developed in this study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The wild-type seeds of Vietnamese soybean cultivar DT26 were obtained from Legumes Research and Development Center, Field Crops Research Institute, Vietnam Academy of Agricultural Sciences. All plant materials and all the experiments in this study complied with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tyug TS, Prasad KN, Ismail A. Antioxidant capacity, phenolics and isoflavones in soybean by-products. Food Chem. 2010;123:583–589. doi: 10.1016/j.foodchem.2010.04.074. [DOI] [Google Scholar]
- 2.Braun U, Cook RTA. Taxonomic manual of the Erysiphales (Powdery mildews). CBS Biodiversity Series 11. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2012.
- 3.McTaggart AR, Ryley MJ, Shivas RG. First report of the powdery mildew Erysiphe diffusa on soybean in Australia. Australasian Plant Dis Notes. 2012;7:127–129. doi: 10.1007/s13314-012-0065-7. [DOI] [Google Scholar]
- 4.Tam LTT, Dung PN, Liem NV. First report of powdery mildew caused by Erysiphe cruciferarum on Brassica juncea in Vietnam. Plant Dis. 2016;100:856. doi: 10.1094/PDIS-06-15-0678-PDN. [DOI] [Google Scholar]
- 5.Takamatsu S, Taguchi Y, Shin H-D, Paksiri U, Limkaisang S, Thi Binh N, et al. Two Erysiphe species associated with recent outbreak of soybean powdery mildew: results of molecular phylogenetic analysis based on nuclear rDNA sequences. Mycoscience. 2002;43:333–341. doi: 10.1007/S102670200049. [DOI] [Google Scholar]
- 6.Dunleavy JM. Yield losses in soybeans induced by powdery mildew. Plant Dis. 1980;64:291–292. doi: 10.1094/PD-64-291. [DOI] [Google Scholar]
- 7.Wrather JA, Anderson TR, Arsyad DM, Tan Y, Ploper LD, Porta-Puglia A, et al. Soybean disease loss estimates for the top ten soybean-producing countries in 1998. Can J Plant Pathol. 2001;23:115–121. doi: 10.1080/07060660109506918. [DOI] [PubMed] [Google Scholar]
- 8.Gonçalves ECP, Di Mauro AO, Centurion MA. Genetics of resistance to powdery mildew (Microsphaera diffusa) in Brazilian soybean populations. Genet Mol Biol. 2002;25:339–42. doi: 10.1590/S1415-47572002000300015. [DOI] [Google Scholar]
- 9.Xian P, Cai Z, Jiang B, Xia Q, Cheng Y, Yang Y, et al. GmRmd1 encodes a TIR-NBS-BSP protein and confers resistance to powdery mildew in soybean. Plant Communications. 2022;3:100418. doi: 10.1016/j.xplc.2022.100418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Schie CCN, Takken FLW. Susceptibility Genes 101: How to be a good host. Annu Rev Phytopathol. 2014;52:551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- 11.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, et al. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 12.Acevedo-Garcia J, Kusch S, Panstruga R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014;204:273–281. doi: 10.1111/nph.12889. [DOI] [PubMed] [Google Scholar]
- 13.Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416:447–451. doi: 10.1038/416447a. [DOI] [PubMed] [Google Scholar]
- 14.Reddy VS, Ali GS, Reddy ASN. Characterization of a pathogen-induced calmodulin-binding protein: mapping of four Ca2+-dependent calmodulin-binding domains. Plant Mol Biol. 2003;52:143–159. doi: 10.1023/A:1023993713849. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen IH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. doi: 10.1007/BF00023919. [DOI] [Google Scholar]
- 16.Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Gen Genet. 1993;239:122–128. doi: 10.1007/BF00281610. [DOI] [PubMed] [Google Scholar]
- 17.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 18.Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, et al. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol Plant-Microbe Interact. 2008;21:30. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]
- 19.Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol. 2011;12:866–878. doi: 10.1111/j.1364-3703.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun S, Fu H, Wang Z, Duan C, Zong X, Zhu Z. Discovery of a novel er1 allele conferring powdery mildew resistance in Chinese pea (Pisum sativum L.) landraces. PLoS One. 2016;11:e0147624. doi: 10.1371/journal.pone.0147624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Z, Nonomura T, Appiano M, Pavan S, Matsuda Y, Toyoda H, et al. Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS ONE. 2013;8:e70723. doi: 10.1371/journal.pone.0070723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 23.Acevedo-Garcia J, Spencer D, Thieron H, Reinstädler A, Hammond-Kosack K, Phillips AL, et al. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol J. 2017;15:367–378. doi: 10.1111/pbi.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu X, Wang Q, Zhang H, Jian H, Zhou N, Ji C, et al. Antisense RhMLO1 gene transformation enhances resistance to the powdery mildew pathogen in rosa multiflora. Plant Mol Biol Rep. 2015;33:1659–1665. doi: 10.1007/s11105-015-0862-1. [DOI] [Google Scholar]
- 25.Pessina S, Angeli D, Martens S, Visser RGF, Bai Y, Salamini F, et al. The knock-down of the expression of MdMLO19 reduces susceptibility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica) Plant Biotechnol J. 2016;14:2033–2044. doi: 10.1111/pbi.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pessina S, Lenzi L, Perazzolli M, Campa M, Dalla Costa L, Urso S, et al. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic Res. 2016;3:16016. doi: 10.1038/hortres.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan D-Y, Guo Y, Cheng Y, Hu Y, Xiao S, Wang Y, et al. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera) Hortic Res. 2020;7:116. doi: 10.1038/s41438-020-0339-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusch S, Panstruga R. mlo-based resistance: An apparently universal “weapon” to defeat powdery mildew disease. Mol Plant Microbe Interact. 2017;30:179–189. doi: 10.1094/MPMI-12-16-0255-CR. [DOI] [PubMed] [Google Scholar]
- 29.Zhang D, Zhang Z, Unver T, Zhang B. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J Adv Res. 2021;29:207–221. doi: 10.1016/j.jare.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7:482. doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pramanik D, Shelake RM, Park J, Kim MJ, Hwang I, Park Y, et al. CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. IJMS. 2021;22:1878. doi: 10.3390/ijms22041878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B, et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature. 2022;602:455–460. doi: 10.1038/s41586-022-04395-9. [DOI] [PubMed] [Google Scholar]
- 33.Shen Q, Zhao J, Du C, Xiang Y, Cao J, Qin X. Genome-scale identification of MLO domain-containing genes in soybean (Glycine max L. Merr.) Genes Genet Syst. 2012;87:89–98. doi: 10.1266/ggs.87.89. [DOI] [PubMed] [Google Scholar]
- 34.Deshmukh R, Singh VK, Singh BD. Comparative phylogenetic analysis of genome-wide Mlo gene family members from Glycine max and Arabidopsis thaliana. Mol Genet Genomics. 2014;289:345–359. doi: 10.1007/s00438-014-0811-y. [DOI] [PubMed] [Google Scholar]
- 35.Malnoy M, Viola R, Jung M-H, Koo O-J, Kim S, Kim J-S, et al. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Do PT, Nguyen CX, Bui HT, Tran LTN, Stacey G, Gillman JD, et al. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2–1A and GmFAD2–1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 2019;19:311. doi: 10.1186/s12870-019-1906-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le H, Nguyen NH, Ta DT, Le TNT, Bui TP, Le NT, et al. CRISPR/Cas9-mediated knockout of galactinol synthase-encoding genes reduces raffinose family oligosaccharide levels in soybean seeds. Front Plant Sci. 2020;11:612942. doi: 10.3389/fpls.2020.612942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen DV, Hoang TT-H, Le NT, Tran HT, Nguyen CX, Moon Y-H, et al. An efficient hairy root system for validation of plant transformation vector and CRISPR/Cas construct activities in cucumber (Cucumis sativus L.) Front Plant Sci. 2022;12:770062. doi: 10.3389/fpls.2021.770062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu BXH, St. Onge RP, Fire AZ, Smith JD. Distinct patterns of Cas9 mismatch tolerance in vitro and in vivo. Nucleic Acids Res. 2016;44:5365–77.. doi: 10.1093/nar/gkw417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu G, Wang X, Chen Q, Cui N, Yu Y, Fan H. Cucumber mildew resistance locus O interacts with calmodulin and regulates plant cell death associated with plant immunity. IJMS. 2019;20:2995. doi: 10.3390/ijms20122995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Z, Appiano M, Pavan S, Bracuto V, Ricciardi L, Visser RGF, et al. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front Plant Sci. 2016;7:380. doi: 10.3389/fpls.2016.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Cai Y, Liu X, Guo C, Sun S, Wu C, et al. Soybean hairy roots produced in vitro by Agrobacterium rhizogenes-mediated transformation. Crop J. 2018;6:162–171. doi: 10.1016/j.cj.2017.08.006. [DOI] [Google Scholar]
- 44.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: Version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 45.Margie MP, Shou H, Guo Z, Zhang Z, Banerjee AK, Wang K. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica. 2004;136:167–179. doi: 10.1023/B:EUPH.0000030670.36730.a4. [DOI] [Google Scholar]
- 46.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White T, Bruns T, Lee S, Taylor J, Innis M, Gelfand D, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide Methods Appl. 1990;18:315–22. [Google Scholar]
- 48.Takamatsu S, Kano Y. PCR primers useful for nucleotide sequencing of rDNA of the powdery mildew fungi. Mycoscience. 2001;42:135–139. doi: 10.1007/BF02463987. [DOI] [Google Scholar]
- 49.Kang S-T, Rouf Mian MA. Powdery mildew resistance in soybean PI 243540 is controlled by a single dominant gene. Can J Plant Sci. 2010;90:939–942. doi: 10.4141/cjps09070. [DOI] [Google Scholar]
- 50.Tran TT, Nguyen TT, Trinh XH, Nguyen DT. The research results of powdery mildew (Microphaera diffusa) on soybean in the North of Vietnam. J Vietnam Agric Sci Technol. 2015;3:94–101. [Google Scholar]
- 51.Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Transcriptomics analysis of GmMLO genes in different tissues of soybean plant. Data were obtained from RNA-Seq Atlas of Glycine max. Fig. S2. Induced mutation analysis of hairy roots. A Gel electrophoresis (agarose 1%) of GmMLO20 edited region in wild-type (WT) and hairy root samples (HR1, HR2, HR3) with large deletions. M: 1 kb DNA marker. Shifted bands in lines HR1, HR2 and HR3 indicated the induced mutations of targeted genes. B Sequencing result of the HR1 line for the edited regions of GmMLO20 compared to wild-type allele. Target sequences and PAMs are indicated in red and blue, respectively. Δ indicates targeted sequence changes: - for deletion. Clones indicate number of colonies with the respective alleles out of total of clones sequenced. Fig. S3. Soybean transformation procedure. A Cotyledons at 5 days on the co-cultivation medium. B, C Shoot induction at 14 and 28 days on the selection medium. D Shoot elongation. E Rooted plants on the rooting medium. F, G Leaf painting using 200 mg/L glufosinate. Fig. S4. Gel electrophoresis of PCR-amplicons of GmMLO target expanding regions at T1 generation. WT: Non transgenic wild-type plant; 3.1-2 to 3.1-5: T1 plants from 3.1 line; 15.1-1 to 15.1-6: T1 plants from 15.1 line; M: 1 kb DNA ladder. PCR products amplified by specific primers for extended regions of GmMLO02, GmMLO10, GmMLO18, GmMLO19, GmMLO20 and GmMLO23 genes. Fig. S5. Inheritance of induced mutations in GmMLO02 (A), GmMLO19 (B), GmMLO20 (C) and GmMLO23 (D) genes in T2 plants. Target sequences and PAMs are indicated in red and blue color, respectively. Inserted nucleotides are shown in yellow. Δ indicates targeted sequence changes: 0 for no change, - for deletion, + for insertion. Fig. S6. E. diffusa isolation and characterization. A Conidiophore and conidia of E. diffusa isolated from infected leaves. Scale bar = 10 μm. B Internal transcribed spacer (ITS) sequences of the collected E. diffusa. C Nucleotide BLAST result of the E. diffusa ITS sequence on GenBank, NCBI. Fig. S7. Powdery mildew resistant assessment of T2 offspring Gmmlo mutant lines under the net- house conditions. Infection levels were recorded using a 0 to 5 scale (described by Tran et al., 2015), which according to strong resistance to severe infection. Infection levels were calculated as the average of 20-30 biological replicates and three experiments. Statistical analysis was done using one-way ANOVA followed by a post hoc Turkey’s test. Significant difference was considered at P < 0.05. Fig. S8. Soybean seed production under the net-house conditions. A The total number of pods per plant. B The frequency of pods with 3 seeds. WT: Control line (DT26 cultivar); 3.1-3-41, 3.1-5-87 44, 15.1-2-2, 15.1-6-4: T2 offspring Gmmlo soybean mutant lines. Error bars indicate standard deviations, n = 9-14. Statistical analysis was done using one-way ANOVA followed by a post hoc Turkey’s test. Significant difference was considered at P < 0.05. Fig. S9. Transgene inheritance and segregation at different generations. A Representative results of herbicide leaf painting with glufosinate solution (200 mg/L) on wild-type (WT), herbicide resistant line (3.1) and herbicide susceptible line (3.1-3-41). B Gel electrophoresis of PCR amplicons of transgenes at T0, T1 and T2 generations. M: 1 kb DNA ladder; WT: non transgenic wild-type plant; (+): positive control (CRISPR/Cas9 vector); bar: herbicide resistance gene; 35S:pFGC: transgene region spanning pFGC vector and 35SPPDK promoter of pcoCas9. Table S1. Sequences of oligonucleotides and primer sets used in this study. Table S2. Genotypes of T0 mutant lines. Table S3. Inheritance and segregation of transgenes at different transgenic soybean generations.
Data Availability Statement
All the data generated or analyzed during this study are included in this published article and its supplementary information files. The ITS sequence of E. diffusa was deposited into GenBank with accession number OQ933656. The partial nucleotide sequences of GmMLO genes were deposited into GenBank with accession numbers OQ945362-OQ945367. The materials developed in this study are available from the corresponding author on reasonable request.