Abstract
A peptidase gene expressing l-proline-β-naphthylamide-hydrolyzing activity was cloned from a gene library of Lactobacillus rhamnosus 1/6 isolated from cheese. Peptidase-expressing activity was localized in a 1.5-kb SacI fragment. A sequence analysis of the SacI fragment revealed the presence of one complete open reading frame (ORF1) that was 903 nucleotides long. The ORF1-encoded 34.2-kDa protein exhibited 68% identity with the PepR protein from Lactobacillus helveticus. Additional sequencing revealed the presence of another open reading frame (ORF2) following pepR; this open reading frame was 459 bp long. Northern (RNA) and primer extension analyses indicated that pepR is expressed both as a monocistronic transcriptional unit and as a dicistronic transcriptional unit with ORF2. Gene replacement was used to construct a PepR-negative strain of L. rhamnosus. PepR was shown to be the primary enzyme capable of hydrolyzing Pro-Leu in L. rhamnosus. However, the PepR-negative mutant did not differ from the wild type in its ability to grow and produce acid in milk. The cloned pepR expressed activity against dipeptides with N-terminal proline residues. Also, Met-Ala, Leu-Leu, and Leu-Gly-Gly and the chromogenic substrates l-leucine-β-naphthylamide and l-phenylalanine-β-naphthylamide were hydrolyzed by the PepR of L. rhamnosus.
Lactic acid bacteria are auxotrophic for many amino acids, and in order to grow to high cell densities in milk, they utilize a complex proteolytic system that degrades the milk protein casein. Extensive studies of the proteolytic system of lactococci have revealed that this system is composed of a cell envelope-associated proteinase, membrane-bound transport systems, and several cytoplasmic peptidases (for a recent review, see reference 28). Most of the genetic studies of the proteolytic system of lactic acid bacteria have focused on Lactococcus lactis, Lactobacillus delbrueckii, and Lactobacillus helveticus that are used in the production of a broad range of food products. However, during cheese maturation mesophilic nonstarter lactic acid bacteria, such as Lactobacillus plantarum, Lactobacillus casei, and Lactobacillus brevis, are frequently found in large numbers during the late ripening period (8, 34). In recent years, several peptidolytic enzymes have been purified from L. casei strains that were originally isolated from cheeses (2, 3, 14, 15, 19, 20), and those enzymes have been characterized biochemically. However, the only component of the proteolytic system of mesophilic lactobacilli that has been characterized at the gene level is the proteinase of L. casei NCDO151 (23). Milk casein contains a high level of proline (16), which results in the generation of proline-rich peptides during proteinase action (27). Peptides containing an N-terminal proline residue are usually not hydrolyzed by general-purpose aminopeptidases, dipeptidases, or tripeptidases. Thus, several proline-specific peptidases having distinct substrate specificities have evolved in lactic acid bacteria. These enzymes may play an important role in cheese ripening because proline-containing peptides are often bitter (20). Some proline-specific peptidases, including X-prolyl-dipeptidyl aminopeptidase (dipeptidyl-peptidase IV; EC 3.4.14.5), proline iminopeptidase (prolyl aminopeptidase; EC 3.4.11.5), and prolidase (imidodipeptidase; EC 3.4.13.9), have been purified from L. casei (13, 15, 19, 20).
We have started to genetically characterize the peptidolytic system of mesophilic lactobacilli by cloning genes encoding proline-specific peptidases in Lactobacillus rhamnosus (formerly L. casei subsp. rhamnosus). In this paper we describe the cloning, expression, and inactivation of a gene encoding prolinase (Pro-X dipeptidase; EC 3.4.13.8) in L. rhamnosus.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. L. rhamnosus 1/6 was isolated from cheese and was identified by using the API 50 CH system (bioMérieux, Marcy l’Etoile, France) and L. rhamnosus-specific PCR primers described by Tilsala-Timisjärvi and Alatossava (40). L. rhamnosus was routinely grown in MRS (Lab M, Bury, England) or whey broth at 37°C without shaking. Whey broth contained (per liter) 50 g of whey permeate (Valio Ltd., Helsinki, Finland), 20 g of casein hydrolysate (Valio Ltd.), and 10 g of yeast extract (Difco Laboratories, Detroit, Mich.). For growth experiments whey broth was inoculated with 1% exponentially growing cells. Growth was monitored by measuring the turbidity with a Klett-Summerson colorimeter. Erythromycin (5 μg/ml) was added when appropriate. Growth experiments in milk were carried out by using 10% reconstituted skim milk (Valio Ltd.) which had been autoclaved for 10 min at 105°C. Cells grown in MRS were pelleted by centrifugation, washed twice with 0.85% NaCl, and used to inoculate 10% reconstituted skim milk to a final concentration of 106 CFU/ml. Colony counts were determined by plating samples onto MRS agar at 1-h intervals, and acid production was monitored by neutralizing preparations with 0.1 N NaOH. Escherichia coli XL1-Blue and CM89 were grown in Luria broth and in Luria broth supplemented with 0.3 mM thymine and 0.05 mM thiamine, respectively. Zeocin (Invitrogen, De Schelp, The Netherlands), an antibiotic belonging to the bleomycin family, or ampicillin was added at a concentration of 50 μg/ml when required. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a concentration of 1 mM.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype(s) or genotype(s) | Source or reference |
|---|---|---|
| L. rhamnosus strains | ||
| 1/6 | Wild type | Valio Ltd. |
| 1/6::pVS101 | Derivative of 1/6 with integrated pVS101, Emr | This study |
| 1/6ΔpepR | Derivative of 1/6 containing 227-bp ClaI-NdeI chromosomal deletion in pepR gene, Ems | This study |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB laclqZΔM15 Tn10 (Tetr)]c | Stratagene |
| CM89 | leu-9 Δ(pro-lac) met thyA pepN102 pepA11 pepB1 pepQ10 | 31 |
| Plasmids | ||
| pZErO | Zeor | Invitrogen |
| pLS19 | Amr Emr, pUC19 carrying the erm gene of pE194 in the NdeI site | K. Leenhouts, University of Groningen |
| pIL252 | Emr, ori+ of pAMβ1 | 39 |
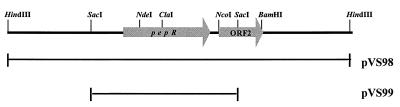
| pVS98 | Zeor PepR+, pZErO carrying 3.5-kb HindIII fragment from L. rhamnosus 1/6 chromosomal DNA with pepR gene | This study |
| pVS99 | Zeor PepR+, pZErO carrying 1.5-kb SacI fragment from pVS98, contains L. rhamnosus pepR gene | This study |
| pVS101 | Amr Emr PepR−, pLS19 with the internally deleted pepR of L. rhamnosus 1/6 | This study |
| pVS102 | Emr PepR+, pIL252 with 1.5-kb SacI fragment from pVS98 | This study |
General DNA techniques and transformation.
Molecular cloning techniques and electrotransformation of E. coli were performed as described by Sambrook et al. (36). Restriction enzymes, the Klenow enzyme, T4 DNA ligase, and deoxynucleotides were obtained from Boehringer Mannheim or New England Biolabs and were used according to the instructions of the suppliers. Chromosomal DNA was isolated from L. rhamnosus by a modification of the method of Anderson and McKay (1), as follows. The mid-log-phase cells in 3 ml of MRS supplemented with 1% glycine were pelleted and resuspended in 380 μl of 6.7% sucrose–50 mM Tris–1 mM EDTA (pH 8.0). Next, 100 μl of a 50-mg/ml lysozyme solution (in 25 mM Tris, pH 8.0) and 100 U of mutanolysin (in 100 mM potassium phosphate buffer, pH 6.2) were added, and the cells were incubated at 37°C for 1 h. After 50 μl of 0.25 M EDTA–50 mM Tris (pH 8.0) was added, the cells were lysed by adding 30 μl of 20% sodium dodecyl sulfate–50 mM Tris–20 mM EDTA (pH 8.0). The proteins were digested by adding 20 μl of proteinase K (20 mg/ml) and incubating the preparation for 1 h at 50°C. Depending on the viscosity of the sample, approximately 300 μl of sterile water was added prior to phenol extraction. Phenol extraction was repeated once and was followed by phenol-chloroform extraction, chloroform-isoamyl alcohol extraction, and ethanol precipitation.
L. rhamnosus was transformed by electroporation with a gene pulser (Bio-Rad Laboratories, Richmond, Calif.) as follows. Cells were grown in 100 ml of MRS supplemented with 2% glycine to an optical density at 600 nm of 0.3 to 0.4 and were harvested by centrifugation at room temperature. The cells were washed twice at room temperature with electroporation buffer (0.5 M sucrose, 7 mM potassium phosphate [pH 7.4], 1 mM MgCl2) (5), resuspended in 1 ml of the same buffer, and placed on ice. A mixture containing 100 μl of cooled cell suspension and 200 ng of DNA was transferred into a precooled electroporation cuvette (with a 0.2-cm electrode gap) and electroporated immediately by using the following settings: 1.5 kV, 25 μF, and 200 Ω. After electroporation, the cells were immediately diluted with 5 ml of MRS containing 2 mM CaCl2 and 20 mM MgCl2 and incubated at 37°C for 3 h before they were plated onto MRS agar containing the appropriate antibiotic.
DNA synthesis.
The oligonucleotides were synthesized with an Applied Biosystems model 392 DNA-RNA synthesizer and were purified by ethanol precipitation or with NAP-10 columns (Pharmacia). For DNA synthesis by PCR amplification, the reaction conditions recommended by the manufacturer of DynaZyme DNA polymerase (Finnzymes) were used. For PCR screening of erythromycin-resistant L. rhamnosus 1/6 colonies after transformation with pVS101, the following pepR-specific primers were used: P1 (5′-GCCATTTGGAGTCGTTACC-3′) and P2 (5′-ATCTCGGCGTTCAAGTCC-3′).
Construction and screening of an L. rhamnosus genomic library.
Chromosomal DNA was partially digested with HindIII, and a 3- to 8-kb fragment pool was selected for a L. rhamnosus genomic library. The L. rhamnosus DNA fragments were ligated into pZErO and transformed into E. coli XL1-Blue by electroporation. The transformant colonies were screened for enzymatic activity against l-proline-β-naphthylamide (Pro-βNA) by using the method originally described by Miller and Mackinnon (30). The colonies expressing Pro-βNA-hydrolyzing activity could be identified with a red, nondiffusible azo dye as a result of the reaction of β-naphthylamine with fast garnet GBC (Sigma).
Nucleotide sequencing and sequence analysis.
Sequencing was performed with a model A.L.F. DNA sequencer (Pharmacia). The dideoxy sequencing reactions (37) were performed by using the methods recommended in the AutoRead sequencing kit manual (Pharmacia). Both DNA strands were sequenced with pUC19-specific primers and sequence-specific oligonucleotides for primer walking. DNA sequences were assembled and analyzed with the PC/GENE set of programs (release 6.85; IntelliGenetics). The PROSITE program of PC/GENE was used to detect specific sites and signatures in protein sequences. Hydropathy analyses were performed by the method of Kyte and Doolittle (29) with the SOAP program of PC/GENE. Protein homology searches were carried out with the SwissProt database by E-mail with the EMBL BLITZ and EMBL FASTA servers.
RNA methods.
Total RNA was isolated from L. rhamnosus cells as described previously (33, 45). RNA gel electrophoresis and Northern blotting were performed as described previously (21). A pepR-specific 0.7-kb PCR fragment, which was synthesized with primers P1 and P2, was used as a probe in Northern hybridizations. The 0.4-kb NcoI-BamHI fragment of pVS98 was used as an ORF2-specific probe. Hybridization probes were labeled with digoxigenin-dUTP, and a digoxigenin luminescent detection kit (Boehringer Mannheim) was used to detect hybrids. Primer extension of pepR was performed with total RNA by using a model A.L.F. DNA sequencer as described previously (32, 44) and 10 pmol of fluorescein-labeled oligonucleotide 5′-ACGACCCGAGTTGATCGTAC-3′, which was complementary to nucleotides at positions 285 to 304 (Fig. 1).
FIG. 1.
Nucleotide and deduced amino acid sequences of L. rhamnosus pepR. The predicted −35 and −10 hexanucleotides are underlined. The 5′ end of the pepR transcript, identified by primer extension, is indicated by a vertical arrow. RBS is the predicted ribosome binding site. The translation stop codon is indicated by boldface type, and the putative transcription terminator is indicated by dashed arrows. The conserved residues of the active site region of prolyl oligopeptidases are indicated by boldface type and asterisks.
Peptidase activity assays.
The peptidase activities in L. rhamnosus and E. coli were determined with liquid cultures as follows. Cells were harvested by centrifugation at 7 000 × g for 15 min, washed once with sterile 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.0), resuspended in one-third the original volume of the same buffer, and then sonicated on ice with a model Ultrasonic 2000 sonicator (B. Braun) by using 10-s bursts until more than 90% of the cells were disrupted. The proline-liberating activity against di- and tripeptides was determined by using a modification of the method of Troll and Lindsley (41) as described by Baankreis and Exterkate (6). Hydrolysis of peptides that did not contain proline as the amino-terminal residue was assayed by using the Cd-ninhydrin method as described by Doi et al. (10). Hydrolysis of chromogenic β-naphthylamide substrates was studied by performing a plate assay with E. coli CM89 as described previously (24, 26).
Construction of a pepR mutant of L. rhamnosus 1/6.
A deletion in pepR was prepared by removing the internal 0.2-kb NdeI-ClaI fragment from the 1.5-kb SacI insert of pVS99 (Fig. 2). An integration vector was constructed by introducing the SacI fragment with an internal deletion into plasmid pLS19, which is a nonreplicative plasmid in L. rhamnosus. The resulting construct, designated pVS101, did not express prolinase activity in E. coli (data not shown). The replacement recombination technique (7, 17) was used to replace the pepR on the chromosome of L. rhamnosus 1/6 with a pepR gene containing the internal deletion.
FIG. 2.
Partial restriction map of the L. rhamnosus 1/6 pepR region. The positions and orientations of pepR and ORF2 are indicated by arrows. The 3.5- and 1.5-kb inserts of plasmids pVS98 and pVS99, respectively, are shown below the map.
Nucleotide sequence accession number.
The nucleotide sequence of pepR has been deposited under EMBL and GenBank accession no. AJ003247.
RESULTS
Cloning of the prolinase gene from L. rhamnosus 1/6.
A genomic library was constructed by inserting HindIII fragments of L. rhamnosus 1/6 chromosomal DNA into the pZErO vector. The ligation mixture was used to transform E. coli XL1-Blue cells. A total of 2 of the 3,000 Zeocin-resistant transformant colonies screened turned red in the enzymatic plate assay performed with Pro-βNA as the substrate. Restriction analysis revealed that the two clones carried plasmids with overlapping inserts that were 3.5 and 8 kb long (data not shown). The plasmid with the 3.5-kb insert was designated pVS98 and was used for further characterization. Subcloning of a 1.5-kb SacI fragment of pVS98 in pZErO resulted in plasmid pVS99, which still expressed Pro-βNA-hydrolyzing activity. Both pVS98 and pVS99 were used for sequencing. A partial restriction map of the 3.5-kb HindIII fragment of pVS98 is shown in Fig. 2.
Nucleotide and amino acid sequence analyses.
DNA sequencing of the 3.5-kb HindIII-fragment revealed two open reading frames (ORF1 and ORF2), which were 903 and 459 bp long, respectively. ORF1 had the capacity to encode a 301-amino-acid protein with a calculated molecular mass of 34.2 kDa. A putative promoter region (TTGTCA-15 nucleotides-TACAAT) and a ribosome binding site (AAGGTG) were found 30 and 5 nucleotides upstream of the start codon (ATG), respectively (Fig. 1). An inverted repeat structure with a ΔG of −81 kJ/mol was detected six nucleotides downstream of the stop codon; this structure represents a putative transcription terminator.
The predicted amino acid sequence encoded by ORF1 exhibited 68% identity with the PepR of L. helveticus (12, 43). At the nucleic acid level the corresponding level of identity was 67%. A search for homologous proteins revealed a level of amino acid sequence identity of 28% between the ORF1-encoded protein and proline iminopeptidases from L. helveticus and L. delbrueckii (4, 25, 42). The amino acid sequence of PepR includes the conserved amino acid residues (GQSWGG) of the active-site region typical of prolyl oligopeptidases (35); identical sequences occur in L. helveticus PepR (12, 43) and L. helveticus and L. delbrueckii PepIs (4, 25, 42). Further analysis of the putative pepR-encoded protein with the PC/GENE set of programs revealed that the protein did not possess any transmembrane or membrane-associated helices or hydrophobic segments likely to be part of a signal peptide, which suggested that PepR is a cytoplasmic protein. A search for proteins homologous to the protein encoded by ORF2 in the SwissProt data bank revealed no significant similarities.
Substrate specificity of L. rhamnosus PepR.
Enzyme activity assays performed with E. coli CM89(pVS99) lysates revealed an activity that liberated proline from the dipeptides Pro-Ala, Pro-Ile, Pro-Leu, Pro-Phe, and Pro-Val but not from the dipeptide Phe-Pro or the tripeptide Pro-Gly-Gly. The cloned pepR also expressed activity against Met-Ala, Leu-Leu, and Leu-Gly-Gly. In addition to activity against Pro-βNA, pVS99 expressed l-phenylalanine-β-naphthylamide (Phe-βNA)- and l-leucine-β-naphthylamide (Leu-βNA)-hydrolyzing activities in E. coli CM89 when the enzymatic plate assay was used.
mRNA analyses.
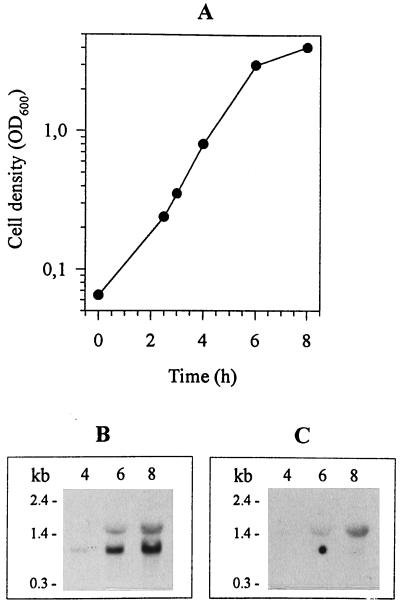
The size of the pepR mRNA was analyzed by using exponentially growing L. rhamnosus cells, Northern blotting, and the pepR-specific 0.7-kb PCR fragment as the hybridization probe. The probe detected a 1.0-kb transcript and also a 1.5-kb transcript (Fig. 3). The size of the 1.0-kb transcript is consistent with the size of the pepR gene sequenced. Northern hybridization performed with the 0.4-kb NcoI-BamHI fragment of ORF2 as the probe resulted in detection of a 1.5-kb transcript that comigrated exactly like the longer transcript detected with the pepR-specific probe. The size of the 1.5-kb transcript is consistent with the size of a read-through transcript containing pepR and ORF2 in the same mRNA. So far, it is not known whether the function of the protein encoded by ORF2 is related to PepR activity. Primer extension mapping of the 5′ end of the pepR mRNA from exponentially growing cells indicated that the transcription initiation site of the pepR gene is located 24 nucleotides upstream of the start codon (data not shown). Thus, mapping of the 5′ end of the pepR transcript confirmed the location of the promoter region predicted on the basis of the DNA sequence. These mRNA analyses revealed that the L. rhamnosus pepR gene is expressed both as a monocistronic transcriptional unit and as a dicistronic transcriptional unit.
FIG. 3.
Expression of the pepR gene and ORF2. (A) L. rhamnosus 1/6 grown in MRS at 37°C. The cell density is shown as a function of growth. OD600, optical density at 600 nm. (B) Northern blot analysis performed with the 0.7-kb pepR-specific hybridization probe. (C) Northern blot analysis performed with the 0.4-kb ORF2-specific hybridization probe. Samples for total RNA isolation were taken 4, 6, and 8 h after cell inoculation. The numbers on the left indicate the sizes of RNA molecular mass markers (Gibco BRL).
Construction and analyses of a chromosomal pepR deletion mutant.
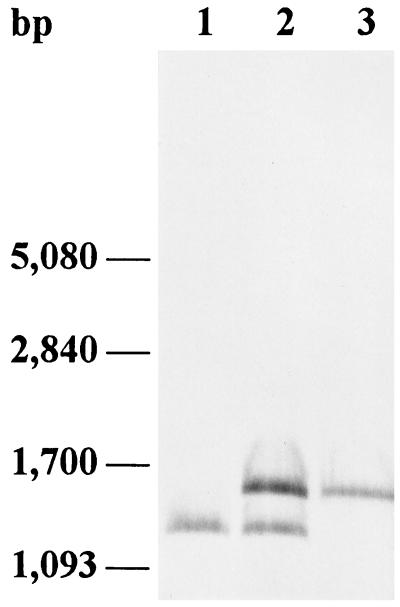
To investigate the function of PepR in L. rhamnosus 1/6, a deletion was introduced into the chromosomal pepR gene by replacement recombination. After transformation of L. rhamnosus 1/6 with pVS101, which includes an erythromycin resistance gene and a pepR gene with an internal 227-bp deletion, the erythromycin-resistant colonies were examined by PCR performed with primers P1 and P2 (data not shown). Integration of pVS101 into the chromosome of L. rhamnosus 1/6 resulted in 661- and 434-bp PCR products, which corresponded to an intact version and a deleted version of pepR. One of the integrants (1/6::pVS101) was used in additional experiments and was analyzed by Southern hybridization with a pepR-specific probe (Fig. 4). We established that excision of the integrated plasmid occurred after nonselective growth for approximately 105 generations in MRS. Cells were plated onto MRS agar, and colonies were replica plated onto MRS agar with and without erythromycin (5 μg/ml). Erythromycin-sensitive colonies were examined by PCR performed with primers P1 and P2 (data not shown), and from one of the colonies examined, only the 434-bp fragment was amplified, indicating that the original intact pepR gene had been excised from the L. rhamnosus 1/6 genome (data not shown). Chromosomal DNA from this strain was isolated, digested with SacI, and analyzed by Southern hybridization (Fig. 4). It was predicted that strain 1/6ΔpepR carrying only the deleted version of pepR contained one SacI fragment (1,284 bp), whereas it was predicted that strain 1/6::pVS101 contained two fragments (1,511 and 1,284 bp) which hybridized with a pepR-specific probe. It was predicted that a hybridization signal corresponding to a 1,511-bp fragment would be obtained from the wild-type strain. The presence of only the 1,284-bp fragment in 1/6ΔpepR (Fig. 4) suggests that a crossover event resulted in excision of pLS19 and the wild-type pepR gene, leaving only the deleted version of pepR. The pepR deletion was complemented with plasmid pVS102, which was constructed by ligating the 1.5-kb SacI fragment of pVS98 containing the pepR gene into pIL252. Plasmid pVS102 was electroporated into L. rhamnosus 1/6ΔpepR and into the wild-type strain.
FIG. 4.
Analysis of L. rhamnosus 1/6 with a 227-bp chromosomal deletion in the pepR gene by Southern hybridization with a pepR-specific probe. Lanes 1, L. rhamnosus 1/6ΔpepR chromosomal DNA digested with SacI; 2, L. rhamnosus 1/6::pVS101 chromosomal DNA digested with SacI; 3, L. rhamnosus 1/6 chromosomal DNA digested with SacI.
PepR activity against Pro-Leu was determined with cell extracts of L. rhamnosus 1/6 and 1/6ΔpepR and their derivatives grown in whey medium (Table 2). The cell extract from strain 1/6ΔpepR was found to have 5% of the Pro-Leu-hydrolyzing activity of the wild-type strain. The effect of the prolinase deficiency of the mutant strain on the ability of this strain to grow in whey medium and milk was examined. There was no difference in growth rate or acid production between 1/6 and 1/6ΔpepR (data not shown).
TABLE 2.
Pro-Leu-hydrolyzing activities of cell extracts from L. rhamnosus 1/6 and its derivatives
| Strain | Relative activity (%)a |
|---|---|
| 1/6 | 100 |
| 1/6ΔpepR | 5 ± 0.5 |
| 1/6(pIL252) | 106 ± 1 |
| 1/6ΔpepR(pIL252) | 7 ± 0.5 |
| 1/6(pVS102) | 325 ± 4 |
| 1/6ΔpepR(pVS102) | 288 ± 4 |
The Pro-Leu-hydrolyzing activity of L. rhamnosus 1/6 was defined as 100%.
DISCUSSION
In this work we cloned the gene encoding prolinase (PepR) in L. rhamnosus by using the chromogenic substrate Pro-βNA to screen a gene library constructed in E. coli. Previously, this substrate has been used to clone a Lactobacillus proline iminopeptidase (pepI) gene (25). The PepR of L. helveticus has been characterized as a relatively broad-specificity dipeptidase with prolinase activity (12, 38, 43). However, in addition to hydrolyzing Pro-βNA, the L. rhamnosus PepR hydrolyzed Leu-βNA and Phe-βNA, which are substrates for aminopeptidases and are not usually cleaved by dipeptidases from lactic acid bacteria (28). Thus, the use of chromogenic substrates to divide peptidases of lactic acid bacteria into different classes is somewhat questionable, and at the least, these substrates are not suitable for distinguishing between Lactobacillus PepI and PepR. On the other hand, the inability of the Lactobacillus PepR to liberate the N-terminal proline from the tripeptide Pro-Gly-Gly distinguishes it from PepI (references 18, 38, 42, and 43 and this study). The suggested serine proteinase nature of L. helveticus PepR (12, 43) has been confirmed by site-directed mutagenesis (38). The active site region of L. helveticus PepR (GQSWGG), which is also found in Lactobacillus PepI proteins (4, 25, 42), is also present in L. rhamnosus PepR (Fig. 1). The PepR of L. helveticus has been purified and biochemically analyzed by Shao et al. (38). This enzyme did not hydrolyze Leu-Gly-Gly. Our results showed that the cloned PepR of L. rhamnosus hydrolyzes Leu-Gly-Gly. Also, the ability of the cloned L. rhamnosus PepR to hydrolyze Pro-βNA, Phe-βNA, and Leu-βNA strongly suggest that the substrate specificity of L. rhamnosus PepR is different from that of L. helveticus PepR. However, this suggestion has yet to be confirmed by purification and biochemical characterization of L. rhamnosus PepR. The pepR gene of the mesophilic organism L. rhamnosus is highly homologous to the pepR gene of the thermophilic organism L. helveticus (12, 43). However, the genetic organizations of the pepR locus are different in these two bacteria. The L. rhamnosus pepR is expressed both as a monocistronic transcriptional unit and as a dicistronic transcriptional unit, whereas L. helveticus pepR is solely a monocistronic transcriptional unit (43). Furthermore, a gene encoding a putative ABC transporter has been located upstream of the L. helveticus pepR and in the opposite orientation (43). No divergent open reading frames were found in the upstream region of the pepR gene described in this paper.
In order to analyze the significance of PepR during growth of L. rhamnosus 1/6 in milk, the gene encoding this peptidase was inactivated. There were no differences in growth rate or acid production between the wild-type and mutant strains. Apparently, PepR is not necessary for the liberation of essential amino acids from casein. This is consistent with results obtained from inactivation of L. helveticus pepR (38). So far, the peptidase genes pepDA, pepX, pepC, pepN, and pepR have been inactivated as single mutations from L. helveticus (9, 11, 38, 46), and only the pepX gene has been shown to be necessary for optimum growth of L. helveticus in milk (46). In lactococci a single mutation of the dipeptidase gene pepV led to a significantly reduced growth rate in milk (22).
Two enzymes, PepR and PepI, that effectively hydrolyze Pro-Leu and other Pro-X dipeptides have been obtained and characterized from lactic acid bacteria (18, 38, 42; this study). Inactivation of the pepR gene abolished most (95%) of the Pro-Leu-hydrolyzing activity of L. rhamnosus, indicating either that the level of PepI activity is low or that the pepI gene is not present in L. rhamnosus. However, a peptidase with PepI activity has been purified from a closely related L. casei strain (20). It has been demonstrated that both genes are present in L. helveticus (12, 42, 43), and inactivation of PepR still resulted in a strain with only 6.7% of the Pro-Leu-hydrolyzing activity of the wild-type strain (38). These results suggest that hydrolysis of dipeptides with N-terminal proline residues by members of the genus Lactobacillus is performed primarily by PepR under the growth conditions used.
ACKNOWLEDGMENTS
We are grateful to Anneli Virta for the running the A.L.F. sequencer and to Juha Laukonmaa and Tuula Vähäsöyrinki for technical assistance.
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora G, Lee B H. Purification and characterization of aminopeptidase from Lactobacillus casei subsp. casei LLG. J Dairy Sci. 1992;75:700–710. [Google Scholar]
- 3.Arora G, Lee B H. Purification and characterization of an aminopeptidase from Lactobacillus casei subsp. rhamnosus S93. Biotechnol Appl Biochem. 1994;19:179–192. [PubMed] [Google Scholar]
- 4.Atlan D, Gilbert C, Blanc B, Portalier R. Cloning, sequencing and characterization of the pepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397. Microbiology. 1994;140:527–535. doi: 10.1099/00221287-140-3-527. [DOI] [PubMed] [Google Scholar]
- 5.Aymerich M T, Hugas M, Garriga M, Vogel R F, Monfort J M. Electrotransformation of meat lactobacilli. Effect of several parameters on their efficiency of transformation. J Appl Bacteriol. 1993;75:320–325. [Google Scholar]
- 6.Baankreis R, Exterkate F. Characterization of a peptidase from Lactococcus lactis ssp. cremoris HP that hydrolyses di- and tripeptides containing proline or hydrophobic residues as the aminoterminal amino acid. Syst Appl Microbiol. 1991;14:317–323. [Google Scholar]
- 7.Bhowmik T, Fernández L, Steele J L. Gene replacement in Lactobacillus helveticus. J Bacteriol. 1993;175:6341–6344. doi: 10.1128/jb.175.19.6341-6344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromme M C, Krause D A, Hickey M W. The isolation and characterization of lactobacilli from cheddar cheese. Aust J Dairy Technol. 1990;45:60–66. [Google Scholar]
- 9.Christensen J E, Steele J L. Abstracts of the Fifth Symposium on Lactic Acid Bacteria. Glasgow, United Kingdom: Federation of European Microbiological Societies, Publications Office; 1996. Characterization of peptidase-deficient Lactobacillus helveticus CNRZ32 derivatives, abstr. K24. [Google Scholar]
- 10.Doi E, Shibata D, Matoba T. Modified colorimetric ninhydrin methods for peptidase assay. Anal Biochem. 1981;118:173–184. doi: 10.1016/0003-2697(81)90175-5. [DOI] [PubMed] [Google Scholar]
- 11.Dudley E G, Husgen A C, He W, Steele J L. Sequencing, distribution, and inactivation of the dipeptidase A gene (pepDA) from Lactobacillus helveticus CNRZ32. J Bacteriol. 1996;178:701–704. doi: 10.1128/jb.178.3.701-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudley E G, Steele J L. Nucleotide sequence and distribution of the pepPN gene from Lactobacillus helveticus CNRZ32. FEMS Microbiol Lett. 1994;119:41–46. doi: 10.1111/j.1574-6968.1994.tb06864.x. [DOI] [PubMed] [Google Scholar]
- 13.El Abboudi M, El Soda M, Pandian S, Simard R E, Olson N F. Purification of X-prolyl dipeptidyl aminopeptidase from Lactobacillus casei subspecies. Int J Food Microbiol. 1992;15:87–98. doi: 10.1016/0168-1605(92)90138-s. [DOI] [PubMed] [Google Scholar]
- 14.Fernández-Esplá M D, Martín-Hernández M C. Purification and characterization of a dipeptidase from Lactobacillus casei ssp. casei IFPL 731 isolated from goat cheese made from raw milk. J Dairy Sci. 1997;80:1497–1504. doi: 10.3168/jds.S0022-0302(97)76078-8. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Esplá M D, Martín-Hernández M C, Fox P F. Purification and characterization of a prolidase from Lactobacillus casei subsp. casei IFPL 731. Appl Environ Microbiol. 1997;63:314–316. doi: 10.1128/aem.63.1.314-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox P. Proteolysis during cheese manufacture and ripening. J Dairy Sci. 1989;72:1379–1400. [Google Scholar]
- 17.Gasson M J, Fitzgerald G F. Gene transfer systems and transposition. In: Gasson M, De Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Blackie Academic and Professional; 1994. pp. 1–51. [Google Scholar]
- 18.Gilbert C, Atlan D, Blanc B, Portalier R. Proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397: purification and characterization. Microbiology. 1994;140:537–542. doi: 10.1099/00221287-140-3-537. [DOI] [PubMed] [Google Scholar]
- 19.Habibi-Najafi M B, Lee B H. Purification and characterization of X-prolyl dipeptidyl peptidase from Lactobacillus casei subsp. casei LLG. Appl Microbiol Biotechnol. 1994;42:280–286. doi: 10.1007/BF00902729. [DOI] [PubMed] [Google Scholar]
- 20.Habibi-Najafi M B, Lee B H. Purification and characterization of proline iminopeptidase from Lactobacillus casei subsp. casei LLG. J Dairy Sci. 1995;78:251–259. [Google Scholar]
- 21.Hames B, Higgins S. Nucleic acid hybridization: a practical approach. Oxford, United Kingdom: IRL Press; 1985. [Google Scholar]
- 22.Hellendoorn M A, Franke-Fayard B M D, Mierau I, Venema G, Kok J. Cloning and analysis of the pepV dipeptidase gene of Lactococcus lactis MG1363. J Bacteriol. 1997;179:3410–3415. doi: 10.1128/jb.179.11.3410-3415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holck A, Naes H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO151. J Gen Microbiol. 1992;138:1353–1364. doi: 10.1099/00221287-138-7-1353. [DOI] [PubMed] [Google Scholar]
- 24.Klein J R, Klein U, Schad M, Plapp R. Cloning, DNA sequence analysis and partial characterization of pepN, a lysyl aminopeptidase from Lactobacillus delbrueckii subsp. lactis DSM7290. Eur J Biochem. 1993;217:105–114. doi: 10.1111/j.1432-1033.1993.tb18224.x. [DOI] [PubMed] [Google Scholar]
- 25.Klein J R, Schmidt U, Plapp R. Cloning, heterologous expression, and sequencing of a novel proline iminopeptidase gene, pepI, from Lactobacillus delbrueckii subsp. lactis DSM7290. Microbiology. 1994;140:1133–1139. doi: 10.1099/13500872-140-5-1133. [DOI] [PubMed] [Google Scholar]
- 26.Klein J R, Schick J, Henrich B, Plapp R. Lactobacillus delbrueckii subsp. lactis DSM7290 pepG gene encodes a novel cysteine aminopeptidase. Microbiology. 1997;143:527–537. doi: 10.1099/00221287-143-2-527. [DOI] [PubMed] [Google Scholar]
- 27.Kok J, De Vos W M. The proteolytic system of lactic acid bacteria. In: Gasson M, De Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Blackie Academic and Professional; 1994. pp. 169–210. [Google Scholar]
- 28.Kunji E R S, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic system of lactic bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Miller C G, Mackinnon K. Peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974;120:355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller C G, Schwartz G. Peptidase-deficient mutants of Escherichia coli. J Bacteriol. 1978;135:603–611. doi: 10.1128/jb.135.2.603-611.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myöhänen S, Wahlfors J. Automated fluorescent primer extension. BioTechniques. 1993;14:16–17. [PubMed] [Google Scholar]
- 33.Palva A, Nyberg K, Palva I. Quantitation of α-amylase mRNA in Bacillus subtilis by nucleic acid sandwich hybridization. DNA. 1988;7:135–142. doi: 10.1089/dna.1988.7.135. [DOI] [PubMed] [Google Scholar]
- 34.Peterson S D, Marshall R T. Nonstarter lactobacilli in cheddar cheese: a review. J Dairy Sci. 1990;73:1395–1410. [Google Scholar]
- 35.Rawlings N, Polgar L, Barret A. A new family of serine type peptidases related to prolyl oligopeptidase. Biochem J. 1991;279:907–911. doi: 10.1042/bj2790907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao W, Yüksel G, Dudley E G, Parkin K L, Steele J L. Biochemical and molecular characterization of PepR, a dipeptidase, from Lactobacillus helveticus CNRZ32. Appl Environ Microbiol. 1997;63:3438–3443. doi: 10.1128/aem.63.9.3438-3443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Bichimie. 1988;70:559–567. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 40.Tilsala-Timisjärvi A, Alatossava T. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int J Food Microbiol. 1997;35:49–56. doi: 10.1016/s0168-1605(97)88066-x. [DOI] [PubMed] [Google Scholar]
- 41.Troll W, Lindsley J. A photometric method for the determination of proline. J Biol Chem. 1955;215:655–661. [PubMed] [Google Scholar]
- 42.Varmanen P, Rantanen T, Palva A. An operon from Lactobacillus helveticus composed of a proline iminopeptidase gene (pepI) and two genes coding for putative members of the ABC transporter family of proteins. Microbiology. 1996;142:3459–3468. doi: 10.1099/13500872-142-12-3459. [DOI] [PubMed] [Google Scholar]
- 43.Varmanen P, Steele J, Palva A. Characterization of prolinase gene and its product and an adjacent ABC transporter gene from Lactobacillus helveticus. Microbiology. 1996;142:809–816. doi: 10.1099/00221287-142-4-809. [DOI] [PubMed] [Google Scholar]
- 44.Vesanto E, Savijoki K, Rantanen T, Steele J L, Palva A. An X-prolyl dipeptidyl aminopeptidase (pepX) gene from Lactobacillus helveticus. Microbiology. 1995;141:3067–3075. doi: 10.1099/13500872-141-12-3067. [DOI] [PubMed] [Google Scholar]
- 45.Vesanto E, Varmanen P, Steele J L, Palva A. Characterization and expression of the Lactobacillus helveticus pepC gene encoding a general aminopeptidase. Eur J Biochem. 1994;224:991–997. doi: 10.1111/j.1432-1033.1994.00991.x. [DOI] [PubMed] [Google Scholar]
- 46.Yüksel G, Steele J L. DNA sequence analysis, expression, distribution and the physiological role of the Xaa-prolyldipeptidyl aminopeptidase gene from Lactobacillus helveticus CNRZ32. Appl Microbiol Biotechnol. 1996;44:766–773. doi: 10.1007/BF00178616. [DOI] [PubMed] [Google Scholar]