This study demonstrates that mRNA targeting and local translation at the midbody play an important role in regulating abscission during cytokinesis. It also identifies localization elements within 3′UTR on midbody-associated mRNAs as mediators of mRNA targeting the midbody.
Abstract
Midbodies function during telophase to regulate the abscission step of cytokinesis. Until recently, it was thought that abscission-regulating proteins, such as ESCRT-III complex subunits, accumulate at the MB by directly or indirectly binding to the MB resident protein, CEP55. However, recent studies have shown that depletion of CEP55 does not fully block ESCRT-III targeting the MB. Here, we show that MBs contain mRNAs and that these MB-associated mRNAs can be locally translated, resulting in the accumulation of abscission-regulating proteins. We demonstrate that localized MB-associated translation of CHMP4B is required for its targeting to the abscission site and that 3′ UTR-dependent CHMP4B mRNA targeting to the MB is required for successful completion of cytokinesis. Finally, we identify regulatory cis-elements within RNAs that are necessary and sufficient for mRNA trafficking to the MB. We propose a novel method of regulating cytokinesis and abscission by MB-associated targeting and localized translation of selective mRNAs.
Introduction
The cell cycle is a key event during the development, growth, and reproduction of all organisms. Cytokinesis is the final stage of the cell cycle that results in the physical separation of two daughter cells. The mother cell divides by the formation of the cleavage furrow, leaving two daughter cells connected by a microtubule-rich intercellular bridge. Additionally, during the ingression of the cleavage furrow, the central spindle microtubules are compacted to form a structure known as the midbody (MB). To complete cytokinesis, the intercellular bridge must be cleaved on one or both sides of the MB via the process known as the abscission (Fig. 1 A). Abscission is a highly regulated event and it is now well established that the MB plays a key role in regulating the completion of cytokinesis by recruiting and activating abscission-mediating proteins, such as the endosomal sorting complex required for transport (ESCRT-III) complex, as well as several regulators of the abscission checkpoint (Carlton, 2010; Schöneberg et al., 2017). While targeting and activation of abscission regulators at the MB is required for the completion of cytokinesis, the mechanisms governing this process remain to be fully defined.
Figure 1.
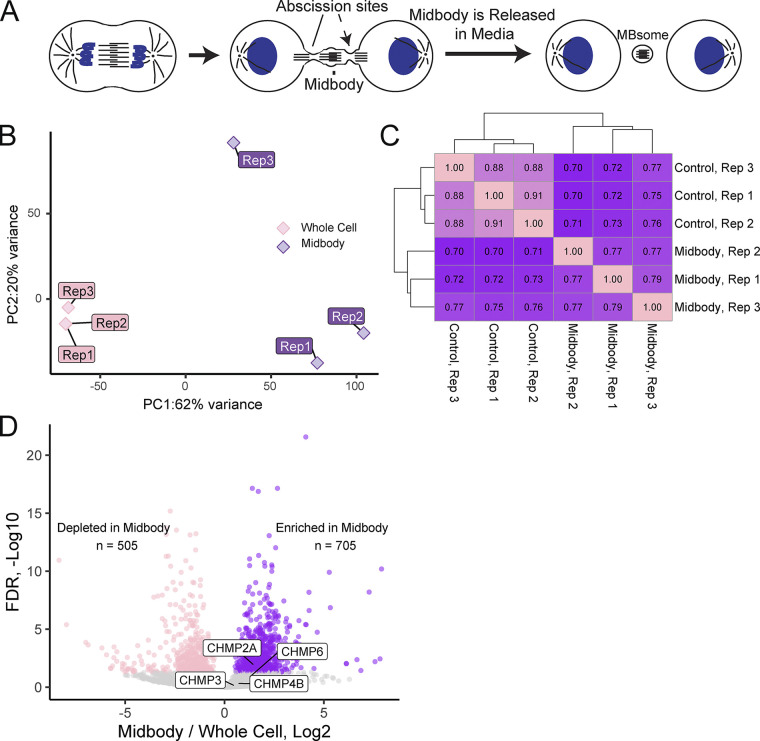
MBsomes contain a distinct subset of mRNAs. (A) Schematic representing MB formation and MBsome release into the media. MBsomes from the media were isolated for RNAseq analysis presented in this study. (B) Principal component analysis representing the difference between the three control and the three MB RNAseq datasets. (C) Correlation analysis representing the correlation between the various control and MB RNAseq datasets. (D) Volcano plot showing the significantly depleted (pink) and significantly enriched (purple) mRNAs found within MBsomes from three RNAseq datasets.
Previous studies have shown that several members of the ESCRT-III machinery are recruited to the MB just before initiation of the abscission (Carlton and Martin-Serrano, 2007; Morita et al., 2007; Carlton et al., 2008; Guizetti et al., 2011). Specifically, it was shown that TSG101 recruits charged multivesicular body proteins (CHMPs, components of the ESCRT-III complex) to the MB during telophase (Goliand et al., 2014). In addition to TSG101, ALIX (also known as PDCD6IP) also contributes to the direct recruitment of the ESCRT-III complex to the MB (Carlton, 2010; Morita et al., 2007; Addi et al., 2020). Upon recruitment to the MB, the ESCRT-III components oligomerize (CHMP4B and CHMP2A or CHMP3 and CHMP6) into spiral structures with progressively smaller diameters toward the abscission site (Elia et al., 2011; Goliand et al., 2018; Christ et al., 2017), a process that is thought to be responsible for the eventual abscission of the intercellular bridge to complete cytokinesis and form two daughter cells (Guizetti et al., 2011; Elia et al., 2011; Goliandi et al., 2018). Inhibiting MB recruitment of ESCRT-III results in abscission defects, demonstrating the importance of appropriately regulating ESCRT-III protein accumulation and activation at the MB (Carlton et al., 2008; Guizetti et al., 2011; Agromayor et al., 2009; Christ et al., 2016; Morita et al., 2010; Carlton et al., 2012).
Until recently, it has been thought that TSG101 accumulates at the MB by binding to the MB resident protein, CEP55 (Carlton and Martin-Serrano, 2007; Morita et al., 2007; Guizetti et al., 2011; Elia et al., 2011; Goliand et al., 2018; Mierzwa and Gerlich, 2014; Addi et al., 2018). This binding was presumed to tether the TSG101 at the MB, gradually leading to ESCRT-III accumulation before initiation of abscission. However, recent studies have shown that depletion of CEP55 does not fully block ESCRT-III targeting to the MB, and CEP55 knock-out mice are able to grow and develop (Tedeschi et al., 2020). Finally, CEP55 is a protein present only in vertebrates; thus, while CEP55 clearly contributes to ESCRT-III recruitment to the MB in vertebrates, it does not appear to be absolutely required for abscission. Indeed, some studies suggested that other mechanisms may also mediate ESCRT-III recruitment. For example, it has been shown that centralspindlin may also mediate this process, at least in Drosophila embryos (Lie-Jensen et al., 2019).
Recent work suggested that in addition to regulating abscission, secreted post-mitotic MBs (also known as MBsomes, MB remnants, or Flemmingsomes) can be internalized by other cells where they regulate cell proliferation and differentiation (Addi et al., 2020; Dionne et al., 2015; Kuo et al., 2011; Crowell et al., 2014; Chen et al., 2013; Peterman and Prekeris, 2019; Rai et al., 2021). It was proposed that these MBsomes may mediate cell-to-cell transfer of various signaling molecules in a manner similar to exosomes and extracellular vesicles (Peterman and Prekeris, 2019; Rai et al., 2021). Consequently, several laboratories, including ours, have analyzed the proteome of secreted MBsomes. Interestingly, these studies have shown that, in addition to various signaling proteins, MBs are also enriched in ribosomal and translation initiation factor proteins (Addi et al., 2020; Peterman and Prekeris, 2019; Rai et al., 2021; Skop et al., 2004), raising an intriguing possibility that MB-associated mRNA targeting and the localized translation may mediate various mitotic and post-mitotic MB functions.
In this study, we show, for the first time, that a specific subset of mRNAs is indeed targeted to and enriched at the MBs. We also demonstrate that MBs contain active ribosomes and that MB-associated protein translation is needed for the successful completion of the abscission. Importantly, by using a combination of RNAseq, qPCR, and smFISH, we demonstrate that mRNA encoding ESCRT-III components, specifically CHMP4B, CHMP2A, CHMP3, and CHMP6, are all enriched at the MB, and that MB-associated translation is needed for CHMP4B accumulation at the MB during telophase. Additionally, the characterization of CHMP4B mRNA revealed that the 3′ UTR is critical for efficient CHMP4B mRNA targeting to the MB. Finally, we identify GA-rich localization elements within 3′ UTR that are necessary and sufficient to target mRNA to the MB. Collectively, this study proposes a novel mechanism for regulating cytokinesis and abscission by MB-associated targeting and localized translation of selective mRNAs that include, but are not limited to, the members of ESCRT-III complex. These findings lay a foundation for future studies in determining the molecular mechanisms regulating mRNA targeting and translation at the MB and future analyses of the roles that this mRNA targeting plays in regulating abscission and post-mitotic MBsome functions.
Results
Identification of midbody-associated mRNAs
Recent studies have shown that MBs are enriched in RNA-binding proteins (Addi et al., 2020; Peterman and Prekeris, 2019; Rai et al., 2021; Skop et al., 2004), leading to the intriguing idea that MBs may contain mRNAs and that local translation at the MB may regulate various MB functions. To test this hypothesis, we first set out to determine whether specific mRNAs actually accumulate at the MBs and began by isolating MBsomes from tissue culture media as previously described (Fig. 1 A and Fig. S2 A; Peterman et al., 2019). This type of MB purification has previously been used in at least three different laboratories, including ours, and has been shown to be distinct from conventional exosome and microparticle preparations (Peterman et al., 2019; Addi et al., 2020; Rai et al., 2021). We then prepared oligo-dT-based mRNA libraries from MBsomes and matched whole-cell RNA samples, analyzing them using RNAseq (Table S1). To assess the reproducibility of the RNA content of our MBsome preparations and their relationship to whole-cell RNA samples, we used principal components analysis and hierarchical clustering (Fig. 1, B and C). With both approaches, we found that the MB RNA samples were highly similar to each other, indicating the reproducible nature of their RNA contents. Importantly, they were well separated from the whole-cell RNA samples, suggesting that the MB RNA samples contained an mRNA composition distinct from that found in the whole-cell samples. Overall, the RNAseq analysis identified 16,922 mRNAs that can be detected in the MBsome. From those, 505 mRNAs were significantly depleted and 705 mRNAs were significantly enriched in the MBsomes as compared to the whole-cell RNA samples (Fig. 1 D and Table S1).
Figure S2.
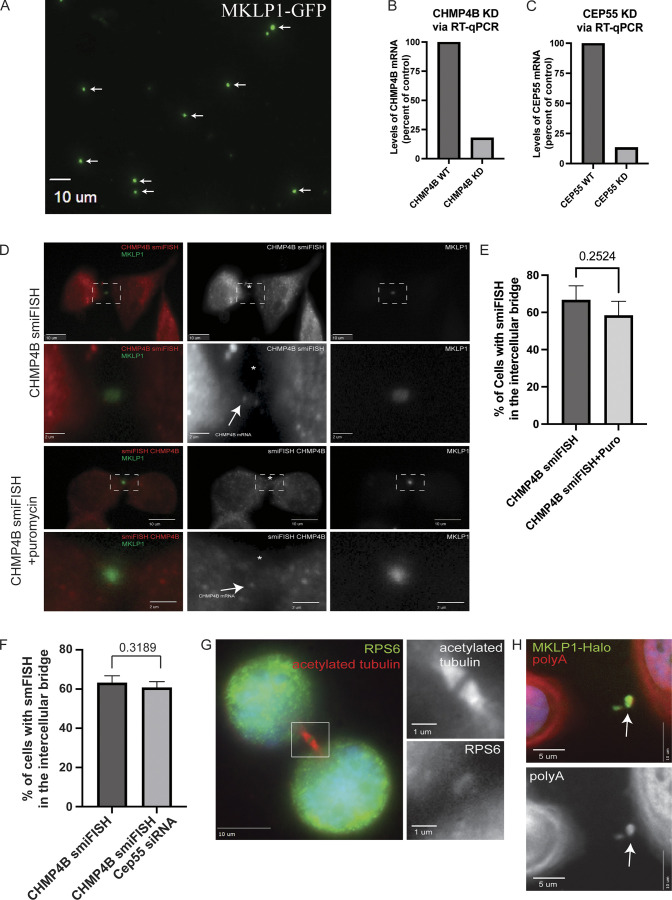
Validation of CHMP4B mRNA localization at the MB. (A) Midbodies were purified from HeLa cells stably expressing GFP-MKLP1. Purified midbodies were plated on poly-L-lysine coated coverslips and analyzed by fluorescence microscopy. Arrows point to GFP-MKLP1 positive midbodies. Scale bar: 10 μm. (B and C) GFP-MKLP1 expressing HeLa cells were either mock-treated or treated with CHMP4B (B) or CEP55 (C) siRNAs. Cells were then incubated for 72 h, followed by mRNA isolation and RT-qPCR analysis using CHMP4B, CEP55, and GAPDH (control) specific primers. Data shown are the CHMP4B or CEP55 mRNA levels normalized against GAPDH mRNA derived from one qPCR analysis. (D and E) GFP-MKLP1 expressing HeLa cells were either mock treated (top two panels) or treated with puromycin for 1 h (bottom two panels) and subjected to staining with smFISH probes against CHMP4B mRNA. CHMP4B mRNA particles are marked by the arrows in the inset and the black dashed square represents the region of the image used for the inset (D). Scale bars: 10 μm. Inset scale bars: 2 μm. The number of cells with CHMP4B smFISH signal was then counted. The data shown in E represents the means and SD derived from three independent experiments. (F) GFP-MKLP1 expressing HeLa cells were either mock treated or treated with CEP55 siRNA for 72 h. Cells were then fixed and subjected to staining with smFISH probes against CHMP4B mRNA. The number of cells with CHMP4B smFISH signal in the MB were then counted and expressed as means and SD derived from three independent experiments. (G) HeLa cells were fixed and stained with anti-RPS6 and anti-acetylated-tubulin antibodies. Box in the image on the left indicates areas that are shown in zoomed-in images on the right. Scale bars: 10 μm. Inset scale bars: 1 μm. (H) HeLa cells expressing MKLP1-Halo (green) were fixed and stained with polyA probes (red). Arrows point to the midbody. Scale bars: 5 μm.
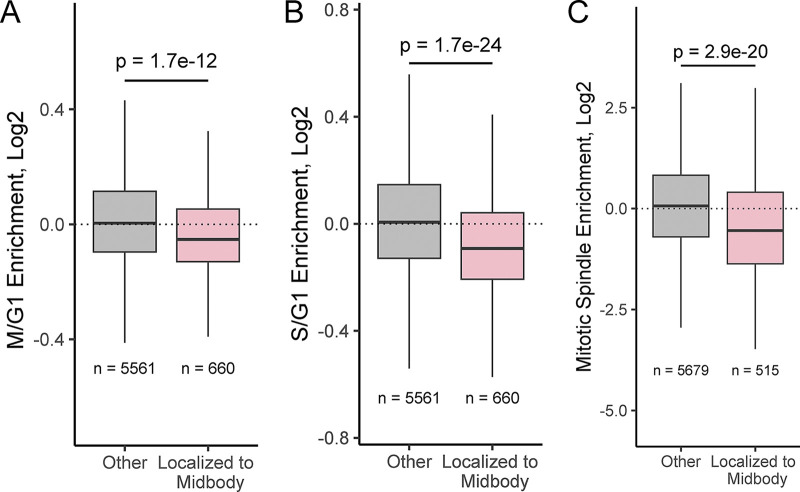
MBsomes are produced exclusively by mitotic cells. Our observed MBsome-enriched RNAs, then, could therefore simply reflect the RNA content of cells that produced MBsomes (i.e., cells undergoing mitosis) rather than RNAs specifically trafficked to the midbody. To assess whether MBsome RNAs detected by RNAseq reflected MBsome-localized RNAs or the general RNA content of mitotic cells, we compared our data to RNAseq data from unsynchronized HeLa cells sorted for cell cycle phase using Fucci reporter expression (Boström et al., 2017). For all RNAs, we calculated their relative enrichment in cells in the G2/M phase compared with cells in G1. We found that our MBsome-enriched RNAs were significantly less abundant in G2/M cells compared with G1 cells (Fig. S1 A). Similarly, MBsome-localized RNAs were less abundant in cells in the S phase compared to G1 cells (Fig. S1 B). From these data, we conclude that RNA enrichments in MBsomes are not due to their increased abundance in mitotic cells.
Figure S1.
Comparative analysis of MB-Associated mRNAs. (A) Comparison of MB-associated mRNA with RNA contents of HeLa cells in G2/M and G1 cell cycle phases. (B) Comparison of MB-associated mRNA with RNA contents of HeLa cells in S and G1 cell cycle phases. (C) Comparison of MB-associated mRNA with spindle-associated RNA.
Given the MB’s connection to the mitotic spindle, we also wondered whether MBsome-localized RNAs were generally localized to the mitotic spindle or were specific to the MB. To test this, we analyzed RNAseq data in which spindle-associated RNAs were identified through differential centrifugation (Hassine et al., 2020). We found that MBsome-localized RNAs were not spindle enriched, indicating that these two RNA populations are likely distinct (Fig. S1 C).
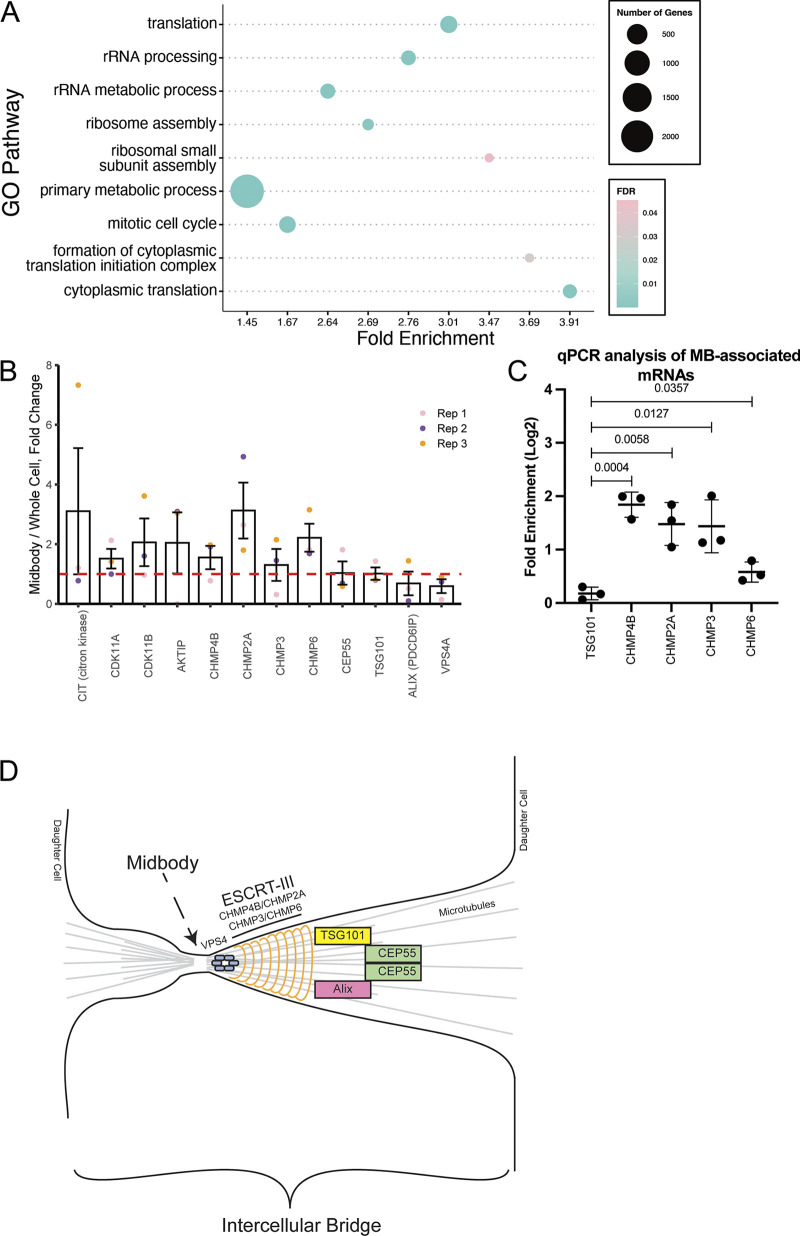
To determine the possible function of these MB-localized mRNAs, we performed a Gene Ontology (GO) Enrichment Analysis (Fig. 2 A) and found that mRNAs encoding for proteins that regulate cell cycle and microtubule dynamics are enriched at the MBsomes (Fig. 2 A). We also determined that mRNAs of several abscission-regulators, such as Citron Kinase, CDK11A, CDK11B, and AKTIP, are also enriched in the MBsomes (Fig. 2 B). Importantly, we found that mRNAs of the ESCRT-III complex proteins, CHMP4B, 2A, 3, and 6, were all enriched at the MBsome (Fig. 2, B–D). This is especially noteworthy as CHMP4B, CHMP2A, CHMP3, and CHMP6 are known to copolymerize and drive the final abscission step (Fig. 2 D; Elia et al., 2011; Goliand et al., 2018; Christ et al., 2017), suggesting that localized translation of ESCRT-III proteins may drive their accumulation at the MB and may also be required for the abscission step of cytokinesis.
Figure 2.
MBsomes are enriched in mRNAs encoding proteins that regulate translation and cell cycle. (A) The Gene Ontology (GO) pathway analysis tool was used to determine the pathways associated with the enriched MB-associated mRNAs. (B) MB enrichment of mRNAs encoding known abscission regulators. Data shown present means and SD derived from three separate RNAseq analyses. Dashed line marks the mRNA levels present in the whole cell transcriptome. (C) Whole-cell or MBsome-associated mRNAs were isolated from GFP-MKLP1-expressing HeLa cell line. The cDNA was subject to RT-qPCR using TSG101, CHMP4B, CHMP2A, CHMP3, CHMP6, or GAPDH (control) specific primers and represented as normalized values. The means and SD were calculated from three independent experimental repeats. Statistical analysis is represented with a P value. (D) Schematic representing key regulators of abscission and how they relate to each other in the intercellular bridge.
Interestingly, not all mRNAs of genes associated with abscission were found to be enriched at the MBsomes. For example, CEP55, TSG101, ALIX, and VPS4 mRNAs, while present in MBsomes, were not enriched as compared to the whole-cell transcriptome (Fig. 2, B–D). This suggests that only a subset of abscission-regulating proteins may be controlled by local translation. All ESCRT-III components are recruited during late telophase, just before completion of the abscission, suggesting an intriguing possibility that the MB targeting of proteins during late telophase may depend on localized translation of specific mRNAs after MB formation. In the rest of the study, we will test this hypothesis by focusing on the machinery mediating MB targeting and translation of CHMP4B mRNA.
Midbodies contain the molecular machinery required for protein translation
Our GO analysis (Fig. 2 A) demonstrated that numerous MB-associated mRNAs encode proteins that regulate ribosomal assembly and function. This further supports the idea that MBs contain ribosomal machinery, which in turn can mediate local translation. Due to this finding, and the fact that CHMP4B mRNA is enriched in the MBs, we hypothesized that local translation of CHMP4B and other ESCRT-III subunits may contribute to ESCRT-III accumulation at the MB, therefore regulating the abscission. For this hypothesis to be correct, MBs must contain CHMP4B mRNA and translation-associated ribosomal machinery, as well as be sites of active protein translation.
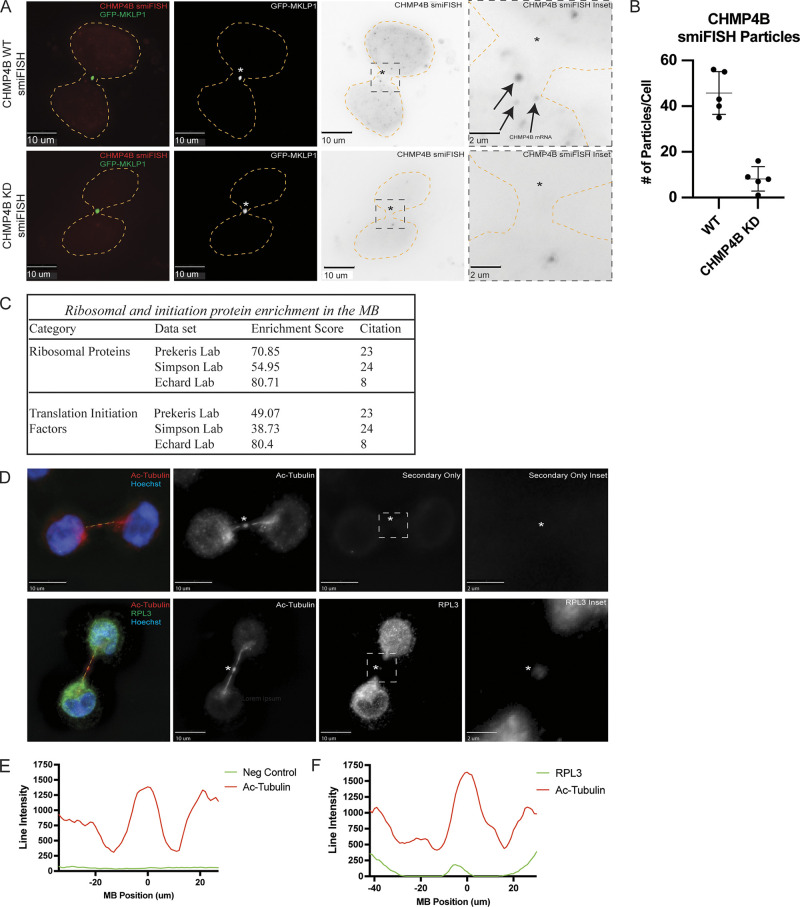
To test this idea, we first visualized CHMP4B mRNA using single-molecule inexpensive RNA in situ hybridization (smiFISH; Tsanov et al., 2016). Consistent with our hypothesis that CHMP4B may be translated at the MB, we found that CHMP4B mRNA is present within the intercellular bridge during telophase (see arrows in Fig. 3 A) in over 60% of dividing HeLa cells (Fig. S2, D and E). To validate the specificity of our CHMP4B smFISH probes, we depleted CHMP4B RNA using siRNA and observed a corresponding decrease in the number of observed CHMP4B RNA molecules per cell (Fig. 3 B).
Figure 3.
Midbodies contain molecular machinery required for protein translation. (A) GFP-MKLP1 expressing HeLa cells were either mock-treated or treated with CHMP4B siRNA for 72 h and subjected to staining with smFISH probes against CHMP4B. Arrows in the inset point to CHMP4B mRNA particles. The cell outline is marked in yellow with the asterisks marking the MB. The black dashed square represents the region of the image used for the inset. Scale bars: 10 μm. (B) ImageJ was used to count the number of CHMP4B smFISH particles in the mock or CHMP4B siRNA-treated cells. Each dot represents a single cell. The means and SD were calculated from five randomly chosen cells in telophase. (C) The Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used to determine whether ribosomal proteins and translation initiation factors are enriched in all three published MB proteomes. (D) HeLa cells were fixed and subjected to immunostaining with anti-acetylated-tubulin (red) and anti-RPL3 (green) antibodies. The line used for the line intensity graphs is marked in yellow and the asterisks mark the MB. The white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (E and F) Line intensity graphs representing the intensity of acetylated-tubulin (red) and RPL3 (green). Negative control was derived from samples that were treated with secondary antibodies but without anti-RPL3 antibodies.
There are multiple mechanisms that can drive mRNA accumulation at specific subcellular compartments. One is direct targeting of mRNA that is independent of translation, and another is cotranslational targeting of mRNA that relies on the targeting of the nascent peptides (rather than mRNA) that emerge from the ribosome during translation. In this case, mRNA is indirectly accumulated in the specific subcellular compartment as a result of protein-dependent targeting of the entire mRNA-associated ribosome. To differentiate between these two possibilities, we briefly treated the cells with puromycin, a known inhibitor of protein translation that dissociates ribosomes from their mRNA substrates. As shown in Fig. S2, B, D, and E, puromycin treatment did not block CHMP4B mRNA accumulation at the MB, suggesting that localized protein translation is not required for the mRNA targeting to the MB.
Next, we set out to determine if ribosomal components that are needed for translation are also present at the MB. Initially, we analyzed three different proteomic datasets of purified MBsomes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; Addi et al., 2020; Peterman and Prekeris, 2019; Rai et al., 2021). Consistent with the possibility of localized MB-associated translation, there are numerous ribosomal and translation initiation factor proteins present in the MB proteome (Fig. 3 C). Not only were there ribosomal and translation initiation factor proteins present in the proteomic datasets but they were also highly enriched according to the DAVID analysis enrichment score (Fig. 3 C). Finally, upon staining HeLa cells with antibodies against Ribosomal Protein L3 (RPL3) and Ribosomal Protein S6 (RPS6), we observed the presence of both RPL3 and RPS6 in the MB and intercellular bridge (marked by anti-acetylated-tubulin antibodies; Fig. 3, D–F and Fig. S2 G). While this does not prove that local translation occurs, it supports the idea that the ribosomal translation machinery is present at the MB and can potentially perform active translation of MB-associated mRNAs.
Midbodies are putative sites for mRNA translation
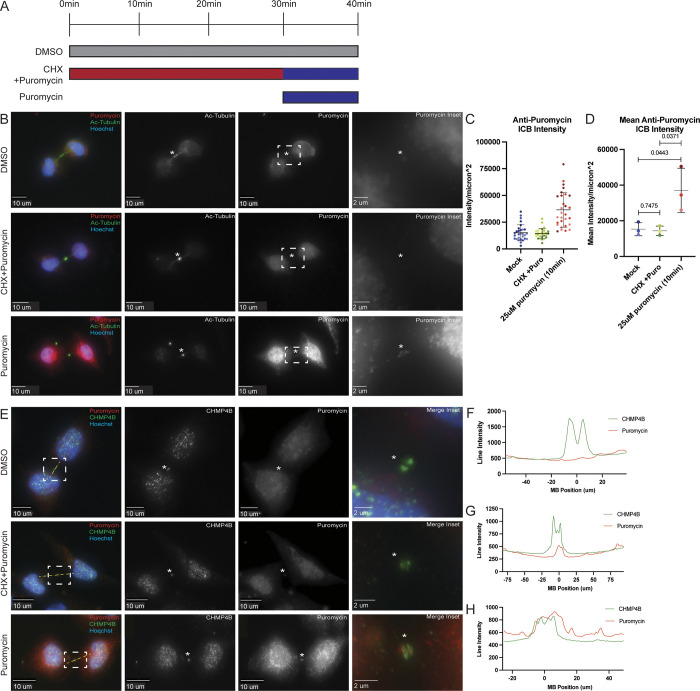
While the presence of ribosomal machinery at the MB supports the idea of local MB-associated protein translation, it is yet to be shown that active translation actually occurs at the MB and intercellular bridge during late telophase. To test this possibility, we used puromycin as a marker for active translation. Puromycin inhibits translation by causing the formation of a puromycylated nascent peptide chain and leading to dissociation of ribosomes and premature chain release. Importantly, these puromycylated peptides accumulate at sites of active translation and can be detected using anti-puromycin antibodies. To test whether translation occurs at the MB, we treated HeLa cells for 10 min with either DMSO or puromycin. We found that an anti-puromycin signal in MBs can be observed in puromycin-treated cells (Fig. 4, A–D), and, importantly, pretreatment of cells with cycloheximide (Fig. 4 A) blocked the anti-puromycin signal (Fig. 4, A–D). Cycloheximide functions by interfering with the translocation step in protein synthesis, thus preventing ribosomes from incorporating puromycin in nascent peptides. Finally, anti-CHMP4B and anti-puromycin signals colocalized at the MB and intercellular bridge (Fig. 4, E–H). The presence of anti-puromycin signal in the MB and intercellular bridge is consistent with our hypothesis that MB-enriched mRNAs are locally translated during telophase. However, it is important to note that our experiments do not rule out the possibility that puromycylated nascent peptides could be generated in the cell body and then diffuse into the MB. Considering that MBs are diffusion-limited structures (due to the high crosslinking of MB microtubules) and short treatment time (only 10 min), we consider that unlikely. However, further experiments will be needed to fully demonstrate that CHMP4B is translated at the MB (also see Fig. 8).
Figure 4.
Active protein translation occurs in the midbody. (A) Schematic representing the three treatment conditions and times used for the experiments represented in B–E. (B) HeLa cells were treated with either DMSO, CHX+Puromycin, or Puromycin and subjected to immunostaining with anti-acetylated-tubulin and anti-puromycin antibodies. The asterisks mark the MB and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (C and D) 3i imaging software was used to measure the anti-puromycin intensity/μm2 in the intercellular bridge of 60 cells from three separate experiments (C; each dot represents one cell). Statistical analysis was done on means and SD derived from the three individual experiments (D). Each experiment is represented by a different shade of color and is the mean derived from all cells analyzed in that particular experiment. Statistical analysis is represented with a P value. (E) HeLa cells were treated with either DMSO, CHX+Puromycin, or Puromycin and subjected to immunostaining with anti-puromycin (to mark the puromycylated peptides) and anti-CHMP4B antibodies. The line used for the line intensity graphs is marked in yellow and the asterisks mark the MB. The white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (F–H) Line intensity graphs representing the intensity of Puromycin (red) and CHMP4B (green) signals after DMSO (F), CHX+Puromycin (G), or Puromycin (H) treatments.
Figure 8.
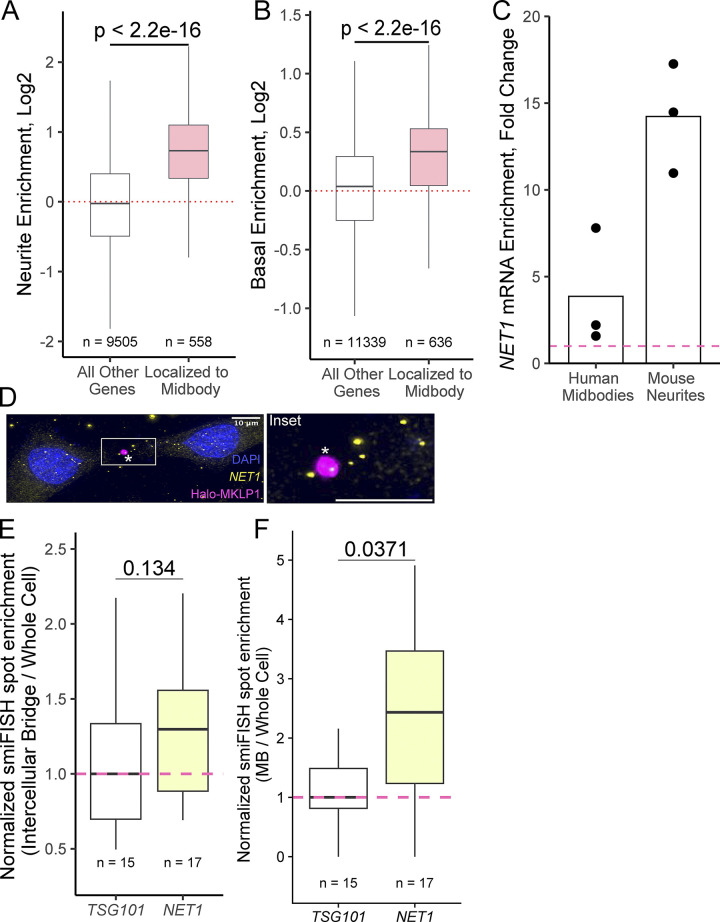
Transcripts associated with the plus-ends of microtubules are enriched in the MB transcriptome. (A) Comparison of MB-associated mRNA with RNAs enriched in neuronal projections of mouse neuronal cells. Genes were binned into MB-localized RNAs and non-MB-localized RNAs. Neurite RNA enrichment values (neurite RNA/soma RNA, log2) from previously published data were then calculated for the mouse orthologs of all genes, and the distributions of enrichment values were compared between bins. P values were calculated using a Wilcoxon rank-sum test. (B) As in A, genes were binned according to their MB RNA enrichment. Using a previous published comparison of the apical and basal compartments of epithelial cells, basal enrichment values (basal RNA/apical RNA, log2) were calculated for all genes and the distributions of enrichment values were compared between bins. P values were calculated using a Wilcoxon rank-sum test. (C) NET1 RNA enrichment in human midbodies compared with whole cells (left) and mouse neurites compared to cell bodies (right). Scale bars: 10 μm. Inset scale bars: 2 μm. (D) smiFISH for endogenous NET1 mRNA localization (yellow) in cells stably expressing Halo-MKLP1 (pink). Midbody is marked with an asterisk. The box indicates the area shown in a higher magnification inset. Scale bar: 5 μm. (E) Quantification of NET1 smiFISH. Transcript counts were quantified in the intercellular bridge and whole-cell areas. For each cell, the number of counts in the intercellular bridge was normalized by the number of counts in the whole cell. The median ratio for this value across all cells for the control TSG101 transcript was set to one. P values were calculated using a t test. (F) As in E, except that transcript counts in the midbody and whole cell regions were compared. P values were calculated using a t test.
Active translation contributes to the accumulation of CHMP4B at the MB during late telophase
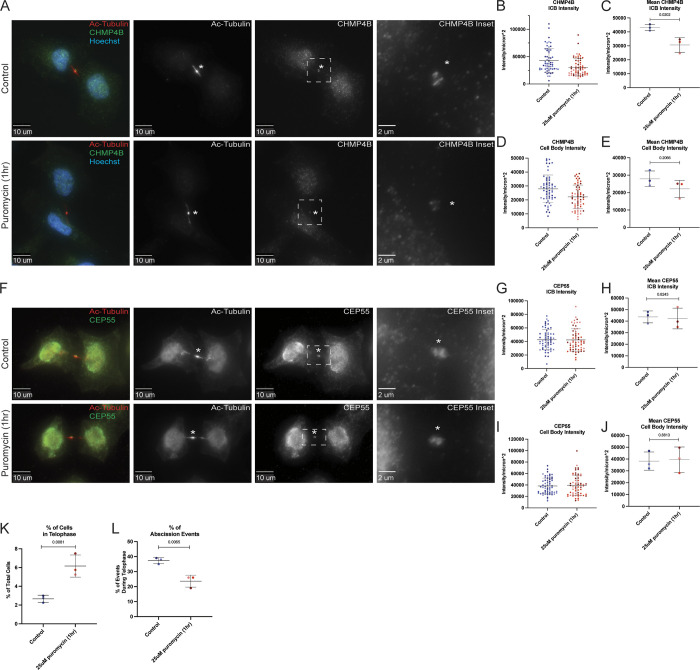
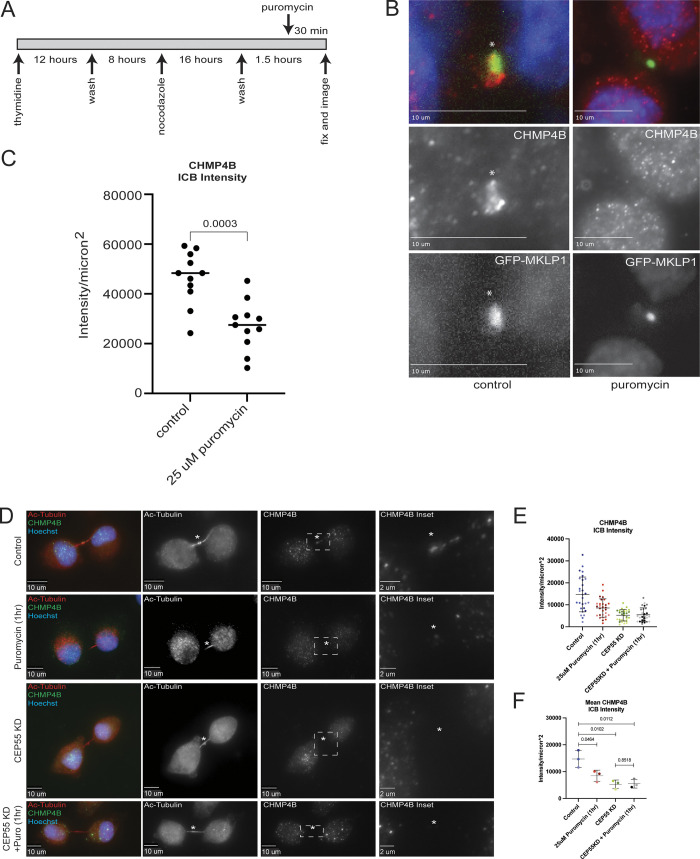
Since our data suggest that local translation may occur in the MB, and that the anti-puromycin signal colocalizes with CHMP4B, we hypothesized that inhibiting protein translation at the MB should affect CHMP4B accumulation at the MB during late telophase. To test this hypothesis, we briefly (1 h) treated HeLa cells with puromycin to block translation and analyzed endogenous CHMP4B levels at the MB by immunofluorescence microscopy. Consistent with our hypothesis, puromycin treatment decreased CHMP4B intensity in the MB (Fig. 5, A–C). CHMP4B levels were also slightly reduced in the cell body, although to a much lesser (and statistically not significant) extent than seen in the MB (Fig. 5, A, D, and E).
Figure 5.
Inhibition of translation inhibits CHMP4B accumulation at the midbody. (A) HeLa cells were either untreated or treated with puromycin for 1 h and subjected to immunostaining with anti-acetylated-tubulin and anti-CHMP4B antibodies. The asterisks mark the MB and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (B and C) Shown data represent CHMP4B intensity/μm2 in the intercellular bridge of 60 cells from three separate experiments. B shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in C was done on the means and SD derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (D and E) Shown data represent CHMP4B intensity/μm2 in the cell body of 60 cells from three separate experiments. D shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in E was done on the means, and SD were derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (F) HeLa cells were either untreated or treated with puromycin for 1 h and subjected to immunostaining with anti-acetylated-tubulin and anti-CEP55 antibodies. The asterisks mark the MB and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (G and H) Shown data represents CEP55 intensity/μm2 in the intercellular bridge of 60 cells from three separate experiments. G shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in H was done on the means and SD were derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (I and J) Shown data represents CEP55 intensity/μm2 in the cell body of 60 cells from three separate experiments. I show the distributions derived from individual cells (each dot represents a single cell). Statistical analysis in J was done on the means, and SD were derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (K) HeLa cells after puromycin treatment were fixed and stained with anti-acetylated-tubulin antibodies. The percentage of cells in telophase was then counted. Shown data are the means and SD derived from three independent experiments. Statistical analysis is represented with a P value. (L) HeLa cells after puromycin treatment were fixed and stained with anti-acetylated-tubulin antibodies. The percentage of cells that just completed abscission was then counted. Shown data are the means and SD derived from three independent experiments. Statistical analysis is represented with a P value.
Puromycin blocks all protein translation. Consequently, even a short puromycin treatment will impact many proteins, thus, it is possible that inhibition of translation also affects other MB proteins. To further bolster this idea, we chose to look at CEP55, a protein that is recruited to the MB during its formation and is required for MB-accumulation of TSG101 and ESCRT-III complex (including CHMP4B; Carlton and Martin-Serrano, 2007; Morita et al., 2007; Guizetti et al., 2011; Elia et al., 2011; Goliand et al., 2018). Since CEP55 mRNA was not found to be enriched in our RNAseq analysis (Fig. 2 B), it presumably does not require MB-associated translation for its targeting. Consistent with this hypothesis, puromycin treatment did not have any effect on CEP55 intensity in the MB or cell body (Fig. 5, F–J).
The experiments described above used non-synchronized cells. Consequently, we cannot discount the possibility that 1 h puromycin treatment affected CHMP4B mRNA translation during metaphase and anaphase before the formation of the MB. If that was the case, the differences we observed in CHMP4B accumulation at the MB would be the result of translation inhibition in the cell body (before telophase) rather than MB. To discount this possibility, we next synchronized HeLa cells stably expressing GFP-MKLP1 using a thymidine/nocodazole block (Fig. S3 A). Cells in metaphase were then washed to release from the block and incubated in serum-supplemented media for 90 min at 37°C followed by fixation and staining with anti-CHMP4B antibodies (Fig. S3 A). We have previously shown that within this 90-min incubation, cells progress from metaphase to telophase (Schiel et al., 2012), and, consistent with our previous work, we observed that over 90% of cells were in late telophase with clearly visible CHMP4B present at the MB (Fig. S3 B). Importantly, the addition of puromycin for the last 30 min of the 90-min incubation significantly decreased the accumulation of endogenous CHMP4B at the MB (Fig. S3, B and C).
Figure S3.
The effect of puromycin treatment on CHMP4B targeting to the MB in synchronized cells. (A) Synchronization and puromycin treatment schematics. (B and C) Synchronized HeLa cells were incubated for 90 min in the presence or absence of puromycin (during last 30 min of incubation). Cells were then foxed and stained with anti-CHMP4B antibodies (red). Asterisk marks the midbody. Scale bars: 10 μm. C shows quantification of CHMP4B localization in the midbody. Data shown are the means and SD calculated from individual cell values. Dots represent localization in individual cells. (D) HeLa cells were either untreated, treated with puromycin, CEP55 siRNA, or CEP55 siRNA plus puromycin and subjected to immunostaining anti-acetylated-tubulin and anti-CHMP4B antibodies. The asterisks mark the MB, and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (E and F) Shown data represents CHMP4B intensity/μm2 in intercellular of 60 cells from three separate experiments. E shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in F was done on the means and SD were derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value.
If localized translation of CHMP4B, and presumably other members of the ESCRT-III complex, is required for their accumulation at the MB, we would expect that inhibition of translation during telophase would affect the cells’ ability to undergo abscission by arresting them at late telophase. To test this hypothesis, we counted the number of cells in telophase and found that 1 h of treatment with puromycin significantly increased the number of cells in telophase (Fig. 5 K), while decreasing the number of cells that have just undergone abscission (Fig. 5 L and Fig. S4, A–D), which are all indications of defects in cell division.
Figure S4.
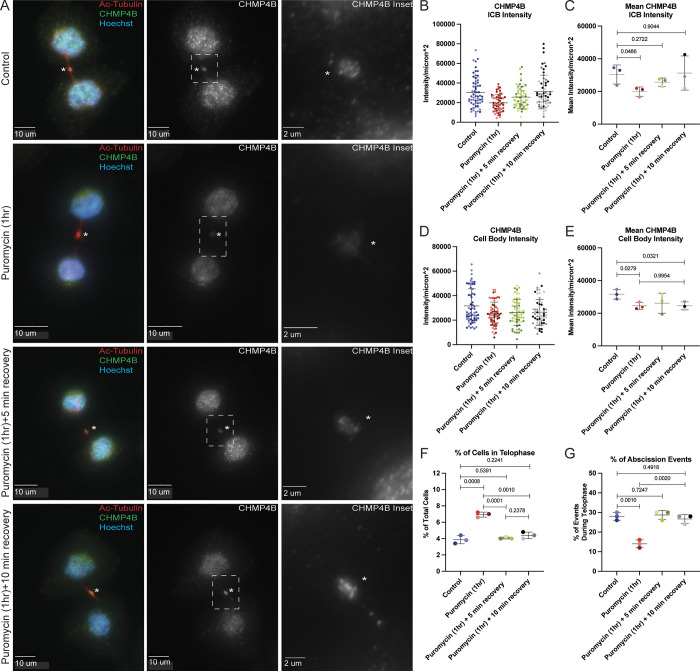
Puromycin wash-out leads to fast recovery of CHMP4B levels in MB. (A) HeLa cells were either untreated or treated for 1 h with puromycin. Cells were then washed and incubated with complete media for either 5 min or 10 min. Cells were then fixed and subjected to immunostaining with anti-acetylated-tubulin and anti-CHMP4B antibodies. The asterisks mark the MB and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (B and C) Shown data represents CHMP4B intensity/μm2 in the intercellular bridge of 60 cells from three separate experiments. B shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in C was done on the means and SD were derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (D and E) Shown data represents CHMP4B intensity/μm2 in the cell body of 60 cells from three separate experiments. D shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in E was done on the means and SD were derived from the three individual experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (F and G) HeLa cells that were untreated or treated for 1 h with puromycin. Cells were then washed and incubated with media for an additional 5 or 10 min. Cells were then fixed and stained with anti-acetylated-tubulin antibodies. The percentage of cells in either telophase (F) or just after abscission (G) was then counted. Shown data are means and SD derived from three independent experiments. Statistical analysis is represented with a P value.
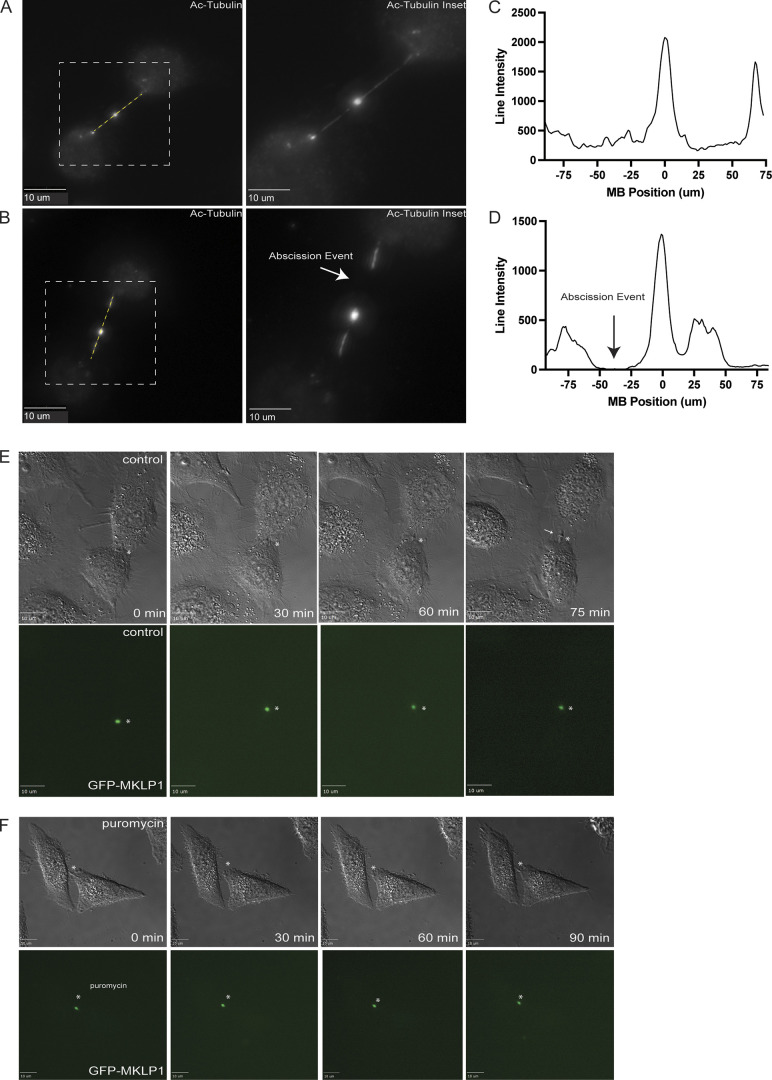
To further test whether puromycin can delay cell abscission, we next imaged telophase cells expressing GFP-MKLP1 for 90 min by time-lapse microscopy (Fig. S4, E and F). In all cases, we selected cells that were already in telophase, as indicated by the flattened cell, round nucleus, and presence of a GFP-MKLP1-labeled MB. Consistent with several previous studies, all cells (5 out of 5 imaged) in telophase successfully divided within 75 min (Fig. S4 E; asterisk marks the MB; arrow points to abscission site). In contrast, none of the telophase cells (0 out of 5 imaged) completed abscission within 90 min if puromycin was added at the beginning of the time-lapse analysis.
While the previous experiments support the idea that local translation of CHMP4B is at least partially responsible for the accumulation of CHMP4B during telophase, it does not exclude other mechanisms for CHMP4B accumulation at the MB. Since it has been proposed that CEP55 is the key initial recruiter of abscission-related proteins in mammalian cells, it is plausible that both pathways are required for efficient recruitment of ESCRT-III during abscission. To further test this idea, we used puromycin, CEP55 siRNA, or CEP55 siRNA/puromycin treatments and tested their effect on CHMP4B accumulation in the MB. As we already have shown, puromycin treatment alone caused a significant reduction in CHMP4B intensity at the MB (Fig. S3, D–F). Similarly, consistent with previous reports, CEP55 knock-down (Fig. S2 C) also decreased CHMP4B intensity at the MB (Fig. S3, D–F). This decrease was not due to a decrease in CHMP4B mRNA accumulation at the MB (Fig. S2 F), but likely due to a lack of CEP55-dependent tethering of Alix and TSG101 at the MB (Fig. 2 D). Surprisingly, a combination of CEP55 KD and puromycin treatment did not further reduce CHMP4B intensity at the MB, suggesting that local MB-associated translation and CEP55 may work together to allow efficient accumulation (by localized translation) and retention (by tethering CHMP4B at the MB) of CHMP4B during late telophase.
Short puromycin wash-out recovers CHMP4B levels at the MB but not cell body
While our previous data shows that MB-localized protein translation contributes to CHMP4B accumulation at the MB, the data are somewhat difficult to interpret because puromycin also resulted in a slight decrease of CHMP4B levels in the cell body. Thus, we could not fully differentiate between two possibilities: (1) CHMP4B targeting relies on MB-associated translation during telophase; (2) CHMP4B is translated at the cell body and then simply diffuses back to the MB. To help differentiate between those two possibilities, we performed a puromycin wash-out experiment (Fig. S4). Briefly, we treated cells with puromycin for 1 h, followed by a quick puromycin wash-out and incubation of cells for 5 or 10 min, before analyzing the levels of CHMP4B in the MB and the cell body. As we already have shown, puromycin treatment resulted in decreased CHMP4B levels in both the MB and the cell body (Fig. S4, A–E). Importantly, after puromycin wash-out, CHMP4B levels at the MB started to increase within 5 min, with complete recovery after 10 min (Fig. S4, A–C). On the contrary, CHMP4B intensity in the cell body did not recover even after 10 min (Fig. S4, A, D, and E), suggesting that CHMP4B accumulation at the MB is primarily driven by localized translation rather than by diffusion of CHMP4B from the cell body.
Since CHMP4B levels rapidly recover at the MB after the puromycin wash-out, this should also lead to a rapid recovery of the cell’s ability to undergo abscission. Consistent with this idea, 5 min after the puromycin wash-out, cells fully recovered their ability to undergo abscission, as determined by a decrease in the number of cells in telophase and an increase in the number of cells that have just undergone abscission (Fig. S4 F). All this data suggests that CHMP4B accumulation at the MB depends on local translation rather than diffusion from the cell body and that MB-associated translation of CHMP4B is required for successful completion of the abscission.
The 3′ UTR of CHMP4B mRNA enhances CHMP4B protein accumulation at the MB
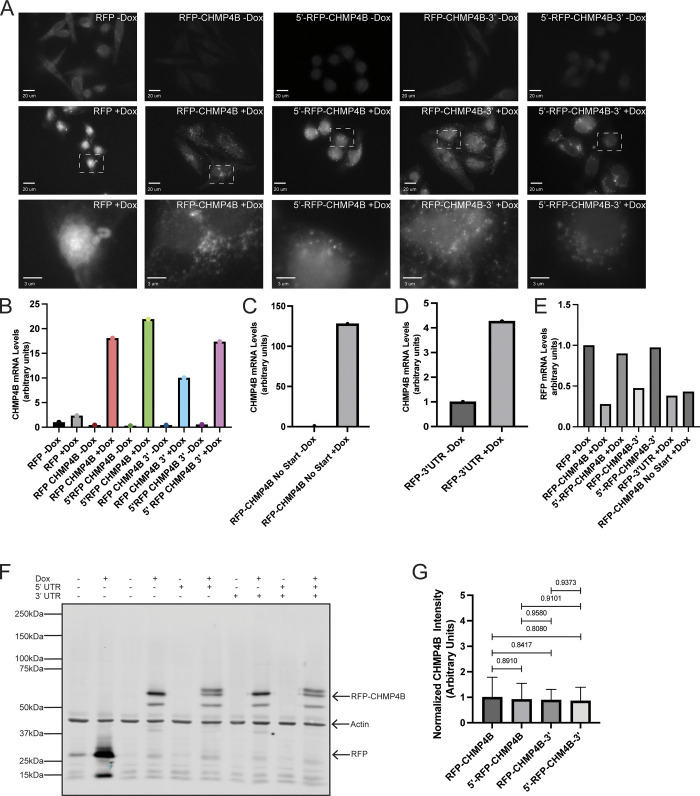
Up to this point, we have been able to show that CHMP4B mRNA is present in the MB, local translation may occur at the MB, and that CHMP4B accumulation at the MB is at least partially regulated by local translation. The next step was to determine how the CHMP4B mRNA is targeted to the MB. RNA trafficking is often driven by cis-regulatory localization elements (LEs) that are commonly found in the 5′- or 3′ UTRs (Holt and Bullock, 2009). To that end, we generated clones containing the 5′- and/or 3′ UTR of CHMP4B fused to RFP-CHMP4B cDNA. One major issue with transient expression is the high variation in expression levels in different cells hindering our ability to tell whether differences in MB targeting of various CHMP4B constructs are due to UTRs or due to the differences in expression levels. To minimize this issue, we used a HeLa LoxP recombination system (Khandelia et al., 2011). Briefly, this system uses cre recombination to allow for a single copy, site-specific recombination of our constructs into the HeLa genome (Khandelia et al., 2011). This allowed us to obtain virtually homogenous cell pools containing various doxycycline-inducible CHMP4B constructs. Importantly, each cell in these pools will have one copy of the construct that is inserted at the same genomic locus. Upon making cell lines containing various CHMP4B constructs, we first used immunofluorescence microscopy, Western blotting, and qPCR to confirm that doxycycline does induce the expression of these constructs to approximately the same level (Fig. S5, A–G). Interestingly, we found that the presence of CHMP4B 3′ UTR without the 5′ UTR slightly decreased CHMP4B mRNA levels, raising the possibility that the 3′ UTR of CHMP4B may also regulate mRNA stability (Fig. S5, B and E). Additionally, please note that the insertion of the 5′ UTR generated an alternative start codon that was in frame with the RFP, resulting in two CHMP4B bands on Western blots (Fig. S5 F).
Figure S5.
The effect of puromycin treatment on cell abscission. (A and B) HeLa cells were fixed and subjected to immunostaining using anti-acetylated-tubulin. A represents a cell in telophase. B represents a cell that had just undergone abscission (see arrow). The lines used for the line intensity graphs are marked in yellow, and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. (C and D) Line intensity graphs representing the intensity of acetylated-tubulin in telophase and abscission cells. The abscission site is marked by an arrow (D). (E and F) Telophase HeLa cells stably expressing GFP-MKLP1 were analyzed by time-lapse microscopy. Cells were imaged for 120 min with 15 min time-lapse. Scale bars: 10 μm.
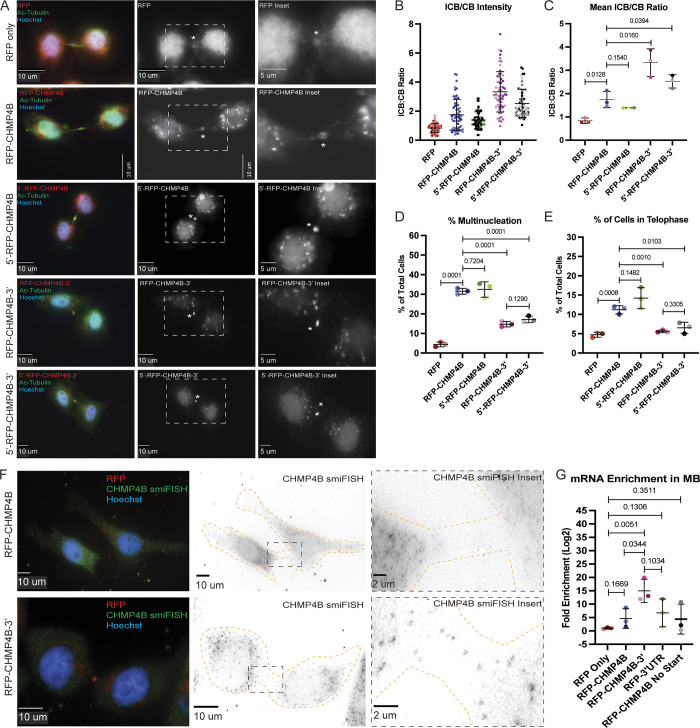
To determine the role of UTRs in CHMP4B targeting, we first tested whether having the 5′- and/or 3′ UTR of CHMP4B resulted in more efficient RFP-CHMP4B accumulation in the MB. To that end, we calculated the ratio of RFP-CHMP4B protein intensity at the MB and cell body. As shown in Fig. 6, A–C, the presence of the 3′ UTR significantly enhanced RFP-CHMP4B enrichment at the MB as compared with RFP-CHMP4B alone (Fig. 6, A–C). Interestingly, the 5′ UTR alone did not enhance RFP-CHMP4B targeting to the MB nor did it enhance the effect of 3′ UTR (Fig. 6, A–C). All these data suggest that 3′ UTR plays an important role in mediating CHMP4B protein accumulation in the MB during telophase, presumably due to targeting and localized translation of RFP-CHMP4B mRNA. Consistent with previous reports, RFP-CHMP4B without UTRs is still able to accumulate in the MB, although at much lower levels as compared with RFP-CHMP4B-3′UTR, potentially showing that there are multiple mechanisms, such as CEP55 binding, that drives ESCRT-III targeting during abscission.
Figure 6.
The 3′ UTR of CHMP4B is important for protein accumulation and mRNA translocation. (A) HeLa cells expressing various RFP-tagged dox-inducible constructs were incubated with 2 μg/ml doxycycline for 48 h and then subjected to immunostaining with anti-acetylated-tubulin. The asterisks mark the MB and the white dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 5 μm. (B and C) Ratio between RFP fluorescence in the intercellular bridge and cell body was calculated from 60 randomly chosen cells in telophase. B shows distributions derived from individual cells (each dot represents a single cell). Statistical analysis in C was done on the means, and SD were derived from the three independent experiments where experimental means were calculated by averaging values from all the cells from each experiment. Each experiment is represented by a different shade of color. Statistical analysis is represented with a P value. (D) HeLa cells expressing various RFP-tagged dox-inducible constructs were incubated with 2 μg/ml doxycycline for 48 h and then subjected to immunostaining with anti-acetylated-tubulin. The percentage of cells with multinucleation was then counted. Shown data are the means and SD derived from three independent experiments. Statistical analysis is represented with a P value. (E) HeLa cells expressing various RFP-tagged dox-inducible constructs were incubated with 2 μg/ml doxycycline for 48 h and then subjected to immunostaining with anti-acetylated-tubulin. The percentage of cells in telophase was then counted. Shown data are the means and SD derived from three independent experiments. Statistical analysis is represented with a P value. (F) HeLa cells expressing dox-inducible RFP-CHMP4B or RFP-CHMP4B-3′UTR constructs were incubated with 2 μg/ml doxycycline for 48 h and subjected to staining with smFISH probes against CHMP4B. The cell outline is marked in yellow with the asterisks marking the MB. The black dashed square represents the region of the image used for the inset. Scale bars: 10 μm. Inset scale bars: 2 μm. (G) Whole-cell or MB-associated mRNA was isolated from these cell lines expressing various RFP-tagged constructs and subject to RT-qPCR analysis using RFP or GAPDH (control) specific primers. RFP mRNA levels were then normalized against GAPDH mRNA and expressed as a ratio between MB and whole-cell RFP mRNA levels. Statistical analysis is represented with a P value and was done on the means and SD derived from three independent experiments.
It has been previously reported that overexpression of exogenous tagged ESCRT-III members inhibits abscission (Christ et al., 2016). Consistent with those studies, we also see an increase in multinucleation and the number of telophase-arrested cells in the cells overexpressing RFP-CHMP4B (Fig. 6, D and E). While the cause of this inhibition remains to be fully understood, it has been suggested that CHMP4B overexpression may lead to the accumulation of cytosolic ESCRT-III aggregates, thus, sequestering the rest of the ESCRT-III subunits away from the MB. We wondered whether the inhibitory effect of exogenous CHMP4B may be due to the fact that its mRNA lacks localization elements mediating the targeting of CHMP4B to the MB. Consistent with this hypothesis, the effect of over-expression on multinucleation or telophase arrest was reversed in cell lines expressing RFP-CHMP4B constructs containing 3′ UTR (Fig. 6, D and E).
The 3′ UTR of CHMP4B mRNA is required for its targeting to the MB
Since the 3′ UTR of CHMP4B RNA is important for CHMP4B protein accumulation at the MB during abscission, it might be expected that the 3′ UTR is also important for CHMP4B mRNA targeting the MB. To determine that, we first analyzed RFP-CHMP4B mRNA localization using smiFISH probes against CHMP4B. Consistent with the requirement of the 3′ UTR for RFP-CHMP4B accumulation, we observed smiFISH signal in the MB of cells only expressing the RFP-CHMP4B-3′UTR construct (Fig. 6 F). One drawback of using smiFISH in the MB is that it is only semiquantifiable due to not knowing how efficiently the probes are able to penetrate the protein-rich region. To better quantify the difference between RFP-CHMP4B mRNA targeting to the MB, we purified MBsomes from cell lines expressing various constructs and analyzed mRNA levels by performing RT-qPCR on purified MBsomes and then compared them to whole cell mRNA levels. To measure only exogenous CHMP4B RNA, we used primers designed to target the RFP sequence. As shown in Fig. 6 G, the presence of the 3′ UTR was required for efficient RFP-CHMP4B mRNA targeting to the MB, as compared with the amount of RFP mRNA alone or RFP-CHMP4B mRNA without 3′ UTRs (Fig. 6 G). This data suggests that the 3′ UTR is required for efficient RFP-CHMP4B mRNA targeting to the MB. Interestingly, RFP-CHMP4B RNA without UTRs was also slightly enriched at the MB as compared with RFP RNA alone. This slight mRNA enrichment could be due to cotranslational RFP-CHMP4B mRNA accumulation at the MB. To test this possibility, we removed the start codon and generated a HeLa cell line expressing doxycycline-inducible RFP-CHMP4B-NoStart construct (Fig. S5, C and E) and tested whether this mRNA also accumulates at the MBs. As shown in Fig. 6 E, the removal of the start codon did not affect the ability of RFP-CHMP4B mRNA to accumulate at the MB, although it was clearly still much lower than RFP-CHMP4B-3′UTR. All this data suggests that the 3′ UTR is required for efficient CHMP4B mRNA targeting, although the coding sequence also appears to contribute to mRNA accumulation at the MB.
To test whether the 3′ UTR of CHMP4B is sufficient for the targeting of any mRNA to the MB, we generated a doxycycline-inducible RFP-only construct that contained the 3′ UTR from CHMP4B (RFP-3′UTR; Fig. S5, D and E) and tested the mRNA levels at the MB. As shown in Fig. 7 E, 3′ UTR of CHMP4B did slightly enhance the levels of RFP mRNA at the MB, although it did not reach the levels of RFP-CHMP4B-3′UTR mRNA. This suggests that while CHMP4B 3′ UTR is essential for mRNA targeting, it may require other targeting elements that are possibly present in the CHMP4B coding region or secondary structure of the coding region to get the full targeting of mRNA to the MB.
Figure 7.
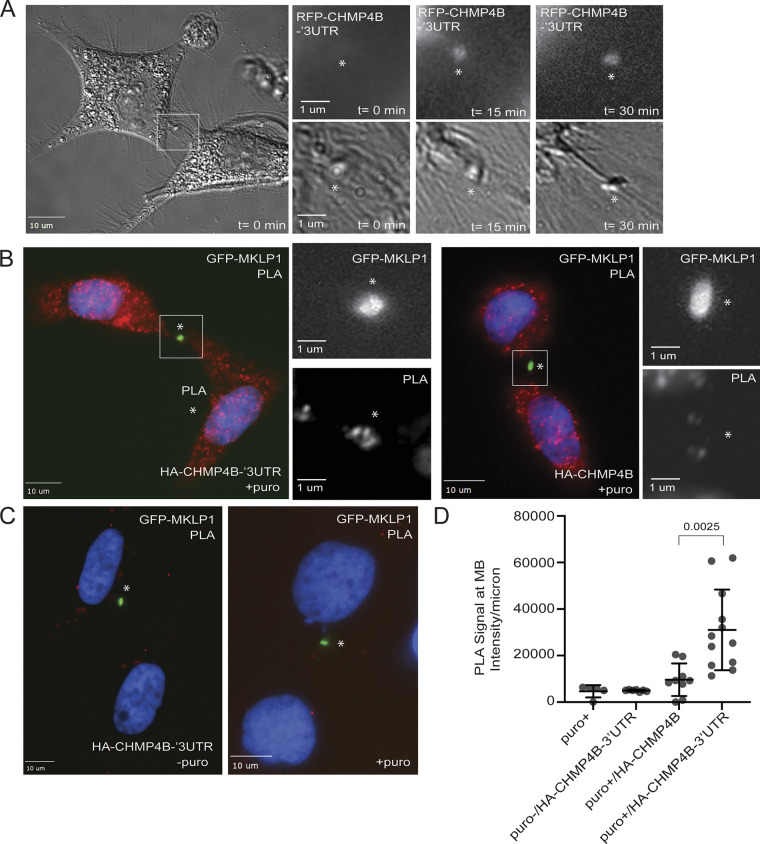
The 3′ UTR is required for CHMP4B translation at the MB during telophase. (A) HeLa cells were transfected with RFP-CHMP4B-3′UTR and plated on glass-bottom dishes. The accumulation of RFP-CHMP4B at the MB during telophase was then imaged using time-lapse microscopy for 120 min with 15 min time-lapse. Asterisk marks the midbody. Scale bars: 10 μm. Inset scale bars: 1 μm. (B) HeLa cells stably expressing GFP-MKLP1 were transfected with either HA-CHMP4B-3′UTR (images on the left) or HA-CHMP4B (images on the right). Cells were then fixed and incubated with anti-HA and anti-puromycin antibodies followed by proximity ligation assay (PLA). Asterisk marks the midbody. Boxes mark area shown in higher magnification insets. Scale bars: 10 μm. Inset scale bars: 1 μm. (C) Negative controls for the PLA experiment are shown in B. Cells in the image on the left were not treated with puromycin but transfected with HA-CHMP4B-3′UTR. Cells in the image on the right were treated with puromycin but not transfected with HA-CHMP4B-3′UTR. Asterisk marks the midbody. Scale bars: 10 μm. (D) Quantification of the levels of PLA signal in the midbody area. Data shown are the means and SEM derived from all cells analyzed. Each circle indicates the data derived from one dividing cell.
The 3′ UTR of CHMP4B mRNA is required for its localized translation at the MB
Since the 3′ UTR is required for CHMP4B mRNA targeting the MB, we wondered whether it may also be required for localized CHMP4B translation at the MB. If CHMP4B is locally translated at the MB, it would be expected to start accumulating at the MB during late telophase after the formation of the MB. Indeed, it has been previously shown that endogenous CHMP4B accumulates at the MB just before the abscission, while CEP55, TSG101, and Alix are known to be present at the MB from the very beginning of MB formation. To understand the timing and dynamics of RFP-CHMP4B accumulation at the MB, we next performed a time-lapse analysis of cells stably expressing RFP-CHMP4B-3′UTR mRNA (Fig. 7 A). To minimize the possibility that RFP-CHMP4B diffuses from the cell body, we chose cells in late telophase (as determined by round nuclei and flattened body) that were still connected with a long intercellular bridge (Fig. 7 A, MB marked by asterisk). As shown in Fig. 7 A, RFP-CHMP4B was initially absent in the MB (see time point 0 min), but then started accumulating with the MB over 10–30 min, the dynamics consistent with translation of CHMP4B at the MB during late telophase.
While the time-lapse analysis of cells expressing RFP-CHMP4B-3′UTR mRNA is consistent with the possibility that RFP-CHMP4B is locally translated at the MB, it does not directly demonstrate that CHMP4B is locally translated and that the 3′ UTR is required for this process. To more directly investigate that, we overexpressed an N-terminally HA-tagged CHMP4B construct with or without CHMP4B’s 3′ UTR. Cells were then treated for 10 min with puromycin, followed by fixation and incubation with anti-HA and anti-puromycin antibodies. We next analyzed fixed cells with proximity ligation assay (PLA). The idea of this experiment is that the accumulation of nascent peptides containing HA (at the N-terminus) and puromycin (more toward the C-terminus) will be an indication of local translation and can be detected by HA/puro PLA. Importantly, since the 3′ UTR of CHMP4B is required for HA-CHMP4B mRNA targeting to the MB, we should not detect PLA signal in cells overexpressing HA-CHMP4B lacking the 3′ UTR. This experiment also allowed us to test whether diffusion from the cell body of puromycylated nascent peptides is sufficient to generate PLA signal in the MB (also see experiments shown in Fig. 4). As shown in Fig. 7, B–D, we could indeed detect HA/puro PLA signal in the cells expressing HA-CHMP4B-3′UTR. This signal was significantly diminished in cells expressing HA-CHMP4B without 3′UTR. Importantly, the PLA signal was essentially absent in cells not treated with puromycin or cells not expressing the HA-tagged CHMP4B construct (Fig. 7, C and D). Taken together, all these data demonstrate that 3′ UTR is required for CHMP4B mRNA targeting to the MB and is also needed for localized CHMP4B translation at the MB during late telophase.
The MBsome transcriptome is similar to other localized transcriptomes associated with the plus-ends of microtubules
We next sought to identify specific cis-elements (LEs) within MBsome-localized RNAs that regulated their trafficking to the MB. We recently identified RNA elements that were necessary and sufficient for transport to both the neurites of neuronal cells and the basal pole of epithelial cells (Goering et al., 2023). Both of these subcellular locations are associated with the plus ends of microtubules and we showed that these RNA elements directed RNAs to their destinations by targeting them to microtubule plus ends. Given that MBs are associated with the plus-ends of microtubules emanating from each daughter cell, we reasoned that perhaps the same regulatory elements and mechanisms that drive RNAs to neurites and the basal pole of epithelial cells would also drive mRNAs to MBs.
If this were true, we would expect the same sets of mRNAs to be enriched at all three subcellular locations. To test this, we asked whether our MBsome-localized RNAs were more neurite-enriched than other RNAs. For this comparison, we used a previously published dataset in which mouse neuronal cells were mechanically fractionated into soma and neurite fractions followed by an analysis of their RNA contents using RNAseq (Goering et al., 2020). We compared the neurite enrichments of the mouse orthologs of our MBsome-enriched RNAs to the neurite enrichments of all other mouse RNAs. We found that the mouse orthologs of the MBsome-enriched RNAs were significantly more neurite-enriched than all other RNAs, suggesting that neurites and MBsomes contain similar mRNA contents (Fig. 8 A).
Using data from human intestinal epithelial cells (Goering et al., 2023), we then compared enrichments across the apicobasal axis for MBsome RNAs and all other detected RNAs. Again, we found that MBsome-enriched RNAs were significantly enriched at the basal pole of these cells compared to non-MBsome-enriched RNAs (Fig. 8 B). Taken together with the neurite RNA enrichments observed above, this indicates that neurites, the basal pole of epithelial cells, and MBsomes are enriched for similar sets of mRNAs, implying that similar mechanisms (e.g., plus-end directed transport along microtubules) may be populating their transcriptomes.
Identification of 3′ UTR localization elements that are necessary and sufficient to target mRNA to the MB
Our previous studies that identified mechanisms behind RNA transport along microtubules focused on the Net1 mRNA (Arora et al., 2022a; Goering et al., 2023). These studies identified a GA-rich 260 nt region within the 3′ UTR of NET1 that was both necessary and sufficient to direct RNA to the plus ends of microtubules in both neuronal and epithelial cells. We, therefore, wondered if the same element had similar abilities to direct RNA transport to the MB and MBsomes.
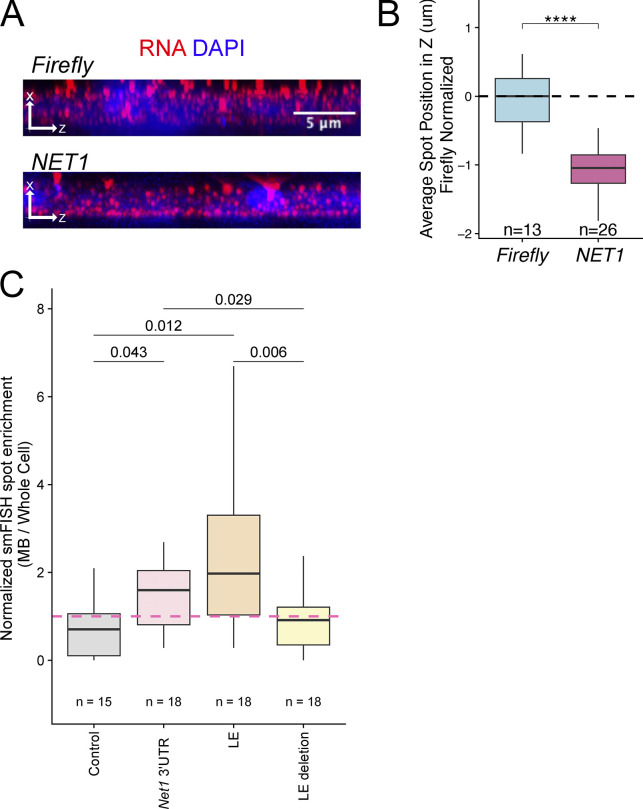
We started by asking if endogenous NET1 mRNA was enriched in our MBsome RNA samples (Fig. 1 D) since our previous studies had found NET1 mRNA enriched in the neurites (Arora et al., 2022a), and we observed NET1 mRNA enriched at the basal pole of intestinal epithelial cells (Fig. S6; and Fig. S7, A and B). We found that NET1 mRNA was approximately fourfold enriched in MBsomes relative to whole cells (Fig. 8 C). This observation was confirmed with smiFISH, showing NET1 mRNA puncta in close proximity to MBs in dividing cells (Fig. 8 D). After quantifying the smiFISH results across many cells, we found that NET1 mRNA was more likely to be enriched at both the intercellular bridge (Fig. 8 E) and the midbody (Fig. 8 F) compared with the control transcript TSG101 (that is not enriched in MBs as measured by RNAseq and qPCR; see Fig. 2).
Figure S6.
Characterization of HeLa LoxP cells expressing doxycycline-inducible RFP-tagged constructs. (A) HeLa LoxP cells expressing various RFP-tagged dox-inducible constructs cells were untreated (−Dox) or treated with 2 μg/ml doxycycline for 48 h (+Dox). RFP was visualized by fluorescent microscopy to test for dox-dependent expression. The white dashed square represents the region of the image used for the inset. Scale bars: 20 μm. Inset scale bars: 3 μm. Scale bars: 5 μm. (B–D) HeLa LoxP cells expressing various RFP-tagged dox-inducible constructs cells were untreated (−Dox) or treated with 2 μg/ml doxycycline for 48 h (+Dox). The construct expression levels were then determined by RT-qPCR using CHMP4B-specific primers (single qPCR run with three technical replicates). (E) HeLa LoxP cells expressing various RFP-tagged dox-inducible constructs cells were treated with 2 μg/ml doxycycline for 48 h. The levels of expression among various constructs were the n compared using RT-qPCR with RFP specific primers (single qPCR run with three technical replicates). (F and G) HeLa LoxP cells expressing various RFP-tagged dox-inducible constructs cells were untreated (−Dox) or treated with 2 μg/ml doxycycline for 48 h (+Dox). The levels of RFP-tagged protein expressions were visualized via Western blot using anti-RFP and anti-Actin (loading control) antibodies. G shows Western blot quantification from three independent experiments. Statistical analysis is represented with a P value. Source data are available for this figure: SourceData FS6.
Figure S7.
Localization analysis of Net1 mRNA. (A) smiFISH for endogenous NET1 mRNA localization (red) in human epithelial cells. As a nonlocalized control, transcripts encoding an exogenous Firefly luciferase are also visualized. (B) Quantification of NET1 RNA localization along the apicobasal axis of epithelial cells. (C) Quantification of MB-localized Net1 UTR-containing reporter transcripts. In all samples, reporter transcript counts in midbodies and whole cells were quantified and the ratio of counts between the two locations is reported. These ratios were normalized by setting the value for the control reporter transcript lacking 3′ UTR additions to one. P values were calculated using a t test.
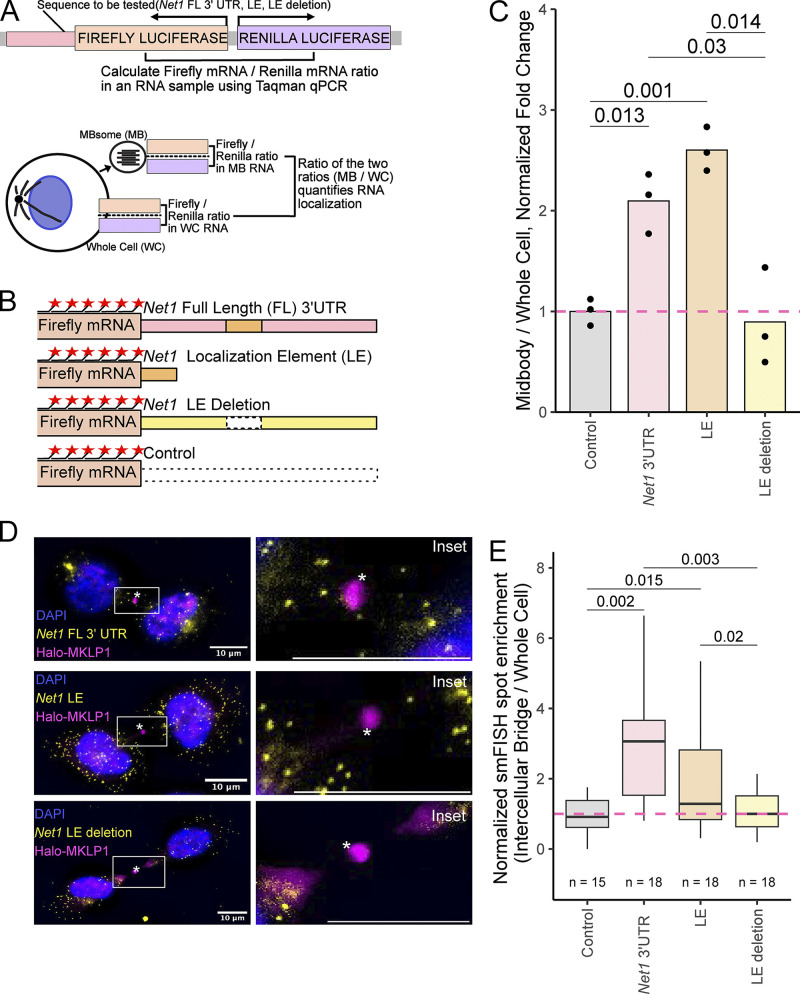
We next asked if the same RNA cis-regulatory element within mouse Net1 mRNA previously shown to promote RNA trafficking to neurites and the basal pole of epithelial cells also controlled RNA localization to MBs and MBsomes. To test this, we devised an RT-qPCR-based reporter assay that quantified the enrichment of designed reporter transcripts in MBsomes (Fig. 9 A). In this assay, Firefly and Renilla luciferases are coexpressed from a bidirectional promoter with the Renilla luciferase RNA serving as an unchanging internal control. RNA from MBsomes and whole cells was collected as described before, and the relative amounts of Firefly and Renilla luciferase transcripts in each fraction were determined using Taqman RT-qPCR. By comparing how the ratios of these transcripts vary between cell fractions depending on the 3′ UTR appended to the Firefly luciferase RNA, we are able to quantify the ability of the 3′ UTR to drive the Firefly luciferase transcript to MBsomes.
Figure 9.
NET1 3′ UTR localization elements are necessary and sufficient to target mRNA to the MB. (A) Schematic RT-qPCR-based reporter RNA approach for assaying MB RNA localization. Plasmids expressing Firefly and Renilla luciferases from a bidirectional reporter are integrated into the genome. Sequences to be tested for MB localization activity (NET1 full length 3′ UTR, LE and LE deletion) are fused to the 3′ UTR of Firefly luciferase. Using Taqman qPCR, the ratio of Firefly to Renilla luciferase mRNA is measured in MB and whole-cell samples. The ratio of these ratios (MB/WC) quantifies the MB enrichment of the Firefly luciferase transcript. By asking how this value changes upon appending additional sequences to the Firefly luciferase RNA, the effects of these additional sequences on MB RNA enrichment can be tested. (B) Diagram of reporter constructs used to interrogate NET1 localization activity in MBs. smFISH probes against the Firefly luciferase coding sequence are shown as red stars. (C) Localization of the reporter RNAs described in B to the MB as quantified using the strategy outlined in A. The value for the control reporter lacking additional sequence elements was set to one. P values were calculated using a t test. (D) smFISH visualizing the reporter RNAs described in B (yellow) in cells stably expressing Halo-MKLP1 (pink). Midbody is marked with an asterisk. The box indicates the area shown in a higher magnification inset. Scale bars: 10 μm. (E) Quantification of MB-localized Net1 UTR-containing reporter transcripts visualized in D. In all samples, reporter transcript counts in intercellular bridges and whole cells were quantified and the ratio of counts between the two locations is reported. These ratios were normalized by setting the value for the control reporter transcript lacking 3′ UTR additions to one. P values were calculated using a t test.
We created four Firefly luciferase reporter RNAs (Fig. 9 B). These reporters contained 3′ UTRs consisting of the entire Net1 3′ UTR, the previously identified 260 nt element within the Net1 UTR that was sufficient to drive RNA transport to neurites and the basal pole of epithelial cells, the entire Net1 UTR lacking this element, and as a control, an unrelated plasmid-encoded UTR. These constructs were site-specifically incorporated into HeLa cells using cre/loxP recombination, and their expression was controlled using a doxycycline-on system.
We then measured the relative MBsome-enrichment of each of these constructs using the RT-qPCR strategy outlined in Fig. 9 A, setting the enrichment of the reporter with the control 3′ UTR to one. We found that the addition of the entire Net1 UTR to the reporter construct increased its MBsome-enrichment by approximately twofold (Fig. 9 C). The reporter containing the previously identified 260 nt subset of the Net1 UTR was similarly enriched in MBsomes, indicating that this cis-element is sufficient to drive RNAs to MBsomes. Deletion of this element completely abrogated the ability of the Net1 3′ UTR to regulate MBsome RNA localization, demonstrating that the element is also necessary for RNA transport to MBsomes.
Next, we confirmed these results using an orthogonal smFISH assay that visualized Firefly luciferase reporter RNAs in situ. The fluorescent probes used in this assay targeted the Firefly luciferase coding sequence that was shared across all reporter RNAs. As with the RT-qPCR approach, we found that the 260-nt cis-element within the Net1 UTR was both necessary and sufficient for reporter RNA targeting to both MBs and the intercellular bridge of dividing cells (Fig. 9, D and E; and Fig. S7 C).
These results demonstrate that discrete RNA cis-regulatory elements control the RNA content of MBs and MBsomes and suggest that previously defined mechanisms that regulate microtubule-based RNA transport in other systems likely also act to populate the MBsome transcriptome (Goering et al., 2023).
Discussion
Our study shows that MBs contain mRNAs encoding for several ESCRT-III components, namely CHMP2A, CHMP3, CHMP4B, and CHMP6. What makes this discovery especially exciting is that all these ESCRT-III complex proteins are known to co-oligomerize and drive the abscission step of cytokinesis. While the role of the ESCRT-III proteins in mediating abscission was first described over a decade ago (Guizetti et al., 2011; Elia et al., 2011; Carlton et al., 2012), how ESCRT-III subunits are delivered and targeted to the MB remains somewhat controversial. Until recently, it has been thought that TSG101 and the ESCRT-III complex accumulate at the MB by directly or indirectly binding to the MB resident protein, CEP55 (Morita et al., 2007; Addi et al., 2018). However, recent studies have shown that depletion of CEP55 does not fully block ESCRT-III targeting to the MB, and CEP55 knock-out mice are able to grow and develop with limited defects (Tedeschi et al., 2020). Additionally, CEP55 is only present in vertebrates, raising a question of how the ESCRT-III may be targeted in other organisms, although CEP55 clearly contributes to ESCRT-III targeting in vertebrate cells. Indeed, recent studies have suggested alternative mechanisms of ESCRT-III recruitment, such as ALIX binding to cetralspindlin (Lie-Jensen et al., 2019) or delivery of ESCRT-III complexes to the MB on the transport vesicles (Pust et al., 2023). The discovery that ESCRT-III mRNAs are enriched at the MB suggests another intriguing possibility that regulated localized translation of ESCRT-III components contribute to the accumulation of ESCRT-III at the MB during abscission. This hypothesis is supported by results in other systems in which precise RNA localization is critical for the successful association of the resulting translated protein with its proper interaction partners (Norris and Mendell, 2023).
Consistent with the hypothesis that localized protein translation contributes to ESCRT-III targeting the MB, we show that the MB has all the components needed for translation. We also use anti-puromycin antibodies to suggest that active translation is occurring in the MB. Finally, we show that inhibition of protein translation during late telophase blocks the completion of cytokinesis. Based on all these findings, we chose to further investigate one of the most important regulators of late-stage abscission, CHMP4B. There are several pieces of evidence suggesting that CHMP4B is being actively translated in the MB during telophase. First, puromycylated CHMP4B nascent peptides are present at the MB. Second, inhibition of protein translation during telophase leads to a decrease in CHMP4B accumulation at the MB. Third, wash-out of the translation inhibitor puromycin results in a very fast recovery of CHMP4B levels at the MB, which could not be explained by simple diffusion of CHMP4B protein that was translated in the cell body. Importantly, the ability of the cell to complete abscission also recovers within a similar timescale. All these data suggest that CHMP4B can be locally translated and that it is at least partially responsible for the accumulation and function of CHMP4B at the MB. Importantly, the mechanisms using CEP55 and local translation to target ESCRT-III to the MB are not necessarily mutually exclusive. Indeed, both of these mechanisms appear to contribute to ESCRT-III localization in vertebrate cells, suggesting that while localized translation may be required to accumulate ESCRT-III components within the MB, CEP55 is likely needed to tether and maintain their MB enrichment during telophase.
Local translation of CHMP4B at the MB depends on the targeting of CHMP4B mRNA to the MB during telophase. What remains unclear is the molecular machinery governing mRNA delivery and accumulation at the MB. It has been previously shown that the subcellular targeting of mRNAs is usually driven by cis-regulatory elements that are commonly found in the 5′ or 3′ UTRs (Holt and Bullock, 2009). Consequently, we tested whether the 5′ and -3′ UTRs drive the translocation of CHMP4B mRNA to the MB. Our data demonstrates that the 3′ UTR is required for translocation and accumulation of CHMP4B at the MB. To test whether the 3′ UTR is sufficient for mRNA accumulation at the MB, we added CHMP4B 3′ UTR to RFP and analyzed its ability to drive RFP mRNA accumulation at the MB. Interestingly, while we did see an increase in the levels of MB-associated RFP mRNA, the amount of RFP-3′UTR mRNA did not fully recapitulate the levels of RFP-CHMP4B-3′UTR mRNA, suggesting that the CHMP4B coding region may also play a role in CHMP4B mRNA targeting to the MB.
It has been previously shown that GA-rich motifs mediate the targeting of RAB13 mRNA to the actin-rich cellular protrusions (Costa et al., 2020; Moissoglu et al., 2020). Similarly, we have shown that GA-rich localization elements (LEs) in mouse Net1 mRNA can mediate Net1 mRNA targeting neurites and the basal pole of epithelial cells (Moissoglu et al., 2020). Our RNAseq analysis of purified MBs showed that there is a significant overlap between mRNAs targeted to the MB, neurite, and basolateral domains of epithelial cells. Furthermore, RAB13 and NET1 mRNAs are also both enriched in the MB, suggesting that their 3′ UTR GA-rich motifs may also mediate mRNA targeting to the MBs. Consistent with this hypothesis, we show that the LE present in the 3′ UTR of mouse Net1 mRNA is necessary and sufficient to target mRNA to the MB. We recently found that these GA-rich motifs likely target RNAs to neurites and the basal pole of epithelial cells by promoting their transport to the plus ends of microtubules in a kinesin 1–dependent manner (Goering et al., 2023). This type of transport would be perfect for targeting mRNAs to the MB as plus-ends of microtubules are oriented toward the MB, and kinesin 1 has already been implicated in transporting endosomes to the MB during cell division (Montagne and Gonzalez-Gaitan, 2014). All these findings hint at a common mechanism that regulates the subcellular targeting of mRNAs across a variety of cell types and subcellular destinations.
While this paper was being reviewed, Skop and colleagues published a study demonstrating that RNA is present in the midbody of CHO cells (Park et al., 2023). This study also suggested that active mRNA translation occurs at the MB during late telophase, although they did not investigate the translation of specific mRNAs, such as CHMP4B. Importantly, this study purified MBs from cells in telophase rather than MBsomes (as we did in this study), suggesting that the presence of MB-associated mRNA is not an artifact of MB purification, and that mRNA is likely targeted to the MB during telophase. Taken together, this work is fully consistent with the hypothesis proposed in this study that mRNA targeting and translation at the MB may play an important role in mediating mitotic cell division.
Taken together, this study demonstrates that a specific subset of mRNAs is targeted and enriched at the MB during cell division and plays a role in MB function during abscission (Fig. 8). At least some of these mRNAs, such as CHMP4B mRNA, appear to be translated at the MB, and this translation plays a key role in the accumulation of ESCRT-III components at the MB, thus regulating the abscission step of cytokinesis. While working out the details governing mRNA targeting and translation at the MB will require future studies, and this work lays a foundation for the identification of a novel mechanism regulating abscission. Finally, this study will not only advance our understanding of the machinery governing ESCRT-III translocation and accumulation at the MB but could also provide novel insight into the mechanisms of RNA targeting to other subcellular compartments and other cell types, as well as provide a framework for studying the role of mRNAs in the post-mitotic MB function.
Recent studies have shown that during cell division MBs can be released in the extracellular milieu where they are called MBsomes or Flemmingsomes (Addi et al., 2020; Peterman and Prekeris, 2019). Uptake of these MBsomes by other cells can lead to changes in cell fate, differentiation, and proliferation capacity, presumably by mediating the lateral transfer of specific signaling molecules (Peterman and Prekeris, 2019; Rai et al., 2021; Farmer and Prekeris, 2022). However, what remains unclear is what these signaling molecules are and how they may affect cells after MBsome internalization. In this study, we demonstrate that MBsomes contain a specific subset of mRNAs, many of them encoding proteins regulating cell division and differentiation. That raises a very intriguing possibility that delivery of specific mRNAs to the acceptor cells is what mediates post-mitotic MBsome functions, although further research will be needed to fully define the role of MBsome-dependent mRNA delivery.
Materials and methods
Cell culture
All HeLa cell lines were kept in a 37°C humidified incubator at 5% CO2, routinely tested for mycoplasma, and maintained in DMEM with 10% FBS and 1% penicillin/streptomycin.
Cell synchronization
HeLa cells were grown to ∼50% confluency and incubated with 5 mM thymidine for 16 h. Cells were then washed and incubated with regular serum-containing media and filled by incubation with 0.1 μg/ml of nocodazole for 16 h. Mitotic cells were then isolated by gently tapping tissue culture plates to dislodge loosely attached cells arrested in prometaphase. Cells were then washed, seeded on collagen-coated coverslips, and incubated in serum-containing media for 90 min in the presence or absence of puromycin (for last 30 min).
Reagents and antibodies
iTaq Universal SYBR Green Supermix was purchased from Bio-Rad. TRIzol, Puromycin, Doxycycline, Lipofectamine 2000, and Lipofectamine RNAiMax were purchased from Thermo Fisher Scientific. Stellaris Hybridization, Wash A, and Wash B Buffers were purchased from Biosearch Technologies. Hoechst stain was purchased from AnaSpec. Antibodies against the following proteins were used: RPL3 (#A305-007A, 1:100 for immunofluorescence; Bethyl Laboratories), acetylated-tubulin (#D20G3, 1:500 for immunofluorescence; Cell Signaling), acetylated-tubulin (#T7451, 1:500 for immunofluorescence; Millipore Sigma), CHMP4B (#16639-I-AP, 1:300 for immunofluorescence; Proteintech), CEP55 (#H00055165-A01, 1:500 for immunofluorescence; Abnova), RFP (#6g6-20, 1:2,000 for immunoblotting; Chromotek), β-actin (#926-42210, 1:2,000 for immunoblotting; LI-COR), donkey anti-mouse-IgG conjugated to Alexa Fluor 488 (#715-545-150, 1:100 dilution for immunofluorescence; Jackson), donkey anti-mouse-IgG conjugated to Alexa Fluor 594 (#715-585-1500, 1:100 dilution for immunofluorescence; Jackson), donkey anti-rabbit-IgG Alexa Fluor 488 (#711-585-152, 1:100 dilution for immunofluorescence; Jackson), donkey anti-rabbit-IgG conjugated to Alexa Fluor 594 (#711-585-152, 1:100 dilution for immunofluorescence; Jackson), donkey anti-mouse IRDye 680CW (#926-32212, 1:5,000 for immunoblotting; LI-COR), and donkey anti-rabbit IRDye 800CW (#926-68072, 1:5,000 for immunoblotting; LI-COR).
RNA isolation
RNA was extracted using Trizol (#15596026; Thermo Fisher Scientific) per the manufacturer’s instructions. The RNA was reverse transcribed using SuperScript IV (#18091050; Thermo Fisher Scientific) per the manufacturer’s instructions.
RT-qPCR
Quantitative PCR was performed using iTaq Universal SYBR Green Supermix (#1725121; Bio-Rad). Primer sequences can be found in Table S2. To quantify the RT-qPCR, each cDNA sample was normalized to GAPDH. If the experiment was looking at enrichment in MBsomes, the data is represented as a ratio of the normalized MB/normalized total cell. Taqman qPCR reactions were performed using Taqman Fast Advanced Master Mix (Life Technologies) with labeled probe sets (Table S3). The quality of each Midbody isolation was quantified by the enrichment of NET1 in MBsomes using the ratio of NET1/TSG101 in MBsomes compared with NET1/TSG101 in whole cells. Reporter enrichment in MBsomes was quantified using Renilla luciferase as an internal control using the ratio of Firefly luciferase/Renilla luciferase RNA in MBsomes compared with the ratio of Firefly luciferase/Renilla luciferase RNA in whole cells.
MB purification and RNAseq
Midbodies were isolated as previously described (Peterman and Prekeris, 2017). Briefly, media from the various HeLa cells was collected and subjected to a series of centrifugation spins (300 × g, 10,000 × g). Sucrose gradient fractionation was performed at 3,000 × g and the interphase between 40% glycerol and 2 M sucrose was collected and spun at 10,000 × g to pellet the midbodies. RNA was extracted using TRIzol (#15596026; Thermo Fisher Scientific) per the manufacturer’s instructions. For control and MB group 1, the library prep and RNAseq were performed by the University of Colorado Anschutz Medical Campus Genomics facility. The Genomics facility generated a poly A enriched library from the total RNA extraction and performed RNAseq on an Illumina HiSeq2000 platform, utilizing 1 lane, single read 125 cycles. For the control group and MB group 2 and 3, the library prep and RNAseq were performed by Novogene. Briefly, Novogene generated a poly A enriched library from the total RNA extraction and performed RNAseq on a NovaSeq PE150 platform with 6 G raw data per sample.
RNAseq data processing
Transcript abundances were calculated using Salmon v1.9.0 (Patro et al., 2017) using the flags −seqBias and −gcBias. Transcript abundances were collapsed to gene level using txImport (Soneson et al., 2015). To determine the enrichment of genes in the midbody, DEseq2 was used to calculate the log2 ratio of midbody/whole cell counts and identify significantly enriched RNAs (Love et al., 2014). Genes with a basemean (average of normalized count values) <250 were excluded. Midbody enriched RNAs have an FDR <0.05 and a midbody enrichment (Log2 fold change) >0. Plots were generated in Rstudio using R version 4.1.1.
siRNA transfections
Custom CHMP4B siRNA oligonucleotides (5′-AUCGAUAAAGUUGAUGAGUUAUU-3′; Dharmacon) and custom CEP55 siRNA oligonucleotides (5′-AGGCAUGUACUUUAGACUU-3′; Millipore Sigma) were transfected into HeLa cells using Liptofectamine RNAiMAX with 40 nM oligonucleotide. The efficiency of the knock-down was measured 72 h after transfection by RT-qPCR (previously described above).
Immunofluorescence and quantification
HeLa cells were treated as indicated in the text and then fixed in 1% paraformaldehyde in PBS for 10 min at room temperature. Cells were rinsed three times in PBS. The cells were then incubated with a primary antibody in PBS containing 0.5% BSA and 0.2% saponin for 1 h at room temperature, washed three times in PBS, and then incubated with the appropriate fluorochrome-conjugated secondary antibodies diluted in PBS containing 0.5% BSA and 0.2% saponin for 30 min. Cells were washed three times in PBS and mounted in Fluoromount (Southern Biotech).
Fixed cells were imaged with an inverted Axiovert 200M microscope (Zeiss) with a ×63 oil immersion lens and QE charge-coupled device camera (Sensicam). Z-stack images were taken at a step size of 500–1,000 nm. Image processing and quantification were performed using 3i Slidebook 6 software (Intelligent Imaging Innovations). Briefly, masks (ROI) were made to calculate the total intensity of the cell body or the intercellular bridge as well as the total area for each mask. The numbers were then used to calculate the intensity/µm2. Unless otherwise stated, all images are represented as a maximal projection of the z-stack.
Time-lapse imaging
HeLa cells stably expressing GFP-MKLP1 were plated on 6-cm glass-bottom dishes and imaged at 37°C for 120 min with a time lapse of 15 min (to minimize phototoxicity). To ensure that cells are in mid-late telophase, cells were selected based on the following criteria: cells were flattened with a round nucleus (a kidney-shaped nucleus is a feature of cells in early telophase) containing identifiable MB (labeled by GFP-MKLP1). Where indicated, 25 μM puromycin was added at the beginning of the time-lapse analysis.
Proximal ligation assay
HeLa cells stably expressing GFP-MKLP1 were transfected with either HA-CHMP4B or HA-CHMP4B-3′UTR constructs. Non-transfected HeLa cells were used as one of the negative controls. Cells were then treated with puromycin for 10 min (non-treated cells were used as another negative control), followed by fixation with 4% paraformaldehyde and permeabilization with 0.1% Triton X-100. Cells were incubated with anti-HA and anti-puromycin antibodies, followed by Duallink proximal ligation assay (PLA), as described in the manufacturers protocol (Sigma-Aldrich). For imaging, cells in mid-to-late telophase were randomly picked using the following criteria: (1) cells had a clearly identifiable MB as determined by GFP-MKLP1 signal; (2) cells had fully reformed round nuclei; (3) cell bodies were flattened; (4) daughter cells were connected by an extended intercellular bridge of at least 2 μm in length.
smiFISH probe design
The smiFISH protocol was adopted from Tsanov et al. (2016). Briefly, the probes were designed by inputting the gene sequences (CHMP4B, NET1, and TSG101) into the Oligostan software (Table S4). smiFISH probes then had a Y flap (5′-TTACACTCGGACCTCGTCGACATGCATT-3′) added to have a location where we could hybridize the fluorescent molecules to the probe. The sequences can be found in Table S4. The fluorescent probe was designed by taking the reverse complement of the Y flap (5′-AATGCATGTCGACGAGGTCCGAGTGTAA-3′) and adding Alexa488, Alexa594, or Cy3 to both ends.
smFISH probe design
smFISH probes for Firefly luciferase were obtained from Biosearch Technologies. The probes were labeled with Quasar 570 dye. The sequences of these probes are proprietary.
smiFISH/smFISH visualization
HeLa cells were plated on collagen-coated coverslips and allowed to grow to 70–75% confluency. The media was aspirated and cells were washed once with 1× PBS. For Halo-tag visualization, HaloTag Oregon Green Ligand was added 4–6 h prior to fixing cells. Cells were fixed in 1% PFA for 10 min at room temperature and then washed twice with 1× PBS. Cells were permeabilized with 0.1% Triton X-100 for 5 min at room temperature. For Halo-tag visualization, cells were not permeabilized to preserve Halo-tag fluorescence. The cells were washed with Wash Buffer A (Stellaris) at room temperature for 5 min. In the meantime, the CHMP4B probes and Y flap were hybridized using the protocol from Tsanov et al. (2016). For smFISH, this hybridization was not necessary. 2 μl of the probe/flap hybridization product (0.833 µM) or smFISH probe was added to 100 μl of smFISH hybridization buffer (Biosearch Technologies). A hybridization chamber was prepared using an empty 15-cm cell culture plate, wrapped in tinfoil with wet paper towels surrounding the rim on the inside and parafilm covering the bottom of the plate. 100 μl of the probe-containing hybridization solution was added to the parafilm. The coverslip was then placed on top of this droplet of hybridization buffer with the cell side down. The hybridization chamber with the coverslips was incubated at 37°C overnight. On the following day, the glass coverslips were transferred to a fresh six-well plate with the cell side up and incubated with Wash Buffer A (Biosearch Technologies) and Hoechst stain (1:2,000; AnaSpec) at room temperature in the dark for 30 min. Hoechst stain was washed with Wash Buffer B (Biosearch Technologies) at room temperature in the dark for 5 min. Coverslips were then mounted onto slides with Fluoromount G (Southern Biotech) and sealed with clear nail polish. Slides were imaged using an inverted Axiovert 200M microscope (Zeiss) with a 63X oil immersion lens and QE charge-coupled device camera (Sensicam).
smFISH particle counting
ImageJ was used to count the total number of smiFISH particles. First, the channels were separated, and an outline of the cell was drawn using the freehand tool in the smFISH channel. Next, a threshold was set using a control cell. The threshold set with the control cell was used for all cells after that. Finally, the particles were analyzed to get the total number of particles in each cell.
FISH-quant
FISH-quant was used to quantify midbody or intercellular bridge enrichment of smFISH spots as previously described (Arora et al., 2022b). Briefly, outlines were drawn in the FITC channel visualizing Halo-MKLP1. Four outlines were drawn per dividing cell: two cells on each side of the bridge, the intercellular bridge, and the midbody marked by Halo-MKLP1. Prior to quantification, identified smFISH spots were thresholded for intensity, sphericity, amplitude, and position. Reporter enrichment was quantified by the total number of spots in the intercellular bridge or midbody of cells over a total number of spots in the whole cell normalized to the control.
Puromycin treatment
HeLa cells were plated on collagen-coated coverslips and allowed to grow to 70–75% confluency. At this time, cells were exposed to 25 μM puromycin and incubated at 37°C for 1 h. Cells were then fixed, stained, and imaged as previously described.
Anti-puromycin labeling
HeLa cells were plated on collagen-coated coverslips and allowed to grow to 70–75% confluency. At this time, the cells were either treated with DMSO (volume equal to that of the other treatments) for 40 min at 37°C, 50 μM cycloheximide for 40 min plus the last 10 min with 25 μM puromycin at 37°C, or only 25 μM puromycin for 10 min at 37°C. Cells were then fixed, stained, and imaged as previously described.
Puromycin wash-out assay
HeLa cells were plated on collagen-coated coverslips and allowed to grow to 70–75% confluency. At this time, the cells were either untreated, treated with 25 μM puromycin at 37°C for 1 h, treated with 25 μM puromycin at 37°C for 1 h, washed with 1× PBS and allowed to recover for 5 min in complete media at 37°C, or treated with 25 μM puromycin at 37°C for 1 h, washed with 1× PBS and allowed to recover for 10 min in complete media at 37°C. Cells were then fixed, stained, and imaged as previously described.
Generation of doxycycline-inducible constructs
Due to the GC-rich nature, we were unable to PCR the full-length 3′-UTR. Therefore, we used Twist Bioscience to generate a modified sequence that we could use for a template (5′-TGGGGTCCAGCGCTGGCTGGGCCCAGACAGACTGTGGTGGCCTGCGCAGCGAGCAGGCGTGTGCGTGTGTGGGGCAGGCAGGATGTGGTGCAGGCAGGTTCCATCGCTTTCGACTCTCACTCCAAAGCAGTAGGGCCGCGTTGCTGCTCACTCTCTGCATAGCATGGTCTGGAAGTGTGCTGTTTATAATGTTGAATTTCTGTAAAATAAACTGTATTTGCAAATCCAACATTGAGCTTCTGGACTACGCTGACTCCACTGCTGAATCCTCAATGGAAAGGGTCGACTGGTTGCAGTTGAAATGACCTGAAATGTAGCCTCTGTCCTTGTAAGTCAGTTGACTTGCCGCACATCTCTTTGTGTACTTGTACGGTACTGGCAGAAAAGTCATTTTTCAAAAGCCATAGGCTTTTCCTTGCCCTTAGCTGTAATAATGCATCTGATTTTGATTTCCTCCAGAGCTGTGTTTCTGTCCATCACCTGTGTATTGGCCCTGTGTTTACCACTCTGGCCCACTCCTCACCCCCTTGCTCCCCTGGTCTTCTGGAGTTTGTGACATTGATTTGAAATGGATGGTGTTCTCTTGAGAGCAAGTGAGATTGTTAGAATTAAGTTCCAACTATACAGTTTTCTAACATAGCTATAAGGTCCTTGTTGCTGTTTGTGATAACTGATAGATAACTCATTGGAAACGTGCATACATTTATATTCAGATGAAATTATGGTTTGCACTGTCTATTAAATATCTCGATTAATTTTCATA-3′). This sequence maintains the GA-rich motifs thought to be important for translocation.
To insert the various doxycycline-inducible constructs into the backbone (pRD-RIPE), we used the NEBuilder Assembly tool and followed the program’s direction to generate forward and reverse primers for generating PCR products for the insert and the backbone (listed in Table S5). Q5 High-Fidelity 2× Master Mix (New England Biolabs) was used to perform the PCR according to the manufacturer’s protocol. NEBuilder HiFi DNA Assembly Master Mix was used to assemble the inserts and backbones according to the manufacturer’s protocol and transformed into XL10-Gold competent cells (Agilent). Bacteria containing the constructs were then grown overnight at 37°C and the DNA was extracted using the Zyppy Plasmid Miniprep Kit (Zymo Research) according to the manufacturer’s protocol.
For the RFP-3′UTR, RFP-CHMP4B-3′UTR were generated as described above and then using the NEBuilder system, the CHMP4B coding region was removed by PCR and reassembled to get the RFP-3′UTR construct. For the RFP-CHMP4B No Start, the RFP-CHMP4B was generated as described above and then using the NEBuilder system, the start codon was removed by PCR and reassembled to get the RFP-CHMP4B No Start construct.
Transfection of doxycycline-inducible constructs
HeLa acceptor lines containing a HeLa LoxP site were plated in a six-well plate and allowed to grow to 60% confluency. Cells were then cotransfected with a pRD-RIPE construct containing a gene of interest, along with a Cre-encoding plasmid (pBT140) at a 0.5–10% wt/wt ratio. To transfect one well in a six-well plate, a total of 2.1 μg of DNA was mixed with 5.625 μl of Lipofectamine 2000 (Thermo Fisher Scientific) and 246 μl of Opti-MEM (Invitrogen) following the manufacturer’s protocol. Cells were incubated with the transfection mixture overnight, the medium was changed, and the incubation continued for another 24 h before adding puromycin. We used a two-step selection protocol, beginning with half of the maximal puromycin concentration (0.5 μg/ml) for the first 48 h of selection, followed by the maximal puromycin concentration (1 μg/ml) for several days until the puromycin-sensitive cells were eliminated. The cultures were incubated until the appearance of visible puromycin-resistant colonies, which were then pooled together and expanded.
Induction of doxycycline-inducible cell lines
Cells containing a doxycycline-inducible gene of interest were subject to 2 μg/ml of doxycycline and allowed to grow for 48 h unless otherwise stated. After 48 h, the cells were either fixed on collagen-coated coverslips for immunofluorescence, lysed for Western blot analysis, or the media was collected for MBsome isolation.
Cell lysis and Western blot analysis
Cells were lysed on ice in a buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF. After 30 min, lysates were clarified at 15,000 g in a prechilled microcentrifuge. Supernatants were collected and analyzed via Bradford assay (5000006; Bio-Rad Protein Assay). 50-µg lysate samples were prepared (unless otherwise stated) in 4× SDS loading dye, boiled for 5 min at 95°C, and separated via SDS-PAGE. Gels were transferred onto 0.45-µm polyvinylidene difluoride membrane (IPFL00010), followed by blocking for 30 min in Intercept Blocking Buffer diluted in TBST 1:3 (927-60001). Primary antibodies (made in diluted Intercept Blocking Buffer) were incubated overnight at 4°C. The next day, blots were washed in TBST followed by incubation with IRDye fluorescent secondary antibody (diluted Intercept Blocking Buffer) for 30 min at room temperature. Blots were washed once again with TBST before final imaging on a Li-Cor Odyssey CLx.
Statistical analysis
All statistical analyses were performed using GraphPad Prism Software (GraphPad). A Student’s t test was used to determine significance unless otherwise noted. Error bars represent SD unless otherwise noted. For all immunofluorescence experiments, at least 20 randomly chosen image fields from at least three separate experiments were used for data collection. For quantitative immunofluorescence analysis, the same exposure was used for all images in that experiment and was quantified using Intelligent Imaging Innovations software. Data distribution was assumed to be normal, but this was not formally tested. For Western blot experiments, three separate experimental examples were used for data collection. For quantitative Western blot analysis, Image Studio V5.2 software (LI-COR) was used to collect intensities.
Online supplemental material
Fig. S3 shows the effect of puromycin treatment on CHMP4B targeting the MB in synchronized cells. Fig. S4 shows that puromycin wash-out leads to fast recovery of CHMP4B levels in MB. Fig. S6 shows the characterization of HeLa LoxP cells expressing doxycycline-inducible RFP-tagged constructs. Table S1 lists the mRNAs identified in RNAseq analyses of purified MBs. Table S2 shows SYBR RT-qPCR primer sequences. Table S3 shows Taqman RT-qPCR primer sequences. Table S4 shows smiFISH probe sequences. Table S5 lists the primer sequences used for cloning.
Supplementary Material
is a list of mRNAs identified in RNAseq analyses of purified MBs.
shows SYBR RT-qPCR primer sequences.
shows Taqman RT-qPCR primer sequences.
shows smiFISH probe sequences.
lists primer sequences used for cloning.
is the source file for Fig. S6.
Acknowledgments
We would like to thank the University of Colorado Anschutz Medical Campus Genomics Facility and Novogene for performing the RNAseq, and the RNA Biosciences Initiative (RBI) for help in analyzing the RNAseq data and Migle Prekeryte for critically reading and editing the manuscript.
This work was supported by National Institute of General Medical Sciences grant R01-GM143774 to R. Prekeris, R35-GM133385 to J.M. Taliaferro, and RBI Pilot Grant to R. Prekeris and J.M. Taliaferro. Additionally, some of the work was also supported by Diversity Supplement GM143774-02S1 to K.F. Vaeth.
Author contributions: R. Prekeris and J.M. Taliaferro oversaw the entire project and helped with various aspects of experimental design, data interpretation, and manuscript writing. T. Farmer and K.F. Vaeth performed most experiments and were directly involved in data analysis and manuscript writing. T. Farmer also performed two repeats of RNAseq analysis. K.-J. Han performed one repeat of RNAseq analysis and also helped with various qPCR analyses. R. Goering contributed to performing comparisons between various RNAseq data sets and also helped with some of smFISH analyses.
References
- Addi, C., Bai J., and Echard A.. 2018. Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr. Opin. Cell Biol. 50:27–34. 10.1016/j.ceb.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Addi, C., Presle A., Frémont S., Cuvelier F., Rocancourt M., Milin F., Schmutz S., Chamot-Rooke J., Douché T., Duchateau M., et al. 2020. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat. Commun. 11:1941. 10.1038/s41467-020-15205-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agromayor, M., Carlton J.G., Phelan J.P., Matthews D.R., Carlin L.M., Ameer-Beg S., Bowers K., and Martin-Serrano J.. 2009. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 20:1374–1387. 10.1091/mbc.e08-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, A., Castro-Gutierrez R., Moffatt C., Eletto D., Becker R., Brown M., Moor A.E., Russ H.A., and Taliaferro J.M.. 2022a. High-throughput identification of RNA localization elements in neuronal cells. Nucleic Acids Res. 50:10626–10642. 10.1093/nar/gkac763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, A., Goering R., Velez P.T., and Taliaferro J.M.. 2022b. Visualization and quantification of subcellular RNA localization using single-molecule RNA fluorescence in situ hybridization. Methods Mol. Biol. 2404:247–266. 10.1007/978-1-0716-1851-6_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström, J., Sramkova Z., Salašová A., Johard H., Mahdessian D., Fedr R., Marks C., Medalová J., Souček K., Lundberg E., et al. 2017. Comparative cell cycle transcriptomics reveals synchronization of developmental transcription factor networks in cancer cells. PLoS One. 12:e0188772. 10.1371/journal.pone.0188772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J. 2010. The ESCRT machinery: A cellular apparatus for sorting and scission. Biochem. Soc. Trans. 38:1397–1412. 10.1042/BST0381397 [DOI] [PubMed] [Google Scholar]
- Carlton, J.G., and Martin-Serrano J.. 2007. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 316:1908–1912. 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carlton, J.G., Agromayor M., and Martin-Serrano J.. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 105:10541–10546. 10.1073/pnas.0802008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J.G., Caballe A., Agromayor M., Kloc M., and Martin-Serrano J.. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 336:220–225. 10.1126/science.1217180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.T., Ettinger A.W., Huttner W.B., and Doxsey S.J.. 2013. Resurrecting remnants: The lives of post-mitotic midbodies. Trends Cell Biol. 23:118–128. 10.1016/j.tcb.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ, L., Wenzel E.M., Liestøl K., Raiborg C., Campsteijn C., and Stenmark H.. 2016. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 212:499–513. 10.1083/jcb.201507009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ, L., Raiborg C., Wenzel E.M., Campsteijn C., and Stenmark H.. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42:42–56. 10.1016/j.tibs.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Costa, G., Bradbury J.J., Tarannum N., and Herbert S.P.. 2020. RAB13 mRNA compartmentalisation spatially orients tissue morphogenesis. EMBO J. 39:e106003. 10.15252/embj.2020106003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, E.F., Gaffuri A.L., Gayraud-Morel B., Tajbakhsh S., and Echard A.. 2014. Engulfment of the midbody remnant after cytokinesis in mammalian cells. J. Cell Sci. 127:3840–3851. 10.1242/jcs.154732 [DOI] [PubMed] [Google Scholar]
- Dionne, L.K., Wang X.J., and Prekeris R.. 2015. Midbody: From cellular junk to regulator of cell polarity and cell fate. Curr. Opin. Cell Biol. 35:51–58. 10.1016/j.ceb.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, N., Sougrat R., Spurlin T.A., Hurley J.H., and Lippincott-Schwartz J.. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA. 108:4846–4851. 10.1073/pnas.1102714108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, T., and Prekeris R.. 2022. New signaling kid on the block: The role of the postmitotic midbody in polarity, stemness, and proliferation. Mol. Biol. Cell. 33:pe2. 10.1091/mbc.E21-06-0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering, R., Hudish L.I., Guzman B.B., Raj N., Bassell G.J., Russ H.A., Dominguez D., and Taliaferro J.M.. 2020. FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. Elife. 9:e52621. 10.7554/eLife.52621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering, R., Arora A., Pockalny M.C., and Taliaferro J.M.. 2023. RNA localization mechanisms transcend cell morphology. Elife. 12:e80040. 10.7554/eLife.80040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliand, I., Nachmias D., Gershony O., and Elia N.. 2014. Inhibition of ESCRT-II-CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol. Biol. Cell. 25:3740–3748. 10.1091/mbc.e14-08-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliand, I., Adar-Levor S., Segal I., Nachmias D., Dadosh T., Kozlov M.M., and Elia N.. 2018. Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM. Cell Rep. 24:1756–1764. 10.1016/j.celrep.2018.07.051 [DOI] [PubMed] [Google Scholar]
- Guizetti, J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., and Gerlich D.W.. 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 331:1616–1620. 10.1126/science.1201847 [DOI] [PubMed] [Google Scholar]
- Hassine, S., Bonnet-Magnaval F., Benoit Bouvrette L.P., Doran B., Ghram M., Bouthillette M., Lecuyer E., and DesGroseillers L.. 2020. Staufen1 localizes to the mitotic spindle and controls the localization of RNA populations to the spindle. J. Cell Sci. 133:jcs247155. 10.1242/jcs.247155 [DOI] [PubMed] [Google Scholar]
- Holt, C.E., and Bullock S.L.. 2009. Subcellular mRNA localization in animal cells and why it matters. Science. 326:1212–1216. 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia, P., Yap K., and Makeyev E.V.. 2011. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. USA. 108:12799–12804. 10.1073/pnas.1103532108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, T.C., Chen C.T., Baron D., Onder T.T., Loewer S., Almeida S., Weismann C.M., Xu P., Houghton J.M., Gao F.B., et al. 2011. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13:1214–1223. 10.1038/ncb2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie-Jensen, A., Ivanauskiene K., Malerod L., Jain A., Tan K.W., Laerdahl J.K., Liestol K., Stenmark H., and Haglund K.. 2019. Centralspindlin recruits ALIX to the midbody during cytokinetic abscission in Drosophila via a mechanism analogous to virus budding. Curr. Biol. 29:3538–3548.e7. 10.1016/j.cub.2019.09.025 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa, B., and Gerlich D.W.. 2014. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell. 31:525–538. 10.1016/j.devcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Moissoglu, K., Stueland M., Gasparski A.N., Wang T., Jenkins L.M., Hastings M.L., and Mili S.. 2020. RNA localization and co-translational interactions control RAB13 GTPase function and cell migration. EMBO J. 39:e104958. 10.15252/embj.2020104958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, C., and Gonzalez-Gaitan M.. 2014. Sara endosomes and the asymmetric division of intestinal stem cells. Development. 141:2014–2023. 10.1242/dev.104240 [DOI] [PubMed] [Google Scholar]
- Morita, E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., and Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, E., Colf L.A., Karren M.A., Sandrin V., Rodesch C.K., and Sundquist W.I.. 2010. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA. 107:12889–12894. 10.1073/pnas.1005938107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, M.L., and Mendell J.T.. 2023. Localization of Kif1c mRNA to cell protrusions dictates binding partner specificity of the encoded protein. Genes Dev. 37:191–203. 10.1101/gad.350320.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S., Dahn R., Kurt E., Presle A., VanDenHeuvel K., Moravec C., Jambhekar A., Olukoga O., Shepherd J., Echard A., et al. 2023. The mammalian midbody and midbody remnant are assembly sites for RNA and localized translation. Dev. Cell. 58:1917–1932.e6. 10.1016/j.devcel.2023.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman, E., and Prekeris R.. 2017. Understanding post-mitotic roles of the midbody during cell differentiation and polarization. Methods Cell Biol. 137:173–186. 10.1016/bs.mcb.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman, E., and Prekeris R.. 2019. The postmitotic midbody: Regulating polarity, stemness, and proliferation. J. Cell Biol. 218:3903–3911. 10.1083/jcb.201906148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman, E., Gibieža P., Schafer J., Skeberdis V.A., Kaupinis A., Valius M., Heiligenstein X., Hurbain I., Raposo G., and Prekeris R.. 2019. The post-abscission midbody is an intracellular signaling organelle that regulates cell proliferation. Nat. Commun. 10:3181. 10.1038/s41467-019-10871-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pust, S., Brech A., Wegner C.S., Stenmark H., and Haglund K.. 2023. Vesicle-mediated transport of ALIX and ESCRT-III to the intercellular bridge during cytokinesis. Cell Mol Life. 80:235. 10.1007/s00018-023-04864-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, A., Greening D.W., Xu R., Chen M., Suwakulsiri W., and Simpson R.J.. 2021. Secreted midbody remnants are a class of extracellular vesicles molecularly distinct from exosomes and microparticles. Commun. Biol. 4:400. 10.1038/s42003-021-01882-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel, J.A., Simon G.C., Zaharris C., Weisz J., Castle D., Wu C.C., and Prekeris R.. 2012. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat. Cell Biol. 14:1068–1078. 10.1038/ncb2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg, J., Lee I.H., Iwasa J.H., and Hurley J.H.. 2017. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18:5–17. 10.1038/nrm.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A.R., Liu H., Yates J. III, Meyer B.J., and Heald R.. 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 305:61–66. 10.1126/science.1097931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson, C., Love M.I., and Robinson M.D.. 2015. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 4:1521. 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi, A., Almagro J., Renshaw M.J., Messal H.A., Behrens A., and Petronczki M.. 2020. Cep55 promotes cytokinesis of neural progenitors but is dispensable for most mammalian cell divisions. Nat. Commun. 11:1746. 10.1038/s41467-020-15359-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov, N., Samacoits A., Chouaib R., Traboulsi A.M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M., et al. 2016. smiFISH and FISH-quant: A flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 44:e165. 10.1093/nar/gkw784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a list of mRNAs identified in RNAseq analyses of purified MBs.
shows SYBR RT-qPCR primer sequences.
shows Taqman RT-qPCR primer sequences.
shows smiFISH probe sequences.
lists primer sequences used for cloning.
is the source file for Fig. S6.