Abstract
Activation of the mechanistic target of rapamycin complex 1 (mTORC1) contributes to the development of chronic pain. However, the specific mechanisms by which mTORC1 causes hypersensitivity remain elusive. The eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) is a key mTORC1 downstream effector that represses translation initiation. Here, we show that nociceptor-specific deletion of 4E-BP1, mimicking activation of mTORC1-dependent translation, is sufficient to cause mechanical hypersensitivity. Using translating ribosome affinity purification in nociceptors lacking 4E-BP1, we identified a pronounced translational up-regulation of tripartite motif-containing protein 32 (TRIM32), an E3 ubiquitin ligase that promotes interferon signaling. Down-regulation of TRIM32 in nociceptors or blocking type I interferon signaling reversed the mechanical hypersensitivity in mice lacking 4E-BP1. Furthermore, nociceptor-specific ablation of TRIM32 alleviated mechanical hypersensitivity caused by tissue inflammation. These results show that mTORC1 in nociceptors promotes hypersensitivity via 4E-BP1–dependent up-regulation of TRIM32/interferon signaling and identify TRIM32 as a therapeutic target in inflammatory pain.
Translation of specific mRNAs via the mTORC1-4E-BP1 axis in nociceptors controls neuronal excitability and pain hypersensitivity.
INTRODUCTION
Chronic pain is caused by functional and structural reorganization of peripheral and central nociceptive circuits. These processes depend on the production of new proteins via de novo gene expression (1, 2). Regulation of mRNA translation by the mechanistic target of rapamycin complex 1 (mTORC1) is one of the key mechanisms that modulate gene expression, particularly in neurons where local axonal and dendritic translation allows rapid changes in the cellular proteome in response to external stimuli or activity (3, 4). mTORC1 is a conserved serine/threonine kinase that is stimulated by numerous signaling cascades in response to tissue injury and disease (e.g., downstream of TrkA/TrkB, insulin receptor, epidermal growth factor receptor/insulin-like growth factor 1, and N-methyl-d-aspartate receptors) to regulate the translation of a subset of mRNAs (5–7). In addition to mRNA translation, mTORC1 modulates other processes, including lipid metabolism, autophagy, and ribosome biogenesis (5, 8). mTORC1 is activated in sensory neurons in models of chronic pain caused by tissue inflammation and peripheral nerve injury (9–14) and pharmacological inhibition of mTORC1 alleviates the development of pain hypersensitivity (9, 10, 15–22). The contribution of mTORC1-dependent translation in nociceptors, versus other mTORC1-dependent processes, in causing hypersensitivity and the identity of translationally regulated mRNAs mediating the effect on neuronal excitability and hypersensitivity are ill-defined.
The mTORC1 stimulates the rate of mRNA translation by phosphorylating and inhibiting the family of translational repressor proteins, eukaryotic initiation factor 4E (eIF4E)–binding proteins (4E-BPs) (23) (Fig. 1A). 4E-BPs bind the mRNA cap-binding protein eIF4E, preventing its interaction with the large scaffolding protein eIF4G and the RNA helicase eIF4A to form the eIF4F complex, which is required for translation initiation. Deletion of 4E-BP releases eIF4E from suppression and enhances eIF4F complex formation, mimicking increased mRNA translation downstream of mTORC1 (23). There are two main isoforms of 4E-BPs in the nervous system: 4E-BP1 and 4E-BP2 (4E-BP3 expression levels are very low), which have similar function but distinct tissue distribution (24). We previously reported that 4E-BP1 is the main isoform involved in nociception in mice as whole-body ablation of 4E-BP1, but not 4E-BP2, causes mechanical hypersensitivity (24).
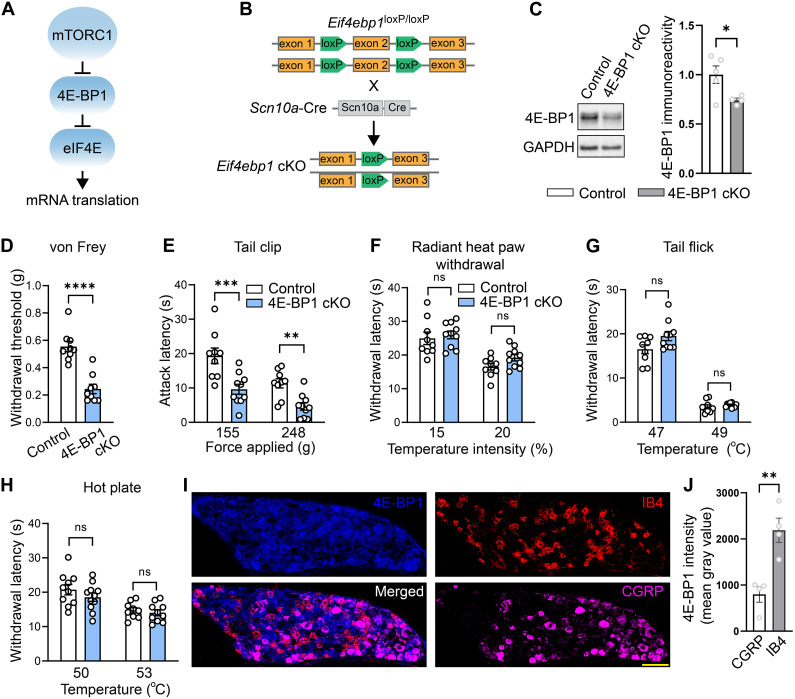
Fig. 1. Nociceptor-specific deletion of 4E-BP1 causes mechanical but not thermal hypersensitivity.
(A) Schematic of the mTORC1 pathway. (B) Generation of 4E-BP1 conditional knockout (cKO) mice. (C) Western blots and quantification showing decreased 4E-BP1 in DRG lysates of 4E-BP1 cKO mice (n = 4 to 5 mice per group, Student’s t test, two-tailed). Baseline testing showed increased mechanical hypersensitivity in the von Frey (D) and tail clip (E) tests but no difference in thermal sensitivity in the radiant heat paw withdrawal (F), tail flick assay (G), and hot plate test (H) (n = 8 to 10 mice per group, Student’s t test, two-tailed). Analysis revealed no main effects of sex or interactions for (D) to (H). (I) Immunostaining of DRG from wild-type mice revealed increased total 4E-BP1 in nonpeptidergic nociceptors compared to peptidergic nociceptors. (J) Quantification (n = 4 mice per group, Student’s t test, two-tailed; scale bar, 100 μm). All data are presented as mean ± SEM. *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001.
Here, we investigated the mechanism by which mTORC1–4E-BP1–dependent translation contributes to mechanical hypersensitivity in nociceptive sensory neurons. We revealed that selective deletion of 4E-BP1 in nociceptors causes robust mechanical hypersensitivity and showed that the effect is mediated via enhanced translation of the E3 ubiquitin ligase, tripartite motif-containing protein 32 (TRIM32), which promotes type I interferon (IFN) signaling. Moreover, down-regulation of TRIM32 or inhibition of type I IFN signaling alleviated inflammation-induced hypersensitivity. Thus, mTORC1–4E-BP1–mediated induction of TRIM32/type I IFN signaling contributes to mechanical hypersensitivity after inflammation.
RESULTS
Deletion of 4E-BP1 in nociceptors induces mechanical hypersensitivity
To study the role of peripheral 4E-BP1–dependent translation in pain, we generated mice lacking 4E-BP1 selectively in nociceptors. Eif4ebp1fl/fl mice were crossed with mice expressing Cre recombinase under the control of the Scn10a (Nav1.8) promoter to generate eif4ebp1fl/fl Scn10aCre mice, hereafter referred to as 4E-BP1 conditional knockout (4E-BP1 cKO) mice (Fig. 1, A and B). As expected, 4E-BP1 cKO mice displayed reduced expression of 4E-BP1 in dorsal root ganglion (DRG) lysates (Fig. 1C). Behavioral analysis demonstrated that 4E-BP1 cKO male and female mice exhibit mechanical hypersensitivity, as measured in the von Frey (Fig. 1D) and tail clip (Fig. 1E) tests, but show no alterations in thermal sensitivity, as assessed in the radiant heat paw-withdrawal (Fig. 1F), tail flick (Fig. 1G), and hot plate (Fig. 1H) assays. Immunostaining of DRG sections revealed that 4E-BP1 is expressed at higher levels in nonpeptidergic IB4+ nociceptors compared to peptidergic CGRP+ nociceptors (Fig. 1, I and J). This finding might partially explain the selective mechanical hypersensitivity phenotype in 4E-BP1 cKO mice, as nonpeptidergic neurons are predominantly involved in mediating mechanical nociception (25, 26).
The translational landscape of nociceptors lacking 4E-BP1
To understand how activation of 4E-BP1–dependent translation, downstream of mTORC1, in nociceptors, induces mechanical hypersensitivity, we used the cell type–specific translating ribosome affinity purification (TRAP) approach. TRAP identifies mRNAs that are bound to ribosomes and therefore undergo active translation in a specific cell population (27, 28) (Fig. 2A). To label ribosomes in nociceptors, an adeno-associated virus (AAV) expressing in a Cre-dependent manner a ribosomal subunit L10a tagged with enhanced green fluorescent protein (eGFP; AAV2/9-DIO-eGFP-L10a) was injected intraperitoneally into eif4ebp1fl/fl Scn10aCre (4E-BP1 cKO) and control Scn10aCre pups at postnatal days 2 to 4. Colabeling of DRG sections with markers of peptidergic (CGRP) and nonpeptidergic (IB4) nociceptors, as well as large-diameter neurons (NF-200) showed that eGFP-L10a is predominantly expressed in nociceptors (31.5% in peptidergic, 52% in nonpeptidergic, and 13.25% in large-diameter neurons; Fig. 2, B and C).
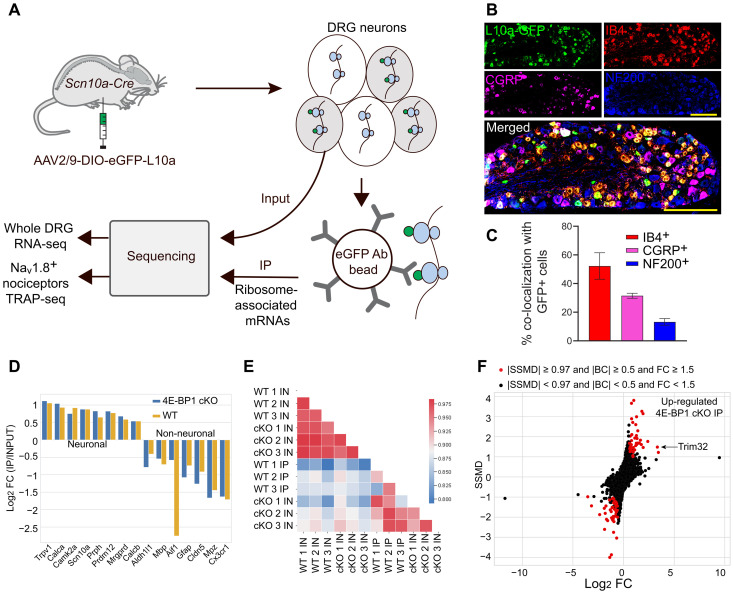
Fig. 2. TRAP RNA-sequencing reveals differentially expressed genes and TRIM32 as translationally up-regulated mRNA.
(A) Schematic of translating ribosome affinity purification approach. (B) Immunostaining of DRG, from Scn10a Cre mice injected with AAV-eGFP-L10a, with IB4, CGRP, and NF200 to confirm tagging of nociceptors but not large-diameter neurons. (C) Quantification (n = 4 mice per group, Student’s t test, two-tailed; scale bar, 200 μm). (D) Neuronal markers are enriched and nonneuronal markers are depleted in IP fractions. (E) Heatmap of the correlation coefficients between different samples (input, IN; immunoprecipitated, IP). (F) Dual-flashlight plot depicting strictly standardized mean difference (SSMD) versus log2 FC for genes in IP samples. Positive Log2 FC indicates increased expression in 4E-BP1 cKO mice. Parameters for defining data as up-regulated or down-regulated in 4E-BP1 KO are indicated on the top.
To capture the eGFP-tagged ribosomes and ribosome-bound mRNAs, we performed immunoprecipitation (IP) with eGFP antibody on DRG lysates from 8-week-old mice (three biological replicates per genotype, DRGs pooled from three mice per replicate), followed by RNA sequencing. IP input samples (IN) from DRG lysates were also sequenced to normalize for mRNA transcription. We observed a high correlation between the biological replicates (r > 0.96 for IN and r > 0.92 for IP; fig. S1, A and B), suggesting high reproducibility across experiments. The immunoprecipitated mRNA populations were enriched for neuronal markers (e.g., Prph, Trpv1, Scn10a, and Calca; Fig. 2D) and depleted for markers of nonneuronal cells (e.g., Gfap, Cx3cr1, Mbp, and Mpz), demonstrating the specificity of the approach.
A heatmap of the correlation coefficients for coding genes in different conditions showed differences between WT and 4E-BP1 cKO mice in TRAP-seq samples, consistent with the role of 4E-B1 in translational regulation (Fig. 2E). Analysis of ribosome-bound mRNA revealed a subset of mRNAs with altered ribosome occupancy in 4E-BP1 cKO nociceptors (Fig. 2F and data S1). The mRNA encoding for the E3 ubiquitin ligase TRIM32 was among the two most up-regulated mRNAs (10.11-fold increase, Fig. 2F). After normalization to mRNA levels, Trim32 showed substantial increase in IP/IN ratio (9.41-fold increase; fig. S1, C and D), demonstrating its translational up-regulation. We further confirmed the increase in TRIM32 protein levels in lumbar DRG lysates of 4E-BP1 cKO mice using Western blotting (Fig. 3A). To study whether increased expression of TRIM32 contributes to the mechanical hypersensitivity of 4E-BP1 cKO mice, we knocked down TRIM32 selectively in nociceptors. To this end, Scn10aCre and eif4ebp1fl/fl Scn10aCre mice were injected (intraperitoneally, postnatal days 2 to 4) with an AAV expressing shRNAmir against TRIM32 in a Cre-dependent manner (AAV9-CAG-DIO-GFP-mTRIM32-shRNAmir, referred to as TRIM32-shRNA) or control scrambled sequence (scr-shRNA, Fig. 3B). We confirmed that this approach corrected the increased levels of TRIM32 in 4E-BP1 cKO mice (Fig. 3C). Down-regulation of TRIM32 reversed the mechanical hypersensitivity in 4E-BP1 cKO animals (Fig. 3D), whereas control animals were not affected. Together, these data show that TRIM32 is translationally up-regulated in 4E-BP1 cKO mice and demonstrate that increased TRIM32 expression contributes to mechanical hypersensitivity.
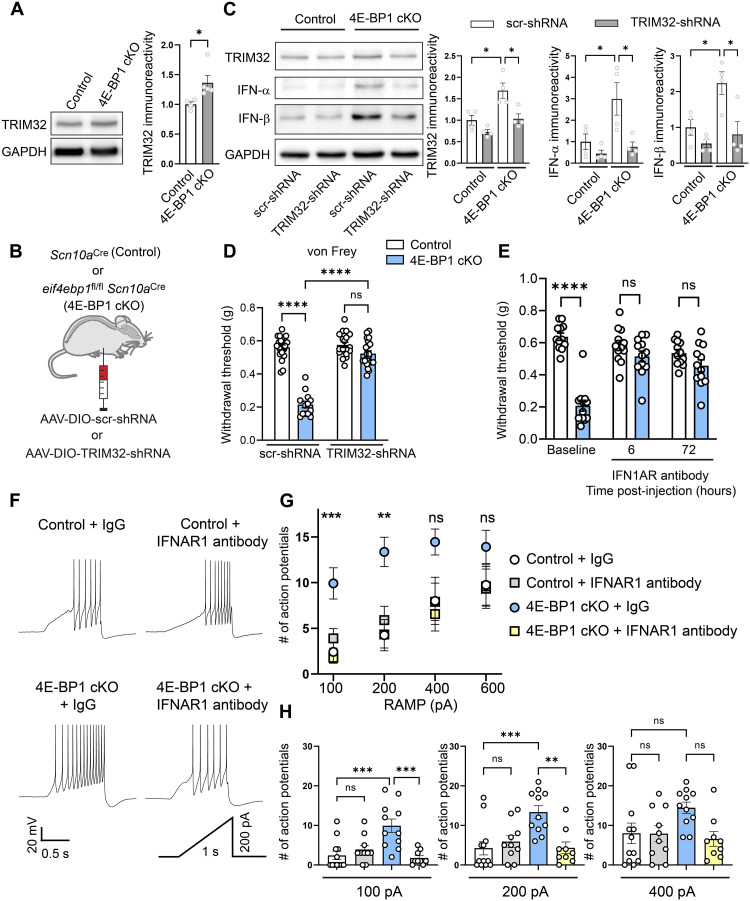
Fig. 3. Down-regulation of TRIM32 alleviates mechanical hypersensitivity in 4E-BP1 cKO.
(A) Western blots and quantification showing increased TRIM32 in DRG lysates of 4E-BP1 cKO mice (n = 4 to 5 mice per group, Student’s t test, two-tailed). (B) Schematic of AAV-DIO-TRIM32-shRNAmir intraperitoneal injections. (C) Western blots and quantification showing corrected TRIM32, IFN-α, and IFN-β in DRG lysates of 8-week-old 4E-BP1 cKO mice that received AAV-DIO-TRIM32-shRNAmir (intraperitoneally, postnatal days 2 to 4, n = 4 mice per group, one-way ANOVA followed by Bonferroni’s post hoc comparison). (D) Mechanical hypersensitivity in the von Frey test was rescued in 8-week-old 4E-BP1 cKO mice that received AAV-DIO-TRIM32-shRNAmir (n = 13 to 21 mice per group, one-way ANOVA followed by Bonferroni’s post hoc comparison). (E) Mechanical hypersensitivity in the von Frey test was corrected in 4E-BP1 cKO mice that received IFNAR1 neutralizing antibody (n = 12 mice per group, two-way ANOVA followed by Bonferroni’s post hoc comparison). Analysis revealed no main effects of sex or interactions for (D) and (E). (F) 4E-BP1 cKO and Scn10a Cre control mice were injected intraperitoneally with AAV-CAG-Flex-tdTomato at P2 to P4 and their DRGs were collected 8 weeks later and cultured for 24 hours. Cultured neurons were stained with IB4 to identify nonpeptidergic nociceptors and incubated in either immunoglobulin G (IgG) control or IFNAR1 neutralizing antibody for 60 min before recordings. Representative traces are shown for 200 pA ramp current injection. (G) 4E-BP1 cKO receiving IgG control had increased number of action potentials at 100 pA and 200 pA ramp intensities (n = 10 to 13 cells from six animals per group, one-way ANOVA followed by Bonferroni’s post hoc comparison). (H) Individual data points for 100-, 200-, and 400-pA stimulations are shown. All data are presented as mean ± SEM. *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001.
TRIM32-mediated type I IFN signaling causes hypersensitivity
TRIM32 plays central roles in innate immunity and antiviral responses by stimulating type I IFN induction via nonproteolytic ubiquitination of the stimulator of interferon genes (STING) (29). Consistent with the elevated levels of TRIM32 in 4E-BP1 cKO mice and the important role of TRIM32 in type I IFN induction, semiquantitative in situ hybridization (RNAScope) showed an increased abundance of interferon-α (IFN-α; Ifnas) and IFN-β (Ifnb1) mRNAs in scn10a+ nociceptors of 4E-BP1 cKO mice (fig. S2). Accordingly, Western blot analysis demonstrated increased protein levels of type I IFN-α and IFN-β in 4E-BP1 cKO DRG lysates compared to control mice (Fig. 3C). The increased protein levels of IFN-α and IFN-β were normalized in 4E-BP1 cKO animals injected with AAV expressing shRNAmir against TRIM32 (Fig. 3C). To assess the physiological role of enhanced type I IFN signaling in mediating mechanical hypersensitivity in 4E-BP1 cKO mice, we used a neutralizing antibody for the interferon alpha and beta receptor subunit 1 (IFNAR1) (30). Inhibition of IFN signaling with IFNAR1 neutralizing antibodies (intraperitoneally) reversed the mechanical hypersensitivity in 4E-BP1 cKO mice (Fig. 3E), indicating that the enhanced activation of this receptor substantially contributes to the hypersensitivity.
4E-BP1 cKO nociceptors exhibit hyperexcitability, which is attenuated by inhibition of IFN signaling
Interferon promotes the excitability of sensory neurons (31). Because IFN signaling is enhanced in 4E-BP1 cKO mice, we assessed the excitability of DRG nociceptors isolated from 4E-BP1 cKO mice. Because 4E-BP1 cKO mice exhibit mechanical but not thermal hypersensitivity, we recorded from nonpeptidergic nociceptors that are thought to predominantly mediate mechanical hypersensitivity (25, 26). To identify nonpeptidergic nociceptors, we injected eif4ebp1fl/fl Scn10aCre and control Scn10aCre mice at postnatal days 2 to 4 with AAV expressing tdTomato in a Cre-dependent manner (AAV2/9-CAG-DIO-tdTomato) to label Nav1.8+ neurons and applied fluorescent IB4 (Alexa Fluor 488), a marker of nonpeptidergic neurons, to the extracellular solution for 2 min before recording. Recording from 4E-BP1 cKO nonpeptidergic nociceptors (positive for tdTomato and IB4) revealed an increase in the number of action potentials in response to ramp current injection (100 pA and 200 pA) as compared to controls (Fig. 3, F to H). Incubation of cultured neurons for 1 hour with IFNAR1 neutralizing antibody reduced the number of stimulation-induced action potentials in 4E-BP1 cKO nociceptors (Fig. 3, F to H). Thus, ablation of 4E-BP1 in nociceptors increases their excitability and enhanced type I IFN signaling contributes to this effect.
Down-regulation of TRIM32 alleviates inflammatory pain
mTORC1 is activated in nociceptors in models of neuropathic and inflammatory pain (9–14) and mTORC1 inhibition alleviates the hypersensitivity in both conditions (9, 10, 15–22). Thus, we tested the prediction that mTORC1–4E-BP1–mediated expression of TRIM32 and type I IFN signaling contribute to pain hypersensitivity in models of chronic pain. We first confirmed previous reports that mTORC1 is activated peripherally in neuropathic and inflammatory pain conditions (9–14). We subjected mice to a model of neuropathic [spared nerve injury (SNI)] and inflammatory [intraplantar injection of complete Freund’s adjuvant (CFA)] pain, and measured mTORC1 activation in DRG by assessing the phosphorylation of S6 (p-S6), which is the mTORC1 downstream effector and most commonly used readout of its activity. Consistent with prior studies showing sustained mTORC1 activation in DRG after peripheral tissue injury (32, 33), p-S6 levels were increased in DRG lysates from SNI animals (day 4 post-surgery, fig. S3A) compared with sham controls, and in CFA-injected animals (day 1 post-injection) compared with vehicle controls (Fig. 4A). Concomitant with the increase in p-S6, phosphorylation of 4E-BP1 at Thr37/46 was also elevated in CFA-injected animals (Fig. 4A). Notably, protein levels of TRIM32, as well as IFN-α and IFN-β, were up-regulated in the DRG of SNI (fig. S3A) and CFA (Fig. 4A) animals.
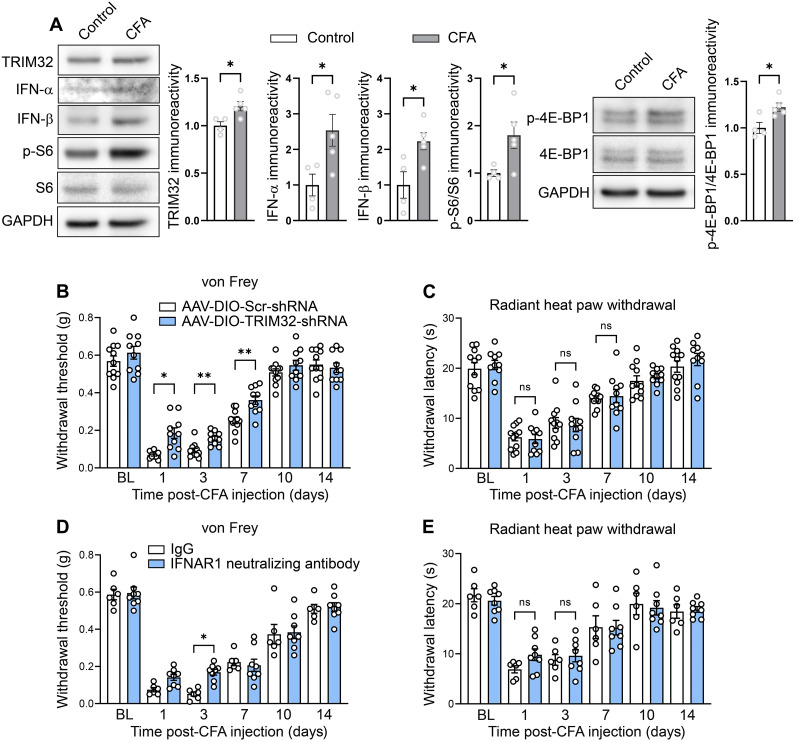
Fig. 4. Down-regulation of TRIM32 alleviates CFA inflammatory pain.
(A) Western blots and quantification showing increased TRIM32, IFN-α, IFN-β, p-S6, and p-4E-BP1 in DRG lysates of animals 1 day after CFA intraplantar injection compared to vehicle control (n = 4 to 5 mice per group, Student’s t test, two-tailed). Animals that received AAV-DIO-TRIM32-shRNAmir showed alleviated mechanical (B) but not thermal (C) hypersensitivity compared to scrambled controls after intraplantar injection of CFA (n = 8 to 10 mice per group, two-way ANOVA followed by Bonferroni’s post hoc comparison). Animals that received IFNAR1 neutralizing antibody showed alleviated mechanical (D) but not thermal (E) hypersensitivity compared to IgG controls after intraplantar injection of CFA (n = 6 to 8 mice per group, two-way ANOVA followed by Bonferroni’s post hoc comparison). All data are presented as mean ± SEM. *P < 0.05; **P < 0.01.
To investigate whether TRIM32 plays a role in mediating pain hypersensitivity, we down-regulated TRIM32 in nociceptors by injecting Scn10aCre pups with AAV9-CAG-DIO-GFP-mTRIM32-shRNAmir or control virus (AAV9-CAG-DIO-GFP-scrambled) at postnatal days 2 to 4 and subjected them to SNI and CFA pain models at 8 weeks of age. We found that whereas mechanical hypersensitivity caused by peripheral nerve injury was not affected by the down-regulation of TRIM32 in nociceptors (fig. S3B), it was alleviated in a model of inflammatory pain (Fig. 4B). CFA intraplantar injection in control mice caused mechanical and thermal hypersensitivity (Fig. 4, B and C). However, in mice with down-regulation of TRIM32 in nociceptors, CFA-induced mechanical hypersensitivity was reduced whereas thermal hyperalgesia was not affected (Fig. 4, B and C). Consistent with this finding, intraperitoneal administration of IFNAR1 neutralizing antibodies selectively alleviated the CFA-induced mechanical hypersensitivity (Fig. 4, D and E). Overall, our results describe a signaling cascade in nociceptors, via mTORC1–4E-BP1–dependent translation of TRIM32 and up-regulation of type I IFN signaling, in mediating mechanical hypersensitivity (Fig. 5).
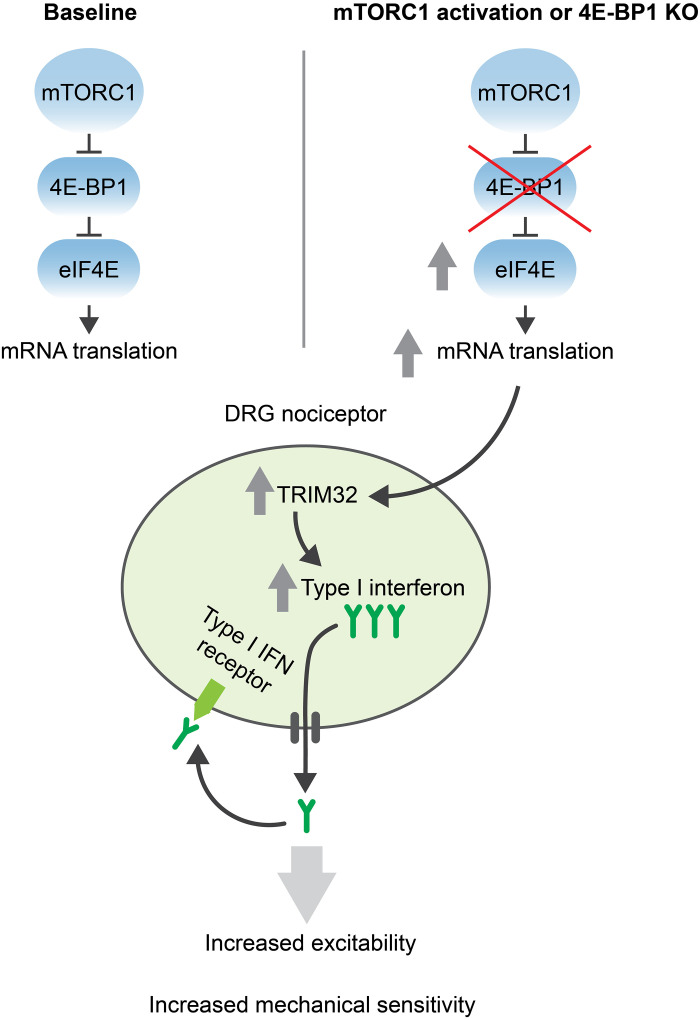
Fig. 5. Proposed model.
Ablation of 4E-BP1, mimicking the hyperactivation of the mTORC1 pathway, in DRG nociceptors, results in increased eIF4E-dependent mRNA translation, including of TRIM32 mRNA. TRIM32 promotes type I IFN signaling by nonproteolytic ubiquitination of STING. Increased production of type I IFN and its autocrine signaling enhances nociceptor excitability, resulting in mechanical hypersensitivity.
DISCUSSION
Here, we found that stimulation of 4E-BP1–dependent translation selectively in nociceptors engenders mechanical hypersensitivity. Furthermore, we showed that 4E-BP1 ablation induces mechanical hypersensitivity via enhanced translation of TRIM32 and increased type I IFN signaling. Targeting TRIM32/type I IFN alleviates mechanical hypersensitivity in a model of inflammation-induced pain.
It is well established that mTORC1 is activated in DRG neurons after tissue injury and its inhibition alleviates pain hypersensitivity; however, the specific cell types and proteins mediating this effect are not well defined. Moreover, previous studies have used mTORC1 pharmacological inhibitors, such as rapamycin and rapalogs, to study its function in pain. Inhibition of mTORC1 affects numerous mTORC1-dependent processes in addition to mRNA translation, such as lipid and nucleotide biosynthesis, autophagy, and glucose metabolism (5). Therefore, using this approach, it is challenging to study the selective contribution and mechanisms of mTORC1-dependent translation in the development of pain, which is the focus of the current study. To address these important questions, we generated mice with specific ablation of 4E-BP1 in nociceptors, mimicking hyperactivation of mTORC1-eIF4E–dependent translation. Activation of mTORC1-eIF4E translation in nociceptors elicited pronounced mechanical hypersensitivity, without affecting heat sensation, consistent with the mechanical hypersensitivity phenotype in whole-body 4E-BP1 KO mice (24). Using the TRAP methodology, we defined the translational landscape in nociceptors lacking 4E-BP1 and identified a substantial translational up-regulation for the mRNA encoding an E3 ubiquitin ligase TRIM32 (9.41-fold increase in IP/IN ratio), which is involved in responses to viral infection. TRIM32 stimulates type I IFN signaling by targeting STING for K63-linked nonproteolytic type of ubiquitination (29) in vitro and in vivo under viral infections characterized by DNA virus–triggered induction of type I IFN (34–36). To study the role of TRIM32 in pain, we down-regulated TRIM32 selectively in nociceptors and found that it corrects mechanical hypersensitivity of 4E-BP1 cKO mice, demonstrating its importance in mediating the mTORC1-eIF4E–dependent mechanical hypersensitivity. Consistent with the role of TRIM32 in inducing type I IFN, we also found that both IFN-α and IFN-β were up-regulated in 4E-BP1 cKO mice and revealed that inhibition of IFN signaling, using IFNAR1 neutralizing antibodies, alleviated the mechanical hypersensitivity in 4E-BP1 cKO animals. Finally, we assessed the role of TRIM32 and IFN in models of chronic pain. Down-regulation of TRIM32 and inhibition of IFNAR1 with neutralizing antibodies alleviated the mechanical but not thermal hypersensitivity in response to peripheral inflammation. Down-regulation of TRIM32, however, had no effect on neuropathic pain, likely because nonnociceptive neurons and spinal mechanisms predominantly mediate the mechanical hypersensitivity caused by peripheral nerve injury.
Translational control via the mTORC1–4E-BP1 axis plays crucial roles in antiviral resistance. Deletion of 4E-BP1 in mouse embryonic fibroblasts enhanced IFN signaling (37) and potentiated viral immunity (29). Our findings of increased levels of IFN-α and IFN-β in 4E-BP1 cKO mice establish the role of the mTORC1–4E-BP1 axis in the regulation of IFN signaling in the pain pathway, demonstrating that this mechanism is shared between antiviral responses and chronic pain.
Our work shows the involvement of TRIM32 in inflammatory pain and reveals TRIM32 as a potential therapeutic target in inflammatory pain conditions. We found that selective down-regulation of TRIM32 in nociceptors alleviates mechanical but not thermal hypersensitivity induced by CFA injection. Although the mechanism underlying the modality specificity of TRIM32 is not completely understood, intraplantar administration of IFN elicited mechanical hypersensitivity by increasing the excitability of DRG neurons, with no effect on thermal sensitivity (31). Our findings are consistent with these results as 4E-BP1 cKO mice demonstrated enhanced IFN levels in DRG and exhibited nociceptor hyperexcitability and selective mechanical hypersensitivity. The selective mechanical hypersensitivity observed in 4E-BP1 cKO mice is likely mediated by both the preferential expression of 4E-BP1 in nonpeptidergic fibers, which mediate noxious mechanical sensation (25, 26), and the specific impact of peripheral type I IFNs on mechanical sensitivity (31). Collectively, this study enhances the understanding of the mechanisms through which mTORC1 promotes pain hypersensitivity, unveiling the role of the mTORC1–4E-BP1–IFN axis in regulating mechanical hypersensitivity after peripheral inflammation.
In our study, we focused on the translational up-regulation of TRIM32 in 4E-BP1 KO nociceptors as it was the second-ranked mRNA exhibiting an increase in ribosome occupancy in 4E-BP1 KO nociceptors, and one of the most highly up-regulated mRNAs after normalization to transcript levels. Furthermore, we took into consideration the established link between TRIM32-mediated IFN signaling and mechanical hypersensitivity (31). However, the roles of other 4E-BP1–dependent mRNAs identified in nociceptors, which could potentially contribute to increased excitability and hypersensitivity, remain to be determined in future studies.
The increase in the IP/IN ratio for TRIM32 mRNA in 4E-BP1 cKO mice in the TRAP analysis (9.41-fold increase) was higher than the increase in TRIM32 protein levels in total DRG lysates (increase of 36 to 65%). The IP/IN ratio was assessed in 4E-BP1 KO nociceptors using IP of tagged ribosomes, whereas protein levels of TRIM32 were evaluated in total DRG lysates, where the presence of nonnociceptive neuronal and nonneuronal cells likely dilutes the effect. Moreover, an increase in translational efficiency typically does not yield an equivalent increase in protein levels, especially in chronic models, due to phenomenon of translational buffering (38).
In summary, our study uncovered the peripheral inflammation-induced and mTORC1–4E-BP1–mediated up-regulation of TRIM32, which acts to promote type I IFN signaling, neuronal excitability, and mechanical hypersensitivity. Our work advances the understanding of translational control mechanisms by which mTORC1 promotes pain and highlights TRIM32 as a potential therapeutic target for conditions characterized by inflammation-induced mechanical hypersensitivity.
MATERIALS AND METHODS
Animals
Eif4ebp1fl/fl mice were crossed with mice expressing Cre recombinase under the control of the Scn10a (Nav1.8) promoter to generate the cKO animals: Eif4ebp1fl/fl Scn10aCre (referred to as 4E-BP1 cKO). Control animals used were Scn10aCre. Mice were weaned at the third postnatal week, and ear samples were collected for genotyping using polymerase chain reaction (PCR). eIF4E-BP1 mutant and wild-type alleles were detected using PCR assay in which primer eIF4E-BP1 F (5′-CACATTTCAGGGAGAGGGTGATG-3′) and primer eIF4E-BP1 R (5′-GCTGGGTTCTAAGAGTGGTACTTT-3′) amplified a 250–base pair (bp) fragment (wild type) and a 347-bp fragment (Exon 2 of the eIF4E-BP1 conditional allele). Cre expression was detected by PCR with primers CreF (5′-GATCTCCGGTATTGAAACTC-3′) and CreR (5′-GCTAAACATGCTTCATCGTC-3′), which amplify a 650-bp fragment. All experiments were conducted on 8- to 12-week-old male and female mice of the C57BL/6 background. To assess for the presence of main effects of sex or interactions of sex with experimental manipulations, key datasets (Figs. 1, D to H, and 3, D and E) were analyzed using sex as a factor. In no cases were any main effects of sex or interactions observed, so data were collapsed for all reported analyses. Food and water were available ad libitum. Mice were kept on a 12-hour light/dark cycle (lights on at 0700). Behavioral experiments were conducted during the light phase of the cycle (0900 to 1700). All procedures complied with the Canadian Council on Animal Care guidelines and International Association for the Study of Pain, and were approved by the McGill University’s downtown Animal Care Committee.
Animal model of neuropathic pain
All animals were 8 weeks of age when undergoing surgical procedures. Neuropathic pain was induced by performing SNI, as described previously (39, 40). The lateral surface of the thigh skin was shaved and incised, the left sciatic nerve was isolated, and the tibial and common peroneal branches were ligated using 7-0 silicone coated silk (Covidien, S-768 K) and a 3-mm portion of each branch was sectioned and removed distal to the ligation point. The muscle and skin were closed with 6-0 Vicryl suture (Ethicon J489G).
Animal model of inflammatory pain
CFA and incomplete Freund’s adjuvant were mixed (1:1; both from Sigma-Aldrich, St. Louis, MO) to produce an emulsion, 20 μl of which was delivered under the skin of the left hind paw (intraplantar).
Reagents and materials
The IFN1AR neutralizing antibody (MAR1-5A3) was purchased from Thermo Fisher Scientific. Animals were injected intraperitoneally with a dose (2.5 mg/kg) consistent with reduced IFN activity in vivo (30).
Western blotting
Samples were homogenized in ice-cold lysis buffer consisting of 200 mM Hepes, 50 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 50 mM NaF, 2 mM Na3VO4, 25 mM β-glycerophosphate, and EDTA-free complete ULTRA tablet. A total of 30 μg of protein per sample was resolved on SDS–polyacrylamide gel electrophoresis (12%), transferred onto nitrocellulose membranes, and imaged using a ChemiDoc Imaging System (Bio-Rad).
Primary antibodies for Western blotting are p-4E-BP1, 4E-BP1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), p-S6 (1:1000; #2855S, #9644S, #2118S, and #4060S, respectively, Cell Signaling and Technology Laboratories, Danvers, MA), TRIM32, IFN-α, and IFN-β (1:1000; #10326-1-AP, #XL3766809, and #XE3598743, respectively, Thermo Fisher Scientific) raised in Rabbit and t-S6 (1:1000; #4691S, Cell Signaling and Technology Laboratories, Danver, MA) raised in mouse. Secondary antibodies for Western blotting are anti-rabbit and anti-mouse horseradish peroxidase (1:5000, #RPN4301, GE Healthcare).
Behavioral testing
The experimenters were blind to the genotype and drug treatments for all behavioral tests. In the von Frey assay, mice were habituated in individual transparent custom Plexiglas cubicles (5 cm by 8.5 cm by 6 cm) set above a perforated steel floor for 1 hour before testing. Nylon monofilaments were applied firmly to the plantar surface of the ipsilateral and contralateral hind paws for 3 s. The up-down method of Dixon (41) was applied to estimate the 50% withdrawal threshold (average of two measurements).
In the tail clip assay, a clip exerting a force of 155g or 248g was applied 1 cm from the base of the tail. Animals were placed in an enclosure (15 cm by 25 cm), and the latency to attacking the clip was measured. The cutoff for responses was set at 60 s.
In the radiant heat paw-withdrawal assay, mice were habituated in individual transparent custom Plexiglas cubicles (5 cm by 8.5 cm by 6 cm) set above a transparent glass floor for 1 hour before testing. A focused beam of high-intensity light was aimed at the plantar surface of both hind paws. An average of four measurements were taken.
In the tail flick assay, animals were restrained in a cloth restrainer, and the tail was placed into a temperature-controlled water bath of either 47° or 49°C. The latency to the animal flicking its tail was measured. The cutoff for response was set at 30 and 15 s, respectively.
In the hot plate test, we used a hot plate apparatus (Columbus Instruments) set at either 50° or 53°C. Animals were placed on the hot plate enclosed within an open Plexiglas cylindrical tube, and latency to demonstrating any nocifensive responses—jumping, hind paw licking, or rapid fluttering/shaking of the hind paw—was measured. The cutoff for response was set at 90 and 60 s, respectively.
Immunohistochemistry, confocal microscopy, and image analysis
Animals were perfused via the left cardiac ventricle with 10 ml of 0.1% NaNO2 perfusion buffer [64 g of NaCl, 2 g of KCl, 4 g of NaHCO3, 400 ml of 0.2 M phosphate buffer (PB) in 8 liters of ddH2O], followed by 100 ml of 4% paraformaldehyde (PFA) in 0.1 M PB. The lumbar 3 to 5 DRGs were extracted and postfixed in the same fixative solution at 4°C. DRGs were transferred to a cryoprotectant solution 24 hours later consisting of 30% sucrose in 0.1 M PB. DRGs were embedded in OCT (optimal cutting temperature compound, Thermo Fisher Scientific), sectioned at 14 μm using a Leica cryostat, and collected directly onto gelatin-subbed histological slides.
Three 10-min washes using 0.2% phosphate-buffered saline (PBS)–Triton X-100 (PBS-T) were conducted on DRG sections and incubated for 24 hours at 4°C in rabbit anti-4E-BP1 (1:500; #2855S, Cell Signaling and Technology Laboratories, Danvers, MA), rabbit anti-CGRP (1:500; #C89198, Sigma-Aldrich, St. Louis, MO), mouse anti-NF200 antibody (1:500; #N5389, Sigma-Aldrich, St. Louis, MO), and IB4 conjugated to Alexa Fluor 568 (1:500; #I21411, Thermo Fisher Scientific) in 0.2% PBS-T. The next day, after several washes with 0.2% PBS-T, sections were incubated in goat secondary antibodies (Alexa Fluor, 1:500; Thermo Fisher Scientific) in 0.2% PBS-T for 2 hours at room temperature. Sections were then washed once for 10 min using 0.2% PBS-T followed by two 10-min washes using PBS and cover-slipped using Aqua Poly/Mount (Polysciences Inc.).
For high-resolution imaging, DRG sections were imaged using an LSM 880 confocal microscope with Airyscan (Zeiss); the AiryScan mode was conducted using Zeiss 63/1.40 Oil DIC f/ELYRA objective and the AiryScan super-resolution module with a 32-channel hexagonal array GaAsP detector for LSM (Zeiss) using 651-nm lasers. Stacks of 20 to 30 optical sections (170-nm step) were acquired and AiryScan super-resolution image stacks were reconstructed using ZEN Black software (Zeiss).
Images were analyzed using ImageJ (NIH) software. For quantification, we analyzed four to six sections per animal, for a total of n = 4 animals.
Translating ribosome affinity purification
4E-BP1 cKO and Cre-positive control postnatal day 2 to 4 pups were injected intraperitoneally with 5 μl of AAV2/9-FLEX-eGFP-L10a (Canadian Neurophotonics Platform Viral Vector Core Facility RRID:SCR_016477), and TRAP-sequencing was performed on 8- to 10-week old mice, as previously described (27, 42).
Briefly, animals were decapitated and the cervical, thoracic, and lumbar DRGs were dissected and transferred into ice-cold dissection buffer [1× Hanks’ balanced salt solution (HBSS), 2.5 mM Hepes-NaOH (pH 7.4), 35 mM glucose, and 5 mM MgCl2; cycloheximide (100 μg/ml) and emetine (0.2 mg/ml) were added just before use]. Once all the DRGs were collected, they were homogenized in lysis buffer [20 mM Hepes-NaOH (pH 7.4), 12 mM MgCl2, and 150 mM KCl in RNase-free water; 0.5 mM dithiothreitol (DTT), Protease Inhibitors Cocktail (1 μl/ml; Roche), cycloheximide (100 μg/ml), emetine (20 μg/ml), RNasin (40 U/ml; Promega), and TURBO DNase (2 U/ml; Invitrogen) were added immediately before use] using a Minilys Personal High Power Tissue Homogenizer (Bertin Technologies) on medium speed for 10 s for a total of eight times at 4°C. DRG homogenates were centrifuged at 2000g for 5 min at 4°C to prepare a post-nuclear fraction (S2). Next, NP-40 (AG Scientific) and DHPC (1,2-diheptanoyl-sn-glycero-3-phosphocholine, Cat# 850306, Avanti Polar lipids) were added to the S2 fraction at a final concentration of 1% and incubated for 5 min at 4°C, followed by centrifugation at 15,000g for 15 min at 4°C to generate a post-mitochondrial fraction (S20). A 200-μl sample of S20 was removed for use as “input”, and 1 μl of TURBO DNase was added to 800 μl of S20 and was incubated with protein-washed G-coated Dynabeads (Invitrogen) bound to 50 μg of anti-GFP antibodies (HtzGFP-19F7 and HtzGFP-19C8 antibodies were acquired from Sloan Memorial Kettering Center) for 3 hours at 4°C on an end-over-end mixer. Subsequently, the beads were collected using a magnetic stand and 200 μl of the “unbound’ fraction was saved for further processing. The beads were washed four times with high salt buffer [20 mM Hepes-NaOH (pH 7.4), 12 mM MgCl2, 0.35 M KCl, and 1% NP-40 (AG Scientific) in RNase-free water; cycloheximide (100 μg/ml) and 0.5 mM DTT were added just before use] to collect the “immunoprecipitated (IP)” fraction. RNA was extracted by adding 300 μl of TRIzol to the beads (“IP” fraction) and 600 μl of TRIzol to the “input” fractions and incubated for 10 min at room temperature. RNA was eluted from all the samples using a Direct-zol RNA kit (Zymo Research) using the manufacturer’s protocol. RNA yield was quantified using a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies Inc.) and RNA quality was determined by a 2100 Bioanalyzer (Agilent Technologies).
Library generation and sequencing
We sequenced the following samples: three immunoprecipitated (IP) replicates paired with three input replicates, which were obtained from pooling three mice per replicate, for each group (4E-BP1 cKO and Cre-positive controls). Each replicate consisted of three mice of the same sex, two replicates are male and one replicate is female for both groups. Total RNA quality check, library generation, library quality check, and sequencing were carried out at the Génome Québec Innovation Centre (McGill University, Montréal, Canada) as per their internal platform protocols. Briefly, rRNA was depleted from total RNA using the QIAseq FastSelect-Human/Mouse/Rat kit (Qiagen) using the manufacturer’s recommended protocol with slight modifications. The shearing time was reduced from 15 to 8 min. Next, cDNA was synthesized using the NEB Next RNA First Strand Synthesis and NEB Next Ultra Directional RNA Second Strand Synthesis Modules (New England BioLabs). The remaining steps of library preparation were done using the NEB Next Ultra II DNA Library Prep Kit for Illumina (New England BioLabs). Adapters and PCR primers were purchased from New England BioLabs. Libraries were quantified using the KAPA Library Quantification Kits - Complete kit (Universal) (Kapa Biosystems). Average size fragment was determined using a LabChip GX (PerkinElmer) instrument.
The normalized and pooled libraries (3 nM) were denatured and neutralized using 0.05 NaOH and HT1 buffer (Illumina) respectively. Subsequently, ExAMP reagent cocktail (Illumina) was added and 360 pM of pooled libraries were loaded on a cBot (Illumina) and the flowcell was run on HiSeq 4000 (Illumina) for 1 × 50 cycles in single-end mode to generate 50-bp, single-end reads and expected sequencing depth of 50 M reads/sample. A phiX library was used as a control and mixed with libraries at the 1% level. The Illumina control software was HCS HD 3.4.0.38, and the real-time analysis program was RTA v. 2.7.7. Program bcl2fastq2 v2.20 was then used to demultiplex samples and generate fastq reads.
Bioinformatics and statistical analysis
Mapping and TPM quantification
The quality of fastq files was checked using FastQC (Babraham Bioinformatics, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were trimmed based on the Phred score and per-base sequence content. Trimmed reads were then aligned against the mouse reference genome and transcriptome (Gencode vM16 and GRCm38.p5) using STAR v2.2.1 (43). We obtained the relative abundances in transcripts per million (TPM) for every gene of every sample using StringTie v1.3.5 (44). We removed the noncoding genes and mitochondrial genes before proceeding with downstream analysis (based on Gencode annotation) and the TPMs for coding genes were re-normalized to sum to 1 million.
Order statistics and re-normalization of expression data
Downstream analysis of the TRAP dataset was done as previously described (42). Briefly, we calculated the percentile ranks for each coding gene for each sample with the aim of identifying a set of consistently expressed genes in the transcriptome (INPUT) samples. We conservatively identified 15,129 genes that were above the 30th percentile in each INPUT sample, for at least one group, to be in the set of genes considered consistently detected in the transcriptome. Quantile normalization was then performed based on the set of all coding genes.
The IP (translatome) analysis was only performed for the 15,129 consistently transcriptome-expressed genes. To identify a set of consistently expressed genes in the translatome (IP) samples, we calculated the percentile ranks on TPMs for each of the 15,129 consistently transcriptome-expressed coding genes for each sample. We then identified 12,553 genes out of those 15,129 genes to be consistently detected in the translatome based on whether their expression was on or above the 15th percentile (out of 15,129 genes) in each IP sample, for at least one group.
The percentile thresholds for choosing consistently transcriptome-expressed and translatome-expressed genes were conservatively estimated by identifying thresholds that would eliminate genes with consistently low or no detected reads.
Differential expression analysis
Differential expression (DE) analysis was done as previously described (42). We first calculated the log2-fold change (based on median TPMs) for each consistently transcriptome-expressed coding gene in the INPUT, and for each consistently translatome-expressed coding gene in the IP. We used strictly standardized mean difference (SSMD) (42, 45) to discover genes with systematically altered expression percentile ranks between the WT and 4E-BP1–cKO groups. SSMD is the difference of means controlled by the variance of the sample measurements. We used SSMD as a measure of effect size because it is appropriate for smaller sample sizes while simultaneously controlling for within-group variability. To determine whether a gene is differentially expressed between WT and cKO, we also calculated the Bhattacharyya distance (46). This measure is used to calculate the amount of overlap in the area under the curve of the two sample distributions (corresponding to each group). In contrast to SSMD, BD does not make assumptions of equal variance in the two compared samples and, thus, is useful for comparing distributions of gene relative abundance (in TPMs). The Bhattacharyya coefficient BC(Q)i ranges between 0 (for totally nonoverlapping distributions) and 1 (for completely identical distributions) and is derived from the Bhattacharyya distance. In our analysis, we used a modified form of the Bhattacharyya coefficient that ranges between 0 (for completely identical distributions) and +1 or −1 (for totally nonoverlapping distributions, sign defined by the log-fold change value). DE genes were identified if the absolute value of SSMD was higher than or equal to 0.97, the absolute value of BC was higher than or equal to 0.5, and fold change is higher than or equal to 1.5. Coding for bioinformatics analysis and data visualization was done in Python (version 3.7 with Anaconda distribution).
Whole-cell patch recording in acutely dissociated mice DRG neurons
DRG cell culture preparation
Before whole-cell recordings, 4E-BP1 cKO and Cre-positive control postnatal day 2 to 4 pups were injected intraperitoneally with AAV2/9-CAG-FLEX-tdTomato (Canadian Neurophotonics Platform Viral Vector Core Facility). DRGs were extracted from male and female 8- to 10-week-old animals and incubated in HBSS containing dispase (1.37 mg/ml, GIBCO) and collagenase II (1.08 mg/ml, GIBCO) for 30 min at 37°C. After two washes in complete medium [Ham’s F-12 Nutrient Mixture, 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, and 1% penicillin-streptomycin], DRGs were mechanically dissociated using three fire-polished Pasteur pipettes with sequentially decreasing diameters. The cells were concentrated by centrifugation (1,000 rpm for 2 min), resuspended in culture medium, and plated on the glass bottom of the 35-mm dish precoated with a mixture of poly-d-lysine (100 μg/ml) and laminin (10 μg/ml) in HBSS.
To identify nonpeptidergic cells, neurons were incubated with IB4 Alexa Fluor 488 (3 μg/ml; Thermo Fisher Scientific) for 5 min and rinsed before recording. For experiments with IFNAR1 neutralizing antibody (Thermo Fisher Scientific) or a mouse IgG1 kappa isotype control antibody (Thermo Fisher Scientific), antibodies were incubated in the bath for at least 45 min at a concentration of 10 μg/ml (47).
Current clamp recordings
Recordings were performed from Nav1.8 tdTomato+, IB4+, and <25-μm-diameter dissociated DRG neurons, up to 48 hours after plating. Whole-cell membrane voltages were recorded using a Multiclamp 700B amplifier (Molecular Devices) at room temperature (24 ± 2°C). Data were sampled at 20 kHz and low pass filtered at 500 Hz. Patch pipettes (4 to 8 megohms) were pulled from borosilicate glass capillaries and fire polished. The intracellular solution contained 130 mM K-gluconate, 10 mM Hepes, 5 mM EGTA, 3 mM Mg-ATP, and 0.4 mM GTP (pH 7.3). The extracellular solution contained 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, and 10 mM d-glucose. pH and osmolarity were adjusted to 7.4 and 300, respectively. Command current protocols were generated with a Digidata 1550B A/D interface (Molecular Devices). Data were digitized using pCLAMP 10.3 (Molecular Devices). Data analysis was performed using Clampfit 10.7 software.
Neurons were recorded under current clamp conditions and held at −60 mV. Resting membrane potential (RMP) was obtained at zero current, and cells with RMP less than −40 mV or those in which the access resistance changed more than 25% from its initial value during the recordings were excluded from analysis. Only cells that responded to the current ramp test (at least one spike at 600 pA) were considered for future analysis. The liquid junction potential (calculated to be +5.6 mV) was not corrected. A series of depolarizing square current injections (500 ms with a 2-s interval and 25-pA increments) were used to measure action potential parameters measured from the first action potential evoked. Input resistance and the sag were measured by separate hyperpolarizing square pulses at −20 pA and −80 pA, respectively. Number of spikes was measured by injecting 1-s ramp currents from baseline up to 600 pA.
Fluorescent RNA in situ hybridization using RNAScope
Animals were perfused via the left cardiac ventricle with 10 ml of 0.1% NaNO2 perfusion buffer, followed by 100 ml of 4% PFA in 0.1 M PB. The lumbar 3 to 5 DRGs were extracted and postfixed in the same fixative solution at 4°C for 24 hours. DRGs were transferred to a cryoprotectant solution 24 hours later consisting of 10% sucrose in PBS for 18 hours, followed by 20% sucrose in PBS for 18 hours, and lastly 30% sucrose in PBS for 18 hours. DRGs were embedded in OCT (Thermo Fisher Scientific), sectioned at 14 μm using a Leica cryostat, and collected directly onto gelatin-subbed histological slides. The RNAScope Multiplex Fluorescent V2 Assay (ACD Biosystems) was performed according to the ACD protocol for fixed-frozen tissue. DRG sections were hybridized with three mRNA probes: Scn10a (426011), Ifnb1 (406531), and Ifnas (471691). The fluorophores TSA Vivid 520, 570, and 650 were used for labeling probes. DAPI (4′,6-diamidino-2-phenylindole) was used for nuclear counterstaining, and slides were mounted with ProLong Gold Antifade mounting medium (Life Technologies). Images were acquired on an LSM 880 confocal microscope with 63/1.40 Oil DIC f/ELYRA. Stacks of 12 to 18 optical sections (0.8-μm step intervals) were reconstructed using ZEN Black software (Zeiss). Images were analyzed using ImageJ (NIH) software. For quantification, the number of puncta clusters corresponding to overlapping mRNA molecules was counted for Ifnb1 and Ifnas probes. We analyzed 8 to 12 cells per animal (using the Scn10a probe marker for nociceptors), for a total of n = 3 animals per group.
Preparation of AAV9-CAG-DIO-GFP-mTRIM32-shRNAmir
AAV9-shRNA viral particles were prepared by Vector Biolabs. The validated sequence targeting TRIM32 was 5′-GCT GTTCAGTGAAGGACTCCATGCAGTTTTGGC CACT GACTGACTGCATGGACCTTCACTGAA CAG-3′.
Statistical analysis
All experimenters and data analysts were blinded to genotype and treatment. No nested data were collected; only one observation per research object. Student’s t test and two-way analysis of variance (ANOVA) were used to analyze behavioral, immunohistochemistry, electrophysiology, and Western blot experiments. All data are reported as mean ± SEM and analyzed using GraphPad Prism 8 (GraphPad Prism Software Inc., USA). The level of significance was set a priori at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We thank J. Wood (UCL) for the Scn10aCre mice.
Funding: This work was supported by Canadian Institutes of Health Research PJT-162412 (A.K.), General Secretariat for Research and Innovation Greece Τ12ΕΡΑ5-00024 (C.G.G.), ERA-NET Neuron Sensory disorders project TRANSMECH (A.K. and C.G.G.), Canadian Institutes of Health Research PJT-183684 (P.S.), NIH NINDS grant NS065926 (T.J.P.), and Louise and Alan Edwards Foundation PhD fellowship (C.W.).
Author contributions: C.W., C.G.G., and A.K. conceived and designed the study, and supervised research; C.W., D.T.-F., C.T.P., B.S., S.U., K.C.L., M.H., and V.N. performed experiments; C.W., D.T.-F., C.T.P., B.S., S.U., M.A., and V.N. analyzed data; P.S., N.S., T.J.P., and C.G.G. helped with interpretation of results; all authors discussed the work and reviewed the manuscript; C.W., C.G.G., and A.K. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data generated in this study have been deposited in the Gene Expression Omnibus under accession no. GSE227978.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.A. Pokhilko, A. Nash, M. Z. Cader, Common transcriptional signatures of neuropathic pain. Pain 161, 1542–1554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. S. Yousuf, S. I. Shiers, J. J. Sahn, T. J. Price, Pharmacological manipulation of translation as a therapeutic target for chronic pain. Pharmacol. Rev. 73, 59–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. E. Holt, K. C. Martin, E. M. Schuman, Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 26, 557–566 (2019). [DOI] [PubMed] [Google Scholar]
- 4.J. Jung, J. Ohk, H. Kim, C. E. Holt, H. J. Park, H. Jung, mRNA transport, translation, and decay in adult mammalian central nervous system axons. Neuron 111, 650–668.e4 (2023). [DOI] [PubMed] [Google Scholar]
- 5.G. Y. Liu, D. M. Sabatini, mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. H. Melick, J. L. Jewell, Regulation of mTORC1 by upstream stimuli. Genes (Basel) 11, 989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. O. Lipton, M. Sahin, The neurology of mTOR. Neuron 84, 275–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. Kim, K. L. Guan, mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71 (2019). [DOI] [PubMed] [Google Scholar]
- 9.S. M. Geranton, L. Jimenez-Diaz, C. Torsney, K. K. Tochiki, S. A. Stuart, J. L. Leith, B. M. Lumb, S. P. Hunt, A rapamycin-sensitive signaling pathway is essential for the full expression of persistent pain states. J. Neurosci. 29, 15017–15027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L. Jimenez-Diaz, S. M. Geranton, G. M. Passmore, J. L. Leith, A. S. Fisher, L. Berliocchi, A. K. Sivasubramaniam, A. Sheasby, B. M. Lumb, S. P. Hunt, Local translation in primary afferent fibers regulates nociception. PLOS ONE 3, e1961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. K. Moy, A. Khoutorsky, M. N. Asiedu, B. J. Black, J. L. Kuhn, P. Barragan-Iglesias, S. Megat, M. D. Burton, C. C. Burgos-Vega, O. K. Melemedjian, S. Boitano, J. Vagner, C. G. Gkogkas, J. J. Pancrazio, J. S. Mogil, G. Dussor, N. Sonenberg, T. J. Price, The MNK–eIF4E signaling axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. Neuroscience 37, 7481–7499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O. K. Melemedjian, M. N. Asiedu, D. V. Tillu, R. Sanoja, J. Yan, A. Lark, A. Khoutorsky, J. Johnson, K. A. Peebles, T. Lepow, N. Sonenberg, G. Dussor, T. J. Price, Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol. Pain 7, 1744-8069-7-70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. Khoutorsky, T. J. Price, Translational control mechanisms in persistent pain. Trends Neurosci. 41, 100–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L. J. Martin, S. B. Smith, A. Khoutorsky, C. A. Magnussen, A. Samoshkin, R. E. Sorge, C. Cho, N. Yosefpour, S. Sivaselvachandran, S. Tohyama, T. Cole, T. M. Khuong, E. Mir, D. G. Gibson, J. S. Wieskopf, S. G. Sotocinal, J. S. Austin, C. B. Meloto, J. H. Gitt, C. Gkogkas, N. Sonenberg, J. D. Greenspan, R. B. Fillingim, R. Ohrbach, G. D. Slade, C. Knott, R. Dubner, A. G. Nackley, A. Ribeiro-da-Silva, G. G. Neely, W. Maixner, D. V. Zaykin, J. S. Mogil, L. Diatchenko, Epiregulin and EGFR interactions are involved in pain processing. J. Clin. Invest. 127, 3353–3366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.I. Obara, S. P. Hunt, Axonal protein synthesis and the regulation of primary afferent function. Dev. Neurobiol. 74, 269–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. J. Price, S. M. Geranton, Translating nociceptor sensitivity: The role of axonal protein synthesis in nociceptor physiology. Eur. J. Neurosci. 29, 2253–2263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O. K. Melemedjian, M. N. Asiedu, D. V. Tillu, K. A. Peebles, J. Yan, N. Ertz, G. O. Dussor, T. J. Price, IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J. Neurosci. 30, 15113–15123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O. Bogen, N. Alessandri-Haber, C. Chu, R. W. Gear, J. D. Levine, Generation of a pain memory in the primary afferent nociceptor triggered by PKCɛ activation of CPEB. J. Neurosci. 32, 2018–2026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. F. Ferrari, O. Bogen, C. Chu, J. D. Levine, Peripheral administration of translation inhibitors reverses increased hyperalgesia in a model of chronic pain in the rat. J. Pain: Off. J. Am. Pain Soc. 14, 731–738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. O. Asante, V. C. Wallace, A. H. Dickenson, Formalin-induced behavioural hypersensitivity and neuronal hyperexcitability are mediated by rapid protein synthesis at the spinal level. Mol. Pain 5, 1744-8069-5-27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.T. J. Price, M. H. Rashid, M. Millecamps, R. Sanoja, J. M. Entrena, F. Cervero, Decreased nociceptive sensitization in mice lacking the fragile X mental retardation protein: Role of mGluR1/5 and mTOR. J. Neurosci. 27, 13958–13967 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Q. Xu, B. Fitzsimmons, J. Steinauer, A. O'Neill, A. C. Newton, X. Y. Hua, T. L. Yaksh, Spinal phosphinositide 3-kinase-Akt-mammalian target of rapamycin signaling cascades in inflammation-induced hyperalgesia. J. Neurosci. 31, 2113–2124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. C. Gingras, Y. Svitkin, G. J. Belsham, A. Pause, N. Sonenberg, Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. U.S.A. 93, 5578–5583 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. Khoutorsky, R. P. Bonin, R. E. Sorge, C. G. Gkogkas, S. A. Pawlowski, S. M. Jafarnejad, M. H. Pitcher, T. Alain, J. Perez-Sanchez, E. W. Salter, L. Martin, A. Ribeiro-da-Silva, Y. De Koninck, F. Cervero, J. S. Mogil, N. Sonenberg, Translational control of nociception via 4E-binding protein 1. eLife 4, e12002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Zhang, D. J. Cavanaugh, M. I. Nemenov, A. I. Basbaum, The modality-specific contribution of peptidergic and non-peptidergic nociceptors is manifest at the level of dorsal horn nociresponsive neurons. J. Physiol. 591, 1097–1110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D. J. Cavanaugh, H. Lee, L. Lo, S. D. Shields, M. J. Zylka, A. I. Basbaum, D. J. Anderson, Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc. Natl. Acad. Sci. U.S.A. 106, 9075–9080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. Megat, P. R. Ray, J. K. Moy, T. F. Lou, P. Barragan-Iglesias, Y. Li, G. Pradhan, A. Wanghzou, A. Ahmad, M. D. Burton, R. Y. North, P. M. Dougherty, A. Khoutorsky, N. Sonenberg, K. R. Webster, G. Dussor, Z. T. Campbell, T. J. Price, Nociceptor translational profiling reveals the Ragulator-Rag GTPase complex as a critical generator of neuropathic pain. J. Neurosci. 39, 393–411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. Sanz, L. Yang, T. Su, D. R. Morris, G. S. McKnight, P. S. Amieux, Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U.S.A. 106, 13939–13944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Zhang, M. M. Hu, Y. Y. Wang, H. B. Shu, TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287, 28646–28655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. S. Diamond, M. Kinder, H. Matsushita, M. Mashayekhi, G. P. Dunn, J. M. Archambault, H. Lee, C. D. Arthur, J. M. White, U. Kalinke, K. M. Murphy, R. D. Schreiber, Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.P. Barragan-Iglesias, U. Franco-Enzastiga, V. Jeevakumar, S. Shiers, A. Wangzhou, V. Granados-Soto, Z. T. Campbell, G. Dussor, T. J. Price, Type I interferons act directly on nociceptors to produce pain sensitization: Implications for viral infection-induced pain. J. Neurosci. 40, 3517–3532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Liang, B. Tao, L. Fan, M. Yaster, Y. Zhang, Y. X. Tao, mTOR and its downstream pathway are activated in the dorsal root ganglion and spinal cord after peripheral inflammation, but not after nerve injury. Brain Res. 1513, 17–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Li, J. Han, D. S. Wang, Q. Yang, B. Feng, W. B. Kang, L. Yang, G. Liu, M. G. Zhao, Sinomenine attenuates chronic inflammatory pain in mice. Metab. Brain Dis. 32, 211–219 (2017). [DOI] [PubMed] [Google Scholar]
- 34.S. Wu, J. Zhang, Q. Xue, J. Liu, B. Huang, Z. He, J. Huang, S. Zu, Z. Chen, B. Zhao, M. Liao, P. Jiao, Duck TRIM32 functions in IFN-β signaling against the infection of H5N6 highly pathogenic avian influenza virus. Front. Immunol. 11, 377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.H. Cui, Y. Liu, Y. Huang, Roles of TRIM32 in corneal epithelial cells after infection with herpes simplex virus. Cell. Physiol. Biochem. 43, 801–811 (2017). [DOI] [PubMed] [Google Scholar]
- 36.L. Carthagena, A. Bergamaschi, J. M. Luna, A. David, P. D. Uchil, F. Margottin-Goguet, W. Mothes, U. Hazan, C. Transy, G. Pancino, S. Nisole, Human TRIM gene expression in response to interferons. PLOS ONE 4, e4894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R. Colina, M. Costa-Mattioli, R. J. Dowling, M. Jaramillo, L. H. Tai, C. J. Breitbach, Y. Martineau, O. Larsson, L. Rong, Y. V. Svitkin, A. P. Makrigiannis, J. C. Bell, N. Sonenberg, Translational control of the innate immune response through IRF-7. Nature 452, 323–328 (2008). [DOI] [PubMed] [Google Scholar]
- 38.E. P. Kusnadi, C. Timpone, I. Topisirovic, O. Larsson, L. Furic, Regulation of gene expression via translational buffering. Biochim. Biophys. Acta Mol. Cell Res. 1869, 119140 (2022). [DOI] [PubMed] [Google Scholar]
- 39.S. D. Shields, W. A. Eckert 3rd, A. I. Basbaum, Spared nerve injury model of neuropathic pain in the mouse: A behavioral and anatomic analysis. J. Pain 4, 465–470 (2003). [DOI] [PubMed] [Google Scholar]
- 40.I. Decosterd, C. J. Woolf, Spared nerve injury: An animal model of persistent peripheral neuropathic pain. Pain 87, 149–158 (2000). [DOI] [PubMed] [Google Scholar]
- 41.S. R. Chaplan, F. W. Bach, J. W. Pogrel, J. M. Chung, T. L. Yaksh, Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994). [DOI] [PubMed] [Google Scholar]
- 42.D. Tavares-Ferreira, P. R. Ray, I. Sankaranarayanan, G. L. Mejia, A. Wangzhou, S. Shiers, R. Uttarkar, S. Megat, P. Barragan-Iglesias, G. Dussor, Sex differences in nociceptor translatomes contribute to divergent prostaglandin signaling in male and female mice. Biol. Psychiatry 91, 129–140 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. Pertea, G. M. Pertea, C. M. Antonescu, T.-C. Chang, J. T. Mendell, S. L. Salzberg, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.X. D. Zhang, S. D. Marine, M. Ferrer, Error rates and powers in genome-scale RNAi screens. J. Biomol. Screen. 14, 230–238 (2009). [DOI] [PubMed] [Google Scholar]
- 46.A. Bhattacharyya, On a measure of divergence between two statistical populations defined by their probability distribution. Bullet. Calc. Math. Soc. 35, 99–110 (1943). [Google Scholar]
- 47.S. Hosseini, K. Michaelsen-Preusse, G. Grigoryan, C. Chhatbar, U. Kalinke, M. Korte, Type I interferon receptor signaling in astrocytes regulates hippocampal synaptic plasticity and cognitive function of the healthy CNS. Cell Rep. 31, 107666 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Legend for data S1
Data S1