Abstract
Inflammation-associated insulin resistance is a key trigger of gestational diabetes mellitus (GDM), but the underlying mechanisms and effective interventions remain unclear. Here, we report the association of placental inflammation (tumor necrosis factor–α) and abnormal maternal glucose metabolism in patients with GDM, and a high fermentable dietary fiber (HFDF; konjac) could reduce GDM development through gut flora–short-chain fatty acid–placental inflammation axis in GDM mouse model. Mechanistically, HFDF increases abundances of Lachnospiraceae and butyrate, reduces placental-derived inflammation by enhancing gut barrier and inhibiting the transfer of bacterial-derived lipopolysaccharide, and ultimately resists high-fat diet–induced insulin resistance. Lachnospiraceae and butyrate have similar anti-GDM and anti–placental inflammation effects, and they can ameliorate placental function and pregnancy outcome effects probably by dampening placental immune dysfunction. These findings demonstrate the involvement of important placental inflammation–related mechanisms in the progression of GDM and the great potential of HFDFs to reduce susceptibility to GDM through gut-flora-placenta axis.
A high fermentable fiber reduces development of gestational diabetes mellitus through gut-flora-placenta axis.
INTRODUCTION
Gestational diabetes mellitus (GDM), the most common metabolic disturbance during pregnancy, affects 14% of pregnancies and is associated with an increased risk of adverse pregnancy outcomes, including prematurity, macrosomia, and placental abruption (1). GDM is characterized by hyperglycemia, hyperinsulinemia, insulin resistance, and abnormal placental development in the second and third trimesters (2). In patients with GDM (GDMs), elevated serum inflammatory cytokines are reported to be associated with insulin resistance and can be used to identify its early onset (3). In general, chronic inflammation is associated with maternal obesity (4). A recent study of a cohort of GDMs controlled in body mass index (BMI) suggested that inflammatory factors may serve as biomarkers of GDM independent of BMI and that other features such as immune-endocrine interactions may play a central role in GDM development (3). Notably, the placenta is a source and target of multiple pathological stimuli in the second and third trimesters in metabolic diseases (5), and a previous study attributed insulin resistance during pregnancy to the increased production of placental tumor necrosis factor–α (TNF-α) (6). However, the role of placental inflammation in GDM remains unclear.
The human gut flora and its derivatives play a key regulatory role in the development of metabolic diseases during pregnancy, such as preeclampsia (3). Several preclinical studies in preeclampsia have demonstrated that the inflammatory response that triggers maternal and placental inflammation involved the disruption of gut barrier and Toll-like receptor (TLR) signaling promoted by bacterial components transferred from gut to placenta (7, 8). Moreover, short-chain fatty acids (SCFAs) produced by gut flora contribute to macrophage differentiation and placental function, which, in turn, inhibit inflammation and resist pre-eclampsia development in preclinical models (9). Emerging clinical studies have indicated that GDM is induced through heightened inflammation initiated by microbial dysbiosis (3). However, the role of gut-placenta axis in GDM progression is unclear. Recent data in pregnant women indicate that a decrease in SCFAs may serve as a potential early biomarker of GDM (3). This suggested that the research focusing on gut flora–derived SCFAs may provide insight for improving the therapeutic effect of GDM.
Dietary fiber, especially high fermentable dietary fiber (HFDF), which can be fermented to produce abundant SCFAs, is promoted as part of a healthy dietary pattern and diabetes management (10). Inadequate dietary fiber intake is an important cause of microflora dysbiosis and GDM (11, 12). In human studies, HFDFs were shown to improve insulin sensitivity and reduce the risk of diabetes because of their protective effects on gut homeostasis and chronic inflammation (13). Our and other previous studies in sows and mice have demonstrated that konjac flour, a source of HFDF, has beneficial effects on glucose metabolism and anti-inflammation during pregnancy (14–16). However, it remains unknown whether HFDF can resist high-fat diet (HFD)–induced maternal insulin resistance, inflammation, and poor pregnancy outcomes via gut-placenta axis.
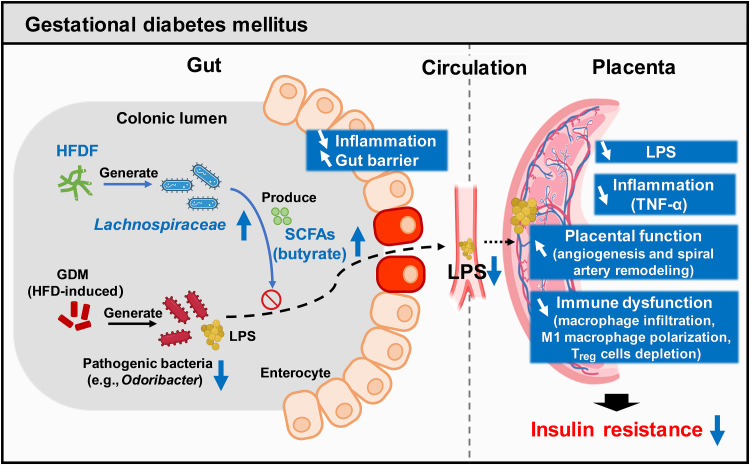
Therefore, this work aimed to investigate the mechanistic link between GDM and placental inflammation and explore how HFDF improves maternal insulin resistance during pregnancy. Here, we report that abnormal glucose metabolism in GDMs is associated with placental inflammation. Konjac flour (hereafter referred to as HFDF) could reduce GDM development through gut flora–SCFA–placental inflammation axis in GDM mouse model. Mechanistically, the HFDF increases the abundances of Lachnospiraceae and butyrate, enables the reduction of placental-derived inflammation by enhancing gut barrier, inhibits the transfer of bacterial components [lipopolysaccharide (LPS)], and ultimately resists HFD-induced insulin resistance. Moreover, butyrate abundances in placentae of patients and mice with GDM are negatively correlated with placental TNF-α. We found that Lachnospiraceae and butyrate have similar anti-GDM and anti–placental inflammation effects, which are associated with dampened macrophage infiltration and M1 macrophage polarization and increased regulatory T (Treg) cells in placenta. All the results indicated that the flora-metabolite-placenta axis may underlie the placental inflammation in GDM and that HFDF intake could be a promising intervention against GDM.
RESULTS
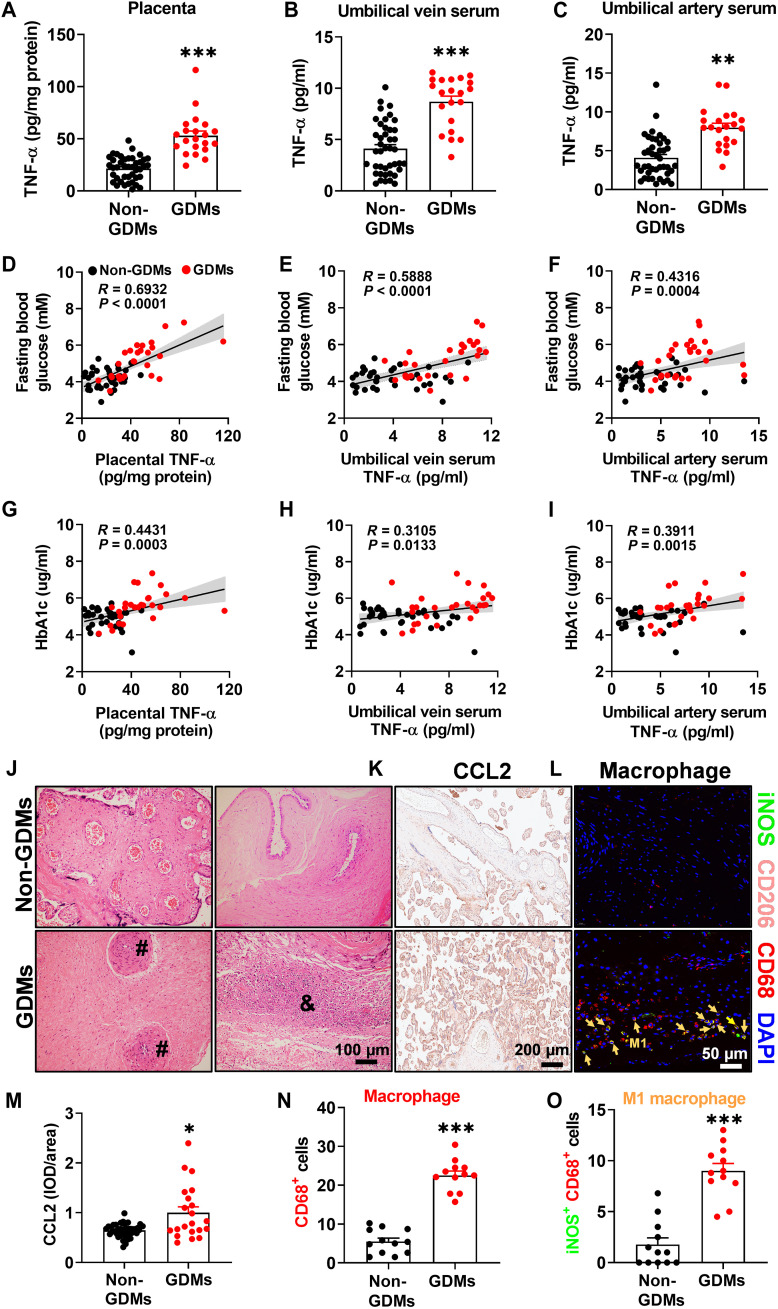
Abnormal glucose metabolism in GDMs is associated with placental inflammation
Abnormal glucose metabolism during pregnancy has been reported to be ascribed to the increased production of TNF-α (6). To examine the association between placental inflammation and abnormal glucose metabolism in GDMs, we included 63 eligible women [21 GDMs and 42 women without GDM (non-GDMs)] in follow-up study. In this cohort, prepregnancy overweight GDMs (average BMI, 24.31 kg/m2) had significantly higher TNF-α levels in the full-term placentae, umbilical vein serum, and artery vein serum compared to non-GDMs (average BMI, 20.58 kg/m2) (Fig. 1, A to C, and table S1), agreeing with previous observations (6). Further linear regression analysis revealed that TNF-α in the full-term placentae, umbilical vein serum, and umbilical artery serum exhibited a significant positive relationship with fasting blood glucose (R = 0.6932, 0.5888, and 0.4316, respectively; P < 0.0001, P < 0.0001, and P = 0.0004, respectively) and hemoglobin A1c (HbA1c; R = 0.4431, 0.3105, and 0.3911, respectively; P = 0.0003, 0.0133, and 0.0015, respectively) (Fig. 1, D to I). Histomorphology showed that, compared to non-GDM placentae, GDM placentae had increases in placental inflammatory cell infiltration, vascular infarction, macrophages numbers (CD68+), C-C motif chemokine ligand 2 (CCL2) expression [one of the key factors in macrophage recruitment (17)], and M1 polarization [inducible nitric oxide synthase (iNOS)+ CD68+; proinflammatory] (Fig. 1, J to O), all of which suggest a higher inflammatory environment in GDM placentae. In human studies, because of the challenge to confirm causation or identify the mechanisms linking GDM and placental inflammation, we established a GDM mouse model, where female C57BL/6J mice were fed the HFD during pregestation (before mating, 4 weeks) and postgestation [embryonic day 0.5 (E0.5) to E18.5, 18.5 days] (fig. S1A). Elevated mRNA levels of placental inflammatory factor (especially TNF-α) indicate the increased placental inflammation in GDM mice (fig. S1B). Notably, there was a characteristic rapid restoration of glucose homeostasis [glucose tolerance tests (GTTs)] and circulating TNF-α in the mother immediately after placental expulsion (within 3 days of delivery) (fig. S1, C to E), reflecting that these changes are mediated by placental factors. In addition, blocking CCL2/CCR2 signaling by intraperitoneal injection of CCR2 inhibitors during gestation was found able to alleviate HFD-induced placental inflammation and GDM phenotype (fig. S1, F to J). These data indicate that abnormal maternal glucose metabolism is associated with placental inflammation in GDMs.
Fig. 1. Relationship between placental inflammation (TNF-α) and abnormal glucose metabolism in GDM women.
(A to C) TNF-α levels in the placenta and umbilical veins and arteries. Non-GDMs, n = 42; GDMs, n = 21. (D to F) Two-tailed Pearson’s correlation coefficient analysis of TNF-α levels in the placenta, umbilical vein serum, and artery serum in terms of fasting glucose levels. (G to I) Two-tailed Pearson’s correlation coefficient analysis of TNF-α levels in the placenta, umbilical vein serum, and artery serum in terms of blood HbA1c levels. (J) Representative hematoxylin and eosin (H&E)–staining images of midsagittal placental tissue sections used in histomorphological analysis. Placental inflammatory cell infiltration (&) and vascular infarction (#). See table S1 for quantification of H&E staining. Non-GDMs, n = 42; GDMs, n = 21. Scale bar, 100 μm. (K and M) Immunohistochemistry and quantification of CCL2 in placenta. Non-GDMs, n = 42; GDMs, n = 21. Scale bar, 200 μm. IOD, integrated optical density. (L, N, and O) Localization of macrophage (CD68+; red), M1 macrophage (iNOS+/CD68+; green/red; yellow arrow), and M2 macrophage (CD206+/CD68+; pink/red; it was not quantified because it was almost nonexistent) in placenta by immunofluorescence. Non-GDMs, n = 12; GDMs, n = 12. Scale bar, 50 μm. DAPI, 4′,6-diamidino-2-phenylindole. Data were analyzed by unpaired Student’s t test (A and M to O) or Mann-Whitney U test (B and C) and represented the means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
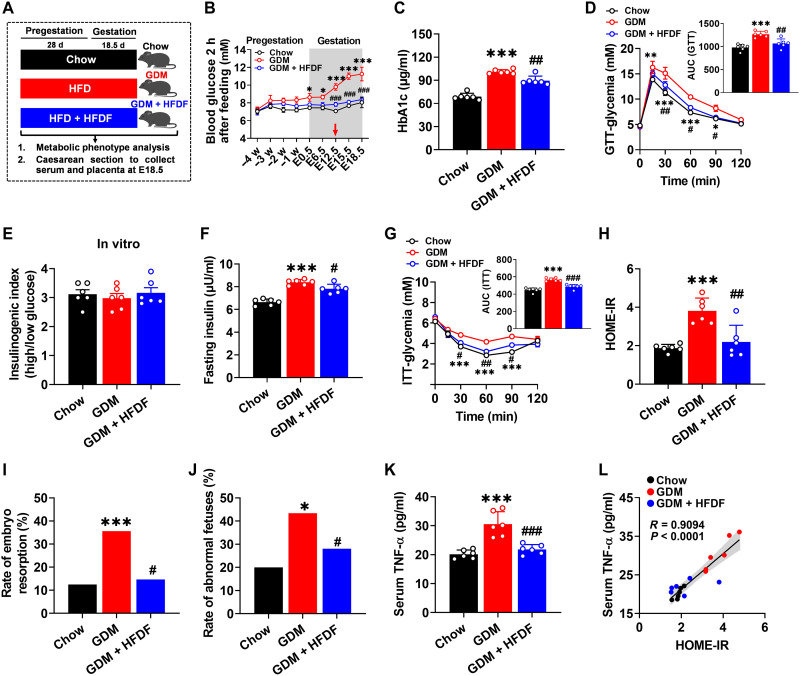
HFDF modulates the phenotypes of GDM, inflammation, and maternal obesity in E18.5 mice
To evaluate the effects of HFDF on GDM phenotypes, we fed female mice chow diet (chow group), HFD (GDM group), or HFD with HFDF (GDM + HFDF group) during pregestation and postgestation (Fig. 2A). In the primary screening tests, we measured circulating glucose and HbA1c and found that HFDF treatment could alleviate hyperglycemia caused by HFD (Fig. 2, B and C). The hypoglycemic effect of HFDF was first observed at E12.5 and maintained through the rest of gestation (Fig. 2B). Further GTTs showed that HFDF treatment could prevent impaired maternal glucose tolerance and hinder GDM development (Fig. 2D). The primary defect in glucose intolerance was determined by measuring insulin sensitivity and insulin secretion, two major features of GDM (18). An in vitro glucose–stimulated insulin secretion test was performed using pancreatic islets isolated from mice, and the stimulation index was shown to be similar between GDM + HFDF group and GDM group (Fig. 2E), implicating a similar insulin secretory capacity. The in vivo results showed that insulin levels in the serum were significantly lower in GDM + HFDF mice than in GDM mice (Fig. 2F), suggesting that compensatory insulin secretion was alleviated in GDM + HFDF mice. Insulin tolerance tests (ITTs) and homeostasis model assessment of insulin resistance (HOME-IR) index also showed significant improvement of insulin tolerance in GDM + HFDF mice relative to GDM mice (Fig. 2, G and H). Moreover, HFDF was seen to reduce the rate of embryo resorption and abnormal fetuses (Fig. 2, I and J), which are typical adverse pregnancy outcomes in GDM (2). Circulating TNF-α levels are positively correlated with insulin resistance (19). Consistent with GDM-associated insulin resistance, HFD increased circulating TNF-α levels, but they were significantly reduced by HFDF (Fig. 2K). Serum TNF-α was seen to have a significant positive relationship with HOME-IR in mice (R = 0.9094, P < 0.0001) (Fig. 2L). These results suggest that HFDF could reduce HFD-induced maternal inflammation and contribute to alleviate insulin resistance by improving insulin sensitivity rather than insulin secretion.
Fig. 2. Effects of HFDF on HFD-induced maternal insulin resistance, inflammation, and poor pregnancy outcomes.
(A) Schematic of the maternal diet regimen. C57BL/6 female mice were fed chow diet (chow group), HFD (GDM group), or HFD supplemented with HFDF (GDM + HFDF group) during pregestation (before mating, 28 days) and postgestation (E0.5 to E18.5, 18 days) (n = 10 mice per group). (B) Blood glucose 2 hours after feeding at different treatment stages (n = 6 mice per group). (C) Serum HbA1c level at E18.5 (n = 6 mice per group). (D) GTT and area under the curve (AUC) at E16.5 (n = 6 mice per group). (E) The stimulation index of insulin release from isolated pancreatic islets of mice at E18.5 (n = 6 mice per group). (F) Serum insulin level (n = 6 mice per group). (G) ITT and AUC (n = 6 mice per group). (H) HOME-IR index (n = 6 mice per group). HOME-IR = [fasting insulin (mIU/liter) × fasting glucose (mM) / 22. 5]. (I and J) Rates of embryo resorption and abnormal fetuses at E18.5 in each group (n = 10 litters per group). (K) Serum TNF-α level at E18.5 (n = 6 mice per group). (L) Two-tailed Pearson’s correlation coefficient analysis of serum TNF-α levels and HOME-IR at E18.5 in mice. Results represent two independent experiments. Data were analyzed by two-way analysis of variance (ANOVA) followed by post hoc Bonferroni tests (B, D, and G), one-way ANOVA followed by post hoc Tukey’s tests [(C), (E), (F), (H), and (K)], and chi-square test [(I) and (J)] and represented the means ± SEM. * or #P < 0.05, ** or ##P < 0.01, and *** or ###P < 0.001, relative to chow and GDM groups, respectively.
HFDF did not affect food and energy intake in GDM mice but significantly reduced HFD-induced prepregnancy body weight after 4 weeks of feeding (fig. S2, A and B). However, the effect of HFDF resistance to maternal weight gain was shown to diminish upon entry into gestation (fig. S2B), probably due to improvement in HFD-induced impaired gestational product development, including placentae and fetus (fig. S2C). After excluding the interference of pregnancy products, HFDF still had a suppressive effect on obesity during pregnancy (fig. S2D). Analysis of organ phenotypes showed that HFDF could resist HFD-induced obesogenic metabolic phenotype, as demonstrated by significant reduction in liver weight and steatosis and a downtrend in fat weight in GDM + HFDF mice versus GDM mice (fig. S2, E to I). Collectively, HFDF intervention can not only suppress the HFD-induced inflammatory phenotype but can also curb maternal obesity, either or both of which may explain the suppressive effect of HFDF on GDM.
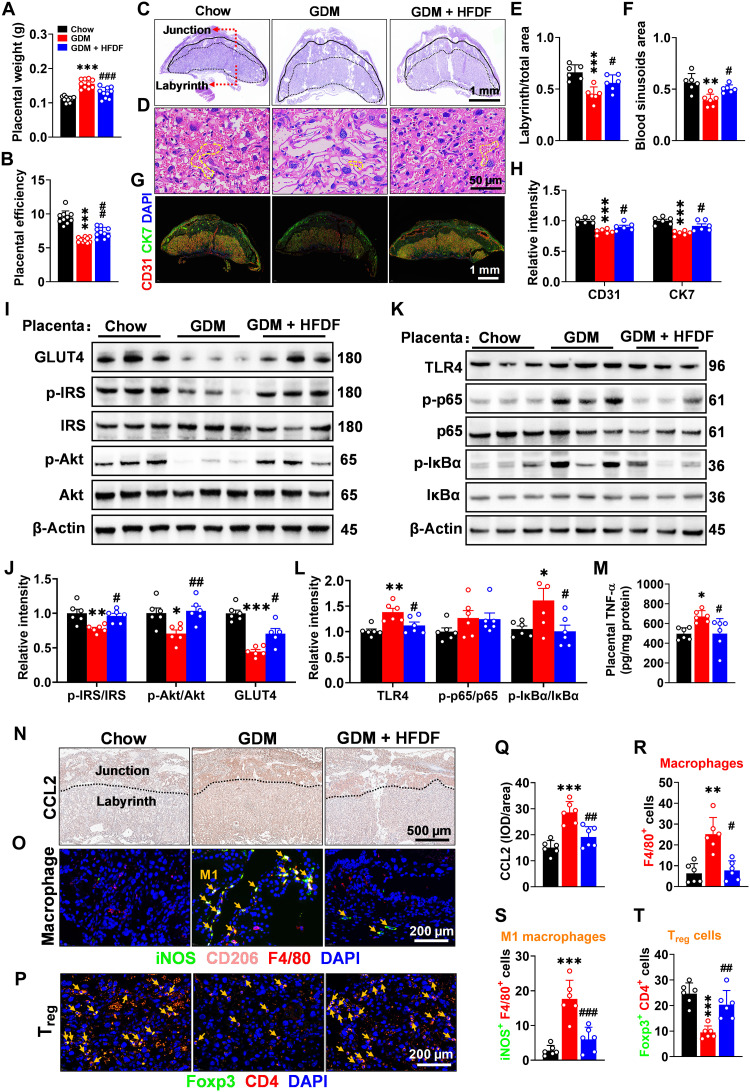
HFDF alleviates placental dysfunction and inflammation in E18.5 GDM mice
We attempted to identify the specific tissue(s) associated with HFDF-induced reduction of inflammation in GDM model, and the changes in HFDF-induced gestational products and liver phenotypes prompted us to investigate their metabolic phenotypes. Compared with GDM group, GDM + HFDF group showed a decrease in placental weight while an increase in placental efficiency (Fig. 3, A and B), suggesting the potential improvement of its function. Hematoxylin and eosin (H&E) staining also showed significant structural changes in GDM + HFDF placentae, with an increase in the proportion of the placental labyrinth/total zones and blood sinusoids area relative to GDM group (Fig. 3, C to F). Moreover, HFDF was seen to up-regulate the expression of platelet endothelial cell adhesion molecule (CD31) and cytokeratin 7 (CK7) in placentae (Fig. 3, G and H), which are the markers of placental angiogenesis and spiral artery remodeling, respectively (20). HFDF was also found able to reduce HFD-induced impairment of insulin signaling pathways [insulin receptor substrate (IRS)–protein kinase B (Akt)–glucose transporter type 4 (GLUT4)] in the placenta and maternal liver (Fig. 3, I and J, and fig. S3, A and B).
Fig. 3. Effects of HFDF on HFD-induced placental endothelial dysfunction, inflammation, and insulin resistance.
Schematic of the maternal diet regimen and breeding is shown in Fig. 2A. (A) Placental weight (n = 10 litters per group). (B) Placental efficiency (ratio of fetal weight to placental weight, n = 10 litters per group). (C to F) Representative H&E images of midsagittal placental tissue sections, with red arrow marking the labyrinth zone/junctional zone and yellow dashed line marking the typical blood sinusoid. Scale bars, 1 mm (C) and 50 μm (D). (G and H) Immunofluorescence of CD31 (marker of placental angiogenesis; red) and CK7 (marker of spiral artery remodeling; green) in placenta. Scale bar, 1 mm. (I to L) Immunoblots and quantification of insulin signaling pathway (GLUT4, p-IRS/IRS, and p-Akt/Akt) and TLR4/NF-κB inflammatory pathway [TLR4, p-p65/p65, and p–inhibitor of NF-κB (IκBα)/IκBα] in placenta. (M) Placental TNF-α level in each group. (N and Q) Immunohistochemistry and quantification of CCL2 (one of the key factors in macrophage recruitment) in placenta. Scale bar, 500 μm. (O, R, and S) Localization of macrophage (F4/80+; red), M1 macrophage (iNOS+/F4/80+; green/red; yellow arrow), and M2 macrophage (CD206+ F4/80+; pink/red; it was not quantified because it was almost nonexistent) in placenta by immunofluorescence. Scale bar, 200 μm. (P and T) Localization of Treg cell (Foxp3+/CD4+; yellow arrow) in placenta by immunofluorescence. Scale bar, 200 μm. (C to T) n = 6 placentae from 6 litters per group. Data were analyzed by one-way ANOVA followed by post hoc Tukey’s tests and represented means ± SEM. * or #P < 0.05, ** or ##P < 0.01, and *** or ###P < 0.001, relative to Chow and GDM group, respectively.
In addition, the inflammatory responses in the placenta and liver were also analyzed. HFDF could inhibit the activation of TLR4/nuclear factor κB (NF-κB) signaling pathway in the placenta and liver in GDM model (Fig. 3, K and L, and fig. S3, C and D). HFDF could also reduce TNF-α levels in the placenta and liver of E18.5 mice (Fig. 3M and fig. S3E). Notably, hepatic inflammatory cell infiltration did not appear to differ between groups (fig. S3F). Immunofluorescence analysis confirmed that the alleviation of placental inflammation by HFDF in E18.5 GDM mice was associated with immune homeostasis, as evidenced by the reduction of CCL2 expression, macrophage infiltration [mouse epidermal growth factor–like module-containing mucin-like hormone receptor–like 1 (F4/80+)] and M1 polarization (iNOS+ F4/80+; proinflammatory) in placenta (Fig. 3, N, O, and Q to S). Macrophage differentiation has been reported to be regulated by Treg cells (21). HFDF could also prevent depletion of Treg cells [forkhead box P3 (Foxp3)+ CD4+] in placenta of GDM mice (Fig. 3, P and T). Collectively, HFDF could reduce placental and hepatic dysfunction at E18.5 in GDM model, particularly the inflammatory responses associated with macrophage infiltration, M1 polarization, and depletion of Treg cells in placenta.
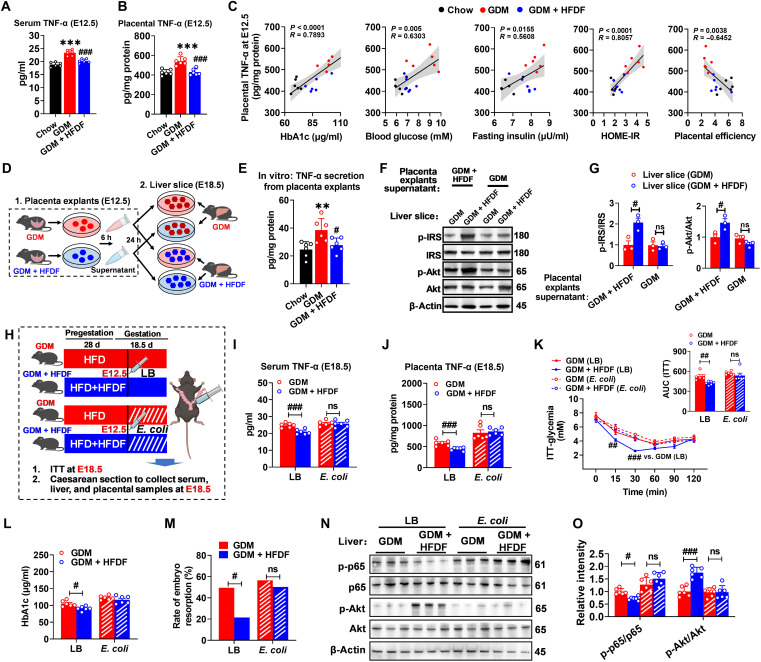
Suppression of placental inflammation in mice during E12.5 to E18.5 is necessary for HFDF to fight HFD-induced GDM
Placenta is the main site of TNF-α production in mid to late gestation (6). In Fig. 2B, HFDF was shown to resist HFD-induced hyperglycemia during E12.5 to E18.5, which is the stage of placental development and secretion of inflammatory factors, inferring that the placenta may be the main source of inflammation affecting the system. We speculate that HFDF may exert anti-GDM effects by reducing placental inflammation at E12.5. HFDF was first found able to reverse maternal and placental TNF-α levels in GDM model as early as E12.5 (Fig. 4, A and B). In Fig. 4C, further Pearson’s correlation analysis showed significant correlations of placental TNF-α with serum HbA1c (R = 0.7893, P < 0.0001), blood glucose (R = 0.6303, P = 0.005), fasting insulin (R = 0.5608, P = 0.0155), HOME-IR (R = 0.8057, P < 0.0001), and placental efficiency (R = −0.6452, P = 0.0038) at E12.5. These results suggest that improved maternal glucose metabolism in GDM + HFDF mice may be associated with reduced placental inflammation.
Fig. 4. The central role of HFDF in hindering GDM development by suppressing placental inflammation.
(A and B) Serum (n = 6 mice per group) and placental (n = 6 placentae from 6 litters per group) TNF-α levels E12.5. Schematic of the maternal diet regimen and breeding is shown in Fig. 2A. (C) Two-tailed Pearson’s correlation coefficient analysis of placental TNF-α levels and GDM parameters in E12.5 mice. (D) Schematic representation of primary liver sections treated with placental explant supernatant. Results represent two independent experiments. (E) TNF-α levels in culture supernatants derived from placental explants (n = 6 placenta from 6 litters per group). (F and G) Immunoblots and quantification of insulin signaling pathway (p-IRS/IRS and p-Akt/Akt) in liver slices (n = 3 liver from 3 mice per group). (H) Schematic representation for placental inflammation induction. (I and J) Serum and placental TNF-α levels of GDM, or GDM + HFDF mice at 6 days after placental inflammation induction treatment (at E18.5) (n = 6 mice per group). (K) ITT and AUC of GDM or GDM + HFDF mice at E18.5 (n = 6 mice per group). (L) Serum HbA1c level of GDM or GDM + HFDF mice at E18.5 (n = 6 mice per group). (M) Rate of embryo resorption in each group (n = 6 litters per group). (N and O) Immunoblots and quantification of p-p65/p65 and p-Akt/Akt in livers of GDM or GDM + HFDF mice at E18.5 (n = 6 placentae from 6 litters per group). Data were analyzed by one-way ANOVA followed by post hoc Tukey’s tests [(A), (B), and (E)] and two-way ANOVA followed by post hoc Bonferroni tests [(G) to (L) and (O)] or chi-square test (M) and represented means ± SEM. #P < 0.05, ** or ##P < 0.01, *** or ###P < 0.001, and ns (not significant), relative to chow and GDM group, respectively.
The causal role of placental inflammation in the systemic effects of HFDF on maternal obesity–related insulin resistance was further explored through an in vitro and an in vivo experiment. For the in vitro experiment, we established a placental explant–liver section model (Fig. 4D), and a higher TNF-α production capacity was found in GDM placental explants than in GDM + HFDF placental explants, indicating a higher inflammation level in the former group at E12.5 (Fig. 4E). Treatment with culture supernatants in placental explants from GDM was shown to eliminate the protective effects of HFDF on insulin resistance and inflammation in the maternal liver (Fig. 4, F and G). For the in vivo experiment, a mouse model was established for the targeted induction of intrauterine inflammation by intrauterine injection of Escherichia coli (Fig. 4H), a typical model of uterine/placental inflammation (22). As expected, intrauterine injection of inactivated E. coli at E12.5 induced maternal and placental TNF-α secretion in E18.5 mice (Fig. 4, I and J). Compared with Luria-Bertani (LB)–injected mice at E18.5, E. coli–injected mice showed an increase in 2-hour postprandial blood glucose, ITT, embryo resorption rate, serum, and placental TNF-α levels (fig. S4, A to E), proving evidence that placental inflammation could induce GDM phenotype independent of HFD or obesity. The up-regulation of placental inflammation levels in the second and third trimesters eliminated the anti-GDM effect of HFDF (Fig. 4, K to M). Besides, consistent with the in vitro placental explant–liver section model, the recovery of placental inflammation abolished HFDF-induced relief of maternal hepatic insulin resistance and inflammation as indicated by the changes of p-65 and Akt protein activities (Fig. 4, N and O). We noted that the effects of HFDF on maternal feed intake, maternal body weight, liver weight, hepatic steatosis, and fat weight were not altered by the short-term placental inflammation induced by inactivated E. coli injection versus LB injection (fig. S4, F to N), but the beneficial effects of HFDF on maternal glucose metabolism and inflammation were eliminated. These results reinforced the notion that HFDF acts on placental inflammation to reduce maternal inflammation and insulin resistance and hinder GDM development, which may be independent of changes in maternal obesity phenotype.
HFDF improves gut barrier and reduces bacterial-derived LPS translocation in GDM model
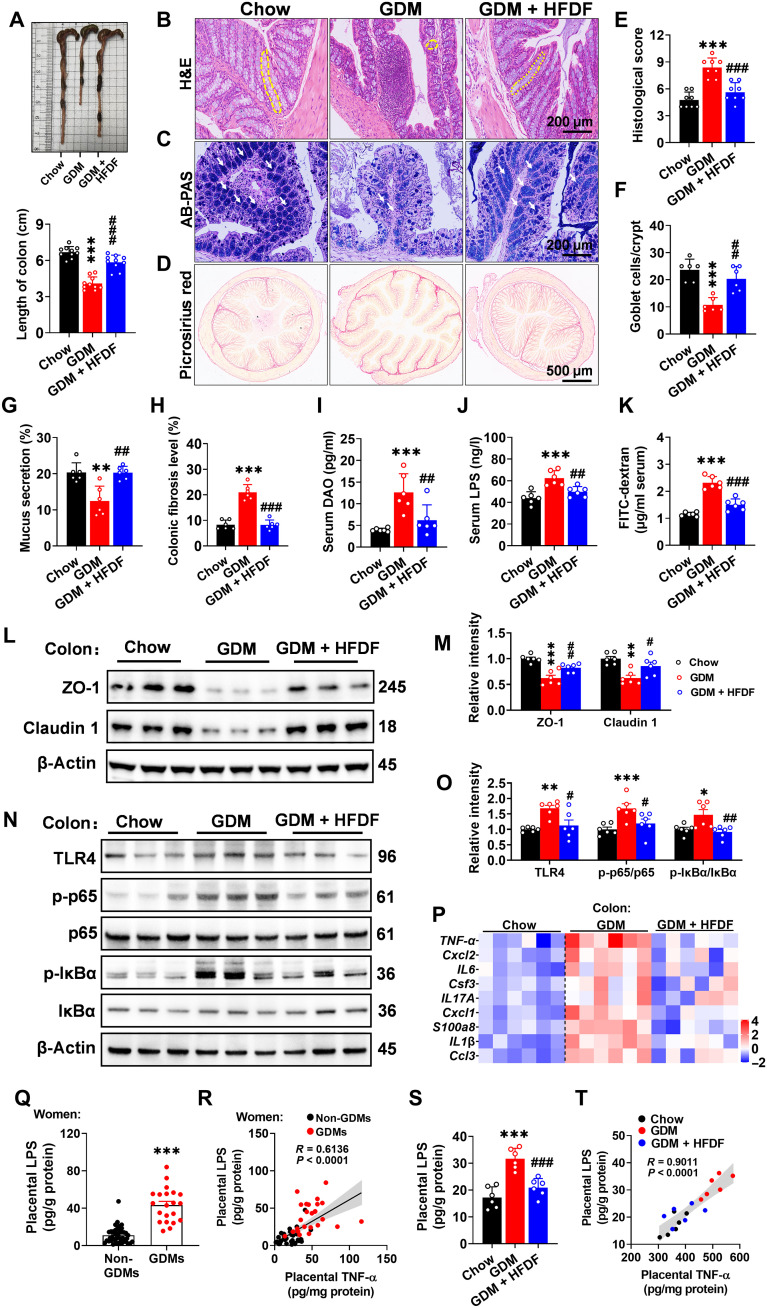
Maternal and placental inflammatory responses are associated with gut barrier dysfunction (23), so we further investigated whether HFDF affects intestinal barrier function in GDM mice. HFD was first found to induce a reduction in colon length in GDM model (Fig. 5A), which is one of the typical features of colonic inflammation (24). Further histological analysis revealed that HFDF could reduce histological damage score, increase the number of goblet cells and mucus secretion, and decrease fibrosis in GDM model (Fig. 5, B to H). These results support that HFDF could alleviate colonic inflammatory effects. Moreover, the serum levels of diamine oxidase (DAO), LPS, and fluorescein isothiocyanate (FITC)–dextran flux were shown to increase significantly in GDM mice but decreased under HFDF treatment (Fig. 5, I to K). Immunoblotting or quantitative real-time polymerase chain reaction (qPCR) results also showed that HFDF treatment could elevate the levels of barrier-forming tight junction markers and down-regulate the expression of inflammatory-related signaling pathways and inflammatory cytokines in the colon (Fig. 5, L to P). These results suggested an overall protective effect of HFDF against HFD-induced gut barrier damage in GDM model.
Fig. 5. Effects of HFDF on placental inflammation associated with impaired gut barrier and placental bacterial components (LPS) translocation in GDM model.
Schematic of the maternal diet regimen and breeding is shown in Fig. 2A. At E18.5, serum and intestinal samples were obtained after execution of mice. (A) Representative colon images and colon length (n = 10 mice per group). (B, E, and F) Representative H&E staining images of colon sections, histological score, and goblet cell (yellow dashed lines) number per colonic crypt (n = 6 mice per group). Scale bar, 200 μm. (C and G) Representative Alcian blue–periodic acid Schiff (AB-PAS) staining images of colon sections and quantification of mucus secretion (white arrows) (n = 6 mice per group). Scale bar, 200 μm. (D and H) Representative Picrosirius red of colon sections and quantification (n = 6 mice per group). (I to K) Serum DAO, LPS, and FITC-dextran flux levels (n = 6 mice per group). FITC-dextran flux was analyzed in another independent experiment. (L to O) Immunoblots and quantification of barrier-forming tight junction markers and TLR4/NF-κB inflammatory pathway in colon (n = 6 mice per group). (P) 4 Expression levels of cytokines and inflammatory-related factors in placenta (n = 6 mice per group). (Q) Placental LPS level in women (non-GDMs, n = 42; GDMs, n = 21). (R) Two-tailed Pearson’s correlation coefficient analysis of placental LPS levels and placental TNF-α in women. (S) Placental LPS level in mice (n = 6 placentae from 6 litters per group). (T) Two-tailed Pearson’s correlation coefficient analysis of placental LPS levels and placental TNF-α in mice. Data were analyzed by one-way ANOVA followed by post hoc Tukey’s tests (A to P and S) or unpaired Student’s t test with Welch’s correction (Q) and represented means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, relative to chow group or non-GDM. #P < 0.05, ##P < 0.01, and ###P < 0.001, relative to GDM group.
Increased gut permeability can lead to the translocation of bacterial components (such as LPS) or gut bacteria into extraintestinal tissues (including placenta) and the subsequent inflammation (25). Intrauterine injection of inactivated E. coli was shown to abolish the protective effect of HFDF on maternal inflammation and GDM (Fig. 4, H to O). Moreover, the GDM and GDM + HFDF placentae showed marked changes in the expression of TLR4 (Fig. 3K), which could act as a receptor for LPS in the outer membrane of Gram-negative bacterial cells (8). On the one hand, we measured the LPS concentration in human placentae, the LPS concentration was significantly higher in the placentae of GDMs than non-GDMs, and placental LPS showed a significant positive correlation with placental TNF-α (R = 0.6136, P < 0.0001) (Fig. 5, Q and R). In GDM mice, HFDF treatment significantly reduced placental LPS concentration, which was significant positively correlated with placental TNF-α (R = 0.9011, P < 0.0001) (Fig. 5, S and T). On the other hand, placental bacterial translocation may play an important role in metabolic disorder of pregnancy (7). In fig. S5A, placentae from each group are separated in a sterile environment, followed by qPCR analysis of bacteria in mouse placentae to evaluate the bacterial 16S rRNA gene, and the levels of total bacterial load were significantly increased in GDM mice while decreased in GDM + HFDF mice. Fluorescence in situ hybridization (FISH) analysis showed that the relative fluorescence intensity was increased in the junction zone of GDM placentae but decreased in that of GDM + HFDF placentae (fig. S5, B and C). Notably, we were still unable to determine whether placental inflammation in pregnant women is associated with the colonization of pathogens in the placenta, as high bacterial loads were present in the placentae of all pregnant women regardless of GDM status (fig. S5, D and E). Collectively, HFDF is more likely to alleviate placental inflammation by reducing gut barrier damage and placental bacterial component (LPS) translocation in GDM model.
HFDF reshapes gut bacteria in GDM model
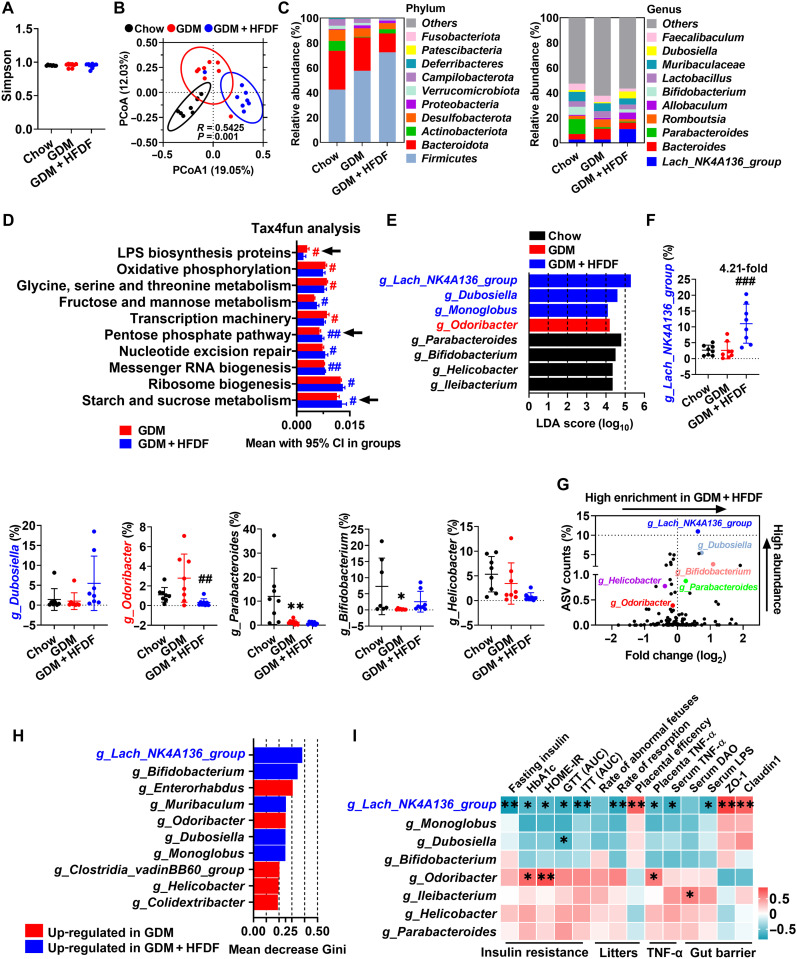
Gut dysbiosis may induce changes in gut barrier function (2), so the effect of HFDF on gut flora was investigated by high-throughput sequencing of 16S rRNA in fresh feces. The α-diversity (Simpson index) was not significantly different between the three groups (Fig. 6A). Notably, we observed a distinct clustering of flora composition for the three groups using Bray-Curtis–based principal coordinates analysis (PCoA) (Fig. 6B). The variations at phylum and genus levels indicated that HFDF still has a unique microbial composition compared to Chow (Fig. 6C), suggesting that HFDF could reshape gut bacteria. The microbial function predicted by Tax4fun revealed that pentose phosphate pathway and starch and sucrose metabolism [related to SCFA synthesis (26)] were enriched in GDM + HFDF mice, but LPS biosynthesis protein pathway was enriched in GDM mice (Fig. 6D). Moreover, the differential bacterial taxa were investigated by linear discriminant analysis (LDA) effect size, and eight core genera were identified (Fig. 6E). Abundance comparison of predominant genera showed the enrichment of g_Lachnospiraceae_NK4A136_group, g_Dubosiella, and g_Monoglobu in GDM + HFDF (Fig. 6F). The volcano plot flagged g_Lachnospiraceae_NK4A136_group as the most enriched and representative bacteria in GDM + HFDF (Fig. 6G). Random forest algorithm identified g_Lachnospiraceae_NK4A136_group as the most important bacterial genus enriched in GDM + HFDF mice, while g_Odoribacter in GDM mice (Fig. 6H). Further Pearson’s correlation analysis of these key genera suggests that g_Lachnospiraceae_NK4A136_group may play a beneficial role in alleviating insulin resistance, reducing placental inflammation, and maintaining gut barrier function, whereas g_Odoribacter may exert a detrimental effect (Fig. 6I). Collectively, HFDF could reshape gut bacteria and avoid microbial dysbiosis, which may play an important role in suppressing inflammation-associated insulin resistance in GDM model. We also proposed that g_Lachnospiraceae_NK4A136_group is crucial for mediating the anti-GDM effect of HFDF.
Fig. 6. Effects of HFDF on gut bacteria in GDM model.
Schematic of the maternal diet regimen and breeding is shown in Fig. 2A. At E12.5, fresh feces of mice were obtained, followed by 16S rRNA analysis (n = 8 mice per group). (A) Simpson index. (B) PCoA based on Bray-Curtis between Chow, GDM, and GDM + HFDF groups. (C) Average relative abundances of predominant taxa at phylum and genus level (n = 8 mice per group). (D) Relative abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (top 10) in gut predicted by Tax4fun. (E) The most differentially abundant taxa between three groups identified by LDA effect size. Only LDA threshold of ≥4 taxa are shown. (F) Relative abundances of g_Lachnospiraceae_NK4A136_group, g_Dubosiella, g_Odoribacter, g_Parabacteroides, g_Bifidobacterium, and g_Helicobacter (n = 8 mice per group). g_Ileibacterium and g_Monoglobu are not shown because they were identified in only about half of the samples. (G) Volcano plot for the relative abundance distribution of microbial amplicon sequence variants (ASVs). Each symbol represents one bacterial taxon. (H) Identification of key bacterial genera by the random forest method using the 16S rRNA gene sequencing data of GDM and GDM + HFDF groups. Higher mean decrease Gini values indicate greater importance of the variable. (I) Pearson’s correlation coefficient analysis between the relative abundance of selected bacterial genera and insulin resistance, litters, TNF-α, or gut barrier parameters in GDM and GDM + HFDF mice. These parameters are based on the data from Figs. 2 to 5. Data were represented means ± SEM or means with 95% confidence interval (CI). P values were analyzed by one-way ANOVA followed by post hoc Tukey’s tests (A), unpaired Student’s t test (D), or one-way ANOVA followed by two-stage step-up false discovery rate (FDR) method of Benjamini, Krieger, and Yekutieli (F). * or #P < 0.05, ** or ##P < 0.01, and ###P < 0.001, relative to chow and GDM group, respectively.
HFDF improves GDM phenotype, bacterial-derived LPS translocation, and placental inflammation of mice through flora mediation
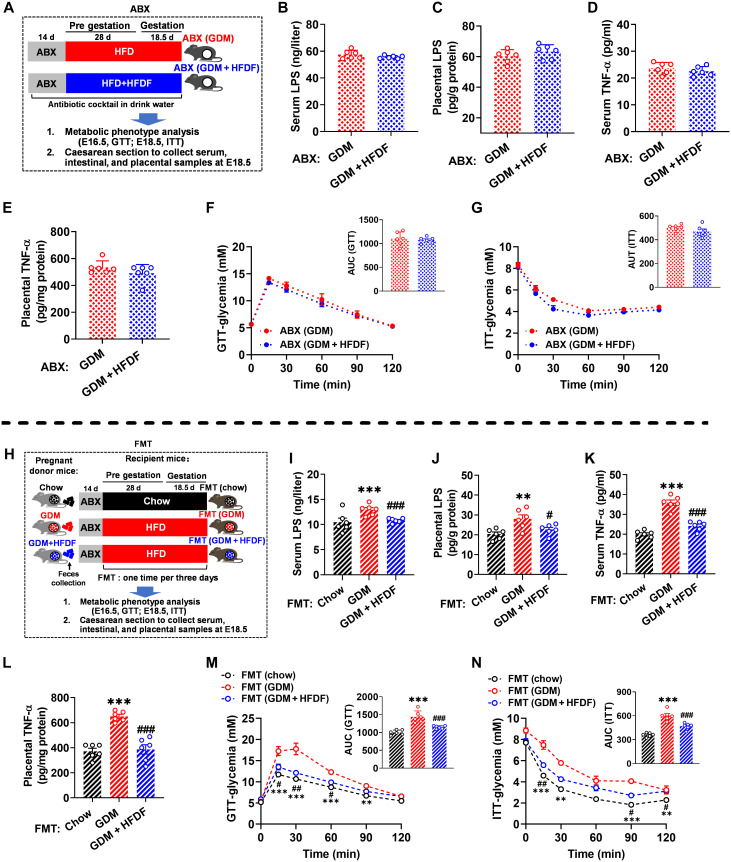
We also examined to what extent the restoration of HFD-induced loss of gut barrier by HFDF requires the presence of a flora. First, we tried to ablate the flora with antibiotics (Fig. 7A). Oral antibiotic mixture was shown to reduce the fecal DNA and total bacterial load in mice to 1/3 and 1/10, respectively (fig. S6, A and B), indicating that their gut flora was mostly cleared. Notably, antibiotic treatment greatly reduced the ameliorative effect of HFDF on bacterial-derived LPS translocation and placental inflammation and suppressed the systemic anti-GDM effect (Fig. 7, B to G, and fig. S6, C to N). The effect of HFDF-induced changes in flora on placental, gut, and metabolic parameters was further investigated by fecal microbiota transplantation (FMT) (Fig. 7H), where the flora was transferred from chow, GDM, or GDM + HFDF donor mice to their respective conventional recipient mice, followed by examination of GDM-related traits. Fecal flora transplantation from GDM or GDM + HFDF donors did not affect α-diversity of HFD-fed recipients but did significantly affect β-diversity on Bray-Curtis (fig. S7, A to D). The phylum- and genus-level variations suggested a different microbial composition among the recipient groups (fig. S7, E and F). From the donors, 810 amplicon sequence variants (ASVs) successfully colonized the intestine of recipient mice (fig. S7G). These results suggested that the fecal flora composition of donors was successfully reproduced in recipient mice. As expected, FMT (GDM + HFDF) mice resisted a series of metabolic syndromes of pregnancy induced by HFD, thus preventing placental LPS translocation and placental inflammation, maintaining gut barrier function, and improving GDM phenotype, adverse pregnancy outcomes, and placental function (Fig. 7, I to N, and fig. S8, A to W). Collectively, these data indicate that HFDF regulates placental inflammation and GDM phenotype via alteration of placental bacterial-derived LPS translocation mediated by flora.
Fig. 7. Effects of flora on the ability of HFDF to prevent placental inflammation and suppress insulin resistance during pregnancy.
(A) Illustration of antibiotic (ABX) experiment. Results represent two independent experiments. (B and I) Serum LPS level of ABX experiment and FMT experiment (n = 6 mice per group). (C and J) Placental LPS level of ABX experiment and FMT experiment (n = 6 placentae from 6 litters per group). (D and K) Serum TNF-α level of ABX experiment and FMT experiment (n = 6 mice per group). (E and L) Placental TNF-α level of ABX experiment and FMT experiment (n = 6 placentae from 6 litters per group). (F and M) GTT of ABX and FMT experiment (n = 6 mice per group). (G and N) ITT of ABX and FMT experiment (n = 6 mice per group). (H) Illustration of FMT experiment. Data were analyzed by unpaired Student’s t test with Welch’s correction [(B) to (E)], one-way ANOVA followed by post hoc Tukey’s tests (I to L), or two-way ANOVA followed by post hoc Bonferroni tests (GTT and ITT) and represented means ± SEM. #P < 0.05, ** or ##P < 0.01, and *** or ###P < 0.001, relative to chow and GDM group, respectively.
Lachnospiraceae-derived butyrate alleviates gut barrier impairment and reduces placental inflammation and GDM phenotype
In Fig. 6 (E to I), g_Lachnospiraceae_NK4A136_group was identified as a key genus enriched under HFDF treatment, which may play a beneficial role in alleviating insulin resistance and maintaining gut barrier function. This pattern was recapitulated in FMT recipient mice. LDA, random forest algorithm, and volcano plot analysis flagged Lachnospiraceae as the most enriched and representative bacterium in GDM + HFDF mice (fig. S7, H to K). At the genus level, the abundance variations in g_Lachnospiraceae_NK4A136_group showed the same trend between recipient and donor mice (fig. S7L). These results further suggest that g_Lachnospiraceae_NK4A136_group could be the key regulator of gut-placenta axis under HFDF treatment.
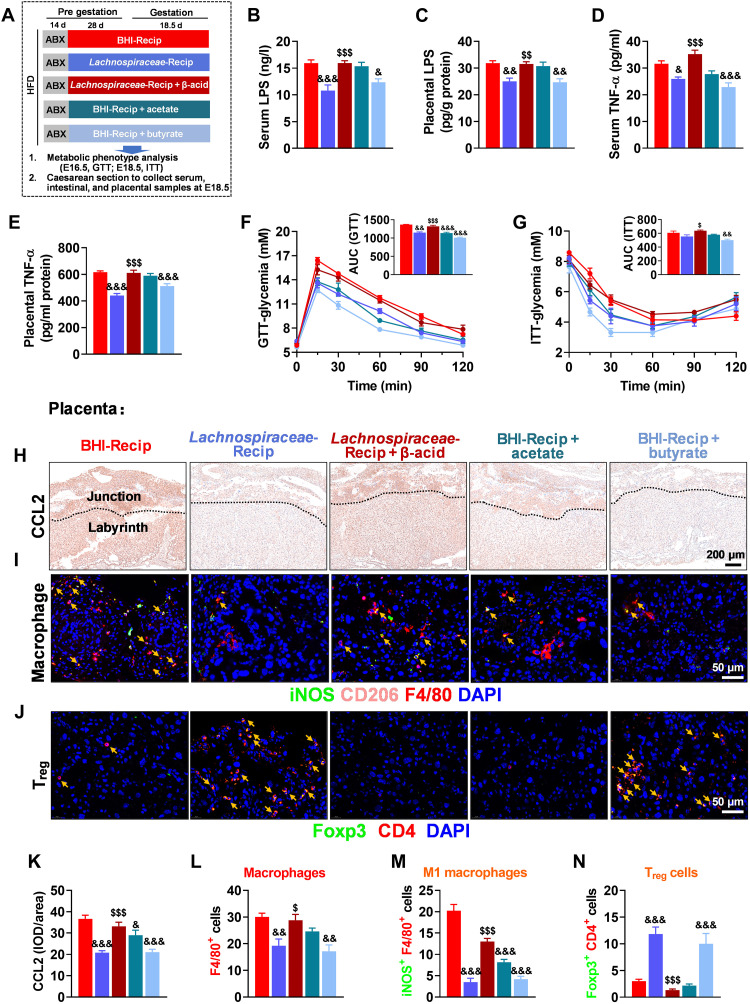
Microbial functions in recipient mice were predicted using Tax4fun, and FMT (GDM + HFDF) recipients showed the enrichment of valine, leucine, and isoleucine degradation and phenylalanine metabolism, related to SCFA metabolism (fig. S7M). Lachnospiraceae and its microbial metabolite butyrate play an ameliorating role in colitis, radiation syndromes, and graft-versus-host disease (27–29). Meanwhile, SCFAs could improve the impaired placental function and inflammatory response induced by intestinal-derived endotoxin (8). We observed increases in acetate and butyrate in colon content and placenta of GDM + HFDF or FMT (GDM + HFDF) mice versus GDM or FMT (GDM) mice (fig. S9, A and B). These SCFAs and metabolic parameters are correlated with g_Lachnospiraceae_NK4A136_group (fig. S9C). Notably, data from pregnant women demonstrated lower butyrate levels in the umbilical vein serum and placenta of GDMs than those of non-GDMs (fig. S9, D and E), and butyrate levels in the umbilical vein serum and placenta were negatively correlated with placental TNF-α (R = −0.3619 and −0.5706, respectively; P = 0.0036 and 0.0230, respectively; fig. S9, F and G). These results suggest that the bacterial metabolite butyrate may enter the placenta through circulation and thus suppress the GDM phenotype associated with placental inflammation. Moreover, the roles of Lachnospiraceae and its microbial metabolite in GDM and placental inflammation were investigated through treatments of live Lachnospiraceae, live Lachnospiraceae + β-acid, sodium acetate, or sodium butyrate in GDM model (Fig. 8A). β-Acid, an ingredient derived from hops, could reduce bacterial-mediated SCFA production (fig. S10A) (30–32). Live Lachnospiraceae and butyrate could reverse HFD-induced placental inflammation (CCL expression, macrophage infiltration, M1 macrophage polarization, and depletion of Treg cell), gut barrier impairment, and GDM phenotype, while β-acid supplementation could eliminate the beneficial effects of live Lachnospiraceae (Fig. 8, B to N, and fig. S10, B to M). The effects of heat-inactivated Lachnospiraceae, which can cause the loss of butyrate production (33), and culture supernatants of Lachnospiraceae were also verified (fig. S10N). Heat-inactivated Lachnospiraceae did not have anti-GDM effects, whereas the culture supernatant of Lachnospiraceae could reproduce the benefits of live Lachnospiraceae or butyrate (fig. S10, O to U). These results suggested that Lachnospiraceae-derived butyrate could alleviate gut barrier impairment and reduce placental inflammation and GDM phenotype.
Fig. 8. Effects of Lachnospiraceae and SCFAs on HFD-induced placental inflammation and GDM development.
(A) Schematic of medium control [brain-heart infusion (BHI)], transfer experiments with bacteria (Lachnospiraceae), Lachnospiraceae + β-acid (an inhibitor of bacterial SCFA production), acetate, or butyrate. At E18.5, serum, intestinal, and placental samples were obtained after execution of mice (n = 6 mice per group). (B and C) Serum and placental LPS level (n = 6 mice per group). (D and E) Serum and placental TNF-α level (n = 6 mice per group). (F) GTT and AUC (n = 6 mice per group). (G) ITT and AUC (n = 6 mice per group). (H and K) Immunohistochemistry and quantification of CCL2 (one of the key factors in macrophage recruitment) in placenta (n = 6 placentae from 6 litters per group). Scale bar, 200 μm. (I, L, and M) Localization of macrophage (F4/80+; red), M1 macrophage (iNOS+/F4/80+; green/red; yellow arrow), and M2 macrophage (CD206+ F4/80+; pink/red; it was not quantified because it was almost nonexistent) in placenta by immunofluorescence (n = 6 placentae from 6 litters per group). Scale bar, 50 μm. (J and N) Localization of Treg cell (Foxp3+/CD4+; yellow arrow) in placenta by immunofluorescence (n = 6 placentae from 6 litters per group). Scale bar, 50 μm. Data were analyzed by one-way ANOVA followed by post hoc Tukey’s tests (B to E, AUC, and K to N) or two-way ANOVA followed by post hoc Bonferroni tests (GTT and ITT) and represented means ± SEM. & and $P < 0.05, && and $$P < 0.01, and &&& and $$$P < 0.001, with & versus BHI-Recip and $ versus Lachnospiraceae-Recip.
Butyrate alleviates gut barrier impairment and placental inflammation in GDM mice partially through GPR109A
G protein–coupled receptor 109A (GPR109A) is an important mediator of butyrate (34), so we hypothesized that GPR109A can mediate the role of butyrate in regulating gut barrier, placental inflammation, and subsequent anti-GDM (fig. S11A). The presence of a specific inhibitor of GPR109A [mepenzolate bromide (MPN)] was shown to partially reverse the effects of butyrate on reducing serum FITC flux, serum LPS, placental LPS levels, and increasing tight junction protein expression (fig. S11, B to E). Moreover, MPN could partially abolish the protective effect of butyrate on placental inflammation associated with placental immune dysfunction (macrophage infiltration and M1 macrophage polarization) (fig. S11, F to M). Notably, MPN could partially abolish the anti-GDM effect of butyrate as shown by GTT and ITT (fig. S11, N to Q). Therefore, butyrate can improve gut barrier, placental inflammation, and the subsequent GDM phenotype at least partially through GPR109A.
DISCUSSION
Although a recent high-profile study reported that inflammation in a human cohort can serve as an early marker of GDM (3), whether placental inflammation is mechanistically involved in the progression of GDM is unclear. This study first focused on circulating and placental TNF-α changes rather than other inflammatory cytokines due to the following considerations: (i) In clinical studies, TNF-α–mediated insulin resistance has been reported as a crucial factor in GDM (6, 35, 36); (ii) TNF-α is mainly synthesized and secreted by the placenta during pregnancy; (iii) most of the placental TNF-α (94%) was released into the maternal circulation during pregnancy (6); and (iv) our results showed that TNF-α had the highest fold change in the placentae of GDM mice. Thus, TNF-α was used to quantify placental inflammation in this study. Here, abnormal glucose metabolism in GDMs was found to be associated with placental TNF-α, and this notion was reinforced by the rapid restoration of glucose homeostasis and circulating TNF-α following placental expulsion at delivery in the mouse model. We found that HFDF treatment could reduce GDM development through the gut-placental inflammation axis. Mechanistically, the HFDF was shown to increase the abundances of Lachnospiraceae and butyrate, enabling the reduction of placental-derived inflammation by enhancing gut barrier and inhibiting the transfer of bacterial-derived LPS, ultimately resisting HFD-induced insulin resistance. Lachnospiraceae and butyrate were also found to have similar anti-GDM and anti–placental inflammation effects, which are associated with dampened macrophage infiltration and M1 macrophage polarization and increased Treg cells in placenta. In addition, butyrate could alleviate gut barrier impairment and placental inflammation in GDM mice partially through GPR109A. These findings demonstrate the central role of placenta in the pathophysiological mechanisms of GDM and the potential of fermentable fibers as a means of reducing GDM susceptibility via the gut flora–placental inflammation axis (Fig. 9).
Fig. 9. GDM is associated with the development of placental hypofunction and inflammation.
A HFDF (konjac) could reduce GDM development through gut-SCFA-placenta axis in the GDM mouse model. Mechanistically, the HFDF could increase the abundances of Lachnospiraceae and butyrate, enabling the reduction of placental-derived inflammation by enhancing gut barrier and inhibiting the transfer of bacterial components (LPS), ultimately resisting HFD-induced insulin resistance. Lachnospiraceae and butyrate have similar anti-GDM and anti–placental inflammation effects, and they can ameliorate placental function and pregnancy outcomes probably by dampening placental immune dysfunction.
Numerous animal-based studies revealed the multiple effects of HFD-induced GDM on placenta, such as endothelial disorders, insulin resistance, or inflammatory responses, among other metabolic phenotypes (37, 38). Although these studies have advanced our knowledge of placental physiological response to maternal glucose metabolism, it is still necessary to elucidate the causal relationship between changes in placental and maternal metabolism for developing strategies against GDM transmission. Here, we demonstrated that HFDF could restore HFD-induced GDM phenotype, including changes in maternal mass, circulating glucose metabolic parameters, and placental and hepatic metabolic phenotypes. Placental inflammation induced by intrauterine E. coli injection was shown to sufficiently reverse the beneficial effects of HFDF on insulin resistance and adverse pregnancy outcomes in the short term without affecting maternal weight, liver, or fat mass. This suggested that weakening the placental inflammatory response, rather than obesity, may be the main mechanism by which HFDF affects maternal insulin resistance. Similar previous studies have shown that TNF-α, which is mainly secreted by placenta during mid to late pregnancy, can cause maternal insulin resistance independent of maternal fat mass (6). This is interesting because it is known that, in the absence of pregnancy, HFD can lead to the secretion of cytokines from various organs, such as the liver and adipose tissue, and these factors play important roles in overall maternal metabolism. We speculate that the widely accepted dogma that increased adiposity equates to increased maternal inflammation and insulin resistance may not be so evident during pregnancy as in nonpregnant state. Consistent with the data in clinical practice, 74% of GDMs did not show obesity during pregnancy (39). Collectively, this study has fortified the view that placental inflammation is a central target for the response and regulation of maternal insulin resistance.
In a healthy pregnancy, macrophages are the second largest leukocytes at the maternal-fetal interface and have been shown to play an important role in remodeling spiral arteries and placental development (40). However, macrophages are also the key regulators of inflammation (8). The deterioration of the placental proinflammatory environment is characterized by placental inflammatory cell infiltration and macrophage phenotype changes, with placental macrophages as the main producers of TNF-α (41). Proinflammatory M1 macrophages secreting TNF-α, interleukin-1β (IL-1β), IL-12, and IL-18 are more abundant in the placenta, decidua, and surrounding uterine spiral arteries of women with metabolic abnormalities than in tissues from healthy pregnancies (40). Dysregulation of macrophage polarization and secretion of proinflammatory factors are responsible for reduced trophoblast cell invasion and inadequate spiral artery remodeling in preeclampsia (40). In the present study, GDM placentae had increases in placental inflammatory cell infiltration, macrophage number, and CCL2 expression. Blocking CCL2/CCR2 signaling [one of the key factors in macrophage recruitment (17)] could alleviate HFD-induced placental inflammation and GDM phenotype in mice. This indicates that macrophage-associated inflammation also plays an important role in GDM. Moreover, placentae in GDM + HFDF, Lachnospiraceae, or butyrate treatments were found to exhibit unpolarized M0 macrophages and reduced macrophage infiltration, whereas GDM placentae exhibited increased macrophage infiltration and proinflammatory M1 macrophage polarization. This suggests that modulating macrophage function may be one of the mechanisms for HFDF and gut flora to reduce placental inflammation.
Maternal glucose metabolism has a potential reciprocal role with gut inflammation. On the one hand, several clinical and preclinical studies have demonstrated that microbial-associated inflammation may contribute to metabolic abnormalities during pregnancy (7, 8, 23). The disruption of intestinal barrier and TLR signaling driven by the transfer of bacterial components such as LPS from gut to placenta could induce an inflammatory response in the maternal placenta in mouse model (7, 8, 42). Previous studies have reported that the gut flora from patients with pre-eclampsia, another metabolic disease of pregnancy, could induce pre-eclampsia in mice by causing damage to intestinal barrier and inflammation and pathogenic bacterial translocation from gut to placenta (7, 8). Similarly, GDMs were reported to have a distinct placental flora profile (43). A recent study on a human cohort reported that GDM was driven by inflammation caused by flora in the months before diagnosis (3). Therefore, we measured the LPS concentration in the placenta in this study, and the LPS concentration was shown to be higher in the placenta of GDMs than that of non-GDMs, and placental LPS showed a positive correlation with placental TNF-α. We also measured the LPS concentration in the placenta of HFDF-treated mice, and HFDF treatment was shown to reduce the LPS concentration in GDM placenta. These results suggested that the entry of bacterial component (LPS) into the placenta through circulation may be an important cause of placental inflammation. With gut flora removed with antibiotics, the subsequent FMT experiment demonstrated an indispensable role of gut flora in HFDF remodeling of gut function and inhibition of placental LPS translocation against HFD-induced placental inflammation and GDM. On the other hand, there is no clear evidence that the development of GDM causes colonic inflammation in pregnant women. Host metabolic health and diabetes status have diet-independent effects on intestinal growth and function in the UC Davis type 2 diabetes mellitus rat (44). Moreover, there is an association between type 2 diabetes and increased risk of multiple gastrointestinal diseases, such as diabetes as an independent risk factor for colon and rectal cancer in human, which highlights the importance of preventing gastrointestinal diseases in patients with diabetes (45, 46). Excessive immune response in GDM has been previously reported to cause chronic low-grade inflammation (3, 4), and our data support the role of the placenta as a source of inflammation in GDM. Therefore, it is interesting to know whether cytokines secreted by the placenta may, in turn, influence the immune and inflammatory response in maternal organs such as the colon.
Maternal dietary intervention in GDM through gut flora is controversial. Despite previous demonstration of marked changes in the gut flora of pregnant women with GDM (47), the effect of dietary modulation of gut flora during pregnancy on GDM, particularly during the intervention window, remains controversial. Several studies have consistently shown that supplementation of probiotics or fish oil in early or mid-pregnancy does not prevent GDM in late pregnancy (47–49). A possible explanation is that the GDM status may disturb maternal gut flora flexibility and thus limit the capacity of GDM women to respond to dietary interventions during gestation (47). A recent systematic review and meta-analysis of prospective studies suggested that higher dietary fiber intake starting before pregnancy was associated with a reduced risk of GDM (50). Therefore, we hypothesize that pregestation and postgestation dietary fiber interventions in mouse model may be more effective in preventing gestational diabetes targeting gut function and flora. The results provide evidence that pregestation and postgestation diet is associated with the susceptibility of mice to GDM, i.e., initiating HFDF supplementation before gestation prevents abnormal glucose metabolism and inflammatory responses to GDM through the gut flora–placenta axis. This also implies that the window period for preventing GDM should start before pregnancy.
Decrease in SCFAs is one of the early biomarkers of GDM (3). SCFAs help maintain glucose homeostasis and suppress inflammatory response (3). Our findings suggest that Lachnospiraceae_NK4A136_group, a member of the Lachnospiraceae family, may be a core genus for GDM prevention under HFDF treatment. Lachnospiraceae is a highly abundant member of the gut bacteria able to synthesize SCFAs by fermenting dietary polysaccharides (27–29). The HFDF used in this study is a hydrogel-like polysaccharide extracted from the tubers of the konjac plant with high microbial availability and SCFA production (14–16). Data from pregnant women demonstrated that butyrate levels in the umbilical vein serum and placenta were lower in GDMs than in non-GDMs, and butyrate levels in the umbilical vein serum and placenta were negatively correlated with placental TNF-α. Moreover, we found that the ameliorative effect of Lachnospiraceae on insulin resistance and inflammation was reduced by β-acid (an inhibitor of bacterial SCFAs production), suggesting the central role of SCFA in anti-GDM. Thus, the circulation of the bacterial metabolite butyrate into the placenta may be another mechanism to alleviate GDM associated with placental inflammation.
Butyrate exerts metabolic protection in metabolic disease models during pregnancy through a variety of mechanisms, including reducing inflammation and improving gut barrier function. On the one hand, butyrate exerts metabolic regulation through activation of GPR109A, the major receptor of butyrate (34). Butyrate could enhance the gut barrier integrity through GPR109A and then regulate the expression of the tight junction protein in an Akt signaling–dependent manner in an enteritis model (51). Moreover, butyrate-GPR109A signaling could induce Treg differentiation and confers an anti-inflammatory phenotype on macrophages in a colon cancer model (34). Our data suggest that the effect of butyric acid in improving the intestinal barrier in GDM mice was partially reversed after pharmacological inhibition of GPR109A. Inhibition of GPR109A also partially restored placental immune dysfunction (increased macrophage infiltration, proinflammatory M1 macrophage polarization, and Treg depletion), placental inflammation, and the GDM cascade in GDM mice. Thus, we proposed that butyrate can ameliorate gut barrier, placental inflammation, and subsequent GDM phenotype at least partially via GPR109A. On the other hand, butyrate exerts effects by inhibiting histone deacetylase (HDAC). Butyrate could promote Treg cell generation by inhibiting HDAC expression, conferring anti-inflammatory effect in rheumatoid arthritis mice (52). In addition, butyrate could constrain neutrophil function and ameliorate mucosal inflammation in inflammatory bowel disease by inhibiting HDAC (53). During pregnancy, Treg cells play an important role in maintaining an anti-inflammatory decidual environment and macrophage differentiation and further regulate implantation and placental development. Insufficient Treg cell numbers or inadequate functional competence is implicated in idiopathic infertility and recurrent miscarriage as well as later-onset pregnancy complications stemming from placental insufficiency, including preeclampsia and fetal growth restriction (54). In the present study, inhibition of GPR109A did not affect the promotion of placental Treg cells by butyrate, suggesting a potential role for butyrate-HDAC in GDM.
Results from the placental inflammation model suggest that HFDF resistance to HFD-induced obesity may be independent of maternal food intake and inflammation. The mechanisms of HFDF resistance to HFD-induced maternal obesity may include, but are not limited to, the following: (i) The FMT trial replicated the inhibition of maternal fat deposition by HFDF, suggesting that microbes may be key to reducing obesity-related traits. HFDF-derived butyrate may increase energy expenditure and thus fight maternal obesity. Butyrate acts on components of the energy balance, i.e., stimulating energy expenditure, thereby reducing obesity and obesity-associated disorders (55). Mechanistically, a previous study indeed showed that butyrate could induce peroxisome proliferator–activated receptor-γ coactivator-1α activity, thereby enhancing mitochondrial function in brown adipose tissue and substantially promoting energy expenditure (56). (ii) Chronic hepatic sympathetic overactivity mediates hepatic steatosis (i.e., excessive hepatic triglyceride accumulation) (57). Monocarboxylic metabolites, as propionate and ketone bodies, have been reported to inhibit hepatic steatosis by modulating the sympathetic nervous system via GPR41 (58), so we speculate that HFDF-derived butyrate may have similar effects in modulating sympathetic nerves. (iii) Obesity in mice can be inhibited by homeostasis in circadian eating rhythms independent of energy intake (59, 60). Mechanistically, the circadian variation in the mechanical sensitivity of the gastric vagus nerve in HFD-fed mice could completely disappear and was replaced by changes in food ingestion behavior that are independent of clock gene expression patterns (61). Our previous study in sows has shown konjac flour as a highly absorbent, swelling, and viscous fiber, which can enhance satiation by increasing gastrointestinal fullness (14). Therefore, HFDF may restore the gastric vagus response to mechanical stimuli through its physical properties, which, in turn, initiates satiety and restores circadian eating patterns, thereby possibly inhibiting HFD-induced maternal obesity. However, these hypotheses need further validation.
There are several limitations to the present study. First, we failed to assess the dietary fiber intake of the subjects by food frequency questionnaire or 24-hour diet recalls in the human trial, so it is necessary to obtain quantified fiber (especially HFDF) intake in relation to GDM and inflammation in future trials. Second, the quantification of gut barrier function and SCFAs in this study was done within colonic region—the putative site of konjac flour fermentation. A recent study demonstrated the limitations of fecal samples in capturing the diversity of microbial communities in the colonic region (62). 16S rRNA analysis of feces may not fully characterize the colonic gut flora. While feces are still a valuable and effective means of characterizing the distal colon microbiota, future work may need to take gut biogeography into account. Last, any study on the presence of bacteria in the placenta is likely to be questioned, but the presence of bacteria in GDM placentae has been previously reported (43). FISH and qPCR analyses showed that bacterial load was elevated in GDM placentae but decreased in GDM + HFDF placentae. Because of a small sample size and limited sampling conditions, these data should be interpreted with caution and only show an association of bacteria with reduced adverse symptoms in GDM model. Notably, we detected high bacteria loads in the placentae of all pregnant women regardless of GDM status, possibly due to contamination from vaginal delivery or cesarean section. Further evidence of the presence and potential colonizers of GDM placental bacterial translocations is required to be demonstrated using sterile model animals to collect placental samples in a stricter environment (sterile, isolated, low-contamination, controlled environment) in further studies. We intend to gradually address these problems step by step and provide further insight into the mechanistic links between placental inflammation and gut flora.
Overall, this study demonstrated that abnormal glucose metabolism in patients with GDM is associated with placental inflammation, and HFDF supplementation can reduce GDM development through gut flora–placenta axis. We identified that the HFDF could increase the abundances of Lachnospiraceae and butyrate, enabling the reduction of placental-derived inflammation by enhancing gut barrier and inhibiting the transfer of LPS, ultimately resisting insulin resistance in an HFD-induced GDM model. These findings show the involvement of important placental inflammation–related mechanisms in the progression of GDM and the great potential of HFDFs to reduce susceptibility to GDM through gut flora–placenta axis.
MATERIALS AND METHODS
Clinical patient data and sample collection
From 2021 to 2023, a total of 24 patients with GDM and 42 non-GDMs were recruited in Shenzhen, Guangdong, China. This observational study was approved by the Ethics Committee of Shenzhen People’s Hospital and followed the Declaration of Helsinki (LL-KY-2021349). All individuals were informed of the nature of the study, and their informed consent was obtained. All procedures conformed to the relevant laws and institutional guidelines. GDM was diagnosed at 24 to 28 weeks of pregnancy according to the criteria of the International Diabetes and Pregnancy Study Group (63). Information about maternal characteristics was obtained from standardized medical records, including age, body weight, first trimester BMI, medical history, and oral GTT at 24 to 28 weeks of gestation, systolic blood pressure, diastolic blood pressure, neonatal weight, and neonatal sex. Moreover, all pregnant women with GDM received only lifestyle modification interventions to avoid any deviations from drug therapy. The exclusion criteria of the sampling were as follows: This study excluded pregnant women with any prenatal infection, high blood pressure, kidney disease, thyroid dysfunction, and any other serious medical condition during pregnancy or receiving medication such as antibiotics and insulin. Initially, 66 women were recruited, but three GDMs did not provide placental samples. Last, 63 women (21 GDMs and 42 non-GDMs) completed the follow-up.
Peripheral blood (5 ml) was collected in the morning after an overnight fast (≥8 hours) 24 to 28 weeks of gestation, and the blood and serum samples were stored in −80°C for glucose and HbA1c analysis. Moreover, venous and arterial blood samples were collected from umbilical cords immediately after delivery, followed by centrifugation at 4000 rpm for 15 min to obtain cord blood serum samples and storage at −80°C for analysis of relevant indicators. Placental samples were collected using a previously reported protocol, with minor modification (64). Briefly, after a vaginal birth or cesarean section, the fresh placentae were washed in a normal saline solution to remove any blood clots. The samples were collected from the placental disc by cutting from the maternal side to the fetal side, and they were divided into two portions. One portion was fixed in 10% neutral buffered formalin for histological and immunological analysis, and the other portion was stored at −80°C for TNF-α analysis. For detailed maternal, neonatal, and placental characteristics, refer to table S1.
HFD-induced GDM mouse model and HFDF treatment
Animal studies were conducted according to the guidelines of Guangdong Province on the Review of Welfare and Ethics of Laboratory Animals and approved by the South China Agricultural University Animal Care and Use Ethics Committee (2022F233). Seven-week-old C57BL/6J mice were purchased from Bestest, China and housed in a temperature/humidity-controlled environment (23° ± 3°C/70 ± 10%) on a 12-hour light/dark cycle with free access to food and water. All mice were stabilized in the same feeding management and environment for a week before initiating any experimental procedures. The same-week-old female mice were completely randomized to different experimental groups based on body weight. The number of samples for each animal trial was specified in the corresponding legend.
GDM mouse model was established as previously described (65), and the experimental procedure is shown in Fig. 2A and fig. S1A. Briefly, 8-week-old female mice were fed standard chow diet (chow, XTADG001, Xietong Shengwu Co. Ltd., China) or a HFD (60% energy from fat; XTHF60, Xietong Shengwu Co. Ltd., China) during pregestation (before mating, 28 days) and postgestation (E0.5 to E18.5, 18 days). For chow and HFD composition, refer to table S2. Female mice were mated with male mice overnight at a proportion of 2:1 per cage. Pregnancy was determined by the presence of vaginal plugs the next morning, which was identified as E0.5. The GDM mice were defined using the International Association of Diabetes and Pregnancy Study Group criteria: a fasting blood glucose of ≥5.1 mM or a blood glucose of 8.5 mM 2 hours after feeding (66). This model is widely used to induce several characteristics resembling human GDM, including maternal obesity and imbalance of glucose metabolism (65).
GDM + HFDF treatment procedure is shown in Fig. 2A. Briefly, 8-week-old female mice received an HFD diet with highly fermentable konjac flour (HFDF) as a source of fiber during pregestation and postgestation. See table S2 for HFD + HFDF diet composition. The concentration of konjac flour in the diet was determined as reported in our previous studies (14). The HFD and HFD + HFDF diets consisted of 60% kcal from fat, 20% kcal from carbohydrates, and 20% kcal from protein. The HFD and HFD + HFDF diets are isocaloric (5243.21 kcal at water fuel energy/kg) and isofiber level (6.5% fiber level in diet). Analysis of mouse metabolic phenotypes was performed at indicated time points. Among them, E6.5, E12.5 to E18.5, and E15.5 to E18.5 are the developmental stages of epiblast, placenta, and fetal weight gain, respectively. After fasting for 4 hours from 8 a.m., mice were euthanized by CO2 inhalation on E12.5 or E18.5 to collect blood and tissues. The placental units were separated from the decidual tissue (maternal origin). All placental tissues were obtained from E12.5 (only in Fig. 4, B to D) or E18.5 mice by cesarean section. The numbers and weight of viable and resorbed pups were counted and recorded. Placental efficiency was indicated by the ratio of fetal weight to placental weight. The weight of the gestational product is the sum of the placenta and fetal weight. Abnormal fetuses were defined as macrosomia and intrauterine growth restriction fetuses. All eligible mice in the same cohort were included in the measurement of litter performance during the experiment, unless they died or miscarried.
Pharmacological blockade of the CCL2/CCR2
The pharmacological blockade of CCL2/CCR2 experimental procedure is shown in fig. S1F. Briefly, HFD-fed mice were intraperitoneally injected with 2 mg/kg of body weight of RS-504393 (CCR2 antagonist, HY-15418, MedChemExpress) or dimethyl sulfoxide once daily during gestation. Intraperitoneal GTT was performed at E18.5. After execution of mice at E18.5, serum and placental samples were obtained after execution of mice.
GTTs and ITTs
Intraperitoneal GTT and ITT were performed at indicated time points (E16.5 or E18.5). For GTT, an injection of glucose (2 g/kg of body weight) was given to overnight (12-hour)–fasted mice, followed by measuring blood glucose levels at different time points. For ITT, 5-hour–fasted mice were intraperitoneally injected with a single dose of insulin (1 U/kg), followed by measuring blood glucose levels in tail vein blood by a blood glucose meter (Sinocare).
Islet isolation and insulin secretion assay
Islet isolation and insulin secretion assay followed a previously reported protocol with minor modification (67). Briefly, E18.5 mice were killed, and the gallbladder region was exposed. After clamping at the confluence of the common bile duct and duodenum, the pancreas was dilated by perfusing the bile duct with collagenase IV (2 mg/ml), followed by excising the pancreas and shaking at 37°C for 30 min. After washing and centrifugation, pancreatic islets were hand-picked under a stereomicroscope and then incubated for 6 hours in RPMI 1640 medium with 5 mM glucose for recovery. Next, the isolated islets were washed and incubated for 1 hour with Krebs Ringer bicarbonate buffer (KRBB) containing 0.1% fatty acid–free bovine serum albumin (BSA) supplemented with 5 mM d-glucose. For stimulated secretion, islets were washed again and incubated in 25 mM d-glucose KRBB for 1 hour, and the stimulation index was calculated as the amount of insulin secreted in 25 mM divided by the amount of insulin secreted in 5 mM. Insulin secretion was determined using commercially available enzyme-linked immunosorbent assay kits as instructed by the manufacturer (MM-0579 M1, MEIMIAN, China).
Inactivated E. coli–induced placental inflammation model
The placental inflammation experiment (Fig. 4H) was performed on E12.5 as previously described (22). Briefly, female mice received HFD or HFD + HFDF during pregestation (before mating, 4 weeks) and postgestation (E0.5 to E18.5, 18 days). E12.5 mice were anesthetized with 0.015 ml/g of body weight of Avertin [2.5% tribromo ethyl alcohol and 2.5% tert-amyl alcohol in phosphate-buffered saline (PBS)], followed by making a 2-cm midline incision in the lower abdomen, then intrauterine injection of a 100-μl solution containing inactivated E. coli bacteria suspended in LB medium (see the following text) or LB medium only (control) in the midsection of the right uterine horn at a site between two adjacent fetuses, taking care not to inject individual fetal sacs. The abdomen was closed in two layers, with 4-0 polyglactin sutures at the peritoneum and wound clips at the skin. Surgical procedures lasted ~15 min. Then, serum, liver, subcutaneous fat, inguinal fat, and placental samples were obtained after execution of E18.5 mice.
E. coli bacteria were prepared as previously described (22). A fresh culture of previously frozen E. coli (number 12014, American Type Culture Collection, Manassas, VA) was grown overnight in 4000 ml of LB broth, followed by concentrating the overnight culture through centrifugation, suspending it in 10 ml of LB broth, and determining its concentration by plating serial dilutions in duplicate. The bacteria within the suspension were inactivated by immersion in a boiling water bath for 30 min and then frozen at −20°C. Inactivation was verified by lack of overnight growth on plates and in broth culture. Once the concentration of the frozen stock was known, it was thawed and diluted to 107 organisms/ml, followed by aliquoting and freezing the latter suspension at −80°C, which was thawed and diluted as needed before each experiment.
Antibiotic treatment
The flora depletion experimental procedure is shown in Fig. 7A. Briefly, 5-week-old C57BL/6J mice were purchased from Bestest, China and housed in a temperature/humidity-controlled environment (23° ± 3°C/70 ± 10%) on a 12-hour light/dark cycle with free access to food and water. Mice were pretreated for 2 weeks with the antibiotic mixture, their sterile drinking water supplemented with an antibiotic solution containing streptomycin (1 g/liter; MB1275, MEILUNBIO, China), ampicillin (1 g/liter; MA0317, MEILUNBIO, China), and neomycin (1 g/liter; MA0312, MEILUNBIO, China), and then randomly divided into two groups (HFD or HFD supplemented with 6.5% konjac flour). Confirmation that each mouse has obtained adequate antibiotics was performed by measuring water consumption (~5 ml/day) in the early morning of each day. Drinking water was prepared daily. The dietary treatment lasted from 4 weeks before breeding to E18.5. The depletion of microbes was confirmed by fecal DNA levels and qPCR using the universal bacteria-specific primers (table S3). At E18.5, serum, intestinal and placental samples were obtained after execution of mice.
FMT experiment
FMT experimental procedure was performed based on established protocols (7, 68, 69) as shown in Fig. 7H. Briefly, diet handling and administration of chow, GDM, and GDM + HFDF donor mice are shown in Fig. 2A. Donor mice were housed in single cages. Feces from each of three pregnant donor mice from chow, GDM, and GDM + HFDF groups were pooled and used as a source of FMT (chow), FMT (GDM), and FMT (GDM + HFDF) recipient mice, respectively. Each diet treatment contained nine donor mice and obtained three sources of fecal transplant material. Donor stool was diluted with saline containing 0.05% cysteine-hydrochloride, homogenized for 1 min using a vortex to obtain a liquid slurry, and then centrifuged at room temperature for 3 min (500g) to remove particulate matter to facilitate administration. Fresh transplant material was prepared under aseptic conditions within 10 min before oral gavage to prevent changes in bacterial composition at 10: 00 a.m. each morning.
For recipient mice, 6-week-old mice housed in single cages were pretreated for 2 weeks with the antibiotic mixture [an antibiotic solution containing streptomycin (1 g/liter), ampicillin (1 g/liter), and neomycin (1 g/liter) in their sterile drinking water]. Water consumption (~5 ml/day) was measured in the early morning of each day to confirm whether each mouth obtained adequate antibiotics, and drinking water was prepared every 3 days. The depletion of microbes was confirmed by fecal DNA levels and qPCR. Eight-week-old mice were then randomly divided into three recipient groups and orally administered 2 ml of respective donor fecal supernatant (100 mg/ml) once every 3 days for 46 days [during pregestation (before mating, 28 days) and postgestation (E0.5 to E18.5, 18 days)]. FMT(chow) mice were fed chow diet, and FMT (GDM) and FMT (GDM + HFDF) mice were fed HFD throughout the experimental period. At E18.5, fresh feces, serum, subcutaneous fat, inguinal fat, intestinal, and placental samples were obtained after execution of recipient mice. The colonization of gut flora from the donors in recipient mice was verified using 16S rRNA gene sequencing.
In vivo permeability assay
Intestinal permeability was assessed with an in vivo FITC-dextran (#46944-4 kDa; Sigma-Aldrich) permeability assay, as described previously (70). Briefly, E18.5 mice fasted for 4 hours were gavaged with 0.6 mg/g of body weight of FITC-dextran solution, and their blood samples were collected by submandibular bleeding 3 hours later. Fluorescence intensity in the serum was measured using a microplate reader. FITC-dextran concentrations were determined from a standard curve generated by serial dilutions of FITC-dextran.
SCFAs analysis in colon content, umbilical vein serum, and placenta
The levels of acetate, propionate, butyrate, isobutyrate, isovalerate, and valerate in colon content, umbilical vein serum, and placenta were measured by gas chromatography. Briefly, colon content (~200 mg), umbilical vein serum (200 μl), or placenta (~100 mg) samples were homogenized in 20 μl of 25% metaphosphoric acid solution and 0.25 g of anhydrous sodium sulfate for acidification and salting out. Subsequently, the samples were homogenized in methyl tert-butyl ether (1 ml), followed by centrifugation at 4°C for 5 min (13,500g), transferring the supernatant into the vial, and storage at −20°C until gas chromatography–mass spectrometry (GC-MS) analysis. The concentrations of SCFAs in colon content were normalized according to stool weight. The concentrations of SCFAs in placenta were normalized according to protein levels (P0012, Beyotime, China). Notably, we used a strategy of 50%/50% males and females to avoid the interference of mice placental sex.
Transfer experiments with live, heat-inactivated Lachnospiraceae cultures, supernatant, β-acid treatment, acetate treatment, and butyrate treatment
The experimental procedure of transfer experiments with live Lachnospiraceae (American Type Culture Collection, TSD-26) cultures, β-acid treatment, acetate treatment, and butyrate treatment is shown in Fig. 8A. Briefly, Lachnospiraceae was cultured in an anaerobic chamber in brain-heart infusion (BHI) broth supplemented with 5% fetal bovine serum (FBS), 0.01% l-cysteine, 1% corn starch, and 0.5% konjac flour. Five-week-old C57BL/6J mice treated with the antibiotic mixture for 2 weeks were randomly divided into five groups, followed by orally gavaging the mice separately once every 3 days (from 4 weeks before mating until day 18.5 of gestation) with 2 ml of BHI medium (control), live Lachnospiraceae cultures [108 colony-forming units (CFU)], and live Lachnospiraceae + β-acid [β-acid (20 mg/liter) in drinking water] (30), acetate (200 mM; #S2889, Sigma-Aldrich), or butyrate (200 mM; #B5887, Sigma-Aldrich). The success of colonization and treatment of Lachnospiraceae, β-acid, and SCFAs in recipient mice was verified using qPCR and GC-MS, respectively. Moreover, the effects of heat-inactivated Lachnospiraceae and culture supernatants of active Lachnospiraceae were verified and shown fig. S10N. Briefly, the supernatant was obtained by centrifugation of live Lachnospiraceae (108 CFU) at 2500g for 10 min and filtered through a 0.2-μm syringe filter. The precipitate (Lachnospiraceae) obtained by centrifugation was rinsed twice and resuspended in BHI medium, followed by pasteurization at 75°C for 30 min and culture to ensure that no active strain remained. Heat inactivation treatment can cause the loss of butyrate production of Lachnospiraceae (33). Fresh transplant material was prepared under aseptic conditions within 10 min before oral gavage to prevent changes in bacterial composition at 10:00 a.m. each morning. At E18.5, serum, intestinal, and placental samples were obtained after execution of mice.
Pharmacological blockade of GPR109A
The pharmacological blockade of GPR109A experimental procedure is shown in fig. S11A. Briefly, to further characterize the role of the GPR109A receptor, HFD-fed mice were supplemented with butyrate in drinking water (200 mM) and intraperitoneally injected with MPN (5 mg/kg per day), a specific inhibitor of GPR109A (71), during pregestation and postgestation. Intraperitoneal GTT and ITT tests were performed at E16.5 and E18.5, respectively. At E18.5, serum, intestinal, and placental samples were obtained after execution of mice.
Histological analysis of colon, placenta, and liver
For histological analysis of the placentae from women, the placentae were fixed in 10% buffered formalin at room temperature, followed by embedding in paraffin, sectioning the tissues into 5-μm-thick slices, and staining them with H&E. Next, two independent observers, unaware of the experimental conditions, scored maternal vasculopathy and placental inflammation associated with pregnancy-induced GDM according to the previously reported criteria (64, 72, 73). Briefly, an important feature of placental inflammation is the presence of large infiltration of lymphocytes, plasma cells, and/or macrophages in the basal plate. Vascular lesions were characterized by the marked thickening of vessel wall, nonfunctioning vessel infarction, and infiltration of inflammatory cells into the vessel wall. Accelerated villous maturation was defined as the presence of small or short hypermature villi during gestation, usually with an increase in syncytial knots.
For histological analysis of mice tissues, mice colons, placentae, and livers were fixed in 10% buffered formalin at room temperature, followed by embedding in paraffin, sectioning the tissues into 5-μm-thick slices, and staining them with Picrosirius red, H&E, or Alcian blue–periodic acid Schiff (AB-PAS). (i) For histological analysis of mice placentae, placental blood sinus area was investigated by selecting six fields in each midsagittal placental tissue section, followed by using ImageJ software to analyze the total areas of placenta, junctional zone, and labyrinth, with the maximum area for the layer of the whole placenta being calculated by two serial sections in each individual. Notably, we used a strategy of 50%/50% males and females to avoid the interference of mice placental sex. (ii) For histological analysis of mice colon, two independent observers blinded to the experimental conditions evaluated the severity of colitis in H&E-stained sections according to the following criteria: 1 and 2, no signs of inflammation; 3 and 4, low leukocyte infiltration; 5 and 6, moderate leukocyte infiltration; 7 and 8, high leukocyte infiltration, moderate fibrosis, high vascular density, thickening of the colon wall, moderate goblet cell loss, and focal loss of crypts; and 9 and 10, transmural infiltrations, massive loss of goblet cells, extensive fibrosis, and diffuse loss of crypts. ImageJ software was used to determine the number of cupped cells in a single crypt from H&E. In addition, one section per colonic sample was stained with AB-PAS to evaluate mucin secretion. The integrated optical density (IOD) of the colon mucus was measured using AB-PAS staining pictures with an image analyzer (Image-pro Plus 6.0). Then, the mucin density (IOD per area) was calculated to show the expression level of colon mucin secretion. For fibrosis analysis, colonic sections were deparaffinized and stained with Picrosirius red dye for 1 hour. Staining quantification was performed by determining the ratio of positively stained (red) pixels to the total pixel number of each section (% fibrosis) using ImageJ. The birefringence of the Picrosirius red–stained colonic sections was also imaged on an Olympus BX41 microscope using the Olympus U-POT drop-in polarizer and U-ANT transmitted light analyzer. (iii) For histological analysis of mouse liver, two independent observers blinded to the experimental conditions evaluated the hepatic steatosis in the H&E-stained sections according to the criteria (ImageJ).
Immunohistochemical analysis of placenta
Briefly, mouse placenta tissues were collected and washed with iced-cold PBS and fixed in 4% paraformaldehyde at 4°C overnight. Next, the fixed samples were dehydrated in graded ethanol, embedded in paraffin, sliced, and blocked by 2% BSA, followed by incubation with appropriate primary and secondary antibodies. The positive areas of each sample were measured using ImageJ software.
Immunofluorescence analysis of placenta
Briefly, the fixed, permeabilized, and sliced placental samples were blocked with Quickblock blocking buffer for Immunol-staining (Beyotime), followed by incubation successively with primary and secondary antibodies. Stained tissues were viewed using a confocal fluorescence microscope (Zeiss).
RNA isolation and qPCR
RNA isolation and qPCR followed the previously reported standard procedures (74, 75). Briefly, total RNA from placenta (whole placenta), liver (~100 mg), or colon (~50 mg) was extracted with the reagent box of total RNA kit as instructed by the manufacturer (EZB-RN001-plus, EZBioscience, China). The high-quality RNA was then reverse-transcribed using the Primer Script RT Kit (A0010CGQ, EZBioscience, China), followed by qPCR in ABI QuantStudio 6 Flex system (Applied Biosystems, Carlsbad, CA) using the iScript One-Step RT-PCR Kit with SYBR Green (A0002, EZBioscience, China) with the primers listed in table S3. Difference in transcript levels was quantified by normalization of each amplicon to β-actin. Notably, we used a strategy of 50%/50% males and females to avoid the interference of mice placental sex.
Western blotting
Immunoblotting analysis was performed following the standard procedures. We used a strategy of 50%/50% males and females to avoid the interference of mice placental sex. The placental (whole placenta), colon (~50 mg), and liver (~100 mg) tissues were washed with ice-cold PBS and lysed in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors (P1048, Beyotime, China). Protein concentration was quantified using a bicinchoninic acid protein assay kit (P0012, Beyotime, China), and an equal amount of protein was subjected to SDS–polyacrylamide gel electrophoresis. Next, proteins were transferred onto polyvinylidene difluoride membranes, followed by incubation with primary and secondary antibodies (AS014, Abclonal, China), and visualized using chemiluminescent reagent (P10300, NCM Biotech, China). The optical density of the signals on the film was quantified by Image Lab software. The primary antibodies used are shown in table. S3. Western blot images are provided in fig. S12.
Biochemical analysis
The levels of TNF-α (MM-0132 M1 and MM-0122H1, MEIMIAN, China), insulin (MM-0579 M1, MEIMIAN), HbA1c (MM-1517H1 and MM-0159 M2, MEIMIAN, China), LPS (CSB-E13066m, Cusabio, China; MM-1309H1, MM-0634 M1, and MEIMIAN, China), and DAO (CSB-E10090m, Cusabio, China) were measured with commercial kits as instructed by the respective manufacturers. Moreover, the concentrations of TNF-α in women and mice placentae were normalized according to protein levels (P0009, Beyotime, China). Notably, we used a strategy of 50%/50% males and females to avoid the interference of mice placental sex.
16S rRNA amplicon sequencing, data processing, and analysis
Fresh feces of mice were obtained, followed by 16S rRNA analysis. Total DNA was extracted from fresh feces (~50 mg) using the DNA Stool Mini Kit (51504, QIAGEN). After determining the DNA quality, the region V3 and V4 of 16S rRNA genes were amplified using the universal primer: forward, 5′-GTGCCAGCMGCCGCGG-TAA-3′; reverse, 5′-GGACTACHVGGGTWTCTAAT-3′. Then, amplicons were extracted from a 2% agarose gel, followed by purification with the GeneJET Gel Extraction Kit (Thermo Fisher Scientific) and quantification using a Quant-iT PicoGreen dsDNA assay kit (6163, Invitrogen, USA). Subsequently, sequencing libraries were generated using NEB Next Ultra II FS DNA PCR-free Library Prep Kit for Illumina (NEB, USA) as directed by the manufacturer, and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific) and Agilent Bioanalyzer 2100 system. Last, the library was sequenced on an Illumina NovaSeq platform, and 250–base pair paired-end reads were generated (raw tags).
The main steps of the bioinformatic analysis are as follows. Sequencing data were processed using QIIME2 (V.2022.02; https://magic.novogene.com/) (76). Briefly, the 16S rRNA gene reads were merged and quality-controlled using fastp (V.0.23.1) to obtain clean tags, followed by aligning the sequences of clean tags with the SILVA species annotation data library (V.138.1; www.arb-silva.de/) to detect and remove the chimeric sequences to obtain the final effective tags. Subsequently, the DADA2 plugin (V.3.11) in QIIME2 was used to reduce the noise of effective tags, and the final amplicon sequence variations and feature table were obtained. The representative sequences of each ASV were annotated based on the SILVA database to obtain the corresponding species information and species-based abundance distribution. R package Vegan (V.4.0.3) was used to analyze rarefaction curve, α-diversity (Shannon, Simpson, Chao1, or observed features), and β-diversity (PCoA) based on Bray-Curtis. One-way analysis of variance (ANOVA) followed by two-stage step-up false discovery rate (FDR) method of Benjamini, Krieger, and Yekutieli was performed to determine whether the flora compositions differed between groups unless otherwise indicated (FDR P < 0.05 was considered as a significant difference). Moreover, the key taxa were identified through the following analysis: (i) LDA effect size (LEfSe, V.1.1.2) was performed to determine the features most likely to explain the differences between groups (77), with an LDA score threshold of ≥4 as an important contributor to the model; (ii) random forest analysis was performed to determine the enrichment of key bacterial genera in GDM or GDM + HFDF group by the OECloud tools at https://cloud.oebiotech.com; (iii) Pearson’s correlation coefficient analysis between the relative abundance of selected bacterial genera and insulin resistance, litters, TNF-α, or gut barrier parameters in mice was performed using the OmicStudio tools at www.omicstudio.cn/tool; (iv) volcano plot was created to display relative abundance distribution of microbial ASVs and fold change; and (v) trend analyses were performed to identify core flora with consistent trends in abundance in recipient and donor mice (7). Moreover, microbial functions were predicted using Tax4fun (V.0.3.1) against Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.kegg.jp/) databases using default parameters (78).
Bacterial quantification in placenta
Bacterial quantification in placenta was performed as previously described (7). Briefly, placentae were isolated in a sterile environment, followed by DNA extraction in an isolated, low-contamination, and controlled environment. We also collected several types of negative controls as follows: (i) Sterile wet swabs that were opened in the sample collection room, waved in the air, and subjected to the same treatment as the placental samples, such as washing with sterile PBS, freezing in cryovials, and storage or transportation of the samples; and (ii) DNA-free water that was processed with the DNA extraction. For quantification of total fecal bacterial load, total DNA was isolated from known amounts of feces using QIAamp DNA Stool Mini Kit (QIAGEN). Next, DNA was subjected to quantitative PCR using QuantiFast SYBR Green PCR kit (A0002, EZBioscience, China) to measure the total bacterial number with universal 16S rRNA primers listed in table S3. Results were expressed as bacterial number per milligram of stool based on a standard curve.
Bacterial localization by FISH staining in mice placenta
The placental samples were handled in an isolated, low-contamination, and controlled environment where surfaces and equipment were treated with ultraviolet radiation to minimize and fragment environmental contaminant DNA. Moreover, personnel wore protective clothing and equipment to cover all exposed human surfaces. The FISH was performed following a modified protocol described by Chen et al. (7). Briefly, paraffin-embedded sections were subjected to deparaffinization, rehydration, and permeabilization. The probes used in this study included: eubacterial probes (EUB338 I-III and BBL-0557, BIOSSCI, China) mixture complementary to a 16S rRNA region specific for most bacteria; NON338, an oligonucleotide complementary to the probe EUB338 as a scrambled control for nonspecific binding. All probes were 5′-labeled with digoxigenin, and antidigoxigenin/horseradish peroxidase antibodies were used as secondary antibodies. Tyramide signal amplification was performed to increase the detection sensitivity of bacterial signals. Six areas of each section were randomly photographed under a fluorescence microscope. The specific probes for FISH are shown in table S3. Notably, we used a strategy of 50%/50% males and females to avoid the interference of mice placental sex.
In vitro tissue culture and placental explant–liver section model
In vitro tissue culture of liver section was measured using a previously reported protocol (79). Briefly, mice liver tissue slices were obtained from mice using sterile surgical techniques, followed by culturing the liver tissue slices in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; 11965175, Thermo Fisher Scientific, USA) at 37°C in a humidified atmosphere with 5% CO2. The high-glucose DMEM was supplemented with 10% FBS (16000044, Thermo Fisher Scientific), streptomycin sulfate (100 mg/liter; 11860038, Thermo Fisher Scientific), and penicillin sodium (100,000 U/liter). The liver was sectioned into 200-μm-thick slices with a vibration slicer (VF-300 Microtome, USA) and then cultured for further treatment. In vitro tissue culture of placental explant was measured using a previously reported protocol with minor modifications (80). Briefly, mice placentae were collected from mice using sterile surgical techniques under 3% isoflurane anesthesia, followed by euthanizing the mice by exsanguination while still under anesthesia. After rinsing with sterile PBS, the placentae were placed in sterile tissue culture medium (45% DMEM/45% Ham’s F12/10% FBS + penicillin/streptomycin), and each placenta was cut into four pieces, followed by placing each of them in an individual well of six-well plates containing 2 ml of the tissue culture medium. Next, placental explant was cultured in sterile tissue culture medium for 6 hours at 37°C in a humidified atmosphere with 5% CO2, followed by collecting the placental explant supernatant to detect TNF-α secretion from isolated placentae or to process liver sections.
Schematic representation of primary liver sections treated with placental explant supernatant is shown in Fig. 4D. Briefly, culture supernatants were collected from placental explants of GDM or GDM + HFDF mice at E12.5, followed by treating the primary liver sections from GDM or GDM + HFDF mice at E18.5 with the two supernatants for 24 hours. Liver sections were then collected for further analysis.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9.5.1 software, and results were presented as means ± SEM unless otherwise noted. Results represent an independent experiment unless indicated. For the blinding to histological analysis and immunofluorescence, samples from each group were named after neutral codes and then given to investigators not involved in the experimental design for quantification. Statistical analysis methods and biological replicates of each experiment were indicated in the corresponding figure legends.
Acknowledgments
Funding: This project was funded by Project of National Natural Science Foundation of China (nos. 32272895 and 32172744) and Double first-class discipline promotion project (2023B10564001).
Author contributions: Conceptualization: C.T. and Y.Y. Methodology: S.H., J.C., Z.C., Q.X., and W.S. Investigation: S.H., J.C., Z.C., K.M., D.W., J.L., F.L., W.X., and S.R. Visualization: S.H. Resources: Q.X., W.S. Supervision: J.D., C.T. Writing—original draft: S.H. Writing—review and editing: C.T., T.S., and S.H.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Materials. The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (GSA) in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA012171, PRJCA018924) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. All source data used to generate plots are available through the Figshare data repository under accession number DOI: 10.6084/m9.figshare.22874480.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 to S3
REFERENCES AND NOTES
- 1.N. H. Cho, J. E. Shaw, S. Karuranga, Y. Huang, J. D. da Rocha Fernandes, A. W. Ohlrogge, B. Malanda, IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 138, 271–281 (2018). [DOI] [PubMed] [Google Scholar]
- 2.E. C. Johns, F. C. Denison, J. E. Norman, R. M. Reynolds, Gestational diabetes mellitus: Mechanisms, treatment, and complications. Trends Endocrinol. Metab. 29, 743–754 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Y. Pinto, S. Frishman, S. Turjeman, A. Eshel, M. Nuriel-Ohayon, O. Shrossel, O. Ziv, W. Walters, J. Parsonnet, C. Ley, E. L. Johnson, K. Kumar, R. Schweitzer, S. Khatib, F. Magzal, E. Muller, S. Tamir, K. Tenenbaum-Gavish, S. Rautava, S. Salminen, E. Isolauri, O. Yariv, Y. Peled, E. Poran, J. Pardo, R. Chen, M. Hod, E. Borenstein, R. E. Ley, B. Schwartz, Y. Louzoun, E. Hadar, O. Koren, Gestational diabetes is driven by microbiota-induced inflammation months before diagnosis. Gut 72, 918–928 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P. M. Catalano, K. Shankar, Obesity and pregnancy: Mechanisms of short term and long term adverse consequences for mother and child. BMJ 356, j1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L. Kang, H. Y. Li, H. Y. Ou, P. Wu, S. H. Wang, C. J. Chang, S. Y. Lin, C. L. Wu, H. T. Wu, Role of placental fibrinogen-like protein 1 in gestational diabetes. Transl. Res. 218, 73–80 (2020). [DOI] [PubMed] [Google Scholar]
- 6.J. P. Kirwan, S. Hauguel-De Mouzon, J. Lepercq, J. C. Challier, L. Huston-Presley, J. E. Friedman, S. C. Kalhan, P. M. Catalano, TNF-alpha is a predictor of insulin resistance in human pregnancy. Diabetes 51, 2207–2213 (2002). [DOI] [PubMed] [Google Scholar]
- 7.X. Chen, P. Li, M. Liu, H. Zheng, Y. He, M. X. Chen, W. Tang, X. Yue, Y. Huang, L. Zhuang, Z. Wang, M. Zhong, G. Ke, H. Hu, Y. Feng, Y. Chen, Y. Yu, H. Zhou, L. Huang, Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut 69, 513–522 (2020). [DOI] [PubMed] [Google Scholar]
- 8.J. Jin, L. Gao, X. Zou, Y. Zhang, Z. Zheng, X. Zhang, J. Li, Z. Tian, X. Wang, J. Gu, C. Zhang, T. Wu, Z. Wang, Q. Zhang, Gut dysbiosis promotes preeclampsia by regulating macrophages and trophoblasts. Circ. Res. 131, 492–506 (2022). [DOI] [PubMed] [Google Scholar]
- 9.L. Zhao, The gut microbiota and obesity: From correlation to causality. Nat. Rev. Microbiol. 11, 639–647 (2013). [DOI] [PubMed] [Google Scholar]
- 10.A. N. Reynolds, A. P. Akerman, J. Mann, Dietary fibre and whole grains in diabetes management: Systematic review and meta-analyses. PLoS Med. 17, e1003053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.W. Ma, L. H. Nguyen, M. Song, D. D. Wang, E. A. Franzosa, Y. Cao, A. Joshi, D. A. Drew, R. Mehta, K. L. Ivey, L. L. Strate, E. L. Giovannucci, J. Izard, W. Garrett, E. B. Rimm, C. Huttenhower, A. T. Chan, Dietary fiber intake, the gut microbiome, and chronic systemic inflammation in a cohort of adult men. Genome Med. 13, 102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.G. J. Burton, E. Jauniaux, Pathophysiology of placental-derived fetal growth restriction. Am. J. Obstet. Gynecol. 218, S745–S761 (2018). [DOI] [PubMed] [Google Scholar]
- 13.L. Zhao, F. Zhang, X. Ding, G. Wu, Y. Y. Lam, X. Wang, H. Fu, X. Xue, C. Lu, J. Ma, L. Yu, C. Xu, Z. Ren, Y. Xu, S. Xu, H. Shen, X. Zhu, Y. Shi, Q. Shen, W. Dong, R. Liu, Y. Ling, Y. Zeng, X. Wang, Q. Zhang, J. Wang, L. Wang, Y. Wu, B. Zeng, H. Wei, M. Zhang, Y. Peng, C. Zhang, Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018). [DOI] [PubMed] [Google Scholar]
- 14.C. Tan, H. Wei, J. Ao, G. Long, J. Peng, Inclusion of konjac flour in the gestation diet changes the gut microbiota, alleviates oxidative stress, and improves insulin sensitivity in sows. Appl. Environ. Microbiol. 82, 5899–5909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R. D. Devaraj, C. K. Reddy, B. Xu, Health-promoting effects of konjac glucomannan and its practical applications: A critical review. Int. J. Biol. Macromol. 126, 273–281 (2019). [DOI] [PubMed] [Google Scholar]
- 16.S. Huang, D. Wu, X. Hao, J. Nie, Z. Huang, S. Ma, Y. Chen, S. Chen, J. Wu, J. Sun, H. Ao, B. Gao, C. Tan, Dietary fiber supplementation during the last 50 days of gestation improves the farrowing performance of gilts by modulating insulin sensitivity, gut microbiota and placental function. J. Anim. Sci. 101, skad021 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.X. Li, W. Yao, Y. Yuan, P. Chen, B. Li, J. Li, R. Chu, H. Song, D. Xie, X. Jiang, H. Wang, Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 66, 157–167 (2017). [DOI] [PubMed] [Google Scholar]
- 18.K. K. Y. Cheng, K. S. L. Lam, D. Wu, Y. Wang, G. Sweeney, R. L. C. Hoo, J. Zhang, A. Xu, APPL1 potentiates insulin secretion in pancreatic β cells by enhancing protein kinase Akt-dependent expression of SNARE proteins in mice. Proc. Natl. Acad. Sci. U.S.A. 109, 8919–8924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P. Plomgaard, A. R. Nielsen, C. P. Fischer, O. H. Mortensen, C. Broholm, M. Penkowa, R. Krogh-Madsen, C. Erikstrup, B. Lindegaard, A. M. W. Petersen, S. Taudorf, B. K. Pedersen, Associations between insulin resistance and TNF-α in plasma, skeletal muscle and adipose tissue in humans with and without type 2 diabetes. Diabetologia 50, 2562–2571 (2007). [DOI] [PubMed] [Google Scholar]
- 20.M. Badran, B. Abuyassin, N. Ayas, I. Laher, Intermittent hypoxia impairs uterine artery function in pregnant mice. J. Physiol. 597, 2639–2650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. Weirather, U. D. W. Hofmann, N. Beyersdorf, G. C. Ramos, B. Vogel, A. Frey, G. Ertl, T. Kerkau, S. Frantz, Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ. Res. 115, 55–67 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Y. Filipovich, J. Klein, Y. Zhou, E. Hirsch, Maternal and fetal roles in bacterially induced preterm labor in the mouse. Am. J. Obstet. Gynecol. 214,386.386.e1–386.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.W. Shin, H. J. Kim, Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. U.S.A. 115, E10539–E10547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Q. Li, Y. Cui, B. Xu, Y. Wang, F. Lv, Z. Li, H. Li, X. Chen, X. Peng, Y. Chen, E. Wu, D. Qu, Y. Jian, H. Si, Main active components of Jiawei Gegen Qinlian decoction protects against ulcerative colitis under different dietary environments in a gut microbiota-dependent manner. Pharmacol. Res. 170, 105694 (2021). [DOI] [PubMed] [Google Scholar]
- 25.S. Manfredo Vieira, M. Hiltensperger, V. Kumar, D. Zegarra-Ruiz, C. Dehner, N. Khan, F. R. C. Costa, E. Tiniakou, T. Greiling, W. Ruff, A. Barbieri, C. Kriegel, S. S. Mehta, J. R. Knight, D. Jain, A. L. Goodman, M. A. Kriegel, Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. Macfarlane, G. T. Macfarlane, Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 62, 67–72 (2003). [DOI] [PubMed] [Google Scholar]
- 27.H. Guo, W.-C. Chou, Y. Lai, K. Liang, J. W. Tam, W. J. Brickey, L. Chen, N. D. Montgomery, X. Li, L. M. Bohannon, A. D. Sung, N. J. Chao, J. U. Peled, A. L. C. Gomes, M. R. M. van den Brink, M. J. French, A. N. Macintyre, G. D. Sempowski, X. Tan, R. B. Sartor, K. Lu, J. P. Y. Ting, Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 370, eaay9097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. Chen, J. E. Wilson, M. J. Koenigsknecht, W.-C. Chou, S. A. Montgomery, A. D. Truax, W. J. Brickey, C. D. Packey, N. Maharshak, G. K. Matsushima, S. E. Plevy, V. B. Young, R. B. Sartor, J. P.-Y. Ting, NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 18, 541–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N. D. Mathewson, R. Jenq, A. V. Mathew, M. Koenigsknecht, A. Hanash, T. Toubai, K. Oravecz-Wilson, S. R. Wu, Y. Sun, C. Rossi, H. Fujiwara, J. Byun, Y. Shono, C. Lindemans, M. Calafiore, T. M. Schmidt, K. Honda, V. B. Young, S. Pennathur, M. van den Brink, P. Reddy, Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 17, 505–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Zou, B. Chassaing, V. Singh, M. Pellizzon, M. Ricci, M. D. Fythe, M. V. Kumar, A. T. Gewirtz, Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microbe 23, 41–53.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.H. Zhang, S. Qin, X. Zhang, P. Du, Y. Zhu, Y. Huang, J. Michiels, Q. Zeng, W. Chen, Dietary resistant starch alleviates Escherichia coli-induced bone loss in meat ducks by promoting short-chain fatty acid production and inhibiting Malt1/NF-κB inflammasome activation. J. Anim. Sci. Biotechnol. 13, 92 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Y. Bai, Y. Li, T. Marion, Y. Tong, M. M. Zaiss, Z. Tang, Q. Zhang, Y. Liu, Y. Luo, Resistant starch intake alleviates collagen-induced arthritis in mice by modulating gut microbiota and promoting concomitant propionate production. J. Autoimmun. 116, 102564 (2021). [DOI] [PubMed] [Google Scholar]
- 33.K. Yang, Y. Hou, Y. Zhang, H. Liang, A. Sharma, W. Zheng, L. Wang, R. Torres, K. Tatebe, S. J. Chmura, S. P. Pitroda, J. A. Gilbert, Y. X. Fu, R. R. Weichselbaum, Suppression of local type I interferon by gut microbiota-derived butyrate impairs antitumor effects of ionizing radiation. J. Exp. Med. 218, e20201915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.N. Singh, A. Gurav, S. Sivaprakasam, E. Brady, R. Padia, H. Shi, M. Thangaraju, P. D. Prasad, S. Manicassamy, D. H. Munn, J. R. Lee, S. Offermanns, V. Ganapathy, Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.U. Kampmann, S. Knorr, J. Fuglsang, P. Ovesen, Determinants of maternal insulin resistance during pregnancy: An updated overview. J. Diabetes Res. 2019, 5320156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.E. A. Ryan, M. J. O'Sullivan, J. S. Skyler, Insulin action during pregnancy. Studies with the euglycemic clamp technique. Diabetes 34, 380–389 (1985). [DOI] [PubMed] [Google Scholar]
- 37.C. Liang, K. DeCourcy, M. R. Prater, High-saturated-fat diet induces gestational diabetes and placental vasculopathy in C57BL/6 mice. Metabolism 59, 943–950 (2010). [DOI] [PubMed] [Google Scholar]
- 38.F. T. Spradley, A. C. Palei, J. P. Granger, Differential body weight, blood pressure and placental inflammatory responses to normal versus high-fat diet in melanocortin-4 receptor-deficient pregnant rats. J. Hypertens. 34, 1998–2007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.P. M. Catalano, H. D. McIntyre, J. K. Cruickshank, D. R. McCance, A. R. Dyer, B. E. Metzger, L. P. Lowe, E. R. Trimble, D. R. Coustan, D. R. Hadden, B. Persson, M. Hod, J. J. N. Oats; HAPO Study Cooperative Research Group , The hyperglycemia and adverse pregnancy outcome study: Associations of GDM and obesity with pregnancy outcomes. Diabetes Care 35, 780–786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. Deer, O. Herrock, N. Campbell, D. Cornelius, S. Fitzgerald, L. M. Amaral, B. LaMarca, The role of immune cells and mediators in preeclampsia. Nat. Rev. Nephrol. 19, 257–270 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. Hoch, M. Gauster, S. Hauguel-de Mouzon, G. Desoye, Diabesity-associated oxidative and inflammatory stress signalling in the early human placenta. Mol. Aspects Med. 66, 21–30 (2019). [DOI] [PubMed] [Google Scholar]
- 42.O. Dowling, P. K. Chatterjee, M. Gupta, H. B. Tam Tam, X. Xue, D. Lewis, B. Rochelson, C. N. Metz, Magnesium sulfate reduces bacterial LPS-induced inflammation at the maternal-fetal interface. Placenta 33, 392–398 (2012). [DOI] [PubMed] [Google Scholar]
- 43.J. Zheng, X. Xiao, Q. Zhang, L. Mao, M. Yu, J. Xu, T. Wang, The placental microbiota is altered among subjects with gestational diabetes mellitus: A pilot study. Front. Physiol. 8, 675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. D. Piccolo, J. L. Graham, P. Kang, C. E. Randolph, K. Shankar, L. Yeruva, R. Fox, M. S. Robeson, B. Moody, T. LeRoith, K. L. Stanhope, S. H. Adams, P. J. Havel, Progression of diabetes is associated with changes in the ileal transcriptome and ileal-colon morphology in the UC Davis type 2 diabetes mellitus rat. Physiol. Rep. 9, e15102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Chen, S. Yuan, T. Fu, X. Ruan, J. Qiao, X. Wang, X. Li, D. Gill, S. Burgess, E. L. Giovannucci, S. C. Larsson, Gastrointestinal consequences of type 2 diabetes mellitus and impaired glycemic homeostasis: A Mendelian randomization study. Diabetes Care 46, 828–835 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Yuhara, C. Steinmaus, S. E. Cohen, D. A. Corley, Y. Tei, P. A. Buffler, Is diabetes mellitus an independent risk factor for colon cancer and rectal cancer? Am. J. Gastroenterol. 106, 1911–1921 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. Mokkala, N. Paulin, N. Houttu, E. Koivuniemi, O. Pellonperä, S. Khan, S. Pietilä, K. Tertti, L. L. Elo, K. Laitinen, Metagenomics analysis of gut microbiota in response to diet intervention and gestational diabetes in overweight and obese women: A randomised, double-blind, placebo-controlled clinical trial. Gut 70, 309–318 (2021). [DOI] [PubMed] [Google Scholar]
- 48.L. K. Callaway, H. D. McIntyre, H. L. Barrett, K. Foxcroft, A. Tremellen, B. E. Lingwood, J. M. Tobin, S. Wilkinson, A. Kothari, M. Morrison, P. O’Rourke, A. Pelecanos, M. D. Nitert, Probiotics for the prevention of gestational diabetes mellitus in overweight and obese women: Findings from the SPRING double-blind randomized controlled trial. Diabetes Care 42, 364–371 (2019). [DOI] [PubMed] [Google Scholar]
- 49.S. J. Davidson, H. L. Barrett, S. A. Price, L. K. Callaway, M. Dekker Nitert, Probiotics for preventing gestational diabetes. Cochrane Database Syst. Rev. 4, CD009951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Y. Cui, M. Liao, A. Xu, G. Chen, J. Liu, X. Yu, S. Li, X. Ke, S. Tan, Z. Luo, Q. Wang, Y. Liu, D. Wang, F. Zeng, Association of maternal pre-pregnancy dietary intake with adverse maternal and neonatal outcomes: A systematic review and meta-analysis of prospective studies. Crit. Rev. Food Sci. Nutr. 63, 3430–3451 (2023). [DOI] [PubMed] [Google Scholar]
- 51.G. Chen, X. Ran, B. Li, Y. Li, D. He, B. Huang, S. Fu, J. Liu, W. Wang, Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine 30, 317–325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. He, Y. Chu, J. Li, Q. Meng, Y. Liu, J. Jin, Y. Wang, J. Wang, B. Huang, L. Shi, X. Shi, J. Tian, Y. Zhufeng, R. Feng, W. Xiao, Y. Gan, J. Guo, C. Shao, Y. Su, F. Hu, X. Sun, J. Yu, Y. Kang, Z. Li, Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci. Adv. 8, eabm1511 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.G. Li, J. Lin, C. Zhang, H. Gao, H. Lu, X. Gao, R. Zhu, Z. Li, M. Li, Z. Liu, Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 13, 1968257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.S. A. Robertson, A. S. Care, L. M. Moldenhauer, Regulatory T cells in embryo implantation and the immune response to pregnancy. J. Clin. Invest. 128, 4224–4235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Z. Li, C. X. Yi, S. Katiraei, S. Kooijman, E. Zhou, C. K. Chung, Y. Gao, J. K. van den Heuvel, O. C. Meijer, J. F. P. Berbée, M. Heijink, M. Giera, K. Willems van Dijk, A. K. Groen, P. C. N. Rensen, Y. Wang, Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut 67, 1269–1279 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Z. Gao, J. Yin, J. Zhang, R. E. Ward, R. J. Martin, M. Lefevre, W. T. Cefalu, J. Ye, Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58, 1509–1517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. Hurr, H. Simonyan, D. A. Morgan, K. Rahmouni, C. N. Young, Liver sympathetic denervation reverses obesity-induced hepatic steatosis. J. Physiol. 597, 4565–4580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.I. Kimura, D. Inoue, T. Maeda, T. Hara, A. Ichimura, S. Miyauchi, M. Kobayashi, A. Hirasawa, G. Tsujimoto, Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl. Acad. Sci. U.S.A. 108, 8030–8035 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Hatori, C. Vollmers, A. Zarrinpar, L. DiTacchio, E. A. Bushong, S. Gill, M. Leblanc, A. Chaix, M. Joens, J. A. J. Fitzpatrick, M. H. Ellisman, S. Panda, Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. Chaix, A. Zarrinpar, P. Miu, S. Panda, Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 20, 991–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.S. J. Kentish, A. D. Vincent, D. J. Kennaway, G. A. Wittert, A. J. Page, High-fat diet-induced obesity ablates gastric vagal afferent circadian rhythms. J. Neurosci. 36, 3199–3207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.J. Folz, R. N. Culver, J. M. Morales, J. Grembi, G. Triadafilopoulos, D. A. Relman, K. C. Huang, D. Shalon, O. Fiehn, Human metabolome variation along the upper intestinal tract. Nat. Metab. 5, 777–788 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.American Diabetes Association , Standards of medical care in diabetes–2014. Diabetes Care 37 (Pt 1), S14–S80 (2014). [DOI] [PubMed] [Google Scholar]
- 64.W. M. Aldahmash, S. H. Alwasel, K. Aljerian, Gestational diabetes mellitus induces placental vasculopathies. Environ. Sci. Pollut. Res. Int. 29, 19860–19868 (2022). [DOI] [PubMed] [Google Scholar]
- 65.T. Tanaka, T. Wada, K. Uno, S. Ogihara, H. Ie, A. Okekawa, A. Ishikawa, T. Ito, Y. Miyazawa, A. Sameshima, Y. Onogi, H. Tsuneki, M. Sasahara, A. Nakashima, S. Saito, T. Sasaoka, Oestrogen receptor α in T cells controls the T cell immune profile and glucose metabolism in mouse models of gestational diabetes mellitus. Diabetologia 64, 1660–1673 (2021). [DOI] [PubMed] [Google Scholar]
- 66.D. R. Coustan, L. P. Lowe, B. E. Metzger, A. R. Dyer; International Assoiciation of Diabetes and Pregnancy Study Groups , The hyperglycemia and adverse pregnancy outcome (HAPO) study: Paving the way for new diagnostic criteria for gestational diabetes mellitus. Am. J. Obstet. Gynecol. 202, 654.e1–654.e6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.B. Chen, Y. R. Du, H. Zhu, M. L. Sun, C. Wang, Y. Cheng, H. Pang, G. Ding, J. Gao, Y. Tan, X. Tong, P. Lv, F. Zhou, Q. Zhan, Z. M. Xu, L. Wang, D. Luo, Y. Ye, L. Jin, S. Zhang, Y. Zhu, X. Lin, Y. Wu, L. Jin, Y. Zhou, C. Yan, J. Sheng, P. R. Flatt, G. L. Xu, H. Huang, Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature 605, 761–766 (2022). [DOI] [PubMed] [Google Scholar]
- 68.S.-L. Zeng, S.-Z. Li, P. T. Xiao, Y.-Y. Cai, C. Chu, B.-Z. Chen, P. Li, J. Li, E.-H. Liu, Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci. Adv. 6, eaax6208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.K. R. Secombe, G. H. Al-Qadami, C. B. Subramaniam, J. M. Bowen, J. Scott, Y. Z. A. Van Sebille, M. Snelson, C. Cowan, G. Clarke, C. E. Gheorghe, J. F. Cryan, H. R. Wardill, Guidelines for reporting on animal fecal transplantation (GRAFT) studies: Recommendations from a systematic review of murine transplantation protocols. Gut Microbes 13, 1979878 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.K. Rahman, C. Desai, S. S. Iyer, N. E. Thorn, P. Kumar, Y. Liu, T. Smith, A. S. Neish, H. Li, S. Tan, P. Wu, X. Liu, Y. Yu, A. B. Farris, A. Nusrat, C. A. Parkos, F. A. Anania, Loss of junctional adhesion molecule a promotes severe steatohepatitis in mice on a diet high in saturated fat, fructose, and cholesterol. Gastroenterology 151, 733–746.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.V. Singh, S. Jamwal, R. Jain, P. Verma, R. Gokhale, K. V. S. Rao, Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe 12, 669–681 (2012). [DOI] [PubMed] [Google Scholar]
- 72.T. Y. Khong, E. E. Mooney, I. Ariel, N. C. Balmus, T. K. Boyd, M. A. Brundler, H. Derricott, M. J. Evans, O. M. Faye-Petersen, J. E. Gillan, A. E. Heazell, D. S. Heller, S. M. Jacques, S. Keating, P. Kelehan, A. Maes, E. M. McKay, T. K. Morgan, P. G. Nikkels, W. T. Parks, R. W. Redline, I. Scheimberg, M. H. Schoots, N. J. Sebire, A. Timmer, G. Turowski, J. P. van der Voorn, I. van Lijnschoten, S. J. Gordijn, Sampling and definitions of placental lesions: Amsterdam placental workshop group consensus statement. Arch. Pathol. Lab. Med. 140, 698–713 (2016). [DOI] [PubMed] [Google Scholar]
- 73.C. J. Kim, R. Romero, P. Chaemsaithong, J. S. Kim, Chronic inflammation of the placenta: Definition, classification, pathogenesis, and clinical significance. Am. J. Obstet. Gynecol. 213, S53–S69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.S. Huang, Z. Wu, Z. Huang, X. Hao, L. Zhang, C. Hu, J. Wei, J. Deng, C. Tan, Maternal supply of cysteamine alleviates oxidative stress and enhances angiogenesis in porcine placenta. J. Anim. Sci. Biotechnol. 12, 91 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.C. Hu, Z. Wu, Z. Huang, X. Hao, S. Wang, J. Deng, Y. Yin, C. Tan, Nox2 impairs VEGF-A-induced angiogenesis in placenta via mitochondrial ROS-STAT3 pathway. Redox Biol. 45, 102051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. B. Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y. X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson 2nd, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight, J. G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett, C. Huttenhower, Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.K. P. Aßhauer, B. Wemheuer, R. Daniel, P. Meinicke, Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Y. Yuan, C. Zhu, Y. Wang, J. Sun, J. Feng, Z. Ma, P. Li, W. Peng, C. Yin, G. Xu, P. Xu, Y. Jiang, Q. Jiang, G. Shu, α-Ketoglutaric acid ameliorates hyperglycemia in diabetes by inhibiting hepatic gluconeogenesis via serpina1e signaling. Sci. Adv. 8, eabn2879 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. Phillippe, S. Adeli, Cell-free DNA release by mouse placental explants. PLOS ONE 12, e0178845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S3