Abstract
A 4.5-kb region of chromosomal DNA carrying the locus responsible for the production of plantaricin S, a two-peptide bacteriocin produced by Lactobacillus plantarum LPCO10 (R. Jiménez-Díaz, J. L. Ruiz-Barba, D. P. Cathcart, H. Holo, I. F. Nes, K. H. Sletten, and P. J. Warner, Appl. Environ. Microbiol. 61:4459–4463, 1995), has been cloned, and the nucleotide sequence has been elucidated. Two genes, designated plsA and plsB and encoding peptides α and β, respectively, of plantaricin S, plus an open reading frame (ORF), ORF2, were found to be organized in an operon. Northern blot analysis showed that these genes are cotranscribed, giving a ca. 0.7-kb mRNA, whose transcription start point was determined by primer extension. Nucleotide sequences of plsA and plsB revealed that both genes are translated as bacteriocin precursors which include N-terminal leader sequences of the double-glycine type. The role of ORF2 is unknown at the moment, although it might be expected to encode an immunity protein of the type described for other bacteriocin operons. In addition, several other potential ORFs have been found, including some which may be responsible for the regulation of bacteriocin production. Two of them, ORF8 and ORF14, show strong homology with histidine protein kinase and response regulator genes, respectively, which have been found to be involved in the regulation of the production of other bacteriocins from lactic acid bacteria. A third ORF, ORF5, shows homology with gene agrB from Staphylococcus aureus, which is involved in the mechanism of regulation of the virulence phenotype in this species. Thus, an agr-like regulatory system for the production of plantaricin S is postulated.
The importance of microbial starters for a variety of fermented foods is now acknowledged worldwide as an efficient means to control these processes and to obtain products of a homogeneous quality which are safely preserved (6). This is particularly true for those products fermented by lactic acid bacteria (LAB) (6, 7, 10). LAB are able to compete and dominate in their environments by means of a very well adapted metabolism which includes the production of a range of antimicrobial substances (10, 22). Among these, the proteinaceous antimicrobial compounds called bacteriocins are undoubtedly the most promising for a biotechnological approach to starter technology (8, 9, 10, 32). Bacteriocins are ribosomally synthesized peptides exhibiting antimicrobial activity directed, in most cases, against bacteria closely related to the producer microorganism (20). Bacteriocins from LAB have been classified by Nes et al. (25) into different groups according to their compositions, sizes, heat-stabilities, modes of action, types of export mechanism, and activity spectra (25). Most of the bacteriocins characterized in recent years belong to the class II bacteriocins, which are small (30 to 100 amino acids), heat-stable, and, usually, not posttranslationally modified (25). As the genetic organization and biosynthesis mechanisms of some of them have been elucidated, a number of common features have become apparent (20, 25).
The genes for bacteriocin production are organized in operons comprising four genes which may or may not be located on the same transcription unit. These genes are the structural gene encoding a prebacteriocin, a dedicated immunity gene (located on the same transcription unit as the structural gene), a gene encoding an ABC transporter (optional), and a gene encoding an accessory protein essential for the externalization of the bacteriocin. The inactive prebacteriocin, which includes an N-terminal leader sequence, is activated by the cleavage of the leader peptide by the proteolytic activity of a transporter protein (usually of the ABC type but occasionally sec dependent), concurrent with the export of the active bacteriocin from the cells. In some cases, a regulatory mechanism for class II bacteriocin production has been described; there is an induction factor (IF), with a bacteriocin-like leader sequence, forming part of a three-component regulatory system, which includes a response regulator (RR) gene and a sensor histidine protein kinase (HPK) gene (5, 12, 16, 25).
The use of bacteriocin-producing strain Lactobacillus plantarum LPCO10, which produces plantaricins S (PLS) and T (PLT) (18), as a starter culture for the fermentation of olives has demonstrated the strain’s suitability for controlling this process and for the preservation of the final product (33). In order to increase our knowledge of the mechanisms by which this bacteriocin-producing strain dominates the microflora in the fermentation and to make use of its properties under optimum conditions, we began studies to biochemically characterize these bacteriocins. Recently we reported the purification to homogeneity and amino acid sequence of PLS (19). This bacteriocin was found to be novel and to require the presence of two different peptide components to achieve its maximum bactericidal activity. It is, therefore, a member of the class IIb (small, heat-stable, two-component) bacteriocins produced by LAB (25), which include lactococcin G (26), plantaricin EF (13), plantaricin JK (13), lactacin F (1), lactococcin M (37), and acidocin J1132 (36).
Here we describe the cloning and sequencing of the genes encoding PLS and show that they are organized as an operon in L. plantarum LPCO10. In addition, several open reading frames (ORFs) have been identified downstream from the PLS operon. Two of these ORFs show strong homology with genes that have been found to be involved in the regulation of the production of other bacteriocins from LAB (25). A third ORF shows homology with a gene from Staphylococcus aureus involved in the regulation of the virulence phenotype (27). A comparison of the DNA sequences and ORF organizations suggests that an agr-like regulatory system may be involved.
MATERIALS AND METHODS
Bacterial strains, culture media, growth conditions, and plasmids.
The PLS and PLT producer L. plantarum LPCO10 and its PLS- and PLT-immune derivative, L. plantarum 55-1, which does not produce bacteriocin, have been described previously (18, 33). Both strains were grown on MRS agar or broth (Oxoid, Basingstoke, England) at 30°C without agitation. Escherichia coli DH5α, JM109, Top10F′, and InvαF′ (Invitrogen, Leek, The Netherlands) were grown in Luria-Bertani broth at 37°C with vigorous agitation. Bacteria were maintained as frozen stocks at −80°C in MRS plus 20% (vol/vol) glycerol. Plasmid pUC19 and the T/A cloning vectors pCR2.1 and pCR3 (Invitrogen) were used as cloning vectors and were selected by adding ampicillin (50 μg/ml) to the culture medium.
DNA isolation.
Total genomic DNA from lactobacilli was prepared as described by Cathcart (7). The extraction of plasmid DNA from E. coli was performed as described previously (34).
Direct and inverse DNA amplification (PCR) techniques.
Degenerate primers used to amplify the genes corresponding to the α and β peptides of PLS are shown in Table 1. They were synthesized by Oswel DNA Services, Southampton, United Kingdom. Where degeneracy was more than twofold for any base, deoxyinosine bases were incorporated into these primers (30). DNA was amplified in 100-μl reaction mixtures containing 4 mM MgCl2, 1× reaction buffer, 100 μM concentrations of each of the deoxynucleoside triphosphates (dNTPs), 100 pmol of each of the primers, 100 ng of genomic DNA as the template, and 2.5 U of AmpliTaq polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) with a Techne PHC-2 Dri-Block thermocycler or a PTC-100 programmable thermal controller (MJ Research, Inc., Watertown, Mass.). Before addition of the enzyme, the tube was heated to 95°C for 5 min (hot start). Amplification with degenerate primers typically proceeded through 30 two-step cycles as follows: annealing at 48°C for 1 min and denaturation at 93°C for 30 s. Amplification with nondegenerate primers αS, αA, βS, and βA (Table 1) was carried out as described previously, except that 10 or 20 pmol of each of the primers was used for the reaction, the annealing temperature was raised to 60°C, and a polymerization step at 72°C for 40 s was performed after the annealing step. All possible pairwise combinations of the nondegenerate primers for the genes encoding the α and β peptides were used in the same PCR procedure.
TABLE 1.
PCR primers used in this study
| Primer | Sequencea |
|---|---|
| Degenerate primers | |
| αF | 5′-AAYAARYTIGCITAYAAYATG-3′ |
| αR | 5′-GCNGCIARICCRAADATNGT-3′ |
| βF | 5′-TRAARCARWSITGGTAYGCIGC-3′ |
| βR | 5′-GCRTTYARRAAICCYTCICC-3′ |
| Nondegenerate primers | |
| αS | 5′-GGGCATTACGCTGGTAAGGC-3′ |
| αA | 5′-GCCTTACCAGCGTAATGCCC-3′ |
| βS | 5′-CTGGTGATGCAATCGTTAGTTT-3′ |
| βA | 5′-AAACTAACGATTGCATCACCAG-3′ |
| Primers for inverse PCR | |
| αINV1 | 5′-CTCTGCAAGCATTGTGAATGG-3′ |
| βINV1 | 5′-CTGACTATATCGTTCACTCAG-3′ |
| α1A | 5′-TTACGGCAGCCCAATTTTATG-3′ |
| α1B | 5′-GARCCCTTTATGGCTTTTTCC-3′ |
| α2A | 5′-GTGCTGAGCAGTATACTACTG-3′ |
| α2B | 5′-GTCATTATGATGTTGACAGCG-3′ |
| α3A | 5′-GCTTAGATTTCACAGCTTCGA-3′ |
| α3B | 5′-GTTCAGGATTTGAGTTGGCAC-3′ |
| β1B | 5′-ATCTCCTTAATTCGACAATCG-3′ |
I, deoxyinosine; R, A/G; W, A/T; S, C/G; Y, C/T; D, A/G/T; N, A/C/G/T.
An inverse PCR strategy (28) was used to determine sequences extending outwards from the known sequences of the genes. Thus, the primer pair αINV1/βINV1, with the 3′ ends facing away from each other, was designed. A 1.3-kb ClaI fragment (Fig. 1) for amplification by inverse PCR was identified by Southern blotting and hybridization (see next section), with primers αINV1 and βINV1 as the labelled probes. Circular template DNA was prepared from ClaI-digested Lactobacillus DNA as described by Ochman et al. (28). The circularized template DNA (100 ng) was used in a 100-μl PCR mixture with the αINV1/βINV1 primer pair (20 pmol of each primer), 200 μM concentrations of each of the dNTPs, 4 mM MgCl2 and 1× reaction buffer. The reaction mixture was heated to 94°C for 5 min before the addition of 5 U of AmpliTaq polymerase. The PCR proceeded through 30 cycles of annealing at 55°C for 30 s, polymerization at 72°C for 2 min, and denaturation at 94°C for 30 s, followed by a final polymerization step for 4 min (20 min when T/A cloning vectors were used). The thermal cycling was performed with a Touchdown thermal cycler (Hybaid, Teddington, United Kingdom). Primers for inverse PCR were obtained from Oswel DNA Services, King’s College, London, United Kingdom, or MWG Biotech, Ebersberg, Germany.
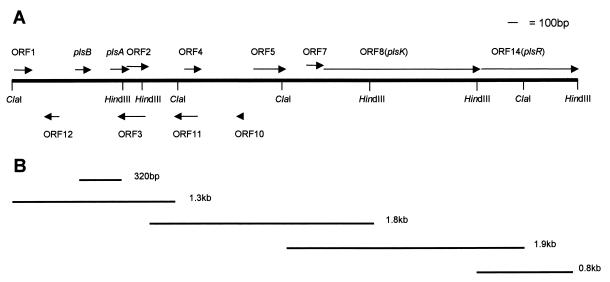
FIG. 1.
Genetic map of the pls locus. (A) Restriction map showing pls genes and putative ORFs. (B) PCR products cloned and sequenced.
Inverse PCR has been used to extend the sequence downstream of the region encoding PLS. For this, new suitable restriction fragments (1.8-kb HindIII, 1.9-kb ClaI, and 0.8-kb HindIII fragments) (Fig. 1) overlapping the existing ones were identified as described above, circularized, and used as templates for new inverse PCRs in which new sets of primers were designed and used: primer set α1A/α1B for the amplification of the 1.8-kb HindIII fragment, primer set α2A/α2B for the amplification of the 1.9-kb ClaI fragment, and primer set α3A/α3B for the amplification of the 0.8-kb HindIII fragment. In all cases, at least two independent PCRs were performed for cloning and sequencing experiments (see below) to guard against any misincorporation errors which may have been introduced by Taq polymerase.
Molecular cloning, Southern hybridization, and DNA sequencing.
Standard DNA cloning techniques were performed as described by Sambrook et al. (34). Enzymes used in the cloning experiments were purchased from Boehringer Mannheim (Indianapolis, Ind.), Sigma (St. Louis, Mo.), Promega Corp. (Madison, Wis.) or Pharmacia LKB (Uppsala, Sweden) and used according to the manufacturers’ recommendations. The T/A cloning vectors pCR2.1 and pCR3 were used as recommended by the manufacturer. Following restriction mapping of the cloned inverse PCR products, subclones in pUC19 were made in order to obtain fragments short enough to completely sequence both strands. Southern hybridizations were carried out as described previously (34, 35). Oligonucleotide probes were 3′-end labelled with fluorescein-11-dUTP by using the ECL 3′-end labelling kit (Amersham Life Science, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer’s instructions. Hybridization conditions were as recommended by Amersham. Nylon membranes (Hybond N+; Amersham) were used throughout. DNA sequencing of the plasmid inserts was performed by King’s College, Department of Molecular Medicine, London, United Kingdom, with an ABI 373A DNA sequencer (Perkin-Elmer Cetus) and Taq DyeDeoxy terminator chemistry or by MWG Biotech, with a LI-COR 4200 sequencer.
RNA isolation, Northern analysis, and primer extension.
RNA was isolated from L. plantarum by the method described by Anba et al. (3) at different growth phases of cultures growing on MRS or MRS plus 4% (wt/vol) NaCl. RNA concentrations were determined spectrophotometrically. Northern blot analysis was performed as described by Ausubel et al. (4). A 320-bp DNA fragment generated by PCR, with the αA/βS primer pair and chromosomal DNA from LPCO10 as the template, was used as the probe after being labelled with [α-32P]CTP by using the Ready-to-Go labelling kit (Pharmacia).
Primer extension analysis was performed as described by Ausubel et al. (4) with the following modifications. Forty micrograms of total RNA from a culture at early stationary phase was annealed with end-labelled (32P; 200,000 cpm) primer B1 (5′-ATAACAGCATTTAATTGATC-3′) in a total volume of 30 μl. The mixture was heat denatured at 80°C for 5 min and then incubated at 47°C for 1 h. Annealed nucleic acids were ethanol precipitated and resuspended in 25 μl of reverse transcriptase mixture, which consisted of 0.56 mM concentrations of each of the dNTPs, 5 μl of reverse transcriptase buffer, 0.5 μl of the RNase inhibitor RNA Guard (Pharmacia), and 25 U of avian myeloblastosis virus reverse transcriptase (Boehringer). The mixture was incubated at 42°C for 1 h to allow polymerization. The products were ethanol precipitated in the presence of 35 μg of glycogen (Appligene, Pleasanton, Calif.). Samples were analyzed on a 6 M urea–6% (wt/vol) polyacrylamide gel. pBluescript II SK(+) (Stratagene, La Jolla, Calif.) with M13 (−20) universal primer (Stratagene) in a standard sequencing reaction mixture was used as the molecular weight ladder.
Computer analysis of DNA and protein sequences.
The PC/Gene program package (version 6.85; IntelliGenetics, Inc., Mountain View, Calif.) and the Genetics Computer Group package (version 8; University of Wisconsin) were used for DNA and protein analyses.
Nucleotide sequence accession number.
The nucleotide sequence presented in this article has been assigned EMBL accession no. Y15127.
RESULTS
PCR with degenerate primers.
On the basis of the available amino acid sequences of the α and β peptides of PLS (19), two sets of degenerate primers were designed and synthesized: αF/αR and βF/βR (Table 1). When LPCO10 genomic DNA was used as the template in PCRs in which the degenerate primer pairs αF/αR and βF/βR were present, products of 61 and 66 bp, respectively, were obtained as predicted from the known amino acid sequences of the α and β peptides of PLS. These PCR products were subsequently isolated and blunt-end ligated into SmaI-linearized pUC19. Thus, recombinant plasmids pSIG201 and pCRU1, containing part of the genes encoding the α and β peptides of PLS, respectively, were obtained and used to transform E. coli DH5α competent cells. DNA sequencing of the inserts in these recombinant plasmids revealed the correspondence to that predicted from the previously known partial amino acid sequence (19). Furthermore, when primers βF and αR were used in a PCR with the same template DNA as that described above, a product of approximately 360 bp resulted, suggesting that the coding regions for the two peptides were colocated on the LPCO10 chromosome, with the gene encoding the β peptide (plsB) upstream of that encoding the α peptide (plsA).
PCR with nondegenerate primers.
Once portions of the DNA sequences for the genes encoding the α and β peptides of PLS were elucidated and once the genes’ proximity in the chromosome of LPCO10 was established, a nondegenerate PCR primer pair was designed: αA/βS (Table 1). These primers were used to amplify a fragment of 320 bp, as predicted from the result with the degenerate primers. Thus, this fragment included the C terminus-encoding region of plsB, the N terminus-encoding region of plsA, and the intergenic region. This PCR product was subsequently cloned into pCR2.1, and the resulting recombinant plasmids, pSAB07 and pSAB045, were used to transform E. coli Top10F′ competent cells. DNA sequencing of the inserts was performed, and the resulting information was used to design a PCR primer pair for inverse PCR: αINV1/βINV1 (Table 1).
Inverse PCR; cloning and sequencing of the PLS operon and surrounding regions.
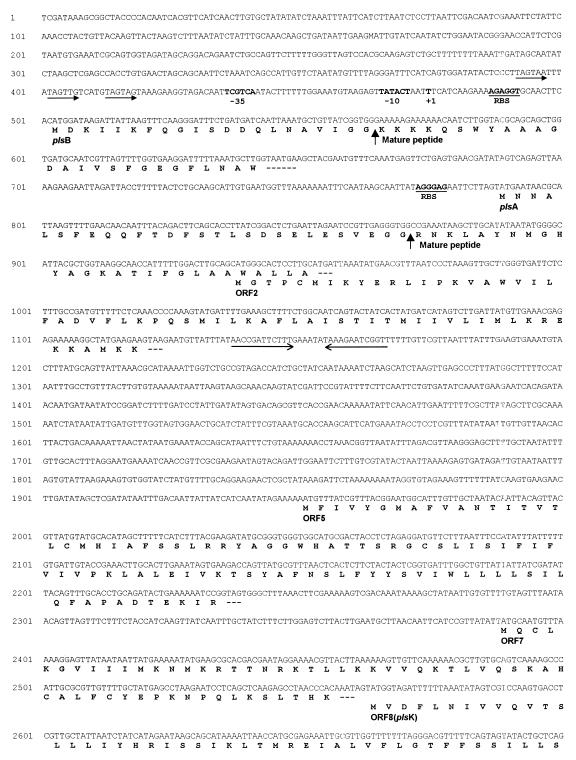
By inverse PCR with primer pair αINV1/βINV1 on circular-template DNA produced from a ClaI digest of LPCO10 genomic DNA, a 1.3-kb region spanning the PLS-encoding region (Fig. 1) was cloned into pCR3, and the resulting recombinant plasmids were used to transform E. coli Top10F′ competent cells. Subsequent DNA sequencing of the 1.3-kb insert revealed the complete sequences of the plsA and plsB genes, a complete ORF, termed ORF2, and an incomplete ORF, termed ORF1 (Fig. 2). DNA sequences of plsA and plsB show that the corresponding two peptides contain leader sequences of the double-glycine type (20). The α peptide consists of 55 amino acids (28 amino acids in the leader sequence and 27 in the mature peptide), and the β peptide consists of 47 amino acids (21 in the leader and 26 in the mature peptide) (Fig. 2). The theoretical pI for the β peptide including the leader is 8.0, and its molecular weight (MW) is 5,117; the pI of the mature peptide is 9.4, and its MW is 2,873. For the α peptide the pI is 4.6 for the native peptide and its MW is 5,987; the pI for the mature peptide is 10.0, and its MW is 2,922. While the α and β leader peptides show strong homology to those reported for other LAB bacteriocins (13, 15), the α and β mature peptides are different from any previously described bacteriocin (Fig. 2). There are −35- and −10-type promoter sequences upstream of plsB, as well as a putative ribosome binding site (RBS) (Fig. 2). Three imperfect direct repeats of 7 nucleotides each, separated by either 3 or 4 nucleotides and having the consensus sequence 5′-TAGTagT-3′, are present 16 nucleotides upstream of the putative −35 sequence (Fig. 2). There is an inverted repeat just downstream of ORF2 which may function as a transcription terminator (Fig. 2). This suggests that the two pls genes plus ORF2 are produced on the same transcript of approximately 0.7 kb (see below). There is a further putative RBS upstream of the plsA sequence, and thus the two peptides are presumed to be translated independently. ORF2, which overlaps the end of plsA and which is in the same reading frame as plsB, has the potential to encode a small protein of 61 amino acids, with a theoretical pI of 10.1 and MW of 7,054.
FIG. 2.
Nucleotide sequence of the pls locus and deduced proteins. Promoter −35 and −10 sites, RBSs, and the transcriptional start (+1) are indicated by boldface letters. Vertical arrows indicate the cleavage sites in the α and β native peptides and the beginnings of the respective mature peptides. Direct repeats upstream of plsB are indicated by arrows. Sequences of dyad symmetry with the potential to serve as transcription terminators are indicated by arrows downstream of ORF2. Underlined amino acid sequences in ORF8 (plsK) indicate putative histidine (H), asparagine (N), and glycine (G) boxes, as defined by O’Connell-Motherway et al. (29). Stop codons are indicated by dashed lines at the ends of the protein sequences.
To further extend the sequence downstream of the PLS operon, the 1.8-kb HindIII, the 1.9-kb ClaI, and the 0.8-kb HindIII fragments amplified by subsequent rounds of inverse PCR (Fig. 1) were cloned into pCR3 or pCR2.1. The resulting recombinant plasmids were sequenced, as described above, revealing several new ORFs (Fig. 1). The sequences of putative proteins encoded by these ORFs were compared with known sequences in the Swiss-Prot and EMBL data banks by using the BLITZ and BLASTP searches. The results of these searches revealed areas of strong homology between ORF8 and several HPKs from LAB, with P values below 10−22 (2) and percentages of identity ranging from 24 to 37% (data not shown). In particular, strong homology was found with HPKs involved in the regulatory mechanisms of the production of several bacteriocins from LAB: the proteins encoded by sppK from Lactobacillus sake Lb674 (16), plnB from L. plantarum C11 (11), sapK from L. sake Lb674 (5), and cbnK from Carnobacterium piscicola LV17B (31). In accordance with this observation, we have tentatively named ORF8 plsK (Fig. 1). In addition, the predicted product of plsK possesses at least three of the motifs which have been found to be common to other HPKs (29): the histidine (H) box, the glycine (G) box, and the asparagine (N) box (Fig. 2). Furthermore, computer-aided analysis of its amino acid sequence predicts the existence of three membrane-associated helices (data not shown) similar to those found in other HPKs associated with the class II bacteriocins from LAB (25). The database searches also identified areas of strong homology between ORF14 and several RR proteins, with P values below 10−8 (2) and percentages of identity ranging from 31 to 54% (data not shown). Again, strong homology was found with RR proteins involved in the regulation of bacteriocins from LAB: the proteins encoded by sppR from L. sake Lb674 (16), plnC and plnD from L. plantarum C11 (11), sapR from L. sake Lb674 (5), and cbnR from C. piscicola LV17B (31). Consequently, we have called ORF14 plsR (Fig. 1). Finally, BLITZ and FASTA searches of the data banks found 33.3% identity, with a P value below 10−8 (2), between the potential product of ORF5 and part of the protein encoded by the agrB gene of S. aureus (27). In S. aureus this gene is involved in the posttranslational modification of the product of agrD, which is part of the mechanism that regulates the virulence phenotype in this bacterium (23, 24).
Transcription analysis of the PLS operon.
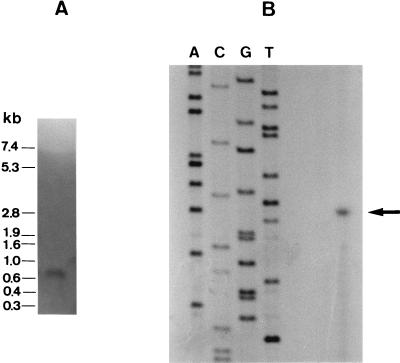
Northern analyses performed on total RNA samples obtained at different times from cultures of LPCO10 growing in MRS broth revealed that a unique transcript of ca. 0.7 kb, which hybridized to the 320-bp probe containing part of the plsA and plsB genes, was produced (Fig. 3A). Detectable levels of this mRNA were obtained when the optical densities at 600 nm of the cultures were over 0.7 and were maintained throughout bacterial growth, with a maximum at the early stationary phase. The same Northern analysis was performed on total RNA from L. plantarum 55-1 (the non-bacteriocin-producing derivative of LPCO10). The results indicated that this strain does not express any of the genes detected by the 320-bp probe.
FIG. 3.
Northern and primer extension analysis of the pls operon. (A) Transcription product of the pls operon in an early-stationary-phase culture of L. plantarum LPCO10, obtained by using a 320-bp fragment containing part of the plsA and plsB genes as the probe. A total of 75 μg of RNA was loaded. Figures on the left indicate the sizes of the RNA standards used. (B) Primer extension analysis of plsB with oligonucleotide B1 (see text) as the primer. The arrow indicates the 84-nucleotide extension product generated. The size of the product was determined by comparison with the sequencing reaction products generated from pBluescript II SK(+) primed with the M13 (−20) universal primer which are shown in lanes A, C, G, and T.
Primer extension analysis of LPCO10 total RNA from a culture at the early stationary phase with oligonucleotide B1 as the primer showed an extension product of 84 nucleotides (Fig. 3B). Further calculations gave nucleotide T at position −28 as the start point of the transcription (+1; Fig. 2). This result was later confirmed by using the PCR primer βA in a similar reaction. Based on these results, putative −10 and −35 sequences are proposed (Fig. 2). Combining Northern analysis and primer extension results showed that the 0.7-kb mRNA would include genes plsA and plsB and could also cover ORF2 (Fig. 2).
Comparison of LPCO10 and 55-1.
The non-bacteriocin-producing mutant 55-1, derived from LPCO10 as described above, was compared with LPCO10 by PCR to determine if the regions of DNA encoding the pls transcripts were still present. A series of PCRs was performed in parallel on both 55-1 and LPCO10 DNA with different sets of primers designed from the known sequence of LPCO10. The following primer pairs were used: βS/αA, β1B/α1A, β1B/βINV1, αINV1/α1A, and α1B/α2A (Table 1). All of the PCR primer pairs amplified fragments of the same size for both strains, and these were of the sizes predicted from the LPCO10 DNA sequence, i.e., 320, 1,200, 700, 700, and 1,800 bp, respectively. Thus, 55-1 was shown to possess the genetic region carrying the pls genes. However, the 0.7-kb transcript seen in LPCO10 was found to be absent from 55-1 (see above). In order to check the possibility of significant mutations in the PLS promoter region, the PCR product amplified by the β1B and βA primers, which covers the promoter region for PLS and part of the plsB gene in LPCO10, was obtained by using 55-1 total DNA as the template, cloned in pCR2.1, and sequenced as described above. The DNA sequence obtained was compared with that from LPCO10, and no significant difference was found between them (data not shown).
DISCUSSION
In this study the genes responsible for the production of PLS in L. plantarum LPCO10 have been identified, cloned, and sequenced. The subsequent analysis of the DNA sequence of this region has revealed two structural genes, plsA and plsB, whose sequences correspond to the amino acid sequences of the two components of PLS, i.e., peptides α and β, respectively (19). Other ORFs which are probably involved in the regulation of the bacteriocin system have been identified.
The DNA sequences of plsA and plsB obtained in this study have confirmed the presence of N-terminal leader sequences of the type recognized by ABC transporters and found in many other bacteriocins (25). The presence of a consensus sequence having homology with all other double-glycine type leader sequences so far described indicates that an ABC transporter system is almost certainly present in LPCO10. Although no ORF whose product shows homology to any of the ABC transport proteins described has been identified in the portion of the LPCO10 chromosome that has been sequenced to date, it is likely that such ORFs are present elsewhere in the genome. As LPCO10 has been shown to produce at least one other bacteriocin, i.e., PLT (18), a single, dedicated ABC transport system may be shared between the two bacteriocins.
According to the model proposed by Nes et al. (25), the gene conferring immunity to the bacteriocin should be located close to the structural gene of the bacteriocin itself and should be expressed as part of the same operon. In our case, ORF2, with the potential to code for a peptide 61 amino acids long, is the best candidate for the PLS immunity gene. It is located in proximity to the structural genes, that is, downstream of plsB and either overlapping or just downstream of plsA, and it could be expressed in LPCO10 concomitant with the expression of both structural genes, as Northern and primer extension experiments have demonstrated (Fig. 3). Furthermore, a hydropathic profile analysis of ORF2 shows several putative transmembrane segments (data not shown), which are thought to be necessary for the integration of immunity proteins into the membrane of the bacteriocin producer (14). No homology between the putative peptide encoded by ORF2 and any of the immunity proteins described for other LAB bacteriocins has been found. However, this is not unexpected, as bacteriocin immunity proteins described so far show little homology with one another (25). The absence of the 0.7-kb transcript in non-bacteriocin-producing strain 55-1 appears to throw doubt on the hypothesis that ORF2 is the immunity gene, since 55-1 is not susceptible to PLS. However, two other possibilities remain: (i) that ORF2 can be transcribed separately from the plsAB genes and (ii) that the lack of susceptibility to PLS results from some mechanism of resistance other than immunity.
The presence of an agr-like system for the regulation of the production of PLS, as described for other bacteriocins from LAB (25), is suggested by the homology found between the potential translation products of plsK and plsR and several HPK and RR proteins, respectively. This is also suggested by the homology between ORF5 and gene agrB from S. aureus (27), which might be involved in the posttranslational modification of the product of agrD. agrD activates transcription from the agr operon promoter (P2), as well as from the divergent RNAIII promoter (P3) (17), and regulates the expression of virulence in S. aureus (27). However, the predicted product of ORF5 is considerably smaller than that of agrB: 95 versus 189 amino acids.
A further element for bacteriocin regulation in other agr-like operons that has been described previously (21), the IF, is not evident at the moment in the putative pls regulatory system. The position of ORF7, i.e., upstream of plsKR and downstream of the putative modification gene ORF5, makes its product the best candidate for the IF. The regulatory operon would then resemble the agrBDCA locus in S. aureus (27). However, the predicted product of ORF7 does not possess a double-glycine processing site, as is the case for other IFs from LAB bacteriocin systems (25), and the protein would be much larger in its native form (57 amino acids) than other IFs, which are typically around 20 amino acids (5, 11, 16, 31). It is possible that, in the PLS system, the posttranslational modification to produce the mature IF is performed by the product of ORF5 (the protein resembling the product of agrB) rather than by cleavage of a leader sequence by the ABC transporter. The presence of three conserved direct repeats in front of the pls operon (Fig. 2) is in agreement with a similar finding for other regulated bacteriocins (13) and suggests that an IF does regulate transcription of the PLS operon.
This study has shown that PLS conforms, at least in part, to the model proposed by Nes et al. (25) regarding the genetic organization of class II bacteriocins from LAB. Further studies are currently being carried out in order to identify either the immunity gene or the existence of a PLS resistance mechanism. Our present efforts are oriented towards the analysis of the expression of those ORFs which are presumably related to the pls operon, such as ORFs 2, 5, 7, 8, and 14. Further DNA sequencing of the region surrounding the PLS operon is also under way in the hope of identifying further related genes such as those involved in the export of the bacteriocin.
ACKNOWLEDGMENTS
This work was supported by contracts from the EU (BIOTECH G-Project on LAB, contract no. BIOT-CT94-3055) and the Spanish government (CICYT Project ALI-94-0980-CO2-01). J.L.R.-B. was the recipient of a grant from the Spanish Ministry of Education and Science and from the European Commission.
We are grateful to Enrique Flores García from the Instituto de Bioquímica Vegetal y Fotosíntesis, C.S.I.C., Seville, Spain, for allowing us to use the radioactive facilities at that center.
REFERENCES
- 1.Allison G E, Fremaux C, Klaenhammer T R. Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J Bacteriol. 1994;176:2235–2241. doi: 10.1128/jb.176.8.2235-2241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. Mol Microbiol. 1990;8:81–91. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Anba J, Bidnenko E, Hillier A, Ehrlich D, Chopin M-C. Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J Bacteriol. 1995;177:3818–3823. doi: 10.1128/jb.177.13.3818-3823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1992. [Google Scholar]
- 5.Axelsson L, Holck A. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Bacteriol. 1995;177:2125–2137. doi: 10.1128/jb.177.8.2125-2137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckenhüskes H J. Selection criteria for lactic acid bacteria to be used as starter cultures for various food commodities. FEMS Microbiol Rev. 1993;12:253–272. [Google Scholar]
- 7.Cathcart D P. Purification, characterization and molecular analysis of plantaricin S, a two-peptide bacteriocin from olive fermenting Lactobacillus plantarum strains. Ph.D. thesis. Bedford, United Kingdom: Cranfield University; 1995. [Google Scholar]
- 8.Daeschel M A, Fleming H P. Selection of lactic acid bacteria for use in vegetable fermentations. Food Microbiol. 1984;1:303–313. [Google Scholar]
- 9.Daeschel M A. Applications and interactions of bacteriocins from lactic acid bacteria in foods and beverages. In: Hoover D G, Steenson L R, editors. Bacteriocins of lactic acid bacteria. New York, N.Y: Academic Press Inc.; 1993. pp. 63–91. [Google Scholar]
- 10.de Vuyst L, Vandamme E J. Antimicrobial potential of lactic acid bacteria. In: de Vuyst L, Vandamme E J, editors. Bacteriocins of lactic acid bacteria: microbiology, genetics and applications. London, United Kingdom: Blackie Academic & Professional; 1994. pp. 91–142. [Google Scholar]
- 11.Diep D B, Håvarstein L S, Nissen-Meyer J, Nes I F. The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcription unit as an agr-like regulatory system. Appl Environ Microbiol. 1994;60:160–166. doi: 10.1128/aem.60.1.160-166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep D B, Håvarstein L S, Nes I F. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol Microbiol. 1995;18:631–639. doi: 10.1111/j.1365-2958.1995.mmi_18040631.x. [DOI] [PubMed] [Google Scholar]
- 13.Diep D B, Håvarstein L S, Nes I F. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fremaux C, Ahn C, Klaenhammer T R. Molecular analysis of the lactacin F operon. Appl Environ Microbiol. 1993;59:3906–3915. doi: 10.1128/aem.59.11.3906-3915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Håvarstein L S, Holo H, Nes I F. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 16.Huehne K, Holck A, Axelsson L, Kroeckel L. Analysis of sakacin P gene cluster from Lactobacillus sake LB674 and its expression in sakacin P negative L. sake strains. Microbiology. 1996;142:1437–1448. doi: 10.1099/13500872-142-6-1437. [DOI] [PubMed] [Google Scholar]
- 17.Ji G Y, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiménez-Díaz R, Ríos-Sánchez R M, Desmazeaud M, Ruiz-Barba J L, Piard J-C. Plantaricins S and T, two new bacteriocins produced by Lactobacillus plantarum LPCO10 isolated from a green olive fermentation. Appl Environ Microbiol. 1993;59:1416–1424. doi: 10.1128/aem.59.5.1416-1424.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiménez-Díaz R, Ruiz-Barba J L, Cathcart D P, Holo H, Nes I F, Sletten K H, Warner P J. Purification and partial amino acid sequence of plantaricin S, a bacteriocin produced by Lactobacillus plantarum LPCO10, the activity of which depends on the complementary action of two peptides. Appl Environ Microbiol. 1995;61:4459–4463. doi: 10.1128/aem.61.12.4459-4463.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 21.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 22.Lindgren S E, Dobrogosz W J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol Rev. 1990;87:149–164. doi: 10.1111/j.1574-6968.1990.tb04885.x. [DOI] [PubMed] [Google Scholar]
- 23.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 25.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 26.Nissen-Meyer J, Holo H, Håvarstein L S, Sletten K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 28.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 219–222. [Google Scholar]
- 29.O’Connell-Motherway M, Fitzgerald G, van Sinderen D. Cloning and sequence analysis of putative histidine protein kinases isolated from Lactococcus lactis MG1363. Appl Environ Microbiol. 1997;63:2454–2459. doi: 10.1128/aem.63.6.2454-2459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patil R V, Dekker E E. PCR amplification of an Escherichia coli gene using mixed primers containing deoxyinosine at ambiguous positions in degenerate amino acid codons. Nucleic Acids Res. 1990;18:3080. doi: 10.1093/nar/18.10.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quadri L E N, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 32.Ray B, Daeschel M A. Bacteriocins of starter culture bacteria. In: Dillon V M, Board R G, editors. Natural antimicrobial systems and food preservation. Wallingford, United Kingdom: CAB International; 1994. pp. 133–165. [Google Scholar]
- 33.Ruiz-Barba J L, Cathcart D P, Warner P J, Jiménez-Díaz R. Use of Lactobacillus plantarum LPCO10, a bacteriocin producer, as a starter culture in Spanish-style green olive fermentations. Appl Environ Microbiol. 1994;60:2059–2064. doi: 10.1128/aem.60.6.2059-2064.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Southern E M. Detection of specific sequences among DNA fragments separated by agarose gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 36.Tahara T, Oshimura M, Umezawa C, Kanatani K. Isolation, partial characterization, and mode of action of acidocin J1132, a two-component bacteriocin produced by Lactobacillus acidophilus JCM 1132. Appl Environ Microbiol. 1996;62:892–897. doi: 10.1128/aem.62.3.892-897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Belkum M J, Hayema B J, Jeeninga R E, Kok J, Venema G. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991;57:492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]