Abstract
The microbiology of the biomass from a nitrite-oxidizing sequencing batch reactor (NOSBR) fed with an inorganic salts solution and nitrite as the sole energy source that had been operating for 6 months was investigated by microscopy, by culture-dependent methods, and by molecular biological methods, and the seed sludge that was used to inoculate the NOSBR was investigated by molecular biological methods. The NOSBR sludge comprised a complex and diverse microbial community containing gram-negative and gram-positive rods, cocci, and filaments. By culture-dependent methods (i.e., micromanipulation and sample dilution and spread plate inoculation), 16 heterotrophs (6 gram positive and 10 gram negative) were identified in the NOSBR sludge (RC), but no autotrophs were isolated. 16S ribosomal DNA clone libraries of the two microbial communities revealed that the seed sludge (GC) comprised a complex microbial community dominated by Proteobacteria (29% beta subclass; 18% gamma subclass) and high G+C gram-positive bacteria (10%). Three clones (4%) were closely related to the autotrophic nitrite-oxidizer Nitrospira moscoviensis. The NOSBR sludge was overwhelmingly dominated by bacteria closely related to N. moscoviensis (89%). Two clone sequences were similar to those of the genus Nitrobacter. Near-complete insert sequences of eight RC and one GC N. moscoviensis clone were determined and phylogenetically analyzed. This is the first report of the presence of bacteria from the Nitrospira phylum in wastewater treatment systems, and it is hypothesized that these bacteria are the unknown nitrite oxidizers in these processes.
Nitrification is the initial step in the removal of nitrogenous compounds from wastewaters. It involves the two-step conversion of ammonia to nitrite (ammonia oxidation) and nitrite to nitrate (nitrite oxidation) (10). Denitrification of the nitrate to nitrogenous gas removes the nitrogen from solution (18). If nitrogen removal fails, the nitrogenous compounds passing into waterways may cause a series of environmental and medical problems (2).
There are a range of autotrophic (11) and heterotrophic bacteria (8) capable of nitrification. Unlike heterotrophic bacteria, autotrophs are dependent on this reaction to generate energy for cell maintenance and growth. In wastewater treatment systems, autotrophs constitute only a small percentage of the mixed liquor microbial community, but they are responsible for the bulk of nitrification (17, 18).
In wastewater treatment systems, the genera Nitrosomonas (an ammonia oxidizer) and Nitrobacter (a nitrite oxidizer) are the two groups of autotrophs presumed to be responsible for nitrification (11). Although ammonia oxidizers have been intensively studied by the use of molecular methods (26, 27), the nitrite oxidizers have not been similarly studied. In one study of activated sludge flocs (15), clusters of Nitrosomonas and Nitrobacter spp. were adjacent to each other as revealed by fluorescent in situ hybridization (FISH) probing. However, in other studies, Nitrobacter could not be detected, and it was speculated that other bacteria were likely responsible for nitrite oxidation (12, 27).
To investigate the identity of the nitrite oxidizers in wastewater treatment plants, a nitrite-oxidizing sequencing batch reactor (NOSBR) was operated. After 6 months of operation of the NOSBR, the developed sludge (RC) was investigated by microscopy, by culture-dependent methods, and by molecular biological methods. In addition, the sludge used to inoculate the NOSBR (GC) was investigated by molecular biological methods and compared with the NOSBR biomass.
MATERIALS AND METHODS
Mixed liquor from the Merrimac Wastewater Treatment Plant at the Gold Coast, Queensland, Australia, was used as inoculum for the NOSBR. The Merrimac plant is a full-scale biological nutrient removal (BNR) plant operating for nitrogen and phosphorus removal. Mixed liquor from the aerobic stage was collected and brought to the laboratory on ice. A volume of 1 liter was used to initiate the NOSBR, while further aliquots were stored at −20°C.
Operation of NOSBR.
The NOSBR was operated according to methods previously reported (6). Briefly, the reactor was a chemostat with an operating volume of 1 liter, and the reactor feed comprised the following (per liter): 400 mg of KNO2, 3.75 g of MgSO4 · 7H2O, 250 mg of CaCl2 · 2H2O, 10 g of KH2PO4, 10 g of KH2PO4, 200 mg of FeSO4 · 7H2O, and 20 g of NaHCO3 (pH 7.2). There were four stages to each cycle and a hydraulic retention time of 12 h. The stages were (i) feed, 500 ml of fresh medium for 30 min (0 to 0.5 h); (ii) aerobic reaction, 4.5 h (0.5 to 5 h); (iii) settle, 40 min (5 to 5.7 h); and (iv) decant, 500 ml of supernatant for 20 min (5.7 to 6 h). The total time per cycle was 6 h.
After the NOSBR was operated for a period of approximately 6 months, a 10-ml grab sample of mixed liquor was removed from the reactor during the middle of the aerobic reaction stage and used immediately for analyses.
Microscopy.
Approximately 50 μl of the NOSBR mixed liquor was Gram stained, and micrographs were taken with a Nikon Microphot FXA microscope.
Culture-dependent methods.
The NOSBR sludge (400 μl) was washed twice with phosphate-buffered saline (PBS; 135 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 1.75 mM K2HPO4 [pH 7.5]), resuspended in 400 μl of PBS, and serially diluted to 10−7. A volume of 50 μl of each dilution was then spread inoculated onto two types of agar media. These were Nutrient Agar (NA; Oxoid, England) and autotrophic nitrite agarose (ANA; composed of the reactor feed [see above] solidified with 10 g of agarose per liter). In addition, a range of the diluted sludge samples was briefly sonicated, and individual cells were isolated by micromanipulation (21) and inoculated onto ANA. Plates were then incubated at 28°C until growth occurred. A range of colonies with different morphologies grew on the NA and ANA inoculated with sludge samples by spread inoculation and on ANA inoculated with micromanipulated cells. These colonies were subcultured to ensure purity. The 16S ribosomal DNA (rDNA) sequence was partially determined and analyzed for a range of these isolates by previously published methods (3, 4).
Molecular biological methods.
The total community DNAs from the NOSBR sludge (RC) and from the sludge used as inoculum for the NOSBR (GC) were isolated, and the 16S rDNAs were PCR amplified and cloned.
DNA extraction.
The biomass (500 μl) was centrifuged at 12,000 × g for 5 min. The supernatant was discarded, and the pellet was resuspended in 500 μl of saline-EDTA (150 mM NaCl, 100 mM EDTA [pH 8.0]). A volume of 100 μl of freshly prepared 100-mg/ml lysozyme was added to the mixture and incubated at 37°C for 1.5 h. The mixture was then subjected to four cycles of freezing and thawing at −20 and 65°C, respectively. Following this, 100 μl of 25% (wt/vol) sodium dodecyl sulfate and 50 μl of 2% (wt/vol) proteinase K were added to the mixture and the mixture was incubated at 60°C for 1.5 h. The DNA was recovered from the tube by phenol-chloroform extraction (19). The nucleic acids from the 0.5-ml aqueous phase were precipitated by adding 0.12 ml of sterile 3 M sodium acetate and 1 ml of ice-cold 100% ethanol and incubating for 1 h at −70°C. The DNA pellet was recovered by centrifuging the solution at 12,000 × g for 20 min at 4°C. The pellet was washed by adding 500 μl of 70% ice-cold ethanol and was recovered by centrifuging at 12,000 × g for 10 min at 4°C. The pellet was then air dried, and the nucleic acids were dissolved in 100 μl of sterile milliQ-purified (mQ) water. Residual RNA was removed from the nucleic acid solution by adding 3 μl of 10-mg/ml RNase and incubating at 37°C for 1 h. The DNA was visualized by agarose gel electrophoresis (19).
Amplification of the 16S rRNA genes (16S rDNA).
Amplification of the near-complete 16S rRNA genes from the extracted DNA was done by employing the bacterial conserved primers 27f and 1492r (14) in a PCR. The components of the PCR were 10 to 100 ng of DNA, 1 U of Tth Plus DNA polymerase (Biotech, Perth, Australia), 10 μl of 10× reaction buffer (Biotech), 200 μM (each) dATP, dCTP, dGTP, and dTTP (deoxynucleoside triphosphates [dNTPs]), and 1 μl of 200-ng/μl (each) primer in a final volume of 100 μl made up with sterile mQ water. The reaction mixture was then overlaid with 80 μl of mineral oil and placed in a thermal cycler (Perkin-Elmer DNA Thermal Cycler 480). A cycling program of 30 cycles of 94°C for 60 s, 48°C for 60 s, and 72°C for 120 s with a final extension of 1 cycle of 48°C for 60 s and 72°C for 300 s was used. The amplicons were visualized by agarose gel electrophoresis and were purified by the Wizard PCR Cleanup Kit (Promega, Sydney, Australia) according to the manufacturer’s instructions.
Cloning of the 16S DNAs.
Amplicons were used immediately in a ligation reaction mixture comprising 1 μl of T4 DNA ligase (1 U/μl), 10× buffer, 1 μl of pGEM-T vector (50 ng), 2 μl of amplicons (75 ng), and 5 μl of sterile mQ water. All components except the amplicons were from the TA Cloning Kit (Invitrogen, Calif.). Ligation occurred at 15°C for 16 h.
Ultracompetent Epicurian Coli XL2-Blue MRF′ cells (Stratagene, Sydney, Australia) were thawed on ice in preparation for the transformation step. A volume of 100 μl of thawed cells was gently placed in a chilled 50-ml Falcon tube, and 1.7 μl of β-mercaptoethanol was added. The mixture was incubated on ice for 10 min with regular gentle swirling. Then, 2 μl of the ligation mixture was added to the cells, and the cells were incubated on ice for 30 min. A heat shock step was done by immersing the Falcon tube in a 42°C water bath for exactly 30 s. Cells were then returned to ice for 2 min. A volume of 900 μl of warm (42°C) sterile SOB (20 g of Bacto Tryptone, 5 g of yeast extract, 0.5 g of NaCl per liter) was added to each tube of transformed cells. These were then shaken at 37°C for 1 h.
A volume of 25 μl of transformed cells was spread inoculated onto Luria-Bertani (LB) agar plates containing ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and isopropyl-β-d-thiogalactopyranoside (LB Ampicillin/X-gal/IPTG), (19) which were incubated at 37°C for 12 to 16 h and then at 4°C for 1 h. Positive clones (those containing 16S rDNA PCR inserts) appeared white and negative clones (no inserts) were blue. Positive clones were picked and patched onto LB Ampicillin/X-gal/IPTG agar plates to ensure that the first screening was correct. Positive clones were picked, homogenized into 300 μl of sterile 50% glycerol, and stored at −20°C until required. These clones constituted the clone libraries.
Amplification of clone inserts.
Stored clones from the library were patched onto LB Ampicillin plates from glycerol stocks and grown overnight at 37°C. A sterile tip from a P200 micropipettor was used to obtain a barely visible amount of overnight growth, which was placed into a microcentrifuge tube containing 63 μl of sterile mQ water and 10 μl of 10× reaction buffer, and the mixture was covered with mineral oil. The tube was placed into the thermal cycler and incubated at 96°C for 10 min. Then, 200 μM (each) dNTP, 1 μl of Tth Plus DNA polymerase, and 1 μl of each of the plasmid primers (SP6 and T7) (200 ng/μl; Invitrogen) were added to each tube. PCR cycling and observation of amplicons were performed as described above.
Restriction enzyme analysis (REA) of clone inserts.
For the NOSBR (RC) library, the amplicons from the SP6-T7 PCR from individual clones were subjected to HaeIII (Sigma, Sydney, Australia) digestion. HaeIII is a restriction enzyme that recognizes and cuts the tetranucleotide sequence 5′ GG-CC 3′, i.e., it is a “4-bp cutter.” The digestion mixture consisted of 0.5 μl of HaeIII enzyme (10 U/μl), 2 μl of NEB Buffer 2 (Sigma), 7 μl of sterile mQ water, and 10 μl of amplicon. The reaction was carried out at 37°C for 3 h. The restriction-digested fragments were visualized by electrophoresis in a 3% Tris-acetate-EDTA agarose gel (19) for 55 min at 80 V.
Clones containing inserts that produced identical restriction patterns were grouped into operational taxonomic units (OTUs), and representatives of each OTU were selected for insert sequencing and analysis.
Partial and near-complete sequencing of clone inserts.
Amplicons from the SP6-T7 PCR from individual clones were purified with the Wizard PCR Cleanup Kit and sequenced with the ABI dideoxy sequencing kit (ABI, Melbourne, Australia) according to the manufacturers’ instructions and with primer 530f (14). For some clones, near-complete insert sequence data were obtained. In this case, PCR of the inserts with the 27f and 1492r primers was employed. A range of bacterial conserved primers (27f, 357f, 530f, 927f, 1114f [4]) were used to determine the sequences. PCR and sequencing were performed as described above.
Analysis of sequence data.
The partial 16S rDNA sequences were compared with those on publicly accessible databases by using the program Basic Local Alignment Search Tool (BLAST [1]). The sequences were also manually aligned, considering secondary structural constraints, with sequences from members of the domain Bacteria. Phylogenetic analysis of aligned data sets was carried out by using the Phylogeny Inference Package (PHYLIP version 3.5) according to previously published methods (4).
RESULTS
Microscopy.
Over the 6-month period of exposure to a very simple medium that favored the growth of autotrophic nitrite oxidizers, a diverse microbial community in terms of morphology (cocci, rods, and filaments) and Gram stain reaction developed.
Culture-dependent methods.
Results for partial 16S rDNA sequences and identities for 16 pure cultures of bacteria obtained from the NOSBR are shown in Table 1. A range of bacteria were able to grow on the ANA medium, although in some cases, the growth took up to 14 days. In addition, prolific growth of a range of bacteria was observed on the NA. Clearly, the NOSBR contained heterotrophs in addition to autotrophic nitrite oxidizers. A range of other bacteria in addition to the 16 reported were isolated. However, none were closely related to known autotrophic nitrite-oxidizing bacteria.
TABLE 1.
Results for isolates obtained from the NOSBR by either sample dilution and spread plate inoculation or micromanipulation of individual cells to ANA
| Isolatea | Information from BLAST comparison
|
Gram stain and cell morphology | ||
|---|---|---|---|---|
| Closest match | No. of nucleotides compared | % Similarity with closest match | ||
| 1-NA-S | Acinetobacter sp. | 422 | 90 | Single, gram-negative rods |
| 2-NA-S | Bacillus firmus | 422 | 98 | Large, long, gram-positive rods; chains |
| 3-NA-S | Pseudomonas mendocina | 500 | 96 | Single, paired, gram-negative rods |
| 4-NA-S | Pseudomonas alcaligenes | 380 | 97 | Long, thin, gram-negative rods |
| 5-NA-S | Acinetobacter sp. | 400 | 96 | Short rods; gram negative |
| 6-NA-S | Acinetobacter sp. | 425 | 100 | Short rods; gram negative |
| 7-NA-S | Bacterial sp. | 375 | 99 | NDb |
| 8-ANA-S | Rhodococcus sp. | 420 | 97 | Gram-positive filaments |
| 9-ANA-S | Rhodococcus rhodochrous | 434 | 98 | Short, fat rods; gram positive |
| 10-ANA-S | Rhodococcus sp. | 218 | 92 | Gram-negative rods |
| 11-ANA-S | Mycobacterium fallax | 280 | 94 | Long, thin, gram-negative rods |
| 12-ANA-S | Staphylococcus epidermidis | 381 | 95 | Gram-positive tetrads |
| 13-ANA-S | Paracoccus aminovorans | 300 | 96 | Medium-length, gram-negative rods |
| 14-ANA-M | Unidentified actinomycete | 328 | 97 | ND |
| 15-ANA-M | Stenotrophomonas sp. | 315 | 96 | ND |
| 16-ANA-M | Comamonas testosteroni | 365 | 97 | ND |
NA, Nutrient Agar; ANA, autotrophic nitrite agarose; S, spread plate inoculation; M, micromanipulation.
ND, not described.
Molecular biological methods.
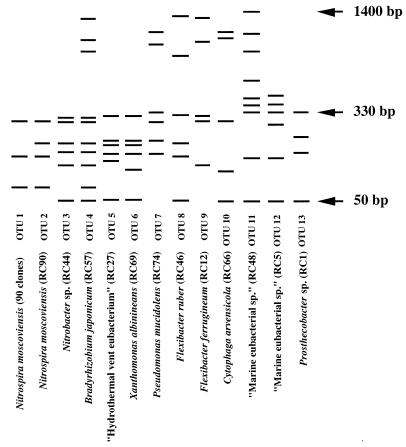
Inserts from a total of 102 clones from the RC library were examined by REA, and they were found to fall into 13 different OTUs (Fig. 1). A total of 90 clones (88%) were grouped into OTU 1, while the remaining 12 OTUs were each composed of individual clones (each of these OTUs was 1% of the total number of clones). Each individual OTU clone insert and six representatives from OTU 1 (RC7, RC11, RC16, RC25, RC73, and RC99) were partially sequenced. Results from BLAST comparisons are given in Fig. 1. According to the BLAST results, the vast bulk of clone inserts in the RC clone library originated from bacteria whose closest relative is Nitrospira moscoviensis. It is recognized that BLAST analysis is a fairly crude way to align sequences from clones with those of specific bacterial genera or species, and a selection of the N. moscoviensis-like clones was analyzed in much more detail. Also according to BLAST analysis, two other clone inserts, RC44 (OTU 3) and RC57 (OTU 4) (Fig. 1), most closely matched sequences from the genera Nitrobacter and Bradyrhizobium, respectively. These genera along with the genera Afipia and Rhodopseudomonas are very closely related according to rRNA comparisons (16, 23). Phylogenetic analysis and direct pairwise comparisons of clones RC44 and RC57 with their closest relatives did not clearly align them with any one of these genera. However, the closest matches from BLAST are given in Fig. 1.
FIG. 1.
Diagrammatic representation of the banding profiles of the 13 OTUs in the RC clone library and their closest matches by BLAST comparisons with partial 16S rDNA sequencing of inserts.
Inserts from a total of 77 clones from the GC library were partially sequenced with the 530f primer and analyzed. The groups to which these clone inserts were affiliated are shown in Table 2. The majority of the clone sequences grouped with the proteobacterial phylum (56%), while 4% (3 clones; GC3, GC86, and GC109) grouped with the phylum Nitrospira. The sequences of GC3 and GC86 were 99% similar, while the sequence similarities between clone GC109 and clones GC3 and GC86 were 91.4 and 92.6%, respectively.
TABLE 2.
Phyla from the domain Bacteria represented in the seed sludge (GC) clone library determined by BLAST comparisons of partial clone insert sequences
| Phylum | % of clone library |
|---|---|
| Proteobacteria | |
| Alpha subclass | 5 |
| Beta subclass | 29 |
| Gamma subclass | 18 |
| Delta subclass | 4 |
| High G+C gram positive | 10 |
| Low G+C gram positive | 7 |
| Flexibacter/Cytophaga/Bacteroides | 5 |
| Nitrospira | 4 |
| Planctomycetales | 9 |
| Unaffiliated | 9 |
Analysis of Nitrospira clones.
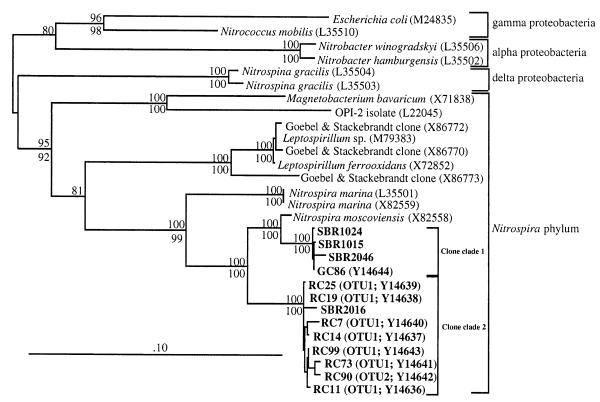
Near-complete insert sequences were determined for eight RC clones (seven from OTU 1 [RC7, RC11, RC14, RC19, RC25, RC73, and RC99] and the one from OTU 2 [RC90]) (Fig. 1), one of the three GC Nitrospira clones (GC86), and four clones (SBR1015, SBR1024, SBR2016, and SBR2046) phylogenetically grouped in the Nitrospira phylum and from a clone library prepared by Bond et al. (5). The data were phylogenetically analyzed as shown in Fig. 2. A similarity matrix of the 13 clone insert sequences and all those from N. moscoviensis and Nitrospira marina is shown in Table 3.
FIG. 2.
Evolutionary distance tree of the Nitrospira phylum and other known nitrite oxidizers in the domain Bacteria based on a comparative analysis of 1,030 nucleotides. Most bootstrap values greater than 92% from 100 resamplings for distance (numbers above branches) and parsimonious (numbers below branches) analyses are presented at the nodes. The outgroup, Bacteroides fragilis, is not shown in the tree. The bar represents 0.1 estimated change per nucleotide.
TABLE 3.
Similarity matrix showing the percent similarities among 16S rDNA sequences of N. moscoviensis, N. marina, and 13 near-complete sequences of clone inserts obtained from biomass from a full-scale BNR activated sludge plant or an NOSBR and clones for which the partial sequences had been previously reporteda
| Strain no. | Species or clone (accession no.) | % Sequence similarity with species of strain no.:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| 1 | N. moscoviensis (X82558) | |||||||||||||||
| 2 | SBR1024 | 96.3 | ||||||||||||||
| 3 | SBR1015 | 96.1 | 99.6 | |||||||||||||
| 4 | GC86 (Y14644) | 96.1 | 99.6 | 99.4 | ||||||||||||
| 5 | SBR2046 | 95.8 | 99.3 | 99.4 | 99.2 | |||||||||||
| 6 | RC25 (Y14639) | 93.4 | 93.4 | 93.6 | 93.6 | 93.1 | ||||||||||
| 7 | RC19 (Y14638) | 93.2 | 93.1 | 93.0 | 93.2 | 92.7 | 98.8 | |||||||||
| 8 | SBR2016 | 93.0 | 92.7 | 92.8 | 92.6 | 92.4 | 99.1 | 98.7 | ||||||||
| 9 | RC7 (Y14640) | 92.9 | 93.1 | 93.2 | 92.9 | 92.8 | 98.7 | 98.7 | 98.5 | |||||||
| 10 | RC14 (Y14637) | 92.8 | 93.0 | 93.1 | 93.1 | 92.7 | 98.7 | 98.9 | 98.5 | 99.3 | ||||||
| 11 | RC99 (Y14643) | 92.7 | 92.9 | 93.0 | 93.0 | 92.6 | 98.5 | 98.7 | 98.4 | 99.2 | 99.6 | |||||
| 12 | RC11 (Y14636) | 92.6 | 92.8 | 93.0 | 92.9 | 92.5 | 98.5 | 98.7 | 98.4 | 99.0 | 99.5 | 99.7 | ||||
| 13 | RC73 (Y14641) | 92.2 | 92.5 | 92.6 | 92.6 | 92.1 | 98.0 | 98.2 | 97.9 | 98.7 | 99.1 | 99.4 | 99.4 | |||
| 14 | RC90 (Y14642) | 92.1 | 92.1 | 92.3 | 92.2 | 91.8 | 98.1 | 98.6 | 98.0 | 98.1 | 98.6 | 98.8 | 98.8 | 99.0 | ||
| 15 | N. marina (X82559) | 88.7 | 88.2 | 88.3 | 88.3 | 87.8 | 88.1 | 87.6 | 87.2 | 87.2 | 87.1 | 87.1 | 87.1 | 86.5 | 86.6 | |
| 16 | N. marina (X35501) | 88.0 | 88.0 | 88.2 | 88.1 | 87.7 | 87.9 | 87.5 | 87.2 | 87.2 | 87.1 | 87.1 | 87.1 | 86.5 | 86.6 | 99.9 |
GC86 was obtained from a BNR activated sludge plant. RC clones were obtained from an NOSBR. Partial sequences for SBR clones are reported in reference 5. Values in boldface indicate the two environmental Nitrospira clone clades.
DISCUSSION
Our goal was to discover the possible nitrite-oxidizing microorganisms in wastewater treatment systems, since Wagner et al. (27) had unequivocally shown that Nitrobacter was not found in sludges by FISH probing. Until then, wastewater treatment personnel had presumed that Nitrobacter was the dominant nitrite oxidizer because it was commonly isolated from sludges. Additional support for this notion came from Mobarry et al. (15) who used FISH to observe clusters of Nitrobacter, closely juxtaposed with clusters of Nitrosomonas, in activated sludge and biofilm samples. However, by quantitative methods of rRNA extraction and slot blot hybridization, it was concluded that the contribution of Nitrobacter to nitrification was minor (15).
Nitrobacter (α subclass of the class Proteobacteria) can grow heterotrophically, while the remaining known nitrite oxidizers, Nitrospina (δ subclass), Nitrococcus (γ subclass), and Nitrospira (Nitrospira phylum), are unable to grow heterotrophically (9). To preclude the selective advantage that Nitrobacter may have gained from heterotrophic growth, we employed strategies that selected chemoautotrophic nitrite oxidizers. We attempted to significantly narrow the microbial community from a complex mixture with multiple functions (carbon, nitrogen, and phosphorus removal from domestic wastewater) to a single function (autotrophic nitrite oxidation) of reduced diversity. Our NOSBR has excellent nitrification capacities (7), and we investigated its microbial community structure.
Microscopy and culture-dependent methods.
The community is composed of complex morphological types and still retains the floccular nature of activated sludge. We were able to isolate a range of heterotrophs on both ANA and NA by classical sample dilution and spread plate inoculation and by micromanipulation. The occurrence in the NOSBR of heterotrophic nitrification and of other reactions such as aerobic denitrification cannot be ruled out, but the paucity of organic carbon in the reactor would slow such reactions. Organic carbon can theoretically come from the extracellular polymers that the bacteria in the flocs are producing and from dead microbial cells. The possibility of heterotrophic nitrification and aerobic denitrification is currently being further investigated. We were unsuccessful in isolating an autotrophic nitrite oxidizer. However, the isolation procedures used are perhaps not likely to favor this since we employed only growth on solid media and other groups have employed liquid media for the isolation of nitrite oxidizers (9).
Molecular biological methods.
Previous studies generating clone libraries with nonselected activated sludges (e.g., that of Bond et al. [5]) indicated that the diversity of the community would be too great to simplify by REA. Consequently, for the seed sludge library (GC), partial insert sequencing was immediately done rather than REA for grouping. As with other sludges, the diversity of the Merrimac sludge was significant. As well, the proteobacterial phylum dominated the library, comprising 56% of clones, with the majority of these being of the beta subclass of the class Proteobacteria (29% of all Bacteria). The next largest group was the high G+C gram-positive bacteria (10%). These findings are similar to those from other researchers where bacteria of the beta subclass and/or high G+C gram-positive bacteria are dominant in BNR systems (5, 13, 24, 25). We did not recover any Nitrobacter clones in the GC library but did identify three clones (4% of the library) most closely related to N. moscoviensis.
We employed REA for the grouping of clones from the NOSBR sludge library (RC) because we hypothesized that the microbial diversity should be reduced in this clone library. Culture-dependent methods had not supported such a hypothesis, but it is well recognized that these methods are heavily biased and the results obtained with them are unrepresentative of the true microbial composition (13). However, because of a range of biases in the methods, clone libraries are not considered adequate for generating quantitative information about the diversity of the microbial community from which the library was prepared (5). Nevertheless, REA proved extremely useful in grouping clones in the RC library because, when 102 clones were examined, one grouping comprised 90 (88% of the total) clones. Of the remaining 12 clones, 11 contained inserts originating from different bacteria with five from the Flexibacter/Cytophaga/Bacteroides phylum and five others from the proteobacterial phylum. Two Nitrobacter-like clones could not be unequivocally aligned with any genus, but more sequence data from these clones could clarify their affiliation. None of the clone inserts came from gram-positive bacteria, but 6 of the 16 isolates reported were gram positive. In addition, gram-positive bacteria were microscopically observed in the sludge. Cell lysis methods may not have been rigorous enough to lyse the gram-positive bacteria or the primers may have preferentially bound to the non-gram-positive templates in the PCR. Bond et al. (5) also found very few gram positives in two clone libraries from sludges, but Wagner et al. (25) hypothesize that gram positives could be responsible for phosphorus removal in BNR systems because increases in this population, as determined by FISH probing, were correlated with initiation of phosphorus removal in activated sludge systems.
The RC clone library was predominantly composed of clones (89% from OTU 1 and OTU 2) with inserts originating from bacteria whose closest relatives are in the Nitrospira phylum and are most similar to Nitrospira moscoviensis. Direct pairwise sequence comparisons between sequences in the two Nitrospira clone clades (see Fig. 2) showed that clone clade 1 (SBR1015, SBR1024, SBR2046, and GC86) had an average 16S rDNA similarity value of 99.4% (Table 3), while for clone clade 2 (RC7, RC11, RC14, RC19, RC25, RC73, RC90, RC99, and SBR2016) this value was 98.7% (Table 3). The average sequence similarity between the two clone clades was 92.8% (Table 3), while those between N. moscoviensis and clone clades 1 and 2 (Fig. 2) were 96.1 and 92.8%, respectively. The highest comparative value between an RC clone sequence and N. moscoviensis was 93.4% for RC25 (Table 3). From the sequence data analysis, the two clone clades would likely represent two separate species. This conclusion is drawn from discussions by Stackebrandt and Goebel (22), who note that organisms with rRNA sequence similarity values of less than 97.5% most likely represent different species.
Conclusions.
From the data presented in this paper, we suggest that the unknown nitrite-oxidizing bacteria in activated sludges belong in the Nitrospira phylum. In the meantime, both Wagner et al. (27) and Schramm et al. (20) have discovered clones originating from Nitrospira in industrial activated sludge and biofilm processes, respectively. Both the seed sludge from the Merrimac plant and the highly selected autotrophic nitrifying bioreactor biomass contain these organisms. Clones with inserts originating from Nitrobacter were not recovered in the GC library, but two RC clones (RC44 and RC57) were closely related to this bacterium and its relatives. In the meantime, we have prepared Nitrospira-specific primers and in preliminary studies involving a PCR test (data not shown) have positively correlated the presence of Nitrospira with excellent nitrification in full-scale activated sludge plants. In addition, in processes where nitrification is poor, these bacteria are absent. These PCR experiments will be complemented with FISH studies to quantify the numbers of Nitrospira in nitrifying systems. However, we will use our PCR test as a screening in advance of FISH probing of sludges to show the presence of nitrospiras. Data from studies on nitrification kinetics from the enhanced nitrite-oxidizing culture in the NOSBR (7) will be combined with numbers of nitrospiras from FISH probing and used in mathematical modelling.
ACKNOWLEDGMENTS
P.C.B. had an Australian Postgraduate Award, and the research was financially supported by the CRC Waste Management & Pollution Control Ltd., a center established by the federal government of Australia.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Argaman Y. Biological nutrient removal. In: Martin A M, editor. Biological degradation of wastes. New York, N.Y: Elsevier Applied Science; 1991. pp. 85–101. [Google Scholar]
- 3.Blackall L L. Molecular identification of activated sludge foaming bacteria. Water Sci Technol. 1994;29:35–42. [Google Scholar]
- 4.Blackall L L, Seviour E M, Cunningham M A, Seviour R J, Hugenholtz P. “Microthrix parvicella” is a novel, deep branching member of the actinomycetes subphylum. Syst Appl Microbiol. 1994;17:513–518. [Google Scholar]
- 5.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrell, P., L. L. Blackall, and J. Keller. Characterisation of the bacterial consortium involved in nitrite oxidation in activated sludge. Water Sci. Technol., in press.
- 7.Burrell, P. C., L. L. Blackall, and J. Keller. Unpublished data.
- 8.Castignetti D, Hollocher T C. Heterotrophic nitrification among denitrifiers. Appl Environ Microbiol. 1984;47:620–623. doi: 10.1128/aem.47.4.620-623.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 10.Halling-Sørensen B. Biological nitrification and denitrification. In: Halling-Sørensen B, Jørgensen S E, editors. The removal of nitrogen compounds from wastewater. Amsterdam, The Netherlands: Elsevier; 1993. pp. 41–53. [Google Scholar]
- 11.Halling-Sørensen B, Jørgensen S E, editors. The removal of nitrogen compounds from wastewater. Amsterdam, The Netherlands: Elsevier; 1993. [Google Scholar]
- 12.Hovanec T A, DeLong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kämpfer P, Erhart R, Beimfohr C, Bohringer J, Wagner M, Amann R. Characterization of bacterial communities from activated sludge—culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb Ecol. 1996;32:101–121. doi: 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- 14.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: Academic Press; 1991. pp. 115–175. [Google Scholar]
- 15.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orso S, Gouy M, Navarro E, Normand P. Molecular phylogenetic analysis of Nitrobacter spp. Int J Syst Bacteriol. 1994;44:83–86. doi: 10.1099/00207713-44-1-83. [DOI] [PubMed] [Google Scholar]
- 17.Randall C W. Introduction and principles of biological nutrient removal. In: Randall C W, editor. Design and retrofit of wastewater treatment plants for biological nutrient removal. Lancaster, Pa: Technomic Publishing Company Inc.; 1992. pp. 7–84. [Google Scholar]
- 18.Robertson L A, Kuenen J G. Physiology of nitrifying and denitrifying bacteria. In: Rogers J E, Whitman W B, editors. Microbial production and consumption of greenhouse gases: methane, nitrogen oxides, and halomethanes. Washington, D.C: American Society for Microbiology; 1991. pp. 189–199. [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Schramm A, De Beer D, van den Heuvel H, Ottengraf S, Amann R. In situ structure/function studies in wastewater treatment systems. 1997. Presented at the Second International Conference on Microorganisms in Activated Sludge and Biofilm Processes, Berkeley, Calif. [Google Scholar]
- 21.Skerman V B D. A new type of micromanipulator and microforge. J Gen Microbiol. 1968;54:287–297. doi: 10.1099/00221287-54-2-287. [DOI] [PubMed] [Google Scholar]
- 22.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 23.Teske A, Alm E, Regan J M, Toze S, Rittmann B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner M, Amann R, Lemmer H, Manz W, Schleifer K H. Probing activated sludge with fluorescently labeled rRNA targeted oligonucleotides. Water Sci Technol. 1994;29:15–23. [Google Scholar]
- 25.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner M, Rath G, Amann R, Koops H P, Schleifer K H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 27.Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]