Abstract
During our whole lifespan, from conception to death, the epigenomes of all tissues and cell types of our body integrate signals from the environment. This includes signals derived from our diet and the uptake of macro- and micronutrients. In most cases, this leads only to transient changes, but some effects of this epigenome programming process are persistent and can even be transferred to the next generation. Both epigenetic programming and redox processes are affected by the individual choice of diet and other lifestyle decisions like physical activity. The nutrient-gene communication pathways have adapted during human evolution and are essential for maintaining health. However, when they are maladaptive, such as in long-term obesity, they significantly contribute to diseases like type 2 diabetes and cancer. The field of nutrigenomics investigates nutrition-related signal transduction pathways and their effect on gene expression involving interactions both with the genome and the epigenomes. Several of these diet-(epi)genome interactions and the involved signal transduction cascades are redox-regulated. Examples include the effects of the NAD+/NADH ratio, vitamin C levels and secondary metabolites of dietary molecules from plants on the acetylation and methylation state of the epigenome as well as on gene expression through redox-sensitive pathways via the transcription factors NFE2L2 and FOXO. In this review, we summarize and extend on these topics as well as those discussed in the articles of this Special Issue and take them into the context of redox biology.
Keywords: Nutrigenomics, Epigenomics, Redox regulation, Lifestyle, Evolution, Epigenetic programming, ROS
Abbreviations
- AKT
AKT serine/threonine kinase
- ATP
adenosine triphosphate
- CNV
copy number variant
- CREBBP
CREB binding protein
- DNMT
DNA methyltransferase
- DOHaD
Developmental Origins of Health and Disease
- FAD
flavin adenine dinucleotide
- FOXO
forkhead box protein, class O
- G6PC1
glucose-6-phosphatase catalytic subunit 1
- G6PD
glucose-6-phosphate dehydrogenase
- GLUT
glucose transporter
- GWAS
genome-wide association study
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- IGF1R
insulin-like growth factor 1 receptor
- IGF2
insulin growth factor 2
- INSR
insulin receptor
- KDM
lysine demethylase
- KEAP1
kelch-like ECH-associated protein 1
- KMT
lysine methyltransferase
- MBD
methyl-DNA-binding domain
- NAD
nicotinamide adenine dinucleotide
- NFE2L2
nuclear factor erythroid-derived 2-like 2, also called NRF2
- NOX
NADPH oxidase
- PCK1
phosphoenolpyruvate carboxykinase 2, cytosolic
- PI3K
phosphoinositide-3-kinase
- PTEN
phosphatase and tensin homolog
- PTPN1
protein tyrosine phosphatase non-receptor type 1
- ROS
reactive oxygen species
- SAM
S-adenosyl-l-methionine
- SIRT
sirtuin
- SLC
solute carrier
- SNV
single nucleotide variant
- T2D
type 2 diabetes
- TET
ten-eleven translocation
- TOR
target of rapamycin
- TP53
tumor protein p53
- TSS
transcription start site
- UDP
uridine diphosphate
- VSC
volatile sulfur compound
- β-OHB
β-hydroxybutyrate
1. Introduction
For thermodynamic reasons, life is impossible without the uptake of nutrients. Macronutrients, i.e. the protein, lipid and carbohydrate components of our diet, are used as providers of building blocks, such as amino acids, fatty acids and glucose as well as for the anabolism of endogenous macromolecules required for the structure and function of organs, tissues and cells. In parallel, macronutrients are used to fuel metabolism in general, as their catabolism provides energy in the form of NADH and ATP [1]. The use of molecular oxygen (O2) as the terminal electron sink in aerobic metabolism, driving the process of oxidative phosphorylation at the inner mitochondrial membrane, renders the extraction of energy from macronutrients far more efficient than anerobic pathways like glycolysis. This also implies that redox processes are at the core of human metabolism. This is further emphasized by the fact that several micronutrients, which in contrast to macronutrients are not used as energy sources but are required as cofactors in metabolism, are redox-active. This includes vitamins like vitamins C and E, but also minerals that participate in cellular redox processes, such as Fe, Cu and Se [2,3].
Another implication of O2 being used in metabolism is that its partial reduction generates reactive oxygen species (ROS), such as superoxide and hydrogen peroxide [4]. While in the early days of research on “oxidative stress” these byproducts were regarded as undesired and potentially damaging, it is now understood that the endogenous generation of ROS contributes to signaling events [5]. This includes nutrient-triggered signal transduction cascades that directly or indirectly affect the genome and epigenome of all cell types by modulating the activity of transcription factors and chromatin-modifying enzymes [6]. This communication of dietary molecules with the genome and epigenome is a central topic of nutrigenomics [7,8]. The purpose of this summarizing review is to provide an overview on the contributions of redox processes to this communication.
2. Nutrients and signaling
2.1. Evolutionary and transgenerational aspects
Macro- and micronutrients, such as polyunsaturated fatty acids [9] and vitamin D [10], directly or indirectly affect the genome and epigenome. Nutrient availability has therefore been a driving factor in evolutionary adaptation. The nutrient-(epi)genome interaction is a lifelong process starting at conception and ending with death, encompassing the adaptation and modulation of gene expression in all organs, including metabolic powerhouses, such as adipose tissue, skeletal muscle, liver and pancreas, but also in brain and the immune system. Thus, for understanding diet-(epi)genome communication one needs to take a whole body's perspective.
Humans differ from one another in properties like eye color [11] but also with respect to their physiological characteristics, such as lactase persistence or non-persistence [12]. The differences that characterize humans as “individuals” relate only to some 0.5 % variations of the genome but are significantly higher on the level of the epigenomes of the >400 different tissues and cell types forming the human body. Accordingly, also the risk of developing most common non-communicable diseases like type 2 diabetes (T2D) or cancer has both a genetic and an epigenetic basis [13]. Some of these differences in these so-called “traits” are the result of an evolutionary adaptation, referred to as “positive selection” [14]. During the past 50,000 years modern humans (Homo sapiens) migrated from Africa to less sunny higher latitudes, such as in Europe, or to high altitudes like in the Andes or the Himalayas, and adapted to these significantly different geographic locations. Moreover, some 10,000 years ago humans started agriculture and domesticated many animal and plant species, so that their diet's content in starch or milk increased [15]. Within this adaption period some 10 % of all protein-coding genes may have undergone positive natural selection [16]. Skin, digestive tract and immune system are in more intensive contact with the environment and therefore more affected by this adaption process than other organs [17]. In addition, there are also randomly occurring changes of the genome sequence, which can cause genetic drifts [18,19], representing approximately 50 bp per individual and generation [20,21]. Nevertheless, it takes hundreds to thousands of years, i.e., dozens to hundreds of generations, until a specific trait gets common within a population. In contrast, the Anthropocene (the human epoch) has started only two to three generations ago [22] and comes not only with geologically measurable alterations of the planet but also with significant changes in human behavior, such as a sedentary lifestyle paired with a surplus of energy-rich foods.
2.2. Missing heritability and nutritional epigenetics
The 1000 Genomes Project (www.internationalgenome.org) [23] indicated that two individuals differ, on average, by 4–5 million single nucleotide variants (SNVs) and some 1000 copy number variants (CNVs). This means that approximately 0.5 % of a person's genome sequence differ from the public reference genome [24]. Interestingly, there are, on average, some 150 SNVs causing protein truncation, 10,000 leading to amino acid changes and 500,000 SNVs affecting transcription factor binding sites [23]. Based on other, even lager whole genome sequencing projects, such as TOPMed [25] and Genomics England [26], there are nearly 500 million known SNVs. Over the past 20 years a large number of genome-wide association studies (GWAS) have described statistically significant associations between SNVs and various traits [27,28]. This resulted in dozens to hundreds of SNVs for various diseases, but, with a few exceptions like type 1 diabetes [29] and macular degeneration [30], the sum of presently known SNVs for a given common disease explain less than 20 % of its risk [31]. This means that having the whole genome sequence of an individual and knowing all SNVs will explain no more than 20 % of the disease risk, which precludes any reliable predictions. The remaining 80 % of the disease risk are often referred to as “missing heritability”. Whole genome sequencing of millions of individuals may identify rare SNVs with large effect size [32], but their contribution to the missing heritability will be minor. It is more likely that environmental exposures, which directly or indirectly lead to epigenetic changes, have the major contribution to most traits. This suggests that, considering all traits establishing the phenotype of an individual, environment and epigenetics plays a larger role than genetics. Whereas the genetic composition of all cells of an individual will largely stay stable over a lifespan as long as the person does not develop cancer, a substantial part of the epigenome dynamically responds to signals from the environment. Many of these signals are related to nutritional molecules or their metabolites [33] (Fig. 1).
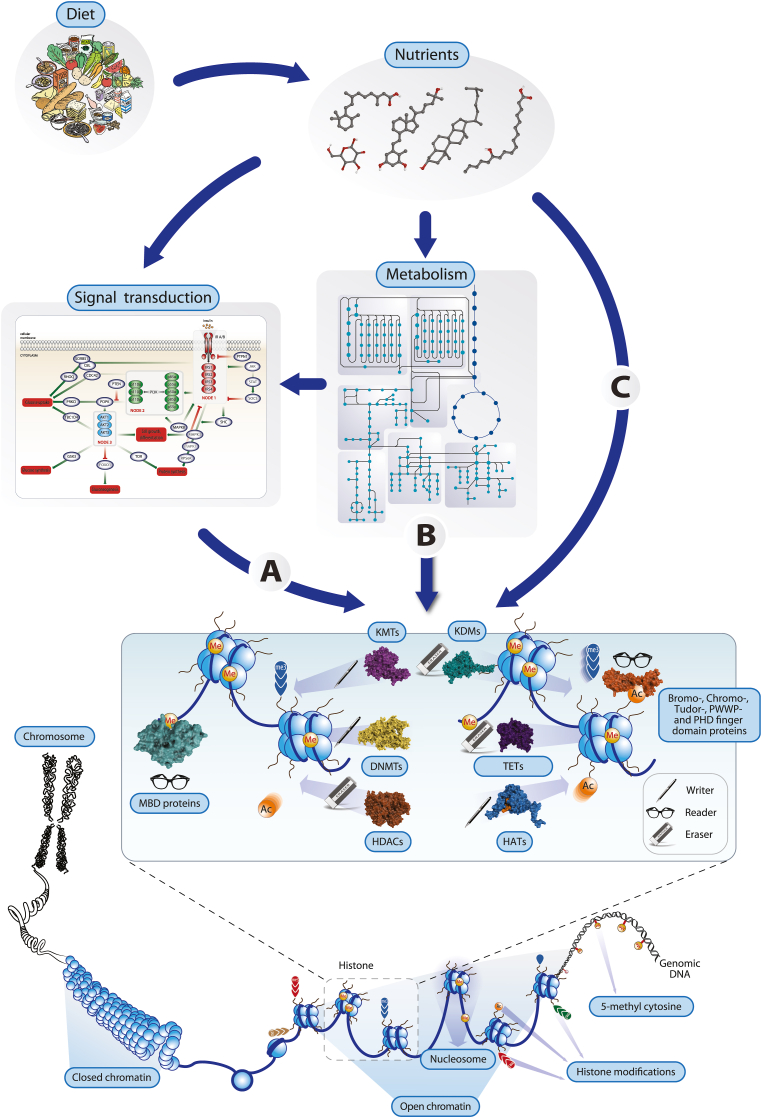
Fig. 1.
Concepts of nutritional epigenetics. Changes in the level of nutrients deriving from diet affect the epigenome of tissues and cell types by affecting chromatin-modifying enzymes such as KMTs, DNMTs and HATs (“writers”), KDMs, TETs and HDACs (“erasers”) and MBD (methyl-DNA-binding domain) proteins (“readers”), via the modulation of signal transduction pathways (A), after their metabolism (B) or through direct binding (C). The activity of these enzymes determine the local chromatin structure through covalent modifications of histones and genomic DNA and mediate a between the active state (euchromatin) and inactive state (heterochromatin). This is essential for controlling gene expression.
Genomic DNA is part of chromatin and covered by proteins. The latter are predominantly nucleosome-forming histone proteins that govern accessibility of DNA. This accessibility is modulated by histone modification, the patterns of which are therefore a physical expression of the epigenome [34]. More specifically, the epigenome is composed of genome-wide patterns of DNA methylation at cytosines, post-translational modifications of histone proteins and the 3D organization of chromatin into loops. These epigenetic patterns can be changed through the action of chromatin-modifying enzymes, such as i) DNA methyltransferases (DNMTs) and TET (ten-eleven translocation) enzymes, which attach or eradicate methyl groups from DNA, ii) histone acetyltransferases (HATs) and histone deacetylases (HDACs) that add or remove acetyl groups from lysines of histone proteins and iii) lysine methyltransferases (KMTs) that provide histones with methyl groups and lysines demethylases (KDMs) that erase them. The approximately 200 chromatin-modifying enzymes encoded by the human genome are often the end point of signal transduction cascades, i.e., their activity is regulated by environmental signals. Interestingly, a wide spectrum of plant-based metabolites, such as epicatechin, genistein, resveratrol, catechin and curcumin from green tea, soy, red grapes, cocoa and curcuma, respectively, are able to modulate the activity of chromatin-modifying enzymes [35]. In addition, intermediary metabolites representing energy metabolism, such as α-ketoglutarate, NAD+, FAD, acetyl-CoA, SAM (S-adenosyl-l-methionine), UDP-N-acetylglucosamine or ATP, act as cofactors or substrates of chromatin-modifying enzymes [36]. Importantly, the redox state of a cell depends on the levels of the oxidized and reduced form of the cofactor NAD, i.e., the NAD+/NADH ratio is inversely proportional to the energy state of a cell [37]. Since HDACs of the sirtuin (SIRT) family use NAD+ as a cofactor, their enzymatic activity and impact on the epigenome increases during fasting [38]. Thus, specific nutritional compounds as well as the energy status of the cell affect the epigenome in multiple ways and induce shifts from active euchromatin via facultative heterochromatin to inactive heterochromatin or vice versa [39].
In summary, the mechanisms of nutritional epigenetics (also sometimes referred to as nutriepigenomics [40]) provide an explanation of how exposure to environmental stimuli, the largest of which is the daily uptake of diet, translate to changes in the epigenome. In terms of evolutionary and adaptive pressure, epigenetics is able to respond faster than genetics and therefore may explain the majority of the missing heritability.
3. Lifelong epigenetic programming
Epigenetics comprises functionally relevant changes of chromatin that do not alter the sequence of the genome [41]. The most prominent example of the impact of epigenetics is embryogenesis, in which totipotent stem cells of the inner cell mass of the blastocyst differentiate into the more than 400 tissues and cell types that form the human body [42]. In the context of this process the cells of the embryo are epigenetically programmed through accurately timed cascades of changes in DNA and histone methylation (Fig. 2A). A similar procedure of epigenetic programming takes place in adult stem cells, e.g., in the bone marrow, colon and skin, and leads to lifelong replenishment with new blood cells, enterocytes and keratinocytes, respectively [43]. In both cases, in particular the patterns of DNA methylation create a so-called epigenetic landscape that preserves terminally differentiated cells in tight borders, so that they keep their cellular identity for the rest of their life [44]. Thus, epigenetic marks that are created during cellular differentiation are stable, i.e., this type of epigenetic programming is irreversible.
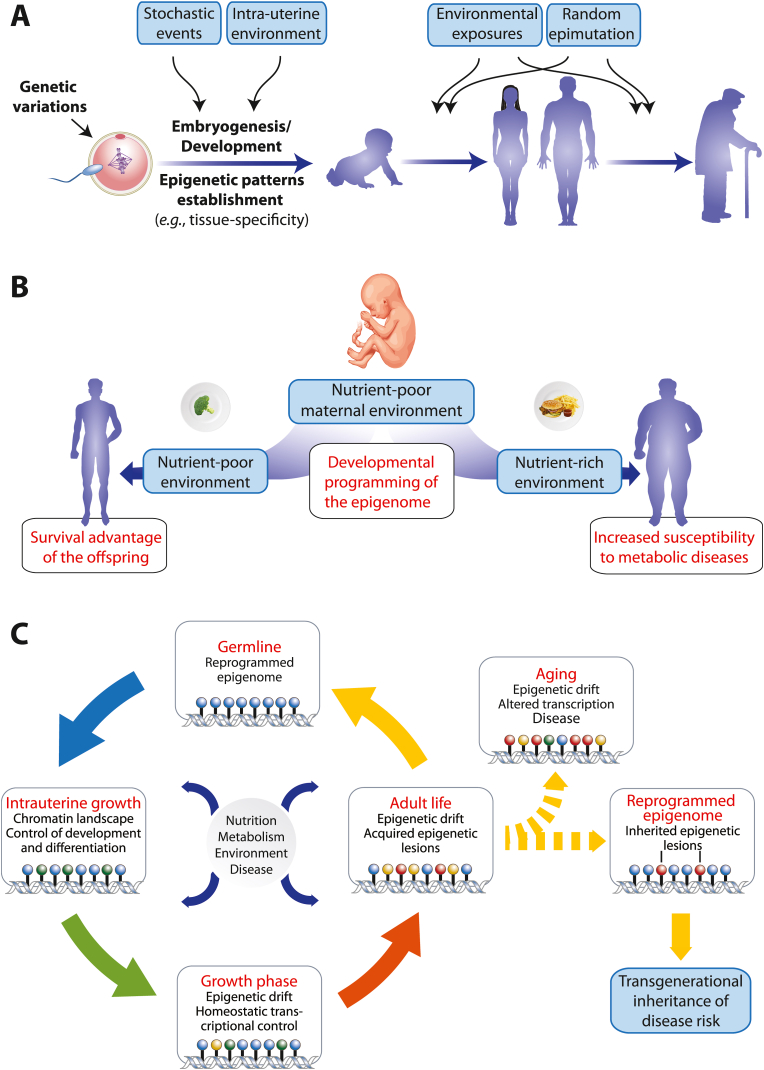
Fig. 2.
Lifelong epigenetic programming. In every phase of life the phenotype of an individual is based on genetics, epigenetics and environment (A). This implies that we have it to a large extent in our own hands to stay healthy and to avoid getting major common diseases. Intrauterine stressors, including maternal undernutrition or placental dysfunction can initiate abnormal patterns of development, histone modifications and DNA methylation (B). During embryogenesis epigenetic marks, such as DNA methylation and histone modifications, are established in order to maintain cell lineage commitment (C). After birth, this epigenetic landscape stays dynamic throughout lifespan and responds to nutritional, metabolic, environmental and noxious signals. Epigenetic drifts are part of homeostatic adaptations and should keep the individual in good health. However, when an adverse epigenetic drift compromises the capacity of metabolic organs to adequately respond to challenges as provided by nutrition and chronic inflammation, the susceptibility to diseases increases. Some of these acquired epigenetic marks can be inherited to subsequent generations when they escape epigenetic reprograming during gametogenesis.
Epigenetic programming can also lead to transient chromatin marks that control short-term effects on the accessibility of TSS (transcription start site) and enhancer regions, which are the binding sites of RNA polymerases and inducible transcription factors, respectively [45]. In addition, extra- and intracellular signals affect via signal transduction cascades the activity of transcription factors and chromatin-modifying enzymes. Most of these signaling molecules are hydrophilic, such as cytokines, growth factors and peptide hormones, and act via membrane receptors, but some lipophilic molecules, such as steroids, oxysterols, polyunsaturated fatty acids and vitamin D3 [10] function via the direct binding to a class of transcription factors referred to as nuclear receptors [46]. Since most of these signals fluctuate in their intensity, they have only short-term effects on transcription factors and chromatin-modifying enzymes, so that gene expression and the local chromatin status are mostly only transiently affected. However, the repeated exposure to the same signal may also lead to more persistent chromatin marks [47]. In this way, our epigenome can memorize environmental events, such as what and how much we have eaten, whether we have been in contact with microbes or whether we have been exposed to stressful stimuli (including oxidative stress) in any other way.
Importantly, the dynamic component of epigenome implies that some epigenetic programming events are reversible [48]. For example, an unhealthy lifestyle, such as food excess paired with physical inactivity, over many years causes epigenetic programming of metabolic tissues that causes insulin resistance. This programming process results in an epigenetic memory, such as histone methylation changes within aortic endothelial cells of T2D patient in response to increased glucose exposure [49]. The “hyperglycemic memory” can lead to an increased risk of macrovascular complications and organ damage even years after initiation of glucose-lowering therapy. Thus, only a significant lifestyle change that leads to the reprogramming of the dynamic component of the epigenome has the potential to reverse the disease condition as long as no irreversible tissue damage, such as β-cell death, has happened. Moreover, the epigenome of metabolic organs, such as skeletal muscle and adipose tissue, is able to memorize nutrient availability and energy status of the body and adapt accordingly, i.e., these tissues “memorize” and integrate via changes of epigenetic patterns in the level of DNA methylation, histone modifications and 3D chromatin structure how much we have eaten and how much physical activity we have had [50]. For example, in caloric restriction, i.e., a dietary regime where only 70–80 % of the recommended dietary (caloric) intake is provided over a longer time period, NAD+ levels and SIRT activity rise and the level of histone acetylation declines [[51], [52], [53]]. In contrast, short-chain fatty acids like the ketone body β-OHB (β-hydroxybutyrate), whose levels are increased after the consumption of fiber, can act as HDAC inhibitors, increase the histone acetylation levels and affect the proliferation of colon cells [40,54]. This suggests that at least part of the beneficial effect of fiber-rich diet, such as the prevention of chronic inflammation and cancer of the colon, is based on epigenetic programming [55].
The memory function of the epigenome can also explain, how under- or overnutrition in the prenatal phase can lead to a postnatal predisposition for obesity, T2D and the metabolic syndrome. Human fetuses can of course not be studied, but there are so-called natural experiments, where the exposure to severe environmental conditions was not under experimental control. For example, in the Dutch Hunger Winter occurring in the Netherlands during the winter of 1944/45 fetuses of their starving mothers were exposed to extreme undernutrition in utero [56,57]. This malnutrition led to impaired growth of the fetuses. The resulting low birth weight favors a thrifty phenotype that is epigenetically programmed to use nutritional energy efficiently. Thus, the children were prepared for a future environment with low resources during adult life. Even many decades after birth the individuals undernourished in utero showed subtle (<10 %) changes in DNA methylation at several loci in adulthood, e.g., at the regulatory region of the imprinted IGF2 (insulin growth factor 2) gene [58]. This epigenetic pattern is associated with an increased risk of obesity, dyslipidemia and insulin resistance, when the respective individuals are exposed to an obesogenic environment. Since genomic DNA gets nearly completely demethylated in early embryogenesis, DNA methylation is particularly sensitive to disturbances of correct epigenetic programming, such as through under- or overnutrition, during the prenatal phase [59].
Similar observations of a rise in T2D prevalence have been made for survivors of the Ukraine Famine (1932-33) [60] and the Chinese Famine (1959-61) [61]. These examples led to the Developmental Origins of Health and Disease (DOHaD) concept indicating that early developmental events, such as perturbations of the nutritional state in utero, have significant effects on disease risk as adult [62] (Fig. 2B). In times of continued famine, this epigenetic sensitizing is a survival advantage, whereas in times of plenty of food it may drive the individual into obesity and T2D. In this way, diseases that are related to the exposure with environmental factors, such as overnutrition causing obesity or microbe infection causing inflammation, have a large epigenetic component [33,63]. Furthermore, diseases that have their onset a long time before the phenotype emerges, such as cancer and T2D, are most susceptible to stepwise epigenetic programming [40].
The epigenomes of all tissues and cell types of our body can record information about their external and internal stimulation in the form of epigenetic drifts, i.e., as changes in the patterns of DNA methylation, histone modifications and 3D organization of chromatin [64,65]. Hypermethylation of CpG islands close to the regulatory regions of key genes, such as TP53 (tumor protein p53), is an example of an epigenetic drift, which significantly contributes to the risk for cancer (Fig. 2C). Avoiding such epimutations, e.g., by dietary polyphenols that affect the activity of DNMTs, will contribute to the prevention of disease onset through a healthy diet [66]. Alterations of histone acetylation patterns are mostly transient, while changes in histone methylation and in particular in DNA methylation are more persistent [67]. In part, this is related to the fact that demethylation occurs via a multistep process that involves an intermediate oxidation requiring enzymes kept active by reductants such as ascorbate (see below).
When epigenetic programming through environmental exposures, including dietary compounds, affects the epigenome of germ cells, there is the chance that epimutations get inherited [68,69]. However, the nearly complete erasure of the DNA methylome during early embryogenesis will filter most epigenetic marks out and only a few selected positions, which may be protected by proteins that bind to methylated DNA, will reach the next generation. Thus, epigenetic inheritance is not as persistent as genetic inheritance [70]. Interestingly, it was observed that populations like Polynesians or Native Americans, such as the Pima, that made a transition from famine to food surplus within just 1–2 generations, are under a significantly higher risk for developing obesity, T2D and the metabolic syndrome, than those that were improving their nutritional conditions gradually over many generations [71]. The ancestors of these populations may have been evolutionarily selected for energetic efficiency, i.e., they went through an evolutionary bottleneck and developed a thrifty metabolism. Thus, populations in countries with rapid changes in urbanization and economic development have an increased risk of the different features of the metabolic syndrome [72].
Taken together, epigenetic programming is an important process that directs the properties and responsiveness of the tissues and cell types forming the human body not only in the prenatal phase but also postnatal and in adult live.
4. Nutrition and aging: from the free radical theory to nutritional epigenetics
Aging is a complex natural phenomenon that applies to everyone, but its rate is individual [73]. Like the risk for common diseases, the rate of aging is a trait that has a genetic and an environmental/epigenetic component. Although longevity has a genetic basis, the process of aging is predominantly (by more than 70 %) determined by non-genetic factors [74]. These are largely lifestyle choices, such as smoking or physical inactivity, but also many environmental factors like air pollution and other stressful stimuli [75]. Aging and metabolism are associated with an oxidative component, giving rise to theories like the “free radical theory of aging” or the “mitochondrial theory of aging” [76,77]. These redox-related theories of aging have been expanded and rephrased, e.g., as “cell signaling disruption theory of aging” [78] by integrating findings [79] like these: (i) antioxidant supplementation has been without beneficial effect with respect to attenuating aging, (ii) signal transduction pathways have been identified that not only respond to nutrient supply, but also regulate aging and (iii) these pathways are part of the cellular redox signaling network, such as insulin signaling via FOXO (forkhead box protein, class O) transcription factors [80].
Animal models suggest that a low basal metabolic rate, which is caused by caloric restriction, paired with increased responsiveness to environmental signals has a major impact on the lifespan of worms, flies and mice [81]. The effect of non-genetic factors on the rate of aging can be explained by the modulation of signal transduction pathways that affect the activity of chromatin-modifying enzymes and transcription factors, i.e., by direct and indirect effects on the epigenome. As already discussed for diseases (Fig. 2C), the process of aging can also result in epigenetic drifts [82]. For example, metals and metalloids like cadmium, arsenic and mercury, air pollutants like carbon black particles and benzene or endocrine disruptors like diethylstilbestrol, bisphenol A and dioxin not only affect the integrity of DNA but also cause epimutations, such as changes in DNA methylation patterns. Thus, epigenetic modifications are a major contributor to the aging process [82].
Alterations in the DNA methylome are found in the context of chronological aging but are also associated with cancer [83]. The methylation status of a few hundred CpG islands, which can be easily measured from skin or white blood cells, were identified as markers for biological age [84]. Correlating the chronological and the biological age with each other indicates significant interindividual variations. For example, at a given chronological age some individuals show a “younger” epigenome, i.e., less than the average extent of epigenetic drift compared to young persons, while that of others is significantly older, i.e., their epigenome exhibits more drift. Since the epigenetic programming of the latter is already progressed, the respective individuals more likely develop age-related diseases and eventually may die at younger chronological age than persons with a younger epigenetic age. Moreover, the different organs of an individual do not age proportionally but respond in a tissue-specific way to environmental challenges, such as nutritional overload or other forms of stress. For example, the liver of an obese person may age faster than his/her skeletal muscles [85]. Thus, the pace of epigenetic programming during the aging process can be considered as an epigenetic clock that serves as a more accurate predictor for mortality by all causes than other biomarkers of aging, such as telomere length [84].
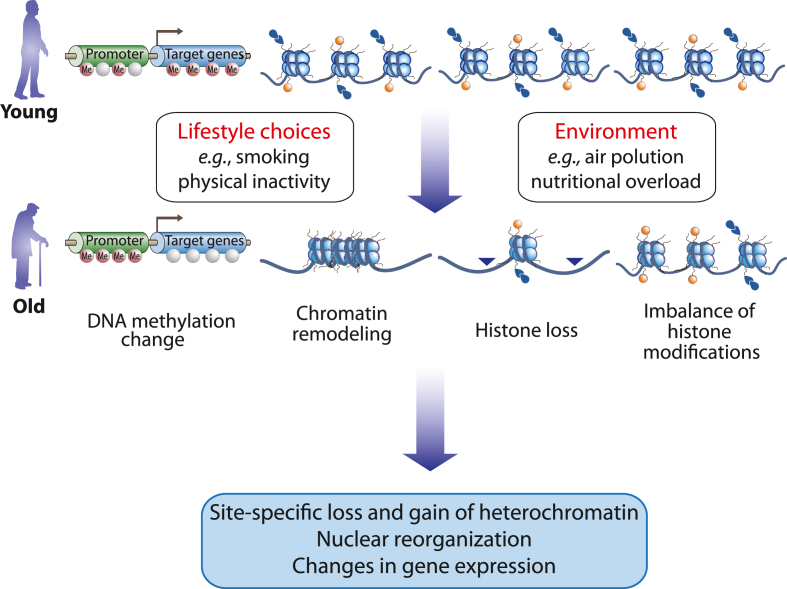
Epigenetic alterations, i.e., epimutations or epigenetic drifts, and genomic instability due to point mutations, deletions, insertions, amplifications and translocations are both hallmarks of aging as well as of cancer [86]. Thus, the processes of aging and tumorigenesis mechanistically overlap. At a cellular level, senescence may be a means of preventing tumorigenesis through attenuation of cell cycle progression. Accordingly, basically all tissues and cell types of the human body age (although at different rates), while only in some 50 % of the population one or the other tissue develop a detectable form of cancer. Cells of young individuals show a robust transcriptome and normal chromatin states, while with aging the transcriptome becomes unstable, and aberrant chromatin states are accumulating. This involves changes in DNA methylation, such as a global hypomethylation primarily lost at repetitive genomic regions and a focal hypermethylation at CpG islands close to TSS regions, a general loss of histones due to local and global chromatin remodeling and an imbalance of activating and repressive histone modifications (Fig. 3). This leads to a site-specific loss and gain in heterochromatin, such as senescence-associated heterochromatin foci, a significant nuclear reorganization and changes in gene expression, such as the upregulation of genes involved in cell adhesion and the downregulation of those related to cell cycle regulation, DNA repair and replication.
Fig. 3.
Epigenetic markers of aging. The hallmarks of aging “epigenetic alterations” includes changes in gene expression and chromatin remodeling, a loss of histones and the imbalance of activating and repressive histone modifications.
Many of the changes in the transcriptome, up- and downregulation, negatively affect longevity [87]. All these changes are heavily dictated by lifestyle choices and environmental stimuli. This includes nutrient availability and levels of intracellular metabolites, such as NAD+, acetyl-CoA or α-ketoglutarate. The peptide hormone insulin and its receptors, the amino acid-sensing kinase TOR (target of rapamycin) and the NAD+-sensing chromatin-modifying enzyme SIRT1 are central elements of nutrient signal transduction pathways that are conserved from worm to human [88]. These pathways inform the epigenome on nutrient availability. They act as proaging, when food is available, and as antiaging under fasting conditions or when one or the other component of the pathway is knocked down [89].
In summary, the process of aging involves epigenetic programming, i.e., it creates epigenetic drifts in response to environmental stimuli. Since many of these stimuli relate to nutritional molecules, their metabolites or intermediary metabolites, the principles of nutritional epigenetics help to understand the molecular basis of aging.
5. Redox regulation in nutritional (epi)genomics
The communication and interaction between dietary molecules and the human genome as well as the epigenome is a central aspect of nutrigenomics. Redox processes govern several of these communication pathways. A relation between diet/nutrition and oxidative stress has long been recognized, e.g., through the obvious liaison between metabolism and the endogenous generation of ROS as well as owing to dietary components resulting in antioxidant metabolites. Urate, the breakdown product of purines in humans, which is present at relatively high concentrations (300–400 μM) in human plasma, is a famous example of the latter: a cellular urate transporter, SLC2A9 (GLUT9), is upregulated by tumor suppressor protein TP53, likely in order to exploit urate antioxidant activity [90].
A free radical component has been described for a plethora of diseases and pathologies, including nutrition-related disorders, such as T2D or cancer, as well as more confined phenomena such as favism, which is an acute hemolytic syndrome that can occur after eating fava beans in individuals with G6PD (glucose-6-phosphate dehydrogenase) deficiency. However, in many instances it is not clear whether the link between disorder and ROS is a cause-effect relationship or rather a mere association [4]. Toning it down from “disease” to “stress”, dietary (or nutritional) oxidative stress was defined as “an imbalance between the prooxidant load and the antioxidant defense as a consequence of excess oxidative load or of inadequate supply of the organism with nutrients” [91]. This rather broad definition was then further broken down by defining postprandial oxidative stress as “characterized by an increased susceptibility of the organism toward oxidative damage after consumption of a meal rich in lipids and/or carbohydrates” [91]. These definitions implicitly refer to stress and nutritional inadequacy, which are potentially damaging. Moreover, they usually refer to what happens with dietary compounds after their absorption into the bloodstream.
It is now clear that the generation of ROS, including superoxide or hydrogen peroxide, per se does not equal damage through oxidative stress, but rather may be a component of cellular signal transduction cascades. In contrast to distress this is referred to as eustress [92], i.e. a beneficial stress that, e.g., causes adaptation to an adverse environment. Moreover, processes prior to absorption, e.g., interactions with the gut microbiome, need to be considered when discussing the relation between diet, redox processes and cellular signaling. An example illustrating both aspects is the recently identified thiol oxidase activity of SELENBP1 (selenium binding protein) [93], which is strongly expressed in differentiated enterocytes [94] and catalyzes the oxidation of methanethiol as well as other volatile sulfur compounds (VSC) to generate H2O2 and the gasotransmitter H2S [3]. As the gut microbiome is a major source of VSCs, it establishes an interaction between ingested food, microbial degradation thereof, and enterocytic generation of H2O2 and H2S (Fig. 4A). This is of significance with respect to the “communication between diet and genome”, because H2O2 and H2S are not necessarily damaging, but also redox-regulatory molecules that are involved in the modulation of cellular signal transduction cascades, including nutrient-responsive cascades.
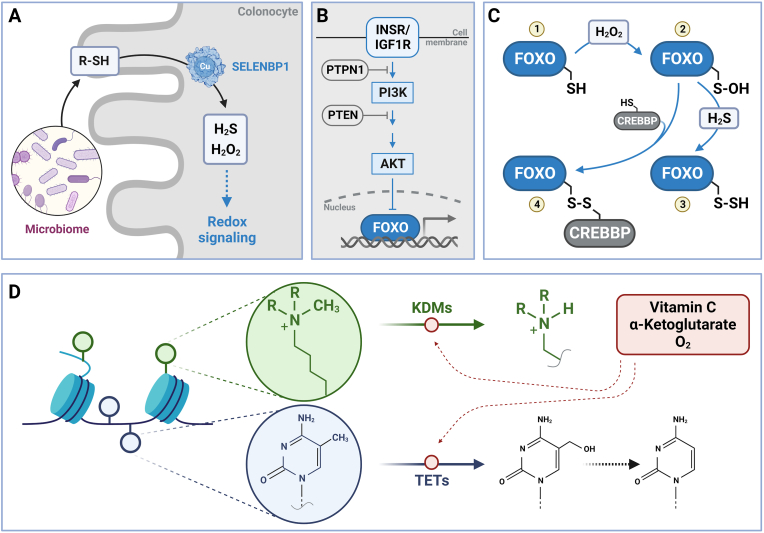
Fig. 4.
Redox regulation in nutritional (epi)genomics. Nutrition-related redox regulation occurs through nutrients involved in redox processes, and redox-regulated signal transduction cascades that control the expression of metabolism-associated genes. Cellular generation of redox regulators, such as hydrogen peroxide and hydrogen sulfide, occurs already at the microbiome/enterocyte (here: colonocyte) interface (A). The gut microbiome is a major source of volatile sulfur compounds which, following absorption by enterocytes, are oxidized by the activity of SELENBP1, which is a copper-dependent thiol oxidase. Insulin signaling following stimulation of INSR (insulin receptor) or IGF1R (insulin-like growth factor 1 receptor) causes attenuation of FOXO-dependent gene expression (B). The INSR/FOXO cascade is redox-sensitive at several stages (see text for details), including FOXOs per se. FOXOs contain conserved cysteine residues (1) that, through an intermediate sulfenic acid stage (2), may be oxidized to disulfides, including persulfides (3) when interacting with hydrogen sulfide, or disulfides (4) with various thiol-containing factors, ranging from glutathione to thiol-containing proteins such as the histone acetyl transferase CREBBP (C). Ascorbic acid (vitamin C) is a cofactor of enzymes catalyzing demethylation of KDMs or TETs, which belong to the family of α-ketoglutarate-dependent dioxygenases (D). Generated with BioRender.com.
Insulin signaling is a classic example of a pathway controlled by a nutrient like glucose, whose blood levels are translated into release of insulin by pancreatic β-cells into the bloodstream. This pathway regulates nutrient and energy metabolism at several levels, including the regulation of gene expression. Insulin signaling is also a pathway under close redox control at multiple stages [95], including both the regulation of glucose-stimulated insulin secretion [96] and the signal transduction cascades triggered in insulin target tissues [95,97] (Fig. 4B). Transient oxidation of phosphatases, such as the tyrosine phosphatase PTPN1 (protein tyrosine phosphatase non-receptor type 1) or the lipid phosphatase PTEN results in their reversible inactivation, which is crucial for cellular insulin-induced signaling. The inactivation of the phosphatases would prevent an interruption of the cascade implying that high levels of antioxidants may have adverse effects under certain conditions [95,98]. However, there are also adverse effects of high levels of ROS that may contribute to the development of insulin resistance [95]. Insulin-induced activation of PI3K (phosphoinositide-3′-kinase) and the downstream serine/threonine kinase AKT results in the phosphorylation and inactivation of FOXO transcription factors (Fig. 4B). In liver, FOXO target genes include those encoding G6PC1 (glucose-6-phosphatase catalytic subunit 1) and PCK1 (phosphoenolpyruvate carboxykinase 1, cytosolic), i.e., enzymes of the gluconeogenesis pathway. Insulin-induced inhibition of FOXOs transcriptionally counteracts gluconeogenesis and insulin resistance [99,100]. A considerable number of FOXO target genes encode proteins with antioxidant activity, ranging from those preventing ROS generation, such as metallothioneins or ceruloplasmin, enzymes intercepting ROS like superoxide dismutases, catalase or peroxiredoxins to others repairing oxidative damage, such as the thioredoxin system [101]. Both stimulation and attenuation of transcriptional activity of FOXOs under the influence of redox regulation has been shown, including the H2O2-induced oxidation resulting in mixed disulfides with regulatory cofactors, such as the histone deacetylase CREBBP (CREB binding protein) [102] (Fig. 4C). Both H2O2 and H2S are able to regulate FOXOs, with H2O2 generating the sulfenic acid form of cysteine, which then interacts with thiols, such as glutathione, resulting in S-glutathionylation, or with protein cysteines, or H2S, altering FOXO activity.
A key redox-sensing mechanism in human cells that is sensing not only oxidants but also electrophilic compounds is the KEAP1 (kelch like ECH associated protein 1)-NFE2L2 (NFE2 like BZIP transcription factor 2) pathway [103] It regulates NFE2L2-dependent expression of genes encoding proteins required for antioxidant defense and adaptation as well as for xenobiotic metabolism. Numerous plant-derived dietary compounds, such as sulforaphane, cardamonin or curcumin are activators of NFE2L2 and stimulate the expression of antioxidant enzymes [104]. Interestingly, endogenous formation of H2O2, such as by NADPH oxidases (NOX), particularly NOX4, modulate NFE2L2-dependent gene expression in relation to metabolism. NOX4 expression as well as NOX4-triggered generation of H2O2 is upregulated in skeletal muscle during exercise. Furthermore, NOX4-derived H2O2 upregulates NFE2L2-dependent expression of protective enzymes, which in skeletal muscle helps to attenuate the decline in insulin sensitivity in obesity and during aging [105].
In addition to signal transduction cascades resulting in the modulation of gene expression at the transcriptional level, the accessibility of the genome is controlled by the redox and nutrient status. This epigenetic control of gene expression was already alluded to above when CREBBP was mentioned in relation to FOXO activity. As also mentioned above (Fig. 1), deacetylation, e.g., by SIRTs, depends on the cellular redox status, particularly the NAD+/NADH ratio [51]. In addition to histone acetylation status being redox-controlled, the methylation status is as well, e.g., through KDMs or TET enzymes that both require ascorbate and α-ketoglutarate as cofactors, i.e., an antioxidant vitamin and an energy metabolism intermediate, respectively, establishing yet another link between diet, metabolism and redox control of epigenetics (Fig. 4D).
6. Conclusions
The communication between dietary molecules and the genome and epigenome is a central topic of nutrigenomics. Redox processes are involved in the modulation of signal transduction cascades mediating this communication. These cascades both regulate nutrient metabolism and are, in turn, controlled by macro- and micronutrients. Nutritional epigenetics is a subdiscipline of nutrigenomics that studies the effects of macro- and micronutrients, their metabolites and/or metabolites of energy metabolism on the programming of the epigenome in all tissues and cell types.
Epigenetic programming plays a role in all phases of life ranging from early embryogenesis to the last moments of life, i.e., it concerns fetuses, children adults and the elderly. Both transcription factors and chromatin-modifying enzymes are the endpoints of signal transduction cascades triggered by environmental signals, of which dietary molecules have a major contribution. While the action of transcription factors is only transient and restricted to the moment of their contact with DNA, chromatin-modifying enzymes leave marks to both parts of the chromatin, genomic DNA and histone proteins, which can last for hours, days or even decades. In this way, epigenetic programming through the action of chromatin-modifying enzymes has not only signaling function but in addition creates short-term to long-term memory.
The insight that for most traits epigenetics has a larger impact than genetics has important consequences for the rapidly changing environment and lifestyle that humans are exposed to. Rapid genetic adaption of the average population to the changes cannot be expected, but epigenetic adaption, e.g., to Western diet, is significantly faster. Moreover, appropriate epigenetic programming can be directed by lifestyle changes, such as healthy diet and sufficient physical activity, and the optimization of the environment, such as stress reduction and better sleep. In this way, not only the process of aging may slow down, but also diseases are prevented. In addition, the fact that humans do not only differ in their genome but even more in their epigenome is important for personalized/precision nutrition [40,106,107]. Thus, personalized dietary advice should be rather directed by the epigenetic than the genetic profile of the individuals. In addition, other levels, such as the individual proteome or metabolome need to be addressed [106].
Financial support and sponsorship
LOK is supported by Deutsche Forschungsgemeinschaft (DFG, Bonn, Germany) through RTG 2155 (ProMoAge). This publication is part of a project that has received funding from the European Union's Horizon2020 research and innovation program under grant agreement no. 952601 and from the David and Amy Fulton Foundation (both to CC).
Declaration of competing interest
There is no conflict of interest.
Data availability
No data was used for the research described in the article.
References
- 1.Chen Y., Michalak M., Agellon L.B. Importance of nutrients and nutrient metabolism on human health. Yale J. Biol. Med. 2018;91(2):95–103. [PMC free article] [PubMed] [Google Scholar]
- 2.Kietzmann T. Vitamin C: from nutrition to oxygen sensing and epigenetics. Redox Biol. 2023;63 doi: 10.1016/j.redox.2023.102753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Philipp T.M., Gernoth L., Will A., Schwarz M., Ohse V.A., Kipp A.P., Steinbrenner H., Klotz L.O. Selenium-binding protein 1 (SELENBP1) is a copper-dependent thiol oxidase. Redox Biol. 2023;65 doi: 10.1016/j.redox.2023.102807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehrer J.P., Klotz L.O. Free radicals and related reactive species as mediators of tissue injury and disease: implications for Health. Crit. Rev. Toxicol. 2015;45(9):765–798. doi: 10.3109/10408444.2015.1074159. [DOI] [PubMed] [Google Scholar]
- 5.Klotz L.O. In: Studies on Experimental Toxicology and Pharmacology. Roberts S.M., Kehrer J.P., Klotz L.O., editors. Springer Int. Publishing; Cham, Switzerland: 2015. Reactive oxygen species as initiators and mediators of cellular signaling processes; pp. 149–171. [Google Scholar]
- 6.Ordovas J.M., Ferguson L.R., Tai E.S., Mathers J.C. Personalised nutrition and health. Bmj. 2018;361 doi: 10.1136/bmj.k2173. bmj k2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller M., Kersten S. Nutrigenomics: goals and strategies. Nat. Rev. Genet. 2003;4(4):315–322. doi: 10.1038/nrg1047. [DOI] [PubMed] [Google Scholar]
- 8.Carlberg C., Ulven S.M., Molnár F. Springer textbook; 2020. Nutrigenomics: How Science Works. [Google Scholar]
- 9.Rundblad A., Sandoval V., Holven K.B., Ordovas J.M., Ulven S.M. Omega-3 fatty acids and individual variability in plasma triglyceride response: a mini-review. Redox Biol. 2023;63 doi: 10.1016/j.redox.2023.102730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlberg C., Raczyk M., Zawrotna N. Vitamin D: a master example of nutrigenomics. Redox Biol. 2023;62 doi: 10.1016/j.redox.2023.102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sturm R.A., Duffy D.L. Human pigmentation genes under environmental selection. Genome Biol. 2012;13(9):248. doi: 10.1186/gb-2012-13-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerbault P., Liebert A., Itan Y., Powell A., Currat M., Burger J., Swallow D.M., Thomas M.G. Evolution of lactase persistence: an example of human niche construction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366(1566):863–877. doi: 10.1098/rstb.2010.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells J.C.K. Body composition and susceptibility to type 2 diabetes: an evolutionary perspective. Eur. J. Clin. Nutr. 2017;71(7):881–889. doi: 10.1038/ejcn.2017.31. [DOI] [PubMed] [Google Scholar]
- 14.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 15.Carlberg C. Nutrigenomics in the context of evolution. Redox Biol. 2023;62 doi: 10.1016/j.redox.2023.102656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Civello D., Adams M.D., Cargill M., Clark A.G. Natural selection on protein-coding genes in the human genome. Nature. 2005;437(7062):1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 17.Dolgova O., Lao O. Evolutionary and medical consequences of archaic introgression into modern human genomes. Genes. 2018;9(7) doi: 10.3390/genes9070358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M. The neutral theory of molecular evolution: a review of recent evidence. Jpn. J. Genet. 1991;66(4):367–386. doi: 10.1266/jjg.66.367. [DOI] [PubMed] [Google Scholar]
- 19.Dudley J.T., Kim Y., Liu L., Markov G.J., Gerold K., Chen R., Butte A.J., Kumar S. Human genomic disease variants: a neutral evolutionary explanation. Genome Res. 2012;22(8):1383–1394. doi: 10.1101/gr.133702.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell C.D., Eichler E.E. Properties and rates of germline mutations in humans. Trends Genet. 2013;29(10):575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasani T.A., Pedersen B.S., Gao Z., Baird L., Przeworski M., Jorde L.B., Quinlan A.R. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. Elife. 2019;8 doi: 10.7554/eLife.46922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steffen W., Grinevald J., Crutzen P., McNeill J. The Anthropocene: conceptual and historical perspectives. Philos. Trans. A Math. Phys. Eng. Sci. 2011;369(1938):842–867. doi: 10.1098/rsta.2010.0327. [DOI] [PubMed] [Google Scholar]
- 23.1000 -Genomes-Project-Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., McPherson J.D., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W.R., de la Bastide M., Dedhia N., Blocker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S., Chen Y.J. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 25.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., Pitsillides A.N., LeFaive J., Lee S.B., Tian X., Browning B.L., Das S., Emde A.K., Clarke W.E., Loesch D.P., Shetty A.C., Blackwell T.W., Smith A.V., Wong Q., Liu X., Conomos M.P., Bobo D.M., Aguet F., Albert C., Alonso A., Ardlie K.G., Arking D.E., Aslibekyan S., Auer P.L., Barnard J., Barr R.G., Barwick L., Becker L.C., Beer R.L., Benjamin E.J., Bielak L.F., Blangero J., Boehnke M., Bowden D.W., Brody J.A., Burchard E.G., Cade B.E., Casella J.F., Chalazan B., Chasman D.I., Chen Y.I., Cho M.H., Choi S.H., Chung M.K., Clish C.B., Correa A., Curran J.E., Custer B., Darbar D., Daya M., de Andrade M., DeMeo D.L., Dutcher S.K., Ellinor P.T., Emery L.S., Eng C., Fatkin D., Fingerlin T., Forer L., Fornage M., Franceschini N., Fuchsberger C., Fullerton S.M., Germer S., Gladwin M.T., Gottlieb D.J., Guo X., Hall M.E., He J., Heard-Costa N.L., Heckbert S.R., Irvin M.R., Johnsen J.M., Johnson A.D., Kaplan R., Kardia S.L.R., Kelly T., Kelly S., Kenny E.E., Kiel D.P., Klemmer R., Konkle B.A., Kooperberg C., Kottgen A., Lange L.A., Lasky-Su J., Levy D., Lin X., Lin K.H., Liu C., Loos R.J.F., Garman L., Gerszten R., Lubitz S.A., Lunetta K.L., Mak A.C.Y., Manichaikul A., Manning A.K., Mathias R.A., McManus D.D., McGarvey S.T., Meigs J.B., Meyers D.A., Mikulla J.L., Minear M.A., Mitchell B.D., Mohanty S., Montasser M.E., Montgomery C., Morrison A.C., Murabito J.M., Natale A., Natarajan P., Nelson S.C., North K.E., O'Connell J.R., Palmer N.D., Pankratz N., Peloso G.M., Peyser P.A., Pleiness J., Post W.S., Psaty B.M., Rao D.C., Redline S., Reiner A.P., Roden D., Rotter J.I., Ruczinski I., Sarnowski C., Schoenherr S., Schwartz D.A., Seo J.S., Seshadri S., Sheehan V.A., Sheu W.H., Shoemaker M.B., Smith N.L., Smith J.A., Sotoodehnia N., Stilp A.M., Tang W., Taylor K.D., Telen M., Thornton T.A., Tracy R.P., Van Den Berg D.J., Vasan R.S., Viaud-Martinez K.A., Vrieze S., Weeks D.E., Weir B.S., Weiss S.T., Weng L.C., Willer C.J., Zhang Y., Zhao X., Arnett D.K., Ashley-Koch A.E., Barnes K.C., Boerwinkle E., Gabriel S., Gibbs R., Rice K.M., Rich S.S., Silverman E.K., Qasba P., Gan W., N.T.-O.f.P.M. Consortium. Papanicolaou G.J., Nickerson D.A., Browning S.R., Zody M.C., Zollner S., Wilson J.G., Cupples L.A., Laurie C.C., Jaquish C.E., Hernandez R.D., O'Connor T.D., Abecasis G.R. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590(7845):290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Investigators G.P.P., Smedley D., Smith K.R., Martin A., Thomas E.A., McDonagh E.M., Cipriani V., Ellingford J.M., Arno G., Tucci A., Vandrovcova J., Chan G., Williams H.J., Ratnaike T., Wei W., Stirrups K., Ibanez K., Moutsianas L., Wielscher M., Need A., Barnes M.R., Vestito L., Buchanan J., Wordsworth S., Ashford S., Rehmstrom K., Li E., Fuller G., Twiss P., Spasic-Boskovic O., Halsall S., Floto R.A., Poole K., Wagner A., Mehta S.G., Gurnell M., Burrows N., James R., Penkett C., Dewhurst E., Graf S., Mapeta R., Kasanicki M., Haworth A., Savage H., Babcock M., Reese M.G., Bale M., Baple E., Boustred C., Brittain H., de Burca A., Bleda M., Devereau A., Halai D., Haraldsdottir E., Hyder Z., Kasperaviciute D., Patch C., Polychronopoulos D., Matchan A., Sultana R., Ryten M., Tavares A.L.T., Tregidgo C., Turnbull C., Welland M., Wood S., Snow C., Williams E., Leigh S., Foulger R.E., Daugherty L.C., Niblock O., Leong I.U.S., Wright C.F., Davies J., Crichton C., Welch J., Woods K., Abulhoul L., Aurora P., Bockenhauer D., Broomfield A., Cleary M.A., Lam T., Dattani M., Footitt E., Ganesan V., Grunewald S., Compeyrot-Lacassagne S., Muntoni F., Pilkington C., Quinlivan R., Thapar N., Wallis C., Wedderburn L.R., Worth A., Bueser T., Compton C., Deshpande C., Fassihi H., Haque E., Izatt L., Josifova D., Mohammed S., Robert L., Rose S., Ruddy D., Sarkany R., Say G., Shaw A.C., Wolejko A., Habib B., Burns G., Hunter S., Grocock R.J., Humphray S.J., Robinson P.N., Haendel M., Simpson M.A., Banka S., Clayton-Smith J., Douzgou S., Hall G., Thomas H.B., O'Keefe R.T., Michaelides M., Moore A.T., Malka S., Pontikos N., Browning A.C., Straub V., Gorman G.S., Horvath R., Quinton R., Schaefer A.M., Yu-Wai-Man P., Turnbull D.M., McFarland R., Taylor R.W., O'Connor E., Yip J., Newland K., Morris H.R., Polke J., Wood N.W., Campbell C., Camps C., Gibson K., Koelling N., Lester T., Nemeth A.H., Palles C., Patel S., Roy N.B.A., Sen A., Taylor J., Cacheiro P., Jacobsen J.O., Seaby E.G., Davison V., Chitty L., Douglas A., Naresh K., McMullan D., Ellard S., Temple I.K., Mumford A.D., Wilson G., Beales P., Bitner-Glindzicz M., Black G., Bradley J.R., Brennan P., Burn J., Chinnery P.F., Elliott P., Flinter F., Houlden H., Irving M., Newman W., Rahman S., Sayer J.A., Taylor J.C., Webster A.R., Wilkie A.O.M., Ouwehand W.H., Raymond F.L., Chisholm J., Hill S., Bentley D., Scott R.H., Fowler T., Rendon A., Caulfield M. 100,000 genomes pilot on rare-disease diagnosis in health care - preliminary report. N. Engl. J. Med. 2021;385(20):1868–1880. doi: 10.1056/NEJMoa2035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cano-Gamez E., Trynka G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uffelmann E., Huang Q.Q., Munung N.S., de Vries J., Okada Y., Martin A.R., Martin H.C., Lappalainen T., Posthuma D. Genome-wide association studies. Nature Reviews Methods Primers. 2021;1(1) [Google Scholar]
- 29.Pociot F. Type 1 diabetes genome-wide association studies: not to be lost in translation. Clin. Transl. Immunol. 2017;6(12):e162. doi: 10.1038/cti.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorin M.B. Genetic insights into age-related macular degeneration: controversies addressing risk, causality, and therapeutics. Mol. Aspect. Med. 2012;33(4):467–486. doi: 10.1016/j.mam.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex ttaits: from polygenic to omnigenic. Cell. 2017;169(7):1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., Cho J.H., Guttmacher A.E., Kong A., Kruglyak L., Mardis E., Rotimi C.N., Slatkin M., Valle D., Whittemore A.S., Boehnke M., Clark A.G., Eichler E.E., Gibson G., Haines J.L., Mackay T.F., McCarroll S.A., Visscher P.M. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breton C.V., Landon R., Kahn L.G., Enlow M.B., Peterson A.K., Bastain T., Braun J., Comstock S.S., Duarte C.S., Hipwell A., Ji H., LaSalle J.M., Miller R.L., Musci R., Posner J., Schmidt R., Suglia S.F., Tung I., Weisenberger D., Zhu Y., Fry R. Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Commun. Biol. 2021;4(1):769. doi: 10.1038/s42003-021-02316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael A.K., Thoma N.H. Reading the chromatinized genome. Cell. 2021;184(14):3599–3611. doi: 10.1016/j.cell.2021.05.029. [DOI] [PubMed] [Google Scholar]
- 35.Reuter S., Gupta S.C., Park B., Goel A., Aggarwal B.B. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 2011;6(2):93–108. doi: 10.1007/s12263-011-0222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Z., Ramesh V., Locasale J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020;21(12):737–753. doi: 10.1038/s41576-020-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C., Wang Z., Qin Y. Connections between metabolism and epigenetics: mechanisms and novel anti-cancer strategy. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.935536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kupkova K., Shetty S.J., Haque R., Petri W.A., Jr., Auble D.T. Histone H3 lysine 27 acetylation profile undergoes two global shifts in undernourished children and suggests altered one-carbon metabolism. Clin. Epigenet. 2021;13(1):182. doi: 10.1186/s13148-021-01173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlberg C., Molnár F. Springer Textbook; 2018. Human Epigenomics. [Google Scholar]
- 40.Malcomson F.C., Mathers J.C. Translation of nutrigenomic research for personalised and precision nutrition for cancer prevention and for cancer survivors. Redox Biol. 2023;62 doi: 10.1016/j.redox.2023.102710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlberg C., Molnár F. Springer textbook; 2019. Human Epigenetics: How Science Works. [Google Scholar]
- 42.Cai S., Quan S., Yang G., Chen M., Ye Q., Wang G., Yu H., Wang Y., Qiao S., Zeng X. Nutritional status impacts epigenetic regulation in early embryo development: a scoping review. Adv. Nutr. 2021;12(5):1877–1892. doi: 10.1093/advances/nmab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bloushtain-Qimron N., Yao J., Shipitsin M., Maruyama R., Polyak K. Epigenetic patterns of embryonic and adult stem cells. Cell Cycle. 2009;8(6):809–817. doi: 10.4161/cc.8.6.7938. [DOI] [PubMed] [Google Scholar]
- 44.Ferrell J.E., Jr. Bistability, bifurcations, and Waddington's epigenetic landscape. Curr. Biol. 2012;22(11):R458–R466. doi: 10.1016/j.cub.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlberg C., Velleuer E. Nutrition and epigenetic programming. Curr. Opin. Clin. Nutr. Metab. Care. 2023;26(3):259–265. doi: 10.1097/MCO.0000000000000900. [DOI] [PubMed] [Google Scholar]
- 46.Carlberg C. Vitamin D in the context of evolution. Nutrients. 2022;14(15):3018. doi: 10.3390/nu14153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badeaux A.I., Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 2013;14(4):211–224. [PubMed] [Google Scholar]
- 48.Machnik M., Oleksiewicz U. Dynamic signatures of the epigenome: friend or foe? Cells. 2020;9(3):653. doi: 10.3390/cells9030653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brasacchio D., Okabe J., Tikellis C., Balcerczyk A., George P., Baker E.K., Calkin A.C., Brownlee M., Cooper M.E., El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58(5):1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomez de Cedron M., Moreno Palomares R., Ramirez de Molina A. Metabolo-epigenetic interplay provides targeted nutritional interventions in chronic diseases and ageing. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1169168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker F., Behrends M.M., Rudolph K.L. Evolution, mechanism and limits of dietary restriction induced health benefits & longevity. Redox Biol. 2023;63 doi: 10.1016/j.redox.2023.102725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y., Daniel M., Tollefsbol T.O. Epigenetic regulation of caloric restriction in aging. BMC Med. 2011;9:98. doi: 10.1186/1741-7015-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green C.L., Lamming D.W., Fontana L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 2022;23(1):56–73. doi: 10.1038/s41580-021-00411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newman J.C., Verdin E. Beta-hydroxybutyrate: a signaling metabolite. Annu. Rev. Nutr. 2017;37:51–76. doi: 10.1146/annurev-nutr-071816-064916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlberg C., Velleuer E. Springer Textbook; 2021. Cancer Biology: How Science Worls. [Google Scholar]
- 56.De Rooij S.R., Bleker L.S., Painter R.C., Ravelli A.C., Roseboom T.J. Lessons learned from 25 Years of research into long term consequences of prenatal exposure to the Dutch famine 1944-45: the Dutch famine birth cohort. Int. J. Environ. Health Res. 2022;32(7):1432–1446. doi: 10.1080/09603123.2021.1888894. [DOI] [PubMed] [Google Scholar]
- 57.Ramirez D., Haas S.A. Windows of vulnerability: consequences of exposure timing during the Dutch Hunger Winter. Popul. Dev. Rev. 2022:959–989. doi: 10.1111/padr.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U. S. A. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breton-Larrivee M., Elder E., McGraw S. DNA methylation, environmental exposures and early embryo development. Anim. Reprod. 2019;16(3):465–474. doi: 10.21451/1984-3143-AR2019-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaiserman A., Lushchak O. Prenatal famine exposure and adult health outcomes: an epigenetic link. Environ. Epigenet. 2021;7(1) doi: 10.1093/eep/dvab013. dvab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou Z., Li C., Patton G.C. Early-life exposure to the Chinese Famine and subsequent T2DM. Nat. Rev. Endocrinol. 2020;16(2):124–125. doi: 10.1038/s41574-019-0299-y. [DOI] [PubMed] [Google Scholar]
- 62.Lacagnina S. The developmental Origins of health and disease (DOHaD) Am. J. Lifestyle Med. 2020;14(1):47–50. doi: 10.1177/1559827619879694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harary D., Akinyemi A., Charron M.J., Fuloria M. Fetal growth and intrauterine epigenetic programming of obesity and cardiometabolic disease. NeoReviews. 2022;23(6):e363–e372. doi: 10.1542/neo.23-6-e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su C., Gao L., May C.L., Pippin J.A., Boehm K., Lee M., Liu C., Pahl M.C., Golson M.L., Naji A., Consortium H., Grant S.F.A., Wells A.D., Kaestner K.H. 3D chromatin maps of the human pancreas reveal lineage-specific regulatory architecture of T2D risk. Cell Metabol. 2022;34(9):1394–1409 e4. doi: 10.1016/j.cmet.2022.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bruno S., Williams R.J., Del Vecchio D. Epigenetic cell memory: the gene's inner chromatin modification circuit. PLoS Comput. Biol. 2022;18(4) doi: 10.1371/journal.pcbi.1009961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y., Tollefsbol T.O. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr. Med. Chem. 2010;17(20):2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J., Zhang J., Hou W., Yang X., Liu X., Zhang Y., Gao M., Zong M., Dong Z., Liu Z., Shen J., Cong W., Ding C., Gao S., Huang G., Kong Q. Metabolic control of histone acetylation for precise and timely regulation of minor ZGA in early mammalian embryos. Cell Discov. 2022;8(1):96. doi: 10.1038/s41421-022-00440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitz-James M.H., Cavalli G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022;23(6):325–341. doi: 10.1038/s41576-021-00438-5. [DOI] [PubMed] [Google Scholar]
- 69.Nilsson E.E., Sadler-Riggleman I., Skinner M.K. Environmentally induced epigenetic transgenerational inheritance of disease. Environ. Epigenet. 2018;4(2):dvy016. doi: 10.1093/eep/dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heard E., Martienssen Robert A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulz L.O., Chaudhari L.S. High-risk populations: the pimas of Arizona and Mexico. Curr. Obes. Rep. 2015;4(1):92–98. doi: 10.1007/s13679-014-0132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ranasinghe P., Mathangasinghe Y., Jayawardena R., Hills A.P., Misra A. Prevalence and trends of metabolic syndrome among adults in the asia-pacific region: a systematic review. BMC Publ. Health. 2017;17(1):101. doi: 10.1186/s12889-017-4041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen A.A., Ferrucci L., Fulop T., Gravel D., Hao N., Kriete A., Levine M.E., Lipsitz L.A., Olde Rikkert M.G.M., Rutenberg A., Stroustrup N., Varadhan R. A complex systems approach to aging biology. Nat Aging. 2022;2(7):580–591. doi: 10.1038/s43587-022-00252-6. [DOI] [PubMed] [Google Scholar]
- 74.Passarino G., De Rango F., Montesanto A. Human longevity: genetics or Lifestyle? It takes two to tango. Immun. Ageing. 2016;13:12. doi: 10.1186/s12979-016-0066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simm A., Klotz L.O. Stress and biological aging: a double-edged sword. Z. Gerontol. Geriatr. 2015;48(6):505–510. doi: 10.1007/s00391-015-0928-6. [DOI] [PubMed] [Google Scholar]
- 76.Vina J., Borras C., Miquel J. Theories of ageing. IUBMB Life. 2007;59(4–5):249–254. doi: 10.1080/15216540601178067. [DOI] [PubMed] [Google Scholar]
- 77.Vina J. The free radical theory of frailty: mechanisms and opportunities for interventions to promote successful aging. Free Radic. Biol. Med. 2019;134:690–694. doi: 10.1016/j.freeradbiomed.2019.01.045. [DOI] [PubMed] [Google Scholar]
- 78.Vina J., Borras C., Abdelaziz K.M., Garcia-Valles R., Gomez-Cabrera M.C. The free radical theory of aging revisited: the cell signaling disruption theory of aging. Antioxidants Redox Signal. 2013;19(8):779–787. doi: 10.1089/ars.2012.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Egea J., Fabregat I., Frapart Y.M., Ghezzi P., Gorlach A., Kietzmann T., Kubaichuk K., Knaus U.G., Lopez M.G., Olaso-Gonzalez G., Petry A., Schulz R., Vina J., Winyard P., Abbas K., Ademowo O.S., Afonso C.B., Andreadou I., Antelmann H., Antunes F., Aslan M., Bachschmid M.M., Barbosa R.M., Belousov V., Berndt C., Bernlohr D., Bertran E., Bindoli A., Bottari S.P., Brito P.M., Carrara G., Casas A.I., Chatzi A., Chondrogianni N., Conrad M., Cooke M.S., Costa J.G., Cuadrado A., My-Chan Dang P., De Smet B., Debelec-Butuner B., Dias I.H.K., Dunn J.D., Edson A.J., El Assar M., El-Benna J., Ferdinandy P., Fernandes A.S., Fladmark K.E., Forstermann U., Giniatullin R., Giricz Z., Gorbe A., Griffiths H., Hampl V., Hanf A., Herget J., Hernansanz-Agustin P., Hillion M., Huang J., Ilikay S., Jansen-Durr P., Jaquet V., Joles J.A., Kalyanaraman B., Kaminskyy D., Karbaschi M., Kleanthous M., Klotz L.O., Korac B., Korkmaz K.S., Koziel R., Kracun D., Krause K.H., Kren V., Krieg T., Laranjinha J., Lazou A., Li H., Martinez-Ruiz A., Matsui R., McBean G.J., Meredith S.P., Messens J., Miguel V., Mikhed Y., Milisav I., Milkovic L., Miranda-Vizuete A., Mojovic M., Monsalve M., Mouthuy P.A., Mulvey J., Munzel T., Muzykantov V., Nguyen I.T.N., Oelze M., Oliveira N.G., Palmeira C.M., Papaevgeniou N., Pavicevic A., Pedre B., Peyrot F., Phylactides M., Pircalabioru G.G., Pitt A.R., Poulsen H.E., Prieto I., Rigobello M.P., Robledinos-Anton N., Rodriguez-Manas L., Rolo A.P., Rousset F., Ruskovska T., Saraiva N., Sasson S., Schroder K., Semen K., Seredenina T., Shakirzyanova A., Smith G.L., Soldati T., Sousa B.C., Spickett C.M., Stancic A., Stasia M.J., Steinbrenner H., Stepanic V., Steven S., Tokatlidis K., Tuncay E., Turan B., Ursini F., Vacek J., Vajnerova O., Valentova K., Van Breusegem F., Varisli L., Veal E.A., Yalcin A.S., Yelisyeyeva O., Zarkovic N., Zatloukalova M., Zielonka J., Touyz R.M., Papapetropoulos A., Grune T., Lamas S., Schmidt H., Di Lisa F., Daiber A. European contribution to the study of ROS: a summary of the findings and prospects for the future from the COST action BM1203 (EU-ROS) Redox Biol. 2017;13:94–162. doi: 10.1016/j.redox.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klotz L.O., Sanchez-Ramos C., Prieto-Arroyo I., Urbanek P., Steinbrenner H., Monsalve M. Redox regulation of FoxO transcription factors. Redox Biol. 2015;6:51–72. doi: 10.1016/j.redox.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lopez-Lluch G., Navas P. Calorie restriction as an intervention in ageing. J. Physiol. 2016;594(8):2043–2060. doi: 10.1113/JP270543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pal S., Tyler J.K. Epigenetics and aging. Sci. Adv. 2016;2(7) doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berben L., Floris G., Wildiers H., Hatse S. Cancer and aging: two tightly interconnected biological processes. Cancers. 2021;13(6):1400. doi: 10.3390/cancers13061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bell C.G., Lowe R., Adams P.D., Baccarelli A.A., Beck S., Bell J.T., Christensen B.C., Gladyshev V.N., Heijmans B.T., Horvath S., Ideker T., Issa J.J., Kelsey K.T., Marioni R.E., Reik W., Relton C.L., Schalkwyk L.C., Teschendorff A.E., Wagner W., Zhang K., Rakyan V.K. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019;20(1):249. doi: 10.1186/s13059-019-1824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salvestrini V., Sell C., Lorenzini A. Obesity may accelerate the aging process. Front. Endocrinol. 2019;10:266. doi: 10.3389/fendo.2019.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frenk S., Houseley J. Gene expression hallmarks of cellular ageing. Biogerontology. 2018;19(6):547–566. doi: 10.1007/s10522-018-9750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan H., Finkel T. Key proteins and pathways that regulate lifespan. J. Biol. Chem. 2017;292(16):6452–6460. doi: 10.1074/jbc.R116.771915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Longo V.D., Anderson R.M. Nutrition, longevity and disease: from molecular mechanisms to interventions. Cell. 2022;185(9):1455–1470. doi: 10.1016/j.cell.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Itahana Y., Han R., Barbier S., Lei Z., Rozen S., Itahana K. The uric acid transporter SLC2A9 is a direct target gene of the tumor suppressor p53 contributing to antioxidant defense. Oncogene. 2015;34(14):1799–1810. doi: 10.1038/onc.2014.119. [DOI] [PubMed] [Google Scholar]
- 91.Sies H., Stahl W., Sevanian A. Nutritional, dietary and postprandial oxidative stress. J. Nutr. 2005;135(5):969–972. doi: 10.1093/jn/135.5.969. [DOI] [PubMed] [Google Scholar]
- 92.Sies H. Oxidative eustress: on constant alert for redox homeostasis. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pol A., Renkema G.H., Tangerman A., Winkel E.G., Engelke U.F., de Brouwer A.P.M., Lloyd K.C., Araiza R.S., van den Heuvel L., Omran H., Olbrich H., Oude Elberink M., Gilissen C., Rodenburg R.J., Sass J.O., Schwab K.O., Schafer H., Venselaar H., Sequeira J.S., Op den Camp H.J.M., Wevers R.A. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 2018;50(1):120–129. doi: 10.1038/s41588-017-0006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Philipp T.M., Will A., Richter H., Winterhalter P.R., Pohnert G., Steinbrenner H., Klotz L.O. A coupled enzyme assay for detection of selenium-binding protein 1 (SELENBP1) methanethiol oxidase (MTO) activity in mature enterocytes. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lennicke C., Cocheme H.M. Redox regulation of the insulin signalling pathway. Redox Biol. 2021;42 doi: 10.1016/j.redox.2021.101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Plecita-Hlavata L., Jaburek M., Holendova B., Tauber J., Pavluch V., Berkova Z., Cahova M., Schroder K., Brandes R.P., Siemen D., Jezek P. Glucose-stimulated insulin secretion fundamentally requires H(2)O(2) signaling by NADPH oxidase 4. Diabetes. 2020;69(7):1341–1354. doi: 10.2337/db19-1130. [DOI] [PubMed] [Google Scholar]
- 97.Klotz L.O., Steinbrenner H. Cellular adaptation to xenobiotics: interplay between xenosensors, reactive oxygen species and FOXO transcription factors. Redox Biol. 2017;13:646–654. doi: 10.1016/j.redox.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klotz L.O. In: Studies on Experimental Toxicology and Pharmacology. Roberts S.M., Kehrer J.P., Klotz L.O., editors. Springer Int. Publishing; Charm, Switzerland: 2015. On the biochemistry of antioxidants: current aspects; pp. 383–396. [Google Scholar]
- 99.Barthel A., Schmoll D., Unterman T.G. FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2005;16(4):183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 100.O-Sullivan I., Zhang W., Wasserman D.H., Liew C.W., Liu J., Paik J., DePinho R.A., Stolz D.B., Kahn C.R., Schwartz M.W., Unterman T.G. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat. Commun. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krafczyk N., Klotz L.O. FOXO transcription factors in antioxidant defense. IUBMB Life. 2022;74(1):53–61. doi: 10.1002/iub.2542. [DOI] [PubMed] [Google Scholar]
- 102.Dansen T.B., Smits L.M., van Triest M.H., de Keizer P.L., van Leenen D., Koerkamp M.G., Szypowska A., Meppelink A., Brenkman A.B., Yodoi J., Holstege F.C., Burgering B.M. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 2009;5(9):664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- 103.Adinolfi S., Patinen T., Jawahar Deen A., Pitkanen S., Harkonen J., Kansanen E., Kublbeck J., Levonen A.L. The KEAP1-NRF2 pathway: targets for therapy and role in cancer. Redox Biol. 2023;63 doi: 10.1016/j.redox.2023.102726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tauber S., Sieckmann M.K., Erler K., Stahl W., Klotz L.O., Steinbrenner H. Activation of nrf2 by electrophiles is largely independent of the selenium status of HepG2 cells. Antioxidants. 2021;10(2):167. doi: 10.3390/antiox10020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xirouchaki C.E., Jia Y., McGrath M.J., Greatorex S., Tran M., Merry T.L., Hong D., Eramo M.J., Broome S.C., Woodhead J.S.T., D'Souza R F., Gallagher J., Salimova E., Huang C., Schittenhelm R.B., Sadoshima J., Watt M.J., Mitchell C.A., Tiganis T. Skeletal muscle NOX4 is required for adaptive responses that prevent insulin resistance. Sci. Adv. 2021;7(51) doi: 10.1126/sciadv.abl4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brennan L., de Roos B. Role of metabolomics in the delivery of precision nutrition. Redox Biol. 2023;65 doi: 10.1016/j.redox.2023.102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Livingstone K.M., Ramos-Lopez O., Pérusse L., Kato H., Ordovas J.M., Martínez J.A. Precision nutrition: a review of current approaches and future endeavors. Trends Food Sci. Technol. 2022;128:253–264. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.