To the Editor:
The FMS-like tyrosine kinase-3 (FLT3) is mutated in ~30% of acute myeloid leukemia (AML) patients. Common FLT3 mutations are internal tandem duplications (FLT3-ITD) and point mutations in its c-terminal tyrosine kinase domain (FLT3-TKD). The resulting active FLT3 receptors promote cell proliferation and resistance to programmed cell death (apoptosis) [1]. The poor prognosis of patients with FLT3-ITD has spurred an intensive search for FLT3 inhibitors (FLT3i) [2]. These include the nanomolar FLT3i quizartinib, which has produced promising benefits in AML patients with FLT3-ITD [3]. TKD mutations in FLT3-ITD arise during the therapy of patients with quizartinib and confer drug resistance. The broad-range tyrosine kinase inhibitors midostaurin and gilteritinib are used with standard chemotherapy to treat AML. Their pleiotropic actions frequently evoke hematological toxicity [1, 2]. Marbotinib is a hybrid FLT3i containing elements of quizartinib and bis(1H-indol-2-yl)methanone. Marbotinib specifically inhibits FLT3-ITD and FLT3-TKD [4, 5].
Epigenetic modifiers of the histone deacetylase (HDAC) family control the development and survival of blood cells. Compared to normal cells, certain leukemic cell types frequently have aberrant expression levels and activities of epigenetic modifiers that belong to the histone deacetylase (HDAC) family. Accordingly, inhibitors of zinc-dependent HDACs (HDACi) are approved drugs for the treatment of subtypes of leukemia and lymphoma. These agents are vorinostat, romidepsin, belinostat, and chidamide. Further HDACi are tested clinically (www.clinicaltrials.org), such as the benzamide entinostat (MS-275) [6, 7]. HDACi decrease FLT3-ITD through ubiquitin-dependent proteasomal degradation and apoptosis-associated caspase activation. Combinations of HDACi and FLT3i synergistically kill FLT3-ITD-positive cells through accelerated elimination of FLT3-ITD and DNA replication stress/DNA damage induction. HDAC1, HDAC2, and particularly HDAC3 maintain the stability of FLT3-ITD [8]. A cellular self-digestion process termed autophagy also eliminates FLT3-ITD [9]. Nonetheless, autophagy can promote FLT3i resistance [10, 11]. HDACs modulate autophagy in leukemic cells [6, 12], but it is unknown if HDACi modulate FLT3-ITD through autophagy and if this has biological relevance.
Despite initial hope in HDACi, their limited clinical efficacy as monotherapies and toxicological issues make additional research necessary. This includes the development of nanomolar HDACi with a low impact on normal cells and the identification of effective drug combinations [7, 13]. KH16 and KH29 are recently developed HDACi, which we have named yanostat-1/yanostat-2 [12, 14]. We reveal that their inhibitory profiles are superior to those of SAHA and MS-275 in AML cells carrying FLT3-ITD. Low doses of HDACi cause hormesis effects through FLT3-ITD. Specific inhibition of FLT3-ITD with the nanomolar FLT3i marbotinib and quizartinib abrogates undesired hormesis and is synergistically lethal in combination with nanomolar doses of KH16.
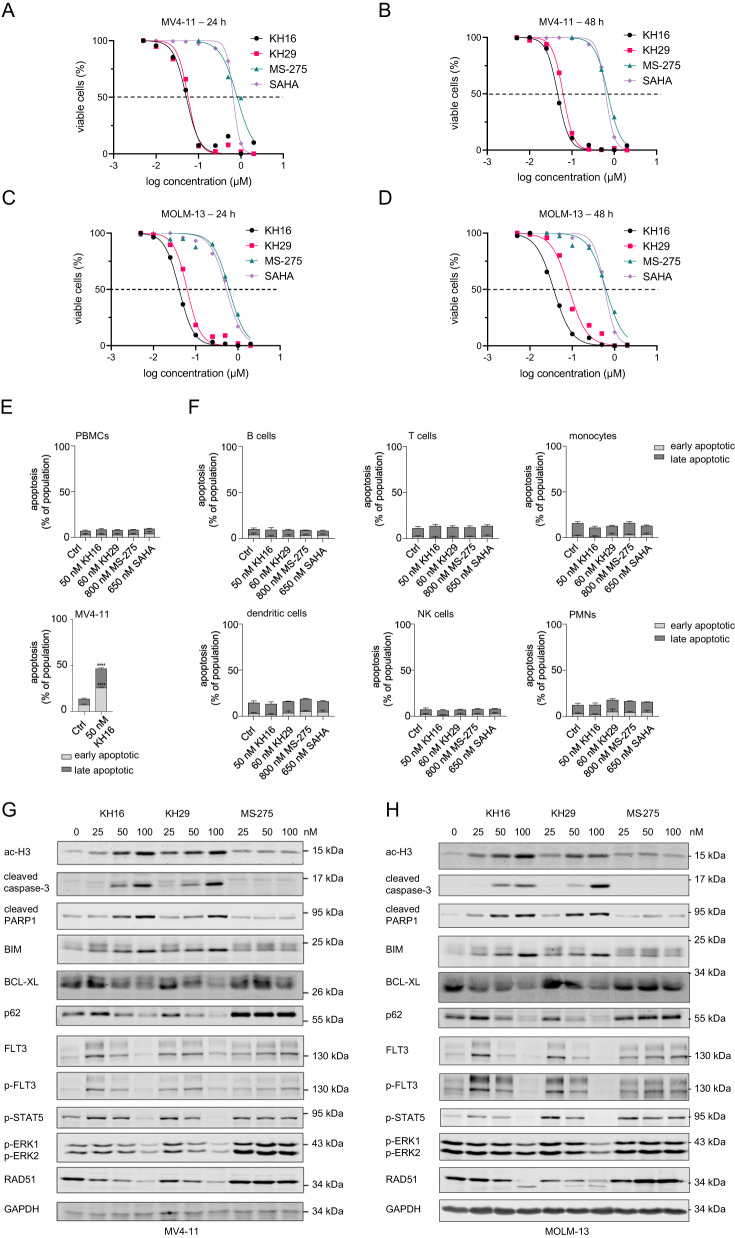
KH16 and KH29 carry the hydroxamic acid as zinc binding moiety being present in SAHA and have nanomolar activity against HDACs [12]. We applied KH16 and KH29 to human FLT3-ITD-positive MV4-11 and MOLM-13 AML cells for 24-48 h. Flow cytometry demonstrated that nanomolar concentrations of KH16 and KH29 induced apoptosis (37 nM < IC50 < 85 nM). KH16 and KH29 were at least 6.3-fold more effective apoptosis inducers than the established HDACi MS-275 and SAHA (533 nM < IC50 < 814 nM) (Fig. 1A–D; Table 1; Fig. S1A, B). Thus, KH16 and KH29 are significantly more potent inducers of apoptosis in FLT3-ITD-positive leukemic cells than the clinical grade HDACi vorinostat and entinostat.
Fig. 1. Cellular and biochemical effects of KH16 and KH29.
MV4-11 and MOLM-13 cells were treated with different concentrations of KH16, KH29, MS-275, or SAHA for 24-48 h and subjected to flow cytometry analyses for annexin-V (indicator of early apoptosis; #130-093-060 from Miltenyi Biotec) and propidium iodide (PI; from Sigma-Aldrich; late apoptosis indicator). A MV4-11 cells were treated with increasing concentrations of KH16, KH29, MS-275, or SAHA from 5 nM to 2 µM for 24 h (n = 3). The x-axis was logarithmically transformed to yield a dose-response curve and to calculate the IC50 using GraphPad Prism 8.4.3. B Same experiments as in A, but for 48 h. Decreased cell viability equals to annexin-V/PI-positivity (n = 3). C MOLM-13 cells were treated with increasing concentrations of KH16, KH29, MS-275, or SAHA (5 nM to 2 µM) and incubated for 24 h (n = 3) or D Same experiments as in C, but for 48 h (n = 3). E Upper panel: PBMCs were isolated from buffy coats of four healthy donors (tested negative for common infections) using Biocoll (Bio&Sell). These were treated with HDACi as indicated for 24 h (Ctrl, untreated) and subjected to flow cytometry assessing apoptosis. To receive and delineate the PBMC populations, debris and doublets were excluded. Lower panel: MV4-11 cells that were treated in parallel to the PBMCs with 50 nM KH16 are a positive control for apoptosis induction (n = 4; mean+SD; two-way ANOVA; ****p ≥ 0.0001). F Untreated and HDACi-treated PBMCs were stained for lineage markers and subjected to flow cytometry (Ctrl, untreated). The cells were defined as: CD3-CD19+ as B cells; CD3+ as T cells, CD3-CD19-CD14+ as monocytes; CD3-CD19-CD1c+ as dendritic cells; CD3-CD19-CD56+ as NK cells; and CD3-CD14-CD19-CD56-CD11b+ as PMNs. The cell populations were analyzed for viability using annexin-V AF647 (#A23204; early apoptosis marker) and FVD eFl780 (#65-0865-18; late apoptosis marker; both from ThermoFisher). The following antibodies were used: CD11b BV510 (#101263), CD1c BV605 (#331538), CD3 BV711 (#344838) from BioLegend; CD14 PE-eFl610 (#61-0149-42), CD56 Pe-Cy7 (#25-0567-42), CD19 AF488 (#53-0199-42) from ThermoFisher. G MV4-11 cells were treated with 25, 50, and 100 nM KH16, KH29, or MS-275, respectively, for 24 h. Immunoblot was carried out for the indicated proteins, ac-H3, acetylated histone H3, GAPDH as representative loading control; n = 2. H MOLM-13 cells were treated with 25, 50, and 100 nM KH16, KH29, or MS-275, respectively for 24 h; n = 2. The n-numbers indicate biological replicates of which some were additionally carried out as technical replicates. Immunoblot was carried out for the indicated proteins, GAPDH as representative loading control; n = 2. Flow cytometry and immunoblot were done as mentioned [4, 12], with the following antibodies: BCL-XL (#ab32370), BIM (#ab32158), GAPDH (#ab128915), RAD51 (#ab6380) from Abcam; cleaved PARP (#552596) from BD Biosciences; FLT3 (#sc-480), SQSTM1/p62 (#sc-25575) from Santa Cruz Biotechnology; cleaved caspase-3 (#cs9661), p-Tyr591-FLT3 (#3461), p-Tyr202/Tyr204-ERK1/ERK2 (#cs9101) from Cell Signaling; p-Tyr694-STAT5 (#MA5-14973) from Thermo Fisher; ac-H3 (#06-599) from Millipore. The protein ladder is the prestained ScientificTM PageRulerTM (#26617) from Thermo Fisher.
Table 1.
KH16 and KH29 are more potent inducers of apoptosis than MS-275 and SAHA in FLT3-ITD-positive leukemic cells.
| KH16 | KH29 | MS-275 | SAHA | |||||
|---|---|---|---|---|---|---|---|---|
| hours | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 |
| MV4-11 (IC50 nM) | 52 | 46 | 57 | 62 | 814 | 706 | 658 | 629 |
| MOLM-13 (IC50 nM) | 41 | 37 | 63 | 85 | 621 | 628 | 533 | 586 |
Shown are the IC50 values for apoptosis induction in MV4-11 and MOLM-13 cells that were treated with KH16, KH29, MS-275, or SAHA for 24-48 h. The values were calculated from the flow cytometry experiments (Fig. 1A–D) using GraphPad Prism 8.4.3; n = 3.
Human embryonic kidney and retinal pigment epithelial (RPE1) cells do not undergo apoptosis when they are exposed to KH16 and KH29 [12, 14]. We extended these analyses. We incubated MV4-11 cells, RPE1 cells, and human keratinocytes (HaCaT cells) with rounded IC50 concentrations of KH16, KH29, and SAHA for 24-72 h. Flow cytometry showed that the HDACi induced apoptosis significantly in MV4-11 cells but not in RPE1 and HaCaT cells (Fig. S2). To additionally consider the responses of normal human blood cells to KH16 and KH29, we isolated peripheral blood mononuclear cells (PBMCs) from healthy donors and treated the cells with rounded IC50 concentrations of KH16, KH29, MS-275, and SAHA for 24 h. None of these HDACi compromised the viability of PBMCs (Fig. 1E). We further distinguished the main leukocyte populations by employing antibodies for lineage-specific surface markers. We found that HDACi did not induce apoptosis in B-cells, T-cells, monocytes, dendritic cells, natural killer (NK) cells, and polymorphonuclear neutrophils (PMNs) (Fig. 1F). Thus, despite being potent inducers of apoptosis in leukemic cells with FLT3-ITD, KH16 and KH29 do not kill normal cells.
Immunoblot analyses with lysates from MV4-11 and MOLM-13 cells that were incubated with equimolar concentrations of KH16, KH29, and MS-275 verified that 25 nM of KH16 and KH29 induced an accumulation of hyperacetylated histone H3 (Fig. 1G, H). This effect increased dose-dependently. Up to 100 nM MS-275 did not induce an accumulation of acetylated histone H3.
We confirmed that KH16 and KH29 induced apoptosis by immunoblots assessing activation of caspase-3 and cleavage of its target PARP1. Moreover, we detected a dose-dependent induction of pro-apoptotic BIM and a reduction of anti-apoptotic BCL-XL. KH16 and KH29 caused these molecular alterations more effectively than MS-275. Whereas 25 nM KH16 and KH29 induced the autophagy protein p62, 100 nM KH16 and KH29 decreased p62. 25–100 nM MS-275 stabilized p62 (Fig. 1G, H).
Unexpectedly, 25–50 nM KH16 and KH29, and 25–100 nM MS-275, evoked an accumulation of total and phosphorylated FLT3-ITD (Fig. 1G, H). At 100 nM concentrations of KH16 and KH29, this effect was lost. This held for the phosphorylation of the kinases ERK1/ERK2, the transcription factor STAT5, and the levels of the DNA repair protein RAD51. Doses of 100 nM KH16 and KH29 attenuated these tumor-promoting targets of FLT3-ITD (Fig. 1G, H).
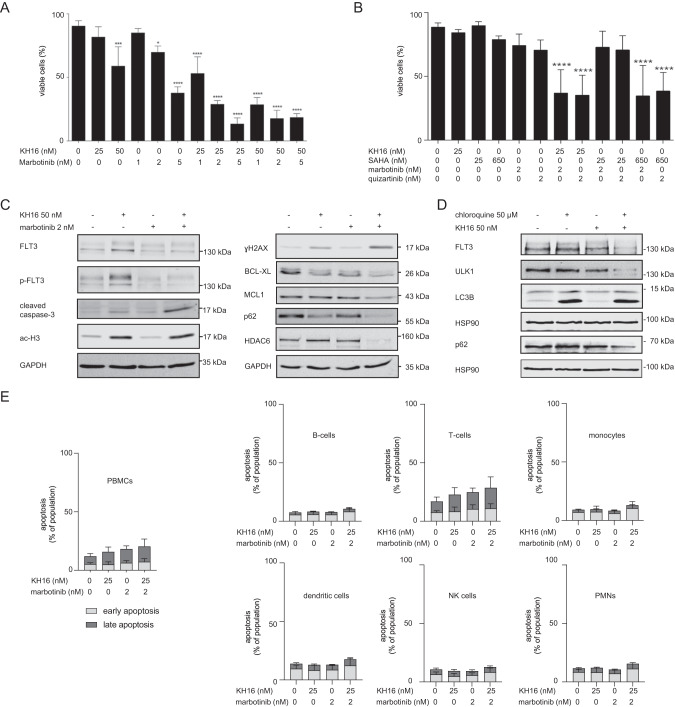
We aimed to define the biological importance of the accumulation of p-FLT3-ITD and FLT3-ITD in response to low doses of KH16 (Fig. 1G, H). We hypothesized that this increase in FLT3-ITD attenuated pro-apoptotic effects of HDACi. We experimentally addressed this with a nanomolar dose of marbotinib. We found that 25 nM KH16 and 2 nM marbotinib combined favorably against MV4-11 cells (Fig. 2A). We also noted this effect using 25 nM KH16 and 2 nM quizartinib. Compared to KH16 plus FLT3i, combinations of 25 nM SAHA and the FLT3i were significantly less effective apoptosis inducers. In such combinations, increasing the dose of SAHA to 650 nM gave similar results as 25 nM KH16 (Fig. 2B). According to these results, combinations of KH16 plus marbotinib or quizartinib are low nanomolar inducers of apoptosis in FLT3-ITD-positive leukemic cell populations.
Fig. 2. Combinations of KH16 and FLT3i efficiently kill FLT3-ITD-positive leukemic cells.
A MV4-11 cells were treated with 25-50 nM KH16 ± 1-5 nM marbotinib for 24 h and subjected to flow cytometry for apoptosis measurement (n = 3; mean+SD; one-way ANOVA; *p ≥ 0.05; ***p ≥ 0.001; ****p ≥ 0.0001). B MV4-11 cells were treated with different combinations of HDACi and FLT3i as indicated for 24 h and subjected to flow cytometry for apoptosis measurement (n = 4; mean+SD; one-way ANOVA; ****p ≥ 0.0001) C Immunoblot shows the levels of the indicated proteins in MV4-11 cells that were treated as indicated for 24 h; GAPDH serves as loading control (n = 2). D MV4-11 cells were treated with 50 nM KH16 ± 50 µM chloroquine for 24 h. Immunoblot was carried out for the indicated proteins; HSP90, loading control (n = 2). The n-numbers indicate biological replicates of which some were additionally carried out as technical replicates. Flow cytometry and immunoblot were done as mentioned [4, 12], with the following antibodies: BCL-XL (#ab32370), GAPDH (#ab128915) from Abcam; FLT3 (#sc-480), HSP90 (#sc-13119), MCL-1 (#sc-12756), SQSTM1/p62 (#sc-25575), ɣH2AX (#sc-101696) from Santa Cruz Biotechnology; cleaved caspase-3 (#cs9661), HDAC6 (#7558), LC3B (#3868), p-Tyr591-FLT3 (#3461), ULK1 (#8054) from Cell Signaling; ac-H3 (#06-599) from Millipore. The protein ladder is the prestained ScientificTM PageRulerTM (#26617) from Thermo Fisher. E PBMCs from four healthy donors were isolated, treated as indicated, separated in cell fractions, and analyzed with flow cytometry (n = 4; mean+SD; see Fig. 1E, F for details).
Immunoblot analyses verified that 50 nM KH16 plus 2 nM marbotinib activated caspase-3, induced the DNA damage marker ɣH2AX, and decreased the anti-apoptotic proteins BCL-XL and MCL1. Histone hyperacetylation was equally evoked by KH16 ± marbotinib (Fig. 2C). These findings suggest that low doses of HDACi cause hormesis through kinase signaling. KH16 plus marbotinib decreased the autophagy regulators p62 and HDAC6 (Fig. 2C), indicating a breakdown of autophagy. Inhibition of autophagy with chloroquine potentiated the ability of 50-100 nM KH16 to attenuate p62 and FLT3-ITD in MV4-11 and MOLM-13 cells. Moreover, this combination treatment caused a reduction and cleavage of the UNC-51-like autophagy-activating kinase-1 (ULK1) (Figs. 2D; S3). Caspase-dependent mechanisms likely conferred this loss and processing of proteins that belong to the autophagy system [12].
We analyzed how the combination of 25 nM KH16 and 2 nM marbotinib affected PBMCs and subpopulations therein. We noted that these drugs alone and in combination did not significantly induce apoptosis of B-cells, T-cells, monocytes, dendritic cells, NK cells, and PMNs (Fig. 2E). These data suggest that these inhibitors selectively kill leukemic cells carrying mutant FLT3 and spare normal blood cells.
In conclusion, the epigenetic drugs KH16 and KH29 are innovative lead compounds that deserve further consideration as treatment options for leukemia. One needs to consider the threshold concentrations at which HDACi evoke caspase-dependent apoptosis and abrogate cytoprotective autophagy. This finding stresses the need for HDACi that are effective at low nanomolar concentrations. Specific FLT3 kinase inhibitors disable undesired hormesis effects that HDACi cause. This allows such drug combinations to favorably combine against leukemic cells that carry the clinically unfavorable marker FLT3-ITD.
Supplementary information
Acknowledgements
Work done in the group of OHK is funded by the German Research Foundation/Deutsche Forschungsgemeinschaft (DFG) KR2291/9-1, project number 427404172; KR2291/12-1, project number 445785155; DFG KR2291/14-1, project number 469954457; KR2291/15-1, project number 495271833; KR2291/16-1, project number 496927074; KR2291/17-1, project number 502534123; KR2291/18-1, project number 528202295; funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 393547839 – SFB 1361; the DAAD Egypt/Germany; the Brigitte und Dr. Konstanze Wegener-Stiftung (projects 65/110); and the Walter Schulz-Stiftung. Work done in the group of WS is funded by the DFG project number 469954457, SI868/22-1, and the Alexander von Humboldt Foundation Project EGY 1191187 (HSI). We thank Prof. Dr. F.-D. Böhmer, Institute of Molecular Cell Biology, CMB, Jena University Hospital, Jena, Germany, for MV4-11 and MOLM-13 cells (originally from the DSMZ, authenticated by DNA fingerprint profiling using eight different and highly polymorphic short tandems repeats, at the Leibniz-Institute DSMZ, Braunschweig, Germany). We thank the Blood transfusion unit of the University Medical Center Mainz for providing us with buffy coats for PBMC isolation.
Author contributions
YZ and MH carried out most biological experiments. MBe supervised YZ and carried out experiments. KH and HSI synthesized and analytically characterized the HDAC inhibitors that were designed by WS. WB contributed material and scientific discussion. SM provided marbotinib and MBr provided PBMCs and analytical tools. OHK designed the experiments. The manuscript was drafted by OHK and approved for submission by all authors.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets that we generated during and analyzed during this study are available from the corresponding authors on reasonable request.
Competing interests
MBe, SM, and OHK declare the patent “Synthesis, pharmacology and use of new and selective FMS-like tyrosine kinase 3 (FLT3) inhibitors, WO2019/034538”; SM and OHK declare “Novel HDAC6 inhibitors and their uses, WO2016020369A1”; OHK declares “The use of molecular markers for the preclinical and clinical profiling of inhibitors of enzymes having histone deacetylase activity, WO/2004/027418”.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanira Zeyn, Kristin Hausmann, Melisa Halilovic.
Contributor Information
Wolfgang Sippl, Email: wolfgang.sippl@pharmazie.uni-halle.de.
Oliver H. Krämer, Email: okraemer@uni-mainz.de
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-023-02036-2.
References
- 1.Daver N, Venugopal S, Ravandi F. FLT3 mutated acute myeloid leukemia: 2021 treatment algorithm. Blood Cancer J. 2021;5:104. doi: 10.1038/s41408-021-00495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antar AI, Otrock ZK, Jabbour E, Mohty M, Bazarbachi A. FLT3 inhibitors in acute myeloid leukemia: ten frequently asked questions. Leukemia. 2020;3:682–96. doi: 10.1038/s41375-019-0694-3. [DOI] [PubMed] [Google Scholar]
- 3.Erba HP, Montesinos P, Kim HJ, Patkowska E, Vrhovac R, Zak P, et al. Quizartinib plus chemotherapy in newly diagnosed patients with FLT3-internal-tandem-duplication-positive acute myeloid leukaemia (QuANTUM-First): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2023;10388:1571–83. doi: 10.1016/S0140-6736(23)00464-6. [DOI] [PubMed] [Google Scholar]
- 4.Beyer M, Henninger SJ, Haehnel PS, Mustafa AM, Gurdal E, Schubert B, et al. Identification of a highly efficient dual type I/II FMS-like tyrosine kinase inhibitor that disrupts the growth of leukemic cells. Cell Chem Biol. 2022;3:398–411.e394. doi: 10.1016/j.chembiol.2021.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Sellmer A, Pilsl B, Beyer M, Pongratz H, Wirth L, Elz S, et al. A series of novel aryl-methanone derivatives as inhibitors of FMS-like tyrosine kinase 3 (FLT3) in FLT3-ITD-positive acute myeloid leukemia. Eur J Med Chem. 2020;193:112232. doi: 10.1016/j.ejmech.2020.112232. [DOI] [PubMed] [Google Scholar]
- 6.Li G, Tian Y, Zhu WG. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front Cell Dev Biol. 2020;8:576946. doi: 10.3389/fcell.2020.576946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mustafa AM, Krämer OH. Pharmacological Modulation of the Crosstalk between Aberrant Janus Kinase Signaling and Epigenetic Modifiers of the Histone Deacetylase Family to Treat Cancer. Pharm Rev. 2023;1:35–61. doi: 10.1124/pharmrev.122.000612. [DOI] [PubMed] [Google Scholar]
- 8.Wachholz V, Mustafa AM, Zeyn Y, Henninger SJ, Beyer M, Dzulko M, et al. Inhibitors of class I HDACs and of FLT3 combine synergistically against leukemia cells with mutant FLT3. Arch Toxicol. 2022;1:177–93. doi: 10.1007/s00204-021-03174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larrue C, Saland E, Boutzen H, Vergez F, David M, Joffre C, et al. Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells. Blood. 2016;7:882–92. doi: 10.1182/blood-2015-05-646497. [DOI] [PubMed] [Google Scholar]
- 10.Zalpoor H, Bakhtiyari M, Akbari A, Aziziyan F, Shapourian H, Liaghat M, et al. Potential role of autophagy induced by FLT3-ITD and acid ceramidase in acute myeloid leukemia chemo-resistance: new insights. Cell Commun Signal. 2022;1:172. doi: 10.1186/s12964-022-00956-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D, Chen Y, Yang Y, Yin Z, Huang C, Wang Q, et al. Autophagy activation mediates resistance to FLT3 inhibitors in acute myeloid leukemia with FLT3-ITD mutation. J Transl Med. 2022;1:300. doi: 10.1186/s12967-022-03498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer MA, Mustafa AM, Hausmann K, Ashry R, Kansy AG, Liebl MC, et al. Novel hydroxamic acid derivative induces apoptosis and constrains autophagy in leukemic cells. J Adv Res. 2023;23:00197–2.. doi: 10.1016/j.jare.2023.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Hai R, Yang D, Zheng F, Wang W, Han X, Bode AM, et al. The emerging roles of HDACs and their therapeutic implications in cancer. Eur J Pharm. 2022;931:175216. doi: 10.1016/j.ejphar.2022.175216. [DOI] [PubMed] [Google Scholar]
- 14.Ashry R, Mustafa AM, Hausmann K, Linnebacher M, Strand S, Sippl W, et al. NOXA Accentuates Apoptosis Induction by a Novel Histone Deacetylase Inhibitor. Cancers. 2023;14:3650. doi: 10.3390/cancers15143650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets that we generated during and analyzed during this study are available from the corresponding authors on reasonable request.