Abstract
Host immunity involves various immune cells working in concert to achieve balanced immune response. Host immunity interacts with tumorigenic process impacting disease outcome. Clusters of different immune cells may reveal unique host immunity in relation to breast cancer progression. CIBERSORT algorithm was used to estimate relative abundances of 22 immune cell types in 3 datasets, METABRIC, TCGA, and our study. The cell type data in METABRIC were analyzed for cluster using unsupervised hierarchical clustering (UHC). The UHC results were employed to train machine learning models. Kaplan–Meier and Cox regression survival analyses were performed to assess cell clusters in association with relapse-free and overall survival. Differentially expressed genes by clusters were interrogated with IPA for molecular signatures. UHC analysis identified two distinct immune cell clusters, clusters A (83.2%) and B (16.8%). Memory B cells, plasma cells, CD8 positive T cells, resting memory CD4 T cells, activated NK cells, monocytes, M1 macrophages, and resting mast cells were more abundant in clusters A than B, whereas regulatory T cells and M0 and M2 macrophages were more in clusters B than A. Patients in cluster A had favorable survival. Similar survival associations were also observed in other independent studies. IPA analysis showed that pathogen-induced cytokine storm signaling pathway, phagosome formation, and T cell receptor signaling were related to the cell type clusters. Our finding suggests that different immune cell clusters may indicate distinct immune responses to tumor growth, suggesting their potential for disease management.
Subject terms: Cancer, Risk factors
Introduction
Host immunity in tumor progression has reemerged as an important focus in cancer research1. The new development offers renewed hopes for novel anti-cancer therapies. Recent breakthrough in cancer immunotherapy, especially in the use of immune checkpoint inhibitors (ICI) to treat solid tumors, has invigorated researchers and oncologists in search for new therapeutic modalities to manage recurrent and metastatic malignancies which are otherwise resistant to available treatment2–4. However, the success in ICI has not been achieved uniformly for all cancer sites as certain types of cancer do not respond well to the new immunotherapy. ICI has shown promising results in treating melanoma, lung cancer (small cell and non-small cell), renal cell carcinoma, and urothelial carcinoma with significant improvement in clinical outcomes5–11, but the efficacy in breast cancer is limited12,13. Hormone receptor-positive tumors which are the most common breast cancer do not respond well to immunotherapy; only triple-negative breast cancer (TNBC) appears to have limited responses14. Thus, to better understand host immunity in breast cancer, we need to know not only the involvement of different immune and tumor cells, but also their interactions and responses to treatment.
Tumor microenvironment (TME) has been recognized to have significant impacts on cancer cell functions and activities and therefore affect tumor progression and metastasis. In addition to tumor cells and stromal components in TME, many local and infiltrating immune cells also play a crucial role in determining tumor growth and disease outcome15–17. Analyzing their configurations and abundances in TME has emerged as important parameters in assessing tumor specimens, predicting disease outcomes, and developing treatment strategies. Studies have shown that infiltrating cytotoxic lymphocytes in TME are associated with the efficacy of immunotherapy17,18. TNBC patients with high tumor infiltrating lymphocytes (TIL) are more responsive to ICI, whereas those with hormone receptor-positive breast tumors and low TIL are less responsive19. This discrepancy in TIL is explained in part by the differences in somatic mutations which not only reprogram cell signal pathways and metabolisms, but also generate tumor-associated and tumor-specific antigens (TAA, TSA)20,21. These altered or mutant molecules induce host immune response by attracting immune cell infiltration and congregation. Characterizing the abundance and composition of immune cell subtypes in tumor samples has shown values in disease prognosis and prediction of treatment responses22,23.
Cell sorting by flow cytometry and tissue staining with immunohistochemistry have been used to assess TIL, but these methods have some limitations with respect to tissue accessibility, processing challenges, and subjective evaluation24. Recently, computational approaches have been developed for in silico prediction of immune cell subtype abundances based on the readily available gene expression data on tissue transcriptomes. To assess if immune cell subtype clusters are useful for breast cancer prognosis, we analyzed transcriptomic data from several breast cancer datasets using the computation algorithm CYBERSORT25. The results of our analyses are presented in this report.
Results
Clusters of immune cell subtypes
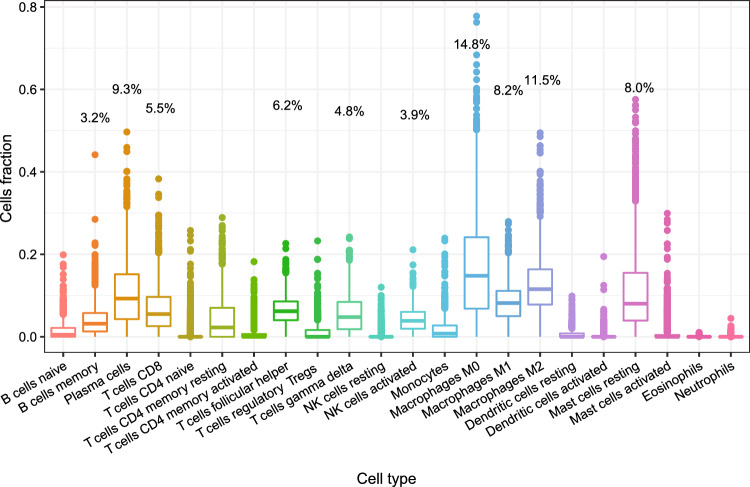
Figure 1 shows the relative abundances of each immune cell subtypes in METABRIC. Over half of the cell subtypes had very low abundances. Cell subtypes with relatively high abundances were M0 macrophages (14.8%), M2 macrophages (11.5%), plasma cells (9.3%), M1 macrophages (8.2%), resting mast cells (8.0%), follicular helper T cells (6.2%), CD8 positive T cells (5.5%), gamma delta T cells (4.8%), activated NK cells (3.9%), and memory B cells (3.2%).
Figure 1.
Distributions of immune cell subtypes in METABRIC.
UHC analysis indicated two clusters of immune cell subtypes in METABRIC (Supplementary Fig. 1). One cluster (hcluster 1 or cluster A) was observed in 1113 patients (83.2%), and another (hcluster 2 or cluster B) was in 224 patients (16.8%). Differences in cell subtypes between the two clusters and their comparisons with normal breast tissues are shown in Table 1. Cell subtypes which were significantly different between the two clusters included memory B cells, plasma cells, CD8 positive T cells, resting memory CD4 T cells, activated NK cells, monocytes, M1 macrophages, and resting mast cells, which showed higher abundances in cluster A than cluster B. Cell subtypes with relative abundances higher in cluster B than cluster A were regulatory T cells and M0 and M2 macrophages.
Table 1.
Two clusters of immune cell subtypes in METABRIC and immune cell subtypes in GTEx breast.
| Immune cell subtype | Median % in Cluster A (n = 1113) | Median % in Cluster B (n = 224) | Median % in GTEx (n = 269) | P value* Cluster A versus Cluster B | P value* Cluster A versus GTEx | P value* Cluster B versus GTEx |
|---|---|---|---|---|---|---|
| B cells naïve | 0.50 | 0.49 | 8.10 | 0.9907 | 4.4E−85 | 1.0E−48 |
| B cells memory | 3.44 | 2.08 | 0.00 | 5.8E−10 | 6.4E−99 | 3.9E−51 |
| Plasma cells | 10.10 | 4.86 | 6.49 | 4.1E−22 | 6.1E−07 | 0.0084 |
| T cells CD8 | 6.33 | 2.71 | 7.41 | 4.7E−29 | 0.0092 | 2.7E−26 |
| T cells CD4 naïve | 0.00 | 0.00 | 0.00 | 0.0922 | 3.1E−13 | 1.7E−07 |
| T cells CD4 memory resting | 2.80 | 0.00 | 9.47 | 3.3E−08 | 1.2E−42 | 2.5E−45 |
| T cells CD4 memory activated | 0.00 | 0.00 | 0.00 | 0.0002 | 7.2E−26 | 1.8E−12 |
| T cells follicular helper | 6.22 | 6.13 | 2.68 | 0.1943 | 1.3E−26 | 3.0E−13 |
| T cells regulatory Tregs | 0.00 | 1.28 | 0.00 | 3.4E−15 | 2.8E−05 | 2.1E−22 |
| T cells gamma delta | 4.86 | 4.25 | 0.00 | 0.1071 | 4.0E−109 | 4.6E−82 |
| NK cells resting | 0.00 | 0.00 | 1.92 | 5.1E−13 | 7.4E−114 | 5.7E−26 |
| NK cells activated | 4.32 | 1.70 | 2.09 | 5.4E−33 | 9.9E−25 | 0.011 |
| Monocytes | 1.03 | 0.00 | 3.16 | 5.5E−13 | 2.8E−32 | 2.4E−40 |
| Macrophages M0 | 12.40 | 36.00 | 0.00 | 8.5E−107 | 8.7E−66 | 2.0E−76 |
| Macrophages M1 | 8.52 | 6.46 | 1.65 | 3.1E−09 | 1.6E−86 | 3.9E−33 |
| Macrophages M2 | 10.90 | 15.40 | 23.79 | 1.5E−14 | 7.2E−65 | 3.5E−26 |
| Dendritic cells resting | 0.00 | 0.00 | 0.00 | 1.1E−15 | 3.4E−22 | 0.29 |
| Dendritic cells activated | 0.00 | 0.00 | 0.00 | 0.8621 | 5.8E−07 | 0.0019 |
| Mast cells resting | 8.98 | 3.93 | 11.72 | 2.3E−27 | 0.0001 | 5.6E−31 |
| Mast cells activated | 0.00 | 0.00 | 0.00 | 2.6E−12 | 0.0074 | 5.2E−14 |
| Eosinophils | 0.00 | 0.00 | 0.00 | 0.0364 | 6.3E−15 | 0.0023 |
| Neutrophils | 0.00 | 0.00 | 0.00 | 0.0605 | 2.5E−21 | 2.7E−06 |
*Mann–Whitney nonparametric test; bold p values < 0.002273 (0.05/22).
Immune cell subtype abundances were very different between normal breasts and breast tumors (Table 1). Compared to normal breasts, less abundant cell types in breast tumors included naïve B cells, resting CD4 memory T cells, resting NK cells, M2 macrophages, and resting mast cells; more abundant cell types in tumor samples were memory B cells, follicular helper T cells, gamma delta T cells, and M0 and M1 macrophages. Different abundances between clusters A and B tumor samples in comparison to normal breasts were plasma cells (higher in A, but lower in B), CD8 T cells (no difference in A, but lower in B), and activated NK cells (higher in A, but no difference in B).
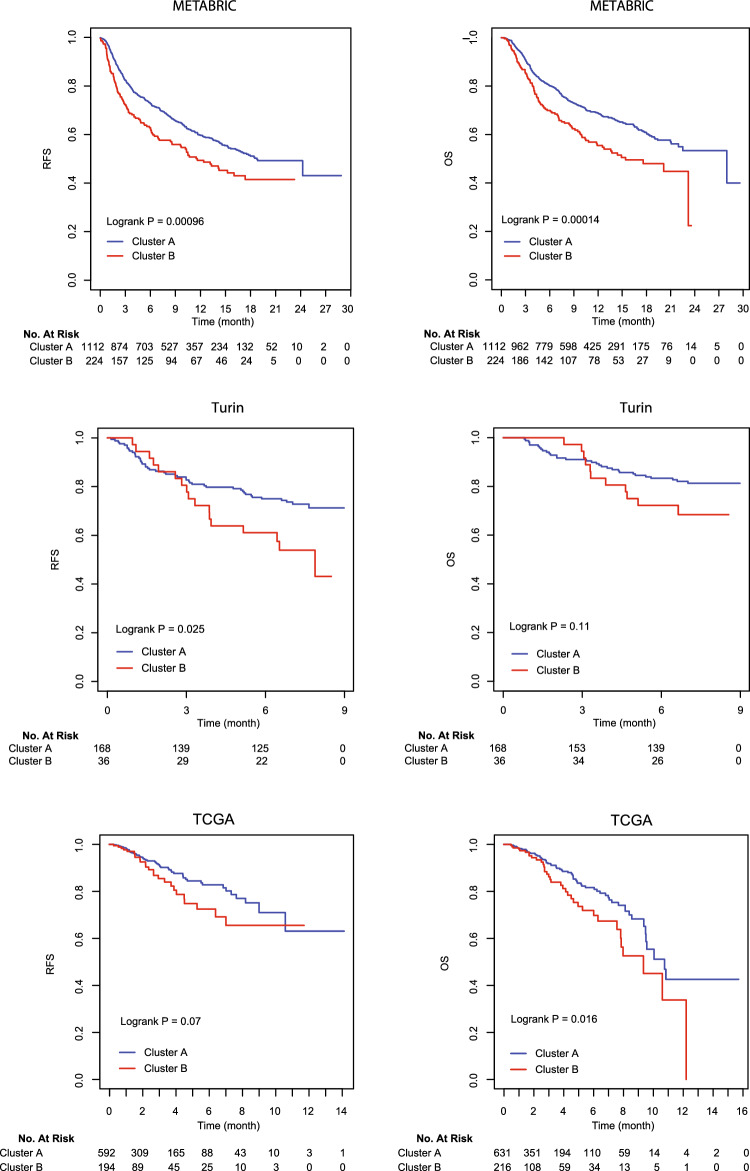
Associations of immune cell clusters with clinical and pathological variables of breast cancer in METABRIC are shown in Table 2. Patients with ER negative tumors or invasive ductal carcinoma were more prevalent in cluster B than in cluster A, and patients in cluster B were also more likely to develop recurrent disease or die. As expected, patients in cluster A had higher immune cytolytic activity or CYT scores compared to those in cluster B. Disease stage, tumor grade, age at diagnosis, PR status, and ERBB2 (HER2) overexpression were not significantly different between the two cell clusters. The cell cluster variable was significantly associated with relapse-free and overall survival (Fig. 2, METABRIC). These associations remained statistically significant in Cox proportional hazards regression models after clinical and pathological variables were adjusted in the analysis, including age at diagnosis, disease stage, tumor grade, tumor histology, and hormone receptor status (Table 3).
Table 2.
Associations between clinicopathological variables in METABRIC and immune cell clusters.
| Clinicopathological variable | Immune cell subtype clusters | p value* | ||
|---|---|---|---|---|
| Cluster A, n = 1113 (83.2%) | Cluster B, n = 224 (16.8%) | Total n = 1337 | ||
| Mean age (SD) | 59.4 (13.1) | 59.8 (13.2) | 59.4 (13.1) | 0.67 |
| Age group | 0.91 | |||
| < 60 years | 556 (50.0) | 111 (49.6) | 667 (49.9) | |
| ≥ 60 years | 557 (50.0) | 113 (50.4) | 670 (50.1) | |
| Stage | 0.41 | |||
| 0 | 2 (0.2) | 1 (0.6) | 3 (0.3) | |
| 1 | 2781 (33.5) | 56 (32.2) | 334 (33.2) | |
| 2 | 464 (55.8) | 105 (60.3) | 569 (56.6) | |
| 3 | 80 (9.6) | 12 (6.9) | 92 (9.2) | |
| 4 | 7 (0.8) | 7 (0.7) | 0 | |
| Grade | 0.11 | |||
| 1 | 89 (8.3) | 13 (6.0) | 102 (7.9) | |
| 2 | 405 (37.7) | 70 (32.4) | 475 (36.8) | |
| 3 | 580 (54.0) | 133 (61.6) | 713 (55.3) | |
| Histology | 0.006 | |||
| Ductal | 830 (75.1) | 191 (86.0) | 1021 (76.9) | |
| Lobular | 96 (8.7) | 10 (4.5) | 106 (8.0) | |
| Mixed | 128 (11.6) | 16 (7.2) | 144 (10.9) | |
| Others | 51 (4.6) | 5 (2.3) | 56 (4.2) | |
| ER | 0.006 | |||
| Positive | 799 (71.8) | 140 (62.5) | 939 (70.2) | |
| Negative | 314 (28.2) | 84 (37.5) | 398 (29.8) | |
| PR | 0.16 | |||
| Positive | 529 (47.5) | 95 (42.4) | 624 (46.7) | |
| Negative | 584 (52.5) | 129 (57.6) | 713 (53.3) | |
| HER2 | 0.24 | |||
| Positive | 168(15.1) | 27(12.1) | 195(14.6) | |
| Negative | 945(84.9) | 197(87.9) | 1,142(85.6) | |
| Relapse | 0.003 | |||
| No | 677(60.9) | 112(50.0) | 789(59.1) | |
| Yes | 435(39.1) | 112(50.0) | 547(40.9) | |
| Death | 0.001 | |||
| No | 522(46.9) | 78(34.8) | 600(44.9) | |
| Yes | 591(53.1) | 146(65.2) | 737(55.1) | |
| Cytolytic activity | < 0.0001 | |||
| CYT score | 6.9 (0.7) | 6.3 (0.6) | 6.8 (0.8) | |
*Student’s T-test, Pearson’s Chi-squared test, or Fisher’s exact test where appropriate.
Significant values are in bold.
Figure 2.
Kaplan–Meier curves on relapse-free survival (RFS) and overall survival (OS) in METABRIC, Turin, and TCGA.
Table 3.
Associations between immune cell subtype cluster and breast cancer survival.
| Dataset | Relapse-free survival | Overall survival | Relapse-free survival* | Overall survival* | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | HR | 95%CI | P | HR | 95%CI | P | |
| METABRIC | 0.001 | < 0.001 | 0.009 | 0.011 | ||||||||
| Cluster A | 1 | 1 | 1 | 1 | ||||||||
| Cluster B | 1.42 | 1.15–1.74 | 1.60 | 1.28–2.00 | 1.38 | 1.08–1.76 | 1.37 | 1.08–1.75 | ||||
| Turin study | 0.028 | 0.114 | 0.154 | 0.358 | ||||||||
| Cluster A | 1 | 1 | 1 | 1 | ||||||||
| Cluster B | 1.87 | 1.07–3.27 | 1.74 | 0.85–3.47 | 1.53 | 0.85–2.73 | 1.40 | 0.68–2.89 | ||||
| TCGA# | 0.072 | 0.017 | 0.002 | 0.004 | ||||||||
| Cluster A | 1 | 1 | 1 | 1 | ||||||||
| Cluster B | 1.54 | 0.96–2.47 | 1.63 | 1.09–2.43 | 2.24 | 1.34–3.74 | 1.93 | 1.24–3.01 | ||||
*Adjusted for age, stage, grade, ER, PR, and histology.
#Tumor grade not included in multivariate analysis.
Significant values are in bold.
Immune cell cluster modeling
We used random forest (RF) to build a prediction model for cell subtype clusters. The RF model was trained with the UHC results in 60% of the METABRIC data, and the model fit well to the UHC clusters with 100% and 98% AUC in the training and testing sets, respectively (Supplementary Fig. 2). Although DNN, elastic net, and stepAIC models were also matched well to UHC, the AUC of RF in the training set was higher than that in other three models. Thus, we used the RF model to predict immune cell clusters in the Turin study and TCGA. The RF predicted cell clusters were analyzed for its associations with patient survival. Similar associations with relapse-free and overall survival were found in the Turin study (Fig. 2), i.e., cluster B associated with poor survival, although the associations were not statistically significant after adjusting for clinicopathological variables (Table 3). Associations between patient survival and immune cell clusters were also observed in TCGA. Patients with immune cell subtypes in cluster B had higher risks for disease recurrence and death compared to those with cell subtypes in cluster A (Fig. 2). The survival associations in TCGA were statistically significant after adjusting for clinicopathological variables (Table 3). No associations between immune cell clusters and ER status or histological types were observed in these validation studies (data not shown).
The importance of each cell type in the RF model was evaluated with mean decreases in accuracy and the Gini coefficient. The top 5 important cell types were M0 and M2 macrophages, CD8 positive T cells, activated NK cells, and resting mast cells (Supplementary Fig. 3). The stepAIC analysis showed a 19-cell model, and the elastic net suggested a 13-cell regression (Supplementary Table 1). Twelve cell types were common in both models, including naïve B cells, memory B cells, plasma cells, CD8 positive T cells, resting memory CD4 T cells, activated memory CD4 T cells, regulatory T cells, activated NK cells, M2 macrophages, resting mast cells, activated mast cells, and neutrophils.
IPA analysis on DEGs
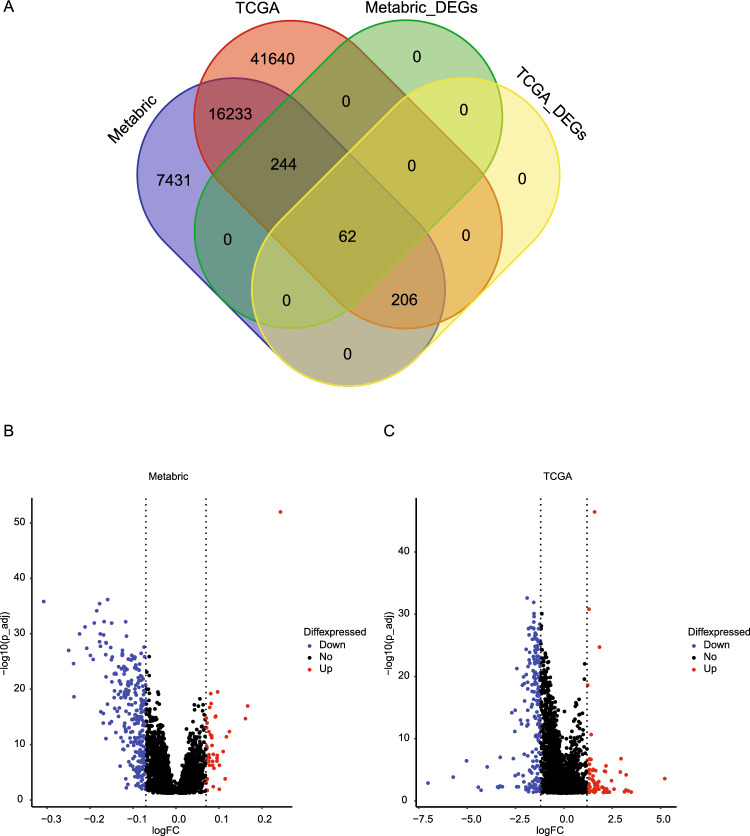
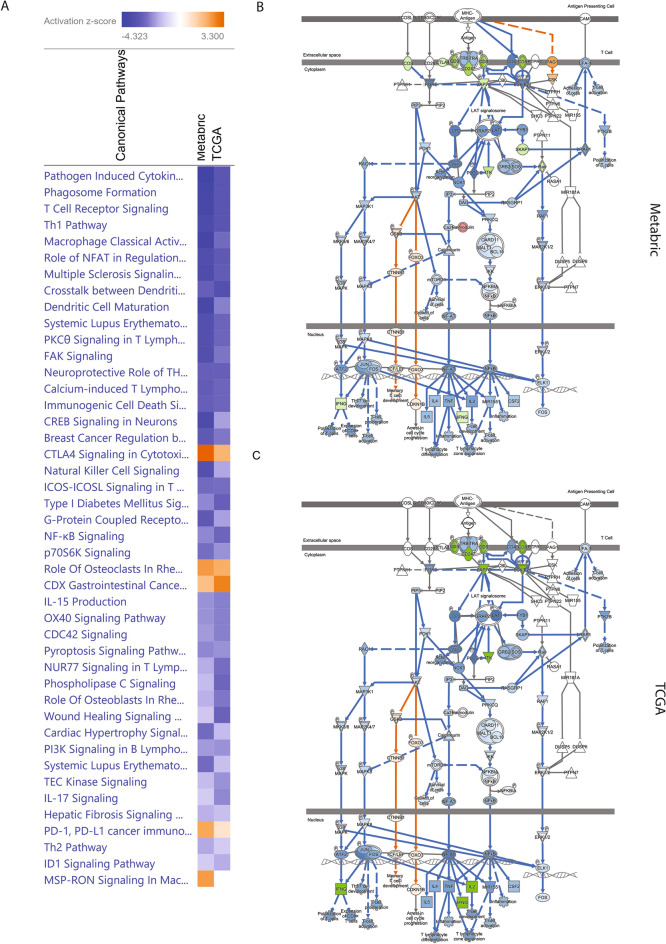
There were 16,621 genes overlapping between the transcriptomic data of METABRIC and TCGA. IPA was performed on the 268 DEGs in TCGA (absolute log2 fold change at 1.2 or larger for cluster B versus cluster A; BH adjusted P < 0.05) (Fig. 3A). Since the expression data in METABRIC had a smaller range and the median fold change was only 1.001 (IQR: 0.996–1.184), we used the absolute log2 fold change at 0.07 as a threshold and selected 306 DEGs for IPA analysis. Volcano plot showed the selected DEGs in METABRIC and TCGA (Fig. 3B,C). Graphical summary of IPA analysis on cell cluster associated DEGs showed that the transcription profiles were similar between METABRIC and TCGA, with most of the signal pathways being downregulated (Supplementary Fig. 4). The top 5 common signal pathways predicted by IPA in METABRIC and TCGA were pathogen induced cytokine storm signaling pathway, phagosome formation, T cell receptor signaling, T helper 1 pathway, and macrophage classical activity, all of which were downregulated (Fig. 4A). The T cell receptor signaling showed the similar patterns of network in METABRIC and TCGA (Fig. 4B,C).
Figure 3.
(A) Venn diagram of total available genes and DEGs in METABRIC and TCGA; (B) Volcano plot of DEGs identified in METABRIC; (C) Volcano plot of DEGs identified in TCGA. FC: fold change.
Figure 4.
(A) Comparison of IPA analysis between METABRIC and TCGA using the ingenuity pathway analysis (IPA, www.qiagen.com/ingenuity); (B) T cell receptor signaling predicted IPA in METABRIC; (C) T cell receptor signaling predicted by IPA in TCGA.
Discussion
We used CIBERSORT to estimate the relative abundances of 22 immune cell subtypes in breast cancer and normal breast tissues and found significant differences in cell types between tumor and normal tissues. The deconvolution results on cell subtypes were further analyzed in breast cancer (METABRIC) with unsupervised hierarchical clustering, and the analysis suggested two distinct clusters of immune cell subtypes associated with different survival outcomes of breast cancer. These survival associations were replicated independently in our study (Turin) and TCGA when using a random forest model which was trained with the UHC classifications in METABRIC. The survival associations with immune cell clusters appeared to be independent from most known clinical and pathological variables of breast cancer, suggesting the importance of host immunity in determining tumor progression and host-tumor interaction. The machine learning-based cell cluster analyses split the tumor samples into large (83%) and small (17%) groups, which appears to match with the general trend of breast cancer outcome where most patients have a favorable prognosis (> 80%).
Previously, Ali et al. performed hierarchical clustering analysis on immune cell subtypes in 10,988 tumor samples from 56 studies26. Their analysis showed 7 clusters in 6071 samples. The authors concluded that there were substantial variations in immune cell subtypes in TME and that tumor characteristics might determine the cell type variability. A recent study by Tekpli et al.27 reported 3 clusters of immune infiltration based on the expression of 509 genes, and the clusters were correlated with lymphoid and myeloid infiltration from low to high, with high and low infiltration clusters associated with favorable survival compared to intermediate infiltration. Since the study used a different method to determine tumor immunity, we cannot directly compare the clustering results between the two studies, but both studies indicate that breast cancer may be classified into immunity-based subtypes which have clinical implications in predicting disease prognosis and treatment response.
Going through the cell types in each cluster, we found that memory B cells, plasma cells, CD8 positive T cells, resting memory CD4 T cells, activated NK cells, monocytes, M1 macrophages, and resting mast cells were significantly higher in the favorable cluster (cluster A), whereas regulatory T cells and M0 and M2 macrophages were substantially higher in unfavorable cluster (cluster B). These differentiating cell types appear to be consistent with the current understanding that hot or immune-inflamed TME, which has favorable prognosis and is responsive to immunotherapy, is infiltrated with cytotoxic T cells (CD8 positive T cells), NK cells, and M1 macrophages, whereas cold TME is filled with immunosuppressive lymphocytes like regulatory T cells and tumor-associated macrophages (TAM), M0 and M228. NK cells and CD8 positive T cells are known to be able to suppress tumor growth through their cytotoxic activities28,29. Furthermore, CD4 memory T cells and M1 microphages facilitate the effects of NK cells and cytotoxic T cells30. Conversely, M2 macrophages and regulatory T cells inhibit the activities of CD4 memory and CD8 cytotoxic T cells, respectively31.
We analyzed the cell type data by focusing on immune cells in clusters instead of individual cells because host immunity is complex and involves different mechanisms and diverse cell lineages which give rise to innate versus adaptive, local versus systemic, and cellular versus humoral immunities. These distinct immune activities are carried out by a variety of cell types which work in concert to mount an appropriate immune response32. Thus, analyzing any single cell or a few cell types may not reveal enough insights into the interplay between tumor immunogenicity and host immune response as well as the potential impact of their interaction on tumor growth and disease outcome33. Ali et al.26 assessed individual immune cell subtypes in relation to breast cancer survival by ER status, and the large study found that only two cell types showed consistent associations with survival outcomes, regulatory T cells and M2 macrophages, both of which were associated with poor survival. Although multiple cell type clusters were found in that study, the survival associations with some cell types were generally consistent with those observed in our cluster analysis. For example, Ali et al.26 found favorable survival associations with monocytes and memory B cells in ER positive tumors and with CD8 positive T cells in ER negative tumors, as well as unfavorable survival associations with M0 macrophages for ER positive tumors.
TCR stimulation is a fundamental step in most T cell responses. TCR signaling is important for many aspects of T cell regulation, including development, differentiation, activation, proliferation, and survival. Dysregulation of TCR signaling can result in allergy and autoimmune diseases34. The molecular mechanism of TCR suppression underlying the link between immune cells in cluster B and breast cancer progression remains to be elucidated.
One limitation of our study is that we cannot assess the temporal and spatial variations of immune cell subtypes in tumor specimens, which is known to play an important role in determining the effect of host immunity and host-immune interplay in addition to cell types35,36. Anti-cancer therapies are known to have significant impacts on TME and immune cell infiltration37. Our analysis of immune cell subtypes in cluster only reflects the cell composition at the time of initial mastectomy which may be considered as a baseline status of TME that is different from those of post-surgery and during systemic anti-cancer treatment. The other limitation is that our deconvolution was not based on the entire 547 reference genes in LM22. Although not all signature matrix genes are required for deconvolution, the algorithm’s performance is improved with the presence of more signature genes38.
Conclusions
This study applied different machine learning methods to analyze immune cell subtypes in clusters and found two distinct clusters in breast cancer associated with survival outcomes. The survival associations were replicated independently in two additional datasets. Immune cell subtypes which were more abundant in the cluster of favorable prognosis included memory B cells, plasma cells, CD8 positive T cells, resting memory CD4 T cells, activated NK cells, monocytes, M1 macrophages, and resting mast cells, and those less abundant were regulatory T cells, and M0 and M2 macrophages. The immune cell clusters associated with breast cancer progression may involve suppression of pathogen induced cytokine storm signaling pathway, phagosome formation, T cell receptor signaling, T helper 1 cell pathway, and macrophage classical activity pathways. Our finding suggests that immune cell clusters in primary breast cancer may be an important parameter to consider, in addition to individual cell types, when predicting disease outcome and planning treatment strategy.
Methods
Study design and participants
Two online datasets on transcriptome, METABRIC and TCGA39,40, were used for analysis together with their clinical and follow-up information. METABRIC, downloaded from cBioPortal (https://www.cbioportal.org/)41,42, has 1903 breast tumor samples with gene expression data on 24,368 genes measured by a microarray chip from Illumine (Illumina HT-12 v3). The log2 intensity values were used for cell type deconvolution. Clinical data and survival information available for analysis in METABRIC include age at diagnosis, disease stage, tumor grade, histological type, estrogen receptor (ER) status, progesterone receptor (PR) status, ERBB2 (HER2) overexpression, disease recurrence, death, and follow-up time. TCGA RNA-seq data, expressed as fragments per kilobase of exon per million mapped fragments (FPKM), on 1075 breast tumor samples were downloaded from the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/)43. The corresponding clinical information was downloaded from cBioPortal.
An independent dataset of tumor transcriptomes from 204 breast cancer patients was available from a previous study (Turin) of ours described in detail elsewhere44. In brief, we recruited 348 patients who were diagnosed with primary breast cancer and underwent mastectomies in the University Hospital at University of Turin in Italy45. Fresh tumor samples were collected during surgery and snap-frozen in liquid nitrogen immediately after resection. Total RNA was extracted, of which 205 were selected for microarray analysis using the Illumina Expression BeadChip (HumanRef-8 v1). The raw expression data (~ .idat) generated by the Illumina microarray assay were processed using GenomeStudio V2011.1. Data was normalized using the function neqc() in R package limma, This function performs normexp background correction using negative controls, then quantile normalizes and finally log2 transforms. The normalized data was ready for CIBERSORT deconvolution of 22 immune cell types24,46. Transcriptomic data on normal breast tissues were downloaded from the GTEx Portal (https://www.gtexportal.org/home/datasets) which contains the transcripts per million (TPM) of RNA-seq data on 459 tissue specimens (GTEx Analysis V8).
CIBERSORT estimates the relative abundances of immune cell subtypes in tissue samples. The computation algorithm deconvolutes 22 immune cell subtypes from tumor transcriptomes using reference LM22. LM22 includes the expression of 547 reference genes, of which 475 were available in METABRIC, 444 in the Turin study, 537 in TCGA, and 527 in the GTEx data. CIBERSORT interrogates tumor transcriptome for immune cell subtypes based on the assumption that tissue samples contain mixed cell populations38. To evaluate the validity of cell type deconvolution in METABRIC and TCGA, we selected 100 permutations as recommended to achieve statistical rigor without applying quantile normalization. Tumor samples with deconvolution results not significantly different from the null hypothesis (p > 0.05) were excluded from final analysis. The null hypothesis assumes no immune cell subtypes present in a tumor sample based on LM22. After removing the samples without significance, we obtained 1337 samples from METABRIC, 848 samples from TCGA, and 269 samples from GTEx qualified for cell type analysis.
Model development and statistical analysis
We performed unsupervised hierarchical clustering (UHC) analysis on the immune cell subtypes from METABRIC using the ‘hclust’ function with ‘complete’ selection in R. Based on the UHC results, we created a dichotomous variable on cell subtype clusters. Differences in immune cell subtypes between clusters were compared using the Mann–Whitney nonparametric U test. Associations of cell subtype clusters with clinical and pathological variables were analyzed with the Chi-square test. Kaplan–Meier survival curves and log-rank test were used to evaluate survival differences between patients in different immune cell clusters. Cox proportional hazards regression analysis was performed to determine survival associations with immune cell clusters while adjusting for clinicopathological variables. Two-side p values < 0.05 were considered statistical significance. All the analyses were performed using R (version 4.0.5).
To predict cell subtype clusters, we tested 4 machine learning models, including random forest (RF), deep neural network (DNN), stepAIC, and elastic net. The models were initially trained based on the UHC results in 60% of METABRIC and then tested in the remaining 40% of the data. METABRIC data were randomly split into training and testing sets. The RF model was developed using the ‘randomForest’ package in R with 500-tree selection. The importance of immune cell subtypes in the model was evaluated by mean decrease in accuracy and the Gini coefficient. The DNN model was trained using the CPU implementation of TensorFlow (version 1.14.0) for 2000 steps with a 7 × 7 hidden layer in Python (version 3.6.13). Regression models of stepAIC and elastic net were developed using the “MASS” and “glmnet” packages in R (version 4.0.3)47. Model comparison was made between UHC and each of the 4 machine learning methods using the “pROC” package in R which calculates the receiver operating characteristic (ROC) curves and area under the curve (AUC)48. DeLong’s test was used for AUC comparison between models. We also evaluated immune cytolytic activity by calculating the CYT score49.
Wilcoxon test was performed for the differentially expressed genes (DEG) analysis between cluster A and cluster B (cluster B vs. cluster A) in METABRIC and TCGA. P values were adjusted for the Benjamin-Hochberg correction (BH). The ingenuity pathway analysis (IPA) (www.qiagen.com/ingenuity) was performed on the significant DEGs to explore the signal pathways enriched in cell clusters.
Supplementary Information
Author contributions
Conceptualization, Z.W. and H.Y.; methodology, Z.W. and H.Y.; data curation, Z.W., D.K., J.W., and N.B.; data analysis, Z.W. and H.Y.; validation, Z.W. and H.Y.; writing the original manuscript, Z.W. and H.Y.; Review and editing, Z.W., D.K., J.W., N.B., B.H., P.F., L.L., H.R. and H.Y.; supervision, H.Y.; All authors reviewed the manuscript.
Data availability
The TCGA, METABRIC, and Transcriptomic data on normal breast tissues are available in the following website: https://portal.gdc.cancer.gov; https://www.cbioportal.org; https://www.gtexportal.org/home/datasets, respectively. All additional information including Turin data required to reproduce our results is available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-45932-4.
References
- 1.Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat. Rev. Cancer. 2021;21(6):345–359. doi: 10.1038/s41568-021-00347-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vesely MD, Schreiber RD. Cancer immunoediting: Antigens, mechanisms, and implications to cancer immunotherapy. Ann. N. Y. Acad. Sci. 2013;1284:1–5. doi: 10.1111/nyas.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaddepally RK, Kharel P, Pandey R, Garje R, Chandra AB. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers (Basel) 2020;12(3):738. doi: 10.3390/cancers12030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mencoboni M, Ceppi M, Bruzzone M, Taveggia P, Cavo A, Scordamaglia F, Gualco M, Filiberti RA. Effectiveness and safety of immune checkpoint inhibitors for patients with advanced non small-cell lung cancer in real-world: Review and meta-analysis. Cancers (Basel) 2021;13(6):1388. doi: 10.3390/cancers13061388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggermont AMM, Blank CU, Mandala M, Long GV, Atkinson VG, Dalle S, Haydon AM, Meshcheryakov A, Khattak A, Carlino MS, Sandhu S, Larkin J, Puig S, Ascierto PA, Rutkowski P, Schadendorf D, Koornstra R, Hernandez-Aya L, Di Giacomo AM, van den Eertwegh AJM, Grob JJ, Gutzmer R, Jamal R, Lorigan PC, van Akkooi ACJ, Krepler C, Ibrahim N, Marreaud S, Kicinski M, Suciu S, Robert C. Longer follow-up confirms recurrence-free survival benefit of adjuvant pembrolizumab in high-risk stage III melanoma: Updated results from the EORTC 1325-MG/KEYNOTE-054 trial. J. Clin. Oncol. 2020;38(33):3925–3936. doi: 10.1200/JCO.20.02110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber J, Mandala M, Del Vecchio M, Gogas HJ, Arance AM, Cowey CL, Dalle S, Schenker M, Chiarion-Sileni V, Marquez-Rodas I, Grob JJ, Butler MO, Middleton MR, Maio M, Atkinson V, Queirolo P, Gonzalez R, Kudchadkar RR, Smylie M, Meyer N, Mortier L, Atkins MB, Long GV, Bhatia S, Lebbe C, Rutkowski P, Yokota K, Yamazaki N, Kim TM, de Pril V, Sabater J, Qureshi A, Larkin J, Ascierto PA, CheckMate C. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med. 2017;377(19):1824–1835. doi: 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- 9.Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthelemy P, Porta C, George S, Powles T, Donskov F, Neiman V, Kollmannsberger CK, Salman P, Gurney H, Hawkins R, Ravaud A, Grimm MO, Bracarda S, Barrios CH, Tomita Y, Castellano D, Rini BI, Chen AC, Mekan S, McHenry MB, Wind-Rotolo M, Doan J, Sharma P, Hammers HJ, Escudier B, CheckMate I. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018;378(14):1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ready NE, Ott PA, Hellmann MD, Zugazagoitia J, Hann CL, de Braud F, Antonia SJ, Ascierto PA, Moreno V, Atmaca A, Salvagni S, Taylor M, Amin A, Camidge DR, Horn L, Calvo E, Li A, Lin WH, Callahan MK, Spigel DR. Nivolumab monotherapy and nivolumab plus ipilimumab in recurrent small cell lung cancer: Results from the CheckMate 032 randomized cohort. J. Thorac. Oncol. 2020;15(3):426–435. doi: 10.1016/j.jtho.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Hellmann MD, Ramalingam SS. Nivolumab plus ipilimumab in non-small-cell lung cancer. N. Engl. J. Med. 2020;382(9):875. doi: 10.1056/NEJMc1916859. [DOI] [PubMed] [Google Scholar]
- 12.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Dieras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA, Investigators IMT. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018;379(22):2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 13.Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, Gallardo C, Lipatov O, Barrios CH, Holgado E, Iwata H, Masuda N, Otero MT, Gokmen E, Loi S, Guo Z, Zhao J, Aktan G, Karantza V, Schmid P, for the KEYNOTE-355 Investigators Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396(10265):1817–1828. doi: 10.1016/S0140-6736(20)32531-9. [DOI] [PubMed] [Google Scholar]
- 14.Kearney MR, McGuinness JE, Kalinsky K. Clinical trial data and emerging immunotherapeutic strategies: Hormone receptor-positive, HER2- negative breast cancer. Breast Cancer Res. Treat. 2021;189(1):1–13. doi: 10.1007/s10549-021-06291-8. [DOI] [PubMed] [Google Scholar]
- 15.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen M, Louise JJ. Jekyll and Hyde: The role of the microenvironment on the progression of cancer. J. Pathol. 2011;223(2):162–176. doi: 10.1002/path.2803. [DOI] [PubMed] [Google Scholar]
- 17.Salmon H, Remark R, Gnjatic S, Merad M. Host tissue determinants of tumour immunity. Nat. Rev. Cancer. 2019;19(4):215–227. doi: 10.1038/s41568-019-0125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo K, Yoshie O, Nakayama T. Multifaceted roles of chemokines and chemokine receptors in tumor immunity. Cancers (Basel) 2021;13(23):6132. doi: 10.3390/cancers13236132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Althobiti M, Aleskandarany MA, Joseph C, Toss M, Mongan N, Diez-Rodriguez M, Nolan CC, Ashankyty I, Ellis IO, Green AR, Rakha EA. Heterogeneity of tumour-infiltrating lymphocytes in breast cancer and its prognostic significance. Histopathology. 2018;73(6):887–896. doi: 10.1111/his.13695. [DOI] [PubMed] [Google Scholar]
- 20.Haen SP, Loffler MW, Rammensee HG, Brossart P. Towards new horizons: Characterization, classification and implications of the tumour antigenic repertoire. Nat. Rev. Clin. Oncol. 2020;17(10):595–610. doi: 10.1038/s41571-020-0387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janelle V, Rulleau C, Del Testa S, Carli C, Delisle JS. T-cell immunotherapies targeting histocompatibility and tumor antigens in hematological malignancies. Front. Immunol. 2020;11:276. doi: 10.3389/fimmu.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safi S, Yamauchi Y, Rathinasamy A, Stamova S, Eichhorn M, Warth A, Rauch G, Dienemann H, Hoffmann H, Beckhove P. Functional T cells targeting tumor-associated antigens are predictive for recurrence-free survival of patients with radically operated non-small cell lung cancer. Oncoimmunology. 2017;6(11):e1360458. doi: 10.1080/2162402X.2017.1360458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, Villaflor V, Giles F. Association of tumor mutational burden with DNA repair mutations and response to anti-PD-1/PD-L1 therapy in non-small-cell lung cancer. Clin. Lung Cancer. 2019;20(2):88–96 e6. doi: 10.1016/j.cllc.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Liu CC, Steen CB, Newman AM. Computational approaches for characterizing the tumor immune microenvironment. Immunology. 2019;158(2):70–84. doi: 10.1111/imm.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: A gene-expression-based retrospective study. PLoS Med. 2016;13(12):e1002194. doi: 10.1371/journal.pmed.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tekpli X, Lien T, Rossevold AH, Nebdal D, Borgen E, Ohnstad HO, Kyte JA, Vallon-Christersson J, Fongaard M, Due EU, Svartdal LG, Sveli MAT, Garred O, Osbreac, Frigessi A, Sahlberg KK, Sorlie T, Russnes HG, Naume B, Kristensen VN. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat. Commun. 2019;10(1):5499. doi: 10.1038/s41467-019-13329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadeghalvad M, Mohammadi-Motlagh HR, Rezaei N. Immune microenvironment in different molecular subtypes of ductal breast carcinoma. Breast Cancer Res. Treat. 2021;185(2):261–279. doi: 10.1007/s10549-020-05954-2. [DOI] [PubMed] [Google Scholar]
- 30.Wein L, Savas P, Luen SJ, Virassamy B, Salgado R, Loi S. Clinical validity and utility of tumor-infiltrating lymphocytes in routine clinical practice for breast cancer patients: Current and future directions. Front. Oncol. 2017;7:156. doi: 10.3389/fonc.2017.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desmedt C, Salgado R, Fornili M, Pruneri G, Van den Eynden G, Zoppoli G, Rothe F, Buisseret L, Garaud S, Willard-Gallo K, Brown D, Bareche Y, Rouas G, Galant C, Bertucci F, Loi S, Viale G, Di Leo A, Green AR, Ellis IO, Rakha EA, Larsimont D, Biganzoli E, Sotiriou C. Immune infiltration in invasive lobular breast cancer. J. Natl. Cancer Inst. 2018;110(7):768–776. doi: 10.1093/jnci/djx268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaplin DD. Overview of the immune response. J. Allergy Clin. Immunol. 2010;125(2 Suppl 2):S3–23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang JR, Byeon Y, Kim D, Park SG. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 2020;52(5):750–761. doi: 10.1038/s12276-020-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Zhai J, Sun B, Parra ER, Jiang M, Ma W, Wang J, Kang AM, Kannan K, Pandurengan R, Zhang S, Solis LM, Haymaker CL, Raso MG, Mendoza Perez J, Sahin AA, Wistuba II, Yam C, Litton JK, Yang F. Effector memory cytotoxic CD3(+)/CD8(+)/CD45RO(+) T cells are predictive of good survival and a lower risk of recurrence in triple-negative breast cancer. Mod. Pathol. 2021 doi: 10.1038/s41379-021-00973-w. [DOI] [PubMed] [Google Scholar]
- 36.Hammerl D, Martens JWM, Timmermans M, Smid M, Trapman-Jansen AM, Foekens R, Isaeva OI, Voorwerk L, Balcioglu HE, Wijers R, Nederlof I, Salgado R, Horlings H, Kok M, Debets R. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat. Commun. 2021;12(1):5668. doi: 10.1038/s41467-021-25962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter JM, Polley MC, Leon-Ferre RA, Sinnwell J, Thompson KJ, Wang X, Ma Y, Zahrieh D, Kachergus JM, Solanki M, Boughey JC, Liu MC, Ingle JN, Kalari KR, Couch FJ, Thompson EA, Goetz MP. Characteristics and spatially defined immune (micro)landscapes of early-stage PD-L1-positive triple-negative breast cancer. Clin. Cancer Res. 2021;27(20):5628–5637. doi: 10.1158/1078-0432.CCR-21-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, METABRIC Group. Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, Tsui DW, Liu B, Dawson SJ, Abraham J, Northen H, Peden JF, Mukherjee A, Turashvili G, Green AR, McKinney S, Oloumi A, Shah S, Rosenfeld N, Murphy L, Bentley DR, Ellis IO, Purushotham A, Pinder SE, Borresen-Dale AL, Earl HM, Pharoah PD, Ross MT, Aparicio S, Caldas C. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen MA, Ferretti V, Grossman RL, Staudt LM. The NCI genomic data commons as an engine for precision medicine. Blood. 2017;130(4):453–459. doi: 10.1182/blood-2017-03-735654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu L, Tuck D, Katsaros D, Lu L, Schulz V, Perincheri S, Menato G, Scarampi L, Harris L, Yu H. Favorable outcome associated with an IGF-1 ligand signature in breast cancer. Breast Cancer Res. Treat. 2012;133(1):321–331. doi: 10.1007/s10549-012-1952-5. [DOI] [PubMed] [Google Scholar]
- 45.Mu L, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Yu H. TGF-beta1 genotype and phenotype in breast cancer and their associations with IGFs and patient survival. Br. J. Cancer. 2008;99(8):1357–1363. doi: 10.1038/sj.bjc.6604689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 2010;38(22):e204. doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33(1):1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 49.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA, METABRIC, and Transcriptomic data on normal breast tissues are available in the following website: https://portal.gdc.cancer.gov; https://www.cbioportal.org; https://www.gtexportal.org/home/datasets, respectively. All additional information including Turin data required to reproduce our results is available from the corresponding author upon request.