Abstract
Microorganisms present in mosquitoes and their interactions are key factors affecting insect development. Among them, Wolbachia is closely associated with the host and affects several fitness parameters. In this study, the bacterial and fungal microbiota from two laboratory Culex quinquefasciatus isolines (wild type and tetracycline-cured) were characterized by metagenome amplicon sequencing of the ITS2 and 16S rRNA genes at different developmental stages and feeding conditions. We identified 572 bacterial and 61 fungal OTUs. Both isolines presented variable bacterial communities and different trends in the distribution of diversity among the groups. The lowest bacterial richness was detected in sugar-fed adults of the cured isoline, whereas fungal richness was highly reduced in blood-fed mosquitoes. Beta diversity analysis indicated that isolines are an important factor in the differentiation of mosquito bacterial communities. Considering composition, Penicillium was the dominant fungal genus, whereas Wolbachia dominance was inversely related to that of Enterobacteria (mainly Thorsellia and Serratia). This study provides a more complete overview of the mosquito microbiome, emphasizing specific highly abundant components that should be considered in microorganism manipulation approaches to control vector-borne diseases.
Subject terms: Microbial communities, Applied microbiology, Computational biology and bioinformatics
Introduction
Mosquitoes (Diptera: Culicidae) are considered the most significant medically important insects because of their impact on global public health, as they can vectorize a wide range of pathogens, including viruses and parasites, that affect millions of people each year1. Effective, cost-efficient, and safe control of these diseases requires the implementation of mosquito population control measures. However, the development of appropriate control strategies is a complex process that depends on multiple interrelated factors, including the specific mosquito species to be targeted, as well as biological, environmental, social, political, and economic considerations2. Microbiota-based approaches are currently being explored as novel mosquito biocontrol alternatives. Insect-associated microbiota include a variety of microorganisms, from free-living to parasites/pathogens and true symbionts, engaging in close and long-term biological interactions3. They can be part of the host environment and are hosted in specific structures within the body4. In mosquitoes, they occur at high loads in the midgut and may also colonize other organs or tissues, such as salivary glands and, in a few specific genera, the reproductive organs5. These microorganisms interact with each other to regulate development and population density3. On the other hand, mosquito microbiota interacts with their host, being important modulators of their phenotype, having a strong influence on multiple aspects of their life cycle, as well as affecting their nutrition, immunity, development, and even vector competence5–7. Different strategies have been explored for the use of native microbiota in the control of vector-borne diseases, including the selection of strains with better spreading or enhancing immune responses. In addition, genetic engineering has been proposed to produce effectors that interfere with pathogens5. Many microorganisms, such as the bacterial genera Aeromonas, Asaia, Pseudomonas, and Serratia; filamentous fungi, such as Aspergillus and Penicillium; and yeasts, such as Candida or Pichia, are commonly associated with mosquitoes. However, the mosquito mycobiome and its interaction with other microorganisms have been poorly studied compared with the bacteriome7, 8.
One of the most studied arthropod-associated bacteria, Wolbachia pipientis (Rickettsiales), has the ability to manipulate host reproduction. In mosquitoes, cytoplasmic incompatibility (CI) is induced, producing non-viable offspring in specific mating combinations. In general, CI allows Wolbachia to spread within insect populations through maternal transmission, effectively invading uninfected host populations and resulting in a selective reproductive advantage over uninfected females9. Wolbachia strains are capable of inducing variable effects on the phenotypes of different host mosquito species. In natural carriers, it has been observed to increase larval survival and longevity. In addition, reproductive fitness parameters, such as fertility, fecundity, and egg viability, can be positively or negatively affected10–12. In transinfected mosquito populations, this bacterium can reduce lifespan and alter locomotor and blood-feeding behavior13. Overall, vectorial competence may be affected by the regulation of the mosquito immune response, with mosquitoes becoming refractory to viral or parasitic infections13, 14.
In this work, the microbiota of the Wolbachia carrier Culex quinquefasciatus population maintained under laboratory conditions was analyzed and compared with a Wolbachia-cured mosquito isoline treated with tetracycline, which was allowed to recover its microbiota for many generations after treatment. The effect of the presence or absence of Wolbachia on fungal and bacterial diversity in both isolines throughout the development of the mosquito life cycle was studied through metagenomic analyses.
Materials and methods
Mosquito rearing and collection
Culex quinquefasciatus naturally infected with Wolbachia (wPipSJ strain) was originally obtained from natural breeding sites in San Juan Province, Argentina, and has been maintained under insectary conditions since 2016 (Biological Control Laboratory of the INBIOTEC-CONICET, FIBA). From the established mosquito line, wild type (wt), an antibiotic-treated isoline (-tet), was generated by exposing larvae to a tetracycline hydrochloride solution (0.1 mg/mL final concentration, Sigma, St Louis, MO; Cat. No. T33 83) during development until the pupal stage. Pupae were transferred to clean water without antibiotics and reared to adulthood. Adults were allowed to feed ad libitum with 10% sucrose containing 0.05 mg/mL tetracycline hydrochloride solution for three consecutive generations. After that, healthy insects were reared in dechlorinated water without antibiotics for three generations, and the entire process was repeated for three more generations. Finally, both isolines were reared in dechlorinated water at 24 ± 1 °C and a 12:12 h light/dark photoperiod. Larvae were fed on commercial fish food microorganisms free, and adults were provided with 20% (w/v) sucrose solution ad libitum; gravid females were also fed with mice blood. The presence of Wolbachia in both mosquito lines was checked through amplification of a wsp gene fragment using the primer combination wsp-81F and wsp-691R from gDNA, as previously described15. Isoline -tet has remained stable free of Wolbachia, without the addition of antibiotics, for more than 6 years, being checked monthly by PCR amplification of the wsp gene.
Mosquito DNA extraction
A pool of 20 individuals from each of four groups was used for total genomic DNA extraction: (1) third instar larvae, (2) sucrose-fed males, (3) sucrose-fed females and (4) blood-fed females. Adults were first killed by freezing, and wings and legs were removed under a stereoscope. Dissected bodies were surface disinfected for 1 min with 70% ethanol, followed by 3 to 4 washes (1 min each) with sterile phosphate-buffered saline solution (PBS). Sucrose-fed adults were processed 48 h post emergence, and blood-fed females 96 h post emergence after blood digestion. For DNA extraction, DNeasy Blood & Tissue kit (QIAGEN) lysis buffer was added to insect samples in 1.5 mL tubes, frozen in liquid nitrogen and homogenized using a mechanical cell lyser and glass beads. Homogenates were incubated with 2 mg/mL proteinase K for 3 h for protein degradation and centrifuged at 12,000 rpm for 5 min. After that, the lysate was transferred to the kit column, following DNA extraction according to the manufacturer’s protocol16.
Metagenome amplicon sequencing
Amplicon sequencing was performed using extracted gDNA previously quantified by the PicoGreen method through Victor 3 fluorometry (Invitrogen, Waltham, MA, USA) and quality checked by gel electrophoresis. From each mosquito group, libraries were constructed with primers targeting the nuclear ribosomal internal transcribed spacer (ITS2) and hypervariable regions V3–V4 of the 16S ribosomal DNA. Primer pairs used were ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′)17 and 341F: (5′-CCTACGGGNGGCWGCAG-3′) and 805R: (5′-GACTACHVGGGTATCTAATCC-3′)18. Amplicons were sequenced on an Illumina MiSeq 300 bp PE platform in accordance with the manufacturer’s protocols. Library construction and amplicon sequencing were performed by Macrogen Inc. (Korea).
Sequence analysis and taxonomic assignment
Paired-end reads were quality-checked by FastQC. Reads longer than 250 bp with an expected error < 1 were clustered into operational taxonomic units (OTUs) at 97% similarity using a combination of UPARSE pipeline19 and Mothur software, applying quality and chimera removal filters. Taxonomic assignment for fungi was performed on representative sequences from each OTU using massBLASTer and SH mappings of the UNITE database20. Bacterial OTUs were classified using Mothur against the SILVA database. Unclassified OTUs were taxonomically identified by BLAST searches against the GenBank standard database, nucleotide collection (nr/nt).
Bioinformatic analysis
Using the raw reads for each sample, rarefaction curves were constructed in R, and bacterial and fungal filtered reads were used to estimate alpha-diversity indices in each community using vegan package 2.521. For ITS2 samples, the reads of replicates were merged prior to diversity analysis. Relative abundance and heatmaps were graphed in GraphPad Prism 8.0.1. Principal component analysis (PCA) of all samples was performed with a combination of ggplot2 and ggfortify using the Galaxy web platform, usegalaxy.org22. Bray–Curtis dissimilarity indices for pairwise communities comparison were computed in the vegan package. Based on those values, a distance matrix was made to construct nonmetric dimensional scaling (NMDS) plots.
Ethical approval
All the experiments were reviewed and approved by the Animal Experimental Committee at the Faculty of Exact and Natural Sciences, Mar del Plata University (Institutional Committee on Care and Use of Experimental Animals (CICUAL) N° 2555-04-14 and RD-2021-623). The animal experiment complies with the ARRIVE guidelines in strict accordance with National Health Service and Food Quality (SENASA) guidelines (Argentina), following the 2011 revised form of The Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health.
Results
Metagenomic sequencing from 16S V3–V4 and ITS2 ribosomal DNA regions revealed a total of 1,765,410 bacterial and 3,049,778 eukaryotic paired-end raw reads (Tables S1 and S2).
Rarefaction curves confirmed that the sequencing depth was sufficient to cover the bacterial diversity in all mosquito groups (Fig. S1). Eukaryotic ITS2 reads from all stages reached a plateau, indicating that the fungal community was well represented after two rounds of sequencing (Fig. S2).
Bacterial alpha diversity
Five hundred and seventy-two (572) bacterial OTUs were clustered from the 16S rRNA amplicon reads. Richness values in larval samples were similar between wt and -tet isolines. In sugar-fed adults, tetracycline-treated richness decreased by at least half in comparison to wt. In blood-fed females, a contrary tendency was determined, displaying higher richness than the -tet line. In the wt, Shannon and InvSimpson indices values were highest in the early stage and decreased in the adult samples. In particular, blood-fed females showed over fivefold lower diversity than larvae. Similarly, the -tet line also presented a differential behavior: females displayed greater diversity than larvae, especially after blood-feeding (Table 1).
Table 1.
Indices of richness and diversity in bacterial communities per Culex quinquefasciatus group in two isolines.
| 16S rRNAa | OTUs | Reads | Dominance | Margalef | Fisher_alpha | Shannon | InvSimpson |
|---|---|---|---|---|---|---|---|
| L | 153 | 23,104 | 0.13 | 15.13 | 21.90 | 2.5 | 7.72 |
| L-tet | 171 | 22,280 | 0.56 | 16.98 | 25.20 | 1.38 | 1.79 |
| M | 192 | 30,000 | 0.44 | 18.53 | 27.44 | 1.47 | 2.30 |
| M-tet | 72 | 26,478 | 0.72 | 6.97 | 9.02 | 0.81 | 1.38 |
| SF | 176 | 44,164 | 0.85 | 16.36 | 23.32 | 0.62 | 1.17 |
| SF-tet | 73 | 31,917 | 0.26 | 6.94 | 8.92 | 1.65 | 3.87 |
| BF | 139 | 53,049 | 0.87 | 12.69 | 17.31 | 0.45 | 1.14 |
| BF-tet | 179 | 30,538 | 0.24 | 17.24 | 25.21 | 2.22 | 4.25 |
aL larvae, M sucrose-fed male, SF sucrose-fed females, BF blood-fed females. Tetracycline treated isoline is indicated with the suffix (-tet).
Fungal alpha diversity
Sixty-one (61) different fungal OTUs resulted from the total samples, with great variation between them, from 2 OTUs in wt blood-fed females to 32 in larvae. For this line, richness decreased during development from larvae to adult stage, while -tet line larvae and sugar-fed adults did not show great differences. Remarkably, blood-fed females of both isolines showed a drastic reduction in the total mycobiota (2 and 3 OTUs) (Table 2). Shannon and InvSimpson indices computed for each sample revealed the highest diversity in -tet sugar-fed females, in contrast to its counterpart in the wt line presenting the lowest values of all samples.
Table 2.
Summary of fungal community richness and alpha diversity per Culex quinquefasciatus group in two isolines.
| ITS2a | OTUs | Reads | Dominance | Margalef | Fisher_alpha | Shannon | InvSimpson |
|---|---|---|---|---|---|---|---|
| L | 32 | 9685 | 0.43 | 3.14 | 4.12 | 1.09 | 2.33 |
| L-tet | 18 | 34,629 | 0.61 | 1.53 | 1.83 | 0.70 | 1.65 |
| M | 19 | 4231 | 0.82 | 1.99 | 2.56 | 0.55 | 1.22 |
| M-tet | 21 | 8651 | 0.86 | 2.05 | 2.59 | 0.41 | 1.16 |
| SF | 13 | 1855 | 0.90 | 1.46 | 1.89 | 0.32 | 1.11 |
| SF-tet | 16 | 1164 | 0.31 | 1.94 | 2.62 | 1.69 | 3.22 |
| BF | 2 | 21 | 0.64 | 0.35 | 0.59 | 0.55 | 1.56 |
| BF-tet | 3 | 78 | 0.44 | 0.55 | 0.76 | 0.90 | 2.25 |
aL larvae, M sucrose-fed male, SF sucrose-fed females, BF blood-fed females. Tetracycline treated isoline is indicated with the suffix (-tet).
Considering all mosquito developmental stages, in wt, the greatest diversity was observed in larvae and decreased in adults independent of their food source. On the other hand, blood-fed samples showed very low richness but displayed higher diversity as a consequence of a more even distribution. -tet isoline sugar-fed females had the highest diversity, while wt had the lowest diversity among all mosquito stages.
Bacterial communities composition in Culex quinquefasciatus
Bacterial communities were largely dominated by the phylum Proteobacteria, with a major relative abundance of more than 70% in all samples (Fig. 1A, Table S3). Wolbachia (Anaplasmataceae) had a high relative abundance in adults of the wt line (60.6–93.4%), although it represented 15% of the larval reads. Instead, in the -tet isoline, this OTU was absent in larvae, while in adults, it occurred in a lower proportion than in wt (5.6–17.4%). In all tetracycline-treated mosquito groups, the wsp gene was never detected by PCR amplification, while it was amplified in the wt groups (results not shown) (Fig. 1B, Table S3).
Figure 1.
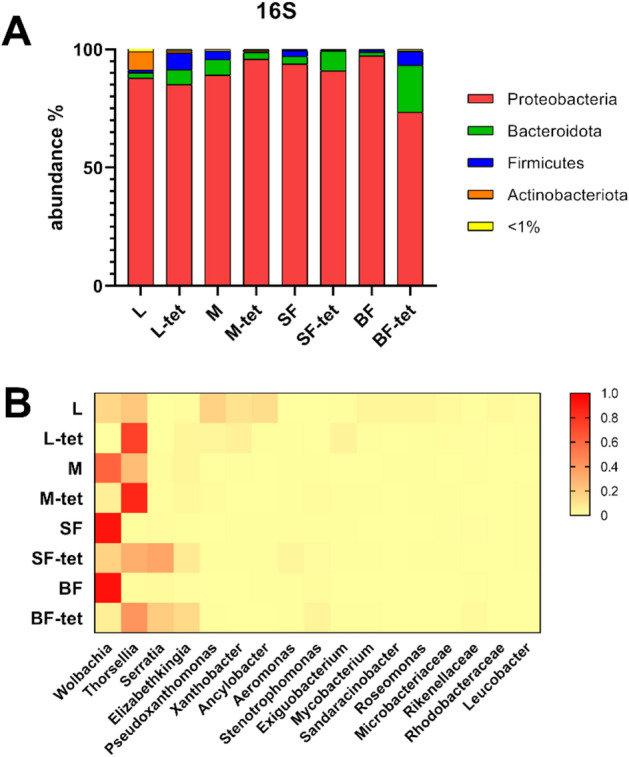
Bacterial relative abundance in wild-type and tetracycline-treated Culex quinquefasciatus isolines. (A) Bacterial community at the phylum level. (B) Heatmap of the relative abundance distribution of the most abundant bacteria in mosquito stages. Red color indicates higher relative abundance. L larvae, M sucrose-fed male, SF sucrose-fed females, BF blood-fed females. Tetracycline treated isoline is indicated with the suffix (-tet).
Another highly represented taxon was Thorselliaceae, with the highest values in larvae and males in wt (21.3 and 2.9%, respectively), even though this family was absent in females. In contrast, in the -tet isoline, Thorselliaceae was dominant in all mosquito stages. Remarkably, Serratia (Yersiniaceae) was the only genus present in both sucrose- and blood-fed females (20%). Xanthobacteraceae was prominent in wt larvae, with an abundance greater than 20%, distributed between Ancylobacter and Xanthobacter. Another remarkable OTU corresponded to Elizabethkingia (Weeksellaceae), which was more abundant in the -tet isoline, especially for females, reaching 14.2% in blood-fed and 7.61% in sugar-fed. Several bacterial genera were found to be considerably abundant in larvae samples, whereas they occurred at low frequency or were absent in adults.
Fungal communities composition in Culex quinquefasciatus
Samples in the wt isoline were dominated by Ascomycota (Fig. 2A). In particular, Penicillium had an abundance greater than 70% in all adult samples, although in larvae, one OTU of Mortierellomycotina (phylum Mucoromycota) showed equal relative abundance (Fig. 2B). Analysis of communities composition in the -tet isoline revealed a drastic reduction in Penicillium occurrence. Despite maintaining its dominance in sugar-fed mosquitoes, it was almost absent in larvae and blood-fed females. Particularly, in blood-fed females, Basidiomycota genera such as Sterigmatomyces and Trametes were identified, while the most abundant larval OTU was assigned to the basidiomycete order Geastrales. Two OTUs of filamentous fungi were more abundant in sugar-fed adults than in larvae samples, one belonging to the order Capnodiales and the other in the genus Dydimella. Both were absent in blood-fed females. Additionally, some genera, such as Erysiphe and Rhodocollybia, were unique to sugar-fed adults of the -tet isoline. Yeasts and yeast-like genera, such as Sterigmatomyces, Debaryomyces, Rhodotorula, Candida and Malassezia, were also identified (Fig. 2, Table S4).
Figure 2.
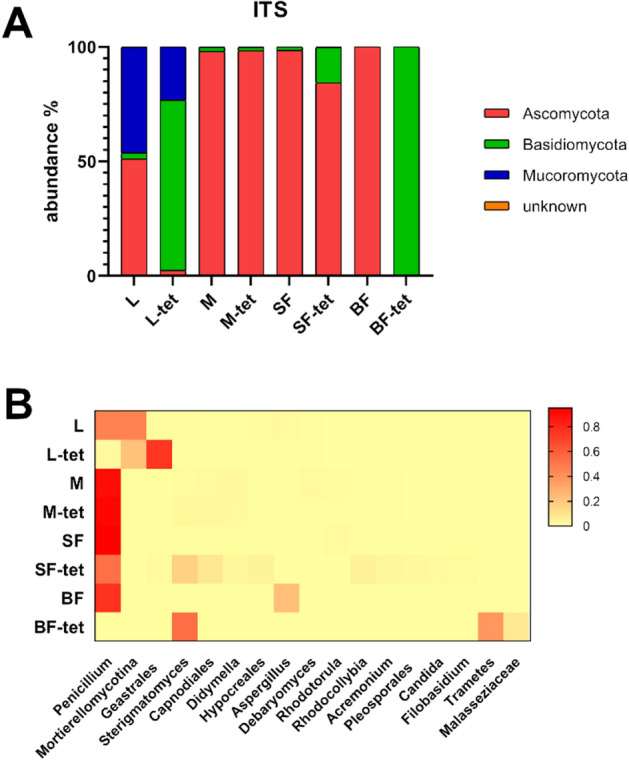
Fungal relative abundance in wild-type and tetracycline-treated Culex quinquefasciatus isolines. (A) Fungal community at the genus level. (B) Heatmap of the relative abundance distribution of the most abundant fungi in mosquito stages. Red color indicates higher relative abundance. L larvae, M sucrose-fed male, SF sucrose-fed females, BF blood-fed females. Tetracycline treated isoline is indicated with the suffix (-tet).
Beta diversity
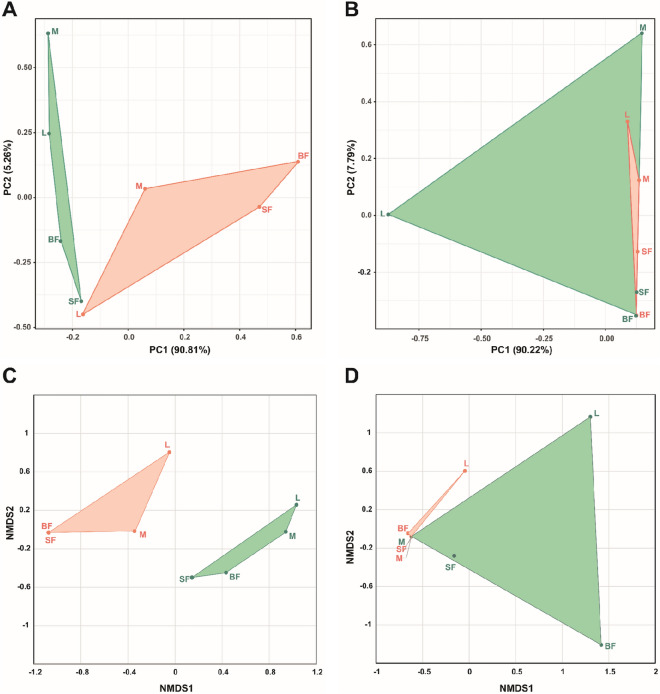
The principal component analysis (PCA) plot for bacterial communities displayed both isolines as separate groups, with PC1 explaining that almost 91% of the variance was due to the differences between isolines (colored groups), while PC2 was associated with variance between stages (Fig. 3A). The Bray–Curtis-based NMDS showed a similar pattern, explaining the dissimilarities in the bacteriome between both populations as distant groups (Fig. 3C). In contrast, the PCA performed for fungal communities assigned most of the variation to PC1, in which samples were close-distanced. Only the wt larval sample was highly separated from the rest. NMDS showed an overlap in points representing samples from both isolines, supporting the PCA in which there was no separation of groups (Fig. 3B, D). In both microbial sets, larval samples were separated from the adults of their own isoline, indicating the presence of distinct and abundant OTUs. In addition, blood-fed female samples tended to have different communities from most samples in pairwise comparisons (Fig. 3, Tables S5 and S6).
Figure 3.
Principal component analysis (PCA) score plots and nonmetric multidimensional scale ordination (NMDS) plots of microorganism communities. Both analyses were performed for bacterial (A, C) and fungal (B, D) communities. In PCA score plots, based on OTU raw counts, X and Y axes show principal component 1 and principal component 2, which together explain 96.07% of the total variance (PC1: 90.81% and PC2: 5.26%) in 16S rRNA samples (A) and 98.01% of the total variance (PC1: 90.22% and PC2: 7.79%) in ITS2 samples (B). The results of the tetracycline-treated samples are represented in green, and the untreated samples are shown in red. L larvae, M sucrose-fed male, SF sucrose-fed females, BF blood-fed females.
Discussion
In this work, bacterial and fungal microbiome from two laboratory Cx. quinquefasciatus isolines (wt and -tet) were compared throughout mosquito development, including changes in female feeding. Bacterial communities in the wt line presented higher Shannon diversity in larvae than adults, as has been demonstrated in a Wolbachia carrier Ae. albopictus population23. On the other hand, the -tet isoline showed a variable diversity distribution between developmental stages, detecting less diversity in larvae than in females. Meanwhile, in previous research, Cx. tarsalis naturally free of Wolbachia exhibited a similar diversity index among all stages24, and Wolbachia-free An. atroparvus population presented greater diversity in larvae than in females25. All these different examples highlight the diverse stage-specific interactions between microbes and their host.
In this study, the bacterial composition at the phylum level showed a dominance of Proteobacteria in all samples. In particular, OTUs such as Thorsellia, Serratia, Aeromonas and Elizabethkingia have been identified, some of which are commonly found in mosquitoes of the Culex genus26–28. The relative abundance data showed a different pattern between the most abundant genera when comparing the -tet and wt isolines. Samples with a high abundance of Wolbachia presented a reduced load of the enterobacteria Thorsellia and Serratia, and vice versa. As reported by Hegde et al.26, interaction network analysis based on metagenomic data from mosquitoes indicated that some bacteria could interact with each other, producing exclusion patterns in which the colonization of the microbiota by one OTU can inhibit the presence of another; finding that Serratia and Aeromonas would be co-excluded by Wolbachia in adult Cx. quinquefasciatus and Ae. albopictus. Additionally, an exclusion effect of the enterobacterium Serratia by Cedecea was experimentally demonstrated in Cx. quinquefasciatus and gnotobiotic Ae. aegypti systems29. Competition for ovary colonization by the acetobacterium Asaia has also been reported in natural Wolbachia carrier mosquito species30. Furthermore, in Anopheles, antibiotic treatment for Asaia reduction allows the vertical transmission of Wolbachia31. Interestingly, in Cx. tarsalis (naturally lacking Wolbachia endosymbiont) Thorsellia was present across all developmental stages in field-collected samples24. In this work, Thorsellia was detected in all stages of development, showing a considerable abundance increase in the -tet isoline. This would suggest that the pattern of dominant bacteria could be explained by an exclusion effect due to competition between Wolbachia and the enterobacteria.
Additionally, the influence of blood intake on the microbiome composition was also analyzed. It has been described that in the Cx. quinquefasciatus females dominated by Wolbachia, blood ingestion produced a drastic reduction in this genus in the midgut, accompanied by an increase in enterobacteria, indicating that Wolbachia would not participate in the digestion of blood32. Our data showed that in the wt isoline, the Wolbachia load remained at similar values, whereas the abundance of Enterobacteriaceae increased after blood ingestion. Serratia increased twice in blood-fed females than in sugar-fed females of the untreated isoline. However, the opposite trend was observed for the -tet isoline. Gaio et al.33 conducted in vivo experiments using antibiotic-treated Ae. aegypti and found that the microbiota actively participates in blood digestion. Furthermore, Enterobacter and Serratia showed the strongest hemolytic activity in vitro. Similarly, in the Cx. pipiens midgut, fluorescently labeled Serratia was shown to increase 700-fold after blood ingestion34. Elizabethkingia (Weeksellaceae) and Stenotrophomonas (Xanthomonadaceae) were increased after the blood meal in both isolines, in agreement with Telang and Skinner32, who observed in Cx. quinquefasciatus females, an increase in the relative abundance of Elizabethkingia after being fed pig blood. In particular, E. anophelis, previously found in the midgut of Anopheles stephensi, would play a role in blood metabolism35. In addition, Stenotrophomonas has been reported as an indicator species of Ae. aegypti gut microbiome after being fed chicken blood36.
Considering the fungal communities, the highest richness was observed in wt larvae; as we have suggested, this biota might be acquired during feeding and reduced after metamorphosis16. Studies on Aedes larvae have revealed that microorganism communities are more similar to the microbiota present in the breeding water than between mosquito populations from different breeding sites, suggesting that the environment is the dominant factor shaping the mycobiota present in the larvae37, 38.
Regarding fungal composition, the genus Penicillium was dominant in the sugar-fed mosquitoes, suggesting acquisition via sucrose solution. An inverse situation was observed in the blood-fed treated females, in which not only this OTU but also all fungal reads were extremely scarce. In engorged females, changes in the physiological state of the gut induce a selective environment for the bacterial microbiota33, 39, suggesting that gut conditions could possibly affect the fungal communities in the same way. As has been reported in Ae. triseriatus females, mycobiota abundance and composition change in the midgut as a response to blood intake40.
Multivariate analysis indicated that there were greater differences between both isolines for bacteria and lesser differences for fungal communities, suggesting that bacterial community structure in -tet isoline was affected by the antibiotic treatment (Fig. 3) due to the broad spectral action of tetracycline41. In mosquitoes42 and other insect hosts43, Wolbachia infection was found to affect the levels of reactive oxygen species (ROS), which are essential players in the immune system42, 43. Additionally, the presence of Wolbachia may influence the expression of phenoloxidase genes, antimicrobial effectors, and the load of bacteria and fungi44. Therefore, the effect of the antibiotic treatment in the initial mosquito generations, in addition to the reduction of Wolbachia influence in the restabilization of microbial communities, could be the cause of dominance/exclusion of certain bacterial genera.
The elimination of Wolbachia in mosquitoes using tetracycline has been identified as a successful method for its cure, possibly because of its bacteriostatic effect on bacterial populations, which would allow the reconstitution of the microbiota after suspending its application41. In this work, Wolbachia was detected in adults of the -tet isoline at a much lower abundance than in wt. Detection of genomic sequences by NGS has much higher sensitivity than endpoint PCR amplification45. Nevertheless, in previous work, CI was detected by crossing the two lines15.
In the case of fungi, the multivariate analysis did not produce a distinctive pattern between groups; therefore, no definitive role could be assigned to them in the differentiation between isolines. However, the high dissimilarity values of the larvae and blood-fed females of both isolines with the rest of the samples suggest that developmental stage and feeding are important factors in mycobiome differentiation.
Here, we demonstrated that richness, diversity, and microbial composition of Cx. quinquefasciatus depends on the characteristics of the initial microbiota, developmental stage, and female feeding. Furthermore, we suggest that the transmission and establishment of the found microbiota could be due to many factors, such as occupation of an ecological niche or competitive exclusion by dominant OTUs such as Wolbachia, Thorsellia, Serratia or Penicillium. Moreover, differential effects on vertical, transstage, and horizontal transmission, environmental influence, or changes in intestinal homeostasis by antibiotics or blood intake could be involved.
The role of the dominant microorganisms in the development and fitness of this mosquito should be further investigated, in order to analyze their potential for biotechnological applications aimed at managing insect populations or reducing their vectorial capacity.
Supplementary Information
Author contributions
G.A.M.F., R.P.L., V.F.C., and C.M.B. designed the research plan. G.A.M.F. and R.P.L. biological material preparation. G.A.M.F., R.P.L., and C.S.C. performed the bioinformatics analysis. V.F.C. and C.M.B. acquired funding and supervised the work. All authors contributed to the result analysis, wrote and edited the manuscript, read and approved the submitted version.
Funding
This work was partially supported by ANPCyT (PICT 2020-SERIE A-02641 and PICT 2020-03165), CONICET (PUE 2017-0101 and PIP 2021-2023 1012) and Universidad Nacional de Mar del Plata (15/E984-EXA1026/21).
Data availability
Data supporting the current investigation and the results reported in this article can be found at the NCBI repository (https://www.ncbi.nlm.nih.gov/). SRA database has been successfully processed with the BioProject name "16S Metagenome amplicon from Culex quinquefasciatus" (PRJNA925532, SUB12534211) with SRA accession numbers SRR23209995, SRR23209996, SRR23209999, SRR23210000, SRR23210001, SRR23209994, SRR23209997, and SRR23209998. Also, BioProject "Tetracycline-treated Culex quinquefasciatus ITS2 Fungi amplicon" (PRJNA925520, SUB12521670) with SRA accession numbers SRR23209969, SRR23209970, SRR23209971, SRR23209972, SRR23209966, SRR23209967, and SRR23209968. The SRA databases will be released once published.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guillermo A. M. Flores and Rocio P. Lopez.
Contributor Information
V. Fabiana Consolo, Email: faconsolo@inbiotec-conicet.gob.ar.
Corina M. Berón, Email: corina.beron@inbiotec-conicet.gob.ar
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-46067-2.
References
- 1.Duval P, Antonelli P, Aschan-Leygonie C, Valiente Moro C. Impact of human activities on disease-spreading mosquitoes in urban areas. J. Urban Health. 2023 doi: 10.1007/s11524-023-00732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker N, Petrić D, Zgomba M, et al. Mosquitoes: Identification, Ecology and Control. Springer; 2020. [Google Scholar]
- 3.Brinker P, Fontaine MC, Beukeboom LW, Salles JF. Host, Symbionts, and the microbiome: The missing tripartite interaction. Trends Microbiol. 2019;27:480–488. doi: 10.1016/j.tim.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit. Vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao H, Cui C, Wang L, et al. Mosquito microbiota and implications for disease control. Trends Parasitol. 2020;36:98–111. doi: 10.1016/j.pt.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cansado-Utrilla C, Zhao SY, McCall PJ, et al. The microbiome and mosquito vectorial capacity: Rich potential for discovery and translation. Microbiome. 2021;9:111. doi: 10.1186/s40168-021-01073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng R, Wang Q, Wu R, et al. Holobiont perspectives on tripartite interactions among microbiota, mosquitoes, and pathogens. ISME J. 2023;1:1–10. doi: 10.1038/s41396-023-01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malassigné S, Valiente Moro C, Luis P. Mosquito mycobiota: An overview of non-entomopathogenic fungal interactions. Pathogens. 2020;9:564. doi: 10.3390/pathogens9070564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinkins SP. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem. Mol. Biol. 2004;34:723–729. doi: 10.1016/j.ibmb.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Dobson SL, Rattanadechakul W, Marsland EJ. Fitness advantage and cytoplasmic incompatibility in Wolbachia single- and superinfected Aedes albopictus. Heredity. 2004;93:135–142. doi: 10.1038/sj.hdy.6800458. [DOI] [PubMed] [Google Scholar]
- 11.de Almeida F, Moura AS, Cardoso AF, et al. Effects of Wolbachia on fitness of Culex quinquefasciatus (Diptera; Culicidae) Infect. Genet. Evol. 2011;11:2138–2143. doi: 10.1016/j.meegid.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 12.Alomar AA, Pérez-Ramos DW, Kim D, et al. Native Wolbachia infection and larval competition stress shape fitness and West Nile virus infection in Culex quinquefasciatus mosquitoes. Front. Microbiol. 2023;14:1138476. doi: 10.3389/fmicb.2023.1138476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caragata EP, Dutra HLC, Moreira LA. Exploiting intimate relationships: Controlling mosquito-transmitted disease with Wolbachia. Trends Parasitol. 2016;32:207–218. doi: 10.1016/j.pt.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Allman MJ, Fraser JE, Ritchie SA, et al. Wolbachia’s deleterious impact on Aedes aegypti egg development: The potential role of nutritional parasitism. Insects. 2020;11:735. doi: 10.3390/insects11110735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Nieto LM, Gil MF, Lazarte JN, et al. Culex quinquefasciatus carrying Wolbachia is less susceptible to entomopathogenic bacteria. Sci. Rep. 2021;11:1094. doi: 10.1038/s41598-020-80034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores GAM, Lopez RP, Cerrudo CS, et al. Culex quinquefasciatus holobiont: A fungal metagenomic approach. Front. Fungal Biol. 2022;3:918052. doi: 10.3389/ffunb.2022.918052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellemain E, Carlsen T, Brochmann C, et al. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson RH, Anslan S, Bahram M, et al. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019;17:95–109. doi: 10.1038/s41579-018-0116-y. [DOI] [PubMed] [Google Scholar]
- 21.Oksanen, J. et al. vegan: Community Ecology Package (Version 2.5-6, R package, 2019).
- 22.Afgan E, Baker D, Batut B, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scolari F, Sandionigi A, Carlassara M, et al. exploring changes in the microbiota of Aedes albopictus: Comparison among breeding site water, larvae, and adults. Front. Microbiol. 2021;12:624170. doi: 10.3389/fmicb.2021.624170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duguma D, Hall MW, Rugman-Jones P, et al. Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol. 2015;15:140. doi: 10.1186/s12866-015-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birnberg L, Climent-Sanz E, Codoñer FM, Busquets N. Microbiota variation across life stages of european field-caught Anopheles atroparvus and during laboratory colonization: New insights for malaria research. Front. Microbiol. 2021;12:775078. doi: 10.3389/fmicb.2021.775078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde S, Khanipov K, Albayrak L, et al. Microbiome interaction networks and community structure from laboratory-reared and field-collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquito vectors. Front. Microbiol. 2018;9:2160. doi: 10.3389/fmicb.2018.02160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C, Beller L, Wang L, et al. Bidirectional interactions between arboviruses and the bacterial and viral microbiota in Aedes aegypti and Culex quinquefasciatus. mBio. 2022;13:e01021–e1022. doi: 10.1128/mbio.01021-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Meng H, Hu X, Yuan Z. Diversity and functional profile of gut symbiotic bacteria between Lysinibacillus sphaericus C3–41 susceptible and resistant Culex quinquefasciatus Say as revealed by 16S rRNA gene high-throughput sequencing. Front. Microbiol. 2022;13:991105. doi: 10.3389/fmicb.2022.991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozlova EV, Hegde S, Roundy CM, et al. Microbial interactions in the mosquito gut determine Serratia colonization and blood-feeding propensity. ISME J. 2021;15:93–108. doi: 10.1038/s41396-020-00763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi P, Ricci I, Cappelli A, et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit. Vectors. 2015;8:278. doi: 10.1186/s13071-015-0888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes GL, Dodson BL, Johnson RM, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. 2014;111:12498–12503. doi: 10.1073/pnas.1408888111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telang A, Skinner J. Effects of host blood meal source on reproductive output, nutrient reserves and gut microbiome of West Nile virus vector Culex quinquefasciatus. J. Insect. Physiol. 2019;114:15–22. doi: 10.1016/j.jinsphys.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Gaio AO, Gusmão DS, Santos AV, et al. Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (diptera: culicidae) (L.) Parasit. Vectors. 2011;4:105. doi: 10.1186/1756-3305-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koosha M, Vatandoost H, Karimian F, et al. Effect of Serratia AS1 (Enterobacteriaceae: Enterobacteriales) on the fitness of Culex pipiens (Diptera: Culicidae) for paratransgenic and RNAi approaches. J. Med. Entomol. 2019;56:553–559. doi: 10.1093/jme/tjy183. [DOI] [PubMed] [Google Scholar]
- 35.Ganley JG, D’Ambrosio HK, Shieh M, Derbyshire ER. Coculturing of mosquito-microbiome bacteria promotes heme degradation in Elizabethkingia anophelis. ChemBioChem. 2020;21:1279–1284. doi: 10.1002/cbic.201900675. [DOI] [PubMed] [Google Scholar]
- 36.Muturi EJ, Njoroge TM, Dunlap C, Cáceres CE. Blood meal source and mixed blood-feeding influence gut bacterial community composition in Aedes aegypti. Parasit. Vectors. 2021;14:83. doi: 10.1186/s13071-021-04579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tawidian P, Coon KL, Jumpponen A, et al. Host-environment interplay shapes fungal diversity in mosquitoes. mSphere. 2021;6:e00646–e721. doi: 10.1128/mSphere.00646-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zouache K, Martin E, Rahola N, et al. Larval habitat determines the bacterial and fungal microbiota of the mosquito vector Aedes aegypti. FEMS Microbiol. Ecol. 2022;98:016. doi: 10.1093/femsec/fiac016. [DOI] [PubMed] [Google Scholar]
- 39.Cui Y, Franz AWE. Heterogeneity of midgut cells and their differential responses to blood meal ingestion by the mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2020;127:103496. doi: 10.1016/j.ibmb.2020.103496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muturi EJ, Bara JJ, Rooney AP, Hansen AK. Midgut fungal and bacterial microbiota of Aedes triseriatus and Aedes japonicus shift in response to La Crosse virus infection. Mol. Ecol. 2016;25:4075–4090. doi: 10.1111/mec.13741. [DOI] [PubMed] [Google Scholar]
- 41.Daghrir R, Drogui P. Tetracycline antibiotics in the environment: A review. Environ. Chem. Lett. 2013;11:209–227. doi: 10.1007/s10311-013-0404-8. [DOI] [Google Scholar]
- 42.Brennan LJ, Keddie BA, Braig HR, Harris HL. The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLOS ONE. 2008;3:e2083. doi: 10.1371/journal.pone.0002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zug R, Hammerstein P. Wolbachia and the insect immune system: what reactive oxygen species can tell us about the mechanisms of Wolbachia-host interactions. Front. Microbiol. 2015;6:1201. doi: 10.3389/fmicb.2015.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez JL, Schumacher MK, Ower G, et al. Impacts of fungal entomopathogens on survival and immune responses of Aedes albopictus and Culex pipiens mosquitoes in the context of native Wolbachia infections. PLoS Negl. Trop. Dis. 2021;15:e0009984. doi: 10.1371/journal.pntd.0009984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y-Y, Floate KD, Fields PG, Pang B-P. Review of treatment methods to remove Wolbachia bacteria from arthropods. Symbiosis. 2014;62:1–15. doi: 10.1007/s13199-014-0267-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the current investigation and the results reported in this article can be found at the NCBI repository (https://www.ncbi.nlm.nih.gov/). SRA database has been successfully processed with the BioProject name "16S Metagenome amplicon from Culex quinquefasciatus" (PRJNA925532, SUB12534211) with SRA accession numbers SRR23209995, SRR23209996, SRR23209999, SRR23210000, SRR23210001, SRR23209994, SRR23209997, and SRR23209998. Also, BioProject "Tetracycline-treated Culex quinquefasciatus ITS2 Fungi amplicon" (PRJNA925520, SUB12521670) with SRA accession numbers SRR23209969, SRR23209970, SRR23209971, SRR23209972, SRR23209966, SRR23209967, and SRR23209968. The SRA databases will be released once published.