Abstract
Purpose
COVID-19 strangely kills some youth with no history of physical weakness, and in addition to the lungs, it may even directly harm other organs. Its complex mechanism has led to the loss of any significantly effective drug, and some patients with severe forms still die daily. Common methods for identifying disease mechanisms and drug design are often time-consuming or reductionist. Here, we use a novel holistic systems biology approach to predict its molecular mechanisms (in vitro), significant molecular relations with SARS, and repurpose drugs.
Methods
We have utilized its relative phylogenic similarity to SARS. Using the available omics data for SARS and the fewer data for COVID-19 to decode the mechanisms and their significant relations, We applied the Cytoscape analyzer, MCODE, STRING, and DAVID tools to predict the topographically crucial molecules, clusters, protein interaction mappings, and functional analysis. We also applied a novel approach to identify the significant relations between the two infections using the Fischer exact test for MCODE clusters. We then constructed and analyzed a drug-gene network using PharmGKB and DrugBank (retrieved using the dgidb).
Results
Some of the shared identified crucial molecules, BPs and pathways included Kaposi sarcoma-associated herpesvirus infection, Influenza A, and NOD-like receptor signaling pathways. Besides, our identified crucial molecules specific to host response against SARS-CoV-2 included FGA, BMP4, PRPF40A, and IFI16.
Conclusion
We also introduced seven new repurposed candidate drugs based on the drug-gene network analysis for the identified crucial molecules. Therefore, we suggest that our newly recommended repurposed drugs be further investigated in Vitro and in Vivo against COVID-19.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s40199-023-00471-1.
Keywords: COVID-19, SARS-CoV, PPI, Host Response, DEG, Drug Repurposing
Introduction
Disease pandemics usually spread to humans due to the human-to-human infection transition. To date, several lethal outbreaks have been recorded in the human medical history of human societies. They usually have left bitter memories of deaths and organ injuries, including Ebola, Zika, SARS, H7N9, Spanish Flu, and Hong Kong Flu [1, 2]. The recent outbreak of COVID-19 has had a devastating impact on health systems worldwide, affecting almost all aspects of human life. A vial agent called SARS-CoV-2 causes the disease (Severe Acute Respiratory Syndrome Coronavirus type 2) [3].
Coronaviruses (CoVs) are an extended family of viruses leading to various diseases ranging from the common cold to severe respiratory tract infections [4]. Viral sequencing projects have shown that SARS‐CoV‐2 belongs to the betacoronavirus (βCoV) as SARS‐CoV and MERS‐CoV do. SARS‐CoV‐2, in particular, shares a highly similar sequence to SARS‐CoV [5]. SARS-CoV-2 genome analysis has indicated that it has about ninety percent sequence proximity with the SARS-CoV [6]. Given the high phylogenetic similarity, identifying the underlying molecular mechanisms of each of these two evolutionary relatives may contribute to predicting the unknown mechanisms of the other virus. However, compared to COVID-19, more high-throughput information is available for the SARS-CoV molecular mechanisms in omics databases.
Systems biology science has recently boosted our knowledge of the protein interactions mediating many illnesses [7]. Different biological networks are used to decipher the molecular pathology of diseases and drug side effects [8, 9]. Various protein–protein interaction networks (PPI) have contributed to predicting the molecular mechanisms underlying different diseases and even repurposing drugs and their side effects [9–12]
Some researchers have applied PPI networks to elucidate underlying molecular mechanisms of diverse human diseases. New omics techniques have provided high-throughput genomics and proteomics data. The high output data have contributed to gaining insight into various molecular mechanisms mediated in the pathogenesis of illnesses. Shared proteins among some related diseases have recently been applied to heighten our perception and insights into the disease's molecular mechanisms in more detail [13]. The new insights may clarify the crucial proteins that may presumably be applied to identify new prophylactic treatments, diagnosis methods, and even new drug designs. To gain a better insight into the biochemical pathways and molecular mechanisms altered by SARS-CoV-2 in human host cells, we analyze the host response molecular mechanism to SARS-CoV through PPINs. Construction and analysis of Protein − protein Interaction Networks (PPINs) are usually considered valuable tools to map out biological processes, biochemical pathways, and molecular functions that can be considered biological functions of the intermediating protein complexes of the disease [14]. PPI networks could be applied in deciphering the molecular pathology underlying diseases [15].
Drug design methods employ a variety of in silico techniques to facilitate and expedite the discovery of effective treatments. Applying docking techniques (global energy scores (Kcal/mol)) and molecular dynamics is a typical strategy. In the relatively short period since SARS-CoV-2 first emerged, some research has sought to offer therapeutic candidates against its essential proteins, such as focusing on SARS-CoV-2 3Clpro and RdRp [16] or the affinity of spike protein and its receptor [17]. Drug-gene network analysis is another in silico strategy for introducing drug candidates, and various researchers have used it to suggest drug candidates for disorders [17, 18].
In the present study, we attempt to achieve two principal goals. First, we focus on analyzing the molecular mechanism of the host response in SARS and COVID-19 diseases. We first focus on determining the SARS host response molecular mechanisms by deciphering SARS and COVID-19 protein–protein interaction clusters. Considering the varieties of clinical manifestations between the two Coronaviruses (SARS-CoV and SARS-CoV-2) as the second goal, we then evaluate the significance of the relationships between both networks of SARS and COVID-19 using comparative statistical methods. Based on the shared proteins between clusters of each disease network and the other disease, we will predict the shared molecular mechanisms that are probably the essential elements of the molecular pathogenesis in the two infections. Applying the novel approach will likely benefit us in gaining a more in-depth insight into the biological processes and molecular mechanisms mediating the two viral pathogenesis. We will also focus on the non-shared crucial molecules and their functions to predict the molecular mechanisms only specific to each infection. |Using this method, we will identify the molecular mechanisms responsible for partially different manifestations of the two infections. Deciphering the molecules and shared signaling underlying both viral infections may also help other researchers repurpose new drug candidates or design new drugs against the newly identified targets of COVID-19 or its symptoms. Therefore, based on the crucial genes, we will repurpose some new medications to possibly prevent the severe consequences of the host response to SARS-CoV-2 in some patients.
Methods
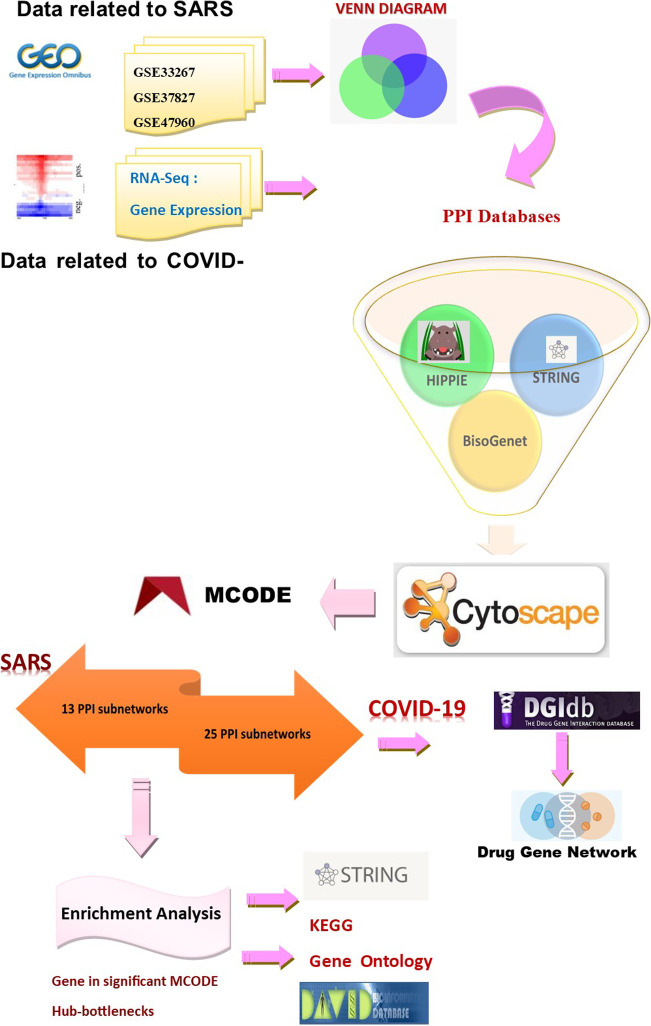
Three separate microarray datasets for respiratory cell lines infected with SARS-CoV were initially identified to identify differentially expressed genes (DEGs). The collected RNA-Seq data from two separate respiratory cell lines were used for COVID-19 data. P-value and fold change (p-value < 0.05 and |Log FC|> 0.5) filtered the up-and-down-regulated DEGs.
Data gathering for COVID-19
Because COVID-19 is a relatively novel infection, omics data on it is still scarce in omics databases. Daniel Blanco-Melo et al.'s released research on respiratory infections was used to extract the COVID-19 data evaluated. This investigation used two epithelial cell lines from the respiratory system of the lungs (NHBE and A549) as in-vitro study models [19].
SARS-CoV-2 was infected into A549 cells at a multiplicity of infection (MOI) of 0.2 virus particles per cell for 24 h. Furthermore, NHBE cells were infected with SARS-CoV-2 at an MOI 2 for 24 h. The TruSeq RNA Library Prep Kit v2 was used to extract total RNA from infected and mock cells and create RNAseq poly-adenylated RNA libraries (Illumina).
The Illumina NextSeq 500 platform was used to sequence the cDNA libraries (www.illumina.com). Each cell line's raw data was examined independently to identify differentially expressed genes (DEGs), then sorted up and down. The filtering criteria in this study were p-value 0.05 and |Log FC|> 0.5. The findings of our research concern respiratory epithelial cells, not immune cells.
Data gathering for SARS
We searched the NCBI Gene Expression Omnibus (GEO) and the European Array Express databank for differentially expressed gene data (DEGs). Compared to data accessible for other illnesses, SARS omics data was scarce in databanks.
One probable explanation is that, compared to other epidemics, the SARS disease was quickly contained and did not last long in communities. Three datasets were discovered and chosen for further analysis (GSE33267, GSE37827, and GSE47960) [20–22].
Calu-3 cells (a lung cancer cell line) were infected with icSARS-CoV in GSE33267 and GSE37827 (Mitchell et al., 2013). In addition, HAE cells (human airway epithelium cell line) were infected with SARS-CoV in GSE47960 [20].
Analysis of differentially expressed genes for SARS
The 60-h early post-infection time point was studied, which was common throughout the three SARS datasets. The ge-Workbench 2.6.0 software was used to filter, standardize, and analyze (t-Test) the three datasets. P-value and fold change criteria (p-value 0.05 and |Log FC|> 0.5) were used to filter DEGs. We then used the online Venn diagram tool (http://bioinformatics.psb.ugent.be) to find the DEGs shared among the three dataset outcomes for the early time-point (60 h post-infection).
Here, We identified the DEGs related to 24 h post-infection, comparing the infected state (24 h post-infection) and control situation (mock-infected 24 h post-infection), and determined the Up downregulated DEGs in both diseases. Since the number of shared DEGs between the diseases was low and insufficient to construct the network for 24 h post-infection, we used raw data from the early 60 h after infection, which was shared among the SARS datasets. We chose the 60-h post-infection data since it was available in all three SARS-related datasets, and it also showed the most significant infection impact (among the shared time points of the available datasets) and had the highest pick in Sims et al. graph.'s curves [20].
Note that, among the three hypothetical stages, the 60-h and 24-h post-infection time points are both considered very early in viral infection with SARS-CoV-2. Moreover, patients may show clinical manifestations even after 14 days post-infection and continue in the first stage for around five days [16, 23]
PPI network data sources
The protein–protein interaction maps were retrieved from three protein interaction databases and then merged. We only mapped the proteins available within our lists. First, our protein lists were mapped using the STRING tool (https://string-db.org/) [24]. The Human Integrated Protein–Protein Interaction rEference database (HIPPIE) (http://cbdm.uni-mainz.de/hippie/) was used to map the interactions. The interactions with confidence scores of more than 0.7 were selected to ensure the reliability of interactions between proteins [25]. BisoGenet, the Cytoscape plugin, was also used (http://bio.cigb.edu.cu/bisogenet-cytoscape/) to retrieve the protein interactions available in the Human Protein Reference Database (HPRD) for the list [26].
PPI network construction
To construct the PPI networks, up and down-regulated data for each disease were merged to construct the SARS and COVID-19 PPIs separately. The three mapped interactions (integrated scores > 0.7 in STRING and HIPPIE) were visualized using Cytoscape (version 3.8.0) and then merged using Cytoscape [27]. The edges representing associations (without direction) were considered interactions between different proteins in both PPI networks.
Topological analysis
Cytoscape 3.8.0 was used to obtain degree and betweenness scores for each disease. The top 10% of nodes with the highest degree were nominated as hubs, and the top 10% with the highest betweenness centrality were considered bottlenecks for each disease. We used the Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to identify the shared and non-shared nodes between the two sets of hubs-bottlenecks in SARS-CoV and COVID-19. The shared proteins probably play essential roles in both infections' pathogenesis since they are considered hubs or bottlenecks in both PPIs. Further functional analysis and studies of the non-shared hub bottlenecks can probably help us predict the molecular mechanisms that are only specific to each infection. It will probably contribute to identifying the molecular mechanisms responsible for partially different manifestations of the two infections.
PPI molecular complex detection (MCODE)
The MCODE algorithm with default parameters (node score cut-off = 0.2, degree cut-off = 2, k-core = 2, maximum depth set at 100) [28] was applied to identify highly connected regions in both disease PPIs. The MCODE algorithm helps find densely interconnected regions of a network. Each region density is the ratio between the number of edges (|E|) and the possible uppermost edges in that region. (|E| max =|n| *(|n|- 1)/2, n is the number of nodes in the region). The product of the number of nodes is considered the MCODE score and is used to detect the dense subnetworks of the network called MCODE clusters. (Score = density * number of nodes) [29].
Enrichment analysis for MCODE clusters
Two different tools were used to perform the functional analysis for MCODE clusters deciphered from SARS and COVID-19 PPI networks, including the DAVID tool [30] and the STRING tool functional analyzer [24]. They were used to enrich the nodes participating in each MCODE cluster separately. We conducted the enrichment analysis for each cluster of nodes separately for GO terms (Gene Ontology terms, including biological process (BP) and KEGG biochemical pathways.
PPI network relations identification between SARS and COVID-19
To decode the shared essential mechanisms underlying both diseases, we first implemented the Chi-square/Fischer exact test analysis [31] between each disease's MCODE cluster and the other disease's PPI separately. The chi-squared test (considering the prerequisites of the Chi-square test) or the alternative Fisher's exact test were applied to detect significant relations between the COVID-19 and the SARS PPI MOCDE subnetworks (p-value < 0.05) and vice versa. The chi-square test only works when the variable under study is categorical. Moreover, in more than 80% of the variables, the expected value of the number of sample observations should be at least 5. (Zero is not permitted for any variable when applying chi-square). Fisher's exact test was applied as an alternative test if the chi-square prerequisites were not met.
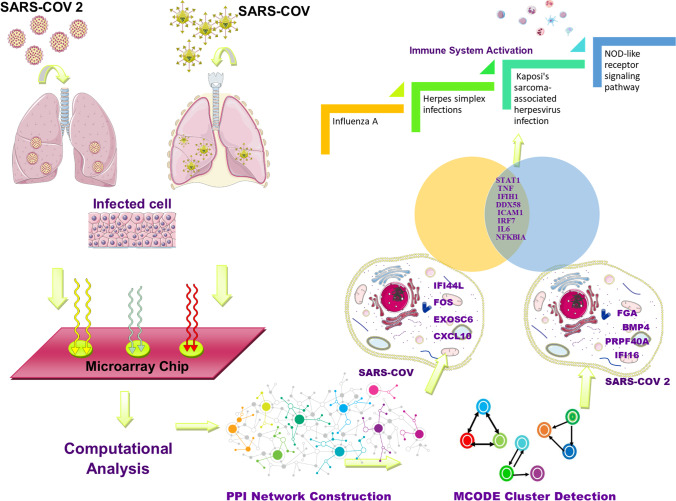
A workflow representing data analysis, PPIN construction, and further survey steps are depicted in Fig. 1.
Fig. 1.
Represents a graphical workflow of the study
Drug–target interaction network construction
Nodes of the three COVID-19 MCODE clusters (significantly) related to SARS shared between the COVID-19 clusters and the SARS network were selected to construct the drug-target interaction network for further drug screenings. Besides, the top 10% of hub bottlenecks shared between SARS and COVID-19 were also selected as drug targets for constructing the drug-gene network. We then used the DGidb database to retrieve the medications with possible interactions with the selected genes [32]. Afterward, the Cytoscape (3.8.0) was used for network visualization. The network was analyzed, and the drugs with higher Degrees were nominated to construct a new drug-gene sub-network. The new repurposed drug candidates were then validated using the ClinicalTrials.gov databank for COVID-19 clinical trials.
Results
This study's primary purpose was to investigate the critical respiratory system genes responsible for host response to the SARS-CoV and SARS-CoV-2 viruses in vitro. We also attempted to determine the shared and non-shared crucial genes and molecular mechanisms between them. Besides, we have repurposed some new medications for further investigations to prevent the severe consequences of the host response to SARS-CoV-2 in some patients based on the identified crucial genes.
Data gathering for COVID-19 and SARS
We studied the SARS data for 24 h post-infection to compare the two disease networks in the 24 h because the omics data for SARS-CoV-2 was only available for 24 h post-infection. First, we used ge-Workbench software to filter, standardize, and analyze the raw data from three SARS-related datasets in the two available shared early time points (24 h and also 60 h, as mentioned in the technique section) (p-value 0.05 and |Log FC|> 0.5).
We looked for DEGs shared between the three data analysis outcomes and identified them as DEGs associated with the SARS-CoV host response in vitro for the distinct time points. Twenty-four hours after infection, we identified seven up-regulated DEGs as SARS-related DEGs. Furthermore, no DEG was identified as a down-regulated DEG in any of the three SARS-related datasets after 24 h. (See Fig. S1 in the Supplementary Materials).
We compared the two disease networks in the two available early time points using SARS data from 60 h after infection (the available shared early point data). First, we used ge-Workbench software to filter, normalize, and analyze the raw data from three SARS-related datasets (p-value < 0.05 and |Log FC|> 0.5). We looked for DEGs shared between the three data analysis outcomes and identified them as DEGs associated with the SARS-CoV host response in vitro for the distinct time points. In 60 h, we found 329 up-regulated and 228 down-regulated DEGs that were SARS-related (data in Supplementary Fig. S1 and Supplementary Table S1). We identified 572 up-regulated and 291 down-regulated genes as COVID-19-related DEGs for two COVID-19 datasets (data is shown in Supplementary Table S2).
We looked for DEGs that were common to both illnesses. We also looked for shared DEGs between COVID-19 and DEGs associated with 60 h SARS-related networks. The findings were limited to respiratory epithelial cells, not immune cells.
Results of topological analysis
To construct the PPI networks, up and down-regulated data for each disease were merged to construct the SARS and COVID-19 PPIs separately. The top 10% node with the highest degree and betweenness centrality was considered hub bottlenecks for SARS and COVID-19 separately (Supplementary Table S3). As visualized in Fig. 2, STAT1, TNF, IFIH1, DDX58, ICAM1, IRF7, IL6, and NFKBIA were the shared nodes between the top 10% hub-bottleneck in SARS and COVIID-19 PPINs. Besides, as represented in S3, 52 nodes were identified exclusively in SARS, and 90 nodes were only related to COVID-19.
Fig. 2.
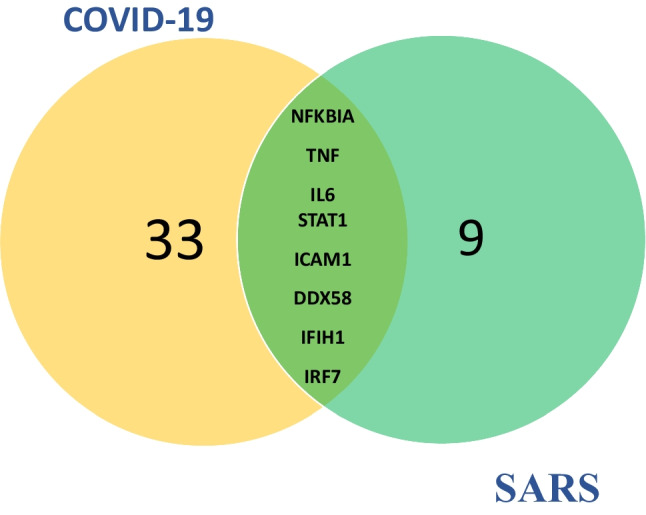
Represents the Venn diagram of the shared DEGs between the top 10% of hubs and bottlenecks of the SARS and COVID-19 PPI networks
Detection of MCODE clusters and assessing the statistical relationship between diseases
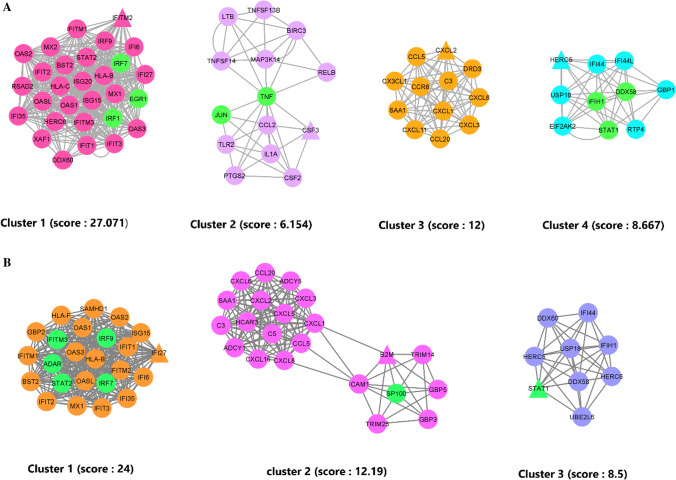
We identified 13 and 25 PPI MCODE cluster subnetworks for SARS and COVID-19. As shown in Supplementary Table S4, to evaluate the relationships between each disease's MCODE clusters and the other disease's PPI, the Chi-square/Fischer exact test analysis was applied. Four clusters of the SARS PPI network had significant statistical relations with COVID-19 PPIN (Fig. 3A). We also identified three COVID-19 clusters having a significant relationship with the SARS PPIN, as shown in Fig. 3B (Supplementary Table S5).
Fig. 3.
MCODE clusters. A represents the significantly scored MCODE clusters in SARS PPI. B represents the significantly scored MCODE clusters in COVID-19 PPI. Nodes with the highest degree score (seeds) are represented in triangle form in each cluster, and nodes with a dual role (TF/Gene) are in green color
Hub-bottleneck functional enrichment
STRING and DAVID Tools were applied to identify the Gene Ontology (GO—biological processes (BP)) and KEGG biochemical pathways as functional enrichment analysis. Tables 1 and S7 show that responses to other organisms and defense responses were the two top biological processes. Besides, as available in Table 2 and S8, Influenza A, and Herpes simplex infections were determined as the two top biochemical pathways shared between SARS and COVID-19 by enriching the top 10% of genes in the PPI network in SARS and COVID-19. As available in S9, positive regulation of the biosynthetic process, biological regulation, and membrane depolarization regulation were SARS' top three biological processes. Also, as represented in Supplementary Table S10, we identified 24 exclusively SARS-related pathways, of which Carbon metabolism, Glycine, serine and threonine metabolism, and Metabolic pathways were the top three.
Table 1.
Top 10% of the COVID-19 and SARS PPIN nodes with the highest degree/betweenness were considered hub bottlenecks
| No | Term (Biological Process) | False discovery rate | Matching proteins in the network |
|---|---|---|---|
| 1 | response to other organism | 3.03E-21 | ADAR,B2M,C3,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,HERC5,HERC6,ICAM1,IFI44,IFIH1,IFIT1,IFIT3,IL1B,IL6,IRF7,NFKB2,NFKBIA,PTGS2,STAT1,TNF,TRIM25 |
| 2 | defense response | 6.21E-18 | ADAR,B2M,C3,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,HERC5,ICAM1,IFIH1,IFIT1,IFIT3,IL1A,IL1B,IL6,IRF7,ITGB2,NFKB2,PTGS2,SP100,STAT1,TNF,TRIM25 |
| 3 | multi-organism process | 2.12E-17 | ADAR,B2M,C3,CDH1,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,FBXW7,HERC5,HERC6,ICAM1,IFI44,IFIH1,IFIT1,IFIT3,IL1B,IL6,IRF7,NFKB2,NFKBIA,PSMB9,PTGS2,SP100,STAT1,TNF,TRIM25 |
| 4 | immune response | 3.74E-17 | ADAR,B2M,C3,CEP290,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,HERC5,ICAM1,IFIH1,IFIT1,IFIT3,IL1A,IL1B,IL6,IRF7,ITGB2,NFKB2,PLAUR,SP100,STAT1,TNF,TRIM25 |
| 5 | immune system process | 8.52E-17 | ADAR,B2M,C3,CEP290,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,HERC5,HERC6,ICAM1,IFIH1,IFIT1,IFIT3,IL1A,IL1B,IL6,IRF7,ITGB2,NFKB2,NFKBIA,PLAUR,PSMB9,SP100,STAT1,TNF,TRIM25 |
| 6 | response to external stimulus | 8.52E-17 | ADAR,B2M,C3,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,HERC5,HERC6,ICAM1,IFI44,IFIH1,IFIT1,IFIT3,IL1B,IL6,IRF7,ITGB2,NFKB2,NFKBIA,PLAUR,PTGS2,STAT1,TNF,TRIM25 |
| 7 | cytokine-mediated signaling pathway | 1.45E-16 | ADAR,B2M,CXCL1,CXCL8,ICAM1,IFIT1,IFIT3,IL1A,IL1B,IL6,IRF7,ITGB2,NFKBIA,PTGS2,SOCS3,SP100,STAT1,TNF,TRIM25 |
| 8 | response to cytokine | 1.05E-15 | ADAR,B2M,CXCL1,CXCL8,EIF2AK2,ICAM1,IFIT1,IFIT3,IL1A,IL1B,IL6,IRF7,ITGB2,NFKB2,NFKBIA,PTGS2,SOCS3,SP100,STAT1,TNF,TRIM25 |
| 9 | response to virus | 7.32E-15 | ADAR,DDX58,DDX60,EIF2AK2,HERC5,IFI44,IFIH1,IFIT1,IFIT3,IL6,IRF7,STAT1,TNF,TRIM25 |
| 10 | response to stress | 1.39E-14 | ADAR,B2M,C3,CXCL1,CXCL8,DDX58,DDX60,EIF2AK2,FBXW7,HERC5,ICAM1,IFIH1,IFIT1,IFIT3,IL1A,IL1B,IL6,IRF7,ITGB2,NFKB2,NFKBIA,PLAUR,PTGS2,SP100,STAT1,TNF,TRIM25,UBE2L6,UBE2V2 |
The shared hub bottlenecks between the COVID-19 and SARS PPI networks were enriched using two different WEB Tools for Biological Processes. (DAVID and STRING). The table describes the top 10 shared biological terms reported by both analyzer tools. (P-values are retrieved from STRING)
Table 2.
The shared hub bottlenecks between the COVID-19 and SARS PPI networks were enriched using two different WEB Tools for KEGG Biochemical Pathways
| No | Term (KEGG pathway) | False discovery rate | Matching proteins in the network |
|---|---|---|---|
| 1 | Influenza A | 1.14E-19 | ADAR,CXCL8,DDX58,EIF2AK2,ICAM1,IFIH1,IL1A,IL1B,IL6,IRF7,NFKBIA,SOCS3,STAT1,TNF,TRIM25 |
| 2 | Herpes simplex infection | 6.73E-16 | C3,DDX58,EIF2AK2,IFIH1,IFIT1,IL1B,IL6,IRF7,NFKBIA,SOCS3,SP100,STAT1,TNF |
| 3 | Legionellosis | 7.21E-14 | C3,CXCL1,CXCL8,IL1B,IL6,ITGB2,NFKB2,NFKBIA,TNF |
| 4 | Measles | 2.05E-12 | ADAR,DDX58,EIF2AK2,IFIH1,IL1A,IL1B,IL6,IRF7,NFKBIA,STAT1 |
| 5 | NF-kappa B signaling pathway | 3.99E-12 | CXCL8,DDX58,ICAM1,IL1B,NFKB2,NFKBIA,PTGS2,TNF,TRIM25 |
| 6 | Leishmaniasis | 2.44E-11 | C3,IL1A,IL1B,ITGB2,NFKBIA,PTGS2,STAT1,TNF |
| 7 | Hepatitis C | 4.55E-11 | CXCL8,DDX58,EIF2AK2,IFIT1,IRF7,NFKBIA,SOCS3,STAT1,TNF |
| 8 | Rheumatoid arthritis | 6.36E-11 | CXCL1,CXCL8,ICAM1,IL1A,IL1B,IL6,ITGB2,TNF |
| 9 | TNF signaling pathway | 3.79E-10 | CXCL1,ICAM1,IL1B,IL6,NFKBIA,PTGS2,SOCS3,TNF |
| 10 | RIG-I-like receptor signaling pathway | 9.23E-10 | CXCL8,DDX58,IFIH1,IRF7,NFKBIA,TNF,TRIM25 |
(DAVID and STRING) The table describes the top 10 shared Biochemical Pathways reported by both analyzer tools. (P-values are retrieved from STRING)
As shown in S11, cell communication, signaling, and viral process regulation were the three top biological processes, and Complement and coagulation cascades, Platelet activation, and Staphylococcus aureus infection were the three top pathways that were exclusively related to the COVID-19 network (Supplementary Table S12).
Functional analysis of the significantly related MCODE clusters between the infections
As shown in Supplementary Table S13 and Table 3, the top five biological processes for cluster 1 were related to immune response. Also, as shown in Table 4, the top five KEGG pathways were related to the host response to a viral infection, such as Herpes simplex infection, Hepatitis C, and Influenza A. Enriching cluster No.2 of SARS showed that the chemokine-mediated signaling pathway was this cluster's most significant biological process. The Chemokine signaling pathway was also the most significant biochemical pathway enriched in cluster No.2, shown in Table 4.
Table 3.
Gene enrichment analysis for biological process (GO-BP) was performed separately in all four MCODE clusters of the SARS PPI network
| Term (Biological Process) |
False discovery rate | Matching proteins in the network | |
|---|---|---|---|
| Cluster 1 | type I interferon signaling pathway | 6.81E-60 |
BST2,EGR1,HLAB,HLAC,IFI27,IFI35,IFI6,IFIT1,IFIT2,IFIT3,IFITM1,IFITM2,I FITM3,IRF1,IRF7,IRF9,ISG15,ISG20,MX1,MX2,OAS1,OAS2,OAS3,OASL,RSAD 2,STAT2,XAF1 |
| innate immune response | 1.25E-37 |
BST2,DDX60,EGR1,HLA-B,HLA-C,IFI27,IFI35,IFI6,IFIT1,IFIT2,IFIT3,IFITM1,IFITM 2,IFITM3,IRF1,IRF7,IRF9,ISG15 ,ISG20,MX1,MX2,OAS1,OAS2,OAS3,OASL,RSAD2,STAT2,XAF1 |
|
| defense response to virus | 3.69E-34 |
BST2,DDX60,IFIT1,IFIT2,IFIT3,IFITM1,IFITM2,IFITM3,IRF1,IRF7,IRF9,ISG15, ISG20,MX1,MX2,OAS1,OAS2,OAS3,OASL,RSAD2,STAT2 |
|
| immune system process | 1.99E-25 |
BST2,DDX60,EGR1,HERC6,HLA-B,HLA-C,IFI27,IFI35,IFI6,IFIT1, IFIT2,IFIT3,IFITM1,IFITM2, IFITM3,IRF1,IRF7,IRF9,ISG15,ISG20,MX1,MX2,OAS1,OAS2,OAS3 ,OASL,RSAD2,STAT2,XAF1 |
|
| response to other organism | 9.51E-25 |
BST2,DDX60,HERC6,HLA-B,IFIT1,IFIT2,IFIT3,IFITM1, IFITM2,IFITM3,IRF1,IRF7,IRF9,ISG15,ISG20,MX1,MX2, OAS1,OAS2,OAS3,OASL,RSAD2,STAT2 |
|
| Cluster 2 | chemokine-mediated signaling pathway | 6.16E-17 | CCL20,CCL5,CCR8,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6 |
| cell chemotaxis | 3.33E-14 | CCL20,CCL5,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6,SAA1 | |
| G protein-coupled receptor signaling pathway | 8.50E-13 | C3,CCL20,CCL5,CCR8,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6,DRD3,SAA1 | |
| inflammatory response | 8.62E-13 | C3,CCL20,CCL5,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6,SAA1 | |
| chemotaxis | 8.87E-13 | CCL20,CCL5,CCR8,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6,SAA1 | |
| Cluster 3 | response to virus | 1.33E-13 | DDX58,EIF2AK2,GBP1,HERC5,IFI44,IFI44L,IFIH1,RTP4,STAT1 |
| defense response to virus | 9.07E-13 | DDX58,EIF2AK2,GBP1,HERC5,IFI44L,IFIH1,RTP4,STAT1 | |
| regulation of cytokine production | 1.07E-05 | DDX58,EIF2AK2,GBP1,HERC5,IFIH1,STAT1 | |
| regulation of type I interferon production | 1.26E-05 | DDX58,HERC5,IFIH1,STAT1 | |
| positive regulation of interferon-alpha production | 1.26E-05 | DDX58,IFIH1,STAT1 | |
| Cluster 4 | response to cytokine | 2.29E-15 |
BIRC3,CCL2,CSF2,CSF3,IL1A,JUN,LTB,MAP3K14,PTGS2,RELB, TLR2,TNF,TNFSF13B,TNFSF14 |
| cytokine-mediated signaling pathway | 1.24E-11 | BIRC3,CCL2,CSF2,CSF3,IL1A,LTB,MAP3K14,PTGS2,TNF,TNFSF13B,TNFSF14 | |
| cellular response to cytokine stimulus | 1.24E-11 |
BIRC3,CCL2,CSF2,CSF3,IL1A,LTB,MAP3K14,PTGS2,TLR2, TNF,TNFSF13B,TNFSF14 |
|
| cell surface receptor signaling pathway | 2.06E-11 |
BIRC3,CCL2,CSF2,CSF3,IL1A,JUN,LTB,MAP3K14,PTGS2, RELB,TLR2,TNF,TNFSF13B,TNFSF14 |
|
| response to tumor necrosis factor | 2.36E-10 | BIRC3,CCL2,LTB,MAP3K14,PTGS2,TNF,TNFSF13B,TNFSF14 |
The top five terms of Biological processes for each cluster have been reported separately (sorted by P-values < 0.05)
Table 4.
Gene enrichment analysis for KEGG biochemical pathways was performed separately in all four MCODE clusters of the SARS PPI network
| Term (KEGG pathway) | False discovery rate | Matching proteins in network | |
|---|---|---|---|
| Custer 1 | Herpes simplex infection | 1.91E-10 | HLA-B,HLA-C,IFIT1,IRF7,IRF9,OAS1,OAS2,OAS3,STAT2 |
| Hepatitis C | 3.56E-10 | IFIT1,IRF1,IRF7,IRF9,OAS1,OAS2,OAS3,STAT2 | |
| Influenza A | 1.58E-09 | IRF7,IRF9,MX1,OAS1,OAS2,OAS3,RSAD2,STAT2 | |
| Measles | 1.01E-08 | IRF7,IRF9,MX1,OAS1,OAS2,OAS3,STAT2 | |
| Human papillomavirus infection | 1.20E-07 | HLA-B,HLA-C,IRF1,IRF9,ISG15,MX1,OASL,STAT2 | |
| Cluster 2 | Chemokine signaling pathway | 3.79E-15 | CCL20,CCL5,CCR8,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6 |
| Cytokine-cytokine receptor interaction | 5.02E-14 | CCL20,CCL5,CCR8,CX3CL1,CXCL1,CXCL11,CXCL2,CXCL3,CXCL6 | |
| TNF signaling pathway | 2.86E-10 | CCL20,CCL5,CX3CL1,CXCL1,CXCL2,CXCL3 | |
| IL-17 signaling pathway | 1.45E-08 | CCL20,CXCL1,CXCL2,CXCL3,CXCL6 | |
| Legionellosis | 1.89E-07 | C3,CXCL1,CXCL2,CXCL3 | |
| Cluster 3 | Measles | 1.40E-05 | DDX58,EIF2AK2,IFIH1,STAT1 |
| Influenza A | 1.74E-05 | DDX58,EIF2AK2,IFIH1,STAT1 | |
| Herpes simplex infection | 1.74E-05 | DDX58,EIF2AK2,IFIH1,STAT1 | |
| Hepatitis C | 0.00027 | DDX58,EIF2AK2,STAT1 | |
| Hepatitis B | 0.00028 | DDX58,IFIH1,STAT1 | |
| Cluster 4 | Rheumatoid arthritis | 5.26E-14 | CCL2,CSF2,IL1A,JUN,LTB,TLR2,TNF,TNFSF13B |
| NF-kappa B signaling pathway | 5.70E-14 | BIRC3,LTB,MAP3K14,PTGS2,RELB,TNF,TNFSF13B,TNFSF14 | |
| TNF signaling pathway | 2.28E-11 | BIRC3,CCL2,CSF2,JUN,MAP3K14,PTGS2,TNF | |
| Cytokine-cytokine receptor interaction | 8.74E-11 | CCL2,CSF2,CSF3,IL1A,LTB,TNF,TNFSF13B,TNFSF14 | |
| IL-17 signaling pathway | 8.06E-10 | CCL2,CSF2,CSF3,JUN,PTGS2,TNF |
The top five terms of KEGG biochemical pathways for each cluster have been reported separately (sorted by P-values < 0.05)
As available in Table 3, response to the virus and defense response to the virus are the top two significant biological processes, and besides, as illustrated in Table 4, Measles and Influenza A are the top two significant pathways. Table 3 shows the cytokine response was the most significant BP in cluster No.4, and also, as demonstrated in Table 4, Rheumatoid arthritis and NF-kappa B signaling pathways were the top two significant pathways in cluster No.4. All of the identified significant BP and KEGG pathways (p-value < 0.05) for all the four significantly related clusters of SARS (to COVID-19) are available in Supplementary Table S13 and Supplementary Table S14, respectively.
Three clusters of COVID-19 related to SARS PPIN were identified as significantly related; hence, the biological process enrichment for the three clusters of COVID-19 was also performed, illustrated in Tables 5 and S15. It shows that the type I interferon signaling pathway, defense response, and response to the virus was the most significant BPs in each cluster. Besides, as shown in Table 6 and S16, Herpes simplex infection, Chemokine signaling pathway, and Hepatitis B were distinctly the top three in Cluster No.1, 2, and 3.
Table 5.
Gene enrichment analysis for biological process (GO-BP) was performed separately in all three MCODE clusters of the COVID-19 PPI network
| Term (Biological Process) | False discovery rate | Matching proteins in the network | ||
|---|---|---|---|---|
| Cluster 1 | 1 | type I interferon signaling pathway | 2.85E-46 |
BST2,HLA-B,IFI27,IFI35,IFI6,IFIT1,IFIT2,IFIT3,IFITM1, IFITM2,IFITM3, IRF7,IRF9,ISG15,MX1,OAS1,OAS2,OAS3,OASL,STAT2 |
| 2 | defense response to virus | 2.63E-27 |
BST2,IFIT1,IFIT2,IFIT3,IFITM1,IFITM2,IFITM3, IRF7,IRF9,ISG15,MX1,OAS1,OAS2,OAS3,OASL,STAT2 |
|
| 3 | response to other organism | 2.71E-19 |
BST2,HLA-B,IFIT1,IFIT2,IFIT3,IFITM1,IFITM2,IFITM3,IRF7 ,IRF9, ISG15,MX1,OAS1,OAS2,OAS3,OASL,STAT2 |
|
| 4 | negative regulation of viral genome replication | 3.23E-19 |
BST2,IFIT1,IFITM1,IFITM2,IFITM3,ISG15,MX1,OAS1,OAS3, OASL |
|
| 5 | immune effector process | 1.19E-18 |
BST2,HLA-B,IFIT1,IFIT2,IFIT3,IFITM1,IFITM2,IFITM3,I RF7,IRF9,ISG15,MX1,OAS1,OAS2,OAS3,OASL,STAT2 |
|
| Cluster 2 | 1 | defense response | 2.80E-17 |
B2M,C3,C5,CCL20,CCL5,CXCL1,CXCL16,CXCL2,CXCL3,CXCL5, CXCL6,CXCL8,GBP3,GBP5,ICAM1,SAA1,SP100,TRIM14,TRIM25 |
| 2 | cell chemotaxis | 2.62E-14 | C5,CCL20,CCL5,CXCL1,CXCL16,CXCL2,CXCL3,CXCL5,CXCL6,CXCL8,SAA1 | |
| 3 | immune response | 4.02E-14 | B2M,C3,C5,CCL20,CCL5,CXCL1,CXCL16,CXCL2,CXCL3,CXCL5,CXCL6,CXCL8,GBP5,ICAM1,SAA1,SP100,TRIM14,TRIM25 | |
| 4 | response to cytokine | 7.03E-14 | B2M,CCL20,CCL5,CXCL1,CXCL16,CXCL2,CXCL3,CXCL5,CXCL6,CXCL8,GBP3,GBP5,ICAM1,SAA1,SP100,TRIM25 | |
| 5 | inflammatory response | 1.44E-13 | C3,C5,CCL20,CCL5,CXCL1,CXCL2,CXCL3,CXCL5,CXCL6,CXCL8,GBP5,ICAM1,SAA1 | |
| Cluster 3 | 1 | response to virus | 2.70E-07 | DDX58,DDX60,HERC5,IFI44,IFIH1,STAT1 |
| 2 | regulation of type I interferon production | 2.70E-07 | DDX58,HERC5,IFIH1,STAT1,UBE2L6 | |
| 3 | negative regulation of type I interferon production | 5.43E-07 | DDX58,HERC5,IFIH1,UBE2L6 | |
| 4 | defense response to virus | 9.92E-07 | DDX58,DDX60,HERC5,IFIH1,STAT1 | |
| 5 | response to other organism | 9.92E-07 | DDX58,DDX60,HERC5,HERC6,IFI44,IFIH1,STAT1 |
The top five terms of Biological processes for each cluster have been reported separately (sorted by P-values < 0.05)
Table 6.
Gene enrichment analysis for KEGG biochemical pathways was performed separately in all three MCODE clusters of the COVID-19 PPI network
| Term (KEGG pathway) | False discovery rate | Matching proteins in the network | ||
|---|---|---|---|---|
| Cluster 1 | 1 | Herpes simplex infection | 2.29E-10 | HLA-B,IFIT1,IRF7,IRF9,OAS1,OAS2,OAS3,STAT2 |
| 2 | Hepatitis C | 8.23E-10 | IFIT1,IRF7,IRF9,OAS1,OAS2,OAS3,STAT2 | |
| 3 | Measles | 8.23E-10 | IRF7,IRF9,MX1,OAS1,OAS2,OAS3,STAT2 | |
| 4 | Influenza A | 2.20E-09 | IRF7,IRF9,MX1,OAS1,OAS2,OAS3,STAT2 | |
| 5 | NOD-like receptor signaling pathway | 9.12E-08 | IRF7,IRF9,OAS1,OAS2,OAS3,STAT2 | |
| Cluster 2 | 1 | Chemokine signaling pathway | 3.67E-15 |
ADCY1,ADCY5,CCL20,CCL5,CXCL1,CXCL16,CXCL2,CXCL3, CXCL5,CXCL6,CXCL8 |
| 2 | Cytokine-cytokine receptor interaction | 2.83E-10 |
CCL20,CCL5,CXCL1,CXCL16,CXCL2,CXCL3,CXCL5,CXCL6, CXCL8 |
|
| 3 | IL-17 signaling pathway | 2.83E-10 | CCL20,CXCL1,CXCL2,CXCL3,CXCL5,CXCL6,CXCL8 | |
| 4 | Rheumatoid arthritis | 2.83E-10 | CCL20,CCL5,CXCL1,CXCL5,CXCL6,CXCL8,ICAM1 | |
| 5 | TNF signaling pathway | 6.02E-10 | CCL20,CCL5,CXCL1,CXCL2,CXCL3,CXCL5,ICAM1 | |
| Cluster 3 | 1 | Hepatitis B | 0.0008 | DDX58,IFIH1,STAT1 |
| 2 | Measles | 0.0008 | DDX58,IFIH1,STAT1 | |
| 3 | Influenza A | 0.0008 | DDX58,IFIH1,STAT1 | |
| 4 | Herpes simplex infection | 0.0008 | DDX58,IFIH1,STAT1 | |
| 5 | RIG-I-like receptor signaling pathway | 0.0028 | DDX58, IFIH1 |
The top 5 terms of KEGG biochemical pathways for each cluster have been reported separately (sorted by P-values < 0.05)
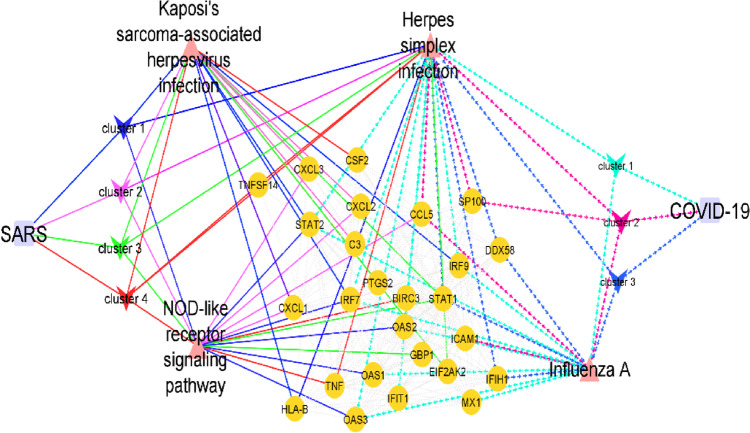
Figure 4 represents the genes shared between SARS and COVID-19 in the PPI network, and the shared pathways between each cluster and disease are represented. Kaposi's sarcoma-associated herpesvirus infection and NOD-like receptor signaling pathway were two shared significant pathways between the SARS clusters and COVID-19. Influenza A was the pathway shared between the COVID-19 clusters and SARS. Furthermore, Herpes simplex infection was also the pathway shared between them.
Fig. 4.
Represents the shared DEGs that relate the MCODE clusters of each infection to the other infection PPI and their significantly enriched corresponding biochemical pathways. DEGs are depicted in orange circles, pathways in triangles, and diseases with purple circles. Edges related to SARS are shown with continuous lines, and COVID-19 with dashed lines. Clusters are labeled distinctly
Drug–target interaction network results
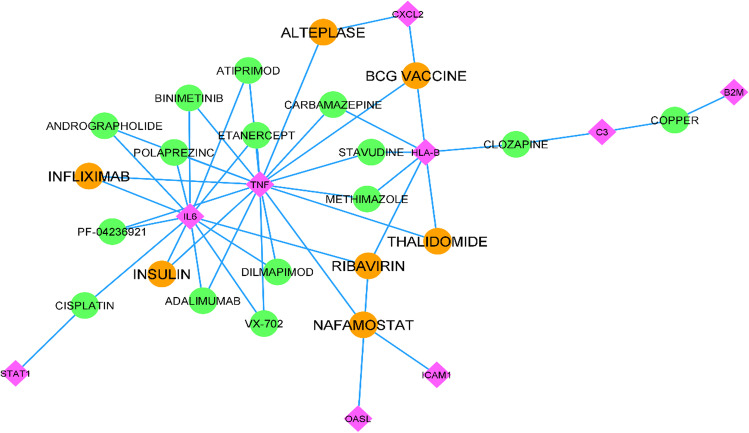
As available in Supplementary Table S6, 13 unique nodes were considered the selected genes, and 169 unique medicines were nominated as the related drugs to the selected genes in the Drug–Target Interaction Network. Among the 169 drugs, 22 drugs with a Degree between three and two were selected to create a new sub-network, as visualized in Fig. 5. To validate the new repurposed drugs, we have shown in Table 7 that seven of the drugs were previously registered in ClinicalTrrials.gov to be evaluated as possible treatments for COVID-19 (Table 7).
Fig. 5.
Represents the Drug-Gene interaction network. Purple diamonds show the crucial identified Genes, and circles depict our repurposed medications. The medicines previously registered for clinical trials against COVID-19 are distinct, using orange color
Table 7.
The table represents the top seven repurposed drugs interacting with the identified crucial genes in the Drug-Gene interaction network sorted by degree value
| Drug name | Gene target | Degree |
|---|---|---|
| BCG vaccine | TNF, CXCL2, HLA-B | 3 |
| Ribavirin | HLA-B, IL6, OASL | 3 |
| Nafamostat | ICAM1, TNF | 2 |
| Infliximab | TNF, IL6 | 2 |
| Alteplase | CXCL2, TNF | 2 |
| Thalidomide | TNF, HLA-B | 2 |
| Insulin | TNF, IL6 | 2 |
Discussion
Altered gene expression is essential to COVID-19 and SARS pathogenesis, affecting proteins in various functional classes. Thus, identifying the host response critical molecular mechanisms to SARS-CoV-2 and SARS-CoV is essential for developing effective management and treatment strategies.
Biological networks probably consist of several sub-network or functional modules contributing to different biological processes. A node may have little effect on the global network or global properties, but it does affect a sub-network with a specific function [33]. Thus, in the present study, we constructed two PPI networks for each disease separately to examine the crucial molecular mechanisms of the host response in respiratory cells in COVID-19 and SARS. Afterward, the MCODE algorithm was used to create cluster sub-networks for each PPI network. Each disease's MCODE clusters were then evaluated for their possible significant relation with the other disease using chi-square/Fisher's exact test. Four SARS MCODE Clusters and three COVID-19 Clusters were significantly related to COVID-19 and SARS PPI networks. We first discuss the molecular mechanisms and critical molecules rooted in the seven clusters to clarify the pathogenesis of the two diseases and their shared molecular mechanisms. Then we will discuss the critical mechanisms which were predicted to be specific to only one of the two diseases.
Functional analysis for pathways in the first significantly related cluster of SARS to COVID-19 (SARS cluster No.1) showed that Herpes simplex infection, Hepatitis C and Influenza A were the top three significant pathways. The Type I interferon signaling pathway and innate immune response were the most significant biological processes that confirm the innate immune system's essential role and interferon in the host response to viral infections [34]. IFITM2 had the highest degree score among SARS cluster No.1 nodes and was considered the seed in this cluster. Previous studies have reported that IFITM2 is vital in different enveloped virus entry, especially in the SARS-CoV entrance mediated by the SARS-CoV spike (S) protein [35]. Furthermore, IFITM2 is also considered a significant node in one of the COVID-19 clusters; hence, it could be hypothesized that this protein also may play a role in SARS-CoV-2 entry into the host cell, similar to SARS-CoV. Besides, IFI27 (interferon Alpha Inducible Protein 27) was nominated as the seed in COVID-19 cluster No.1, which has already been introduced to have antiviral activity in other viruses like hepatitis C [36]. It can inhibit virus replication and potentiate the anti-HCV activity of IFN-α through induced production of type I IFNs and activation of the Jak/STAT signaling pathway independent of autophagy and cell apoptosis [37]. Since HCV is a positive-sense single-stranded RNA virus similar to SARS-CoV-2, we can presume that IFI27 may activate the innate immune system in COVID-19. However, further experimental investigations seem necessary to confirm the hypothesis fully.
CXCL2 (C-X-C Motif Chemokine Ligand 2) is another node in cluster No.3 of SARS-CoV considered a seed and a significant node in COVID-19 cluster No.2. This protein is produced by activated monocytes and neutrophils and expressed at sites of inflammation [38]. Our results indicate that two drugs, BCG Vaccine, and ALTEPLASE, can target this gene. Some previous studies have reported that the BCG Vaccine can reduce the initial virus spread in cells, leading to less severe symptoms [39, 40]. The BCG vaccination, primarily used to prevent tuberculosis, has demonstrated some promise for enhancing the immune response against several diseases, including COVID-19. According to several studies, the BCG vaccine may offer some protection against severe symptoms or lower the risk of infection. To establish its effectiveness, however, more research is necessary [41, 42].
Besides, some experimental and in-silico studies have shown the role of ALTEPLASE as an anticoagulant element [43–45]. The thrombolytic drug called alteplase dissolves blood clots, such as those associated with stroke or pulmonary embolism. According to studies, alteplase medication may benefit COVID-19 individuals who exhibit significant respiratory distress and aberrant blood coagulation markers. It has demonstrated potential for increasing oxygenation and decreasing clot load in these patients. The available evidence is, however, limited [46, 47]
B2M in this COVID-19 cluster was considered a seed that could induce the expression of various interleukins, such as 6 (IL-6), 8, and 10, in several cell types [48]. Several studies have shown the increase of IL-6 in COVID-19 as a driver of the inflammation that causes cytokine storm [49, 50]. The blockage of IL-6 can decrease cytokine storm symptoms [51, 52]. In our in-silico analysis, Insulin, Ribavirin, and Infliximab were three drugs interacting with IL-6 in the drug-target interaction network. Therefore their possible effects on cytokine storms are recommended for further experimental investigations.
Insulin has been investigated for its potential role in COVID-19 treatment. According to studies, administering insulin to COVID-19 individuals who already have diabetes may improve results. However, others contend that people with COVID-19 and diabetes who use insulin may see a rise in severe/critical consequences [53–55].
A variety of viral infections are treated with ribavirin, an antiviral medication. Some research has looked into its possible effectiveness against COVID-19. While some studies point to a potential advantage, others report no appreciable clinical improvement. It might play some part in antiviral combination therapy with other medications [56, 57].
An immunosuppressant medication called infliximab is prescribed to treat autoimmune diseases like rheumatoid arthritis and Crohn's disease. Mixed outcomes have been seen in studies looking at its use in severe COVID-19 cases. While some claim there may be advantages in lowering inflammation and enhancing respiratory function, others claim no appreciable clinical improvement exists. Infliximab may modify the immunological response, although whether it can be used to treat COVID-19 is still controversial [58–60].
The pathway enrichment analysis of COVID-19 and SARS clusters demonstrated four shared pathways, including the NOD-like receptor signaling pathway, Kaposi's sarcoma-associated herpesvirus infection, Influenza A, and Herpes simplex infection pathway, which contain 26 shared proteins between COVID-19 and SARS networks. Among these 26 proteins, DDX58, IFIH1, STAT1, IRF7, ICAM1, and TNF are among the top 10% shared hub and bottlenecks between SARS and COVID-19 PPI networks. Among these genes, TNF and ICAM1 were significant in our drug-target network. ICAM1 and TNF interacted with Nafamostat. One previous study has indicated the role of Nafamostat in the blockage of MERS-CoV infection [61]. Besides, various research has confirmed the role of Nafamostat as an anticoagulant in COVID-19 patients [62, 63]. The serine protease inhibitor, Nafamostat, is primarily used as an anticoagulant. Nafamostat may prevent the SARS-CoV-2 virus from entering and infecting human cells. Recent research suggests that it might prevent the SARS-CoV-2 virus from replicating and lessen inflammation, thereby improving COVID-19 patient outcomes. More research is necessary to determine its clinical efficacy against COVID-19 [64, 65]. The present study showed that TNF and ICAM1 are two crucial network nodes interacting with this drug. Some studies have shown that increased TNF levels in COVID-19 patients correlate with or even lead to ARDS [66]. We hypothesize that Nafamostat could play a role as an anti-inflammatory agent in COVID-19 patients.
TNF and HLA-B were the two target genes interacting with Thalidomide. Thalidomide is registered in two clinical trial experiments against COVID-19 [67, 68]. It has been investigated for its immunomodulatory qualities and potential antiviral and anti-inflammatory actions against COVID-19, while it was commonly used to treat several malignancies and skin diseases. According to recent research, it could lessen lung harm in serious situations and modify immune responses. It might enhance clinical results in severe COVID-19 instances. More study is needed, nevertheless, to prove its efficacy and safety [69, 70].
Thalidomide acts as an anti-inflammatory element due to its ability to accelerate the degradation of messenger RNAs in blood cells and reduce the tumor necrosis factor α (TNFα). It could be an anti-inflammatory drug in COVID-19 patients and is recommended as a repurposed drug candidate to be investigated in alleviating the inflammatory phase in COVID-19 patients. It has been discovered that Thalidomide boosts the production of natural killer, T, and B cells, which in turn stimulates the immunological response [71] Since it has been proposed to suppress the generation of pro-inflammatory cytokines responsible for the cytokine storm seen in severe instances of COVID-19, it may also inhibit the cytokine storm [71, 72]. Additionally, in COVID-19 patients at high risk for thrombotic events, Thalidomide may have anti-thrombotic effects and lower the risk of blood clots [73]. Interestingly, Thalidomide has been demonstrated to prevent SARS-CoV-2 replication [74].
Thalidomide seems a promising candidate medication for treating COVID-19 since it may have numerous positive benefits for COVID-19 patients. Also, the interaction of Thalidomide with the HLA-B gene in our drug-gene network suggests the hypothesis that maybe the effect of it on the treatment of COVID-19 patients is affected by the subtype of this gene; however, additional clinical studies are required to ascertain the hypothesis and also its security and effectiveness in treating COVID-19.
DDX58 (DExD/H-Box Helicase 58) encodes RIG-I, a vital commencer protein in the immune system. RIG-I is reported to be responsible for responding to some negative-strand RNA viruses, such as influenza viruses, and positive-strand RNA viruses, like the Coronaviridae family [75–77]. Coronaviruses are giant RNA viruses containing the RNA genome that encodes for various proteins interacting with different factors in the innate immune signaling pathway, such as the 5'cap segment to block RIG-I recognition of RNA [77]. For example, in SARS-CoV, deubiquitination of RIG-I, STING, TRAF, and TBK1 are reported to cause inhibition of IRF3 activation through encoding a Papin-like protease (PLpro) [78, 79]. In this in-silico study, DDX58 was nominated as a shared hub and bottleneck protein between SARS and COVID-19, which explains that it might also have an essential role in the pathogenesis of SARS-CoV-2 infection.
Another shared hub-bottleneck protein was IFIH1 (Interferon Induced with Helicase C Domain 1), which encodes MDA5 (Melanoma Differentiation-Associated protein 5) [80]. Other studies have reported that infection by various positive-strand RNA viruses, such as picornaviruses [81], arteriviruses [82], and also Kaposi sarcoma-associated herpesvirus (KSHV) [83], trigger MDA5 activation [84]. Indeed, MDA5 activation causes different immune cascades in the cell, leading to the expression of type 1 interferon genes (IFN1: IFNα and IFNβ). Production of IFN1 causes the recruitment of macrophages, neutrophils, and dendritic cells to the infection site with chemokine gradients aid [85]. Therefore, it seems logical that it probably also has an essential role in the pathogenesis of COVID-19 entry. However, further experimental assays are required to confirm the conclusion fully.
One of the other well-known players of the innate immune response against viral infections is IRF7 (Interferon Regulatory Factor 7), induced by type I interferon (INF) in many cell types. Type 1 interferon (IFNs) seem to act as a human first defense line against viral infections and could sometimes be considered the starting point of innate immunity and an essential inducer of adaptive immune responses [86, 87]. Some mouse studies have shown that the lack of IRF-7 in IRF-7 knockout mice could impair the induction of antigen-specific CD8 + T cell responses [86]. In line with them, we hypothesize that IRF7 also plays a critical role in the host response to both SARS and COVID-19.
In conclusion, this in-silico study utilized the phylogenic similarity of SARS-CoV-2 and SARS to predict the molecular mechanisms behind SARS-CoV-2 pathogenesis and the host response against it. Using a systems biology approach and network analysis, we revealed the shared crucial molecules behind the host response to both disease and specific mechanisms of each disease. We recommended some repurposed drug candidates against COVID-19 using the identified target seeds. We revealed that Cell communication, Signaling, and Regulation of the viral process were the top three significant BPs. Complement/coagulation cascades and Platelet activation were the top two significant pathways mediating COVID-19 pathogenesis. The top three crucial molecules specific to the host response against SARS-CoV-2 included FGA, BMP4, and PRPF40A. We have suggested BCG VACCINE, RIBAVIRIN, NAFAMOSTAT, INFLIXIMAB, ALTEPLASE, and THALIDOMIDE as new repurposed candidates drugs by the drug-gene network analysis for further investigations against COVID-19 or its symptoms.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study is related to project NO. 1400/65279 from the Student Research Committee, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We also appreciate the Student Research Committee and Research & Technology Chancellor at Shahid Beheshti University of Medical Sciences for their financial support of this study.
Data availability
Readers may have access to the raw data, details of the analyzed data, and issues in the represented supplementary files.
Code availability
The software used was free (Cytoscape).
Declarations
Ethics approval
The approval was issued by the ethical committee of the SBMU (project NO. 1400/65279).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest/competing interests
Authors declare that they have no conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rewar S, Mirdha D, Rewar P. Treatment and prevention of pandemic H1N1 influenza. Ann Glob Health. 2015;81(5):645–653. doi: 10.1016/j.aogh.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Maurice J. Cost of protection against pandemics is small. Lancet. 2016;387(10016):e12. doi: 10.1016/S0140-6736(16)00156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohrabi C, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Lienau J, Witzenrath M. Coronaviruses as the cause of respiratory infections. Internist (Berl) 2019;60(11):1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F, et al. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22(2):74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen TP, Liu WC, Jordan F. Inferring pleiotropy by network analysis: linked diseases in the human PPI network. BMC Syst Biol. 2011;5:179. doi: 10.1186/1752-0509-5-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farahani M, et al. Deciphering the transcription factor-microRNA-target gene regulatory network associated with graphene oxide cytotoxicity. Nanotoxicology. 2018;12(9):1014–1026. doi: 10.1080/17435390.2018.1513090. [DOI] [PubMed] [Google Scholar]
- 9.Ghani S, et al. Specific regulatory motifs network in SARS-CoV-2-Infected Caco-2 Cell Line, as a model of gastrointestinal infections. Cell Reprogram. 2022;24(1):26–37. doi: 10.1089/cell.2021.0055. [DOI] [PubMed] [Google Scholar]
- 10.Ma J, et al. A comparative study of cluster detection algorithms in protein-protein interaction for drug target discovery and drug repurposing. Front Pharmacol. 2019;10:109. doi: 10.3389/fphar.2019.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sameni M, et al. Deciphering molecular mechanisms of SARS-CoV-2 pathogenesis and drug repurposing through GRN motifs: a comprehensive systems biology study. 3 Biotech. 2023;13(4):117. doi: 10.1007/s13205-023-03518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehghan Z, et al. A motif-based network analysis of regulatory patterns in Doxorubicin effects on treating breast cancer, a systems biology study. Avicenna J Med Biotechnol. 2022;14(2):137. doi: 10.18502/ajmb.v14i2.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramly B, Afiqah-Aleng N, Mohamed-Hussein Z-A. Protein–protein interaction network analysis reveals several diseases highly associated with polycystic ovarian syndrome. Int J Mol Sci. 2019;20(12):2959. doi: 10.3390/ijms20122959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King AD, Przulj N, Jurisica I. Protein complex prediction via cost-based clustering. Bioinformatics. 2004;20(17):3013–3020. doi: 10.1093/bioinformatics/bth351. [DOI] [PubMed] [Google Scholar]
- 15.Dadashkhan S, et al. Deciphering crucial genes in multiple sclerosis pathogenesis and drug repurposing: A systems biology approach. J Proteomics. 2023;280:104890. doi: 10.1016/j.jprot.2023.104890. [DOI] [PubMed] [Google Scholar]
- 16.Molavi Z, et al. Identification of FDA approved drugs against SARS-CoV-2 RNA dependent RNA polymerase (RdRp) and 3-chymotrypsin-like protease (3CLpro), drug repurposing approach. Biomed Pharmacother. 2021;138:111544. doi: 10.1016/j.biopha.2021.111544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solo P. Potential inhibitors of SARS-CoV-2 (COVID 19) spike protein of the delta and delta plus variant: in silico studies of medicinal plants of North-East India. Curr Res Pharmacol Drug Discov. 2021;2:100065. doi: 10.1016/j.crphar.2021.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dehghan Z, et al. Repurposing new drug candidates and identifying crucial molecules underlying PCOS Pathogenesis Based On Bioinformatics Analysis. DARU J Pharm Sci. 2021;29:353–366. doi: 10.1007/s40199-021-00413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco-Melo D, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181(5):1036–45. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims AC, et al. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol. 2013;87(7):3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aevermann BD, et al. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci Data. 2014;1:140033. doi: 10.1038/sdata.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell HD, et al. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS ONE. 2013;8(7):e69374. doi: 10.1371/journal.pone.0069374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romagnoli S, et al. SARS-CoV-2 and COVID-19: from the bench to the bedside. Physiol Rev. 2020;100(4):1455–1466. doi: 10.1152/physrev.00020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alanis-Lobato G, Andrade-Navarro MA, Schaefer MH. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017;45(D1):D408–D414. doi: 10.1093/nar/gkw985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin A, et al. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brohee S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao X, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludbrook J. Analysis of 2 x 2 tables of frequencies: matching test to experimental design. Int J Epidemiol. 2008;37(6):1430–1435. doi: 10.1093/ije/dyn162. [DOI] [PubMed] [Google Scholar]
- 32.Cotto KC, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 34.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 35.Huang IC, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue B, et al. ISG12a restricts hepatitis C virus infection through the ubiquitination-dependent degradation pathway. J Virol. 2016;90(15):6832–6845. doi: 10.1128/JVI.00352-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, et al. ISG12a inhibits HCV replication and potentiates the anti-HCV activity of IFN-alpha through activation of the Jak/STAT signaling pathway independent of autophagy and apoptosis. Virus Res. 2017;227:231–239. doi: 10.1016/j.virusres.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 38.King AG, et al. Identification of unique truncated KC/GRO beta chemokines with potent hematopoietic and anti-infective activities. J Immunol. 2000;164(7):3774–3782. doi: 10.4049/jimmunol.164.7.3774. [DOI] [PubMed] [Google Scholar]
- 39.Escobar LE, Molina-Cruz A, Barillas-Mury C. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19) Proc Natl Acad Sci U S A. 2020;117(30):17720–17726. doi: 10.1073/pnas.2008410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis N, et al. Considering BCG vaccination to reduce the impact of COVID-19. Lancet. 2020;395(10236):1545–1546. doi: 10.1016/S0140-6736(20)31025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong W, et al. BCG vaccination: a potential tool against COVID-19 and COVID-19-like Black Swan incidents. Int Immunopharmacol. 2022;108:108870. doi: 10.1016/j.intimp.2022.108870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parmar K, Siddiqui A, Nugent K. Bacillus Calmette-Guerin vaccine and nonspecific immunity. Am J Med Sci. 2021;361(6):683–689. doi: 10.1016/j.amjms.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore HB, et al. STudy of alteplase for respiratory failure in SARS-Cov2/COVID-19: study design of the phase IIa STARS trial. Res Pract Thromb Haemost. 2020;4(6):984–996. doi: 10.1002/rth2.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loi M, et al. COVID-19 anticoagulation recommendations in children. Pediatr Blood Cancer. 2020;67(9). [DOI] [PMC free article] [PubMed]
- 45.Wang J, et al. Tissue plasminogen activator (tPA) treatment for COVID-19 associated acute respiratory distress syndrome (ARDS): A case series. J Thromb Haemost. 2020;18(7):1752–1755. doi: 10.1111/jth.14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaffe MB. Study of alteplase for respiratory failure in SARS-Cov2/COVID-19: study design of the phase IIa STARS Trial. 2020;161(3):710–727.
- 47.Price LC, et al. Rescue therapy with thrombolysis in patients with severe COVID-19 ARDS. 2020;10(4):1–5. [DOI] [PMC free article] [PubMed]
- 48.Tsai CY, et al. Increased excretions of beta2-microglobulin, IL-6, and IL-8 and decreased excretion of Tamm-Horsfall glycoprotein in urine of patients with active lupus nephritis. Nephron. 2000;85(3):207–214. doi: 10.1159/000045663. [DOI] [PubMed] [Google Scholar]
- 49.Chen X, et al. Detectable serum severe acute respiratory syndrome Coronavirus 2 Viral Load (RNAemia) is closely correlated with drastically elevated Interleukin 6 Level in Critically Ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71(8):1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz M, Fatima R, Assaly R. Elevated interleukin-6 and severe COVID-19: A meta-analysis. J Med Virol. 2020;92(11):2283. doi: 10.1002/jmv.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B, et al. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crisafulli S, et al. Potential role of Anti-interleukin (IL)-6 drugs in the treatment of COVID-19: rationale. Clin Evid Risks BioDrugs. 2020;34(4):415–422. doi: 10.1007/s40259-020-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hartmann-Boyce J, et al. Diabetes and COVID-19: risks, management, and learnings from other national disasters. Diabetes Care. 2020;43(8):1695–1703. doi: 10.2337/dc20-1192. [DOI] [PubMed] [Google Scholar]
- 54.Gupta R, Hussain A, Misra A. Diabetes and COVID-19: evidence, current status and unanswered research questions. Eur J Clin Nutr. 2020;74(6):864–870. doi: 10.1038/s41430-020-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riahi S, et al. Insulin use, diabetes control, and outcomes in patients with COVID-19. Endocr Res. 2021;46(2):45–50. doi: 10.1080/07435800.2020.1856865. [DOI] [PubMed] [Google Scholar]
- 56.Chu C, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings Thorax. 2004;59(3):252–6. [DOI] [PMC free article] [PubMed]
- 57.Gong W-J, et al. A retrospective analysis of clinical efficacy of ribavirin in adults hospitalized with severe COVID-19. J Infect Chemother. 2021;27(6):876–881. doi: 10.1016/j.jiac.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrokhpour M, et al. Infliximab and intravenous gammaglobulin in hospitalized severe COVID-19 patients in intensive care unit. Arch Iran Med. 2021;24(2):139–143. doi: 10.34172/aim.2021.22. [DOI] [PubMed] [Google Scholar]
- 59.Velez MP, McCarthy MW. Infliximab as a potential treatment for COVID-19. Expert Rev Anti Infect Ther. 2023;21(1):1–5. doi: 10.1080/14787210.2023.2151438. [DOI] [PubMed] [Google Scholar]
- 60.Honore PM, et al. Infliximab can reduce mortality from 35 to 14% in critically ill patients with COVID-19: perhaps some potential confounders to consider. Crit Care. 2020;24:1–2. doi: 10.1186/s13054-020-03294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto M, et al. Identification of Nafamostat as a potent inhibitor of Middle East respiratory syndrome Coronavirus S Protein-Mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi W, et al. Potential mechanisms of nafamostat therapy for severe COVID-19 pneumonia with disseminated intravascular coagulation. Int J Infect Dis. 2020;102:529–31. doi: 10.1016/j.ijid.2020.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osawa I, et al. Dynamic changes in fibrinogen and D-dimer levels in COVID-19 patients on nafamostat mesylate. J Thromb Thrombolysis. 2020;51:649–56. doi: 10.1007/s11239-020-02275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briand S, et al. Managing epidemics: key facts about major deadly diseases. World health organization; 2018.
- 65.Takahashi W, et al. Potential mechanisms of nafamostat therapy for severe COVID-19 pneumonia with disseminated intravascular coagulation. Int J Infect Dis. 2021;102:529–531. doi: 10.1016/j.ijid.2020.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perlin DS, et al. Levels of the TNF-Related cytokine LIGHT increase in hospitalized COVID-19 patients with cytokine release syndrome and ARDS. mSphere. 2020;5(4):10–128. doi: 10.1128/mSphere.00699-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park SH, et al. Highly efficient editing of the β-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Res. 2019;47(15):7955–7972. doi: 10.1093/nar/gkz475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, et al. Thalidomide combined with short-term low-dose glucocorticoid therapy for the treatment of severe COVID-19: A case-series study. Int J Infect Dis. 2021;103:507–513. doi: 10.1016/j.ijid.2020.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dastan F, et al. Thalidomide against coronavirus disease 2019 (COVID-19): a medicine with a thousand faces. Iran J Pharm Res: IJPR. 2020;19(1):1–2. doi: 10.22037/ijpr.2020.113369.14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morgulchik N, et al. Potential therapeutic approaches for targeted inhibition of inflammatory cytokines following COVID-19 infection-induced cytokine storm. Interface Focus. 2021;12(1):20210006. doi: 10.1098/rsfs.2021.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elkhodary MSM. Treatment of COVID-19 by controlling the activity of the nuclear factor-kappa B. CellBio. 2020;9(2):109–121. doi: 10.4236/cellbio.2020.92006. [DOI] [Google Scholar]
- 73.Hermans C, Lambert C. Impact of the COVID-19 pandemic on therapeutic choices in thrombosis-hemostasis. J Thromb Haemost. 2020;18(7):1794–1795. doi: 10.1111/jth.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sundaresan L, et al. Repurposing of thalidomide and its derivatives for the treatment of SARS-coV-2 infections: Hints on molecular action. Br J Clin Pharmacol. 2021;87(10):3835–3850. doi: 10.1111/bcp.14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 76.Weber-Gerlach M, Weber F. Standing on three legs: antiviral activities of RIG-I against influenza viruses. Curr Opin Immunol. 2016;42:71–75. doi: 10.1016/j.coi.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 77.Kell AM, Gale M., Jr RIG-I in RNA virus recognition. Virology. 2015;479–480:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X, et al. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun L, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7(2):e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang DC, et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci U S A. 2002;99(2):637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deddouche S, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Kasteren PB, et al. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J Virol. 2012;86(2):773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao Y, et al. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat Commun. 2018;9(1):4841. doi: 10.1038/s41467-018-07314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4(3):a006049. [DOI] [PMC free article] [PubMed]
- 86.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 87.Honda K, et al. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17(11):1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Readers may have access to the raw data, details of the analyzed data, and issues in the represented supplementary files.
The software used was free (Cytoscape).