Abstract
An enrichment of the neuston bacterium Nevskia ramosa was investigated by the cultivation-independent rRNA approach. N. ramosa was first described by Famintzin in 1892 as a rod-shaped, slightly bent bacterium forming typical flat rosettes on the surface of shallow freshwater habitats by unilateral slime formation. PCR in combination with cloning and sequencing was used for retrieving 21 partial and 5 nearly full-length 16S rRNA sequences forming three tight clusters. In situ hybridization with rRNA-targeted oligonucleotide probes allowed us to assign the three sequence clusters to three distinct bacterial populations abundant in the enrichment. The two probes that unambiguously identified the N. ramosa morphotype were derived from a 16S rRNA sequence that had similarities of 87.9 to 88.9% to the rRNA sequences of the most closely related group in the database, Xanthomonas sp. and relatives. N. ramosa currently is the only representative of an independent, deep branch of the gamma subclass of the class Proteobacteria. The two other populations abundant in the enrichment were affiliated with the alpha subclass of the class Proteobacteria. They were most closely related to Blastobacter sp. (97.2% similarity) and Mycoplana bullata (97.6% similarity) and might represent new species in the respective genera.
By definition, neuston is the biocenosis at the air-water interphase. The physicochemical conditions in the neuston layer are very different from those of the remaining water body and are characterized, e.g., by strong UV irradiance and relatively high nutrient concentrations (14). It is the habitat of specialized organisms, including hitherto-uncultured bacteria (4). The rRNA approach to microbial ecology and evolution (2, 18) today allows us to analyze the phylogeny of microorganisms without prior cultivation and to construct nucleic acid probes for their in situ identification. Nevskia ramosa, first described in the late 19th century by Famintzin (8), is a morphologically conspicuous neuston bacterium. It is a rod-shaped, slightly bent bacterium with cell sizes ranging from 0.4 to 1.0 by 1.0 to 5.8 μm (12). In the original publication (8), cell dimensions of 2 to 6 by 12 μm were described. Despite the uncertainties regarding the cell size, there is one idiosyncrasy that definitely allows us to relate later observations to the original description: N. ramosa grows in characteristic flat rosettes on the surface of aquatic systems (Fig. 1). More precisely, N. ramosa forms dichotomously branched stalks based on binary fission of the cells and unilateral excretion of slime. N. ramosa has frequently been observed (4, 8, 10, 12) on the surface of various shallow aquatic habitats as a member of the neuston, the community at the air-water interphase. Only two reports are known in which N. ramosa was seen in subsurface water (9, 12). Although it is apparently widespread and locally abundant, no pure culture had been obtained. Since the taxonomy of N. ramosa was based solely on morphological data, it has undergone several changes in the last 100 years. Henrici and Johnson in 1935 (10) created a new order called Caulobacteriales (from Greek kaulos, stalk) with the family Nevskiaceae, the genus Nevskia, and the species N. ramosa. In 1959, Krassilnikov (13) affiliated N. ramosa with the genus Gallionella (family, Ferribacteriaceae; order, Ferribacteriales). For Prèvot in 1961 (19), it was the only member of the family Nevskiaceae (familia incertae sedis), order Beggiatoales, and in 1959, Skerman (20) supposed a relationship with the genus Thiobacterium Janke (family, Thiobacteriaceae; order, Pseudomonadales). In the current edition of The Prokaryotes, N. ramosa is consequently found in the section on “phylogenetically unaffiliated bacteria” (12).
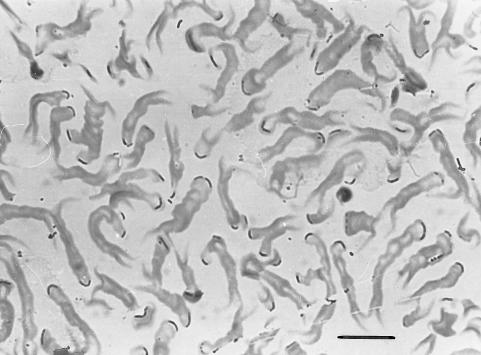
FIG. 1.
Photomicrograph of a formaldehyde-fixed and toluidine blue-stained enrichment of N. ramosa. Thin bent rods are located at the tips of dichotomously branched slime stalks. Bar, 15 μm.
It was the aim of this study to analyze the phylogeny of N. ramosa by the rRNA approach and to construct oligonucleotide probes for its unambiguous in situ identification.
MATERIALS AND METHODS
Enrichment of N. ramosa.
Surface contact slides were prepared from Lake Soelkensee (Greifswald, Germany). They were transferred to petri dishes containing sterile water from the same site. Under microscopic control, conspicuous microcolonies from the surface were transferred to Erlenmeyer flasks containing sterile water from the habitat supplemented with 0.1% sodium lactate. After incubation for 1 to 3 weeks in the dark at room temperature, a film became visible on the surface. This film was maintained by one of us (H.-D.B.) for more than 30 years by repeated transfers of the surface pellicle to sterile supplemented lake water (4).
Oligonucleotide probes.
Fluorescently labeled oligonucleotides were purchased from Interactiva (Ulm, Germany). Probe sequences, hybridization conditions, and references are given in Table 1.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′ to 3′) | Target site; rRNA positionsa | % FAb in situ | Reference |
|---|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S; 338–355 | 0–35 | 1 |
| NON338 | ACTCCTACGGGAGGCAGC | 16S; 338–355 | 0–35 | 24 | |
| ALF968 | Alpha subclass of Proteobacteria | GGTAAGGTTCTGCGCGTT | 16S; 968–986 | 35 | 17 |
| ALF4-1322 | Alpha4 group of Proteobacteria | TCCGCCTTCATGCTCTCG | 16S; 1322–1339 | 35 | 17 |
| BET42a | Beta subclass of Proteobacteria | GCCTTCCCACTTCGTTT | 23S; 1027–1043 | 35 | 15 |
| GAM42a | Gamma subclass of Proteobacteria | GCCTTCCCACATCGTTT | 23S; 1027–1043 | 35 | 15 |
| CF319a | Cytophaga-flavobacterium cluster of CFBc phylum | TGGTCCGTGTCTCAGTAC | 16S; 319–336 | 35 | 16 |
| NEV656 | N. ramosa | CGCCTCCCTCTACCGTTT | 16S; 656–674 | 0–50 | This study |
| NEV177 | N. ramosa | GCTCTTGCGAGATCATGC | 16S; 177–195 | 0–50 | This study |
| NAL657 | Thin bent rods | CCACACACCTCTCTCATA | 16S; 657–675 | 35 | This study |
| NAL1208 | Thin bent rods, M. bullata, Brevundimonas diminuta | TAGCCCACCCTGTAAGGG | 16S; 1208–1226 | 0 | This study |
| ALF921 | Star-like microcolonies | CTTGTGCAGGCCCCGTCA | 16S; 921–939 | 35 | This study |
E. coli numbering (5).
Percent formamide (FA) in in situ hybridization buffer.
CFB, cytophaga-flavobacterium-bacteroides.
In situ hybridization.
For in situ hybridization a 10-day-old enrichment with a surface film that was only slightly visible was used. This film was carefully transferred with a loop to gelatin-coated slides (Paul Marienfeldt KG, Bad Mergentheim, Germany) and air dried at 46°C. The slides were subsequently overlaid with 4% paraformaldehyde solution, fixed at room temperature for 1 h, washed with phosphate-buffered saline (130 mM sodium chloride, 10 mM sodium phosphate [pH 7.2]), and finally dehydrated in 50, 80, and 96% (vol/vol) ethanol for 3 min each. The slides were stored in the dark at room temperature until processed further.
Each well of the slides was overlaid with 10 μl of hybridization solution containing 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 0 to 35% formamide (depending on the probe [Table 1]), 0.01% sodium dodecyl sulfate (SDS), and 5 ng of labeled oligonucleotide/μl and incubated at 46°C for 90 min in an equilibrated humid chamber. Hybridization mixtures were removed, and the slides were transferred to a prewarmed (48°C) vial containing 50 ml of washing solution (80 to 900 mM NaCl, 20 mM Tris-HCl [pH 7.4], 5 mM EDTA, and 0.01% SDS) and incubated without shaking for 15 min. The stringency of the washing step was adjusted by changing the NaCl concentration in the washing buffer. The concentrations were 900, 225, and 80 mM for probes hybridized at 0, 20, and 35% formamide, respectively. The slides were briefly rinsed with distilled water, air dried, and mounted in Citifluor (Citifluor Ltd., London, United Kingdom).
The slides were observed with an Axioplan epifluorescence microscope (Zeiss, Jena, Germany) equipped with a 50-W high-pressure mercury bulb and specific filter sets (fluorescein, Zeiss 09; carboxytetramethylrhodamine, Chroma HQ 41007 [Chroma Technology Corp., Brattleboro, Vt.]). Color photomicrographs were done on Kodak (Rochester, N.Y.) Panther 1600 film. For phase-contrast micrographs, the automatic exposure system was used; for epifluorescence micrographs, the exposure times were manually set to 8 or 15 s.
Composition of a 16S rDNA clone library.
The total surface film in a 3-liter Erlenmeyer flask was collected and subsequently harvested by centrifugation (15 min; 14,000 × g). The pellet was washed several times with distilled water. Total cellular nucleic acids were isolated by lysis with high-salt extraction buffer (1.5 M NaCl), extended heating (2 to 3 h), proteinase K, and SDS, followed by phenol-chloroform extraction as described by Zhou et al. (25). Almost full-length bacterial 16S rRNA gene fragments were amplified from genomic DNA by PCR with two general bacterial 16S ribosomal DNA (rDNA) primers (21). The nucleotide sequences of the primers were 5′-AGAGTTTGATYMTGGCTCAG-3′ (Escherichia coli 16S rDNA positions 8 to 27 [5]) and 5′-CAKAAAGGAGGTGATCC-3′ (E. coli 16S rDNA positions 1529 to 1546 [5]). Amplification was performed with a capillary PCR cycler (Idaho Technologies, Idaho Falls, Idaho). In a final volume of 50 μl, the reaction mixture contained 25 pmol of each of the amplification primers, 200 μM [each] deoxynucleoside triphosphate, 50 mM Tris, 250 μg of bovine serum albumin, 1 mM tartrazine, 2 mM MgCl2, 5 μl of template, and 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.). After initial heating of the mixture to 94°C for 45 s, 35 cycles were performed consisting of 94°C (15 s), 49°C (20 s), and 72°C (30 s). Following the final cycle, the reaction was extended at 72°C for 1 min.
The amplification products were analyzed by agarose gel electrophoresis. The 1.5-kb band was excised, and the DNA was purified with an extraction kit (Boehringer, Mannheim, Germany) and subsequently ligated into the pCR 2.1 vector with the TA cloning kit (Invitrogen, Carlsbad, Calif.). Transformants were plated on dYT agar plates (1.6% tryptone, 1.0% yeast extract, 0.5% NaCl, 0.2% glucose, 1.5% Bacto Agar) containing the antibiotic ampicillin (100 μg ml−1), spread evenly with 40 μl of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 mg ml−1) and 40 μl of IPTG (isopropyl-β-d-thiogalactopyranoside) (100 mM), and incubated at 37°C overnight. Forty-eight white clones were picked on dYT agar plates for further examination. Plasmid DNA was extracted with the QIAprep-spin kit (Qiagen, Hilden, Germany). The presence of right-sized inserts was determined by linearization of recombinant plasmids with NotI followed by agarose gel electrophoresis.
16S rDNA sequencing.
16S rDNA clones were sequenced with a LICOR DNA 4000 automated sequencer (MWG-Biotech, Ebersberg, Germany). Cycle sequencing protocols of the chain termination technique (7) were applied according to the manufacturer’s instructions (Amersham-Buchler, Braunschweig, Germany) with infrared-dye-labeled primers.
Data analysis.
The sequences were added to the 16S rRNA sequence data base of the Technical University Munich with the program package ARB (22). The tool ARB EDIT was used for sequence alignment. The alignment was checked by eye and corrected manually. 16S rRNA-based phylogenetic trees were reconstructed based on distance matrix analyses of all available 16S rRNA primary structures of gamma, beta, and alpha Proteobacteria. Tree topologies were evaluated by performing maximum-parsimony, neighbor-joining, and maximum-likelihood analyses of the full data set and subsets. Only sequences that were at least 90% complete were used for treeing. Alignment positions at which less than 50% of the sequences of the entire data set shared the same residues were excluded from the calculations. Partial sequences were inserted into the reconstructed tree by applying the parsimony criteria without allowing changes in the overall tree topology.
Nucleotide sequence accession numbers.
The three almost full-length 16S rRNA sequences obtained have been deposited with EMBL (accession no. AJ001343 [NO4 and -10], AJ001344 [NO22], and AJ001345 [NO2 and -11]).
RESULTS
Enrichment.
Phase-contrast microscopy of the enrichment (Fig. 2A, C, and E) showed that the following two distinct morphotypes were abundant: (i) a phase-dense rod with a cell diameter of approximately 1.0 μm and a length of 2 to 3 μm forming star-like microcolonies and (ii) a thinner, slightly bent rod with cell dimensions of 0.7 by 3 to 4 μm. The fully developed two-dimensional flat rosettes typical for N. ramosa were not observed. However, numerous early stages of dichotomously branched microcolonies consisting of 2 to 4 cells could be seen. In these cells, spherical bodies of various diameters were also visible, as described by Famintzin (8).
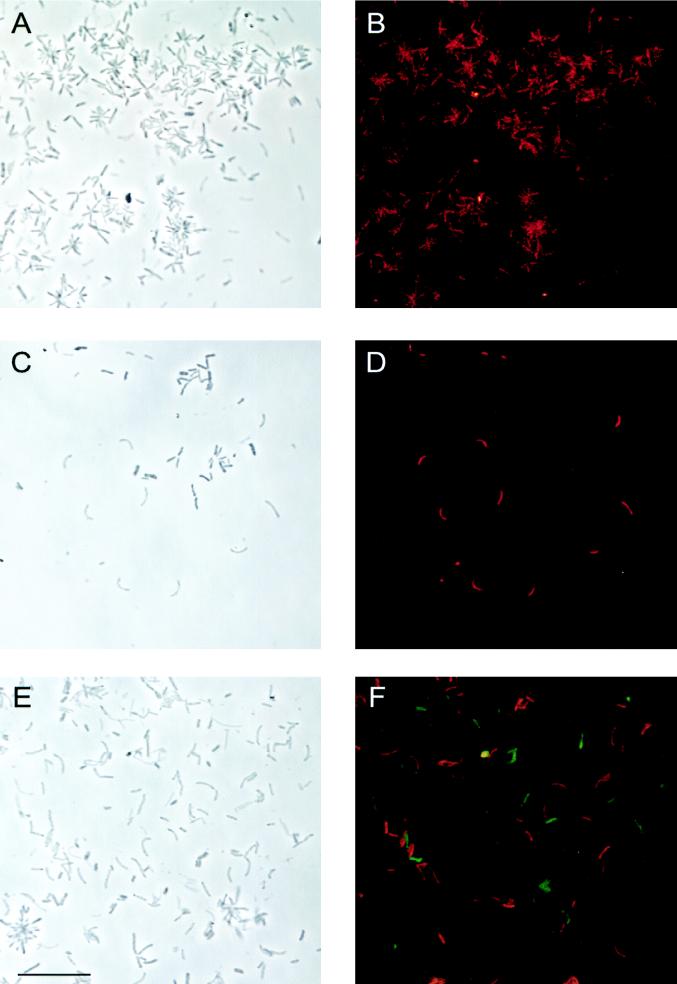
FIG. 2.
In situ hybridization of N. ramosa enrichment. Phase contrast (A, C, and E) and epifluorescence (B, D, and F) micrographs are shown for identical microscopic fields. (A and B) Probe ALF921 (CT labeled; red). (C and D) Probe NEV656 (CT labeled; red). (E and F) Simultaneous hybridization with probes NEV656 (CT labeled; red) and NAL657 (fluorescein labeled; green) distinguishes two populations of thin bent rods. Bar, 10 μm (all panels).
Preliminary phylogenetic characterization of the enrichment by in situ hybridization with group-specific probes.
A set of domain- and group-specific rRNA-targeted probes was used for initial characterization. All cells in the enrichment could be detected with probe EUB338, specific for bacteria. Probing on the group level revealed three distinct populations. The phase-dense rods forming star-like microcolonies consistently hybridized with the probes ALF968 (17) and ALF4-1322 (17), specific for the alpha subclass and alpha4 group of the class Proteobacteria, respectively. The thin bent rods consisted of at least two genotypes. One population hybridized with probe ALF968, and the other hybridized only with EUB338. No signals could be detected by applying the probes GAM42a, BET42a, and CF319a, specific for the beta and gamma subclasses of the class Proteobacteria and the cytophaga-flavobacterium group, respectively, or with the oligonucleotide NON338, routinely applied to check for nonspecific probe binding and autofluorescence (Table 2).
TABLE 2.
Hybridization results and affiliation of clones
| Probe | Hybridization witha:
|
||
|---|---|---|---|
| Star-like microcoloniesb | Thin bent rodsc | N. ramosad | |
| EUB338 | + | + | + |
| NON338 | − | − | − |
| ALF968 | + | + | − |
| ALF4-1322 | + | − | − |
| BET42a | − | − | − |
| GAM42a | − | − | − |
| CF319a | − | − | − |
| ALF921 | + | − | − |
| NAL657 | − | + | − |
| NAL1208 | − | + | − |
| NEV656 | − | − | + |
| NEV177 | − | − | + |
+, hybridized; −, did not hybridize.
Represented by clone NO22; closest-matching sequence (97.2% similarity), Blastobacter sp.
Represented by clones NO2 and -11; closest-matching sequence (97.6% similarity), M. bullata.
Represented by clones NO4 and -10; closest-matching sequences (87.9 to 88.9% similarity), Xanthomonas group.
Phylogenetic analysis of a 16S rDNA clone library.
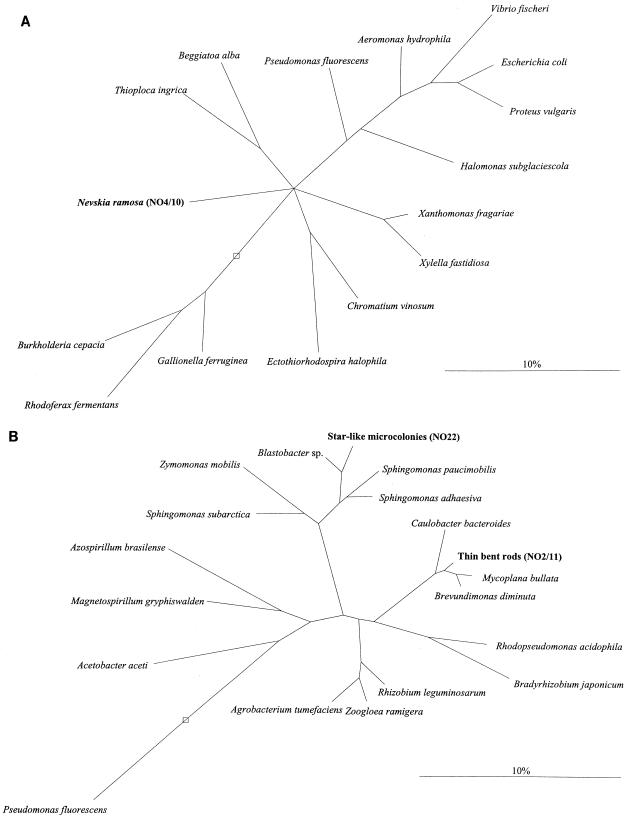
Twenty-one 16S rDNA clones of a library constructed from the enrichment were randomly chosen for partial sequencing (400 to 1,000 bases) in order to get at least one sequence from each of the three populations. One sequence (NO22) was found to be affiliated with the alpha4 group of the class Proteobacteria. A cluster of five sequences (NO2, NO6, NO8, NO11, and NO21) was clearly of alpha-proteobacterial origin, but not from the alpha4 group of the class Proteobacteria. A second coherent cluster of 15 sequences (NO1, NO4 and -5, NO7, NO9 and -10, NO12 to -14, NO16 to -20, and NO24) branched deep in the gamma subclass of the class Proteobacteria. Comparative sequence analysis within the two clusters revealed very high similarity values, between 98.3 and 99.9%. Clone NO22 and two clones from each of the two clusters (NO2 and -11, and NO4 and -10) were fully sequenced. No sequence differences could be seen within the two pairs NO2 and -11 and NO4 and -10. Comparative analysis of the nearly full-length sequences showed similarities of 97.2% between NO22 and Blastobacter sp., 97.6% between NO2 and -11 and Mycoplana bullata, and 87.9 to 88.9% between NO4 and -10 and the Xanthomonas group (Table 2). Based on these data, two trees were reconstructed showing the relationship of the newly determined sequences to selected alpha, beta, and gamma sequences (Fig. 3). The tree calculated with the NO4 and -10 clone sequence (Fig. 3B) was corrected by taking into consideration the differing results of the various tree reconstruction algorithms. Bifurcations indicate branchings which appeared stable and well separated from neighboring branchings in all cases. Multifurcations indicate tree topologies which could not be significantly resolved based on the available data set.
FIG. 3.
Phylogenetic trees based on comparative analysis of 16S rDNA retrieved from the enrichment. The bars indicate 10% estimated sequence divergence. (A) Relationships of N. ramosa, represented by the clones NO4 and -10, and its closest relatives among the gamma subclass of the class Proteobacteria. (B) Relationships of the thin bent rods and the star-like microcolonies, represented by the clones NO2 and -11 and NO22, respectively, and their closest relatives among the alpha subclass of the class Proteobacteria. The open boxes indicate the root.
In situ identification of N. ramosa.
Based on the full-length sequences, five probes were constructed to assign the three different 16S rRNA sequences to cell populations in the enrichment. ALF921 was designed to be specific for the NO22 clone sequence and was successfully used for in situ hybridization showing the typical star-like microcolonies in the enrichment (Fig. 2A and B). Probes NEV656 and NEV177 both hybridized with the population of thin bent rods that had before only hybridized with probe EUB338 but with none of the group-specific probes. These two probes, targeted to the most frequent 16S rDNA sequence type, stained all of the characteristically arranged cells (Fig. 1 and 2C and D), and therefore we assigned the NO4 and -10 sequence to N. ramosa. Two probes were designed to be specific for NO2 and -11. Simultaneous in situ hybridization with differently labeled probes revealed that NAL657 and NAL1208 bound specifically to the probe ALF968-positive population of thin bent rods. Simultaneous hybridization with probes NEV656 labeled with carboxytetramethylrhodamine (red) and NAL657 labeled with fluorescein (Fig. 2E and F) demonstrated that, despite similar morphologies, the second population of thin bent rods in the enrichment was distinct from N. ramosa.
For ALF921, NAL657, and NAL1208, probe specificities at different hybridization stringencies were only evaluated for the other bacteria in the enrichment of interest. ALF921 and NAL657 showed specific signals at 35% formamide, and NAL1208 showed specific signals at 0% formamide. No further testing was done, since the focus of the study was N. ramosa.
The optimal hybridization conditions for the two probes targeting N. ramosa, NEV656 and NEV177, were tested in more detail. Both probes showed specific in situ hybridization signals at stringencies between 0 and 50% formamide. The probes did not hybridize to several organisms of the beta and gamma subclasses of the class Proteobacteria tested (data not shown), which reflects the presence of at least three mismatches between the probes and any of the other entries in the publicly available rRNA databases.
DISCUSSION
In this study, the phylogeny of N. ramosa was revealed more than 100 years after its initial description in 1892 (8). At the time we started, N. ramosa was not available as a pure culture. Therefore, an enrichment was used which, based on microscopic investigation, was thought to consist of only two populations, a contaminant showing star-like microcolonies of phase-dense rods, and the thin bent rods typical for N. ramosa. Since individual thin bent rods without unilateral slime formations could be detected outside the characteristic colonies consisting of dichotomously branched stalks with a single cell at the tip, the working hypothesis was that N. ramosa might be present in different growth stages (3). However, the in situ hybridization with group-specific probes had already revealed that the enrichment still contained three major constituents, two of those being separate populations of thin bent rods. Considering the low complexity of bacterial shapes, this is not surprising. On this level, the group-specific probing not only revealed genetic heterogeneity behind morphological similarity but it also predicted phylogenetic affiliations for the different population members. After comparative 16S rDNA sequence analysis it is now possible to retrospectively judge the quality of these predictions. The contaminating thick rod, clearly distinct from N. ramosa in its morphology, was indeed shown to be a member of the alpha4 group of the class Proteobacteria, as originally indicated by hybridization with probes ALF968 and ALF4-1322. Also, the assignment of one of the two populations of thin bent rods to the alpha subclass of the class Proteobacteria outside the alpha4 group was shown to be correct. The true N. ramosa, however, now known to branch deep in the gamma subclass of the class Proteobacteria, did not hybridize with any of the group-specific probes, including GAM42a, designed to be specific for the gamma subclass. Recently, it has been reported that another deep-branching genus of the gamma subclass of the class Proteobacteria, Xanthomonas sp., is also not detected by this probe (6). These results should be considered when using the 23S rRNA-targeted probe GAM42a. Even though so far no cases are known in which this probe binds to bacteria other than those from the gamma subclass of the class Proteobacteria, it clearly does not bind to all members of this group and might miss some of the deep-branching members. In the light of the huge natural rRNA sequence diversity and the constantly increasing rRNA data set it should be kept in mind that oligonucleotide probes are tools that have to be checked regularly. It might be time to supplement GAM42a with one or several new 16S rRNA-targeted probes for the gamma subclass of the class Proteobacteria.
Evolutionary consequences.
With an overall 16S rRNA similarity of only 87.9 to 88.9% to its nearest cultured relatives, the Xanthomonas group, N. ramosa currently is the sole representative of a deep branch of the gamma subclass of the class Proteobacteria. With this low level of relatedness, it is almost impossible to deduce anything about the physiological potential of N. ramosa from its location in the phylogenetic tree. The large evolutionary distance from its nearest characterized relative, however, provides ample potential for specific adaptation to the habitat of the neuston layer, which is characterized, e.g., by a high level of radiation with its damaging influence. The current lack of additional taxa on the Nevskia branch of the phylogenetic tree might be due simply to problems in cultivation that are shared by the members of this group. From a taxonomic point of view it is now clear that N. ramosa should not be affiliated with either the Caulobacteriales (10), which are indeed alpha-subclass proteobacteria, or Gallionella sp. (13), which are beta-subclass proteobacteria. There is also no close relationship with the Beggiatoales (19) or the genus Thiobacterium (20).
The situation for the other two bacteria present in the enrichment is different. Closely related cultured species have been sequenced before. The 16S rRNA of M. bullata has a similarity of 97.6% with the second population of thin bent rods, and the 16S rRNA of Blastobacter sp. is 97.2% similar to that of the phase-dense thick rods forming star-like microcolonies. The morphology of the latter, including cell shape and dimensions and polar attachment resulting in the formation of rosettes, is indeed typical for Blastobacter sp. (11). Both sequences are sufficiently different from the closest entries in the rRNA databases that they could originate from new species within the respective genera. However, a final taxonomic placement of the two organisms to which the sequences belong has to await cultivation and the examination of DNA-DNA homology with their closest relatives. Blastobacter sp. is known to be a widespread member of the neuston (11), but the natural habitat of the M. bullata-related organism remains unclear. It obviously was able to establish itself in quite high abundance in the particular laboratory enrichment investigated in this study. However, whether it is also abundant in natural neuston communities remains to be evaluated. The specific nucleic acid probes, facilitating tracing of both organisms in situ, will be a helpful tool for further ecological studies.
The oligonucleotide probes.
Considering the various possibilities for getting false-positive or -negative hybridization results (2), two specific probes were constructed for each of the two clusters of 16S rDNA clone sequences, whereas only one probe was designed, based on clone NO22, for the easily distinguishable phase-dense rods. Under the hybridization conditions used in this study, each of the five probes, ALF921, NAL657, NAL1208, NEV177, and NEV656, hybridized specifically to only one of the three populations present in the enrichment. Probes ALF921, NAL657, and NEV656 yielded brighter signals than NAL1208 and NEV177. The differences in signal strength within the two pairs of probes targeted to the same cells, NAL657 and NAL1208 and NEV656 and NEV177, can only be attributed to different in situ accessibilities of the respective 16S rRNA target sites (2). On several occasions, the region between positions 650 and 680 proved to be ideally suited for in situ probing. This site, like the target site of probe NEV177, is not one of the most variable on the 16S rRNA, and it remains to be seen whether the two probes designed for N. ramosa have species or genus specificity. With only one species on a deep phylogenetic branch, this question is currently not relevant. It should, however, be kept in mind when more sequences or organisms related to N. ramosa are found.
With our new knowledge it is now possible to evaluate whether N. ramosa is indeed widely distributed in aquatic habitats, as suggested before (12). This, together with the possibility of obtaining pure cultures of N. ramosa (23), should stimulate further research in the physiology and ecology of the bacterial community found in the neuston layer.
During this study, two more sequences from pure cultures of N. ramosa became available (23). The strain Soe1 (DSMZ 11499T) was isolated from the same enrichment used in this study (Lake Soelkensee), whereas strain OL1 (DSMZ 11500) came from a freshwater ditch in Oldenburg. Both sequences showed high similarity to the NO4 and -10 clone sequence: 99.2 and 98.0% for Soe1 and OL1, respectively. Also, the two specific probes NEV177 and NEV656, developed in this study for N. ramosa, had no mismatches and hybridized specifically with the pure cultures.
ACKNOWLEDGMENTS
This work has been supported by a grant from the Deutsche Forschungsgemeinschaft (Am73/2-4).
We thank Sibylle Schadhauser and Kirsten Meissner for expert technical assistance, Alexander Neef for making available probes ALF968 and ALF4-1322 before publication, and Wolfgang Ludwig for ARB and help in the reconstruction of phylogenetic trees.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babenzien H-D. Genus Nevskia Famintzin 1892, 484. In: Buchanan R E, Gibbons N E, editors. Bergey’s manual of determinative bacteriology. 8th ed. Baltimore, Md: Williams and Wilkins; 1974. pp. 161–162. [Google Scholar]
- 4.Babenzien H-D. Genus Nevskia Famintzin 1892, 484AL. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams and Wilkins; 1989. pp. 1979–1981. [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz-Cleven B E E, Rattunde B, Straub K L. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. System Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 7.Chen E Y, Seeburg P H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 8.Famintzin A. Eine neue Bacterienform: Nevskia ramosa. Arb Bot Lab Kaiserlichen Akad Wiss St-Petersburg. 1892;2:481–487. [Google Scholar]
- 9.Heldal M, Tumyr O. Morphology and content of dry matter and some elements in cells and stalks of Nevskia from an eutrophic lake. Can J Microbiol. 1986;32:89–92. [Google Scholar]
- 10.Henrici A T, Johnson D E. Studies of freshwater bacteria. II. Stalked bacteria, a new order of Schizomycetes. J Bacteriol. 1935;30:61–93. doi: 10.1128/jb.30.1.61-93.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch P. The genus Blastobacter. In: Balows A, Trüper H G, Dworkin M, Harder W, Schlegel H G, editors. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. New York, N.Y: Springer; 1991. pp. 2171–2175. [Google Scholar]
- 12.Hirsch P. The genus Nevskia. In: Balows A, Trüper H G, Dworkin M, Harder W, Schlegel H G, editors. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. New York, N.Y: Springer; 1991. pp. 4090–4092. [Google Scholar]
- 13.Krassilnikov N A. Diagnostik der Bacterien und Actionomyceten. Jena, Germany: Fischer Verlag; 1959. [Google Scholar]
- 14.Lambert W, Sommer U. Limnoecology. Oxford, United Kingdom: Oxford University Press; 1997. [Google Scholar]
- 15.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 16.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 17.Neef A. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Munich, Germany: Technical University Munich; 1997. [Google Scholar]
- 18.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 19.Prèvot A-R. Traité de systématique bactérienne. Paris, France: Dunod; 1961. [Google Scholar]
- 20.Skerman V B. A guide to the identification of the genera of bacteria. Baltimore, Md: Williams & Wilkins; 1959. [Google Scholar]
- 21.Springer N, Ludwig W, Amann R, Schmidt H J, Görtz H-D, Schleifer K-H. Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc Natl Acad Sci USA. 1993;90:9892–9895. doi: 10.1073/pnas.90.21.9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckman, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. http://www.mikro.biologie.tu-muenchen.de. Department of Microbiology, Technische Universität München, Munich, Germany.
- 23.Stürmeyer H, Overmann J, Babenzien H-D, Cypionka H. Ecophysiological and phylogenetic studies of Nevskia ramosa in pure culture. Appl Environ Microbiol. 1998;64:1890–1894. doi: 10.1128/aem.64.5.1890-1894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization of suspended cells with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]