Abstract
Conjugal plasmid transfer was examined on the phylloplane of bean (Phaseolus vulgaris) and related to the spatial distribution pattern and metabolic activity of the bacteria. The donor (Pseudomonas putida KT2442) harbored a derivative of the TOL plasmid, which conferred kanamycin resistance and had the gfp gene inserted downstream of a lac promoter. A chromosomal insertion of lacIq prevented expression of the gfp gene. The recipient (P. putida KT2440) had a chromosomal tetracycline resistance marker. Thus, transconjugants could be enumerated by plating and visualized in situ as green fluorescent cells. Sterile bean seedlings were inoculated with donors and recipients at densities of approximately 105 cells per cm2. To manipulate the density and metabolic activity (measured by incorporation of [3H]leucine) of the inoculated bacteria, plants were grown at various relative humidities (RH). At 100% RH, the transconjugants reached a density of 3 × 103 CFU/cm2, corresponding to about one-third of the recipient population. At 25% RH, numbers of transconjugants were below the detection limit. Immediately after inoculation onto the leaves, the per-cell metabolic activity of the inocula increased by up to eight times (100% RH), followed by a decrease to the initial level after 96 h. The metabolic activity of the bacteria was not rate limiting for conjugation, and no correlation between the two parameters was observed. Apparently, leaf exudates insured that the activity of the bacteria was above a threshold value for transfer to occur. Transconjugants were primarily observed in junctures between epidermal cells and in substomatal cavities. The distribution of the transconjugants was similar to the distribution of indigenous bacteria on nonsterile leaves. Compared to polycarbonate filters, with cell densities equal to the overall density on the leaves, transfer ratios on leaves were up to 30 times higher. Thus, aggregation of the bacteria into microhabitats on the phylloplane had a great stimulatory effect on transfer.
Genetic exchange by conjugal plasmid transfer has been observed in diverse aquatic (2, 3, 38, 43, 47) and terrestrial (28, 32, 49, 51, 52) environments and has been suggested to be an important mechanism in the adaptation of microbial communities to changing environmental conditions (4, 31).
An important habitat in the terrestrial environment is the phyllosphere. Gene transfer by conjugation between epiphytic bacteria is, however, poorly investigated. Lacy and Leary (30), Knudsen et al. (25), and Björklöf et al. (5) studied conjugation on the phylloplane of bean. Transfer ratios up to 3 × 10−1 (number of transconjugants per recipients [T/R]) were observed at humidities close to 100% (30). In other studies, Lilley and Bailey (31) demonstrated transfer of natural mercury resistance plasmids from indigenous bacteria of the sugar beet phylloplane to an added pseudomonad.
The phylloplane can under many environmental conditions be considered a hostile habitat as the epiphytic bacteria are exposed to desiccation and solar UV radiation (8, 33, 45). On the other hand, leaf exudates, such as carbohydrates, amino acids, and organic acids (37) may support bacterial densities of up to 5 × 107 CFU/g (fresh weight) under humid conditions (23). In addition, the structurally complex leaf surface, consisting of epidermal cells, interstitial spaces, trichomes, and stomata (7, 22), may provide bacteria with survival habitats. Both availability of growth substrates, a high bacterial density, and the presence of solid surfaces are believed to stimulate conjugal transfer (15, 20, 35, 50).
An understanding of the factors that influence genetic exchange by conjugation is pertinent for assessing the significance of conjugation in the evolution of microbial communities as well as for more pragmatic reasons, such as risk assessment of released genetically engineered bacteria. The aim of the present study was to investigate the significance of bacterial distribution and metabolic activity on conjugation on the phylloplane. To the best of our knowledge, this is the first report which relates conjugal transfer on the phylloplane to the bacterial metabolic activity, and it is the first study in which the effect of cell distribution on transfer is directly assessed. To accomplish these objectives, bean plants were grown at various relative humidities (RH) to simultaneously manipulate the density and activity of the inoculated bacteria. In situ metabolic activity and distribution of transconjugant cells were determined by incorporation of tritiated leucine (Leu) and by using green fluorescent protein as plasmid reporter gene, respectively.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Characteristics of the strains and plasmids used are listed in Table 1. Pseudomonas putida KT2442::lacIq served as donor strain in biparental mating experiments. The strain harbored a derivative of the TOL plasmid which conferred kanamycin resistance and had the gfpmut3b reporter gene cloned downstream of the lac promoter, PA1/O4/O3 (constructions are described below). As recipient strain, P. putida KT2440 with a chromosomal tetracycline resistance marker was used. Donors were grown in Luria broth (LB) (36) supplemented with 50 μg of kanamycin per ml (KM50), while recipients were grown in LB with 15 μg of tetracycline per ml (TC15). Transconjugants (P. putida KT2440/TOL) were enumerated on LB plates containing both KM50 and TC15. Plates were incubated at 30°C.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristic | Source or reference |
|---|---|---|
| Bacteria | ||
| P. putida KT2440 | TOL plasmid-cured derivative of P. putida mt-2 | (1) |
| P. putida KT2440-Tc | KT2440 with mini-Tn5 insertion of Tcr | (29) |
| P. putida KT2442 | Rifr mutant of KT2440 | (1) |
| P. putida KT2442::lacIq | KT2442 with mini-Tn5 insertion of lacIq, Rifr | This study |
| P. fluorescens AS12 | arg mutant of P. fluorescens R2f, Rifr | M. Givskov |
| E. coli MC1061 | Strr | ATCC 53338 |
| Plasmids | ||
| TOL | Self-transmissible 117-kb plasmid from P. putida mt-2 | (51) |
| TOL::gfpmut3b | TOL with mini-Tn5 insertion of PA1/O4/O3::gfpmut3b, Kmr | This study |
| RP4 | Self-transmissible 60-kb broad-host-range IncP plasmid, Kmr, Tcr, Ampr | (6) |
In studies of transfer to indigenous epiphytic bacteria, the auxotrophic (arg mutant) Pseudomonas fluorescens AS12 containing plasmid RP4 was used. P. fluorescens AS12 was chosen as donor in these experiments because it, unlike for P. putida KT2442, is possible to counterselect this strain on transconjugant selective plates (see below). RP4 was the plasmid of choice because it is a promiscuous plasmid that can be transferred to a large variety of bacterial species (26). The TOL plasmid, on the other hand, has a more narrow host range (40).
Donors were enumerated on LB supplemented with KM50 and TC15, indigenous recipients on minimal medium (16) amended with 0.2% glucose, and indigenous transconjugants on minimal medium amended with 0.2% glucose with KM50 and TC15. To avoid overgrowth by fungi, media were supplemented with 25 μg of natamycin (Delvocid; Gist-Brocades, Delft, Holland) per ml. Plates were incubated at 25°C.
Construction of strains and plasmids.
The lacIq gene (48) was inserted into the chromosome of P. putida KT2442 by triparental mating (14) by using a modified pUT vector with resolvase sites flanking the npt gene (27). Subsequently, the npt gene was deleted by a second round of triparental mating and a Kms transconjugant was picked.
To construct a PA1/O4/O3::gfpmut3b gene cassette, the gfpmut3b gene (12) was amplified by PCR as a 0.7-kb SphI-HindIII fragment. The gfpmut3b gene is a variant of the wild-type gfp gene in which two amino acids have been substituted. These substitutions result in an enhanced fluorescent signal (12). To introduce a SphI restriction site in the start codon of gfpmut3b, the sequence was changed during PCR so that the gfpmut3b contained an Arg instead of a Ser residue at position 2. The gfpmut3b fragment was cloned downstream from the promoter PA1/O4/O3 (34) in an optimal distance from the ribosome binding site of phage T5 (RBSII) and upstream of a region with translational stop codons in all three reading frames, as well as two strong transcriptional terminators, T0 (from phage lambda) and T1 (from the rrnB operon of Escherichia coli). The NotI fragment from the resulting plasmid (pJBA27), containing RBSII, gfpmut3b, the translational stop codons, and the transcriptional terminators, was inserted into the NotI site of pUT-Km (13), resulting in a transposon delivery vector (pJBA28) containing the PA1/O4/O3::gfpmut3b and npt gene cassette.
Insertion of the PA1/O4/O3::gfpmut3b cassette into the TOL plasmid was performed in two steps. First, pJBA28 was transferred to P. putida KT2440 by triparental mating. Isolation on AB minimal plates (11), containing KM50 and 10 mM citrate, resulted in KT2440 derivatives carrying the PA1/O4/O3::gfpmut3b cassette either on the chromosome or on the TOL plasmid. To isolate clones with the cassette integrated on the plasmid, a second round of conjugation was performed. All colonies from the selective plates (>1,000 per plate) were scraped off and suspended in 1 ml of 0.9% NaCl. Cells were then mixed with the Kms P. putida KT2442::lacIq. Isolation on plates containing KM50 and 50 μg of rifampin per ml (RIF50) resulted in different Kmr derivatives carrying the modified TOL plasmid. The clone chosen for the gene transfer experiments was able to grow on AB minimal plates supplemented with either 5 mM benzyl alcohol or 5 mM benzoate (53); it showed green fluorescence upon addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and illumination with blue light, and the conjugation frequency of the gfp-tagged plasmid was similar to that of the wild-type TOL plasmid as tested on agar plates.
Sterilization and growth of plants.
Seeds of bush bean (Phaseolus vulgaris cv. Montana) were sterilized in a solution of 0.25% benzalkoniumchloride and 25% H2SO4 for 2 h followed by careful rinsing in sterile MilliQ-water. The sterilized seeds were pregerminated on LB plates (to test for sterility) for 3 to 4 days in the dark after which they were transferred aseptically to sterile rock wool cubes with 5 ml of autoclaved Hoagland’s plant nutrient solution (18) in 30-ml plastic pots. The pots were incubated in a growth chamber at 26 to 28°C and a 23:1-h light-dark cycle. The plants were watered with autoclaved Hoagland’s plant nutrient solution when needed. Untreated bean plants were grown in pots with soil from an uncultivated field at Risø, near Roskilde, Denmark. In this case, pots were kept in the dark for 4 to 5 days, after which they were transferred to the growth chamber. Prior to inoculation, all plants were incubated for 24 h at the RH to be used in the specific experiment.
The growth chamber was equipped with two halogen-quartz-iodine-tungsten lamps (Osram Daylight HQI-T 250 W/D). Light intensities were 240 to 270 μmol/m2/s. RH was controlled by a vaporizer and measured by a Kane May 8004 RH sensor and time logged by a Tinytalk datalogger (Orion Components [Chichester] Ltd., United Kingdom). Both sensors had an accuracy of ±2% RH and an upper limit of 95% RH. An RH of approximately 100% was obtained by incubating plants in plastic containers (15 to 20 liters) covered with polyethylene film and with water added to the bottom of the containers.
Inoculation of plants.
Overnight cultures were washed twice in 10 mM phosphate buffer (pH 7.0) (7,740 g, 8 min in a Beckman JA20 rotor), starved for 24 h at room temperature and adjusted to approximately 108 cells/ml according to predetermined optical density curves. The starvation period was used to reduce intracellular energy resources.
Leaves of sterile 12- to 14-day-old plants were inoculated by carefully immersing the green parts of the plants in a 1:1 mixture of the P. putida donor and recipient suspensions or in the P. fluorescens AS12/RP4 suspension for 10 to 15 s. Excess drops of liquid were removed by gentle shaking of the plants. Densities of approximately 108 CFU per g (dry weight) or 105 CFU per cm2 were achieved (Fig. 1). In some instances seed inoculation was used. This was done by inoculating the Hoagland solution of the sterile rock wool cubes (see above) with 107 CFU/ml of the cells.
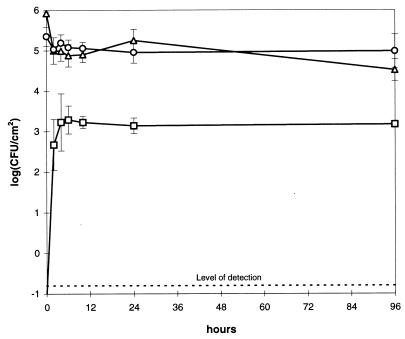
FIG. 1.
Survival of P. putida KT2442/TOL (donors, ▵) and P. putida KT2440-Tc (recipients, ○) and appearance of KT2440-Tc/TOL (transconjugants, □) on bean leaves at 100% RH (A) and 90% ± 2% RH (B). Error bars are ± SD of triplicate samples.
In the biparental mating experiments, the metabolic activity of the inocula was determined by incorporation of tritiated Leu (24, 28). One milliliter of a 0.01 mM Leu solution containing 0.40 μCi of [4,5-3H]Leu (139 Ci/mmol; Amersham Life Science) was added to 4-ml aliquots of the bacterial suspensions to give a final Leu concentration of 2,000 nM. Killed controls were set up by addition of 500 μl of 37% formaldehyde. The incorporation was terminated after 30 min by addition of 500 μl of 37% formaldehyde. Suspensions were filtered through 0.2-μm-pore-size cellulose-nitrate filters (Sartorius GmbH, Göttingen, Germany). Filters were rinsed with 5 ml of 10 mM phosphate buffer and counted on a Beckman LS1801 scintillation counter. The concentration of Leu required to reach the saturation level with respect to bacterial assimilation had been determined in a preliminary experiment.
Sampling procedure.
At each sampling time, both the metabolic activity and bacterial population size were determined. One leaf was excised from each of three replicate plants, and leaves were submerged individually in 5 ml of phosphate buffer (10 mM, pH 7.0) containing 250 nM Leu and 1 μCi of [4,5-3H]Leu (139 Ci/mmol; Amersham Life Science). Controls were set up by addition of 500 μl of 37% formaldehyde. Incorporation of Leu was stopped after 30 min by transferring the leaves to new phosphate buffer without Leu. The bacteria were then extracted by sonication for 7 min in a Branson 5210 ultrasonic bath followed by 15 to 20 s of vortexing. Aliquots (4 ml) of the extracts were filtered through 0.2-μm-pore-size cellulose-nitrate filters. Filters were rinsed and counted as described above.
Numbers of donors, recipients, and transconjugants were determined by serially diluting the remaining extract and plating on selective medium. To improve the detection limit of transconjugants, aliquots of 400 μl were mixed with 1.6 ml of 10 mM phosphate buffer and filtered through 0.2-μm-pore-size polycarbonate membrane filters (Poretics Products, Livermore, Calif.). Filters were placed on transconjugant selective media. Parallel to sampling, the significance of mating on the transconjugant selective media was assessed. This was done by combining extracts of leaves, inoculated with donors and recipients separately, and plating on transconjugant selective media as described above. Plate mating constituted less than 5% of the observed transconjugants. Reported numbers of transconjugants (see Results) are corrected for plate mating.
Leaves were dried for 24 h at 110°C, and the dry weight was determined. Conversion of dry weight to surface area (both sides) was performed according to the following equation: surface area (cm2) = 0.747 × dry weight (mg) (n = 22; P < 0.0001). The equation was determined by measuring the dry weight of 1- by 1-cm squares of the leaves.
Filter matings.
Two different filter-mating experiments were performed. In one experiment, starved donor and recipient suspensions were filtered onto 0.2-μm-pore-size polycarbonate filters (Poretics Products) to a density of 107 CFU/cm2. A monolayer of cells was formed (verified by microscopy), which insured cell-to-cell contact. Filters were presoaked for 10 min in 10% (vol/vol) Suprapur HCl (Merck, Darmstadt, Germany) and washed three times in 0.9% NaCl (solid purity, 99.5%; Merck) in UV-treated MilliQ-water. The filters with the bacteria were floated on saline in acid-rinsed petri dishes and incubated in the dark at 26°C. In the other experiment, starved donors and recipients were filtered into the polycarbonate membranes to a density similar to that on the leaves, i.e., ca. 105 CFU/cm2. The filters were placed on agarose plates. In both experiments, cell numbers and metabolic activity were determined at regular intervals as described above for the leaves. The concentration of dissolved organic carbon in the saline (<0.25 ppm) was measured on a Shimadzu TOC-5000 analyzer.
Verification of transconjugants and identification of indigenous epiphytic bacteria.
Putative transconjugants were either tested for green fluorescence, to show the presence of the TOL plasmid, or tested for their ability to act as donors of RP4 to E. coli MC1061 (Table 1).
Natural epiphytic isolates possessing different cell and/or colony morphology were gram-identified by the KOH method (39). Subsequently, gram-negative isolates were characterized by the API 20E and API 20NE test kits (Biomerieux SA, Marcy l’Etoile, France).
In situ detection of bacteria on leaves.
Epiphytic indigenous bacteria were stained with 0.2 μm-pore-size-filtered (Nalgene sterilization filter; Nalge Company, Rochester, N.Y.) phenolic aniline blue (PAB) according to Jones et al. (21) and Hossell and Baker (19). Basically, a leaf was submerged in PAB for 1 to 2 min. A square of approximately 5 by 5 mm was excised and placed on a microscope slide mounted in a drop of PAB. A Zeiss Axioplan microscope fitted with a 12-V tungsten lamp was used for transmitted illumination. Digital images were recorded with a 12-bit cooled slow-scan charge-coupled device camera (KAF 1400 chip; Photometrics Ltd., Tucson, Ariz.).
The spatial distribution of transconjugant cells was determined by examining a 5- by 5-mm leaf square with a Zeiss Axioplan microscope equipped with an HBO-100 mercury lamp and Zeiss filter set 10 (BP 470- to 490-nm exitation filter, 510-nm dichroic mirror, and BP 515- to 565-nm emission filter). Plan-Neofluar 40× and 63× oil immersion lenses and 20×, 40×, and 100× dry lenses were used.
Three-dimensional images were obtained by a Leica Lasertechnik TCS 4D confocal scanning laser microscope equipped with a 15-mW argon-krypton ion laser (excitation wavelength, 488 nm). To discriminate between the green fluorescence emitted by the cells and the red fluorescence emitted by the leaf, BP-510 and LP-515 emission filters (Leica) were used. Series of monochrome 2-D sections along the optical axis were recorded and combined to create a 3-D image by use of the simulated fluorescent projection technique provided by the Scanware 1.02 software (Leica). Stereo-pairs of 3-D images of the green fluorescent cells and the leaf surface were colored and combined within a red-green-blue display by using Adobe Photoshop for Windows 95 (Adobe Systems Inc., San Jose, Calif.).
Data analysis.
Plasmid transfer was calculated as T/R and T/D (number of transconjugants per number of donors) ratios and as the time- and density-independent transfer coefficient, kt1 (44). kt1 was calculated for two data points as (ΔT/Δt)/(D × R) under the assumption that D≫T, R≫T, and D and R were constant (35).
One-way analyses of variance on log10-transformed data and linear regression analysis were performed by using the SigmaStat software for Windows (Jandel Corp., Erkrath, Germany).
RESULTS
Effect of RH on survival and conjugal transfer.
Survival of the bacteria on the leaves depended on RH. At 100% RH, numbers of CFU of P. putida KT2442/TOL almost doubled during the 96-h incubation (Fig. 1A), whereas at 90% RH, numbers declined by a factor of 200 within the first 24 h (Fig. 1B). At lower humidities (80, 55, and 25%), population densities of the donor were reduced further (Fig. 2). The P. putida KT2440 recipient did not survive as well as the donor (P < 0.003) as numbers were reduced by a factor of 8 during the incubation at 100% RH (Fig. 1A). At 90% RH and lower humidities, survival rates of donors and recipients were comparable (P > 0.17) (Fig. 1B and 2).
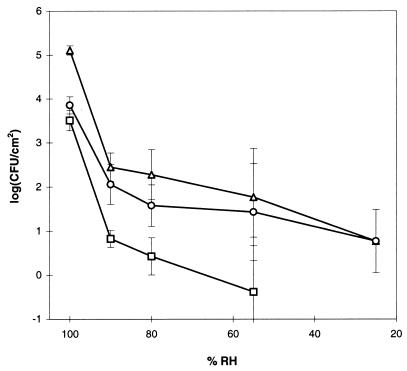
FIG. 2.
Density of donors (▵), recipients (○) and transconjugants (□) after 96 h of incubation as function of RH. No transconjugants were observed after 96 h of incubation at 25% RH. Error bars are ± SD of triplicate samples.
RH also affected the appearance of transconjugants. At 100% RH, numbers of transconjugants reached a level of 3 × 103 CFU/cm2 within 24 h, corresponding to about one-third of the recipient population (Fig. 1A). Relative to 100% RH, transconjugant densities were approximately 500, 1,200 and 8,000 times lower at 90, 80, and 55% RH, respectively (Fig. 2). At 25% RH, no transconjugants were detected.
Incubation at low RH did not result in an irreversible decline in the population size of the bacteria. In the experiment performed at 55% RH, some plants were transferred to 100% RH after 96 h. After an additional 72 h at this humidity, densities of donors, recipients, and transconjugants increased 70, 10, and 100 times, respectively (data not shown).
Maximal transfer ratios, calculated as T/D, ranged from 0.007 to 0.026 whereas ratios calculated as T/R ranged from 0.01 to 0.343 (Table 2). Ratios did not appear to be directly related to RH. However, by exerting an effect on cell survival, RH did have an effect on transfer, especially on T/R which was fivefold lower at 90% than at 100% RH.
TABLE 2.
Metabolic activity and maximal transfer ratios
| Experimenta | Metabolic activityb (fmol of Leu × 103/CFU/h) | Maximal transfer ratios
|
|
|---|---|---|---|
| T/D | T/R | ||
| 100% RH | 15.8 → 2.60 | 0.019 | 0.343 |
| 90% RH | 8.66 → BDc | 0.026 | 0.064 |
| 80% RH | 10.4 → BD | 0.017 | 0.088 |
| 55% RH | NDd | 0.007 | 0.010 |
| 25% RH | 8.18 → BD | BD | BD |
| Low inoculum density (100% RH; 103–104 CFU/cm2) | ND | 0.004 | 0.009 |
| Nonsterile leaves (100% RH) | ND | 0.045 | 0.015 |
| Filter on agarose (100% RH) | ND | 0.007 | 0.010 |
| Filter on saline (100% RH; 107 CFU/cm2) | 0.87 → 0.24 | BD | BD |
Inoculum density was 105 CFU/cm2 unless otherwise specified.
Maximal (0 h) and minimal (96 h) activities.
BD, below detection.
ND, not determined.
In situ distribution of cells on leaves.
Green fluorescent transconjugants were observable 5 to 6 h after inoculation of the plants (100% RH). Thus, detection by microscopy was delayed about 4 h relative to detection by plating, due to an approximately 4-h processing time of the fluorophore (17). After 24 h of incubation, numerous green fluorescent cells were found. Highest numbers were observed in the epidermal interstices (Fig. 3A and B), but transconjugants were also seen in 5 to 10% of the ca. 300 stomata investigated on five leaves. From 1 up to more than 100 cells per stoma were observed (Fig. 3C). Occasionally, transconjugants were observed at the base of trichomes. Leaves inoculated directly with transconjugants showed an identical distribution. The distribution of the cells did not depend upon the inoculation procedure, i.e., transconjugants were distributed as described above, when sterile seeds were inoculated with either transconjugants or a 1:1 mixture of donors and recipients.
FIG. 3.
Confocal scanning laser microscopy photographs showing green fluorescent transconjugant cells in the interstices of epidermal cells (A), in the interstices of vein cells (B), and inside a stoma (C). Photographs (D) and (E) show charge-coupled device-recorded images of indigenous bacteria in the interstices of epidermal cells and in the interstices of vein cells, respectively. Bars represent 10 μm.
The fluorescent signal was reduced if cells on microscope slides were exposed to desiccation. Similarly, the signal disappeared after a few hours if the plants were incubated at 60% RH. The signal persisted the longest time in the substomatal cavities followed by the interstitial spaces. Hence, relative to the surface of the epidermal cells, these habitats most likely protected the bacteria against desiccation.
Examination of unsterile leaves grown at 100% RH revealed a distribution of the indigenous bacteria similar to that of the transconjugants (Fig. 3D and E). Microcolonies consisting of 100 to 1,000 cells were often found associated with the epidermal interstices. Cells were observed in about 10% of the stomata. Plants incubated at 60% RH showed a similar distribution of the bacteria; however, fewer cells were observed. Densities of CFU were 6.1 ± 5.0 × 102 CFU/cm2 and 1.8 ± 1.1 × 105 CFU/cm2 (± standard deviation [SD]) (n = 6) at 60 and 100% RH, respectively.
In situ metabolic activity.
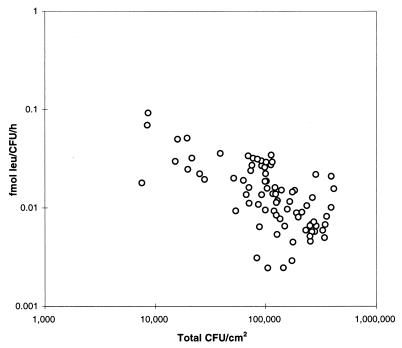
Immediately after inoculation onto the leaves, the per-cell metabolic activity increased four to eight times relative to the activity of the cells when in the inoculation buffer. For instance at 100% RH, the activity increased significantly (P < 0.0005), from 0.2 × 10−2 to 1.6 × 10−2 fmol of Leu/CFU/h (Fig. 4). Through the incubation, the activity decreased and approached the level of the inocula after 96 h. At lower RHs, metabolic activities on the leaves decreased to the level of the inocula after 4 h, following which activity could no longer be detected (Table 2). The metabolic activity was inversely correlated with cell density (r2 = 0.218; P < 0.0001) (Fig. 5).
FIG. 4.
Metabolic activity of the donor prior to inoculation (▵), metabolic activity of the recipient prior to inoculation (○), and the mean activity of donors and recipients after inoculation onto leaves at 100% RH (□). Error bars are ± SD of triplicate samples.
FIG. 5.
Relationship between bacterial density and metabolic activity at 100% RH. Data points represent single leaves.
Effect of metabolic activity and density on conjugal transfer.
Metabolic activity and conjugal transfer on the leaves were not correlated (r2 = 0.267; P > 0.05) (Fig. 6). Calculated kt1 values ranged between 6.4 × 10−11 and 1.4 × 10−7 cm2/CFU/h and metabolic activities ranged between 0.0034 and 0.030 fmol of Leu/CFU/h (Fig. 6).
FIG. 6.
kt1 as a function of metabolic activity (100% RH). Open symbols (○) represent individual leaves, while filled symbols (•) represent individual filters floating on saline (see text for details).
Per-cell metabolic activities on filters placed on saline were about six times lower than activities measured on the leaves, ranging between 2.6 × 10−4 to 5.5 × 10−4 fmol of Leu/CFU/h (Fig. 6). Although cell densities on the filters were 100 times higher than on the leaves, no transconjugants were observed by plating or microscopical examination for fluorescent cells.
Transfer ratios were not correlated to cell density (P > 0.11) on leaves with densities around 106 CFU/cm2 (not shown). However, maximal numbers of transconjugants and maximal transfer ratios were about 100 and 35 times lower, respectively, at densities between 103 and 104 CFU/cm2 (Table 2).
Transfer to indigenous epiphytic bacteria.
The highest numbers of indigenous bacteria that had received the RP4 plasmid were attained after 6 h of incubation, after which the population size remained stable at 1.5 × 103 CFU/cm2 (Fig. 7). Under the conditions employed here, more than 95% of the culturable indigenous bacteria were prototrophic and thus were potential recipients of RP4. The T/R ratio, however, was 23 times lower than the maximal ratio for the biparental mating with the TOL plasmid (Table 2). RP4 was transferred to six different indigenous Pseudomonas spp., to Stenotrophomonas maltophilia, and to four unidentified gram-negative isolates.
FIG. 7.
Survival of P. fluorescens AS12/RP4 (▵), indigenous recipients (○), and appearance of transconjugants (□) on bean leaves at 100% RH. Error bars are ± SD of triplicate samples.
DISCUSSION
This is the first study on effects of bacterial distribution and metabolic activity on conjugal gene transfer on the phylloplane. The experiments demonstrated that the phylloplane of bean is a habitat conducive to conjugal transfer. Transfer primarily took place in the interstitial spaces and stomata (Fig. 3), and numbers of transconjugants were positively related to RH and inoculum concentration (Fig. 2 and Table 2). The metabolic activity of the bacteria inoculated onto the leaf surface was stimulated, possibly due to leaf exudates (Fig. 4). No correlation, however, between conjugal transfer on the leaves and metabolic activity was observed (Fig. 6).
The observed T/Rs of up to 0.34 in the biparental mating experiments (Table 2) are similar to results of earlier studies of the phylloplane (5, 30). A literature comparison of transfer to indigenous bacteria is not feasible, however, as only one study has been published and no transfer ratios were reported (5). Relative to the biparental mating experiment at 100% RH, maximal transfer ratios to indigenous bacteria were 23 times lower (Table 2). Although RP4 is transmissible to a wide range of gram-negative and a few gram-positive bacteria (26), transfer to all epiphytic bacteria would not be expected. Furthermore, only 95% of the indigenous bacteria were prototrophic and would be scored as transconjugant on the selective media. Compared to results of the rhizosphere, however, the maximal T/R (0.02) was high. For instance, Smit et al. (46) and Richaume et al. (41) reported T/R values of RP4 in the range of 10−6 to 10−4 between added pseudomonad donors and indigenous soil or wheat rhizosphere bacteria.
In order to estimate the in situ activity of the donors and recipients it was necessary to use sterilized plants. Although this gnotobiotic model system does not completely reproduce the complexity of the natural situation, it allowed us to specifically address the importance of metabolic activity by eliminating the large numbers of uncontrolled parameters of a more complex system.
Possibly as the result of growing the plants aseptically, the metabolic activity of the bacteria increased upon inoculation onto the leaves (Fig. 4). Most likely, accumulated exudates initially stimulated the bacterial activity. During incubation, however, the surplus exudates were used up and the bacterial activity approached the level of the starved inocula (Fig. 4). A negative correlation between density and metabolic activity was observed at 100% RH (Fig. 5). The relatively low numbers of CFU at RHs below 100% should, according to Fig. 5, result in an elevated activity of the surviving cells. This, however, was not the case (Table 2). Possibly, the Leu uptake was impeded by the lower water potential.
The physiological state of the bacteria has been suggested to be important for conjugal transfer due to the energy required for synthesis of a pilus and replication of the plasmid DNA (35, 42). For instance, the kt1 for transfer of RP4 from E. coli to Rhodobacter capsulatus in batch cultures was found to be proportional to substrate concentration (35). van Elsas et al. (51) suggested that root exudates in the rhizosphere of wheat stimulated conjugal transfer, and Björklöf et al. (5) proposed that availability of nutrients could be responsible for the high transfer ratios on the phylloplane.
No relationship between metabolic activity and transfer on the phylloplane was found in the present study (Fig. 6); i.e., metabolic activity was not rate limiting. However, a minimum level of activity appeared to be necessary for transfer to occur. This was demonstrated by the filter mating experiment in which the bacteria were kept at low activity on saline (<0.9 × 10−3 fmol of Leu/CFU/h). In this case, no transfer was observed. Since a monolayer of cells was present on the filters, the required cell-to-cell contact was achieved. Our data suggest that the threshold level of metabolic activity must have been somewhere between 0.9 and 3 × 10−3 fmol of Leu/CFU/h (Fig. 6).
The hypothesis that metabolic activity is not limiting for conjugation in planta is supported by recent evidence by Kroer et al. (28), who reported a lack of causal relationship between transfer and Leu uptake in the rhizosphere of water grass (Echinochlora crusgalli). In their study, measured metabolic activities were in the interval of 8 × 10−3 to 16 × 10−3 fmol of Leu/CFU/h and, hence, above the estimated threshold activity level observed for the phylloplane in this study.
It may be argued that accumulated exudates on sterile leaves supported bacterial activity at levels that were not limiting for transfer, whereas on nonsterile leaves, where exudates could not accumulate due to consumption by the resident microflora, a correlation between activity and transfer may have been observed. Transfer, however, occurred immediately on the nonsterile leaves (Fig. 7). Thus, an experimental bias was not introduced by using sterile plants.
Since metabolic activity did not appear to determine the rate of conjugal transfer, other factors must have been playing that role. Recently it has been hypothesized that leaf and rhizosphere habitats support conjugation by increasing the local density of the bacteria (5, 28). In the present study, cells were applied to the leaves at a density of approximately 105 CFU/cm2. But the clustering of the bacteria into interstices and stomata resulted in densities that locally were much higher. A comparison of transfer ratios between the phylloplane and the filters placed on agarose shows that transfer ratios on the phylloplane (100% RH) were more than 30 times higher (Table 2). Since the inoculated densities were the same in both cases, the spatial aggregation of the bacteria in microhabitats on the phylloplane probably was responsible for the high transfer ratios.
The clustering of bacteria in the interstices and stomata could have been an experimental artifact of the inoculation procedure. This however, was not the case because a similar distribution was observed when seed inoculation was applied. Furthermore, the location of indigenous bacteria was similar to that of inoculated bacteria. Also, scanning electron microscopy studies of the phylloplane of potato (7) and the rhizosphere of tomato (9) showed that bacteria were clustered in interstitial spaces. Due to the hydrophobicity of some parts of the leaf and condensation of water (33), the bacteria probably passively end up and proliferate in the most hydrophillic environments, such as the interstices and stomata.
Wilson and Lindow (54) argued that cells, upon reduction in RH, survive in protected habitats, whereas they die in unprotected habitats. Thus, cells in protected habitats are less subject to changes in humidity. Our data support this hypothesis, as the bacteria persisted the longest time in the substomatal cavities and epidermal interstices. Persistence of the bacteria in the microhabitats may explain why transfer occurred at RHs lower than 100%, despite the fact that the bacteria were highly sensitive to desiccation.
It is generally observed that transconjugants primarily appear during the first day of an experiment (10, 25, 28, 35, 49). In the present study, transconjugants appeared within the first 10 h after which their population size stabilized (Fig. 1). Since metabolic activity only appears to be limiting for transfer under extreme conditions, transfer probably takes place whenever a donor and a recipient are in contact. Thus, while a plasmid may quickly be spread among all recipients within a microhabitat, further transfer is less likely because of the physical separation of the microhabitats. Consequently, the distribution of the cells (on, for instance, the phylloplane), may initially stimulate transfer but at a later stage be the limiting factor.
Conclusion.
Aggregation of the bacteria into microhabitats on the leaf surface greatly stimulated survival and transfer. Metabolic activity, on the other hand, was not rate limiting for conjugal transfer in this habitat. Most likely, very little energy is required for completion of transfer, and the leaf exudates insured that the activity of the bacteria was well above the threshold value.
ACKNOWLEDGMENTS
This research was partly financed by grants from the Danish Environmental Protection Agency and The Plasmid Foundation.
We thank Tamar Barkay for critically reviewing the manuscript.
REFERENCES
- 1.Bagdasarian M B, Lurz B, Ruckert F C, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 2.Bale M J, Fry J C, Day M J. Plasmid transfer between strains of Pseudomonas aeruginosa on membrane filters attached to river stones. J Gen Microbiol. 1987;133:3099–3107. doi: 10.1099/00221287-133-11-3099. [DOI] [PubMed] [Google Scholar]
- 3.Barkay T, Kroer N, Rasmussen L D, Sørensen S. Conjugal transfer at natural population densities in a microcosm simulating an estuarine environment. FEMS Microbiol Ecol. 1995;16:43–54. [Google Scholar]
- 4.Barkay T, Liebert C, Gillman M. Conjugal gene transfer to aquatic bacteria detected by the generation of a new phenotype. Appl Environ Microbiol. 1993;59:807–814. doi: 10.1128/aem.59.3.807-814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björklöf K, Suoniemi A, Haahtela K, Romantschuk M. High frequency of conjugation versus plasmid segregation of RP1 in epiphytic Pseudomonas syringae populations. Microbiology. 1995;141:2719–2727. doi: 10.1099/13500872-141-10-2719. [DOI] [PubMed] [Google Scholar]
- 6.Black W A, Girdwood R W A. Carbenicillin resistance in Pseudomonas aeruginosa. Br Med J. 1969;4:234. doi: 10.1136/bmj.4.5677.234-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blakeman J P. Ecological succession of leaf surface microorganisms in relation to biological control. In: Windels C E, Lindow S E, editors. Biological control on the phylloplane. St. Paul, Minn: The American Phytopathological Society; 1985. pp. 6–30. [Google Scholar]
- 8.Burrage S W. The micro-climate at the leaf surface. In: Preece T F, Dickinson C H, editors. Ecology of leaf surface micro-organisms. London, United Kingdom: Academic Press; 1971. pp. 89–101. [Google Scholar]
- 9.Chin-A-Woeng T F C, de Priester W, van der Bij A J, Lugtenberg B J J. Description of the colonization of a gnotobiotic tomato rhizosphere by Pseudomonas fluorescens biocontrol strain WCS365, using scanning electron microscopy. Mol Plant-Microbe Interact. 1997;10:79–86. [Google Scholar]
- 10.Christensen B B, Sternberg C, Molin S. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (Gfp) from Aequorea victoria as marker. Gene. 1996;173:59–65. doi: 10.1016/0378-1119(95)00707-5. [DOI] [PubMed] [Google Scholar]
- 11.Clark J D, Maaloe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;21:99–112. [Google Scholar]
- 12.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived mini-transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Astorga A, Muela A, Cisterna R, Iriberri J, Barcina I. Biotic and abiotic factors affecting plasmid transfer in Escherichia coli strains. Appl Environ Microbiol. 1992;58:392–398. doi: 10.1128/aem.58.1.392-398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hareland W A, Crawford R L, Chapman P J, Daley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975;121:272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heim R, Prasher D C, Tsien R Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoagland D R, Arnon D I. The water culture method of growing plants without soil. California agricultural experimental station circular 347. Berkeley, Calif. 1938. [Google Scholar]
- 19.Hossel J C, Baker J H. A note on the enumeration of epiphytic bacteria by microscopic methods with particular reference to two freshwater plants. J Appl Bacteriol. 1979;46:87–92. [Google Scholar]
- 20.Ippen-Ihler K. Bacterial conjugation. In: Levy S B, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill Publishing Company; 1989. pp. 33–72. [Google Scholar]
- 21.Jones P C T, Mollison J E, Quenouille M H. A technique for the quantitative estimation of soil micro-organisms. J Gen Microbiol. 1948;2:54–69. doi: 10.1099/00221287-6-3-4-261. [DOI] [PubMed] [Google Scholar]
- 22.Juniper B E. The leaf from the inside and the outside: a microbe’s perspective. In: Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 21–42. [Google Scholar]
- 23.Kinkel L L, Wilson M, Lindow S E. Utility of microcosm studies for predicting phylloplane bacterium population sizes in the field. Appl Environ Microbiol. 1996;62:3413–3423. doi: 10.1128/aem.62.9.3413-3423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchman D L, K’Nees E, Hodson R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knudsen G R, Walter M V, Porteous L A, Prince V J, Armstrong J L, Seidler R J. Predictive model of conjugative plasmid transfer in the rhizosphere and phyllosphere. Appl Environ Microbiol. 1988;54:343–347. doi: 10.1128/aem.54.2.343-347.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnapillai V. Molecular genetic analysis of bacterial plasmid promiscuity. FEMS Microbiol Rev. 1988;54:223–238. doi: 10.1111/j.1574-6968.1988.tb02744.x. [DOI] [PubMed] [Google Scholar]
- 27.Kristensen C S, Eberl L, Sanchez-Romero J M, Molin S, Givskov M, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroer N, Barkay T, Sørensen S, Weber D. Effect of root exudates and bacterial metabolic activity on conjugal gene transfer in the rhizosphere of a marsh plant. FEMS Microbiol Ecol. 1998;25:375–384. [Google Scholar]
- 29.Kroer N, Normander B, Molin S. Factors affecting conjugal gene transfer on the phylloplane of bean. Poster presented at the Fallen Leaf Lake Conference on “Horizontal Gene Transfer: Implications & Consequences”, South Lake Tahoe, Calif. 1996. [Google Scholar]
- 30.Lacy G H, Leary J V. Transfer of antibiotic resistance plasmid RP1 into Pseudomonas glycinea and Pseudomonas phaseolicola in vitro and in planta. J Gen Microbiol. 1975;88:49–57. doi: 10.1099/00221287-88-1-49. [DOI] [PubMed] [Google Scholar]
- 31.Lilley A K, Bailey M J. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol. 1997;63:1577–1583. doi: 10.1128/aem.63.4.1577-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilley A K, Fry J C, Day M J, Bailey M J. In situ transfer of an exogenously isolated plasmid between Pseudomonas spp. in sugar beet rhizosphere. Microbiology. 1994;140:27–33. [Google Scholar]
- 33.Lindow S E. Determinants of epiphytic fitness in bacteria. In: Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 295–314. [Google Scholar]
- 34.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacDonald J A, Smets B F, Rittmann B E. The effects of energy availability on the conjugative-transfer kinetics of plasmid RP4. Water Res. 1992;26:461–468. [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 37.Morgan J V, Tukey H B., Jr Characterization of leachate from plant foliage. Plant Physiol. 1964;39:590–593. doi: 10.1104/pp.39.4.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muela A, Pocino M, Arana I, Justo J I, Iriberri J, Barcina I. Effect of growth phase and parental cell survival in river water on plasmid transfer between Escherichia coli strains. Appl Environ Microbiol. 1994;60:4273–4278. doi: 10.1128/aem.60.12.4273-4278.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Leary W M. Practical handbook of microbiology. Boca Raton, Fla: CRC Press Inc.; 1989. [Google Scholar]
- 40.Ramos-Gonzalez M-I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richaume A, Smit E, Faurie G, van Elsas J D. Influence of soil type on the transfer of plasmid-RP4(p) from Pseudomonas fluorescens to introduced recipient and to indigenous bacteria. FEMS Microbiol Ecol. 1992;101:281–292. [Google Scholar]
- 42.Rittmann B E, Smets B F, Stahl D A. The role of genes in biological processes. Part 1 of a two-part article. Environ Sci Technol. 1990;24:23–29. [Google Scholar]
- 43.Sandaa R, Enger Ø. Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl Environ Microbiol. 1994;60:3430–3437. doi: 10.1128/aem.60.12.4234-4238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonsen L, Gordon D M, Stewart F M, Levin B R. Estimating the rate of plasmid transfer: an end-point method. J Gen Microbiol. 1990;136:2319–2325. doi: 10.1099/00221287-136-11-2319. [DOI] [PubMed] [Google Scholar]
- 45.Sleesman J P, Leben C. Bacterial desiccation: effects of temperature, relative humidity, and culture age on survival. Ecol Epidemiol. 1976;66:1334–1338. [Google Scholar]
- 46.Smit E, van Elsas J D, van Veen J A, de Vos W M. Detection of plasmid transfer from Pseudomonas fluorescens to indigenous bacteria in soil using bacteriophage φR2f for donor counterselection. Appl Environ Microbiol. 1991;57:3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sørensen S J. Transfer of plasmid RP4 from Escherichia coli K-12 to indigenous bacteria of seawater. Microb Releases. 1993;2:135–141. [PubMed] [Google Scholar]
- 48.Stark M J R. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression. Gene. 1987;51:255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 49.Sudarshana P, Knudsen G R. Effect of parental growth on dynamics of conjugative plasmid transfer in the pea spermosphere. Appl Environ Microbiol. 1995;61:3136–3141. doi: 10.1128/aem.61.8.3136-3141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Elsas J D, Trevors J T, Starodub M E. Bacterial conjugation between pseudomonads in the rhizosphere of wheat. FEMS Microbiol Ecol. 1988;53:299–306. [Google Scholar]
- 52.Weinberg S R, Stotzky G. Conjugation and genetic recombination of Escherichia coli in soil. Soil Biol Biochem. 1972;4:171–180. [Google Scholar]
- 53.Williams P A, Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence of a TOL plasmid. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson M, Lindow S E. Inoculum density-dependent mortality and colonization of the phyllosphere by Pseudomonas syringae. Appl Environ Microbiol. 1994;60:2232–2237. doi: 10.1128/aem.60.7.2232-2237.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]