Abstract
The response of the bacterial strains Comamonas acidovorans PX54 (β subclass of the class Proteobacteria) and Vibrio strain CB5 (γ subclass of the class Proteobacteria) to grazing by the bacterivorous flagellate Ochromonas sp. was examined in one-stage chemostat experiments under conditions of low growth rates with a complex carbon source. The two bacterial strains were cultured together; they were cultured without flagellates in the first phase of the experiments and in the presence of the flagellates in the second phase. Monoclonal and polyclonal antibodies were used to determine the numbers and sizes of C. acidovorans PX54 and Vibrio strain CB5 cells. The flagellates caused strong changes in total bacterial cell numbers, in the relative abundances of the individual bacterial strains, and in bacterial cell size distribution. Vibrio strain CB5 dominated the total bacterial cell numbers during the flagellate-free phase of the experiments with a relative abundance of 93%, but this declined to 33% after inoculation with the flagellate. In contrast to Vibrio strain CB5, C. acidovorans PX54 responded to grazing with a strong expansion of cell length distribution toward large, filamentous cells. These changes in cell morphology resulted in a high percentage of inedible cells in the C. acidovorans PX54 population but not in the Vibrio strain CB5 population, which caused the observed change in the relative abundances of the strains. Batch culture experiments without the flagellate demonstrated that the elongation of C. acidovorans PX54 cells was dependent on their growth rate. This indicates that the occurrence of filamentous C. acidovorans PX54 cells is not a direct response to chemical stimuli released by the flagellates but rather a response to increased growth rates due to flagellate grazing.
Predator-prey interactions of coexisting, free-living aquatic bacteria and bacterivorous protozoa have coevolved for more than a billion years (28). This enormous time span and the short generation times of both groups of microorganisms should have resulted in a high degree of evolutionary adaptation on both sides. Bacteria may have developed defense strategies to prevent themselves from being ingested (preingestional strategies) or digested (postingestional strategies) by their protozoan predators, which, expectedly, adaptated to circumvent the bacterial defense mechanisms. Information about the strategies involved in these predator-prey interactions is scarce. Recently, Jürgens and Güde (20) reviewed the strategies of bacteria and stressed the lack of knowledge in this field.
Studies on size-selective ingestion (grazing) of bacterivorous protozoa (6, 10, 25) indicate that very small and large bacteria are partly or totally protected from protozoan grazing (12, 20). This finding is supported by field and experimental observations showing the occurrence and persistence of large bacterial filaments and aggregates during times of high grazing pressure (11, 21, 29, 41). The experimental evidence for protection and the increasing number of reports on the presence of filamentous bacteria in freshwater ecosystems (12, 13, 19, 35, 39, 41) indicate that this bacterial morphotype exhibits an ecologically significant defense strategy against protozoan grazing. It is not known to which species these protected forms belong. Additionally, it is unclear if the filamentous bacteria grow permanently with these, with respect to grazing, advantageous morphological properties or if they express these characteristic features only under strong grazing pressure.
In a recent study, Pernthaler et al. (30) demonstrated that a slow-growing bacterial community reacted to the addition of bacterivorous flagellates within 1 day: one group produced filamentous, grazing-resistant forms, and another group of bacteria reacted with a massive growth rate increase. Similarly, Jürgens et al. (21) observed in enclosure studies, after experimentally increasing the protozoan grazing pressure, that there was a rapid and strong change in the morphological structure of the bacterial community. After 3 days, mainly filamentous and other inedible bacterial cells dominated the bacterial biomass, with a prevalence of 80 to 90%.
Different mechanisms are conceivable for such changes in the morphological structure of bacterial communities. First, nonfilamentous, edible strains may simply be replaced after some time by inedible, permanently filamentous strains. In situations with bacterial generation times longer than 1 day and undetectably low abundances of filamentous cells (30), such an indirect selection mechanism can hardly cause visible changes in community structure within 24 h. But the possibility cannot be ruled out that this mechanism is of relevance in natural ecosystems. Second, medium-size, edible cells may become elongated and thus form filaments. This type of response to strong protistan grazing might be controlled by two different mechanisms: (i) elongation of the cells due to grazing-mediated changes in bacterial growth conditions (indirect induction of filament formation) or (ii) direct induction of morphological changes by chemical stimuli. Such chemical stimuli might be produced and released by the protozoan predators (predator kairomone) or produced by the prey bacteria and set free by the predators during digestion. The second type of stimuli would act as an alarm substance. It is not known if selection or one of the induction mechanisms triggers the observed reactions of bacterial communities. Pernthaler et al. (30) speculated that a chemical stimulus caused the observed changes in their experiments, since they found an immediate response upon addition of a flagellate grazer.
Detailed information on the interactions of bacteria with protozoan grazers and the resulting bacterial defense strategies are necessary for a comprehensive understanding of a number of important issues in microbial ecology. This includes questions about the influence of protozoa on (i) the bacterial species composition of natural communities, (ii) the regulation of bacterial production and mineralization in aquatic systems, and (iii) the survival and behavior of allochthonous bacteria such as pathogenic members of the family Enterobacteriaceae or genetically engineered microorganisms in the environment.
In this study, we used a model system to investigate the interactions of two bacterial strains with the bacterivorous nanoflagellate Ochromonas sp. The bacterium Vibrio sp. strain CB5 originated from the pelagic zone of Lake Constance (southern Germany) and was isolated from a chemostat inoculated with a water sample from that lake (14). The other strain, Comamonas acidovorans PX54, represents a member of a phylogenetic group which is abundant in Lake Plußsee (located near Plön, northern Germany) and in other lakes in the same area (9).
In this study, we investigated mechanisms that control the observed changes in the composition of the model community and investigated possible defense strategies of pelagic bacteria against protozoan grazing.
MATERIALS AND METHODS
Microbial strains, culture conditions, and media.
Two bacterial strains were used as a defined, simple bacterial community. C. acidovorans PX54 (β subclass of the class Proteobacteria) was isolated from Lake Plußsee, Germany (9). Vibrio strain CB5 (γ subclass of the class Proteobacteria) was obtained from a chemostat which was inoculated with a mixed bacterial culture from Lake Constance, Germany (14). Both strains were identified by low-molecular-weight RNA profiling (9, 16, 17) and sequencing of the 16S rRNA gene (26).
Bacterial strains were stored at −70°C and cultured on nutrient broth-soyotone-yeast extract medium (NSY medium) consisting of a mineral medium (modified Chu medium; 14, 27) plus equal amounts of nutrient broth, soyotone, and yeast extract (all from Difco) as a complex substrate. Different substrate concentrations were used for the chemostat and the other media. Liquid NSY medium for growth curve determination in batch culture and solid NSY medium contained a total of 9 g of the complex substrate per liter (3 g of each source per liter); the chemostat NSY medium and liquid medium used as an inoculum contained 9 mg of the same complex substrate per liter (3 mg of each source per liter). The solid medium contained 15 g of agar per liter.
A facultatively mixotrophic Ochromonas sp. strain isolated from Lake Constance by D. Springmann served as a model organism for bacterivorous flagellates. The flagellate (4 to 6 μm in diameter) is able to grow entirely heterotrophically (bacterivorously) in the dark. In the light, the flagellate is unable to maintain growth based on photosynthesis alone. During mixotrophic growth under optimal light intensities, the heterotrophic production of the Ochromonas sp. dominates and less than 10% of its biomass production results from primary production (15). Under the experimental conditions used in this study, the flagellate behaves like a typical heterotrophic interception feeder. Flagellates were grown in the mineral medium enriched with an autoclaved wheat grain (nonaxenic cultures with living bacteria present) or heat-killed bacteria (axenic cultures without living bacteria) as a food source.
Axenic Ochromonas sp. cultures.
Axenic flagellate cultures (free of living bacteria) were produced by adding antibiotics (tetracycline, streptomycin, and chloramphenicol, each at 40 mg/liter plus 5 mg of nutrient broth per liter) to a culture with a bacterium-flagellate cell ratio of 3:1. After 12 h, the culture was diluted to a concentration of 0.8 flagellate ml−1. Samples (100 μl) of this dilution were then pipetted into 24-well cell culture plates containing mineral medium and about 107 heat-killed bacteria ml−1 (70°C, 2 h). After 1 week, a sample from each well (total, 96) was inspected for flagellate growth by phase-contrast microscopy. The presence of bacterial contamination was confirmed by plating subsamples on NSY agar, by culturing subsamples in liquid NSY medium, and by inspection of 4′,6-diamidino-2-phenylindole (DAPI)-stained subsamples by epifluorescence microscopy (see below). With this screening procedure, six cultures of Ochromonas sp. clones were found to contain no living bacteria. These Ochromonas sp. cultures were kept in six-well cell culture plates with heat-killed bacteria in the light. The axenic cultures were tested routinely and just prior to use for bacterial contamination with the procedures described above.
Chemostat cultures.
Chemostat experiments were run in a one-stage chemostat with a 2-liter reactor at a dilution rate of 0.5 day−1 (doubling times, 33.3 h) at 15°C in the dark. Chemostat cultures were mixed by aeration with sterile air. NSY chemostat medium was sterilized by filtering through a sandwich filter consisting of a glass fiber prefilter and a 0.2-μm-pore-size cellulose-acetate filter (both from Sartorius, Göttingen, Germany). The absence of bacterial contaminants during the chemostat experiments was verified by epifluorescence microscopy for DAPI-stained cells not labelled with one of the antibodies (see below) and plating on NSY agar.
Three chemostat experiments with the two bacterial strains were run (Table 1). Each experiment consisted of two phases. In the first phase, the bacteria were cultured alone or together; the second phase started with inoculation of the flagellate (designated the Flag1 and Flag2 experiments, respectively). In the third experiment, no flagellate was introduced (Table 1); instead, the dilution rate was increased from 0.5 to 2.0 day−1 (designated the Dilut experiment). At the beginning of the Flag1 and Dilut experiments, the chemostat was inoculated with both bacterial strains simultaneously. In the Flag2 experiment, the chemostat was inoculated initially with C. acidovorans PX54 and 15 days later with Vibrio strain CB5. Samples (100 ml) were taken at 24- to 48-h intervals. Additional samples (10 ml) were taken in the transient stages after the inoculations with the flagellate. In the Flag1 and Flag2 chemostat experiments, different Ochromonas sp. clones were used because the axenic stock culture used for inoculation in the Flag1 experiment was contaminated with bacteria before the Flag2 experiment began.
TABLE 1.
Overview of the three chemostat experiments
| Expt | Day of inoculation with:
|
Day of:
|
|||
|---|---|---|---|---|---|
| C. acidovorans PX54 | Vibrio strain CB5 | Ochromonas sp. | Increase in dilution rate | End of expt | |
| Flag1 | 1 | 1 | 14 | 38 | |
| Flag2 | 1 | 15 | 34 | 62 | |
| Dilut | 1 | 1 | 10 | 17 | |
Microbial abundance, bacterial cell size, and biomass.
Ten-milliliter chemostat subsamples were taken and fixed with formaldehyde (2% final concentration) for determination of total bacterial abundance, flagellate abundance, the abundances of the two bacterial strains, and bacterial cell size. Samples were stored at 4°C until analysis. Fixed subsamples were stained with 0.1% (wt/vol) DAPI, filtered on 0.2-μm-pore-size black polycarbonate filters (Nuclepore), and enumerated by epifluorescence microscopy for determination of bacterial and flagellate abundances (31). At least 500 bacterial and 100 protistan cells were counted per sample. For determination of bacterial cell size and biomass (whole bacterial population), the lengths and widths of at least 1,000 DAPI-stained cells were measured by using digitized images. These images of bacterial cells were produced by an epifluorescence microscope (Zeiss Axiovert 135TV) equipped with a charge-coupled device camera (MIT Dage) at a magnification of ×1,000. Sizing was done with the Image I image analysis system (Brock & Michelsen, Birkerod, Denmark). The system was calibrated with fluorescently stained latex beads of known diameter (Polysciences Inc., Warrington, Pa.). For latex beads 0.5, 0.75, 1.0, and 2.0 μm in diameter, a linear relationship between the measured mean diameter (dm) and mean real diameter (dr) was found (dm = 1.50 · dr; r = 0.996; n = 4). This relationship was used to calculate bacterial cell sizes from measured data for all cells up to 2.0 μm in length. For the minority of cells longer than 2.0 μm, a constant length overestimation of 0.66 μm per cell was assumed. Bacterial cells longer than 10 μm were termed filamentous. Cell volume was calculated by using the formula of Andersson et al. (2). For calculation of bacterial biomass, a conversion factor of 220 fg of C μm−1 was used (40).
Abundance of C. acidovorans PX54 and Vibrio strain CB5 and specific cell size.
The two bacterial strains were distinguished by using specific antibodies and indirect immunofluorescence microscopy. Vibrio strain CB5 cells were recognized by a polyclonal rabbit antiserum, and C. acidovorans PX54 cells were recognized by a monoclonal antibody (9). Both strains showed a characteristic ring fluorescence and no cross-reactivity. Immunofluorescent staining of bacterial cells was done with the primary antibodies plus dichlorotriazinylaminofluorescein (DTAF)- or Texas red-labelled secondary antibodies (Dianova, Hamburg, Germany) in accordance with the protocol of Faude and Höfle (9). In addition, cells were stained with 0.1% (wt/vol) DAPI before filtration on 0.2-μm-pore-size polycarbonate filters. Due to the heterogeneity of the distribution of cells on the filter, relative abundances of the labelled cells were determined instead of absolute abundances. DAPI-stained cells (at least 500) and antibody-labelled cells were counted on the same areas. The resulting mean percentage of labelled cells was used to calculate the absolute abundance of Vibrio strain CB5 and C. acidovorans PX54 with the help of separate determination of the total bacterial cell number (see above).
For determination of strain-specific cell sizes, DTAF-antibody-labelled cells were sized as described above for DAPI-stained cells. To compensate for overestimation due to fluorescent labels at the surface of the bacterial cells, a correction factor was established. To obtain a wide range of cell lengths and widths, pure cultures of C. acidovorans PX54 and Vibrio strain CB5 were grown in batch cultures at 15 and 30°C and harvested at several stages. Specimens of each subsample stained with either DTAF-labelled antibodies or DAPI were sized. Linear regression analysis of the resulting mean cell lengths and widths gave the following relationship of measured length or width between DAPI- and DTAF-antibody-stained cells: dDAPI = (dDTAF − 0.1986)/1.0036 (n = 11; r = 0.997), where dDTAF is the measured size (length or width) of DTAF-antibody-labelled cells and dDAPI is the measured size of DAPI-stained cells of the same population. All specific size or biomass data were corrected for overestimation due to antibody labelling and overestimation due to DAPI staining. An exception is the size class distribution of the Vibrio strain CB5 and C. acidovorans PX54 populations shown in Fig. 5. The size data presented in that figure were only corrected for size overestimation caused by antibody labelling. This was done to avoid a strong distortion of bacterial size class distribution due to different handling of cells smaller and larger than 2.0 μm in correcting for DAPI-caused size overestimation. This resulted in a slight overestimation of measured cell lengths, but size class distribution was not distorted.
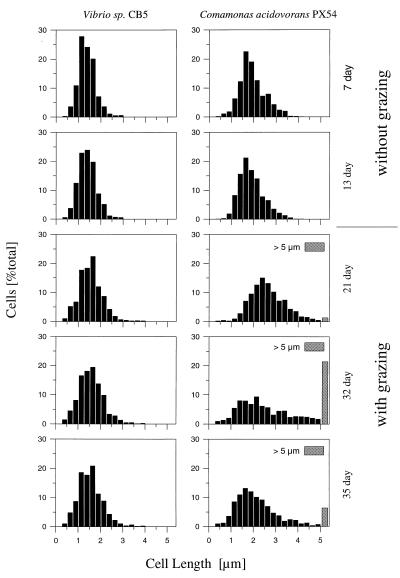
FIG. 5.
Size class distribution of Vibrio strain CB5 cells (left panels) and C. acidovorans PX54 cells (right panels) in corresponding chemostat samples from flagellate-free and flagellate-controlled phases (Flag1 experiment). Cells longer than 5 μm were pooled in one size class (>5 μm). To avoid distortion of size class distribution, measured cell lengths were corrected for size overestimation caused by cell staining with fluorescently labelled antibodies but not for size overestimation caused by DAPI staining (for details, see Materials and Methods).
Determination of specific CFU from chemostat samples.
Chemostat samples were diluted with sterile mineral medium in 10-fold steps up to dilutions yielding 30 to 100 colonies per agar plate, plated on solid NSY medium, incubated at room temperature for 4 days, and inspected by eye for the total number of colonies, the percentage of colonies from the two bacterial strains, and possible contamination. The colonies of the two strains were easily distinguishable by morphology.
Batch culture growth studies.
Batch cultures were used to study the dependence of bacterial cell size on growth stages. Triplicate 150-ml Erlenmeyer flasks were enriched with NSY medium (9 g of substrate per liter) and rotated at 150 rpm and 15°C. Optical density was measured at 578 nm, and bacterial cell size was measured as outlined above. Pure cultures of the bacterial strains were grown in the absence of the flagellate.
Simulation of the effect of nonselective grazing by increasing the chemostat dilution rate (Dilut experiment).
During steady-state growth in chemostat cultures, the bacterial growth rate (μ) is identical to the dilution rate of the chemostat (D). If bacterivorous flagellates are introduced, a nonselectively grazed bacterial population grows under steady-state conditions at the following rate (per hour):
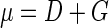 |
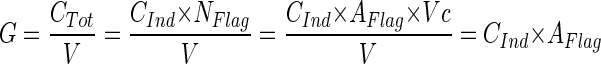 |
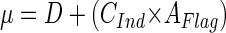 |
where G is the grazing rate of the flagellate population, CTot is the community clearance rate of the flagellate population, CInd is the mean individual flagellate clearance rate, NFlag is the total flagellate number in the chemostat, V is the volume of the chemostat reactor, and AFlag is the abundance of flagellates. To simulate the effect of grazing by a flagellate population with a given abundance and clearance rate in a bacterial community growing at dilution rate D, this rate has to be increased to Ds: D + (CInd × AFlag) = μ ≅ Ds (per hour). In the Dilut experiment, the dilution rate was increased from 0.5 to 2.0 day−1. This increase simulates the effect of nonselective grazing by a flagellate population of 8.8 × 103 ml−1 at a clearance rate of 7.1 nl flagellate−1 h−1 (15).
RESULTS
Abundance of C. acidovorans PX54 and Vibrio strain CB5 in flagellate-free and flagellate-controlled phases of the Flag1 and Flag2 experiments.
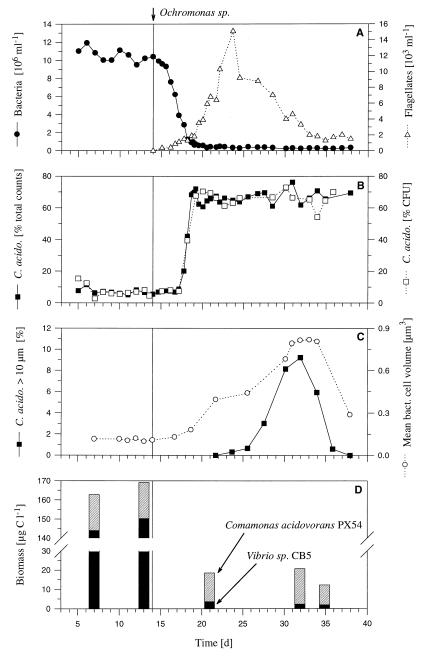
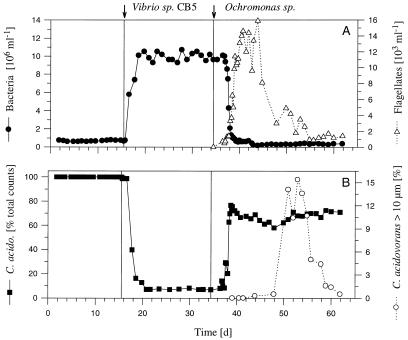
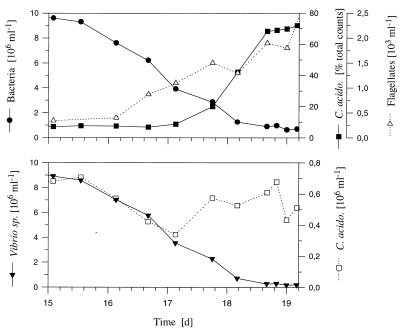
Average total bacterial numbers were (10.3 ± 0.5) × 106 ml−1 (Flag1 experiment) and (9.9 ± 0.4) × 106 ml−1 (Flag2 experiment, after inoculation with Vibrio strain CB5) in the flagellate-free phase (Table 2 and Fig. 1 and 2). After inoculation with Ochromonas sp., bacterial abundance dropped to mean steady-state values of (0.34 ± 0.07) × 106 ml−1 (Flag1 experiment) and (0.32 ± 0.06) × 106 ml−1 (Flag2 experiment). Kinetics of flagellate increase and bacterial decrease in the transient stage differed considerably between the two experiments, but steady-state bacterial abundances before and after inoculation with the flagellate showed only slight differences (Table 2). After inoculation, the numbers of flagellates increased exponentially over periods of 10 days (Flag1 experiment) and 6 days (Flag2 experiment), to maximum abundances of 15.1 × 103 ml−1 (Flag1 experiment) and 14.0 × 103 ml−1 (Flag2 experiment). Thereafter, abundances decreased slowly and reached steady-state levels of (1.7 ± 0.3) × 103 ml−1 (Flag1 experiment) and (1.2 ± 0.3) × 103 ml−1 (Flag2 experiment) toward the end of the experiments (Fig. 1 and 2). During the steady state of the flagellate-free phase of the Flag2 experiment, the C. acidovorans PX54 abundance showed no significant change (P > 0.1) after inoculation with Vibrio strain CB5 and only slight differences (P < 0.05) from the steady state of the flagellate-free phase of the Flag1 experiment (Table 3). During the flagellate-free phase, Vibrio strain CB5 dominated, with mean relative abundances of 93.5% (Flag1 experiment) and 92.7% (Flag2 experiment). During the flagellate-controlled steady state, C. acidovorans PX54 constituted 67.0% of the total bacterial abundance in the Flag1 and Flag2 experiments (Fig. 1, 2, and 3). In the Flag1 experiment, a grazing-induced decrease in total bacterial abundance started 1.5 days after inoculation with the flagellate while changes in the relative abundances of the two strains followed 1.2 days later (Fig. 4). In the Flag2 experiment, the total bacterial numbers decreased first and an increase in the relative abundance of C. acidovorans PX54 followed with a delay of 0.6 day. The shorter delay in the Flag2 experiment corresponded to the faster kinetics of changes during the transient stage of this experiment (Table 2).
TABLE 2.
Steady-state parameters of the flagellate-free and flagellate-controlled phases and parameters of the transient stages (after flagellate introduction until establishment of a new steady state) of the Flag1 and Flag2 chemostat experimenta
| Parameter | Flag1 expt | Flag2 exptb |
|---|---|---|
| Steady state | ||
| Total bacterial abundance (106 ml−1) | ||
| Without flagellates | 10.3 ± 0.5 | 9.9 ± 0.4 |
| With flagellates | 0.34 ± 0.07 | 0.32 ± 0.06 |
| Relative abundance of C. acidovorans PX54 (%) | ||
| Without flagellates | 6.5 ± 1.0 | 7.3 ± 0.4 |
| With flagellates | 67.0 ± 3.8 | 67.0 ± 1.1 |
| Dynamic state (from transient stage; day−1) | ||
| Rate of increase in flagellate no.b | 0.51 | 1.15 |
| Rate of bacterial cell no.c decrease | 0.65 | 1.14 |
Data on the flagellate-free phase of the Flag2 experiment represent the situation after inoculation with Vibrio strain CB5. Steady-state parameters represent mean values ± standard deviations, and transient-stage values represent rates of exponential changes following flagellate inoculation.
After inoculation with Vibrio strain CB5.
After inoculation with the flagellate.
FIG. 1.
Influence of grazing by the bacterivorous flagellate Ochromonas sp., inoculated on day 14 into C. acidovorans PX54 and Vibrio strain CB5 chemostat cultures (Flag1 experiment). (A) Total bacterial abundance and flagellate abundance. (B) Relative abundance of C. acidovorans PX54 determined either by immunofluorescence microscopy or by distinguishing colony types on agar plates. (C) Mean cell volume of the total bacterial community and percentage of C. acidovorans PX54 cells larger than 10 μm (filamentous cells). (D) Biomasses of C. acidovorans PX54 and Vibrio strain CB5 populations in the flagellate-free and flagellate-controlled phases. Inoculation of the bacterivorous flagellate Ochromonas sp. is indicated by vertical lines.
FIG. 2.
Influence of grazing by Ochromonas sp. on the model community consisting of C. acidovorans PX54 and Vibrio strain CB5 (Flag2 experiment). (A) Bacterial abundance before and after inoculation with Vibrio strain CB5 and after introduction of the predator Ochromonas sp. and abundance of this flagellate. (B) Relative abundance of C. acidovorans PX54 and percentage of C. acidovorans PX54 cells larger than 10 μm (filamentous cells).
TABLE 3.
Abundance of C. acidovorans PX54 and Vibrio strain CB5 in the steady state of the flagellate-free phases of the three chemostat experimentsa
| Expt | Period (days) | Mean abundance ± SD (106 ml−1)
|
|
|---|---|---|---|
| C. acidovorans PX54 | Vibrio strain CB5 | ||
| 1 | 4–14 | 0.67 ± 0.08 | 9.63 ± 0.55 |
| 2b | 2–15 | 0.68 ± 0.06 | |
| 2c | 20–34 | 0.72 ± 0.07 | 9.18 ± 0.51 |
| 3 | 6–10 | 1.00 ± 0.17 | 6.30 ± 0.13 |
In each experiment, the growth rate during the first phase was 0.5 day−1. In contrast to the other experiments, C. acidovorans PX54 grew initially in the Flag2 experiment without Vibrio strain CB5. This species was introduced at day 15 of the experiment, and 4 days later, a second steady state was established. Note that C. acidovorans PX54 abundance was not significantly (P > 0.1) influenced by the Vibrio strain CB5 population.
Steady state before inoculation with Vibrio strain CB5.
Steady state after inoculation with Vibrio strain CB5.
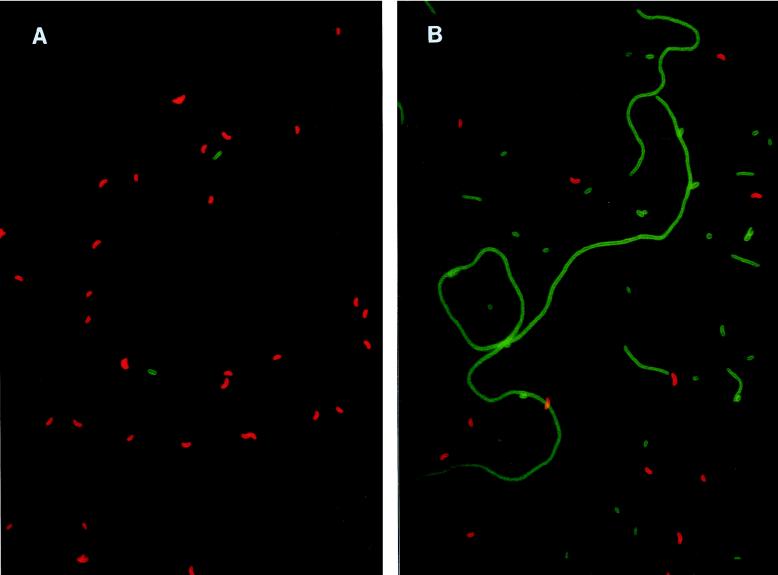
FIG. 3.
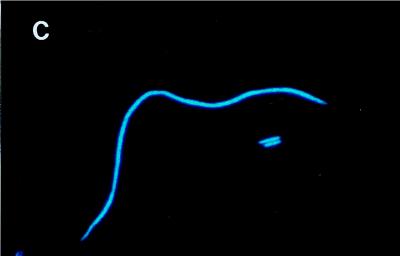
(A and B) Microphotographs of C. acidovorans PX54 (green) and Vibrio strain CB5 (red) stained with fluorescently labelled antibodies (Flag1 experiment). (A) Before flagellate inoculation (day 8). (B) After flagellate inoculation (day 32). (C) Filamentous C. acidovorans PX54 cells grown in a flagellate-free batch culture (late exponential growth stage; cells stained with DAPI).
FIG. 4.
Transient stage after introduction of the flagellate (on day 14) into a chemostat culture of C. acidovorans PX54 and Vibrio strain CB5 (Flag1 experiment). In the beginning, the total bacterial numbers of both species decreased at similar rates, indicating nonselective grazing by the flagellate (lower panel). After day 16, Vibrio strain CB5 abundance continued to decrease but C. acidovorans PX54 abundance increased again and the relative species composition changed (upper panel).
Bacterial biomass, cell size, and filament formation.
After inoculation with the flagellate, the abundance of the Vibrio strain CB5 populations was reduced by 98.8% in both experiments and the biomass decreased by 98.1% (Fig. 1D). In contrast, the C. acidovorans PX54 population decreased by 66.0% (Flag1 experiment) and 70.3% (Flag2 experiment) in abundance but only by 23.3% in biomass (Fig. 1D). Differences in changes in abundance and biomass between the two species are due to the different relative changes in mean cell size and cell size distribution (Fig. 3 and 5). The cell sizes of both bacterial strains remained unchanged during the flagellate-free phase of the experiments (Fig. 1C and 5). After introduction of the flagellate, the mean bacterial cell size increased in both experiments but the increase in Vibrio strain CB5 cell size was less than that of C. acidovorans PX54. The mean cell length of Vibrio strain CB5 increased only during the transient stage, and during the following flagellate-controlled phase, it showed a higher but stable mean value (Fig. 5). The cell size of C. acidovorans PX54 increased during the transient stage too, but in contrast to that of Vibrio strain CB5, the increase in cell size continued during the first part of the steady state in the flagellate-controlled phase. At 9 (Flag1 experiment) and 10 (Flag2 experiment) days after inoculation with the predator, the first filamentous C. acidovorans PX54 cells (larger than 10 μm) were observed (Fig. 1C and 2B). Eight (Flag1 experiment) and 9 (Flag2 experiment) days later, the numbers of filamentous cells peaked. The number of large C. acidovorans PX54 cells decreased thereafter, and at the end of both experiments, no filamentous cells were observed. The measured C. acidovorans PX54 cell length ranged from 0.3 to 4.2 μm (flagellate-free phase) and from 0.3 to 49.8 μm (flagellate-controlled phase) (Fig. 5). During both experiments, no Vibrio strain CB5 cells larger than 5 μm occurred (Flag1 experiment; Fig. 3) and the Vibrio strain CB5 cell size ranges were 0.3 to 2.5 μm (flagellate-free phase) and 0.3 to 4.2 μm (flagellate-controlled phase) (Fig. 5).
Due to the different cell size distributions, the two bacterial populations showed different percentages of cells smaller than 1.2 μm, which are presumably easily ingestible (29). During the flagellate-controlled phase of the Flag1 experiment, 72 to 75% of the Vibrio strain CB5 cells but only 14 to 41% of the C. acidovorans PX54 cells were smaller than 1.2 μm.
The observed strong changes in the relative abundances of the two strains occurred in both experiments several days before the occurrence of filamentous C. acidovorans PX54 cells. Flagellate control of the composition of the bacterial community remained unchanged in both cases until the end of the experiments and lasted longer than the occurrence of filaments (Fig. 1B and C and 2B).
Increase in dilution rate to simulate nonselective grazing pressure (Dilut experiment).
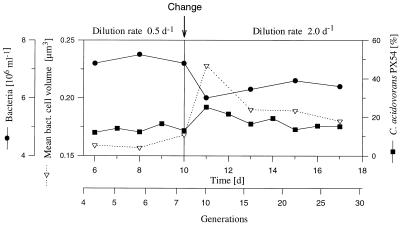
To simulate nonselective loss rates, the dilution rate was increased to 2.0 day−1 (about 50% of μmax for both strains) after the chemostat was run at 0.5 day−1 for 10 days (Fig. 6). This increase in dilution rate was supposed to simulate mortality equivalent to a grazing rate of 1.5 day−1 on a total bacterial community growing at a rate of 0.5 day−1. Theoretically, such a grazing rate could be caused by an Ochromonas sp. population of 8.8 × 103 ml−1 with a clearance rate of 7.1 nl flagellum−1 h−1. Such a clearance rate was measured for Ochromonas sp. under comparable conditions (15). The first phase of the experiment with a dilution rate of 0.5 day−1 was identical to the other two chemostat experiments. However, the initial steady-state total bacterial number (7.3 × 106 ml−1) was significantly (P < 0.01 for both experiments) lower and the relative C. acidovorans PX54 abundance (13.7% ± 1.8%) was slightly but significantly (P < 0.01 for both experiments) higher than in the respective first phases of the two other experiments (Fig. 1, 2, and 6). The lower total bacterial number was balanced by a higher mean cell volume (0.161 ± 0.006 μm3), so that the total bacterial biomass of 204.9 ± 5.6 μg of C liter−1 showed no significant difference (P > 0.1 for both of the other experiments) from the other chemostat studies (Flag1 experiment, 199 ± 17 μg of C liter−1; Flag2 experiment, 186 ± 27 μg of C liter−1).
FIG. 6.
Influence of growth rate on the cell size and abundance of C. acidovorans PX54 and Vibrio strain CB5 in chemostat cultures (Dilut experiment). The change in dilution rate (growth rate) from 0.5 to 2.0 day−1 at day 10 simulated nonselective grazing similar to size-selective grazing by Ochromonas sp. in flagellate-controlled phases of the Flag1 and Flag2 experiments.
The dilution rate increase initially resulted in changes in total bacterial abundance, mean cell volume, and the relative abundance of the strains (Fig. 6). After 3 days, a new steady state was established with a significantly higher mean cell volume (P < 0.01) and a significantly lower total bacterial abundance (P < 0.01) but no significant difference (P > 0.1) in the relative abundance of C. acidovorans PX54. Filamentous cells (>10 μm) were not observed in the first or second phase, i.e., after the fourfold increase in the dilution rate.
Dependence of Vibrio strain CB5 and C. acidovorans PX54 cell size on growth stage in flagellate-free batch cultures.
The cell sizes of both species changed with the growth stage of batch cultures, and in both cases, they reached a maximum during the exponential growth stage. In each growth stage, the mean cell volume of C. acidovorans PX54 was larger than that of Vibrio strain CB5 (Table 4). Additionally, the variation in the mean cell volume between the exponential growth stage and the stationary stage was twofold stronger for C. acidovorans PX54 than for Vibrio strain CB5. During the late exponential growth stage, C. acidovorans PX54 cells larger than 10 μm developed with a relative abundance of less than 1% (Table 4). However, the measured maximum growth rate of Vibrio strain CB5 was higher than that of C. acidovorans PX54 (Table 4).
TABLE 4.
Effect of growth phase on cell size in batch culture experimentsa
| Bacterium | Mean cell vol (μm3)
|
% of cells >10 μm long | μmax (h−1) | |
|---|---|---|---|---|
| Log phase | Stationary phase | |||
| C. acidovorans PX54 | 0.76 | 0.09 | <1 | 0.17 |
| Vibrio strain CB5 | 0.31 | 0.07 | 0.20 | |
Mean cell volume in the late logarithmic and stationary phases (35 h after the end of the log phase), percentage of filamentous cells (late logarithmic phase), and the maximum growth rates of the two bacterial strains are shown. Batch cultures were grown in NSY medium at 15°C and 150 rpm.
DISCUSSION
We observed strong changes in the cell morphology and the absolute and relative abundances of the two bacterial strains after introduction of a bacterivorous flagellate in our chemostat experiments (Fig. 1, 2, 3, and 5). Similar changes caused by protozoan grazing, especially the occurrence of filamentous bacteria, were observed by others in complex continuous cultures (30, 38) and in field experiments (21). The methods used in these studies, however, did not allow the analysis of species-specific bacterial responses to protozoan grazing. In the former two studies, the investigators examined the influence of protozoan predators on different phylogenetic groups of bacteria (α, β, and γ subclasses of the class Proteobacteria). Each of these groups covers a wide range of bacterial species and includes members which differ drastically in the type of metabolism. In the latter experiments, Jürgens et al. (21) analyzed the effect of protozoan grazing on morphologically defined groups of natural bacteria, which may also cover a wide range of species. In contrast to these studies, we investigated the influence of flagellate grazing on a well-defined bacterial community consisting of only two species. This allowed us to examine the impact of grazing on single bacterial species and to study species- or strain-specific strategies against grazing losses.
Mechanisms controlling the changes after introduction of the flagellate.
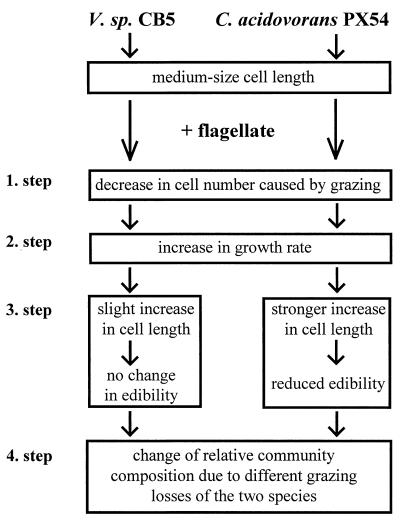
We suggest that the observed changes in the relative abundances of the two strains and the occurrence of filamentous C. acidovorans PX54 cells resulted from increasing growth rates caused by flagellate grazing combined with differences in the species’ ability to increase cell length with growth rate and size-selective grazing of the flagellate. During the compositional changes, several steps could be distinguished (Fig. 7).
FIG. 7.
Four-step mechanism explaining the observed changes in relative species composition of binary chemostat cultures after introduction of the flagellate Ochromonas sp. Changes were caused by a combination of a grazing-controlled increase in the bacterial growth rate with the different growth rate-dependent elongation of cells of the two species and size-selective grazing by the flagellate.
First, grazing of the introduced flagellate reduced bacterial numbers (Fig. 1A, 2A, and 4). Since the cell sizes of the two strains were similar, both populations were grazed at similar rates from the beginning and thus decreased at similar rates (Fig. 4). Due to similar loss rates, their relative abundances remained stable (Fig. 4).
Second, the growth rate of the surviving bacteria increased due to the reduction of bacterial cell numbers and biomass. The reduction of inter- and intraspecific competitors increased the amount of substrate available for every single cell, thereby enabling higher growth rates. The dependence of growth rate on the chemostat dilution rate and grazing rate is defined above (see Materials and Methods). Strains which were not able to balance grazing losses with higher growth rates would have been washed out of the system. Higher activity of the bacterial population under grazing pressure in comparison to the uninfluenced population was also demonstrated in other chemostat experiments (4, 42). In addition, bacterial growth rate may be positively influenced by release of dissolved and particulate organic matter and recycling of nutrients by the phagotrophic flagellates (1, 18, 36).
Third, the growth rate increase paralleled an increase in cell size for both species (Fig. 1C and 5) but the extents of cell elongation were very different between the two species (Fig. 5). A dependence of cell size on the growth rate was demonstrated for both strains in the chemostat Dilut experiment (Fig. 6) and the batch culture experiments (Table 4) and is also known for other bacterial species (5, 7, 34). This dependence is partly caused by an increasing need for space for the ribosomes, which increase in number as the growth rate increases (5, 32, 33). In contrast to Vibrio strain CB5, in both continuous and batch cultures, C. acidovorans PX54 showed a strong increase in mean cell length with growth rate and a strong expansion of the cell length distribution toward long filamentous cells.
Fourth, the strong increase in cell length made a large percentage of the C. acidovorans PX54 cells less edible or completely inedible. Despite an increase in size, the majority of Vibrio strain CB5 cells remained in the edible size range. The resulting differences in edibility between the two populations resulted in different specific loss rates and then caused the observed changes in the species compositions of the communities.
Exclusion of other conceivable mechanisms.
The possibility of species-specific grazing based on characteristics other than cell size can be excluded, because of the initially similar rates of decrease of the two bacterial species (Fig. 4). The observation that the absence or presence of Vibrio strain CB5 in the flagellate-free stage did not influence the steady-state abundance of C. acidovorans PX54 (Table 3) indicated that reduction of the possible competitor Vibrio strain CB5 by the flagellate was not the reason for the lesser decrease of the C. acidovorans PX54 population. Low C. acidovorans PX54 abundances and biomasses during flagellate-free growth indicate that this strain was able to use only a small part of the supplied complex carbon source. During the flagellate-controlled phase, the growth of both strains may have been enhanced by the additional substrate supply due to possible release of dissolved and particulate organic material by the flagellates (1, 18). C. acidovorans PX54 might have profited more than the other strain by such a substrate release.
The Dilut experiment demonstrated that a change in growth rate is not enough to change the relative composition of the bacterial community studied (Fig. 6). This experiment simulated a grazing pressure assumed to occur in the flagellate-controlled phases of the other two chemostat experiments. In contrast to real flagellate grazing, the simulated grazing pressure is nonselective. Both bacterial populations reacted to the treatment with an increase in cell size, but without size-selective grazing, no compositional changes occurred. The observed absence of filamentous C. acidovorans PX54 cells may be caused by physiological disadvantages of strongly elongated cells. For example, filamentous cells have a smaller surface-to-volume ratio than medium-size cells. When filamentous cells grow under limited-substrate conditions (e.g., chemostat culture) and have to compete with much smaller cells, then they have a disadvantage in terms of substrate uptake, resulting in lower growth rates. In chemostat experiments with grazing pressure, this disadvantage may be balanced due to protection against grazing losses (a lower growth rate than the competitors but also lower loss rates). Growth during the exponential phase of batch cultures was, in contrast to growth in the chemostat Dilut experiment, not substrate limited. Therefore, filamentous growth of C. acidovorans PX54 was enabled by lack of intraspecific competition with smaller cells.
The observed differences in flagellate increase and bacterial decrease kinetics during the transient stage between the Flag1 and Flag2 experiments (Table 2) may reflect the different growth abilities of the two different Ochromonas sp. clones used in the two studies. It is surprising that despite different kinetics during the transient stage, mostly insignificant differences in the overall percentage of bacterial strains and total bacterial abundance occurred. It seems that the two clones have different maximum growth rates (transient stage) but grow at lower rates (i.e., during the steady state of chemostat experiments) with comparable growth efficiencies.
Role of filamentous bacteria in the natural environment and in chemostat experiments.
In general, filament formation by bacteria is considered to be a protection mechanism against protozoan grazing (11, 20, 21, 30, 38). To have a refuge from bacterivorous nanoflagellates like Ochromonas sp. (4 to 6 μm in diameter), it is not advantageous to form filaments longer than 10 μm. It is sufficient if cells exceed the maximum ingestion size of these flagellates. In bacterivorous flagellates, this size is normally smaller than the cell diameter. Therefore, bacteria larger than 4 to 6 μm should be fully protected against grazing by Ochromonas sp. Both chemostat experiments with the flagellate-controlled phase confirm this presumption. In each experiment, compositional changes started before filament formation (Fig. 1B, 1C, and 2B). Additionally, the stable flagellate-controlled community composition lasted until the end of each experiment and, thus, longer than the filaments were present in the chemostats (Fig. 1B, 1C, and 2B). We assume that the observed occurrence of C. acidovorans PX54 filaments longer than necessary for full protection is caused by an overshooting reaction in response to the increasing growth rate. For bacteria from ecosystems like meso- to hypertrophic lakes, it is uncertain that filament formation is such an overshooting reaction. It is, rather, conceivable that formation of long filaments is a protection strategy against larger protozoan predators like large heterotrophic flagellates (3) or ciliates (37). However, in an investigation of Lake Plußsee bacterioplankton, Weinbauer and Höfle (43) found that viral lysis is size specific and can affect the cell size distribution of bacterial communities. In oxic waters, cells larger than 2.4 μm were not infected with viruses, suggesting that filamentous bacteria have advantages other than grazing resistance.
Delay of filament formation.
In contrast to other experimental studies which demonstrated that grazing or the presence of a protistan grazer caused the formation and dominance of filamentous bacteria (21, 30, 38), filament formation in our chemostat experiments occurred after 9 and 10 days and not within 1 to 3 days as described by others. This may have been caused by the initially low flagellate abundances in our studies and the following slow increase in abundance and grazing pressure (Fig. 1A and 2A). Thus, grazing pressure during the first days after inoculation with the flagellate was much lower than in the studies mentioned above.
Conclusions.
The main conclusion of this study is that changes in the morphological structure of bacterial communities like development of filamentous cells under high grazing pressure are not necessarily a result of chemical stimuli released by the protozoan predators. This was demonstrated by the development of filaments in the absence of any protozoan predator by fast-growing C. acidovorans PX54 cells (Fig. 3). Furthermore, the ability of C. acidovorans PX54 to grow as normal-size cells and as filaments larger than 10 μm shows that shifts in the morphological structure of natural bacterial communities towards filamentous cells do not necessarily lead to the replacement of permanently medium-size bacteria by permanently filamentous bacteria. There may be other species that change their cell morphology by indirect or direct induction mechanisms. We observed that influence of grazing caused an indirect induction of filament formation via an increase in growth rate of C. acidovorans PX54. This is one possible mechanism, but it is conceivable that other indirect or direct mechanisms exist. Chemical stimuli released by protozoan predators are likely to be another (direct) induction mechanism, because interactions between other groups of planktonic organisms are controlled by such stimuli (8, 22, 23). Intraspecific communication of bacterial populations by chemical stimuli (pheromones) has been documented for several bacterial species in different phylogenetic positions (24, 44). However, chemical induction of defense strategies in bacteria has not previously been proven.
In natural environments, a single protozoan predator species is in contact with a wide range of bacterial species and each bacterial species or strain might have a different grazing defense strategy. To gain more insights into these complex interactions between bacterivorous protozoa and their prey, more studies with single predator species and single or a limited number of bacterial strains under controlled conditions are necessary. We cannot exclude the possibility that bacteria possess grazing defense strategies similar in diversity to their metabolic abilities.
ACKNOWLEDGMENTS
We thank D. Springmann for providing the original Ochromonas sp. culture, E. R. B. Moore for identification of the bacterial strains by 16S rDNA analysis, and K. Jürgens, M. G. Weinbauer, and T. Weisse for discussions and help in improving the manuscript.
This study was supported by funds from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (grant BEO-0319433B).
REFERENCES
- 1.Andersson A, Lee C, Azam F, Hagström Å. Release of amino acids and inorganic nutrients by heterotrophic marine microflagellates. Mar Ecol Prog Ser. 1985;23:99–106. [Google Scholar]
- 2.Andersson A, Larsson U, Hagström Å. Size-selective grazing by a microflagellate on pelagic bacteria. Mar Ecol Prog Ser. 1986;33:51–57. [Google Scholar]
- 3.Arndt H, Mathes J. Large heterotrophic flagellates form a significant part of protozooplankton biomass in lakes and rivers. Ophelia. 1991;33:225–234. [Google Scholar]
- 4.Bloem J, Starink M, Bär-Gilissen M-J B, Cappenberg T E. Protozoan grazing, bacterial activity, and mineralization in two-stage continuous cultures. Appl Environ Microbiol. 1988;54:3113–3121. doi: 10.1128/aem.54.12.3113-3121.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 6.Chrzanowski T H, Šimek K. Prey-size selection by freshwater flagellated protozoa. Limnol Oceanogr. 1990;35:1424–1436. [Google Scholar]
- 7.Chrzanowski T H, Crotty R D, Hubbard G J. Seasonal variation in cell volume of epilimnetic bacteria. Microb Ecol. 1988;16:155–163. doi: 10.1007/BF02018911. [DOI] [PubMed] [Google Scholar]
- 8.Dodson S I. The ecological role of chemical stimuli for the zooplankton: predator-induced morphology in Daphnia. Oecologia. 1989;78:361–367. doi: 10.1007/BF00379110. [DOI] [PubMed] [Google Scholar]
- 9.Faude U C, Höfle M G. Development and application of monoclonal antibodies for in-situ detection of indigenous bacterial strains in aquatic ecosystems. Appl Environ Microbiol. 1997;63:4534–4542. doi: 10.1128/aem.63.11.4534-4542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González J M, Sherr E B, Sherr B F. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol. 1990;56:583–589. doi: 10.1128/aem.56.3.583-589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Güde H. Grazing by protozoa as selection factor for activated sludge bacteria. Microb Ecol. 1979;5:225–237. doi: 10.1007/BF02013529. [DOI] [PubMed] [Google Scholar]
- 12.Güde H. The role of grazing on bacteria in plankton succession. In: Sommer U, editor. Plankton ecology. Succession in plankton communities. Berlin, Germany: Springer-Verlag KG; 1989. pp. 337–369. [Google Scholar]
- 13.Güde H, Haibel B, Müller H. Development of planktonic bacterial populations in a water column of Lake Constance (Bodensee-Obersee) Arch Hydrobiol. 1985;105:59–77. [Google Scholar]
- 14.Hahn M W. Experimentelle Untersuchungen zur Interaktion von bakterivoren Nanoflagellaten mit planktischen Bakterien. Ph.D. thesis. Braunschweig, Germany: University of Braunschweig; 1996. [Google Scholar]
- 15.Hahn, M. W., and T. Weisse. Unpublished data.
- 16.Höfle M G. RNA chemotaxonomy of bacterial isolates and natural microbial communities. In: Overbeck J, Chrost R J, editors. Aquatic microbial ecology—biochemical and molecular approaches. Berlin, Germany: Springer-Verlag KG; 1990. pp. 129–159. [Google Scholar]
- 17.Höfle M G. Transfer RNAs as genotypic fingerprints of eubacteria. Arch Microbiol. 1990;153:299–304. [Google Scholar]
- 18.Jumars D, Penry L, Baross J A, Perry M J, Frost B W. Closing the microbial loop: dissolved carbon pathway to heterotrophic bacteria from incomplete ingestion, digestion and absorption in animals. Deep-Sea Res. 1989;36:483–495. [Google Scholar]
- 19.Jürgens K, Stolpe G. Seasonal dynamics of crustacean zooplankton, heterotrophic nanoflagellates and bacteria in a shallow, eutrophic lake. Freshwater Biol. 1995;33:27–38. [Google Scholar]
- 20.Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 21.Jürgens K, Arndt H, Rothhaupt K O. Zooplankton-mediated changes of bacterial community structure. Microb Ecol. 1994;27:27–42. doi: 10.1007/BF00170112. [DOI] [PubMed] [Google Scholar]
- 22.Kusch J. Induction of defensive morphological changes in ciliates. Oecologia. 1993;94:571–575. doi: 10.1007/BF00566974. [DOI] [PubMed] [Google Scholar]
- 23.Lampert W, Rothhaupt K O, von Elert E. Chemical induction of colony formation in a green alga (Scenedesmus acutus) by grazers (Daphnia) Limnol Oceanogr. 1994;39:1543–1550. [Google Scholar]
- 24.Magnuson R, Solomon J, Grossman A D. Biochemical and genetic characterisation of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 25.Monger B C, Landry M R. Prey-size dependency of grazing by free-living marine flagellates. Mar Ecol Prog Ser. 1991;74:239–248. [Google Scholar]
- 26.Moore, E. R. B. Unpublished data.
- 27.Müller H. Wachstum und Phosphatbedarf von Nitzschia actinastroides (LEMM.) v. GOOR in statischer und homokontinuierlicher Kultur unter Phosphatlimitierung. Arch Hydrobiol Suppl. 1972;38:399–484. [Google Scholar]
- 28.Patterson D J. Protozoa: evolution and systematics. In: Hausmann K, Hülsmann N, editors. Progress in protozoology. Stuttgart, Germany: Gustav Fischer Verlag; 1994. pp. 1–14. [Google Scholar]
- 29.Pernthaler J, Sattler B, Šimek K, Schwarzenbacher A, Psenner R. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat Microb Ecol. 1996;10:255–263. [Google Scholar]
- 30.Pernthaler J, Posch T, Šimek K, Vrba J, Amann R, Psenner R. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol. 1997;63:596–601. doi: 10.1128/aem.63.2.596-601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 32.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosset R, Julien J, Monier R. Ribonucleic acid composition of bacteria as a function of growth rate. J Mol Biol. 1966;18:308–320. doi: 10.1016/s0022-2836(66)80248-6. [DOI] [PubMed] [Google Scholar]
- 34.Schaechter M, Maaløe O, Kjeldgaard N O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 35.Schmaljohann R, Pollingher U, Berman T. Natural populations of bacteria in Lake Kinneret: observations with scanning electron and epifluorescence microscopy. Microb Ecol. 1987;13:1–12. doi: 10.1007/BF02014959. [DOI] [PubMed] [Google Scholar]
- 36.Sherr B F, Sherr E B, Berman T. Decomposition of organic detritus: a selective role for microflagellate protozoa. Limnol Oceanogr. 1982;37:765–769. [Google Scholar]
- 37.Shikano S, Luckinbill L S, Kurihara Y. Changes of traits in a bacterial population associated with protozoan predation. Microb Ecol. 1990;20:75–84. doi: 10.1007/BF02543868. [DOI] [PubMed] [Google Scholar]
- 38.Šimek K, Vrba J, Pernthaler J, Posch T, Hartman P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sime-Ngando T, Bouredier G, Amblard C, Pinel-Alloul B. Short-term variations in specific biovolumes of different bacterial forms in aquatic ecosystems. Microb Ecol. 1991;21:211–226. doi: 10.1007/BF02539155. [DOI] [PubMed] [Google Scholar]
- 40.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 41.Sommaruga R, Psenner R. Permanent presence of grazing-resistant bacteria in a hypertrophic lake. Appl Environ Microbiol. 1995;61:3457–3459. doi: 10.1128/aem.61.9.3457-3459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhagen F J M, Laanbroek H J. Effects of grazing by flagellates on competition for ammonium between nitrifying and heterotrophic bacteria in a chemostat. Appl Environ Microbiol. 1992;58:1962–1969. doi: 10.1128/aem.58.6.1962-1969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinbauer, M. G., and M. G. Höfle. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat. Microbial Ecol., in press.
- 44.Wirth R, Muscholl A, Wanner G. The role of pheromones in bacterial interactions. Trends Microbiol. 1996;4:96–103. doi: 10.1016/0966-842X(96)81525-3. [DOI] [PubMed] [Google Scholar]