Abstract
There are thousands of unannotated translated open reading frames (ORFs) in the Saccharomyces cerevisiae genome. Previous investigation into one such unannotated ORF, which was systemically labeled YGR016C-A based on its genomic coordinates, showed that replacing the ORF’s ATG start codon with AAG led to a change in cellular fitness under different stress conditions (Wacholder et al., 2023). This suggested translation of YGR016C-A plays a role in cellular fitness. Here, we investigate Ygr016c-a’s subcellular localization to gain insight into its cellular function. Computational prediction tools, co-expression analysis and fluorescence microscopy suggest that the Ygr016c-a protein localizes to the endoplasmic reticulum.
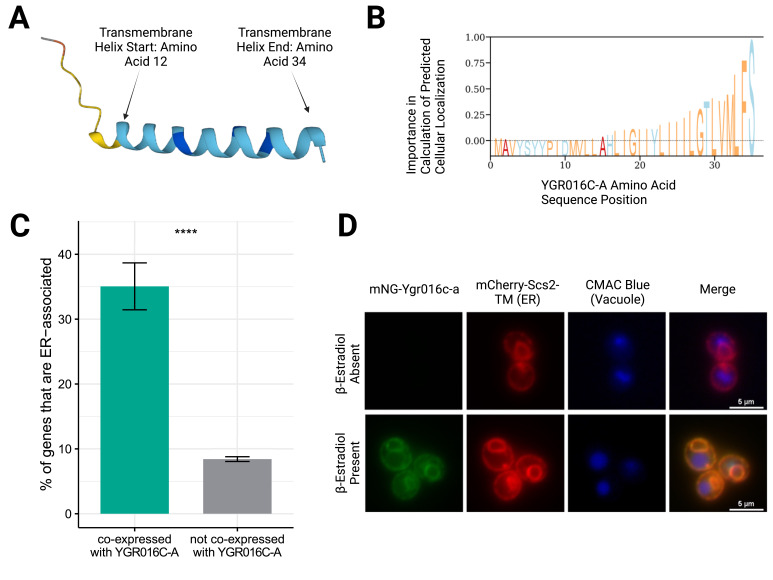
Figure 1. Ygr016c-a protein localizes to the endoplasmic reticulum .
(A) The protein structure for Ygr016c-a was predicted using ESMFold (Lin et al., 2022). The colors (orange, yellow, light blue, and dark blue) represent predicted-LDDT confidence intervals for the modelled residues (<0.5, 0.5-0.7, 0.7-0.9, and 0.9-1.0 confidence, respectively). From amino acid position 11-34, an alpha helix is predicted with a confidence of at least 0.7. Ygr016c-a is predicted to possess a transmembrane domain from amino acid position 12-34 using Phobius (Käll et al., 2004, 2007), which is the basis for the transmembrane helix Start and End annotations on the structure prediction.
(B) The DeepLoc 2.0 prediction tool (Thumuluri et al., 2022) reported Ygr016c-a to localize to the endoplasmic reticulum (ER) with a probability of 0.7518, which is higher than the background threshold of 0.6090. Represented here is the emphasis DeepLoc placed on each residue, with higher importance on the residues toward the C-terminus.
(C) Analysis of co-expression data curated by Rich et al. (2023) compares the percentage of genes co-expressed with YGR016C-A whose encoded proteins localize to the ER (n=61 ER-localizing, 35.1% of 174 total co-expressed; green bar) to the percentage of genes not co-expressed with YGR016C-A whose encoded proteins localize to the ER (n=483 ER-localizing, 8.4% of 5734 total non-co-expressed; gray bar). The error bars represent the standard error of the percentage (see methods). The four stars (****) represent the high significance conferred by a p-value of 2.2e-16, calculated with Fisher’s exact test.
(D) A yeast strain containing the construct Z 3 EVpr-mNG-YGR016C-A:HYG at the HIS3 locus was imaged on an epifluorescence microscope without (top row) and with (bottom row) induction by β-estradiol. The first column (left to right) shows the localization of the fusion protein mNG-Ygr016c-a, while the second column represents the localization of the ER marker, Scs2TM fused with mCherry. The CMAC blue dye was used to dye the vacuole as seen in the third column. The last column shows the merge of all channels, confirming the co-localization of the fused protein mNG-Ygr016c-a with the mCherry-Scs2TM ER marker. Cells shown are representative of the entire field.
Description
The Saccharomyces cerevisiae genome contains approximately 6,000 annotated open reading frames (ORFs) that are translated into proteins (Goffeau et al., 1996) . Recently, a large-scale meta-analysis of ribosome profiling results demonstrated that in addition to these annotated ORFs, there are 18,953 unannotated ORFs that are also translated (Wacholder et al., 2023) . A subset of this group of ORFs was further studied, with some showing experimental evidence of affecting cellular phenotype and fitness (Wacholder et al., 2023) . Among the unannotated translated ORFs exhibiting cellular effects in deletion screenings is an ORF that encodes for a 35 amino acid microprotein of unknown function (Wacholder et al., 2023) which was given the systematic name YGR016C-A based on its genomic coordinates, chrVII:523246-523353(-). This unannotated ORF was shown to cause a change in cell fitness under different stress conditions when its normal ATG start codon was mutated to an AAG (Wacholder et al., 2023) . This suggested translation of YGR016C-A plays a role in cellular phenotype and fitness. To further characterize this ORF, we combined computational prediction tools and fluorescence microscopy to determine the cellular localization of Ygr016c-a.
ESMFold, a deep learning model that has demonstrated high levels of accuracy pertaining to protein structure prediction based on amino acid sequences, was used to predict structure (Lin et al., 2022) . ESMFold predicted that Ygr016c-a would exhibit a large α-helix between residues 11 to 34 with a confidence score of at least 0.7 ( Figure 1A ). Next, Phobius (Käll et al., 2004, 2007) , a computational tool used for predicting the probability of transmembrane domains for a given amino acid sequence, reported Ygr016c-a to have a singular transmembrane helix ( Figure 1A ) between amino acids 12-34, which aligns with the α-helix reported by ESMFold. Since transmembrane helices interact with either the plasma membrane or the membranes of specific organelles, this result suggests a non-cytoplasmic localization of Ygr016c-a. To predict the localization of Ygr016c-a, DeepLoc 2.0 (Thumuluri et al., 2022) , a protein language model relying on amino-acid sequence input, was used. Notably, this tool outputs the probability of the protein localizing to distinct subcellular localizations and a corresponding threshold probability for each localization. If the calculated probability of a protein’s localization is less than the threshold probability, the probability is considered not significant. The only localization where Ygr016c-a had a calculated probability of localization (0.752) above the threshold (0.609) was the endoplasmic reticulum (ER; Figure 1B ). Additionally, DeepLoc placed higher importance on residues towards the C-terminus when calculating this localization ( Figure 1B ). Gene co-expression networks can be used for function and localization inferences (van Dam et al., 2018) . Considering the predicted ER membrane localization, we checked to see if genes that are known to be associated with the ER have similar transcript expression patterns as YGR016C-A using co-expression data (Rich et al., 2023) and cellular component annotations from the Gene Ontology (GO) database (Ashburner et al., 2000; The Gene Ontology Consortium et al., 2023) . 35.1% of genes co-expressed with YGR016C-A localize to the ER, while only 8.4% of genes that aren’t co-expressed with YGR016C-A localize to the ER ( Figure 1C ). We calculated that annotated ORFs that are co-expressed with YGR016C-A are 5.9 times more likely to have an ‘ER’ GO cellular component annotation than genes that are not co-expressed with YGR016C-A (Odds Ratio, 95% confidence interval: 4.2-8.2, p-value < 2.2e-16, Fisher’s Exact test; Figure 1C ). Holistically, these computational analyses provided evidence that Ygr016c-a may localize to the ER.
Wacholder et al., 2023 tagged YGR016C-A at the endogenous locus under its native promoter with a fluorescent protein at the C-terminus and reported inconclusive localization, likely due to low native expression levels. In our study, to test our computational predictions, we genetically engineered a S. cerevisiae strain to overexpress the ORF YGR016C-A fused with a mNeonGreen (mNG) tag at the N-terminus. This overexpression was driven by the Z 3 EV promoter, a β - estradiol inducible promoter (Ohira et. al, 2017) . This construct was integrated at the HIS3 locus. Furthermore, these cells were also transformed with a plasmid expressing mCherry-Scs2TM, a well-known ER marker (Zhou et al., 2014) . Fluorescence microscopy showed co-localization of the mNG-Ygr016c-a fusion and the mCherry-Scs2TM fusion ( Figure 1D ), confirming the predicted ER localization for Ygr016c-a.
Localization of a protein can provide insight into its potential function (Pan et al., 2021) . Here, we have shown ER localization and a predicted C-terminal transmembrane helix for Ygr016c-a. A protein with a lone transmembrane domain at the C-terminus of the peptide chain is considered a tail anchored protein (Schuldiner et al., 2008). The Guided Entry of Tail-anchored proteins (GET) pathway transports tail anchored proteins to the ER (Schuldiner et al., 2008). Therefore, Ygr016c-a may localize to the ER via the GET pathway. This is a possible avenue for future research.
Methods
Sequence and chromosome location of unannotated ORF known as YGR016C-A
The name YGR016C-A given to the unannotated ORF was chosen using the naming system based on SGD conventions.
The sequence of YGR016C-A is: ATGGCGGTTTATTCATACTATCCAATTGACATGGTTTTGCTCGCTCACCTCATTGGCATCATCTACTTAATTATAATTCTAGGTACATTGGTCATGTTGTTTTCTTGA
Furthermore, the ORF is located on the minus strand of chromosome VII between coordinates 523246 and 523353.
Structure, TM and localization predictions
The protein structure of Ygr016c-a was predicted by inputting the amino acid sequence to the online version of ESMFold (Lin et al., 2022) . The website was accessed at: https://esmatlas.com/resources?action=fold on 26 May 2023.
The prediction of transmembrane helices was computed by inputting the amino acid sequence to the online version of Phobius (Käll et al., 2004, 2007). The website was accessed at: https://phobius.sbc.su.se/ on 11 August 2023.
The predicted cellular localization was determined by inputting the amino acid sequence to the online version of DeepLoc 2.0 (Thumuluri et al., 2022) . The website was accessed at: https://services.healthtech.dtu.dk/services/DeepLoc-2.0/ on 29 May 2023.
Co-expression and ER enrichment analysis
Co-expressed was defined as a value greater than the 99th percentile for all co-expression values in the Rich et al. coexpression data; i.e., 99th percentile of all pairwise combinations for the 11,630 ORFs in the Rich et al. co-expression data (co-expression value > 0.836). The coexpression matrix was then subdivided to only include genes that have at least one GO cellular component annotation using the GO slim annotation file downloaded from SGD on 20 January 2021 (Saccharomyces Genome Database). This resulted in n = 5,908 genes. Of this subset, 174 genes were co-expressed with YGR016C-A and 5,734 genes were not co-expressed with YGR016C-A. Any gene with at least one GO cellular compartment annotation with the words ‘endoplasmic reticulum’ was labeled as ‘ER’ associated (n= 544) and all other genes were labeled as ‘not ER’ associated (n= 5,364). Fisher’s exact test was subsequently used to quantify the significance of enrichment using the R function fisher.test(). Standard error of the percentage was calculated using the following equation: root-square (percentage*(100-percentage)/n)
Saccharomyces cerevisiae strain
2x Master mix Q5 Hot Start polymerase (New England BioLabs) was used to amplify the construct containing Z 3 EVpr-mNG-YGR016C-A:HYG, using plasmid pARC0400 as template. Plasmid pARC0400 was made by LR recombinase (ThermoFisher) between an Entry Clone containing the YGR016C-A ORF and a destination plasmid containing Z 3 EVpr-mNG-ccdB:HYG. The PCR product containing homology to HIS3 locus was used to transform the strain yARC0085 following the LiAc/PEG/ssDNA transformation protocol (Dunham, 2015) . Positive transformants were selected on YPD+Hygromycin (200μg/ml) and used for a subsequent transformation with the plasmid pARC0006 (containing the mCherry-SCS2TM). The final transformants were selected on SC-LEU+GLU+Hygromycin (200μg/ml).
Microscopy methods
Yeast cells were inoculated into overnight cultures of SC-LEU+GLU+Hygromycin. The following day the culture was diluted to an optical density of 0.2 at 600nm, and after 1 hour of growth at 30°C with constant shaking at 120rpm, half of the culture was inoculated with β-estradiol (to a final concentration of 10μM) to induce the expression of mNG-YGR016C-A. In parallel, 100% ethanol was added to the other half of the culture to serve as negative control for the induction.
After 3 hours of induction, the vacuolar dye CMAC Blue (ThermoFisher) was added to the cells to a final concentration of 1μM and incubated at room temperature for 15-minutes prior to imaging. Cells were transferred to glass bottom culture dishes and imaged on a NIKON Ts2R-FL Epifluorescence microscope (Nikon) with a camera ORCA-Flash 4.0. All images were taken using 100x oil immersion objective and analyzed with the software NIS Elements (Nikon).
Reagents
Table 1 | Plasmids, strains and reagents used in this study
|
Plasmids | ||||
|
Name |
Insert |
Description |
Source |
|
|
pARC0006 |
mCherry-SCS2TM |
Plasmid containing mCherry-SCS2TM . LEU2 gene for yeast selection. |
Zhou et al. 2014 |
|
|
pARC0400 |
Z 3 EVpr-mNG- YGR016C-A:HYG |
Expression plasmid used to amplify the construct Z 3 EV-mNG-YGR016C-A:HYG to integrate at HIS3 locus |
This study |
|
|
Strains | ||||
|
Name |
Genotype |
Source |
||
|
yARC0085 |
MAT α ura3Δ0 leu2Δ ::ACT1pr-Z 3 EV:NatMX |
McIsaac et al. 2013 |
||
|
yARC0988 |
MAT α ura3Δ0 leu2Δ ::ACT1pr-Z 3 EV:NaTMX his3Δ : Z 3 EVpr-mNG-YGR016C-A:HYG + plasmid pARC0006 (mCherry-SCS2TM) |
This study |
||
|
Reagents | ||||
|
Name |
Final Concentration |
Reference |
Company |
|
|
2x Q5 Master Mix |
1x |
M0492L |
NEB BioLabs |
|
|
LiAc |
10mM |
L4158 |
Millipore Sigma |
|
|
PEG |
37% |
P4338 |
Millipore Sigma |
|
|
ssDNA |
2mg/ml |
15632-011 |
Invitrogen |
|
|
Hygromycin B |
200μg/ml |
Research Products International |
||
|
CMAC Blue |
1μM |
C2110 |
ThermoFisher |
|
|
β -estradiol |
10μM |
E8875 |
Millipore Sigma |
|
|
Primers | ||||
|
Name |
Sequence |
|||
|
HIS3 integration primer Fw |
5’-TCTATATTTTTTTATGCCTCGGTAATGATTTTCATTTTTTTTTTTCCACCTAGCGGATGACTCTTTTTTTTTCTTAGCGATTGGCATTATCACATAATGAATTATACATTATATAAAGTAATGTGATTTCTTCGAAGAATATACTAAAAAATGAGCAGGCAAGATAAACGAAGGCAAAGacaaaagctggagctctagta-3’ |
|||
|
HIS3 integration primer Rv |
5’-AAAGAAAAAGCAAAAAGAAAAAAGGAAAGCGCGCCTCGTTCAGAATGACACGTATAGAATGATGCATTACCTTGTCATCTTCAGTATCATACTGTTCGTATACATACTTACTGACATTCATAGGTATACATATATACACATGTATATATATCGTATGCTGCAGCTTTAAATAATCGGTGTCAgcgaattgggtaccggcc-3’ |
|||
Acknowledgments
Acknowledgments
The authors thank the Carvunis Lab members, especially Carly Houghton, Lin Chou, Saurin Parikh and Aaron Wacholder at the Department of Computational and Systems Biology at the University of Pittsburgh School of Medicine for the lectures, insightful discussions and support during the “Adopt a Proto-gene” Summer internship during which we generated the results described here.
References
- Ashburner Michael, Ball Catherine A., Blake Judith A., Botstein David, Butler Heather, Cherry J. Michael, Davis Allan P., Dolinski Kara, Dwight Selina S., Eppig Janan T., Harris Midori A., Hill David P., Issel-Tarver Laurie, Kasarskis Andrew, Lewis Suzanna, Matese John C., Richardson Joel E., Ringwald Martin, Rubin Gerald M., Sherlock Gavin. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000 May 1;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, M.J., Gartenberg, M.R., and Brown, G.W. (2015). Methods in Yeast Genetics and Genomics: a Cold Spring Harbor Laboratory Course Manual (Cold Spring Harbor Laboratory Press)
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., Oliver S. G. Life with 6000 Genes. Science. 1996 Oct 25;274(5287):546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Käll Lukas, Krogh Anders, Sonnhammer Erik L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. Journal of Molecular Biology. 2004 May 1;338(5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E. L.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Research. 2007 May 8;35(Web Server):W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Zeming, Akin Halil, Rao Roshan, Hie Brian, Zhu Zhongkai, Lu Wenting, Smetanin Nikita, Verkuil Robert, Kabeli Ori, Shmueli Yaniv, dos Santos Costa Allan, Fazel-Zarandi Maryam, Sercu Tom, Candido Salvatore, Rives Alexander. Evolutionary-scale prediction of atomic level protein structure with a language model. 2022 Jul 21; doi: 10.1101/2022.07.20.500902. [DOI] [PubMed]
- McIsaac R. Scott, Oakes Benjamin L., Wang Xin, Dummit Krysta A., Botstein David, Noyes Marcus B. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Research. 2012 Dec 25;41(4):e57–e57. doi: 10.1093/nar/gks1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira Makoto J., Hendrickson David G., Scott McIsaac R., Rhind Nicholas. An estradiol‐inducible promoter enables fast, graduated control of gene expression in fission yeast. Yeast. 2017 Jun 8;34(8):323–334. doi: 10.1002/yea.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Xiaoyong, Li Hao, Zeng Tao, Li Zhandong, Chen Lei, Huang Tao, Cai Yu-Dong. Identification of Protein Subcellular Localization With Network and Functional Embeddings. Frontiers in Genetics. 2021 Jan 20;11 doi: 10.3389/fgene.2020.626500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich April, Acar Omer, Carvunis Anne-Ruxandra. Massively integrated coexpression analysis reveals transcriptional regulation, evolution and cellular implications of the noncanonical translatome. 2023 Mar 17; doi: 10.1101/2023.03.16.533058. [DOI] [PMC free article] [PubMed]
- Saccharomyces Genome Database | SGD n.d. https://www.yeastgenome.org/ (accessed January 20, 2021).
- The Gene Ontology Consortium. Aleksander Suzi A, Balhoff James, Carbon Seth, Cherry J Michael, Drabkin Harold J, Ebert Dustin, Feuermann Marc, Gaudet Pascale, Harris Nomi L, Hill David P, Lee Raymond, Mi Huaiyu, Moxon Sierra, Mungall Christopher J, Muruganugan Anushya, Mushayahama Tremayne, Sternberg Paul W, Thomas Paul D, Van Auken Kimberly, Ramsey Jolene, Siegele Deborah A, Chisholm Rex L, Fey Petra, Aspromonte Maria Cristina, Nugnes Maria Victoria, Quaglia Federica, Tosatto Silvio, Giglio Michelle, Nadendla Suvarna, Antonazzo Giulia, Attrill Helen, dos Santos Gil, Marygold Steven, Strelets Victor, Tabone Christopher J, Thurmond Jim, Zhou Pinglei, Ahmed Saadullah H, Asanitthong Praoparn, Luna Buitrago Diana, Erdol Meltem N, Gage Matthew C, Ali Kadhum Mohamed, Li Kan Yan Chloe, Long Miao, Michalak Aleksandra, Pesala Angeline, Pritazahra Armalya, Saverimuttu Shirin C C, Su Renzhi, Thurlow Kate E, Lovering Ruth C, Logie Colin, Oliferenko Snezhana, Blake Judith, Christie Karen, Corbani Lori, Dolan Mary E, Drabkin Harold J, Hill David P, Ni Li, Sitnikov Dmitry, Smith Cynthia, Cuzick Alayne, Seager James, Cooper Laurel, Elser Justin, Jaiswal Pankaj, Gupta Parul, Jaiswal Pankaj, Naithani Sushma, Lera-Ramirez Manuel, Rutherford Kim, Wood Valerie, De Pons Jeffrey L, Dwinell Melinda R, Hayman G Thomas, Kaldunski Mary L, Kwitek Anne E, Laulederkind Stanley J F, Tutaj Marek A, Vedi Mahima, Wang Shur-Jen, D’Eustachio Peter, Aimo Lucila, Axelsen Kristian, Bridge Alan, Hyka-Nouspikel Nevila, Morgat Anne, Aleksander Suzi A, Cherry J Michael, Engel Stacia R, Karra Kalpana, Miyasato Stuart R, Nash Robert S, Skrzypek Marek S, Weng Shuai, Wong Edith D, Bakker Erika, Berardini Tanya Z, Reiser Leonore, Auchincloss Andrea, Axelsen Kristian, Argoud-Puy Ghislaine, Blatter Marie-Claude, Boutet Emmanuel, Breuza Lionel, Bridge Alan, Casals-Casas Cristina, Coudert Elisabeth, Estreicher Anne, Livia Famiglietti Maria, Feuermann Marc, Gos Arnaud, Gruaz-Gumowski Nadine, Hulo Chantal, Hyka-Nouspikel Nevila, Jungo Florence, Le Mercier Philippe, Lieberherr Damien, Masson Patrick, Morgat Anne, Pedruzzi Ivo, Pourcel Lucille, Poux Sylvain, Rivoire Catherine, Sundaram Shyamala, Bateman Alex, Bowler-Barnett Emily, Bye-A-Jee Hema, Denny Paul, Ignatchenko Alexandr, Ishtiaq Rizwan, Lock Antonia, Lussi Yvonne, Magrane Michele, Martin Maria J, Orchard Sandra, Raposo Pedro, Speretta Elena, Tyagi Nidhi, Warner Kate, Zaru Rossana, Diehl Alexander D, Lee Raymond, Chan Juancarlos, Diamantakis Stavros, Raciti Daniela, Zarowiecki Magdalena, Fisher Malcolm, James-Zorn Christina, Ponferrada Virgilio, Zorn Aaron, Ramachandran Sridhar, Ruzicka Leyla, Westerfield Monte, Aleksander Suzi A, Balhoff James, Carbon Seth, Cherry J Michael, Drabkin Harold J, Ebert Dustin, Feuermann Marc, Gaudet Pascale, Harris Nomi L, Hill David P, Lee Raymond, Mi Huaiyu, Moxon Sierra, Mungall Christopher J, Muruganugan Anushya, Mushayahama Tremayne, Sternberg Paul W, Thomas Paul D, Van Auken Kimberly, Ramsey Jolene, Siegele Deborah A, Chisholm Rex L, Fey Petra, Aspromonte Maria Cristina, Nugnes Maria Victoria, Quaglia Federica, Tosatto Silvio, Giglio Michelle, Nadendla Suvarna, Antonazzo Giulia, Attrill Helen, dos Santos Gil, Marygold Steven, Strelets Victor, Tabone Christopher J, Thurmond Jim, Zhou Pinglei, Ahmed Saadullah H, Asanitthong Praoparn, Luna Buitrago Diana, Erdol Meltem N, Gage Matthew C, Ali Kadhum Mohamed, Li Kan Yan Chloe, Long Miao, Michalak Aleksandra, Pesala Angeline, Pritazahra Armalya, Saverimuttu Shirin C C, Su Renzhi, Thurlow Kate E, Lovering Ruth C, Logie Colin, Oliferenko Snezhana, Blake Judith, Christie Karen, Corbani Lori, Dolan Mary E, Drabkin Harold J, Hill David P, Ni Li, Sitnikov Dmitry, Smith Cynthia, Cuzick Alayne, Seager James, Cooper Laurel, Elser Justin, Jaiswal Pankaj, Gupta Parul, Jaiswal Pankaj, Naithani Sushma, Lera-Ramirez Manuel, Rutherford Kim, Wood Valerie, De Pons Jeffrey L, Dwinell Melinda R, Hayman G Thomas, Kaldunski Mary L, Kwitek Anne E, Laulederkind Stanley J F, Tutaj Marek A, Vedi Mahima, Wang Shur-Jen, D’Eustachio Peter, Aimo Lucila, Axelsen Kristian, Bridge Alan, Hyka-Nouspikel Nevila, Morgat Anne, Aleksander Suzi A, Cherry J Michael, Engel Stacia R, Karra Kalpana, Miyasato Stuart R, Nash Robert S, Skrzypek Marek S, Weng Shuai, Wong Edith D, Bakker Erika, Berardini Tanya Z, Reiser Leonore, Auchincloss Andrea, Axelsen Kristian, Argoud-Puy Ghislaine, Blatter Marie-Claude, Boutet Emmanuel, Breuza Lionel, Bridge Alan, Casals-Casas Cristina, Coudert Elisabeth, Estreicher Anne, Livia Famiglietti Maria, Feuermann Marc, Gos Arnaud, Gruaz-Gumowski Nadine, Hulo Chantal, Hyka-Nouspikel Nevila, Jungo Florence, Le Mercier Philippe, Lieberherr Damien, Masson Patrick, Morgat Anne, Pedruzzi Ivo, Pourcel Lucille, Poux Sylvain, Rivoire Catherine, Sundaram Shyamala, Bateman Alex, Bowler-Barnett Emily, Bye-A-Jee Hema, Denny Paul, Ignatchenko Alexandr, Ishtiaq Rizwan, Lock Antonia, Lussi Yvonne, Magrane Michele, Martin Maria J, Orchard Sandra, Raposo Pedro, Speretta Elena, Tyagi Nidhi, Warner Kate, Zaru Rossana, Diehl Alexander D, Lee Raymond, Chan Juancarlos, Diamantakis Stavros, Raciti Daniela, Zarowiecki Magdalena, Fisher Malcolm, James-Zorn Christina, Ponferrada Virgilio, Zorn Aaron, Ramachandran Sridhar, Ruzicka Leyla, Westerfield Monte. The Gene Ontology knowledgebase in 2023. GENETICS. 2023 Mar 3;224(1) doi: 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumuluri Vineet, Almagro Armenteros José Juan, Johansen Alexander Rosenberg, Nielsen Henrik, Winther Ole. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Research. 2022 Apr 30;50(W1):W228–W234. doi: 10.1093/nar/gkac278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam Sipko, Võsa Urmo, van der Graaf Adriaan, Franke Lude, de Magalhães João Pedro. Gene co-expression analysis for functional classification and gene–disease predictions. Briefings in Bioinformatics. 2017 Jan 10;:bbw139–bbw139. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacholder A, Parikh SB, Coelho NC, Acar O, Houghton C, Chou L, Carvunis AR. A vast evolutionarily transient translatome contributes to phenotype and fitness. Cell Syst. 2023 May 17;14(5):363-381.e8. [DOI] [PMC free article] [PubMed]
- Zhou Chuankai, Slaughter Brian D., Unruh Jay R., Guo Fengli, Yu Zulin, Mickey Kristen, Narkar Akshay, Ross Rhonda Trimble, McClain Melainia, Li Rong. Organelle-Based Aggregation and Retention of Damaged Proteins in Asymmetrically Dividing Cells. Cell. 2014 Oct 1;159(3):530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]