Abstract
arfI encoded the 57.7-kDa subunit of Cytophaga xylanolytica arabinofuranosidase I (ArfI). arfII encoded a 59.2-kDa subunit of ArfII. Products of both cloned genes liberated arabinose from arabinan and arabinoxylan. The deduced amino acid sequences of ArfI and ArfII revealed numerous regions that were identical to each other and to regions of homologous proteins from Bacteroides ovatus, Bacillus subtilis, and Clostridium stercorarium.
As was reported previously (7), oat spelt arabinoxylan-grown Cytophaga xylanolytica XM3 produced up to 15 electrophoretically separable endoxylanases but only a single α-l-arabinofuranosidase (ARAF) activity, which was purified, characterized, and referred to as ArfI.
During the initial stages of purification of ArfI, there was a parallel effort to clone and sequence the gene encoding it. The first approach used was to shotgun clone (8) the ArfI-encoding gene into Escherichia coli, a strategy that was seemingly successful as it readily yielded an E. coli clone expressing ARAF activity. However, when ArfI was finally purified (7), it became apparent that the gene that had been cloned did not encode ArfI, because none of the amino acid sequences of the four internal peptide fragments of ArfI matched the amino acid sequence deduced from the clone. This indicated that C. xylanolytica contained more than one ARAF-encoding gene, including the gene that was initially cloned and expressed by E. coli but not expressed by C. xylanolytica under our growth conditions.
In this paper, we describe the cloning and sequencing of two ARAF-encoding genes from C. xylanolytica, the ARAF-encoding gene cloned initially (which we refer to as arfII) and the authentic ArfI-encoding gene (designated arfI) that was subsequently obtained by a PCR walking technique.
Bacterial strains and growth conditions.
C. xylanolytica XM3 (= DSM 6779) was grown anaerobically at 30°C on oat spelt arabinoxylan as previously described (7). E. coli strains were routinely grown in Luria-Bertani (LB) broth at 37°C with shaking (8). E. coli DH5αF′ and TOP10F′ were used as the recipient strains for recombinant plasmids pUC19 (4) and pCR2.1 (TA cloning kit; Invitrogen, Carlsbad, Calif.), respectively. To select for plasmid-containing transformants of E. coli, ampicillin was included in media at a concentration of 100 μg · ml−1. Solid media contained 15 g of agar per liter.
Cell extracts and enzyme assays.
Soluble cell extracts of E. coli were prepared by sonication and centrifugation. These extracts were used either without further treatment (to determine the specific activities of the arabinofuranosidases with p-nitrophenylarabinoside as the substrate) or after concentration by ultrafiltration and dialysis (to determine arabinose-releasing ability with sugar beet arabinan or rye or wheat arabinoxylans as the substrates). The methods used for these procedures have been described previously (7).
Isolation and manipulation of DNA.
C. xylanolytica XM3 genomic DNA and E. coli plasmid DNAs were isolated by using genomic and plasmid DNA isolation kits (Qiagen, Valencia, Calif.) according to the manufacturer’s instructions. All PCR amplicons were purified with a QIAquick gel extraction kit (Qiagen) used according to the manufacturer’s instructions. Standard procedures were used to digest DNA with restriction enzymes, to separate the fragments by gel electrophoresis, and to transfer the fragments to nylon membrane filters (8). Southern blots were prepared by using probe DNA labeled with a digoxigenin DNA labeling and detection kit (Boehringer Mannheim, Indianapolis, Ind.) used according to the manufacturer’s instructions.
Amino acid sequences of ArfI peptides.
To determine the partial amino acid sequence of ArfI, purified ArfI (ca. 15 μg per lane) was electrophoresed on a 16% (wt/vol) sodium dodecyl sulfate–polyacrylamide gel electrophoresis resolving gel with a 4% (wt/vol) polyacrylamide stacking gel and blotted onto an Immobilon PSQ membrane as described previously (7). After blotting, the membrane was rinsed with H2O, soaked in 100% methanol, stained with 0.2% amido black in 40% methanol for 30 s, and destained with multiple changes of H2O. The band corresponding to the position of ArfI in each lane of the membrane was excised with a clean, sterile razor blade and placed in a sterile Eppendorf tube. The individual ArfI-containing membrane fragments were sent to the Worcester Foundation for Biomedical Research (Shrewsbury, Mass.) for sequence determination. Upon receipt, ArfI was digested with trypsin, and the oligopeptide fragments that were released were purified by reversed-phase high-performance liquid chromatography. The N-terminal amino acid sequences of three such fragments were then determined by the Edman degradation method.
Separate 48-μg samples of ArfI were also digested with Endoproteinase Lys-C as recommended by the manufacturer (Boehringer Mannheim), and one of the resulting peptides was sequenced at the Michigan State University Macromolecular Sequence Facility.
Cloning of arfI and arfII.
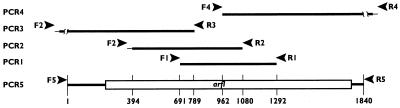
arfI was cloned by the PCR walking technique (Table 1 and Fig. 1). To do this, the amino acid sequences of three trypsin-generated fragments of ArfI were aligned with similar regions in the deduced amino acid sequence of an ARAF from Bacteroides ovatus V975 (asdII gene product; GenBank accession no. U15179 [10]), a protein to which the trypsin fragments were similar as determined by BLASTp analysis (1). Based on this alignment, the two trypsin fragments (peptides 1 and 3) (Fig. 2) presumed to be farthest apart in ArfI were used to design degenerate primers F1 and R1 for PCR 1, in which genomic DNA from C. xylanolytica was used as the template (all primer sequences are shown in Table 1). The resulting 599-bp amplified product (amplicon) was inserted into cloning vector pCR2.1 and sequenced. Based on the nucleotide sequence of the first amplicon, a new primer (primer R2) was designed and used in PCR 2, in which a pUC19 library of EcoRI-restricted C. xylanolytica genomic DNA was used as the template. The forward primer for this reaction (primer F2) corresponded to a region of pUC19 about 70 bp away from its own EcoRI restriction site, and the sequence of the resulting amplicon was aligned with the sequence of the homologous region from the previous PCR. Analogous procedures were used for PCR 3 and 4; in these PCR pUC19 libraries of C. xylanolytica DNA digested with KpnI and HindIII, respectively, were used as the templates.
TABLE 1.
Templates and primers used in the PCR walking technique to clone and sequence arfI
| PCR | Template | PCR primers
|
|
|---|---|---|---|
| Namea | Sequenceb | ||
| 1 | C. xylanolytica genomic | F1 | 5′-GAR GCN GCN CAR TGG GT-3′ (forward) |
| DNA | R1 | 5′-GCR TTR TTR TTR AAD ATR TT-3′ (reverse) | |
| 2 | pUC19-EcoRI libraryc | F2 | 5′-CAG TCA CGA CGT TGT AAA ACG ACG GC-3′ (forward) |
| R2d | 5′-CCA AGT TTG ATG CCA CGA CTG TTC-3′ (reverse) | ||
| 3 | pUC19-KpnI libraryc | F2 | 5′-CAG TCA CGA CGT TGT AAA ACG ACG GC-3′ (forward) |
| R3 | 5′-GAT ATC CCA GGG TTT ATC ACG ACC G-3′ (reverse) | ||
| 4 | pUC19-HindIII libraryc | F4 | 5′-CGC AGC ATC ACA AGT TCC TCA TGC-3′ (forward) |
| R4 | 5′-TCA CAC AGG AAA CAG CTA TGA CCA TG-3′ (reverse) | ||
| 5 | C. xylanolytica genomic | F5 | 5′-GGT CGT TGG TGA AAT ACA GCG G-3′ (forward) |
| DNA | R5 | 5′-GAC AAA TCG CTC CCA CCG AAC AC-3′ (reverse) | |
The priming sites of F2 and R4 complemented regions within the multiple cloning site of pUC19.
A, adenosine; T, thymidine; G, guanosine; C, cytosine; R, adenosine or guanosine; N, adenosine, thymidine, cytosine, or guanosine; D, adenosine, thymidine, or guanosine.
Library of C. xylanolytica genomic DNA created by restriction with an enzyme and ligated into pUC19.
The second and fourth nucleotides of primer R2 (cytosine and adenosine, respectively) were identified incorrectly in the amplicon from PCR 1 and did not correspond to the homologous nucleotides in amplicons from PCR 4 and 5. However, their distances from the 3′ end did not compromise the efficacy of R2 as a primer for PCR 2.
FIG. 1.
PCR walking procedure used to clone and sequence arfI. Amplicons from PCR 1 through 5 were generated by using primer sets F1-R1 through F5-R5 (arrowheads), whose sequences (Table 1) were complementary to the ends of the amplicons indicated. The thick lines represent nucleotide sequences of C. xylanolytica DNA; the thin lines represent pUC19 vector DNA sequences. Breaks in the amplicons from PCR 3 and 4 represent portions of C. xylanolytica DNA that were sequenced but lie outside the PCR 5 amplicon. The putative ORF corresponding to arfI is represented by the box in the amplicon from PCR 5. The numbers at the bottom (drawn to scale) indicate nucleotide positions in amplicon PCR 5.
FIG. 2.
CLUSTAL W comparison of the deduced amino acid sequences of the arfI gene product (ArfI) and the arfII gene product (ArfII) with the amino acid sequences of homologous enzymes from B. ovatus V975 (asdII gene product), C. stercorarium (C. sterc.) (arfB gene product), and B. subtilis (putative arabinosidase). The boxes indicate regions where the level of sequence identity was ≥80%. Dashes in a sequence indicate gaps. The numbers to the left and right of each row indicate the numbers of the first and last amino acids, respectively, in the row. The shaded areas in the ArfI sequence correspond to internal peptides 1 (positions 155 to 175), 2 (positions 188 to 212), and 3 (positions 339 to 356) generated by digestion of ArfI with trypsin and internal peptide 4 (positions 269 to 288) generated by digestion with Endoproteinase Lys-C.
After PCR 3 and 4, transcription initiation and termination codons for the putative open reading frame (ORF) encoding ArfI (i.e., arfI) were recognizable in each amplicon. Therefore, one final PCR (PCR 5) was performed by using unrestricted C. xylanolytica genomic DNA as the template and forward primer F5 and reverse primer R5 corresponding to portions of the regions flanking the putative arfI gene. The resulting 1,839-bp amplicon (with an additional A overhang at each 3′ end) was cloned into pCR2.1 with the TA cloning kit (see above) and was transformed into E. coli TOP10F′, which then expressed ARAF activity. Both strands of this cloned fragment were then sequenced.
All PCR mixtures (total volume, 100 μl) contained 2 mM MgCl2, each deoxynucleotide triphosphate at a concentration of 0.2 mM, 24 pmol of each primer, 380 ng of template DNA, and 2.5 U of Taq DNA polymerase (Gibco BRL, Grand Island, N.Y.). PCR were performed for 30 cycles, with each cycle consisting of denaturation at 94°C for 2 min (initial cycle) or 30 s (remaining 29 cycles), annealing at 53°C (PCR 1) or 60°C (PCR 2 to 5) for 30 s, and extension at 72°C for 45 s (PCR 1), 1 min (PCR 2 to 4), or 3 min (PCR 5).
Shotgun cloning of arfII was initiated by partially digesting C. xylanolytica genomic DNA with EcoRI and then ligating the resulting DNA fragments with T4 DNA ligase into EcoRI-restricted, dephosphorylated pUC19. The ligation products were used to transform competent cells of E. coli DH5αF′ (8). Transformants were screened on LB agar plates containing ampicillin and 20 μg of 4-methylumbelliferyl-α-l-arabinofuranoside (MU-AF) per ml. Three colonies of ARAF-positive transformants (out of ca. 6,200 transformants examined) were identified by their ability to release methylumbelliferone from MU-AF, which gave them a UV-fluorescent halo. These organisms were restreaked onto LB agar to ensure that they were pure and then rescreened by growing them overnight in microtiter plates containing (per well) 300 μl of LB broth supplemented with ampicillin and MU-AF. Positive clones were found to contain a 9-kb insert, which could be more completely digested with EcoRI to yield fragments of about 7 kb and 1,940 bp (see below), the latter of which (when subcloned into pUC19) still conferred ARAF activity on E. coli DH5α F′ transformants. Both strands of the 1,940-bp insert containing the ARAF-encoding gene were sequenced.
All nucleotide sequencing was done with an automated fluorescence sequencer by personnel at the Michigan State University DNA Sequencing Facility. DNA sequences were assembled and edited by using Sequencher (Gene Codes Corporation, Ann Arbor, Mich.). The arfI and arfII nucleotide sequences and the deduced amino acid sequences were compared to sequences from appropriate databases by using BLAST (1). The amino acid sequences which were determined and deduced were aligned and compared by using the CLUSTAL W program (9).
Sequences of arfI and arfII and their translation products.
The putative ORF of arfI begins with the initiation codon ATG at position 231 and ends at position 1,763 with the stop codon TAA. The 509-amino-acid sequence deduced from this 1,533-bp ORF is shown in Fig. 2 (as ArfI) and corresponds to a 57.7-kDa polypeptide. This size agrees well with the subunit size of ArfI determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (56 kDa) (7) and suggests that the native ArfI enzyme is a tri- or tetramer consisting of similar, if not identical, subunits. Consistent with this interpretation were the amino sequences of three trypsin-generated fragments of ArfI (two of which [peptides 1 and 3] were used to design primers for PCR 1) and a fourth peptide fragment (peptide 4) generated by Endoproteinase Lys-C digestion of ArfI, all of which were present in the arfI gene product (Fig. 2). The amino acid sequence of each peptide fragment determined by Edman degradation was identical to the amino acid sequence deduced from the corresponding nucleotide sequence, except for the following amino acids in peptide 4: the deduced T at position 282, which was reported as I; and W and T at positions 279 and 286, respectively, which were not unambiguously resolved after Edman degradation.
The 1,940-bp EcoRI restriction fragment containing the putative arfII gene was verified to originate from C. xylanolytica DNA by using Southern hybridization, which showed that the digoxigenin-labeled fragment hybridized as a single band to an EcoRI digest of C. xylanolytica genomic DNA (data not shown). The putative ORF of arfII begins with the initiation codon ATG at position 293 and ends at position 1,897 with the stop codon TGA. The 534 amino acids encoded by this 1,605-bp ORF are also shown in Fig. 2 (as ArfII) and correspond to a 59.2-kDa polypeptide. It is noteworthy that there is a 48-amino-acid sequence at the N terminus of ArfII, which resembles a standard signal peptide of secreted proteins in having basic amino acids at the N terminus (in this case, K or R at positions 2 to 4, 9, and 10), followed by a domain (residues 11 to 27) in which 14 of 17 amino acids are hydrophobic (5). The proline residue at position 28 may represent the beginning of a “C domain,” which is ultimately cleaved by a peptidase between the glycine and proline residues at positions 47 and 48, respectively. If this interpretation is correct, it suggests that when and if conditions which permit expression of arfII by C. xylanolytica are found, ArfII is likely to be found in the periplasm of cells and/or the extracellular growth medium.
Also present on the 1,940-bp fragment was a GAAA sequence just upstream from arfII at positions −8 to −5; this sequence represents a potential Shine-Dalgarno sequence. Moreover, a TATAAAT sequence at positions −16 to −10 and a TTGATG sequence at positions −37 to −32 resembled the −10 and −35 consensus sequences observed at RNA polymerase binding sites (6).
When arfI and arfII were expressed by E. coli, they conferred ARAF activity capable of releasing both p-nitrophenol and methylumbelliferone from the respective α-l-arabinofuranoside derivatives. The specific activities of cell extracts on p-nitrophenyl-α-l-arabinoside were relatively low (1.96 and 1.61 nmol hydrolyzed · min−1 · mg of protein−1 for the arfI and arfII gene products, respectively); however, they were 80- to 100-fold greater than the specific activities of cell extracts from E. coli hosts containing no cloning vector or containing a vector without an insert. Moreover, the arfI and arfII gene products made in E. coli were capable of liberating arabinose from sugar beet arabinan and rye and wheat arabinoxylans.
Relationship of ArfI and ArfII to other ARAFs.
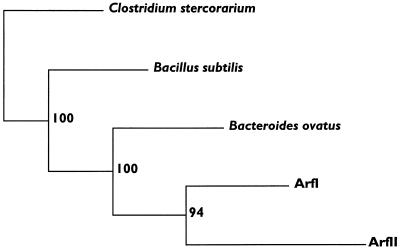
BLASTp analysis of the deduced amino acid sequences of ArfI and ArfII revealed significant similarities (smallest sum probability, ≤10−13) to ARAFs from (in order of similarity) B. ovatus V975 (asdII gene product; GenBank accession no. U15179), Clostridium stercorarium (arfB gene product; GenBank accession no. AF002664), Bacillus subtilis (putative arabinosidase and putative ARAF; GenBank accession no. Z75208 and X89810, respectively), Streptomyces lividans (AbfA; GenBank accession no. U04630), and B. ovatus (asdI gene product; GenBank accession no. U15178). A CLUSTAL W alignment of ArfI and ArfII with the first three proteins mentioned above revealed numerous regions of conserved amino acids (Fig. 2). Based on this alignment, a consensus tree of these ARAFs was generated by parsimony analysis (Fig. 3). The bootstrap values indicate that the evolutionary relationships shown in the tree are strongly supported by the data. Not surprisingly, ArfI and ArfII are most closely related evolutionarily to the asdII gene product of B. ovatus, a member of the same 16S rRNA phylogenetic group as C. xylanolytica (i.e., the “Bacteroides group” of the Flexibacter-Cytophaga-Bacteroides phylum [3]). However, compared to ArfI, ArfII has diverged further from the hypothetical common ancestor. This may reflect its more specialized purpose in C. xylanolytica, a notion supported by its probable export out of the cellular compartment (i.e., cytoplasm) occupied by ArfI.
FIG. 3.
Consensus tree showing the evolutionary relationships among the sequences shown in Fig. 2. One hundred randomly sampled replications of the data set were created with SEQBOOT and were subjected to parsimony analysis with PROTPARS. A majority rule consensus tree was generated from the 100 output trees with CONSENSE. Bootstrap values that indicate the number of times that a given cluster was formed are to the right of each node. The lengths of the horizontal lines represent the relative rates of divergence. All of the programs used for this analysis were part of the PHYLIP package (2).
The results of this study expand the limited database of prokaryotic genes relevant to xylan degradation, and the genes examined in this study are the first such genes to be cloned from any species belonging to the genus Cytophaga, a group of gliding bacteria widely known for (but poorly studied with respect to) biopolymer degradation. The results of this study also underscore the potential danger of making conclusions about gene-enzyme relationships unless both entities are examined individually. Were it not for the efforts to purify and characterize the ArfI protein (7), it might have been concluded that the ARAF activity of C. xylanolytica was due to expression of arfII, the first gene which was cloned and sequenced but a gene which could conceivably be silent in this bacterium.
Nucleotide sequence accession numbers.
The sequence of the 1,839-bp clone containing arfI has been deposited in the GenBank database under accession no. AF028018. The nucleotide sequence of the 1,940-bp EcoRI restriction fragment containing the putative arfII gene has been deposited in the GenBank database under accession no. AF028019.
Acknowledgments
This research was supported by grant DE-FG02-94ER20141 from the U.S. Department of Energy and by grant BIR91-20006 from the National Science Foundation to the Michigan State University Center for Microbial Ecology.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1989. [Google Scholar]
- 3.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messing J F. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 5.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Record M T J, Reznikoff W S, Craig M L, McQuade K I, Schlax P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss R I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 7.Renner M J, Breznak J A. Purification and properties of ArfI, an α-l-arabinofuranosidase from Cytophaga xylanolytica. Appl Environ Microbiol. 1998;64:43–52. doi: 10.1128/aem.64.1.43-52.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 9.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehead T R. Nucleotide sequences of xylan-inducible xylanase and xylosidase/arabinosidase genes from Bacteroides ovatus V975. Biochim Biophys Acta. 1995;1244:239–241. doi: 10.1016/0304-4165(95)00051-c. [DOI] [PubMed] [Google Scholar]