Abstract
Current literature suggests PFAS carbon chain length may be a predictive variable of toxicity. If so, statistical modeling may be used to help predict toxicity, thus improving the efficiency of PFAS regulation development. Data were analyzed using one-way ANOVAs, Tukey’s HSD post hoc tests, and simple linear regressions. A dataset was predicted using modeling from this data. Analysis indicated that 11 of 15 health outcomes showed significant differences in mean values. Two of 15 health outcomes were analyzed using simple linear regressions, with statistically significant results. After predictive modeling generated a theoretical dataset, unpaired t-tests comparing the results of an actual dataset indicated no significant differences among the mean values of the two health outcomes. Therefore, predictive statistical modeling may be used to predict health outcomes for PFAS exposure.
Keywords: PFAS toxicity, predictive toxicity, computational toxicology, PFAS carbon chain length, PFAS modeling, toxicity modeling, QSAR
Introduction
Perfluoroalkyl and polyfluoroalkyl substances (PFAS) are a class of chemicals exhibiting similar molecular structures (i.e., at least with one aliphatic per-fluorocarbon moiety), environmental properties, and biological hazards (Kwiatkowski et al. 2020). The exact number of PFAS is unknown, although 4,730 have currently been identified (Organisation for Economic Co-operation and Development 2018). PFAS can be described as either “long-chained” or “short-chained”, although these descriptions vary by type of PFAS and the individual chemical structure. For example, perfluoroalkyl carboxylic acids with at least seven carbon atoms and perfluoroalkane sulfonic acids with at least six carbon atoms are considered long-chained (Buck et al. 2011; Wang et al. 2017). Used mainly as industrial chemicals, flame retardants, and in food packaging, PFAS are ubiquitous (Ding et al. 2020; Pelch et al. 2019). Due to their strong carbon-fluorine bonds, they pose a concern to the environment (Cousins et al. 2020; Death et al. 2021) and for potential toxicity to humans (Ding et al. 2020; Cousins et al. 2020; Pelch et al. 2019; Fenton et al. 2020).
Human health threats associated with PFAS exposure have been found from both epidemiological and toxicological studies. Some detrimental health effects associated with PFAS exposure include impaired immune function, impaired thyroid function, including thyroid disease and increased thyroid stimulating hormone (TSH), liver disease and cancer, insulin dysregulation, kidney disease and cancer, impaired sperm mobility, testicular cancer, and developmental effects, including reduced birth weight and neurodevelopmental effects (Fenton et al. 2020). Furthermore, PFAS are associated with endocrine disturbing effects and may affect female fertility and reproductive health outcomes (Ding et al. 2020).
Although they are a chemical class, currently, PFAS have historically been often regulated on a substance-by-substance basis (Kwiatkowski et al. 2020). There is much debate surrounding the current approaches to PFAS regulation, with arguments favoring a regulatory model in which PFAS are evaluated as a chemical class, or possible subclasses (Kwiatkowski et al. 2020). Due to the high quantity of PFAS in existence and their potential to cause detrimental effects on the environment and humans, there is a demonstrated need for fast and accurate methods to produce data to assist in the regulation of PFAS.
In advocacy towards the need for a class-based regulatory model, in December 2019, a document, ‘Elements for an EU-Strategy for PFASs’, was published by the European Union. The document highlights the premises that there are far too many PFAS to implement a substance-by-substance regulatory model that is both temporally and fiscally effective, most PFAS lack a Chemical Abstract Service (CAS) number and have unclear composition, and there is insufficient information about their chemical properties. While it is suggested that some PFAS may be less toxic than others, because many PFAS exhibit toxicity and the full extent and scope of potential effects on human health from the combination of exposure to multiple PFAS are unknown, the entire chemical class is of concern. Based on these premises, the EU argues that regulation as a class avoids regrettable substitution and favors the protection of human health (European Union 2019).
Although, others argue that regulation of PFAS as a chemical class or chemical subclasses may not be feasible. For example, the Vermont General Assembly instructed the Vermont Department of Environmental Conservation to complete an assessment of the feasibility of regulating PFAS as a class of chemicals in public water systems. This action was based on the following alleged issues: 1) a purportedly inadequate amount of toxicological research, which is needed to create a framework for developing a class-based regulatory standard; 2) the existence of thousands of PFAS , while the ability of current laboratory methods purportedly to can only reliably identify ≈30 of the thousands of PFAS , leading to the inability to comprehensively quantify PFAS in drinking water; 3) the concern that treatment options to remove PFAS from drinking water could present new health risks; and 4) lack of regional and national coordination (Vermont Official State 2021). The Vermont General Assembly ultimately decided that it is not feasible for them to regulate PFAS as a chemical class at this time. (Vermont Official State 2021).
As such, it is vital to consider alternative approaches that enhance the ability to regulate these compounds. One such approach includes building statistical models to predict health outcomes from PFAS exposure. The number and structure of elements in chemical compounds are frequently used to predict many physical characteristics, such as boiling point, melting point, environmental fate, and toxicity (Wu and Wang, 2018).
Quantitative Structure-Activity Relationships (QSAR) use chemical structure parameters to predict several chemical properties (including toxicity) of families of compounds. The earliest of these QSAR methods, the Hansch approach (proposed in 1962) used linear regressions as its main statistical approach to create a predictive model (Wu and Wang 2018). The Free-Wilson method (1964) used the molecular structure as a predictive variable for regression analysis of physiological activity (Wu and Wang 2018).
While international regulatory bodies have typically relied upon in vivo and in vitro data, there is a recent movement towards the use of QSAR in regulation. The United States Environmental Protection Agency (USEPA) has released documentation detailing the consideration and possible advancements of the use of QSAR for pesticide regulation (Manibusan, et al. 2012). Furthermore, the EU’s European Chemical Agency (ECHA) welcomes the use of QSAR and other computational techniques, under strict restrictions including the scientific validity, reliability, and quality of the model (ECHA 2016).
The toxicity of PFAS is a result of their chemical structure. PFAS are organic fluorine compounds, which are characterized by the carbon-fluorine bond, the strongest bond found in organic chemistry, due largely to the high electronegativity of fluorine, (Cousins, et al. 2020). Additionally, in general, the longer the length of the carbon-chain, the less water-soluble is the PFAS (Gagliano et al. 2020). These properties result in PFAS demonstrating extreme persistence. Importantly, because the longer-chained PFAS have an increased number of these persistent C-F bonds, and lower rates of water solubility, they exhibit longer half-lives in living organisms (Chambers et al. 2021).
Due to these properties, there is a widely held belief that longer chained PFAS are more toxic (Cousins et al. 2020; Chambers et al. 2021). Using the concept of QSAR, there may be a mathematical basis between PFAS carbon chain length and toxicity. This basis could be used to build statistical models to predict adverse health outcomes, information that could supplement toxicological and epidemiological data in the evaluation and regulation of new and existing PFAS . This additional approach could increase the speed, reliability, and accuracy of future PFAS regulation. The present study describes an attempt to develop and test one such statistical model using the number of carbon atoms in PFAS to predict adverse health outcomes. The development and use of statistical modeling and QSAR in the advancement of the understanding of PFAS toxicity and their regulation is not yet well understood, making this a novel and important analysis.
The current analysis compares mean values of health outcomes between groups in exposed laboratory rats, with the hypothesis that mean values will differ significantly between groups. To advance the understanding of statistical modeling as a means of supplementing current methods of regulation, a predictive model is used to generate a dataset predicting health outcomes in rats exposed to a theoretical PFAS. The mean values of this generated dataset will be compared to mean values of an observed dataset of health outcomes of rats exposed to a PFAS of the same carbon chain length. The hypothesis is that the generated dataset will not statistically significantly differ between mean values of health outcomes, when compared to the observed dataset. These analyses will help provide evidence for the use of statistical modeling based on the chemical structure in the regulation of PFAS.
Clinical chemistry health outcomes serve as the health outcomes which will be used as the dependent variables in these analyzes. Clinical chemistry health outcomes are measured by analyzing bodily fluids, such as urine or blood. These measures are useful in epidemiological research, as they are continuous variables and often associated with adverse health outcomes (NTP 2022 a). Many clinical chemistry health outcomes are used to monitor and diagnosis adverse health outcomes. For example, cholesterol levels in the blood may be used to monitor risk of cardiovascular disease (Gjuladin-Hellon 2018), and total protein in urine may be used as a means to help diagnosis kidney disease (Bökenkamp 2020). Effects of PFAS exposure have been associated with changes in clinical chemistry levels in humans (NTP 2022 a).
Methods & Materials
Materials
Datasets sourced from the National Toxicology Program (NTP) contain data from 28-day long rat toxicity studies. Studies include TOX-96 of Perfluoroalkyl Sulfonates, which include (PFBS) and (PFOS) (NTP 2002 b). As well as TOX-97 of Perfluoroalkyl Carboxylates, which include (PFHxA), (PFOA), (PFNA, and (PFDA) (NTP 2022 c). Additional datasets from the TOX-96 study concerning PFHSKslt were used in the analyses (NTP 2022 b).
The rat species used for these experiments were Harlan Sprague Dawley rats. The animals were exposed to the substances by gavage, at five separate doses. Only the first three doses were used in this analysis, due to the excessive deaths leading to missing data in doses four and five. Rats were exposed to the substances for 28 days and then sacrificed and health outcomes were measured. 10 rats were included in each study at each dose. Only male rats were included in this analysis.
Health outcomes that were assessed in male rats during the initial study and included in the secondary analysis include alanine aminotransferase levels in IU/L, albumin levels in g/dL, alkaline phosphatase levels in IU/L, aspartate aminotransferase levels in IU/L, bile salts/acids levels in μmol/L, cholesterol levels in mg/dL, creatine kinase levels in IU/L, creatinine levels in mg/dL, direct bilirubin levels in mg/dL, serum glucose levels in mg/dL, sorbitol dehydrogenase levels in IU/L, total bilirubin levels in mg/dL, total protein levels in g/dL, triglycerides levels in mg/dL and urea nitrogen levels in mg/dL.
Methods
The analysis began with the use of one-way ANOVAs to compare group mean values between PFAS. Here type of PFAS is the independent variable, and clinical chemistry health outcome is the dependent variable. One-Way ANOVA was chosen as the hypothesis test because there is one categorical independent variable, one numeric dependent variable, there were more than 2 (6) groups, and data were normally distributed. A test was completed for each dose and each health outcome.
The statistical significance threshold, alpha, was set to .05 (5%). Bonferroni correction was applied to the .05 threshold as a multiple hypothesis correction. 45 ANOVAs were completed. .05/45= .001, therefore if p<.001 between all three doses for a health outcome, the difference between the mean values was considered statistically significant and a post hoc analysis was conducted. The post hoc analysis used for this analysis was Tukey’s Honestly Significant Difference (HSD) test. Tukey’s HSD post hoc test was completed to assess which pairs of PFAS have statistically significantly different mean values from one another. Results were considered statistically significant at p<.001. One test was completed for each statistically significant health outcome for each dose.
Furthermore, simple linear regression analyses were conducted to form a predictive model for health outcomes. A simple linear regression approach was used because the number of carbon atoms per PFAS served as the only predictor variable for these models, due to issues of multicollinearity in other variables considered as covariates. Additionally, both the response and predictor variables are quantitative. The statistical significance threshold, alpha, for simple linear regression models is set to <.05.
Linear regression is the most used model for identifying possible relationships between a response variable and predicator variables, as it is a simple model (Degregory et al. 2018). Therefore, the simple linear regression serves as a reliable starting point for the generation of a predictive model. There are a few assumptions that must be met before the implementing the use of a linear regression analysis including:
Linearity: The relationship between the independent variable and the mean of the dependent variable is linear.
Homoscedasticity: The variance of residual is the same for any value of the independent variable
Independence: The dependent variable has no relationship with the residuals.
Normality: The residuals must be approximately normally distributed..
Linearity of the variables were tested by calculating the correlation coefficient in R. This study held a requirement of at least a moderate linear relationship between variables at all doses, which is defined as a correlation coefficient of .40. Three of the 15 health outcomes met this assumption: alanine aminotransferase, creatine kinase, and sorbitol dehydrogenase. Homoscedasticity was tested by applying a Breusch-Pagan test to the three remaining health outcomes, with p<.05 indicating heteroscedasticity. Homoscedasticity was seen in two of the three health outcomes: creatine kinase and sorbitol hydrogenase. Values for these variables were normally distributed and it is assumed that there is no relationship between observations.
If statistical significance was found in all three doses for a health outcome, a dataset for a theoretical C6 PFAS was predicted. This was done by using the ‘predict’ function in R. The predict function is “a generic function for predictions from the results of various model fitting functions” (RStudio Team 2021). The function implements data previously inputted to produce predictive results. For linear regression models, the function uses the Y-intercept and coefficient to create predicted values for an unmeasured dependent value. In this case, linear regressions were conducted with clinical chemistry health outcomes as the response variable, and the number of carbon atoms per PFAS as the predictive variable. These analyzes produce a y-intercept and coefficient for each health outcome. Therefore, the model uses the value of the response variable at the y-intercept, the value of the coefficient, and a value for the predictive variable for which a prediction is to be made. In this case, 6 is used for the predictive variable, which is the carbon chain length of the theoretical PFAS , to project a value for the health outcome.
Linear regression formula:
Unpaired T-Tests were used to compare the mean values of the predicted health outcomes from the generated datasets to the mean values of health outcomes from existing datasets. The existing datasets came from experiments exposing rats to an actual C6 PFAS , PFHSKslt, sourced from the National Toxicology Program (NTP 2022 b).
Under the Safe Drinking Water Act, every five years, the United States Environmental Protection Agency (USEPA), releases a document, ‘Unregulated Contaminant Monitoring Rule (UCMR)’, which contains a list of unregulated drinking water contaminants that are to be monitored closely by public water systems. The fifth and most recent UCMR contained a list of 29 PFAS . Based on current technologies, this list purportedly contains each and every PFAS the USEPA is able to detect in drinking water (USEPA 2021; USEPA 2019; USEPA 2020). PFHSKslt is currently not listed on the UCMR because the USEPA claims to not have the technology to detect PFHSKslt in drinking water. Therefore, this molecule was chosen as the PFAS to be compared to a predicted value, as this purportedly cannot be detected in drinking water with current methods.
If the T-Test resulted in a p-value of more than .05, in all three doses, there was no statistically significant difference in the mean values, indicating the statistical model was successful at predicting a health outcome. An unpaired t-test was used because variables were continuous, data were normally distributed, and the mean values were collected from two independent groups. R software was used for every analysis.
Results
45 one-way ANOVAs were conducted to compare the effects of different PFAS on 15 male clinical chemistry health outcomes at three doses among laboratory rats. Differences in the mean values were observed in all three doses in male rats for the following health outcomes: alanine aminotransferase levels; albumin levels; alkaline phosphatase levels; aspartate aminotransferase levels; bile salts/acids levels; cholesterol levels; creatine kinase levels; direct bilirubin levels; sorbitol dehydrogenase levels; total protein levels; and urea nitrogen levels. Statistically significant differences were found in over 70% of the tested health outcomes. Given this information, the null hypothesis that there will be no statistically significant differences can be rejected.
The one-way ANOVAs revealed that at dose 1 results were not statistically significant for creatine levels (F (5, 54) = [1.75], p= [.14]), glucose levels (F (5, 54) = [2.54], p= [.04]), and total bilirubin levels (F (5, 54) = [1.87], p= [.12]), and at dose 2 for creatine levels (F (5, 53) = [4.54], p= [.002]), glucose levels (F (5, 53) = [4.24], p= [.002]), and triglycerides levels (F (5, 53) = [4.07], p= [.003]).
Moreover, one-way ANOVAs indicated that results were statistically significant at dose 1 for triglycerides levels (F (5, 54) = [6.88], p= [<.001]), dose 2 for total bilirubin levels (F (5, 53) = [11.36], p= [<.01]). Results at dose 3 for statistically significant for creatine levels (F (5, 54) = [10.06], p= [<.01]), glucose levels (F (5, 54) = [9.41], p= [<.001]), triglycerides levels (F (5, 54) = [7.91], p= [<.001]) and total bilirubin levels (F (5, 54) = [40.54], p= [<.01]).
A Tukey’s HSD post hoc test was conducted for each health outcome indicating statistically significant results between all 3 doses. Results of the post hoc analyses can be found in Table 5.
Table 5.
Results of Tukey’s Post Hoc Tests, p-values
| PFBS (C4) 62.6, 125, 250 |
PFOS (C8) 0.31, 0.63, 1.30 |
PFHxA (C6) 62.6, 150, |
PFOA (C8) 0.63, 1.3, 2.5 |
PFNA (C9) 0.63, 1.3, 2.5 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| PFOS (C8) |
AA | .57 | .38 | .52 | ||||||||||||
| A | 1.0 | 1.0 | .79 | |||||||||||||
| AK | .89 | 1.0 | .93 | |||||||||||||
| AsA | .76 | .67 | 1.0 | |||||||||||||
| B | 1.0 | 1.0 | .98 | |||||||||||||
| C | .05 | .35 | .85 | |||||||||||||
| CK | 1.0 | <.01 | 1.0 | |||||||||||||
| DB | 1.0 | .60 | 1.0 | |||||||||||||
| SD | 1.0 | .39 | 1.0 | |||||||||||||
| TP | 1.0 | .99 | <.05 | |||||||||||||
| U | 1.0 | .16 | .99 | |||||||||||||
|
| ||||||||||||||||
| PFHxA (C6) |
AA | .22 | 1.0 | .15 | .99 | .27 | .98 | |||||||||
| A | <.01 | <.001 | <.001 | <.001 | <.001 | <.01 | ||||||||||
| AK | .87 | .80 | .58 | 1.0 | .87 | .98 | ||||||||||
| AsA | .51 | .98 | .99 | 1.0 | .24 | 1.0 | ||||||||||
| B | .87 | 1.0 | 1.0 | .99 | 1.0 | 1.0 | ||||||||||
| C | .30 | .09 | <.001 | .96 | <.001 | <.05 | ||||||||||
| CK | .99 | .97 | 1.0 | .99 | .5 | .95 | ||||||||||
| DB | .61 | .96 | 1.0 | .37 | .16 | 1.0 | ||||||||||
| SD | .99 | 1.0 | .05 | .91 | .19 | .13 | ||||||||||
| TP | .68 | <.05 | .27 | .57 | <.05 | <.001 | ||||||||||
| U | 1.0 | .65 | .63 | .97 | .96 | .95 | ||||||||||
|
| ||||||||||||||||
| PFOA (C8) 0.63, 1.3, 2.5 |
AA | .33 | .46 | .62 | <.05 | 1.0 | <.05 | <.001 | .34 | <.01 | ||||||
| A | .38 | .54 | .26 | .38 | .28 | .94 | <.001 | <.001 | <.001 | |||||||
| AK | .37 | .93 | <.001 | <.05 | .89 | <.001 | .03 | .26 | <.001 | |||||||
| AsA | .59 | .74 | .20 | .05 | 1.0 | .09 | <.05 | .30 | .05 | |||||||
| B | .83 | .11 | 1.0 | .98 | .05 | .87 | 1.0 | .13 | .96 | |||||||
| C | <.001 | <.05 | 1.0 | <.001 | .85 | .92 | <.001 | <.001 | <.01 | |||||||
| CK | <.05 | .10 | .94 | <.05 | .88 | .99 | <.01 | .44 | .71 | |||||||
| DB | .86 | 1.0 | 1.0 | .97 | .76 | 1.0 | .09 | .88 | 1.0 | |||||||
| SD | <.001 | <.05 | <.05 | <.001 | .78 | <.01 | <.001 | <.05 | <.001 | |||||||
| TP | 1.0 | <.001 | .08 | <.05 | <.001 | <.001 | .37 | .82 | .99 | |||||||
| U | .26 | .21 | 1.0 | 0.14 | <.001 | 1.00 | .53 | <.01 | 0.81 | |||||||
|
| ||||||||||||||||
| PFNA (C9) |
AA | <.05 | <.001 | .86 | <.001 | .05 | .09 | <.001 | <.001 | .77 | .65 | .03 | .99 | |||
| A | <.01 | .45 | <.001 | <.01 | 0.74 | <.001 | <.001 | <.001 | <.001 | .38 | .01 | <.001 | ||||
| AK | <.05 | <.001 | .62 | <.01 | <.001 | .99 | <.01 | <.001 | 1.00 | .90 | <.001 | <.001 | ||||
| AsA | 0.16 | <.001 | <.001 | <.01 | <.001 | <.001 | <.01 | <.001 | <.001 | .96 | <.001 | <.001 | ||||
| B | <.01 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | ||||
| C | <.001 | 1.0 | <.001 | 0.11 | 0.64 | <.01 | <.05 | <.05 | .99 | <.05 | .09 | <.001 | ||||
| CK | .15 | <.01 | <.01 | 0.17 | 0.49 | <.001 | <.05 | <.001 | <.001 | .97 | .06 | .97 | ||||
| DB | <.05 | <.001 | <.001 | <.001 | <.001 | <.001 | .29 | <.001 | <.001 | <.01 | <.001 | <.001 | ||||
| SD | <.001 | <.01 | <.01 | <.001 | 0.34 | <.001 | <.001 | <.001 | <.001 | 1.0 | .98 | 1.0 | ||||
| TP | <.01 | <.001 | <.001 | <.01 | <.001 | <.001 | .21 | <.001 | <.001 | 1.0 | <.01 | <.001 | ||||
| U | <.001 | <.001 | <.001 | <.01 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | ||||
|
| ||||||||||||||||
| PFDA (C10) 0.16, 0.31, 0.63 |
AA | .16 | <.001 | <.05 | <.01 | .12 | <.001 | <.001 | <.001 | <.001 | 1.0 | .09 | .35 | .87 | 1.0 | <.001 |
| A | .75 | .64 | .91 | .75 | .36 | .21 | <.001 | <.001 | <.01 | .99 | 1.00 | .03 | .12 | .02 | <.001 | |
| AK | 1.0 | .89 | .56 | .65 | .84 | .11 | .62 | .21 | <.05 | .64 | 1.00 | .46 | .12 | <.001 | <.05 | |
| AsA | .12 | <.05 | .26 | <.01 | .35 | .13 | <.001 | <.01 | .07 | .93 | .29 | 1.0 | 1.0 | <.01 | <.001 | |
| B | .57 | 1.0 | .99 | .84 | .98 | .82 | 1.0 | 1.0 | .93 | 1.0 | .22 | 1.0 | <.001 | <.001 | <.001 | |
| C | <.001 | <.001 | 1.0 | <.001 | <.01 | .97 | <.001 | <.001 | <.01 | .67 | .06 | 1.0 | .43 | <.001 | <.001 | |
| CK | <.05 | <.05 | .52 | <.05 | 1.0 | .77 | <.05 | .11 | .24 | 1.0 | .06 | <.01 | .97 | .31 | <.05 | |
| DB | .99 | .83 | 1.0 | .94 | .07 | 1.0 | .90 | 1.0 | 1.0 | .55 | .68 | 1.0 | <.01 | <.001 | <.001 | |
| SD | <.001 | <.001 | .70 | <.001 | .06 | .43 | <.001 | <.001 | <.001 | .98 | .60 | .38 | .99 | .95 | .16 | |
| TP | .33 | .09 | <.001 | .25 | <.05 | <.001 | .52 | 1.0 | .03 | .73 | .50 | .66 | .52 | <.001 | <.001 | |
| U | .67 | <.05 | .98 | .46 | .82 | 1.0 | .99 | .33 | .95 | .98 | <.001 | <.01 | <.001 | <.001 | <.001 | |
Note: Parameters measured included creatinine kinase (CK), and sorbitol dehydrogenase (SD).
Furthermore, simple linear regressions were conducted for 2 clinical chemistry health outcomes, creatine kinase levels and sorbitol dehydrogenase levels at all three doses. Results were statistically significant at all three doses for both creatine kinase levels and sorbitol dehydrogenase levels.
Results were not significant for both creatine kinase levels and sorbitol dehydrogenase levels, indicating that the statistical models were successful in predicting these health outcomes.
There was no statistically significant difference t (68) =0.92, p=.36 between the mean values of creatine kinase levels of the rats exposed to PFHSKslt at dose 1, (M =111.20, SD = 15.52) compared to the predicted value (M = 107.52, SD = 11.06).
There was no statistically significant difference t (67) =0.60, p=.55 between the mean values of creatine kinase levels of the rats exposed to PFHSKslt at dose 2, (M = 108.50, SD = 16.32) compared to the predicted value (M = 105.47, SD = 14.60).
There was no statistically significant difference t (10.30) =−0.18, p=.86 between the mean values of creatine kinase levels of the rats exposed to PFHSKslt at dose 3, (M = 104.00, SD = 25.16) compared to the predicted value (M = 105.52, SD = 16.35).
There was no statistically significant difference t (68) =1.62, p=.11 between the mean values of sorbitol dehydrogenase levels of the rats exposed to PFHSKslt at dose 1, (M =9.80, SD = 2.04) compared to the predicted value (M = 8.30, SD = 2.80).
There was no statistically significant difference t (67) =0.21, p=.84 between the mean values of sorbitol dehydrogenase levels of the rats exposed to PFHSKslt at dose 2, (M = 10.10, SD = 2.33) compared to the predicted value (M = 9.85, SD = 3.72).
There was no statistically significant difference t (68) =0.87, p=.39 between the mean values of sorbitol dehydrogenase levels of the rats exposed to PFHSKslt at dose 3, (M = 9.80, SD = 2.44) compared to the predicted value (M = 9.23, SD = 1.81).
Discussion
The findings of the one-way ANOVAs indicate that there are statistically significant differences in the mean values of several PFAS pairs, among several health outcomes. Tukey’s HSD post hoc test serves the purpose of comparing every possible pair of mean values that were originally compared in the one-way ANOVA, to determine which pairs are statistically significantly different from one another. Surprisingly, there were the highest difference (proportionately) in means among pair groups with two long chained PFAS, compared to paired groups of one short chained PFAS or two short chained PFAS. This may be because, as PFAS increase in chain length, they become more toxic (Chambers et al. 2021), thus resulting in more extreme mean values and larger differences among the means. This is further supported by the fact that there are highest frequencies of differences between paired groups involving the PFAS PFNA (C9) and PFDA (C10), and the longest chained PFAS included in the analysis.
Predictive models were developed for two of 15 clinical chemistry health outcomes in this study: creatine kinase and sorbitol dehydrogenase. Increased levels of serum creatine kinase levels are associated with thyroid (Hekimsoy and Oktem 2005) and musculoskeletal conditions (Johannsen et al. 2013) and increased levels of sorbitol dehydrogenase levels are associated with diabetes (Harvey et al. 2011). Models were useful in predicating the theorical health outcomes, indicating the models can be successful. These models are certainly not perfect and need refinement before being utilized as regulatory tools. As more data is inputted into these models, they may become more accurate. Therefore, statistical modeling should not be considered as the sole basis for regulation, but rather to supplement both toxicological and epidemiological data as they become more accurate.
To ensure the most comprehensive analysis of toxicity, epidemiological studies will continue to be important to identify PFAS of particular concern, sites of contamination, and specific health outcomes of concern (Bartlett and Judge 1997). It is impossible to collect experimental data for every PFAS , and therefore these epidemiological studies serve as crucial guidance to toxicological studies. They help to inform which PFAS to test, for what health outcomes, and for how long of an exposure. From toxicological studies, experimental data is acquired. Experimental data is crucial for toxicity assessment, as it helps to eliminate confounding variables and establish causal inferences (Adami et al. 2011). This data can then be used to refine statistical modeling and to predict health outcomes, which is crucial for chemical classes such as PFAS, where there is simply not enough time or resources to test every chemical. Together, data from epidemiological and toxicological studies, as well as modeling techniques should be considered when making important decisions regarding the regulation of PFAS , whether as a class, a subclass, or as a substance-by-substance basis.
Limitations
Like all studies, this study has limitations. First, the National Toxicology Program currently has data available for only seven PFAS . Of these seven PFAS , only six were used in the one-way ANOVAs, the post hoc tests, and the simple linear regression analyses. There are thousands of PFAS in global circulation (Organization for Economic Co-operation and Development 2018). Six PFAS may, therefore, not be enough to form a fully accurate, comprehensive, predictive model for toxicity health outcomes. More data will help to produce more accurate models in the future.
Furthermore, each of the datasets analyzed, contained data from only 10 rats. This is not a large enough sample size. The rats used in these models also are assumed to be healthy, adult rats. This is not necessarily representative of the total human population in which there are infants, the elderly, and people with pre-existing health conditions which may worsen health outcomes when exposed to PFAS. Although, toxicological research relies heavily on the 3r principles, which enforce the rule that only the smallest number of animals may be used for research involving animals. Statistically, larger sample sizes are desirable, but on the grounds of animal welfare, researchers must rely on smaller sizes, and a sample size of 10 rats is not unique to this study (DFG, 2019). Therefore, any statistical modelling utilizing animal data from toxicological studies will inevitably face the reality of small sample sizing and lack of statical power.
Additionally, animal modeling will have certain limitations when attempting to demonstrate toxicological effects in humans. We cannot be sure that the effects seen in rat models will always be seen in humans. At present, however, it is crucial to use animal models, as it is unethical to conduct these experiments on humans. Furthermore, dose adjustments are always applied to animal toxicity models, to account for these issues (Nair and Jacob 2016).
An additional limitation is that the health outcomes selected for analysis in this study were arbitrary. It was not determined in this analysis whether the health outcomes were either positive or negative to overall health. Only the numerical value of health outcomes was considered when analyzing differences in health outcomes, not what these numerical values mean for health. The question being explored here was whether the numerical value is different, regardless of the real-life applicable value.
This study utilized data from male rats only. While the use of data from both sexes is crucial to health research, it was not feasible to use data from female rats in this study. Male and female rats differ significantly in their height, weights and production of hormones (NTP 2022 a). Due to this reasoning, toxicological data from male and female rats should not be combined to one sample. For the purpose of brevity and to reduce complexity, researchers from this study choose to analyze data from one sex only. Reviewing data from multiple studies on multiple health outcomes conducted at the NTP, indicted male rats were more significantly affected by exposure to PFAS, compared to female rats, across several health outcomes (NTP 2022 a). Therefore, male rats were chosen for this study. This is a limitation, as health research should be inclusive to all sexes and genders. Future studies should consider this, and more research is needed to account for these discrepancies.
Furthermore, the only aspect of chemical structure considered in this study was the number of carbon atoms per PFAS . All PFAS have fluorine, which are contained in the carbon-fluorine bonds making up PFAS chains (Kwiatkowski et al. 2020). However, the number of fluorine atoms per PFAS was not considered during this analysis, due to the issue of multicollinearity. It is plausible that the number of fluorine atoms in PFAS is a predictive factor of health outcomes. Consideration of this variable in future models may help to produce more accurate predictions. Additionally, many PFAS contain a wide range of other elements such as nitrogen, oxygen, sulfur etc., and it is not known whether these elements may also affect health outcomes. Several other chemical properties, such as branching, terminal moiety, chemical weight, chemical density, and pKa value, may affect PFAS toxicity and effects of exposure on health outcomes. There are several types of models which can consider multiple independent covariates such as multilinear regressions, random forest, extreme gradient boosting, and neural networks. Future research seeking to understand the relationship between chemical structures and properties of PFAS and health outcomes will have to implement these models.
For example, in the instances of number of carbon atoms per PFAS, number of fluorine atoms per PFAS, and molecular weight, these properties cannot be analyzed together utilizing multilinear regressions, due to high levels of multicollinearity. It may be useful in the future to utilize a random forest model, as this model is not affected by multicollinearity (Yilmazerab and Kocamana 2020). As research into QSAR and modelling for regulation advances and there is more experimental toxicological data available for PFAS, it is expected that a variety of different models will be utilized. This study did not capacity for these advanced models, which is a limitation that we predict will be addressed in future research.
In addition to PFAS having several chemical structures not considered, the subclasses of PFAS were not considered in this analysis. This analysis included both carboxylic acids and sulfonic acid PFAS , but they were treated the same and data was combined to develop the model. It is likely that health outcomes differ by subclass, and analyzing them separately will produce different results, which may be important to consider in future research.
Conclusions
This study helps to provide evidence to suggest that there may be differences in the health outcomes of short chained versus long chained PFAS . This study also provides evidence that by using the number of carbon atoms per PFAS as a predictive variable, it may be possible to generate datasets predicting health outcomes of PFAS exposure. Based on current legislation, knowledge, technology, time constraints and resources, the possibly of efficiently regulating PFAS based on epidemiology and toxicology studies does not seem feasible. There are not enough resources to regulate PFAS on either a molecule-by-molecule basis or using a class-based approach. Development of new approaches to predict PFAS adverse health outcomes are important to address the problems in current regulatory approaches. Our study shows that statistical approaches may be able to predict some, but not all, of the adverse health outcomes associated with PFAS exposure. The approach taken in this study was rudimentary, implementing carbon chain length as a predictive factor. There are many other important features in PFAS such as fluorine, acid moiety, chemical structure, chemical weight, etc. which can be used in these models. We are hoping our positive, yet preliminary results can motivate other investigators to apply similar approaches using different variables to develop more advances and accurate models for the use of PFAS toxicity predication.
Figure 1.

Linear Regression Best Fit Lines, creatine kinase. (A) Dose 1 creatine kinase, (B) Dose 2 creatine kinase, (C) Dose 3 creatine kinase.
Figure 2.
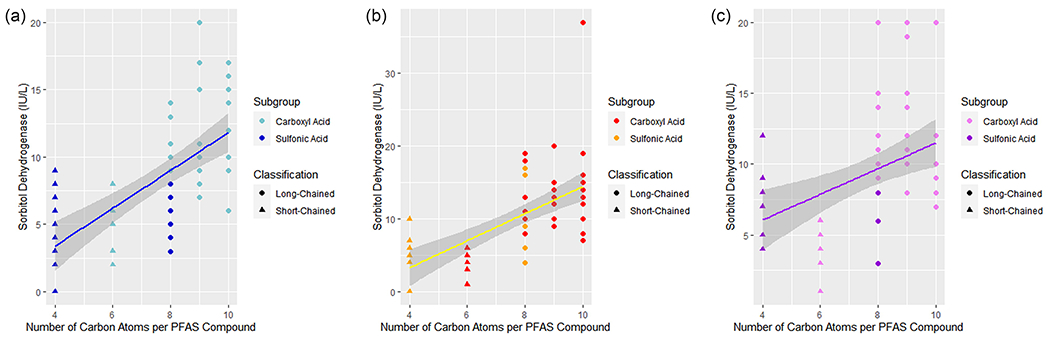
Linear Regression Best Fit Lines, sorbitol dehydrogenase. (A) Dose 1 sorbitol dehydrogenase, (B) Dose 2 sorbitol dehydrogenase, (C) Dose 3 sorbitol dehydrogenase
Table 1.
PFAS Physical Characteristics and Classifications
| PFAS Name | Molecular Formula | Molecular Weight | Classification |
|---|---|---|---|
| Perfluorobutanesulfonic Acid (PFBS) | C4HF9O3S | 300.01 | Short Chained, C4 |
| Perfluorooctanesulfonic Acid (PFOS) | C8HF17O3S | 500.13 | Long chained, C8 |
| Perfluorononanoic Acid (PFNA) | C9HF17O2 | 464.08 | Long chained, C9 |
| Perfluorohexanoic Acid (PFHxA) | C6HF11O2 | 314.05 | Short Chained, C6 |
| Perfluorodecanoic Acid (PFDA) | C10HF19O2 | 514.08 | Long Chained, C10 |
| Perfluorooctanoic Acid (PFOA) | C8HF15O2 | 414.07 | Long Chained, C8 |
| Perfluorohexanesulfonic acid (PFHSKslt) | C6HF13O3S | 400.12 | Long chained, C6 |
Table 2.
Dose per PFAS in mg/kg/day
| PFAS | Dose 1 | Dose 2 | Dose 3 |
|---|---|---|---|
| PFBS | 62.6 | 125 | 250 |
| PFOS | 0.312 | 0.625 | 1.25 |
| PFDA | 0.156 | 0.312 | 0.625 |
| PFHxA | 62.6 | 125 | 250 |
| PFNA | 0.625 | 1.25 | 2.5 |
| PFOA | 0.625 | 1.25 | 2.5 |
| PFHSKslt | 0.625 | 1.25 | 2.5 |
Table 3.
Correlation Coefficient for Strength of Linear Relationship and Breusch-Pagan test for Homoscedasticity
| Health Outcome | R | Strength | R | Strength | r | Strength | Breusch-Pagan p-value | ||
|---|---|---|---|---|---|---|---|---|---|
| AA | .42 | moderate | .61 | strong | .31 | weak | |||
| A | .42 | moderate | .27 | weak | −.20 | weak | |||
| AK | .25 | weak | .38 | weak | .16 | weak | |||
| AsA | .40 | moderate | .55 | moderate | .45 | moderate | .44 | <.01 | .16 |
| B | .07 | weak | .34 | weak | .35 | weak | |||
| C | .66 | moderate | .54 | moderate | .07 | weak | |||
| CK | .45 | moderate | .57 | moderate | .40 | moderate | .39 | .05 | .17 |
| Cr | −.18 | weak | −.04 | weak | −.47 | moderate | |||
| DB | .12 | weak | .29 | weak | .30 | weak | |||
| G | .26 | weak | .27 | weak | −.10 | weak | |||
| SD | .63 | moderate | .62 | moderate | .41 | moderate | .24 | .16 | .71 |
| TB | −.30 | weak | −.10 | weak | .28 | weak | |||
| TP | −.33 | weak | −.38 | weak | −.41 | moderate | |||
| Tg | −.24 | weak | −.30 | weak | −.33 | weak | |||
| U | .07 | weak | .10 | weak | .23 | weak | |||
Note: Parameters measured included alanine aminotransferase (AA), albumin (A), alkaline phosphatase (Ak), aspartate aminotransferase (AsA), bile salts/acids (B), cholesterol (C), creatinine kinase (CK), direct bilirubin (DB), sorbitol dehydrogenase (SD), total protein (TP), and urea nitrogen (U). Abbreviations indicate the parameter in which a difference (p<.001) was identified and the dose levels (1, 2 and/or 3, shown as superscript) at which a difference occurred.
Table 4.
Results of One Way ANOVAs
| Dose 1 | Dose 2 | Dose 3 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Health Outcome | df | SS | MS | F | p | df | SS | MS | F | p | df | SS | MS | F | p |
| AA | Between | 5 | 5,064 | 1,012 | 11.22 | <.001 | 5 | 11,434 | 2,286 | 9.35 | <.001 | 5 | 7,370 | 1,473 | 10.25 | <.001 |
| Within Groups | 54 | 4,874 | 90.30 | 53 | 12,958 | 244 | 54 | 7,764 | 143 | ||||||
|
| |||||||||||||||
| A| Between | 5 | 1.46 | 0.29 | 13.36 | <.001 | 5 | 1.93 | 0.39 | 18.25 | <.001 | 5 | 10.33 | 2.07 | 68.37 | <.001 |
| Within Groups | 54 | 1.18 | 0.02 | 53 | 1.12 | 0.02 | 54 | 1.63 | 0.03 | ||||||
|
| |||||||||||||||
| AK| Between | 5 | 25,440 | 5,088 | 5.58 | <.001 | 5 | 260,061 | 53,212 | 29.98 | <.001 | 5 | 100,973 | 20,195 | 8.33 | <.001 |
| Within Groups | 54 | 49,223 | 912 | 53 | 94,085 | 1,775 | 54 | 130,911 | 2,424 | ||||||
|
| |||||||||||||||
| AsA | Between | 5 | 2,207 | 441 | 7.18 | <.001 | 5 | 9,263 | 1,852 | 18.29 | <.001 | 5 | 23,235 | 4,647 | 18.23 | <.001 |
| Within Groups | 54 | 3,321 | 61.50 | 54 | 5,470 | 101 | 54 | 13,764 | 255 | ||||||
|
| |||||||||||||||
| B| Between | 5 | 2,589 | 517 | 7.82 | <.001 | 5 | 31,211 | 6,242 | 43.69 | <.001 | 5 | 349,411 | 69,882 | 139.60 | <.001 |
| Within Groups | 54 | 3,574 | 66.20 | 53 | 7,572 | 143 | 54 | 27,032 | 501 | ||||||
|
| |||||||||||||||
| C| Between | 5 | 10,513 | 2,1020 | 22.93 | <.001 | 5 | 8,772 | 1,754 | 17.57 | <.001 | 5 | 11,217 | 2,244 | 10.06 | <.001 |
| Within Groups | 54 | 4,951 | 91.70 | 53 | 5,290 | 99.80 | 54 | 12,041 | 223 | ||||||
|
| |||||||||||||||
| CK| Between | 5 | 13,104 | 2,6200 | 6.15 | <.001 | 5 | 16,774 | 3,355 | 8.23 | <.001 | 5 | 41,036 | 8,207 | 7.50 | <.001 |
| Within Groups | 54 | 23,021 | 426 | 53 | 21,611 | 408 | 54 | 59,081 | 1,094 | ||||||
|
| |||||||||||||||
| DB| Between | 5 | <.005 | <.001 | 5.96 | <.001 | 5 | 0.01 | <.005 | 23.12 | <.001 | 5 | 0.75 | 0.15 | 47.78 | <.001 |
| Within Groups | 54 | <.005 | <.001 | 54 | <.005 | <.001 | 54 | 0.17 | <.005 | ||||||
|
| |||||||||||||||
| SD| Between | 5 | 720 | 144 | 17.45 | <.001 | 5 | 960 | 192 | 9.01 | <.001 | 5 | 632 | 126 | 13.20 | <.001 |
| Within Groups | 54 | 446 | 8.26 | 53 | 1,129 | 21.31 | 54 | 517 | 9.59 | ||||||
|
| |||||||||||||||
| TP| Between | 5 | 2.06 | 0.41 | 5.58 | <.001 | 5 | 6.41 | 1.28 | 22.04 | <.001 | 5 | 33.31 | 6.66 | 93.33 | <.001 |
| Within Groups | 54 | 3.90 | 0.07 | 53 | 3.08 | 0.06 | 54 | 3.85 | 0.07 | ||||||
|
| |||||||||||||||
| U| Between | 5 | 199 | 39.90 | 10.42 | <.001 | 5 | 521 | 104 | 38.68 | <.001 | 5 | 616 | 123 | 15.03 | <.001 |
| Within Groups | 54 | 206 | 3.83 | 53 | 142 | 2.69 | 54 | 442 | 8.20 | ||||||
Note: Parameters measured included alanine aminotransferase (AA), albumin (A), alkaline phosphatase (Ak), aspartate aminotransferase (AsA), bile salts/acids (B), cholesterol (C), creatinine kinase (CK), creatine (CR), direct bilirubin (DB), glucose (G), sorbitol dehydrogenase (SD), total bilirubin (TB), total protein (TP), Triglycerides (Tg), and urea nitrogen (U).
Table 6.
Results of Linear Regression Analyses
| Dose 1 | Dose 2 | Dose 3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Health Outcome | β | Se | t | p | β | Se | t | p | β | Se | t | p |
| CK | 65.95 | 11.30 | 5.84 | <.01 | 50.62 | 10.90 | 4.64 | <.01 | 44.07 | 19.30 | 2.28 | <.05 |
| Carbon Chain as Predictor | 5.54 | 1.46 | 3.81 | <.01 | 7.29 | 1.40 | 5.20 | <.01 | 8.19 | 2.49 | 3.29 | <.01 |
|
| ||||||||||||
| SD | −2.23 | 1.76 | −1.28 | .21 | −4.15 | 2.42 | −1.71 | .09 | 2.44 | 2.06 | 1.19 | .24 |
|
| ||||||||||||
| Carbon Chain as Predictor | 1.40 | 0.23 | 6.18 | <.01 | 1.86 | 0.31 | 5.97 | <.01 | 0.91 | 0.27 | 3.42 | <.01 |
Note: Parameters measured included alanine aminotransferase (AA), albumin (A), alkaline phosphatase (Ak), aspartate aminotransferase (AsA), bile salts/acids (B), cholesterol (C), creatinine kinase (CK), direct bilirubin (DB), sorbitol dehydrogenase (SD), total protein (TP), and urea nitrogen (U). Abbreviations indicate the parameter in which a difference (p<.001) was identified and the dose levels (1, 2 and/or 3, shown as superscript) at which a difference occurred.
Table 7.
Differences between pairs of the PFAS for each dosing level determined by Tukey’s HSD post hoc tests
| PFBS 62.6, 125, 250 |
PFOS 0.31, 0.63, 1.3 |
PFHxA 62.6, 125, 250 |
PFOA 0.63, 1.3, 2.5 |
PFNA 0.63, 1.3, 2.5 |
PFDA 0.16, 0.31, 0.63 |
|
|---|---|---|---|---|---|---|
|
| ||||||
| Dose (mg/kg) | ||||||
|
| ||||||
| # Carbons | 4 | 8 | 6 | 8 | 9 | 10 |
|
| ||||||
| PFBS | ||||||
|
| ||||||
| PFOS | ||||||
|
| ||||||
| PFHxA | A23, C3 | A23, C2, TP3 | ||||
|
| ||||||
| PFOA | Ak3, C1, SD1, TP2 | Ak3, C1, SD1, TP23, U2 | AA1, A123, Ak3, C12, SD13, | |||
|
| ||||||
| PFNA | AA2, A3, Ak2, AsA23, B23, C13, CK3, DB23,SD1, TP23,U123 | AA1, A3, Ak2, AsA23, B123CK3, , DB123,SD13, TP23,U23 | AA12, A123, Ak2, AsA23, B123, CK23, DB23,SD123, TP23,U123 | A3, Ak23,
AsA23, B123, C3,
DB23,TP3 U123 |
||
|
| ||||||
| PFDA | AA2, C12, SD12, TP3 | AA3, C1, SD1, TP3 | AA123, A12, AsA1, C12,SD123 | U2 | AA3, A3, Ak2, AsA3, B123, C23, DB23,, TP23,U123 | |
Acknowledgments
We gratefully thank the Yale School of Public Health, Department of Environmental Health Sciences for the resources and mentorship provided by the facility, faculty, and students. We graciously thank the National Toxicology Program for the open access data they have provided, as well as for the continued commitment to toxicology and public health.
Funding Details
This work was supported in part by P42ES033815-01.
Footnotes
Declaration of interest statement
Robert A. Bilott, J.D., reports serving as counsel for various plaintiffs in on-going litigation involving exposure and damage from PFAS . No other declarations of interest have been reported.
Data Availability Statement
The data that support the findings of this study are available in the National Toxicology Program at [https://cebs.niehs.nih.gov/cebs/publication/TOX-96] and [https://cebs.niehs.nih.gov/cebs/publication/TOX-97]
References
- Adami HO, Berry SCL, Breckenridge CB, Smith LL, Swensberg JA, Trichopoulos D, Weiss NS, Pastoor TP. 2011. Toxicology and epidemiology: improving the science with a framework for combining toxicological and epidemiological evidence to establish causal inference. Toxicol Sci. 122(2):223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett PC, Judge LJ. 1997. The role of epidemiology in public health. Rev Sci Tech.16(2):3316. [DOI] [PubMed] [Google Scholar]
- Benfenati E. 2012. Theory, Guidance, Applications on QSAR and REACH. Milano, Italy. Istituto di Ricerche Farmacologiche “Mario Negri.” [Google Scholar]
- Bökenkamp A. 2020. Proteinuria-take a closer look! Pediatr Nephrol. Apr;35(4):533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck RC, Franklin J, Berger U, Conder JM, Cousins IT, de Voogt P, Jensen AA, Kannan K, Mabury SA, van Leeuwen SP. 2011. Perfluoroalkyl and polyfluoroalkyl substances in the environment: terminology, classification, and origins. Integr Environ Assess Manag. 7: 513–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers WS, Hopkins JG, Richards SM. 2021. A review of per- and polyfluorinated alkyl substance impairment of reproduction. Frontiers in Toxicology, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins IT, DeWitt JC, Gluge J, Goldenman G, Herzke D, Lohmann R, Ng CA, Scheringer M, Wang Z. 2020. The High Persistence of PFAS is sufficient for their management as a chemical class. Environ. Sci.: Processes Impacts, 22:2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Harlow SD, Randolph JF Jr, Loch-Caruso R, Park SK. 2020. Perfluoroalkyl and polyfluoroalkyl substances (PFAS) and their effects on the ovary. Hum Reprod Update. 1;26(5):724–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Death C, Bell C, Champness D, Milne C, Reichman S, Hagen T. 2021.Per- and polyfluoroalkyl substances (PFAS) in livestock and game species: A review. Science of The Total Environment. 774 (144795): 0048–9697 [DOI] [PubMed] [Google Scholar]
- DeGregory KW, Kuiper P, DeSilvio T, Pleuss JD, Miller R, Roginski JW, Fisher CB, Harness D, Viswanath S, Heymsfield SB, et al. 2018. A review of machine learning in obesity. Obes Rev. May 19(5): 668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DFG. 2019. Germeny. Deutsche Forschungsgemeinschaft; [November 7, 2022]. https://www.dfg.de/download/pdf/dfg_im_profil/geschaeftsstelle/publikationen/handreichung_sk_tierversuche_en.pdf [Google Scholar]
- European Chemical Agency. 2016. “Practical Guide How to Use and Report (Q)SARS - Europa.” [Google Scholar]
- European Union. 2019. Elements for an EU-Strategy for PFASs. [Google Scholar]
- Fenton SE, Ducatman A, Boobis A, DeWitt JC, Lau C, Ng C, Smith JS and Roberts SM 2021. Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ Toxicol Chem, 40: 606–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano E, Sgroi M, Falciglia PP, Vagliasindi FGA, Roccaro P. 2020. Removal of poly- and perfluoroalkyl substances (PFAS) from water by adsorption: Role of PFAS chain length, effect of organic matter and challenges in adsorbent regeneration. Water Research, 171: (115381):0043–1354. [DOI] [PubMed] [Google Scholar]
- Gjuladin-Hellon T, Davies IG, Penson P, Baghbadorani RA. 2018. Effects of carbohydrate-restricted diets on low-density lipoprotein cholesterol levels in overweight and obese adults: a systematic review and meta-analysis. Nutr Rev. 77(3): 161–180. [DOI] [PubMed] [Google Scholar]
- Harvey RA, Ferrier DR. 2011. Biochemistry. Philadelphia (PA). Lippincott Williams & Wilkins. [Google Scholar]
- Hekimsoy Z, Oktem IK. 2005. Serum creatine kinase levels in overt and subclinical hypothyroidism. Endocr Res. 31(3):171–5. [DOI] [PubMed] [Google Scholar]
- Johannsen S, Berberich C, Metterlein T, Roth C, Reiners K, Roewer N, Schuster F. 2013. Screening test for malignant hyperthermia in patients with persistent hyperCKemia: a pilot study. Muscle Nerve.47(5):677–81. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski CF, Andrews DQ, Birnbaum LS, Bruton TA, DeWitt JC, Knappe DRU, Maffini MV, Miller MF, Pelch KE, Reade A, et al. 2020. Scientific basis for managing PFAS as a chemical class. Environ Sci Technol Lett. 7(8):532–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manibusan M, Paterson J, Kent R, Chen J, Tao J, Scollon E, Olinger C, Schmieder P, Russom C, Mayo K. 2012. (Q)QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIP [(Q)SAR] GUIDANCE DOCUMENT. United States Environmental Protection Agency. [Google Scholar]
- Nair AB, Jacob S. 2016. A simple practice guide for dose conversion between animals and humans. J Basic Clin Pharmacy. 7(2), 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program. 2022. NTP Technical Report on the Toxicity Studies of Perfluoroalkyl Sulfonates (Perfluorobutane Sulfonic Acid, Perfluorohexane Sulfonate Potassium Salt, and Perfluorooctane Sulfonic Acid) Administered by Gavage to Sprague Dawley (Hsd: Sprague Dawley SD) Rats (Revised). Research Triangle Park, NC. National Toxicology Program. 96. [Google Scholar]
- NTP. TOX-96: 1-Perfluorobutanesulfonic acid (375-73-5), Potassium perfluorohexanesulfonate (3871-99-6), Perfluorooctane sulfonate (1763-23-1), WY-14643 (50892-23-4). Chemical Effects in Biological Systems (CEBS). Research Triangle Park, NC (USA): National Toxicology Program (NTP). [Google Scholar]
- NTP. TOX-97: Perfluorohexanoic acid (307-24-4), Perfluorooctanoic acid (335-67-1), Perfluorononanoic acid (375-95-1), Perfluorodecanoic acid (335-76-2), WY-14643 (50892-23-4). Chemical Effects in Biological Systems (CEBS). Research Triangle Park, NC (USA): National Toxicology Program (NTP). [Google Scholar]
- RStudio Team. 2021. RStudio: Integrated development environment for R. RStudio, PBC, Boston, MA: URL http://www.rstudio.com/ [Google Scholar]
- Organization for Economic Co-operation and Development: OECD. 2018. Toward a new comprehensive global database of per- and polyfluoroalkyl substances (PFASs): Summary report on updating the OECD 2007 list of per- and polyfluoroalkyl substances (PFASs). Paris, France. Report No.: ENV/JM/MONO(2018)7 [Google Scholar]
- United State Environmental Protection Agency. 2019. Method 533: determination of per- and polyfluoroalkyl substances in drinking water by Isotope dilution anion exchange solid phase Extraction and liquid chromatography/tandem Mass spectrometry, Cinninnati (OH): Office of Water. Report No.: EPA/815-B-19-020 [Google Scholar]
- United State Environmental Protection Agency. 2020. Method 537.1 determination of selected per- and Polyfluorinated alkyl substances in drinking Water by solid phase extraction and liquid Chromatography/tandem mass spectrometry (lc/ms/ms), Cincinnati (OH), Office of Research and Development. Report No.: EPA/600/R-20/006 [Google Scholar]
- United State Environmental Protection Agency. 2021. Revisions to the Unregulated Contaminant Monitoring Rule (UCMR 5) for Public Water Systems and Announcement of Public Meetings, Cincinnati (OH), Office of Ground Water and Drinking Water. Report No.: EPA–HQ–OW–2020–0530 [Google Scholar]
- Vermont Official State. 2021. Notice of decision not to adopt a maximum contaminant level regulating PFAS as a class in public drinking water systems. Vermont Agency of Natural Resources. https://dec.vermont.gov/sites/dec/files/PFAS/PFAS-MCL-Final-Decision.pdf [Google Scholar]
- Wang Z, DeWitt JC, Higgins CP, Cousins IT. 2017. A never ending story of per- and polyfluoroalkyl substances (PFASs?). Environ. Sci. Technol. 51(5):2508–2518. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang G. (2018). Machine learning based toxicity prediction: from chemical structural description to transcriptome analysis. International journal of molecular sciences. 19(8):2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmazerab S, Kocamana S. 2020. A mass appraisal assessment study using machine learning based on multiple regression and random forest. Land Use Pol. 99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in the National Toxicology Program at [https://cebs.niehs.nih.gov/cebs/publication/TOX-96] and [https://cebs.niehs.nih.gov/cebs/publication/TOX-97]


