Abstract
The transcripts of structurally related cellobiohydrolase genes in Phanerochaete chrysosporium-colonized wood chips were quantified. The transcript patterns obtained were dramatically different from the transcript patterns obtained previously in defined media. Cellobiose dehydrogenase transcripts were also detected, which is consistent with the hypothesis that such transcripts play an important role in cellulose degradation.
In submerged cultures, the white rot basidiomycete Phanerochaete chrysosporium secretes an array of hydrolytic cellulases, including multiple isozyme forms of endoglucanase (13), cellobiohydrolase (CBH) (14, 33), and β-glucosidase (11, 28) (for reviews see references 15 and 31).
On the basis of structural similarities to corresponding Trichoderma reesei cellulase genes, the genes for a single CBHII-like clone (32) and six CBHI-like clones (8–10, 26) were isolated from P. chrysosporium. Except for cbh1-1, all of these genes have the tripartite architecture common to microbial cellulase genes (i.e., catalytic and cellulose-binding domains separated by a glycosylated linker region) (reviewed in references 16 and 37). The cbh1-1 gene lacks a binding domain and hinge region (9), and similarly truncated cbh1 genes have now been identified in the non-wood-decaying fungi Cochiobolus carbonum and Cryphonectria parasitica (29, 35). No β-glucosidase or endoglucanase-like genes have been cloned from P. chrysosporium, and it has been suggested that cbh1-like genes may encode proteins with endoglucanase activity (27).
The multiple CBHI genes of P. chrysosporium are transcriptionally regulated. With cellulose induction, the levels of the dominant transcript, cbh1-4, exceed the levels of the closely related genes cbh1-2 and cbh1-1 by more than 1,000-fold (9, 34). These results are consistent with the results of protein purification, which identified the cbh1-4 gene product as the dominant isozyme (33, 34). The transcript levels for cbh1-1 and cbh1-2 are relatively low in cellulose-supplemented medium. However, both cbh1-1 and cbh1-2 transcripts can be detected in minimal media containing glucose as the sole carbon source, while cbh1-3, cbh1-4, cbh1-5, and cbh1-6 cannot be detected under these conditions (9). The precise roles and interactions of individual genes in cellulose deg-radation are unclear, particularly in complex substrates, such as wood.
In addition to the hydrolytic cellulases, extracellular oxidative enzymes apparently are involved in cellulose degradation by P. chrysosporium and, presumably, other white rot fungi (3, 15). Cellobiose dehydrogenase (CDH) is an extracellular enzyme containing two domains; one domain contains flavin adenine dinucleotide, and the other domain contains a heme (18, 19). CDH is produced by several fungi, including P. chrysosporium, and has been shown to oxidize cellobiose and various oligosaccharides. Production of CDH is stimulated by the presence of cellulose as a carbon source, and, like the cellulases, CDH strongly binds to the substrate. The exact role of CDH is uncertain, but removal of cellobiose by CDH removes a powerful inhibitor of CBHs (4, 36). Igarashi et al. (20) showed that CDH adsorbed to the surfaces of cellulose particles in stationary cultures and suggested that CDH plays a role in cellulose degradation in cooperation with the hydrolytic cellulases.
cDNAs encoding P. chrysosporium CDH have been characterized previously (24, 25). The flavin- and heme-binding domains were recognized in the predicted amino acid sequence, but no cellulase-like cellulose-binding domain was observed. The results of Southern blotting suggested that CDH is encoded by a single gene, and the results of Northern blotting identified CDH transcripts in cultures containing cellulose but not in glucose- or cellobiose-containing cultures (24). Like expression of the CBH genes, nothing is known about CDH gene expression in solid wood. To address this issue, magnetic capture and reverse transcriptase PCR (RT-PCR) techniques were used to quantify CBH and CDH transcripts in wood chips.
Approximately 2.5 kg of fresh aspen wood chips that were not amended with any nutrients were steam sterilized and inoculated by standard biopulping methods (1). Ten-gram samples were collected every 2 weeks, snap frozen in liquid nitrogen, and stored at −80°C. Frozen samples were ground in a clean coffee grinder in the presence of dry ice. Each frozen powder sample was suspended in 20 ml of a solution containing 4 M guanidinium thiocyanate, 100 mM Tris-HCl (pH 8.0), 1% dithiothreitol, and 0.5% lauryl sarcosinate in a 50-ml conical tube. The contents of the tube were incubated on ice for 30 min and mixed by gently inverting the tube every 5 to 10 min. Following centrifugation at 2,000 × g for 10 min, the supernatant was filtered through a Miracloth filter (Calbiochem, San Diego, Calif.) and incubated on ice for 30 min with 1.5 mg of Dynabeads oligo(dT)25 (Dynal, Inc., Great Neck, N.Y.). Dynabead-poly(A) hybrids were isolated with a model MPC-1 magnetic concentrator (Dynal, Inc.) and washed repeatedly in a 1.5-ml Eppendorf tube as previously described (6, 7). The eluted mRNA was stored as an ethanol precipitate at −20°C. Aliquots (2 μl) were dried under a vacuum, and reverse transcriptions were carried out in 20-μl reaction mixtures containing 50 U of Moloney murine leukemia virus reverse transcriptase (GIBCO BRL, Gaithersburg, Md.), 15 pmol of oligo (dT)15, and 20 U of RNasin (Promega Biotech, Madison, Wis.). Reactions were performed at 23°C for 10 min, at 42°C for 45 min, and at 95°C for 5 min.
Competitive PCRs were performed as reported previously (6, 7, 17, 30) with 10−13 to 10−8 μg of genomic template. The competitive templates consisted of full-length genomic copies of the genes which had been PCR amplified and subcloned into pKSII (Stratagene, La Jolla, Calif.). Gene-specific primers were prepared based on multiple alignments (Fig. 1) and previously published sequences (24, 32) (Table 1). As described previously (6, 7), the reaction mixtures contained 21 pmol of each primer (Table 1).
FIG. 1.
Partial sequence alignment of the six known cbh1 genes of P. chrysosporium BKM-F-1767. The positions of PCR primers are indicated by arrows and underlining. The conserved intron is enclosed in a box, as are individual residues which differ from the residues in the consensus sequence.
TABLE 1.
Oligonucleotides used as primers for competitive RT-PCR
| Gene | 5′ Primer (5′→3′) | 3′ Primer (5′→3′) |
|---|---|---|
| cbh1-1a | AAGATCGTACTAGACGCTAACCG | GGTGACAACGGTGAAGGTCTTCGTC |
| cbh1-2 | AAAGTCGTCCTCGACGTGAACTG | GGTGACGACAGTGAAGAGTTTGGTG |
| cbh1-3 | AAGATCGTGCTCGACGCGAA | GTGACAACGGTGAAGGGCTT |
| cbh1-4 | AAGGTCGTCCTCGACTCGAA | CGACGGTGAAGGGCTTGGAG |
| cbh1-5 | ACCGTCGTGCTCGACTCCAA | CGACGGTGAAGGGCTTGCTG |
| cbh1-6 | AAGATCGTGCTCGACGCCAA | GTGACGACCGTGAACGGCTT |
| cbh2b | AGCGCCAACTACCAGAACTACCT | CTTCGGGGTCACGGGGATGAAAC |
| cdhb | CAGTGCGGTGGCATTGGCTG | CTGGTCGACGATGAAGGTCG |
The specificity of cbh1 primer pairs (Fig. 1 and Table 1) was established experimentally by performing PCR amplifications with separate clones containing different cbh1 genes. No cross amplification was observed for cbh1-1-, cbh1-2-, cbh1-3-, cbh1-4-, and cbh1-6-specific primers. Primers designed for cbh1-5 sometimes yielded minor products with other plasmid templates, but direct sequencing of the RT-PCR products revealed only the cbh1-5 sequence (data not shown).
The PCR products were size fractionated in 2% agarose containing 1% NuSieve (FMC) in 0.5× TBE buffer. Ethidium bromide-stained agarose gels were analyzed by using NIH Image 1.58 (National Institutes of Health). Image data was analyzed with Cricket Graphic (version 1.53).
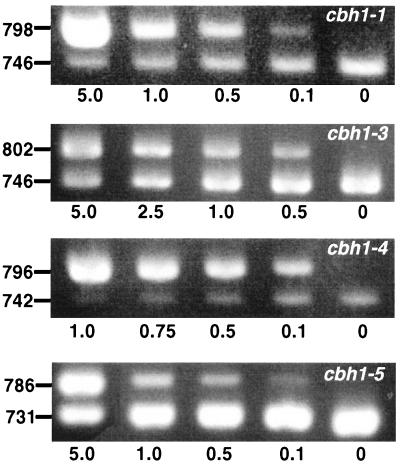
The transcript patterns of P. chrysosporium cbh1-like genes on aspen wood chips differed markedly from the transcript patterns observed previously on defined media supplemented with cellulose or glucose. For example, after 4 weeks of incubation, the cbh1-5 transcript levels were highest and the cbh1-4 transcripts were barely detectable in wood chips (Fig. 2 and Table 2). In contrast, cbh1-4 encodes the dominant transcript and isozyme found in submerged cultures (33, 34). Transcripts of cbh1-6 and cbh1-2 were not detected, although both transcripts are present in cellulose-amended cultures. These results provide a framework for strain improvement, particularly in biomechanical pulping processes in which active cellulase genes must be targeted (5, 12, 22).
FIG. 2.
Competitive PCRs for 4-week aspen chip samples comparing the detectable cbh1 transcripts. The gene-specific PCR primers used are shown in Fig. 1 and Table 1. PCR mixtures contained the amounts of the competitive templates indicated below the gels in picograms of plasmid. As described previously (17), the levels of transcripts in samples were based on estimated equivalence points between competitive product and target cDNAs. The sizes of PCR products in base pairs are indicated on the left. Ethidium bromide-stained gels were photographed with a Foto/Analyst Visionary system (Fotodyne, Hartland, Wis.) and then scanned with a Microtek ScanMaker III and Adobe Photoshop 3.0.
TABLE 2.
Transcript levels in aspen wood chips as estimated by competitive PCR analysis
| Gene | cDNA concn (pg) after:
|
||||
|---|---|---|---|---|---|
| 2 weeks | 4 weeks | 6 weeks | 8 weeks | 10 weeks | |
| cbh1-1 | 0.36 | 0.47 | 0.14 | 0.32 | 0.12 |
| cbh1-3 | 1.87 | 2.43 | 0.26 | 1.81 | 0.25 |
| cbh1-4 | NDa | 0.10 | ND | 0.08 | 0.03 |
| cbh1-5 | 0.44 | 3.30 | 0.33 | 2.35 | 1.61 |
| cbh2 | 2.68 | 5.96 | 0.67 | 3.51 | 5.62 |
| cdh | 0.20 | 1.55 | 0.10 | 0.45 | 0.29 |
ND, not detected.
The relative cbh1 transcript levels were similar throughout the 10-week time course, and the levels of the cbh1-3 and/or cbh1-5 transcripts were consistently highest (Table 2). All cellulase transcript levels decreased at 6 weeks, although the reasons for this are unclear. The levels of lignin peroxidase transcripts were not similarly depressed (21), suggesting that variability in mRNA yields was not responsible.
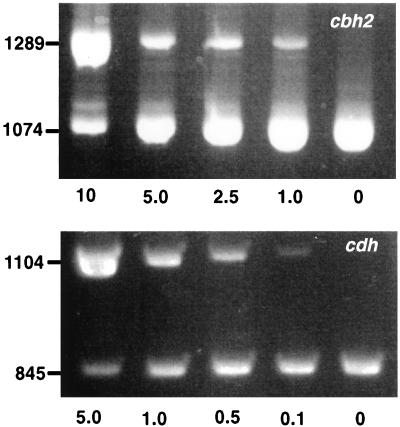
In marked contrast to submerged cultures (9, 34), substantial levels of the transcript of the truncated CBH gene, cbh1-1, were detected in all wood samples. This result supports the hypothesis that cbh1-1 contributes to the degradation of native cellulose. In submerged cultures, the levels of cbh1-1 transcripts are exceedingly low (more than 1,000-fold less than the levels of cbh1-3, cbh1-4, cbh1-5, and cbh1-6 transcripts) (9, 34). Similarly, the consistent presence of cbh2 and cdh transcripts (Fig. 3 and Table 2) strongly supports the hypothesis that these transcripts play a role in cellulose degradation.
FIG. 3.
Competitive PCRs for 4-week aspen chip samples showing amplification of cbh2 and cdh with the gene-specific PCR primers listed in Table 1. The amounts of the competitive templates used are indicated below the gels in picograms. The sizes of PCR products in base pairs are indicated on the left.
It is interesting that the same P. chrysosporium-colonized wood chips also contained transcripts of certain lignin peroxidase and manganese peroxidase genes (21). The appearance of cdh and peroxidase transcripts together in wood may support the hypothesis that there is a physiological connection (reviewed in reference 2) in which CDH regulates lignin depolymerization through reduction of peroxidase products (phenoxy radicals) and thereby prevents their repolymerization.
Acknowledgments
This research was supported by grant DE-FG02-87ER13712 from the Department of Energy to D.C. and by a grant from Conselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq, Brazil.
We thank Masood Akhtar and Eric Horn for preparation of the bioreactors.
REFERENCES
- 1.Akhtar, M. April 1997. U.S. patent 5,620,564.
- 2.Ander P. The cellobiose-oxidizing enzymes CBQ and CBO as related to lignin and cellulose degradation—a review. FEMS Microbiol Rev. 1994;13:297–312. [Google Scholar]
- 3.Ander P, Daniel G, Pettersson B, Westermark U. Possible applications of cellobiose oxidizing and other flavin adenine dinucleotide enzymes in the pulp and paper industry. In: Jeffries T, Viikari L, editors. Enzymes for pulp and paper processing. Washington, D.C: American Chemical Society; 1996. pp. 297–307. [Google Scholar]
- 4.Ayers A R, Ayers S B, Eriksson K-E. Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur J Biochem. 1978;90:171–181. doi: 10.1111/j.1432-1033.1978.tb12588.x. [DOI] [PubMed] [Google Scholar]
- 5.Blanchette R A, Burnes T A, Eerdmans M M, Akhtar M. Evaluating isolates of Phanerochaete chrysosporium and Ceriporiopsis subvermispora for use in biological pulping processes. Holzforschung. 1992;46:109–115. [Google Scholar]
- 6.Bogan B, Schoenike B, Lamar R, Cullen D. Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:3697–3703. doi: 10.1128/aem.62.10.3697-3703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogan B, Schoenike B, Lamar R, Cullen D. Expression of mnp genes during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covert S, Bolduc J, Cullen D. Genomic organization of a cellulase gene family in Phanerochaete chrysosporium. Curr Genet. 1992;22:407–413. doi: 10.1007/BF00352442. [DOI] [PubMed] [Google Scholar]
- 9.Covert S, Vanden Wymelenberg A, Cullen D. Structure, organization, and transcription of a cellobiohydrolase gene cluster from Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:2168–2175. doi: 10.1128/aem.58.7.2168-2175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covert S F. The molecular cloning and chromosomal organization of a cellulase gene family from Phanerochaete chrysosporium. Ph.D thesis. University of Wisconsin, Madison; 1990. [Google Scholar]
- 11.Deshpande V, Eriksson K-E, Pettersson B. Production, purification and partial characterization of 1,4-β-glucosidase enzymes from Sporotrichum pulverulentum. Eur J Biochem. 1978;90:191–198. doi: 10.1111/j.1432-1033.1978.tb12590.x. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson K-E, Kirk T K. Biopulping, biobleaching, and treatments of kraft bleaching effluents with white rot fungi. New York, N.Y: Pergamon Press; 1985. [Google Scholar]
- 13.Eriksson K-E, Pettersson B. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 1. Separation, purification, and physio-chemical characterization of five endo-1,4-β-glucanases. Eur J Biochem. 1975;51:193–206. doi: 10.1111/j.1432-1033.1975.tb03919.x. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson K-E, Pettersson B. Extracellular enzyme system used by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 3. Purification and physiochemical characterization of an exo-1,4-β-glucanase. Eur J Biochem. 1975;51:213–218. doi: 10.1111/j.1432-1033.1975.tb03921.x. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson K-E, Blanchette R A, Ander P. Microbial and enzymatic degradation of wood and wood components. Berlin, Germany: Springer-Verlag; 1990. [Google Scholar]
- 16.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilliland G, Perrin S, Bunn H. Competitive PCR for quantitation of mRNA. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols. New York, N.Y: Academic Press; 1990. pp. 60–69. [Google Scholar]
- 18.Habu N, Samejima M, Dean J F D, Eriksson K-E. Release of the FAD domain from cellobiose oxidase by proteases from cellulolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1993;327:101–106. doi: 10.1016/0014-5793(93)80162-n. [DOI] [PubMed] [Google Scholar]
- 19.Henriksson G, Pettersson G, Johansson G, Ruiz A, Uzcategui E. Cellobiose oxidase from Phanerochaete chrysosporium can be cleaved by papain into two domains. Eur J Biochem. 1991;196:101–106. doi: 10.1111/j.1432-1033.1991.tb15791.x. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi K, Samejima M, Saburi Y, Habu N, Eriksson K. Localization of cellobiose dehydrogenase in cellulose-grown cultures of Phanerochaete chrysosporium. Fungal Genet Biol. 1997;21:214–222. doi: 10.1006/fgbi.1996.0954. [DOI] [PubMed] [Google Scholar]
- 21.Janse, B. J. H., J. Gaskell, and D. Cullen. 1997. Unpublished data.
- 22.Kirk T K, Koning J W, Burgess R R, Akhtar M, Blanchette R A, Cameron D C, Cullen D, Kersten P J, Lightfoot E N, Myers G C, Sykes M, Wall M B. Biopulping: a glimpse of the future? USDA Forest Service research publication FPL-RP-523. Madison, Wis: USDA Forest Service; 1993. [Google Scholar]
- 23.Li B, Nagalla S, Renganathan V. Cellobiose dehydrogenase from Phanerochaete chrysosporium is encoded by two allelic variants. Appl Environ Microbiol. 1997;63:796–799. doi: 10.1128/aem.63.2.796-799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Nagalla S, Renganathan V. Cloning of a cDNA encoding cellobiose dehydrogenase, a hemoflavoenzyme from Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:1329–1335. doi: 10.1128/aem.62.4.1329-1335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raices M, Paifer E, Cremata J, Montesino R, Stahlberg J, Divne C, Szabo I, Henriksson G, Johansson G, Pettersson G. Cloning and characterization of a cDNA encoding a cellobiose dehydrogenase from the white rot fungus Phanerochaete chrysosporium. FEBS Lett. 1995;369:233–238. doi: 10.1016/0014-5793(95)00758-2. [DOI] [PubMed] [Google Scholar]
- 26.Sims P, James C, Broda P. The identification, molecular cloning and characterisation of a gene from Phanerochaete chrysosporium that shows strong homology to the exo-cellobiohydrolase I gene from Trichoderma reesei. Gene. 1988;74:411–422. doi: 10.1016/0378-1119(88)90174-6. [DOI] [PubMed] [Google Scholar]
- 27.Sims P, Soares-Felipe M, Wang Q, Gent M, Tempelaars C, Broda P. Differential expression of multiple exo-cellobiohydrolase I-like genes in the lignin-degrading fungus Phanerochaete chrysosporium. Mol Microbiol. 1994;12:209–216. doi: 10.1111/j.1365-2958.1994.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 28.Smith M H, Gold M H. Phanerochaete chrysosporium β-glucosidase: induction, cellular localization, and physical characterization. Appl Environ Microbiol. 1979;37:938–942. doi: 10.1128/aem.37.5.938-942.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sposato P, Ahn J, Walton J. Characterization and disruption of a gene in the maize pathogen Cochiobolus carbonum encoding a cellulase lacking a cellulose binding domain and hinge region. Mol Plant Microbe Interact. 1995;8:602–609. doi: 10.1094/mpmi-8-0602. [DOI] [PubMed] [Google Scholar]
- 30.Stewart P, Kersten P, Vanden Wymelenberg A, Gaskell J, Cullen D. The lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation, and the identification of a second dimorphic chromosome. J Bacteriol. 1992;174:5036–5042. doi: 10.1128/jb.174.15.5036-5042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teeri T T. Crystalline cellulose degradation: new insight into the function of cellobiohydrolase. Trends Biotechnol. 1997;15:160–167. [Google Scholar]
- 32.Tempelaars C, Birch P, Sims P, Broda P. Isolation, characterization, and analysis of the cbhII gene of Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:4387–4393. doi: 10.1128/aem.60.12.4387-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uzcategui E, Ruiz A, Montesino R, Johansson G, Pettersson G. The 1,4-β-d-glucan cellobiohydrolase from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. J Biotechnol. 1991;19:271–286. doi: 10.1016/0168-1656(91)90064-3. [DOI] [PubMed] [Google Scholar]
- 34.Vanden Wymelenberg A, Covert S, Cullen D. Identification of the gene encoding the major cellobiohydrolase of the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:3492–3494. doi: 10.1128/aem.59.10.3492-3494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Nuss D. Induction of a Cryphonectria parasitca cellobiohydrolase I gene is suppressed by hypovirus infection and regulated by a GTP-binding-protein-linked signaling pathway involved in fungal pathogenesis. Proc Natl Acad Sci USA. 1995;92:11529–11533. doi: 10.1073/pnas.92.25.11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westermark U, Eriksson K-E. Carbohydrate-dependent enzymatic quinone reduction during lignin degradation. Acta Chem Scand. 1974;28:204–208. [Google Scholar]
- 37.Wood T M. Fungal cellulases. Biochem Soc Trans. 1992;20:46–52. doi: 10.1042/bst0200046. [DOI] [PubMed] [Google Scholar]