Abstract
Methylobacterium sp. strain CM4 metabolized chloromethane quantitatively with a molar yield of 2.8 g of whole-cell protein/mol of C. This value was similar to that observed after growth with methanol (2.9 g of protein/mol of C) and about three times larger than the yield with formate (0.94 g of protein/mol of C). Chloromethane dehalogenation activity was inducible. MiniTn5 transposon insertion mutants with altered growth characteristics with chloromethane and other C1 compounds were isolated and characterized. Nine of these were unable to grow with chloromethane but were able to grow with methanol, methylamine, or formate. Seventy-three transposon mutants that were defective in the utilization of either methanol, methylamine, methanol plus methylamine, or formate could still grow with chloromethane. Based on the protein yield data and the properties of the transposon mutants, we propose a pathway for chloromethane metabolism that depends on methyltransferase and dehydrogenase activities.
Chloromethane is of environmental concern since it is believed to be responsible for about 15% of the destruction of the stratospheric ozone layer (14). The primary sources of the compound are from biological and nonbiological processes that occur in nature (5). Chloromethane can be cometabolized both oxidatively (15, 16) and hydrolytically (7) by bacteria. In addition, several methylotrophic bacteria have been characterized which are able to utilize chloromethane as a growth substrate. These include the strictly anaerobic homoacetogenic bacterium strain MC (12) and several aerobic methylotrophs of the genera Hyphomicrobium and Methylobacterium (3, 6). Anoxic dehalogenation of chloromethane by strain MC has been shown to be catalyzed by an enzyme that transfers the methyl group of chloromethane onto tetrahydrofolate and thereby releases inorganic chloride and methyl tetrahydrofolate (13). It is not known how the aerobic methylotrophs metabolize chloromethane, although closely related species are known to dehalogenate dichloromethane by a glutathione-S-transferase-dependent mechanism (9). Therefore, we have initiated physiological and genetic studies to explore the mechanism of chloromethane metabolism in Methylobacterium sp. strain CM4, a recently isolated chloromethane-utilizing methylotrophic bacterium (3). This organism was chosen since in preliminary experiments it has proven accessible to transposon insertion mutagenesis (17).
Growth and cell yields with C1 substrates.
Methylobacterium sp. strain CM4 (3) was kindly provided by Y. A. Trotsenko (Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Pushchino, Russia). It was grown in minimal mineral medium which contained (per liter of distilled water) KH2PO4 (1.9 g), Na2HPO4 (5.1 g), (NH4)2SO4 (2.0 g), and MgSO4 · 7H2O (0.1 g) at a pH of 7.2. After sterilization, 1 ml of a trace element solution containing (in grams/liter) FeSO4 · 7H2O (1.0), MnSO4 · H2O (1.0), Na2MoO4 · H2O (0.25), H3BO3 (0.10), CuCl2 · H2O (0.25), ZnCl (0.25), NH4VO3 (0.10), Co(NO3)2 · 6H2O (0.25), NiSO4 · 6H2O (0.10), H2SO4 (9.2), and 1 ml of a 25-g/liter solution of Ca(NO3)2 was added to 1 liter of medium. Cultures (1 liter) were grown at 30°C on a rotary shaker (180 rpm) in 5-liter Erlenmeyer flasks sealed with rubber stoppers. For growth with chloromethane, the chloromethane gas was added directly to the headspace through the rubber stopper with a syringe to a final concentration of 2% (vol/vol) and was further added during growth as needed. Sterile NaOH (5 M) was also added periodically to maintain the pH at 7.2. For growth on other carbon sources, methanol, methylamine, or formate was added to a final concentration of 40 mM from sterile filtered solutions. The cells were harvested (10,000 × g, 15 min) in the late-exponential-growth phase, washed with minimal medium twice, and either used immediately or frozen at −20°C as a cell pellet. These cells were then used to measure O2 uptake activities (10), chloride liberation (1), and whole-cell protein as described previously (11).
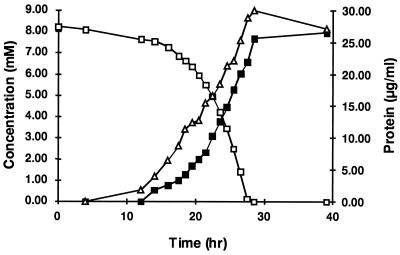
During growth, Methylobacterium sp. strain CM4 metabolized chloromethane with the concurrent production of chloride and protein. The production of bacterial protein precedes chloromethane utilization and chloride formation, and this lack of proportionality may be due to initial growth at the expense of endogenous reserve material (Fig. 1). When 2% (vol/vol) chloromethane was supplied in the headspace, the specific growth rate (μ) was 0.12 h−1. At higher concentrations of chloromethane the growth rate decreased. Nitromethane, dichloromethane, trichloromethane, and n-haloalkanes were not growth substrates, and no chloride or nitrite was released from these compounds by the cells. At a concentration of 2% (vol/vol), bromomethane was rapidly dehalogenated by resting cells grown with chloromethane. Attempts to use dibromomethane (2% [vol/vol]) as the sole substrate failed, probably due to toxicity. Iodomethane was also dehalogenated by these cells, but it was not tested as a growth substrate since it was readily hydrolyzed to methanol in the medium used. For resting cells of Methylobacterium sp. strain CM4, the molar ratio of chloride produced to chloromethane utilized was 1.03 ± 0.08, whereas 1.52 ± 0.06 mol of oxygen was consumed per mol of chloromethane. These values are in agreement with 1 mol of chloromethane being metabolized to produce 1 mol of CO2 and 1 mol of hydrochloric acid while consuming 1.5 mol of O2.
FIG. 1.
Metabolism of chloromethane by Methylobacterium sp. strain CM4. Chloromethane disappearance in the headspace was analyzed by gas chromatography. Chloride and protein appearance in the solution were measured colorimetrically. Symbols: □, chloromethane (mM); ▪, chloride (mM); ▵, protein (μg/ml).
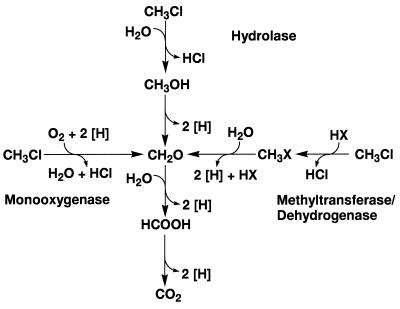
With a methylotrophic bacterium such as Methylobacterium sp. strain CM4, the observed reaction stoichiometry is consistent with three possible mechanisms that involve either a hydrolase, a monooxygenase, or a methyltransferase-dehydrogenase in the chloromethane metabolic pathway (Fig. 2). In the first step of a putative hydrolytic pathway, methanol and chloride are produced from chloromethane in an oxygen-independent manner. Methanol is then oxidized to formaldehyde and further to formate and to carbon dioxide by methanol, formaldehyde, and formate dehydrogenases, respectively, yielding a total of six reducing equivalents. For this pathway, the growth yield with chloromethane would therefore be the same as that for growth with methanol. As is evident from Fig. 2, the methyltransferase-dehydrogenase pathway would also yield a total of six reducing equivalents per chloromethane metabolized and have a growth yield with chloromethane that would be the same as that for growth with methanol. We observed growth yields of 2.8 ± 0.1 g of protein/mol of C with chloromethane and 2.9 ± 0.1 g of protein/mol of C with methanol and thus cannot discriminate between the hydrolase and the methyltransferase-dehydrogenase pathways.
FIG. 2.
Possible mechanisms for the metabolism of chloromethane by methylotrophs. Each of the possible pathways, i.e., hydrolytic, monooxygenase dependent, and methyltransferase-dehydrogenase dependent, follows a stoichiometry of CH3Cl + 3/2 O2 → CO2 + H2O + HCl.
In contrast, from growth yield data for Methylobacterium sp. strain CM4, it is unlikely that chloromethane is metabolized via the putative monooxygenase pathway (Fig. 2). In the first step of this pathway, chloromethane is oxidized to chloromethanol, which then breaks down abiotically to form formaldehyde and chloride in an oxygen-dependent manner. The formaldehyde is then further oxidized to formate and to carbon dioxide, yielding a total of four reducing equivalents. Because the initial monooxygenase-catalyzed step consumes two of these four reducing equivalents, a net yield of only two reducing equivalents is expected, and the growth yield with chloromethane should be the same as that for growth with formate. With formate, however, we have observed a growth yield of 0.94 ± 0.02 g of protein/mol of C, a value amounting to one-third of that obtained with chloromethane. A degradative pathway including a monooxygenase reaction hence appears to be highly unlikely.
The growth yield values measured for Methylobacterium sp. strain CM4 are comparable to the values reported for different bacteria which assimilate C1 compounds via the serine pathway. Yield constants (given as grams of cell dry weight/mol of C) range from 9.8 to 13.1 on methanol, 7.2 to 9.6 on formaldehyde, and 3.35 to 6.95 on formate (4).
Inducibility of chloromethane utilization.
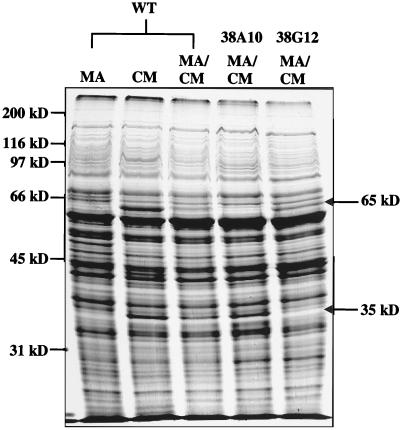
Resting cells of Methylobacterium sp. strain CM4 grown with chloromethane oxidized methanol, formaldehyde, and formate at rates comparable to those of cells grown with methanol (data not shown). However, cells grown with methanol were unable to metabolize chloromethane, indicating that chloromethane metabolism was inducible. Since all attempts to demonstrate chloromethane dehalogenase activity with extracts of chloromethane-grown cells failed, we examined the protein pattern in crude extracts by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), looking for proteins that were specifically synthesized during growth with chloromethane. Cells grown with methanol, chloromethane, or a mixture of these two carbon sources were suspended (20% wet weight/volume of phosphate buffer [50 mM, pH 7.2]) and disrupted by three passages through a French pressure cell (55 mPa). After centrifugation to remove cell debris (10,000 × g, 15 min) the cell extracts were subjected to SDS-PAGE according to the method of Laemmli (8) on 12% gels, and the proteins were visualized by staining with Coomassie blue R-250. As shown in Fig. 3, extracts of cells grown with chloromethane and extracts of chloromethane-methanol-grown cells contained a 65-kDa protein and a 35-kDa protein, indicating that growth with chloromethane involves specific induction of at least two proteins.
FIG. 3.
SDS-PAGE (12%) of chloromethane-induced proteins from Methylobacterium sp. strain CM4. Lanes: WT, 10 μg of protein from extracts of wild-type; MA, cells grown on methanol; CM, cells grown on chloromethane; MA/CM, cells grown on methanol and chloromethane; 38A10 and 38G12, 10 μg of protein from extracts of Cmu− mutant cells, i.e., 38A10 (a group 1 mutant) and 38G12 (a group 2 mutant), grown with methanol in the presence of chloromethane. The arrows indicate the 65- and 35-kDa chloromethane-induced proteins.
Phenotypic characterization of chloromethane utilization-negative mutants.
The data described so far show that chloromethane metabolism proceeds by an inducible pathway based on either a hydrolytic or a methyltransferase-dehydrogenase dehalogenation mechanism. To discriminate between these two mechanisms, chloromethane utilization-negative (Cmu−) transposon insertion mutants were isolated and phenotypically characterized. Transposon mutagenesis of Methylobacterium sp. strain CM4 was carried out by using the miniTn5 system (2). The transposon was introduced into the methylotroph, with a transposition frequency of 10−7, by plate conjugation on Luria-Bertani medium at 30°C for 24 h with a donor/recipient ratio of 1:4. Escherichia coli S17-1 λpir (pUT-miniTn5Km) was used as the donor strain. Exconjugants were selected at 30°C on minimal agar plates containing 10 mM methylamine and 10 mM formate plus 25 μg of kanamycin per ml and were incubated in a closed desiccator containing chloromethane and methanol in the headspace. Kanamycin-resistant colonies of Methylobacterium sp. strain CM4 appeared after 7 days and were inoculated into microtiter plate wells containing 100 μl of minimal medium with methanol, methylamine, and formate (10 mM each) and 25 μg of kanamycin per ml. After overnight growth at 30°C, a multipin replicator was used to transfer each mutant onto four separate agar plates, each containing kanamycin and only one of the four growth substrates (in the plate or in the headspace). Mutants unable to grow with one or more of the C1 compounds were repurified on plates and tested further in liquid media.
From a total of 4,032 kanamycin-resistant Methylobacterium exconjugants tested, 53 were unable to grow with methanol, 7 did not utilize methylamine, 11 were defective in the utilization of both methanol and methylamine, 9 were Cmu−, and 2 could not grow with formate and accumulated formate when grown with chloromethane. The Cmu− mutants were all able to grow with the other C1 substrates tested and all the mutants unable to grow with the other C1 substrates were Cmu+. This shows that methanol, the product of a putative hydrolytic dehalogenation mechanism, is not an intermediate of chloromethane metabolism (Fig. 2). Therefore, the most likely route for chloromethane metabolism is a pathway based on a methyltransferase-dehydrogenase dehalogenation mechanism. Although the first step of a putative methyltransferase-dehydrogenase pathway is oxygen independent, the second step is dependent on oxygen as a terminal electron acceptor for the reducing equivalents produced by the dehydrogenase. This may explain why resting cells grown with chloromethane did not liberate chloride from chloromethane in the absence of oxygen but instead immediately upon the addition of air to the incubation mixture began to release chloride at a constant rate (data not shown). Under anoxic conditions, the methylated intermediate produced in the first step of the methyltransferase-dehydrogenase pathway may accumulate, preventing further dehalogenation and limiting the amount of chloride produced to levels below the detection limit of the chloride assay.
The nine Cmu− mutants could be divided phenotypically into two groups. Group 1 mutants (Table 1) could not grow with or dehalogenate chloromethane. Group 2 mutants, by contrast, released chloride from chloromethane but were also unable to use it as a growth substrate. All group 1 mutants produced the 35-kDa chloromethane-induced protein, whereas three mutants of this class were defective in the formation of the 65-kDa protein (Fig. 3). Southern blot analysis (15) of genomic DNA isolated from the mutants indicated that the transposon insertions of two group 1 mutants, 22B3 and 38A10, were probably in the same DNA region (Table 1). The five group 2 Cmu− mutants produced the 65-kDa protein but two of them, 30F5 and 38G12, did not form the 35-kDa chloromethane-induced protein (Fig. 3). From Southern blot analysis, these two mutants probably had transposon insertions in the same DNA region. Growth with chloromethane and the ability to dehalogenate this compound therefore seem to be encoded at several different loci. Thus, the two chloromethane-induced proteins observed represent only part of the chloromethane utilization system that is present in this bacterium.
TABLE 1.
Phenotypes of Methylobacterium sp. strain CM4 chloromethane utilization-negative mutants
| Strain | Growth substratea | Production of chloride:
|
Presence of chloromethane-induced proteins on SDS-PAGEc:
|
DNA restriction fragments (kb) on Southern blotting withd:
|
|||
|---|---|---|---|---|---|---|---|
| During growth | In resting cells (μmol/min/mg of protein)b | 65 kDa | 35 kDa | ClaI | KpnI | ||
| Wild type | CM | + | 1.16 | +++ | +++ | − | − |
| Wild type | MA | − | <0.05 | − | − | − | − |
| Wild type | MA-CM | + | 0.29 | + | +++ | − | − |
| Group 1 mutants | |||||||
| 19D10 | MA-CM | − | <0.05 | + | +++ | 3.9 | 7.8 |
| 22B3 | MA-CM | − | <0.05 | − | +++ | 8.2 | 7.1 |
| 38A10 | MA-CM | − | <0.05 | − | +++ | 8.2 | 7.1 |
| 27C10 | MA-CM | − | <0.05 | − | +++ | 12.4 | 11.7 |
| Group 2 mutants | |||||||
| 36D3 | MA-CM | + | 0.86 | + | +++ | 4.2 | 7.8 |
| 30F5 | MA-CM | + | 0.32 | + | − | 8.2 | 6.9 |
| 38G12 | MA-CM | + | 0.37 | + | − | 8.2 | 6.9 |
| 27B11 | MA-CM | + | 0.67 | + | +++ | 12.4 | 12.4 |
| 11G7 | MA-CM | + | 0.28 | + | +++ | 9.9 | 15.7 |
None of the mutants grew with chloromethane (CM) as the sole growth substrate. MA, methanol.
Average of triplicate runs.
+++, CM-grown wild-type levels; +, MA-CM-grown wild-type levels; −, not detectable.
Probes were synthesized from MiniTn5Km sequence.
In summary, the growth experiments and the mutant characterization reported here suggest that Methylobacterium sp. strain CM4 metabolizes chloromethane by initial dehalogenation via a methyl transfer reaction, followed by a series of dehydrogenase-based steps which are different from those involved in the downstream steps of methanol or methylamine metabolism in the same organism. Our studies thus provide a testable hypothesis for the as-yet-unknown dehalogenation reaction involved in aerobic chloromethane metabolism. So far the nature of the dehalogenating enzyme which is thought to transfer the methyl group of chloromethane onto an acceptor has remained unknown. Addition of glutathione, which is involved in the dehalogenation of dichloromethane by Methylobacterium sp. strain DM4 (9), did not lead to stable chloromethane dehalogenase activity in cell extracts of Methylobacterium sp. strain CM4, so another nucleophilic cofactor may be necessary for the dehalogenation of chloromethane. Analysis of the genes inactivated by transposon insertion in the Cmu− mutants is in progress and may shed light on the biochemistry of the chloromethane degradative pathway.
Acknowledgments
This work was supported by a grant from the Swiss Federal Institute of Technology, Zürich, Switzerland.
We thank Yuri A. Trotsenko who, in the course of INTAS project 94-3122, made Methylobacterium sp. strain CM4 available to us.
REFERENCES
- 1.Bergmann J G, Sanik J. Determination of trace amounts of chlorine in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 2.De Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 3.Doronina N V, Sokolov A P, Trotsenko Y A. Isolation and initial characterization of aerobic chloromethane-utilizing bacteria. FEMS Microbiol Lett. 1996;142:179–183. [Google Scholar]
- 4.Goldberg I, Rock J S, Ben-Bassat A, Mateles R I. Bacterial yields on methanol, methylamine, formaldehyde, and formate. Biotechnol Bioeng. 1976;18:1657–1668. doi: 10.1002/bit.260181202. [DOI] [PubMed] [Google Scholar]
- 5.Harper D B. Biosynthesis of halogenated methanes. Biochem Soc Trans. 1994;22:1007–1011. doi: 10.1042/bst0221007. [DOI] [PubMed] [Google Scholar]
- 6.Hartmans S, Schmuckle A, Cook A M, Leisinger T. Methyl chloride: naturally occurring toxicant and C-1 growth substrate. J Gen Microbiol. 1986;132:1139–1142. [Google Scholar]
- 7.Keuning S, Janssen D B, Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Leisinger T, La Roche S, Bader R, Schmid-Appert M, Braus-Stromeyer S, Cook A M. Chlorinated methanes as carbon sources for aerobic and anaerobic bacteria. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, United Kingdom: Intercept Ltd.; 1993. pp. 351–363. [Google Scholar]
- 10.Locher H H, Leisinger T, Cook A M. 4-Sulphobenzoate 3,4-dioxygenase. Purification and properties of a desulphonative two-component enzyme system from Comamonas testosteroni T-2. Biochem J. 1991;274:833–842. doi: 10.1042/bj2740833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mägli A, Rainey F A, Leisinger T. Acetogenesis from dichloromethane by a two-component mixed culture comprising a novel bacterium. Appl Environ Microbiol. 1995;61:2943–2949. doi: 10.1128/aem.61.8.2943-2949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messmer M, Wohlfarth G, Diekert G. Methyl chloride metabolism of the strictly anaerobic methyl chloride-utilizing homoacetogen strain MC. Arch Microbiol. 1993;160:383–387. [Google Scholar]
- 13.Messmer M, Reinhardt S, Wohlfarth G, Diekert G. Studies on methyl chloride dehalogenase and O-demethylase in cell extracts of the homoacetogen strain MC based on a newly developed coupled enzyme assay. Arch Microbiol. 1996;165:12–25. [Google Scholar]
- 14.Montzka S A, Butler J H, Myers R C, Thompson T M, Swanson T H, Clarke A D, Lock L T, Elkins J W. Decline in the tropospheric abundance of halogen from halocarbons: implications for stratospheric ozone depletion. Science. 1996;272:1318–1322. doi: 10.1126/science.272.5266.1318. [DOI] [PubMed] [Google Scholar]
- 15.Rasche M E, Hicks R E, Hyman M R, Arp D J. Oxidation of monohalogenated ethanes and n-chlorinated alkanes by whole cells of Nitrosomonas europaea. J Bacteriol. 1990;172:5368–5373. doi: 10.1128/jb.172.9.5368-5373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stirling D I, Dalton H. The fortuitous oxidation and cometabolism of various carbon compounds by whole-cell suspensions of Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1979;5:315–318. [Google Scholar]
- 17.Vannelli, T., and T. Leisinger. Unpublished data.