Keywords: hypertrophy, mechanical overload, myofiber, resistance training, skeletal muscle
Abstract
Mechanisms underlying mechanical overload-induced skeletal muscle hypertrophy have been extensively researched since the landmark report by Morpurgo (1897) of “work-induced hypertrophy” in dogs that were treadmill trained. Much of the preclinical rodent and human resistance training research to date supports that involved mechanisms include enhanced mammalian/mechanistic target of rapamycin complex 1 (mTORC1) signaling, an expansion in translational capacity through ribosome biogenesis, increased satellite cell abundance and myonuclear accretion, and postexercise elevations in muscle protein synthesis rates. However, several lines of past and emerging evidence suggest that additional mechanisms that feed into or are independent of these processes are also involved. This review first provides a historical account of how mechanistic research into skeletal muscle hypertrophy has progressed. A comprehensive list of mechanisms associated with skeletal muscle hypertrophy is then outlined, and areas of disagreement involving these mechanisms are presented. Finally, future research directions involving many of the discussed mechanisms are proposed.
CLINICAL HIGHLIGHTS.
Loss of muscle mass with aging and certain noncommunicable diseases (e.g., cancer, COPD, and others) is associated with increased mortality. Thus, understanding the mechanisms controlling skeletal muscle hypertrophy can help determine the most effective interventions to preserve or enhance muscle mass.
Studies in animals and humans suggest that mechanical overload (e.g., resistance training) best achieves skeletal muscle hypertrophy. Bouts of mechanical overload induce transient increases in mammalian/mechanistic target of rapamycin complex 1 (mTORC1) signaling leading to elevations in muscle protein synthesis rates. With repeated bouts of mechanical overload, these events contribute to skeletal muscle hypertrophy.
An expansion in translational capacity through ribosome biogenesis and increases in satellite cell abundance and myonuclear accretion also contribute to skeletal muscle hypertrophy following days to weeks of repeated mechanical overload bouts.
Aside from these three aforementioned mechanisms, several lines of past, current, and emerging research suggest that other mechanisms may also contribute to mechanical overload-induced skeletal muscle hypertrophy (e.g., mTORC1-independent signaling, microRNAs, genetic polymorphisms, and enhanced angiogenesis among others).
There are also potential manners in which epigenetic alterations in myonuclear and mitochondrial DNA, extracellular matrix remodeling, cytoskeletal remodeling, mitochondrial biogenesis, bioenergetic adaptations, and other mechanisms can contribute to mechanical overload-induced skeletal muscle hypertrophy.
The current and rapidly emerging molecular tools available to researchers as well as rodent and human studies being performed in tandem will continue to provide insight into novel mechanisms that are needed for mechanical overload-induced skeletal muscle hypertrophy to occur.
1. INTRODUCTION
Hypertrophy (hy·per·tro·phy)
/hīˈpərtrəfē/
Noun PHYSIOLOGY
Definition: Increase in the size of a tissue or organ as a result of an increase in cell size rather than increased numbers of cells (hyperplasia).
Source: Oxford Dictionary of Sports Science and Medicine
Skeletal muscle hypertrophy occurs in response to various loading paradigms over prolonged periods, and these stimuli have been deemed as providing “mechanical overload” to the involved musculature. There are various methods to achieve mechanical overload in animals including the surgical removal of synergist muscles (i.e., synergist ablation), simulated resistance training through electrical hindlimb stimulation, loaded wheel running, weighted ladder climbing, weighted sled pulling, weighted limb stretches, and resistance-loaded devices to challenge animals as they obtain food. Mechanical overload in humans is most adequately achieved through progressive resistance training. When performed consistently, resistance training over an 8- to 16-wk period can lead to a 5–20% increase in skeletal muscle volume or mass in younger to middle-aged adults (1).
Over the past 40 years researchers have sought to identify the mechanisms that are associated with mechanical overload-induced skeletal muscle hypertrophy. These investigations have led to a plethora of comprehensive reviews on this topic (2–52), and the word cloud sizing in the graphical abstract is representative of these viewpoints.
Although sections of the present review rearticulate excellent perspectives from these reviews, the broader aims here are to 1) provide a historical perspective of the early discoveries in the field (∼late 1800s to 1940s), discuss animal and human studies from the 1960s to 2000s that were the first to mechanistically interrogate mechanical overload-induced skeletal muscle hypertrophy, and highlight how these studies have guided current-day research efforts; 2) discuss highly investigated mechanisms that are thought to promote skeletal muscle hypertrophy, as well as opposing evidence when applicable; and 3) posit mechanisms that may be involved in promoting skeletal muscle hypertrophy but have little to no evidence and warrant further investigation. Other sections of the present review include discussions on skeletal muscle architecture, methodological considerations with skeletal muscle hypertrophy research, and an abbreviated discussion focused on how sex, race, and aging affect hypertrophic outcomes.
2. BRIEF OVERVIEW: MUSCLE AS TISSUE, MYOFIBERS, THE EXTRACELLULAR MATRIX, AND OTHER RESIDENT CELLS
Mammalian muscle cells (a.k.a. myofibers) are typically considered postmitotic (or nondividing) and possess a unique structure in that they are multinucleated and the largest mammalian cells with a tubular morphology (53). It has been posited that myofibers require multiple nuclei to regulate cellular homeostasis (i.e., the myonuclear domain theory) (51, 54). Most of the intracellular area in myofibers is occupied by myofibrils (∼70–85%) (55–57), and these specialized organelles consist of thick filaments, thin filaments, and other associated proteins. These proteins contribute to active and passive force generation as well as sarcomere structure for muscle contraction. A cytoskeletal network is also present within myofibers and consists of actin, microtubules, microfilaments, and other associated proteins that anchor nuclei and myofibrils within the cell, while also serving as a scaffold for force transmission (58). Proteins enriched in myofibers have been subclassified into different categories including (59) 1) contractile proteins (e.g., myosin, actin, tropomyosin, troponins), 2) sarcomeric-associated proteins (e.g., titin, myosin binding protein C, α-actinin, myomesin, and M protein), 3) cytoskeletal proteins (e.g., tubulin, desmin, and actin), and 4) membrane-associated proteins (e.g., dystrophin, spectrin, talin, vinculin, and ankyrin). Although the ultrastructural characteristics of myofibers have been largely limited to two-dimensional analyses, the Glancy laboratory (60) has recently used three-dimensional ion beam scanning electron microscopy (FIB-SEM) to show that myofibers contain interconnected myofibrils whereby branching is higher in slow-twitch versus fast-twitch myofibers in adult mice.
Some of the intracellular space within myofibers (∼5–10%) is also occupied by the mitochondrial reticulum and sarcoplasmic reticulum, and these organelles are primarily responsible for supporting muscle contraction through adenosine triphosphate (ATP) replenishment and calcium handling, respectively (61). Again, the Glancy laboratory (62) has also provided excellent insight into muscle mitochondrial structure, using FIB-SEM to develop the hypothesis that membrane potential conduction is the prominent pathway for skeletal muscle energy distribution. The cytoplasm (a.k.a. sarcoplasm) is an aqueous medium that facilitates the exchange of ions and metabolites to and from different organelles (8). Several enzymes, proteins, and protein complexes that facilitate anabolic and catabolic reactions also reside in the sarcoplasm. The cell membrane of myofibers is termed the sarcolemma, and this structure contains transmembrane proteins that aid in ion transport, nutrient transport, ligand-receptor signaling, and the anchoring of intracellular cytoskeletal proteins to the extracellular matrix (8). The latter of these functions is responsible for force transmission during muscle contraction from the Z disk to the basal lamina via the dystrophin-glycoprotein and integrin adhesion complexes (63).
The basal lamina is a thin layer of connective tissue that sheaths myofibers, is linked to the sarcolemma through protein-protein interactions, and is considered part of the extracellular matrix (64). Proteins enriched in the basal lamina include fibronectin, laminin, α-dystroglycan, and other proteins that participate in the sarcolemmal protein-protein interactions discussed above (65). The thick fibrillar extracellular matrix (a.k.a. the interstitial matrix) is mainly comprised of collagen proteins and various extracellular growth factors (65). In muscle tissue, a variety of cell types reside in the extracellular matrix and include resident immune cells, fibro-adipogenic progenitor cells, fibroblasts, adipocytes, endothelial cells, and pericytes (66). At the interface between the sarcolemma of myofibers and the basal lamina are satellite cells. Microscopic evidence supports that myofibers, rather than stromal cells of the extracellular matrix, spatially occupy ∼85–90% of muscle tissue (67). This is largely due to myofiber cross-sectional areas in adults typically averaging between 5,000 and 6,000 µm2 and the cell bodies of stromal cells only being slightly larger than their nuclei, which (although not commonly measured) average to be <100 µm2 (68, 69). However, it should be noted that the extracellular matrix contains numerous stromal cell types that outnumber the presence of myofibers. In this regard, histological examinations of muscles from young healthy adults suggest that per 100 myofibers there are ∼10 satellite cells (70), ∼2 lymphocytes and ∼20 macrophages (71, 72), ∼30 fibro-adipogenic progenitor cells (73), ∼13 fibroblasts (74), ∼35 pericytes (75), and ∼200 capillaries, which are comprised of endothelial cells (76, 77). Thus, cells residing in the extracellular matrix potentially outnumber myofibers at a ratio of ∼3:1, and this estimate does not consider age-related changes or the influx of cells into muscle tissue following exercise bouts or injury (e.g., neutrophils and macrophages) (71, 78).
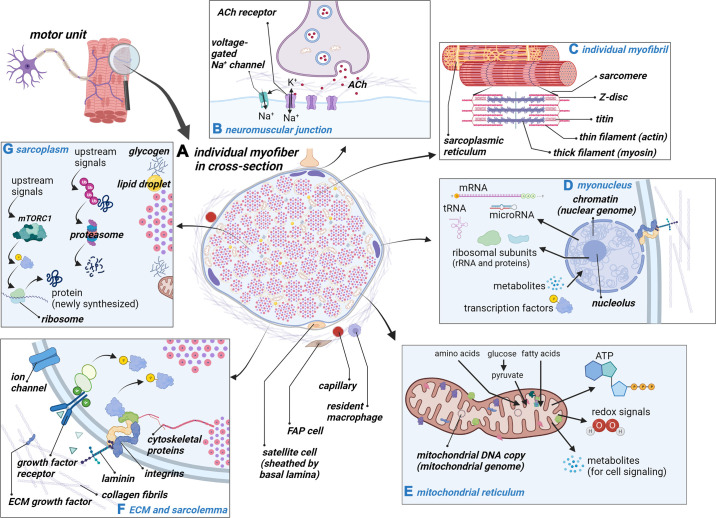
Single-nucleus sequencing studies in rodents have recently provided estimates of the cell types contained in muscle tissue (66, 79). These investigations suggest that of the total nuclear pool in skeletal muscle tissue ∼50–70% are associated with myofibers, 20% are from fibro-adipogenic progenitor cells, 17% are from endothelial cells, 4% are from pericytes, 3% are neuronal, 3% are from macrophages, 2% are from satellite cells, and 1% are from neutrophils. Notably, these estimates are not constant, since some reports suggest that slow-twitch myofibers contain more nuclei per fiber than fast-twitch fibers (80, 81), and immune cell abundance is also higher in slow-twitch versus fast-twitch muscles (82). FIGURE 1 provides a summary diagram of content discussed in this section of the review, and readers are referred to other recent reviews providing related schematics (1, 48, 58, 65, 83–85).
FIGURE 1.
Skeletal muscle fiber components and biological processes. This schematic (constructed with BioRender.com, with permission) illustrates the molecular attributes and processes that occur in a myofiber. A represents an individual myofiber in cross section as well as some of the stromal cells that exist in the extracellular matrix. B depicts the neuromuscular junction and how the ligand binding of acetylcholine (ACh) can lead to myofiber activation through voltage-gated sodium (Na+) channels. C shows an individual myofibril and some of the prominent proteins that make up the structure of the sarcomere. D shows a single myonucleus, some of its key structures (e.g., chromatin), and some of its functions (e.g., RNA transcription and output). E depicts a portion of the mitochondrial reticulum, some of its key structures (e.g., mitochondrial DNA), and some of its key functions (e.g., producing ATP and metabolites). F shows the interface of the extracellular matrix (ECM) and muscle cell membrane (or sarcolemma), and signaling through growth factor receptors and laminin-integrin complexes are also summarized. G shows a portion of the sarcoplasm (which makes up <10% of the myofiber spatially), some of the features between myofibrils (e.g., glycogen granules and lipid droplets), and some of the many reactions that can occur in this region (note that protein synthesis can also occur at ribosomes localized in close proximity to myofibrils). FAP, fibro-adipogenic progenitor cell; K+, potassium; mTORC1, mammalian target of rapamycin complex 1.
A final topic of discussion in this section is myofiber type classification and some of the characteristic differences that exist between myofiber types. As described by Schiaffino and Reggiani (86), mammalian skeletal muscle contains different myofiber types that can be differentiated by either myosin isoforms and contraction speed or metabolic characteristics such as oxidative capacity. Whereas the authors explained the history of past methods used for fiber type classification (e.g., red vs. white appearance and histological classifications using succinate dehydrogenase and myosin ATPase staining), monoclonal antibodies against different myosin heavy chain isoforms developed in the 1980s have been widely used via immunohistochemistry to report myofiber type adaptations to mechanical overload (87). The four predominant mammalian myosin heavy chain isoforms include the slow-twitch type I isoform (encoded by the MYH7 gene) and fast-twitch isoforms including IIA (encoded by the MYH2 gene), IIX (encoded by the MYH1 gene), and IIB (encoded by the MYH4 gene) (86). Although most fibers express a prominent myosin heavy chain isoform, hybrid myofibers coexpressing multiple isoforms in humans have been reported with histochemical and electrophoresis-based techniques (88–93). Notwithstanding, several studies suggest that the commonly biopsied vastus lateralis (VL) muscle in men and women contains a high percentage of type I myofibers (∼30–50%) and type IIA + IIA/X hybrid fibers (∼40–50%) and ∼5% of I/II-coexpressing hybrid myofibers and ∼2% of type IIX myofibers (88, 91, 94, 95). Moreover, a common adaptation to resistance training (and endurance training) in humans is the rapid downregulation of IIX gene expression and shift of IIX + IIA/X to IIA myofibers (96, 97).
Intracellular morphology may differ between type I and IIA/X myofibers, albeit this may depend on species. Beyond myosin typing, the characterization of metabolic features within muscle fibers gets more complex (93), so again it is important to consider the muscle analyzed and species. In rats, Schiaffino et al. (98) used transmission electron microscopy (TEM) to report that slow-twitch soleus myofibers (which possess >90% type I fibers) contain more mitochondria in cross section relative to extensor digitorum longus (EDL) myofibers (which possess >90% type II fibers). These authors also reported that myofibril diameters are larger in fast-twitch EDL myofibers, although Z disks are slightly thicker, and sarcomeres are slightly longer in slow-twitch soleus myofibers. These myofiber type morphology differences in rats are not as dramatic in humans. For instance, Alway and colleagues (57) reported that type I and II myofibers from gastrocnemius and soleus biopsies in men spatially possess ∼5% and ∼3% mitochondria, respectively, whereas myofibril area occupies similar intracellular spacing in both fiber types (∼80%). Ruple et al. (55) more recently used immunohistochemistry to report that type I and II myofibers from the VL muscle in men spatially possess ∼5–6% mitochondria, whereas myofibril area occupied similar intracellular spacing in both fiber types (∼80%), and this largely agrees with a prior study by Wang et al. (99), who used TEM to interrogate type I and II myofiber characteristics from VL muscle tissue in 12 women. Also notable are the data from Wang and colleagues suggesting that myofibril size (∼0.70 µm2) and density (1.06 myofibrils per µm2) are similar in type I and IIA myofibers in humans. Interestingly, recent protein expression profiling between type I and II myofibers in humans indicates that, of the ∼3,800 proteins detected, ∼400 (or 10%) show significant fiber type-specific differences. Hence, these proteome profiles between type I and II myofibers, rather than robust morphology differences, likely drive the divergence in cellular phenotypes (i.e., oxidative potential, force generation, and excitation-contraction coupling characteristics) (100).
3. A HISTORICAL ACCOUNT OF RESISTANCE TRAINING RESEARCH, MECHANICAL OVERLOAD STUDIES IN RODENTS, AND THE MOLECULAR INTERROGATION OF MUSCLE HYPERTROPHY
Research into skeletal muscle hypertrophy has flourished over the past 50 years. However, a general interest in this topic has existed for several millennia. Milo of Croton (∼6th century B.C.) is considered by most to be the first documented practitioner of progressive resistance training. According to anecdote and written history, Milo hauled a newborn calf (which developed into a full-grown bull) over his shoulders daily for nearly 4 years, leading to enhancements in muscle mass and strength (101). Despite the clear implications of progressive overload, scientists would not intensively research resistance training for another two and a half millennia. Much of the current mainstream interest in resistance training and skeletal muscle hypertrophy was largely driven by Eugen Sandow (102), a Prussian bodybuilder and showman (∼1890s), and Canadian Louis Cyr, who was an avid weightlifter and strongman (∼1880s to 1890s) (103).
The notion that tissue could grow via cellular hypertrophy can be traced back to classic work of the German pathologist Rudolf Virchow. In 1858, Virchow (104) published a study detailing the morphology of lymph nodes through the use of microscopy. Virchow reported greater cell counts in enlarged versus normal-appearing lymph nodes and reported that other organs could grow without increasing cell number. These observations led to the current-day definitions of “hyperplasia” and “hypertrophy” and inspired Morpurgo’s landmark study published in 1897 showing that skeletal muscle hypertrophy occurs in response to exercise training (105). Interestingly, Morpurgo reported that 2 mo of treadmill training increased sartorius myofiber diameters by ∼50% without increasing myofiber number or length, and he termed this phenomenon “work-induced hypertrophy.” This study was the first scientific documentation of skeletal muscle hypertrophy in response to exercise training. Nevertheless, research from the early 1900s to 1930s that examined work-induced hypertrophy neglected skeletal muscle in favor of cardiac muscle adaptations (reviewed in Ref. 106), with these studies utilizing regimens that were endurance training by today’s standards.
Scientific writings on progressive resistance training surfaced around the time of Morpurgo’s work. Wilhelm Roux and Willi Lange authored perspectives between 1895–1917 suggesting that skeletal muscle hypertrophy occurred when bouts of work intensity routinely exceeded that which was performed during normal daily activities (107). Dr. Theodor Hettinger, a research fellow at the Max Planck Institute from 1950 to 1960, attributed the beginnings of resistance training research to studies published by Petow, Siebert, and Eyster between 1925 and 1927 that documented the strength adaptations to weightlifting (107). Viewpoints on the hypertrophic and strength adaptations to resistance training were also provided in a 1933 commentary by Steinhaus (108) and in MacFadden’s 1940 Encyclopedia of Health and Physical Education. In the mid-1930s through the 1940s entrepreneurs Bob Hoffman (founder of York Barbell Company) and Joe Weider (founder of the International Federation of Bodybuilding as well as several mainstream fitness magazines) largely stoked public interest in resistance training (109). Skeletal muscle hypertrophy research was also published in the late 1940s and early 1950s (110–112), albeit these observational human studies examined masseter muscle hypertrophy due to excessive chewing or clinching of the jaws under stressful conditions. As written by Bompa and Haff (113), theories regarding recovery and adaptation from exercise training were also published in the 1940s by Folbrot. The triphasic stress response termed the “general adaptation syndrome” (or “GAS”) by Hans Selye (114) was published in this same era, and the exercise physiology field has largely adopted this model to explain the stress response to mechanical overload.
In 1945, US Army Captain Dr. Thomas Delorme (115) reported that progressive resistance training promoted skeletal muscle hypertrophy and a restoration of muscle function in rehabilitating soldiers. This publication led to a flurry of human research into how resistance training affected strength and local muscle endurance in diseased and rehabilitating patients (116–119). Around the same time (circa 1949), Novikov and Ozolin published papers detailing the implementation of complex training methods (i.e., strength and endurance training) (120). Although these papers were informative, there would be a 25-year lapse between these reports and mechanistic human investigations since skeletal muscle biopsies were not widely adopted in the research setting until the late 1960s.
Although sparse research in the 1950s and early 1960s utilized exercise paradigms in livestock to examine meat quality outcomes (121, 122), Geoffrey Goldspink (1964) (123) published the first mechanistic interrogation of mechanical overload-induced skeletal muscle hypertrophy. The author indicated that myofiber and myofibril diameters are generally larger in mice trained with a resistance-loaded pulley apparatus versus untrained mice, albeit it is unclear how this interpretation was formulated given that the light microscopy that was utilized did not provide clear resolution of myofibrils. Notwithstanding, this publication, in part, led to a widely adopted mindset in the field that resistance training increases myofiber hypertrophy through increasing myofibril size rather than number. Three years after Goldspink’s report, Goldberg (124) used tenotomy to elicit hindlimb skeletal muscle hypertrophy in rats (i.e., functional overload). This technique was later refined in mice (125), and these seminal investigations led to the widespread utilization of the synergist ablation model to study hypertrophic mechanisms. The synergist ablation model surgically excises a portion of the gastrocnemius muscle, which imposes continuous mechanical overload on the remaining muscle(s). Numerous rodent studies have utilized various forms of synergist ablation (126), and although it is generally viewed as a nonphysiological form of rapid hypertrophy, these studies were foundational for many of the anabolic signaling mechanisms that are discussed here. Interestingly, a 1975 review by Goldberg and colleagues (127) summarizing findings of studies that utilized these models indicated that overload-induced skeletal muscle hypertrophy was largely independent of endocrine factors (e.g., growth hormone, insulin, testosterone, and thyroid hormones). This viewpoint has since been supported by research suggesting that intrinsic signaling mechanisms, such as mechanotransduction-based signaling, are prominently responsible for mechanical overload-induced skeletal muscle hypertrophy (128, 129).
In the 1960s, researchers utilized biochemical assays to determine how postnatal muscle growth affects tissue protein, deoxyribonucleic acid (DNA), and ribonucleic acid (RNA) concentrations in animals (130, 131). First-generation tracer studies were performed using “hot” radioisotopes in isolated muscles and cell-free preparations in the late 1950s and early 1960s to determine the fate of amino acids exposed to cellular environments (132, 133). These studies inspired subsequent research in the late 1960s and early 1970s by Goldberg, Millward, and others, who administered radioisotope tracers to live rodents to determine muscle protein, DNA, and RNA synthesis rates during different loading paradigms (134–137). The collective evidence from these studies supported that mechanical overload increased the synthesis rates of these macromolecules. Human research examining skeletal muscle tissue adaptations to resistance training also surfaced in this same decade because of skeletal muscle biopsy sampling, pioneered for research purposes by Jonas Bergstrom in 1962 (138). Penman (1969) (139) provided the first report in humans that used TEM to describe the ultrastructural myofiber adaptations in response to 8 wk of knee extensor resistance training. In 1970, Penman (140) published a similar report with three college-aged male participants who underwent 10 wk of resistance training. Although Penman’s investigations were limited in scope, his scientific approach of obtaining skeletal muscle tissue from humans who performed resistance training was soon adopted by other scientists in the field.
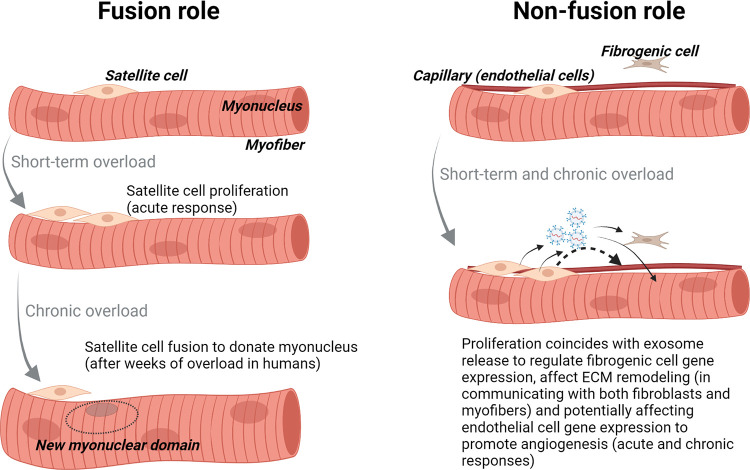
In the late 1960s and early 1970s, histological staining methods were used to characterize the metabolic phenotypes of slow- and fast-twitch myofibers. Animal work by Ogata and Mori (141), Edgerton et al. (142), Barnard et al. (143), and Brooke and Kaiser (144) and human work by Edström and Nyström (145) were foundational in establishing several of these techniques. A 1972 paper by Gollnick et al. (146) built upon Penman’s previous work given that it was adequately powered from a statistical perspective to compare fiber type characteristics between weightlifters, endurance-trained athletes, and untrained participants. The investigators subjected muscle sections to specialized reagents to assess glycogen content, myosin ATPase activity (for myofiber type), succinate dehydrogenase and reduced diphosphopyridine nucleotide-diaphorase (DPNH-diaphorase) activity (for oxidative capacity), and phosphofructokinase activity (for glycolytic capacity). Compared with the other participants, weightlifters were reported to possess larger myofibers as well as a lower percentage of oxidative myofibers. In 1973, Schiaffino and Bormioli (147) utilized similar histological techniques to support that synergist ablation in rats elicits myofiber growth accompanied by a shift toward a more oxidative phenotype. Additional work performed by Schiaffino and colleagues (148) in the early 1970s, which involved [3H]thymidine administration and TEM-based autoradiography, indicated that satellite cell proliferation occurs days after mechanical overload induced by synergist ablation in rats. These authors published a separate report supporting the idea that satellite cells become incorporated into the myofibers as myonuclei (149). These studies provided the first evidence that satellite cells have a role in mechanical overload-induced skeletal muscle hypertrophy and inspired work in this area of muscle biology described below in this review.
The first study to formally assess whether myofiber hypertrophy occurred in humans with resistance training was published by Thorstensson and colleagues (150) in 1976. Although it was reported that 8 wk of resistance training increased strength outcomes, slow- and fast-twitch myofiber cross-sectional area (fCSA) values were not significantly altered. The authors hypothesized that the duration of training was not long enough to observe the myofiber hypertrophy reported in the weightlifters that Gollnick and colleagues examined (146). Contrary to this report, 1979 reports by Dons et al. (151) and Costill et al. (152) indicated that myofiber hypertrophy occurred after 7 wk of resistance training. That same year, Moritani and deVries (153) published a landmark electromyography paper indicating that neural factors accounted for the initial strength gains during the first few weeks of resistance training. The authors also posited that muscle hypertrophy (as assessed through limb circumference measurements) became a more influential factor for continued strength gains thereafter. MacDougall et al. (154) published a paper in 1980 showing that 6 mo of resistance training increased slow- and fast-twitch fCSA values, and the authors utilized TEM to support a mechanism of myofiber growth primarily occurring through the expansion of the sarcoplasmic space. Although these findings were provocative, several studies published in later years would challenge this mode of hypertrophy (1, 155), and this area of the literature remains controversial. In the 1980s, several research groups continued to detail the histological, biochemical, and ultrastructural differences of biopsied muscle between weightlifters and nonweightlifting participants (56, 57, 156–162). Research by Staron, Hikida, Dudley, Kraemer, Gonyea, and others in the 1990s documented how weeks to months of resistance training in previously untrained participants affected fCSA as assessed by the myosin ATPase staining technique (99, 163–166). Research by Tesch, Costill, and associated colleagues, which employed biochemical assays and other staining techniques, was also published in this same era detailing metabolic adaptations in muscle tissue following months of resistance training (152, 167, 168). Although hyperplasia has been largely dismissed as a significant contributor to mammalian skeletal muscle hypertrophy (1), it is notable that the Gonyea laboratory performed experiments on this topic during the 1980s and 1990s by chronically stretching and loading the anterior latissimus dorsi muscle in quails (169–172) or performing various forms of resistance training in cats (173, 174). Indeed, much of the resistance training research performed during the 1970s and 1980s provided information on myofiber size and metabolic adaptations. However, aside from sparse TEM reports by MacDougall and colleagues and others through the early 1990s (56, 67, 99, 175), researchers have since largely neglected examination of the ultrastructural adaptations that occur in myofibers in response to mechanical overload.
Seminal molecular work in the late 1980s, the 1990s, and the early 2000s led to a research breakthrough focused on mechanisms associated with skeletal muscle hypertrophy. One line of research spanning from the early to late 1990s involved infusing stable isotope tracers into human participants to assess the muscle protein synthesis and breakdown kinetics to single or multiple bouts of resistance exercise (176–180). The collective evidence from these studies indicated that changes in muscle protein synthetic and breakdown rates were significantly elevated for hours to days after resistance exercise bouts. These findings led to the widely adopted hypothesis that resistance training facilitates myofiber hypertrophy through intracellular protein accretion and that this process is largely driven by pulsatile postexercise increases in muscle protein synthesis that eventually supersede muscle protein breakdown rates as individuals become more trained (181). This initial human tracer research was innovative and foundational in establishing the more accessible and less invasive methodology of administering deuterium oxide (D2O) to humans and rodents through drinking water to assess longer-term (or integrated) mixed-muscle or myofibrillar protein synthesis rates (182, 183); notably, these reports provide cumulative protein synthetic responses ranging from days to months into training interventions. What is less appreciated, however, is that the rodent radioisotope tracer work discussed above in this section largely inspired this line of research in humans.
Another line of innovative molecular research during this era involved the elucidation of skeletal muscle mRNA and phosphosignaling responses to mechanical loading, and again studies in rodents predated human investigations. In 1990, the Booth laboratory was the first to document skeletal muscle RNA expression responses to acute and chronic mechanical overload. In short, these authors reported that one bout of concentric exercise via hindlimb muscle stimulation against load in rats did not affect relative expression levels of α-actin mRNA, cytochrome c mRNA, 18S rRNA, or 28S rRNA in the gastrocnemius muscle (184). However, 10 wk of training increased the overall abundance of these RNAs when considering training-induced increases in gastrocnemius masses. These authors published a second paper detailing the RNA responses that occurred in rat tibialis muscles that were eccentrically targeted during the acute and chronic stimulation models (185). This work largely guided subsequent research examining the acute and chronic RNA responses to mechanical overload and resistance training in rodents and humans, respectively. Additionally, this work inspired research into how concentric- versus eccentric-only training affects the molecular milieu in skeletal muscle (186).
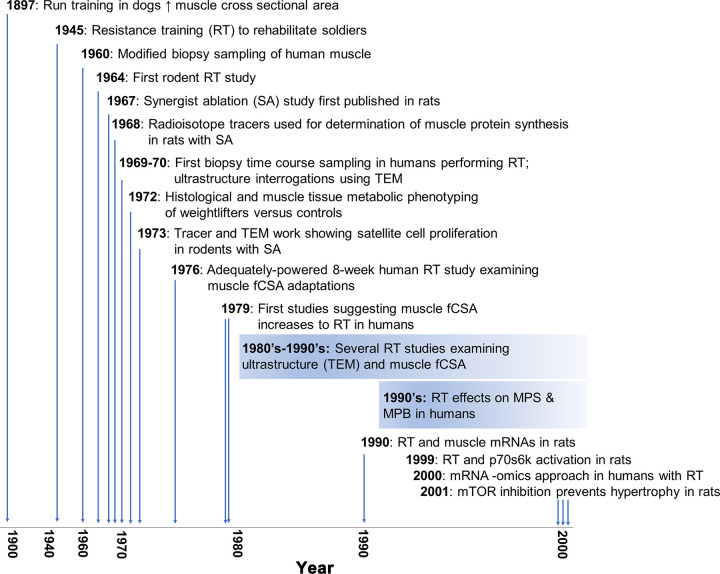
In 1999, Baar and Esser (187) published a landmark paper on muscle signaling responses to mechanical overload. In short, the authors used Western blotting to demonstrate that the phosphorylation status of the 70-kDa S6 protein kinase (p70S6K) protein 6 h after stimulated lengthening contractions was associated with the degree of muscle hypertrophy in various rat hindlimb muscles after 6 wk of training using the same stimulation protocol. This finding was confirmed by Nader and Esser (2001) (188), who reported that the prolonged increase in p70S6K and a transient increase in protein kinase B phosphorylation occur in response to a hypertrophy-inducing stimulus but not low-frequency stimulation or running exercise. Notably, studies in humans published 5–7 years later indicated that similar signaling responses occur in response to a resistance exercise bout (189, 190). In 2001, Bodine and colleagues (191) published a landmark study that built upon Baar and Esser’s work showing that rapamycin, a mammalian/mechanistic target of rapamycin (mTOR) inhibitor, blunted plantaris hypertrophy following 14 days of mechanical overload induced by synergist ablation in rats. This response was also reported to coincide with the diminished phosphorylation of p70S6K, which is now appreciated as being a downstream kinase that is phosphorylated and activated by mammalian/mechanistic target of rapamycin complex 1 (mTORC1) (192). These findings were, in part, validated in humans 5 years later by the Rasmussen laboratory, who reported that rapamycin administration blocks the early (1–2 h after exercise) increases in muscle protein synthesis and mTORC1 signaling after a resistance exercise bout (190). Another notable milestone publication around this time (2000) was the first human muscle mRNA-omics dataset published by the Peterson laboratory (193). These authors isolated muscle RNA from 12 older and 11 younger participants before and 24 h after a resistance exercise bout and used [32P]ATP labeling during cDNA construction (which preceded the currently used fluorometric technology) before chip hybridization reactions. Of the 588 annotated targets the array provided probes for, the authors reported that vascular endothelial growth factor (VEGF) mRNA, inflammatory mRNAs (IL-1β and RANTES), and immediate-early response mRNAs (c-jun, EGR-1) were dynamically altered at the postexercise time point in both cohorts. Indeed, this publication led the way to current high-density microarray and RNA-sequencing (RNA-seq) investigations whereby load-induced changes in all annotated muscle mRNAs and several annotated miRNAs and small RNAs can be interrogated (194–196). A timeline of studies discussed in this section is summarized in FIGURE 2.
FIGURE 2.
An overview of landmark studies. A timeline of landmark studies investigating skeletal muscle adaptations to mechanical overload in rodents and subsequent resistance training studies in humans. fCSA, myofiber cross-sectional area; MPB, muscle protein breakdown; MPS, muscle protein synthesis; mTOR, mammalian/mechanistic target of rapamycin; p70S6K, 70-kDa S6 protein kinase; TEM, transmission electron microscopy.
From the early 2000s to the present day, independent research groups around the world have utilized assays to determine the transient RNA, phosphosignaling, and protein synthetic responses to mechanical overload in animals and resistance exercise bouts in humans (197–217). The widespread availability of antibody-conjugated chromagens and fluorophores to label proteins in myofibers, or cells in the extracellular matrix, has also led to a greater understanding of the cellular and molecular signaling responses to resistance training (218–230). The advancement of genetic mouse models has enabled the determination of genes that may be critically involved in promoting load-induced skeletal muscle hypertrophy (231–233). Also notable is the advent and utilization of various -omics-based techniques in human and rodent resistance training and mechanical overload studies. These investigations have included chip-based genomics (234), chip- and sequencing-based transcriptomics (194, 195, 235–252), DNA methylomics (195, 253–258), and mass spectrometry-based proteomics (100, 195, 259–265), phosphoproteomics (217, 264, 266–268), and metabolomics (269–271). The democratization of these techniques has led to a rapid expansion of molecular data in the field, and current-day research now involves analyzing large-scale multi-omics-based datasets. Examples of such efforts include 1) the implementation of the MetaMEx interactive database by the Zierath laboratory to elucidate changes in mRNA expression across 66 exercise studies that contained muscle transcriptome information (272); 2) work from the Phillips laboratory that used a variety of bioinformatics approaches to validate a gene signature responsive to mechanical loading in humans that was associated with hypertrophy and in vitro experiments indicating that this signature is functionally is associated with protein synthesis (250); and 3) the broader goal of the Molecular Transducers of Physical Activity Consortium (MoTrPAC) to overlay multiple -omics-based datasets and generate a molecular map that is triggered by single and multiple bouts of resistance training (7, 273). These efforts have and will continue to greatly expand the amount of information related to molecular signaling events that are associated with load-induced skeletal muscle hypertrophy. However, the need for research utilizing innovative genetic rodent models will also persist so that novel signaling mechanisms can be validated through loss- and gain-of-function studies. The utilization of higher-throughput in vitro contraction models (see Ref. 274 for example) is also needed to validate or unveil novel contraction-induced signaling mechanisms associated with myofiber hypertrophy. In silico analyses as described by Rupert et al. (275) can also be performed to develop novel hypotheses in this area of muscle physiology by leveraging online rodent muscle phenotype, genotype, and transcriptomic databases. Finally, refining -omics-based pathway analyses approaches will be instrumental in examining novel mechanisms associated with mechanical overload-induced skeletal muscle hypertrophy. Although this topic is beyond the scope of this review, an excellent review by Stokes et al. (276) provides guidance in performing pathway analyses and modeling of -omics data.
4. METHODOLOGICAL CONSIDERATIONS WITH SKELETAL MUSCLE HYPERTROPHY RESEARCH
To fully appreciate the content of this review, readers should be aware of various methodological aspects involved in skeletal muscle hypertrophy research including tissue processing, limitations to whole tissue lysate analysis, the interpretation of molecular data, time course considerations, the general lack of agreement between surrogate measures of skeletal muscle hypertrophy, and the human translatability of rodent studies.
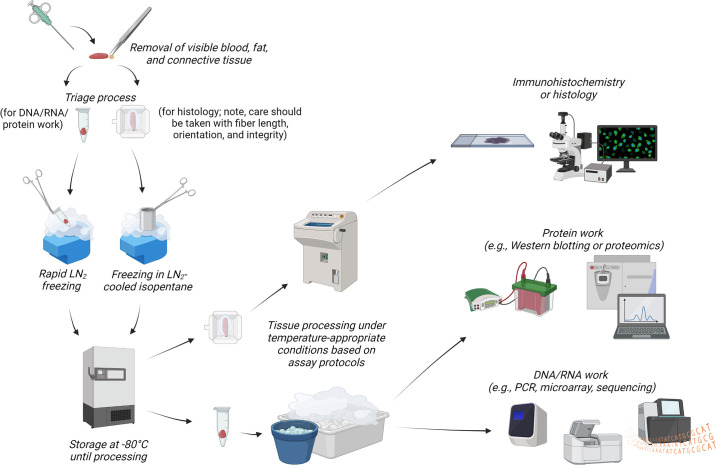
First, muscle-molecular outcomes can be affected by tissue collection, preservation, and processing methods. Although many studies indicate that tissue is “immediately processed and frozen for future analysis,” there is often little to no description of the time taken to preserve tissue for the different analyses. Oftentimes researchers collect animal or human skeletal muscle specimens for multiple assays, which requires more time to triage and preserve samples before freezing. A significant time lapse in tissue processing (e.g., 5 s vs. 10 min) may result in biomarker quality issues (277), and evidence in rodents indicates that postmortem delays in tissue processing cause a linear decay in RNA quality and an exponential decay in phosphoprotein status (278, 279). Tissue retrieval from deep freeze storage and thawing to isolate RNA or protein can also have deleterious effects on RNA and phosphoprotein quality (280, 281). Finally, histological artifacts due to freeze fracture can arise if tissue is not properly mounted and/or is frozen improperly (282). To mitigate some of these issues, researchers are encouraged to preserve tissue after excision as quickly as possible (e.g., snap-freeze in liquid nitrogen within a minute after extraction). Additionally, tissue can be preserved in specialized reagents to preserve RNA integrity (283). Finally, there are published protocols detailing the process of proper muscle tissue preservation for histology (282, 284). A brief illustrative summary of proper muscle tissue processing is provided in FIGURE 3.
FIGURE 3.
General muscle tissue processing steps for histology and molecular analyses. Muscle tissue procurement from human and animal studies involves either a biopsy (humans) or dissections (rodents; not pictured). It is advised that the removal of visible blood, fat, and connective tissue, tissue triage, and liquid nitrogen (LN2) tissue preservation occur as rapidly as possible (e.g., between 1 and 3 min). Noted in the diagram are different preservation methods when sampling tissue for histology vs. nucleic acid or protein work. Researchers are advised to consult with published literature based on the assays desired to be performed to ensure that tissues are placed in adequate buffers (if needed) before cold storage and/or LN2 freezing and deep freeze storage. Upon tissue removal from deep freeze storage, care should be taken in most circumstances to ensure that the tissue is kept in a frozen state. As illustrated in the schematic, tissue processing for nucleic acid and protein work involves keeping tissue on dry ice, LN2-cooled stages, and/or ice throughout several of the processing steps to prevent macromolecule degradation. Tissue processing for immunohistochemistry or histology on nonfixed tissue typically involves sectioning in a cryostat at approximately −20°C. Again, researchers are encouraged to consult with published literature to obtain the desired conditions based on the assay(s) desired to be performed. This schematic was constructed with BioRender.com, with permission.
Second, much of the protein, RNA, DNA, and tracer work in the field provides information on muscle tissue lysates. There is an appreciable presence of stromal cells in the extracellular matrix, as mentioned above (see FIGURE 1). Although it is commonly assumed that information acquired from whole muscle lysates represents phenomena occurring within myofibers, a certain level of non-myofiber-specific signaling exists and must be considered. Researchers are beginning to circumvent this issue by labeling, isolating, and analyzing myonuclei with specialized genetic mouse models (254, 285, 286). The utilization of immunohistochemical techniques is also becoming more common to decipher protein localization responses to mechanical overload paradigms. Notwithstanding, DNA, RNA, and protein data from crude muscle lysates is still largely prevalent in much of the research discussed in this review. Also notable, buffer formulation is critical when working with muscle tissue, and lysates yielded from nonoptimal buffers can contribute to the signal-to-noise issue discussed in this paragraph. Specialized buffers and centrifugation protocols can be used to isolate myofibrils, nonmyofibrillar proteins, mitochondria, nuclear proteins, and extracellular matrix proteins (287–290). However, researchers commonly allocate general cell lysis buffers when analyzing muscle tissue without considering optimal buffer alternatives relative to the research question. Some researchers also use precleared lysates (i.e., removal of insoluble proteins) whereas others use whole muscle lysates, and this methodological difference likely leads to different outcomes being reported. Hence, researchers should attempt to best determine buffer selection according to the research question. Furthermore, although whole muscle lysates are more difficult to work with given the poor solubility of large contractile proteins, working with muscle-specific lysis buffers that solubilize most proteins is ideal in studies that seek to examine how loading paradigms affect certain aspects of the muscle-molecular milieu (e.g., enzyme activities or protein-protein interactions). Finally, whole tissue analysis does not reflect motor unit recruitment changes, which is one of the earliest physiological adaptations to resistance training (291). Whole tissue analysis collectively assays recruited and nonrecruited myofibers together, which has the potential to dilute signals in myofibers that are recruited during training. Single-myofiber analyses provide a more nuanced view of the myofiber size/function and signaling dynamics, albeit there can still be contamination of adherent stromal cells (254). Although single-fiber analysis is cumbersome, this analysis has been performed by several independent laboratories (265, 292–295), and recent work from Murgia et al. (100) indicates that single-fiber preparations from humans are suitable for proteomic analysis.
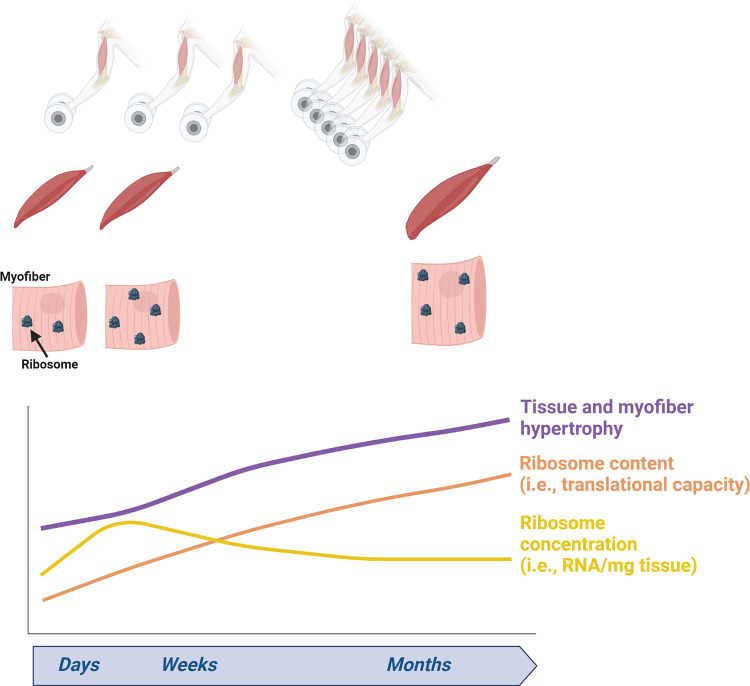
Third, molecular data interpretation can be challenging in the context of skeletal muscle hypertrophy. Some assays (e.g., qPCR and Western blotting) normalize molecular targets to housekeeping genes or proteins (296, 297), both of which can be altered during or after periods of mechanical overload (298, 299). Western blotting normalization for protein expression can be achieved through Ponceau or stain-free signals, which represents the total solubilized protein pool (296, 300). Phosphorylated proteins are commonly normalized to pan (or total) protein levels for a given target. Omics-based assays have specialized normalization procedures as well. Chip-based RNA and DNA assays are normalized to a global fluorescent intensity (301), mass spectrometry-based proteomic data are normalized as a percentage of total spectra (261), and RNA-seq data are commonly normalized to read counts (302). Finally, there are commonly interrogated variables such as total muscle RNA (a surrogate of ribosome content) and muscle citrate synthase (CS) activity (a surrogate of mitochondrial volume density). These variables can be normalized to wet or dry muscle weights, albeit CS activity can also be normalized to muscle or mitochondrial protein content. It is critical to appreciate that muscle tissue and myofiber hypertrophy is accompanied by an absolute increase in muscle protein and macromolecule content as discussed above. Thus, in models that induce skeletal muscle hypertrophy, researchers should ensure that their normalization variable (e.g., normalizer protein or housekeeping gene mRNA) is not altered. Slight nonsignificant changes in opposing directions from the target and normalization marker could show significant differences in the target-normalized outcome. Likewise, it is important to conceptualize that modest changes, no changes, or even a decrease in the relative abundance or concentration of a target molecule during tissue hypertrophy can indicate an increase in the overall abundance (and thus an upregulation) of the molecule (184). As a contextual example, Roberts et al. (80) reported that rat plantaris total RNA concentrations (µg/g wet tissue) are 19% higher in hindlimbs subjected to 14 days of synergist ablation compared with sham-treated legs. When considering that plantaris masses are also 25% higher in the surgical versus sham-treated legs, plantaris total RNA content in the surgical versus sham-treated legs is estimated to be 47% higher by multiplying RNA concentration in micrograms per gram of wet tissue by wet tissue weights. The Booth laboratory (184, 185) adopted a similar approach when reporting rRNA and mRNA content differences between nonexercised rats and rats that performed hindlimb resistance-like training; specifically, targets were presented in relative (% of total extracted RNA) and absolute (RNA content adjusted for muscle mass) terms. Hence, although not commonly adopted, it is recommended that researchers reporting protein or RNA expression changes during chronic periods of mechanical overload discuss (or even report) how the degree of hypertrophy potentially alters the relative versus total content of assayed biomarkers. Finally, non-steady-state differential equation models have been championed when using D2O during atrophy models to calculate integrated protein synthetic rates since calculations are contingent on muscle protein pool size (303), and the same could be argued for muscle hypertrophy models that elicit increases in the total muscle protein pool.
Fourth, the timing of tissue sampling after a bout or period of mechanical overload can be critical relative to the research question. For example, the temporal pattern of changes in protein synthesis following exercise does not directly track with intracellular signaling responses (17, 26, 304–306), and this is likely related to the timing of tissue sampling that will require innovative approaches to address. The RNA profile of muscle can vastly differ when sampled minutes, hours, or days after a resistance exercise bout (307, 308). Incongruent findings between muscle protein turnover rates and hypertrophy during the earlier periods of resistance training have also been noted (304, 309, 310), albeit this relationship is more coupled as subjects become trained (311). These findings indicate that exercise-induced protein synthesis rates may be more of a hypertrophic stimulus as these signals become more “refined” with training. Unfortunately, several studies that have examined the muscle protein synthesis response to naive bouts of resistance exercise likely interrogated a damage-synthesis response versus the hypertrophy-synthesis response that occurs later into training (310). Hence, the timing of tissue sampling is critically important to consider depending on the research question (e.g., examining peak mRNA responses to mechanical overload vs. phosphosignaling responses, etc.).
Fifth, it is often underappreciated that the size and percentage of slow- and fast-twitch myofibers differ depending on depth and proximo-distal location in a muscle group in humans and rodents (312, 313). This issue is difficult to mitigate in humans given the invasiveness of obtaining multiple biopsies. In rodents, however, a common practice is to examine most (if not all) myofibers at the midbelly of excised muscle. Also notable, data from multiple methods used to assess whole muscle versus myofiber size changes to mechanical overload in rodents and humans yield weak-to-moderate correlations. This observation was first noted by Gordon (314) in animals in the 1960s when comparing changes in mean fCSA and muscle weights following a period of treadmill training. Similar findings have since been reported in human resistance training studies that have compared MRI-derived VL muscle volume changes to ultrasound-derived VL muscle thickness changes (315), MRI-derived midthigh muscle CSA changes to dual-energy X-ray absorptiometry (DXA)-derived leg lean mass changes (316), and MRI- and ultrasound-based tissue level changes to mean fCSA changes (317). Why these discrepancies exist is not discussed here, and interested readers are encouraged to refer to Haun et al. (83) for more details. However, readers should be aware that this is still a salient issue that has not been resolved.
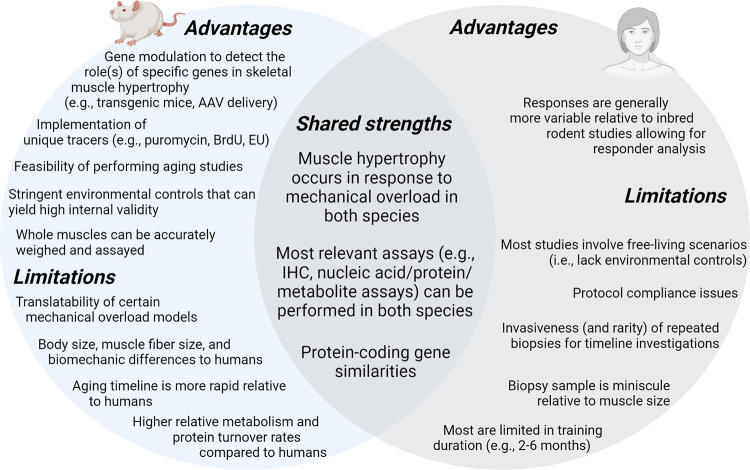
Finally, there are strengths and weaknesses with rodent models. Other than the clear discrepancies in myofiber size (81) and oxidative phenotype (93), metabolic and protein turnover rates are appreciably different between humans and rodents (318). Muscle protein synthesis rates have been reported to be 1.3- to 2-fold greater in type I versus type II fibers in rodents (319), and similar evidence exists (295). Conversely, type I versus II fiber differences in muscle protein synthesis rates are less dramatic in the resting and postexercise states in humans (∼10–30%) (295, 320). Mechanical overload models in rodents vary in duration and stimulus, and the advantages and disadvantages of these models have been more thoroughly described by Lowe and Alway (321) and Booth and Thomason (322) and more recently by Murach et al. (232). Although studies using this procedure have yielded insightful information, it is a surgical model in which the intact muscle(s) is exposed to persistent load and exhibits rapid hypertrophy. Hence, despite the discussion of several studies using the synergist ablation model, the unfamiliar reader should be aware that this model does not resemble the physiological stimulus provided through progressive resistance training. Finally, it is common for researchers to examine rodents between the ages of 2 and 4 mo, and this can yield incongruent results between studies because this time frame is a formative stage of muscle maturation in the animal (323). There are, however, several strengths with rodent models. For instance, although it has been reported that human and mouse genomes on the whole show ∼40% sequence overlap (324), there are similarities between genomes such as genome size (human [GRCh30]: 3,088,269,832; mouse [GRCm38]: 2,725,521,370) and the number of protein-coding genes (human: 19,950; mouse: 22,018), and both species possess ∼70% sequence similarities in protein coding gene sequences (325, 326). Genetic mouse models have also been developed to determine loss or gain of function in relation to signaling mechanisms involved with skeletal muscle hypertrophy, and this approach is not possible in humans. Commonly interrogated hindlimb rodent muscles such as the soleus and plantaris predominantly consist of type I or type II fibers (80), whereas human muscles that are commonly biopsied contain a mixture of fibers as discussed in sect. 2. Hence, unless single-fiber approaches are used in humans, examining fiber type-specific mechanisms associated with hypertrophy may be more fruitful in rats given that dual overload via synergist ablation differentially affects muscle protein synthesis, ribosome biogenesis, proteasome activity levels, satellite cell counts, and the magnitude of hypertrophy in the type I fiber-prominent soleus versus type II fiber-prominent plantaris muscle (80). The use of adeno-associated virus (AAV) vectors is becoming more widespread in rodents given the high uptake efficiency across most myofibers within a muscle (275). Genes delivered through AAV-based vectors can also be coupled with a muscle-specific promoter to transduce muscle-specific gene expression across virtually all muscles in the animal. Gene delivery is also possible through the electroporation of plasmids containing genes encoding proteins or shRNAs for gene knockdown (327), and this methodology also has good utility in examining mechanisms. The ability to control environmental factors more stringently in rodents, such as food administration and the light-dark (sleep) cycle, is also advantageous in reducing variability in outcome measures. Finally, a prominent theme in this review is that several mechanistic rodent studies predated (and were confirmed) by human discoveries (see FIGURE 3). Thus, although limitations exist, the discussed strengths and general human translatability of rodent models in skeletal muscle hypertrophy research illustrate their utility in examining associated mechanisms. FIGURE 4 provides a summary of advantages, limitations, and shared strengths of rodent and human studies in the literature.
FIGURE 4.
Advantages, limitations, and shared strengths of rodent and human studies. This figure (created with BioRender.com, with permission) summarizes the advantages and limitations of using rodent models. Additionally, limitations of human studies are presented. Finally, shared strengths of both models are displayed in the overlap region of the Venn diagram. AAV, adeno-associated virus; EU, 5-ethynyluridine; IHC, immunohistochemistry.
4.1. Mechanisms Commonly Associated with Skeletal Muscle Hypertrophy
4.1.1. The involvement of mTORC1 and its upstream activators.
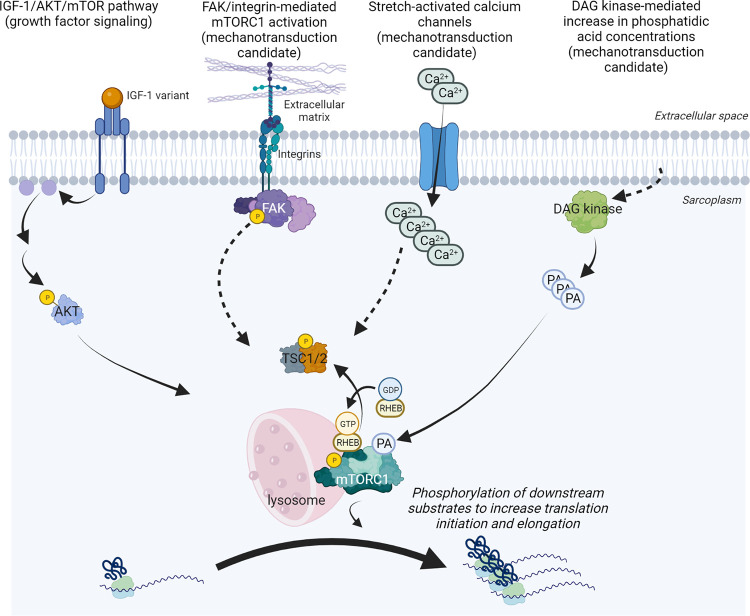
mTOR is a 289-kDa serine/threonine protein kinase in the phosphatidylinositol 3-kinase (PI3K)-related protein kinase (PI3K) family (328). In mammals, mTOR acts as a catalytic subunit of two distinct complexes known as mTOR complex 1 (previously defined as mTORC1) and complex 2 (mTORC2). These complexes differ in their accessory proteins, differential sensitivity to rapamycin, downstream substrates, and functions. Specific to this review, mTORC1 phosphorylates substrates that increase the synthesis of proteins, lipids, nucleotides, and ATP while limiting the autophagic breakdown of cellular components (329). Two and a half decades since the discovery of rapamycin (330, 331) and mTOR (332), mTORC1 has been the most investigated mechanism linked to skeletal muscle hypertrophy. mTORC1 contains six accessory proteins including (333) 1) mTOR, which possesses kinase activity, 2) mammalian lethal with sec-13 (mLST8), 3) DEP-domain containing mTOR-interacting protein (DEPTOR), 4) the Tti1/Tel2 complex, 5) regulatory-associated protein of mammalian target of rapamycin (RAPTOR), and 6) proline-rich Akt substrate 40 kDa (PRAS40). Providing a more expanded discussion of mTORC1 signaling is beyond the scope of this review, and these details are provided elsewhere (10, 13, 334). However, the reader should appreciate that active mTORC1 complexes enhance muscle protein synthesis by regulating the phosphorylation of downstream substrates involved in translation initiation (e.g., p70S6K and 4EBP1) and elongation (e.g., eEF2) (192, 335). Although various cellular conditions are needed to stimulate increases in mTORC1 activity, one that has gained recent notoriety is the interaction of the mTORC1 complex with the lysosome (221, 311, 336–338). Increased translocation of mTORC1 to the periphery of myofibers following mechanical overload has also been reported, and future investigations will likely unveil the relevance of this event (218, 221, 339).
As mentioned in sect. 3, Baar and Esser (187) and Bodine et al. (191) published landmark studies outlining the involvement of p70S6K and mTOR, respectively, in mechanical overload-induced skeletal muscle hypertrophy, and these findings were subsequently validated in other rodent studies (340, 341). Goodman et al. (342) also reported that mTOR within myofibers is the rapamycin-sensitive element that confers the hypertrophic response to mechanical overload in mice, and follow-up mouse studies provided compelling evidence to suggest that mTORC1 is critical in this process (343, 344). This animal work led to other rodent and human investigations reporting enhanced mTORC1 signaling hours to days after a bout (or bouts) of mechanical overload (189, 206, 218, 221, 225, 345–371). Although a large body of research provides strong evidence suggesting that mTORC1 signaling is involved with skeletal muscle hypertrophy, the upstream activators of mTORC1 that are responsive to mechanical loading have not been fully elucidated. Over the years, various signals have been posited to be responsible for mTORC1 activation during overload stimuli including growth factor signaling (namely insulin-like growth factor 1, or IGF1), membrane-associated proteins involved with mechanotransduction, proteins involved with amino acid sensing that converge to activate mTORC1, and other proteins that act as upstream activators and inhibitors of mTOR. These topics are the crux of discussion in this section of the review.
AKT is a protein kinase that acts as an upstream activator of mTORC1 (372). IGF1, and its muscle-specific mechano-growth factor (MGF) variant, are upregulated at the mRNA and protein levels in rodent and human skeletal muscle subjected to mechanical overload (197, 373–382). These observations largely inspired a hypothesis that was pervasive in the literature from the late 1990s through ∼2010 suggesting that postloading increases in localized IGF1 isoforms are largely responsible for mTORC1 activation via ligand binding to the IGF1 receptor, IGF1 receptor autophosphorylation, and increased AKT kinase activity (383–387). However, the IGF1 hypothesis has been rigorously challenged. Hornberger et al. (388) reported that the stretch-induced activation of mTOR signaling ex vivo is not abrogated in Akt1-knockout mice. Spangenburg et al. (389) reported that synergist ablation-induced mTORC1 signaling and plantaris hypertrophy are not perturbed in dominant-negative IGF1 receptor (Igf1r) mice. Maruyama et al. (390) used an AKT inhibitor (MK2206) in rodents to show that mTORC1 activation via hindlimb electrical stimulation occurs independently of AKT1/2 phosphorylation. Miyazaki et al. (391) reported that synergist ablation can still lead to the activation of mTORC1 in mice treated with a PI3K/AKT inhibitor. However, recent reports indicate that Akt1/2 double-knockout mice present stark impairments in muscle mass and protein synthesis during the rapid growth phase (8–12 wk old) (392), and this extends into adulthood (393). Hence, AKT may be indispensable for muscle maturation and growth, although it does not appear to have a central role in mTORC1 activation in response to mechanical overload.
The conflicting IGF1 and AKT findings presented above have, in part, shifted emphasis to the current-day mechanotransduction hypothesis of mechanical overload-induced skeletal muscle hypertrophy. This hypothesis was pioneered by Goldberg et al. (127) (1975), expanded by Vandenburgh (394) (1987), and further refined by Flück and colleagues (395–397) as well as Hornberger and colleagues (398, 399). Mechanotransduction in myofibers occurs when mechanical perturbations of the basal lamina, sarcolemma, and cytoskeleton catalyze downstream signaling events. It is thought that these biochemical signaling events, in turn, activate mTORC1 and upregulate protein synthesis in an AKT-independent manner. Hornberger and colleagues (400, 401) established that various forms of mechanical overload transiently increase myocellular concentrations of phosphatidic acid (PA) and mTORC1 signaling by stimulating the membrane-associated activity of diacylglycerol (DAG) kinases. This group has also reported that PA can directly bind to and activate mTOR (402, 403). Collectively, these studies established a working model in which muscle contractions upregulate mTORC1 activity by promoting an increase in membrane-associated DAG kinase activity and subsequent increases in intracellular PA. This model is further strengthened by research from Hornberger’s laboratory (404) showing that the knockout of the zeta DAG kinase isoform (DGKζ) significantly attenuates increases in fCSA, protein accretion, and plantaris mass after 7 days of synergist ablation. In humans, Thalacker-Mercer et al. (238) reported that human participants who exhibited the greatest muscle hypertrophy after a resistance training program (i.e., termed “extreme responders”) exhibited heightened DGKζ mRNA expression before training. Although this latter human report does not provide a cause-and-effect relationship, it further supports the involvement of DGKζ as an anabolic signal during resistance training. However, the upstream mechanisms through which mechanical stimuli increase DGKζ activity have yet to be defined, and this mechanism will likely continue to be investigated.
Another mode of skeletal muscle mechanotransduction potentially involves transmembrane integrins and accessory proteins localized to the internal portion of the sarcolemma that propagate signals to activate mTORC1 (14). This model has been largely inspired by the findings of the Booth laboratory (405) reporting that focal adhesion complex-associated proteins, specifically focal adhesion kinase (FAK), are upregulated in rat soleus muscle 1 day and 8 days after synergist ablation. In this same publication, these authors reported increased FAK autophosphorylation in rooster muscle subjected to chronic loaded stretch. The Flück laboratory (395) later reported that FAK autophosphorylation preceded increases in p70S6K activity during an unloading and reloading paradigm in mice whose hindlimb muscles are transfected with a pCMV-FAK plasmid. Crossland et al. (406) reported in vitro data in this area showing that shRNA-mediated FAK knockdown reduced IGF1-stimulated increases in myotube protein synthesis and hypertrophy. Chaillou et al. (407) reported that mRNAs related to the integrin-linked kinase pathway are upregulated during the earlier phases of synergist ablation in mice, and these authors speculated that transmembrane integrins may signal an upregulation in other genes that coordinate the anabolic response.
The notion of mechanotransduction operating through integrins or FAK signaling is scientifically grounded given the evidence discussed above. As well, FAK is localized with integrins on the interior portion of the sarcolemma (408), and FAK autophosphorylation associates with mTORC1 signaling in myotubes and other cell types (395, 409–411). However, several studies have challenged whether mechanotransduction operates through integrins or FAK to promote downstream anabolic signaling and skeletal muscle hypertrophy. Relative to wild-type mice, mice overexpressing the α7BX2-integrin subunit in skeletal muscle (α7Tg mice) exhibit reduced mTORC1 signaling after one bout of downhill running (412) despite these same mice exhibiting rapid myofiber hypertrophy after multiple bouts of the same stimulus (413). Boppart and Mahmassani (14) have also discussed unpublished findings from their laboratory showing that mTORC1 signaling trended downward in α7Tg mice compared with wild-type mice after 1 day of synergist ablation. Interestingly, Petrosino et al. (414) more recently reported that synergist ablation-induced plantaris hypertrophy is impaired in Ccn2-knockout mice and noted that the CCN2 gene (also known as connective tissue growth factor) encodes a matricellular protein that exists in the extracellular matrix. Synergist ablation-induced elevations in muscle protein synthesis are also reduced in Ccn2-knockout mice, which presented lower basal levels of pan and phosphorylated FAK concentrations in skeletal muscle. The authors hypothesized that CCN2 might stimulate mechanical overload-induced muscle protein synthesis and hypertrophy through FAK signaling. However, contrary to the authors’ own hypotheses, mechanical overload-induced increases in pan and phosphorylated FAK are not impaired in Ccn2-knockout mice 3 and 7 days after synergist ablation despite muscle protein synthesis and hypertrophy being dampened. In humans, Glover et al. (415) reported that muscle FAK phosphorylation is not transiently altered after a bout of resistance exercise despite an upregulation in p70S6K phosphorylation being observed, and similar evidence exists in rats subjected to a bout of eccentric contractions (416). Franchi et al. (417) demonstrated that FAK phosphorylation is upregulated with 8 wk of eccentric-only versus concentric-only resistance training in humans. However, both forms of training elicit similar increases in thigh lean mass values, midthigh thickness values, and 8 wk integrated muscle protein synthesis responses. It is also notable that FAK phosphorylation is prevented with mTORC1 inhibition in vitro, which suggests that FAK could be a downstream target of mTORC1 signaling rather than an upstream activator (410). Finally, sarcomere-based mechanotransduction has been shown to contribute to skeletal muscle hypertrophy independently of FAK involvement. Specifically, van der Pijl et al. (418) used a unilateral diaphragm denervation hypertrophy model in genetic mouse models in which titin stiffness is increased (TtnΔIAjxn) and decreased (RBM20ΔRRM), respectively, and reported that RBM20ΔRRM mice (decreased titin stiffness) presented significant impairments in hypertrophy whereas TtnΔIAjxn mice presented exaggerated increases in hypertrophy. Collectively, these conflicting reports make it difficult to determine whether integrin or FAK signaling is involved with load-induced increases in skeletal muscle hypertrophy.
A final mechanotransduction candidate discussed here is stretch-activated channels (SACs), which permit the influx of calcium and sodium ions into myofibers (16). There are various lines of evidence to support that contraction-induced increases in intracellular calcium increase mTORC1 signaling and muscle protein synthesis. For instance, the pharmacological blockade of SACs in rats with streptomycin has been shown to blunt eccentric contraction-induced increases in p70S6K phosphorylation (419). Others have reported that mTORC1 signaling and muscle protein synthesis are attenuated in rats administered a SAC inhibitor after eccentric contractions (420). However, there are also several independent lines of conflicting evidence in this area. The landmark paper by Bodine et al. (191) also reported that the calcium-mediated calcineurin pathway is not affected during periods of mechanical overload and calcineurin inhibition does not impair mechanical overload-induced skeletal muscle hypertrophy. It is also difficult to disentangle how calcium release from organelles in myofibers, rather than the influx of calcium into myofibers via SACs, affects mTORC1 signaling. For instance, Li et al. (421) performed in vitro experiments to show that the inhibition of calcium release from lysosomes reduces mTORC1 activity. Calcium transients from the sarcoplasmic reticulum during muscle contractions also presumably have a role in intracellular calcium signaling (422), and this mechanism operates independently of SAC-mediated calcium influx. Ito et al. (423) reported that synergist ablation-induced hypertrophy is abrogated in Nnos1-null mice. These researchers attributed this effect to a mechanism involving neuronal nitric oxide synthase (nNOS)-mediated nitric oxide formation, the subsequent formation of peroxynitrite, sarcoplasmic reticulum Trpv1 channel activation via increased peroxynitrite concentrations, and the increased influx of calcium into sarcoplasm from the sarcoplasmic reticulum to enhance mTORC1 signaling. Subsequent work by this group strengthened this mechanism (424), and a more recent paper suggests that the stimulation of the P2Y2 receptors promotes increased intracellular calcium concentrations to enhance mTORC1 signaling in the type I myofiber-rich soleus muscle (425). Again, although these data support the role of calcium in propagating anabolic signaling, these calcium-mediated mechanisms do not involve SAC-mediated mechanotransduction. Finally, the manner in which calcium activates mTOR has not been well resolved, and the involvement of calcium-mediated signaling in skeletal muscle hypertrophy has been challenged. A 1999 study supported a mechanism in which calcium-mediated calcineurin activation promotes mechanical overload-induced skeletal muscle hypertrophy in rodents (426). A separate study published the same year suggested that calcineurin acted downstream of IGF1 to elicit the nuclear translocation of the NF-ATc1 transcription factor and drive transcriptional processes that resulted in myofiber hypertrophy (427). However, subsequent research rigorously challenged the notion that calcineurin activation is involved in IGF1-mediated and/or mechanical overload-induced skeletal muscle hypertrophy (428–430). More recently, Ferey et al. (431) demonstrated that the overexpression of calcium/calmodulin-dependent protein kinase kinase-α (CaMKKα/CAMKK1), which is a prominent signaling mediator for intracellular calcium, stimulated mTORC1 signaling and muscle protein synthesis in mice. As with much of the data presented above, this finding supports the notion that calcium signaling (via CaMKKα activation) may act as an upstream activator of mTORC1. What strikingly opposes this paradigm, however, is data in this same paper showing that Camkk1-knockout mice exhibited 15% greater muscle hypertrophy and enhanced mTORC1 signaling relative to wild-type mice after synergist ablation. Prior work by Hornberger et al. (402) has also shown that the chelation of intracellular calcium with BAPTA-AM has no effect on the stretch-induced activation of mTORC1. Thus, although various lines of evidence have linked increases in intracellular calcium concentrations to enhanced mTORC1 signaling, the role that SACs (and calcium signaling at large) exhibit during load-induced increases in mTORC1 activity is riddled with conflicting data and needs further clarity.
Aside from the discussed mechanotransduction mechanisms, various upstream activators of mTORC1 signaling may be affected during periods of mechanical overload including amino acid-sensing and amino acid transport proteins. It is generally recognized that dietary proteins and essential amino acids increase mTORC1 signaling and muscle protein synthesis in the basal state (432). It is also recognized that dietary proteins and essential amino acids additively enhance anabolic signaling in skeletal muscle after a resistance exercise bout (433–435). However, preliminary data suggest that skeletal muscle upregulates the activity and content of proteins involved in the transport and sensing of amino acids in a load-dependent and nutrient-independent manner. For instance, electrically simulated hindlimb contractions have been shown to increase the activity of mammalian Vps34 (mVps34), an amino acid-sensing protein, 3 h after contractions (436). Others have reported in humans that chronic resistance training increases the expression and sarcolemmal enrichment of the L-type amino acid transporter 1 (LAT1) protein (219), which is responsible for transporting several essential amino acids into myofibers (437). Although this area is limited, the available data support that load-dependent increases in proteins that promote amino acid transport and sensing may be partially responsible for enhanced mTORC1 activation and skeletal muscle hypertrophy during periods of mechanical overload.
Other upstream activators and inhibitors of mTORC1 signaling are also altered during periods of mechanical overload. TSC2 inhibits mTORC1 by acting as a GTPase-activating protein that converts active GTP-Rheb into inactive GDP-Rheb (438), and TSC2, Rheb, and mTOR are enriched at the lysosome (438). Jacobs et al. (337) demonstrated that eccentric contractions in mice reduced the localization of TSC2 with the lysosome, coinciding with a hyperphosphorylation of TSC2. A subsequent investigation by these same researchers indicated that the inducible and skeletal muscle-specific knockout of Rheb led to a reduction in the eccentric contraction-induced activation of mTORC1 signaling (439). These studies lend support for a model in which the load-induced phosphorylation of TSC2 causes it to dissociate from the lysosome and these events enable Rheb to obtain its active GTP-bound state to upregulate mTORC1 signaling. In humans, Song et al. (221) reported that a bout of resistance exercise transiently leads to similar postloading events (e.g., dissociation of TSC2 from Rheb), lending further credibility to this model. The Regulated in DNA damage and development 1 (REDD1) protein, which is an inhibitor of mTORC1 signaling (440), may also be affected during periods of mechanical overload. Gordon et al. (352) reported that skeletal muscle REDD1 protein levels are transiently reduced after an overload stimulus in mice. These authors published a follow-up study showing that load-induced hypertrophy is enhanced in Redd1-knockout mice (350), coinciding with heightened mTORC1 activity and a reduction in autophagy. Drummond et al. (441) partially confirmed these findings in humans by showing that REDD1 mRNA is transiently downregulated 3 h after a low-intensity resistance exercise bout with blood flow restriction. However, skeletal muscle REDD1 mRNA and protein levels have been reported to be elevated 1 and 3 h after a bout of resistance exercise (203). Others have also shown that resistance exercise does not transiently affect REDD1 protein levels (442). Thus, additional research into REDD1 and its role during mechanical overload-induced skeletal muscle hypertrophy is warranted.
Finally, the increased myocellular concentrations of certain substrates linked to enhanced mTORC1 signaling may also be involved with skeletal muscle hypertrophy. For instance, polyamine synthesis enzymes are upregulated in skeletal muscle by synergist ablation in an mTORC1-dependent fashion (443). Polyamines are small compounds containing two or more amino groups (e.g., spermidine, spermine), and various polyamines are required for cellular homeostasis and protein synthesis (444). However, these data are relatively new to the field, and the function(s) that polyamines exhibit during skeletal muscle hypertrophy remains to be determined.
In summary, the collective evidence suggests that enhanced mTORC1 signaling promotes skeletal muscle hypertrophy during various loading paradigms in animals and humans. However, it is critical to note that this signaling likely needs to be pulsatile given that mTOR hyperactivity in TSC1-knockout mice increases oxidative stress, elicits myofiber damage, and causes myofiber loss over the life span (445). It is also apparent that increased mTORC1 activity during these scenarios does not require certain upstream signals such as an upregulation in IGF1 signaling, enhanced AKT activity, or calcineurin activation. Instead, several lines of evidence support that a DAG kinase-mediated increase in PA and a dissociation of TSC2 from the mTOR-lysosome complex are involved with load-induced increases in mTORC1 activity. The other upstream mTORC1 signals discussed (e.g., integrins and FAK signaling, an upregulation in amino acid transport and sensing proteins, a downregulation in REDD1) have limited supporting evidence or are confounded by inconsistent data and require further clarity. FIGURE 5 summarizes several of the mTORC1-associated mechanisms discussed in this section.
FIGURE 5.
Signals associated with mechanical overload posited to upregulate mammalian/mechanistic target of rapamycin complex 1 (mTORC1) activity in skeletal muscle. This schematic (constructed with BioRender.com, with permission) provides an overview of content discussed in the review related to signals associated with mechanical overload that have been posited to upregulate mTORC1 activity in skeletal muscle. Notably, 1 of these signals [insulin-like growth factor 1 (IGF1)] operates through canonical ligand-receptor binding, whereas the other 3 signals are thought to operate through mechanotransduction. Single or multiple solid arrows indicate the pathway (or a portion of the pathway) has been relatively well defined and/or extensively investigated. Dashed arrows indicate that not much is known about how the upstream signal operates in skeletal muscle during periods of mechanical overload. DAG, diacylglycerol; FAK, focal adhesion kinase; PA, phosphatidic acid.
4.1.2. A brief discussion of mTORC1-independent mechanisms.
Although a high level of emphasis has been placed on mTORC1 signaling, several lines of evidence support mTORC1-independent signaling being involved with load-induced anabolic outcomes or skeletal muscle hypertrophy (36). For instance, West et al. (446) demonstrated that mTOR inhibition through rapamycin inhibited 6 h postexercise muscle protein synthesis after sciatic stimulation of the hindlimb muscles in rats. However, rapamycin only partially inhibited protein synthesis 18 h after exercise, and this was attributed to the mTORC1-independent phosphorylation of ERK1/2 (a mitogen-activated protein kinase, or MAPK), eEF2 (which regulates translation elongation), and UBF (a transcriptional regulator of ribosome biogenesis) as well as alterations in the mRNA expression patterns of Akirin1/Mighty, Myc, and other genes involved in ribosome biogenesis. Ogasawara and Suginohara (447) similarly demonstrated that early hypertrophic signaling (<3 h after exercise) following an electrical stimulation bout in rats is sensitive to rapamycin, whereas later increases in protein synthesis (>6 h after contraction) occurred despite mTOR inhibition. You et al. (343) demonstrated that load-induced increases in protein synthesis are not impaired by the muscle-specific, inducible knockout of Raptor or when mice are treated with rapamycin (342). Since contraction-induced muscle protein synthesis was subsequently shown to be completely inhibited by an ATP-competitive mTOR inhibitor (447), it is reasonable to speculate that mTORC2 may act in tandem with mTORC1 to regulate muscle protein synthesis. However, mTORC2 inhibition via muscle-specific Rictor knockout in mice does not affect contraction-induced muscle protein synthesis (448). Goodman et al. (449) reported that synergist ablation increases muscle Yes-Associated Protein (YAP) protein concentrations, a transcriptional coactivator of the TEA domain transcription factors and constituent of the Hippo signaling pathway (450, 451). An increase in YAP phosphorylation also occurs, and skeletal muscle YAP overexpression in vivo induces hypertrophy in an mTORC1-independent fashion. Through a series of in vivo transfection experiments, it was shown that increased YAP protein expression enhances the promoter activities of Myc and Myod1 while reducing the promoter activity of Trim63/Murf1. Hence, the YAP-induced increase in Myc expression could drive hypertrophy through enhanced ribosome biogenesis in a mTOR-independent fashion. Contrary to these findings, however, is work by the Wackerhage laboratory (452) showing that the constitutive overexpression of Yap1 in mice leads to muscle atrophy. Given the novelty of this target, as well as conflicting data, the involvement of YAP in skeletal muscle hypertrophy needs to be further explored. Steinert and colleagues (267) more recently demonstrated that S473 phosphorylation of the Tripartite Motif-Containing 28 (TRIM28) protein is transiently elevated after a bout of maximal hindlimb contractions in mice and this occurs independent of mTORC1 signaling. In addition, TRIM28 phosphorylation confers myofiber hypertrophy in mice transfected with a TRIM28 phosphomimetic plasmid construct. The authors posited that TRIM28 phosphorylation likely occurs through upstream MAPK signaling and, once phosphorylated, the protein could enhance the expression of the muscle specific MYOD and MEF2 transcription factors to promote hypertrophy.
As alluded to in the prior paragraph, MAPK signaling has been commonly cited as an mTORC1-independent signaling mechanism involved with skeletal muscle hypertrophy. Three MAPKs (ERK1/2, JNK1/2, and p38) have been extensively examined with in vitro and in rodent synergist ablation models (354, 453–455). More recent data suggest that activation of MAPKs occurs through mechanotransduction (e.g., the MAP3K ZAKβ localizing to Z disks) (456), and there is evidence that certain aspects of MAPK signaling converge to activate downstream mTORC1 targets in skeletal muscle (351). Several groups have reported that elevated MAPK signaling occurs after one or multiple resistance exercise bouts in humans (363, 457–462), and in some cases these signaling events coincide with elevated mTORC1 signaling and increases in muscle protein synthesis. The Goodyear laboratory (463) reported that mechanical overload-induced increases in fCSA and muscle mass are impaired in inducible and muscle-specific Mapk8/Jnk1-knockout mice after 14 days of synergist ablation, which again underscores the importance of MAPK signaling in promoting skeletal muscle hypertrophy. There is evidence from a recent human study suggesting that β2-adrenergic signaling operates in an mTORC1-independent fashion to stimulate myofibrillar protein synthesis following resistance exercise (464). And perhaps the most compelling example of non-mTORC1 signaling being involved with overload-induced hypertrophy comes from Ogasawara and colleagues (465), who reported that chronic rapamycin treatments dampened, but did not prevent, increases in hindlimb muscle masses and fCSA in rats following 8 wk of electrically evoked hindlimb contractions. Although the mTORC1-independent mechanisms associated with this response were not determined, this was the first resistance training-like loading paradigm in rodents to show such an effect. This is very important to note because all prior evidence of rapamycin preventing skeletal muscle hypertrophy came from surgical models of chronic mechanical overload.
Indeed, several lines of evidence support that mTORC1 inhibition inhibits skeletal muscle hypertrophy during chronic mechanical overload (191, 340, 341, 343, 466). Notwithstanding, several studies have also suggested that non-mTORC1 signals are involved (e.g., MAPKs, YAP, TRIM28, UBF, MYC, and others), and the importance of these mechanisms during physiologically relevant forms of mechanical overload-induced hypertrophy warrants further consideration.
4.1.3. The involvement of ribosome biogenesis in mechanical overload-induced skeletal muscle hypertrophy.
Millward and colleagues (467) published a report in 1973 indicating that muscle protein synthesis in rat skeletal muscle scaled linearly with changes in ribosomal (r) muscle RNA content. They noted that
“More than 80% of muscle RNA is ribosomal and this proportion appears to be maintained during protein depletion, so that a change in RNA also reflects a change in ribosome content. If alteration in ribosomal content affects control, then this alteration may be termed a change in the ribosomal capacity for protein synthesis. If, however, a change in synthesis is brought about by alterations in the other factors modulating each phase of translation, then it is a change in the ribosomal efficiency.”
This study provided the basis for the current-day definitions of translational capacity, the concentration of ribosomes in myofibers, and translational efficiency, the ability of existing ribosomes to catalyze protein synthesis per unit of RNA. Additionally, rRNA accounts for ∼80% of the total RNA content of cells (468); this work implied that the determination of total RNA concentrations reliably reflects alterations in skeletal muscle ribosome content. Increases in translational or ribosomal capacity occur through a process termed ribosome biogenesis. Describing the process of ribosome biogenesis is beyond the scope of this review and has been detailed elsewhere (20, 469–472). Subsequent rodent studies have shown that mechanical overload-induced increases in tissue total RNA and rRNA concentrations are associated with skeletal muscle hypertrophy, as reviewed by Goldberg et al. (127). Using an intermittent loading paradigm, Wong and Booth (473) confirmed the increase in RNA content after 10 wk of hindlimb loading. Several studies have since indicated that loading paradigms increase rRNA or total RNA concentrations after one bout or with chronic loading to amplify the ribosomal capacity of the muscle (80, 380, 382, 474–480).
A series of in vitro studies from 1989–2005 suggested that an increase in translational capacity through rRNA synthesis is involved in cardiomyocyte and myotube hypertrophy (481–483). In skeletal myotubes specifically, Nader et al. (481) reported that serum/growth factor stimulation increased rRNA concentrations and that this effect is abrogated by rapamycin, which implicated that ribosome biogenesis in myotubes is largely stimulated through mTORC1 signaling. Although mTORC1 converges at the ribosome to promote increased translational efficiency, this was the first evidence suggesting that mTORC1 signaling also promotes ribosome biogenesis in skeletal muscle. More recently, these same researchers reported that mTORC1 can undergo nuclear localization to bind to rRNA gene promoters, and this process can be inhibited by rapamycin (484). Furthermore, the release of mTOR from ribosomal gene promoters with rapamycin treatment correlated with chromatin marks indicative of transcriptional silencing.
To understand the mechanisms responsible for ribosome biogenesis in skeletal muscle, von Walden et al. (478) investigated the transcriptional response of rRNA genes during the initial stages of mechanical overload. rRNA transcription peaked at 3 days and preceded rRNA accumulation and hypertrophy. A transcriptional burst involved the enrichment of specific transcription factors at the rDNA promoter including the Upstream Binding Factor (UBF), c-Myc (Myc), and the Williams Syndrome Transcription Factor (WSTF), a component of the B-WHICH chromatin remodeling complex. This was consistent with the increase in 45S pre-rRNA and suggests both transcriptional and epigenetic regulation of ribosome biogenesis in skeletal muscle hypertrophy. A recent study by Murach et al. (485) expanded upon the role that Myc transcription factor has in driving ribosome biogenesis. Specifically, in silico analysis of several datasets suggested that Myc gene regulation is evident at the onset of mechanical overload and that certain genes related to ribosome biogenesis (e.g., Bop1, Polr3g, and Rps19) are likely driven by Myc.
Studies in humans have shown results consistent with these data. For example, a bout of resistance exercise stimulates rRNA gene transcription as early as 4 h after exercise (486, 487), and this response can persist for at least 48–72 h after exercise (472, 488). Figueiredo (17) also authored a recent review describing the process of ribosome biogenesis, and a summary table discusses several studies that have reported an increase in muscle total RNA or rRNA concentrations days to weeks into resistance training (353, 489–495). Since myofiber hypertrophy is typically detected after 15–20 training sessions in humans (223, 310, 496), the increase in ribosomal production appears to precede hypertrophy (see FIGURE 6). A recent human study by Figueiredo et al. (497) indicates that ribosomal DNA (rDNA) copy number, which can range between hundreds and thousands of copies on an individual-to-individual basis (498), is positively associated with ribosome biogenesis markers in response to an acute bout of resistance exercise. These authors also reported that a mechanical overload stimulus in mice transiently alters the promoter methylation status of genes associated with ribosome biogenesis, confirming earlier findings that load-dependent epigenetic mechanisms, in part, modulate ribosome biogenesis.
FIGURE 6.
Timeline of ribosome biogenesis during load-induced skeletal muscle hypertrophy. This schematic (constructed with Biorender.com, with permission) provides a general timeline of ribosome biogenesis during periods of mechanical overload. Importantly, researchers have shown that ribosome biogenesis precedes skeletal muscle (and myofiber) hypertrophy, and this can result in increases in both ribosome content as well as ribosome concentrations. However, ribosome concentrations renormalize after longer-term training periods where myofiber hypertrophy is evident.
Strengthening the case for ribosome biogenesis being a critical mechanism for load-induced skeletal muscle hypertrophy, other research has indicated that the magnitude of ribosome biogenesis in response to different loading paradigms is associated with hypertrophic outcomes. For instance, Kirby and colleagues (477) reported that older mice present impairments in load-induced skeletal muscle hypertrophy compared with younger mice and this corresponded with a diminished ribosomal response in the old mouse cohort. Nakada et al. (476) utilized various forms of synergist ablation in rats to produce four different levels of plantaris hypertrophy. Fourteen days after overload, plantaris masses increased by 8% in the first cohort, 22% in the second, 32% in the third, and 45% in the fourth. rRNA content increased by 80%, 120%, and 150% in the latter three groups 5 days after overload, and the 5 day increase in translational capacity was strongly correlated to 14 day muscle weight data. These rodent findings have been validated in humans, in part, by Hammarström et al. (499), who reported that the degree of muscle hypertrophy following 12 wk of resistance training is associated with increased rRNA concentrations. A more recent study by Hammarström et al. (500) in humans also expanded these findings through a time course examination of resistance training-induced changes in rRNA levels. The authors reported that muscle rRNA concentrations increased in response to the first four training sessions, and this was followed by a plateau and peak in concentrations after eight sessions. Furthermore, the increases in muscle total RNA concentrations correlated with the magnitude of resistance training-induced skeletal muscle hypertrophy. This latter finding was recently confirmed by Figueiredo et al. (501), who reported that 2 wk of ambulatory recovery from cast immobilization followed by 2 wk of resistance training increases midthigh muscle CSA and rRNA concentrations.
Despite the strong evidence suggesting that an increase in ribosomal capacity plays a central role in muscle hypertrophy, a few reports are inconsistent with this notion. For instance, Goodman et al. (342) reported that ribosome biogenesis occurs in the absence of myofiber hypertrophy after 7 days of synergist ablation in mice administered rapamycin, but whether this occurs during longer-term mechanical overload was not reported. Others have also shown that AAV-mediated skeletal muscle Myc overexpression during a 2-wk period does not stimulate muscle hypertrophy despite upregulating ribosome biogenesis markers and muscle protein synthesis (502). However, this may have been due to the persistent expression of Myc, and as discussed with mTORC1 signaling, pulsatile Myc responses to mechanical overload may be needed to contribute to the hypertrophic response. Although these limited data challenge the importance of ribosome biogenesis, most of the studies discussed here support that increases in translational capacity through ribosome biogenesis are associated with the magnitude of skeletal muscle hypertrophy in response to mechanical loading.
An additional theme gaining traction, despite limited evidence to date, is the notion of ribosome specialization being involved with load-induced skeletal muscle hypertrophy. A review by Chaillou (18) suggested that ribosome heterogeneity exists within myofibers in that each ribosome likely contains a unique profile of ribosomal proteins and rRNA spliced variants that act to modulate ribosome function. Ribosomal proteins can also be subjected to posttranslational modifications (e.g., phosphorylation, methylation, and acetylation), which may increase ribosome heterogeneity within myofibers (18). Perhaps most intriguing is the notion put forth by Chaillou suggesting that ribosome specialization during skeletal muscle hypertrophy may lead to enhanced translational fidelity and selection events whereby certain mRNAs are prioritized for translation. Potential evidence of ribosome specialization comes from the McCarthy laboratory (503) showing that the mRNA expression of Rpl3 and Rpl3l is differentially affected in skeletal muscle after synergist ablation (+400% and −82%, respectively), whereas the mRNA levels of all other genes encoding ribosomal proteins are modestly affected or not affected at all. These authors performed additional experiments demonstrating that the induction of Rpl3l expression in vitro impaired myotube growth and protein accretion by −23% and −14%, respectively, compared with a control cell line. Beyond these data, no published research to date has determined whether ribosome specialization occurs during periods of resistance training in humans and/or plays an appreciable role in skeletal muscle hypertrophy as implicated in vitro. Thus, like other mechanisms discussed in this review with limited or incongruent data, the research potential in this area is high.
4.1.4. The involvement of satellite cells in myofiber hypertrophy.
Satellite cells were first observed in frog muscle by Mauro (504) and Katz (505) in 1961 through TEM. As noted above, rodent work by Schiaffino et al. (148) indicated that satellite cell proliferation occurs in rodents after mechanical overload and that the fate of some of these satellite cells is to become incorporated into overloaded myofibers (149). Since these landmark studies, human investigations have provided evidence to support a mechanism in which satellite cell-derived myoblasts fuse to myofibers in response to resistance training (70, 223, 480, 493, 506–516). A meta-analysis by Conceição et al. (23) examined 27 resistance training studies totaling 903 participants. The authors reported that myofiber hypertrophy of ≤10% induces a modest increase in myonuclear content and that a significantly higher increase is observed when muscle hypertrophy is ∼22%; notably, these effects are independent of age, sex, and myofiber type composition. Increased satellite cell abundance following either a resistance exercise bout or longer-term resistance training has also been shown to be correlated with the magnitude of skeletal muscle hypertrophy (70, 222, 517, 518). Moreover, although some evidence to the contrary exists (519), increased satellite cell activation during periods of resistance training has been reported to coincide with skeletal muscle hypertrophy in older participants (520, 521).
Genetic mouse models have yielded tremendous insight in this area as well. The Pax7-DTA mouse uses the Cre-loxP system to kill satellite cells by driving the expression of a diphtheria toxin A fragment in a cell-specific fashion through tamoxifen administration. In 2011, McCarthy et al. (522) were the first to use the Pax7-DTA mouse model to remove ∼90% of all satellite cells in adult mice (4 mo of age). In short, the authors reported that plantaris growth in response to 14 days of synergist ablation is not impaired relative to control mice, thus providing the first evidence that satellite cell-mediated myonuclear accretion is not obligatory for load-induced skeletal muscle hypertrophy during this shorter time frame. However, Egner et al. (523) replicated the experimental approach utilized by McCarthy and colleagues to show that satellite cells are necessary for load-induced plantaris hypertrophy in juvenile Pax7-DTA mice (2–3 mo of age). These studies suggested that earlier maturation phases likely influence the requirement for satellite cells in overload-induced skeletal muscle hypertrophy. Murach et al. (323) confirmed the age-dependent requirement of satellite cells in reporting that the plantaris muscle of 2-mo-old Pax7-DTA did not hypertrophy in response to overload when satellite cells were ablated. Goh and Millay prevented myonuclear accretion in response to mechanical loading by inactivating the Myomaker (Tmem8c) gene in satellite cells, this being a gene required for satellite cell fusion (524). In agreement with Egner and colleagues, these authors found that satellite cell fusion is required for increasing myofiber size following 14 days of synergist ablation (525). Similarly, Englund et al. (526) reported that muscle hypertrophy induced by 8 wk of resistance-loaded wheel running is blunted in Pax7-DTA mice in which satellite cells had been depleted. Finally, a recent study by Kobayashi et al. (527) utilized inducible satellite cell-specific Cdk1-knockout mice, which show impaired satellite cell proliferation, to demonstrate that myonuclear accretion is blunted and increases in fCSA are limited after 14 days of synergist ablation. Although some conflicting evidence exists, these mouse data form a collective paradigm agreeing with the human data to suggest that satellite cells are needed for optimizing load-induced skeletal muscle hypertrophy in maturing rodents and/or during longer periods of mechanical loading in adult rodents. Furthermore, although several of these studies used the synergist ablation model, the data from Englund and colleagues support that satellite cells are needed to optimize hypertrophy induced by progressive resistance-loaded wheel running, which is a more physiological model.
Although data discussed in the prior paragraphs provide strong support to satellite cells assuming a critical role in resistance training-induced skeletal muscle hypertrophy, hypotheses in this area are being rapidly refined given that this is an intensely studied topic in muscle biology. For instance, Murach et al. (25) authored a recent review citing human resistance training studies that show that myofiber hypertrophy occurs either before or without myonuclear accretion (223, 528, 529), and similar data were published thereafter (512, 530–533). These eight studies reported mixed and type II myofiber radial size increases that averaged ∼15–20%; thus it is likely that some participants from these studies did not obtain the ∼22% fCSA increase threshold proposed by Conceição et al. (23), thereby explaining the lack of myonuclear accretion. Murach and colleagues also provide evidence to suggest that type II myofibers exhibit the ability to hypertrophy with less myonuclear accretion relative to type I myofibers, which results in larger type II fiber-specific myonuclear domains. Type II myofiber myonuclei have also been shown to compensate for the loss of myonuclear accretion via satellite cell depletion by significantly increasing their transcriptional output in response to mechanical overload induced by synergist ablation (534); alternatively stated, the myonuclei of type II myofibers appear to possess a transcriptional reserve to support myofiber growth in the absence of myonuclear accretion. One of the challenges in understanding myonuclear dynamics is that the relationship between myonuclear content and domain size is not constant for different-sized myofibers. For example, in opposition to the concept of a constant myonuclear domain with changing myofiber size, smaller myofibers have markedly smaller domains, and myofiber perimeter (rather than myofiber CSA) per myonucleus is constant across a fCSA range from 2,000 to 8,000 µm2 (520, 535). Additionally, there are recent data from the Miller laboratory in mice showing increased DNA synthesis of myonuclei in vivo using D2O, and this process is enhanced with synergist ablation-induced mechanical overload in the plantaris muscle (536). Other reports have indicated that bone marrow-derived cells and other stromal cells (e.g., Twist2 positive and Hox11 positive) provide additional sources of myonuclei (537–539), but more research is needed to definitely determine whether these cells contribute myonuclei in response to mechanical overload. Hence, although satellite cells are seemingly critical for optimizing load-induced skeletal muscle hypertrophy, these data imply that much remains unknown regarding myonuclear dynamics, and satellite cell fusion may not be the sole source of newly acquired myonuclei. As an interesting aside, a consequence of the satellite cell depletion studies using the Pax7-DTA model is the revelation of nonfusion roles that satellite cells seemingly exhibit during different loading paradigms. Independent lines of evidence support that satellite cells interact with myofibers and fibroblasts to promote extracellular matrix remodeling during tissue repair (74, 540–542). This cell-to-cell communication also appears to be bidirectional, with evidence of muscle fibroblasts stimulating satellite cell fusion (74). Mice depleted of fibroblasts have altered satellite cell dynamics and smaller regenerating myofibers after injury (540). In the absence of muscle damage (i.e., a state of strong extracellular matrix adhesion), satellite cells remain quiescent. However, satellite cells rapidly proliferate after a bout of unaccustomed resistance exercise with associated tissue damage, and this occurs without myonuclear accretion and coincides with an upregulation in genes involved with extracellular matrix remodeling (543). The Pax7-DTA genetic mouse model has been used to demonstrate that longer-term (8 wk) loading blunts skeletal muscle hypertrophy, and this coincides with a significant increase in fibrosis (541, 544). In vitro experiments from these studies have provided evidence showing that primary myogenic progenitor cells communicate with primary fibroblasts via miRNA-containing exosomes to downregulate Rrbp1, a master regulator of collagen biosynthesis, as well as collagen-related mRNAs. These studies by the Peterson laboratory support a mechanism in which satellite cells assume a nonfusion role in regulating extracellular matrix remodeling during muscle growth as summarized by Murach et al. (545). The findings of Roberts et al. (80), Moro et al. (546), and Damas et al. (533) showing that different loading paradigms increase type II myofiber hypertrophy and satellite cell number in the absence of myonuclear accretion suggest that this mechanism may also be operative in humans; however, these studies did not examine extracellular matrix markers or miRNAs. Beyond fibroblasts, satellite cells communicate with other stromal cells during the early stages of hypertrophy (e.g., endothelial cells, fibro-adipogenic progenitors, and mesenchymal progenitors) (542, 547, 548), as shown in the context of muscle regeneration (549). Hence, the nonfusion roles that satellite cells assume during mechanical overload may be involved in skeletal muscle hypertrophy, and this area of research is ripe for further investigation. Content in this section review is summarized in FIGURE 7.
FIGURE 7.
Summary of the fusion and nonfusion roles of satellite cells. This schematic (constructed with BioRender.com, with permission) provides a general overview of how satellite cells can respond to mechanical overload. The fusion role has been well defined, and this involves satellite cell proliferation (acute response) followed by the fusion of a subpopulation of satellite cells to increase myonuclear number. The nonfusion role involves satellite cells secreting exosomes containing microRNA (and presumably other cargo). Exosomes can transport this cargo to myofiber and nonmyofiber cell types in the interstitial space to regulate gene expression. In the example pictured, satellite cells are regulating gene expression in myofibers and fibrogenic cells, and this may affect extracellular matrix remodeling during myofiber hypertrophy (as discussed in main text). Satellite cells are likely to communicate with other cell types, and this is also illustrated via cell-cell communication with vascular endothelial cells. ECM, extracellular matrix.
4.2. Other Mechanisms Involved with Skeletal Muscle Hypertrophy
4.2.1. Genetic variants.
Genetic polymorphisms likely play a role in the hypertrophic response to resistance training in humans. Heterogeneous responses in hypertrophic outcomes exist with weeks to months of training (27), and genetic differences between individuals are commonly touted as being partially responsible for this effect. A recent meta-analysis including 24 heritability studies indicates that strength adaptations to resistance training possess ∼50% genetic component (550), and this likely holds true for hypertrophic outcomes. In a larger-scale study, Stokes et al. (250) recently reported that a strong genetic component exists for resistance training and limb immobilization adaptations. Angleri et al. (531) more recently reported that two different unilateral leg training paradigms in 20 resistance-trained subjects led to statistically similar increases in mean fCSA (within-subject r value = 0.89 for this measure, P < 0.05). Authors from both studies suggested that intrinsic biological factors (i.e., genetic factors leading to transcriptome-wide responses that promote training adaptations) are likely responsible for these observed effects. Kilikevicius et al. (551) reported that soleus and plantaris hypertrophy elicited by 28 days of synergist ablation differs between eight strains of laboratory mice, and the authors concluded that this is likely mitigated by genetic differences between strains.
Despite these data suggesting that a genetic component exists for skeletal muscle hypertrophy, candidate polymorphisms that affect hypertrophic outcomes have varied and tempered enthusiasm in this area. The Functional Single Nucleotide Polymorphisms Associated with Human Muscle Size and Strength (FAMuSS) multicenter trial was a targeted analysis that provided novel insight into polymorphisms that affect the hypertrophic response to 12 wk of single-arm resistance training (552). In short, the authors examined ∼500 gene variants in 1,300 younger adult men and women and reported that polymorphisms in 17 genes (ACE, ACTN3, ANKRD6, BMP2, CCL2, CCR2, CNTF, FST, MSTN, IGF1, IL15, IL15Rα, LEP, LEPR, NOS3, RETN, SPP1) are associated with muscle size changes during resistance training. Contrary to these findings, Vann et al. (234) recently used a DNA microarray to examine whether any of the ∼315,000 polymorphism targets are associated with changes in whole body lean mass or mean myofiber fCSA with 12 wk of resistance training in 109 males. In short, none of the assayed polymorphisms, including many of those from the FAMuSS trial, was significantly associated with hypertrophic outcomes. Although the FAMuSS trial and the study published by Vann and colleagues employed similar-length training interventions, discrepancies between studies could have been due to differences in training modality (single-arm vs. full-body training, respectively), methods to quantify hypertrophy (MRI vs. DXA and VL myofiber histology, respectively), and the lower number of participants in the study by Vann and colleagues. However, one insightful finding by Vann and colleagues was that one annotated intronic gene variant (GLI3; rs10263647) is significantly associated with mean fCSA changes. GLI3 encodes a transcription factor that regulates Sonic hedgehog signaling (553), and Vann and colleagues reported that the GLI3 T/C and C/C genotypes achieved myonuclear addition in response to training, whereas the T/T cohort did not. The Gli3 gene has been shown to regulate satellite cell differentiation and fusion in mice by affecting the expression of myogenic regulatory factors (i.e., Myf5, Myog, and Myod1) (554). Hence, it is possible that those with the T/T genotype have impairments in satellite cell fusion. However, the significance threshold (P < 1 × 10−5) utilized by Vann and colleagues was adjusted for exploratory purposes and differs from the commonly utilized significance threshold in genome-wide association studies (GWAS) (P < 1 × 10−8). Thus, more research is needed to validate these findings. Additionally, a notable limitation with single-gene candidate studies and GWAS is the lack of resolution in detecting novel polymorphism candidates, and this issue has been illustrated in other research disciplines. For instance, Rivas et al. (555) utilized deep DNA sequencing to interrogate 56 genes and gene regions previously associated with Crohn’s disease. These authors identified 70 novel protein-altering variants that likely contribute to the disease phenotype. Novel polymorphisms related to insulin secretion and glucose tolerance have also been recently identified with deep DNA sequencing (556). Findings from both studies imply that unidentified gene variants related to hypertrophy may exist, and future deep DNA sequencing efforts in this area will likely lead to fruitful discoveries. FIGURE 8 provides a summary of what has been performed to date as well as future directions that could be pursued to increase the knowledge base in this area.
FIGURE 8.
Past research and future directions regarding the delineation of gene polymorphisms associated with hypertrophic outcomes. This schematic (constructed with BioRender.com, with permission) provides a summary of past efforts examining genetic polymorphisms associated with the skeletal muscle hypertrophic response to resistance training in humans. As mentioned in main text, future methods using deep DNA sequencing and bioinformatics are needed to garner additional information in this area. FAMuSS, Functional Single Nucleotide Polymorphisms Associated with Human Muscle Size and Strength; RT, resistance training; SNP, single-nucleotide polymorphism.
It is important to reiterate that genetic mouse models are useful tools whereby genes can be overexpressed or knocked out to yield a hypertrophic phenotype (231). These models, however, do not support that genetics is the prominent mechanism involved in load-mediated skeletal muscle hypertrophy. As a contextual example, the myostatin (MSTN) gene received considerable attention in the late 1990s and early 2000s as a gene that limits muscle growth. McPherron and Lee (557) reported that Belgian Blue cattle harbored an 11-nucleotide deletion in the MSTN gene that led to a doubling in muscle mass relative to normal cattle. Around the same time McPherron and colleagues developed Mstn-knockout mice, and these mice exhibit robust hypertrophy in the absence of mechanical overload (558). Seven years later, a case report in a child indicated that a rare MSTN mutation led to a hypermuscular phenotype (559). A proliferation of myostatin-related research ensued in the field, and various human studies sought to examine whether MSTN-related polymorphisms are associated with muscle mass or skeletal muscle hypertrophy in response to resistance training (234, 560–562). Notably, most of these studies have revealed few to no appreciable effects regarding MSTN genotype-phenotype (hypertrophy) outcomes (234, 561, 562). Hence, readers should appreciate that genetic mouse models (and case reports regarding rare human mutations) can be useful in identifying proteins involved with skeletal muscle hypertrophy, albeit this process is a complex trait that likely involves still-to-be resolved polymorphisms as well as the other mechanisms discussed here.
4.2.2. Epigenetic alterations, with an emphasis on DNA methylation.
Geneticist Adrian Bird was a pioneer in the scientific discipline of epigenetics, or changes in gene activity that do not involve DNA sequence alterations. In a landmark review, Bird (563) discussed research in the 1980s, much of which was from his laboratory, detailing the presence and postulating the significance of DNA methylation in eukaryotes. Through decades of subsequent research, it is now well appreciated that gene expression is regulated, in large part, through DNA methylation. Approximately 98% of DNA methylation occurs on cytosine residues present in cytosine guanine dinucleotide pairing sites (CpG sites), and DNA methylation is catalyzed by DNA methyltransferase (DNMT) enzymes whereas demethylation is catalyzed by ten-eleven translocation (TET) enzymes (564–567). Increased methylation levels in a promoter or enhancer region of a gene generally downregulate RNA transcription by either impairing transcription factor binding or compacting DNA and making it transcriptionally inaccessible (567). Additionally, although data in skeletal muscle are lacking, in vitro work in budding yeast suggests that alterations in genome-wide DNA methylation patterns can cause chromatin remodeling events that may indirectly impact the expression of genes by allowing certain DNA regions access to transcriptional machinery (568).
More recent enthusiasm has surrounded how exercise alters the collective skeletal muscle DNA methylome, and several reviews have been published on this topic (37, 39, 569, 570). Barrès et al. (571) provided the first evidence showing that changes in skeletal muscle DNA methylation transiently occur across various metabolic genes after a single high-intensity aerobic exercise session in humans. These authors also reported that the postexercise DNA demethylation patterns across various metabolic genes correspond with the mRNA expression patterns of these genes. Subsequent genome-wide methylation (or methylome) studies in humans have indicated that resistance training elicits the demethylation and upregulation of genes related to actin/cytoskeletal, extracellular matrix, growth-related, and/or metabolic pathways (255, 257). Moreover, some genes retain a demethylated signature after an earlier period of resistance training and detraining, and the mRNA expression of some of these genes is enhanced during the retraining period. These data have led Sharples and colleagues (572) to hypothesize that skeletal muscle possesses an epigenetic memory following periods of resistance training (or “epi-memory”), which could mechanistically explain why an increase in muscle mass occurs more rapidly during retraining periods following weeks to months of detraining. Also compelling are two recent collaborative studies from the Roberts and Sharples laboratories. The first study by Ruple et al. (258) indicated that 6 wk of resistance training in 65-yr-old men causes a robust demethylation of the mitochondrial genome and these methylation changes correspond with an increased mRNA expression of numerous mitochondrion-specific genes in the presence of skeletal muscle hypertrophy. The second study by Sexton et al. (573) in previously trained college-aged men indicated that global skeletal muscle DNA methylation patterns are more robustly altered 3 h versus 6 h after a resistance exercise bout (239,951 vs. 12,419 CpG site methylation changes, respectively; FIGURE 9). Like the aforementioned endurance exercise data discussed by Barrès and colleagues, these data suggest that alterations in skeletal muscle DNA methylation occur rapidly after a loading stimulus. Moreover, these authors used bioinformatics to report that genes related to “focal adhesion,” “MAPK signaling,” and “PI3K-AKT signaling” are significantly affected at both the DNA methylation and transcriptome-wide levels.
FIGURE 9.
Skeletal muscle genome-wide DNA methylation and transcriptome responses to a bout of resistance exercise. This schematic (constructed with BioRender.com, with permission) summarizes recent data [Sexton et al. 2023 (573)] demonstrating alterations in skeletal muscle tissue DNA methylation status after a bout of resistance exercise in humans. The researchers concluded that 1) alterations in DNA methylation statuses occur very rapidly (i.e., 3 h vs. 6 h after exercise); 2) contrary to past hypotheses suggesting that exercise generally elicits a reduction in DNA methylation, more hypermethylation events occurred 3 h after exercise relative to hypomethylation events; and 3) alterations in DNA methylation patterns likely precede and are, in part, responsible for altered mRNA expression patterns. HIF, hypoxia-inducible factor; PI3K, phosphatidylinositol 3-kinase.
Rodent data also exist supporting dynamic alterations in myonuclear DNA methylation accompanies skeletal muscle hypertrophy. Figueiredo et al. (497) reported that a mechanical overload stimulus in mice alters the promoter methylation status rDNA to favor transcription. von Walden et al. (254) used the HSA-GFP (HSA-rtTA;Rosa26-H2B-GFP) genetic mouse model, as well as myonuclear capture and bisulfite sequencing techniques, to show that 11,210 CpG sites are hypomethylated and 3,491 sites are hypermethylated with plantaris hypertrophy induced by synergist ablation. Several CpG sites in proximity to genes involved in mTORC1 signaling, autophagy, and ribosome biogenesis are also hypomethylated, and an upregulation in several corresponding mRNAs also occurs. Wen et al. (286) used this same mouse model to show that enhanced muscle growth (relative to naive hypertrophy) occurs after detraining-retraining. Notably, detraining-retraining hypertrophy corresponds with a myonuclear methylome “memory” signature, which resonates with the human data from Sharples’s group. Murach et al. (285) subsequently used the HSA-GFP genetic mouse model, as well as myonuclear capture and bisulfite sequencing techniques like von Walden and colleagues, to elucidate epigenetic adaptations that occur after 8 wk of progressive resistance-loaded wheel running in two groups of mice. The first group of mice had resident myonuclei labeled before training to allow for their capture for DNA methylation analysis after training. The second group of mice had both resident myonuclei and newly acquired myonuclei via satellite cell-mediated myonuclear addition analyzed after training. Bioinformatics indicated that genes related to the PI3K-AKT and FOXO signaling pathways favored hypomethylation in the first group of mice. In contrast, genes associated with RNA polymerase II-mediated transcription and cell-to-cell adhesion pathways favored hypomethylation in the second group of mice (285). Additionally, multiple CpG sites near the 28S rDNA transcription termination area and one CpG site in the rDNA enhancer region favored hypomethylation in the second versus the first group of mice. These data suggest that resident myonuclei may utilize DNA hypomethylation to upregulate genes associated with protein turnover in response to mechanical overload, whereas newly acquired nuclei may utilize this process to upregulate genes associated with satellite cell proliferation, fusion, and ribosome biogenesis. The Murach laboratory (574) more recently reported that loaded wheel running in older mice alters muscle DNA methylation suggestive of reduced biological aging (i.e., the Horvath and developmental muscle clocks) and used a skeletal muscle-specific Myc-inducible mouse to suggest that mechanical overload operates through MYC to elicit these changes.
These collective data from independent research groups demonstrate that myonuclear DNA, DNA from cells in the extracellular matrix, and mitochondrial DNA display altered methylation profiles in response to mechanical overload in rodents or resistance training in humans. However, as with other data discussed here, much of the epigenetics work to date has been descriptive and more data are needed to confirm that mechanical overload-induced alterations in DNA methylation result in appreciable mRNA and protein expression changes. It is also difficult to determine the impact that DNA methylation has on downstream processes given that some data in this area indicate a smaller than anticipated overlap of DNA methylation changes and mRNA expression changes. In this regard, Sharples’s group (575) recently compared methylome and transcriptome data in muscle samples collected from humans 30 min and 24 h after various high-intensity running protocols. Only 431 of 879 genes were reported to exhibit exercise-induced mRNA expression patterns corresponding to DNA methylation patterns. More striking are data published by Laker et al. (576) where skeletal muscle transcriptome and methylome data were obtained in humans before and after three resistance exercise bouts over a 9-day period. In short, RNA-sequencing indicated that training alters the expression of 2,616 mRNAs, and methylome analysis using reduced representation bisulfite sequencing (RRBS) indicates that 474 genomic regions are differentially methylated. Surprisingly, only 54 genes exhibited DNA methylation changes that correspond to mRNA expression changes following training, indicating that DNA methylation changes are only associated with only ∼2% of the genes whose mRNA abundances are altered. Indeed, this discordance may be due to the timing of tissue acquisition (307). For instance, many of the load-induced alterations in DNA methylation patterns are seemingly transient as discussed above (i.e., within 3 h after a resistance exercise bout), and these events may affect mRNA expression patterns hours to days thereafter. In support of this hypothesis, Telles et al. (577) examined DNA methylation and mRNA expression time course changes for select myogenic regulatory factors (MYOD1, MYF5, and MYF6) immediately after and 4 h and 8 h after a single bout of resistance, high-intensity interval, and concurrent exercise. Although the methylation and mRNA expression responses were reported to be interrelated, the respective profiles were not synchronized at the postexercise time points; specifically, mRNA responses to promoter methylation events seemingly occur after ∼8 h. The aforementioned study from Sexton and colleagues (573) also indicates that global DNA methylation patterns 3 h after a resistance exercise bout exhibit a high association with global mRNA expression patterns 6 h after exercise. Thus, there is still much to learn about how dynamic changes in DNA methylation temporally affect gene expression.
Finally, unlike other mechanisms discussed in this review where genetic mouse models have provided cause-and-effect relationships, no study to date has examined how genetic mouse models harboring modified genes that affect skeletal muscle DNA methylation impact overload-induced hypertrophy. Interestingly, Wang et al. (578) reported that Tet2-knockout mice do not exhibit impairments in postnatal muscle development, albeit these mice presented fewer newly formed myofibers in response to cardiotoxin-induced injury compared to wild-type mice. These authors also generated tamoxifen-inducible Tet2-knockout mice (i.e., Pax7CreERT2:Tet2 flox/flox mice where Tet2 knockout occurs specifically in satellite cells), and similar effects were noted. Given that muscle DNA hypomethylation is commonly reported during periods of overload and TET2 catalyzes DNA demethylation, using similar mouse models with mechanical overload paradigms will provide more compelling evidence as to whether explicit DNA hypomethylation events at certain promoter regions are required for skeletal muscle hypertrophy.
4.2.3. Muscle proteolysis.
As discussed in other sections of this review, many molecular-based studies in the field have focused on examining mTORC1 signaling and/or either mixed or myofibrillar protein synthesis rates. The reasons for such interrogations are twofold. First, data in the field indicate that muscle protein synthesis (rather than muscle proteolysis) is more responsive to mechanical loading during well-fed states (579, 580), and Brook et al. (581) have similarly suggested that “…the measurement of [muscle protein synthesis] remains a cornerstone for understanding the control of hypertrophy—mainly because it is the underlying driving force behind skeletal muscle hypertrophy.” Second, there are logistical issues that preclude assessing muscle protein breakdown rates and related mechanisms. Unlike protein synthesis in which mTORC1 acts as a central signaling hub that converges at the ribosome, there are several proteolytic mechanisms operative in skeletal muscle including the ATP-dependent ubiquitin proteasome pathway, calpain-mediated proteolysis, and lysosome-mediated autophagy (582, 583). Given the increased complexity of muscle proteolysis regulation, it is inherently more difficult to ascribe a particular proteolytic mechanism as being involved (rather than coinciding) with mechanical overload-induced skeletal muscle hypertrophy. Examining proteolysis rates in humans can also be technically challenging and invasive (e.g., with tracer infusions or using the invasive arterial-venous balance method requiring arterial and venous cannulations), and other methods require additional analysis and expertise (e.g., assessing isotope dilution in the free muscle amino acid pool for the extrapolation of breakdown rates) (583). Additionally, there are several more assumptions that are applied when calculating muscle protein breakdown versus synthetic rate kinetics. Several studies have assessed how proteolysis-related biomarkers (e.g., poly-ubiquitinated proteins, proteasome activity, and atrogene mRNAs) are transiently and chronically affected during periods of resistance training in humans (308, 369, 492, 493, 584–589) or during periods of mechanical overload in rodents (80, 474, 590). The overactivation of muscle proteolytic pathways in genetic mouse models or during longer-term fasting, prolonged unloading, or diseased states mechanistically contributes to muscle atrophy (591–594). Under normal physiological circumstances, however, a dysregulation in one or more of the proteolytic systems in skeletal muscle may impair hypertrophy. This theme subtly emerged in human tracer studies in muscle protein synthesis, and breakdown rates were shown to be tightly coupled days after exercise (178, 595). Several human studies have since shown that a bout of resistance exercise transiently upregulates mRNAs, proteins, or protein signaling involved with muscle proteolysis (e.g., TRIM63 and FBXO32 mRNAs, calpain mRNAs, and autophagy markers) (308, 345, 369, 584–587, 596–600), and similar data exist after chronic training (589, 600, 601). Baehr et al. (590) demonstrated that proteasome activity, as well as the mRNA expression of Trim63 and Fbxo32, increase after 2 wk of synergist ablation in mice. Abou Sawan et al. (41) also contend that, although muscle lysosomes are generally viewed as cellular “garbage cans” that rid the cell of damaged organelles, research into their emerging roles includes nutrient sensing, regulation of protein synthesis, and cell growth.
Genetic mouse models have also provided additional evidence to suggest that a dysregulation of components in proteolytic mechanisms may impair hypertrophy. For instance, ATG7 is involved in autophagosome formation, and Masiero and Sandri (602) have shown that robust muscle atrophy occurs in Atg7-null mice. PSMC4 is a subunit of the 20S proteasome, and a similar atrophy phenotype has been reported in Psmc4-knockout mice (603). Visual inspections of myofibers by TEM were provided in both papers, and both reports indicate that these knockout mouse lines possess disorganized myofibrils, abnormal intracellular vacuoles, disruptions to the sarcoplasmic reticulum, and mitochondria with abnormal appearances. Hence, a complete disruption in one or multiple proteolytic mechanisms likely leads to an inability of myofibers to purge damaged macromolecules or organelles, which in turn leads to a broader catabolic (or antianabolic) signaling cascades. Work by Steinert and colleagues (267) further adds to this working hypothesis. As mentioned, a phosphoproteomic approach was used to demonstrate that the S473 phosphorylation of the TRIM28 protein transiently occurs after maximal hindlimb contractions in mice. In addition to showing in mice that myofibers hypertrophy when transfected with a plasmid encoding a phosphomimetic version of TRIM28, the authors reported that tamoxifen-inducible Trim28-knockout mice exhibit myofiber atrophy as well as an impairment in hypertrophy in response to mechanical overload induced by synergist ablation. The authors posited that TRIM28 phosphorylation may stimulate the E3 ligase activity of the protein (note that E3 ligases act to transfer ubiquitin from an E2 ubiquitin-conjugating enzyme to targeted proteins) and, subsequently, accelerate proteasome-mediated proteolysis to enhance protein turnover and promote muscle hypertrophy. However, these authors did not extensively pursue this mechanism. Hughes et al. (604) recently demonstrated that the knockdown of an E3 ligase (Ubr5) in murine skeletal muscle leads to myofiber atrophy. Notable in vitro data also exist in this area of research. Osburn et al. (605) demonstrated that the pharmacological blockade of the proteasome and calpains in murine myotubes abrogates leucine-induced increases in muscle protein synthesis. Lewis et al. (606) demonstrated that autophagy inhibition blocks myofibrillar protein synthesis rates in L6 myotubes. Although kinetic interrogations were not performed in either investigation, both groups speculated that proteolysis serves to provide an intracellular pool of free amino acids that is sufficient to support muscle protein synthesis (i.e., intracellular recycling of amino acids).
Although data in the paragraph above support enhanced proteolytic activity being involved with skeletal muscle hypertrophy, there is counterevidence suggesting that these mechanisms are downregulated as well. For instance, Roberts et al. (80) reported a significant reduction in proteasome activity in hypertrophied plantaris and soleus muscles following 14 days of dual overload induced by synergist ablation. Human data from these same researchers indicates that 6 wk of high-volume resistance training reduces muscle polyubiquitinated protein levels (an indirect measure of E3 ligase and proteasome activities) in what were defined as higher and lower hypertrophic responders (492). Human data that have linked changes in protein turnover (i.e., the balance of muscle protein synthesis and muscle protein breakdown) to the hypertrophic response to resistance training are also lacking. Only two studies have utilized tracer techniques to concurrently assess protein turnover in humans after resistance training (607, 608), and one of these studies demonstrated improved muscle net balance in the postabsorptive state and that pre- to posttraining changes in basal muscle protein synthesis rates significantly correlate with changes in VL muscle thickness. Improvements in the intracellular recycling of amino acids may occur in response to resistance training or be a feature of higher responders to promote a more efficient protein turnover and muscle hypertrophy. However, no research has directly measured the intracellular recycling of amino acids from proteolysis to protein synthesis in this context. Further analysis of this hypothesis with utilization of isotope tracers, three-compartment modeling (609), and non-steady-state equations (303) could provide more insight into the role(s) that proteolysis has in intracellular amino acid recycling for muscle protein synthesis and anabolism during hypertrophy. Depending on the method used and duration and form of tracer labeling, researchers should consider adjusting for possible changes in the protein pool size, use steady-state conditions, or, when this is not possible, adjust with non-steady-state equations appropriate to the chosen tracer model.
These collective data indicate that fully operational proteolytic mechanisms are likely integral for mechanical overload-induced skeletal muscle hypertrophy. What remains less understood, however, is how the coupling between protein synthesis and muscle protein breakdown is modulated during periods of overload to regulate skeletal muscle hypertrophy. Emerging methods that provide insight at the level of the single protein (610), single fiber, and even maintaining the spatial dimensions with emerging technologies such as nanoscale secondary ion mass spectrometry (nanoSIMS) (611) have the potential to greatly advance our understanding of this area of muscle physiology.
4.2.4. A reduction in myostatin mRNA expression and signaling markers.
As mentioned in sect. 4.2.1, myostatin (MSTN) has received considerable attention as a negative regulator of muscle growth since the landmark reports by McPherron and colleagues. MSTN is a putative myokine that is part of the transforming growth factor (TGF)-β superfamily, and signaling pathways associated with MSTN have been described in detail (612–615). Briefly, MSTN mRNA is translated into a propeptide secreted into the extracellular matrix. Once the latent propeptide is cleaved by extracellular matrix proteases, MSTN forms an active dimer that binds to the activin receptor type-2B (ACVRIIB). Ligand binding promotes the recruitment and activation of the activin receptor-like kinase-4 and -5 (ALK4 and ALK5) serine/threonine type 1 receptor kinases, which phosphorylate the SMAD2/3 heterodimer transcription factor (616). The SMAD2/3 complex interacts with SMAD4, and this heterotrimer enters the nucleus to transcriptionally regulate numerous genes that contain SMAD binding sites. It is difficult to determine which MSTN-regulated genes affect hypertrophic outcomes given that in vitro data indicate that 1,787 genes contain SMAD2/3 binding sites, 925 genes contain SMAD4 binding sites, and the TGF-β-mediated activation of SMAD2/3 can either upregulate or downregulate a fraction of these genes (617). However, MSTN signaling can affect the expression of genes without putative SMAD binding elements. For instance, proteolysis-related mRNAs are upregulated in myotubes treated with MSTN in vitro (618), in rat muscles incubated with MSTN (619), and in mice that express a constitutively active mutant of ALK5 (620). Furthermore, rat muscles subjected to MSTN gene electrotransfer display an atrophy phenotype coincident with a downregulation in structural genes (myosin heavy chain IIb, troponin I, and desmin) and myogenic transcription factors (MyoD and myogenin) (621). Data in Mstn-knockout mice suggest that MSTN signaling upregulates collagen-related genes (622) and genes that inhibit canonical Wnt signaling (623) in skeletal muscle.
Nongenomic MSTN signaling events have also been documented in skeletal muscle. For instance, MSTN-treated myotubes exhibit a reduction in AKT activity and mTORC1 signaling (624). Sartori et al. (620) reported that SMAD2/3 inhibition through shRNA knockdown promotes muscle hypertrophy in mice through enhanced mTORC1 signaling. Hulmi et al. (625) similarly demonstrated that blocking MSTN signaling in mice through intraperitoneal injections of the soluble activin receptor IIb increases mTORC1 signaling and muscle protein synthesis after 1–2 days and myofiber size after 2 wk of treatment. Also mentioned previously, Mstn-knockout mice exhibit a robust hypertrophic phenotype (558), and other genetic mouse models have been used to show that the conditional overexpression of follistatin (Fst), a MSTN inhibitor, and a mutated form of Acvr2b increases whole body muscle mass (612). More recent data in mice indicate that the overexpression of Fst increases the protein expression of several mTORC1 signaling-related proteins (e.g., AKT and p70S6k) as well as basal-state and insulin-stimulated muscle protein synthesis rates (626). Recent clinical trials in overweight and insulin-resistant participants have also indicated that 10 or 48 wk of bimagrumab administration, which blocks the activin type II receptor, significantly increases muscle mass and reduces fat mass (627, 628). Although resistance training was not employed in either study, these data reiterate that the blockade of MSTN signaling increases muscle mass in humans. Collectively, these data suggest that MSTN operates through genomic and nongenomic signaling mechanisms to upregulate proteolytic genes, downregulate structural genes and muscle-specific transcription factors, and reduce AKT-mTORC1 signaling in myofibers (FIGURE 10). Additionally, independent lines of evidence in mice indicate that longer-term MSTN inhibition does not operate through satellite cells to affect myofiber hypertrophy and, instead, likely acts through the other mechanisms discussed above (629–631).
FIGURE 10.
Summary of myostatin signaling and how mechanical overload affects signaling outcomes. This schematic (constructed with BioRender.com, with permission) provides a summary of myostatin (MSTN) signaling in skeletal muscle and how mechanical overload in rodents or humans affects signaling outcomes based on the research cited in-text. Upon ligand binding, MSTN leads to the phosphorylation of SMAD2/3, which causes the formation of the SMAD2/3/4 complex. This complex can then enter nuclei and affect the expression of genes that negatively impact muscle size and growth (i.e., genomic effects). A putative nongenomic effect of MSTN signaling includes the inhibition of AKT phosphorylation, and this can lead to diminished protein synthesis rates. Notably, blue arrows and inhibitor indicators illustrate how resistance exercise affects aspects of MSTN signaling. As discussed in main text, acute and/or chronic resistance training has been documented to upregulate myostatin inhibitors (follistatin and FSTL3), reduce SMAD2/3/4 nuclear translocation through JNK1/2 activation, and reduce MSTN mRNA levels through decreasing transcription rates.
Mechanical overload can act in various manners to downregulate MSTN signaling. Multiple studies in rats have indicated that one to four bouts of concentric, eccentric, and/or isometric contractions downregulate skeletal muscle Mstn mRNA levels (381, 382, 632). In mice, Lessard et al. (463) reported that 14 days of synergist ablation-induced mechanical overload enhances JNK/MAPK signaling, which acts to alter SMAD2/3/4 binding activity to gene promoter regions. In humans, Pillon et al. (272) analyzed transcriptome-wide array data from eight resistance exercise studies and reported that MSTN mRNA is downregulated across multiple studies hours to days after exercise. Hulmi and colleagues (633–635) published several reports using qPCR to show that MSTN mRNA is downregulated in response to one bout of resistance exercise, and Dalbo and colleagues (587, 636), Louis et al. (308), and Fernandez-Gonzalo et al. (370) have published similar reports. Hulmi et al. (635) also reported that correlations existed between MSTN mRNA levels 48 h after a naive resistance exercise bout and changes in whole body muscle mass (r = −0.88, P = 0.002) and VL muscle thickness (r = −0.51, P = 0.12) after prolonged resistance training. Other reports have also indicated that reductions in serum MSTN concentrations and skeletal muscle MSTN mRNA occur after longer-term resistance training (493, 637–641), and a recent study by McIntosh et al. (642) shows that resistance exercise downregulates MSTN mRNA levels 3 and 6 h after exercise and upregulates muscle follistatin protein levels 6 h after exercise. Collectively, these findings provide the basis for a model in which MSTN signaling is inhibited via mechanical overload through the downregulation of MSTN mRNA and upregulation in muscle follistatin protein, and through other signaling mechanisms such as increased MAPK activation. Additionally, the load-induced reduction in MSTN mRNA appears to be both transient (i.e., after a single exercise bout) and also persisting weeks/months into training.
Although the abovementioned reports are compelling, data are not consistent in this area. Willoughby (643) reported that 12 wk of resistance training in humans upregulates MSTN mRNA and protein expression concomitant with muscle hypertrophy. Kim et al. (644) reported that MSTN mRNA expression is downregulated 24 h after a single bout of resistance exercise and 16 wk after resistance training in humans. However, this pattern is similar in participants classified as hypertrophic nonresponders (i.e., participants who did not experience increases in mean fCSA with training), moderate responders (change in mean fCSA = +1,111 µm2 on average), or extreme responders (change in mean fCSA = +2,475 µm2 on average). More strikingly, muscle tissue concentrations of the active MSTN peptide were reported to increase by 44%, and, again, this pattern is evident across responder cohorts. Mobley et al. (493) reported that MSTN mRNA is not altered in lower, moderate, and higher hypertrophic responders after 12 wk of resistance training despite serum MSTN concentrations equally decreasing across responder cohorts. Data from certain rodent studies have not yielded consistent results, either. Minderis et al. (645) reported that Berlin high (BEH) mice, which are homozygous for a mutation causing a dysfunction in Mstn, present larger muscle masses versus wild-type mice. However, these authors also reported that soleus mass increases in response to synergist ablation are paradoxically impaired in the BEH line. Aoki et al. (646) reported that muscle Mstn mRNA rapidly increases in response to stretch overload in rats, and MacKenzie et al. (647) reported that hindlimb stimulations in rats result in a rapid postbout increase in muscle Mstn mRNA. Eight weeks of ladder climbing has been shown to elicit similar increases in flexor hallucis longus muscle masses and fCSA value in rats treated with either a placebo or SB431542, a myostatin inhibitor (648).
Most of the data support that MSTN mRNA is downregulated in skeletal muscle during periods of mechanical overload in rodents and humans, and some human studies suggest that a similar response occurs with circulating MSTN concentrations. There are also various animal models that suggest that Mstn gene mutations affect muscular phenotype, and pharmacological studies in rodents indicate that anabolic signaling is enhanced in skeletal muscle with MSTN inhibition. However, whether a downregulation in MSTN gene expression is needed for load-induced skeletal muscle hypertrophy requires further investigation given the conflicting data discussed here. Moreover, although a plethora of human MSTN mRNA data exist, more human resistance training studies are needed to determine how MSTN protein levels, the protein levels of endogenous MSTN inhibitors (e.g., FST and FSTL3), and SMAD signaling are affected, and whether these outcomes are predictive of hypertrophic outcomes.
4.2.5. Extracellular matrix remodeling.
The extracellular matrix has long been recognized as more than a cellular scaffold. Skeletal muscle extracellular matrix components participate in a vast array of molecular processes (649, 650), from acting as a growth factor reservoir to orchestrating fundamental cell behavior in response to loading and injury (651, 652). Given that myofibers are encapsulated by the extracellular matrix, and fCSA increases occur in response to mechanical overload, extracellular matrix adaptations likely coincide with myofiber hypertrophy. In support of this theory, a 1995 review by Millward (653) presented a “bag theory” for intramyofiber protein accretion whereby the extracellular matrix of myofibers acts like a bag and, when filled, the “bag enlargement” (i.e., the extracellular matrix remodeling) is needed for myofiber growth to continue. Several lines of rodent evidence support the idea that extracellular matrix adaptations occur during periods of mechanical overload. For instance, work by the Peterson laboratory (541, 544) supports a fusion-independent role of satellite cells during hypertrophy involving an exosome-mediated downregulation in collagen-related genes in fibroblasts. Mendias et al. (654) reported that the concentrations of a collagen cross-link marker (pyridinoline) increase and concentrations of a collagen structural stability marker (hydroxyproline) decrease in skeletal muscle 3, 7, and 28 days after synergist ablation in rats. Several mRNAs for collagen proteins and matrix metalloproteases (MMPs) were also reported to be higher at one or multiple postsurgical time points. Other rodent synergist ablation studies (414, 485, 654, 655) provide further evidence that extracellular matrix adaptations occur in skeletal muscle subjected to mechanical overload. Likewise, a recent review by Brightwell et al. (48) discusses mouse data showing that several collagen-related mRNAs are upregulated 3–15 days after mechanical overload and collagen I-expressing cells increase in abundance 4–7 days after mechanical overload (541).
Studies in humans also support the notion that extracellular matrix remodeling, via increases in collagen synthesis rates or increased gene/protein expression of extracellular matrix genes (241, 531, 656–659), occurs in response to one bout or weeks of resistance training. A recent study by the Peterson laboratory sought to extensively examine how resistance training mechanistically affects extracellular matrix remodeling mechanisms. In this study, Peck et al. (660) reported that several mRNAs associated with extracellular matrix remodeling are upregulated after 14 wk of resistance training in older adults. These authors also performed elegant in vitro experiments to show that electrically stimulated myotubes secrete leukemia inhibitory factor (LIF) to stimulate the production and secretion of MMP-14 from resident macrophages. Moreover, bone-derived macrophages in vitro treated with media isolated from electrically stimulated myotubes increased type I collagen degradation, which is abrogated with an anti-LIF neutralizing antibody. Given that others have reported that resident macrophages are needed for mechanical overload-induced hypertrophy (661), the data by Peck and colleagues suggest that resident macrophages promote skeletal muscle hypertrophy, in part, through the secretion of MMPs and extracellular matrix remodeling. It should be noted, however, that although the total content of extracellular matrix components likely increases with skeletal muscle hypertrophy via remodeling mechanisms, the relative content is likely not disproportionally altered. In this regard, MacDougall et al. (67) used trichrome staining to show that the proportion of connective tissue is similar (∼13% of imaged tissue) in the biceps brachii muscle of bodybuilders and nontrained control subjects despite bodybuilders demonstrating greater fCSA values. These data were expanded upon by a recent study performed by the Roberts laboratory (73) indicating that 10 wk of resistance training in 38 younger adult men does not affect VL muscle fascial thickness, protein and histology markers of extracellular matrix content, tissue MMP activity, or the protein expression of MMP-2/9 with concomitant VL hypertrophy.
When considering the collective evidence, it seems plausible that mechanical overload initially enhances collagen protein turnover and reduces collagen content to allow for myofiber hypertrophy, and this may occur days after the initial stimulus in rodents or days to weeks into a resistance training program in humans. These events may be promoted, in part, by stromal cells in the extracellular matrix secreting factors (e.g., exosomes from satellite cells) that inhibit the expression of collagen genes from fibroblasts and myofibers and other factors that stimulate collagen breakdown (e.g., MMP14 from macrophages). However, after myofiber and tissue growth, the extracellular matrix may favor collagen synthesis over breakdown to establish a strengthened scaffold. Although data supporting this hypothesis are limited, this notion is partially supported by transcriptome-wide investigations showing that skeletal muscle extracellular matrix-related mRNAs are more dynamically altered in the earlier (rather than later) phases of resistance training (243, 662).
Finally, it is becoming more evident that excessive fibrotic tissue deposition and disorganized collagen orientation in the extracellular matrix can constrain myofiber growth and impair hypertrophic outcomes, respectively. Regarding the former, Fry et al. (541) utilized the genetic Pax7-DTA mouse model to demonstrate that satellite cell depletion before 8 wk of synergist ablation substantially increases extracellular matrix collagen deposition and impairs myofiber hypertrophy. Long et al. (663) more recently reported that pretraining collagen characteristics (e.g., total content and packing density) are negatively associated with hypertrophic outcomes in older participants who performed 14 wk of resistance training. These data, along with the data provided by Peck and colleagues in an older population (660), raise an intriguing possibility that extracellular matrix adaptations in older adults becomes more critical in promoting myofiber hypertrophy.
4.2.6. Involvement of angiogenesis in mechanical overload-induced skeletal muscle hypertrophy.
Angiogenesis, or the formation of new capillaries, is seemingly critical for mechanical overload-induced skeletal muscle hypertrophy to occur. Synergist ablation studies in rodents indicate that angiogenesis occurs in skeletal muscle in response to mechanical overload (664–666), and Degens et al. (667) reported that plantaris hypertrophy (induced via denervation of synergist muscles) is accompanied by an increase in the capillary-to-myofiber ratio. Vascular endothelial growth factor (VEGF) promotes angiogenesis (668), and plantaris hypertrophy has been reported to be impaired in skeletal muscle-specific Vegf-knockout mice because of a reduction in the capillary-to-myofiber ratio (669). A recent report by Ato et al. (670) demonstrated that Heyl-knockout mice, which show a blunted hypertrophic response to synergist ablation (671), also present impairments in mechanical overload-induced angiogenesis. In humans, VEGF has been reported to be upregulated in skeletal muscle at the mRNA and protein levels 2 and 4 h after a resistance exercise bout (672). Longer-term resistance training studies have also indicated that increased muscle capillarization occurs (673–676). Studies in older adults have indicated that lower pretraining muscle capillary density is associated with limited hypertrophic outcomes (663, 677, 678), and a 12-yr longitudinal study in older men indicates that a reduction in capillary number per fiber accompanies leg extensor muscle atrophy (679). Nederveen et al. (680) reported that satellite cells reside near capillaries in younger men, and the capillary‐to‐fiber perimeter exchange index is associated with the satellite cell proliferation response 24 h after an eccentric exercise bout. Thomas et al. (77) recently examined how 6 wk of unilateral leg aerobic training affected subsequent 10-wk resistance training outcomes in both legs. These authors reported that aerobic training increased type I and type II myofiber capillary number and augmented subsequent resistance training-induced increases in fCSA and satellite cell abundance. Collectively, these studies support a model in which angiogenesis during mechanical overload affects satellite cell dynamics to potentially enhance skeletal muscle hypertrophy. Furthermore, the age-related loss in muscle capillaries seemingly reduces muscle plasticity to loading paradigms and may be an involved mechanism with muscle atrophy.
Pericytes, or mural cells that encapsulate and support the microvasculature and exist in the extracellular matrix, have also received attention as a cell type that adapts to mechanical overload. Dvoretskiy et al. (681) recently demonstrated that muscle-resident pericytes upregulate genes associated with angiogenesis and extracellular matrix remodeling following an acute bout of resistance exercise in mice. Additionally, pericyte transplantation in combination with exercise training resulted in significant enhancements in the capillary-to-myofiber ratio and collagen turnover. Interestingly, the Boppart laboratory (682) recently reported that pericyte transplantation in mice recovered fCSA losses and the capillary-to-fiber ratio after 14 days of hindlimb immobilization and remobilization. These findings suggest that pericytes function to promote angiogenesis and, in agreement with the literature cited above, suggest that enhanced capillarization promoted the hypertrophy observed during reloading. In agreement with such a mechanism, Hansen-Smith et al. (665) reported that synergist ablation in rats promoted angiogenesis, an increase in pericyte abundance, and muscle hypertrophy.
Collectively, a large body of evidence suggests that angiogenesis is needed to optimize skeletal muscle hypertrophy induced by mechanical loading. As this is a consistently researched area, more research will undoubtedly examine the stromal cells and signaling mechanisms involved with this process.
4.2.7. Alterations in muscle-specific and circulating microRNAs.
MicroRNAs (miRNAs) are ∼20 nucleotides in length, are noncoding, and act to inhibit the translation of mRNAs in a sequence-specific fashion via the miRNA-induced silencing complex (miRISC) (683, 684). In 1993, Lee et al. (685) reported that Caenorhabditis elegans express a miRNA (lin-4) during postembryonic development to negatively regulate the LIN-14 protein. Since this foundational discovery, there has been widespread research interest into how miRNA profiles are altered in different tissues during diseased states (686, 687). Alterations in skeletal muscle miRNAs to exercise training have also received considerable attention, and several reviews have been written on the topic (44, 688–691). Various reports have detailed the skeletal muscle miRNA responses to mechanical overload in rodents and humans. McCarthy and Esser (692) reported that 7 days of mechanical overload induced by synergist ablation increased plantaris mass by 45% while reducing muscle-enriched miR-1 and miR-133a levels by ∼50%. Several human studies have since provided miRNA targets that are altered acutely or after chronic training interventions (239, 248, 693–696), and recent in vitro and rodent evidence suggests that miR-16 is lower in mechanically overloaded muscle and that this may lead to the derepression of genes involved in myoblast differentiation and ribosome biogenesis (697).
Although these data have been informative, a 2019 study by Vechetti et al. (698) tempers enthusiasm in this area. To determine the role of microRNAs in muscle hypertrophy, the authors generated a skeletal muscle-specific Dicer knockout to globally deplete skeletal muscle miRNAs; notably, Dicer is responsible for producing mature miRNAs from pre-miRNAs (683). Despite an 80% knockdown of Dicer expression, miRNAs levels were only reduced by ∼50% and do not appear to affect mechanical overload-induced skeletal muscle hypertrophy, atrophy induced by hindlimb unloading, or age-related muscle loss. In agreement with this finding, Oikawa et al. (699) found that Dicer inactivation only reduced myomiR levels by 30–50% in this same genetic mouse model and did not affect endurance exercise adaptations following 2 wk of voluntary wheel running. Although these studies suggest that the regulation of skeletal muscle miRNA levels is maintained through a yet-to-be described Dicer-independent mechanism, they also question the involvement of miRNAs in mechanical overload-induced skeletal muscle hypertrophy and endurance training adaptations, respectively.
4.2.8. Involvement of sex hormone signaling.
Androgens, which include testosterone and its metabolites, exert downstream effects on skeletal muscle through binding to androgen receptors localized in the sarcoplasm (700). Upon activation through ligand binding, androgen receptors undergo nuclear translocation and act as transcription factors to alter the mRNA expression of hundreds to thousands of genes (701). The administration of supraphysiological levels of anabolic steroids promotes appreciable skeletal muscle hypertrophy in males (702–707) and females (532, 708). Human and rodent studies imply that these effects are due to satellite cell-mediated myonuclear accretion (703, 709), enhanced mTORC1 signaling (presumably through noncanonical androgen signaling) (710), increased ribosome biogenesis (711), and heightened muscle protein synthesis (705, 712–714). However, a more recent study has shown that Pax7-DTA mice depleted of satellite cells experience a similar magnitude of skeletal muscle hypertrophy relative to control mice with testosterone administration (715), thus implying that satellite cells may not be needed for androgens to elicit anabolic effects. It has also been demonstrated that male androgen receptor knockout (ARKO) mice, but not female ARKO mice, exhibit impairments in muscle mass during adulthood (716), and similar data have been reported in male muscle-specific ARKO mice (717). Male muscle-specific ARKO mice have also been shown to exhibit impairments in plantaris hypertrophy after 28 days of synergist ablation versus wild-type mice (+54% vs. +115%) (718). The pharmacological blockade of the androgen receptor has been shown to slightly (albeit significantly) impair skeletal muscle growth in the gastrocnemius muscle of male rats that underwent 2 wk of hindlimb stimulation (719). Likewise, Yin et al. (720) reported that androgen receptor blockade via flutamide prevents gastrocnemius hypertrophy in male rats that underwent 3 wk of ladder climbing. In men, longer-term testosterone suppression with a gonadotropin-releasing hormone analog has been shown to impair resistance training-induced skeletal muscle hypertrophy (713). Hence, there is a robust body of evidence to suggest that androgen signaling is involved with skeletal muscle hypertrophy, albeit it is notable that most of these data are prominently derived from testosterone administration studies or the blockade of androgen secretion and/or signaling.
Although the data above link androgen signaling to skeletal muscle hypertrophy, it is still unclear how androgen signaling under physiological circumstances affects this process. The necessity of circulating hormones in mechanical overload-induced hypertrophy was originally challenged by Goldberg who, as stated above, indicated that their role is minor in driving synergist ablation-induced hypertrophy in hypophysectomized rats (i.e., removal of the pituitary gland) (127). A series of studies published in the 1990s and 2000s suggested that transient elevations in circulating anabolic hormones [testosterone, growth hormone (GH), and IGF1] following bouts of resistance exercise are significant contributors to hypertrophic adaptations (721–726). However, several human studies have since indicated that the postexercise endocrine responses to bouts of resistance exercise do not correlate with transient anabolic signaling or long-term hypertrophic outcomes (727–731). Moreover, although there is limited evidence to suggest that androgen receptor protein content in skeletal muscle is associated with hypertrophic outcomes (732, 733), the most practical evidence against skeletal muscle androgen receptor signaling being a significant contributor to overload-induced hypertrophy comes from a comprehensive meta-analysis by Roberts et al. (734). These authors reported 12 hypertrophy outcomes from numerous resistance training studies involving male and female participants that spanned from 7 to 24 wk in duration (166, 735–742). The pooled effect size favoring relative (or body mass corrected) hypertrophy in males is small and not statistically significant (effect size: 0.07, P = 0.31). In explaining their findings, the authors rejected the notion that low circulating androgen levels, and presumably lower androgen signaling in skeletal muscle, are barriers to skeletal muscle hypertrophy in females.
Although some studies have examined the relationship between muscle androgen receptor protein content and hypertrophic outcomes, no study to date has extensively investigated canonical androgen receptor signaling events that occur with different loading paradigms. Assessing mTORC1 signaling in muscle tissue is straightforward given that there are relatively well-defined downstream targets that can be assayed for phosphorylation status. However, assessing whether canonical androgen receptor signaling is altered with mechanical overload is inherently more difficult given that nuclear lysates must be obtained from muscle tissue, the interaction of the androgen receptor to consensus DNA binding elements should be assessed, and transcriptional targets should be assayed. Given that the androgen receptor affects the mRNA expression of dozens to hundreds of genes, these collective endeavors can be cumbersome. Even if these assays are performed, extensive genetic screening would be needed to determine which androgen-sensitive mRNAs affected by mechanical overload promote skeletal muscle hypertrophy. Cardaci et al. (743) reported that androgen receptor DNA binding affinity is enhanced 3 h after a resistance exercise bout in men. Hence, replicating this approach with human time course resistance training studies and more downstream analyses may yield insightful information as to whether alterations in androgen receptor DNA binding affinity, as well as changes in nuclear androgen receptor concentrations and downstream mRNAs, correlate with hypertrophic outcomes. The transgenic androgen response element luciferase (ARE-Luc) mouse would also be an excellent model to utilize to further enhance our knowledge in this area (744). Specifically, this mouse model allows researchers to monitor androgen receptor DNA binding, and time course mechanical overload studies that examine muscle luciferase activity could yield insightful associations between androgen DNA binding activity and hypertrophy. Given these knowledge gaps and conflicting data in this area, more research is needed to determine the degree to which enhanced androgen signaling during periods of mechanical overload contributes to skeletal muscle hypertrophy.
As with testosterone, female sex hormones such as estrogen and progesterone operate through canonical nuclear receptor DNA binding and noncanonical protein kinase signaling (32). However, unlike exogenous testosterone administration, oral contraceptives (which consist of estrogens and/or progestins) have no meaningful impact on muscle hypertrophy in younger female participants who resistance train (512, 745–749). Furthermore, there is evidence to suggest that estrogen replacement in older women diminishes the myofibrillar protein synthesis response to a resistance exercise bout (750), and a recent meta-analysis by Javed et al. (751) indicated that estrogen-progesterone or estrogen-only replacement therapy does not prevent muscle mass loss with aging in women over the age of 50 yr. Hence, most evidence to date suggests that factors aside from sex hormone signaling are response for mechanical overload-induced skeletal muscle hypertrophy in females.
4.2.9. Involvement of inflammation via prostaglandin signaling.
Prostaglandins are lipid mediators formed through a multicatalytic reaction involving cyclooxygenase 1/2 (COX-1/2) enzymes and cell-specific prostaglandin synthases (752). Rodemann and Goldberg (753) were the first to report that prostaglandin F2α (PGF2α) increases protein synthesis in isolated rat hindlimb muscles. Two decades later, Trappe et al. (754) demonstrated in humans that 1,200 mg of ibuprofen, which inhibits the COX enzymes and blunts PGF2α levels in skeletal muscle (755), prevents increases in muscle protein synthesis during a 24-h period following a high-volume eccentric resistance exercise bout. PGF2α operates through the G protein-coupled prostanoid FP receptor to stimulate mTORC1 signaling in myotubes in vitro (756). There are also four known E-prostanoid (EP) receptors (EP1, EP2, EP3, and EP4), and EP4 may be especially relevant for skeletal muscle hypertrophy (237, 757, 758). Ho et al. (759) reported that prostaglandin E2 (PGE2) stimulates satellite cell proliferation through the EP4 receptor and that the genetic ablation of satellite cell EP4 receptor in mice impairs muscle repair after various forms of injury. These authors additionally reported that the acute administration of PGE2 after muscle injury improves the morphology and function of muscle tissue. Hence, there are multiple lines of evidence indicating that prostaglandins likely contribute to anabolic signaling events in skeletal muscle.
Studies that have inhibited prostaglandin synthesis during periods of mechanical overload have yielded intriguing results. For instance, daily ibuprofen administration reduces plantaris hypertrophy in rats by ∼50% after synergist ablation (760), and the daily administration of a COX-2-specific inhibitor in mice almost completely abrogates plantaris hypertrophy and muscle protein accretion 14 days after synergist ablation (761). In younger adults, Markworth et al. (762) reported that higher-dose ibuprofen administration (1,200 mg/day) blunts certain aspects of MAPK and mTORC1 signaling 3 and 24 h after a single resistance exercise bout, and others have reported that 1,200 mg/day of ibuprofen blunts increases in leg muscle volume by ∼50% after 8 wk of resistance training compared to a group receiving 75 mg/day of acetylsalicylic acid (763). COX inhibition via ibuprofen infusion into muscle during and hours after an eccentric training bout prevents satellite cell proliferation 8 days after exercise (764). COX-2 inhibition in mice also prevents stromal cell proliferation up to 14 days after synergist ablation (761), and Mackey and colleagues (765) have reported similar findings in humans after a 36-km run. Peterson and Fyfe (28) authored a recent review citing multiple studies that examined the efficacy of cold-water immersion (CWI) as a means to potentially enhance recovery aspects during resistance training. Notably, this practice is popular in the athletic sphere given that certain lines of research indicate that CWI reduces postexercise soreness and promotes a more rapid restoration of muscle strength after rigorous exercise (766). However, limited research in this area indicates that CWI blunts certain aspects of mTORC1 signaling and satellite cell proliferation (767, 768). Although not explicitly stated, the potential contributions of reduced prostaglandin signaling cannot be discounted, given that cryotherapy has been shown to reduce tissue prostaglandins in other models of inflammation (769, 770). Hence, a reduction in exercise-induced inflammation via anti-inflammatory drugs or CWI may interfere with muscle repair or remodeling, and mechanisms that promote skeletal muscle hypertrophy may be subsequently impaired.
Like several mechanisms discussed in this review, there are also incongruent data in this area. Mikkelsen et al. (659) reported that indomethacin (a COX inhibitor) infusion into muscle during and after a resistance exercise bout did not affect myofibrillar or collagen protein synthesis rates during the 24–28 h postexercise period. Krentz et al. (771) reported that lower-dose ibuprofen administration (400 mg/day) in younger adults did not affect increases in biceps muscle thickness following 6 wk of resistance training, and Candow et al. (772) reported similar findings in postmenopausal women who consumed 400 mg/day of ibuprofen over a 9-week period. Lilja et al. (480) reported that higher-dose ibuprofen administration (1,200 mg/day) did not impair acute or chronic hypertrophy mechanisms (i.e., mTOR signaling, ribosome biogenesis, satellite cell content, myonuclear accretion, and muscle capillarization) in younger adults who partook in 8 wk of resistance training. Trappe et al. (773) reported that higher-dose ibuprofen administration (1,200 mg/day) enhanced hypertrophic outcomes in older participants, which contrasts with the data discussed above in younger participants indicating that higher-dose ibuprofen administration either does not affect or inhibits the hypertrophic response to resistance training. Damas and colleagues (310, 774) conducted a series of studies with an experimental design to test the relationship between changes in muscle damage and inflammation, myofibrillar protein synthesis, and muscle hypertrophy in previously untrained participants. Correlational analysis revealed that a greater magnitude of muscle damage and inflammation after the first four resistance exercise bouts does not confer a significantly greater hypertrophic response to 10 wk of resistance training. In addition, myofibrillar protein synthesis does not significantly correlate with muscle hypertrophy when damage and inflammation are highest (i.e., in response to the first resistance exercise session). After a progressive attenuation of muscle damage and inflammation throughout resistance training, however, myofibrillar protein synthesis is strongly correlated with muscle hypertrophy induced by 10 wk of resistance training (r = ∼0.90). As an interesting aside, Damas et al. (243) utilized a microarray approach to report that mRNAs related to inflammation and proteolysis are upregulated after the first bout of resistance exercise. However, a subsequent bout that occurred after 10 wk of resistance training in these same participants resulted in an upregulation in mRNAs related to muscle structure and contractile function, suggesting that increased training status results in a refined transcriptomic response.
Several lines of evidence published to date suggest that prostaglandin signaling in skeletal muscle is elevated in response to mechanical overload. Additionally, the human studies discussed in this section indicate that a greater reduction in prostaglandin synthesis occurs with higher doses of nonsteroidal anti-inflammatory drugs (1,200 mg/day), and this may partially abrogate resistance training-induced skeletal muscle hypertrophy compared with lower doses (i.e., 400 mg/day). However, findings are mixed and seemingly age dependent. Moreover, the necessity of inflammation, in general, for skeletal muscle hypertrophy to occur is confounded by the lack of associations discussed above by Damas and colleagues. Thus, more time course studies are needed to further elucidate the relevance of mechanical overload-induced inflammation in the hypertrophic process.
4.2.10. Involvement of β-adrenergic signaling in myofibers.
β2-Adrenergic receptor signaling operates through a canonical signaling pathway that involves 1) ligand (catecholamine) binding to the G-coupled protein receptor, 2) the intracellular activation of adenylyl cyclase, 3) the production of cyclic adenosine monophosphate (cAMP), and 4) the activation of protein kinases (e.g., protein kinase A, or PKA) (775). Active PKA phosphorylates and activates the cAMP response element binding protein (CREB) transcription factor (50). Several studies have shown that the administration of β-adrenergic receptor agonists to animals enhances skeletal muscle hypertrophy (776–786), which occurs independently of mechanical overload. Hinkle et al. (787) also reported that administration of the β-adrenergic receptor agonist clenbuterol to mice lacking β1-adrenergic receptors enhances skeletal muscle hypertrophy, whereas mice lacking β1/2-adrenergic receptors did not show this response. Woodall et al. (788) used genetic mouse models to determine that clenbuterol enhances muscle Akt activity, and transgenic mice conditionally overexpressing CREB-regulated transcriptional coregulators (Crtc) show a hypertrophic phenotype (789). Jessen et al. (790) reported that clenbuterol transiently upregulates skeletal muscle mTORC1 signaling markers in humans, and these researchers also reported that the selective β2-agonist salbutamol augments type II myofiber hypertrophy in college-aged men after 11 wk of resistance training (791). These studies, and others reporting similar findings (792), indicate that β2-adrenergic signaling can enhance mTORC1 activity in skeletal muscle. However, the administration of salbutamol (a short-acting β2-adrenergic receptor agonist) has been shown to concomitantly enhance the phosphorylation of CREB, AKT2, and the myofibrillar protein synthetic response to a single bout of resistance exercise in humans without affecting mTORC1 signaling markers (464).Thus, along with activating mTORC1 signaling, β2-adrenergic signaling seemingly stimulates skeletal muscle hypertrophy in an mTORC1-independent manner.
Select reviews such as those by Glass (793), Schiaffino et al. (10), and Sartori et al. (9) have highlighted the role that β2-adrenergic receptor signaling may have in promoting skeletal muscle hypertrophy. However, although research on this topic blossomed in the 1980s, it is perplexing that this area of the hypertrophy literature has been largely overlooked in recent years. Several recent hypertrophy reviews have prioritized the involvement of mTORC1 signaling, satellite cells, and ribosome biogenesis over β2-adrenergic signaling, which also reflects the low number of original articles on this mechanism in recent years. Although reasons as to why diminished interest in this area has occurred are difficult to posit, we speculate that certain discoveries in the field provide plausible explanations. First, clenbuterol administration was largely researched as a pharmacological means to increase meat yields in livestock, and this approach was abandoned shortly after food poisoning outbreaks in Europe linked to clenbuterol accumulation in bovine liver and meat products (794–796). Additionally, findings from the Bodine laboratory (797) indicating that mTOR inhibition in mice that were administered clenbuterol blunts, but does not completely abrogate, the hypertrophic effects of overload likely dampened enthusiasm in this area. The authors rightfully concluded that the anabolic effects of clenbuterol are mediated, in large part, through the activation of the mTOR signaling pathway. However, the authors also reported that canonical β2-adrenergic signaling might reduce atrophic signaling in an mTOR-independent manner. Indeed, more recent human findings by Jessen and colleagues (790, 791) indicating that β2-adrenergic agonists operate in an mTORC1-independent manner to enhance the protein synthesis response to resistance exercise are provocative, and these studies may instigate future research in this area.
Although the evidence regarding the anabolic effects of pharmacological β2-adrenergic agonists has been informative, virtually no data exist on the potential anabolic effects of endogenous catecholamines during periods of mechanical overload. Several studies have shown that circulating epinephrine concentrations transiently increase during and after resistance exercise bouts (798–801), and epinephrine is a well-known β2-adrenergic receptor ligand (49, 802). Moreover, one report indicates that β2-adrenergic receptor blockade via propranolol does not affect strength outcomes following strength training interventions (789). However, mechanistic studies seeking to determine whether inhibiting β2-adrenergic receptor signaling during periods of mechanical overload affects anabolic signaling and/or skeletal muscle hypertrophy are lacking, and performing such studies will provide much needed insight in this area.
4.2.11. Involvement of angiotensin II signaling.
Research interest in angiotensin II signaling and hypertrophy was initially rooted in cardiac hypertrophy research (803–805). These findings motivated the Booth laboratory to investigate the role of angiotensin II signaling in skeletal muscle hypertrophy. Gordon et al. (806) reported that the pharmacological blockade of angiotensin II signaling through the angiotensin II type 1 (AT1) receptor reduced overload-induced hindlimb hypertrophy in rats 28 days after synergist ablation. Subsequent studies have demonstrated similar phenomena such as that the inhibition of angiotensin II production with an angiotensin-converting enzyme (ACE) inhibitor blunts the satellite cell proliferation response to synergist ablation in rat soleus muscles (807) and AT1 receptor blockade through losartan prevented skeletal muscle hypertrophy in rats following 4 wk of eccentric training (808). Notwithstanding, follow-up research in this area has been relatively sparse and conflicting. For instance, Zempo et al. (809) paradoxically reported that AT1 receptor global-knockout mice exhibited a similar magnitude of skeletal muscle hypertrophy relative to wild-type mice in response to 14 days of synergist ablation. Heisterberg et al. (810) reported that losartan administration did not affect various hypertrophy indexes after 4 mo of resistance training in older men. Another study by this group indicated that losartan generally did not affect the acute satellite cell or mRNA expression responses to one bout of exercise (811). Additionally, mouse studies suggest that heightened circulating angiotensin induces muscle atrophy through the hepatic production of proinflammatory mediators, which in turn leads to chronic elevations in muscle proteolysis (812). Hence, this collective evidence suggests that angiotensin II signaling through the AT1 receptor in skeletal muscle may coincide with overload-induced hypertrophy, albeit the conflicting evidence makes it difficult to conclude whether this hormonal mechanism exerts an appreciable role in the process.
4.3. Emerging Mechanisms That May Be Involved with Skeletal Muscle Hypertrophy
4.3.1. Mitochondrial biogenesis.
In vitro microbial research suggests that energy harnessed from the catabolism of four ATP molecules is required per peptide bond synthesized (813). Muscle proteolysis also requires ATP (583). Thus, although no formal estimates have been made, the bioenergetic requirement to support enhanced myofiber protein turnover and subsequent protein accretion during periods of resistance training, where mean fCSA values can increase ∼15–30% in size on average (814), is appreciable. Indeed, the dysregulation of mitochondrial biogenesis and function leads to muscle loss. For instance, mice expressing a proofreading-deficient version of mtDNA polymerase gamma (PolG) show heightened mitochondrial fission and autophagy levels that coincide with muscle atrophy (815, 816). Moreover, mice overexpressing the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), which is a key regulator of mitochondrial biogenesis (817), show a reduction in muscle atrophy induced by denervation, fasting, and hindlimb unloading (818, 819). Conversely, muscle-specific PGC-1α-knockout mice have been reported to show modest impairments in mechanical overload-induced plantaris hypertrophy after 14 days of synergist ablation (820). Uemichi et al. (466) demonstrated that 14 days of synergist ablation increased plantaris markers of mitochondrial remodeling to presumably enhance mitochondrial expansion and function. The investigators also used TEM to demonstrate that the area in myofibers occupied by mitochondria increased approximately fivefold after synergist ablation despite plantaris muscle mass only increasing approximately twofold. Thus, these several independent observations in rodents suggest that a critical mass of normally functioning mitochondria is needed within myofibers to maintain muscle mass and an expansion of the mitochondria may be needed to optimize load-induced skeletal muscle hypertrophy.
Although resistance training affects mitochondrial markers in humans, the data are mixed and this may be due to differences in training paradigms as well as different methodologies used to detect these markers. Costill et al. (152) were the first to report that resistance training increases mitochondrial enzyme activity markers, albeit this was reported to occur with higher-volume training. Groennebaek and Vissing (821) authored a review including 16 human studies examining how chronic resistance training affected mitochondrial volume density and function. It should be noted that several studies cited in the review assayed muscle tissue CS activity as a surrogate of mitochondrial volume density given the findings of Larsen et al. (822) indicating that this metric exhibits a strong correlation with myofiber mitochondrial content as assessed through TEM imaging (r value = 0.84, P < 0.05). Additionally, older studies mentioned in the Groennebaek and Vissing review used TEM to assess the percentage of intramyofiber space occupied by mitochondria. Only two of the cited studies in the Groennebaek and Vissing review (195, 823) reported increases in mitochondrial volume density after 12 wk of resistance training, whereas the other 14 studies reported no changes or decreases. Three of the five studies that assessed markers of mitochondrial function (e.g., a tighter coupling of oxidative phosphorylation) reported improvements (824–826). These data, along with other studies (263, 827), motivated Parry, Roberts, and Kavazis (45) to author a review in 2020 positing that myofiber hypertrophy during resistance training may occur more rapidly than the expansion of the mitochondrial reticulum. However, subsequent findings by Ruple et al. (55) in college-aged males who partook in 10 wk of resistance training challenged this notion. In short, the investigators reported that mitochondrial volume density, assessed by immunostaining the outer mitochondrial TOMM20 protein, increased in type I and II myofibers and these increases outpaced fCSA increases in both fiber types. The authors also reported that CS activity values were not significantly altered with resistance training and that CS activity change score values exhibit a poor association with changes in type I and II myofiber mitochondrial area assessed by TOMM20 immunostaining.
The aforementioned reports have multiple implications. First, more research is needed to determine the veracity of using CS activity as a marker to track changes in mitochondrial volume density with resistance training. Second, increases in mitochondrial volume density in humans may precede load-induced increases in myofiber hypertrophy as observed by Uemichi et al. (466) in rodents. Pillon et al. (272), who analyzed numerous studies that examined the transcriptomic responses to resistance exercise, reported that PGC1-α mRNA is transiently upregulated after single bouts of resistance exercise. Although these data provide further indirect support for mitochondrial biogenesis being a response to resistance training, these microarray investigations may have captured an upregulation in the PGC-1α4 mRNA variant rather than the common PGC-1α1 variant. Notably, various studies in humans support that an upregulation in the PGC-1α4 isoform occurs with resistance training and that this could lead to hypertrophic signaling (e.g., downregulation in MSTN and upregulation in IGF1) while also upregulating glycolytic (rather than mitochondrion related) genes (828, 829). As a final note in this section, an expansion or remodeling of the mitochondrial reticulum during periods of mechanical overload may serve nonenergetic roles such as calcium buffering (and thus refining calcium signaling) (812), and the mitochondrial propagation of redox signaling may assist with myotube hypertrophy in vitro (813) and in rodents (814). Thus, although it appears that mitochondria play various roles in mechanical overload-induced skeletal muscle hypertrophy, much remains to be determined in deciphering all these roles.
4.3.2. Other bioenergetic adaptations in myofibers.
An emerging research theme in muscle biology involves the necessity of metabolic reprogramming and an enhanced uptake and utilization of glucose to provide substrates for myofiber growth during periods of mechanical overload. A seminal 1972 paper by Gollnick et al. (146) supported that weightlifters possessed a lower percentage of oxidative myofibers versus endurance-trained and untrained participants. Tesch and colleagues (167) indicated that 6 mo of conventional resistance training did not alter muscle enzyme activities related to the ATP-PCr or glycolytic systems, albeit earlier work by Costill et al. (152) reported that several glycolytic enzyme markers were affected with 7 wk of higher- versus lower-volume resistance training. In agreement with Costill et al., two proteomic investigations by the Roberts laboratory indicate that higher-volume resistance training induces a significant elevation in glycolytic proteins relative to lower-volume resistance training (262, 263). Verbrugge et al. (830) reported that muscle pyruvate kinase 2 (PKM2) is preferentially upregulated over the PKM1 isoform after 6 wk of resistance training in humans, and the same group found that the knockdown of both Pkm1 and Pkm2 blunts myotube hypertrophy in vitro. Valentino et al. (831) used microarrays to show that mechanical overload in mice activates the pentose phosphate pathway (PPP) leading to enhanced NADPH synthesis, and this mechanism was proposed to be necessary for heightened redox regulation during the early stage of hypertrophy. Other in vitro evidence suggests that glycolysis inhibition through 2-deoxy-d-glucose reduced murine and primary myotube size by ∼40% (832), and the Ogasawara laboratory (833) reported that the inhibition of glycolysis reduces skeletal muscle mTORC1 signaling in rats following isometric contractions induced by hindlimb stimulations. Several of these findings prompted Wackerhage et al. (834) to posit that metabolic adaptations accompanying mechanical overload-induced myofiber growth parallel metabolic reprogramming events that occur in cancer cells; specifically, both cell types increase the uptake and utilization of glucose for cell growth as posited by Otto Warburg in the 1920s (835). Others have similarly hypothesized that glucose and downstream metabolites are shunted to metabolic pathways in myofibers during hypertrophic growth to provide macromolecules (e.g., nucleotides, amino acids, and lipids) necessary for the anabolic processes (830, 831, 836). Thus, determining the roles these metabolic adaptations have in skeletal muscle hypertrophy represents an exciting new area of research for the field and will provide a more comprehensive description of the metabolism underlying muscle growth.
4.3.3. The muscle circadian clock.
Virtually all cells possess a circadian clock mechanism that is characterized by a ∼24-h transcriptional-translational feedback loop (837). The circadian clock transcriptional program is commonly referred to as clock output, and this has been characterized in both mouse and human muscle (838–840). Clock gene expression patterns are similar between nocturnal rodents and diurnal humans when viewed in the context of rest-active cycle rather than the light- dark cycle. Beyond the commonality in core clock genes, comparisons of clock output have identified >400 common mRNAs that cycle in human and mouse skeletal muscle. Functional analysis of the common clock output mRNAs highlights the links between the circadian clock and substrate metabolism (e.g., PDK4), transcription factors (e.g., MYOD1), and proteostasis (e.g., TFEB) (841). Studies using genetic mouse models to disrupt the muscle circadian clock indicate that metabolism, mitochondrial function, and muscle function are impaired (842, 843).
Despite evidence demonstrating that clock function in muscle is critical for homeostasis, it is still unclear what the role of the muscle clock is in response to resistance exercise. In humans, Zambon and colleagues (844) provided the first and most extensive investigation as to how resistance exercise modulates the expression of circadian-regulated genes in skeletal muscle. The researchers had participants perform a session of unilateral leg resistance exercise at 1:30 PM after 8 days of controlling diet and physical activity. Muscle biopsies were obtained 6 h and 18 h after resistance exercise in both the exercised and nonexercised legs. Microarray results revealed that 704 genes and 1,479 genes are differentially expressed at 6 h and 18 h after the resistance exercise session, respectively. In addition, 40% of circadian rhythm-related genes are significantly altered 6 h after resistance exercise. Three of the core circadian clock genes (CRY1, PER2, and BMAL1) as well as the muscle-specific transcription factor MYOG were reported to also be upregulated 6 h after resistance exercise in the exercised leg. Other human data indicate that one bout of resistance exercise affects the muscle mRNA expression of core clock genes (844). However, the single-bout nature of these studies has not allowed investigators to determine whether exercise-induced alterations in the muscle circadian transcriptome are related to longer-term hypertrophic outcomes. Furthermore, the researchers did not vary the time of exercise, which would have provided stronger evidence that resistance exercise modulates circadian-related mRNA expression patterns.
It is also worth noting that a meta-analysis by Grgic and colleagues (845) indicates that the magnitude of muscle hypertrophy in humans is similar regardless of the time of day at which the resistance training is conducted. This can be viewed in one of two ways, including 1) resistance training induces muscle circadian clock gene adjustments to better align metabolism and other cell functions around the training stimulus to better optimize skeletal muscle hypertrophy or 2) although certain muscle circadian clock genes are responsive to resistance exercise as indicated above, resistance training has no appreciable influence on the muscle circadian transcriptome and this is not an involved mechanism in skeletal muscle hypertrophy. To test these hypotheses in humans requires the application of randomly timed exercise bouts throughout training, which is difficult to execute. Thus, the use of mouse genetic models will allow for direct testing of the requirement of the clock in adaptations to mechanical overload. Additionally, there are very few studies of resistance exercise or models of muscle hypertrophy that include true circadian design strategies, and this too warrants further investigation.
4.3.4. Microtubules and myonuclear and RNA trafficking.
The microtubule network in myofibers serves as an intracellular cytoskeletal scaffold that structurally harnesses myofibrils and other organelles. Elegant TEM work in the 1980s indicated that microtubules exist in the intermyofibrillar space and appear to wrap around myofibrils in a helical fashion (846). Boudriau et al. (847) subsequently used immunohistochemistry to show that slow- and fast-twitch myofibers possess extensive microtubule networks surrounding myofibrils and myonuclei.
Until recently, interest in the role myofiber microtubules have in exercise adaptations has been relatively subdued. However, three recent studies have detailed critical roles that microtubules possess in myofibers, and there are stark implications regarding how the microtubule network may participate in overload-induced skeletal muscle hypertrophy. In 2021, Denes et al. (848) used an ex vivo RNA fluorescence in situ hybridization (FISH) visualization strategy to show that various mRNAs colocalize with microtubules in adult mouse myofibers, and this agrees with a 2021 report by Pinheiro et al. (849) indicating that mRNA distribution away from myonuclei in myofibers is dependent upon microtubule-mediated transport. Denes and colleagues also reported that the pharmacological inhibition of microtubule assembly leads to an aggregation of RNAs around nuclear envelopes and a robust downregulation in mRNA translation at the Z disks. These and other findings from this study led the authors to posit that microtubule-dependent RNA transport from myonuclei to ribosomes is essential for properly localizing muscle protein synthesis at the sarcomeres. The same month, Roman et al. (850) published a report demonstrating that the microtubule network transports myonuclei to contraction-induced muscle injury sites hours after exercise in mice. Additionally, these investigators performed in vitro experiments to illustrate that myotubes treated with compounds that slowed myonuclear migration exhibit a delay in sarcomere repair following laser-induced damage. On the basis of results from these experiments, the authors concluded that (contrary to the satellite cell-centric view of muscle repair) myonuclear migration is likely critical for the local delivery of mRNAs required for protein production and repair of damaged sarcomeres transiently following exercise bouts. An excellent review by Bagley, Denes, Wang, and others (51) discusses the implications of these papers for interested readers.
As an interesting aside, several investigations in cardiomyocytes have indicated that microtubule reorganization coincides with overload-induced cardiac hypertrophy (851–854), and Scarborough et al. (855) recently reported that that microtubules are indispensable for cardiac growth via spatiotemporal control of the translational machinery. However, no studies have directly sought to determine how mechanical overload in rodents or humans affects the expression or spatial orientation of microtubule proteins (e.g., α-tubulin and β-tubulin) or other proteins present in the microtubule network in skeletal muscle (e.g., nuclear lamins and desmin). Interestingly, α-tubulin is commonly used as a housekeeping protein, which is widely assumed not to be altered by various exercise stressors. In a 2016 article titled “Housekeeping proteins: how useful are they in skeletal muscle diabetes studies and muscle hypertrophy models?”, Fortes et al. (856) reported that α-tubulin and γ-tubulin protein levels more than double in the extensor digitorum longus muscle of rats after 7 days of synergist ablation. Human data from the Bamman laboratory indicate that moderate and higher hypertrophic responders to 16 wk of resistance training exhibit an upregulation in muscle lysate α-tubulin protein levels, whereas lower responders do not (238). Both studies imply that myofiber hypertrophy may rely on the expansion or reorganization of the microtubule network given its putative role in RNA trafficking, protein synthesis regulation, and myofiber repair. However, this is highly speculative, and innovative investigations are needed to provide additional insight.
4.3.5. The gut microbiome-skeletal muscle signaling axis.
Several trillion bacteria inhabit the gastrointestinal tract, and these microbes affect physiological processes ranging from immune function to nutrient absorption (857). The continued development of ever more powerful sequencing technology underlies the heightened interest in the gut microbiome by allowing for the identification of individual bacterial species via metagenomic analysis. Various reviews have summarized the few studies that have investigated how exercise is able to alter the bacterial composition of the gut microbiome (858–860). A review by Mailing et al. (858) detailing how exercise affects the microbiome in humans indicates that 1) 6 wk of endurance training increases the abundance of short-chain fatty acid (SCFA)-producing taxa and these effects are reversed after detraining (861), 2) a trend for increased bacterial diversity occurs with 8 wk of endurance training (862), and 3) a higher Firmicutes-to-Bacteroidetes ratio is associated with a higher aerobic capacity (863). Although informative, less research exists detailing the gut microbiome responses to resistance training. Cronin et al. (862) performed an 8-wk combined aerobic and resistance training intervention in which 90 participants were randomized to one of three groups including exercise training alone, exercise training with whey protein supplementation, and whey protein supplementation only. In short, the authors reported no significant changes in fecal taxonomic composition following the exercise interventions. Bycura et al. (864) reported how 8 wk of endurance training versus resistance training affected the gut microbiome in healthy, younger adults. Endurance training elicited more robust microbiome alterations compared to resistance training, indicating that resistance training either does not appreciably affect the microbiome or does so in a more subtle manner. Moore et al. (865) also examined fecal samples in older participants after 6 wk of resistance training. The authors reported that biome diversity metrics were not significantly altered despite this shorter period of resistance training causing a significant increase in strength and skeletal muscle hypertrophy. Recent data in mice agree, in principle, with the two aforementioned human trials and also indicate that 4 wk of ladder climbing does not appreciably alter gut microbiome diversity metrics (866).
Although this preliminary evidence suggests that shorter-term resistance training does not appreciably impact the composition of the gut microbiome in humans, a preclinical study found that a healthy gut microbiome is necessary for skeletal muscle adaptation to exercise (867). Specifically, these authors reported that antibiotic-induced gut dysbiosis impairs soleus and plantaris muscle hypertrophy in mice subjected to 8 wk of loaded voluntary wheel running and this occurs despite drug-treated and untreated mice running similar distances. On the basis of these findings, the authors speculated that there are yet-to-be identified microbially derived metabolites that are required for optimal muscle adaptation to exercise training, which may include amino acids, bile acids, or SCFAs. Also notable are the earlier findings of Bäckhed et al. (868), who reported that germ-free mice, which completely lack commensal bacteria, display an atrophy phenotype. Finally, Castro et al. (869) recently determined that 12 wk of weighted ladder climbing in rats decreases the relative phyla abundance of Pseudomonas, Serratia, and Comamonas, while increasing Coprococcus. Although it remains to be determined, the authors speculated that the change in the composition of the gut microbiome with mechanical overload reduces inflammation, which improves metabolic and hypertrophic outcomes. This area of study is still in its infancy, and the goal of future studies will be to identify specific bacterial species and their respective metabolites that have direct or indirect roles in regulating muscle hypertrophy in response to mechanical loading.
5. A BRIEF DISCUSSION ON HOW SEX, RACE, AND AGE AFFECT OVERLOAD-INDUCED SKELETAL MUSCLE HYPERTROPHY
Skeletal muscle hypertrophy during periods of resistance training appears to be conserved between sexes. Evidence supporting this contention comes from recent meta-analyses showing that the degree of muscle hypertrophy in response to resistance training (when considering relative or body mass-adjusted values) is similar between males and females (734, 870), and similar data have been published since these meta-analyses (871). Moreover, mechanisms such as increased muscle protein synthesis, mTORC1 signaling, and the satellite cell response to resistance exercise are similar between sexes (222, 366, 731), which has been confirmed in preclinical models. However, these findings do not consider the fact that females are an understudied population in sports sciences (872, 873). As mentioned above, more data are needed in females to characterize how estrogen receptor signaling, among other inherent aspects of female physiology, may affect hypertrophic outcomes.
Although much more limited, there is evidence suggesting that race does not significantly affect the hypertrophic response to resistance training (874). Notwithstanding, more research is needed on diverse races, given that younger adult Caucasians have been the commonly examined population in many studies cited here.
Finally, although aging may impair the hypertrophic responses to mechanical overload, this is a more nuanced topic in the literature. Multiple studies have indicated that resistance training can lead to skeletal muscle hypertrophy in older adults (68, 722, 737, 740, 875–886), albeit some studies have indicated that aging impairs acute anabolic signaling and longer-term hypertrophic responses (880, 887–889). A recent meta-analysis by Straight et al. (890) suggests that increase in myofiber size with resistance training is impaired in older participants, which supports the notion that aging blunts hypertrophic outcomes. Studies that have obtained muscle biopsies have also indicated that participants >80 yr old show limited muscle plasticity in response to resistance training (520, 891) (e.g., limited increases in fCSA or satellite cell abundance). There are a variety of mechanisms that may be responsible for these age-related responses including, but not limited to, heightened low-grade inflammation with aging that blunts anabolic signaling in skeletal muscle (892, 893), older individuals showing a dampened anabolic response to protein and amino acid ingestion (894), a loss in higher-threshold motor units and myofibers (895), and impairments in skeletal muscle ribosome biogenesis and proteostasis in response to one or multiple bouts of resistance training (362, 519, 896).
As an interesting aside related to age-related responses to mechanical overload, recent rodent work has utilized senolytic cocktails (i.e., dasatinib and quercetin, or D + Q) to enhance skeletal muscle hypertrophy in older mice in response to overload. Specifically, Dungan et al. (897) reported that two gavage feedings of D + Q increases the plantaris hypertrophic response to 14 days of synergist ablation in older mice, which coincides with a blunted increase in senescence-associated beta-galactosidase-positive cells during the overload period. These researchers also reported that older mice present more senescent cells in the extracellular matrix in response to overload (but not in the basal state) relative to younger adult mice, and this is independent of D + Q administration. Hence, the hypertrophic response may be impaired in older participants because of a heightened senescent cell accumulation in the extracellular matrix during resistance training. Likewise, more research is needed to determine whether senolytic cocktails enhance skeletal muscle hypertrophy in older participants who perform longer-term resistance training.
While the effect of aging on mechanical overload-induced skeletal muscle hypertrophy remains a salient issue in the field, it is practically undebatable that resistance training increases strength and functional outcomes in older individuals (898–902). Furthermore, a recent meta-analysis indicated that muscle-strengthening activities are inversely associated with the risk of all-cause mortality and diseases including cardiovascular disease, diabetes, cancer (overall), and lung cancer (903). Position stands by the National Strength and Conditioning Association (899) and the American College of Sports Medicine (904) provide resistance training recommendations in older persons for interested readers.
6. MOVING TOWARD A UNIFIED PERSPECTIVE ON A DEFINITION OF AND MECHANISMS INVOLVED WITH MECHANICAL OVERLOAD-INDUCED SKELETAL MUSCLE HYPERTROPHY
Several attempts in the literature have been made to define mechanical overload-induced hypertrophy in adult skeletal muscle. Russell and colleagues (905) suggest that hypertrophy is an increase in muscle mass and cross-sectional area at the whole tissue and cellular levels, and this largely agrees with Oxford’s definition presented above in this review. However, more complex definitions exist in attempts to describe molecular nuance. Glass (906) defined skeletal muscle hypertrophy in adults as an increase in muscle mass, which manifests as an increase in the size, as opposed to the number, of preexisting skeletal myofibers. Roberts et al. (155) speculated that various forms of myofiber hypertrophy may occur, including conventional hypertrophy, or the proportional increase in contractile protein as myofibers increase in diameter, the disproportional increase in (or packing of) contractile protein as myofibers increase in diameter, or the disproportional increase in myofiber diameter relative to contractile protein accretion. Jorgenson et al. (1) suggest that conventional myofiber hypertrophy persists during various loading paradigms, and in some cases increases in fiber length can coincide to interactively promote tissue cross-sectional area changes. Damas, Libardi, and Ugrinowitsch (26) suggest that “true” hypertrophy occurs when there is an increase in the cross-sectional area of the myofibers or whole muscle, without the presence of exercise-induced muscle swelling. Finally, there is evidence to support that longitudinal myofiber hypertrophy may mechanistically differ from radial myofiber hypertrophy, and this is a continued area of investigation (1).
Each definition implies that muscle tissue and myofiber growth occur in tandem with contractile protein accretion. However, several points of contention exist regarding involved mechanisms. For instance, Jorgenson and colleagues (1) and Roberts and colleagues (155) argue that the current evidence is weak regarding mechanical overload-induced myofibril hypertrophy. There are also opposing viewpoints regarding whether myonuclear accretion via satellite cell fusion is obligatory for myofiber hypertrophy (545), whether myonuclei that are gained during resistance training demonstrate permanence or are lost during detraining (907–911), or whether hyperplasia contributes to skeletal muscle hypertrophy during extreme loading (912, 913). Ribosome biogenesis, rather than enhanced translational efficiency following bouts of overload, has been posited to be just as, if not more, critical in promoting muscle hypertrophy (5, 17). Researchers have exchanged viewpoints regarding whether edema is a significant contributor to the early stages of muscle hypertrophy (774, 914, 915), and the evidence is mixed concerning whether myofiber length increases appreciably contribute to hypertrophy with conventional resistance training (913, 916). Recent preliminary data in humans indicate that individuals may exhibit different morphological adaptations to the same resistance training program (917): specifically, some individuals may show tissue-level hypertrophy predominantly through fascicle length changes, whereas others may show tissue-level hypertrophy predominantly through fCSA increases. One of the more provocative questions related to myofiber hypertrophy was posed in 1982 by J. D. MacDougall and colleagues who pondered (56) whether
“…skeletal muscle fibers possess an unlimited capacity for protein synthesis and enlargement, or is there a maximal or optimal size which can be attained?”
Despite the remarkable discoveries that have been made during the past four decades, this fundamental question remains to be answered.
The present authors agree that skeletal muscle hypertrophy in response to mechanical overload generally involves cross-sectional (or radial) growth at the tissue and myofiber levels and that this coincides with a proportional expansion of the extracellular matrix. Moreover, although less resolved, limited literature discussed here (55, 99) [and in other recent reviews (1, 155)] supports the idea that myofibril and mitochondrial content mostly scale with myofiber hypertrophy induced by resistance training in humans. However, there are limited data or knowledge gaps that require further investigation into how different modes of mechanical overload affect 1) longitudinal tissue and myofiber growth, especially since most investigations examine radial growth; 2) type I versus type II myofiber hypertrophy, which seems to be load dependent in humans albeit not well delineated (814); 3) alterations in the size and number of myofibrils; 4) the three-dimensional properties of myofibrils, the mitochondrial and sarcoplasmic reticula, and the cytoskeletal network in type I and II myofibers; and 5) the time courses of 1–4 listed here. Additionally, a central tenet of this review is that several mechanisms are required for mechanical overload-induced skeletal muscle hypertrophy as shown in TABLE 1, and much remains to be learned in these areas as well.
Table 1.
Summary of mechanisms discussed in this review
| Mechanism | Responses to Mechanical Overload | Knowledge of Current Role(s) | Knowledge Gaps |
|---|---|---|---|
| mTORC1 signaling | ↑↑↑ | mTORC1 signaling is critically involved with skeletal muscle hypertrophy through increased translation initiation and/or elongation. | Further interrogation of the upstream activating signals during mechanical overload |
| mTORC1-independent signaling | ↑↑↑ | MAPK signaling and other mTORC1-independent signals are transiently activated after a bout of resistance exercise to presumably affect aspects of transcription and translation. | Further elucidating the role MAPK signaling and other mTORC1-independent signals (e.g., YAP and TRIM28 phosphorylation) have in promoting skeletal muscle hypertrophy |
| Ribosome biogenesis | ↑↑↑ | Increased translational capacity | Determining whether ribosome specialization occurs with overload and, if so, determining whether this is a critical aspect of hypertrophy |
| Satellite cells | ↑↑↑ | Myonuclear accretion via fusion, muscle repair, and nonfusion roles | In humans, validating preliminary animal findings suggesting that satellite cells coordinate extracellular matrix adaptations during overload; also examining whether hypertrophy can proceed in the absence of satellite cell-mediated myonuclear accretion in humans with certain diseases where satellite cell counts are reduced (e.g., MYMK mutations); finally, determining how satellite cell fusion alters molecular processes in myofibers (single-cell studies) or myofiber morphology |
| Genetic variants | Inherently present; no changes to overload | Various single-candidate polymorphism studies show small hypertrophic advantages with certain genotypes. | Performing deep sequencing efforts to identify novel variants and adopting statistical approaches to examine the combinatorial effects of multiple variants |
| Epigenetic alterations | ↑↑↑ and ↓↓↓ | Methylation changes occur across hundreds of genes in the nuclear genome, and preliminary evidence suggests demethylation of mitochondrial genome with resistance training in humans. | Determining whether gene-specific methylation responses to overload are needed for hypertrophy to occur; further determining whether prolonged DNA demethylation during periods of mechanical overload confers more robust skeletal muscle hypertrophy |
| Muscle proteolysis | ↑↑↑ early into training, but response subsides with increased training status. | Potentially needed for removing damaged proteins and organelles after initial stages of resistance training | Determining which proteolytic system(s) is primarily responsible for adaptive responses early (i.e., weeks) and later (i.e., months to years) into training; additionally, determining whether these proteolytic systems are required for muscle hypertrophy to occur in response to loading paradigms and/or whether synchronization between synthesis and proteolysis directs the degree of hypertrophy |
| Myostatin markers | ↓↓↓ | Numerous lines of evidence suggest that resistance training acutely and transiently decreases muscle MSTN mRNA levels. | Elucidating how MSTN pathway signaling (e.g., SMAD2/3 phosphorylation and the mRNA expression of downstream targets) is transiently affected during the initial and later stages of overload and whether these events are critically involved in the hypertrophic response |
| Extracellular matrix remolding | ↑↑↑ | Markers of extracellular matrix remodeling are altered during periods of resistance training, but much of this work has been confined to mRNAs. | Broadening the scope of extracellular matrix remodeling markers during resistance training studies to determine whether remodeling is required or merely coincides with skeletal muscle hypertrophy |
| Angiogenesis | ↑↑↑ | Preliminary evidence suggests that capillary number per fiber prior to resistance training is associated with hypertrophic response to training. | Determining whether the magnitude of angiogenesis induced by resistance training (and/or enhanced microvessel function) enhances muscle hypertrophy |
| Muscle microRNA expression | ↑↑↑ and ↓↓↓ | Genes involved with IGF1/PI3K/AKT/mTOR signaling are directly and/or indirectly regulated by various miRNAs that are altered in response to overload. | Moving beyond microRNA-omics-based studies in humans to show a core set of microRNAs involved with or needed for skeletal muscle hypertrophy |
| Testosterone signaling | ??? | Transient postexercise responses in circulating testosterone concentrations do not correlate with intracellular anabolic signaling events (e.g., mTORC1 signaling or muscle protein synthesis) and hypertrophy. However, muscle hormone receptor protein content modestly correlates with anabolic outcomes in some studies. | Determining whether androgen DNA binding is altered during periods of overload and which of the identified hormone receptor-affected genes are involved with skeletal muscle hypertrophy |
| Inflammation through prostaglandin signaling | ↑↑↑ | Coincides with robust elevations in protein synthesis and satellite cell proliferation during in the initial phases of resistance training. | Determining whether certain aspects of inflammation (e.g., EP and FP receptor signaling) are needed for, or merely coincide with, skeletal muscle hypertrophy |
| β-Adrenergic signaling through endogenous catecholamines | ??? | The administration of β-adrenergic receptor agonists promotes skeletal muscle hypertrophy. | Determining whether intrinsic β-adrenergic receptor signaling via endogenous catecholamines, in part, promotes skeletal muscle hypertrophy during periods of mechanical overload |
| Angiotensin II signaling | ??? | Preliminary animal evidence suggested angiotensin II signaling is involved with overload-induced skeletal muscle hypertrophy. However, follow-up animal studies suggest angiotensin II signaling may blunt hypertrophic responses, and human data in this area are mixed. | Determining whether intrinsic angiotensin II signaling blunts, enhances, or does not affect hypertrophic outcomes in humans |
| Mitochondrial biogenesis | ↑, ↓, or ↔ | The increase in mitochondrial volume density may precede or concomitantly occur with muscle hypertrophy. | Demonstrating whether an expansion of the mitochondria is required for myofiber hypertrophy, or whether mitochondrial biogenesis, mitochondrial expansion, and myofiber hypertrophy merely coincide with one another |
| Other bioenergetic adaptations | ↑↑↑ | Resistance training can promote differential metabolic adaptations in skeletal muscle. | Determining whether metabolic adaptations (e.g., enhanced glycolytic flux) provide skeletal muscle with substrates needed for cell growth |
| Muscle circadian transcriptome | ??? | The oscillation of core clock genes in muscle transcriptionally regulates hundreds of genes related to metabolism, protein turnover, ribosome biogenesis, and other processes. | Determining whether the muscle circadian transcriptome is altered (or disrupted) during periods of overload; if so, is this an involved mechanism with molecular adaptations (e.g., ribosome biogenesis or the altered expression of metabolic genes)? |
| Microtubule networks, and myonuclear and RNA trafficking | ??? | Studies suggest that 1) microtubule-dependent RNA transport from myonuclei to ribosomes is essential for the translation process to occur and 2) myonuclear trafficking to focal injury sites occurs through microtubules. | Determining whether microtubule network expansion scales with hypertrophy and if this process is needed to support RNA and myonuclear trafficking |
| Microbiome alterations | Minimal changes in bacterial species diversity | Preliminary evidence suggests that resistance training may not drastically alter diversity metrics in the gut microbiome. | Determining whether metabolites produced by the gut microbiome change in response to training; if so, do they act as signals to affect anabolic signaling pathways? |
↑↑↑ or ↓↓↓, several independent laboratories have shown increases or decreases in markers associated with mechanism; ↑, ↓, or ↔, less evidence supports the mechanism being involved in mechanical overload-induced skeletal muscle hypertrophy; ???, involvement in mechanical overload-induced skeletal muscle hypertrophy has not been well elucidated relative to other discussed mechanisms. IGF1, insulin-like growth factor 1; PI3K, phosphatidylinositol 3-kinase; MSTN, myostatin; mTOR, mammalian/mechanistic target of rapamycin; mTORC1, mammalian/mechanistic target of rapamycin complex 1.
7. CONCLUSIONS
Skeletal muscle hypertrophy research has rapidly evolved since the landmark report by Morpurgo in 1897. Pioneering discoveries in the field have motivated others to adopt innovative methodologies and drive the research boundaries in meaningful ways. Given the rapid advancements in molecular-based research techniques, investigations in upcoming years will continue to confirm or refute which of the discussed mechanisms are obligatory for (rather than coinciding with) load-induced skeletal muscle hypertrophy. More importantly, these efforts will likely unveil novel mechanisms that continue to reshape our thinking in this area of muscle biology.
GRANTS
T.A.H. is supported by the National Institutes of Health (NIH) (R01AR074932). S.M.P. is supported by the Canada Research Chairs Programme, National Science and Engineering Research Council (NSERC) of Canada, and the Canadian Institutes for Health Research (CIHR). S.M.P. also reports grants or research contracts, current or recent, from the US National Dairy Council, Dairy Farmers of Canada, Roquette Freres, Nestle Health Sciences, Myos, NSERC, and NIH during the execution of studies; and personal fees from the US National Dairy Council and nonfinancial support from Enhanced Recovery outside the submitted work. G.A.N. is supported by NIH (AR-078430). M.D.B. is supported by NIH (R01AR072735). P.T.R. is supported by Miami University College of Education, Health and Society Summer Research Funding. R.O. is supported by Japan Society for the Promotion of Science (no. 22H03465). C.A.L. is supported by the São Paulo Research Foundation (no. 2020/13613-4) and the National Council for Scientific and Technological Development (no. 311387/2021-7). Article processing charges were provided in full by Auburn University’s School of Kinesiology.
DISCLOSURES
M.D.R. has received funding in the form of contracts, gifts, and grants from industry sources, Auburn University (Intramural Grants Program), and the Peanut Institute (commodities) for work in certain areas discussed in this article. S.M.P. has patent (Canadian) 3052324 assigned to Exerkine and patent (US) 20200230197 pending to Exerkine but reports no financial gains from any patent or related work. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
M.D.R. conceived and designed research; M.D.R., J.J.M., T.A.H., G.A.N., and A.N.K. prepared figures; M.D.R., J.J.M., T.A.H., S.M.P., A.L.M., G.A.N., M.D.B., A.N.K., P.T.R., R.O., C.A.L., C.U., and F.W.B. drafted manuscript; M.D.R., J.J.M., T.A.H., S.M.P., A.L.M., G.A.N., M.D.B., P.T.R., R.O., C.A.L., C.U., F.W.B, and K.A.E., edited and revised manuscript; M.D.R., J.J.M., T.A.H., S.M.P., A.L.M., G.A.N., M.D.B., A.N.K., P.T.R., R.O., C.A.L., C.U., F.W.B, and K.A.E. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors extend gratitude to Dr. Sue Bodine (University of Iowa) for critiques and assistance in the publication process.
REFERENCES
- 1. Jorgenson KW, Phillips SM, Hornberger TA. Identifying the structural adaptations that drive the mechanical load-induced growth of skeletal muscle: a scoping review. Cells 9: 1658, 2020. doi: 10.3390/cells9071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Attwaters M, Hughes SM. Cellular and molecular pathways controlling muscle size in response to exercise. FEBS J 289: 1428–1456, 2022. doi: 10.1111/febs.15820. [DOI] [PubMed] [Google Scholar]
- 3. Bamman MM, Roberts BM, Adams GR. Molecular regulation of exercise-induced muscle fiber hypertrophy. Cold Spring Harb Perspect Med 8: a029751, 2018. doi: 10.1101/cshperspect.a029751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wackerhage H, Schoenfeld BJ, Hamilton DL, Lehti M, Hulmi JJ. Stimuli and sensors that initiate skeletal muscle hypertrophy following resistance exercise. J Appl Physiol (1985) 126: 30–43, 2019. doi: 10.1152/japplphysiol.00685.2018. [DOI] [PubMed] [Google Scholar]
- 5. Brook MS, Wilkinson DJ, Smith K, Atherton PJ. It’s not just about protein turnover: the role of ribosomal biogenesis and satellite cells in the regulation of skeletal muscle hypertrophy. Eur J Sport Sci 19: 952–963, 2019. doi: 10.1080/17461391.2019.1569726. [DOI] [PubMed] [Google Scholar]
- 6. Fukada SI, Ito N. Regulation of muscle hypertrophy: involvement of the Akt-independent pathway and satellite cells in muscle hypertrophy. Exp Cell Res 409: 112907, 2021. doi: 10.1016/j.yexcr.2021.112907. [DOI] [PubMed] [Google Scholar]
- 7. Lavin KM, Coen PM, Baptista LC, Bell MB, Drummer D, Harper SA, Lixandrão ME, McAdam JS, O’Bryan SM, Ramos S, Roberts LM, Vega RB, Goodpaster BH, Bamman MM, Buford TW. State of the knowledge on molecular adaptations to exercise in humans: historical perspectives and future directions. Compr Physiol 12: 3193–3279, 2022. doi: 10.1007/978-3-031-05164-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mukund K, Subramaniam S. Skeletal muscle: a review of molecular structure and function, in health and disease. Wiley Interdiscip Rev Syst Biol Med 12: e1462, 2020. doi: 10.1002/wsbm.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sartori R, Romanello V, Sandri M. Mechanisms of muscle atrophy and hypertrophy: implications in health and disease. Nat Commun 12: 330, 2021. doi: 10.1038/s41467-020-20123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schiaffino S, Reggiani C, Akimoto T, Blaauw B. Molecular mechanisms of skeletal muscle hypertrophy. J Neuromuscul Dis 8: 169–183, 2021. doi: 10.3233/JND-200568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Solsona R, Sanchez AM. Exercise and ribosome biogenesis in skeletal muscle hypertrophy: impact of genetic and epigenetic factors. J Physiol 599: 3803–3805, 2021. doi: 10.1113/JP281984. [DOI] [PubMed] [Google Scholar]
- 12. Vainshtein A, Sandri M. Signaling pathways that control muscle mass. Int J Mol Sci 21, 2020. doi: 10.3390/ijms21134759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodman CA. Role of mTORC1 in mechanically induced increases in translation and skeletal muscle mass. J Appl Physiol (1985) 127: 581–590, 2019. doi: 10.1152/japplphysiol.01011.2018. [DOI] [PubMed] [Google Scholar]
- 14. Boppart MD, Mahmassani ZS. Integrin signaling: linking mechanical stimulation to skeletal muscle hypertrophy. Am J Physiol Cell Physiol 317: C629–C641, 2019. doi: 10.1152/ajpcell.00009.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirby TJ. Mechanosensitive pathways controlling translation regulatory processes in skeletal muscle and implications for adaptation. J Appl Physiol (1985) 127: 608–618, 2019. doi: 10.1152/japplphysiol.01031.2018. [DOI] [PubMed] [Google Scholar]
- 16. Olsen LA, Nicoll JX, Fry AC. The skeletal muscle fiber: a mechanically sensitive cell. Eur J Appl Physiol 119: 333–349, 2019. doi: 10.1007/s00421-018-04061-x. [DOI] [PubMed] [Google Scholar]
- 17. Figueiredo VC. Revisiting the roles of protein synthesis during skeletal muscle hypertrophy induced by exercise. Am J Physiol Regul Integr Comp Physiol 317: R709–R718, 2019. doi: 10.1152/ajpregu.00162.2019. [DOI] [PubMed] [Google Scholar]
- 18. Chaillou T. Ribosome specialization and its potential role in the control of protein translation and skeletal muscle size. J Appl Physiol (1985) 127: 599–607, 2019. doi: 10.1152/japplphysiol.00946.2018. [DOI] [PubMed] [Google Scholar]
- 19. Mesquita PH, Vann CG, Phillips SM, McKendry J, Young KC, Kavazis AN, Roberts MD. Skeletal muscle ribosome and mitochondrial biogenesis in response to different exercise training modalities. Front Physiol 12: 725866, 2021. doi: 10.3389/fphys.2021.725866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. von Walden F. Ribosome biogenesis in skeletal muscle: coordination of transcription and translation. J Appl Physiol (1985) 127: 591–598, 2019. doi: 10.1152/japplphysiol.00963.2018. [DOI] [PubMed] [Google Scholar]
- 21. Francaux M, Deldicque L. Exercise and the control of muscle mass in human. Pflugers Arch 471: 397–411, 2019. doi: 10.1007/s00424-018-2217-x. [DOI] [PubMed] [Google Scholar]
- 22. Joanisse S, Lim C, McKendry J, Mcleod JC, Stokes T, Phillips SM. Recent advances in understanding resistance exercise training-induced skeletal muscle hypertrophy in humans. F1000Res 9: F1000 Faculty Rev-141, 2020. doi: 10.12688/f1000research.21588.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conceição MS, Vechin FC, Lixandrão M, Damas F, Libardi CA, Tricoli V, Roschel H, Camera D, Ugrinowitsch C. Muscle fiber hypertrophy and myonuclei addition: a systematic review and meta-analysis. Med Sci Sports Exerc 50: 1385–1393, 2018. doi: 10.1249/MSS.0000000000001593. [DOI] [PubMed] [Google Scholar]
- 24. Murach KA, Fry CS, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Fusion and beyond: satellite cell contributions to loading-induced skeletal muscle adaptation. FASEB J 35: e21893, 2021. doi: 10.1096/fj.202101096R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murach KA, Englund DA, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. myonuclear domain flexibility challenges rigid assumptions on satellite cell contribution to skeletal muscle fiber hypertrophy. Front Physiol 9: 635, 2018. doi: 10.3389/fphys.2018.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Damas F, Libardi CA, Ugrinowitsch C. The development of skeletal muscle hypertrophy through resistance training: the role of muscle damage and muscle protein synthesis. Eur J Appl Physiol 118: 485–500, 2018. doi: 10.1007/s00421-017-3792-9. [DOI] [PubMed] [Google Scholar]
- 27. Roberts MD, Haun CT, Mobley CB, Mumford PW, Romero MA, Roberson PA, Vann CG, McCarthy JJ. Physiological differences between low versus high skeletal muscle hypertrophic responders to resistance exercise training: current perspectives and future research directions. Front Physiol 9: 834, 2018. doi: 10.3389/fphys.2018.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petersen AC, Fyfe JJ. Post-exercise cold water immersion effects on physiological adaptations to resistance training and the underlying mechanisms in skeletal muscle: a narrative review. Front Sports Act Living 3: 660291, 2021. doi: 10.3389/fspor.2021.660291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alix-Fages C, Del Vecchio A, Baz-Valle E, Santos-Concejero J, Balsalobre-Fernandez C. The role of the neural stimulus in regulating skeletal muscle hypertrophy. Eur J Appl Physiol 122: 1111–1128, 2022. doi: 10.1007/s00421-022-04906-6. [DOI] [PubMed] [Google Scholar]
- 30. Lavin KM, Roberts BM, Fry CS, Moro T, Rasmussen BB, Bamman MM. The importance of resistance exercise training to combat neuromuscular aging. Physiology (Bethesda) 34: 112–122, 2019. doi: 10.1152/physiol.00044.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lundberg TR, Howatson G. Analgesic and anti-inflammatory drugs in sports: Implications for exercise performance and training adaptations. Scand J Med Sci Sports 28: 2252–2262, 2018. doi: 10.1111/sms.13275. [DOI] [PubMed] [Google Scholar]
- 32. Gharahdaghi N, Phillips BE, Szewczyk NJ, Smith K, Wilkinson DJ, Atherton PJ. Links between testosterone, oestrogen, and the growth hormone/insulin-like growth factor axis and resistance exercise muscle adaptations. Front Physiol 11: 621226, 2020. doi: 10.3389/fphys.2020.621226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cornish SM, Bugera EM, Duhamel TA, Peeler JD, Anderson JE. A focused review of myokines as a potential contributor to muscle hypertrophy from resistance-based exercise. Eur J Appl Physiol 120: 941–959, 2020. doi: 10.1007/s00421-020-04337-1. [DOI] [PubMed] [Google Scholar]
- 34. Fukada SI. The roles of muscle stem cells in muscle injury, atrophy and hypertrophy. J Biochem 163: 353–358, 2018. doi: 10.1093/jb/mvy019. [DOI] [PubMed] [Google Scholar]
- 35. Shamim B, Hawley JA, Camera DM. Protein availability and satellite cell dynamics in skeletal muscle. Sports Med 48: 1329–1343, 2018. doi: 10.1007/s40279-018-0883-7. [DOI] [PubMed] [Google Scholar]
- 36. Ogasawara R, Jensen TE, Goodman CA, Hornberger TA. Resistance exercise-induced hypertrophy: a potential role for rapamycin-insensitive mTOR. Exerc Sport Sci Rev 47: 188–194, 2019. doi: 10.1249/JES.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Widmann M, Nieß AM, Munz B. Physical exercise and epigenetic modifications in skeletal muscle. Sports Med 49: 509–523, 2019. doi: 10.1007/s40279-019-01070-4. [DOI] [PubMed] [Google Scholar]
- 38. Jacques M, Hiam D, Craig J, Barrès R, Eynon N, Voisin S. Epigenetic changes in healthy human skeletal muscle following exercise—a systematic review. Epigenetics 14: 633–648, 2019. doi: 10.1080/15592294.2019.1614416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seaborne RA, Sharples AP. The interplay between exercise metabolism, epigenetics, and skeletal muscle remodeling. Exerc Sport Sci Rev 48: 188–200, 2020. doi: 10.1249/JES.0000000000000227. [DOI] [PubMed] [Google Scholar]
- 40. Graham ZA, Lavin KM, O’Bryan SM, Thalacker-Mercer AE, Buford TW, Ford KM, Broderick TJ, Bamman MM. Mechanisms of exercise as a preventative measure to muscle wasting. Am J Physiol Cell Physiol 321: C40–C57, 2021. doi: 10.1152/ajpcell.00056.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abou Sawan S, Mazzulla M, Moore DR, Hodson N. More than just a garbage can: emerging roles of the lysosome as an anabolic organelle in skeletal muscle. Am J Physiol Cell Physiol 319: C561–C568, 2020. doi: 10.1152/ajpcell.00241.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huey KA. Potential roles of vascular endothelial growth factor during skeletal muscle hypertrophy. Exerc Sport Sci Rev 46: 195–202, 2018. doi: 10.1249/JES.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 43. Kraemer WJ, Ratamess NA, Hymer WC, Nindl BC, Fragala MS. Growth hormone(s), testosterone, insulin-like growth factors, and cortisol: roles and integration for cellular development and growth with exercise. Front Endocrinol (Lausanne) 11: 33, 2020. doi: 10.3389/fendo.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Domańska-Senderowska D, Laguette MJ, Jegier A, Cięszczyk P, September AV, Brzeziańska-Lasota E. MicroRNA profile and adaptive response to exercise training: a review. Int J Sports Med 40: 227–235, 2019. doi: 10.1055/a-0824-4813. [DOI] [PubMed] [Google Scholar]
- 45. Parry HA, Roberts MD, Kavazis AN. Human skeletal muscle mitochondrial adaptations following resistance exercise training. Int J Sports Med 41: 349–359, 2020. doi: 10.1055/a-1121-7851. [DOI] [PubMed] [Google Scholar]
- 46. Lustgarten MS. The role of the gut microbiome on skeletal muscle mass and physical function: 2019 update. Front Physiol 10: 1435, 2019. doi: 10.3389/fphys.2019.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolff CA, Esser KA. Exercise timing and circadian rhythms. Curr Opin Physiol 10: 64–69, 2019. doi: 10.1016/j.cophys.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brightwell CR, Latham CM, Thomas NT, Keeble AR, Murach KA, Fry CS. A glitch in the matrix: the pivotal role for extracellular matrix remodeling during muscle hypertrophy. Am J Physiol Cell Physiol 323: C763–C771, 2022. doi: 10.1152/ajpcell.00200.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hostrup M, Onslev J. The beta2-adrenergic receptor—a re-emerging target to combat obesity and induce leanness? J Physiol 600: 1209–1227, 2022. doi: 10.1113/JP281819. [DOI] [PubMed] [Google Scholar]
- 50. Berdeaux R, Hutchins C. Anabolic and pro-metabolic functions of CREB-CRTC in skeletal muscle: advantages and obstacles for type 2 diabetes and cancer cachexia. Front Endocrinol (Lausanne) 10: 535, 2019. doi: 10.3389/fendo.2019.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bagley JR, Denes LT, McCarthy JJ, Wang ET, Murach KA. The myonuclear domain in adult skeletal muscle fibres: past, present and future. J Physiol 601: 723–741, 2023. doi: 10.1113/JP283658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith JA, Murach KA, Dyar KA, Zierath JR. Exercise metabolism and adaptation in skeletal muscle. Nat Rev Mol Cell Biol. In Press. doi: 10.1038/s41580-023-00606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roman W, Gomes ER. Nuclear positioning in skeletal muscle. Semin Cell Dev Biol 82: 51–56, 2018. doi: 10.1016/j.semcdb.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 54. Allen DL, Roy RR, Edgerton VR. Myonuclear domains in muscle adaptation and disease. Muscle Nerve 22: 1350–1360, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 55. Ruple BA, Godwin JS, Mesquita PH, Osburn SC, Sexton CL, Smith MA, Ogletree JC, Goodlett MD, Edison JL, Ferrando AA, Fruge AD, Kavazis AN, Young KC, Roberts MD. Myofibril and mitochondrial area changes in type I and II fibers following 10 weeks of resistance training in previously untrained men. Front Physiol 12: 728683, 2021. doi: 10.3389/fphys.2021.728683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. MacDougall JD, Sale DG, Elder GC, Sutton JR. Muscle ultrastructural characteristics of elite powerlifters and bodybuilders. Eur J Appl Physiol Occup Physiol 48: 117–126, 1982. doi: 10.1007/BF00421171. [DOI] [PubMed] [Google Scholar]
- 57. Alway SE, MacDougall JD, Sale DG, Sutton JR, McComas AJ. Functional and structural adaptations in skeletal muscle of trained athletes. J Appl Physiol (1985) 64: 1114–1120, 1988. doi: 10.1152/jappl.1988.64.3.1114. [DOI] [PubMed] [Google Scholar]
- 58. Henderson CA, Gomez CG, Novak SM, Mi-Mi L, Gregorio CC. Overview of the muscle cytoskeleton. Compr Physiol 7: 891–944, 2017. doi: 10.1002/cphy.c160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mado K, Chekulayev V, Shevchuk I, Puurand M, Tepp K, Kaambre T. On the role of tubulin, plectin, desmin, and vimentin in the regulation of mitochondrial energy fluxes in muscle cells. Am J Physiol Cell Physiol 316: C657–C667, 2019. doi: 10.1152/ajpcell.00303.2018. [DOI] [PubMed] [Google Scholar]
- 60. Willingham TB, Kim Y, Lindberg E, Bleck CK, Glancy B. The unified myofibrillar matrix for force generation in muscle. Nat Commun 11: 3722, 2020. doi: 10.1038/s41467-020-17579-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Glancy B, Balaban RS. Energy metabolism design of the striated muscle cell. Physiol Rev 101: 1561–1607, 2021. doi: 10.1152/physrev.00040.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Glancy B, Hartnell LM, Malide D, Yu ZX, Combs CA, Connelly PS, Subramaniam S, Balaban RS. Mitochondrial reticulum for cellular energy distribution in muscle. Nature 523: 617–620, 2015. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lapidos KA, Kakkar R, McNally EM. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res 94: 1023–1031, 2004. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 64. Sanes JR. The basement membrane/basal lamina of skeletal muscle. J Biol Chem 278: 12601–12604, 2003. doi: 10.1074/jbc.R200027200. [DOI] [PubMed] [Google Scholar]
- 65. Csapo R, Gumpenberger M, Wessner B. Skeletal muscle extracellular matrix—what do we know about its composition, regulation, and physiological roles? A narrative review. Front Physiol 11: 253, 2020. doi: 10.3389/fphys.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Englund DA, Zhang X, Aversa Z, LeBrasseur NK. Skeletal muscle aging, cellular senescence, and senotherapeutics: current knowledge and future directions. Mech Ageing Dev 200: 111595, 2021. doi: 10.1016/j.mad.2021.111595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. MacDougall JD, Sale DG, Alway SE, Sutton JR. Muscle fiber number in biceps brachii in bodybuilders and control subjects. J Appl Physiol Respir Environ Exerc Physiol 57: 1399–1403, 1984. doi: 10.1152/jappl.1984.57.5.1399. [DOI] [PubMed] [Google Scholar]
- 68. Snijders T, Holwerda AM, van Loon LJ, Verdijk LB. Myonuclear content and domain size in small versus larger muscle fibres in response to 12 weeks of resistance exercise training in older adults. Acta Physiol (Oxf) 231: e13599, 2021. doi: 10.1111/apha.13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang S, Stoops E, Cp U, Markus B, Reuveny A, Ordan E, Volk T. Mechanotransduction via the LINC complex regulates DNA replication in myonuclei. J Cell Biol 217: 2005–2018, 2018. doi: 10.1083/jcb.201708137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Petrella JK, Kim JS, Mayhew DL, Cross JM, Bamman MM. Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: a cluster analysis. J Appl Physiol (1985) 104: 1736–1742, 2008. doi: 10.1152/japplphysiol.01215.2007. [DOI] [PubMed] [Google Scholar]
- 71. Malm C, Sjödin TL, Sjöberg B, Lenkei R, Renström P, Lundberg IE, Ekblom B. Leukocytes, cytokines, growth factors and hormones in human skeletal muscle and blood after uphill or downhill running. J Physiol 556: 983–1000, 2004. doi: 10.1113/jphysiol.2003.056598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Walton RG, Kosmac K, Mula J, Fry CS, Peck BD, Groshong JS, Finlin BS, Zhu B, Kern PA, Peterson CA. Human skeletal muscle macrophages increase following cycle training and are associated with adaptations that may facilitate growth. Sci Rep 9: 969, 2019. doi: 10.1038/s41598-018-37187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Godwin JS, Sexton CL, Kontos NJ, Ruple BA, Willoughby DS, Young KC, Mobley CB, Roberts MD. Extracellular matrix content and remodeling markers do not differ in college-aged men classified as higher and lower responders to resistance training. J Appl Physiol (1985) 134: 731–741, 2023. doi: 10.1152/japplphysiol.00596.2022. [DOI] [PubMed] [Google Scholar]
- 74. Mackey AL, Magnan M, Chazaud B, Kjaer M. Human skeletal muscle fibroblasts stimulate in vitro myogenesis and in vivo muscle regeneration. J Physiol 595: 5115–5127, 2017. doi: 10.1113/JP273997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. De Lisio M, Farup J, Sukiennik RA, Clevenger N, Nallabelli J, Nelson B, Ryan K, Rahbek SK, de Paoli F, Vissing K, Boppart MD. The acute response of pericytes to muscle-damaging eccentric contraction and protein supplementation in human skeletal muscle. J Appl Physiol (1985) 119: 900–907, 2015. doi: 10.1152/japplphysiol.01112.2014. [DOI] [PubMed] [Google Scholar]
- 76. Nielsen JL, Frandsen U, Jensen KY, Prokhorova TA, Dalgaard LB, Bech RD, Nygaard T, Suetta C, Aagaard P. Skeletal muscle microvascular changes in response to short-term blood flow restricted training-exercise-induced adaptations and signs of perivascular stress. Front Physiol 11: 556, 2020. doi: 10.3389/fphys.2020.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Thomas AC, Brown A, Hatt AA, Manta K, Costa-Parke A, Kamal M, Joanisse S, McGlory C, Phillips SM, Kumbhare D, Parise G. Short-term aerobic conditioning prior to resistance training augments muscle hypertrophy and satellite cell content in healthy young men and women. FASEB J 36: e22500, 2022. doi: 10.1096/fj.202200398RR. [DOI] [PubMed] [Google Scholar]
- 78. Fielding RA, Manfredi TJ, Ding W, Fiatarone MA, Evans WJ, Cannon JG. Acute phase response in exercise. III. Neutrophil and IL-1 beta accumulation in skeletal muscle. Am J Physiol Regul Integr Comp Physiol 265: R166–R172, 1993. doi: 10.1152/ajpregu.1993.265.1.R166. [DOI] [PubMed] [Google Scholar]
- 79. Dos Santos M, Backer S, Saintpierre B, Izac B, Andrieu M, Letourneur F, Relaix F, Sotiropoulos A, Maire P. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat Commun 11: 5102, 2020. doi: 10.1038/s41467-020-18789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roberts MD, Mobley CB, Vann CG, Haun CT, Schoenfeld BJ, Young KC, Kavazis AN. Synergist ablation-induced hypertrophy occurs more rapidly in the plantaris than soleus muscle in rats due to different molecular mechanisms. Am J Physiol Regul Integr Comp Physiol 318: R360–R368, 2020. doi: 10.1152/ajpregu.00304.2019. [DOI] [PubMed] [Google Scholar]
- 81. Liu JX, Höglund AS, Karlsson P, Lindblad J, Qaisar R, Aare S, Bengtsson E, Larsson L. Myonuclear domain size and myosin isoform expression in muscle fibres from mammals representing a 100,000-fold difference in body size. Exp Physiol 94: 117–129, 2009. doi: 10.1113/expphysiol.2008.043877. [DOI] [PubMed] [Google Scholar]
- 82. Reidy PT, McKenzie AI, Mahmassani ZS, Petrocelli JJ, Nelson DB, Lindsay CC, Gardner JE, Morrow VR, Keefe AC, Huffaker TB, Stoddard GJ, Kardon G, O’Connell RM, Drummond MJ. Aging impairs mouse skeletal muscle macrophage polarization and muscle-specific abundance during recovery from disuse. Am J Physiol Endocrinol Metab 317: E85–E98, 2019. doi: 10.1152/ajpendo.00422.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Haun CT, Vann CG, Roberts BM, Vigotsky AD, Schoenfeld BJ, Roberts MD. A critical evaluation of the biological construct skeletal muscle hypertrophy: size matters but so does the measurement. Front Physiol 10: 247, 2019. doi: 10.3389/fphys.2019.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int 96: 183–195, 2015. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 85. Zhang J, Han X, Lin Y. Dissecting the regulation and function of ATP at the single-cell level. PLoS Biol 16: e3000095, 2018. doi: 10.1371/journal.pbio.3000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev 91: 1447–1531, 2011. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 87. Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gundersen K, Lømo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil 10: 197–205, 1989. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- 88. Andersen JL, Klitgaard H, Saltin B. Myosin heavy chain isoforms in single fibres from m. vastus lateralis of sprinters: influence of training. Acta Physiol Scand 151: 135–142, 1994. doi: 10.1111/j.1748-1716.1994.tb09730.x. [DOI] [PubMed] [Google Scholar]
- 89. Bagley JR, McLeland KA, Arevalo JA, Brown LE, Coburn JW, Galpin AJ. Skeletal muscle fatigability and myosin heavy chain fiber type in resistance trained men. J Strength Cond Res 31: 602–607, 2017. doi: 10.1519/JSC.0000000000001759. [DOI] [PubMed] [Google Scholar]
- 90. Machek SB, Hwang PS, Cardaci TD, Wilburn DT, Bagley JR, Blake DT, Galpin AJ, Willoughby DS. Myosin heavy chain composition, creatine analogues, and the relationship of muscle creatine content and fast-twitch proportion to Wilks coefficient in powerlifters. J Strength Cond Res 34: 3022–3030, 2020. doi: 10.1519/JSC.0000000000003804. [DOI] [PubMed] [Google Scholar]
- 91. Williamson DL, Gallagher PM, Carroll CC, Raue U, Trappe SW. Reduction in hybrid single muscle fiber proportions with resistance training in humans. J Appl Physiol (1985) 91: 1955–1961, 2001. doi: 10.1152/jappl.2001.91.5.1955. [DOI] [PubMed] [Google Scholar]
- 92. Murach KA, Dungan CM, Kosmac K, Voigt TB, Tourville TW, Miller MS, Bamman MM, Peterson CA, Toth MJ. Fiber typing human skeletal muscle with fluorescent immunohistochemistry. J Appl Physiol (1985) 127: 1632–1639, 2019. doi: 10.1152/japplphysiol.00624.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bloemberg D, Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One 7: e35273, 2012. doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Williamson DL, Godard MP, Porter DA, Costill DL, Trappe SW. Progressive resistance training reduces myosin heavy chain coexpression in single muscle fibers from older men. J Appl Physiol (1985) 88: 627–633, 2000. doi: 10.1152/jappl.2000.88.2.627. [DOI] [PubMed] [Google Scholar]
- 95. Parcell AC, Sawyer RD, Craig Poole R. Single muscle fiber myosin heavy chain distribution in elite female track athletes. Med Sci Sports Exerc 35: 434–438, 2003. doi: 10.1249/01.MSS.0000053735.99344.C0. [DOI] [PubMed] [Google Scholar]
- 96. Plotkin DL, Roberts MD, Haun CT, Schoenfeld BJ. Muscle fiber type transitions with exercise training: shifting perspectives. Sports (Basel) 9: 127, 2021. doi: 10.3390/sports9090127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Andersen JL, Gruschy-Knudsen T. Rapid switch-off of the human myosin heavy chain IIX gene after heavy load muscle contractions is sustained for at least four days. Scand J Med Sci Sports 28: 371–380, 2018. doi: 10.1111/sms.12914. [DOI] [PubMed] [Google Scholar]
- 98. Schiaffino S, Hanzlíková V, Pierobon S. Relations between structure and function in rat skeletal muscle fibers. J Cell Biol 47: 107–119, 1970. doi: 10.1083/jcb.47.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang N, Hikida RS, Staron RS, Simoneau JA. Muscle fiber types of women after resistance training–quantitative ultrastructure and enzyme activity. Pflugers Arch 424: 494–502, 1993. doi: 10.1007/BF00374913. [DOI] [PubMed] [Google Scholar]
- 100. Murgia M, Nogara L, Baraldo M, Reggiani C, Mann M, Schiaffino S. Protein profile of fiber types in human skeletal muscle: a single-fiber proteomics study. Skelet Muscle 11: 24, 2021. doi: 10.1186/s13395-021-00279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Atha J. Strengthening muscle. Exerc Sport Sci Rev 9: 1–73, 1981. [PubMed] [Google Scholar]
- 102. Kraemer WJ, Ratamess NA, Flanagan SD, Shurley JP, Todd JS, Todd TC. Understanding the science of resistance training: an evolutionary perspective. Sports Med 47: 2415–2435, 2017. doi: 10.1007/s40279-017-0779-y. [DOI] [PubMed] [Google Scholar]
- 103. Buck J. Louis Cyr and Charles Sampson: archetypes of vaudevillian strongmen. Iron Game History 5: 18–28, 1998. [Google Scholar]
- 104. Virchow R. Die Cellularpathologie in ihrer Begründung auf physiologische und pathologische Gewebelehre. Berlin: Hirschwald, 1858. [PubMed] [Google Scholar]
- 105. Morpurgo B. Ueber Activitäts-Hypertrophie der willkürlichen Muskeln. Arch Pathol Anat 150: 522–554, 1897. doi: 10.1007/BF01924319. [DOI] [Google Scholar]
- 106. Donaldson HH. Summary of data for the effects of exercise on the organ weights of the albino rat: comparison with similar data from the dog. Am J Anat 56: 57–70, 1935. doi: 10.1002/aja.1000560104. [DOI] [Google Scholar]
- 107. Hettinger T. Physiology of Strength. Springfield, IL: Thomas Books, 1961, p. 48. [Google Scholar]
- 108. Steinhaus AH. Chronic effects of exercise. Physiol Rev 13: 103–147, 1933. doi: 10.1152/physrev.1933.13.1.103. [DOI] [Google Scholar]
- 109. Shurley JP, Todd J, Todd T. Building the barbell athlete: Bob Hoffman, Joe Weider, and the promotion of strength training for sport, 1932–1969. In: Strength Coaching in America. Austin, TX: University of Texas Press, 2021. p. 39–66. [Google Scholar]
- 110. Hersh JH. Hypertrophy of the masseter muscle. Arch Otolaryngol (1925) 43: 593–596, 1946. doi: 10.1001/archotol.1946.00680050619005. [DOI] [PubMed] [Google Scholar]
- 111. Maxwell JH, Waggoner RW. Hypertrophy of the masseter muscles. Trans Am Laryngol Assoc 53: 193–204, 1952. [PubMed] [Google Scholar]
- 112. Neal RD. Benign hypertrophy of the masseter muscle. Bull Charlotte Meml Hosp 3: 53–55, 1948. [PubMed] [Google Scholar]
- 113. Bompa TO, Buzzichelli C. Periodization: Theory and Methodology of Training. Champaign, IL: Human Kinetics, 2018. [Google Scholar]
- 114. Selye H. Stress and the general adaptation syndrome. Br Med J 1: 1383–1392, 1950. doi: 10.1136/bmj.1.4667.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. DeLorme TL. Restoration of muscle power by heavy-resistance exercises. J Bone Joint Surg 27: 645–667, 1945. [Google Scholar]
- 116. Mead S. Intermittent treatment of poliomyelitis with progressive resistance exercise. J Am Med Assoc 144: 458–460, 1950. doi: 10.1001/jama.1950.02920060020006. [DOI] [PubMed] [Google Scholar]
- 117. Watkins AL. Practical applications of progressive resistance exercises. J Am Med Assoc 148: 443–446, 1952. doi: 10.1001/jama.1952.02930060025008. [DOI] [PubMed] [Google Scholar]
- 118. Delorme TL, Schwab RS, Watkins AL. The response of the quadriceps femoris to progressive-resistance exercises in poliomyelitic patients. J Bone Joint Surg Am 30A: 834–847, 1948. [PubMed] [Google Scholar]
- 119. Anderson EH. Heavy resistance, low repetition exercises in the restoration of function in the knee joint. Treat Serv Bull 2: 22–26, 1947. [PubMed] [Google Scholar]
- 120. Myslinski T. The Development of the Russian Conjugate Sequence System. 2003. EliteFTS com.
- 121. Briskey EJ, Bray RW, Hoekstra WG, Grummer RH, Phillips PH. The effect of various levels of exercise in altering the chemical and physical characteristics of certain pork ham muscles. J Anim Sci 18: 153–157, 1959. doi: 10.2527/jas1959.181153x. [DOI] [Google Scholar]
- 122. Skjervold H, Standal N, Bruflot R. Effect of one form of exercise on the body development in pigs. J Anim Sci 22: 458–462, 1963. doi: 10.2527/jas1963.222458x. [DOI] [Google Scholar]
- 123. Goldspink G. The combined effects of exercise and reduced food intake on skeletal muscle fibers. J Cell Comp Physiol 63: 209–216, 1964. doi: 10.1002/jcp.1030630211. [DOI] [PubMed] [Google Scholar]
- 124. Goldberg AL. Work-induced growth of skeletal muscle in normal and hypophysectomized rats. Am J Physiol 213: 1193–1198, 1967. doi: 10.1152/ajplegacy.1967.213.5.1193. [DOI] [PubMed] [Google Scholar]
- 125. James NT. Compensatory muscular hypertrophy in the extensor digitorum longus muscle of the mouse. J Anat 122: 121–131, 1976. [PMC free article] [PubMed] [Google Scholar]
- 126. Terena SM, Fernandes KP, Bussadori SK, Deana AM, Mesquita-Ferrari RA. Systematic review of the synergist muscle ablation model for compensatory hypertrophy. Rev Assoc Med Bras (1992) 63: 164–172, 2017. doi: 10.1590/1806-9282.63.02.164. [DOI] [PubMed] [Google Scholar]
- 127. Goldberg AL, Etlinger JD, Goldspink DF, Jablecki C. Mechanism of work-induced hypertrophy of skeletal muscle. Med Sci Sports 7: 185–198, 1975. [PubMed] [Google Scholar]
- 128. Schoenfeld BJ. Postexercise hypertrophic adaptations: a reexamination of the hormone hypothesis and its applicability to resistance training program design. J Strength Cond Res 27: 1720–1730, 2013. doi: 10.1519/JSC.0b013e31828ddd53. [DOI] [PubMed] [Google Scholar]
- 129. West DW, Phillips SM. Anabolic processes in human skeletal muscle: restoring the identities of growth hormone and testosterone. Phys Sportsmed 38: 97–104, 2010. doi: 10.3810/psm.2010.10.1814. [DOI] [PubMed] [Google Scholar]
- 130. Winick M, Noble A. Quantitative changes in DNA, RNA, and protein during prenatal and postnatal growth in the rat. Dev Biol 12: 451–466, 1965. doi: 10.1016/0012-1606(65)90009-6. [DOI] [PubMed] [Google Scholar]
- 131. Enesco M, LeBlond CP. Increase in cell number as a factor in the growth of the organs of the young male rat. J Embryol Exp Morph 10: 530–562, 1962. doi: 10.1242/dev.10.4.530. [DOI] [Google Scholar]
- 132. Florini JR. Incorporation of labeled amino acids into interior sites of protein by a cell-free system from rat skeletal muscle. Biochem Biophys Res Commun 8: 125–130, 1962. doi: 10.1016/0006-291x(62)90249-8. [DOI] [PubMed] [Google Scholar]
- 133. McLean JR, Cohn GL, Brandt IK, Simpson MV. Incorporation of labeled amino acids into the protein of muscle and liver mitochondria. J Biol Chem 233: 657–663, 1958. doi: 10.1016/S0021-9258(18)64722-2. [DOI] [PubMed] [Google Scholar]
- 134. Goldberg AL, Goodman HM. Amino acid transport during work-induced growth of skeletal muscle. Am J Physiol 216: 1111–1115, 1969. doi: 10.1152/ajplegacy.1969.216.5.1111. [DOI] [PubMed] [Google Scholar]
- 135. Jablecki CK, Heuser JE, Kaufman S. Autoradiographic localization of new RNA synthesis in hypertrophying skeletal muscle. J Cell Biol 57: 743–759, 1973. doi: 10.1083/jcb.57.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Goldberg AL. Protein synthesis during work-induced growth of skeletal muscle. J Cell Biol 36: 653–658, 1968. doi: 10.1083/jcb.36.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Millward DJ. Protein turnover in skeletal muscle. I. The measurement of rates of synthesis and catabolism of skeletal muscle protein using (14C)Na2CO3 to label protein. Clin Sci 39: 577–590, 1970. doi: 10.1042/cs0390577. [DOI] [PubMed] [Google Scholar]
- 138. Bergstrom J. Muscle electrolytes in man determined by neutron activation analysis on needle biopsy specimens. Scand J Clin Lab Invest (Engl) 14: 68, 1962. [Google Scholar]
- 139. Penman KA. Ultrastructural changes in human striated muscle using three methods of training. Res Q 40: 764–772, 1969. [PubMed] [Google Scholar]
- 140. Penman KA. Human striated muscle ultrastructural changes accompanying increased strength without hypertrophy. Res Q 41: 418–424, 1970. [PubMed] [Google Scholar]
- 141. Ogata T, Mori M. Histochemical study of oxidative enzymes in vertebrate muscles. J Histochem Cytochem 12: 171–182, 1964. doi: 10.1177/12.3.171. [DOI] [PubMed] [Google Scholar]
- 142. Edgerton VR, Gerchman L, Carrow R. Histochemical changes in rat skeletal muscle after exercise. Exp Neurol 24: 110–123, 1969. doi: 10.1016/0014-4886(69)90009-0. [DOI] [PubMed] [Google Scholar]
- 143. Barnard RJ, Edgerton VR, Peter JB. Effect of exercise on skeletal muscle. I. Biochemical and histochemical properties. J Appl Physiol 28: 762–766, 1970. doi: 10.1152/jappl.1970.28.6.762. [DOI] [PubMed] [Google Scholar]
- 144. Brooke MH, Kaiser KK. Muscle fiber types: how many and what kind? Arch Neurol 23: 369–379, 1970. doi: 10.1001/archneur.1970.00480280083010. [DOI] [PubMed] [Google Scholar]
- 145. Edström L, Nyström B. Histochemical types and sizes of fibres in normal human muscles. A biopsy study. Acta Neurol Scand 45: 257–269, 1969. doi: 10.1111/j.1600-0404.1969.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 146. Gollnick PD, Armstrong RB, Saubert CW, Piehl K, Saltin B. Enzyme activity and fiber composition in skeletal muscle of untrained and trained men. J Appl Physiol 33: 312–319, 1972. doi: 10.1152/jappl.1972.33.3.312. [DOI] [PubMed] [Google Scholar]
- 147. Schiaffino S, Bormioli SP. Adaptive changes in developing rat skeletal muscle in response to functional overload. Exp Neurol 40: 126–137, 1973. doi: 10.1016/0014-4886(73)90129-5. [DOI] [PubMed] [Google Scholar]
- 148. Schiaffino S, Bormioli SP, Aloisi M. Cell proliferation in rat skeletal muscle during early stages of compensatory hypertrophy. Virchows Arch B Cell Pathol 11: 268–273, 1972. doi: 10.1007/BF02889406. [DOI] [PubMed] [Google Scholar]
- 149. Schiaffino S, Bormioli SP, Aloisi M. The fate of newly formed satellite cells during compensatory muscle hypertrophy. Virchows Arch B Cell Pathol 21: 113–118, 1976. doi: 10.1007/BF02899148. [DOI] [PubMed] [Google Scholar]
- 150. Thorstensson A, Hultén B, von Döbeln W, Karlsson J. Effect of strength training on enzyme activities and fibre characteristics in human skeletal muscle. Acta Physiol Scand 96: 392–398, 1976. doi: 10.1111/j.1748-1716.1976.tb10207.x. [DOI] [PubMed] [Google Scholar]
- 151. Dons B, Bollerup K, Bonde-Petersen F, Hancke S. The effect of weight-lifting exercise related to muscle fiber composition and muscle cross-sectional area in humans. Eur J Appl Physiol Occup Physiol 40: 95–106, 1979. doi: 10.1007/BF00421155. [DOI] [PubMed] [Google Scholar]
- 152. Costill DL, Coyle EF, Fink WF, Lesmes GR, Witzmann FA. Adaptations in skeletal muscle following strength training. J Appl Physiol Respir Environ Exerc Physiol 46: 96–99, 1979. doi: 10.1152/jappl.1979.46.1.96. [DOI] [PubMed] [Google Scholar]
- 153. Moritani T, deVries HA. Neural factors versus hypertrophy in the time course of muscle strength gain. Am J Phys Med 58: 115–130, 1979. [PubMed] [Google Scholar]
- 154. MacDougall JD, Elder GC, Sale DG, Moroz JR, Sutton JR. Effects of strength training and immobilization on human muscle fibres. Eur J Appl Physiol Occup Physiol 43: 25–34, 1980. doi: 10.1007/BF00421352. [DOI] [PubMed] [Google Scholar]
- 155. Roberts MD, Haun CT, Vann CG, Osburn SC, Young KC. Sarcoplasmic hypertrophy in skeletal muscle: a scientific “unicorn” or resistance training adaptation? Front Physiol 11: 816, 2020. doi: 10.3389/fphys.2020.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Tesch PA, Larsson L. Muscle hypertrophy in bodybuilders. Eur J Appl Physiol Occup Physiol 49: 301–306, 1982. doi: 10.1007/BF00441291. [DOI] [PubMed] [Google Scholar]
- 157. Thorstensson A, Larsson L, Tesch P, Karlsson J. Muscle strength and fiber composition in athletes and sedentary men. Med Sci Sports 9: 26–30, 1977. [PubMed] [Google Scholar]
- 158. Tesch PA, Thorsson A, Kaiser P. Muscle capillary supply and fiber type characteristics in weight and power lifters. J Appl Physiol Respir Environ Exerc Physiol 56: 35–38, 1984. doi: 10.1152/jappl.1984.56.1.35. [DOI] [PubMed] [Google Scholar]
- 159. Apple FS, Tesch PA. CK and LD isozymes in human single muscle fibers in trained athletes. J Appl Physiol (1985) 66: 2717–2720, 1989. doi: 10.1152/jappl.1989.66.6.2717. [DOI] [PubMed] [Google Scholar]
- 160. Staron RS, Hikida RS, Hagerman FC, Dudley GA, Murray TF. Human skeletal muscle fiber type adaptability to various workloads. J Histochem Cytochem 32: 146–152, 1984. doi: 10.1177/32.2.6229571. [DOI] [PubMed] [Google Scholar]
- 161. Prince FP, Hikida RS, Hagerman FC, Staron RS, Allen WH. A morphometric analysis of human muscle fibers with relation to fiber types and adaptations to exercise. J Neurol Sci 49: 165–179, 1981. doi: 10.1016/0022-510x(81)90076-9. [DOI] [PubMed] [Google Scholar]
- 162. Lüthi JM, Howald H, Claassen H, Rösler K, Vock P, Hoppeler H. Structural changes in skeletal muscle tissue with heavy-resistance exercise. Int J Sports Med 7: 123–127, 1986. doi: 10.1055/s-2008-1025748. [DOI] [PubMed] [Google Scholar]
- 163. Staron RS, Malicky ES, Leonardi MJ, Falkel JE, Hagerman FC, Dudley GA. Muscle hypertrophy and fast fiber type conversions in heavy resistance-trained women. Eur J Appl Physiol Occup Physiol 60: 71–79, 1990. doi: 10.1007/BF00572189. [DOI] [PubMed] [Google Scholar]
- 164. Staron RS, Leonardi MJ, Karapondo DL, Malicky ES, Falkel JE, Hagerman FC, Hikida RS. Strength and skeletal muscle adaptations in heavy-resistance-trained women after detraining and retraining. J Appl Physiol (1985) 70: 631–640, 1991. doi: 10.1152/jappl.1991.70.2.631. [DOI] [PubMed] [Google Scholar]
- 165. Staron RS, Karapondo DL, Kraemer WJ, Fry AC, Gordon SE, Falkel JE, Hagerman FC, Hikida RS. Skeletal muscle adaptations during early phase of heavy-resistance training in men and women. J Appl Physiol (1985) 76: 1247–1255, 1994. doi: 10.1152/jappl.1994.76.3.1247. [DOI] [PubMed] [Google Scholar]
- 166. Alway SE, Grumbt WH, Stray-Gundersen J, Gonyea WJ. Effects of resistance training on elbow flexors of highly competitive bodybuilders. J Appl Physiol (1985) 72: 1512–1521, 1992. doi: 10.1152/jappl.1992.72.4.1512. [DOI] [PubMed] [Google Scholar]
- 167. Tesch PA, Komi PV, Häkkinen K. Enzymatic adaptations consequent to long-term strength training. Int J Sports Med 8, Suppl 1: 66–69, 1987. doi: 10.1055/s-2008-1025706. [DOI] [PubMed] [Google Scholar]
- 168. Tesch PA, Thorsson A, Essén-Gustavsson B. Enzyme activities of FT and ST muscle fibers in heavy-resistance trained athletes. J Appl Physiol (1985) 67: 83–87, 1989. doi: 10.1152/jappl.1989.67.1.83. [DOI] [PubMed] [Google Scholar]
- 169. Winchester PK, Davis ME, Alway SE, Gonyea WJ. Satellite cell activation in the stretch-enlarged anterior latissimus dorsi muscle of the adult quail. Am J Physiol Cell Physiol 260: C206–C212, 1991. doi: 10.1152/ajpcell.1991.260.2.C206. [DOI] [PubMed] [Google Scholar]
- 170. Alway SE, Gonyea WJ, Davis ME. Muscle fiber formation and fiber hypertrophy during the onset of stretch-overload. Am J Physiol Cell Physiol 259: C92–C102, 1990. doi: 10.1152/ajpcell.1990.259.1.C92. [DOI] [PubMed] [Google Scholar]
- 171. Alway SE, Winchester PK, Davis ME, Gonyea WJ. Regionalized adaptations and muscle fiber proliferation in stretch-induced enlargement. J Appl Physiol (1985) 66: 771–781, 1989. doi: 10.1152/jappl.1989.66.2.771. [DOI] [PubMed] [Google Scholar]
- 172. Antonio J, Gonyea WJ. Progressive stretch overload of skeletal muscle results in hypertrophy before hyperplasia. J Appl Physiol (1985) 75: 1263–1271, 1993. doi: 10.1152/jappl.1993.75.3.1263. [DOI] [PubMed] [Google Scholar]
- 173. Gonyea WJ, Sale DG, Gonyea FB, Mikesky A. Exercise induced increases in muscle fiber number. Eur J Appl Physiol Occup Physiol 55: 137–141, 1986. doi: 10.1007/BF00714995. [DOI] [PubMed] [Google Scholar]
- 174. Giddings CJ, Neaves WB, Gonyea WJ. Muscle fiber necrosis and regeneration induced by prolonged weight-lifting exercise in the cat. Anat Rec 211: 133–141, 1985. doi: 10.1002/ar.1092110204. [DOI] [PubMed] [Google Scholar]
- 175. MacDougall JD, Sale DG, Moroz JR, Elder GC, Sutton JR, Howald H. Mitochondrial volume density in human skeletal muscle following heavy resistance training. Med Sci Sports 11: 164–166, 1979. [PubMed] [Google Scholar]
- 176. MacDougall JD, Gibala MJ, Tarnopolsky MA, MacDonald JR, Interisano SA, Yarasheski KE. The time course for elevated muscle protein synthesis following heavy resistance exercise. Can J Appl Physiol 20: 480–486, 1995. doi: 10.1139/h95-038. [DOI] [PubMed] [Google Scholar]
- 177. MacDougall JD, Tarnopolsky MA, Chesley A, Atkinson SA. Changes in muscle protein synthesis following heavy resistance exercise in humans: a pilot study. Acta Physiol Scand 146: 403–404, 1992. doi: 10.1111/j.1748-1716.1992.tb09437.x. [DOI] [PubMed] [Google Scholar]
- 178. Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol Endocrinol Metab 273: E99–E107, 1997. doi: 10.1152/ajpendo.1997.273.1.E99. [DOI] [PubMed] [Google Scholar]
- 179. Chesley A, MacDougall JD, Tarnopolsky MA, Atkinson SA, Smith K. Changes in human muscle protein synthesis after resistance exercise. J Appl Physiol (1985) 73: 1383–1388, 1992. doi: 10.1152/jappl.1992.73.4.1383. [DOI] [PubMed] [Google Scholar]
- 180. Yarasheski KE, Zachwieja JJ, Bier DM. Acute effects of resistance exercise on muscle protein synthesis rate in young and elderly men and women. Am J Physiol Endocrinol Metab 265: E210–E214, 1993. doi: 10.1152/ajpendo.1993.265.2.E210. [DOI] [PubMed] [Google Scholar]
- 181. Phillips SM. Short-term training: when do repeated bouts of resistance exercise become training? Can J Appl Physiol 25: 185–193, 2000. doi: 10.1139/h00-014. [DOI] [PubMed] [Google Scholar]
- 182. Wilkinson DJ, Franchi MV, Brook MS, Narici MV, Williams JP, Mitchell WK, Szewczyk NJ, Greenhaff PL, Atherton PJ, Smith K. A validation of the application of D2O stable isotope tracer techniques for monitoring day-to-day changes in muscle protein subfraction synthesis in humans. Am J Physiol Endocrinol Metab 306: E571–E579, 2014. doi: 10.1152/ajpendo.00650.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Gasier HG, Fluckey JD, Previs SF, Wiggs MP, Riechman SE. Acute resistance exercise augments integrative myofibrillar protein synthesis. Metabolism 61: 153–156, 2012. doi: 10.1016/j.metabol.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 184. Wong TS, Booth FW. Protein metabolism in rat gastrocnemius muscle after stimulated chronic concentric exercise. J Appl Physiol (1985) 69: 1709–1717, 1990. doi: 10.1152/jappl.1990.69.5.1709. [DOI] [PubMed] [Google Scholar]
- 185. Wong TS, Booth FW. Protein metabolism in rat tibialis anterior muscle after stimulated chronic eccentric exercise. J Appl Physiol (1985) 69: 1718–1724, 1990. doi: 10.1152/jappl.1990.69.5.1718. [DOI] [PubMed] [Google Scholar]
- 186. Franchi MV, Reeves ND, Narici MV. Skeletal Muscle remodeling in response to eccentric vs. concentric loading: morphological, molecular, and metabolic adaptations. Front Physiol 8: 447, 2017. doi: 10.3389/fphys.2017.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Baar K, Esser K. Phosphorylation of p70S6k correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol Cell Physiol 276: C120–C127, 1999. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- 188. Nader GA, Esser KA. Intracellular signaling specificity in skeletal muscle in response to different modes of exercise. J Appl Physiol (1985) 90: 1936–1942, 2001. doi: 10.1152/jappl.2001.90.5.1936. [DOI] [PubMed] [Google Scholar]
- 189. Karlsson HK, Nilsson PA, Nilsson J, Chibalin AV, Zierath JR, Blomstrand E. Branched-chain amino acids increase p70S6k phosphorylation in human skeletal muscle after resistance exercise. Am J Physiol Endocrinol Metab 287: E1–E7, 2004. doi: 10.1152/ajpendo.00430.2003. [DOI] [PubMed] [Google Scholar]
- 190. Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol 576: 613–624, 2006. doi: 10.1113/jphysiol.2006.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019, 2001. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 192. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976, 2017. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Jozsi AC, Dupont-Versteegden EE, Taylor-Jones JM, Evans WJ, Trappe TA, Campbell WW, Peterson CA. Aged human muscle demonstrates an altered gene expression profile consistent with an impaired response to exercise. Mech Ageing Dev 120: 45–56, 2000. doi: 10.1016/s0047-6374(00)00178-0. [DOI] [PubMed] [Google Scholar]
- 194. Lim C, Shimizu J, Kawano F, Kim HJ, Kim CK. Adaptive responses of histone modifications to resistance exercise in human skeletal muscle. PLoS One 15: e0231321, 2020. doi: 10.1371/journal.pone.0231321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Robinson MM, Dasari S, Konopka AR, Johnson ML, Manjunatha S, Esponda RR, Carter RE, Lanza IR, Nair KS. Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metab 25: 581–592, 2017. doi: 10.1016/j.cmet.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Nair VD, Ge Y, Li S, Pincas H, Jain N, Seenarine N, Amper MA, Goodpaster BH, Walsh MJ, Coen PM, Sealfon SC. Sedentary and trained older men have distinct circulating exosomal microRNA profiles at baseline and in response to acute exercise. Front Physiol 11: 605, 2020. doi: 10.3389/fphys.2020.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Psilander N, Damsgaard R, Pilegaard H. Resistance exercise alters MRF and IGF-I mRNA content in human skeletal muscle. J Appl Physiol (1985) 95: 1038–1044, 2003. doi: 10.1152/japplphysiol.00903.2002. [DOI] [PubMed] [Google Scholar]
- 198. Zhu WG, Hibbert JE, Lin KH, Steinert ND, Lemens JL, Jorgenson KW, Newman SM, Lamming DW, Hornberger TA. Weight pulling: a novel mouse model of human progressive resistance exercise. Cells 10: 2459, 2021. doi: 10.3390/cells10092459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kotani T, Takegaki J, Tamura Y, Kouzaki K, Nakazato K, Ishii N. Repeated bouts of resistance exercise in rats alter mechanistic target of rapamycin complex 1 activity and ribosomal capacity but not muscle protein synthesis. Exp Physiol 106: 1950–1960, 2021. doi: 10.1113/EP089699. [DOI] [PubMed] [Google Scholar]
- 200. Kotani T, Takegaki J, Tamura Y, Kouzaki K, Nakazato K, Ishii N. The effect of repeated bouts of electrical stimulation-induced muscle contractions on proteolytic signaling in rat skeletal muscle. Physiol Rep 9: e14842, 2021. doi: 10.14814/phy2.14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Camera DM, Edge J, Short MJ, Hawley JA, Coffey VG. Early time course of Akt phosphorylation after endurance and resistance exercise. Med Sci Sports Exerc 42: 1843–1852, 2010. doi: 10.1249/MSS.0b013e3181d964e4. [DOI] [PubMed] [Google Scholar]
- 202. Apró W, Blomstrand E. Influence of supplementation with branched-chain amino acids in combination with resistance exercise on p70S6 kinase phosphorylation in resting and exercising human skeletal muscle. Acta Physiol (Oxf) 200: 237–248, 2010. doi: 10.1111/j.1748-1708.2010.02151.x. [DOI] [PubMed] [Google Scholar]
- 203. Apró W, Wang L, Pontén M, Blomstrand E, Sahlin K. Resistance exercise induced mTORC1 signaling is not impaired by subsequent endurance exercise in human skeletal muscle. Am J Physiol Endocrinol Metab 305: E22–E32, 2013. doi: 10.1152/ajpendo.00091.2013. [DOI] [PubMed] [Google Scholar]
- 204. Mazo CE, D’Lugos AC, Sweeney KR, Haus JM, Angadi SS, Carroll CC, Dickinson JM. The effects of acute aerobic and resistance exercise on mTOR signaling and autophagy markers in untrained human skeletal muscle. Eur J Appl Physiol 121: 2913–2924, 2021. doi: 10.1007/s00421-021-04758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Camera DM, West DW, Burd NA, Phillips SM, Garnham AP, Hawley JA, Coffey VG. Low muscle glycogen concentration does not suppress the anabolic response to resistance exercise. J Appl Physiol (1985) 113: 206–214, 2012. doi: 10.1152/japplphysiol.00395.2012. [DOI] [PubMed] [Google Scholar]
- 206. Burd NA, Holwerda AM, Selby KC, West DW, Staples AW, Cain NE, Cashaback JG, Potvin JR, Baker SK, Phillips SM. Resistance exercise volume affects myofibrillar protein synthesis and anabolic signalling molecule phosphorylation in young men. J Physiol 588: 3119–3130, 2010. doi: 10.1113/jphysiol.2010.192856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Wette SG, Birch NP, Soop M, Zügel M, Murphy RM, Lamb GD, Smith HK. Expression of titin-linked putative mechanosensing proteins in skeletal muscle after power resistance exercise in resistance-trained men. J Appl Physiol (1985) 130: 545–561, 2021. doi: 10.1152/japplphysiol.00711.2020. [DOI] [PubMed] [Google Scholar]
- 208. Vissing K, Overgaard K, Nedergaard A, Fredsted A, Schjerling P. Effects of concentric and repeated eccentric exercise on muscle damage and calpain-calpastatin gene expression in human skeletal muscle. Eur J Appl Physiol 103: 323–332, 2008. doi: 10.1007/s00421-008-0709-7. [DOI] [PubMed] [Google Scholar]
- 209. Lamas L, Aoki MS, Ugrinowitsch C, Campos GE, Regazzini M, Moriscot AS, Tricoli V. Expression of genes related to muscle plasticity after strength and power training regimens. Scand J Med Sci Sports 20: 216–225, 2010. doi: 10.1111/j.1600-0838.2009.00905.x. [DOI] [PubMed] [Google Scholar]
- 210. Santos AR, Lamas L, Ugrinowitsch C, Tricoli V, Miyabara EH, Soares AG, Aoki MS. Different resistance-training regimens evoked a similar increase in myostatin inhibitors expression. Int J Sports Med 36: 761–768, 2015. doi: 10.1055/s-0035-1547219. [DOI] [PubMed] [Google Scholar]
- 211. Witard OC, Tieland M, Beelen M, Tipton KD, van Loon LJ, Koopman R. Resistance exercise increases postprandial muscle protein synthesis in humans. Med Sci Sports Exerc 41: 144–154, 2009. doi: 10.1249/MSS.0b013e3181844e79. [DOI] [PubMed] [Google Scholar]
- 212. Damas F, Angleri V, Phillips SM, Witard OC, Ugrinowitsch C, Santanielo N, Soligon SD, Costa LA, Lixandrão ME, Conceição MS, Libardi CA. Myofibrillar protein synthesis and muscle hypertrophy individualized responses to systematically changing resistance training variables in trained young men. J Appl Physiol (1985) 127: 806–815, 2019. doi: 10.1152/japplphysiol.00350.2019. [DOI] [PubMed] [Google Scholar]
- 213. Mayhew DL, Hornberger TA, Lincoln HC, Bamman MM. Eukaryotic initiation factor 2B epsilon induces cap-dependent translation and skeletal muscle hypertrophy. J Physiol 589: 3023–3037, 2011. doi: 10.1113/jphysiol.2010.202432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Farrell PA, Fedele MJ, Vary TC, Kimball SR, Lang CH, Jefferson LS. Regulation of protein synthesis after acute resistance exercise in diabetic rats. Am J Physiol Endocrinol Metab 276: E721–E727, 1999. doi: 10.1152/ajpendo.1999.276.4.E721. [DOI] [PubMed] [Google Scholar]
- 215. Hasten DL, Pak-Loduca J, Obert KA, Yarasheski KE. Resistance exercise acutely increases MHC and mixed muscle protein synthesis rates in 78-84 and 23-32 yr olds. Am J Physiol Endocrinol Metab 278: E620–E626, 2000. doi: 10.1152/ajpendo.2000.278.4.E620. [DOI] [PubMed] [Google Scholar]
- 216. Terzis G, Spengos K, Mascher H, Georgiadis G, Manta P, Blomstrand E. The degree of p70 S6k and S6 phosphorylation in human skeletal muscle in response to resistance exercise depends on the training volume. Eur J Appl Physiol 110: 835–843, 2010. doi: 10.1007/s00421-010-1527-2. [DOI] [PubMed] [Google Scholar]
- 217. Blazev R, Carl CS, Ng YK, Molendijk J, Voldstedlund CT, Zhao Y, Xiao D, Kueh AJ, Miotto PM, Haynes VR, Hardee JP, Chung JD, McNamara JW, Qian H, Gregorevic P, Oakhill JS, Herold MJ, Jensen TE, Lisowski L, Lynch GS, Dodd GT, Watt MJ, Yang P, Kiens B, Richter EA, Parker BL. Phosphoproteomics of three exercise modalities identifies canonical signaling and C18ORF25 as an AMPK substrate regulating skeletal muscle function. Cell Metab 34: 1561–1577.e9, 2022. doi: 10.1016/j.cmet.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 218. Hodson N, McGlory C, Oikawa SY, Jeromson S, Song Z, Rüegg MA, Hamilton DL, Phillips SM, Philp A. Differential localization and anabolic responsiveness of mTOR complexes in human skeletal muscle in response to feeding and exercise. Am J Physiol Cell Physiol 313: C604–C611, 2017. doi: 10.1152/ajpcell.00176.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Roberson PA, Mobley CB, Romero MA, Haun CT, Osburn SC, Mumford PW, Vann CG, Greer RA, Ferrando AA, Roberts MD. LAT1 protein content increases following 12 weeks of resistance exercise training in human skeletal muscle. Front Nutr 7: 628405, 2020. doi: 10.3389/fnut.2020.628405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Mazzulla M, Hodson N, Lees M, Scaife PJ, Smith K, Atherton PJ, Kumbhare D, Moore DR. LAT1 and SNAT2 protein expression and membrane localization of LAT1 are not acutely altered by dietary amino acids or resistance exercise nor positively associated with leucine or phenylalanine incorporation in human skeletal muscle. Nutrients 13: 3906, 2021. doi: 10.3390/nu13113906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Song Z, Moore DR, Hodson N, Ward C, Dent JR, O’Leary MF, Shaw AM, Hamilton DL, Sarkar S, Gangloff YG, Hornberger TA, Spriet LL, Heigenhauser GJ, Philp A. Resistance exercise initiates mechanistic target of rapamycin (mTOR) translocation and protein complex co-localisation in human skeletal muscle. Sci Rep 7: 5028, 2017. doi: 10.1038/s41598-017-05483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Abou Sawan S, Hodson N, Babits P, Malowany JM, Kumbhare D, Moore DR. Satellite cell and myonuclear accretion is related to training-induced skeletal muscle fiber hypertrophy in young males and females. J Appl Physiol (1985) 131: 871–880, 2021. doi: 10.1152/japplphysiol.00424.2021. [DOI] [PubMed] [Google Scholar]
- 223. Kadi F, Schjerling P, Andersen LL, Charifi N, Madsen JL, Christensen LR, Andersen JL. The effects of heavy resistance training and detraining on satellite cells in human skeletal muscles. J Physiol 558: 1005–1012, 2004. doi: 10.1113/jphysiol.2004.065904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Snijders T, Verdijk LB, Smeets JS, McKay BR, Senden JM, Hartgens F, Parise G, Greenhaff P, van Loon LJ. The skeletal muscle satellite cell response to a single bout of resistance-type exercise is delayed with aging in men. Age (Dordr) 36: 9699, 2014. doi: 10.1007/s11357-014-9699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Koopman R, Zorenc AH, Gransier RJ, Cameron-Smith D, van Loon LJ. Increase in S6K1 phosphorylation in human skeletal muscle following resistance exercise occurs mainly in type II muscle fibers. Am J Physiol Endocrinol Metab 290: E1245–E1252, 2006. doi: 10.1152/ajpendo.00530.2005. [DOI] [PubMed] [Google Scholar]
- 226. Bjørnsen T, Wernbom M, Kirketeig A, Paulsen G, Samnøy L, Bækken L, Cameron-Smith D, Berntsen S, Raastad T. Type 1 muscle fiber hypertrophy after blood flow-restricted training in powerlifters. Med Sci Sports Exerc 51: 288–298, 2019. doi: 10.1249/MSS.0000000000001775. [DOI] [PubMed] [Google Scholar]
- 227. Crameri RM, Langberg H, Magnusson P, Jensen CH, Schrøder HD, Olesen JL, Suetta C, Teisner B, Kjaer M. Changes in satellite cells in human skeletal muscle after a single bout of high intensity exercise. J Physiol 558: 333–340, 2004. doi: 10.1113/jphysiol.2004.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Bjørnsen T, Wernbom M, Paulsen G, Markworth JF, Berntsen S, D’Souza RF, Cameron-Smith D, Raastad T. High-frequency blood flow-restricted resistance exercise results in acute and prolonged cellular stress more pronounced in type I than in type II fibers. J Appl Physiol (1985) 131: 643–660, 2021. doi: 10.1152/japplphysiol.00115.2020. [DOI] [PubMed] [Google Scholar]
- 229. Bjørnsen T, Wernbom M, Løvstad A, Paulsen G, D’Souza RF, Cameron-Smith D, Flesche A, Hisdal J, Berntsen S, Raastad T. Delayed myonuclear addition, myofiber hypertrophy, and increases in strength with high-frequency low-load blood flow restricted training to volitional failure. J Appl Physiol (1985) 126: 578–592, 2019. doi: 10.1152/japplphysiol.00397.2018. [DOI] [PubMed] [Google Scholar]
- 230. Gaulton N, Wakelin G, Young LV, Wotherspoon S, Kamal M, Parise G, Nederveen JP, Holwerda A, Verdijk LB, van Loon LJ, Snijders T, Johnston AP. Twist2-expressing cells reside in human skeletal muscle and are responsive to aging and resistance exercise training. FASEB J 36: e22642, 2022. doi: 10.1096/fj.202201349RR. [DOI] [PubMed] [Google Scholar]
- 231. Verbrugge SA, Schönfelder M, Becker L, Yaghoob Nezhad F, Hrabě de Angelis M, Wackerhage H. Genes whose gain or loss-of-function increases skeletal muscle mass in mice: a systematic literature review. Front Physiol 9: 553, 2018. doi: 10.3389/fphys.2018.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Murach KA, McCarthy JJ, Peterson CA, Dungan CM. Making mice mighty: recent advances in translational models of load-induced muscle hypertrophy. J Appl Physiol (1985) 129: 516–521, 2020. doi: 10.1152/japplphysiol.00319.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Steinert ND, Jorgenson KW, Lin KH, Hermanson JB, Lemens JL, Hornberger TA. A novel method for visualizing in-vivo rates of protein degradation provides insight into how TRIM28 regulates muscle size. iScience 26: 106526, 2023. doi: 10.1016/j.isci.2023.106526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Vann CG, Morton RW, Mobley CB, Vechetti IJ, Ferguson BK, Haun CT, Osburn SC, Sexton CL, Fox CD, Romero MA, Roberson PA, Oikawa SY, McGlory C, Young KC, McCarthy JJ, Phillips SM, Roberts MD. An intron variant of the GLI family zinc finger 3 (GLI3) gene differentiates resistance training-induced muscle fiber hypertrophy in younger men. FASEB J 35: e21587, 2021. doi: 10.1096/fj.202100113RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol 545: 27–41, 2002. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Chen YW, Hubal MJ, Hoffman EP, Thompson PD, Clarkson PM. Molecular responses of human muscle to eccentric exercise. J Appl Physiol (1985) 95: 2485–2494, 2003. doi: 10.1152/japplphysiol.01161.2002. [DOI] [PubMed] [Google Scholar]
- 237. Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S. Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol (1985) 112: 1625–1636, 2012. doi: 10.1152/japplphysiol.00435.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Thalacker-Mercer A, Stec M, Cui X, Cross J, Windham S, Bamman M. Cluster analysis reveals differential transcript profiles associated with resistance training-induced human skeletal muscle hypertrophy. Physiol Genomics 45: 499–507, 2013. doi: 10.1152/physiolgenomics.00167.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Ogasawara R, Akimoto T, Umeno T, Sawada S, Hamaoka T, Fujita S. MicroRNA expression profiling in skeletal muscle reveals different regulatory patterns in high and low responders to resistance training. Physiol Genomics 48: 320–324, 2016. doi: 10.1152/physiolgenomics.00124.2015. [DOI] [PubMed] [Google Scholar]
- 240. Willis CR, Deane CS, Ames RM, Bass JJ, Wilkinson DJ, Smith K, Phillips BE, Szewczyk NJ, Atherton PJ, Etheridge T. Transcriptomic adaptation during skeletal muscle habituation to eccentric or concentric exercise training. Sci Rep 11: 23930, 2021. doi: 10.1038/s41598-021-03393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Khan Y, Hammarström D, Rønnestad BR, Ellefsen S, Ahmad R. Increased biological relevance of transcriptome analyses in human skeletal muscle using a model-specific pipeline. BMC Bioinformatics 21: 548, 2020. doi: 10.1186/s12859-020-03866-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Gordon PM, Liu D, Sartor MA, IglayReger HB, Pistilli EE, Gutmann L, Nader GA, Hoffman EP. Resistance exercise training influences skeletal muscle immune activation: a microarray analysis. J Appl Physiol (1985) 112: 443–453, 2012. doi: 10.1152/japplphysiol.00860.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Damas F, Ugrinowitsch C, Libardi CA, Jannig PR, Hector AJ, McGlory C, Lixandrão ME, Vechin FC, Montenegro H, Tricoli V, Roschel H, Phillips SM. Resistance training in young men induces muscle transcriptome-wide changes associated with muscle structure and metabolism refining the response to exercise-induced stress. Eur J Appl Physiol 118: 2607–2616, 2018. doi: 10.1007/s00421-018-3984-y. [DOI] [PubMed] [Google Scholar]
- 244. Melov S, Tarnopolsky MA, Beckman K, Felkey K, Hubbard A. Resistance exercise reverses aging in human skeletal muscle. PLoS One 2: e465, 2007. doi: 10.1371/journal.pone.0000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Murton AJ, Billeter R, Stephens FB, Des Etages SG, Graber F, Hill RJ, Marimuthu K, Greenhaff PL. Transient transcriptional events in human skeletal muscle at the outset of concentric resistance exercise training. J Appl Physiol (1985) 116: 113–125, 2014. doi: 10.1152/japplphysiol.00426.2013. [DOI] [PubMed] [Google Scholar]
- 246. Phillips BE, Williams JP, Gustafsson T, Bouchard C, Rankinen T, Knudsen S, Smith K, Timmons JA, Atherton PJ. Molecular networks of human muscle adaptation to exercise and age. PLoS Genet 9: e1003389, 2013. doi: 10.1371/journal.pgen.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Stepto NK, Coffey VG, Carey AL, Ponnampalam AP, Canny BJ, Powell D, Hawley JA. Global gene expression in skeletal muscle from well-trained strength and endurance athletes. Med Sci Sports Exerc 41: 546–565, 2009. doi: 10.1249/MSS.0b013e31818c6be9. [DOI] [PubMed] [Google Scholar]
- 248. Telles GD, Libardi CA, Conceição MS, Vechin FC, Lixandrão ME, DE Andrade AL, Guedes DN, Ugrinowitsch C, Camera DM. Time course of skeletal muscle miRNA expression after resistance, high-intensity interval, and concurrent exercise. Med Sci Sports Exerc 53: 1708–1718, 2021. doi: 10.1249/MSS.0000000000002632. [DOI] [PubMed] [Google Scholar]
- 249. Lundberg TR, Fernandez-Gonzalo R, Tesch PA, Rullman E, Gustafsson T. Aerobic exercise augments muscle transcriptome profile of resistance exercise. Am J Physiol Regul Integr Comp Physiol 310: R1279–R1287, 2016. doi: 10.1152/ajpregu.00035.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Stokes T, Timmons JA, Crossland H, Tripp TR, Murphy K, McGlory C, Mitchell CJ, Oikawa SY, Morton RW, Phillips BE, Baker SK, Atherton PJ, Wahlestedt C, Phillips SM. Molecular transducers of human skeletal muscle remodeling under different loading states. Cell Rep 32: 107980, 2020. doi: 10.1016/j.celrep.2020.107980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Chapman MA, Arif M, Emanuelsson EB, Reitzner SM, Lindholm ME, Mardinoglu A, Sundberg CJ. Skeletal muscle transcriptomic comparison between long-term trained and untrained men and women. Cell Rep 31: 107808, 2020. doi: 10.1016/j.celrep.2020.107808. [DOI] [PubMed] [Google Scholar]
- 252. Gautvik KM, Olstad OK, Raue U, Gautvik VT, Kvernevik KJ, Utheim TP, Ravnum S, Kirkegaard C, Wiig H, Jones G, Pilling LC, Trappe S, Raastad T, Reppe S. Heavy-load exercise in older adults activates vasculogenesis and has a stronger impact on muscle gene expression than in young adults. Eur Rev Aging Phys Act 19: 23, 2022. doi: 10.1186/s11556-022-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Bagley JR, Burghardt KJ, McManus R, Howlett B, Costa PB, Coburn JW, Arevalo JA, Malek MH, Galpin AJ. Epigenetic responses to acute resistance exercise in trained vs. sedentary men. J Strength Cond Res 34: 1574–1580, 2020. doi: 10.1519/JSC.0000000000003185. [DOI] [PubMed] [Google Scholar]
- 254. Von Walden F, Rea M, Mobley CB, Fondufe-Mittendorf Y, McCarthy JJ, Peterson CA, Murach KA. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics 15: 1151–1162, 2020. doi: 10.1080/15592294.2020.1755581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Seaborne RA, Strauss J, Cocks M, Shepherd S, O’Brien TD, van Someren KA, Bell PG, Murgatroyd C, Morton JP, Stewart CE, Sharples AP. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep 8: 1898, 2018. doi: 10.1038/s41598-018-20287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Turner DC, Seaborne RA, Sharples AP. Comparative transcriptome and methylome analysis in human skeletal muscle anabolism, hypertrophy and epigenetic memory. Sci Rep 9: 4251, 2019. doi: 10.1038/s41598-019-40787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Seaborne RA, Strauss J, Cocks M, Shepherd S, O’Brien TD, Someren KA, Bell PG, Murgatroyd C, Morton JP, Stewart CE, Mein CA, Sharples AP. Methylome of human skeletal muscle after acute & chronic resistance exercise training, detraining & retraining. Sci Data 5: 180213, 2018. doi: 10.1038/sdata.2018.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Ruple BA, Godwin JS, Mesquita PH, Osburn SC, Vann CG, Lamb DA, Sexton CL, Candow DG, Forbes SC, Frugé AD, Kavazis AN, Young KC, Seaborne RA, Sharples AP, Roberts MD. Resistance training rejuvenates the mitochondrial methylome in aged human skeletal muscle. FASEB J 35: e21864, 2021. doi: 10.1096/fj.202100873RR. [DOI] [PubMed] [Google Scholar]
- 259. de Sousa Neto IV, Carvalho MM, Marqueti RC, Almeida JA, Oliveira KS, Barin FR, Petriz B, de Araújo HS, Franco OL, Durigan JL. Proteomic changes in skeletal muscle of aged rats in response to resistance training. Cell Biochem Funct 38: 500–509, 2020. doi: 10.1002/cbf.3497. [DOI] [PubMed] [Google Scholar]
- 260. Tibana RA, Franco OL, Cunha GV, Sousa NM, Sousa Neto IV, Carvalho MM, Almeida JA, Durigan JL, Marqueti RC, Navalta JW, Lobo MO, Voltarelli FA, Prestes J. The effects of resistance training volume on skeletal muscle proteome. Int J Exerc Sci 10: 1051–1066, 2017. [PMC free article] [PubMed] [Google Scholar]
- 261. Vann CG, Roberson PA, Osburn SC, Mumford PW, Romero MA, Fox CD, Moore JH, Haun CT, Beck DT, Moon JR, Kavazis AN, Young KC, Badisa VL, Mwashote BM, Ibeanusi V, Singh RK, Roberts MD. Skeletal muscle myofibrillar protein abundance is higher in resistance-trained men, and aging in the absence of training may have an opposite effect. Sports (Basel) 8: 7, 2020. doi: 10.3390/sports8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Vann CG, Osburn SC, Mumford PW, Roberson PA, Fox CD, Sexton CL, Johnson MR, Johnson JS, Shake J, Moore JH, Millevoi K, Beck DT, Badisa VL, Mwashote BM, Ibeanusi V, Singh RK, Roberts MD. Skeletal muscle protein composition adaptations to 10 weeks of high-load resistance training in previously-trained males. Front Physiol 11: 259, 2020. doi: 10.3389/fphys.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Haun CT, Vann CG, Osburn SC, Mumford PW, Roberson PA, Romero MA, Fox CD, Johnson CA, Parry HA, Kavazis AN, Moon JR, Badisa VL, Mwashote BM, Ibeanusi V, Young KC, Roberts MD. Muscle fiber hypertrophy in response to 6 weeks of high-volume resistance training in trained young men is largely attributed to sarcoplasmic hypertrophy. PLoS One 14: e0215267, 2019. doi: 10.1371/journal.pone.0215267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Deane CS, Phillips BE, Willis CR, Wilkinson DJ, Smith K, Higashitani N, Williams JP, Szewczyk NJ, Atherton PJ, Higashitani A, Etheridge T. Proteomic features of skeletal muscle adaptation to resistance exercise training as a function of age. Geroscience. In Press. doi: 10.1007/s11357-022-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Deshmukh AS, Steenberg DE, Hostrup M, Birk JB, Larsen JK, Santos A, Kjøbsted R, Hingst JR, Schéele CC, Murgia M, Kiens B, Richter EA, Mann M, Wojtaszewski JF. Deep muscle-proteomic analysis of freeze-dried human muscle biopsies reveals fiber type-specific adaptations to exercise training. Nat Commun 12: 304, 2021. doi: 10.1038/s41467-020-20556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Potts GK, McNally RM, Blanco R, You JS, Hebert AS, Westphall MS, Coon JJ, Hornberger TA. A map of the phosphoproteomic alterations that occur after a bout of maximal-intensity contractions. J Physiol 595: 5209–5226, 2017. doi: 10.1113/JP273904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Steinert ND, Potts GK, Wilson GM, Klamen AM, Lin KH, Hermanson JB, McNally RM, Coon JJ, Hornberger TA. Mapping of the contraction-induced phosphoproteome identifies TRIM28 as a significant regulator of skeletal muscle size and function. Cell Rep 34: 108796, 2021. doi: 10.1016/j.celrep.2021.108796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, Stöckli J, Burchfield JG, Jensen TE, Jothi R, Kiens B, Wojtaszewski JF, Richter EA, James DE. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab 22: 922–935, 2015. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Berton R, Conceição MS, Libardi CA, Canevarolo RR, Gáspari AF, Chacon-Mikahil MP, Zeri AC, Cavaglieri CR. Metabolic time-course response after resistance exercise: a metabolomics approach. J Sports Sci 35: 1211–1218, 2017. doi: 10.1080/02640414.2016.1218035. [DOI] [PubMed] [Google Scholar]
- 270. Valério DF, Berton R, Conceição MS, Canevarolo RR, Chacon-Mikahil MP, Cavaglieri CR, Meirelles GV, Zeri AC, Libardi CA. Early metabolic response after resistance exercise with blood flow restriction in well-trained men: a metabolomics approach. Appl Physiol Nutr Metab 43: 240–246, 2018. doi: 10.1139/apnm-2017-0471. [DOI] [PubMed] [Google Scholar]
- 271. Gehlert S, Weinisch P, Römisch-Margl W, Jaspers RT, Artati A, Adamski J, Dyar KA, Aussieker T, Jacko D, Bloch W, Wackerhage H, Kastenmüller G. Effects of acute and chronic resistance exercise on the skeletal muscle metabolome. Metabolites 12: 445, 2022. doi: 10.3390/metabo12050445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Pillon NJ, Gabriel BM, Dollet L, Smith JA, Sardon Puig L, Botella J, Bishop DJ, Krook A, Zierath JR. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat Commun 11: 470, 2020. doi: 10.1038/s41467-019-13869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Sanford JA, Nogiec CD, Lindholm ME, Adkins JN, Amar D, Dasari S, Drugan JK, Fernández FM, Radom-Aizik S, Schenk S, Snyder MP, Tracy RP, Vanderboom P, Trappe S, Walsh MJ; Molecular Transducers of Physical Activity Consortium. Molecular Transducers of Physical Activity Consortium (MoTrPAC): mapping the dynamic responses to exercise. Cell 181: 1464–1474, 2020. doi: 10.1016/j.cell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Valero-Breton M, Warnier G, Castro-Sepulveda M, Deldicque L, Zbinden-Foncea H. Acute and chronic effects of high frequency electric pulse stimulation on the Akt/mTOR pathway in human primary myotubes. Front Bioeng Biotechnol 8: 565679, 2020. doi: 10.3389/fbioe.2020.565679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Rupert JE, Jengelley DH, Zimmers TA. In vitro, in vivo, and in silico methods for assessment of muscle size and muscle growth regulation. Shock 53: 605–615, 2020. doi: 10.1097/SHK.0000000000001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Stokes T, Cen HH, Kapranov P, Gallagher IJ, Pitsillides AA, Volmar CH, Kraus WE, Johnson JD, Phillips SM, Wahlestedt C, Timmons JA. Transcriptomics for clinical and experimental biology research: hang on a seq. Adv Genet (Hoboken) 4: 2200024, 2023. doi: 10.1002/ggn2.202200024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, Banks S, Deng J, VanMeter AJ, Geho DH, Pastore L, Sennesh J, Petricoin EF 3rd, Liotta LA. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics 7: 1998–2018, 2008. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Bonadio RS, Nunes LB, Moretti PN, Mazzeu JF, Cagnin S, Pic-Taylor A, de Oliveira SF. Insights into how environment shapes post-mortem RNA transcription in mouse brain. Sci Rep 11: 13008, 2021. doi: 10.1038/s41598-021-92268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Wang Y, Zhang Y, Hu W, Xie S, Gong CX, Iqbal K, Liu F. Corrigendum: Rapid alteration of protein phosphorylation during postmortem: implication in the study of protein phosphorylation. Sci Rep 6: 17241, 2016. doi: 10.1038/srep17241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Botling J, Edlund K, Segersten U, Tahmasebpoor S, Engström M, Sundström M, Malmström PU, Micke P. Impact of thawing on RNA integrity and gene expression analysis in fresh frozen tissue. Diagn Mol Pathol 18: 44–52, 2009. doi: 10.1097/PDM.0b013e3181857e92. [DOI] [PubMed] [Google Scholar]
- 281. Rountree CB, Van Kirk CA, You H, Ding W, Dang H, VanGuilder HD, Freeman WM. Clinical application for the preservation of phospho-proteins through in-situ tissue stabilization. Proteome Sci 8: 61, 2010. doi: 10.1186/1477-5956-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Meng H, Janssen PM, Grange RW, Yang L, Beggs AH, Swanson LC, Cossette SA, Frase A, Childers MK, Granzier H, Gussoni E, Lawlor MW. Tissue triage and freezing for models of skeletal muscle disease. J Vis Exp 89: 51586, 2014. doi: 10.3791/51586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Mutter GL, Zahrieh D, Liu C, Neuberg D, Finkelstein D, Baker HE, Warrington JA. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics 5: 88, 2004. doi: 10.1186/1471-2164-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Kumar A, Accorsi A, Rhee Y, Girgenrath M. Do’s and don’ts in the preparation of muscle cryosections for histological analysis. J Vis Exp 99: e52793, 2015. doi: 10.3791/52793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Murach KA, Dungan CM, von Walden F, Wen Y. Epigenetic evidence for distinct contributions of resident and acquired myonuclei during long-term exercise adaptation using timed in vivo myonuclear labeling. Am J Physiol Cell Physiol 322: C86–C93, 2022. doi: 10.1152/ajpcell.00358.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Wen Y, Dungan CM, Mobley CB, Valentino T, von Walden F, Murach KA. Nucleus type-specific DNA methylomics reveals epigenetic “memory” of prior adaptation in skeletal muscle. Function (Oxf) 2: zqab038, 2021. doi: 10.1093/function/zqab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Roberts MD, Young KC, Fox CD, Vann CG, Roberson PA, Osburn SC, Moore JH, Mumford PW, Romero MA, Beck DT, Haun CT, Badisa VL, Mwashote BM, Ibeanusi V, Kavazis AN. An optimized procedure for isolation of rodent and human skeletal muscle sarcoplasmic and myofibrillar proteins. J Biol Methods 7: e127, 2020. doi: 10.14440/jbm.2020.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Yates LD, Greaser ML. Quantitative determination of myosin and actin in rabbit skeletal muscle. J Mol Biol 168: 123–141, 1983. doi: 10.1016/s0022-2836(83)80326-x. [DOI] [PubMed] [Google Scholar]
- 289. Wang K, McClure J, Tu A. Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci USA 76: 3698–3702, 1979. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Garcia-Cazarin ML, Snider NN, Andrade FH. Mitochondrial isolation from skeletal muscle. J Vis Exp 49: 2452, 2011. doi: 10.3791/2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Sale DG. Neural adaptation to resistance training. Med Sci Sports Exerc 20: S135–S145, 1988. doi: 10.1249/00005768-198810001-00009. [DOI] [PubMed] [Google Scholar]
- 292. Tobias IS, Lazauskas KK, Arevalo JA, Bagley JR, Brown LE, Galpin AJ. Fiber type-specific analysis of AMPK isoforms in human skeletal muscle: advancement in methods via capillary nanoimmunoassay. J Appl Physiol (1985) 124: 840–849, 2018. doi: 10.1152/japplphysiol.00894.2017. [DOI] [PubMed] [Google Scholar]
- 293. Galpin AJ, Raue U, Jemiolo B, Trappe TA, Harber MP, Minchev K, Trappe S. Human skeletal muscle fiber type specific protein content. Anal Biochem 425: 175–182, 2012. doi: 10.1016/j.ab.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Kristensen DE, Albers PH, Prats C, Baba O, Birk JB, Wojtaszewski JF. Human muscle fibre type-specific regulation of AMPK and downstream targets by exercise. J Physiol 593: 2053–2069, 2015. doi: 10.1113/jphysiol.2014.283267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Dickinson JM, Lee JD, Sullivan BE, Harber MP, Trappe SW, Trappe TA. A new method to study in vivo protein synthesis in slow- and fast-twitch muscle fibers and initial measurements in humans. J Appl Physiol (1985) 108: 1410–1416, 2010. doi: 10.1152/japplphysiol.00905.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Bass JJ, Wilkinson DJ, Rankin D, Phillips BE, Szewczyk NJ, Smith K, Atherton PJ. An overview of technical considerations for Western blotting applications to physiological research. Scand J Med Sci Sports 27: 4–25, 2017. doi: 10.1111/sms.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Kuang J, Yan X, Genders AJ, Granata C, Bishop DJ. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS One 13: e0196438, 2018. doi: 10.1371/journal.pone.0196438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, Tarnopolsky MA. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics 18: 226–231, 2004. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- 299. Sunderland KL, Roberts MD, Dalbo VJ, Kerksick CM. Aging and sequential resistance exercise bout effects on housekeeping gene messenger RNA expression in human skeletal muscle. J Strength Cond Res 27: 1–7, 2013. doi: 10.1519/JSC.0b013e3182779830. [DOI] [PubMed] [Google Scholar]
- 300. Vigelso A, Dybboe R, Hansen CN, Dela F, Helge JW, Guadalupe Grau A. GAPDH and beta-actin protein decreases with aging, making Stain-Free technology a superior loading control in Western blotting of human skeletal muscle. J Appl Physiol (1985) 118: 386–394, 2015. doi: 10.1152/japplphysiol.00840.2014. [DOI] [PubMed] [Google Scholar]
- 301. Qin S, Kim J, Arafat D, Gibson G. Effect of normalization on statistical and biological interpretation of gene expression profiles. Front Genet 3: 160, 2012. doi: 10.3389/fgene.2012.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25, 2010. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Kobak KA, Lawrence MM, Pharaoh G, Borowik AK, Peelor FF 3rd, Shipman PD, Griffin TM, Van Remmen H, Miller BF. Determining the contributions of protein synthesis and breakdown to muscle atrophy requires non-steady-state equations. J Cachexia Sarcopenia Muscle 12: 1764–1775, 2021. doi: 10.1002/jcsm.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Witard OC, Bannock L, Tipton KD. Making sense of muscle protein synthesis: a focus on muscle growth during resistance training. Int J Sport Nutr Exerc Metab 32: 49–61, 2022. doi: 10.1123/ijsnem.2021-0139. [DOI] [PubMed] [Google Scholar]
- 305. Damas F, Phillips S, Vechin FC, Ugrinowitsch C. A review of resistance training-induced changes in skeletal muscle protein synthesis and their contribution to hypertrophy. Sports Med 45: 801–807, 2015. doi: 10.1007/s40279-015-0320-0. [DOI] [PubMed] [Google Scholar]
- 306. Mitchell CJ, Churchward-Venne TA, Cameron-Smith D, Phillips SM. What is the relationship between the acute muscle protein synthesis response and changes in muscle mass? J Appl Physiol (1985) 118: 495–497, 2015. doi: 10.1152/japplphysiol.00609.2014. [DOI] [PubMed] [Google Scholar]
- 307. Kuang J, McGinley C, Lee MJ, Saner NJ, Garnham A, Bishop DJ. Interpretation of exercise-induced changes in human skeletal muscle mRNA expression depends on the timing of the post-exercise biopsies. PeerJ 10: e12856, 2022. doi: 10.7717/peerj.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Louis E, Raue U, Yang Y, Jemiolo B, Trappe S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol (1985) 103: 1744–1751, 2007. doi: 10.1152/japplphysiol.00679.2007. [DOI] [PubMed] [Google Scholar]
- 309. Mitchell CJ, Churchward-Venne TA, Parise G, Bellamy L, Baker SK, Smith K, Atherton PJ, Phillips SM. Acute post-exercise myofibrillar protein synthesis is not correlated with resistance training-induced muscle hypertrophy in young men. PLoS One 9: e89431, 2014. doi: 10.1371/journal.pone.0089431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Damas F, Phillips SM, Libardi CA, Vechin FC, Lixandrão ME, Jannig PR, Costa LA, Bacurau AV, Snijders T, Parise G, Tricoli V, Roschel H, Ugrinowitsch C. Resistance training-induced changes in integrated myofibrillar protein synthesis are related to hypertrophy only after attenuation of muscle damage. J Physiol 594: 5209–5222, 2016. doi: 10.1113/JP272472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Abou Sawan S, Hodson N, Malowany JM, West DW, Tinline-Goodfellow C, Brook MS, Smith K, Atherton PJ, Kumbhare D, Moore DR. Trained integrated postexercise myofibrillar protein synthesis rates correlate with hypertrophy in young males and females. Med Sci Sports Exerc 54: 953–964, 2022. doi: 10.1249/MSS.0000000000002878. [DOI] [PubMed] [Google Scholar]
- 312. Wang LC, Kernell D. Proximo-distal organization and fibre type regionalization in rat hindlimb muscles. J Muscle Res Cell Motil 21: 587–598, 2000. doi: 10.1023/a:1026584307999. [DOI] [PubMed] [Google Scholar]
- 313. Horwath O, Envall H, Röja J, Emanuelsson EB, Sanz G, Ekblom B, Apró W, Moberg M. Variability in vastus lateralis fiber type distribution, fiber size, and myonuclear content along and between the legs. J Appl Physiol (1985) 131: 158–173, 2021. doi: 10.1152/japplphysiol.00053.2021. [DOI] [PubMed] [Google Scholar]
- 314. Gordon EE. Anatomical and biochemical adaptations of muscle to different exercises. JAMA 201: 755–758, 1967. [PubMed] [Google Scholar]
- 315. Franchi MV, Longo S, Mallinson J, Quinlan JI, Taylor T, Greenhaff PL, Narici MV. Muscle thickness correlates to muscle cross-sectional area in the assessment of strength training-induced hypertrophy. Scand J Med Sci Sports 28: 846–853, 2018. doi: 10.1111/sms.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Tavoian D, Ampomah K, Amano S, Law TD, Clark BC. Changes in DXA-derived lean mass and MRI-derived cross-sectional area of the thigh are modestly associated. Sci Rep 9: 10028, 2019. doi: 10.1038/s41598-019-46428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Ruple BA, Smith MA, Osburn SC, Sexton CL, Godwin JS, Edison JL, Poole CN, Stock MS, Fruge AD, Young KC, Roberts MD. Comparisons between skeletal muscle imaging techniques and histology in tracking midthigh hypertrophic adaptations following 10 weeks of resistance training. J Appl Physiol (1985) 133: 416–425, 2022. doi: 10.1152/japplphysiol.00219.2022. [DOI] [PubMed] [Google Scholar]
- 318. Kummitha CM, Kalhan SC, Saidel GM, Lai N. Relating tissue/organ energy expenditure to metabolic fluxes in mouse and human: experimental data integrated with mathematical modeling. Physiol Rep 2: e12159, 2014. doi: 10.14814/phy2.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Garlick PJ, Maltin CA, Baillie AG, Delday MI, Grubb DA. Fiber-type composition of nine rat muscles. II. Relationship to protein turnover. Am J Physiol Endocrinol Metab 257: E828–E832, 1989. doi: 10.1152/ajpendo.1989.257.6.E828. [DOI] [PubMed] [Google Scholar]
- 320. Koopman R, Gleeson BG, Gijsen AP, Groen B, Senden JM, Rennie MJ, van Loon LJ. Post-exercise protein synthesis rates are only marginally higher in type I compared with type II muscle fibres following resistance-type exercise. Eur J Appl Physiol 111: 1871–1878, 2011. doi: 10.1007/s00421-010-1808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Lowe DA, Alway SE. Animal models for inducing muscle hypertrophy: are they relevant for clinical applications in humans? J Orthop Sports Phys Ther 32: 36–43, 2002. doi: 10.2519/jospt.2002.32.2.36. [DOI] [PubMed] [Google Scholar]
- 322. Booth FW, Thomason DB. Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol Rev 71: 541–585, 1991. doi: 10.1152/physrev.1991.71.2.541. [DOI] [PubMed] [Google Scholar]
- 323. Murach KA, White SH, Wen Y, Ho A, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Differential requirement for satellite cells during overload-induced muscle hypertrophy in growing versus mature mice. Skelet Muscle 7: 14, 2017. doi: 10.1186/s13395-017-0132-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, , et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562, 2002. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 325. Breschi A, Gingeras TR, Guigó R. Comparative transcriptomics in human and mouse. Nat Rev Genet 18: 425–440, 2017. doi: 10.1038/nrg.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, , et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364, 2014. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Hughes DC, Hardee JP, Waddell DS, Goodman CA. CORP: gene delivery into murine skeletal muscle using in vivo electroporation. J Appl Physiol (1985) 133: 41–59, 2022. doi: 10.1152/japplphysiol.00088.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270: 50–51, 1995. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 329. Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21: 183–203, 2020. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol 55: 48–51, 1977. doi: 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- 331. Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 28: 721–726, 1975. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 332. Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78: 35–43, 1994. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 333. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 149: 274–293, 2012. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Yoon MS. mTOR as a key regulator in maintaining skeletal muscle mass. Front Physiol 8: 788, 2017. doi: 10.3389/fphys.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Marabita M, Baraldo M, Solagna F, Ceelen JJ, Sartori R, Nolte H, Nemazanyy I, Pyronnet S, Kruger M, Pende M, Blaauw B. S6K1 is required for increasing skeletal muscle force during hypertrophy. Cell Rep 17: 501–513, 2016. doi: 10.1016/j.celrep.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 336. Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 14: 133–139, 2013. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Jacobs BL, You JS, Frey JW, Goodman CA, Gundermann DM, Hornberger TA. Eccentric contractions increase the phosphorylation of tuberous sclerosis complex-2 (TSC2) and alter the targeting of TSC2 and the mechanistic target of rapamycin to the lysosome. J Physiol 591: 4611–4620, 2013. doi: 10.1113/jphysiol.2013.256339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. D’Lugos AC, Patel SH, Ormsby JC, Curtis DP, Fry CS, Carroll CC, Dickinson JM. Prior acetaminophen consumption impacts the early adaptive cellular response of human skeletal muscle to resistance exercise. J Appl Physiol (1985) 124: 1012–1024, 2018. doi: 10.1152/japplphysiol.00922.2017. [DOI] [PubMed] [Google Scholar]
- 339. Hodson N, Mazzulla M, Holowaty MN, Kumbhare D, Moore DR. RPS6 phosphorylation occurs to a greater extent in the periphery of human skeletal muscle fibers, near focal adhesions, after anabolic stimuli. Am J Physiol Cell Physiol 322: C94–C110, 2022. doi: 10.1152/ajpcell.00357.2021. [DOI] [PubMed] [Google Scholar]
- 340. Hornberger TA, McLoughlin TJ, Leszczynski JK, Armstrong DD, Jameson RR, Bowen PE, Hwang ES, Hou H, Moustafa ME, Carlson BA, Hatfield DL, Diamond AM, Esser KA. Selenoprotein-deficient transgenic mice exhibit enhanced exercise-induced muscle growth. J Nutr 133: 3091–3097, 2003. doi: 10.1093/jn/133.10.3091. [DOI] [PubMed] [Google Scholar]
- 341. Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci USA 99: 9213–9218, 2002. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS, Hornberger TA. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol 589: 5485–5501, 2011. doi: 10.1113/jphysiol.2011.218255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. You JS, McNally RM, Jacobs BL, Privett RE, Gundermann DM, Lin KH, Steinert ND, Goodman CA, Hornberger TA. The role of raptor in the mechanical load-induced regulation of mTOR signaling, protein synthesis, and skeletal muscle hypertrophy. FASEB J 33: 4021–4034, 2019. doi: 10.1096/fj.201801653RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Bentzinger CF, Lin S, Romanino K, Castets P, Guridi M, Summermatter S, Handschin C, Tintignac LA, Hall MN, Ruegg MA. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skelet Muscle 3: 6, 2013. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Haun CT, Mumford PW, Roberson PA, Romero MA, Mobley CB, Kephart WC, Anderson RG, Colquhoun RJ, Muddle TW, Luera MJ, Mackey CS, Pascoe DD, Young KC, Martin JS, DeFreitas JM, Jenkins ND, Roberts MD. Molecular, neuromuscular, and recovery responses to light versus heavy resistance exercise in young men. Physiol Rep 5: e13457, 2017. doi: 10.14814/phy2.13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Mitchell CJ, Churchward-Venne TA, West DW, Burd NA, Breen L, Baker SK, Phillips SM. Resistance exercise load does not determine training-mediated hypertrophic gains in young men. J Appl Physiol (1985) 113: 71–77, 2012. doi: 10.1152/japplphysiol.00307.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Reidy PT, Walker DK, Dickinson JM, Gundermann DM, Drummond MJ, Timmerman KL, Fry CS, Borack MS, Cope MB, Mukherjea R, Jennings K, Volpi E, Rasmussen BB. Protein blend ingestion following resistance exercise promotes human muscle protein synthesis. J Nutr 143: 410–416, 2013. doi: 10.3945/jn.112.168021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Gundermann DM, Walker DK, Reidy PT, Borack MS, Dickinson JM, Volpi E, Rasmussen BB. Activation of mTORC1 signaling and protein synthesis in human muscle following blood flow restriction exercise is inhibited by rapamycin. Am J Physiol Endocrinol Metab 306: E1198–E1204, 2014. doi: 10.1152/ajpendo.00600.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Blomstrand E, Eliasson J, Karlsson HK, Köhnke R. Branched-chain amino acids activate key enzymes in protein synthesis after physical exercise. J Nutr 136: 269S–273S, 2006. doi: 10.1093/jn/136.1.269S. [DOI] [PubMed] [Google Scholar]
- 350. Gordon BS, Liu C, Steiner JL, Nader GA, Jefferson LS, Kimball SR. Loss of REDD1 augments the rate of the overload-induced increase in muscle mass. Am J Physiol Regul Integr Comp Physiol 311: R545–R557, 2016. doi: 10.1152/ajpregu.00159.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Martin TD, Dennis MD, Gordon BS, Kimball SR, Jefferson LS. mTORC1 and JNK coordinate phosphorylation of the p70S6K1 autoinhibitory domain in skeletal muscle following functional overloading. Am J Physiol Endocrinol Metab 306: E1397–E1405, 2014. doi: 10.1152/ajpendo.00064.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Gordon BS, Steiner JL, Lang CH, Jefferson LS, Kimball SR. Reduced REDD1 expression contributes to activation of mTORC1 following electrically induced muscle contraction. Am J Physiol Endocrinol Metab 307: E703–E711, 2014. doi: 10.1152/ajpendo.00250.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Szewczyk NJ, Greenhaff PL, Smith K, Atherton PJ. Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide-derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J 29: 4485–4496, 2015. doi: 10.1096/fj.15-273755. [DOI] [PubMed] [Google Scholar]
- 354. Chalé-Rush A, Morris EP, Kendall TL, Brooks NE, Fielding RA. Effects of chronic overload on muscle hypertrophy and mTOR signaling in young adult and aged rats. J Gerontol A Biol Sci Med Sci 64: 1232–1239, 2009. doi: 10.1093/gerona/glp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Hamilton DL, Philp A, MacKenzie MG, Patton A, Towler MC, Gallagher IJ, Bodine SC, Baar K. Molecular brakes regulating mTORC1 activation in skeletal muscle following synergist ablation. Am J Physiol Endocrinol Metab 307: E365–E373, 2014. doi: 10.1152/ajpendo.00674.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. May AK, Russell AP, Della Gatta PA, Warmington SA. Muscle adaptations to heavy-load and blood flow restriction resistance training methods. Front Physiol 13: 837697, 2022. doi: 10.3389/fphys.2022.837697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Bolster DR, Kubica N, Crozier SJ, Williamson DL, Farrell PA, Kimball SR, Jefferson LS. Immediate response of mammalian target of rapamycin (mTOR)-mediated signalling following acute resistance exercise in rat skeletal muscle. J Physiol 553: 213–220, 2003. doi: 10.1113/jphysiol.2003.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bepsilon mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem 280: 7570–7580, 2005. doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- 359. Agergaard J, Bülow J, Jensen JK, Reitelseder S, Drummond MJ, Schjerling P, Scheike T, Serena A, Holm L. Light-load resistance exercise increases muscle protein synthesis and hypertrophy signaling in elderly men. Am J Physiol Endocrinol Metab 312: E326–E338, 2017. doi: 10.1152/ajpendo.00164.2016. [DOI] [PubMed] [Google Scholar]
- 360. Eliasson J, Elfegoun T, Nilsson J, Köhnke R, Ekblom B, Blomstrand E. Maximal lengthening contractions increase p70 S6 kinase phosphorylation in human skeletal muscle in the absence of nutritional supply. Am J Physiol Endocrinol Metab 291: E1197–E1205, 2006. doi: 10.1152/ajpendo.00141.2006. [DOI] [PubMed] [Google Scholar]
- 361. Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol 587: 1535–1546, 2009. doi: 10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Kumar V, Selby A, Rankin D, Patel R, Atherton P, Hildebrandt W, Williams J, Smith K, Seynnes O, Hiscock N, Rennie MJ. Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. J Physiol 587: 211–217, 2009. doi: 10.1113/jphysiol.2008.164483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Moore D, Atherton P, Rennie M, Tarnopolsky M, Phillips S. Resistance exercise enhances mTOR and MAPK signalling in human muscle over that seen at rest after bolus protein ingestion. Acta Physiol (Oxf) 201: 365–372, 2011. doi: 10.1111/j.1748-1716.2010.02187.x. [DOI] [PubMed] [Google Scholar]
- 364. Edman S, Söderlund K, Moberg M, Apró W, Blomstrand E. mTORC1 signaling in individual human muscle fibers following resistance exercise in combination with intake of essential amino acids. Front Nutr 6: 96, 2019. doi: 10.3389/fnut.2019.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Deldicque L, Atherton P, Patel R, Theisen D, Nielens H, Rennie MJ, Francaux M. Decrease in Akt/PKB signalling in human skeletal muscle by resistance exercise. Eur J Appl Physiol 104: 57–65, 2008. doi: 10.1007/s00421-008-0786-7. [DOI] [PubMed] [Google Scholar]
- 366. Dreyer HC, Fujita S, Glynn EL, Drummond MJ, Volpi E, Rasmussen BB. Resistance exercise increases leg muscle protein synthesis and mTOR signalling independent of sex. Acta Physiol (Oxf) 199: 71–81, 2010. doi: 10.1111/j.1748-1716.2010.02074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Sedliak M, Zeman M, Buzgó G, Cvečka J, Hamar D, Laczo E, Zelko A, Okuliarová M, Raastad T, Nilsen TS, Kyröläinen H, Häkkinen K, Ahtiainen JP, Hulmi JJ. Effects of time of day on resistance exercise-induced anabolic signaling in skeletal muscle. Biol Rhythm Res 44: 756–770, 2013. doi: 10.1080/09291016.2012.740314. [DOI] [Google Scholar]
- 368. Fujita S, Abe T, Drummond MJ, Cadenas JG, Dreyer HC, Sato Y, Volpi E, Rasmussen BB. Blood flow restriction during low-intensity resistance exercise increases S6K1 phosphorylation and muscle protein synthesis. J Appl Physiol (1985) 103: 903–910, 2007. doi: 10.1152/japplphysiol.00195.2007. [DOI] [PubMed] [Google Scholar]
- 369. Mascher H, Tannerstedt J, Brink-Elfegoun T, Ekblom B, Gustafsson T, Blomstrand E. Repeated resistance exercise training induces different changes in mRNA expression of MAFbx and MuRF-1 in human skeletal muscle. Am J Physiol Endocrinol Metab 294: E43–E51, 2008. doi: 10.1152/ajpendo.00504.2007. [DOI] [PubMed] [Google Scholar]
- 370. Fernandez‐Gonzalo R, Lundberg TR, Tesch PA. Acute molecular responses in untrained and trained muscle subjected to aerobic and resistance exercise training versus resistance training alone. Acta Physiol (Oxf) 209: 283–294, 2013. doi: 10.1111/apha.12174. [DOI] [PubMed] [Google Scholar]
- 371. Langer HT, West D, Senden J, Spuler S, van Loon LJ, Baar K. Myofibrillar protein synthesis rates are increased in chronically exercised skeletal muscle despite decreased anabolic signaling. Sci Rep 12: 7553, 2022. doi: 10.1038/s41598-022-11621-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem 280: 32081–32089, 2005. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- 373. Roberts MD, Dalbo VJ, Sunderland KL, Poole CN, Hassell SE, Bemben D, Cramer J, Stout J, Kerksick CM. IGF-1 splice variant and IGF-1 peptide expression patterns in young and old human skeletal muscle prior to and following sequential exercise bouts. Eur J Appl Physiol 110: 961–969, 2010. doi: 10.1007/s00421-010-1588-2. [DOI] [PubMed] [Google Scholar]
- 374. Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J Physiol 547: 247–254, 2003. doi: 10.1113/jphysiol.2002.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Greig CA, Hameed M, Young A, Goldspink G, Noble B. Skeletal muscle IGF-I isoform expression in healthy women after isometric exercise. Growth Horm IGF Res 16: 373–376, 2006. doi: 10.1016/j.ghir.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 376. Philippou A, Papageorgiou E, Bogdanis G, Halapas A, Sourla A, Maridaki M, Pissimissis N, Koutsilieris M. Expression of IGF-1 isoforms after exercise-induced muscle damage in humans: characterization of the MGF E peptide actions in vitro. In Vivo 23: 567–575, 2009. [PubMed] [Google Scholar]
- 377. Bamman MM, Petrella JK, Kim JS, Mayhew DL, Cross JM. Cluster analysis tests the importance of myogenic gene expression during myofiber hypertrophy in humans. J Appl Physiol (1985) 102: 2232–2239, 2007. doi: 10.1152/japplphysiol.00024.2007. [DOI] [PubMed] [Google Scholar]
- 378. Adams GR, Haddad F. The relationships among IGF-1, DNA content, and protein accumulation during skeletal muscle hypertrophy. J Appl Physiol (1985) 81: 2509–2516, 1996. doi: 10.1152/jappl.1996.81.6.2509. [DOI] [PubMed] [Google Scholar]
- 379. Wilborn CD, Taylor LW, Greenwood M, Kreider RB, Willoughby DS. Effects of different intensities of resistance exercise on regulators of myogenesis. J Strength Cond Res 23: 2179–2187, 2009. doi: 10.1519/JSC.0b013e3181bab493. [DOI] [PubMed] [Google Scholar]
- 380. Adams GR, Haddad F, Baldwin KM. Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. J Appl Physiol (1985) 87: 1705–1712, 1999. doi: 10.1152/jappl.1999.87.5.1705. [DOI] [PubMed] [Google Scholar]
- 381. Garma T, Kobayashi C, Haddad F, Adams GR, Bodell PW, Baldwin KM. Similar acute molecular responses to equivalent volumes of isometric, lengthening, or shortening mode resistance exercise. J Appl Physiol (1985) 102: 135–143, 2007. doi: 10.1152/japplphysiol.00776.2006. [DOI] [PubMed] [Google Scholar]
- 382. Haddad F, Adams GR. Aging-sensitive cellular and molecular mechanisms associated with skeletal muscle hypertrophy. J Appl Physiol (1985) 100: 1188–1203, 2006. doi: 10.1152/japplphysiol.01227.2005. [DOI] [PubMed] [Google Scholar]
- 383. Philippou A, Maridaki M, Halapas A, Koutsilieris M. The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In Vivo 21: 45–54, 2007. [PubMed] [Google Scholar]
- 384. Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle 1: 4, 2011. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Schoenfeld BJ. The mechanisms of muscle hypertrophy and their application to resistance training. J Strength Cond Res 24: 2857–2872, 2010. doi: 10.1519/JSC.0b013e3181e840f3. [DOI] [PubMed] [Google Scholar]
- 386. Adams GR. Role of insulin-like growth factor-I in the regulation of skeletal muscle adaptation to increased loading. Exerc Sport Sci Rev 26: 31–60, 1998. [PubMed] [Google Scholar]
- 387. Philp A, Hamilton DL, Baar K. Signals mediating skeletal muscle remodeling by resistance exercise: PI3-kinase independent activation of mTORC1. J Appl Physiol (1985) 110: 561–568, 2011. doi: 10.1152/japplphysiol.00941.2010. [DOI] [PubMed] [Google Scholar]
- 388. Hornberger TA, Stuppard R, Conley KE, Fedele MJ, Fiorotto ML, Chin ER, Esser KA. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem J 380: 795–804, 2004. doi: 10.1042/BJ20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Spangenburg EE, Le Roith D, Ward CW, Bodine SC. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. J Physiol 586: 283–291, 2008. doi: 10.1113/jphysiol.2007.141507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Maruyama Y, Ikeda C, Wakabayashi K, Ato S, Ogasawara R. High-intensity muscle contraction-mediated increases in Akt1 and Akt2 phosphorylation do not contribute to mTORC1 activation and muscle protein synthesis. J Appl Physiol (1985) 128: 830–837, 2020. doi: 10.1152/japplphysiol.00578.2019. [DOI] [PubMed] [Google Scholar]
- 391. Miyazaki M, McCarthy JJ, Fedele MJ, Esser KA. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. J Physiol 589: 1831–1846, 2011. doi: 10.1113/jphysiol.2011.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Jaiswal N, Gavin M, Loro E, Sostre-Colón J, Roberson PA, Uehara K, Rivera-Fuentes N, Neinast M, Arany Z, Kimball SR, Khurana TS, Titchenell PM. AKT controls protein synthesis and oxidative metabolism via combined mTORC1 and FOXO1 signalling to govern muscle physiology. J Cachexia Sarcopenia Muscle 13: 495–514, 2022. doi: 10.1002/jcsm.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Sasako T, Umehara T, Soeda K, Kaneko K, Suzuki M, Kobayashi N, Okazaki Y, Tamura-Nakano M, Chiba T, Accili D, Kahn CR, Noda T, Asahara H, Yamauchi T, Kadowaki T, Ueki K. Deletion of skeletal muscle Akt1/2 causes osteosarcopenia and reduces lifespan in mice. Nat Commun 13: 5655, 2022. doi: 10.1038/s41467-022-33008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Vandenburgh HH. Motion into mass: how does tension stimulate muscle growth? Med Sci Sports Exerc 19: S142–S149, 1987. [PubMed] [Google Scholar]
- 395. Klossner S, Durieux AC, Freyssenet D, Flueck M. Mechano-transduction to muscle protein synthesis is modulated by FAK. Eur J Appl Physiol 106: 389–398, 2009. doi: 10.1007/s00421-009-1032-7. [DOI] [PubMed] [Google Scholar]
- 396. Flück M, Hoppeler H. Molecular basis of skeletal muscle plasticity–from gene to form and function. Rev Physiol Biochem Pharmacol 146: 159–216, 2003. doi: 10.1007/s10254-002-0004-7. [DOI] [PubMed] [Google Scholar]
- 397. Durieux AC, Desplanches D, Freyssenet D, Flück M. Mechanotransduction in striated muscle via focal adhesion kinase. Biochem Soc Trans 35: 1312–1313, 2007. doi: 10.1042/BST0351312. [DOI] [PubMed] [Google Scholar]
- 398. Hornberger TA. Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle. Int J Biochem Cell Biol 43: 1267–1276, 2011. doi: 10.1016/j.biocel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Hornberger TA, Esser KA. Mechanotransduction and the regulation of protein synthesis in skeletal muscle. Proc Nutr Soc 63: 331–335, 2004. doi: 10.1079/PNS2004357. [DOI] [PubMed] [Google Scholar]
- 400. You JS, Lincoln HC, Kim CR, Frey JW, Goodman CA, Zhong XP, Hornberger TA. The role of diacylglycerol kinase zeta and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy. J Biol Chem 289: 1551–1563, 2014. doi: 10.1074/jbc.M113.531392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. O’Neil TK, Duffy LR, Frey JW, Hornberger TA. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J Physiol 587: 3691–3701, 2009. doi: 10.1113/jphysiol.2009.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402. Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci USA 103: 4741–4746, 2006. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. You JS, Frey JW, Hornberger TA. Mechanical stimulation induces mTOR signaling via an ERK-independent mechanism: implications for a direct activation of mTOR by phosphatidic acid. PLoS One 7: e47258, 2012. doi: 10.1371/journal.pone.0047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404. You JS, Dooley MS, Kim CR, Kim EJ, Xu W, Goodman CA, Hornberger TA. A DGKzeta-FoxO-ubiquitin proteolytic axis controls fiber size during skeletal muscle remodeling. Sci Signal 11: eaao6847, 2018. doi: 10.1126/scisignal.aao6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Flück M, Carson JA, Gordon SE, Ziemiecki A, Booth FW. Focal adhesion proteins FAK and paxillin increase in hypertrophied skeletal muscle. Am J Physiol Cell Physiol 277: C152–C162, 1999. doi: 10.1152/ajpcell.1999.277.1.C152. [DOI] [PubMed] [Google Scholar]
- 406. Crossland H, Kazi AA, Lang CH, Timmons JA, Pierre P, Wilkinson DJ, Smith K, Szewczyk NJ, Atherton PJ. Focal adhesion kinase is required for IGF-I-mediated growth of skeletal muscle cells via a TSC2/mTOR/S6K1-associated pathway. Am J Physiol Endocrinol Metab 305: E183–E193, 2013. doi: 10.1152/ajpendo.00541.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol (1985) 115: 1065–1074, 2013. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci 116: 1409–1416, 2003. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 409. Paul R, Luo M, Mo X, Lu J, Yeo SK, Guan JL. FAK activates AKT-mTOR signaling to promote the growth and progression of MMTV-Wnt1-driven basal-like mammary tumors. Breast Cancer Res 22: 59, 2020. doi: 10.1186/s13058-020-01298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Lee FY, Zhen YY, Yuen CM, Fan R, Chen YT, Sheu JJ, Chen YL, Wang CJ, Sun CK, Yip HK. The mTOR-FAK mechanotransduction signaling axis for focal adhesion maturation and cell proliferation. Am J Transl Res 9: 1603–1617, 2017. [PMC free article] [PubMed] [Google Scholar]
- 411. Gan B, Yoo Y, Guan JL. Association of focal adhesion kinase with tuberous sclerosis complex 2 in the regulation of s6 kinase activation and cell growth. J Biol Chem 281: 37321–37329, 2006. doi: 10.1074/jbc.M605241200. [DOI] [PubMed] [Google Scholar]
- 412. Boppart MD, Burkin DJ, Kaufman SJ. Alpha7beta1-integrin regulates mechanotransduction and prevents skeletal muscle injury. Am J Physiol Cell Physiol 290: C1660–C1665, 2006. doi: 10.1152/ajpcell.00317.2005. [DOI] [PubMed] [Google Scholar]
- 413. Zou K, Meador BM, Johnson B, Huntsman HD, Mahmassani Z, Valero MC, Huey KA, Boppart MD. The alpha7beta1-integrin increases muscle hypertrophy following multiple bouts of eccentric exercise. J Appl Physiol (1985) 111: 1134–1141, 2011. doi: 10.1152/japplphysiol.00081.2011. [DOI] [PubMed] [Google Scholar]
- 414. Petrosino JM, Longenecker JZ, Angell CD, Hinger SA, Martens CR, Accornero F. CCN2 participates in overload-induced skeletal muscle hypertrophy. Matrix Biol 106: 1–11, 2022. doi: 10.1016/j.matbio.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415. Glover EI, Oates BR, Tang JE, Moore DR, Tarnopolsky MA, Phillips SM. Resistance exercise decreases eIF2Bepsilon phosphorylation and potentiates the feeding-induced stimulation of p70S6K1 and rpS6 in young men. Am J Physiol Regul Integr Comp Physiol 295: R604–R610, 2008. doi: 10.1152/ajpregu.00097.2008. [DOI] [PubMed] [Google Scholar]
- 416. Eftestøl E, Franchi MV, Kasper S, Flück M. JNK activation in TA and EDL muscle is load-dependent in rats receiving identical excitation patterns. Sci Rep 11: 16405, 2021. doi: 10.1038/s41598-021-94930-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Franchi MV, Ruoss S, Valdivieso P, Mitchell KW, Smith K, Atherton PJ, Narici MV, Flück M. Regional regulation of focal adhesion kinase after concentric and eccentric loading is related to remodelling of human skeletal muscle. Acta Physiol (Oxf) 223: e13056, 2018. doi: 10.1111/apha.13056. [DOI] [PubMed] [Google Scholar]
- 418. van der Pijl R, Strom J, Conijn S, Lindqvist J, Labeit S, Granzier H, Ottenheijm C. Titin-based mechanosensing modulates muscle hypertrophy. J Cachexia Sarcopenia Muscle 9: 947–961, 2018. doi: 10.1002/jcsm.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Spangenburg EE, McBride TA. Inhibition of stretch-activated channels during eccentric muscle contraction attenuates p70S6K activation. J Appl Physiol (1985) 100: 129–135, 2006. doi: 10.1152/japplphysiol.00619.2005. [DOI] [PubMed] [Google Scholar]
- 420. Tyganov S, Mirzoev T, Shenkman B. An anabolic signaling response of rat soleus muscle to eccentric contractions following hindlimb unloading: a potential role of stretch-activated ion channels. Int J Mol Sci 20: 1165, 2019. doi: 10.3390/ijms20051165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Li RJ, Xu J, Fu C, Zhang J, Zheng YG, Jia H, Liu JO. Regulation of mTORC1 by lysosomal calcium and calmodulin. Elife 5: e19360, 2016. doi: 10.7554/eLife.19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Bravo-Sagua R, Parra V, Muñoz-Cordova F, Sanchez-Aguilera P, Garrido V, Contreras-Ferrat A, Chiong M, Lavandero S. Sarcoplasmic reticulum and calcium signaling in muscle cells: Homeostasis and disease. Int Rev Cell Mol Biol 350: 197–264, 2020. doi: 10.1016/bs.ircmb.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 423. Ito N, Ruegg UT, Kudo A, Miyagoe-Suzuki Y, Takeda S. Activation of calcium signaling through Trpv1 by nNOS and peroxynitrite as a key trigger of skeletal muscle hypertrophy. Nat Med 19: 101–106, 2013. doi: 10.1038/nm.3019. [DOI] [PubMed] [Google Scholar]
- 424. Ito N, Ruegg UT, Kudo A, Miyagoe-Suzuki Y, Takeda S. Capsaicin mimics mechanical load-induced intracellular signaling events: involvement of TRPV1-mediated calcium signaling in induction of skeletal muscle hypertrophy. Channels (Austin) 7: 221–224, 2013. doi: 10.4161/chan.24583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Ito N, Ruegg UT, Takeda S. ATP-induced increase in intracellular calcium levels and subsequent activation of mTOR as regulators of skeletal muscle hypertrophy. Int J Mol Sci 19: 2804, 2018. doi: 10.3390/ijms19092804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Dunn SE, Burns JL, Michel RN. Calcineurin is required for skeletal muscle hypertrophy. J Biol Chem 274: 21908–21912, 1999. doi: 10.1074/jbc.274.31.21908. [DOI] [PubMed] [Google Scholar]
- 427. Semsarian C, Wu MJ, Ju YK, Marciniec T, Yeoh T, Allen DG, Harvey RP, Graham RM. Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature 400: 576–581, 1999. doi: 10.1038/23054. [DOI] [PubMed] [Google Scholar]
- 428. Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilson JR, Liberatore CM, Yutzey KE, Crabtree GR, Tsika RW, Molkentin JD. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. J Biol Chem 279: 26192–26200, 2004. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- 429. Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lømo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci USA 98: 13108–13113, 2001. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430. Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI3K/Akt/mTOR and PI3K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009–1013, 2001. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 431. Ferey JL, Brault JJ, Smith CA, Witczak CA. Constitutive activation of CaMKKalpha signaling is sufficient but not necessary for mTORC1 activation and growth in mouse skeletal muscle. Am J Physiol Endocrinol Metab 307: E686–E694, 2014. doi: 10.1152/ajpendo.00322.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 21: 362–369, 2006. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 433. Yang Y, Breen L, Burd NA, Hector AJ, Churchward-Venne TA, Josse AR, Tarnopolsky MA, Phillips SM. Resistance exercise enhances myofibrillar protein synthesis with graded intakes of whey protein in older men. Br J Nutr 108: 1780–1788, 2012. doi: 10.1017/S0007114511007422. [DOI] [PubMed] [Google Scholar]
- 434. Tang JE, Moore DR, Kujbida GW, Tarnopolsky MA, Phillips SM. Ingestion of whey hydrolysate, casein, or soy protein isolate: effects on mixed muscle protein synthesis at rest and following resistance exercise in young men. J Appl Physiol (1985) 107: 987–992, 2009. doi: 10.1152/japplphysiol.00076.2009. [DOI] [PubMed] [Google Scholar]
- 435. D’Souza RF, Markorth JF, Figueiredo VC, Della Gatta PA, Petersen AC, Mitchell CJ, Cameron-Smith D. Dose-dependent increases in p70S6K phosphorylation and intramuscular branched-chain amino acids in older men following resistance exercise and protein intake. Physiol Rep 2: e12112, 2014. doi: 10.14814/phy2.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436. MacKenzie MG, Hamilton DL, Murray JT, Taylor PM, Baar K. mVps34 is activated following high-resistance contractions. J Physiol 587: 253–260, 2009. doi: 10.1113/jphysiol.2008.159830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. Hodson N, Brown T, Joanisse S, Aguirre N, West DWD, Moore DR, Baar K, Breen L, Philp A. Characterisation of L-Type Amino Acid Transporter 1 (LAT1) expression in human skeletal muscle by immunofluorescent microscopy. Nutrients 10: 23, 2017., doi: 10.3390/nu10010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 412: 179–190, 2008. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Jacobs BL, McNally RM, Kim KJ, Blanco R, Privett RE, You JS, Hornberger TA. Identification of mechanically regulated phosphorylation sites on tuberin (TSC2) that control mechanistic target of rapamycin (mTOR) signaling. J Biol Chem 292: 6987–6997, 2017. doi: 10.1074/jbc.M117.777805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440. Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, Tanti JF, Giorgetti-Peraldi S, Bost F. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res 71: 4366–4372, 2011. doi: 10.1158/0008-5472.CAN-10-1769. [DOI] [PubMed] [Google Scholar]
- 441. Drummond MJ, Fujita S, Abe T, Dreyer HC, Volpi E, Rasmussen BB. Human muscle gene expression following resistance exercise and blood flow restriction. Med Sci Sports Exerc 40: 691–698, 2008. doi: 10.1249/MSS.0b013e318160ff84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442. Etheridge T, Atherton PJ, Wilkinson D, Selby A, Rankin D, Webborn N, Smith K, Watt PW. Effects of hypoxia on muscle protein synthesis and anabolic signaling at rest and in response to acute resistance exercise. Am J Physiol Endocrinol Metab 301: E697–E702, 2011. doi: 10.1152/ajpendo.00276.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443. Tabbaa M, Ruz Gomez T, Campelj DG, Gregorevic P, Hayes A, Goodman CA. The regulation of polyamine pathway proteins in models of skeletal muscle hypertrophy and atrophy: a potential role for mTORC1. Am J Physiol Cell Physiol 320: C987–C999, 2021. doi: 10.1152/ajpcell.00078.2021. [DOI] [PubMed] [Google Scholar]
- 444. Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc Natl Acad Sci USA 110: 2169–2174, 2013. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445. Tang H, Inoki K, Brooks SV, Okazawa H, Lee M, Wang J, Kim M, Kennedy CL, Macpherson PC, Ji X, Van Roekel S, Fraga DA, Wang K, Zhu J, Wang Y, Sharp ZD, Miller RA, Rando TA, Goldman D, Guan KL, Shrager JB. mTORC1 underlies age-related muscle fiber damage and loss by inducing oxidative stress and catabolism. Aging Cell 18: e12943, 2019. doi: 10.1111/acel.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446. West DW, Baehr LM, Marcotte GR, Chason CM, Tolento L, Gomes AV, Bodine SC, Baar K. Acute resistance exercise activates rapamycin-sensitive and -insensitive mechanisms that control translational activity and capacity in skeletal muscle. J Physiol 594: 453–468, 2016. doi: 10.1113/JP271365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Ogasawara R, Suginohara T. Rapamycin-insensitive mechanistic target of rapamycin regulates basal and resistance exercise-induced muscle protein synthesis. FASEB J 32: 5824–5834, 2018., doi: 10.1096/fj.201701422R. [DOI] [PubMed] [Google Scholar]
- 448. Ogasawara R, Knudsen JR, Li J, Ato S, Jensen TE. Rapamycin and mTORC2 inhibition synergistically reduce contraction-stimulated muscle protein synthesis. J Physiol 598: 5453–5466, 2020. doi: 10.1113/JP280528. [DOI] [PubMed] [Google Scholar]
- 449. Goodman CA, Dietz JM, Jacobs BL, McNally RM, You JS, Hornberger TA. Yes-Associated Protein is up-regulated by mechanical overload and is sufficient to induce skeletal muscle hypertrophy. FEBS Lett 589: 1491–1497, 2015. doi: 10.1016/j.febslet.2015.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450. Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal 7: re4, 2014. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- 451. Gnimassou O, Francaux M, Deldicque L. Hippo pathway and skeletal muscle mass regulation in mammals: a controversial relationship. Front Physiol 8: 190, 2017. doi: 10.3389/fphys.2017.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452. Judson RN, Gray SR, Walker C, Carroll AM, Itzstein C, Lionikas A, Zammit PS, De Bari C, Wackerhage H. Constitutive expression of Yes-associated protein (Yap) in adult skeletal muscle fibres induces muscle atrophy and myopathy. PLoS One 8: e59622, 2013. doi: 10.1371/journal.pone.0059622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453. Haddad F, Adams GR. Inhibition of MAP/ERK kinase prevents IGF-I-induced hypertrophy in rat muscles. J Appl Physiol (1985) 96: 203–210, 2004. doi: 10.1152/japplphysiol.00856.2003. [DOI] [PubMed] [Google Scholar]
- 454. Winter JN, Jefferson LS, Kimball SR. ERK and Akt signaling pathways function through parallel mechanisms to promote mTORC1 signaling. Am J Physiol Cell Physiol 300: C1172–C1180, 2011. doi: 10.1152/ajpcell.00504.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455. Carlson CJ, Fan Z, Gordon SE, Booth FW. Time course of the MAPK and PI3-kinase response within 24 h of skeletal muscle overload. J Appl Physiol (1985) 91: 2079–2087, 2001. doi: 10.1152/jappl.2001.91.5.2079. [DOI] [PubMed] [Google Scholar]
- 456. Nordgaard C, Vind AC, Stonadge A, Kjøbsted R, Snieckute G, Antas P, , et al. ZAKbeta is activated by cellular compression and mediates contraction-induced MAP kinase signaling in skeletal muscle. EMBO J 41: e111650, 2022. doi: 10.15252/embj.2022111650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457. Burd NA, West DW, Staples AW, Atherton PJ, Baker JM, Moore DR, Holwerda AM, Parise G, Rennie MJ, Baker SK, Phillips SM. Low-load high volume resistance exercise stimulates muscle protein synthesis more than high-load low volume resistance exercise in young men. PLoS One 5: e12033, 2010. doi: 10.1371/journal.pone.0012033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458. Galpin AJ, Fry AC, Chiu LZ, Thomason DB, Schilling BK. High-power resistance exercise induces MAPK phosphorylation in weightlifting trained men. Appl Physiol Nutr Metab 37: 80–87, 2012. doi: 10.1139/h11-131. [DOI] [PubMed] [Google Scholar]
- 459. Taylor LW, Wilborn CD, Kreider RB, Willoughby DS. Effects of resistance exercise intensity on extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase activation in men. J Strength Cond Res 26: 599–607, 2012. doi: 10.1519/JSC.0b013e318242f92d. [DOI] [PubMed] [Google Scholar]
- 460. Gonzalez AM, Hoffman JR, Townsend JR, Jajtner AR, Boone CH, Beyer KS, Baker KM, Wells AJ, Mangine GT, Robinson EH, Church DD, Oliveira LP, Fukuda DH, Stout JR. Intramuscular MAPK signaling following high volume and high intensity resistance exercise protocols in trained men. Eur J Appl Physiol 116: 1663–1670, 2016. doi: 10.1007/s00421-016-3417-8. [DOI] [PubMed] [Google Scholar]
- 461. Nicoll JX, Fry AC, Mosier EM, Olsen LA, Sontag SA. MAPK, androgen, and glucocorticoid receptor phosphorylation following high-frequency resistance exercise non-functional overreaching. Eur J Appl Physiol 119: 2237–2253, 2019. doi: 10.1007/s00421-019-04200-y. [DOI] [PubMed] [Google Scholar]
- 462. Nicoll JX, Fry AC, Galpin AJ, Sterczala AJ, Thomason DB, Moore CA, Weiss LW, Chiu LZ. Changes in resting mitogen-activated protein kinases following resistance exercise overreaching and overtraining. Eur J Appl Physiol 116: 2401–2413, 2016. doi: 10.1007/s00421-016-3492-x. [DOI] [PubMed] [Google Scholar]
- 463. Lessard SJ, MacDonald TL, Pathak P, Han MS, Coffey VG, Edge J, Rivas DA, Hirshman MF, Davis RJ, Goodyear LJ. JNK regulates muscle remodeling via myostatin/SMAD inhibition. Nat Commun 9: 3030, 2018. doi: 10.1038/s41467-018-05439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464. Hostrup M, Reitelseder S, Jessen S, Kalsen A, Nyberg M, Egelund J, Kreiberg M, Kristensen CM, Thomassen M, Pilegaard H, Backer V, Jacobson GA, Holm L, Bangsbo J. Beta2 -adrenoceptor agonist salbutamol increases protein turnover rates and alters signalling in skeletal muscle after resistance exercise in young men. J Physiol 596: 4121–4139, 2018. doi: 10.1113/JP275560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465. Ogasawara R, Fujita S, Hornberger TA, Kitaoka Y, Makanae Y, Nakazato K, Naokata I. The role of mTOR signalling in the regulation of skeletal muscle mass in a rodent model of resistance exercise. Sci Rep 6: 31142, 2016. doi: 10.1038/srep31142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Uemichi K, Shirai T, Hanakita H, Takemasa T. Effect of mechanistic/mammalian target of rapamycin complex 1 on mitochondrial dynamics during skeletal muscle hypertrophy. Physiol Rep 9: e14789, 2021. doi: 10.14814/phy2.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467. Millward DJ, Garlick PJ, James WP, Nnanyelugo DO, Ryatt JS. Relationship between protein synthesis and RNA content in skeletal muscle. Nature 241: 204–205, 1973. doi: 10.1038/241204a0. [DOI] [PubMed] [Google Scholar]
- 468. Hirsch CA. Quantitative determination of the ribosomal ribonucleic acid content of liver and Novikoff hepatoma from fed and from fasted rats. J Biol Chem 242: 2822–2827, 1967. doi: 10.1016/S0021-9258(18)99580-3. [DOI] [PubMed] [Google Scholar]
- 469. Figueiredo VC, McCarthy JJ. Regulation of ribosome biogenesis in skeletal muscle hypertrophy. Physiology (Bethesda) 34: 30–42, 2019. doi: 10.1152/physiol.00034.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Chaillou T, Kirby TJ, McCarthy JJ. Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol 229: 1584–1594, 2014. doi: 10.1002/jcp.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471. Wen Y, Alimov AP, McCarthy JJ. Ribosome biogenesis is necessary for skeletal muscle hypertrophy. Exerc Sport Sci Rev 44: 110–115, 2016. doi: 10.1249/JES.0000000000000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472. Kim HG, Guo B, Nader GA. Regulation of ribosome biogenesis during skeletal muscle hypertrophy. Exerc Sport Sci Rev 47: 91–97, 2019. doi: 10.1249/JES.0000000000000179. [DOI] [PubMed] [Google Scholar]
- 473. Wong TS, Booth FW. Skeletal muscle enlargement with weight-lifting exercise by rats. J Appl Physiol (1985) 65: 950–954, 1988. doi: 10.1152/jappl.1988.65.2.950. [DOI] [PubMed] [Google Scholar]
- 474. Mobley CB, Holland AM, Kephart WC, Mumford PW, Lowery RP, Kavazis AN, Wilson JM, Roberts MD. Progressive resistance-loaded voluntary wheel running increases hypertrophy and differentially affects muscle protein synthesis, ribosome biogenesis, and proteolytic markers in rat muscle. J Anim Physiol Anim Nutr (Berl) 102: 317–329, 2018. doi: 10.1111/jpn.12691. [DOI] [PubMed] [Google Scholar]
- 475. Kotani T, Takegaki J, Takagi R, Nakazato K, Ishii N. Consecutive bouts of electrical stimulation-induced contractions alter ribosome biogenesis in rat skeletal muscle. J Appl Physiol (1985) 126: 1673–1680, 2019. doi: 10.1152/japplphysiol.00665.2018. [DOI] [PubMed] [Google Scholar]
- 476. Nakada S, Ogasawara R, Kawada S, Maekawa T, Ishii N. Correlation between ribosome biogenesis and the magnitude of hypertrophy in overloaded skeletal muscle. PLoS One 11: e0147284, 2016. doi: 10.1371/journal.pone.0147284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477. Kirby TJ, Lee JD, England JH, Chaillou T, Esser KA, McCarthy JJ. Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J Appl Physiol (1985) 119: 321–327, 2015. doi: 10.1152/japplphysiol.00296.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478. von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol 302: C1523–C1530, 2012. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- 479. West DW, Marcotte GR, Chason CM, Juo N, Baehr LM, Bodine SC, Baar K. Normal ribosomal biogenesis but shortened protein synthetic response to acute eccentric resistance exercise in old skeletal muscle. Front Physiol 9: 1915, 2018. doi: 10.3389/fphys.2018.01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480. Lilja M, Moberg M, Apró W, Martínez-Aranda LM, Rundqvist H, Langlet B, Gustafsson T, Lundberg TR. Limited effect of over-the-counter doses of ibuprofen on mechanisms regulating muscle hypertrophy during resistance training in young adults. J Appl Physiol (1985) 134: 753–765, 2023. doi: 10.1152/japplphysiol.00698.2022. [DOI] [PubMed] [Google Scholar]
- 481. Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol 289: C1457–C1465, 2005. doi: 10.1152/ajpcell.00165.2005. [DOI] [PubMed] [Google Scholar]
- 482. McDermott PJ, Carl LL, Conner KJ, Allo SN. Transcriptional regulation of ribosomal RNA synthesis during growth of cardiac myocytes in culture. J Biol Chem 266: 4409–4416, 1991. doi: 10.1016/S0021-9258(20)64337-X. [DOI] [PubMed] [Google Scholar]
- 483. McDermott PJ, Rothblum LI, Smith SD, Morgan HE. Accelerated rates of ribosomal RNA synthesis during growth of contracting heart cells in culture. J Biol Chem 264: 18220–18227, 1989. doi: 10.1016/S0021-9258(19)84700-2. [DOI] [PubMed] [Google Scholar]
- 484. von Walden F, Liu C, Aurigemma N, Nader GA. mTOR signaling regulates myotube hypertrophy by modulating protein synthesis, rDNA transcription, and chromatin remodeling. Am J Physiol Cell Physiol 311: C663–C672, 2016. doi: 10.1152/ajpcell.00144.2016. [DOI] [PubMed] [Google Scholar]
- 485. Murach KA, Liu Z, Jude B, Figueiredo VC, Wen Y, Khadgi S, Lim S, Morena da Silva F, Greene NP, Lanner JT, McCarthy JJ, Vechetti IJ, von Walden F. Multi-transcriptome analysis following an acute skeletal muscle growth stimulus yields tools for discerning global and MYC regulatory networks. J Biol Chem 298: 102515, 2022. doi: 10.1016/j.jbc.2022.102515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486. Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, Gordon PM. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol (1985) 116: 693–702, 2014. doi: 10.1152/japplphysiol.01366.2013. [DOI] [PubMed] [Google Scholar]
- 487. Fyfe JJ, Bishop DJ, Bartlett JD, Hanson ED, Anderson MJ, Garnham AP, Stepto NK. Enhanced skeletal muscle ribosome biogenesis, yet attenuated mTORC1 and ribosome biogenesis-related signalling, following short-term concurrent versus single-mode resistance training. Sci Rep 8: 560, 2018. doi: 10.1038/s41598-017-18887-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488. Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol (1985) 98: 482–488, 2005. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- 489. Brook MS, Wilkinson DJ, Mitchell WK, Lund JL, Phillips BE, Szewczyk NJ, Kainulainen H, Lensu S, Koch LG, Britton SL, Greenhaff PL, Smith K, Atherton PJ. A novel D2O tracer method to quantify RNA turnover as a biomarker of de novo ribosomal biogenesis, in vitro, in animal models, and in human skeletal muscle. Am J Physiol Endocrinol Metab 313: E681–E689, 2017. doi: 10.1152/ajpendo.00157.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490. Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron-Smith D, Blazevich AJ. Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. Am J Physiol Endocrinol Metab 309: E72–E83, 2015. doi: 10.1152/ajpendo.00050.2015. [DOI] [PubMed] [Google Scholar]
- 491. Haddad F, Baldwin KM, Tesch PA. Pretranslational markers of contractile protein expression in human skeletal muscle: effect of limb unloading plus resistance exercise. J Appl Physiol (1985) 98: 46–52, 2005. doi: 10.1152/japplphysiol.00553.2004. [DOI] [PubMed] [Google Scholar]
- 492. Haun CT, Vann CG, Mobley CB, Osburn SC, Mumford PW, Roberson PA, Romero MA, Fox CD, Parry HA, Kavazis AN, Moon JR, Young KC, Roberts MD. Pre-training skeletal muscle fiber size and predominant fiber type best predict hypertrophic responses to 6 weeks of resistance training in previously trained young men. Front Physiol 10: 297, 2019. doi: 10.3389/fphys.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493. Mobley CB, Haun CT, Roberson PA, Mumford PW, Kephart WC, Romero MA, Osburn SC, Vann CG, Young KC, Beck DT, Martin JS, Lockwood CM, Roberts MD. Biomarkers associated with low, moderate, and high vastus lateralis muscle hypertrophy following 12 weeks of resistance training. PLoS One 13: e0195203, 2018. doi: 10.1371/journal.pone.0195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494. Sieljacks P, Wang J, Groennebaek T, Rindom E, Jakobsgaard JE, Herskind J, Gravholt A, Møller AB, Musci RV, de Paoli FV, Hamilton KL, Miller BF, Vissing K. Six weeks of low-load blood flow restricted and high-load resistance exercise training produce similar increases in cumulative myofibrillar protein synthesis and ribosomal biogenesis in healthy males. Front Physiol 10: 649, 2019. doi: 10.3389/fphys.2019.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495. Stec MJ, Kelly NA, Many GM, Windham ST, Tuggle SC, Bamman MM. Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. Am J Physiol Endocrinol Metab 310: E652–E661, 2016. doi: 10.1152/ajpendo.00486.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496. Goreham C, Green HJ, Ball-Burnett M, Ranney D. High-resistance training and muscle metabolism during prolonged exercise. Am J Physiol Endocrinol Metab 276: E489–E496, 1999. doi: 10.1152/ajpendo.1999.276.3.E489. [DOI] [PubMed] [Google Scholar]
- 497. Figueiredo VC, Wen Y, Alkner B, Fernandez-Gonzalo R, Norrbom J, Vechetti IJ Jr, Valentino T, Mobley CB, Zentner GE, Peterson CA, McCarthy JJ, Murach KA, von Walden F. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J Physiol 599: 3363–3384, 2021. doi: 10.1113/JP281244. [DOI] [PubMed] [Google Scholar]
- 498. Nelson JO, Watase GJ, Warsinger-Pepe N, Yamashita YM. Mechanisms of rDNA copy number maintenance. Trends Genet 35: 734–742, 2019. doi: 10.1016/j.tig.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499. Hammarström D, Øfsteng S, Koll L, Hanestadhaugen M, Hollan I, Apró W, Whist JE, Blomstrand E, Ronnestad BR, Ellefsen S. Benefits of higher resistance-training volume are related to ribosome biogenesis. J Physiol 598: 543–565, 2020. doi: 10.1113/JP278455. [DOI] [PubMed] [Google Scholar]
- 500. Hammarström D, Øfsteng SJ, Jacobsen NB, Flobergseter KB, Rønnestad BR, Ellefsen S. Ribosome accumulation during early phase resistance training in humans. Acta Physiol (Oxf) e13806, 2022. doi: 10.1111/apha.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501. Figueiredo VC, D’Souza RF, Van Pelt DW, Lawrence MM, Zeng N, Markworth JF, Poppitt SD, Miller BF, Mitchell CJ, McCarthy JJ, Dupont-Versteegden EE, Cameron-Smith D. Ribosome biogenesis and degradation regulate translational capacity during muscle disuse and reloading. J Cachexia Sarcopenia Muscle 12: 130–143, 2021. doi: 10.1002/jcsm.12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502. Mori T, Ato S, Knudsen JR, Henriquez-Olguin C, Li Z, Wakabayashi K, Suginohara T, Higashida K, Tamura Y, Nakazato K, Jensen TE, Ogasawara R. c-Myc overexpression increases ribosome biogenesis and protein synthesis independent of mTORC1 activation in mouse skeletal muscle. Am J Physiol Endocrinol Metab 321: E551–E559, 2021. doi: 10.1152/ajpendo.00164.2021. [DOI] [PubMed] [Google Scholar]
- 503. Chaillou T, Zhang X, McCarthy JJ. Expression of muscle-specific ribosomal protein L3-like impairs myotube growth. J Cell Physiol 231: 1894–1902, 2016. doi: 10.1002/jcp.25294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504. Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9: 493–495, 1961. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505. Katz B. The termination of the afferent nerve fibre in the muscle spindle of the frog. Philos Trans R Soc Lond Ser B Biol Sci 243: 221–240, 1961. [Google Scholar]
- 506. Mackey AL, Esmarck B, Kadi F, Koskinen SO, Kongsgaard M, Sylvestersen A, Hansen JJ, Larsen G, Kjaer M. Enhanced satellite cell proliferation with resistance training in elderly men and women. Scand J Med Sci Sports 17: 34–42, 2007. doi: 10.1111/j.1600-0838.2006.00534.x. [DOI] [PubMed] [Google Scholar]
- 507. Lundberg TR, Martínez-Aranda LM, Sanz G, Hansson B, von Walden F, Tesch PA, Fernandez-Gonzalo R. Early accentuated muscle hypertrophy is strongly associated with myonuclear accretion. Am J Physiol Regul Integr Comp Physiol 319: R50–R58, 2020. doi: 10.1152/ajpregu.00061.2020. [DOI] [PubMed] [Google Scholar]
- 508. Olsen S, Aagaard P, Kadi F, Tufekovic G, Verney J, Olesen JL, Suetta C, Kjaer M. Creatine supplementation augments the increase in satellite cell and myonuclei number in human skeletal muscle induced by strength training. J Physiol 573: 525–534, 2006. doi: 10.1113/jphysiol.2006.107359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509. Kadi F, Thornell LE. Concomitant increases in myonuclear and satellite cell content in female trapezius muscle following strength training. Histochem Cell Biol 113: 99–103, 2000. doi: 10.1007/s004180050012. [DOI] [PubMed] [Google Scholar]
- 510. Snijders T, Smeets JS, van Kranenburg J, Kies AK, van Loon LJ, Verdijk LB. Changes in myonuclear domain size do not precede muscle hypertrophy during prolonged resistance-type exercise training. Acta Physiol (Oxf) 216: 231–239, 2016. doi: 10.1111/apha.12609. [DOI] [PubMed] [Google Scholar]
- 511. Reidy PT, Fry CS, Igbinigie S, Deer RR, Jennings K, Cope MB, Mukherjea R, Volpi E, Rasmussen BB. Protein supplementation does not affect myogenic adaptations to resistance training. Med Sci Sports Exerc 49: 1197–1208, 2017. doi: 10.1249/MSS.0000000000001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512. Oxfeldt M, Dalgaard LB, Jørgensen EB, Johansen FT, Dalgaard EB, Ørtenblad N, Hansen M. Molecular markers of skeletal muscle hypertrophy following 10 wk of resistance training in oral contraceptive users and nonusers. J Appl Physiol (1985) 129: 1355–1364, 2020. doi: 10.1152/japplphysiol.00562.2020. [DOI] [PubMed] [Google Scholar]
- 513. Mackey AL, Holm L, Reitelseder S, Pedersen TG, Doessing S, Kadi F, Kjaer M. Myogenic response of human skeletal muscle to 12 weeks of resistance training at light loading intensity. Scand J Med Sci Sports 21: 773–782, 2011. doi: 10.1111/j.1600-0838.2010.01178.x. [DOI] [PubMed] [Google Scholar]
- 514. Hanssen KE, Kvamme NH, Nilsen TS, Rønnestad B, Ambjørnsen IK, Norheim F, Kadi F, Hallèn J, Drevon CA, Raastad T. The effect of strength training volume on satellite cells, myogenic regulatory factors, and growth factors. Scand J Med Sci Sports 23: 728–739, 2013. doi: 10.1111/j.1600-0838.2012.01452.x. [DOI] [PubMed] [Google Scholar]
- 515. Herman-Montemayor JR, Hikida RS, Staron RS. Early-phase satellite cell and myonuclear domain adaptations to slow-speed vs. traditional resistance training programs. J Strength Cond Res 29: 3105–3114, 2015. doi: 10.1519/JSC.0000000000000925. [DOI] [PubMed] [Google Scholar]
- 516. Kadi F, Eriksson A, Holmner S, Butler-Browne GS, Thornell LE. Cellular adaptation of the trapezius muscle in strength-trained athletes. Histochem Cell Biol 111: 189–195, 1999. doi: 10.1007/s004180050348. [DOI] [PubMed] [Google Scholar]
- 517. Bellamy LM, Joanisse S, Grubb A, Mitchell CJ, McKay BR, Phillips SM, Baker S, Parise G. The acute satellite cell response and skeletal muscle hypertrophy following resistance training. PLoS One 9: e109739, 2014. doi: 10.1371/journal.pone.0109739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518. Smith MA, Sexton CL, Smith KA, Osburn SC, Godwin JS, Beausejour JP, Ruple BA, Goodlett MD, Edison JL, Fruge AD, Robinson AT, Gladden LB, Young KC, Roberts MD. Molecular predictors of resistance training outcomes in young untrained female adults. J Appl Physiol (1985) 134: 491–507, 2023. doi: 10.1152/japplphysiol.00605.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519. Brook MS, Wilkinson DJ, Tarum J, Mitchell KW, Lund JL, Phillips BE, Szewczyk NJ, Kadi F, Greenhaff PL, Smith K, Atherton PJ. Neither myonuclear accretion nor a myonuclear domain size ceiling is a feature of the attenuated hypertrophic potential of aged human skeletal muscle. Geroscience 45: 451–462, 2023. doi: 10.1007/s11357-022-00651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520. Karlsen A, Bechshøft RL, Malmgaard-Clausen NM, Andersen JL, Schjerling P, Kjaer M, Mackey AL. Lack of muscle fibre hypertrophy, myonuclear addition, and satellite cell pool expansion with resistance training in 83-94-year-old men and women. Acta Physiol (Oxf) 227: e13271, 2019. doi: 10.1111/apha.13271. [DOI] [PubMed] [Google Scholar]
- 521. Karlsen A, Soendenbroe C, Malmgaard-Clausen NM, Wagener F, Moeller CE, Senhaji Z, Damberg K, Andersen JL, Schjerling P, Kjaer M, Mackey AL. Preserved capacity for satellite cell proliferation, regeneration, and hypertrophy in the skeletal muscle of healthy elderly men. FASEB J 34: 6418–6436, 2020. doi: 10.1096/fj.202000196R. [DOI] [PubMed] [Google Scholar]
- 522. McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, Van Zant G, Campbell KS, Esser KA, Dupont-Versteegden EE, Peterson CA. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138: 3657–3666, 2011. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523. Egner IM, Bruusgaard JC, Gundersen K. Satellite cell depletion prevents fiber hypertrophy in skeletal muscle. Development 143: 2898–2906, 2016. doi: 10.1242/dev.134411. [DOI] [PubMed] [Google Scholar]
- 524. Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499: 301–305, 2013. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525. Goh Q, Millay DP. Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. Elife 6: e20007, 2017. doi: 10.7554/eLife.20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526. Englund DA, Figueiredo VC, Dungan CM, Murach KA, Peck BD, Petrosino JM, Brightwell CR, Dupont AM, Neal AC, Fry CS, Accornero F, McCarthy JJ, Peterson CA. Satellite cell depletion disrupts transcriptional coordination and muscle adaptation to exercise. Function (Oxf) 2: zqaa033, 2021. doi: 10.1093/function/zqaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527. Kobayashi Y, Tanaka T, Mulati M, Ochi H, Sato S, Kaldis P, Yoshii T, Okawa A, Inose H. Cyclin-dependent kinase 1 is essential for muscle regeneration and overload muscle fiber hypertrophy. Front Cell Dev Biol 8: 564581, 2020. doi: 10.3389/fcell.2020.564581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528. Verney J, Kadi F, Charifi N, Féasson L, Saafi MA, Castells J, Piehl-Aulin K, Denis C. Effects of combined lower body endurance and upper body resistance training on the satellite cell pool in elderly subjects. Muscle Nerve 38: 1147–1154, 2008. doi: 10.1002/mus.21054. [DOI] [PubMed] [Google Scholar]
- 529. Dirks ML, Tieland M, Verdijk LB, Losen M, Nilwik R, Mensink M, de Groot L, van Loon LJ. Protein supplementation augments muscle fiber hypertrophy but does not modulate satellite cell content during prolonged resistance-type exercise training in frail elderly. J Am Med Dir Assoc 18: 608–615, 2017. doi: 10.1016/j.jamda.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 530. Shamim B, Camera DM, Whitfield J. Myofibre hypertrophy in the absence of changes to satellite cell content following concurrent exercise training in young healthy men. Front Physiol 12: 625044, 2021. doi: 10.3389/fphys.2021.625044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Angleri V, Damas F, Phillips SM, Selistre-de-Araujo HS, Cornachione AS, Stotzer US, Santanielo N, Soligon SD, Costa LA, Lixandrão ME, Conceição MS, Vechin FC, Ugrinowitsch C, Libardi CA. Resistance training variable manipulations are less relevant than intrinsic biology in affecting muscle fiber hypertrophy. Scand J Med Sci Sports 32: 821–832, 2022. doi: 10.1111/sms.14134. [DOI] [PubMed] [Google Scholar]
- 532. Horwath O, Apró W, Moberg M, Godhe M, Helge T, Ekblom M, Hirschberg AL, Ekblom B. Fiber type-specific hypertrophy and increased capillarization in skeletal muscle following testosterone administration in young women. J Appl Physiol (1985) 128: 1240–1250, 2020. doi: 10.1152/japplphysiol.00893.2019. [DOI] [PubMed] [Google Scholar]
- 533. Damas F, Libardi CA, Ugrinowitsch C, Vechin FC, Lixandrão ME, Snijders T, Nederveen JP, Bacurau AV, Brum P, Tricoli V, Roschel H, Parise G, Phillips SM. Early- and later-phases satellite cell responses and myonuclear content with resistance training in young men. PLoS One 13: e0191039, 2018. doi: 10.1371/journal.pone.0191039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534. Kirby TJ, Patel RM, McClintock TS, Dupont-Versteegden EE, Peterson CA, McCarthy JJ. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Mol Biol Cell 27: 788–798, 2016. doi: 10.1091/mbc.E15-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535. Karlsen A, Couppé C, Andersen JL, Mikkelsen UR, Nielsen RH, Magnusson SP, Kjaer M, Mackey AL. Matters of fiber size and myonuclear domain: does size matter more than age? Muscle Nerve 52: 1040–1046, 2015. doi: 10.1002/mus.24669. [DOI] [PubMed] [Google Scholar]
- 536. Borowik AK, Davidyan A, Peelor FF 3rd, Voloviceva E, Doidge SM, Bubak MP, Mobley CB, McCarthy JJ, Dupont-Versteegden EE, Miller BF. Skeletal muscle nuclei in mice are not post-mitotic. Function 4: zqac059, 2022. doi: 10.1093/function/zqac059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537. Dreyfus PA, Chretien F, Chazaud B, Kirova Y, Caramelle P, Garcia L, Butler-Browne G, Gherardi RK. Adult bone marrow-derived stem cells in muscle connective tissue and satellite cell niches. Am J Pathol 164: 773–779, 2004. doi: 10.1016/S0002-9440(10)63165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538. Liu N, Garry GA, Li S, Bezprozvannaya S, Sanchez-Ortiz E, Chen B, Shelton JM, Jaichander P, Bassel-Duby R, Olson EN. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol 19: 202–213, 2017. doi: 10.1038/ncb3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539. Flynn CG, Ginkel PR, Hubert KA, Guo Q, Hrycaj SM, McDermott AE, Madruga A, Miller AP, Wellik DM. Hox11-expressing interstitial cells contribute to adult skeletal muscle at homeostasis. Development 150: dev201026, 2023. doi: 10.1242/dev.201026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540. Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138: 3625–3637, 2011. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541. Fry CS, Kirby TJ, Kosmac K, McCarthy JJ, Peterson CA. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 20: 56–69, 2017. doi: 10.1016/j.stem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542. Murach KA, Vechetti IJ Jr, Van Pelt DW, Crow SE, Dungan CM, Figueiredo VC, Kosmac K, Fu X, Richards CI, Fry CS, McCarthy JJ, Peterson CA. Fusion-independent satellite cell communication to muscle fibers during load-induced hypertrophy. Function (Oxf) 1: zqaa009, 2020. doi: 10.1093/function/zqaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543. Hyldahl RD, Nelson B, Xin L, Welling T, Groscost L, Hubal MJ, Chipkin S, Clarkson PM, Parcell AC. Extracellular matrix remodeling and its contribution to protective adaptation following lengthening contractions in human muscle. FASEB J 29: 2894–2904, 2015. doi: 10.1096/fj.14-266668. [DOI] [PubMed] [Google Scholar]
- 544. Fry CS, Lee JD, Jackson JR, Kirby TJ, Stasko SA, Liu H, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Regulation of the muscle fiber microenvironment by activated satellite cells during hypertrophy. FASEB J 28: 1654–1665, 2014. doi: 10.1096/fj.13-239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545. Murach KA, Fry CS, Kirby TJ, Jackson JR, Lee JD, White SH, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Starring or supporting role? Satellite cells and skeletal muscle fiber size regulation. Physiology (Bethesda) 33: 26–38, 2018. doi: 10.1152/physiol.00019.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546. Moro T, Brightwell CR, Volpi E, Rasmussen BB, Fry CS. Resistance exercise training promotes fiber type-specific myonuclear adaptations in older adults. J Appl Physiol (1985) 128: 795–804, 2020. doi: 10.1152/japplphysiol.00723.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547. Murach KA, Peck BD, Policastro RA, Vechetti IJ, Van Pelt DW, Dungan CM, Denes LT, Fu X, Brightwell CR, Zentner GE, Dupont-Versteegden EE, Richards CI, Smith JJ, Fry CS, McCarthy JJ, Peterson CA. Early satellite cell communication creates a permissive environment for long-term muscle growth. iScience 24: 102372, 2021. doi: 10.1016/j.isci.2021.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548. Kaneshige A, Kaji T, Zhang L, Saito H, Nakamura A, Kurosawa T, Ikemoto-Uezumi M, Tsujikawa K, Seno S, Hori M, Saito Y, Matozaki T, Maehara K, Ohkawa Y, Potente M, Watanabe S, Braun T, Uezumi A, Fukada SI. Relayed signaling between mesenchymal progenitors and muscle stem cells ensures adaptive stem cell response to increased mechanical load. Cell Stem Cell 29: 265–280.e6, 2022. doi: 10.1016/j.stem.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 549. Abou-Khalil R, Mounier R, Chazaud B. Regulation of myogenic stem cell behavior by vessel cells: the “ménage à trois” of satellite cells, periendothelial cells and endothelial cells. Cell Cycle 9: 892–896, 2010. doi: 10.4161/cc.9.5.10851. [DOI] [PubMed] [Google Scholar]
- 550. Zempo H, Miyamoto-Mikami E, Kikuchi N, Fuku N, Miyachi M, Murakami H. Heritability estimates of muscle strength-related phenotypes: a systematic review and meta-analysis. Scand J Med Sci Sports 27: 1537–1546, 2017. doi: 10.1111/sms.12804. [DOI] [PubMed] [Google Scholar]
- 551. Kilikevicius A, Bunger L, Lionikas A. Baseline muscle mass is a poor predictor of functional overload-induced gain in the mouse model. Front Physiol 7: 534, 2016. doi: 10.3389/fphys.2016.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552. Pescatello LS, Devaney JM, Hubal MJ, Thompson PD, Hoffman EP. Highlights from the functional single nucleotide polymorphisms associated with human muscle size and strength or FAMuSS study. Biomed Res Int 2013: 643575, 2013. doi: 10.1155/2013/643575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553. Fu L, Wu H, Cheng SY, Gao D, Zhang L, Zhao Y. Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. Elife 5: e15690, 2016. doi: 10.7554/eLife.15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554. Renault MA, Vandierdonck S, Chapouly C, Yu Y, Qin G, Metras A, Couffinhal T, Losordo DW, Yao Q, Reynaud A, Jaspard-Vinassa B, Belloc I, Desgranges C, Gadeau AP. Gli3 regulation of myogenesis is necessary for ischemia-induced angiogenesis. Circ Res 113: 1148–1158, 2013. doi: 10.1161/CIRCRESAHA.113.301546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555. Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, , et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43: 1066–1073, 2011. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556. Bailetti D, Sentinelli F, Prudente S, Cimini FA, Barchetta I, Totaro M, Di Costanzo A, Barbonetti A, Leonetti F, Cavallo MG, Baroni MG. Deep resequencing of 9 candidate genes identifies a role for ARAP1 and IGF2BP2 in modulating insulin secretion adjusted for insulin resistance in obese Southern Europeans. Int J Mol Sci 23: 1221, 2022. doi: 10.3390/ijms23031221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557. McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94: 12457–12461, 1997. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558. Lin J, Arnold HB, Della-Fera MA, Azain MJ, Hartzell DL, Baile CA. Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochem Biophys Res Commun 291: 701–706, 2002. doi: 10.1006/bbrc.2002.6500. [DOI] [PubMed] [Google Scholar]
- 559. Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, Kömen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688, 2004. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 560. Li X, Wang SJ, Tan SC, Chew PL, Liu L, Wang L, Wen L, Ma L. The A55T and K153R polymorphisms of MSTN gene are associated with the strength training-induced muscle hypertrophy among Han Chinese men. J Sports Sci 32: 883–891, 2014. doi: 10.1080/02640414.2013.865252. [DOI] [PubMed] [Google Scholar]
- 561. Thomis MA, Huygens W, Heuninckx S, Chagnon M, Maes HH, Claessens AL, Vlietinck R, Bouchard C, Beunen GP. Exploration of myostatin polymorphisms and the angiotensin-converting enzyme insertion/deletion genotype in responses of human muscle to strength training. Eur J Appl Physiol 92: 267–274, 2004. doi: 10.1007/s00421-004-1093-6. [DOI] [PubMed] [Google Scholar]
- 562. Kostek MA, Angelopoulos TJ, Clarkson PM, Gordon PM, Moyna NM, Visich PS, Zoeller RF, Price TB, Seip RL, Thompson PD, Devaney JM, Gordish-Dressman H, Hoffman EP, Pescatello LS. Myostatin and follistatin polymorphisms interact with muscle phenotypes and ethnicity. Med Sci Sports Exerc 41: 1063–1071, 2009. doi: 10.1249/MSS.0b013e3181930337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563. Bird AP. CpG-rich islands and the function of DNA methylation. Nature 321: 209–213, 1986. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 564. Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev 4: 255–259, 1994. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 565. Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem 12: 206–222, 2011. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 566. Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18: 517–534, 2017. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 567. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 38: 23–38, 2013. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568. Buitrago D, Labrador M, Arcon JP, Lema R, Flores O, Esteve-Codina A, Blanc J, Villegas N, Bellido D, Gut M, Dans PD, Heath SC, Gut IG, Brun Heath I, Orozco M. Impact of DNA methylation on 3D genome structure. Nat Commun 12: 3243, 2021. doi: 10.1038/s41467-021-23142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569. Sellami M, Bragazzi N, Prince MS, Denham J, Elrayess M. Regular, intense exercise training as a healthy aging lifestyle strategy: preventing DNA damage, telomere shortening and adverse DNA methylation changes over a lifetime. Front Genet 12: 652497, 2021. doi: 10.3389/fgene.2021.652497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570. Swiatowy WJ, Drzewiecka H, Kliber M, Sasiadek M, Karpinski P, Plawski A, Jagodzinski PP. Physical activity and DNA methylation in humans. Int J Mol Sci 22: 12989, 2021. doi: 10.3390/ijms222312989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571. Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15: 405–411, 2012. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 572. Sharples AP, Stewart CE, Seaborne RA. Does skeletal muscle have an ‘epi’-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell 15: 603–616, 2016. doi: 10.1111/acel.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573. Sexton CL, Godwin JS, McIntosh MC, Ruple BA, Osburn SC, Hollingsworth BR, Kontos NJ, Agostinelli PJ, Kavazis AN, Ziegenfuss TN, Lopez HL, Smith R, Young KC, Dwaraka VB, Frugé AD, Mobley CB, Sharples AP, Roberts MD. Skeletal muscle DNA methylation and mRNA responses to a bout of higher versus lower load resistance exercise in previously trained men. Cells 12: 263, 2023. doi: 10.3390/cells12020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574. Jones RG 3rd, Dimet-Wiley A, Haghani A, da Silva FM, Brightwell CR, Lim S, Khadgi S, Wen Y, Dungan CM, Brooke RT, Greene NP, Peterson CA, McCarthy JJ, Horvath S, Watowich SJ, Fry CS, Murach KA. A molecular signature defining exercise adaptation with ageing and in vivo partial reprogramming in skeletal muscle. J Physiol 601: 763–782, 2023. doi: 10.1113/JP283836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575. Maasar MF, Turner DC, Gorski PP, Seaborne RA, Strauss JA, Shepherd SO, Cocks M, Pillon NJ, Zierath JR, Hulton AT, Drust B, Sharples AP. The comparative methylome and transcriptome after change of direction compared to straight line running exercise in human skeletal muscle. Front Physiol 12: 619447, 2021. doi: 10.3389/fphys.2021.619447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576. Laker RC, Garde C, Camera DM, Smiles WJ, Zierath JR, Hawley JA, Barrès R. Transcriptomic and epigenetic responses to short-term nutrient-exercise stress in humans. Sci Rep 7: 15134, 2017. doi: 10.1038/s41598-017-15420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577. Telles GD, Libardi CA, Conceição MS, Vechin FC, Lixandrão ME, Mangone FR, Pavanelli AC, Nagai MA, Camera DM, Hawley JA, Ugrinowitsch C. Interrelated but not time-aligned response in myogenic regulatory factors demethylation and mRNA expression after divergent exercise bouts. Med Sci Sports Exerc 55: 199–208, 2023. doi: 10.1249/MSS.0000000000003049. [DOI] [PubMed] [Google Scholar]
- 578. Wang H, Huang Y, Yu M, Yu Y, Li S, Wang H, Sun H, Li B, Xu G, Hu P. Muscle regeneration controlled by a designated DNA dioxygenase. Cell Death Dis 12: 535, 2021. doi: 10.1038/s41419-021-03817-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579. Burd NA, Tang JE, Moore DR, Phillips SM. Exercise training and protein metabolism: influences of contraction, protein intake, and sex-based differences. J Appl Physiol (1985) 106: 1692–1701, 2009. doi: 10.1152/japplphysiol.91351.2008. [DOI] [PubMed] [Google Scholar]
- 580. Kumar V, Atherton P, Smith K, Rennie MJ. Human muscle protein synthesis and breakdown during and after exercise. J Appl Physiol (1985) 106: 2026–2039, 2009. doi: 10.1152/japplphysiol.91481.2008. [DOI] [PubMed] [Google Scholar]
- 581. Brook MS, Wilkinson DJ, Smith K, Atherton PJ. The metabolic and temporal basis of muscle hypertrophy in response to resistance exercise. Eur J Sport Sci 16: 633–644, 2016. doi: 10.1080/17461391.2015.1073362. [DOI] [PubMed] [Google Scholar]
- 582. Tipton KD, Hamilton DL, Gallagher IJ. Assessing the role of muscle protein breakdown in response to nutrition and exercise in humans. Sports Med 48: 53–64, 2018. doi: 10.1007/s40279-017-0845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583. Pasiakos SM, Carbone JW. Assessment of skeletal muscle proteolysis and the regulatory response to nutrition and exercise. IUBMB Life 66: 478–484, 2014. doi: 10.1002/iub.1291. [DOI] [PubMed] [Google Scholar]
- 584. Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J Gerontol A Biol Sci Med Sci 62: 1407–1412, 2007. doi: 10.1093/gerona/62.12.1407. [DOI] [PubMed] [Google Scholar]
- 585. Dalbo VJ, Roberts MD, Hassell SE, Brown RD, Kerksick CM. Effects of age on serum hormone concentrations and intramuscular proteolytic signaling before and after a single bout of resistance training. J Strength Cond Res 25: 1–9, 2011. doi: 10.1519/JSC.0b013e3181fc5a68. [DOI] [PubMed] [Google Scholar]
- 586. Yang Y, Jemiolo B, Trappe S. Proteolytic mRNA expression in response to acute resistance exercise in human single skeletal muscle fibers. J Appl Physiol (1985) 101: 1442–1450, 2006. doi: 10.1152/japplphysiol.00438.2006. [DOI] [PubMed] [Google Scholar]
- 587. Dalbo VJ, Roberts MD, Hassell S, Kerksick CM. Effects of pre-exercise feeding on serum hormone concentrations and biomarkers of myostatin and ubiquitin proteasome pathway activity. Eur J Nutr 52: 477–487, 2013. doi: 10.1007/s00394-012-0349-x. [DOI] [PubMed] [Google Scholar]
- 588. Stefanetti RJ, Lamon S, Wallace M, Vendelbo MH, Russell AP, Vissing K. Regulation of ubiquitin proteasome pathway molecular markers in response to endurance and resistance exercise and training. Pflugers Arch 467: 1523–1537, 2015. doi: 10.1007/s00424-014-1587-y. [DOI] [PubMed] [Google Scholar]
- 589. Stefanetti RJ, Lamon S, Rahbek SK, Farup J, Zacharewicz E, Wallace MA, Vendelbo MH, Russell AP, Vissing K. Influence of divergent exercise contraction mode and whey protein supplementation on atrogin-1, MuRF1, and FOXO1/3A in human skeletal muscle. J Appl Physiol (1985) 116: 1491–1502, 2014. doi: 10.1152/japplphysiol.00136.2013. [DOI] [PubMed] [Google Scholar]
- 590. Baehr LM, Tunzi M, Bodine SC. Muscle hypertrophy is associated with increases in proteasome activity that is independent of MuRF1 and MAFbx expression. Front Physiol 5: 69, 2014. doi: 10.3389/fphys.2014.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591. Zamir O, Hasselgren PO, Higashiguchi T, Frederick JA, Fischer JE. Tumour necrosis factor (TNF) and interleukin-1 (IL-1) induce muscle proteolysis through different mechanisms. Mediators Inflamm 1: 247–250, 1992. doi: 10.1155/S0962935192000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592. Baehr LM, Hughes DC, Lynch SA, Van Haver D, Maia TM, Marshall AG, Radoshevich L, Impens F, Waddell DS, Bodine SC. Identification of the MuRF1 skeletal muscle ubiquitylome through quantitative proteomics. Function (Oxf) 2: zqab029, 2021. doi: 10.1093/function/zqab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593. Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol 185: 1083–1095, 2009. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594. Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 595. Phillips SM, Tipton KD, Ferrando AA, Wolfe RR. Resistance training reduces the acute exercise-induced increase in muscle protein turnover. Am J Physiol Endocrinol Metab 276: E118–E124, 1999. doi: 10.1152/ajpendo.1999.276.1.E118. [DOI] [PubMed] [Google Scholar]
- 596. Dickinson JM, Reidy PT, Gundermann DM, Borack MS, Walker DK, D’Lugos AC, Volpi E, Rasmussen BB. The impact of postexercise essential amino acid ingestion on the ubiquitin proteasome and autophagosomal-lysosomal systems in skeletal muscle of older men. J Appl Physiol (1985) 122: 620–630, 2017. doi: 10.1152/japplphysiol.00632.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597. Hentilä J, Ahtiainen JP, Paulsen G, Raastad T, Häkkinen K, Mero AA, Hulmi JJ. Autophagy is induced by resistance exercise in young men, but unfolded protein response is induced regardless of age. Acta Physiol (Oxf) 224: e13069, 2018. doi: 10.1111/apha.13069. [DOI] [PubMed] [Google Scholar]
- 598. Kerksick CM, Roberts MD, Dalbo VJ, Kreider RB, Willoughby DS. Changes in skeletal muscle proteolytic gene expression after prophylactic supplementation of EGCG and NAC and eccentric damage. Food Chem Toxicol 61: 47–52, 2013. doi: 10.1016/j.fct.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 599. Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Volpi E, Rasmussen BB. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci 68: 599–607, 2013. doi: 10.1093/gerona/gls209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600. Zeng N, D’Souza RF, Figueiredo VC, Markworth JF, Roberts LA, Peake JM, Mitchell CJ, Cameron-Smith D. Acute resistance exercise induces Sestrin2 phosphorylation and p62 dephosphorylation in human skeletal muscle. Physiol Rep 5: e13526, 2017. doi: 10.14814/phy2.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601. Léger B, Cartoni R, Praz M, Lamon S, Dériaz O, Crettenand A, Gobelet C, Rohmer P, Konzelmann M, Luthi F, Russell AP. Akt signalling through GSK-3beta, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J Physiol 576: 923–933, 2006. doi: 10.1113/jphysiol.2006.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602. Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6: 307–309, 2010. doi: 10.4161/auto.6.2.11137. [DOI] [PubMed] [Google Scholar]
- 603. Kitajima Y, Tashiro Y, Suzuki N, Warita H, Kato M, Tateyama M, Ando R, Izumi R, Yamazaki M, Abe M, Sakimura K, Ito H, Urushitani M, Nagatomi R, Takahashi R, Aoki M. Proteasome dysfunction induces muscle growth defects and protein aggregation. J Cell Sci 127: 5204–5217, 2014. doi: 10.1242/jcs.150961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604. Hughes DC, Turner DC, Baehr LM, Seaborne RA, Viggars M, Jarvis JC, Gorski PP, Stewart CE, Owens DJ, Bodine SC, Sharples AP. Knockdown of the E3 ubiquitin ligase UBR5 and its role in skeletal muscle anabolism. Am J Physiol Cell Physiol 320: C45–C56, 2021. doi: 10.1152/ajpcell.00432.2020. [DOI] [PubMed] [Google Scholar]
- 605. Osburn SC, Vann CG, Church DD, Ferrando AA, Roberts MD. Proteasome- and calpain-mediated proteolysis, but not autophagy, is required for leucine-induced protein synthesis in C2C12 myotubes. Physiologia 1: 22–33, 2021. doi: 10.3390/physiologia1010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606. Lewis MH, Ryan PJ, Stanelle ST, O’Reilly C, Cardin JM, Fluckey JD. Autophagy, but not proteolysis, may aid in muscle protein synthesis. Int J Exer Sci 2: 66, 2022. [Google Scholar]
- 607. Reidy PT, Borack MS, Markofski MM, Dickinson JM, Fry CS, Deer RR, Volpi E, Rasmussen BB. Post-absorptive muscle protein turnover affects resistance training hypertrophy. Eur J Appl Physiol 117: 853–866, 2017. doi: 10.1007/s00421-017-3566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608. Phillips SM, Parise G, Roy BD, Tipton KD, Wolfe RR, Tamopolsky MA. Resistance-training-induced adaptations in skeletal muscle protein turnover in the fed state. Can J Physiol Pharmacol 80: 1045–1053, 2002. doi: 10.1139/y02-134. [DOI] [PubMed] [Google Scholar]
- 609. Biolo G, Fleming RY, Maggi SP, Wolfe RR. Transmembrane transport and intracellular kinetics of amino acids in human skeletal muscle. Am J Physiol Endocrinol Metab 268: E75–E84, 1995. doi: 10.1152/ajpendo.1995.268.1.E75. [DOI] [PubMed] [Google Scholar]
- 610. Camera DM, Burniston JG, Pogson MA, Smiles WJ, Hawley JA. Dynamic proteome profiling of individual proteins in human skeletal muscle after a high-fat diet and resistance exercise. FASEB J 31: 5478–5494, 2017. doi: 10.1096/fj.201700531R. [DOI] [PubMed] [Google Scholar]
- 611. Nuñez J, Renslow R, Cliff JB 3rd, Anderton CR. NanoSIMS for biological applications: Current practices and analyses. Biointerphases 13: 03B301, 2017. doi: 10.1116/1.4993628. [DOI] [PubMed] [Google Scholar]
- 612. Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA 98: 9306–9311, 2001. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613. Chen MM, Zhao YP, Zhao Y, Deng SL, Yu K. Regulation of myostatin on the growth and development of skeletal muscle. Front Cell Dev Biol 9: 785712, 2021. doi: 10.3389/fcell.2021.785712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614. Kollias HD, McDermott JC. Transforming growth factor-beta and myostatin signaling in skeletal muscle. J Appl Physiol (1985) 104: 579–587, 2008. doi: 10.1152/japplphysiol.01091.2007. [DOI] [PubMed] [Google Scholar]
- 615. Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J 280: 4294–4314, 2013. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 616. Goodman CA, Hornberger TA. New roles for Smad signaling and phosphatidic acid in the regulation of skeletal muscle mass. F1000Prime Rep 6: 20, 2014. doi: 10.12703/P6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617. Koinuma D, Tsutsumi S, Kamimura N, Imamura T, Aburatani H, Miyazono K. Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci 100: 2133–2142, 2009. doi: 10.1111/j.1349-7006.2009.01299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618. Wang DT, Yang YJ, Huang RH, Zhang ZH, Lin X. Myostatin activates the ubiquitin-proteasome and autophagy-lysosome systems contributing to muscle wasting in chronic kidney disease. Oxid Med Cell Longev 2015: 684965, 2015., doi: 10.1155/2015/684965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619. Manfredi LH, Paula-Gomes S, Zanon NM, Kettelhut IC. Myostatin promotes distinct responses on protein metabolism of skeletal and cardiac muscle fibers of rodents. Braz J Med Biol Res 50: e6733, 2017. doi: 10.1590/1414-431X20176733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620. Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol 296: C1248–C1257, 2009. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- 621. Durieux AC, Amirouche A, Banzet S, Koulmann N, Bonnefoy R, Pasdeloup M, Mouret C, Bigard X, Peinnequin A, Freyssenet D. Ectopic expression of myostatin induces atrophy of adult skeletal muscle by decreasing muscle gene expression. Endocrinology 148: 3140–3147, 2007. doi: 10.1210/en.2006-1500. [DOI] [PubMed] [Google Scholar]
- 622. Welle S, Cardillo A, Zanche M, Tawil R. Skeletal muscle gene expression after myostatin knockout in mature mice. Physiol Genomics 38: 342–350, 2009. doi: 10.1152/physiolgenomics.00054.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623. Steelman CA, Recknor JC, Nettleton D, Reecy JM. Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J 20: 580–582, 2006. doi: 10.1096/fj.05-5125fje. [DOI] [PubMed] [Google Scholar]
- 624. Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol 296: C1258–C1270, 2009. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 625. Hulmi JJ, Oliveira BM, Silvennoinen M, Hoogaars WM, Ma H, Pierre P, Pasternack A, Kainulainen H, Ritvos O. Muscle protein synthesis, mTORC1/MAPK/Hippo signaling, and capillary density are altered by blocking of myostatin and activins. Am J Physiol Endocrinol Metab 304: E41–E50, 2013. doi: 10.1152/ajpendo.00389.2012. [DOI] [PubMed] [Google Scholar]
- 626. Han X, Møller LL, De Groote E, Bojsen-Møller KN, Davey J, Henríquez-Olguin C, Li Z, Knudsen JR, Jensen TE, Madsbad S, Gregorevic P, Richter EA, Sylow L. Mechanisms involved in follistatin-induced hypertrophy and increased insulin action in skeletal muscle. J Cachexia Sarcopenia Muscle 10: 1241–1257, 2019. doi: 10.1002/jcsm.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627. Heymsfield SB, Coleman LA, Miller R, Rooks DS, Laurent D, Petricoul O, Praestgaard J, Swan T, Wade T, Perry RG, Goodpaster BH, Roubenoff R. Effect of bimagrumab vs placebo on body fat mass among adults with type 2 diabetes and obesity: a phase 2 randomized clinical trial. JAMA Netw Open 4: e2033457, 2021. doi: 10.1001/jamanetworkopen.2020.33457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628. Garito T, Roubenoff R, Hompesch M, Morrow L, Gomez K, Rooks D, Meyers C, Buchsbaum MS, Neelakantham S, Swan T, Filosa LA, Laurent D, Petricoul O, Zakaria M. Bimagrumab improves body composition and insulin sensitivity in insulin-resistant individuals. Diabetes Obes Metab 20: 94–102, 2018. doi: 10.1111/dom.13042. [DOI] [PubMed] [Google Scholar]
- 629. Wang Q, McPherron AC. Myostatin inhibition induces muscle fibre hypertrophy prior to satellite cell activation. J Physiol 590: 2151–2165, 2012. doi: 10.1113/jphysiol.2011.226001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630. Amthor H, Otto A, Vulin A, Rochat A, Dumonceaux J, Garcia L, Mouisel E, Hourdé C, Macharia R, Friedrichs M, Relaix F, Zammit PS, Matsakas A, Patel K, Partridge T. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc Natl Acad Sci USA 106: 7479–7484, 2009. doi: 10.1073/pnas.0811129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631. Lee SJ, Huynh TV, Lee YS, Sebald SM, Wilcox-Adelman SA, Iwamori N, Lepper C, Matzuk MM, Fan CM. Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc Natl Acad Sci USA 109: E2353–E2360, 2012. doi: 10.1073/pnas.1206410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632. Heinemeier KM, Olesen JL, Schjerling P, Haddad F, Langberg H, Baldwin KM, Kjaer M. Short-term strength training and the expression of myostatin and IGF-I isoforms in rat muscle and tendon: differential effects of specific contraction types. J Appl Physiol (1985) 102: 573–581, 2007. doi: 10.1152/japplphysiol.00866.2006. [DOI] [PubMed] [Google Scholar]
- 633. Hulmi JJ, Kovanen V, Lisko I, Selänne H, Mero AA. The effects of whey protein on myostatin and cell cycle-related gene expression responses to a single heavy resistance exercise bout in trained older men. Eur J Appl Physiol 102: 205–213, 2008. doi: 10.1007/s00421-007-0579-4. [DOI] [PubMed] [Google Scholar]
- 634. Hulmi JJ, Tannerstedt J, Selänne H, Kainulainen H, Kovanen V, Mero AA. Resistance exercise with whey protein ingestion affects mTOR signaling pathway and myostatin in men. J Appl Physiol (1985) 106: 1720–1729, 2009. doi: 10.1152/japplphysiol.00087.2009. [DOI] [PubMed] [Google Scholar]
- 635. Hulmi JJ, Ahtiainen JP, Kaasalainen T, Pöllänen E, Häkkinen K, Alen M, Selänne H, Kovanen V, Mero AA. Postexercise myostatin and activin IIb mRNA levels: effects of strength training. Med Sci Sports Exerc 39: 289–297, 2007. doi: 10.1249/01.mss.0000241650.15006.6e. [DOI] [PubMed] [Google Scholar]
- 636. Dalbo VJ, Roberts MD, Sunderland KL, Poole CN, Stout JR, Beck TW, Bemben M, Kerksick CM. Acute loading and aging effects on myostatin pathway biomarkers in human skeletal muscle after three sequential bouts of resistance exercise. J Gerontol A Biol Sci Med Sci 66: 855–865, 2011. doi: 10.1093/gerona/glr091. [DOI] [PubMed] [Google Scholar]
- 637. Bagheri R, Rashidlamir A, Motevalli MS, Elliott BT, Mehrabani J, Wong A. Effects of upper-body, lower-body, or combined resistance training on the ratio of follistatin and myostatin in middle-aged men. Eur J Appl Physiol 119: 1921–1931, 2019. doi: 10.1007/s00421-019-04180-z. [DOI] [PubMed] [Google Scholar]
- 638. Bagheri R, Moghadam BH, Church DD, Tinsley GM, Eskandari M, Moghadam BH, Motevalli MS, Baker JS, Robergs RA, Wong A. The effects of concurrent training order on body composition and serum concentrations of follistatin, myostatin and GDF11 in sarcopenic elderly men. Exp Gerontol 133: 110869, 2020. doi: 10.1016/j.exger.2020.110869. [DOI] [PubMed] [Google Scholar]
- 639. Saremi A, Gharakhanloo R, Sharghi S, Gharaati MR, Larijani B, Omidfar K. Effects of oral creatine and resistance training on serum myostatin and GASP-1. Mol Cell Endocrinol 317: 25–30, 2010. doi: 10.1016/j.mce.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 640. Laurentino GC, Ugrinowitsch C, Roschel H, Aoki MS, Soares AG, Neves M Jr, Aihara AY, Fernandes R, Tricoli V. Strength training with blood flow restriction diminishes myostatin gene expression. Med Sci Sports Exerc 44: 406–412, 2012. doi: 10.1249/MSS.0b013e318233b4bc. [DOI] [PubMed] [Google Scholar]
- 641. Roth SM, Martel GF, Ferrell RE, Metter EJ, Hurley BF, Rogers MA. Myostatin gene expression is reduced in humans with heavy-resistance strength training: a brief communication. Exp Biol Med (Maywood) 228: 706–709, 2003. doi: 10.1177/153537020322800609. [DOI] [PubMed] [Google Scholar]
- 642. McIntosh MC, Sexton CL, Godwin JS, Ruple BA, Michel JM, Plotkin DL, Ziegenfuss TN, Lopez HL, Smith R, Dwaraka VB, Sharples AP, Dalbo VJ, Mobley CB, Vann CG, Roberts MD. Different resistance exercise loading paradigms similarly affect skeletal muscle gene expression patterns of myostatin-related targets and mTORC1 signaling markers. Cells 12: 898, 2023. doi: 10.3390/cells12060898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643. Willoughby DS. Effects of heavy resistance training on myostatin mRNA and protein expression. Med Sci Sports Exerc 36: 574–582, 2004. doi: 10.1249/01.mss.0000121952.71533.ea. [DOI] [PubMed] [Google Scholar]
- 644. Kim JS, Petrella JK, Cross JM, Bamman MM. Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: a cluster analysis. J Appl Physiol (1985) 103: 1488–1495, 2007. doi: 10.1152/japplphysiol.01194.2006. [DOI] [PubMed] [Google Scholar]
- 645. Minderis P, Kilikevicius A, Baltusnikas J, Alhindi Y, Venckunas T, Bunger L, Lionikas A, Ratkevicius A. Myostatin dysfunction is associated with reduction in overload induced hypertrophy of soleus muscle in mice. Scand J Med Sci Sports 26: 894–901, 2016. doi: 10.1111/sms.12532. [DOI] [PubMed] [Google Scholar]
- 646. Aoki MS, Soares AG, Miyabara EH, Baptista IL, Moriscot AS. Expression of genes related to myostatin signaling during rat skeletal muscle longitudinal growth. Muscle Nerve 40: 992–999, 2009. doi: 10.1002/mus.21426. [DOI] [PubMed] [Google Scholar]
- 647. MacKenzie MG, Hamilton DL, Pepin M, Patton A, Baar K. Inhibition of myostatin signaling through Notch activation following acute resistance exercise. PLoS One 8: e68743, 2013. doi: 10.1371/journal.pone.0068743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648. Nikooie R, Jafari-Sardoie S, Sheibani V, Nejadvaziri Chatroudi A. Resistance training-induced muscle hypertrophy is mediated by TGF-beta1-Smad signaling pathway in male Wistar rats. J Cell Physiol 235: 5649–5665, 2020. doi: 10.1002/jcp.29497. [DOI] [PubMed] [Google Scholar]
- 649. Jacobson KR, Lipp S, Acuna A, Leng Y, Bu Y, Calve S. Comparative analysis of the extracellular matrix proteome across the myotendinous junction. J Proteome Res 19: 3955–3967, 2020. doi: 10.1021/acs.jproteome.0c00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650. Karlsen A, Gonzalez-Franquesa A, Jakobsen JR, Krogsgaard MR, Koch M, Kjaer M, Schiaffino S, Mackey AL, Deshmukh AS. The proteomic profile of the human myotendinous junction. iScience 25: 103836, 2022. doi: 10.1016/j.isci.2022.103836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651. Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol 49: 10–24, 2016. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652. Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev 84: 649–698, 2004. doi: 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- 653. Millward DJ. A protein-stat mechanism for regulation of growth and maintenance of the lean body mass. Nutr Res Rev 8: 93–120, 1995. doi: 10.1079/NRR19950008. [DOI] [PubMed] [Google Scholar]
- 654. Mendias CL, Schwartz AJ, Grekin JA, Gumucio JP, Sugg KB. Changes in muscle fiber contractility and extracellular matrix production during skeletal muscle hypertrophy. J Appl Physiol (1985) 122: 571–579, 2017. doi: 10.1152/japplphysiol.00719.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655. White JP, Reecy JM, Washington TA, Sato S, Le ME, Davis JM, Wilson LB, Carson JA. Overload-induced skeletal muscle extracellular matrix remodelling and myofibre growth in mice lacking IL-6. Acta Physiol (Oxf) 197: 321–332, 2009. doi: 10.1111/j.1748-1716.2009.02029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656. Moore DR, Phillips SM, Babraj JA, Smith K, Rennie MJ. Myofibrillar and collagen protein synthesis in human skeletal muscle in young men after maximal shortening and lengthening contractions. Am J Physiol Endocrinol Metab 288: E1153–E1159, 2005. doi: 10.1152/ajpendo.00387.2004. [DOI] [PubMed] [Google Scholar]
- 657. Mackey AL, Donnelly AE, Turpeenniemi-Hujanen T, Roper HP. Skeletal muscle collagen content in humans after high-force eccentric contractions. J Appl Physiol (1985) 97: 197–203, 2004. doi: 10.1152/japplphysiol.01174.2003. [DOI] [PubMed] [Google Scholar]
- 658. Mackey AL, Brandstetter S, Schjerling P, Bojsen-Moller J, Qvortrup K, Pedersen MM, Doessing S, Kjaer M, Magnusson SP, Langberg H. Sequenced response of extracellular matrix deadhesion and fibrotic regulators after muscle damage is involved in protection against future injury in human skeletal muscle. FASEB J 25: 1943–1959, 2011. doi: 10.1096/fj.10-176487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659. Mikkelsen UR, Schjerling P, Helmark IC, Reitelseder S, Holm L, Skovgaard D, Langberg H, Kjaer M, Heinemeier KM. Local NSAID infusion does not affect protein synthesis and gene expression in human muscle after eccentric exercise. Scand J Med Sci Sports 21: 630–644, 2011. doi: 10.1111/j.1600-0838.2010.01170.x. [DOI] [PubMed] [Google Scholar]
- 660. Peck BD, Murach KA, Walton RG, Simmons AJ, Long DE, Kosmac K, Dungan CM, Kern PA, Bamman MM, Peterson CA. A muscle cell-macrophage axis involving matrix metalloproteinase 14 facilitates extracellular matrix remodeling with mechanical loading. FASEB J 36: e22155, 2022. doi: 10.1096/fj.202100182RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661. DiPasquale DM, Cheng M, Billich W, Huang SA, van Rooijen N, Hornberger TA, Koh TJ. Urokinase-type plasminogen activator and macrophages are required for skeletal muscle hypertrophy in mice. Am J Physiol Cell Physiol 293: C1278–C1285, 2007. doi: 10.1152/ajpcell.00201.2007. [DOI] [PubMed] [Google Scholar]
- 662. Vechin FC, Libardi CA, Conceição MS, Damas F, Cavaglieri CR, Chacon-Mikahil MPT, Coutinho LL, Andrade SC, Neves MT Jr, Roschel H, Tricoli V, Baptista IL, Moriscot AA, Ugrinowitsch C. Low-intensity resistance training with partial blood flow restriction and high-intensity resistance training induce similar changes in skeletal muscle transcriptome in elderly humans. Appl Physiol Nutr Metab 44: 216–220, 2019. doi: 10.1139/apnm-2018-0146. [DOI] [PubMed] [Google Scholar]
- 663. Long DE, Peck BD, Lavin KM, Dungan CM, Kosmac K, Tuggle SC, Bamman MM, Kern PA, Peterson CA. Skeletal muscle properties show collagen organization and immune cell content are associated with resistance exercise response heterogeneity in older persons. J Appl Physiol (1985), 2022. doi: 10.1152/japplphysiol.00025.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664. Egginton S, Hudlická O, Brown MD, Walter H, Weiss JB, Bate A. Capillary growth in relation to blood flow and performance in overloaded rat skeletal muscle. J Appl Physiol (1985) 85: 2025–2032, 1998. doi: 10.1152/jappl.1998.85.6.2025. [DOI] [PubMed] [Google Scholar]
- 665. Hansen-Smith F, Egginton S, Zhou AL, Hudlicka O. Growth of arterioles precedes that of capillaries in stretch-induced angiogenesis in skeletal muscle. Microvasc Res 62: 1–14, 2001. doi: 10.1006/mvre.2001.2308. [DOI] [PubMed] [Google Scholar]
- 666. Ballak SB, Busé-Pot T, Harding PJ, Yap MH, Deldicque L, de Haan A, Jaspers RT, Degens H. Blunted angiogenesis and hypertrophy are associated with increased fatigue resistance and unchanged aerobic capacity in old overloaded mouse muscle. Age (Dordr) 38: 39, 2016. doi: 10.1007/s11357-016-9894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667. Degens H, Turek Z, Hoofd LJ, Van’t Hof MA, Binkhorst RA. The relationship between capillarisation and fibre types during compensatory hypertrophy of the plantaris muscle in the rat. J Anat 180: 455–463, 1992. [PMC free article] [PubMed] [Google Scholar]
- 668. Wagner PD. The critical role of VEGF in skeletal muscle angiogenesis and blood flow. Biochem Soc Trans 39: 1556–1559, 2011. doi: 10.1042/BST20110646. [DOI] [PubMed] [Google Scholar]
- 669. Huey KA, Smith SA, Sulaeman A, Breen EC. Skeletal myofiber VEGF is necessary for myogenic and contractile adaptations to functional overload of the plantaris in adult mice. J Appl Physiol (1985) 120: 188–195, 2016. doi: 10.1152/japplphysiol.00638.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670. Ato S, Fukada SI, Kokubo H, Ogasawara R. Implication of satellite cell behaviors in capillary growth via VEGF expression-independent mechanism in response to mechanical loading in HeyL-null mice. Am J Physiol Cell Physiol 322: C275–C282, 2022. doi: 10.1152/ajpcell.00343.2021. [DOI] [PubMed] [Google Scholar]
- 671. Fukuda S, Kaneshige A, Kaji T, Noguchi YT, Takemoto Y, Zhang L, Tsujikawa K, Kokubo H, Uezumi A, Maehara K, Harada A, Ohkawa Y, Fukada SI. Sustained expression of HeyL is critical for the proliferation of muscle stem cells in overloaded muscle. Elife 8: e48284, 2019. doi: 10.7554/eLife.48284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672. Gavin TP, Drew JL, Kubik CJ, Pofahl WE, Hickner RC. Acute resistance exercise increases skeletal muscle angiogenic growth factor expression. Acta Physiol (Oxf) 191: 139–146, 2007. doi: 10.1111/j.1748-1716.2007.01723.x. [DOI] [PubMed] [Google Scholar]
- 673. McCall GE, Byrnes WC, Dickinson A, Pattany PM, Fleck SJ. Muscle fiber hypertrophy, hyperplasia, and capillary density in college men after resistance training. J Appl Physiol (1985) 81: 2004–2012, 1996. doi: 10.1152/jappl.1996.81.5.2004. [DOI] [PubMed] [Google Scholar]
- 674. Holloway TM, Snijders T, Van Kranenburg J, Van Loon LJ, Verdijk LB. Temporal response of angiogenesis and hypertrophy to resistance training in young men. Med Sci Sports Exerc 50: 36–45, 2018. doi: 10.1249/MSS.0000000000001409. [DOI] [PubMed] [Google Scholar]
- 675. Verdijk LB, Snijders T, Holloway TM, Van Kranenburg J, Van Loon LJ. Resistance training increases skeletal muscle capillarization in healthy older men. Med Sci Sports Exerc 48: 2157–2164, 2016. doi: 10.1249/MSS.0000000000001019. [DOI] [PubMed] [Google Scholar]
- 676. Campos GE, Luecke TJ, Wendeln HK, Toma K, Hagerman FC, Murray TF, Ragg KE, Ratamess NA, Kraemer WJ, Staron RS. Muscular adaptations in response to three different resistance-training regimens: specificity of repetition maximum training zones. Eur J Appl Physiol 88: 50–60, 2002. doi: 10.1007/s00421-002-0681-6. [DOI] [PubMed] [Google Scholar]
- 677. Moro T, Brightwell CR, Phalen DE, McKenna CF, Lane SJ, Porter C, Volpi E, Rasmussen BB, Fry CS. Low skeletal muscle capillarization limits muscle adaptation to resistance exercise training in older adults. Exp Gerontol 127: 110723, 2019. doi: 10.1016/j.exger.2019.110723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678. Snijders T, Nederveen JP, Joanisse S, Leenders M, Verdijk LB, van Loon LJ, Parise G. Muscle fibre capillarization is a critical factor in muscle fibre hypertrophy during resistance exercise training in older men. J Cachexia Sarcopenia Muscle 8: 267–276, 2017. doi: 10.1002/jcsm.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679. Frontera WR, Hughes VA, Fielding RA, Fiatarone MA, Evans WJ, Roubenoff R. Aging of skeletal muscle: a 12-yr longitudinal study. J Appl Physiol (1985) 88: 1321–1326, 2000. doi: 10.1152/jappl.2000.88.4.1321. [DOI] [PubMed] [Google Scholar]
- 680. Nederveen JP, Joanisse S, Snijders T, Thomas AC, Kumbhare D, Parise G. The influence of capillarization on satellite cell pool expansion and activation following exercise-induced muscle damage in healthy young men. J Physiol 596: 1063–1078, 2018. doi: 10.1113/JP275155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681. Dvoretskiy S, Garg K, Munroe M, Pincu Y, Mahmassani ZS, Coombs C, Blackwell B, Garcia G, Waterstradt G, Lee I, Drnevich J, Rhodes JS, Boppart MD. The impact of skeletal muscle contraction on CD146+Lin- pericytes. Am J Physiol Cell Physiol 317: C1011–C1024, 2019. doi: 10.1152/ajpcell.00156.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682. Munroe M, Dvoretskiy S, Lopez A, Leong J, Dyle MC, Kong H, Adams CM, Boppart MD. Pericyte transplantation improves skeletal muscle recovery following hindlimb immobilization. FASEB J 33: 7694–7706, 2019. doi: 10.1096/fj.201802580R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9: 402, 2018. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684. Biasini A, Abdulkarim B, de Pretis S, Tan JY, Arora R, Wischnewski H, Dreos R, Pelizzola M, Ciaudo C, Marques AC. Translation is required for miRNA-dependent decay of endogenous transcripts. EMBO J 40: e104569, 2021. doi: 10.15252/embj.2020104569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 686. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM, Voinea SC. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells 9: 276, 2020. doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687. Herrera-Espejo S, Santos-Zorrozua B, Álvarez-González P, Lopez-Lopez E, Garcia-Orad A. A systematic review of microRNA expression as biomarker of late-onset Alzheimer’s disease. Mol Neurobiol 56: 8376–8391, 2019. doi: 10.1007/s12035-019-01676-9. [DOI] [PubMed] [Google Scholar]
- 688. Silva GJ, Bye A, El Azzouzi H, Wisløff U. MicroRNAs as important regulators of exercise adaptation. Prog Cardiovasc Dis 60: 130–151, 2017. doi: 10.1016/j.pcad.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 689. Sapp RM, Shill DD, Roth SM, Hagberg JM. Circulating microRNAs in acute and chronic exercise: more than mere biomarkers. J Appl Physiol (1985) 122: 702–717, 2017. doi: 10.1152/japplphysiol.00982.2016. [DOI] [PubMed] [Google Scholar]
- 690. Russell AP, Lamon S. Exercise, skeletal muscle and circulating microRNAs. Prog Mol Biol Transl Sci 135: 471–496, 2015. doi: 10.1016/bs.pmbts.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 691. Hitachi K, Tsuchida K. Role of microRNAs in skeletal muscle hypertrophy. Front Physiol 4: 408, 2013. doi: 10.3389/fphys.2013.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692. McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol (1985) 102: 306–313, 2007. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 693. Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab 295: E1333–E1340, 2008. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694. Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, Timmons JA, Phillips SM. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J Appl Physiol (1985) 110: 309–317, 2011. doi: 10.1152/japplphysiol.00901.2010. [DOI] [PubMed] [Google Scholar]
- 695. Zacharewicz E, Della Gatta P, Reynolds J, Garnham A, Crowley T, Russell AP, Lamon S. Identification of microRNAs linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PLoS One 9: e114009, 2014. doi: 10.1371/journal.pone.0114009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696. Rivas DA, Peng F, Benard T, Ramos da Silva AS, Fielding RA, Margolis LM. miR-19b-3p is associated with a diametric response to resistance exercise in older adults and regulates skeletal muscle anabolism via PTEN inhibition. Am J Physiol Cell Physiol 321: C977–C991, 2021. doi: 10.1152/ajpcell.00190.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697. Lim S, Lee DE, Morena da Silva F, Koopmans PJ, Vechetti IJ Jr, von Walden F, Greene NP, Murach KA. MicroRNA control of the myogenic cell transcriptome and proteome: the role of miR-16. Am J Physiol Cell Physiol 324: C1101–C1109, 2023. doi: 10.1152/ajpcell.00071.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698. Vechetti IJ Jr, Wen Y, Chaillou T, Murach KA, Alimov AP, Figueiredo VC, Dal-Pai-Silva M, McCarthy JJ. Life-long reduction in myomiR expression does not adversely affect skeletal muscle morphology. Sci Rep 9: 5483, 2019. doi: 10.1038/s41598-019-41476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699. Oikawa S, Lee M, Motohashi N, Maeda S, Akimoto T. An inducible knockout of Dicer in adult mice does not affect endurance exercise-induced muscle adaptation. Am J Physiol Cell Physiol 316: C285–C292, 2019. doi: 10.1152/ajpcell.00278.2018. [DOI] [PubMed] [Google Scholar]
- 700. Vingren JL, Kraemer WJ, Ratamess NA, Anderson JM, Volek JS, Maresh CM. Testosterone physiology in resistance exercise and training: the up-stream regulatory elements. Sports Med 40: 1037–1053, 2010. doi: 10.2165/11536910-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 701. Jiang M, Ma Y, Chen C, Fu X, Yang S, Li X, Yu G, Mao Y, Xie Y, Li Y. Androgen-responsive gene database: integrated knowledge on androgen-responsive genes. Mol Endocrinol 23: 1927–1933, 2009. doi: 10.1210/me.2009-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702. Bhasin S, Storer TW, Berman N, Callegari C, Clevenger B, Phillips J, Bunnell TJ, Tricker R, Shirazi A, Casaburi R. The effects of supraphysiologic doses of testosterone on muscle size and strength in normal men. N Engl J Med 335: 1–7, 1996. doi: 10.1056/NEJM199607043350101. [DOI] [PubMed] [Google Scholar]
- 703. Sinha-Hikim I, Artaza J, Woodhouse L, Gonzalez-Cadavid N, Singh AB, Lee MI, Storer TW, Casaburi R, Shen R, Bhasin S. Testosterone-induced increase in muscle size in healthy young men is associated with muscle fiber hypertrophy. Am J Physiol Endocrinol Metab 283: E154–E164, 2002. doi: 10.1152/ajpendo.00502.2001. [DOI] [PubMed] [Google Scholar]
- 704. Forbes GB, Porta CR, Herr BE, Griggs RC. Sequence of changes in body composition induced by testosterone and reversal of changes after drug is stopped. JAMA 267: 397–399, 1992. [PubMed] [Google Scholar]
- 705. Griggs RC, Kingston W, Jozefowicz RF, Herr BE, Forbes G, Halliday D. Effect of testosterone on muscle mass and muscle protein synthesis. J Appl Physiol (1985) 66: 498–503, 1989. doi: 10.1152/jappl.1989.66.1.498. [DOI] [PubMed] [Google Scholar]
- 706. Young NR, Baker HW, Liu G, Seeman E. Body composition and muscle strength in healthy men receiving testosterone enanthate for contraception. J Clin Endocrinol Metab 77: 1028–1032, 1993. doi: 10.1210/jcem.77.4.8408450. [DOI] [PubMed] [Google Scholar]
- 707. Bhasin S, Woodhouse L, Casaburi R, Singh AB, Bhasin D, Berman N, Chen X, Yarasheski KE, Magliano L, Dzekov C, Dzekov J, Bross R, Phillips J, Sinha-Hikim I, Shen R, Storer TW. Testosterone dose-response relationships in healthy young men. Am J Physiol Endocrinol Metab 281: E1172–E1181, 2001. doi: 10.1152/ajpendo.2001.281.6.E1172. [DOI] [PubMed] [Google Scholar]
- 708. Huang G, Basaria S, Travison TG, Ho MH, Davda M, Mazer NA, Miciek R, Knapp PE, Zhang A, Collins L, Ursino M, Appleman E, Dzekov C, Stroh H, Ouellette M, Rundell T, Baby M, Bhatia NN, Khorram O, Friedman T, Storer TW, Bhasin S. Testosterone dose-response relationships in hysterectomized women with or without oophorectomy: effects on sexual function, body composition, muscle performance and physical function in a randomized trial. Menopause 21: 612–623, 2014. doi: 10.1097/GME.0000000000000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709. Sinha-Hikim I, Roth SM, Lee MI, Bhasin S. Testosterone-induced muscle hypertrophy is associated with an increase in satellite cell number in healthy, young men. Am J Physiol Endocrinol Metab 285: E197–E205, 2003. doi: 10.1152/ajpendo.00370.2002. [DOI] [PubMed] [Google Scholar]
- 710. White JP, Gao S, Puppa MJ, Sato S, Welle SL, Carson JA. Testosterone regulation of Akt/mTORC1/FoxO3a signaling in skeletal muscle. Mol Cell Endocrinol 365: 174–186, 2013. doi: 10.1016/j.mce.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711. Mobley CB, Mumford PW, Kephart WC, Conover CF, Beggs LA, Balaez A, Yarrow JF, Borst SE, Beck DT, Roberts MD. Effects of testosterone treatment on markers of skeletal muscle ribosome biogenesis. Andrologia 48: 967–977, 2016. doi: 10.1111/and.12539. [DOI] [PubMed] [Google Scholar]
- 712. Ferrando AA, Tipton KD, Doyle D, Phillips SM, Cortiella J, Wolfe RR. Testosterone injection stimulates net protein synthesis but not tissue amino acid transport. Am J Physiol Endocrinol Metab 275: E864–E871, 1998. doi: 10.1152/ajpendo.1998.275.5.E864. [DOI] [PubMed] [Google Scholar]
- 713. Gharahdaghi N, Rudrappa S, Brook MS, Farrash W, Idris I, Aziz MHA, Kadi F, Papaioannou K, Phillips BE, Sian T, Herrod PJ, Wilkinson DJ, Szewczyk NJ, Smith K, Atherton PJ. Pharmacological hypogonadism impairs molecular transducers of exercise-induced muscle growth in humans. J Cachexia Sarcopenia Muscle 13: 1134–1150, 2022. doi: 10.1002/jcsm.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714. Urban RJ, Bodenburg YH, Gilkison C, Foxworth J, Coggan AR, Wolfe RR, Ferrando A. Testosterone administration to elderly men increases skeletal muscle strength and protein synthesis. Am J Physiol Endocrinol Metab 269: E820–E826, 1995. doi: 10.1152/ajpendo.1995.269.5.E820. [DOI] [PubMed] [Google Scholar]
- 715. Englund DA, Peck BD, Murach KA, Neal AC, Caldwell HA, McCarthy JJ, Peterson CA, Dupont-Versteegden EE. Resident muscle stem cells are not required for testosterone-induced skeletal muscle hypertrophy. Am J Physiol Cell Physiol 317: C719–C724, 2019. doi: 10.1152/ajpcell.00260.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716. MacLean HE, Chiu WS, Notini AJ, Axell AM, Davey RA, McManus JF, Ma C, Plant DR, Lynch GS, Zajac JD. Impaired skeletal muscle development and function in male, but not female, genomic androgen receptor knockout mice. FASEB J 22: 2676–2689, 2008. doi: 10.1096/fj.08-105726. [DOI] [PubMed] [Google Scholar]
- 717. Ophoff J, Van Proeyen K, Callewaert F, De Gendt K, De Bock K, Vanden Bosch A, Verhoeven G, Hespel P, Vanderschueren D. Androgen signaling in myocytes contributes to the maintenance of muscle mass and fiber type regulation but not to muscle strength or fatigue. Endocrinology 150: 3558–3566, 2009. doi: 10.1210/en.2008-1509. [DOI] [PubMed] [Google Scholar]
- 718. Ferry A, Schuh M, Parlakian A, Mgrditchian T, Valnaud N, Joanne P, Butler-Browne G, Agbulut O, Metzger D. Myofiber androgen receptor promotes maximal mechanical overload-induced muscle hypertrophy and fiber type transition in male mice. Endocrinology 155: 4739–4748, 2014. doi: 10.1210/en.2014-1195. [DOI] [PubMed] [Google Scholar]
- 719. Inoue K, Yamasaki S, Fushiki T, Okada Y, Sugimoto E. Androgen receptor antagonist suppresses exercise-induced hypertrophy of skeletal muscle. Eur J Appl Physiol Occup Physiol 69: 88–91, 1994. doi: 10.1007/BF00867933. [DOI] [PubMed] [Google Scholar]
- 720. Yin L, Lu L, Lin X, Wang X. Crucial role of androgen receptor in resistance and endurance trainings-induced muscle hypertrophy through IGF-1/IGF-1R- PI3K/Akt- mTOR pathway. Nutr Metab (Lond) 17: 26, 2020. doi: 10.1186/s12986-020-00446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721. McCall GE, Byrnes WC, Fleck SJ, Dickinson A, Kraemer WJ. Acute and chronic hormonal responses to resistance training designed to promote muscle hypertrophy. Can J Appl Physiol 24: 96–107, 1999. doi: 10.1139/h99-009. [DOI] [PubMed] [Google Scholar]
- 722. Kraemer WJ, Häkkinen K, Newton RU, Nindl BC, Volek JS, McCormick M, Gotshalk LA, Gordon SE, Fleck SJ, Campbell WW, Putukian M, Evans WJ. Effects of heavy-resistance training on hormonal response patterns in younger vs. older men. J Appl Physiol (1985) 87: 982–992, 1999. doi: 10.1152/jappl.1999.87.3.982. [DOI] [PubMed] [Google Scholar]
- 723. Häkkinen K, Pakarinen A, Kraemer WJ, Newton RU, Alen M. Basal concentrations and acute responses of serum hormones and strength development during heavy resistance training in middle-aged and elderly men and women. J Gerontol A Biol Sci Med Sci 55: B95–B105, 2000. doi: 10.1093/gerona/55.2.b95. [DOI] [PubMed] [Google Scholar]
- 724. Kraemer WJ, Staron RS, Hagerman FC, Hikida RS, Fry AC, Gordon SE, Nindl BC, Gothshalk LA, Volek JS, Marx JO, Newton RU, Häkkinen K. The effects of short-term resistance training on endocrine function in men and women. Eur J Appl Physiol Occup Physiol 78: 69–76, 1998. doi: 10.1007/s004210050389. [DOI] [PubMed] [Google Scholar]
- 725. Kraemer WJ, Volek JS, Bush JA, Putukian M, Sebastianelli WJ. Hormonal responses to consecutive days of heavy-resistance exercise with or without nutritional supplementation. J Appl Physiol (1985) 85: 1544–1555, 1998. doi: 10.1152/jappl.1998.85.4.1544. [DOI] [PubMed] [Google Scholar]
- 726. Kraemer WJ, Ratamess NA. Hormonal responses and adaptations to resistance exercise and training. Sports Med 35: 339–361, 2005. doi: 10.2165/00007256-200535040-00004. [DOI] [PubMed] [Google Scholar]
- 727. Morton RW, Oikawa SY, Wavell CG, Mazara N, McGlory C, Quadrilatero J, Baechler BL, Baker SK, Phillips SM. Neither load nor systemic hormones determine resistance training-mediated hypertrophy or strength gains in resistance-trained young men. J Appl Physiol (1985) 121: 129–138, 2016. doi: 10.1152/japplphysiol.00154.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728. West DW, Burd NA, Tang JE, Moore DR, Staples AW, Holwerda AM, Baker SK, Phillips SM. Elevations in ostensibly anabolic hormones with resistance exercise enhance neither training-induced muscle hypertrophy nor strength of the elbow flexors. J Appl Physiol (1985) 108: 60–67, 2010. doi: 10.1152/japplphysiol.01147.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729. West DW, Kujbida GW, Moore DR, Atherton P, Burd NA, Padzik JP, De Lisio M, Tang JE, Parise G, Rennie MJ, Baker SK, Phillips SM. Resistance exercise-induced increases in putative anabolic hormones do not enhance muscle protein synthesis or intracellular signalling in young men. J Physiol 587: 5239–5247, 2009. doi: 10.1113/jphysiol.2009.177220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730. Morton RW, Sato K, Gallaugher MPB, Oikawa SY, McNicholas PD, Fujita S, Phillips SM. Muscle androgen receptor content but not systemic hormones is associated with resistance training-induced skeletal muscle hypertrophy in healthy, young men. Front Physiol 9: 1373, 2018. doi: 10.3389/fphys.2018.01373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731. West DW, Burd NA, Churchward-Venne TA, Camera DM, Mitchell CJ, Baker SK, Hawley JA, Coffey VG, Phillips SM. Sex-based comparisons of myofibrillar protein synthesis after resistance exercise in the fed state. J Appl Physiol (1985) 112: 1805–1813, 2012. doi: 10.1152/japplphysiol.00170.2012. [DOI] [PubMed] [Google Scholar]
- 732. Ahtiainen JP, Hulmi JJ, Kraemer WJ, Lehti M, Nyman K, Selänne H, Alen M, Pakarinen A, Komulainen J, Kovanen V, Mero AA, Häkkinen K. Heavy resistance exercise training and skeletal muscle androgen receptor expression in younger and older men. Steroids 76: 183–192, 2011. doi: 10.1016/j.steroids.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 733. Mitchell CJ, Churchward-Venne TA, Bellamy L, Parise G, Baker SK, Phillips SM. Muscular and systemic correlates of resistance training-induced muscle hypertrophy. PLoS One 8: e78636, 2013. doi: 10.1371/journal.pone.0078636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734. Roberts BM, Nuckols G, Krieger JW. Sex differences in resistance training: a systematic review and meta-analysis. J Strength Cond Res 34: 1448–1460, 2020. doi: 10.1519/JSC.0000000000003521. [DOI] [PubMed] [Google Scholar]
- 735. Abe T, DeHoyos DV, Pollock ML, Garzarella L. Time course for strength and muscle thickness changes following upper and lower body resistance training in men and women. Eur J Appl Physiol 81: 174–180, 2000. doi: 10.1007/s004210050027. [DOI] [PubMed] [Google Scholar]
- 736. Cureton KJ, Collins MA, Hill DW, McElhannon FM Jr.. Muscle hypertrophy in men and women. Med Sci Sports Exerc 20: 338–344, 1988. doi: 10.1249/00005768-198808000-00003. [DOI] [PubMed] [Google Scholar]
- 737. Häkkinen K, Newton RU, Gordon SE, McCormick M, Volek JS, Nindl BC, Gotshalk LA, Campbell WW, Evans WJ, Häkkinen A, Humphries BJ, Kraemer WJ. Changes in muscle morphology, electromyographic activity, and force production characteristics during progressive strength training in young and older men. J Gerontol A Biol Sci Med Sci 53: B415–B423, 1998. doi: 10.1093/gerona/53a.6.b415. [DOI] [PubMed] [Google Scholar]
- 738. Hurlbut DE, Lott ME, Ryan AS, Ferrell RE, Roth SM, Ivey FM, Martel GF, Lemmer JT, Fleg JL, Hurley BF. Does age, sex, or ACE genotype affect glucose and insulin responses to strength training? J Appl Physiol (1985) 92: 643–650, 2002. doi: 10.1152/japplphysiol.00499.2001. [DOI] [PubMed] [Google Scholar]
- 739. Ivey FM, Roth SM, Ferrell RE, Tracy BL, Lemmer JT, Hurlbut DE, Martel GF, Siegel EL, Fozard JL, Jeffrey Metter E, Fleg JL, Hurley BF. Effects of age, gender, and myostatin genotype on the hypertrophic response to heavy resistance strength training. J Gerontol A Biol Sci Med Sci 55: M641–M648, 2000. doi: 10.1093/gerona/55.11.m641. [DOI] [PubMed] [Google Scholar]
- 740. Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J Appl Physiol (1985) 101: 531–544, 2006. doi: 10.1152/japplphysiol.01474.2005. [DOI] [PubMed] [Google Scholar]
- 741. O’Hagan FT, Sale DG, MacDougall JD, Garner SH. Response to resistance training in young women and men. Int J Sports Med 16: 314–321, 1995. doi: 10.1055/s-2007-973012. [DOI] [PubMed] [Google Scholar]
- 742. Weiss LW, Clark FC, Howard DG. Effects of heavy-resistance triceps surae muscle training on strength and muscularity of men and women. Phys Ther 68: 208–213, 1988. doi: 10.1093/ptj/68.2.208. [DOI] [PubMed] [Google Scholar]
- 743. Cardaci TD, Machek SB, Wilburn DT, Heileson JL, Willoughby DS. High-load resistance exercise augments androgen receptor-DNA binding and Wnt/beta-catenin signaling without increases in serum/muscle androgens or androgen receptor content. Nutrients 12: 3829, 2020. doi: 10.3390/nu12123829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744. Dart DA, Waxman J, Aboagye EO, Bevan CL. Visualising androgen receptor activity in male and female mice. PLoS One 8: e71694, 2013. doi: 10.1371/journal.pone.0071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745. Dalgaard LB, Jørgensen EB, Oxfeldt M, Dalgaard EB, Johansen FT, Karlsson M, Ringgaard S, Hansen M. Influence of second generation oral contraceptive use on adaptations to resistance training in young untrained women. J Strength Cond Res 36: 1801–1809, 2022. doi: 10.1519/JSC.0000000000003735. [DOI] [PubMed] [Google Scholar]
- 746. Romance R, Vargas S, Espinar S, Petro JL, Bonilla DA, Schöenfeld BJ, Kreider RB, Benítez-Porres J. Oral contraceptive use does not negatively affect body composition and strength adaptations in trained women. Int J Sports Med 40: 842–849, 2019. doi: 10.1055/a-0985-4373. [DOI] [PubMed] [Google Scholar]
- 747. Dalgaard LB, Dalgas U, Andersen JL, Rossen NB, Møller AB, Stødkilde-Jørgensen H, Jorgensen JO, Kovanen V, Couppe C, Langberg H, Kjaer M, Hansen M. Influence of oral contraceptive use on adaptations to resistance training. Front Physiol 10: 824, 2019. doi: 10.3389/fphys.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748. Ruzić L, Matković BR, Leko G. Antiandrogens in hormonal contraception limit muscle strength gain in strength training: comparison study. Croat Med J 44: 65–68, 2003. [PubMed] [Google Scholar]
- 749. Riechman SE, Lee CW. Oral contraceptive use impairs muscle gains in young women. J Strength Cond Res 36: 3074–3080, 2022. doi: 10.1519/JSC.0000000000004059. [DOI] [PubMed] [Google Scholar]
- 750. Hansen M, Skovgaard D, Reitelseder S, Holm L, Langbjerg H, Kjaer M. Effects of estrogen replacement and lower androgen status on skeletal muscle collagen and myofibrillar protein synthesis in postmenopausal women. J Gerontol A Biol Sci Med Sci 67: 1005–1013, 2012. doi: 10.1093/gerona/gls007. [DOI] [PubMed] [Google Scholar]
- 751. Javed AA, Mayhew AJ, Shea AK, Raina P. Association between hormone therapy and muscle mass in postmenopausal women: a systematic review and meta-analysis. JAMA Netw Open 2: e1910154, 2019. doi: 10.1001/jamanetworkopen.2019.10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752. Nørregaard R, Kwon TH, Frøkiær J. Physiology and pathophysiology of cyclooxygenase-2 and prostaglandin E2 in the kidney. Kidney Res Clin Pract 34: 194–200, 2015. doi: 10.1016/j.krcp.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753. Rodemann HP, Goldberg AL. Arachidonic acid, prostaglandin E2 and F2 alpha influence rates of protein turnover in skeletal and cardiac muscle. J Biol Chem 257: 1632–1638, 1982. doi: 10.1016/S0021-9258(19)68084-1. [DOI] [PubMed] [Google Scholar]
- 754. Trappe TA, White F, Lambert CP, Cesar D, Hellerstein M, Evans WJ. Effect of ibuprofen and acetaminophen on postexercise muscle protein synthesis. Am J Physiol Endocrinol Metab 282: E551–E556, 2002. doi: 10.1152/ajpendo.00352.2001. [DOI] [PubMed] [Google Scholar]
- 755. Trappe TA, Fluckey JD, White F, Lambert CP, Evans WJ. Skeletal muscle PGF2alpha and PGE2 in response to eccentric resistance exercise: influence of ibuprofen acetaminophen. J Clin Endocrinol Metab 86: 5067–5070, 2001. doi: 10.1210/jcem.86.10.7928. [DOI] [PubMed] [Google Scholar]
- 756. Markworth JF, Cameron-Smith D. Prostaglandin F2alpha stimulates PI3K/ERK/mTOR signaling and skeletal myotube hypertrophy. Am J Physiol Cell Physiol 300: C671–C682, 2011. doi: 10.1152/ajpcell.00549.2009. [DOI] [PubMed] [Google Scholar]
- 757. Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem 282: 11613–11617, 2007. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- 758. Trappe TA, Standley RA, Jemiolo B, Carroll CC, Trappe SW. Prostaglandin and myokine involvement in the cyclooxygenase-inhibiting drug enhancement of skeletal muscle adaptations to resistance exercise in older adults. Am J Physiol Regul Integr Comp Physiol 304: R198–R205, 2013. doi: 10.1152/ajpregu.00245.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759. Ho AT, Palla AR, Blake MR, Yucel ND, Wang YX, Magnusson KE, Holbrook CA, Kraft PE, Delp SL, Blau HM. Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc Natl Acad Sci USA 114: 6675–6684, 2017. doi: 10.1073/pnas.1705420114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760. Soltow QA, Betters JL, Sellman JE, Lira VA, Long JH, Criswell DS. Ibuprofen inhibits skeletal muscle hypertrophy in rats. Med Sci Sports Exerc 38: 840–846, 2006. doi: 10.1249/01.mss.0000218142.98704.66. [DOI] [PubMed] [Google Scholar]
- 761. Novak ML, Billich W, Smith SM, Sukhija KB, McLoughlin TJ, Hornberger TA, Koh TJ. COX-2 inhibitor reduces skeletal muscle hypertrophy in mice. Am J Physiol Regul Integr Comp Physiol 296: R1132–R1139, 2009. doi: 10.1152/ajpregu.90874.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762. Markworth JF, Vella LD, Figueiredo VC, Cameron-Smith D. Ibuprofen treatment blunts early translational signaling responses in human skeletal muscle following resistance exercise. J Appl Physiol (1985) 117: 20–28, 2014. doi: 10.1152/japplphysiol.01299.2013. [DOI] [PubMed] [Google Scholar]
- 763. Lilja M, Mandić M, Apró W, Melin M, Olsson K, Rosenborg S, Gustafsson T, Lundberg TR. High doses of anti-inflammatory drugs compromise muscle strength and hypertrophic adaptations to resistance training in young adults. Acta Physiol (Oxf) 222: e12948, 2018. doi: 10.1111/apha.12948. [DOI] [PubMed] [Google Scholar]
- 764. Mikkelsen UR, Langberg H, Helmark IC, Skovgaard D, Andersen LL, Kjaer M, Mackey AL. Local NSAID infusion inhibits satellite cell proliferation in human skeletal muscle after eccentric exercise. J Appl Physiol (1985) 107: 1600–1611, 2009. doi: 10.1152/japplphysiol.00707.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 765. Mackey AL, Kjaer M, Dandanell S, Mikkelsen KH, Holm L, Døssing S, Kadi F, Koskinen SO, Jensen CH, Schrøder HD, Langberg H. The influence of anti-inflammatory medication on exercise-induced myogenic precursor cell responses in humans. J Appl Physiol (1985) 103: 425–431, 2007. doi: 10.1152/japplphysiol.00157.2007. [DOI] [PubMed] [Google Scholar]
- 766. Versey NG, Halson SL, Dawson BT. Water immersion recovery for athletes: effect on exercise performance and practical recommendations. Sports Med 43: 1101–1130, 2013. doi: 10.1007/s40279-013-0063-8. [DOI] [PubMed] [Google Scholar]
- 767. Roberts LA, Raastad T, Markworth JF, Figueiredo VC, Egner IM, Shield A, Cameron-Smith D, Coombes JS, Peake JM. Post-exercise cold water immersion attenuates acute anabolic signalling and long-term adaptations in muscle to strength training. J Physiol 593: 4285–4301, 2015. doi: 10.1113/JP270570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 768. Fyfe JJ, Broatch JR, Trewin AJ, Hanson ED, Argus CK, Garnham AP, Halson SL, Polman RC, Bishop DJ, Petersen AC. Cold water immersion attenuates anabolic signaling and skeletal muscle fiber hypertrophy, but not strength gain, following whole-body resistance training. J Appl Physiol (1985) 127: 1403–1418, 2019. doi: 10.1152/japplphysiol.00127.2019. [DOI] [PubMed] [Google Scholar]
- 769. Guillot X, Tordi N, Laheurte C, Pazart L, Prati C, Saas P, Wendling D. Local ice cryotherapy decreases synovial interleukin 6, interleukin 1beta, vascular endothelial growth factor, prostaglandin-E2, and nuclear factor kappa B p65 in human knee arthritis: a controlled study. Arthritis Res Ther 21: 180, 2019. doi: 10.1186/s13075-019-1965-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770. Zhang J, Pan T, Wang JH. Cryotherapy suppresses tendon inflammation in an animal model. J Orthop Translat 2: 75–81, 2014. doi: 10.1016/j.jot.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771. Krentz JR, Quest B, Farthing JP, Quest DW, Chilibeck PD. The effects of ibuprofen on muscle hypertrophy, strength, and soreness during resistance training. Appl Physiol Nutr Metab 33: 470–475, 2008. doi: 10.1139/H08-019. [DOI] [PubMed] [Google Scholar]
- 772. Candow DG, Chilibeck PD, Weisgarber K, Vogt E, Baxter-Jones AD. Ingestion of low-dose ibuprofen following resistance exercise in postmenopausal women. J Cachexia Sarcopenia Muscle 4: 41–46, 2013. doi: 10.1007/s13539-012-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773. Trappe TA, Carroll CC, Dickinson JM, LeMoine JK, Haus JM, Sullivan BE, Lee JD, Jemiolo B, Weinheimer EM, Hollon CJ. Influence of acetaminophen and ibuprofen on skeletal muscle adaptations to resistance exercise in older adults. Am J Physiol Regul Integr Comp Physiol 300: R655–R662, 2011. doi: 10.1152/ajpregu.00611.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774. Damas F, Phillips SM, Lixandrão ME, Vechin FC, Libardi CA, Roschel H, Tricoli V, Ugrinowitsch C. An inability to distinguish edematous swelling from true hypertrophy still prevents a completely accurate interpretation of the time course of muscle hypertrophy. Eur J Appl Physiol 116: 445–446, 2016. doi: 10.1007/s00421-015-3287-5. [DOI] [PubMed] [Google Scholar]
- 775. Johnson M. Molecular mechanisms of beta2-adrenergic receptor function, response, and regulation. J Allergy Clin Immunol 117: 18–24, 2006. doi: 10.1016/j.jaci.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 776. Murphy RJ, Beliveau L, Gardiner PF, Calderone A. Nifedipine does not impede clenbuterol-stimulated muscle hypertrophy. Proc Soc Exp Biol Med 221: 184–187, 1999. doi: 10.3181/00379727-221-44402. [DOI] [PubMed] [Google Scholar]
- 777. Beermann DH, Butler WR, Hogue DE, Fishell VK, Dalrymple RH, Ricks CA, Scanes CG. Cimaterol-induced muscle hypertrophy and altered endocrine status in lambs. J Anim Sci 65: 1514–1524, 1987. doi: 10.2527/jas1987.6561514x. [DOI] [PubMed] [Google Scholar]
- 778. Wang SY, Beermann DH. Reduced calcium-dependent proteinase activity in cimaterol-induced muscle hypertrophy in lambs. J Anim Sci 66: 2545–2550, 1988. doi: 10.2527/jas1988.66102545x. [DOI] [PubMed] [Google Scholar]
- 779. Maltin CA, Delday MI, Hay SM, Smith FG, Lobley GE, Reeds PJ. The effect of the anabolic agent, clenbuterol, on overloaded rat skeletal muscle. Biosci Rep 7: 143–149, 1987. doi: 10.1007/BF01121878. [DOI] [PubMed] [Google Scholar]
- 780. Maltin CA, Hay SM, Delday MI, Smith FG, Lobley GE, Reeds PJ. Clenbuterol, a beta agonist, induces growth in innervated and denervated rat soleus muscle via apparently different mechanisms. Biosci Rep 7: 525–532, 1987. doi: 10.1007/BF01116510. [DOI] [PubMed] [Google Scholar]
- 781. Maltin CA, Delday MI, Reeds PJ. The effect of a growth promoting drug, clenbuterol, on fibre frequency and area in hind limb muscles from young male rats. Biosci Rep 6: 293–299, 1986. doi: 10.1007/BF01115158. [DOI] [PubMed] [Google Scholar]
- 782. Carter WJ, Lynch ME. Comparison of the effects of salbutamol and clenbuterol on skeletal muscle mass and carcass composition in senescent rats. Metabolism 43: 1119–1125, 1994. doi: 10.1016/0026-0495(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 783. Claeys MC, Mulvaney DR, McCarthy FD, Gore MT, Marple DN, Sartin JL. Skeletal muscle protein synthesis and growth hormone secretion in young lambs treated with clenbuterol. J Anim Sci 67: 2245–2254, 1989. doi: 10.2527/jas1989.6792245x. [DOI] [PubMed] [Google Scholar]
- 784. Emery PW, Rothwell NJ, Stock MJ, Winter PD. Chronic effects of beta 2-adrenergic agonists on body composition and protein synthesis in the rat. Biosci Rep 4: 83–91, 1984. doi: 10.1007/BF01120827. [DOI] [PubMed] [Google Scholar]
- 785. Reeds PJ, Hay SM, Dorward PM, Palmer RM. The effect of beta-agonists and antagonists on muscle growth and body composition of young rats (Rattus sp.). Comp Biochem Physiol C Comp Pharmacol Toxicol 89: 337–341, 1988. doi: 10.1016/0742-8413(88)90234-4. [DOI] [PubMed] [Google Scholar]
- 786. Kim J, Grotegut CA, Wisler JW, Li T, Mao L, Chen M, Chen W, Rosenberg PB, Rockman HA, Lefkowitz RJ. Beta-arrestin 1 regulates beta2-adrenergic receptor-mediated skeletal muscle hypertrophy and contractility. Skelet Muscle 8: 39, 2018. doi: 10.1186/s13395-018-0184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787. Hinkle RT, Hodge KM, Cody DB, Sheldon RJ, Kobilka BK, Isfort RJ. Skeletal muscle hypertrophy and anti-atrophy effects of clenbuterol are mediated by the beta2-adrenergic receptor. Muscle Nerve 25: 729–734, 2002. doi: 10.1002/mus.10092. [DOI] [PubMed] [Google Scholar]
- 788. Woodall BP, Woodall MC, Luongo TS, Grisanti LA, Tilley DG, Elrod JW, Koch WJ. Skeletal muscle-specific G protein-coupled receptor kinase 2 ablation alters isolated skeletal muscle mechanics and enhances clenbuterol-stimulated hypertrophy. J Biol Chem 291: 21913–21924, 2016. doi: 10.1074/jbc.M116.721282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 789. Stewart KJ, Effron MB, Valenti SA, Kelemen MH. Effects of diltiazem or propranolol during exercise training of hypertensive men. Med Sci Sports Exerc 22: 171–177, 1990. [PubMed] [Google Scholar]
- 790. Jessen S, Solheim SA, Jacobson GA, Eibye K, Bangsbo J, Nordsborg NB, Hostrup M. Beta2-adrenergic agonist clenbuterol increases energy expenditure and fat oxidation, and induces mTOR phosphorylation in skeletal muscle of young healthy men. Drug Test Anal 12: 610–618, 2020. doi: 10.1002/dta.2755. [DOI] [PubMed] [Google Scholar]
- 791. Jessen S, Reitelseder S, Kalsen A, Kreiberg M, Onslev J, Gad A, Ørtenblad N, Backer V, Holm L, Bangsbo J, Hostrup M. beta2-Adrenergic agonist salbutamol augments hypertrophy in MHCIIa fibers and sprint mean power output but not muscle force during 11 weeks of resistance training in young men. J Appl Physiol (1985) 130: 617–626, 2021. doi: 10.1152/japplphysiol.00553.2020. [DOI] [PubMed] [Google Scholar]
- 792. Koopman R, Gehrig SM, Léger B, Trieu J, Walrand S, Murphy KT, Lynch GS. Cellular mechanisms underlying temporal changes in skeletal muscle protein synthesis and breakdown during chronic {beta}-adrenoceptor stimulation in mice. J Physiol 588: 4811–4823, 2010. doi: 10.1113/jphysiol.2010.196725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793. Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol 5: 87–90, 2003. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 794. Sporano V, Grasso L, Esposito M, Oliviero G, Brambilla G, Loizzo A. Clenbuterol residues in non-liver containing meat as a cause of collective food poisoning. Vet Hum Toxicol 40: 141–143, 1998. [PubMed] [Google Scholar]
- 795. Elliott CT, McCaughey WJ, Shortt HD. Residues of the beta-agonist clenbuterol in tissues of medicated farm animals. Food Addit Contam 10: 231–244, 1993. doi: 10.1080/02652039309374145. [DOI] [PubMed] [Google Scholar]
- 796. Mitchell GA, Dunnavan G. Illegal use of beta-adrenergic agonists in the United States. J Anim Sci 76: 208–211, 1998. doi: 10.2527/1998.761208x. [DOI] [PubMed] [Google Scholar]
- 797. Kline WO, Panaro FJ, Yang H, Bodine SC. Rapamycin inhibits the growth and muscle-sparing effects of clenbuterol. J Appl Physiol (1985) 102: 740–747, 2007. doi: 10.1152/japplphysiol.00873.2006. [DOI] [PubMed] [Google Scholar]
- 798. Pullinen T, Mero A, MacDonald E, Pakarinen A, Komi PV. Plasma catecholamine and serum testosterone responses to four units of resistance exercise in young and adult male athletes. Eur J Appl Physiol Occup Physiol 77: 413–420, 1998. doi: 10.1007/s004210050353. [DOI] [PubMed] [Google Scholar]
- 799. Pullinen T, Nicol C, MacDonald E, Komi PV. Plasma catecholamine responses to four resistance exercise tests in men and women. Eur J Appl Physiol Occup Physiol 80: 125–131, 1999. doi: 10.1007/s004210050568. [DOI] [PubMed] [Google Scholar]
- 800. Pullinen T, Huttunen P, Komi PV. Plasma catecholamine responses and neural adaptation during short-term resistance training. Eur J Appl Physiol 82: 68–75, 2000. doi: 10.1007/s004210050653. [DOI] [PubMed] [Google Scholar]
- 801. Stock C, Schaller K, Baum M, Liesen H, Weiss M. Catecholamines, lymphocyte subsets, and cyclic adenosine monophosphate production in mononuclear cells and CD4+ cells in response to submaximal resistance exercise. Eur J Appl Physiol Occup Physiol 71: 166–172, 1995. doi: 10.1007/BF00854975. [DOI] [PubMed] [Google Scholar]
- 802. Steiner JL, Johnson BR, Hickner RC, Ormsbee MJ, Williamson DL, Gordon BS. Adrenal stress hormone action in skeletal muscle during exercise training: an old dog with new tricks? Acta Physiol (Oxf) 231: e13522, 2021. doi: 10.1111/apha.13522. [DOI] [PubMed] [Google Scholar]
- 803. Sadoshima J, Xu Y, Slayter HS, Izumo S. Autocrine release of angiotensin II mediates stretch-induced hypertrophy of cardiac myocytes in vitro. Cell 75: 977–984, 1993. doi: 10.1016/0092-8674(93)90541-w. [DOI] [PubMed] [Google Scholar]
- 804. Baker KM, Aceto JF. Angiotensin II stimulation of protein synthesis and cell growth in chick heart cells. Am J Physiol Heart Circ Physiol 259: H610–H618, 1990. doi: 10.1152/ajpheart.1990.259.2.H610. [DOI] [PubMed] [Google Scholar]
- 805. Neyses L, Vetter H. Action of atrial natriuretic peptide and angiotensin II on the myocardium: studies in isolated rat ventricular cardiomyocytes. Biochem Biophys Res Commun 163: 1435–1443, 1989. doi: 10.1016/0006-291x(89)91139-x. [DOI] [PubMed] [Google Scholar]
- 806. Gordon SE, Davis BS, Carlson CJ, Booth FW. ANG II is required for optimal overload-induced skeletal muscle hypertrophy. Am J Physiol Endocrinol Metab 280: E150–E159, 2001. doi: 10.1152/ajpendo.2001.280.1.E150. [DOI] [PubMed] [Google Scholar]
- 807. Westerkamp CM, Gordon SE. Angiotensin-converting enzyme inhibition attenuates myonuclear addition in overloaded slow-twitch skeletal muscle. Am J Physiol Regul Integr Comp Physiol 289: R1223–R1231, 2005. doi: 10.1152/ajpregu.00730.2004. [DOI] [PubMed] [Google Scholar]
- 808. McBride TA. AT1 receptors are necessary for eccentric training-induced hypertrophy and strength gains in rat skeletal muscle. Exp Physiol 91: 413–421, 2006. doi: 10.1113/expphysiol.2005.032490. [DOI] [PubMed] [Google Scholar]
- 809. Zempo H, Suzuki J, Ogawa M, Watanabe R, Isobe M. A different role of angiotensin II type 1a receptor in the development and hypertrophy of plantaris muscle in mice. J Appl Genet 57: 91–97, 2016. doi: 10.1007/s13353-015-0291-8. [DOI] [PubMed] [Google Scholar]
- 810. Heisterberg MF, Andersen JL, Schjerling P, Lund A, Dalskov S, Jønsson AO, Warming N, Fogelstrøm M, Kjaer M, Mackey AL. Losartan has no additive effect on the response to heavy-resistance exercise in human elderly skeletal muscle. J Appl Physiol (1985) 125: 1536–1554, 2018. doi: 10.1152/japplphysiol.00106.2018. [DOI] [PubMed] [Google Scholar]
- 811. Heisterberg MF, Andersen JL, Schjerling P, Bülow J, Lauersen JB, Roeber HL, Kjaer M, Mackey AL. Effect of losartan on the acute response of human elderly skeletal muscle to exercise. Med Sci Sports Exerc 50: 225–235, 2018. doi: 10.1249/MSS.0000000000001438. [DOI] [PubMed] [Google Scholar]
- 812. Zhang L, Du J, Hu Z, Han G, Delafontaine P, Garcia G, Mitch WE. IL-6 and serum amyloid A synergy mediates angiotensin II-induced muscle wasting. J Am Soc Nephrol 20: 604–612, 2009. doi: 10.1681/ASN.2008060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813. Stouthamer AH. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek 39: 545–565, 1973. doi: 10.1007/BF02578899. [DOI] [PubMed] [Google Scholar]
- 814. Grgic J, Schoenfeld BJ. Are the hypertrophic adaptations to high and low-load resistance training muscle fiber type specific? Front Physiol 9: 402, 2018. doi: 10.3389/fphys.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815. Joseph AM, Adhihetty PJ, Wawrzyniak NR, Wohlgemuth SE, Picca A, Kujoth GC, Prolla TA, Leeuwenburgh C. Dysregulation of mitochondrial quality control processes contribute to sarcopenia in a mouse model of premature aging. PLoS One 8: e69327, 2013. doi: 10.1371/journal.pone.0069327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 816. Ross JM, Coppotelli G, Branca RM, Kim KM, Lehtiö J, Sinclair DA, Olson L. Voluntary exercise normalizes the proteomic landscape in muscle and brain and improves the phenotype of progeroid mice. Aging Cell 18: e13029, 2019. doi: 10.1111/acel.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839, 1998. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 818. Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA 103: 16260–16265, 2006. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 819. Wang J, Wang F, Zhang P, Liu H, He J, Zhang C, Fan M, Chen X. PGC-1alpha over-expression suppresses the skeletal muscle atrophy and myofiber-type composition during hindlimb unloading. Biosci Biotechnol Biochem 81: 500–513, 2017. doi: 10.1080/09168451.2016.1254531. [DOI] [PubMed] [Google Scholar]
- 820. Pérez-Schindler J, Summermatter S, Santos G, Zorzato F, Handschin C. The transcriptional coactivator PGC-1alpha is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc Natl Acad Sci USA 110: 20314–20319, 2013. doi: 10.1073/pnas.1312039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821. Groennebaek T, Vissing K. Impact of resistance training on skeletal muscle mitochondrial biogenesis, content, and function. Front Physiol 8: 713, 2017. doi: 10.3389/fphys.2017.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822. Larsen S, Nielsen J, Hansen CN, Nielsen LB, Wibrand F, Stride N, Schroder HD, Boushel R, Helge JW, Dela F, Hey-Mogensen M. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol 590: 3349–3360, 2012. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823. Tang JE, Hartman JW, Phillips SM. Increased muscle oxidative potential following resistance training induced fibre hypertrophy in young men. Appl Physiol Nutr Metab 31: 495–501, 2006. doi: 10.1139/h06-026. [DOI] [PubMed] [Google Scholar]
- 824. Pesta D, Hoppel F, Macek C, Messner H, Faulhaber M, Kobel C, Parson W, Burtscher M, Schocke M, Gnaiger E. Similar qualitative and quantitative changes of mitochondrial respiration following strength and endurance training in normoxia and hypoxia in sedentary humans. Am J Physiol Regul Integr Comp Physiol 301: R1078–R1087, 2011. doi: 10.1152/ajpregu.00285.2011. [DOI] [PubMed] [Google Scholar]
- 825. Salvadego D, Domenis R, Lazzer S, Porcelli S, Rittweger J, Rizzo G, Mavelli I, Simunic B, Pisot R, Grassi B. Skeletal muscle oxidative function in vivo and ex vivo in athletes with marked hypertrophy from resistance training. J Appl Physiol (1985) 114: 1527–1535, 2013. doi: 10.1152/japplphysiol.00883.2012. [DOI] [PubMed] [Google Scholar]
- 826. Porter C, Reidy PT, Bhattarai N, Sidossis LS, Rasmussen BB. Resistance exercise training alters mitochondrial function in human skeletal muscle. Med Sci Sports Exerc 47: 1922–1931, 2015. doi: 10.1249/MSS.0000000000000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827. Roberts MD, Romero MA, Mobley CB, Mumford PW, Roberson PA, Haun CT, Vann CG, Osburn SC, Holmes HH, Greer RA, Lockwood CM, Parry HA, Kavazis AN. Skeletal muscle mitochondrial volume and myozenin-1 protein differences exist between high versus low anabolic responders to resistance training. PeerJ 6: e5338, 2018. doi: 10.7717/peerj.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828. Koh JH, Pataky MW, Dasari S, Klaus KA, Vuckovic I, Ruegsegger GN, Kumar AP, Robinson MM, Nair KS. Enhancement of anaerobic glycolysis—a role of PGC-1alpha4 in resistance exercise. Nat Commun 13: 2324, 2022. doi: 10.1038/s41467-022-30056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 829. Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, Lanza IR, Rasbach KA, Okutsu M, Nair KS, Yan Z, Leinwand LA, Spiegelman BM. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151: 1319–1331, 2012. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830. Verbrugge SA, Gehlert S, Stadhouders LE, Jacko D, Aussieker T, de Wit G, Vogel IS, Offringa C, Schönfelder M, Jaspers RT, Wackerhage H. PKM2 determines myofiber hypertrophy in vitro and increases in response to resistance exercise in human skeletal muscle. Int J Mol Sci 21: 7062, 2020. doi: 10.3390/ijms21197062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 831. Valentino T, Figueiredo VC, Mobley CB, McCarthy JJ, Vechetti IJ Jr.. Evidence of myomiR regulation of the pentose phosphate pathway during mechanical load-induced hypertrophy. Physiol Rep 9: e15137, 2021. doi: 10.14814/phy2.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832. Stadhouders LE, Verbrugge SA, Smith JAB, Gabriel BM, Hammersen TD, Kolijin D, Vogel IS, Mohamed AD, de Wit GM, Offringa C, Hoogaars WM, Gehlert S, Wackerhage H, Jaspers RT. Myotube hypertrophy is associated with cancer-like metabolic reprogramming and limited by PHGDH (Preprint). bioRxiv 2020.12.01.403949, 2020. doi: 10.1101/2020.12.01.403949. [DOI]
- 833. Suginohara T, Wakabayashi K, Ato S, Ogasawara R. Effect of 2-deoxyglucose-mediated inhibition of glycolysis on the regulation of mTOR signaling and protein synthesis before and after high-intensity muscle contraction. Metabolism 114: 154419, 2021. doi: 10.1016/j.metabol.2020.154419. [DOI] [PubMed] [Google Scholar]
- 834. Wackerhage H, Vechetti IJ, Baumert P, Gehlert S, Becker L, Jaspers RT, de Angelis MH. Does a hypertrophying muscle fibre reprogramme its metabolism similar to a cancer cell? Sports Med 52: 2569–2578, 2022. doi: 10.1007/s40279-022-01676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 835. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 8: 519–530, 1927. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 836. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 837. Chatterjee S, Ma K. Circadian clock regulation of skeletal muscle growth and repair. F1000Res 5: 1549, 2016. doi: 10.12688/f1000research.9076.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838. Perrin L, Loizides-Mangold U, Skarupelova S, Pulimeno P, Chanon S, Robert M, Bouzakri K, Modoux C, Roux-Lombard P, Vidal H, Lefai E, Dibner C. Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol Metab 4: 834–845, 2015. doi: 10.1016/j.molmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 839. McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics 31: 86–95, 2007. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840. Perrin L, Loizides-Mangold U, Chanon S, Gobet C, Hulo N, Isenegger L, Weger BD, Migliavacca E, Charpagne A, Betts JA, Walhin JP, Templeman I, Stokes K, Thompson D, Tsintzas K, Robert M, Howald C, Riezman H, Feige JN, Karagounis LG, Johnston JD, Dermitzakis ET, Gachon F, Lefai E, Dibner C. Transcriptomic analyses reveal rhythmic and CLOCK-driven pathways in human skeletal muscle. Elife 7: e34114, 2018. doi: 10.7554/eLife.34114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841. Harfmann BD, Schroder EA, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. J Biol Rhythms 30: 84–94, 2015. doi: 10.1177/0748730414561638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842. Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA 107: 19090–19095, 2010. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 843. Hodge BA, Wen Y, Riley LA, Zhang X, England JH, Harfmann BD, Schroder EA, Esser KA. The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skelet Muscle 5: 17, 2015. doi: 10.1186/s13395-015-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844. Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, Takahashi JS, Schambelan M, Conklin BR. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol 4: R61, 2003. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845. Grgic J, Lazinica B, Garofolini A, Schoenfeld BJ, Saner NJ, Mikulic P. The effects of time of day-specific resistance training on adaptations in skeletal muscle hypertrophy and muscle strength: A systematic review and meta-analysis. Chronobiol Int 36: 449–460, 2019. doi: 10.1080/07420528.2019.1567524. [DOI] [PubMed] [Google Scholar]
- 846. Cartwright J Jr, Goldstein MA. Microtubules in soleus muscles of the postnatal and adult rat. J Ultrastruct Res 79: 74–84, 1982. doi: 10.1016/s0022-5320(82)90053-3. [DOI] [PubMed] [Google Scholar]
- 847. Boudriau S, Vincent M, Côté CH, Rogers PA. Cytoskeletal structure of skeletal muscle: identification of an intricate exosarcomeric microtubule lattice in slow- and fast-twitch muscle fibers. J Histochem Cytochem 41: 1013–1021, 1993. doi: 10.1177/41.7.8515044. [DOI] [PubMed] [Google Scholar]
- 848. Denes LT, Kelley CP, Wang ET. Microtubule-based transport is essential to distribute RNA and nascent protein in skeletal muscle. Nat Commun 12: 6079, 2021. doi: 10.1038/s41467-021-26383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 849. Pinheiro H, Pimentel MR, Sequeira C, Oliveira LM, Pezzarossa A, Roman W, Gomes ER. mRNA distribution in skeletal muscle is associated with mRNA size. J Cell Sci 134: jcs256388, 2021. doi: 10.1242/jcs.256388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 850. Roman W, Pinheiro H, Pimentel MR, Segalés J, Oliveira LM, García-Domínguez E, Gomez-Cabrera MC, Serrano AL, Gomes ER, Munoz-Canoves P. Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science 374: 355–359, 2021. doi: 10.1126/science.abe5620. [DOI] [PubMed] [Google Scholar]
- 851. Wang X, Li F, Campbell SE, Gerdes AM. Chronic pressure overload cardiac hypertrophy and failure in guinea pigs: II. Cytoskeletal remodeling. J Mol Cell Cardiol 31: 319–331, 1999. doi: 10.1006/jmcc.1998.0885. [DOI] [PubMed] [Google Scholar]
- 852. Monreal G, Nicholson LM, Han B, Joshi MS, Phillips AB, Wold LE, Bauer JA, Gerhardt MA. Cytoskeletal remodeling of desmin is a more accurate measure of cardiac dysfunction than fibrosis or myocyte hypertrophy. Life Sci 83: 786–794, 2008. doi: 10.1016/j.lfs.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 853. Tsutsui H, Ishihara K, Cooper G. Cytoskeletal role in the contractile dysfunction of hypertrophied myocardium. Science 260: 682–687, 1993. doi: 10.1126/science.8097594. [DOI] [PubMed] [Google Scholar]
- 854. Fassett JT, Xu X, Hu X, Zhu G, French J, Chen Y, Bache RJ. Adenosine regulation of microtubule dynamics in cardiac hypertrophy. Am J Physiol Heart Circ Physiol 297: H523–H532, 2009. doi: 10.1152/ajpheart.00462.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 855. Scarborough EA, Uchida K, Vogel M, Erlitzki N, Iyer M, Phyo SA, Bogush A, Kehat I, Prosser BL. Microtubules orchestrate local translation to enable cardiac growth. Nat Commun 12: 1547, 2021. doi: 10.1038/s41467-021-21685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 856. Fortes MA, Marzuca-Nassr GN, Vitzel KF, da Justa Pinheiro CH, Newsholme P, Curi R. Housekeeping proteins: how useful are they in skeletal muscle diabetes studies and muscle hypertrophy models? Anal Biochem 504: 38–40, 2016. doi: 10.1016/j.ab.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 857. Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol 31: 69–75, 2015. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 858. Mailing LJ, Allen JM, Buford TW, Fields CJ, Woods JA. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc Sport Sci Rev 47: 75–85, 2019. doi: 10.1249/JES.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 859. Clauss M, Gérard P, Mosca A, Leclerc M. Interplay between exercise and gut microbiome in the context of human health and performance. Front Nutr 8: 637010, 2021. doi: 10.3389/fnut.2021.637010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860. Clark A, Mach N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J Int Soc Sports Nutr 13: 43, 2016. doi: 10.1186/s12970-016-0155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 861. Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, Holscher HD, Woods JA. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc 50: 747–757, 2018. doi: 10.1249/MSS.0000000000001495. [DOI] [PubMed] [Google Scholar]
- 862. Cronin O, Barton W, Skuse P, Penney NC, Garcia-Perez I, Murphy EF, Woods T, Nugent H, Fanning A, Melgar S, Falvey EC, Holmes E, Cotter PD, O’Sullivan O, Molloy MG, Shanahan F. A prospective metagenomic and metabolomic analysis of the impact of exercise and/or whey protein supplementation on the gut microbiome of sedentary adults. mSystems 3: e00044-18, 2018. doi: 10.1128/mSystems.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 863. Durk RP, Castillo E, Márquez-Magaña L, Grosicki GJ, Bolter ND, Lee CM, Bagley JR. Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int J Sport Nutr Exerc Metab 29: 249–253, 2019. doi: 10.1123/ijsnem.2018-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864. Bycura D, Santos AC, Shiffer A, Kyman S, Winfree K, Sutliffe J, Pearson T, Sonderegger D, Cope E, Caporaso JG. Impact of different exercise modalities on the human gut microbiome. Sports (Basel) 9: 14, 2021. doi: 10.3390/sports9020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865. Moore JH, Smith KS, Chen D, Lamb DA, Smith MA, Osburn SC, Ruple BA, Morrow CD, Huggins KW, McDonald JR, Brown MD, Young KC, Roberts MD, Frugé AD. Exploring the effects of six weeks of resistance training on the fecal microbiome of older adult males: secondary analysis of a peanut protein supplemented randomized controlled trial. Sports (Basel) 10: 65, 2022. doi: 10.3390/sports10050065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 866. Fernández J, Fernández-Sanjurjo M, Iglesias-Gutiérrez E, Martinez-Camblor P, Villar CJ, Tomas-Zapico C, Fernández-Garcia B, Lombo F. Resistance and endurance exercise training induce differential changes in gut microbiota composition in murine models. Front Physiol 12: 748854, 2021. doi: 10.3389/fphys.2021.748854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867. Valentino TR, Vechetti IJ Jr, Mobley CB, Dungan CM, Golden L, Goh J, McCarthy JJ. Dysbiosis of the gut microbiome impairs mouse skeletal muscle adaptation to exercise. J Physiol 599: 4845–4863, 2021. doi: 10.1113/JP281788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868. Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 104: 979–984, 2007. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869. Castro AP, Silva KK, Medeiros CS, Alves F, Araujo RC, Almeida JA. Effects of 12 weeks of resistance training on rat gut microbiota composition. J Exp Biol 224, 2021. doi: 10.1242/jeb.242543. [DOI] [PubMed] [Google Scholar]
- 870. Jones MD, Wewege MA, Hackett DA, Keogh JW, Hagstrom AD. Sex differences in adaptations in muscle strength and size following resistance training in older adults: a systematic review and meta-analysis. Sports Med 51: 503–517, 2021. doi: 10.1007/s40279-020-01388-4. [DOI] [PubMed] [Google Scholar]
- 871. Kojic F, Mandic D, Ilic V. Resistance training induces similar adaptations of upper and lower-body muscles between sexes. Sci Rep 11: 23449, 2021. doi: 10.1038/s41598-021-02867-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 872. Costello JT, Bieuzen F, Bleakley CM. Where are all the female participants in sports and exercise medicine research? Eur J Sport Sci 14: 847–851, 2014. doi: 10.1080/17461391.2014.911354. [DOI] [PubMed] [Google Scholar]
- 873. O’Bryan SM, Connor KR, Drummer DJ, Lavin KM, Bamman MM. Considerations for sex-cognizant research in exercise biology and medicine. Front Sports Act Living 4: 903992, 2022. doi: 10.3389/fspor.2022.903992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 874. Walts CT, Hanson ED, Delmonico MJ, Yao L, Wang MQ, Hurley BF. Do sex or race differences influence strength training effects on muscle or fat? Med Sci Sports Exerc 40: 669–676, 2008. doi: 10.1249/MSS.0b013e318161aa82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 875. Lai X, Bo L, Zhu H, Chen B, Wu Z, Du H, Huo X. Effects of lower limb resistance exercise on muscle strength, physical fitness, and metabolism in pre-frail elderly patients: a randomized controlled trial. BMC Geriatr 21: 447, 2021. doi: 10.1186/s12877-021-02386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 876. Häkkinen K, Pakarinen A, Kraemer WJ, Häkkinen A, Valkeinen H, Alen M. Selective muscle hypertrophy, changes in EMG and force, and serum hormones during strength training in older women. J Appl Physiol (1985) 91: 569–580, 2001. doi: 10.1152/jappl.2001.91.2.569. [DOI] [PubMed] [Google Scholar]
- 877. Charette SL, McEvoy L, Pyka G, Snow-Harter C, Guido D, Wiswell RA, Marcus R. Muscle hypertrophy response to resistance training in older women. J Appl Physiol (1985) 70: 1912–1916, 1991. doi: 10.1152/jappl.1991.70.5.1912. [DOI] [PubMed] [Google Scholar]
- 878. Pyka G, Lindenberger E, Charette S, Marcus R. Muscle strength and fiber adaptations to a year-long resistance training program in elderly men and women. J Gerontol 49: M22–M27, 1994. doi: 10.1093/geronj/49.1.m22. [DOI] [PubMed] [Google Scholar]
- 879. Hurley BF, Redmond RA, Pratley RE, Treuth MS, Rogers MA, Goldberg AP. Effects of strength training on muscle hypertrophy and muscle cell disruption in older men. Int J Sports Med 16: 378–384, 1995. doi: 10.1055/s-2007-973024. [DOI] [PubMed] [Google Scholar]
- 880. Welle S, Totterman S, Thornton C. Effect of age on muscle hypertrophy induced by resistance training. J Gerontol A Biol Sci Med Sci 51: M270–M275, 1996. doi: 10.1093/gerona/51a.6.m270. [DOI] [PubMed] [Google Scholar]
- 881. Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol (1985) 107: 1655–1662, 2009. doi: 10.1152/japplphysiol.91234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 882. Roth SM, Ivey FM, Martel GF, Lemmer JT, Hurlbut DE, Siegel EL, Metter EJ, Fleg JL, Fozard JL, Kostek MC, Wernick DM, Hurley BF. Muscle size responses to strength training in young and older men and women. J Am Geriatr Soc 49: 1428–1433, 2001. doi: 10.1046/j.1532-5415.2001.4911233.x. [DOI] [PubMed] [Google Scholar]
- 883. Ivey FM, Tracy BL, Lemmer JT, NessAiver M, Metter EJ, Fozard JL, Hurley BF. Effects of strength training and detraining on muscle quality: age and gender comparisons. J Gerontol A Biol Sci Med Sci 55: B152–B157, 2000. doi: 10.1093/gerona/55.3.b152. [DOI] [PubMed] [Google Scholar]
- 884. Treuth MS, Ryan AS, Pratley RE, Rubin MA, Miller JP, Nicklas BJ, Sorkin J, Harman SM, Goldberg AP, Hurley BF. Effects of strength training on total and regional body composition in older men. J Appl Physiol (1985) 77: 614–620, 1994. doi: 10.1152/jappl.1994.77.2.614. [DOI] [PubMed] [Google Scholar]
- 885. Bickel CS, Cross JM, Bamman MM. Exercise dosing to retain resistance training adaptations in young and older adults. Med Sci Sports Exerc 43: 1177–1187, 2011. doi: 10.1249/MSS.0b013e318207c15d. [DOI] [PubMed] [Google Scholar]
- 886. Churchward-Venne TA, Tieland M, Verdijk LB, Leenders M, Dirks ML, de Groot LC, van Loon LJ. There are no nonresponders to resistance-type exercise training in older men and women. J Am Med Dir Assoc 16: 400–411, 2015. doi: 10.1016/j.jamda.2015.01.071. [DOI] [PubMed] [Google Scholar]
- 887. Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Dhanani S, Volpi E, Rasmussen BB. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skelet Muscle 1: 11, 2011. doi: 10.1186/2044-5040-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 888. Lee H, Kim IG, Sung C, Jeon TB, Cho K, Ha YC, Park KS, Yoo JI, Kang GH, Kim SJ, Kim JS. Exercise training increases skeletal muscle strength independent of hypertrophy in older adults aged 75 years and older. Geriatr Gerontol Int 19: 265–270, 2019. doi: 10.1111/ggi.13597. [DOI] [PubMed] [Google Scholar]
- 889. Tieland M, Dirks ML, van der Zwaluw N, Verdijk LB, van de Rest O, de Groot LC, van Loon LJ. Protein supplementation increases muscle mass gain during prolonged resistance-type exercise training in frail elderly people: a randomized, double-blind, placebo-controlled trial. J Am Med Dir Assoc 13: 713–719, 2012. doi: 10.1016/j.jamda.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 890. Straight CR, Fedewa MV, Toth MJ, Miller MS. Improvements in skeletal muscle fiber size with resistance training are age-dependent in older adults: a systematic review and meta-analysis. J Appl Physiol (1985) 129: 392–403, 2020. doi: 10.1152/japplphysiol.00170.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 891. Slivka D, Raue U, Hollon C, Minchev K, Trappe S. Single muscle fiber adaptations to resistance training in old (>80 yr) men: evidence for limited skeletal muscle plasticity. Am J Physiol Regul Integr Comp Physiol 295: R273–R280, 2008. doi: 10.1152/ajpregu.00093.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 892. Wang J, Leung KS, Chow SK, Cheung WH. Inflammation and age-associated skeletal muscle deterioration (sarcopaenia). J Orthop Translat 10: 94–101, 2017. doi: 10.1016/j.jot.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 893. Draganidis D, Jamurtas AZ, Chondrogianni N, Mastorakos G, Jung T, Grune T, Papadopoulos C, Papanikolaou K, Papassotiriou I, Papaevgeniou N, Poulios A, Batrakoulis A, Deli CK, Georgakouli K, Chatzinikolaou A, Karagounis LG, Fatouros IG. Low-grade systemic inflammation interferes with anabolic and catabolic characteristics of the aged human skeletal muscle. Oxid Med Cell Longev 2021: 8376915, 2021. doi: 10.1155/2021/8376915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 894. Morton RW, Traylor DA, Weijs PJ, Phillips SM. Defining anabolic resistance: implications for delivery of clinical care nutrition. Curr Opin Crit Care 24: 124–130, 2018. doi: 10.1097/MCC.0000000000000488. [DOI] [PubMed] [Google Scholar]
- 895. Larsson L, Degens H, Li M, Salviati L, Lee YI, Thompson W, Kirkland JL, Sandri M. Sarcopenia: aging-related loss of muscle mass and function. Physiol Rev 99: 427–511, 2019. doi: 10.1152/physrev.00061.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 896. Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Phillips BE, Szewczyk NJ, Greenhaff PL, Smith K, Atherton PJ. Synchronous deficits in cumulative muscle protein synthesis and ribosomal biogenesis underlie age-related anabolic resistance to exercise in humans. J Physiol 594: 7399–7417, 2016. doi: 10.1113/JP272857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 897. Dungan CM, Figueiredo VC, Wen Y, VonLehmden GL, Zdunek CJ, Thomas NT, Mobley CB, Murach KA, Brightwell CR, Long DE, Fry CS, Kern PA, McCarthy JJ, Peterson CA. Senolytic treatment rescues blunted muscle hypertrophy in old mice. Geroscience 44: 1925–1940, 2022. doi: 10.1007/s11357-022-00542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 898. Hunter GR, McCarthy JP, Bamman MM. Effects of resistance training on older adults. Sports Med 34: 329–348, 2004. doi: 10.2165/00007256-200434050-00005. [DOI] [PubMed] [Google Scholar]
- 899. Fragala MS, Cadore EL, Dorgo S, Izquierdo M, Kraemer WJ, Peterson MD, Ryan ED. resistance training for older adults: position statement from the National Strength and Conditioning Association. J Strength Cond Res 33: 2019–2052, 2019. doi: 10.1519/JSC.0000000000003230. [DOI] [PubMed] [Google Scholar]
- 900. Westcott WL. Resistance training is medicine: effects of strength training on health. Curr Sports Med Rep 11: 209–216, 2012. doi: 10.1249/JSR.0b013e31825dabb8. [DOI] [PubMed] [Google Scholar]
- 901. Lopez P, Pinto RS, Radaelli R, Rech A, Grazioli R, Izquierdo M, Cadore EL. Benefits of resistance training in physically frail elderly: a systematic review. Aging Clin Exp Res 30: 889–899, 2018. doi: 10.1007/s40520-017-0863-z. [DOI] [PubMed] [Google Scholar]
- 902. Cruz-Jentoft AJ, Landi F, Schneider SM, Zúñiga C, Arai H, Boirie Y, Chen LK, Fielding RA, Martin FC, Michel JP, Sieber C, Stout JR, Studenski SA, Vellas B, Woo J, Zamboni M, Cederholm T. Prevalence of and interventions for sarcopenia in ageing adults: a systematic review. Report of the International Sarcopenia Initiative (EWGSOP and IWGS). Age Ageing 43: 748–759, 2014. doi: 10.1093/ageing/afu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 903. Momma H, Kawakami R, Honda T, Sawada SS. Muscle-strengthening activities are associated with lower risk and mortality in major non-communicable diseases: a systematic review and meta-analysis of cohort studies. Br J Sports Med 56: 755–763, 2022. doi: 10.1136/bjsports-2021-105061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 904.American College of Sports Medicine, Chodzko-Zajko WJ, Proctor DN, Fiatarone Singh MA, Minson CT, Nigg CR, Salem GJ, Skinner JS. American College of Sports Medicine position stand. Exercise and physical activity for older adults. Med Sci Sports Exerc 41: 1510–1530, 2009. doi: 10.1249/MSS.0b013e3181a0c95c. [DOI] [PubMed] [Google Scholar]
- 905. Russell B, Motlagh D, Ashley WW. Form follows function: how muscle shape is regulated by work. J Appl Physiol (1985) 88: 1127–1132, 2000. doi: 10.1152/jappl.2000.88.3.1127. [DOI] [PubMed] [Google Scholar]
- 906. Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 907. Psilander N, Eftestøl E, Cumming KT, Juvkam I, Ekblom MM, Sunding K, Wernbom M, Holmberg HC, Ekblom B, Bruusgaard JC, Raastad T, Gundersen K. Effects of training, detraining, and retraining on strength, hypertrophy, and myonuclear number in human skeletal muscle. J Appl Physiol (1985) 126: 1636–1645, 2019. doi: 10.1152/japplphysiol.00917.2018. [DOI] [PubMed] [Google Scholar]
- 908. Murach KA, Dungan CM, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Muscle memory” not mediated by myonuclear number? Secondary analysis of human detraining data. Sports Med Health Sci 5: 2–9, 2023. doi: 10.1152/japplphysiol.00506.2019. [DOI] [PubMed] [Google Scholar]
- 909. Dungan CM, Murach KA, Frick KK, Jones SR, Crow SE, Englund DA, Vechetti IJ Jr, Figueiredo VC, Levitan BM, Satin J, McCarthy JJ, Peterson CA. Elevated myonuclear density during skeletal muscle hypertrophy in response to training is reversed during detraining. Am J Physiol Cell Physiol 316: C649–C654, 2019. doi: 10.1152/ajpcell.00050.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 910. Snijders T, Aussieker T, Holwerda A, Parise G, van Loon LJ, Verdijk LB. The concept of skeletal muscle memory: evidence from animal and human studies. Acta Physiol (Oxf) 229: e13465, 2020. doi: 10.1111/apha.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 911. Rahmati M, McCarthy JJ, Malakoutinia F. Myonuclear permanence in skeletal muscle memory: a systematic review and meta-analysis of human and animal studies. J Cachexia Sarcopenia Muscle 13: 2276–2297, 2022. doi: 10.1002/jcsm.13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 912. Murach KA, Dungan CM, Peterson CA, McCarthy JJ. Muscle fiber splitting is a physiological response to extreme loading in animals. Exerc Sport Sci Rev 47: 108–115, 2019. doi: 10.1249/JES.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 913. Jorgenson KW, Hornberger TA. The overlooked role of fiber length in mechanical load-induced growth of skeletal muscle. Exerc Sport Sci Rev 47: 258–259, 2019. doi: 10.1249/JES.0000000000000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 914. DeFreitas JM, Beck TW, Stock MS, Dillon MA, Kasishke PR Jr.. An examination of the time course of training-induced skeletal muscle hypertrophy. Eur J Appl Physiol 111: 2785–2790, 2011. doi: 10.1007/s00421-011-1905-4. [DOI] [PubMed] [Google Scholar]
- 915. DeFreitas JM, Beck TW, Stock MS. The findings of Damas et al. have not influenced the previously proposed time course of skeletal muscle hypertrophy. Eur J Appl Physiol 116: 443–444, 2016. doi: 10.1007/s00421-015-3286-6. [DOI] [PubMed] [Google Scholar]
- 916. Maden-Wilkinson TM, Balshaw TG, Massey GJ, Folland JP. What makes long-term resistance-trained individuals so strong? A comparison of skeletal muscle morphology, architecture, and joint mechanics. J Appl Physiol (1985) 128: 1000–1011, 2020. doi: 10.1152/japplphysiol.00224.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 917. Ruple BA, Mesquita PH, Godwin JS, Sexton CL, Osburn SC, McIntosh MC, Kavazis AN, Libardi CA, Young KC, Roberts MD. Changes in vastus lateralis fiber cross-sectional area, pennation angle, and fascicle length do not predict changes in muscle cross-sectional area. Exp Physiol 107: 1216–1224, 2022. doi: 10.1113/EP090666. [DOI] [PMC free article] [PubMed] [Google Scholar]