Abstract
In recent years, neoantigens have been recognized as ideal targets for tumor immunotherapy. With the development of neoantigen-based tumor immunotherapy, comprehensive neoantigen databases are urgently needed to meet the growing demand for clinical studies. We have built the tumor-specific neoantigen database (TSNAdb) previously, which has attracted much attention. In this study, we provide TSNAdb v2.0, an updated version of the TSNAdb. TSNAdb v2.0 offers several new features, including (1) adopting more stringent criteria for neoantigen identification, (2) providing predicted neoantigens derived from three types of somatic mutations, and (3) collecting experimentally validated neoantigens and dividing them according to the experimental level. TSNAdb v2.0 is freely available at https://pgx.zju.edu.cn/tsnadb/.
Keywords: Neoantigen, Tumor immunotherapy, Human leukocyte antigen, Somatic mutation, Database
Introduction
Tumor neoantigens are tumor-specific antigens derived from somatic mutations in tumor cells, which have been recognized as ideal targets for tumor immunotherapy in recent years [1], [2], [3], [4]. Due to the huge workload for experimental verification, it is preferred to utilize cancer genomics and bioinformatics for neoantigen identification. Numerous prediction tools considering the biological process of neoantigen generation, such as human leukocyte antigen (HLA)–peptide binding [5], [6], [7], have been developed, which have been embedded in neoantigen prediction pipelines such as pVACtools [8], tumor-specific neoantigen detector (TSNAD) [9], [10], and pTuneos [11]. Neoantigen-related databases such as TRON cell line portal (TCLP) [12], the cancer immunome atlas (TCIA) [13], and tumor-specific neoantigen database (TSNAdb) [14] have also been created for better usage of neoantigens in clinical research. In TSNAdb v1.0, we took the complex of mutated peptides and HLA class I molecules (peptide–HLA pairs, pHLAs) as tumor neoantigens and predicted binding affinities between mutated/wild-type pHLAs by NetMHCpan v2.8/v4.0. We then obtained 3,707,562/1,146,961 potential neoantigens derived from single nucleotide variants (SNVs) of 7748 tumor samples from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/). With the development of neoantigen-based tumor immunotherapy, neoantigens from other types of mutations have been identified, and more experimental data have been generated [15], [16]. Therefore, it is urgent to perform system updates for the TSNAdb v1.0.
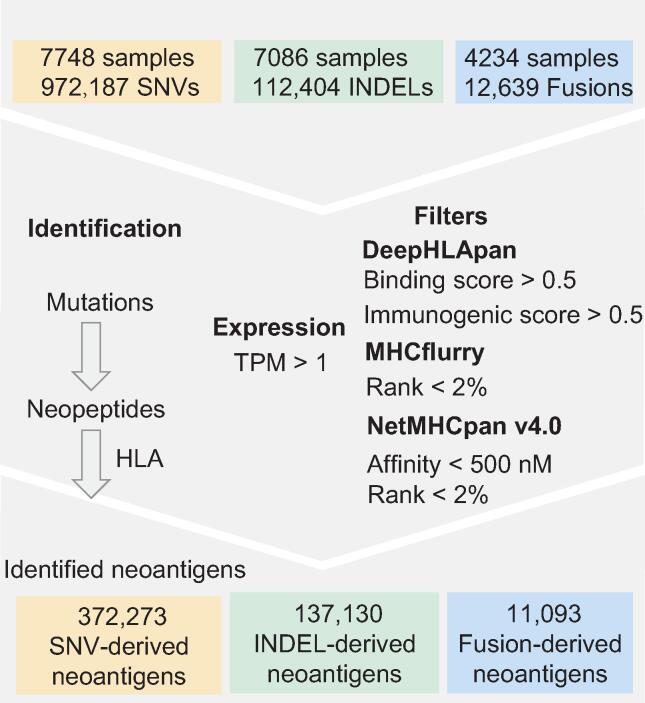
Here, we present an updated version of TSNAdb v1.0 that improves on the following points. (1) More stringent criteria were used for neoantigen identification to reduce the high false-positive rate of neoantigen prediction in practice. Only the pHLAs that met the thresholds of three tools were considered potential neoantigens (Figure 1). The pHLAs would not be considered neoantigens if the mutated genes were not expressed in the tumor cells. (2) We provided predicted neoantigens derived not only from SNVs but also from insertions/deletions (INDELs) and gene fusions (Fusions). In total, 372,273 SNV-derived neoantigens, 137,130 INDEL-derived neoantigens, and 11,093 Fusion-derived neoantigens were obtained. The mean number of neoantigens generated for each SNV (0.38) was lower than each INDEL (1.22) or Fusion (0.88). (3) We collected as many experimentally validated neoantigens from public databases and literature as possible (1856 neoantigens) and divided them into three tiers according to the level of experimental verification. Corresponding genes and mutations of the collected neoantigens were linked to the cancer-driving site profiling database (CandrisDB) [17] since neoantigens derived from driver genes or driver mutations would be ideal targets for tumor immunotherapy [18].
Figure 1.
The neoantigen prediction process of TSNAdb v2.0
SNV, single nucleotide variant; INDEL, insertion/deletion; Fusion, gene fusion; HLA, human leukocyte antigen; TPM, transcripts per million reads.
We believe that the updated database will contribute to neoantigen-based tumor immunotherapy and that the database will continue updating in the aspect of predicting neoantigens from more types of mutations and collecting more experimentally validated neoantigens.
Database content and usage
Data collection and preprocessing
The SNVs, INDELs, and the expression level of corresponding genes were collected from TCGA. Mutated nucleotide sequences generated by SNVs and INDELs are translated into mutated amino acid sequences and have been decomposed into 8 to 11 peptides using the pipeline TSNAD v2.0 [10]. The Fusions were collected from Gao et al. [19], and the mutated proteins were generated by STAR-Fusion [20]. The HLA alleles of corresponding samples were collected from TCIA. Finally, 972,187 SNVs from 7748 samples, 112,404 INDELs from 7086 samples, and 12,639 Fusions from 4234 samples were used for neoantigen prediction.
Stricter criteria for neoantigen identification
Neoantigen-based tumor immunotherapy has shown good application prospects in clinical practice. However, the high false-positive rate of neoantigen prediction limits its usage. How to select high-confidence immunogenic neoantigens remains to be resolved. To reduce the potential false-positive rate in our predicted results, three tools (DeepHLApan, MHCflurry, and NetMHCpan v4.0) were used for neoantigen prediction, and only the pHLAs that met all the criteria of the three tools were considered potential neoantigens (Figure 1). The reason we chose these three tools is as follows: NetMHCpan [7] is the most frequently used tool for neoantigen prediction in clinical practice. MHCflurry [6] obtains the prediction neoantigen efficiently and with high quality. DeepHLApan [5] considers both HLA-peptide binding and immunogenicity of pHLA that the other two tools have not taken into consideration for high-confidence neoantigen prediction. The threshold of each tool is as follows. For NetMHCpan v4.0, pHLA with rank < 2% or affinity < 500 nM is considered binding, and we used both thresholds to select higher quality neoantigens. The output of MHCflurry is rank % and has no specific threshold. We set rank < 2% as the threshold, which is the same as NetMHCpan v4.0. The predicted scores of DeepHLApan are posterior probabilities, so we set the threshold to 0.5. In addition, the pHLAs whose corresponding genes were not expressed [transcripts per million reads (TPM) < 1] were removed.
Neoantigens derived from three types of mutation
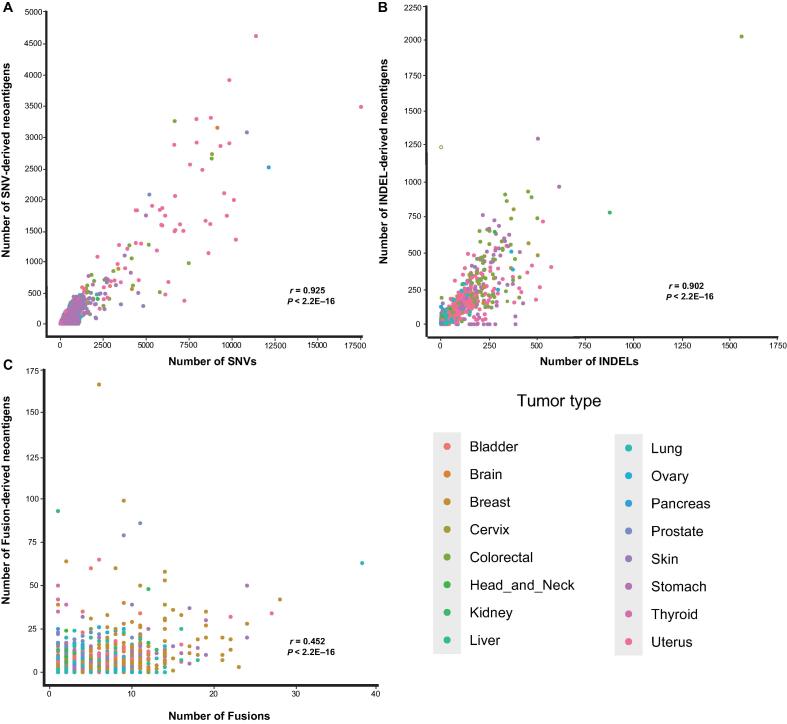
Neoantigens are not only generated from SNVs but also generated from other mutations, such as INDELs and Fusions. Based on the analysis of different types of mutations in TCGA tumor samples, we provided 137,130 INDEL-derived neoantigens and 11,093 Fusion-derived neoantigens into TSNAdb v2.0. The number of predicted neoantigens derived from SNVs was greater than that derived from INDELs and Fusions due to the greatest number of SNVs among the somatic mutations collected. However, the average number of neoantigens derived from each SNV (0.38) was less than that derived from each INDEL (1.22) or each Fusion (0.88) (Table 1). We further explored the relationship between the number of mutations and neoantigens for the three mutation types. The results showed that the numbers of SNV-derived neoantigens and INDEL-derived neoantigens had positive correlations with the numbers of SNVs and INDELs, with the Pearson correlation coefficient r = 0.925 and r = 0.902, respectively (Figure 2A and B). There was no significant correlation between the number of Fusion-derived neoantigens and the number of Fusions (Figure 2C, r = 0.452), which might be attributed to the fact that the number of neopeptides each Fusion generated varies greatly.
Table 1.
The distribution of mutations and neoantigens across 16 tumor types
| Mutation type | Tissue | No. of samples | No. of mutations | No. of neoantigens | No. of neoantigens per mutation |
|---|---|---|---|---|---|
| SNV | Bladder | 408 | 74,707 | 21,070 | 0.38 |
| Brain | 151 | 16,197 | 5227 | ||
| Breast | 982 | 62,211 | 18,238 | ||
| Cervix | 286 | 46,607 | 11,906 | ||
| Colorectal | 531 | 165,293 | 39,116 | ||
| Head and neck | 495 | 55,968 | 13,728 | ||
| Kidney | 681 | 26,170 | 8499 | ||
| Liver | 358 | 28,024 | 5987 | ||
| Lung | 999 | 212,861 | 52,801 | ||
| Ovary | 305 | 28,478 | 7631 | ||
| Pancreas | 166 | 17,221 | 3985 | ||
| Prostate | 493 | 16,424 | 5363 | ||
| Skin | 466 | 206,907 | 43,882 | ||
| Stomach | 411 | 103,103 | 24,497 | ||
| Thyroid | 488 | 4425 | 1218 | ||
| Uterus | 528 | 415,499 | 109,125 | ||
| INDEL | Bladder | 389 | 2890 | 3532 | 1.22 |
| Brain | 130 | 366 | 375 | ||
| Breast | 888 | 5207 | 6123 | ||
| Cervix | 257 | 3095 | 4234 | ||
| Colorectal | 520 | 21,327 | 29,876 | ||
| Head and neck | 478 | 3569 | 4913 | ||
| Kidney | 648 | 5318 | 6395 | ||
| Liver | 353 | 2120 | 2143 | ||
| Lung | 983 | 10,790 | 12,407 | ||
| Ovary | 296 | 4260 | 4441 | ||
| Pancreas | 141 | 457 | 806 | ||
| Prostate | 416 | 1448 | 2119 | ||
| Skin | 430 | 1965 | 2557 | ||
| Stomach | 393 | 19,912 | 24,838 | ||
| Thyroid | 248 | 1746 | 1657 | ||
| Uterus | 516 | 27,934 | 30,714 | ||
| Fusion | Bladder | 269 | 809 | 819 | 0.88 |
| Brain | 92 | 244 | 245 | ||
| Breast | 787 | 3390 | 3457 | ||
| Cervix | 139 | 280 | 215 | ||
| Colorectal | 167 | 328 | 231 | ||
| Head and neck | 285 | 532 | 530 | ||
| Kidney | 171 | 318 | 420 | ||
| Liver | 179 | 513 | 423 | ||
| Lung | 716 | 1992 | 710 | ||
| Ovary | 316 | 983 | 609 | ||
| Pancreas | 72 | 130 | 161 | ||
| Prostate | 381 | 1079 | 1120 | ||
| Skin | 287 | 841 | 918 | ||
| Stomach | 178 | 559 | 486 | ||
| Thyroid | 110 | 140 | 250 | ||
| Uterus | 85 | 501 | 499 | ||
Note: SNV, single nucleotide variant; INDEL, insertion/deletion; Fusion, gene fusion.
Figure 2.
The relationship between the number of mutations and the number of neoantigens across 16 tumor types of three mutation types
A. The relationship between the number of SNVs and SNV-derived neoantigens. B. The relationship between the number of INDELs and INDEL-derived neoantigens. C. The relationship between the number of Fusions and Fusion-derived neoantigens. The Pearson correlation coefficient is used for evaluation.
Shared neoantigens generated from frequent somatic mutations
Currently, most neoantigen-targeted immunotherapies are personalized and expensive, which led us to wonder if we could identify shared neoantigens that can be applied to a wider range of tumor patients. Here, we analyzed the frequency of each neoantigen and obtained 16,913 neoantigens shared in at least two tumor samples (Table S1). Among three SNV-derived neoantigens shared in more than 20 samples, the mutated peptides are generated from BRAF and KRAS, which are well-known cancer driver genes. The most frequent shared neoantigen derived from SNV is the complex of HLA-B57:01 and mutated peptide GLATEKSRW generated by BRAF V600E, which is present in 41 tumor samples. The complex of HLA-A02:01 and the neopeptides RLMAPVGSV and SLLTQPSPA generated by the frameshift mutation XYLT2 G529Afs*78 are the most frequent neoantigens among INDEL-derived neoantigens, which both appear in 31 samples (Table 2). The two Fusion-derived shared neoantigens are the complex of HLA-A02:01 and the neopeptides ALNSEALSVV and ALNSEALSV generated by the fusion of the TMPRSS2 and ERG genes, which both appear in 14 samples (Table S1). We believe that these shared neoantigens are expected to be ideal drug targets for tumor immunotherapy, which might need further experimental validation.
Table 2.
The detailed information of shared neoantigens present in more than 20 samples
| Mutation type | Gene | Mutation | HLA | Peptide | No. of samples |
|---|---|---|---|---|---|
| SNV | BRAF | V600E | HLA-B57:01 | GLATEKSRW | 41 |
| SNV | KRAS | G12D | HLA-B08:01 | DGVGKSAL | 37 |
| INDEL | XYLT2 | G529Afs*78 | HLA-A02:01 | RLMAPVGSV | 31 |
| INDEL | XYLT2 | G529Afs*78 | HLA-A02:01 | SLLTQPSPA | 31 |
| SNV | KRAS | G12V | HLA-A03:01 | VVGAVGVGK | 30 |
| INDEL | SETD1B | H8Tfs*27 | HLA-C07:01 | LRARGGTTI | 29 |
| INDEL | PLEKHA6 | V328Yfs*172 | HLA-A02:01 | IMMSWMPPL | 25 |
| INDEL | PLEKHA6 | V328Yfs*172 | HLA-A02:01 | VLSGCHLAV | 25 |
| INDEL | XYLT2 | G529Afs*78 | HLA-C07:02 | GRTPTTRLM | 25 |
| INDEL | PLEKHA6 | V328Yfs*172 | HLA-A02:01 | FTPLSAHPV | 25 |
| INDEL | PLEKHA6 | V328Yfs*172 | HLA-A02:01 | CLAGSLSTM | 25 |
| INDEL | PLEKHA6 | V328Yfs*172 | HLA-A02:01 | SAMPSAMGV | 25 |
| INDEL | PLEKHA6 | V328Yfs*172 | HLA-A02:01 | SIMMSWMPPL | 25 |
| INDEL | XYLT2 | G529Afs*78 | HLA-B07:02 | RPACTCISM | 23 |
| INDEL | XYLT2 | G529Afs*78 | HLA-B07:02 | HPQWAPHSA | 23 |
| INDEL | XYLT2 | G529Afs*78 | HLA-B07:02 | RPTGRTPTTRL | 23 |
| INDEL | XYLT2 | G529Afs*78 | HLA-B07:02 | SPGACRPAC | 23 |
| INDEL | XYLT2 | G529Afs*78 | HLA-B07:02 | RPTGRTPTT | 23 |
| INDEL | MUC4 | L1937Pfs*1069 | HLA-B07:02 | HPQVTPPL | 23 |
| INDEL | MUC4 | L1937Pfs*1069 | HLA-B07:02 | HPQVTPPLF | 23 |
| INDEL | SETD1B | H8Tfs*27 | HLA-C07:02 | LRARGGTTI | 22 |
| INDEL | MUC4 | L1937Pfs*1069 | HLA-B07:02 | LPQHPQVTPPL | 21 |
Note: fs*78 indicates that 78 amino acids have been changed after the frameshift site. HLA, human leukocyte antigen; fs, frameshift.
Experimentally validated neoantigens
On the “Validation” page of TSNAdb v1.0, we only collected experimental data about wild-type pHLAs that were difficult to identify as neoantigens due to the limited binding data between mutated pHLAs. With the development of clinical studies on neoantigen-based tumor immunotherapy, a large number of experimental results have provided a rich source for the functional confirmation of neoantigens. Here, we collected experimentally validated mutated pHLAs not only from several neoantigen databases (dbPepNeo [21], NeoPeptide [22], NEPdb [23], and Cancer Antigenic Peptide Database [24]) but also from published literature through data mining. For the neoantigens without gene or mutation information, BLAST was used to determine the mutated genes and the positions of somatic mutations at proteins. All collected data were further checked to determine whether the neoantigens were immunogenic or presented to the cell surface, and the collected neoantigens were divided into three tiers according to the experimental level. Neoantigens that have been both validated as immunogenic and to be presented to the cell surface were labeled tier 1, while those only validated as immunogenic were labeled tier 2, and those only validated to be presented to the cell surface were labeled tier 3. We collected 1856 experimental neoantigens, among which 67 neoantigens were classified as tier 1, 1190 neoantigens were classified as tier 2, and 599 neoantigens were classified as tier 3. Among the collected neoantigens, most were SNV-derived (22 were Fusion-derived, 125 were INDEL-derived, 23 were noncoding-derived, 33 were RNA splice-derived, and the remaining were SNV-derived) and enriched in several tumor types (430 belonged to lung cancer, 477 belonged to skin cancer, 361 belonged to B-cell lymphoma, and 123 belonged to colorectal cancer).
The usage of TSNAdb v2.0
The web interface of TSNAdb v2.0 contains seven pages: “Home”, “Browse”, “Search”, “Collected”, “Tools”, “Download”, and “Help”. The pages “Home”, “Download”, and “Help” are similar to those presented in TSNAdb v1.0. The “Tools” page is newly added to provide the links of DeepHLApan and TSNAD, which we developed previously for neoantigen prediction. Major changes (such as more presentation forms, more correlation analysis, and more meaningful links) have been made in the pages “Browse”, “Search”, and “Collected” compared to the TSNAdb v1.0.
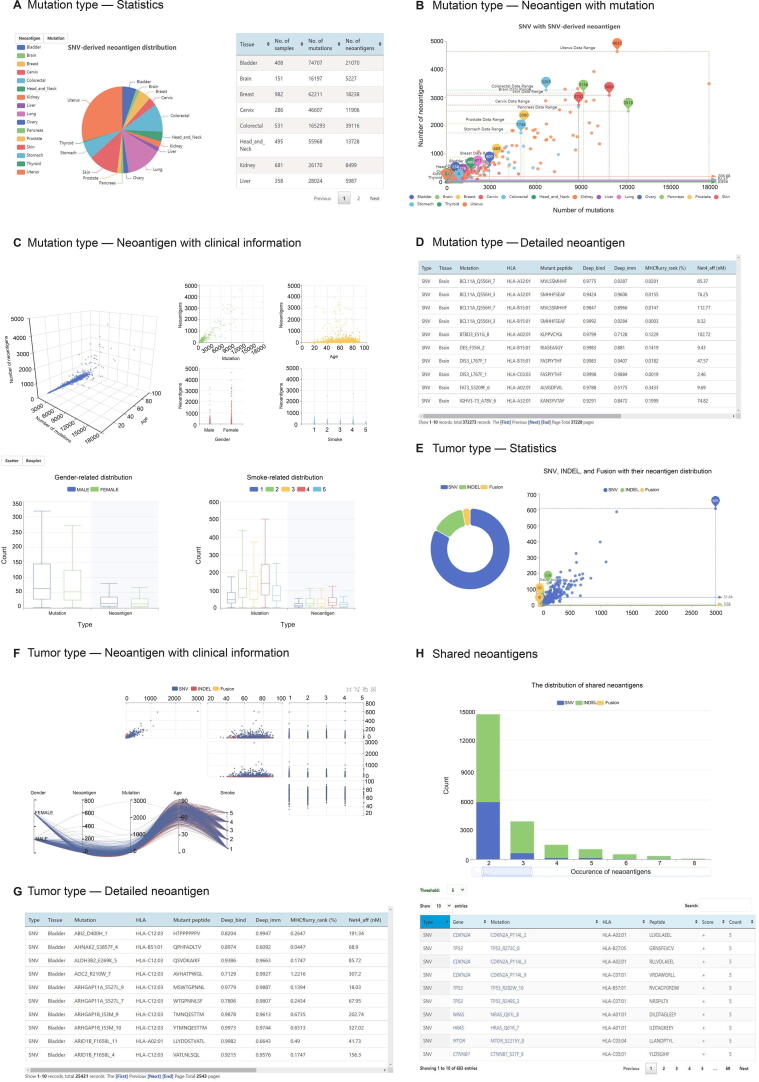
On the “Browse” page, three subpages (“Mutation type”, “Tumor type”, and “Shared neoantigens”) are provided for customized neoantigen browsing. In “Mutation type”, four parts, “Statistics” (Figure 3A), “Neoantigen with mutation” (Figure 3B), “Neoantigen with clinical information” (Figure 3C), and “Detailed neoantigen” (Figure 3D), are displayed. In “Tumor type”, three parts, including the “Statistics” (Figure 3E), “Neoantigen with clinical information” (Figure 3F), and “Detailed neoantigen” (Figure 3G), are displayed. In the “Shared neoantigens”, the distribution of shared neoantigens that occur in at least two tumor samples is displayed. The table below the boxplot displays the shared neoantigens that meet different thresholds. The genes and mutations are linked to CandrisDB [17] to check whether they are driver genes or driver mutations since shared neoantigens derived from driver mutations would be potential ideal targets for tumor immunotherapy (Figure 3H).
Figure 3.
Screenshots of the “Browse” page of TSNAdb v2.0
A. The “Statistics” part of the “Mutation type” page. B. The “Neoantigen with mutation” part of the “Mutation type” page. C. The “Neoantigen with clinical information” part of the “Mutation type” page. D. The “Detailed neoantigen” part of the “Mutation type” page. E. The “Statistics” part of the “Tumor type” page. F. The “Neoantigen with mutation” part of the “Tumor type” page. G. The “Detailed neoantigen” part of the “Tumor type” page. H. The “Shared neoantigens” page.
The “Search” page contains the main page and two subpages “Gene” and “HLA”. On the main page of “Search”, users could search for desired neoantigens by selecting the mutation type, tumor type, and gene. Compared with the “Detailed neoantigen” of the “Browse” page, it could provide more customized functions, such as sorting and searching. In the subpages “Gene” and “HLA”, the detailed neoantigens and their distribution of selected genes or HLAs would be displayed once searching. The displayed pie charts are linked with the bellowed table that the detailed neoantigens would be changed once clicking on the part of the pie charts.
On the “Collected” page, all collected neoantigens are validated by experiments to be presented to the cell surface or immunogenic, which are different from those in TSNAdb v1.0. The corresponding genes and mutations of neoantigens are linked to CandrisDB as those in the “Shared neoantigens”.
Discussion and perspectives
Neoantigens play an important role in tumor immunotherapy. A comprehensive and high-confidence neoantigen database would greatly meet the needs of clinical research. In TSNAdb v2.0, we predict more mutation type-derived neoantigens with stricter criteria, present the tissue-specific and gene-specific distribution of candidate tumor-specific neoantigens of TCGA tumor samples, and collect 1856 neoantigens that have been experimentally validated, which is the most systematic database of tumor-specific neoantigens at present. Compared with other databases, TSNAdb v2.0 has several advantages as follows. First, TSNAdb v2.0 provides both high-quality predicted neoantigens and experimentally validated neoantigens, while most of the other databases except NEPdb only provide one of them. Compared with NEPdb, TSNAdb v2.0 provides more sources of predicted neoantigens and has richer forms of presentation. Second, TSNAdb v2.0 provides the analysis of shared neoantigens and links corresponding genes and mutations to CandrisDB to identify high-quality neoantigens, which other databases do not provide. Finally, TNSAdb will be updated continuously to provide constant service for related researchers and clinicians. We believe that it would certainly contribute to neoantigen-based tumor immunotherapy.
However, neoantigens are derived not only from SNVs, INDELs, and Fusions but also from splice variants [15], the mitochondrial genome [16], and translated unannotated open reading frames [25]. It is necessary to predict all sources of neoantigens to construct a comprehensive neoantigen database. Limited by the difficulty of collecting other mutations and corresponding HLAs, we only chose three sources of neoantigen in this version of the database. In the following update of TNSAdb, we would add neoantigens from more sources and collect more validated neoantigens to construct a more comprehensive neoantigen database.
Data availability
TSNAdb v2.0 is freely available at https://pgx.zju.edu.cn/tsnadb/.
Competing interests
The authors have declared no competing interests.
CRediT authorship contribution statement
Jingcheng Wu: Methodology, Formal analysis, Writing – original draft, Visualization. Wenfan Chen: Formal analysis, Investigation. Yuxuan Zhou: Data curation, Investigation. Ying Chi: Methodology, Resources. Xiansheng Hua: Resources. Jian Wu: Methodology. Xun Gu: Methodology. Shuqing Chen: Conceptualization, Resources. Zhan Zhou: Conceptualization, Supervision, Writing – review & editing. All authors have read and approved the final manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31971371 and U20A20409), the Key R&D Program of Zhejiang Province, China (Grant No. 2020C03010), the Huadong Medicine Joint Funds of the Zhejiang Provincial Natural Science Foundation of China (Grant No. LHDMZ22H300002), and the Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare. We thank the Information Technology Center, State Key Lab of CAD&CG, and Innovation Institute for Artificial Intelligence in Medicine, Zhejiang University, and also Alibaba Cloud for the support of computing resources. We also gratefully acknowledge Prof. Feng Zhu from Zhejiang University for the critical reading of the manuscript, the TCGA Research Network for referencing the TCGA datasets, and the TCIA for referencing HLA-type data of TCGA samples.
Handled by Yu Xue
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2022.09.012.
Supplementary material
The following are the Supplementary material to this article:
The detailed information of neoantigens shared in at least two samples
References
- 1.Zacharakis N., Chinnasamy H., Black M., Xu H., Lu Y.C., Zheng Z., et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24:724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran E., Robbins P.F., Lu Y.C., Prickett T.D., Gartner J.J., Jia L., et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilf N., Kuttruff-Coqui S., Frenzel K., Bukur V., Stevanović S., Gouttefangeas C., et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 5.Wu J., Wang W., Zhang J., Zhou B., Zhao W., Su Z., et al. DeepHLApan: a deep learning approach for neoantigen prediction considering both HLA–peptide binding and immunogenicity. Front Immunol. 2019;10:2559. doi: 10.3389/fimmu.2019.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donnell T.J., Rubinsteyn A., Laserson U. MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst. 2020;11:42–48. doi: 10.1016/j.cels.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hundal J., Kiwala S., McMichael J., Miller C.A., Xia H., Wollam A.T., et al. pVACtools: a computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res. 2020;8:409–420. doi: 10.1158/2326-6066.CIR-19-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z., Lyu X., Wu J., Yang X., Wu S., Zhou J., et al. TSNAD: an integrated software for cancer somatic mutation and tumour-specific neoantigen detection. R Soc Open Sci. 2017;4:170050. doi: 10.1098/rsos.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z., Wu J., Ren J., Chen W., Zhao W., Gu X., et al. TSNAD v2.0: a one-stop software solution for tumor-specific neoantigen detection. Comput Struct. Biotechnol J. 2021;19:4510–4516. doi: 10.1016/j.csbj.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou C., Wei Z., Zhang Z., Zhang B., Zhu C., Chen K., et al. pTuneos: prioritizing tumor neoantigens from next-generation sequencing data. Genome Med. 2019;11:67. doi: 10.1186/s13073-019-0679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., et al. TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression. Genome Med. 2015;7:118. doi: 10.1186/s13073-015-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype–immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Wu J., Zhao W., Zhou B., Su Z., Gu X., Zhou Z., et al. TSNAdb: a database for tumor-specific neoantigens from immunogenomics data analysis. Genomics Proteomics Bioinformatics. 2018;16:276–282. doi: 10.1016/j.gpb.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishto M., Mansurkhodzhaev A., Ying G., Bitra A., Cordfunke R.A., Henze S., et al. An in silico–in vitro pipeline identifying an HLA-A*02:01+ KRAS G12V+ spliced epitope candidate for a broad tumor-immune response in cancer patients. Front Immunol. 2019;10:2572. doi: 10.3389/fimmu.2019.02572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prota G., Lechuga-vieco A.V., De Libero G. Mitochondrial proteins as source of cancer neoantigens. Int J Mol Sci. 2022;23:2627. doi: 10.3390/ijms23052627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao W., Yang J., Wu J., Cai G., Zhang Y., Haltom J., et al. CanDriS: posterior profiling of cancer-driving sites based on two-component evolutionary model. Brief Bioinform. 2021;22:bbab131. doi: 10.1093/bib/bbab131. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W., Wu J., Chen S., Zhou Z. Shared neoantigens: ideal targets for off-the-shelf cancer immunotherapy. Pharmacogenomics. 2020;21:637–645. doi: 10.2217/pgs-2019-0184. [DOI] [PubMed] [Google Scholar]
- 19.Gao Q., Liang W.W., Foltz S.M., Mutharasu G., Jayasinghe R.G., Cao S., et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23:227–238. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas B.J., Dobin A., Li B., Stransky N., Pochet N., Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20:213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan X., Li D., Huang P., Jian X., Wan H., Wang G., et al. dbPepNeo: a manually curated database for human tumor neoantigen peptides. Database. 2020;2020:baaa004. doi: 10.1093/database/baaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W.J., Qu Z., Song C.Y., Sun Y., Lai A.L., Luo M.Y., et al. NeoPeptide: an immunoinformatic database of T-cell-defined neoantigens. Database. 2019;2019:baz128. doi: 10.1093/database/baz128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J., Bai P., Fan W., Li Q., Li Y., Wang D., et al. NEPdb: a database of T-cell experimentally-validated neoantigens and pan-cancer predicted neoepitopes for cancer immunotherapy. Front Immunol. 2021;12:644637. doi: 10.3389/fimmu.2021.644637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigneron N., Stroobant V., Van Den Eynde B.J., Van Der Bruggen P. Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun. 2013;13:15. [PMC free article] [PubMed] [Google Scholar]
- 25.Ouspenskaia T., Law T.E., Clauser K.R., Klaeger S., Keskin D.B., Li B., et al. Abstract 566: Neoantigens from translated unannotated open reading frames in cancer. Annual Meeting of the American-Association-for-Cancer-Research. 2019;79:566. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The detailed information of neoantigens shared in at least two samples
Data Availability Statement
TSNAdb v2.0 is freely available at https://pgx.zju.edu.cn/tsnadb/.