Abstract
Kinesin‐driven intracellular transport is essential for various cell biological events and thus plays a crucial role in many pathological processes. However, little is known about the molecular basis of the specific and dynamic cargo‐binding mechanism of kinesins. Here, an integrated structural analysis of the KIF3/KAP3 and KIF3/KAP3‐APC complexes unveils the mechanism by which KIF3/KAP3 can dynamically grasp APC in a two‐step manner, which suggests kinesin‐cargo recognition dynamics composed of cargo loading, locking, and release. Our finding is the first demonstration of the two‐step cargo recognition and stabilization mechanism of kinesins, which provides novel insights into the intracellular trafficking machinery.
Keywords: Cargo binding, Intracellular transport, KIF3, Kinesin, Protein dynamics
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Membranes & Trafficking; Structural Biology
Kinesin‐2 recognizes and binds its specific cargo in a two‐step manner. The kinesin–cargo interaction can be modulated through stabilization by the coiled‐coil stalk.
Introduction
Molecular mechanisms that produce the remarkable dynamics of the motor protein‐mediated intracellular transport have been a focus of extensive research in the past decades (Vale, 2003; Hirokawa & Takemura, 2005; Hirokawa et al, 2010; Lee Sweeney & Holzbaur, 2018). Kinesin superfamily proteins (KIFs) play key roles in many cellular functions as pivotal components of the microtubule (MT)‐based intracellular trafficking system that specifically recognizes their diverse cargos (Hirokawa et al, 2009). To date, the mechanism of cargo recognition and binding remains one of the most crucial unexplained aspects in the research field of motor proteins, including kinesins (Hirokawa et al, 2009; Lee Sweeney & Holzbaur, 2018). While the mechanism of kinesin motor‐MT association, dynamics and regulation has been well elucidated in the past 25 years, little progress has been achieved regarding how kinesins grasp and transport a variety of cargos. Therefore, biochemical and structural analysis of kinesin‐cargo complexes is necessary to verify their reported functions and elucidate the functional basis. Notably, a few crystal structures of kinesin light chain (KLC) 2 in complex with a cargo peptide have reported evidence of a direct interaction between kinesin‐1 and cargos (Pernigo et al, 2013). However, we still have insufficient information regarding the dynamic events of cargo recognition and the interaction of KIFs. To better understand the functional diversity of kinesin‐cargo recognition mechanisms, it is necessary to elucidate their dynamic interactions based on integrated analyses in solution.
Heterotrimeric kinesin‐2 motors, which exist ubiquitously from algae to vertebrates, were intensively identified in the 1990s in model organisms, including Chlamydomonas FLA10/FLA8/FLA3 (Waither et al, 1994; Cole et al, 1998), Caenorbabditis elegans KLP‐20/KLP‐11/KAP‐1 (Signor et al, 1999), Drosophila KLP64D/KLP68D/KAP (Pesavento et al, 1994), Strongylocentrotus purpuratus KRP85/KRP95/KAP (Cole et al, 1992, 1993; Rashid et al, 1995; Wedaman et al, 1996), Xenopus XKLP3A/B/KAP3 (Vernos et al, 1993; Le Bot et al, 1998), mammalian KIF3A/B/KAP3 (Aizawa et al, 1992; Yamazaki et al, 1995, 1996; Scholey, 1996) and KIF3A/C/KAP3 (Muresan et al, 1998; Garbouchian et al, 2022). Mammalian KIF3/KAP3 is one of the most ubiquitously and abundantly expressed KIFs (Takeda et al, 2000; Scholey, 2013) that forms a kinesin complex consisting of heterodimeric KIF3 motors (KIF3A/KIF3B or KIF3A/KIF3C) and the armadillo repeat (ARM)‐ containing cargo‐binding adaptor KAP3 (Kondo et al, 1994; Yamazaki et al, 1995, 1996). KIF3/KAP3 has been revealed to play diverse critical roles in cells (Morris & Scholey, 1997; Nonaka et al, 1998; Takeda et al, 1999; Haraguchi et al, 2006; Mattie et al, 2010; Gaudin et al, 2012; Guzik‐Lendrum et al, 2015). Notably, in addition to the essential role of KIF3/KAP3 in driving intraflagellar transport (Rosenbaum & Witman, 2002; Hirokawa et al, 2012; Scholey, 2013), recent studies have also identified its crucial functions in powering the movement of diverse intracellular cargos outside cilia (Teng et al, 2005; Lolkema et al, 2007; Carpenter et al, 2015; Ichinose et al, 2015, 2019; Alsabban et al, 2020; Yoshihara et al, 2021; Wang et al, 2022). For instance, the RNA‐binding tumor suppressor protein adenomatous polyposis coli (APC) has been identified as a KIF3/KAP3 cargo to mediate RNA transport (Jimbo et al, 2002; Baumann et al, 2020), which is associated with ~ 12% of KIF3 organelles (Garbouchian et al, 2022). Nevertheless, little is known of how KIF3/KAP3 specifically recognizes its diverse cargos.
In this report, we focused on KIF3/KAP3 with intensive biochemical analyses as a proof‐of‐principle for kinesin‐cargo interaction. We first identified the distinct structural outlines of the KIF3/KAP3 and KIF3/KAP3‐APC complexes by small‐angle X‐ray scattering (SAXS), suggesting the molecular bases of its specific cargo binding. More importantly, by performing further high‐speed atomic force microscopy (HS‐AFM) and cryo‐electron microscopy (cryoEM) analysis, the dynamic complex structure of KIF3/KAP3 with its specific cargo was unveiled, suggesting stepwise cargo recognition dynamics composed of cargo loading, locking, and release. Overall, our findings provide a crucial complement to the current understanding of the kinesin‐cargo system with crucial insights into its dynamic cargo‐recognition mechanism and relevant pathological processes.
Results & Discussion
KIF3/KAP3 exhibits a specific cargo‐binding cavity
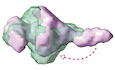
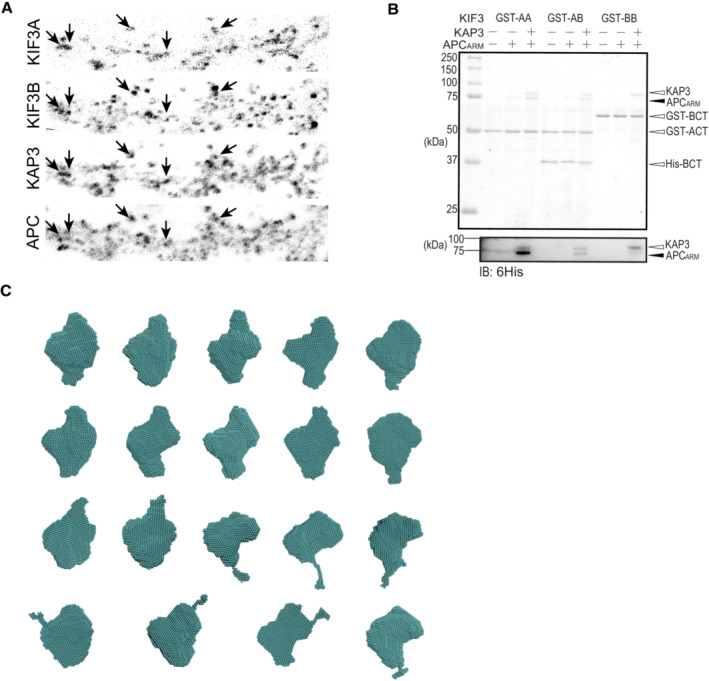
To implement specific cargo transport, how do KIF3 motors configure their specific cargo recognition system? We first focused on the structural arrangement of the cargo‐associating region of the major KIF3 components, KIF3A/B/KAP3. As the KAP3‐bound KIF3 C‐terminus is reportedly responsible for the cargo binding, we constructed and purified the KIF3A/B/KAP3 complex (ABK) consisting of C‐terminal domains of KIF3A (ACT, 481‐701) and 3B (BCT, 472‐741), and KAP3 (1‐693) for structural assessments (Fig 1A). The highly purified ABK proteins were evaluated by the combination of size exclusion chromatography and multiangle light scattering (SEC‐MALS) analysis. As a result, the molecular weight (MW) of the ABK complex was determined to be 145 kDa, corresponding to a 1:1 stoichiometric ratio of the KIF3A/B dimer and KAP3 (Fig 1B).
Figure 1. Reconstruction and biochemical evaluation of KIF3/KAP3 complex.
-
ACartoon representation of the structure of the KIF3/KAP3 complex (upper panel); schematic diagrams showing the major constructs used for biochemical analysis (bottom panel), which are tagged by 6 × His or GST for different uses. ACT: KIF3A residues 481–701; BCT: KIF3B residues 472–747.
-
BSEC‐MALS analysis of the KIF3/KAP3 complex. ABK indicates the reconstructed KIF3/KAP3 (KIF3‐ACT/BCT/KAP3) protein. The MW of the complex was found to be 145 kDa, which is close to the theoretical calculation with a 1:1:1 binding ratio. The SDS–PAGE result of the reconstructed complex is shown on the right side.
-
CRg and I(0) versus number of diffraction frames from SEC‐SAXS experiment measuring the ABK. Rg is indicated by open circles, within which the green spots indicate the data range used for subsequent modeling analysis.
-
DEvaluation of the experimental (colored in green) and theoretical (colored in black) SAXS profiles with the χ2 value of 0.107 for ABK.
-
ESAXS‐based structural outline of ABK. Dmax was examined to be 270 Å. ABK exhibited an elongated conformation with the coiled‐coil domain shown on the lower side and the KIF3 tail‐KAP3 region (Docking domain) shown on the upper side. Potential cargo‐binding pocket was observed in the docking domain.
Next, The ABK protein was further investigated in solution by SEC‐SAXS analysis. The chromatogram results show that the radius of gyration (Rg) between the ascent and descent sides of the peak are different (Figs 1C and EV1A). The shape of the scattering profile on the ascent side of the peak differs from that of the descent side of the peak, where the Rg value is stable (Fig EV1B). In addition, the Guinier plot also shows a rise in the scattering profile on the small angle region (Fig EV1C). The above results indicated the conformational heterogeneity of ABK protein and the possible existence of oligomeric components on the ascent side of the peak, which made it difficult to obtain a single structure. Therefore, we calculated the structure using only the descent side of the peak in this paper (Fig 1C). The real‐space Rg and Dmax were estimated to be ~62.7 and 270 Å for ABK, and ab initio shape analysis was then conducted to obtain the structural outlines. Twenty individual structural models were calculated by the DAMMIF analysis, which were then superimposed and analyzed using cluster analysis. One of the structures was excluded from subsequent calculations based on the normalized spatial discrepancy (NSD) score obtained during the superimposition process. Consequently, the remaining 19 models are presented, and all the structures obtained here exhibited a characteristic formation of a C‐shaped upper part and a rod‐like lower part (Fig EV1D). These structures were averaged, and the excess portions of each structure relative to the average structure were filtered out to create an initial structure. This initial structure was used for the final DAMMIN calculation, and the resulting structure from this process was considered the final model. The final averaged model was established with relatively low χ2 values (Fig 1D). The results indicated that the ABK complex exhibits elongated shapes with the sequential arrangement of the coiled‐coil region, KIF3 tail domains, and KAP3 (Fig 1E). Remarkably, the KIF3 tail‐KAP3 interaction exposes a distinct surface cavity potentially responsible for cargo binding (Fig 1E), where the structure models of the reported KIF3/KAP3 binding partners, ARM domains of APC (APCARM) and β‐catenin (Jimbo et al, 2002; Teng et al, 2005), could be well docked with complementary shapes into the KIF3 tail‐KAP3 region (docking domain; Fig EV1E).
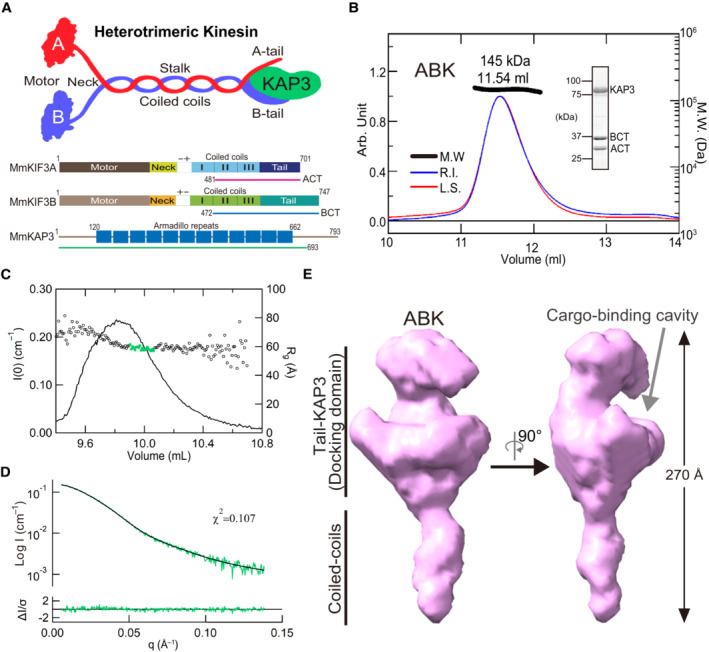
Figure EV1. SEC‐SAXS analysis of ABK complex.
-
A–CExperimental profiles (A), Guinier plots (B) and dimensionless Kratky plots (C) of the SAXS measurements of ABK comparing the ascent and decent sides of the peak. The chromatogram results show that the Rg between the ascent and descent sides of the peak are different. The shape of the scattering profile on the ascent side of the peak differs from that of the descent side of the peak, where the Rg value is stable. Guinier plot indicates a rise of the scattering profile on the small angle region. Based on the above results, the ascent side of the peak contains an oligomeric component, making it difficult to obtain a single structure. Thus, we calculated the structure using only the descent side of the peak.
-
DResults of the DAMMIF modeling of ABK using the data from the descent side of the peak. The DAMMIF process involved calculating 20 individual structural models, which were then superimposed and analyzed using cluster analysis. One of the structures was excluded from subsequent calculations based on the normalized spatial discrepancy (NSD) score obtained during the superimposition process. Consequently, the remaining 19 models are presented. All the structures obtained here exhibited a characteristic formation of a C‐shaped upper part and a rod‐like lower part. These structures were averaged, and the excess portions of each structure relative to the average structure were filtered out to create an initial structure. This initial structure was used for the final DAMMIN calculation. The resulting structure from this process was considered as the final model.
-
EDocking simulation of APC armadillo‐repeat domain and β‐catenin structures (PDB codes: 3NMZ and 1I7X) into the possible cargo‐binding cavity of the ABK SAXS model. ABK: KIF3‐ACT/BCT/KAP3 complex.
KIF3/KAP3 binds APC via the specific surface cleft
Given the potential cargo‐binding cavity of KIF3/KAP determined by the SEC‐SAXS analysis, we next sought to investigate the biochemical and structural bases underlying the cargo‐binding ability and specificity of KIF3/KAP3. To this end, we sought to reconstitute a cargo‐bound KIF3/KAP3 complex with its specific cargo, APC (Jimbo et al, 2002; Baumann et al, 2020), whose colocalization with KIF3/KAP3 was clearly observed in dendrites of primary neuron cultures (Figs 2A and EV2A). APCARM, which reportedly binds KAP3 with a high in vitro affinity (Baumann et al, 2020), was purified (Fig 2B), and its binding with KIF3/KAP was validated by SEC and pull‐down assays (Fig 2C and D). In addition, APCARM did not interact with KIF3 complex in the absence of either KAP3 or KIF3A (Fig EV2B), suggesting that both the KIF3A tail and KAP3 are indispensable for the specific binding of APC with KIF3/KAP3. The reconstituted ABK‐APCARM complex was then purified and assessed by SEC‐MALS analysis, in which ABK‐APCARM exhibited a monodispersed peak with a MW of 274 kDa, which was close to the theoretical MW with a 1:1 stoichiometric ratio (Fig 2E). The complex was subsequently applied to SEC‐SAXS for in‐solution structural analysis. The Rg distribution along the elution peak of ABK‐APCARM is stable compared to ABK (Fig 2F) and the DAMMIF models exhibit approximately spherical structures (Fig EV2C). The final model (Rg: ~ 53.9 Å; Dmax: 195 Å) was obtained with low χ2 values (Fig 2G and Table EV1). ABK‐APCARM exhibits a relatively compacted and globular outline compared to apo ABK (Fig 2H), displaying the docking of APCARM into the binding cleft of ABK. Similar to our results, a cargo‐docking cavity of kinesin‐1 was also observed in the complex structure of cargo peptide‐bound KLC (Pernigo et al, 2013). As KIFs differ majorly in the tail region, and accessory and adaptor proteins that probably constitute the cargo‐specific binding cavities, the discrepancy in the cargo‐binding cavities may underlie the diversity and specificity of kinesin‐cargo recognition.
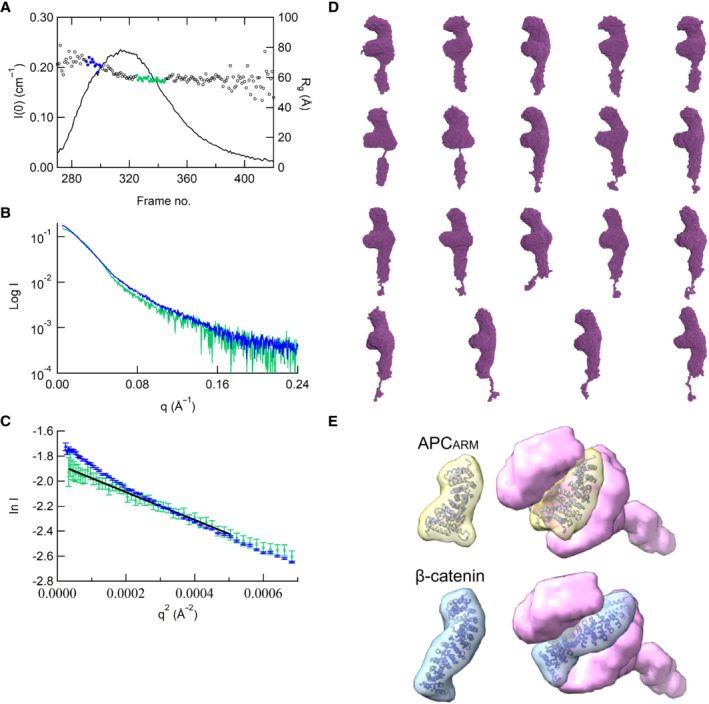
Figure 2. Biochemical validation and reconstruction of the KIF3/KAP3‐cargo complex.
-
AColocalization analysis of KIF3A/B/KAP3 with APC by immunostaining of hippocampal primary neurons. Image of each channel before merging is presented in Fig EV2A.
-
BDiagrams showing functional domains of APC and the construct used in the biochemical assays. The SDS–PAGE result of the purified APCARM protein was shown in the right side.
-
CAnalytic SEC analysis of the ABK‐APCARM binding.
-
DPull‐down analysis of the binding of APCARM with KIF3/KAP3.
-
ESEC‐MALS evaluation of ABK‐APCARM complex.
-
FRg and I(0) versus number of diffraction frames from SEC‐SAXS experiment measuring the ABK‐APCARM. Rg is indicated by open circles, within which the green spots indicate the data range used for subsequent modeling analysis.
-
GEvaluation of the experimental (colored in green) and theoretical (colored in black) SAXS profiles with the χ2 value of 0.075 for ABK‐APCARM.
-
HSAXS‐based structural outline of ABK‐APCARM. Dmax was examined to be 195 Å. APCARM: The armadillo‐repeat domain of APC.
Source data are available online for this figure.
Figure EV2. Binding analysis of KIF3A/B/KAP3 with APC and DAMMIF modeling of ABK‐APCARM complex.
-
ARepresentative images of the immunostaining of KIF3A, KIF3B, KAP3 and APC in the dendrite region of hippocampal primary neurons. Arrows indicate representative colocalizations between KIF3A/B/KAP3 complex and APC. Images shown here are used to generate merged images in Fig 2A.
-
BPull‐down assay assessing the binding capability of KIF3/KAP3 complexes with APCARM. GST‐AA, GST‐BB, and GST‐AB indicate the GST‐tagged homodimeric ACT (KIF3A 481–701), homodimeric BCT (KIF3B 472–747), and heterodimeric ACT/BCT, respectively. The results were evaluated by CBB staining (top panel), and Western blot (bottom panel) using an anti‐6His antibody to probe His‐tagged KAP3 and APCARM.
-
CResults of the DAMMIF modeling of ABK‐APCARM. The DAMMIF process involved calculating 20 individual structural models, which were then superimposed and analyzed using cluster analysis. One of the structures was excluded from subsequent calculations based on the NSD score obtained during the superimposition process. Consequently, the remaining 19 models are presented. The molecular shapes obtained here all exhibit approximately spherical structures. These structures were averaged, and the excess portions of each structure relative to the average structure were filtered out to create an initial structure. This initial structure was used for the final DAMMIN calculation. The resulting structure from this process was considered the final model.
Coiled‐coil region of KIF3 contributes to the cargo binding
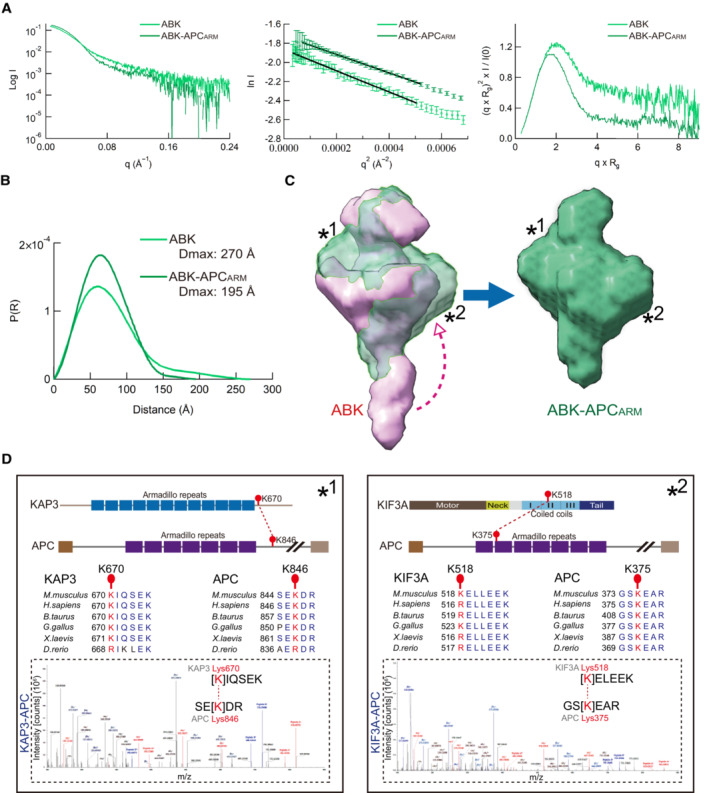
Remarkably, the Guinier plots, dimensionless Kratky plots, and Pair distribution function P(R) calculations from SEC‐SAXS analysis indicated significant structural differences between ABK and ABK‐APCARM (Fig 3A and B). Upon superposition of the SAXS models of ABK and ABK‐APCARM, significant structural changes of ABK upon APCARM binding were observed with the filled and absent cargo‐binding cavity, shortened coiled coils, and swollen intermediate region (Fig 3C), which suggested the involvement of both KIF3 coiled coils and the docking domain in the cargo binding. In addition, to determine the specific interaction sites within the ABK‐APCARM complex, cross‐linking mass spectrometry (XL‐MS) was conducted (Figs 3D and EV3). The cross‐linked peptides were assigned close contact sites not only between the C‐terminal regions of KAP3 (Lys670) and APCARM (Lys846) but also in an unexpected interaction of the N‐terminus of APCARM (Lys375) with the KIF3A coiled‐coil region (Lys518; Fig 3D), suggesting that the KIF3A coiled‐coil region may contribute to enhancing the interaction between ABK and APC by locking on cargo binding. In line with our findings, recent studies on kinesin‐1‐mediated RNA transport indicated the involvement of the coiled coil region of kinesin heavy chain (KHC) in the RNA‐cargo adaptor (aTm1) binding, and suggested the stabilization of the KHC tail‐cargo binding by the coiled‐coil interaction (Cross et al, 2021; Dimitrova‐Paternoga et al, 2021).
Figure 3. Coiled‐coil region of KIF3 participates in the cargo binding.
-
AExperimental profiles, Guinier plots, and dimensionless Kratky plots of the SAXS measurements of ABK and ABK‐APCARM complexes.
-
BPair distribution function P(R) of the ABK and ABK‐APCARM complexes calculated using SAXS data.
-
CSuperposition of SAXS models of ABK and ABK‐APCARM. Asterisks indicate the major structural discrepancy, and the arrowhead indicates the possible conformational change of the KIF3 coiled‐coil region upon APCARM binding.
-
DXL‐MS analysis of the interaction sites between KIF3/KAP3 and APCARM. Two crosslinked site pairs were found to correspond to the possible KIF3/KAP3‐APCARM binding region marked by asterisks in C. Cartoon diagrams, sequence alignment and LC–MS data are shown with crosslinked sites highlighted.
Figure EV3. XL‐MS analysis of ABK‐APCARM complex.
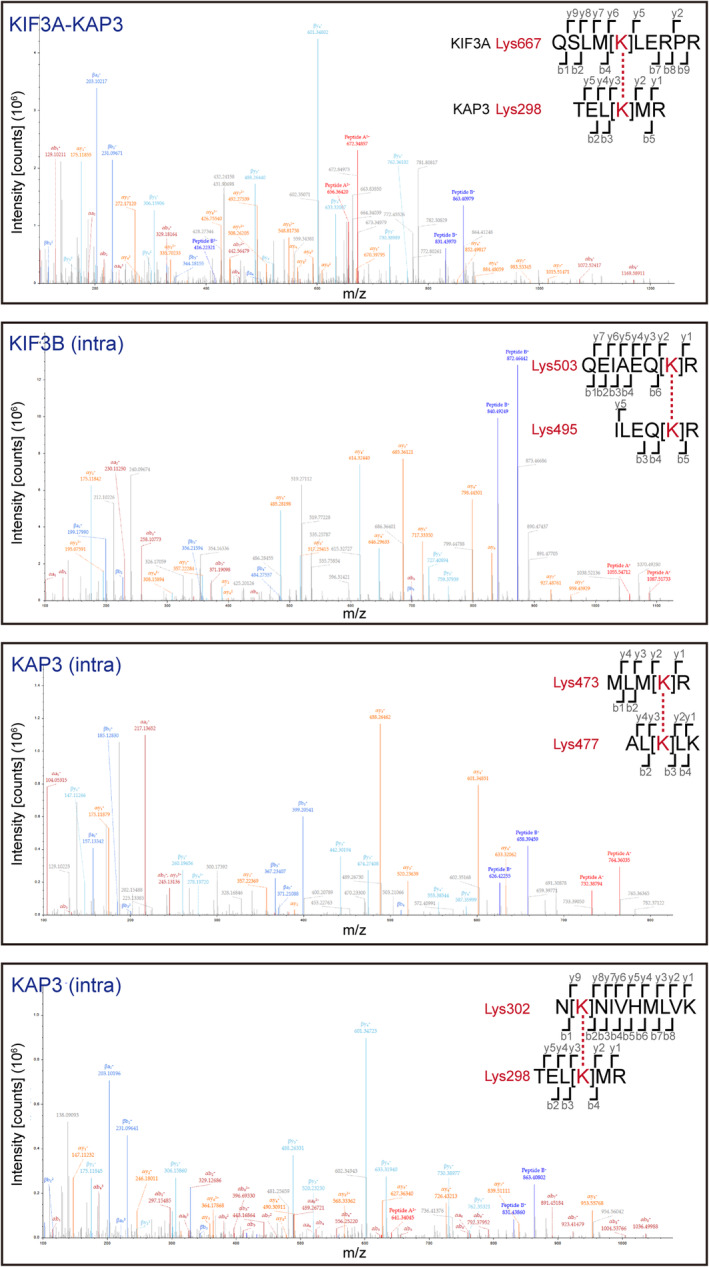
XL‐MS results in addition to those shown in Fig 3D. Intra indicates intramolecular crosslinking.
AFM analysis indicated the conformational dynamics of KIF3/KAP3 complexes
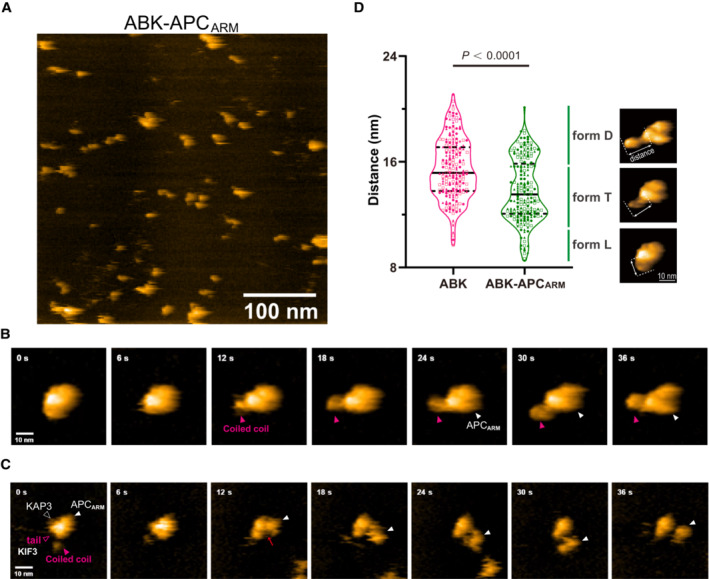
The molecular dynamics of the ABK‐APCARM complex were further examined by single‐molecule real‐time imaging via HS‐AFM. As a result, HS‐AFM images of the protein particles that highly resemble the SAXS models were observed for the complex, and the compacted globular model was validated by methodologically independent analyses in solution (Fig 4A–C). At the same time, a cargo‐free form of ABK was determined by HS‐AFM, which also shares a similar conformation with its SAXS model (Fig EV4A–C). To investigate the transitional conformations through cargo binding between ABK and ABK‐APCARM, we continued the observation of ABK‐APCARM and intensively induced cargo dissociation by continuous tapping of AFM. In the initial stage, ABK‐APCARM exhibited a stable globular conformation with the wound coiled‐coil region, which might interact with bound APCARM to lock and stabilize it (Fig 4B, 0–6 s). Then, when the tapping force was intentionally increased to induce dissociation, the coiled‐coil region was released from binding with APCARM (Fig 4B, 6–24 s) to generate the free and elongated conformation of the coiled‐coil region (Fig 4B, 30 s). Finally, the docking of APCARM into the binding pocket of ABK was clearly visualized (Fig 4B, 30–36 s) compared to cargo‐free ABK (Fig EV4A–C). These distinct transitional conformations of ABK‐APCARM with apparent changes of the coiled‐coil region may largely mimic the dynamics of binding of APCARM to ABK in a reverse manner (Fig 4B and Movie EV1).
Figure 4. HS‐AFM analysis of the KIF3/KAP3‐APC complex indicated its dynamic conformations.
-
AHS‐AFM imaging of KIF3/KAP3‐APC particles.
-
BTime lapse HS‐AFM images of a representative ABK‐APCARM particle with a continuous tapping force.
-
CRepresentative HS‐AFM images of ABK‐APCARM illustrating the dissociation dynamic. Continuous tapping of AFM triggered the dissociation of APCARM from ABK. Red arrows indicate the binding interface between ABK and ABK‐APCARM.
-
DStatistical analysis of the AFM‐based ABK and ABK‐APCARM particle conformations. Two hundred frames extracted from three independent AFM movies of ABK and ABK‐APCARM were used for the analysis. Distance from the coiled coil distal tip to the docking domain (length of the protrusion as shown by the dotted lines) was calculated and shown as violin plots. Data derived from each of the three movies were indicated by circles, triangles and rectangles, respectively. Based on the distal‐center distance, particle conformations were classified into three forms, L (cargo‐locked globular form), T (Transitional curved form) and D (cargo‐docked elongated form). Unpaired t‐test was used to evaluate the statistical significance. The solid and dotted lines indicate the median and quartiles, respectively. The AFM images shown here are re‐used in Fig 5A.
Source data are available online for this figure.
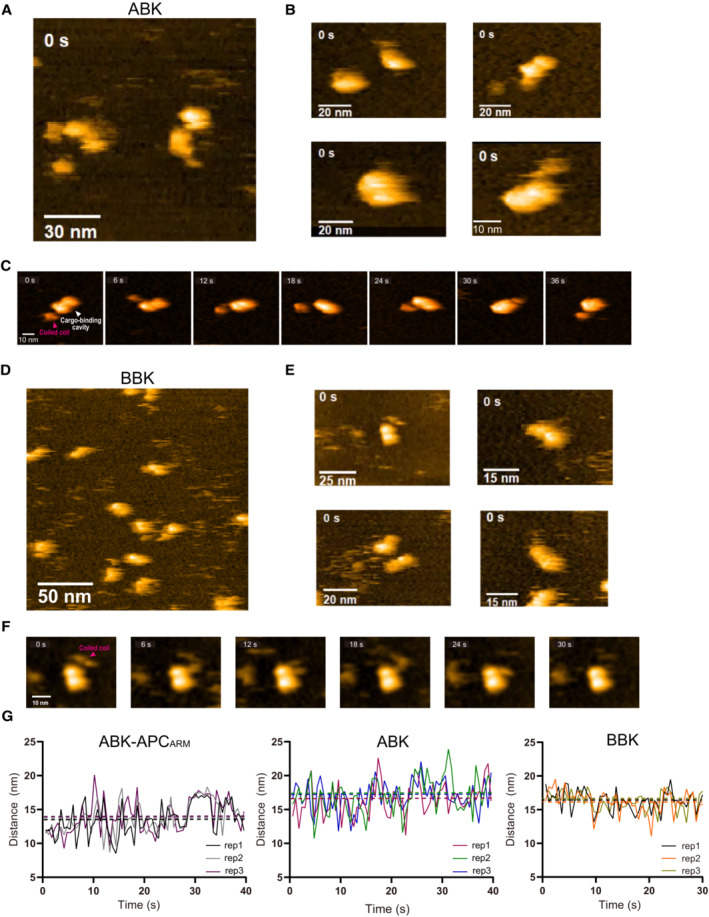
Figure EV4. HS‐AFM analysis of ABK and BBK complexes.
-
A, BHS‐AFM observation and representative particle images of ABK.
-
CTime lapse HS‐AFM images of a representative ABK particle.
-
D, EHS‐AFM observation and representative particle images of BBK.
-
FTime lapse HS‐AFM images of a representative BBK particle.
-
GStatistical analysis of the AFM‐based ABK, BBK and ABK‐APCARM particle conformations. Distance from the coiled coil distal tip to the docking domain (as shown in Fig 4D) was calculated and plotted against time. Data derived from each of the three movies were indicated by different colors. The dotted lines indicate the averages of each group. BBK: KIF3‐BCT/BCT/KAP3 complex.
Meanwhile, the dissociation process of APCARM from the binding pocket of ABK was also visualized via HS‐AFM time series with a continuous tapping force (Fig 4C and Movie EV2), which started from the coiled‐coil‐free state (Fig 4C, 0 s). First, a part of APCARM that needed to be locked by coiled coils became detached from ABK, during which the core binding area of APCARM was still docked in the binding pocket (Fig 4C, 6–12 s). The border of ABK and APCARM was clearly observed at this stage, which also indicated the positions of KAP3 and tail domains of KIF3A and 3B (Fig 4C, 12 s). Subsequently, the KAP3‐binding area of APC was completely released, while the KIF3A tail domain still interacted with APCARM (Fig 4C, 12–30 s). Finally, the binding between APC and the KIF3A tail was also destroyed, and APCARM dissociated from ABK (Fig 4C, 36 s).
In contrast, while the coiled‐coil region also exhibited high flexibility and motility in the HS‐AFM analysis with continuous tapping, cargo‐free ABK displayed less overall conformational flexibility, and particularly globular conformations with coiled coils attached to the docking domain were rarely observed compared to ABK‐APCARM (Fig EV4C). To further quantify the conformational divergence, the distance from the distal point of coiled coils to the docking domain (i.e. length of the protrusion) as indicated in Fig 4D were measured in extracted particle movie frames of ABK and ABK‐APCARM (Figs 4D and EV4G). As a result, ABK exhibited a significantly larger averaged distance than ABK‐APCARM, indicating the smaller frequency of coiled‐coil winding of ABK than ABK‐APCARM. Remarkably, the violin plots of ABK‐APCARM statistically indicated the distribution of the three observed typical conformations, forms D (APC‐Docked elongated form), T (Transitional form) and L (APC‐Locked globular form), corresponding to different distance ranges (Fig 4D).
In addition, the KIF3‐BCT/BCT/KAP3 (BBK) complex that showed no binding with APC (Fig EV2B) was also purified and evaluated by AFM for comparison (Fig EV4D and E). The result suggested that BBK exhibits an average length of protrusion similar to ABK but shows less conformational flexibility than ABK and ABK‐APCARM (Fig EV4F and G). Taken together, AFM data supported the SAXS and XL‐MS findings that the coiled‐coil region participates in KIF3/KAP3‐APC interaction, and illustrated the dynamics of cargo‐free and cargo‐bound ABK particles in solution.
Integrated analysis revealed a two‐step cargo recognition mechanism
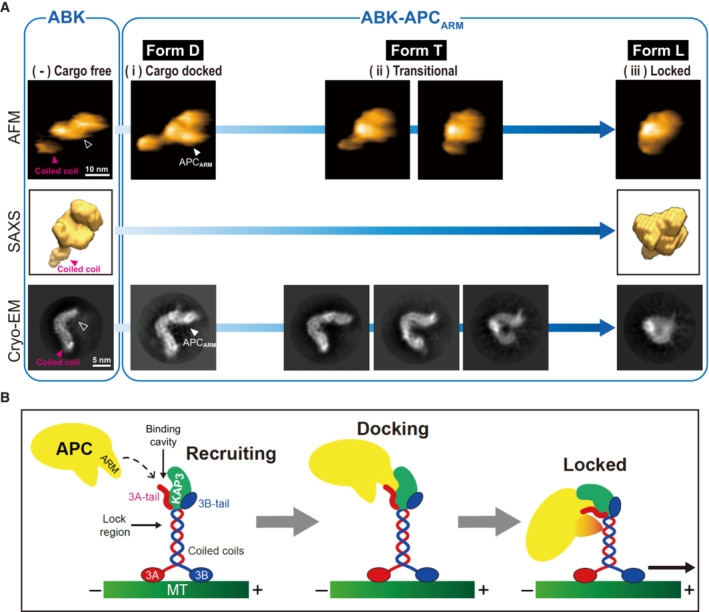
CryoEM analysis was subsequently performed to further evaluate the configurations of ABK and ABKARM complexes, and particle averaging views of high clarity were obtained by 2D classifications. The docking position of APCARM was clearly seen by comparing the major averaging groups of ABK and ABK‐APCARM particles (Figs 5A and EV5A and B). The intermediate conformations of ABK‐APCARM identified by HS‐AFM were also observed in the 2D classification results of cryoEM single particle analysis (Figs 5A and EV5A). Interestingly, while a curved and elongated conformation of the ABK‐APCARM particles was clearly observed by the 2D classification (Fig EV5A, group 1), intermediated (Fig EV5A, groups 2–3) and globular (Fig EV5A, group 4) conformations were also found in the 2D class averages. Consistently, the HS‐AFM and cryoEM results revealed the flexibility and motility of the KIF3 coiled coils and its stabilizing trend after APCARM binding (Figs 4 and 5). These findings on the conformational heterogeneity of kinesin‐2 are in line with the previous rotary shadow EM studies on kinesin‐2 that observed both globular and extended conformations (Wedaman et al, 1996).
Figure 5. The integrated analysis suggests the two‐step APC binding mechanism of KIF3/KAP3.
-
AIntegrated structural analysis of ABK and ABK‐APCARM using SEC‐SAXS, HS‐AFM and cryoEM. Similar structural outlines of the cargo‐free ABK from the three distinct analyses are shown in the left side, and the cargo‐binding cavity is marked by black arrowhead. Paralleled conformations of ABK‐APCARM corresponding to forms D, T and L, which are also presented in Fig 4D, are shown in the right side. Unfilled arrowheads indicate the APC‐binding pocket region of ABK.
-
BModel representation of the potential two‐step APC‐binding process of KIF3A/KAP3 including cargo recruiting by the KIF3 tail, docking into the specific binding cavity formed by KIF3 tail and KAP3, and locking by the KIF3 coiled coil for stabilization and subsequent transport.
Figure EV5. CryoEM analysis of ABK and ABKARM complexes.
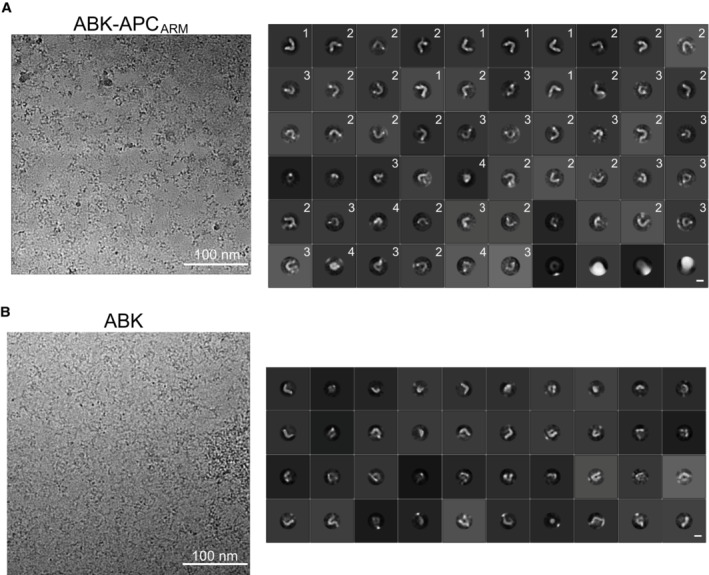
-
ALeft: representative cryoEM image of ABK‐APCARM. Right: 2D class averages of ABK‐APCARM from cryoEM single particle analysis. 2D class averages were classified into four major groups with distinct conformations. HS‐AFM observation and representative particle images of ABK‐APCARM. Scale bars: 100 nm (left), 10 nm (right).
-
BLeft: representative cryoEM image of ABK. Right: 2D class averages of ABK‐APCARM from cryoEM single particle analysis. Scale bars: 100 nm (left), 10 nm (right).
Since the dissociation process often mimics the binding process in a reverse manner, the HS‐AFM and cryoEM results suggested possible conformational changes of KIF3/KAP3 upon APC binding: first, KIF3/KAP3 (Fig 5A, cargo‐free form) binds APC via the KIF3 tail‐KAP3 region with an initially extended APC‐docked form (Fig 5A, form D); second, the KIF3 coiled coil transitionally swings and winds to interact with APCARM (Fig 5A, form T); and last, the wound coiled coil tightly attaches to and binds APC for the final sequestration and stabilization (Fig 5A, form L).
In summary, the specific cargo binding of KIF3/KAP3 was validated in vitro (Figs 2 and EV2), and the biochemical basis of the KIF3‐cargo interaction was revealed by the integrated analysis via SEC‐SAXS, HS‐AFM and cryoEM (Figs 3, EV3, 4, EV4, 5), which supported the previous cell biological observations that KIF3 functionally transported ARM domain‐containing cargos in neurons (Jimbo et al, 2002; Teng et al, 2005; Baumann et al, 2020). Remarkably, the SAXS model of ABK revealed the binding cleft responsible for their specific cargo docking (Figs 1E and EV1E), which was further verified by analysis of ABK‐APCARM (Figs 2 and 3). The above results agree with the previous report that only the heterotrimeric kinesin‐2 was found to be processive rather than other kinesin‐2 subtypes (Brunnbauer et al, 2010), in that the cargo binding essential for autoinhibition release requires the specific binding cavity formed by heterotrimeric KIF3/KAP3 (Figs 1, EV1, 2, EV2, 3). Considering that a scaffold structure was shared by different kinesins (Hirokawa et al, 2009), the specificity and difference mainly resulted from the C‐terminal tail region that form distinguishable docking cavities with adaptor proteins. Interestingly, the coiled‐coil region was also found to contribute to the APCARM binding of KIF3/KAP3 according to the XL‐MS and SAXS results (Fig 3), suggesting a new mechanism by which kinesin–cargo interaction can be modulated through stabilization by the coiled‐coil stalk. Intriguingly, a KIF3B Leu523Pro site mutation in the coiled‐coil region was reported to be associated with retinitis pigmentosa without affecting protein stability (Cogné et al, 2020). Since KIF3B Leu523 localizes closely to the APC‐associated KIF3A Lys518 site in the KIF3A/B coiled‐coil region (Fig 3D), the pathological effect of KIF3B Leu523Pro would likely be the disruption of cargo binding. Moreover, disease‐associated mutations in the coiled‐coil region which potentially affect the cargo interaction have also been reported for kinesin‐1 (KIF5) (Brenner et al, 2018), kinesin‐3 (KIF1) (Pennings et al, 2019) and kinesin‐4 (KIF4) (Wan et al, 2023). Hence, the identified coiled‐coil involvement in cargo association would help clarify the pathogenic mechanism of mutations in kinesin coiled‐coil domains. More importantly, HS‐AFM tapping‐induced dissociation of ABK‐APCARM indicated the temporal roles of the KIF3A tail, KIF3 coiled‐coil region, and KAP3 in APCARM binding, suggesting a stepwise KIF3/KAP3 cargo‐binding mechanism after cargo recruitment: (i) Form D, cargo docking into the binding cleft of KIF3‐tail/KAP3; (ii) Form T, transitional states; and (iii) Form L, subsequent locking and stabilization by wound and attached coiled coils (Fig 5B). Conversely, the cargo‐release process may be in a reverse manner of the binding process suggested above. This binding mechanism might be implicated in the precise regulation of kinesin‐mediated cargo transport by upstream signals such as kinases. In addition, ionic‐strength‐dependent conformational changes of kinesin‐2 were reported that the globular conformer can be converted to the extended conformer with elevated ionic strength, which suggested that ion concentration‐mediated conformational change might also be a regulatory factor for cargo loading and unloading of kinesins in cells (Wedaman et al, 1996). Since motor proteins, including kinesin, dynein and myosin, share a similar elongated C‐terminal structure with a coiled‐coil stalk and a cargo‐binding tail region (Lee Sweeney & Holzbaur, 2018), this two‐step cargo‐binding mechanism might be conserved among motor proteins, and the cargo specificity may depend on their distinct tail regions and mediators. However, this needs to be further investigated on other motor proteins. Moreover, we cannot exclude the possibility that intermediate conformations in the cargo association of kinesin‐2 are different from what we observed in cargo dissociation by AFM, which needs to be further investigated and monitored by advanced techniques in future.
The heterotrimeric kinesin‐cargo interaction is thought to be very flexible as this motor reportedly not only binds ARM domain‐containing cargoes including APC and catenin/cadherins, but also interacts with IFT complexes, GLI and many other cargo proteins which possibly lack high specificity. Our structural analysis suggests that the extended conformation of KIF3 tail‐KAP3 region with a potential binding cavity underlines the specific binding with its high‐affinity cargo such as APC, while the flexibility of the binding cavity and the coiled‐coil region was also indicated by the intermediate and globular conformations observed, which may confer the additional binding capability with other cargoes on kinesin‐2.
Collectively, our integrated analysis of the KIF3/KAP3 and KIF3/KAP3‐APC complexes unveiled the molecular basis of its dynamic cargo‐binding activity of heterotrimeric kinesin‐2 motors, which provides crucial insights into the intracellular trafficking and its relevant pathological processes. Further investigations involving either high‐resolution structural analysis or functional diversity of KIF3/KAP3 are expected.
Materials and Methods
Constructs, protein expression, and purification
All the truncated segments of encoding genes of Mus musculus KIF3A (NCBI accession: NP_032469.2), M. musculus KIF3B (NCBI accession: NP_032470.3), M. musculus KAP3A (NCBI accession: NP_001292572.1) were PCR amplified using their full‐length templates from the previous works (Ichinose et al, 2019) and subsequently cloned into a pET‐21b vector with an N‐terminal 6×His‐tag. In particular, the gene encoding KIF3A C‐terminus (residues 481–701) was also cloned into a pGEX‐6p‐3 vector containing an N‐terminal GST‐tag for the pull‐down assay and into a pETDuet‐1 vector with the gene of KIF3B C‐terminus (residues 475–747) inserted at another MCS for the co‐expression of the C‐terminal part of KIF3A/B heterodimer. The encoding gene of the ARM domain of M. musculus APC (NCBI accession: AAB59632.1; residues 338–1010) was PCR extracted and amplified from mouse cDNA extracts and subsequently cloned into a pET‐21b vector with a 6×His‐tag for biochemical assays and without any tags for complex purification.
The recombinant plasmids were sequenced and transformed into E. coli BL21 (DE3) strains (Novagen) and cells were cultured and harvested as previously described (Ogawa et al, 2017). For reconstruction and purification of the ABK and ABK‐APCARM complexes, the cells expressing each of the complex components were premixed. The lysed cells were centrifuged at 95,000 g for 30 min, and the supernatants were loaded on a Ni‐NTA affinity column (QIAGEN) with a buffer consisting of 20 mM Tris–HCl pH 8.0, 300 mM NaCl, 7 mM β‐ME, 5% (v/v) glycerol, 1 μg/ml DNase I and protein inhibitors, and eluted with 400 mM imidazole. The eluate was further purified by anion exchange chromatography using a Resource Q column (GE Healthcare), followed by size exclusion chromatography (SEC) using a HiLoad 16/60 Superdex™ 200 SEC column (GE Healthcare) with a buffer consisting of 20 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM DTT and 5% (v/v) glycerol. For the ABK and ABK‐APCARM complexes, an additional purification step was performed using the HiRes‐SEC (Ogawa et al, 2017; Toyoshima et al, 2019). The purified proteins were concentrated using ultrafiltration to 5–20 mg/ml and then quick‐frozen in liquid nitrogen and stored at −80°C for biochemical use.
Pull‐down assays and immunoblotting
Escherichia coli cells expressing the bait proteins (GST‐fusion C‐terminal KIF3A or 3B truncations) were premixed with each group of the cells containing overdosed input proteins and lysed by sonication with a buffer of 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 7 mM β‐ME and 5% (v/v) glycerol. After centrifugation, the supernatants were loaded onto the balanced Glutathione Sepharose™ 4B beads (GE Healthcare) and incubated for 30 min on ice; the beads were then washed three times with the buffer to remove unbound proteins. Purified APCARM proteins were further applied to the beads containing each group of KIF3A/B/KAP3 complex, and then washed to remove unbound proteins. The bound proteins were finally eluted with 10 mM reduced glutathione, and the eluates were analyzed using SDS–PAGE and Western blotting. A control group using GST protein as bait was included. For Western blotting analysis, the proteins separated on SDS–PAGE gels were transferred to polyvinylidene difluoride (PVDF) membranes (Thermo Fisher Scientific). The membranes were blocked for 30 min at room temperature with the blocking buffer (3% BSA (m/v) and 0.05% (v/v) Tween in TBS). The blocked membranes were incubated with the rabbit polyclonal anti‐6His antibody (1/1,000, Recenttec) overnight at 4°C followed by incubation with the secondary antibodies (anti‐rabbit IgG antibody, 1/5,000, GE Healthcare) for 30 min at room temperature. Then the membranes were washed and processed with Amersham™ ECL™ Prime Western Blotting Detection Reagent (GE Healthcare) for 2 min, and the signals were detected by chemiluminescence using ImageQuant LAS 4000 (GE Healthcare).
Cell culture and immunocytochemistry
Primary cultures of dissected hippocampal neurons were prepared as previously described (Ichinose et al, 2019). Cells were fixed with 4% paraformaldehyde in PBS for 20 min and then permeabilized with 0.1% Triton X‐100 for 2 min followed by 30 min of blocking with 0.1% BSA in PBS. The proteins were probed with primary antibodies (mouse monoclonal anti‐KIF3A antibody, 1/1,000, RRID:AB_398968; mouse monoclonal anti‐KAP3 antibody, 1/1,000, RRID:AB_397967; rabbit polyclonal anti‐KIF3B antibody, 1/2,000, RRID:AB_2715472; rabbit polyclonal anti‐APC antibody, 1/300, RRID:AB_2057493) overnight at 4°C. Next, the cells were incubated with secondary antibodies (Alexa fluor 568‐conjugated anti rabbit IgG fluorescent antibody, 1/1,000, RRID:AB_1056356; Alexa fluor 488‐conjugated anti mouse IgG fluorescent antibody, 1/1,000, RRID:AB_2534069) for 1 h at room temperature, and then washed three times by PBS for observation. Images of immunostaining were acquired with a confocal laser‐scanning microscope (LSM 880, ZEISS) equipped with 63×/1.4 Plan‐Apochromat oil immersion.
SEC‐MALS
Multi‐angle light scattering (MALS) was performed in‐line with SEC by using an Alliance 2695 high‐performance liquid chromatography system equipped with Dawn Heleos II 18‐angle MALS detectors (Wyatt Technology) and a 2414 Refractive Index (RI) detector (Waters). Purified ABK and ABK‐APCARM samples (30 μl) were applied onto a Superdex™ 200 Increase 10/300 column and examined on the RI‐MALS system under the flow rate of 0.5 ml/min. The obtained data were analyzed by the ASTRA 6.1 software (Wyatt Technology). The data were plotted using GraphPad Prism7.
SEC‐SAXS
SEC‐SAXS data were collected at beamlines 10C and 15A2 in KEK‐PF at 293 K. The SEC‐SAXS experiments were performed using Prominence‐i at 10C and Nexera‐i at 15A2 (SHIMADZU) connected to a Superdex™ 200 increase 10/300 column (GE healthcare) equilibrated with the buffer shown in table EV1. Serial scattering images were taken with 20‐s exposure at 10C and 5‐s exposure at 15A2, respectively. The 15 images measured before the sample fraction were scaled and averaged for a background profile. All scattering images were azimuthally averaged to convert the one‐dimensional scattering intensity data, and data processing was then performed using SAngler (Shimizu et al, 2016). Scattering profiles in the last half of the elution peaks were utilized, and the scattering profiles extrapolated to an infinite dilution condition were calculated from these scattering profiles by using MOLASS (Yonezawa et al, 2023). The radius of gyration (Rg) and forward scattering intensity (I(0)) were calculated from the Guinier approximation by AUTORG (Petoukhov et al, 2007), and the pair distribution function (P(R)) was calculated by GNOM (Svergun, 1992). The absolute intensity calibration was performed using water as a standard. The dummy atom models were calculated using DAMMIF (Franke & Svergun, 2009). Then the 20 dummy atom models were averaged and selected using DAMAVER (Volkov & Svergun, 2003), and the final model calculation using DAMMIN slow mode was performed using the filtered model derived from DAMAVER as the initial starting model. Deposition IDs of SASBDB are SASDMV5 for ABK and SASDMW5 for ABK‐APCARM, respectively (Kikhney et al, 2020).
HS‐AFM
High‐Speed AFM (HS‐AFM) data were collected using a home‐built high‐speed AFM apparatus at Kanazawa University (Ando et al, 2001; Ando, 2019). The cantilevers (AC10, Olympus) were ~ 10 μm long, 2 μm wide, and 90 nm thick with a spring constant of ~ 0.1 N/m. Their resonant frequency and quality factor in aqueous solution were 450–600 kHz and ~ 2, respectively. Mica was used as an observation substrate for adsorbing ABK‐APCARM. 3 μl of buffer solution (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM DTT) containing ABK‐APCARM was placed on a clean mica substrate immediately after cleavage. After 3 min of incubation, the excess unabsorbed molecules were washed away with the buffer, high‐speed AFM observation was performed immediately in the same buffer. For high‐speed AFM imaging, the free vibration amplitude of the cantilever was about 1 nm, and the set‐point amplitude of the feedback operation was about 80–90% of the free amplitude, so that the disturbance from the probe was as small as possible. In some measurements, the set‐point amplitude was decreased to about 70% to intentionally induce dissociation of ABK‐APCARM. We gradually reduced the amplitude setting manually, leading to challenges in determining the exact magnitude of tapping force exerted by the probe during each imaging frame. Nevertheless, we approximated that the tapping force corresponded to approximately 18 pN when the amplitude setting was at 90% of 1 nm free oscillation amplitude, and 22 pN at 70%. The AFM data were analyzed using home‐built analysis software based on IGOR Pro ver. 8 (Wavemetrics). Statistical analysis of the AFM‐based ABK and ABK‐APCARM particle conformations. Two hundred frames extracted from three independent AFM movies of ABK and ABK‐APCARM were used for the analysis. Distance from the coiled coil distal tip to the docking domain (the highest and brightest point with a fixed position) was calculated and shown as violin plots.
XL‐MS
To determine the potential interactions among KIF3A/B, KAP3 and APC, ABK and ABK‐APCARM proteins was crosslinked using the crosslinker disuccinimidyl sulfoxide (DSSO, Thermo Fisher Scientific) for 1 h at room temperature, then trypsinized, desalted, and measured using the LTQ‐Orbitrap Lumos instrument with Easy‐nLC system (Thermo Fisher). Non‐crosslinked peptides were determined through an MS/MS ion search against the amino acid sequences of the recombinant proteins by MASCOT (RRID:SCR_014322). Crosslinked fragments with DSSO were analyzed in XlinkX software (Liu et al, 2017). The MS raw data and results files have been deposited in the ProteomeXchangeConsortium (http://proteomecentral.proteomexchange.org) via the jPOST partner repository (Okuda et al, 2017).
CryoEM
For CryoEM analysis of both ABK and ABK‐APCARM, 3 μl of sample at 1 mg/ml concentration was applied onto a hydrophilized holey carbon grid (Cu, R1.2/1.3, 300 mesh, Quantifoil, Jena, Germany) and was then blotted for 15 s (blot force 15) and flash‐frozen in liquid ethane using Vitrobot Mark IV (FEI) at 18°C and 100% humidity. Images were acquired with a Talos Arctica (FEI, Hillsboro, USA) microscope operating at 200 kV using the EPU software for automated data collection. The movie frames were collected by a 4 k × 4 k Falcon 3 direct electron detector (DED) in linear mode at a nominal magnification of 120,000, yielding a pixel size of 0.88 Å/pixel. Movie frames were recorded at an exposure of 1.51 electrons per Å2 per frame (total exposure: 50 electrons per Å2, number of frames: 33 frames, exposure time: 3.34 s) with the defocus steps of −1.0, −1.5, −2.0, and − 2.5 μm.
For the image processing of ABK dataset, collected frames were aligned, dose‐weighted, and averaged using MotionCor2 (Zheng et al, 2017) on 5 × 5 tiled frames with a B‐factor of 200 applied. The micrographs whose total accumulated motions were larger than 30 Å were discarded. The non‐weighted movie sums were used for CTF estimation via the GCTF (RRID:SCR_016500) with 512‐pixel box size, 30 Å minimum resolution, 4 Å maximum resolution and 0.10 amplitude contrast. The images whose CTF max resolutions were better than 4.5 Å were selected. The particles were picked up using SPHIRE crYOLO with a generalized model (Wagner & Raunser, 2020) using selection threshold of 0.005. The micrographs that contained less than 55 picks were excluded. A stack of 215,256 particle images was extracted from 757 dose‐weighted sum micrographs while rescaling to 3.52 Å/pixel with 64‐pixel box size. Then the stack was subjected to two successive runs of reference‐free 2D classifications (200 expected classes, 213 Å and then 174 Å mask diameter) in RELION3 (RRID:SCR_016274), while selecting the 2D classes with relatively clear protein‐like views after the first run. The selected particle images were rescaled to 1.10 Å/pixel with a 416‐pixel box size. The particle images which became duplicated as the result of alignments and not fully inside of the micrograph boundary anymore due to changing the box size were excluded. 152,484 selected particles were subjected to reference‐free 2D classifications (200 expected classes, 174 Å mask diameter), and the results of this run were reported.
For the image processing of ABK‐APCARM dataset, collected frames were aligned, dose‐weighted, and averaged using RELOIN3.1 own implementation of MotionCor2 on 5 × 5 tiled frames with a B‐factor of 300 applied. The non‐weighted movie sums were used for CTF estimation via the CTFFIND‐4 (RRID:SCR_016732) with 512‐pixel box size, 30 Å minimum resolution, 4 Å maximum resolution and 0.10 amplitude contrast. The particles were picked up using Topaz with the model trained with this dataset (Bepler et al, 2019) where selection threshold of −3.0. A stack of 272,832 particle images was extracted from 844 dose‐weighted sum micrographs while rescaling to 3.52 Å/pixel with 128‐pixel box size. Then, the stack was subjected to two successive runs of reference‐free 2D classifications (300 expected classes, 227 Å and then 199 Å mask diameter) in RELION3.1, while selecting the 2D classes with relatively clear protein‐like views after the first run. The selected particle images were rescaled to 1.10 Å/pixel with a 416‐pixel box size. The particle images which became duplicated as the result of alignments and not fully inside of the micrograph boundary anymore due to changing the box size were excluded. 271,599 selected particles were subjected to reference‐free 2D classifications (300 expected classes, 199 Å mask diameter) and the results of this run were reported.
Sequence and structural analysis
Structure‐based multiple sequence alignments were performed using the programs Clustal Omega (RRID:SCR_001591). The structural coordinates of APCARM (PDB code: 3NMZ) and β‐catenin (PDB code: 1I7X) were downloaded from the PDB. Structural analysis was performed and structural figures were generated using the programs PyMOL (RRID:SCR_000305) and UCSF ChimeraX (RRID:SCR_015872).
Author contributions
Xuguang Jiang: Conceptualization; data curation; investigation; methodology; writing – original draft; writing – review and editing. Tadayuki Ogawa: Conceptualization; data curation; supervision; funding acquisition; validation; investigation; methodology; writing – original draft; project administration; writing – review and editing. Kento Yonezawa: Investigation. Nobutaka Shimizu: Validation; methodology. Sotaro Ichinose: Investigation. Takayuki Uchihashi: Investigation; methodology. Wataru Nagaike: Investigation. Toshio Moriya: Investigation; methodology. Naruhiko Adachi: Methodology. Masato Kawasaki: Investigation. Naoshi Dohmae: Methodology. Toshiya Senda: Validation; writing – review and editing. Nobutaka Hirokawa: Conceptualization; data curation; supervision; funding acquisition; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
PDF+
Source Data for Figure 2
Source Data for Figure 4
Acknowledgements
We thank previous and current members of the N.H. laboratory for their valuable suggestions and assistance. Funding: The Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers JP16H06372 (NH), JP22K06246 (NH), JP20H05499 (TO), JP20K07222 (TO), JP19K06516 (NS); Support from JEOL Ltd. (NH); Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) from AMED under Grant Number JP20am0101071 (TS) (support number 1133 (TO)); the Tokyo Society of Medical Sciences (TO); Joint Research by Exploratory Research Center on Life and Living Systems (ExCELLS) program No 20‐328, 21‐322 (TO); Japanese Government Doctoral Program 161527 (XJ) and the International Postdoctoral Exchange Fellowship Program of China Postdoc Council (Talent‐Introduction Program; YJ20210107) (XJ).
EMBO reports (2023) 24: e56864
Data availability
The MS raw data and results files have been deposited in the ProteomeXchangeConsortium (http://proteomecentral.proteomexchange.org) via the jPOST partner repository (http://jpostdb.org) under the dataset identifiers “ProteomeXchange: PXD027942 and PXD027943” (https://proteomecentral.proteomexchange.org/cgi/GetDataset), and “jPOST: JPST001293 and JPST001294”. The SEC‐SAXS data have been deposited in the SASBDB (https://www.sasbdb.org) under the deposition ID SASDMV5 for ABK and SASDMW5 for ABK‐APCARM, respectively. All other data are available from the corresponding author on request.
References
- Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N (1992) Kinesin family in murine central nervous system. J Cell Biol 119: 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsabban AH, Morikawa M, Tanaka Y, Takei Y, Hirokawa N (2020) Kinesin Kif3b mutation reduces NMDAR subunit NR 2A trafficking and causes schizophrenia‐like phenotypes in mice. EMBO J 39: e101090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T (2019) High‐speed atomic force microscopy. Curr Opin Chem Biol 51: 105–112 [DOI] [PubMed] [Google Scholar]
- Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A (2001) A high‐speed atomic force microscope for studying biological macromolecules. Proc Natl Acad Sci U S A 98: 12468–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Komissarov A, Gili M, Ruprecht V, Wieser S, Maurer SP (2020) A reconstituted mammalian APC‐kinesin complex selectively transports defined packages of axonal mRNAs. Sci Adv 6: eaaz1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, Berger B (2019) Positive‐unlabeled convolutional neural networks for particle picking in cryo‐electron micrographs. Nat Methods 16: 1153–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Yilmaz R, Müller K, Grehl T, Petri S, Meyer T, Grosskreutz J, Weydt P, Ruf W, Neuwirth C et al (2018) Hot‐spot KIF5A mutations cause familial ALS. Brain 141: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnbauer M, Mueller‐Planitz F, Kösem S, Ho TH, Dombi R, Gebhardt JCM, Rief M, Ökten Z (2010) Regulation of a heterodimeric kinesin‐2 through an unprocessive motor domain that is turned processive by its partner. Proc Natl Acad Sci U S A 107: 10460–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BS, Barry RL, Verhey KJ, Allen BL (2015) The heterotrimeric kinesin‐2 complex interacts with and regulates GLI protein function. J Cell Sci 128: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogné B, Latypova X, Senaratne LDS, Martin L, Koboldt DC, Kellaris G, Fievet L, Le Meur G, Caldari D, Debray D et al (2020) Mutations in the kinesin-2 motor KIF3B cause an autosomal-dominant ciliopathy. Am J Hum Genet 106: 893–904. 10.1016/j.ajhg.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM (1992) Isolation of a sea urchin egg kinesin‐related protein using peptide antibodies. J Cell Sci 101: 291–301 [DOI] [PubMed] [Google Scholar]
- Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM (1993) Novel heterotrimeric kinesin‐related protein purified from sea urchin eggs. Nature 366: 268–270 [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998) Chlamydomonas kinesin‐II‐dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JA, Woolfson DN, Dodding MP (2021) Kinesin‐1 captures RNA cargo in its adaptable coils. Genes Dev 35: 937–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova‐Paternoga L, Jagtap PKA, Cyrklaff A, Vaishali V, Lapouge K, Sehr P, Perez K, Heber S, Löw C, Hennig J et al (2021) Molecular basis of mRNA transport by a kinesin‐1–atypical tropomyosin complex. Genes Dev 35: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI (2009) DAMMIF, a program for rapid ab‐initio shape determination in small‐angle scattering. J Appl Cryst 42: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbouchian A, Montgomery A, Gilbert SP, Bentley M (2022) KAP is the neuronal organelle adaptor for Kinesin‐2 KIF3AB and KIF3AC. Mol Biol Cell 33: ar133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin R, de Alencar BC, Jouve M, Bèrre S, le Bouder E, Schindler M, Varthaman A, Gobert FX, Benaroch P (2012) Critical role for the kinesin KIF3A in the HIV life cycle in primary human macrophages. J Cell Biol 199: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik‐Lendrum S, Rank KC, Bensel BM, Taylor KC, Rayment I, Gilbert SP (2015) Kinesin‐2 KIF3AC and KIF3AB can drive long‐range transport along microtubules. Biophys J 109: 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T (2006) Role of the kinesin‐2 family protein, KIF3, during mitosis. J Biol Chem 281: 4094–4099 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R (2005) Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci 6: 201–214 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S (2009) Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10: 682–696 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y (2010) Molecular motors in neurons: Transport mechanisms and roles in brain function, development, and disease. Neuron 68: 610–638 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y (2012) Cilia, KIF3 molecular motor and nodal flow. Curr Opin Cell Biol 24: 31–39 [DOI] [PubMed] [Google Scholar]
- Ichinose S, Ogawa T, Hirokawa N (2015) Mechanism of activity‐dependent cargo loading via the phosphorylation of KIF3A by PKA and CaMKIIa. Neuron 87: 1022–1035 [DOI] [PubMed] [Google Scholar]
- Ichinose S, Ogawa T, Jiang X, Hirokawa N (2019) The spatiotemporal construction of the axon initial segment via KIF3/KAP3/TRIM46 transport under MARK2 signaling. Cell Rep 28: 2413–2426.e7 [DOI] [PubMed] [Google Scholar]
- Jimbo T, Kawasaki Y, Koyama R, Sato R, Takada S, Haraguchi K, Akiyama T (2002) Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol 4: 323–327 [DOI] [PubMed] [Google Scholar]
- Kikhney AG, Borges CR, Molodenskiy DS, Jeffries CM, Svergun DI (2020) SASBDB: towards an automatically curated and validated repository for biological scattering data. Protein Sci 29: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Sato‐Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N (1994) KIF3A is a new microtubule‐based anterograde motor in the nerve axon. J Cell Biol 125: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot N, Antony C, White J, Karsenti E, Vernos I (1998) Role of Xklp3, a subunit of the Xenopus Kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the golgi apparatus. J Cell Biol 143: 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Sweeney H, Holzbaur ELF (2018) Motor proteins. Cold Spring Harb Perspect Biol 10: a021931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lössl P, Scheltema R, Viner R, Heck AJR (2017) Optimized fragmentation schemes and data analysis strategies for proteome‐wide cross‐link identification. Nat Commun 8: 15473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolkema MP, Mans DA, Snijckers CM, van Noort M, van Beest M, Voest EE, Giles RH (2007) The von Hippel‐Lindau tumour suppressor interacts with microtubules through kinesin‐2. FEBS Lett 581: 4571–4576 [DOI] [PubMed] [Google Scholar]
- Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM (2010) Directed microtubule growth, +TIPs, and Kinesin‐2 are required for uniform microtubule polarity in dendrites. Curr Biol 20: 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Scholey JM (1997) Heterotrimeric kinesin‐II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J Cell Biol 138: 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Abramson T, Lyass A, Winter D, Porro E, Hong F, Chamberlin NL, Schnapp BJ (1998) KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol Biol Cell 9: 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N (1998) Randomization of left‐right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95: 829–837 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Saijo S, Shimizu N, Jiang X, Hirokawa N (2017) Mechanism of catalytic microtubule depolymerization via KIF2‐tubulin transitional conformation. Cell Rep 20: 2626–2638 [DOI] [PubMed] [Google Scholar]
- Okuda S, Watanabe Y, Moriya Y, Kawano S, Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa AC et al (2017) JPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res 45: D1107–D1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings M, Schouten MI, van Gaalen J, Meijer RPP, de Bot ST, Kriek M, Saris CGJ, van den Berg LH, van Es MA, Zuidgeest DMH et al (2019) KIF1A variants are a frequent cause of autosomal dominant hereditary spastic paraplegia. Eur J Hum Genet 28: 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernigo S, Lamprecht A, Steiner RA, Dodding MA (2013) Structural basis for kinesin‐1:cargo recognition. Science 340: 356–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento PA, Stewart RJ, Goldstein LSB (1994) Characterization of the KLP68D kinesin‐like protein in Drosophila: Possible roles in axonal transport. J Cell Biol 127: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI (2007) ATSAS 2.1—towards automated and web‐supported small‐angle scattering data analysis. J Appl Cryst 40: 223–228 [Google Scholar]
- Rashid DJ, Wedaman KP, Scholey JM (1995) Heterodimerization of the two motor subunits of the heterotrimeric kinesin, KRP85/95. J Mol Biol 252: 157–162 [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825 [DOI] [PubMed] [Google Scholar]
- Scholey JM (1996) Kinesin‐II, a membrane traffic motor in axons, axonemes, and spindles. J Cell Biol 133: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM (2013) Kinesin‐2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol 29: 443–469 [DOI] [PubMed] [Google Scholar]
- Shimizu N, Yatabe K, Nagatani Y, Saijyo S, Kosuge T, Igarashi N (2016) Software development for analysis of small‐angle x‐ray scattering data. AIP Conf Proc 1471: 050017 [Google Scholar]
- Signor D, Wedaman KP, Rose LS, Scholey JM (1999) Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans . Mol Biol Cell 10: 345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameter in indirect‐transform methods using perceptual criteria. J Appl Cryst 25: 495–503 [Google Scholar]
- Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N (1999) Left‐right asymmetry and kinesin superfamily protein KIF3a: new insights in determination of laterality and mesoderm induction by KIF3A(−/−) mice analysis. J Cell Biol 145: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Yamazaki H, Seog DH, Kanai Y, Terada S, Hirokawa N (2000) Kinesin superfamily protein 3 (KIF3) motor transports fodrin‐associating vesicles important for neurite building. J Cell Biol 148: 1255–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J, Rai T, Tanaka Y, Takei Y, Nakata T, Hirasawa M, Kulkarni AB, Hirokawa N (2005) The KIF3 motor transports N‐cadherin and organizes the developing neuroepithelium. Nat Cell Biol 7: 474–482 [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Jiang X, Ogawa T, Ohnishi T, Yoshihara S, Balan S, Yoshikawa T, Hirokawa N (2019) Enhanced carbonyl stress induces irreversible multimerization of CRMP2 in schizophrenia pathogenesis. Life Sci Alliance 2: e201900478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- Vernos I, Heasman J, Wylie C (1993) Multiple kinesin‐like transcripts in xenopus oocytes. Dev Biol 157: 232–239 [DOI] [PubMed] [Google Scholar]
- Volkov V v, Svergun DI (2003) Uniqueness of ab initio shape determination in small‐angle scattering. J Appl Cryst 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Raunser S (2020) The evolution of SPHIRE‐crYOLO particle picking and its application in automated cryoEM processing workflows. Commun Biol 3: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waither Z, Vashishtha M, Hall JL (1994) The Chlamydomonas kinesin‐like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol 126: 175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Morikawa M, Morikawa M, Iwata S, Naseer MI, Ahmed Chaudhary AG, Tanaka Y, Hirokawa N (2023) KIF4 regulates neuronal morphology and seizure susceptibility via the PARP1 signaling pathway. J Cell Biol 222: e202208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tanaka Y, Xu Y, Takeda S, Hirokawa N (2022) KIF3B promotes a PI3K signaling gradient causing changes in a Shh protein gradient and suppressing polydactyly in mice. Dev Cell 57: 2273–2289.e11 [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Meyer DW, Rashid DJ, Cole DG, Scholey JM (1996) Sequence and submolecular localization of the 115‐kD accessory subunit of the heterotrimeric kinesin‐II (KRP85/95) complex. J Cell Biol 132: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N (1995) KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end‐directed motor for membrane organelle transport. J Cell Biol 130: 1387–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N (1996) Cloning and characterization of KAP3: a novel kinesin superfamily‐associated protein of KIF3A/3B. Proc Natl Acad Sci U S A 93: 8443–8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa K, Takahashi M, Yatabe K, Nagatani Y, Shimizu N (2023) MOLASS: software for automatic processing of matrix data obtained from small‐angle X‐ray scattering and UV‐visible spectroscopy combined with size‐exclusion chromatography. Biophys Physicobiol 20: e200001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara S, Jiang X, Morikawa M, Ogawa T, Ichinose S, Yabe H, Kakita A, Toyoshima M, Kunii Y, Yoshikawa T et al (2021) Betaine ameliorates schizophrenic traits by functionally compensating for KIF3‐based CRMP2 transport. Cell Rep 35: 108971 [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]


