Abstract
Background:
Minimum inhibitory concentration (MIC) of slow growing mycobacteria (SGM) often do not correlate with the treatment response. Among the challenges is the identification of MIC of drugs that degrade in solution faster than the doubling time of the SGM.
Methods:
First, we identified the rate of omadacycline degradation in solution, and its effect on the rapidly growing methicillin resistant Staphylococcus aureus (MRSA). We then identified doubling times versus MICs for Mycobacterium abscessus, M. avium, and M. kansasii, with and without supplementation for degraded drug.
Results:
Omadacycline concentration in solution declined ~50% over 24hr. In the MRSA experiments, omadacycline demonstrated 66.48 ± 19.38% loss in potency over 24hr, confirming the degradation rate in solution. M. abscessus had a doubling time of 8.75 ± 1.23hr and the omadacycline MIC after 24hr of incubation was 2mg/L with and without 50% daily drug supplementation, indicating that drug degradation had no effect on this rapid grower. The doubling time for M. avium was 29.52hr (95% confidence interval (CI): 23.18–33.89hr) and 31.15hr (95%CI: 19.45–38.49 hr) for M. kansasii. The M. avium MICs ±50% daily omadacycline supplementation were 1mg/L and 0.5mg/L on day 7, whereas the M. kansasii MICs ±50% daily supplementation were >128mg/L and 32mg/L on day 7.
Conclusion:
Omadacycline degradation in solution leads to falsely high MICs when SGM doubling time exceed the drug degradation rates in solution. The challenge could be overcome by daily drug supplementation to account for the loss of potency, which is laborious, or perhaps stabilizing the drug from degradation.
Keywords: Susceptibility testing, Nontuberculous mycobacteria, Trailing effect
1. Introduction
Nontuberculous mycobacteria (NTM) cause pulmonary disease in humans that is recalcitrant to treatment. The role of drug susceptibility testing in the choice of anti-infective agents for the treatment of NTM disease, especially slowly growing mycobacteria (SGM) is debatable because of discrepancies between in vitro minimum inhibitory concentration (MIC) of the drug and the efficacy of the same drug observed in patients [1,2]. Therefore, there is need to determine the potential reasons for these discrepancies and develop better methods to perform reproducible drug susceptibility (DST) results. Based on our previous work with omadacycline and ertapenem, we hypothesize that some laboratory technical issues could partly explain these discrepancies [3, 4]. In addition, such drugs often demonstrate an MIC “trailing effect” which makes it further difficult to establish the precise MIC [5]. The danger of relying on MICs is that potentially useful drugs could be discarded based on their falsely high MICs; thus, the technical challenges need to be overcome. Tetracyclines are known to have solubility and stability problem across temperature range [6,7]. Therefore, various agents including ascorbic acid have been tested to stabilize tetracyclines in the solution [8]. Here we present the technical challenges with performing drug susceptibility testing with the novel tetracycline, omadacycline, which recently demonstrated efficacy against both SGM and rapidly growing mycobacteria (RGM) despite high MICs [5,9–12].
2. Materials and methods
Omadacycline (BOC Sciences, NY, USA) was freshly prepared by dissolving the drug in 100% dimethyl sulfoxide (DMSO) followed by further dilution in sterile water. Final DMSO concentration was <0.01% (v/v) with no effect on the growth of the bacteria. The culture media used in the experiments were cation-adjusted Mueller Hinton broth (CAMHB), Mueller Hinton agar (MHA), Middlebrook 7H9 broth supplemented with 10% oleic acid, albumin, dextrose, catalase (OADC) (herein termed 7H9 broth), or Middlebrook 7H10 agar supplemented with 10% OADC (herein termed 7H10 agar), depending upon the bacterial species used in the experiment. All experiments were performed twice with two replicates per condition, unless reported otherwise. GraphPad Prism (v9) was used for data analysis and graphing.
Firstly, to determine the degree of drug degradation, four aliquots of 1mL fresh drug were prepared (final concentration 0.5mg/L). One aliquot was immediately stored at −80°C for drug concentration measurement using a validated LC-MS/MS assay [3]. The remaining three tubes were stored at 37°C. One tube was removed at 24hr, 48hr, and 72hr, respectively, and stored at −80°C for drug concentration measurement.
Secondly, methicillin-resistant Staphylococcus aureus (MRSA; ATCC 29213) was grown to logarithmic phase in CAMHB, turbidity of the cultures was adjusted to 0.5 McFarland standard, followed by 100-fold dilution of the turbidity adjusted cultures to prepare the inoculum with a bacterial density of ~105 CFU/mL. The cultures were co-incubated with freshly made omadacycline at a concentration of 0.5mg/L. After 24hr of incubation at 37°C under shaking conditions at ambient environment, cultures were washed to remove carry-over drug, 10-fold serially diluted, and inoculated on MHA for estimation of bacterial burden as colony forming unit per milliliter (CFU/mL). The experiment was repeated, but this time instead of using the freshly prepared drug, the MRSA was co-incubated with omadacycline solution that was stored at 37°C for 24hr. After 24hr of incubation, samples were processed for enumeration of bacterial burden. The CFU/mL data was used to calculate the percent MRSA growth inhibition with fresh drug versus the drug solution stored at 37°C. In addition, from both set of the experiments, the culture supernatant was collected in a fresh tube, filtered sterilized using 0.22μM filters and stored at −80°C for drug concentration measurement.
Thirdly, we performed broth micro-dilution MIC experiments, using 7H9 broth with the rapidly growing Mycobacterium abscessus (ATCC 19977), with and without 50% drug supplementation every 24 hr. The 50% drug supplementation volume was 1μL (100x concentration) per well. The non-treated control wells were treated in the same manner with addition of 1μL media to keep the volume constant in all wells. The final omadacycline concentrations tested were 0.06, 0.12, 0.25, 0.5, 1, 2, 4, and 8mg/L. At the end of 48hr of incubation at 30°C under ambient environment, 96-well plates were visually inspected and the drug concentration completely inhibiting the bacterial growth was recorded as MIC. In addition, after recording the visual MIC, the samples were collected from each well, washed twice with normal saline to remove the carryover drug, serially diluted, and inoculated on 7H10 agar for enumeration of bacterial burden.
Fourthly, we performed broth macro-dilution MIC studies with the SGM Mycobacterium avium (ATCC 700898) and M. kansasii (ATCC 12478) in 7H9 broth, in a total volume of 5mL. The final omadacycline concentrations tested ranged from 0.5mg/L to 128mg/L, in a 2-fold serial dilution. For each NTM, one set of cultures was co-incubated for 7-days without any drug supplementation, while the second set of the cultures received 50% daily supplementation for each drug concentration. The drug supplementation volume was 25μL (100x concentration) per tube. After 7 days of incubation at 37°C under ambient environment, the cultures were washed twice with normal saline to remove the carryover drug, 10-fold serially diluted, and inoculated on 7H10 agar for enumeration of bacterial burden. After 10 days of incubation at 37°C, the colonies were recorded and used to determine the drug concentration associated with >99.9% bacterial kill compared to the nontreated controls as well as the relationship between the bacterial burden and drug concentrations using the inhibitory sigmoid maximal effect model. A secondary objective to determine the drug concentration with >99.9% reduction was to identify which endpoint by visual inspection (i.e., 80% versus 100% pellet size reduction) correlate better with visual MIC to overcome the issue of the trailing effect.
3. Results
The omadacycline concentration measured in the freshly prepared drug solution as well as the samples incubated at 37°C (from the MRSA experiment) is shown in Fig. 1A. The omadacycline decline rate had a half-life of 21.70hr (r2 = 1), based on an exponential decline model. The omadacycline concentration fell by 45.2% over the first 24hrs. Similarly, in Fig. 1A the omadacycline decline in CAMHB at 37°C had a half-life of 31.37hr (95% confidence interval (CI): 19.76–60.22 hr) (r2 = 0.999). The omadacycline concentration fell by approximately 49.8% over the first 24hrs. Fig. 1B shows the % MRSA growth inhibition compared to the non-treated control. We did not include the results beyond 24hr as there was virtually no difference between the non-treated control versus omadacycline treated cultures, indicating loss in efficacy due to drug degradation. The MRSA kill rate with freshly made omadacycline was −0.047 log10 CFU/mL/hr, and −0.005 log10 CFU/mL/hr with drug solution stored at 37°C for 24hrs. Thus, these results reveal loss of omadacycline efficacy, in solution, over time when stored or longer incubation at 37°C, as seen in the MRSA experiment.
Fig. 1. Effect of incubation temperature and time on omadacycline potency.
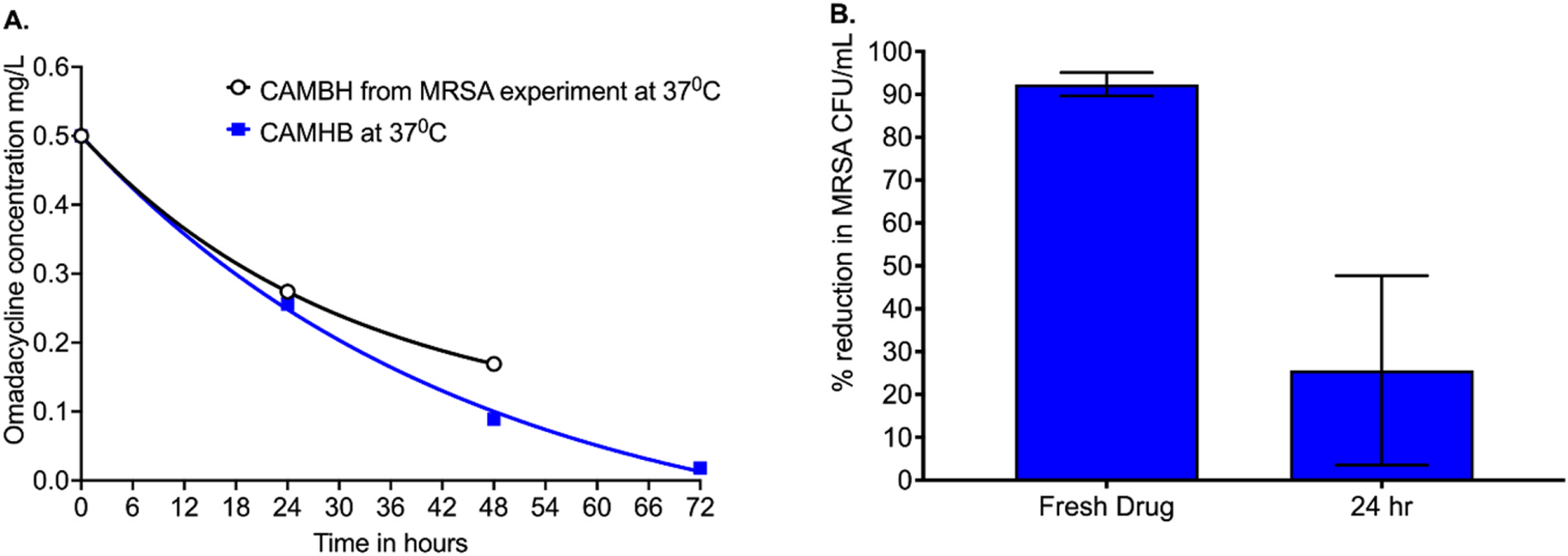
(A) Omadacycline concentrations versus time were modeled using an exponential decline model to identify the half-life of decay under the two different conditions. The concentration of drug solution was observed to be reduced by ~50% in the first 24hrs. (B) Results shown are % reduction in methicillin-resistant Staphylococcus aureus (MRSA) incubated with freshly prepared omadacycline at 0.5mg/L and the same omadacycline preparation after incubation at 37°C. Microbial kill was reduced by 66.45 + 19.38% over the 24hrs.
Fig. 2 shows the results of the M. abscessus experiments. The doubling time of M. abscessus in cultures without omadacycline exposure (non-treated control) was 8.75 ± 1.23hr, in the same range as observed by others in the past [13], but considerably lower than the omadacycline decline half-life in CAMHB. The omadacycline MICs were done with and without supplementation of 50% drug to compensate for drug degradation, with results shown in Fig. 2A. The was no difference in MIC based on the visual inspection with and without drug supplementation. There was a trailing effect observed. The CFU/mL from each well culture were analyzed using the inhibitory sigmoid Emax model (Fig. 2B and Table 1). Table 1 shows that neither the potency (EC50 or effective concentration mediating 50% of the maximal effect) nor the efficacy (Emax or maximal kill compared to the non-treated control) were altered by the 50% drug supplementation. Omadacycline failed to show M. abscessus kill below stasis (initial bacterial burden in the inoculum on day 0) at any concentration.
Fig. 2. M. abscessus and omadacycline.
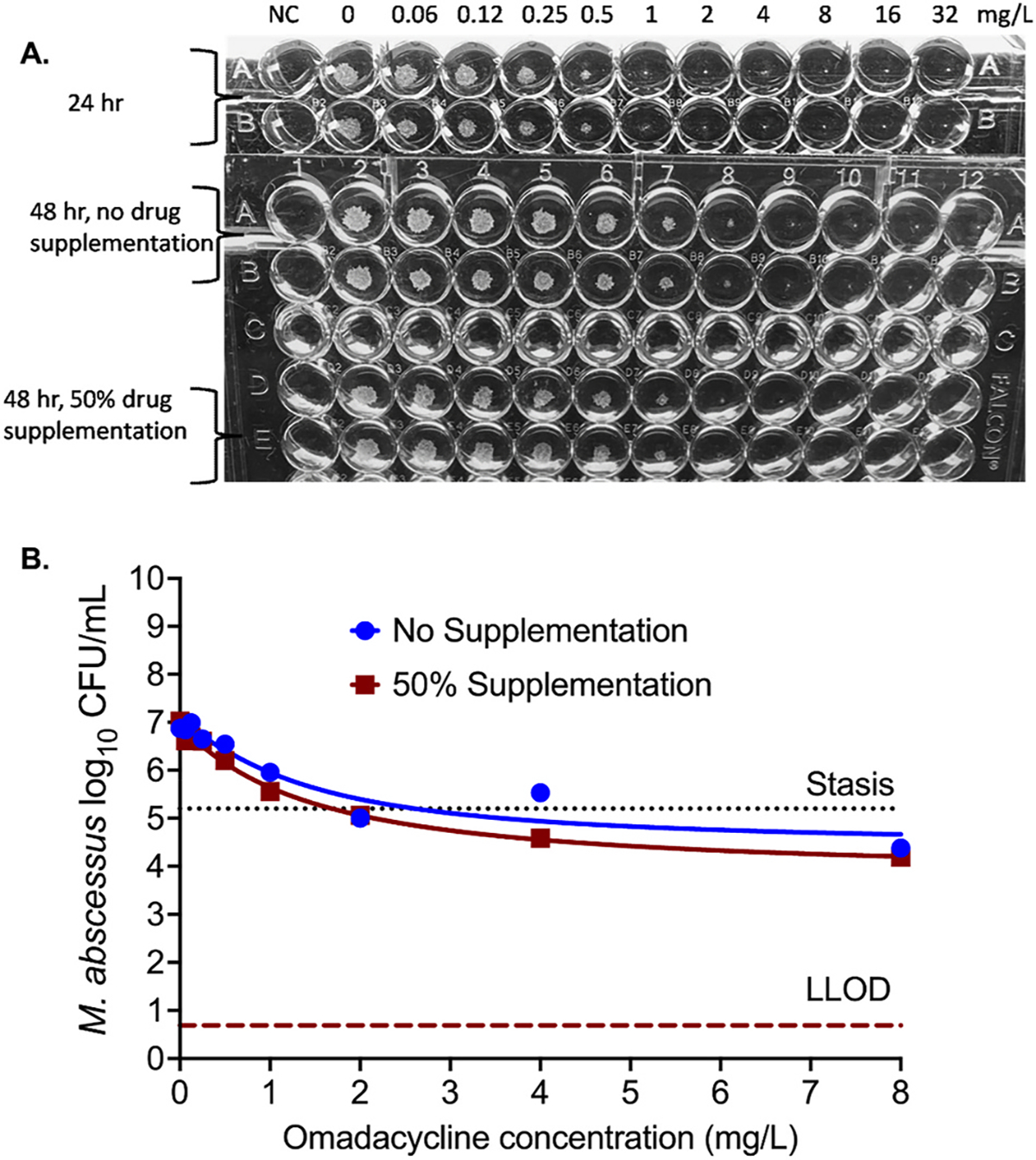
(A) After 24 hr of incubation, considering the trailing effect the MIC would be 0.5mg/L, whereas 100% growth inhibition was seen with 1mg/L concentration. However, in the repeat experiment in which drug was supplemented after 24hr of incubation and cultures were incubated for additional 24hr, the omadacycline MIC, accounting for the trailing effect was recorded as 1mg/L with and without the 50% drug supplementation. Thus, longer time of incubation would result in higher MIC. (B) There was no statistically significant difference in the bacterial kill with difference omadacycline concentration, irrespective of the 50% daily drug supplementation versus no drug supplementation. (Stasis, bacterial burden in the inoculum; LLOD, lower limit of detection by CFU/mL).
Table 1.
Inhibitory sigmoid Emax model for M. abscessus versus omadacycline concentration after 2 days of incubation.
| Parameter | No Supplementation |
50% Daily Drug Supplementation |
|---|---|---|
| Estimate (95% confidence intervals) | Estimate (95% confidence intervals) | |
|
| ||
| Econ (log10 CFU/mL) | 6.95 (6.28–7.62) | 6.91 (6.66–7.17) |
| Emax (log10 CFU/mL) | 2.53 (0.16–4.89) | 3.12 (2.10–4.13) |
| H | 1.29(0.82–3.51) | 1.09 (0.53–1.65) |
| EC50 (mg/L] | 1.39 (−0.93 to 3.71) | 1.41 (0.50–2.32) |
| R2 | 0.91 | 0.99 |
Regarding the omadacycline MIC of SGM, the doubling time for M. avium was 29.52hr (95% CI: 23.18–33.89hr); longer than the omadacycline decline half-life of 21.70hrs. Fig. 3A show the MIC results, highlighting the trailing effect in cultures with or without 50% daily drug supplementation to keep the concentrations constant as in the beginning of the experiment. The omadacycline MIC against M. avium with 100% growth inhibition or absence of bacterial pellet was recorded as 128mg/L when no drug was supplemented, versus 64mg/L MIC with daily 50% drug supplementation. However, if we consider 80% reduction in the pellet size and the trailing effect, the MICs would be 1 mg/L and 0.5mg/L, respectively. Fig. 3B shows the M. avium inhibitory sigmoid Emax model results for the day 7 cultures. Parameter estimates with and without 50% daily drug supplementation were compared for the null hypothesis that one curve fits both sets of data, which was rejected and demonstrated 7.98-fold differences in potency (p < 0.0001) as shown in Table 2. Thus, 50% daily drug supplementation did improve omadacycline potency against M. avium. Another interesting observation, shown in Fig. 3C, was >99% growth inhibition was with 4mg/L concentration with daily drug supplementation and 16mg/L without supplementation. Thus, with drugs showing trailing effect, precise MIC should be determined using the CFU/mL which, however, will make it labor intensive and difficult to perform in a routine clinical laboratory.
Fig. 3. Omadacycline and M. avium.
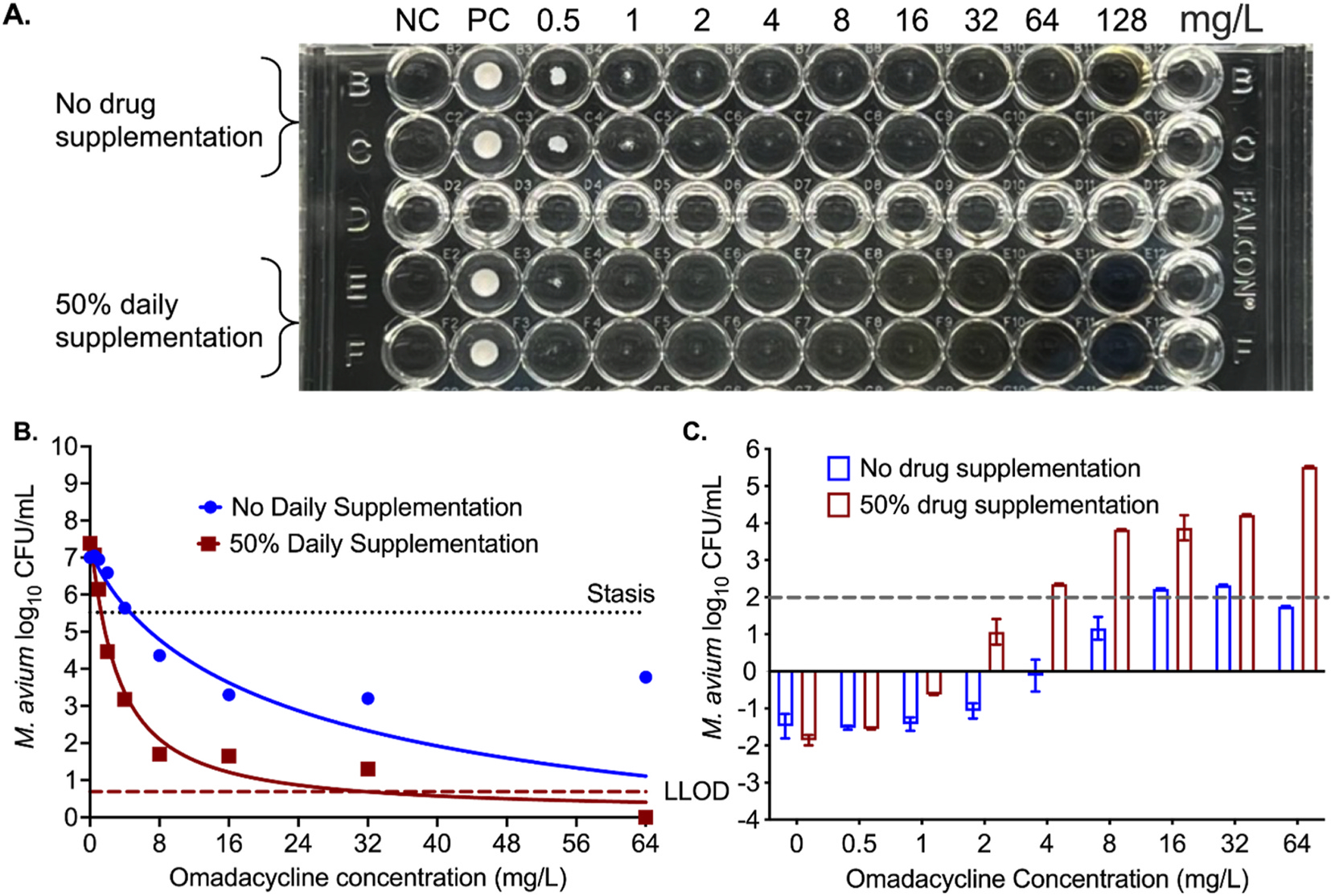
(A) Trailing effects can be seen in wells with or without drug supplementation. Considering 80% growth inhibition, 1mg/L concentration was recorded as MIC when no drug was supplemented versus 0.5mg/L MIC with daily 50% drug supplementation. (NC = negative control). (B) Relationship between the bacterial burden and omadacycline concentration using the 4-parameter inhibitory sigmoidal maximal effect model. (Stasis, bacterial burden in the inoculum; LLOD, lower limit of detection by CFU/mL). (C) Bacterial kill below stasis with different omadacycline concentrations after 7 days of incubation at 37°C. Negative numbers on the y-axis indicate bacterial growth compared to stasis, whereas positive numbers mean kill below stasis.
Table 2.
Inhibitory sigmoid Emax model for Mycobacterium avium versus omadacycline concentration after 7 days of incubation.
| Parameter | No Supplementation |
50% Daily Drug Supplementation |
|---|---|---|
| Estimate (95% confidence intervals) | Estimate (95% confidence intervals) | |
|
| ||
| Econ (log10 CFU/mL) | 7.28 (6.80–7.82) | 7.56 (6.95–8.19) |
| Emax (log10 CFU/mL) | 9.01 (6.89–19.27) | 7.45 (6.48–8.64) |
| H | 0.84 (0.51–1.25) | 1.08 (0.76–1.59) |
| EC50 (mg/L] | 25.30 (12.30–292.0) | 3.17 (2.21–5.07) |
| R 2 | 0.98 | 0.98 |
As regards to M. kansasii, the doubling time was calculated as 31.15hr (95% CI: 19.45–38.49 hr), which was longer than the omadacycline decline half-life. Fig. 4A shows the omadacycline MICs for M. kansasii with and without 50% daily drug supplementation. The figure shows that the MIC was >128mg/L without drug supplementation and 32mg/L with 50% daily drug supplementation. This is at least a 3-tube dilution difference and does not consider the trailing effect that might interpret a lower MIC. However, if we consider the trailing effect and 80% reductions in pellet size, the MIC would be 32mg/L and 4 mg/L, respectively. Fig. 4B and Table 3 shows the inhibitory sigmoid Emax models with and without 50% daily drug supplementation. Parameter estimates with and without 50% daily drug supplementation were compared for the null hypothesis that one curve fits both sets of data, which was rejected and demonstrated 4.3-fold differences in potency (p < 0.0001). Fig. 4C shows that the drug concentration required for 2 log10 CFU/mL kill below stasis would be 32mg/L with supplementation and >128mg/L without supplementation, matching the visual inspection readouts for 100% cut-off instead of the currently used 80% reduction in the pellet size.
Fig. 4. Omadacycline and M. kansasii.
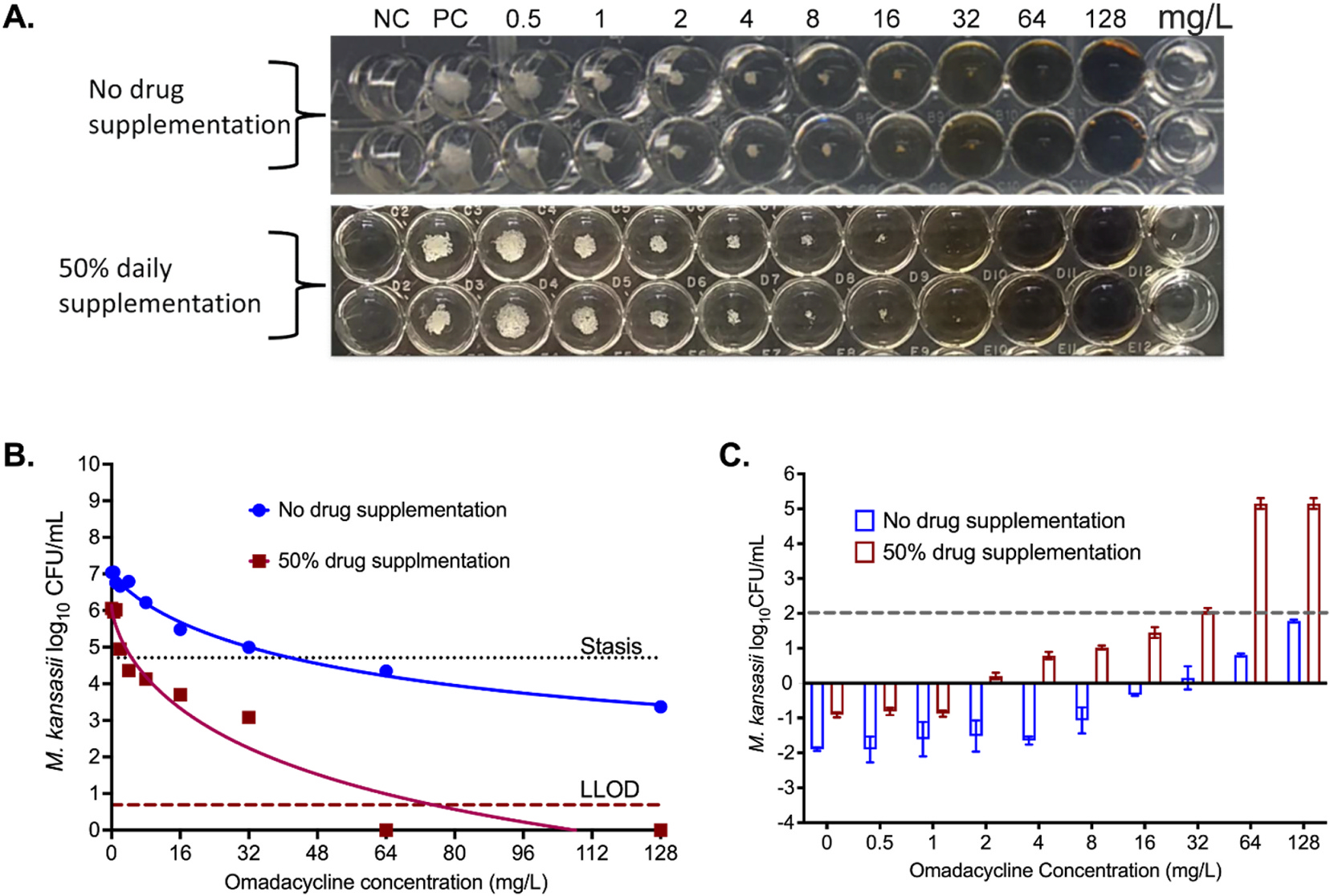
(A) Picture of MIC plates, showing trailing effects. Considering trailing effect, 100% growth inhibition was not achieved with the highest concentration of 128 mg/L concentration when no drug was supplemented to account for the drug degradation. However, 100% growth inhibition was recorded to 32mg/L with daily 50% drug supplementation. (B) The inhibitory sigmoidal maximal effect model showed better kill and lower EC50 with daily drug supplementation; data points for omadacycline 16mg/L and 32mg/L in wells with supplementation were automatically eliminated by program as outliers and are not shown. (Stasis, bacterial burden in the inoculum; LLOD, lower limit of detection by CFU/mL). C) With daily drug supplementation, the drug concentration to reduce the bacterial burden 2 log10 CFU/mL was 4-fold lower. Negative numbers on the y-axis indicate bacterial growth compared to stasis, whereas positive numbers mean kill below stasis
Table 3.
Inhibitory sigmoid Emax model for M. kansasii burden versus omadacycline concentration after 7 days of incubation.
| Parameter | No Supplementation |
50% Daily Drug Supplementation |
|---|---|---|
| Estimate (95% confidence intervals) | Estimate (95% confidence intervals) | |
|
| ||
| Econ (log10 CFU/mL) | 7.07 (6.72–7.54) | 6.08 (5.65–6.58) |
| Emax (log10 CFU/mL) | 5.70 (3.21 to imprecise) | 6.97 (5.81–9.76) |
| H | 0.83 (Imprecise to 1.68) | 1.05 (0.68–1.68) |
| EC50 (mg/L] | 63.39 (18.50 to imprecise) | 14.79 (9.44–39.22) |
| R 2 | 0.93 | 0.98 |
4. Discussion
Our first message is that for SGM (M. avium and M. kansasii) whose doubling times are longer compared to the rate of omadacycline degradation in solution, visual readouts using the broth micro-dilution method will result in several tube dilutions higher MICs, leading to the erroneous assumption that drugs such as omadacycline may not work for SGM. Yet, in vivo model data [11], hollow fiber data [3], and limited number of case series demonstrate good omadacycline efficacy [12,14]. Here, using M. abscessus as model organism, we show that when the bacterial doubling-time is lower than the omadacycline degradation half-life, the MICs are the same with and without 50% daily drug supplementation, so that MICs are not artefactual. However, once the bacterial doubling-time approaches or exceeds the omadacycline degradation half-life, supplementation that takes into account the 50% degradation every 24hrs leads to MIC reduction of several-tube dilution. The pharmacokinetics/pharmacodynamics explanation is straight forward, there is loss of drug potency (EC50) due to the drug degradation. Our solution here was daily drug supplementation that is easy to perform in research laboratories. However, this would be laborious in clinical laboratories and is difficult to automate. Thus, a more permanent solution might be to fortify test compounds that degrade rapidly with chemicals that could slow test compound’s degradation. However, specific adjuvants/compounds may alter drug activity in vitro. Thus, more works needs to be done regarding optimization of drug susceptibility methods for drugs that degrade in solution.
The data presented here highlight some of the challenges with drug susceptibility testing in NTM. The technical challenges include difficulties in inoculum calibration, drug instability, as well as a lack of standardized interpretive criteria (visual MIC) due to the trailing effect [5,15]. In addition, Nicklas et al. have also shown differences in MICs of RGM (M. abscessus) by culture media: CAMHB gave lower omadacycline MICs than Middlebrook 7H9 broth [11]. This adds further complexity to susceptibility testing, on top of drug degradation, and on clinical interpretation of the antimicrobial susceptibility results. Therefore, there is a need for the development and implementation of quality-controlled reliable and reproducible drug susceptibility testing techniques for NTM, especially for the drugs that degrade faster than the doubling time of the bacteria [4,16].
Second, the problem of trailing MICs is as old as clinical antibiotic susceptibility testing, with extensive descriptions by Bauer and Sherris with early sulfonamides and in Mycobacteria described by Wallace et al. in the 1980s [17,18]. We have previously shown that this could be accounted in part, by drug degradation in solution [4]. Here, we identified the CFUs in all wells, including those showing trailing, in three NTM species (M. abscessus, M. avium, and M. kansasii). We specifically wanted to determine which of the endpoints 80% or 100% pellet size reduction by naked eye, correlate with >99% inhibition by CFU/mL. Using SGM results, we show that 99% inhibition gives results more like the 100% cut-off value than 80%.
Third, the MIC, doubling time versus degradation half-life results for SGM, and to a certain extent even the RGM, means that time kill studies with static concentrations conducted over >24hrs with drugs that rapidly degrade might also give erroneous results. Conceivably, one would get rapid regrowth that would be attributed to the emergence of resistance when in fact it may simply be drug being degraded. This means investigators should consider testing the stability of the agents in the culture medium over the conduct of the study under the same temperature conditions.
Among the limitations of our study is that in attempt to explore the reason for MIC discrepancy, our methods may be considered as deviation from the recommended CLSI [19] method of drug susceptibility testing for mycobacteria. We used 7H9 broth that has different composition compared to the CAMHB. In addition, CLSI does not recommend addition of any growth supplement for RGM, whereas 7H9 broth was supplemented with 10% OADC. This protocol deviation was necessary to compare the growth rate/doubling time of the different NTM species between different media commonly used to grow the mycobacteria and how these might affect the overall test results. The hypothesis was if a media supports rapid growth of bacteria, longer incubation time will result in higher MIC. It turned out that indeed the MICs between 7H9 broth were higher compared to CAMHB and this could partially be attributed to faster growth in the given culture medium. Our findings also suggest that during the approval process, the drug manufacturer should also advise the laboratories which growth medium should be used to perform the drug susceptibility testing.
In summary, with the prospect of new drugs, such as omadacycline, to treat difficult-to-treat NTM infections, parallel efforts are needed to develop new drug susceptibility testing methods for drug that degrade faster than the doubling time of the slowly growing mycobacteria.
Funding sources
SS is supported by funding from the department of Pulmonary Immunology, UTHCT [423500/14000], University of Texas System STARS award [250439/39411], and 1R21AI148096 from the National Institute of Allergy and Infectious Diseases. SKH is supported by funding from NIAID R01 AI137080 and U01 AI150508.
Footnotes
Declaration of competing interest
Tawanda Gumbo founded and is president and CEO of Praedicare Inc., a pre-clinical and translational contract research organization, and founded Praedicare Africa, a clinical contract research organization. All other authors have nothing to declare.
Contributor Information
Prem Shankar, Department of Pulmonary Immunology, University of Texas Health Science Centre at Tyler, Tyler, TX, USA.
Sanjay Singh, Department of Pulmonary Immunology, University of Texas Health Science Centre at Tyler, Tyler, TX, USA.
Gunavanthi D. Boorgula, Department of Pulmonary Immunology, University of Texas Health Science Centre at Tyler, Tyler, TX, USA
Tawanda Gumbo, Quantitative Preclinical & Clinical Sciences Department, Praedicare Inc., Dallas, TX, USA; Hollow Fiber System & Experimental Therapeutics Laboratories, Praedicare Inc, Dallas, TX, USA.
Scott K. Heysell, Division of Infectious Diseases and International Health, University of Virginia, USA
Shashikant Srivastava, Department of Pulmonary Immunology, University of Texas Health Science Centre at Tyler, Tyler, TX, USA.
References
- [1].Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ Jr, Andrejak C, Bottger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis 2020;71:e1–36. 10.1093/cid/ciaa241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brown-Elliott BA, Woods GL. Antimycobacterial susceptibility testing of non-tuberculous mycobacteria. J Clin Microbiol 2019;57. 10.1128/JCM.00834-19. e00834-00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chapagain M, Pasipanodya JG, Athale S, Bernal C, Trammell R, Howe D, Gumbo T. Omadacycline efficacy in the hollow fibre system model of pulmonary Mycobacterium avium complex and potency at clinically attainable doses. J Antimicrob Chemother 2022. 10.1093/jac/dkac068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Srivastava S, van Rijn SP, Wessels AM, Alffenaar JW, Gumbo T. Susceptibility testing of antibiotics that degrade faster than the doubling time of slow-growing mycobacteria: ertapenem sterilizing effect versus Mycobacterium tuberculosis. Antimicrob Agents Chemother 2016;60:3193–5. 10.1128/AAC.02924-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brown-Elliott BA, Wallace RJ Jr. In vitro Susceptibility testing of omadacycline against nontuberculous mycobacteria. Antimicrob Agents Chemother 2021;65. 10.1128/AAC.01947-20. e01947-01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Robiyanto R, Zaidi ST, Shastri MD, Castelino RL, Wanandy ST, Jose MD, Patel RP. Stability of tigecycline in different types of peritoneal dialysis solutions. Perit Dial Int 2016;36:410–4. 10.3747/pdi.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kurjogi M, Issa Mohammad YH, Alghamdi S, Abdelrahman M, Satapute P, Jogaiah S. Detection and determination of stability of the antibiotic residues in cow’s milk. PLoS One 2019;14:e0223475. 10.1371/journal.pone.0223475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jitkova Y, Gronda M, Hurren R, Wang X, Goard CA, Jhas B, Schimmer AD. A novel formulation of tigecycline has enhanced stability and sustained antibacterial and antileukemic activity. PLoS One 2014;9:e95281. 10.1371/journal.pone.0095281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tanaka SK, Steenbergen J, Villano S. Discovery, pharmacology, and clinical profile of omadacycline, a novel aminomethylcycline antibiotic. Bioorg Med Chem 2016;24:6409–19. 10.1016/j.bmc.2016.07.029. [DOI] [PubMed] [Google Scholar]
- [10].Kaushik A, Ammerman NC, Martins O, Parrish NM, Nuermberger EL. In vitro activity of new tetracycline analogs omadacycline and eravacycline against drug-resistant clinical Isolates of Mycobacterium abscessus. Antimicrob Agents Chemother 2019;63. 10.1128/AAC.00470-19. e00470-00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nicklas DA, Maggioncalda EC, Story-Roller E, Eichelman B, Tabor C, Serio AW, Keepers TR, Chitra S, Lamichhane G. Potency of omadacycline against Mycobacteroides abscessus clinical isolates in vitro and in a mouse model of pulmonary infection. Antimicrob Agents Chemother 2022;66:e0170421. 10.1128/AAC.01704-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pearson JC, Dionne B, Richterman A, Vidal SJ, Weiss Z, Velasquez GE, Marty FM, Sax PE, Yawetz S. Omadacycline for the treatment of Mycobacterium abscessus disease: a case series. Open Forum Infect Dis 2020;7:ofaa415. 10.1093/ofid/ofaa415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roux AL, Viljoen A, Bah A, Simeone R, Bernut A, Laencina L, Deramaudt T, Rottman M, Gaillard JL, Majlessi L, Brosch R, Girard-Misguich F, Vergne I, de Chastellier C, Kremer L, Herrmann JL. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol 2016;6. 10.1098/rsob.160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morrisette T, Alosaimy S, Philley JV, Wadle C, Howard C, Webb AJ, Veve MP, Barger ML, Bouchard J, Gore TW, Lagnf AM, Ansari I, Mejia-Chew C, Cohen KA, Rybak MJ. Preliminary, real-world, multicenter experience with omadacycline for Mycobacterium abscessus infections. Open Forum Infect Dis 2021;8. 10.1093/ofid/ofab002.ofab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].No_Author_Listed. EUCAST reading guide for broth microdilution. European Committee on Antimicrobial Susceptibility Testing (EUCAST); 2022. [Google Scholar]
- [16].Nikolayevskyy V, Maurer FP, Holicka Y, Taylor L, Liddy H, Kranzer K. Novel external quality assurance scheme for drug susceptibility testing of non-tuberculous mycobacteria: a multicentre pilot study. J Antimicrob Chemother 2019;74:1288–94. 10.1093/jac/dkz027. [DOI] [PubMed] [Google Scholar]
- [17].Bauer AW, Sherris JC. The determination of sulfonamide susceptibility of bacteria. Chemotherapia 1964;9:1–19. 10.1159/000220337. [DOI] [PubMed] [Google Scholar]
- [18].Wallace RJ Jr, Jones DB, Wiss K. Sulfonamide activity against Mycobacterium fortuitum and Mycobacterium chelonei. Rev Infect Dis 1981;3:898–904. 10.1093/clinids/3.5.898. [DOI] [PubMed] [Google Scholar]
- [19].CLSI. Susceptibility testing of mycobacteria, nocardia spp., and other aerobic actinomycetes. In: CLSI standard M24. third ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. [PubMed] [Google Scholar]


