Abstract
Melissococcus pluton is the causative agent of European foulbrood, a disease of honeybee larvae. This bacterium is particularly difficult to isolate because of its stringent growth requirements and competition from other bacteria. PCR was used selectively to amplify specific rRNA gene sequences of M. pluton from pure culture, from crude cell lysates, and directly from infected bee larvae. The PCR primers were designed from M. pluton 16S rRNA sequence data. The PCR products were visualized by agarose gel electrophoresis and confirmed as originating from M. pluton by sequencing in both directions. Detection was highly specific, and the probes did not hybridize with DNA from other bacterial species tested. This method enabled the rapid and specific detection and identification of M. pluton from pure cultures and infected bee larvae.
European foulbrood (EFB) is a disease of honeybee larvae which kills them when they are 4 or 5 days old, mostly in the early summer when bee colonies are rapidly growing. There is often a well-defined seasonal outbreak followed immediately by a spontaneous recovery. When the larvae die, they often turn brown, decompose, and give off a foul odor which is associated with the disease. Several bacteria may be associated with cases of EFB, and most have at one time or another been considered as the primary pathogen (5). At various times the disease has been attributed to Streptococcus apis (12), Bacillus alvei (10), and Bacterium eurydice (8). Today it is accepted that EFB is caused by Melissococcus pluton (5), which is a gram-positive bacterium formerly known as Bacillus pluton (17, 18). The other bacteria previously thought to cause EFB are in fact commonly associated secondary bacteria which rapidly accelerate the death of the infected larvae.
The disease is widespread and has been of economic importance in North and South America, Europe, Japan, Australia, India, and southern Africa (13). The diagnosis of EFB is initially based on typical disease symptoms in the honeybee colony. If the disease is widespread, the comb takes on a pepperbox appearance, with many uncapped and diseased cells mixed with normal capped cells. Another symptom is that caps sealing healthy brood are convex while those of diseased brood are concave and sometimes punctured (16). Infected larvae first turn yellow and then brown and die twisted against the side of the cell or melted at the bottom of the cell. The remains of the larva dry out and form a scalelike protrusion in the cell. Worker, drone, and queen larvae are all susceptible to EFB.
Despite the widespread economic importance of EFB, the routine cultivation and identification of M. pluton remain problematic, as evidenced by the organism’s being poorly described in Bergey’s Manual of Systematic Bacteriology (10a). In addition, EFB is not routinely screened by honeybee disease laboratories worldwide, despite a number of publications comparing techniques for analysis of EFB (1, 11). Instead, these laboratories tend to use the presence of the secondary bacterial invaders Achromabacter eurydice, Bacillus laterosporus, and B. alvei as presumptive evidence of the presence of M. pluton (11, 16). Although selective medium for the cultivation of M. pluton exists (2, 3, 5, 6, 7, 11), involving anaerobic incubation on yeast extract medium supplemented with sugar, starch, cysteine, and potassium, the cultural requirements are fastidious and M. pluton is extremely difficult to isolate. Competition from other bacteria is a problem, and even if isolated, M. pluton is difficult to identify due to its pleomorphic nature (16).
EFB has most commonly been diagnosed by the microscopic identification of M. pluton early in the infection cycle, before the appearance of secondary microflora associated with the disease. Negative staining with nigrosin assists in the identification of bacteria resembling M. pluton (1, 2). The bacterium is a non-spore-forming, lanceolate coccus, occurring singly, in pairs, and in chains and measuring 0.5 to 0.7 by 1.0 μm (1, 16). M. pluton may also be distinguished by scanning electron microscopy (1), by enzyme-linked immunosorbent assays (Pinnock and Featherstone [14]), and with polyclonal antisera by using type strains (2).
No method for the identification of the causative organism of EFB has, however, become established in routine screening. In order to quickly and accurately identify the presence of M. pluton, PCR primers were designed from the 16S rRNA gene of M. pluton. The 16S rRNA genes are essential for the survival of all organisms and are highly conserved in all kingdoms; therefore, it is an ideal molecular marker to use for identification (20). However, to isolate the 16S rRNA gene by PCR and sequence it, in order to identify the organism from which it came, would be too time-consuming and expensive to perform on a routine basis. The PCR primers used here were designed from a region of the M. pluton 16S rRNA gene that was not homologous to other bacterial 16S rRNA gene sequences deposited in the available databases. The primers were designed to amplify a PCR amplicon which was 831 bp and would be unique to M. pluton.
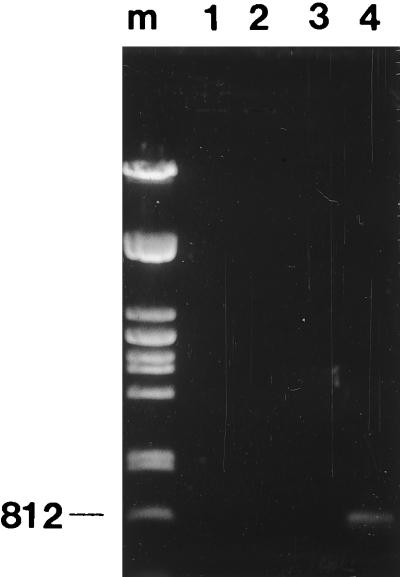
The PCR primers were tested against four bacteria, including the M. pluton type strain (LMG 9003). Escherichia coli, B. alvei, and Staphylococcus aureus were grown in Luria broth, and M. pluton was grown in basal medium (4). Genomic DNAs were prepared according to standard methods (19). The DNA pellets were resuspended in 50 μl of 1× TE buffer (10 mM Tris-HCl, pH 7.5; 1 mM EDTA). Approximately 1 to 3 μg of genomic DNA was amplified in a 50-μl reaction mixture in a Hybaid OMN-E thermocycler. The PCR was optimized with the following concentrations: 2 mM MgCl2, 50 pmol of primers (primer 1, 5′ GAAGAGGAGTTAAAAGGCGC 3′; primer 2, 5′ TTATCTCTAAGGCGTTCAAAGG 3′) per μl, 25 mM (each) deoxyribonucleoside triphosphate, and 1 U of Taq polymerase per μl. The PCR conditions consisted of a cycle of 95°C (1 min); 30 cycles of 93°C (1 min), 55°C (30 s), and 72°C (1 min); and a final cycle of 72°C (5 min). The molecular weights of the PCR products were determined by electrophoresis in a 0.8% agarose gel and staining with ethidium bromide. Under these PCR conditions only the M. pluton type strain produced a PCR product. As expected, this product banded alongside the 812-bp lambda Pst marker (Fig. 1).
FIG. 1.
Agarose gel (0.8%) showing PCR products from genomic DNA from different lysed bacterial species (pure cultures). Lanes: m, Pst-cut lambda DNA marker (the 812-bp band is indicated); 1, E. coli; 2, B. alvei; 3, S. aureus; and 4, M. pluton.
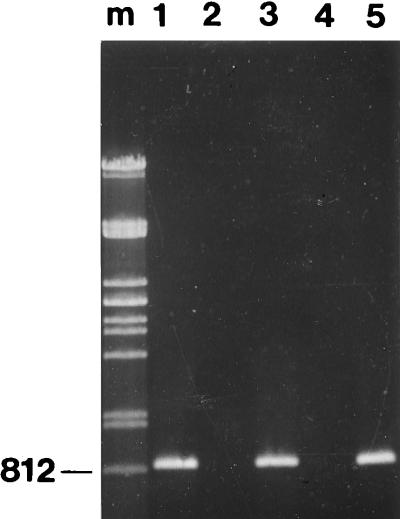
The PCR primers were then used directly with honeybee larvae. Two bee larvae that showed symptoms of EFB disease and two apparently healthy worker larvae from Stellenbosch, South Africa, were tested. Each larva was incubated individually in the basal medium overnight, anaerobically, at 30°C in an anaerobic jar containing hydrogen plus 10% (vol/vol) CO2. Two milliliters of each sample was then centrifuged at 1,000 × g for 2 min, and the supernatant was centrifuged at 10,000 × g for 5 min. The resultant pellets were resuspended in 100 μl of sterile H2O and heated at 95°C for 15 min. One microliter was amplified in a 50-μl PCR mixture. When the resulting PCR products were run on agarose gels only the diseased larvae produced bands of approximately 812 bp, whereas the healthy larvae did not (Fig. 2).
FIG. 2.
Agarose gel (0.8%) showing PCR products from healthy and diseased bee larvae and an M. pluton pure culture. Lanes: m, Pst-cut lambda DNA marker (the 812-bp band is indicated); 1, M. pluton type strain; 2, nondiseased bee larvae; 3, diseased bee larvae; 4, nondiseased bee larvae; and 5, diseased bee larvae.
To confirm that the bee larva PCR product was from M. pluton, the PCR product was cloned into pCR-Script Amp SK(+) cloning vector and transformed with the pCR-Script Amp SK(+) cloning kit (Stratagene). Plasmids were purified with the nucleobond kit (Maherey-Nagel) for plasmid isolation. The PCR product was sequenced in both directions by standard methods (15). A sequence similarity search was done by using the BLAST server at the National Center for Biotechnology Information. The PCR product was found to be 831 bp long, which was the same size as the region on the M. pluton 16S rRNA gene between the two primers. An alignment of the PCR product and the M. pluton 16S rRNA gene sequence showed the two sequences to have base pair mismatches at positions 593 and 824 of the 16S rRNA gene (Fig. 3).
FIG. 3.
Alignment of bases 84 to 913 of the 16S rRNA gene of M. pluton (NCBI accession no. X75752) with the PCR product obtained from enriched bee lysate. The primer-binding regions are indicated in boldface type, and the two mismatches at positions 593 and 824 are boxed.
The high degree of homology between the PCR product and the M. pluton 16S rDNA gene (99.7% [data not shown]) clearly indicates that the PCR product was from M. pluton. Previous studies have shown the 16S rDNA gene of M. pluton to exhibit the highest levels of sequence similarity with members of the Enterococcus genus (range, approximately 94 to 96%) (9). The PCR product used in this study was found to have its closest sequence similarity to the Enterococcus faecium 16S rRNA gene (96.3% [data not shown]). More significantly, the PCR primers used here, compared to E. faecium, had six and five mismatches over the 20-mer regions of primer 1 and primer 2, respectively. Since Enterococcus is the genus most closely related to Melissococcus, it is not possible that these bacteria were detected with these PCR primers.
The technique described here is a quick and accurate method for the identification of M. pluton directly from diseased bee larvae. The time required to complete the procedure, following overnight incubation of the bee larvae, is only 6 h, thus making the method an ideal diagnostic technique. This is a particularly useful technique because of the internationally recognized difficulty in isolating and identifying M. pluton.
Acknowledgments
This work was partly funded by the Foundation for Research Development.
We are grateful for sequencing carried out by Di James from the Department of Microbiology at the University of Cape Town.
REFERENCES
- 1.Alippi A M. A comparison of laboratory techniques for the detection of significant bacteria of the honey bee, Apis mellifera, in Argentina. J Apic Res. 1991;30:75–80. [Google Scholar]
- 2.Allen M F, Ball B V. The cultural characteristics and serological relationships of isolates of Melissococcus pluton. J Apic Res. 1993;32:80–88. [Google Scholar]
- 3.Anderson D L. Pests and pathogens of the honeybee (Apis mellifera L.) in Fiji. J Apic Res. 1990;29:53–59. [Google Scholar]
- 4.Bailey L. An improved method for the isolation of Streptococcus pluton and observations on its distribution and ecology. J Insect Pathol. 1959;1:80–85. [Google Scholar]
- 5.Bailey L. Melissococcus pluton, the cause of European foulbrood of honey bees (Apis spp.) J Appl Bacteriol. 1983;55:65–69. [Google Scholar]
- 6.Bailey L. Melissococcus pluton and European foulbrood. Bee World. 1985;66:134–136. [Google Scholar]
- 7.Bailey L, Collins M D. Reclassification of Streptococcus pluton (White) in a new genus Melissococcus pluton. J Appl Bacteriol. 1982;53:215–217. [Google Scholar]
- 8.Burri R. Neue Untersuchungen über den Erreger der Sauerbrut der Bienen. Beih Schweiz Bienen-Zeitung. 1941;1:1–28. [Google Scholar]
- 9.Cai J, Collins M D. Evidence for a close phylogenetic relationship between Melissococcus pluton, the causative agent of European foulbrood disease, and the genus Enterococcus. Int J Syst Bacteriol. 1994;44:365–367. doi: 10.1099/00207713-44-2-365. [DOI] [PubMed] [Google Scholar]
- 10.Cheshire F R, Cheyne W W. The pathogenic history and history under cultivation of a new bacillus (B. alvei), the cause of a disease of the hive bee hitherto known as foulbrood. J R Microsc Soc Ser 2. 1905;5:581–601. [Google Scholar]
- 10a.Hardie J M. Genus Streptococcus. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1043–1071. [Google Scholar]
- 11.Hornitsky M A Z, Wilson S C. A system for the diagnosis of the major bacterial brood diseases of honeybees. J Apic Res. 1989;28:191–195. [Google Scholar]
- 12.Maasen A. Zur Aetiologie der Sogennanten Faulbrut der Honigbiene. Arb Biol Reichsanstalt Land Fortwirt Berlin-Dahlem. 1908;6:53–70. [Google Scholar]
- 13.Matheson A. World bee health report. Bee World. 1993;74:176–212. [Google Scholar]
- 14.Pinnock D E, Featherstone N E. Detection and quantification of Melissococcus pluton infection in honey bee colonies by means of enzyme-linked immunosorbent assay. J Apic Res. 1984;23:168–170. [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimanuki H, Knox D A, Furgala B, Caron D M, Williams J L. Diseases and pests of honey bees. In: Graham J, editor. The hive and the honey bee. Hamilton, Ill: Dadant and Sons; 1992. pp. 1083–1151. [Google Scholar]
- 17.White G F. The cause of European foulbrood. U.S. Department of Agriculture Bureau of Entomology circular no. 157. U.S. Washington, D.C: Department of Agriculture; 1912. [Google Scholar]
- 18.White G F. European foulbrood. U.S. Department of Agriculture bulletin no. 810. U.S. Washington, D.C: Department of Agriculture; 1920. [Google Scholar]
- 19.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1990. p. 2-4-1. [Google Scholar]
- 20.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]