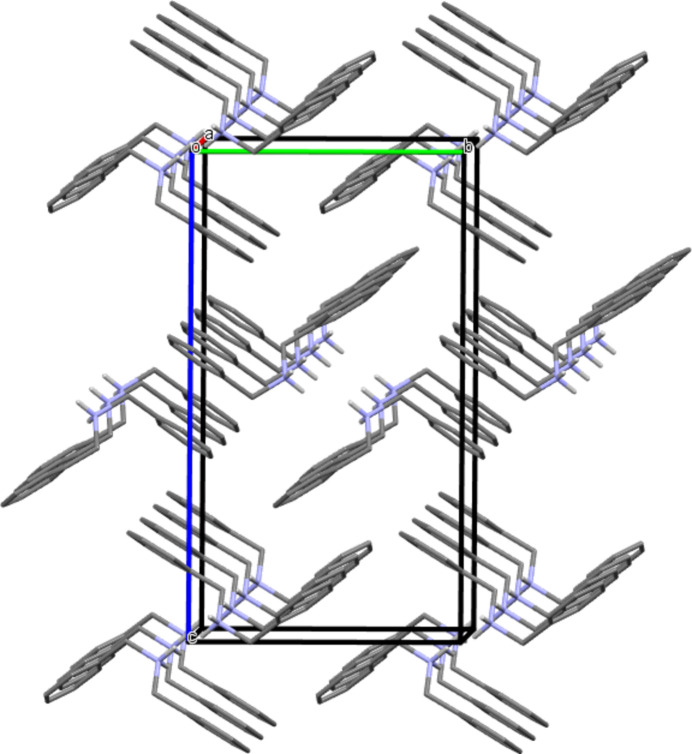
The title salt consists of three components, comprising one dibenzylammonium cation, [(C6H5CH2)2NH2]+, one hydrogen (4-aminophenyl)arsonate anion, [H2NC6H4As(OH)O2]−, and one molecule of water. In the crystal, these components are organized in infinite zigzag chains via intermolecular hydrogen bonds. Weak interactions between the chains lead to a three-dimensional network.
Keywords: crystal structure, group 15 - pnictogen elements, organic salt, phenylarsonic derivatives, hydrogen bonds, infinite chain
Abstract
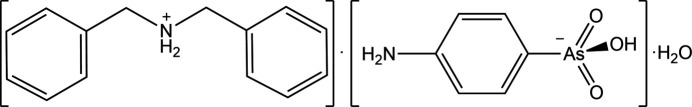
The title salt, C14H16N+·C6H7AsNO3 −·H2O or [(C6H5CH2)2NH2][H2NC6H4As(OH)O2]·H2O, (I), was synthesized by mixing an aqueous solution of (4-aminophenyl)arsonic acid with an ethanolic solution of dibenzylamine at room temperature. Compound I crystallizes in the monoclinic P21/c space group. The three components forming I are linked via N—H⋯O and O—H⋯O intermolecular hydrogen bonds, resulting in the propagation of an infinite zigzag chain. Additional weak interactions between neighbouring chains, such as π–π and N—H⋯O contacts, involving phenyl rings, –NH2 and –As(OH)O3 functions, and H2O, respectively, lead to a three-dimensional network.
1. Chemical context
Organoarsenic compounds have been known for a long time and sparked great interest when they were discovered. Tetramethyldiarsine (Me2As-AsMe2), commonly known as Cacodyl, was isolated in the middle of the 18th century by Cadet de Glaussicourt (Garje & Jain, 1999 ▸). During the next century, in 1859, Antoine Béchamp reported the synthesis of p-arsanilic acid sodium salt (named Atoxyl) by reacting aniline with arsenic acid. This compound was employed for pharmaceutical applications, in particular against trypanosomal infection. Subsequently, in the early 20th century, Paul Ehrlich was inspired by this work to develop a new organoarsenic derivative, called Arsphenamine or Salvarsan (Ehrlich & Bertheim, 1907 ▸). This molecule has proved particularly effective in the treatment of syphilis and sleeping sickness (African Trypanosomiasis) and is considered as being the first chemotherapeutic agent (Williams, 2009 ▸). The use of organoarsenicals as medicines was subsequently abandoned in favour of penicillin, as they were found to be highly toxic to humans, causing significant side effects (including blindness). However, they have continued to be used, until recently, as feed additives and veterinary drugs, particularly in the livestock and poultry breeding industry, but with serious negative effects on the environment. Soil and groundwater contamination resulting from the excessive use of aromatic organoarsenic compounds is now a major environmental concern (Fei et al., 2018 ▸). Current investigations involving academics focus on improving analytical detection (Depalma et al., 2008 ▸; Yang et al., 2018 ▸) and remediation methods (Jun et al., 2015 ▸; Chen et al., 2022 ▸).
From a structural point of view, the crystal structure of phenylarsonic acid was first solved in the early 1960s (refcode ARSACP: Shimada, 1960 ▸). Since then, the X-ray structure for the zwitterionic form of p-arsanilic acid (p-ammoniophenylarsonate) has been determined (CUDSEZ: Shimada, 1961 ▸; CUDSEZ01: Nuttall & Hunter, 1996 ▸) as well as of the hydrated ammonium and sodium salt hydrates of 4-aminophenylarsonic acid (KOKWOY, KOKWUE: Smith & Wermuth, 2014 ▸). We report herein the structure of a new salt of 4-aminophenylarsonate, isolated from a mixture of (4-aminophenyl)arsonic acid and dibenzylamine and characterized as dibenzylammonium hydrogen (4-aminophenyl)arsonate monohydrate, [(C6H5CH2)2NH2][H2NC6H4As(OH)O2]·H2O (I).
2. Structural commentary
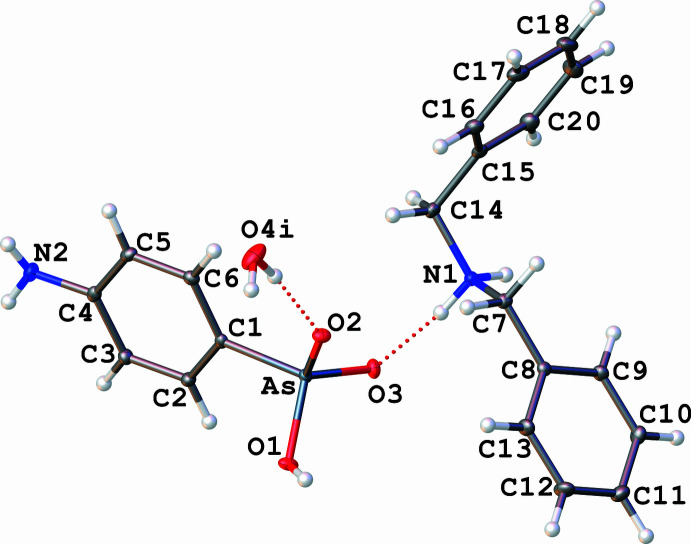
The asymmetric unit of the title salt, which is depicted in Fig. 1 ▸, comprises one dibenzylammonium cation [(C6H5CH2)2NH2]+, one hydrogen (4-aminophenyl)arsonate anion [H2NC6H5As(OH)O2]− and one water molecule of solvation. The three components of I are linked together through intermolecular N—H⋯O and O—H⋯O hydrogen bonds. The As atom of the anion is bonded to three O atoms and one carbon atom of the phenyl ring, describing a slightly distorted tetrahedral geometry [O1—As—C1 = 103.71 (6)°, O2—As—C1 = 110.47 (6)°, O3—As—C1 = 111.73 (6)°, O2—As—O1 = 110.71 (5)°, O3—As—O1 = 108.46 (5)°, O3—As—O2 = 111.48 (5)°]. The As—O bonds exhibit two distinct lengths: As—O1 = 1.7267 (10) Å, and As—O2 = 1.6730 (10) Å and As—O3 =1.6699 (10) Å, which can be considered to be identical. The As—O1 distance is consistent with the presence of a hydroxyl group (Yang et al., 2002 ▸), while the As—O2 and As—O3 distances, which are shorter, reflect rather a double-bond character. In the literature, based on a comparison of structural examples, the average length of the As—O bond is defined as 1.77 Å and that of the As=O bond as 1.67 Å (Nuttall & Hunter, 1996 ▸). The nature of the As=O2 and As=O3 double bonds implies that the negative charge is delocalized on the arsonate. The three oxygen atoms of the arsonate function are engaged in hydrogen bonding, the O1 and O2 atoms being linked head-to-tail [O1—H⋯O2iv, D⋯A = 2.5444 (15) Å; symmetry code: (iv) −x, −y + 1, −z + 1, Table 1 ▸]. The length of the As—C1 bond [1.8955 (13) Å] is within the range of values measured for related compounds such as ammonium 4-nitrophenylarsonate (Yang et al., 2002 ▸) and guanidinium phenylarsonate (Smith & Wermuth, 2010 ▸). An amino group is positioned on the phenyl ring in the para position to the arsonate function. Both functional groups are contained in the plane of the phenyl ring. The negative charge of [H2NC6H4As(OH)O2]− is compensated by the presence of one dibenzylammonium cation, [(C6H5CH2)2NH2]+, whose NH2 + group is hydrogen bonded to the oxygen atom O3 of the arsonate function [N1—H1A⋯O3, D⋯A = 2.6842 (16) Å, N1—H1B⋯O3iii, D⋯A = 2.7260 (15) Å; symmetry code: (iii) −x + 1, −y + 1, −z + 1]. Moreover, the dibenzylammonium cation shows a syn–anti conformation, displaying C—C—N—C torsion angles of 57.65 (16)° and −178.14 (11)°, which are in the range of previous examples of X-ray structures involving [(C6H5CH2)2NH2]+ (Trivedi & Dastidar, 2006 ▸). A water molecule (co-solvent of the reaction) participates in a hydrogen-bond interaction with the oxygen atom O2 of –As(OH)O2 − [O4—H4A⋯O2V, D⋯A = 2.8074 (18) Å; symmetry code: (v) 1 + x, y, z] completes the composition of salt I. From a spectroscopic point of view, the infrared spectrum of I (ATR mode) highlights ν(As—C) and ν(As—O) absorption bands, which are characteristic of the arsonate function (Cowen et al., 2008 ▸), at 1096 cm−1 and between 925–690 cm−1, respectively. The percentages of C, H, N and O determined by elemental analysis support the chemical composition of I, but show that the salt is partially dehydrated (see the Synthesis and crystallization section).
Figure 1.
The molecular structure of I with displacement ellipsoids at the 30% probability level.The water molecule was found to be disordered over two positions, the minor part was omitted and the major part is represented with the following symmetry code: (i): −1 + x, y, z. Dotted lines indicate hydrogen bonds.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2A⋯O4i | 0.84 (2) | 2.37 (2) | 3.165 (2) | 158.0 (18) |
| N2—H2B⋯O1ii | 0.83 (2) | 2.25 (2) | 3.0769 (17) | 175.6 (18) |
| N1—H1A⋯O3 | 0.91 | 1.78 | 2.6842 (16) | 172 |
| N1—H1B⋯O3iii | 0.91 | 1.89 | 2.7260 (15) | 151 |
| O1—H1⋯O2iv | 0.83 (3) | 1.73 (3) | 2.5445 (15) | 170 (3) |
| O4—H4A⋯O2v | 0.87 | 1.95 | 2.8074 (18) | 169 |
Symmetry codes: (i)
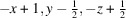 ; (ii)
; (ii)
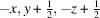 ; (iii)
; (iii)
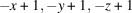 ; (iv)
; (iv)
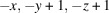 ; (v)
; (v)
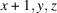 .
.
3. Supramolecular features
At the supramolecular stage, two levels of organization can be observed in the crystal structure of I:
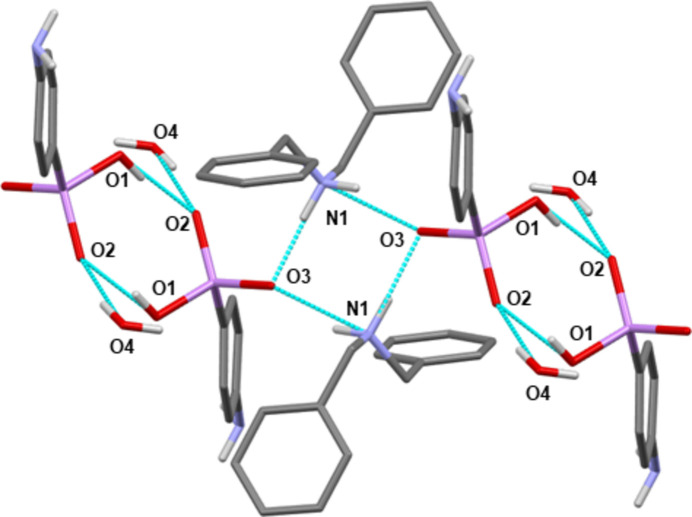
(i) The propagation of one-dimensional zigzag chains along the a-axis direction resulting from the hydrogen-bonding interactions (Fig. 2 ▸). The NH2 groups of two dibenzylammonium cations are involved in two independent hydrogen bonds, oriented perpendicularly [O3⋯N1⋯O3 = 92.63 (5)°], with the oxygen atoms O3 of two arsonate moieties [N1—H1A⋯O3 and N1—H1B⋯O3iii, Table 1 ▸]. This leads to the formation of a tetrameric unit describing a four-membered ring (Fig. 3 ▸). These units are linked together by two additional and parallel hydrogen bonds involving two hydrogen (4-aminophenyl)arsonate anions [O1—H1⋯O2iv, Table 1 ▸]. This creates a six-membered ring. In addition, the water molecule contained in I is also in hydrogen-bonding interaction with the oxygen atom O2 of the arsonate group [O4—H4A⋯O2v, Table 1 ▸]. The 4-aminophenyl groups can be viewed as perpendicular to the chain axis and positioned alternately on either side of it.
Figure 2.
Mercury representation (Macrae et al., 2020 ▸; colour code: C = grey, N = blue, O = red, As = pink, H = white] of the infinite chain structure of I propagating along the a-axis direction via hydrogen bonds (dotted cyan lines).
Figure 3.
Mercury representation (Macrae et al., 2020 ▸; colour code: C = grey, N = blue, O = red, As = pink, H = white) highlighting the hydrogen-bonding network (cyan dotted lines) involving the components of I (the benzyl H atoms have been omitted for clarity).
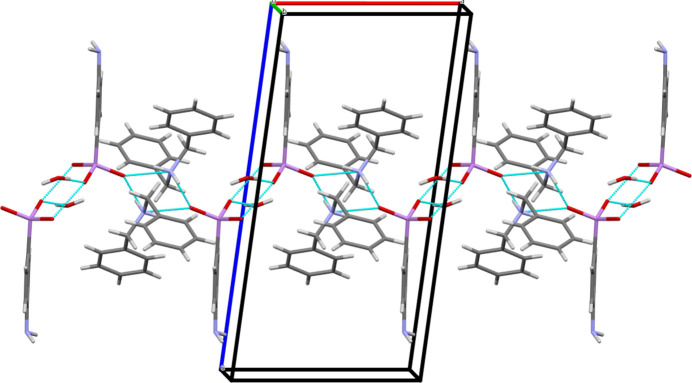
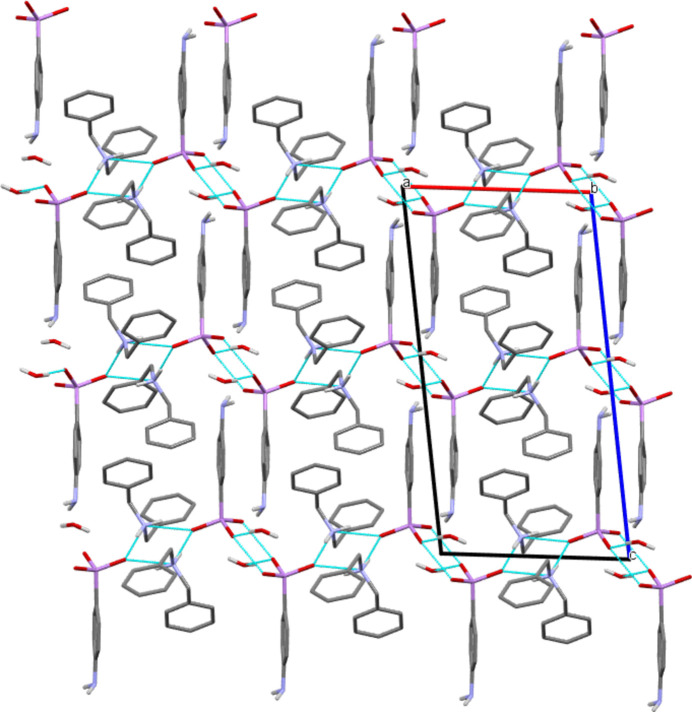
(ii) The association of chains leading to a three-dimensional network and resulting from a combination of weak interactions (Fig. 4 ▸). Two types of π–π stacking interactions involving the phenyl rings of the dibenzylammonium cations can be described (Fig. 5 ▸): (a) centroid(C15–C20)–centroid (C15i–C20i) = 3.9384 (10) Å, interplanar distance = 3.4310 (18) Å, slip angle (angle between the normal to the plane and the centroid–centroid vector) = 29.4, corresponding to a slippage distance of 1.933 Å; symmetry code: (i) 1 − x, 2 − y, 1 − z; (b) centroid(C8–C13)–centroid(C15ii–C20ii) = 4.0178 (10) Å, interplanar distance = 3.5093 (6) Å, slip angle = 29.1°, corresponding to a slippage distance of 1.957 Å; symmetry code: (ii) 1 − x, −
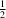 + y,
+ y,
 − z. In addition, the NH2 groups located in the para position of C6H4As(OH)O2, interact via hydrogen bonding with a water molecule [N2—H2A⋯O41 = 3.165 (2) Å] and the O1 oxygen atom of an adjacent –As(OH)O2 function [N2—H2B⋯O1ii = 3.0769 (17) Å] (symmetry codes as in Table 1 ▸).
− z. In addition, the NH2 groups located in the para position of C6H4As(OH)O2, interact via hydrogen bonding with a water molecule [N2—H2A⋯O41 = 3.165 (2) Å] and the O1 oxygen atom of an adjacent –As(OH)O2 function [N2—H2B⋯O1ii = 3.0769 (17) Å] (symmetry codes as in Table 1 ▸).
Figure 4.
Arrangement of the chains in the crystal of I and along the b-axis, leading to a three-dimensional network (Mercury representation; Macrae et al., 2020 ▸; colour code: C = grey, N = blue, O = red, As = pink, H = white). H atoms of phenyl and benzyl groups are omitted for clarity. The hydrogen bonds propagating the infinite chains are represented by dotted cyan lines.
Figure 5.
View of the π–π stacking interactions between phenyl rings of the dibenzylammonium cations of I [along the a-axis, Mercury representation (Macrae et al., 2020 ▸); colour code: C = grey, N = blue, H = white). H atoms of phenyl rings, anions and water molecules have been omitted for clarity.
4. Database survey
A search of the Cambridge Structural Database (WebCSD update 11/2022; Groom et al., 2016 ▸), revealed that, to date, there are relatively few X-ray structures exhibiting the isolated hydrogen phenylarsonate moiety, C6H5As(OH)O2 −. To our knowledge, eleven examples including this fragment have already been identified: ammonium 4-nitrophenylarsonate (AHILAE: Yang et al., 2002 ▸), guanidinium phenylarsonate guanidine dihydrate (DUSCIE: Smith & Wermuth, 2010 ▸), p-aminophenylarsonic acid (CUDSEZ: Shimada, 1961 ▸; CUDSEZ01: Nuttall & Hunter, 1996 ▸), ammonium hydrogen (4-aminophenyl)arsonate monohydrate (KOKWOY: Smith & Wermuth, 2014 ▸), 1-(4-hydroxy-2-methylphenyl)-2,4,6-triphenylpyridinium hydrogen o-arsanilate monohydrate (PAZRIS: Wojtas et al., 2006 ▸), tetrabutylammonium hydrogen phenylarsonate–phenylarsonic acid (QECBEH: Reck & Schmitt, 2012 ▸), 3-ammonio-4-hydroxyphenylarsonate (ROBDAO: Lloyd et al., 2008 ▸), hexaaquamanganese(II) bis[hydrogen (4-aminophenyl)arsonate] tetrahydrate (UBURIV: Smith & Wermuth, 2016a ▸), hexaaqua-magnesium bis(hydrogen (4-aminophenyl)arsonate) tetrahydrate (UDAPIB: Smith & Wermuth, 2017a ▸), 2,3-dimethoxy-10-oxostrychnidin-19-ium hydrogen (4-aminophenyl)arsonate tetrahydrate (ULIROY: Smith & Wermuth, 2016b ▸), 2,4-diamino-5-(3,4,5-trimethoxybenzyl)pyrimidinium 4-hydroxy-3-nitrophenylarsonate monohydrate (XEMZIZ: Pan et al., 2006 ▸). In coordination chemistry, phenylarsonic acid and its derivatives constitute also suitable ligands to generate coordination polymers and heteropolyoxometalates in the presence of transition metals (Lesikar-Parrish et al., 2013 ▸), main-group metals (Xie et al., 2008 ▸), alkali metals (Smith & Wermuth, 2017a ▸) and alkali-earth metal precursors (Smith & Wermuth, 2017b ▸). Regarding the dibenzylammonium cation, [(C6H5CH2)2NH2]+, 117 hits incorporating such an entity were found in the Cambridge Structural Database.
5. Synthesis and crystallization
All chemicals were purchased from Sigma-Aldrich (Germany) and used without any further purification. (4-Aminophenyl)arsonic acid [H2NC6H4As(OH)2O] was prepared according to a previous work (Lewis & Cheetham, 1923 ▸), by reacting aniline (C6H5NH2) and arsenic acid (As(OH)3O). The title salt was obtained by neutralization of an aqueous solution (20 mL) of (4-aminophenyl)arsonic acid (2.15 g, 9.90 mmol) with dibenzylamine ((C6H5CH2)2NH) (3.90 g, 19.80 mmol) dissolved in 20 mL of ethanol. The mixture was stirred for about two h at room temperature (301 K). After three days of slow solvent evaporation, colourless prism-shaped crystals of [(C6H5CH2)2NH2][H2NC6H4As(OH)O2]·H2O (5.25 g, 64% yield), suitable for an X-ray crystallographic analysis, were collected from the solvent (m.p. 393 K). FT–IR (ATR, Bruker Alpha FTIR spectrometer, cm−1): 3447, 3304, 3187, 1595, 1501, 1454, 1096, 923, 878, 825,752, 735, 695. Elemental analysis (Elemental Analyser, ThermoFisher FlashSmart CHNS/O) – analysis calculated for C20H23N2O3As·0.25H2O (418.83), salt I partially dehydrated: C, 57.35; H, 5.66; N, 6.69; O, 12.41; found: C, 57.82; H, 5.61; N, 6.62; O, 12.37%.
6. Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The asymmetric unit contains the dibenzylammonium hydrogen (4-aminophenyl)arsonate monohydrate. The water molecule was found disordered over two main positions with occupancy factors that converged to 0.94:0.06. Hence, the minor part of the water molecule was refined only isotropically and without the hydrogen atoms. The hydrogen atoms for the major component of the water molecule were refined geometrically as a rigid group (O—H = 0.87 Å) with U iso(H) = 1.5U eq(O). C-bound hydrogen atoms were placed at calculated positions [C—H = 0.95 Å (aromatic) or 0.99 Å (methylene group)] and H atoms of the NH2 and OH terminal groups were placed geometrically (N—H = 0.83–0.84 Å, O—H = 0.83 Å) and refined as riding with U iso(H) = 1.2U eq(N, C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C14H16N+·C6H7AsNO3 −·H2O |
| M r | 432.34 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 9.8242 (5), 10.6574 (6), 19.2507 (11) |
| β (°) | 97.7500 (18) |
| V (Å3) | 1997.15 (19) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.73 |
| Crystal size (mm) | 0.5 × 0.25 × 0.18 |
| Data collection | |
| Diffractometer | Bruker D8 VENTURE |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.610, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 67932, 4584, 4119 |
| R int | 0.037 |
| (sin θ/λ)max (Å−1) | 0.650 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.022, 0.054, 1.07 |
| No. of reflections | 4584 |
| No. of parameters | 261 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.42, −0.21 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S205698902300837X/dj2065sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902300837X/dj2065Isup2.hkl
Supporting information file. DOI: 10.1107/S205698902300837X/dj2065Isup3.cml
CCDC reference: 2297206
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful for general and financial support from the University Cheikh Anta Diop-Dakar (Senegal), the University of Bourgogne-Dijon (France) and the Centre National de la Recherche Scientifique (CNRS-France). They would like to thank in particular Ms T. Régnier for elemental analysis measurements.
supplementary crystallographic information
Crystal data
| C14H16N+·C6H7AsNO3−·H2O | F(000) = 896 |
| Mr = 432.34 | Dx = 1.438 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.8242 (5) Å | Cell parameters from 9824 reflections |
| b = 10.6574 (6) Å | θ = 2.8–27.5° |
| c = 19.2507 (11) Å | µ = 1.73 mm−1 |
| β = 97.7500 (18)° | T = 100 K |
| V = 1997.15 (19) Å3 | Prism, clear light colourless |
| Z = 4 | 0.5 × 0.25 × 0.18 mm |
Data collection
| Bruker D8 VENTURE diffractometer | 4584 independent reflections |
| Radiation source: X-ray tube, Siemens KFF Mo 2K-90C | 4119 reflections with I > 2σ(I) |
| TRIUMPH curved crystal monochromator | Rint = 0.037 |
| Detector resolution: 1024 x 1024 pixels mm-1 | θmax = 27.5°, θmin = 2.8° |
| φ and ω scans | h = −12→12 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | k = −13→13 |
| Tmin = 0.610, Tmax = 0.746 | l = −24→25 |
| 67932 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.022 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.054 | w = 1/[σ2(Fo2) + (0.0228P)2 + 1.3431P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max = 0.001 |
| 4584 reflections | Δρmax = 0.42 e Å−3 |
| 261 parameters | Δρmin = −0.21 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| As | 0.15165 (2) | 0.51188 (2) | 0.42563 (2) | 0.01186 (5) | |
| O1 | 0.04372 (11) | 0.38487 (9) | 0.42927 (6) | 0.0184 (2) | |
| O2 | 0.09995 (10) | 0.63317 (9) | 0.47065 (5) | 0.0167 (2) | |
| O3 | 0.31128 (10) | 0.46779 (10) | 0.45696 (5) | 0.0176 (2) | |
| N2 | 0.08041 (14) | 0.62660 (14) | 0.11336 (7) | 0.0208 (3) | |
| H2A | 0.062 (2) | 0.565 (2) | 0.0867 (10) | 0.025* | |
| H2B | 0.043 (2) | 0.694 (2) | 0.1009 (10) | 0.025* | |
| C1 | 0.13469 (13) | 0.55046 (13) | 0.32872 (7) | 0.0126 (3) | |
| C2 | 0.13667 (14) | 0.45347 (13) | 0.27999 (7) | 0.0152 (3) | |
| H2 | 0.149937 | 0.369445 | 0.295952 | 0.018* | |
| C3 | 0.11956 (15) | 0.47831 (14) | 0.20880 (7) | 0.0164 (3) | |
| H3 | 0.121018 | 0.411363 | 0.176326 | 0.020* | |
| C4 | 0.10001 (14) | 0.60216 (14) | 0.18428 (7) | 0.0150 (3) | |
| C5 | 0.10337 (14) | 0.69966 (13) | 0.23332 (8) | 0.0166 (3) | |
| H5 | 0.094757 | 0.784146 | 0.217604 | 0.020* | |
| C6 | 0.11920 (14) | 0.67391 (13) | 0.30476 (7) | 0.0150 (3) | |
| H6 | 0.119468 | 0.740721 | 0.337450 | 0.018* | |
| O4 | 0.94231 (17) | 0.85325 (13) | 0.45149 (7) | 0.0389 (3) | 0.94 |
| H4A | 0.999317 | 0.790435 | 0.455138 | 0.058* | 0.94 |
| H4B | 0.870836 | 0.825432 | 0.468917 | 0.058* | 0.94 |
| N1 | 0.43532 (12) | 0.63057 (11) | 0.55144 (6) | 0.0142 (2) | |
| H1A | 0.391629 | 0.570678 | 0.522995 | 0.017* | |
| H1B | 0.523283 | 0.604491 | 0.564385 | 0.017* | |
| C7 | 0.36565 (15) | 0.64208 (13) | 0.61550 (7) | 0.0156 (3) | |
| H7A | 0.268703 | 0.667011 | 0.601690 | 0.019* | |
| H7B | 0.411155 | 0.708549 | 0.646221 | 0.019* | |
| C8 | 0.37054 (14) | 0.52036 (13) | 0.65533 (7) | 0.0134 (3) | |
| C9 | 0.49603 (15) | 0.46656 (14) | 0.68231 (8) | 0.0167 (3) | |
| H9 | 0.579319 | 0.506225 | 0.674660 | 0.020* | |
| C10 | 0.50021 (15) | 0.35571 (14) | 0.72019 (8) | 0.0194 (3) | |
| H10 | 0.586162 | 0.319539 | 0.738161 | 0.023* | |
| C11 | 0.37894 (16) | 0.29743 (15) | 0.73190 (8) | 0.0227 (3) | |
| H11 | 0.381734 | 0.221469 | 0.757878 | 0.027* | |
| C12 | 0.25392 (16) | 0.35050 (15) | 0.70557 (9) | 0.0251 (3) | |
| H12 | 0.170863 | 0.311054 | 0.713808 | 0.030* | |
| C13 | 0.24933 (15) | 0.46135 (15) | 0.66711 (8) | 0.0200 (3) | |
| H13 | 0.163208 | 0.496855 | 0.648829 | 0.024* | |
| C14 | 0.43754 (16) | 0.75047 (14) | 0.51084 (8) | 0.0194 (3) | |
| H14A | 0.342130 | 0.775473 | 0.493084 | 0.023* | |
| H14B | 0.486851 | 0.736507 | 0.469927 | 0.023* | |
| C15 | 0.50659 (15) | 0.85489 (14) | 0.55510 (8) | 0.0182 (3) | |
| C16 | 0.42794 (16) | 0.94873 (15) | 0.57986 (8) | 0.0220 (3) | |
| H16 | 0.330863 | 0.947490 | 0.568182 | 0.026* | |
| C17 | 0.49027 (19) | 1.04468 (15) | 0.62169 (9) | 0.0277 (4) | |
| H17 | 0.435865 | 1.108628 | 0.638562 | 0.033* | |
| C18 | 0.6313 (2) | 1.04676 (17) | 0.63861 (9) | 0.0322 (4) | |
| H18 | 0.673979 | 1.112232 | 0.667157 | 0.039* | |
| C19 | 0.71081 (18) | 0.95354 (18) | 0.61403 (10) | 0.0323 (4) | |
| H19 | 0.807876 | 0.955423 | 0.625699 | 0.039* | |
| C20 | 0.64919 (16) | 0.85748 (16) | 0.57247 (9) | 0.0249 (3) | |
| H20 | 0.703946 | 0.793541 | 0.555854 | 0.030* | |
| H1 | 0.002 (3) | 0.386 (3) | 0.4638 (14) | 0.062 (8)* | |
| O4B | 0.729 (2) | 0.7346 (18) | 0.4434 (10) | 0.026 (4)* | 0.06 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| As | 0.01149 (7) | 0.01330 (7) | 0.01092 (7) | −0.00011 (5) | 0.00195 (5) | −0.00127 (5) |
| O1 | 0.0235 (5) | 0.0140 (5) | 0.0192 (5) | −0.0066 (4) | 0.0084 (4) | −0.0043 (4) |
| O2 | 0.0200 (5) | 0.0125 (5) | 0.0187 (5) | −0.0005 (4) | 0.0070 (4) | −0.0030 (4) |
| O3 | 0.0143 (5) | 0.0226 (5) | 0.0150 (5) | 0.0041 (4) | −0.0009 (4) | −0.0025 (4) |
| N2 | 0.0233 (7) | 0.0235 (7) | 0.0151 (6) | 0.0019 (6) | 0.0010 (5) | 0.0037 (5) |
| C1 | 0.0096 (6) | 0.0163 (6) | 0.0119 (6) | −0.0005 (5) | 0.0018 (5) | 0.0004 (5) |
| C2 | 0.0157 (7) | 0.0132 (6) | 0.0164 (7) | 0.0004 (5) | 0.0011 (5) | 0.0009 (5) |
| C3 | 0.0179 (7) | 0.0164 (7) | 0.0147 (7) | 0.0000 (5) | 0.0013 (5) | −0.0022 (5) |
| C4 | 0.0097 (6) | 0.0198 (7) | 0.0158 (7) | 0.0003 (5) | 0.0026 (5) | 0.0029 (5) |
| C5 | 0.0149 (7) | 0.0140 (6) | 0.0211 (7) | 0.0016 (5) | 0.0035 (5) | 0.0042 (5) |
| C6 | 0.0127 (6) | 0.0147 (6) | 0.0179 (7) | −0.0005 (5) | 0.0035 (5) | −0.0018 (5) |
| O4 | 0.0569 (10) | 0.0308 (7) | 0.0304 (7) | 0.0169 (7) | 0.0104 (7) | 0.0108 (6) |
| N1 | 0.0149 (6) | 0.0140 (6) | 0.0132 (6) | −0.0003 (4) | 0.0007 (4) | 0.0000 (4) |
| C7 | 0.0170 (7) | 0.0144 (6) | 0.0160 (7) | 0.0009 (5) | 0.0045 (5) | −0.0008 (5) |
| C8 | 0.0143 (6) | 0.0137 (6) | 0.0123 (6) | −0.0003 (5) | 0.0023 (5) | −0.0020 (5) |
| C9 | 0.0129 (6) | 0.0194 (7) | 0.0181 (7) | −0.0015 (5) | 0.0031 (5) | 0.0003 (6) |
| C10 | 0.0160 (7) | 0.0223 (7) | 0.0198 (7) | 0.0047 (6) | 0.0016 (6) | 0.0031 (6) |
| C11 | 0.0253 (8) | 0.0182 (7) | 0.0247 (8) | 0.0005 (6) | 0.0043 (6) | 0.0059 (6) |
| C12 | 0.0172 (7) | 0.0229 (8) | 0.0354 (9) | −0.0053 (6) | 0.0044 (7) | 0.0068 (7) |
| C13 | 0.0119 (7) | 0.0214 (7) | 0.0260 (8) | −0.0005 (5) | −0.0001 (6) | 0.0028 (6) |
| C14 | 0.0230 (8) | 0.0187 (7) | 0.0161 (7) | −0.0018 (6) | 0.0010 (6) | 0.0048 (6) |
| C15 | 0.0204 (7) | 0.0173 (7) | 0.0169 (7) | −0.0042 (6) | 0.0022 (6) | 0.0057 (6) |
| C16 | 0.0226 (8) | 0.0191 (7) | 0.0246 (8) | −0.0032 (6) | 0.0050 (6) | 0.0045 (6) |
| C17 | 0.0409 (10) | 0.0180 (7) | 0.0256 (8) | −0.0041 (7) | 0.0095 (7) | 0.0018 (6) |
| C18 | 0.0434 (10) | 0.0244 (8) | 0.0276 (9) | −0.0172 (8) | 0.0004 (8) | 0.0012 (7) |
| C19 | 0.0237 (8) | 0.0356 (10) | 0.0358 (10) | −0.0118 (7) | −0.0023 (7) | 0.0051 (8) |
| C20 | 0.0207 (8) | 0.0257 (8) | 0.0283 (8) | −0.0022 (6) | 0.0037 (6) | 0.0044 (7) |
Geometric parameters (Å, º)
| As—O1 | 1.7267 (10) | C7—C8 | 1.5044 (19) |
| As—O2 | 1.6730 (10) | C8—C9 | 1.395 (2) |
| As—O3 | 1.6699 (10) | C8—C13 | 1.392 (2) |
| As—C1 | 1.8955 (13) | C9—H9 | 0.9500 |
| O1—H1 | 0.83 (3) | C9—C10 | 1.386 (2) |
| N2—H2A | 0.84 (2) | C10—H10 | 0.9500 |
| N2—H2B | 0.83 (2) | C10—C11 | 1.389 (2) |
| N2—C4 | 1.3776 (19) | C11—H11 | 0.9500 |
| C1—C2 | 1.3978 (19) | C11—C12 | 1.385 (2) |
| C1—C6 | 1.3957 (19) | C12—H12 | 0.9500 |
| C2—H2 | 0.9500 | C12—C13 | 1.392 (2) |
| C2—C3 | 1.384 (2) | C13—H13 | 0.9500 |
| C3—H3 | 0.9500 | C14—H14A | 0.9900 |
| C3—C4 | 1.406 (2) | C14—H14B | 0.9900 |
| C4—C5 | 1.401 (2) | C14—C15 | 1.506 (2) |
| C5—H5 | 0.9500 | C15—C16 | 1.387 (2) |
| C5—C6 | 1.391 (2) | C15—C20 | 1.396 (2) |
| C6—H6 | 0.9500 | C16—H16 | 0.9500 |
| O4—H4A | 0.8696 | C16—C17 | 1.391 (2) |
| O4—H4B | 0.8701 | C17—H17 | 0.9500 |
| N1—H1A | 0.9100 | C17—C18 | 1.380 (3) |
| N1—H1B | 0.9100 | C18—H18 | 0.9500 |
| N1—C7 | 1.4939 (17) | C18—C19 | 1.386 (3) |
| N1—C14 | 1.4995 (18) | C19—H19 | 0.9500 |
| C7—H7A | 0.9900 | C19—C20 | 1.387 (2) |
| C7—H7B | 0.9900 | C20—H20 | 0.9500 |
| O1—As—C1 | 103.71 (6) | C13—C8—C7 | 120.22 (13) |
| O2—As—O1 | 110.71 (5) | C13—C8—C9 | 119.09 (13) |
| O2—As—C1 | 110.47 (6) | C8—C9—H9 | 119.7 |
| O3—As—O1 | 108.46 (5) | C10—C9—C8 | 120.55 (13) |
| O3—As—O2 | 111.48 (5) | C10—C9—H9 | 119.7 |
| O3—As—C1 | 111.73 (5) | C9—C10—H10 | 120.0 |
| As—O1—H1 | 113.0 (19) | C9—C10—C11 | 120.09 (14) |
| H2A—N2—H2B | 116.7 (18) | C11—C10—H10 | 120.0 |
| C4—N2—H2A | 116.7 (13) | C10—C11—H11 | 120.1 |
| C4—N2—H2B | 116.7 (13) | C12—C11—C10 | 119.73 (14) |
| C2—C1—As | 119.52 (10) | C12—C11—H11 | 120.1 |
| C6—C1—As | 121.39 (10) | C11—C12—H12 | 119.8 |
| C6—C1—C2 | 119.08 (13) | C11—C12—C13 | 120.33 (14) |
| C1—C2—H2 | 119.6 | C13—C12—H12 | 119.8 |
| C3—C2—C1 | 120.83 (13) | C8—C13—H13 | 119.9 |
| C3—C2—H2 | 119.6 | C12—C13—C8 | 120.20 (14) |
| C2—C3—H3 | 119.8 | C12—C13—H13 | 119.9 |
| C2—C3—C4 | 120.33 (13) | N1—C14—H14A | 109.2 |
| C4—C3—H3 | 119.8 | N1—C14—H14B | 109.2 |
| N2—C4—C3 | 120.31 (13) | N1—C14—C15 | 111.83 (12) |
| N2—C4—C5 | 120.99 (13) | H14A—C14—H14B | 107.9 |
| C5—C4—C3 | 118.69 (13) | C15—C14—H14A | 109.2 |
| C4—C5—H5 | 119.7 | C15—C14—H14B | 109.2 |
| C6—C5—C4 | 120.62 (13) | C16—C15—C14 | 119.86 (14) |
| C6—C5—H5 | 119.7 | C16—C15—C20 | 119.40 (14) |
| C1—C6—H6 | 119.8 | C20—C15—C14 | 120.73 (14) |
| C5—C6—C1 | 120.37 (13) | C15—C16—H16 | 119.8 |
| C5—C6—H6 | 119.8 | C15—C16—C17 | 120.45 (15) |
| H4A—O4—H4B | 104.5 | C17—C16—H16 | 119.8 |
| H1A—N1—H1B | 107.7 | C16—C17—H17 | 120.1 |
| C7—N1—H1A | 108.8 | C18—C17—C16 | 119.86 (16) |
| C7—N1—H1B | 108.8 | C18—C17—H17 | 120.1 |
| C7—N1—C14 | 113.62 (11) | C17—C18—H18 | 119.9 |
| C14—N1—H1A | 108.8 | C17—C18—C19 | 120.13 (16) |
| C14—N1—H1B | 108.8 | C19—C18—H18 | 119.9 |
| N1—C7—H7A | 109.4 | C18—C19—H19 | 119.9 |
| N1—C7—H7B | 109.4 | C18—C19—C20 | 120.25 (16) |
| N1—C7—C8 | 111.30 (11) | C20—C19—H19 | 119.9 |
| H7A—C7—H7B | 108.0 | C15—C20—H20 | 120.0 |
| C8—C7—H7A | 109.4 | C19—C20—C15 | 119.91 (16) |
| C8—C7—H7B | 109.4 | C19—C20—H20 | 120.0 |
| C9—C8—C7 | 120.67 (13) | ||
| As—C1—C2—C3 | 177.66 (11) | N1—C14—C15—C20 | 74.49 (17) |
| As—C1—C6—C5 | −178.33 (10) | C7—N1—C14—C15 | 57.65 (16) |
| O1—As—C1—C2 | −44.30 (12) | C7—C8—C9—C10 | 178.70 (13) |
| O1—As—C1—C6 | 135.23 (11) | C7—C8—C13—C12 | −178.27 (14) |
| O2—As—C1—C2 | −162.96 (10) | C8—C9—C10—C11 | −0.3 (2) |
| O2—As—C1—C6 | 16.57 (13) | C9—C8—C13—C12 | 0.3 (2) |
| O3—As—C1—C2 | 72.32 (12) | C9—C10—C11—C12 | 0.0 (2) |
| O3—As—C1—C6 | −108.15 (11) | C10—C11—C12—C13 | 0.4 (3) |
| N2—C4—C5—C6 | 178.11 (13) | C11—C12—C13—C8 | −0.6 (2) |
| C1—C2—C3—C4 | 0.1 (2) | C13—C8—C9—C10 | 0.2 (2) |
| C2—C1—C6—C5 | 1.2 (2) | C14—N1—C7—C8 | −178.14 (11) |
| C2—C3—C4—N2 | −178.78 (13) | C14—C15—C16—C17 | 179.01 (14) |
| C2—C3—C4—C5 | 2.3 (2) | C14—C15—C20—C19 | −179.17 (15) |
| C3—C4—C5—C6 | −3.0 (2) | C15—C16—C17—C18 | 0.1 (2) |
| C4—C5—C6—C1 | 1.3 (2) | C16—C15—C20—C19 | −0.1 (2) |
| C6—C1—C2—C3 | −1.9 (2) | C16—C17—C18—C19 | 0.0 (3) |
| N1—C7—C8—C9 | 60.72 (17) | C17—C18—C19—C20 | −0.2 (3) |
| N1—C7—C8—C13 | −120.76 (14) | C18—C19—C20—C15 | 0.2 (3) |
| N1—C14—C15—C16 | −104.55 (16) | C20—C15—C16—C17 | 0.0 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2A···O4i | 0.84 (2) | 2.37 (2) | 3.165 (2) | 158.0 (18) |
| N2—H2B···O1ii | 0.83 (2) | 2.25 (2) | 3.0769 (17) | 175.6 (18) |
| N1—H1A···O3 | 0.91 | 1.78 | 2.6842 (16) | 172 |
| N1—H1B···O3iii | 0.91 | 1.89 | 2.7260 (15) | 151 |
| O1—H1···O2iv | 0.83 (3) | 1.73 (3) | 2.5445 (15) | 170 (3) |
| O4—H4A···O2v | 0.87 | 1.95 | 2.8074 (18) | 169 |
Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) −x, y+1/2, −z+1/2; (iii) −x+1, −y+1, −z+1; (iv) −x, −y+1, −z+1; (v) x+1, y, z.
References
- Bruker (2013). SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2014). APEX2. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, H., Liu, W., Cheng, L., Meledina, M., Meledin, A., Van Deun, R., Leus, K. & Van Der Voort, P. (2022). Chem. Eng. J. 429, 132162.
- Cowen, S., Duggal, M., Hoang, T. & Al-Abadleh, H. A. (2008). Can. J. Chem. 86, 942–950.
- Depalma, S. S. C. O. T. T., Cowen, S., Hoang, T. & Al-Abadleh, H. A. (2008). Environ. Sci. Technol. 42, 1922–1927. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Ehrlich, P. & Bertheim, A. (1907). Ber. Dtsch. Chem. Ges. 40, 3292–3297.
- Fei, J., Wang, T., Zhou, Y., Wang, Z., Min, X., Ke, Y., Hu, W. & Chai, L. (2018). Chemosphere, 207, 665–675. [DOI] [PubMed]
- Garje, S. S. & Jain, V. K. (1999). Main Group Met. Chem. 22, 45–58.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jun, J. W., Tong, M., Jung, B. K., Hasan, Z., Zhong, C. & Jhung, S. H. (2015). Chem. Eur. J. 21, 347–354. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Lesikar-Parrish, L. A., Neilson, R. H. & Richards, A. F. (2013). J. Solid State Chem. 198, 424–432.
- Lewis, W. L. & Cheetham, H. C. (1923). Org. Synth. 3, 13–16.
- Lloyd, N. C., Morgan, H. W., Nicholson, B. K. & Ronimus, R. S. (2008). J. Organomet. Chem. 693, 2443–2450.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Nuttall, R. H. & Hunter, W. N. (1996). Acta Cryst. C52, 1681–1683.
- Pan, T.-T., Liu, B.-X. & Xu, D.-J. (2006). Acta Cryst. E62, m2198–m2199.
- Reck, L. & Schmitt, W. (2012). Acta Cryst. E68, m1212–m1213. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shimada, A. (1960). Bull. Chem. Soc. Jpn, 33, 301–304.
- Shimada, A. (1961). Bull. Chem. Soc. Jpn, 34, 639–643.
- Smith, G. & Wermuth, U. D. (2010). Acta Cryst. E66, o1893–o1894. [DOI] [PMC free article] [PubMed]
- Smith, G. & Wermuth, U. D. (2014). Acta Cryst. C70, 738–741. [DOI] [PubMed]
- Smith, G. & Wermuth, U. D. (2016a). IUCrData, 1, x161985.
- Smith, G. & Wermuth, U. D. (2016b). Acta Cryst. E72, 751–755. [DOI] [PMC free article] [PubMed]
- Smith, G. & Wermuth, U. D. (2017a). Acta Cryst. E73, 203–208. [DOI] [PMC free article] [PubMed]
- Smith, G. & Wermuth, U. D. (2017b). Acta Cryst. C73, 61–67. [DOI] [PubMed]
- Trivedi, D. R. & Dastidar, P. (2006). Cryst. Growth Des. 6, 2115-2121.
- Williams, K. J. (2009). J. R. Soc. Med. 102, 343–348. [DOI] [PMC free article] [PubMed]
- Wojtas, Ł., Milart, P. & Stadnicka, K. (2006). J. Mol. Struct. 782, 157–164.
- Xie, Y.-P., Yang, J., Ma, J.-F., Zhang, L.-P., Song, S.-Y. & Su, Z.-M. (2008). Chem. Eur. J. 14, 4093–4103. [DOI] [PubMed]
- Yang, J., Ma, J.-F., Liu, Y.-C., Zheng, G.-L., Li, L., Liu, J.-F., Hu, N.-H. & Jia, H.-Q. (2002). Acta Cryst. C58, m613–m614. [DOI] [PubMed]
- Yang, T., Wang, L., Liu, Y., Jiang, J., Huang, Z., Pang, S.-Y., Cheng, H., Gao, D. & Ma, J. (2018). Environ. Sci. Technol. 52, 13325–13335. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S205698902300837X/dj2065sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902300837X/dj2065Isup2.hkl
Supporting information file. DOI: 10.1107/S205698902300837X/dj2065Isup3.cml
CCDC reference: 2297206
Additional supporting information: crystallographic information; 3D view; checkCIF report