Abstract
A collection of pediocin AcH amino acid substitution mutants was generated by PCR random mutagenesis of DNA encoding the bacteriocin. Mutants were isolated by cloning mutagenized DNA into an Escherichia coli malE plasmid that directs the secretion of maltose binding protein-pediocin AcH chimeric proteins and by screening transformant colonies for bactericidal activity against Lactobacillus plantarum NCDO955 (K. W. Miller, R. Schamber, Y. Chen, and B. Ray, 1998. Appl. Environ. Microbiol. 64:14–20, 1998). In all, 17 substitution mutants were isolated at 14 of the 44 amino acids of pediocin AcH. Seven mutants (N5K, C9R, C14S, C14Y, G37E, G37R, and C44W) were completely inactive against the pediocin AcH-sensitive strains L. plantarum NCDO955, Listeria innocua Lin11, Enterococcus faecalis M1, Pediococcus acidilactici LB42, and Leuconostoc mesenteroides Ly. A C24S substitution mutant constructed by other means also was inactive against these bacteria. Nine other mutants (K1N, W18R, I26T, M31T, A34D, N41K, H42L, K43N, and K43E) retained from <1% to ∼60% of wild-type activity when assayed against L. innocua Lin11. One mutant, K11E, displayed ∼2.8-fold-higher activity against this indicator. About one half of the mutations mapped to amino acids that are conserved in the pediocin-like family of bacteriocins. All four cysteines were found to be required for activity, although only C9 and C14 are conserved among pediocin-like bacteriocins. Several basic amino acids as well as nonpolar amino acids located within the hydrophobic C-terminal region also were found to be important. The mutations are discussed in the context of structural models that have been proposed for the bacteriocin.
Pediocin AcH (same sequence as pediocin PA-1) is a ribosomally synthesized bacteriocin that is produced by certain strains of Pediococcus acidilactici (24, 27). Synthesis is conferred by the papABCD operon, which encodes the pediocin AcH structural gene (papA) and ancillary genes used for production (4, 26). The bacteriocin is translated as a 62-amino-acid preprotein and is converted by the PapD protein to a 44-amino-acid mature form by enzymatic processing of an 18-amino-acid leader peptide (4, 33). The PapC and PapD proteins belong to the ABC export system family of proteins (11). Although an ABC export system is used for production in Pediococcus, the mature sequence region of pediocin AcH can be secreted via the Escherichia coli sec machinery when its N terminus is fused to the E. coli secretory protein maltose binding protein (MBP) (25). These results indicate that PapD is necessary for recognition and processing of the leader peptide rather than for accommodating the mature region as it passes through the membrane. Once secreted, pediocin AcH becomes fully active after the formation of two intramolecular disulfide bonds (14, 19).
Independent studies performed with pediocins PA-1 and AcH have begun to reveal their mode of action and structural requirements for activity. These bacteriocins kill susceptible bacteria by permeabilizing the cytoplasmic membrane, causing leakage of ions and small molecules (2, 5, 7, 19). The interaction of pediocin PA-1 with membranes is promoted by acidic phospholipids (5), and positively charged amino acids in this cationic peptide appear to be important for membrane binding (6, 19). In fact, both lysines and histidines may mediate membrane binding in the low-pH environment (pH, ≤5.0) in which Pediococcus strains can grow (3, 19, 35). Other pediocin-like bacteriocins, such as sakacin P (32) (same as sakacin 674) (17), leucocin A (13), and curvacin A (31) (same as sakacin A) (16), also are cationic and may rely in part on electrostatic interactions between basic amino acids and negatively charged phospholipid head groups for membrane adsorption.
Pediocins PA-1 and AcH contain two structurally distinct sequence regions (5, 12, 19). The N-terminal 20 amino acids are polar and are highly conserved among pediocin-like bacteriocins. Located within this region is a -Y3-G4-N5-G6-V7- sequence that is present in all family members. While the function of this sequence is unknown, its deletion from pediocin AcH completely inactivates the molecule (25). Based on secondary structure analysis of pediocin PA-1, it has been proposed that the first 18 amino acids fold into a two-strand β hairpin that is stabilized by a β turn at position -G4-N5-G6-V7- (5) and the C9—C14 disulfide bond (5, 14). Solution nuclear magnetic resonance analysis of a related bacteriocin, leucocin A, indicated that its conformation indeed is ordered near the C9—C14 disulfide bond, but specific H-bond interactions expected for a β-hairpin structure were not detected (28). Perhaps due to the short length of these molecules, regions of defined secondary structure may not form until after they have adsorbed to a membrane (20).
The C-terminal sequence region of pediocins PA-1 and AcH (residues A21 to C44) is much less polar and conserved than the N-terminal sequence region (19). The C-terminal region is proposed to contain a hydrophobic membrane interaction domain (12), and analysis of the properties of hybrid peptides constructed from pediocin-like bacteriocins (12) and MBP-pediocin AcH chimeric proteins (25) supports this hypothesis. It also has been shown that the membrane interaction domain becomes amphipathic if folded into an α helix (12). While it is well established that membrane binding is promoted by amphipathic α helices (9, 20), it should be noted that the C24—C44 disulfide bond which is unique to pediocins PA-1 and AcH (19) bends the C-terminal region into a loop, and it is unknown whether an amphipathic structure can be formed in the presence of the loop.
To facilitate analysis of the structure and mode of action of pediocin AcH, we generated a collection of pediocin AcH substitution mutants that display altered bactericidal activity. Mutants were produced and detected with the E. coli MBP-pediocin AcH chimeric protein secretion system and colony overlay screening methods (25). Over one third of the amino acids in pediocin AcH were found to be important for activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Plasmid pMBR1.0 carries the papABCD operon from the P. acidilactici LB42-923 plasmid pSMB74. pMBR1.0 derivatives encoding pediocin AcH cysteine substitution mutants were constructed as described below. E. coli JM109 served as the host for these plasmids and was grown at 37°C in Luria-Bertani (LB) broth or agar containing 30 μg of chloramphenicol per ml. Plasmid pPR682 was used to construct mutant malE-papA translational fusion genes. Transcription of the chimeric genes was controlled by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter. E. coli E609L was used as the host for the malE-papA plasmids, and transformed E609L strains were grown at 37°C in LB broth or agar containing 12.5 μg of tetracycline and 100 μg of ampicillin per ml. MBP-pediocin AcH mutants were tested for activity against the bacterial indicator strains listed in Table 1. Lactic acid-producing bacterial strains were grown in tryptone-glucose-yeast extract (TGE) broth without Tween 80 (3, 35). Listeria innocua Lin11 was grown in tryptic soy broth (Difco) at 30°C.
TABLE 1.
Bacterial strains and plasmids
| Straina or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Enterococcus faecalis M1 | Pediocin AcH sensitive | Our isolate |
| Escherichia coli E609L | E609 lpp::Tn10, periplasmic leaky Tcr host for malE-papA plasmids | Henry C. Wu |
| Escherichia coli JM109 | Host for pMBR1.0-type plasmids | 36 |
| Escherichia coli TG1 | Host used for M13 mutagenesis | Amersham |
| Escherichia coli XL1-Blue | Host used for DNA sequencing | Stratagene |
| Lactobacillus plantarum NCDO955 | Pediocin AcH sensitive | 3 |
| Leuconostoc mesenteroides Ly | Pediocin AcH sensitive | Our isolate |
| Listeria innocua Lin11 | Nonpathogenic, pediocin AcH sensitive | Jean Richard, Institut National de la Recherche Agronomique, Paris, France |
| Pediococcus acidilactici LB42 | Pediocin AcH sensitive | Our isolate |
| Pediococcus acidilactici LB42-923 | Pediocin AcH producer | 26 |
| Plasmids | ||
| pMBR1.0 | papABCD operon in pHPS9, Cmr Emr | 4 |
| pMCS9, pMCS14, pMCS24, and pMCS44 | pMBR1.0 cysteine substitution mutant plasmids, Cmr Emr | This study |
| pPR682 | malE plasmid, Apr | New England BioLabs |
| pPR6821 | malE-papA plasmid, Apr | 25 |
| pKN1, pNK5, and so forth | malE-papA substitution mutant plasmids, Apr | This study |
The wild-type and mutant MBP-pediocin AcH chimeric proteins were tested against 11 other bacterial strains (gram positive and gram negative) that were either sensitive or insensitive to pediocin AcH. None of the mutants acquired activity against resistant strains.
Construction of mutagenized malE-papA plasmids.
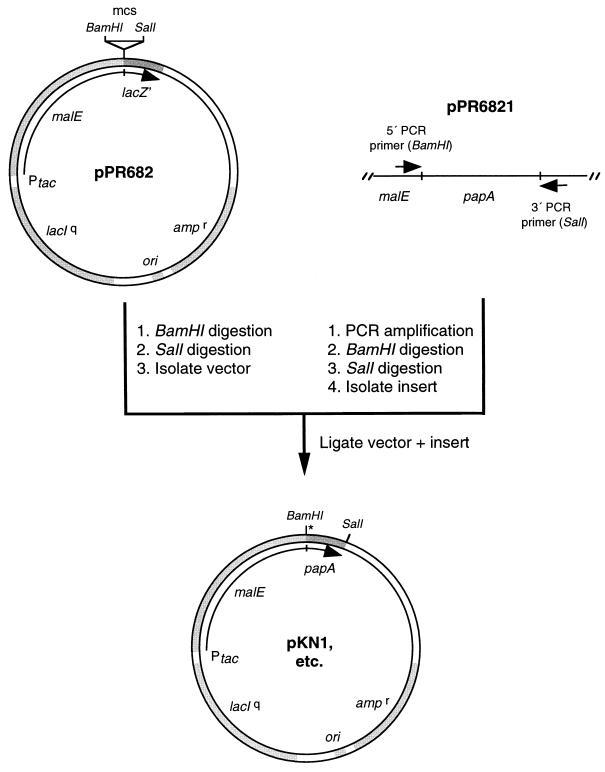
Base substitutions were introduced into the papA gene by PCR random mutagenesis (22) (Fig. 1). Mutagenesis reaction mixtures contained 25 U of Taq DNA polymerase (GIBCO-BRL) per ml, 0.1 μg of pPR6821 template per ml, 5 mM MgCl2, 0.5 mM MnCl2, 10 mM 2-mercaptoethanol, 10% dimethyl sulfoxide, 10 mM each nucleoside triphosphate, and 0.07 μg of each primer per ml. A 30-cycle repeated protocol consisting of 90 s of strand denaturation at 94°C, 60 s of primer annealing at 55°C, and 60 s of primer extension at 72°C was used to amplify papA DNA. The 5′ PCR primer (5′-CGGGGATCCATCGAGGGTAGG-3′) binds immediately upstream of the papA gene residing in pPR6821 and primes DNA synthesis beginning with the K1 codon of the mature sequence region. A BamHI restriction endonuclease digestion site is contained in the primer and was used for cloning purposes. The 3′ PCR primer (5′-CAAGCTTGCCTGCAGGTCGACCTA-3′) binds immediately downstream of codon C44 of the papA gene and contains a SalI restriction endonuclease digestion site. Amplified papA DNA fragments were treated with the Klenow fragment of E. coli DNA polymerase I to repair ragged ends, digested with BamHI and SalI restriction endonucleases, gel purified, and ligated into BamHI-SalI-digested pPR682 (29). Fusion genes in which the malE and mutagenized papA coding sequences were joined in frame were created by ligation.
FIG. 1.
Construction of mutagenized malE-papA plasmids. papA DNA fragments were synthesized by PCR amplification under random mutagenesis conditions with plasmid pPR6821 as a template. Subsequently, papA DNA fragments were digested with BamHI and SalI restriction endonucleases and ligated into plasmid pPR682 to construct malE-papA translational fusion genes. Plasmids containing mutant MBP-pediocin AcH chimeric proteins were identified by colony overlay screening, as explained in the text. Plasmids were named based on the location of the mutation, e.g., plasmid pKN1 contains the K1N mutation (indicated by an asterisk). Abbreviations: ampr, gene encoding β-lactamase; lacIq, gene encoding the lac repressor; lacZ′, gene encoding truncated β-galactosidase; malE, gene encoding MBP; mcs, multiple cloning site; ori, origin of replication; Ptac, tac promoter.
The mutagenized pool of malE-papA plasmids was transformed into strain E609L, and transformants were selected by plating on LB agar containing 12.5 μg of tetracycline and 100 μg of ampicillin per ml. Because the indicator strain Lactobacillus plantarum NCDO955 is sensitive to these antibiotics, the activity of chimeric proteins synthesized by the clones could not be screened directly on the primary transformant plates. Instead, E609L colonies were collected by scraping into LB medium containing antibiotics and were grown overnight at 37°C. On the following day, cells were pelleted by centrifugation and resuspended in LB broth without antibiotics. Aliquots of the suspension containing ∼100 cells were plated on 5 ml of TGE soft agar as described previously (25). The plates were incubated for 24 h at 37°C until colonies had formed and then were overlaid with 5 ml of TGE soft agar containing ∼106 cells of L. plantarum NCDO955 and 1 mM IPTG. Overlaid plates were incubated for an additional 24 h until zones of growth inhibition had formed around E609L producer colonies. Nonproducer mutant colonies were picked from the plates and grown in LB medium containing antibiotics, and plasmid DNA was isolated for transformation into E. coli XL1-Blue to obtain sequencing-quality DNA. Dideoxy terminator double-stranded DNA sequencing was performed with Sequenase T7 DNA polymerase (Amersham). Mutagenized plasmids were designated based on the location of the substitution site, e.g., pKN1, pNK5, and so forth.
Construction of pediocin AcH cysteine substitution mutant plasmids.
Cysteine substitution mutations were introduced into the papA gene of plasmid pMBR1.0 by three different methods. The C9S and C24S codon changes were created by use of a phosphorothioate oligonucleotide-directed in vitro mutagenesis procedure (30). An M13mp18 bacteriophage vector (36) containing the subcloned papA sequence served as the mutagenesis template. The sequences of oligonucleotides used to create cysteine codons were 5′-GGGGTTACTAGTGGCAA-3′ (oligo C9S) and 5′-CTACCACTAGCATAATC-3′ (oligo C24S); bases changed from the wild type are underlined. After in vitro mutagenesis procedures, modified double-stranded bacteriophage DNAs were transformed into E. coli TG1, and cysteine substitution mutants were identified by plaque hybridization with the 32P-labeled mutagenic oligonucleotides that were used to screen nitrocellulose filters (29). The sequences of mutagenized papA inserts were confirmed by single-stranded DNA sequencing with Sequenase T7 DNA polymerase. The C9S and C24S papA genes were excised from the replicative forms of the bacteriophage vectors by digestion with MscI and Bpu1102I restriction endonucleases and were cloned into pMBR1.0, replacing the wild-type papA coding region. The papA sequences of the resulting pMBR1.0 cysteine substitution mutant plasmids (named pMCS9 and pMCS24; Table 1) were subjected to double-stranded DNA sequencing to confirm the swap of mutant for wild-type DNA.
The C44S substitution mutation was created in the papA gene by PCR amplification with pMBR1.0 as a template. The C44S codon change was incorporated at the underlined base into the 3′ PCR primer (5′-CAGCTCAGCATAATGCTAGCTTTTATG-3′) used in the amplification reaction. The primer also contains a Bpu1102I restriction endonuclease digestion site for cloning into pMBR1.0. The 5′ PCR primer (5′-AGAAATGGCCAATATCATTGGTGGTAAA-3′) used in the amplification reaction contains an MscI restriction endonuclease digestion site. The reaction conditions used to synthesize the C44S papA DNA fragment have been described before (25). The C44S DNA product was treated with the Klenow fragment of DNA polymerase I, digested with MscI and Bpu1102I restriction endonucleases, gel purified, and ligated into pMBR1.0. The substitution of the mutant for the wild-type sequence in the resulting plasmid, pMCS44, was confirmed by double-stranded DNA sequencing.
The C14S codon change was introduced into the papA gene by extension overlap PCR mutagenesis (15). The papA coding sequence first was amplified as two DNA fragments in which the 3′ end of the upstream fragment overlapped the 5′ end of the downstream fragment. The targeted C14 codon resided in the overlapping region. Plasmid pMBR1.0 served as a template, and PCR conditions were the same as those used for synthesis of the C44S DNA fragment. The primers used to synthesize the upstream DNA fragment were 5′-AGAAATGGCCAATATCATTGGTGGTAAA-3′ (5′ PCR primer) and 5′-CCAGTCAACAGAGCTGGAATGTTTG-3′ (3′ PCR primer), and the primers used to synthesize the downstream DNA fragment were 5′-CAAACATTCCAGCTCTGTTGACTGG-3′ (5′ PCR primer) and 5′-ATTGATGCCAGCTCAGCATAATGCTA-3′ (3′ PCR primer); the bases changed to create the C14S substitution in the region of overlap are underlined. After the two primary PCR products were synthesized, they were gel purified and combined at a concentration of 0.1 μg/ml each in the extension overlap PCR mixture. Conditions used to synthesize the extension overlap secondary PCR product were identical to those used to synthesize the primary products, except that only the two outermost primers were used. The extension overlap papA DNA fragment was treated with the Klenow fragment of DNA polymerase I, digested with MscI and Bpu1102I restriction enzymes, gel purified, and ligated into pMBR1.0. Again, transfer of the mutant sequence to the resulting plasmid, pMCS14, was confirmed by double-stranded DNA sequencing.
Comparison of MBP-pediocin AcH chimeric protein synthesis levels.
E. coli E609L cells transformed with the mutant malE-papA plasmids were grown at 37°C in LB medium containing tetracycline and ampicillin. At the mid-log phase, IPTG (1 mM final concentration) was added to induce the synthesis of chimeric proteins. After 3 h of continuous growth, culture broths were subjected to trichloroacetic acid precipitation, and precipitated proteins were solubilized in sample loading buffer containing 3% sodium dodecyl sulfate (SDS), boiled, and analyzed on 10% acrylamide-bisacrylamide–SDS gels (21). Proteins were visualized by Coomassie brilliant blue dye staining, and relative levels of synthesis of wild-type and mutant chimeric proteins were compared by scanning densitometry with a Bio-Rad Gel Dock laser densitometer (1, 25).
Measurement of the bactericidal activities of MBP-pediocin AcH chimeric proteins.
Chimeric protein-expressing strains were grown at 37°C in LB medium without antibiotics and induced at the mid-log phase for 3 h by the addition of 1 mM IPTG. Culture broths were boiled, and aliquots were tested against the indicator strains listed in Table 1. Two types of activity tests were performed. The first assay (the well assay) was used to establish if the mutants exhibited any activity. In this assay, 100-μl aliquots of the culture broths were placed into wells cut in TGE agar plates on which soft-agar lawns of the indicator bacteria had been spread. Plates were incubated overnight and examined for zones of growth inhibition around the wells. The second assay (the titration assay) was used to determine the activity levels of mutants that were shown by the well assay to be partially active. In this assay, aliquots of boiled, 3-h IPTG-induced culture broths were applied to plates containing an overlay of L. innocua Lin11, and the minimum volume necessary to produce a zone of growth inhibition in the lawn was determined (3, 35). It should be noted that 1 μl of a boiled, 3-h IPTG-induced culture broth from the wild-type MBP-pediocin AcH-producing strain (E609L/pPR6821) was sufficient to form a small zone of growth inhibition against L. innocua Lin11. After titration of the activities of the broths, the activities of mutant chimeric proteins relative to that of the wild-type MBP-pediocin AcH chimeric protein were calculated. Activities were corrected for differences in culture optical density at 600 nm (OD600) and the relative levels of proteins in the culture broths.
Analysis of pediocin AcH sequence hydrophobicity, amphipathicity, and β-turn potentials.
The average sequence hydrophobicities and average sequence hydrophobic moments of wild-type and mutant pediocin AcH sequence regions were calculated by use of the normalized amino acid hydrophobicity scale and hydrophobic moment calculation method of Eisenberg et al. (9). β-Turn potentials of selected pediocin AcH sequence regions also were calculated by use of published methods (18).
RESULTS
Isolation of MBP-pediocin AcH chimeric protein random mutants.
An MBP-pediocin AcH chimeric protein secretion system (25) was used for the generation and detection of pediocin AcH mutants. Mutations were introduced into DNA encoding the mature sequence region of the papA gene (codons K1 to C44) by PCR random mutagenesis (22). The conditions used typically create about 1 base substitution per 100 bp of DNA. Subsequently, mutagenized papA DNA fragments were cloned into the pPR682 malE expression plasmid (Fig. 1), and the pool of plasmid DNAs was transformed into the periplasmic leaky E. coli host, E609L. About one third of the MBP-pediocin AcH chimeric proteins synthesized by this strain are secreted into the periplasm and released into the culture medium (25). All chimeric protein substitution mutants described in this study were isolated by colony overlay screening against the pediocin AcH-sensitive indicator strain L. plantarum NCDO955 (3, 35).
Approximately 2,500 colonies were obtained from transformation of E609L with the pool of mutagenized plasmid DNAs. Of these transformants, about one half displayed reduced or no activity. Plasmid DNAs from 53 mutant colonies were sequenced, and 17 unique substitution mutations affecting 14 codons in the papA gene were identified (Table 2). Two substitution mutations (G37R and C44W) occurred frequently (>10 times each), perhaps due to a tendency of Taq DNA polymerase to misincorporate bases at these codons under the mutagenesis conditions used. Only a few multiple-substitution mutants were obtained (data not shown), confirming the bias of the technique for generating single base substitutions in short DNA sequences (22). Other mutants might have been recovered if more of the inactive clones in the pool had been sequenced.
TABLE 2.
Substitution sites and bactericidal activities of MBP-pediocin AcH chimeric protein mutants
| Strain | Activitya of mutantsb with the indicated mutation in the following sequence:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K1 Y Y G | N G V T | C9 G | K H S | C14 S V D | W G K A T T C24 I | I N N G A | M A W | A T G | G H Q G | N | H | K | C44 | |
| N1 | K5 | R9 | E11 | S14 Y14 | R18 | T26 | T31 | D34 | E37 R37 | K41 | L42 | E43 N43 | W44 | |
| L. plantarum NCDO955 | + | − | − | + | −c | + | + | + | + | −c | + | + | +c | − |
| P. acidilactici LB42 | + | − | − | + | − | + | + | + | −c | − | + | + | + | − |
| L. mesenteroides Ly | + | − | − | + | − | + | + | + | −c | − | + | + | + | − |
| E. faecalis M1 | + | − | − | + | − | + | + | + | + | − | + | + | + | − |
| L. innocua Lin11 | + | − | − | + | − | + | + | + | + | − | + | + | + | − |
Activity was tested with 100 μl of IPTG-induced (3 h) culture broths from the substitution mutant strains. The five indicator strains are sensitive to wild-type pediocin AcH and to the wild-type MBP-pediocin AcH chimeric protein. The relative activity levels of mutants retaining activity (indicated by +) are presented in Table 3; inactive mutants are indicated by −.
All mutants are single-amino-acid-substitution mutants.
Both mutants at position 14 and position 37 were inactive. The two mutants at position 43 were partially active. The mutant at position 34 showed a loss of activity against two of the five indicator strains.
About one half of the mutations (K1N, N5K, C9R, K11E, C14S/Y, W18R, and A34D) mapped to amino acids that are conserved in the pediocin-like family of bacteriocins (19). Other mutations (I26T, M31T, G37E/R, N41K, H42L, K43N/E, and C44W) affected amino acids that either are not conserved or are minimally conserved. As expected, the side chains of substituted amino acids typically differed substantially from those of wild-type residues. Because mutations often mapped to conserved residues and residues such as cysteines that are known by experimental means to be important for activity (7, 19), we concluded that the mutagenesis and screening procedures were highly effective in identifying important amino acids in the pediocin AcH sequence.
Analysis of the bactericidal activities of MBP-pediocin AcH chimeric protein mutants.
The mutants listed in Table 2 were tested for activity against the gram-positive pediocin AcH-sensitive strains L. plantarum NCDO955, P. acidilactici LB42, Leuconostoc mesenteroides Ly, Enterococcus faecalis M1, and L. innocua Lin11. Five strains were used to determine how broadly the mutations inactivated pediocin AcH. In this regard, a mutation that interferes with a fundamental property such as membrane binding could inactivate pediocin AcH for a variety of bacteria.
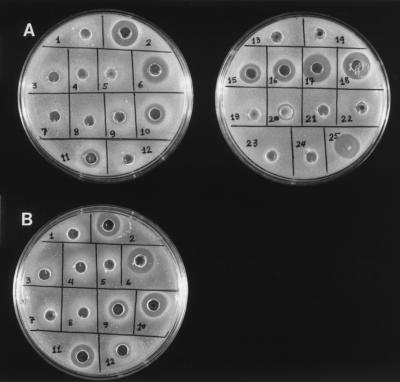
The mutants first were classified as inactive or active by well assay testing against the five indicator strains. Examples of plates on which mutants were tested against L. innocua Lin11 and E. faecalis M1 are presented in Fig. 2. Based on testing against the five indicator strains, 7 mutants (N5K, C9R, C14S, C14Y, G37E, G37R, and C44W) were classified as inactive and 10 mutants (K1N, K11E, W18R, I26T, M31T, A34D, N41K, H42L, K43E, and K43N) were classified as partially to fully active compared to the wild type (Table 2). The test strains were uniformly sensitive or insensitive to all mutants except A34D. The A34D mutant retained some activity against L. plantarum NCDO955, E. faecalis M1, and L. innocua Lin11 but was inactive against P. acidilactici LB42 and L. mesenteroides Ly with the method used. Note that a 100-μl sample of the wild-type E609L/pPR6821 culture broth contained ∼100-fold more chimeric protein than was necessary to inhibit the growth of these bacteria.
FIG. 2.
Well assay analysis of MBP-pediocin AcH substitution mutant and pediocin AcH cysteine substitution mutant antibacterial activities. Screening was performed against L. innocua Lin11 (A) and E. faecalis M1 (B) with 100 μl of boiled, IPTG-induced (3 h) culture broths (chimeric protein strains) or 100 μl of boiled, overnight-grown culture broths (pediocin AcH cysteine substitution mutant strains) per well. The analysis of only culture samples 1 to 12 is shown for E. faecalis M1. Samples: 1, E609L; 2, E609L/pPR6821; 3, E609L/pKN1; 4, E609L/pNK5; 5, E609L/pCR9; 6, E609L/pKE11; 7, E609L/pCS14; 8, E609L/pCY14; 9, E609L/pWR18; 10, E609L/pIT26; 11, E609L/pMT31; 12, E609L/pAD34; 13, E609L/pGE37; 14, E609L/pGR37; 15, E609L/pNK41; 16, E609L/pHL42; 17, E609L/pKE43; 18, E609L/pKN43; 19, E609L/pCW44; 20, JM109/pMBR1.0; 21, JM109/pMCS9; 22, JM109/pMCS14; 23, JM109/pMCS24; 24, JM109/pMCS44; 25, 1 μl of boiled P. acidilactici LB42-923 culture broth containing wild-type pediocin AcH. E609L/pKN1 and E609L/pAD34 had very slight activities against both indicator strains.
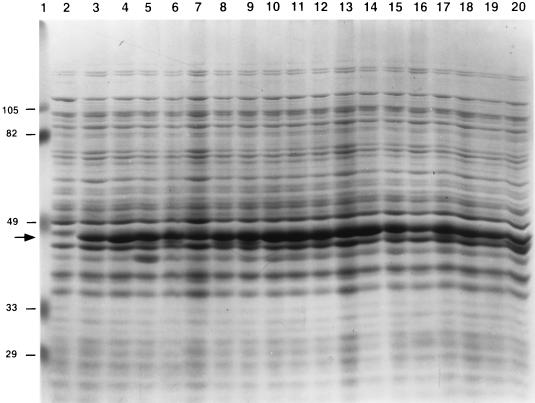
The active mutants subsequently were subjected to titration assays to measure levels of activity relative to that of the wild-type chimeric protein. As shown in Table 3, the active mutants retained from <1% (K1N, W18R, M31T, and A34D) to ∼60% (K43N) wild-type activity against L. innocua Lin11. The K11E mutant actually exhibited 2.75-fold-greater activity than the wild-type protein against this strain. L. innocua Lin11 was used in titration assays because it is relatively more sensitive to pediocin AcH than the other strains. The level of activity of the wild-type culture broth was calculated to be 1,000 activity units/ml because 1 μl was sufficient to produce a small zone of growth inhibition against this indicator strain (Table 3). The activities of the mutants were corrected for differences in the optical densities of cultures and levels of chimeric proteins in culture broths. Relative protein synthesis levels varied by about twofold (Table 3), based on an analysis of SDS-polyacrylamide gels like the one shown in Fig. 3. Protein synthesis levels were lowest for the K11E and H42L mutants and highest for the I26T, M31T, and A34D mutants.
TABLE 3.
Relative bactericidal activities of wild-type and mutant MBP-pediocin AcH chimeric proteins
| Strain | OD600a | Activity units/ml of culture brothb | Protein levelc | Corrected activity units/mld |
|---|---|---|---|---|
| Wild typee | 1.1 | 1,000 | 1.0 | 1,000 |
| K1N | 2.2 | 10 | 1.4 | 4 |
| K11E | 0.5 | 1,000 | 0.8 | 2,750 |
| W18R | 2.3 | ≤10 | 1.4 | ≤3 |
| I26T | 2.4 | 100 | 1.6 | 29 |
| M31T | 2.3 | 20 | 1.6 | 6 |
| A34D | 0.9 | ≤10 | 1.8 | ≤7 |
| N41K | 1.0 | 100 | 1.1 | 100 |
| H42L | 2.3 | 400 | 0.9 | 212 |
| K43N | 1.8 | 1,000 | 1.0 | 611 |
| K43E | 2.4 | 1,000 | 1.1 | 417 |
Determined for culture broths following a 3-h IPTG induction (average of two experiments).
Calculated by (i) determining the minimum volume (microliters) of culture broth required to produce a zone of growth inhibition in a lawn of L. innocua Lin11 cells and (ii) dividing 1,000 AU · μl/ml by this volume (average of four experiments).
Relative protein synthesis levels were determined by laser scanning densitometry of Coomassie brilliant blue-stained gels containing samples of culture broths (average of two experiments).
Corrected for differences in OD600 values and relative chimeric protein levels in wild-type and mutant culture broths.
Wild-type MBP-pediocin AcH chimeric protein expressed by the E609L/pPR6821 strain.
FIG. 3.
SDS-polyacrylamide gel analysis of mutant MBP-pediocin AcH chimeric protein synthesis levels. Proteins were visualized by staining with Coomassie brilliant blue dye. The arrow marks the position of migration of the MBP-pediocin AcH chimeric proteins. Equivalent masses of total culture proteins were analyzed in each lane. Lanes: 1, prestained standards; 2, E609L; 3, E609L/pPR6821; 4, E609L/pKN1; 5, E609L/pNK5; 6, E609L/pCR9; 7, E609L/pKE11; 8, E609L/pCS14; 9, E609L/pCY14; 10, E609L/pWR18; 11, E609L/pIT26; 12, E609L/pMT31; 13, E609L/pAD34; 14, E609L/pGE37; 15, E609L/pGR37; 16, E609L/pNK41; 17, E609L/pHL42; 18, E609L/pKE43; 19, E609L/pKN43; 20, E609L/pCW44. Molecular weights (in thousands) of prestained standards are shown at the left. The levels of synthesis of chimeric proteins varied by about twofold among the strains.
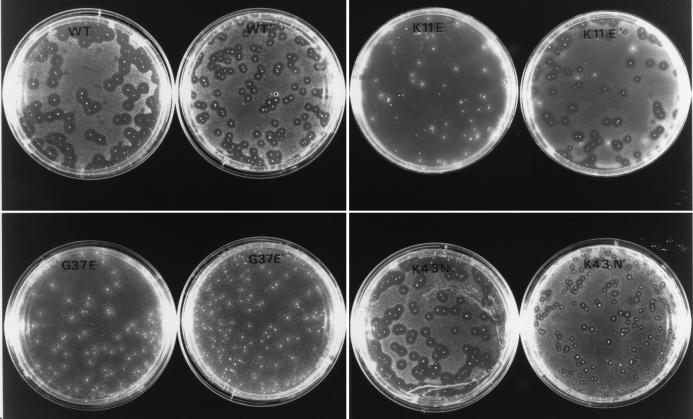
Experiments were performed to determine why the highly active K11E mutant originally was picked up by colony overlay screening as a null mutant. As shown in Fig. 4, zones of growth inhibition typically do not form around E609L/pKE11 colonies when IPTG is added to the primary plating agar instead of to the agar overlay. This result indicates that zones of growth inhibition typically do not form when these colonies are growing at the time at which they are exposed to IPTG. In contrast, colonies of strains that synthesize less potent chimeric proteins, such as the wild-type or K43N mutant proteins, actually show larger zones of growth inhibition when IPTG is added to the primary plating agar (Fig. 4). Taken together, the data indicate that high-level synthesis of the K11E mutant during the growth of colonies causes a null activity phenotype. It seems likely that under these conditions, cells within the colony lose the plasmid and stop synthesizing the K11E protein, which is toxic for the host (data not shown). Possibly, the original K11E mutant colony was recovered from a plate on which it was growing well at the time at which it was overlaid with top agar containing IPTG.
FIG. 4.
Effects of timing of IPTG induction on sizes of zones of growth inhibition formed by MBP-pediocin AcH expression strains against L. innocua Lin11. IPTG (1 mM) was added either in the primary agar containing E. coli colonies or ∼24 h later (indicated by prime symbol) with the agar overlay containing the indicator strain. Strains tested were E609L/pPR6821 (wild type; WT), E609L/pKE11 (K11E mutant), E609L/pGE37 (G37E mutant), and E609L/pKN43 (K43N mutant). The diameters of zones of growth inhibition formed by wild-type and mutant colonies differed depending on the time of induction (see the text).
Analysis of the bactericidal activities of pediocin AcH cysteine substitution mutants.
A serine was substituted for each of the four cysteines in the native pediocin AcH molecule to determine if C24 is important for activity and to verify that C9, C14, and C44 are required in both native and chimeric proteins. Substitution mutations were introduced into the papA gene of plasmid pMBR1.0 by PCR or oligonucleotide-directed site-specific mutagenesis. Mutant plasmids were named pMCS9, pMCS14, pMCS24, and pMCS44 based on the location of the substitution site. Pediocin AcH mutants were expressed in E. coli JM109, in which secretion is directed via the PapC-PapD export machinery carried by pMBR1.0 (4).
All four substitution mutant strains grew well and grew at rates comparable to that of the wild-type JM109/pMBR1.0 strain. However, only the wild-type strain showed zones of growth inhibition in colony overlay screening (data not shown) and in well assay testing of culture broths (Fig. 2). These results showed that C24 is required for activity. They also indicated that substitution of cysteines causes the same inhibitory effects in native and chimeric forms of the bacteriocin. Thus, it appears that the effects of the mutations in chimeric proteins can be extrapolated to the native molecule. It is possible, however, that the extent of inactivation caused by some mutations may vary between native and chimeric molecules due to the presence of the MBP domain.
DISCUSSION
Fourteen amino acids that are important for the activity of pediocin AcH have been identified by random mutagenesis. The amino acids are distributed across the entire peptide chain, indicating that little of its sequence may be dispensable for function. Mutations were obtained at residues anticipated to be required based on sequence conservation as well as at residues that could not be predicted to be necessary. Some mutations within the N-terminal region (N5K, C9R, and C14S/Y) mapped to amino acids that are proposed to stabilize the structure of the molecule (5, 19). Several mutations were obtained at positively charged amino acids (K1N, H42L, and K43N/E) and at amino acids contained within a hydrophobic sequence in the C-terminal region (I26T, M31T, A34D, and G37E/R) that may mediate membrane binding (5, 12, 19). In about one half of the cases, the mutants were completely inactive against several bacterial strains, suggesting that the affected residues play central roles in the mode of action of the bacteriocin. Only one mutation (A34D) may have a species-specific effect on activity.
The experiments showed that all four cysteines in pediocin AcH are necessary for activity. It seems likely that cysteines are required for the formation of disulfide bonds, which stabilize the structure of this short peptide. For example, the C9—C14 disulfide bond may be required to establish the conformation of the intervening -G10-K11-H12-S13- sequence that forms the apex of the putative β hairpin within the N-terminal region (5). As indicated in Table 4, this sequence exhibits weak homology to six known types of consensus β turns (18) and therefore may not be able to maintain an active conformation without the assistance of the C9—C14 disulfide bond. It may be possible to construct an active pediocin AcH molecule lacking the C9—C14 disulfide bond if a strong consensus β-turn sequence is substituted for the G10-to-S13 region. There are consensus-type β turns in which lysine frequently occurs (18). It should be noted that the substitution of glutamate for K11 neither creates nor destroys a strong consensus β turn in this sequence region (Table 4). The possible role of the C24—C44 disulfide bond is discussed below.
TABLE 4.
β-turn potentials of selected pediocin AcH sequence regionsa
| Sequence | % Identity to β-turn consensus sequence of type:
|
||
|---|---|---|---|
| I′b | II′c | VIIId | |
| -G10-K11-H12-S13- | 12 (12) | 25 (24) | 48 (47) |
| -Y2-Y3-G4-N5- | 67 (76) | 30 (30) | |
| -Y3-G4-N5-G6- | 28 (26) | 97 (89) | |
| -G4-N5-G6-V7- | 79 (55) | 20 (21) | |
The percent identity of each sequence to six consensus-type β turns was calculated according to the method of Hutchinson and Thornton (18). Values are shown only for the most relevant types of β turns. Values in parentheses were calculated for the K11E (relevant for the -G10-K11-H12-S13- sequence) and N5K (relevant for the other sequences) substitution mutants.
-Y-N-G-K-.
-Y-G-N-T-.
-P-P-N-P-.
The N5K mutation maps to a putative β-turn sequence (-G4-N5-G6-V7-) (5) within the β hairpin that is 79% identical to a type I′ β turn (Table 4). We have determined that two other sequences in this region (-Y2-Y3-G4-N5- and -Y3-G4-N5-G6-) also have a high potential to form β turns. Of the three sequences, -Y3-G4-N5-G6- exhibits the highest identity (97%) to a known type of β turn, in this case, a type II′ β turn (Table 4). While the calculations do not definitively identify the type of β turn present, they do support the hypothesis that the N-terminal region is folded into a β hairpin, since both type I′ and II′ β turns occur frequently in these structures (18). In addition, the calculations show that the substitution of lysine for asparagine moderately changes the identity scores of the three β turns (Table 4) and perhaps does not prohibit their formation per se. However, the N5K mutation may perturb the structure of this region due to charge repulsion between lysine and other positively charged residues that may be located nearby in the three-dimensional structure.
Experiments performed with magainin (34) and model peptides (23) have indicated that positively charged amino acids are particularly important for the binding of moderately hydrophobic peptides, such as pediocin AcH (see below), to phospholipid bilayers. Of the four lysines and three histidines in the molecule, three residues (K1, H42, and K43) were found to be important for activity. As shown in Table 3, the K1N substitution nearly fully inactivated the molecule, whereas the H42L and K43N/E substitutions decreased activity by ∼40 to 80%. The results indicate that if these residues help mediate membrane binding, then K1 plays an essential role whereas H42 and K43 do not. Instead, H42 and K43 may only augment the binding of pediocin AcH to membranes. It should be possible to define the functions of these positively charged residues by performing membrane binding assays with the mutants. Binding experiments have been used to show that K11 modulates the binding of pediocin PA-1 fragments to membranes (6). While the experiments suggested that K11 participates in membrane binding, the finding that high activity was conserved when glutamate was substituted for K11 indicated that K11 plays another, more important role in the function of the bacteriocin.
The C-terminal sequence region of pediocin AcH was subjected to hydrophobicity analysis to locate nonpolar sequences that could mediate membrane binding. The most hydrophobic stretch of amino acids in this region is the 13-residue I25-to-G37 sequence, for which the average sequence hydrophobicity is 0.45 (Table 5) (9). A 17-amino-acid sequence (A21 to G37) is only slightly less hydrophobic (hydrophobicity, 0.39). The hydrophobicities for both sequences are slightly higher than those exhibited by a number of surface-active peptides and are quite low compared to those exhibited by sequences that are inserted individually into membranes with a transmembrane orientation (9). However, either sequence should be sufficiently hydrophobic to localize to the interior of a phospholipid bilayer if two or more molecules are inserted into the membrane together (9, 10), as may occur during the assembly of bacteriocin pore complexes (19). While many questions remain about the structure and function of the C-terminal region, the mutagenesis results strongly suggest that the hydrophobicity of this region is important for activity. In this regard, all mutations within the I25-to-G37 region increased polarity (Table 5) and resulted in a complete loss of activity (Table 3).
TABLE 5.
Effects of substitution mutations on average hydrophobicities and average hydrophobic moments of selected pediocin AcH sequence regionsa
| Sequence | Mutation | Hydrophobicity | Hydrophobic moments (<μ>) |
|---|---|---|---|
| A21 to G37 | None (wild type) | 0.39 | 0.20 |
| I25 to G37 | None (wild type) | 0.45 | 0.30 |
| I26T | 0.34 | 0.21 | |
| M31T | 0.40 | 0.35 | |
| A34D | 0.34 | 0.36 | |
| G37E | 0.36 | 0.25 | |
| G37R | 0.22 | 0.24 | |
| I25 to H42 | None (wild type) | 0.22 | 0.30 |
| I26T | 0.14 | 0.24 | |
| M31T | 0.18 | 0.34 | |
| A34D | 0.14 | 0.34 | |
| G37E | 0.15 | 0.27 | |
| G37R | 0.05 | 0.24 | |
| N41K | 0.18 | 0.31 | |
| H42L | 0.30 | 0.22 |
Values were calculated by the method of Eisenberg et al. (9). <μ> values are a measure of the amphipathicity of a sequence.
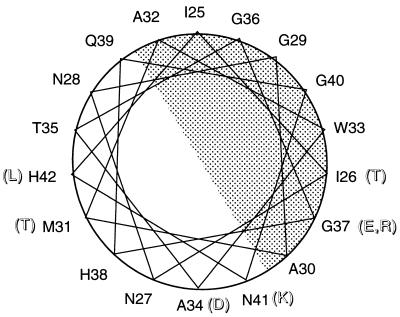
As mentioned above, the nonpolar C-terminal region, which includes the I25-to-G37 sequence, would be amphipathic if it were folded into an α helix (12). Although the C24—C44 disulfide bond may interfere with the formation of an α helix, we nonetheless thought it might be informative to calculate hydrophobic moments (9) for putative α-helical sequences within the C-terminal region (Table 5). The calculations indicated that the wild-type I25-to-G37 sequence and the much less hydrophobic I25-to-H42 sequence are moderately amphipathic if they are folded into α helices. Although the hydrophobic moments of these sequences are fairly low compared to those of many surface-active peptides (18), this fact would not necessarily preclude adsorption of these α helices to a membrane, since the hydrophobicities (0.75 to 0.78) of their nonpolar faces are quite high. Interestingly, all mutations fell on one half of a helical wheel diagram representing this sequence region and would overlap both the nonpolar and the polar faces of the α helices (Fig. 5). In all but one case (N41K), substitutions changed the hydrophobic moments of the sequences (Table 5) and therefore could change the depth or angle of contact at which pediocin AcH adsorbs to the membrane interface. While the calculations and data make it tempting to conclude that the amphipathicity of this region is important for activity, the effect of the C24—C44 disulfide bond on the structure of this region must be determined before conclusions about the role of sequence amphipathicity can be drawn.
FIG. 5.
Schiffer-Edmundsen helical wheel analysis of the I25-to-H42 sequence region of pediocin AcH. Amino acids in the wild-type sequence are shown next to the wheel, and substitutions are indicated in parentheses. Note that all substitution sites are located on the bottom half of the α helix and that the sites fall on both the polar (white) and nonpolar (shaded) faces.
The final mutation to be discussed, W18R, occurred at a site located between the N- and C-terminal regions of the molecule. The tryptophan residue at this position is conserved in several pediocin-like bacteriocins. W18 may penetrate into the acyl-chain region of membrane phospholipids when the bacteriocin adsorbs to the interface (7). We speculate that the W18R mutation could alter the depth to which pediocin AcH adsorbs to the membrane interface. In this regard, tryptophans located near the ends of integral membrane protein transmembrane segments are thought to help establish the interfacial boundaries of these segments (8).
In the future, models for the structure of pediocin AcH will be tested further with these and related mutants. Biochemical analysis of the mutants also should help clarify the mode of action of pediocin AcH and may explain why it has greater potency and spectrum of activity than other pediocin-like bacteriocins (19). In this regard, the structure of pediocin AcH may differ substantially from those of other pediocin-like bacteriocins due to its unique C24—C44 disulfide bond. Finally, the finding that the K11E mutation increased potency demonstrates that it is possible to improve the properties of pediocin AcH and perhaps other bacteriocins by mutagenesis.
ACKNOWLEDGMENTS
We thank investigators for providing bacterial strains.
Financial support was provided by the National Science Foundation, the state of Wyoming, and The Michigan Biotechnology Institute.
REFERENCES
- 1.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Bhunia A K, Johnson M C, Ray B, Kalchayanand N. Mode of action of pediocin AcH from Pediococcus acidilactici H on sensitive bacterial strains. J Appl Bacteriol. 1991;70:25–30. [Google Scholar]
- 3.Biswas S R, Ray P, Johnson M C, Ray B. Influence of growth conditions on the production of bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol. 1991;57:1265–1267. doi: 10.1128/aem.57.4.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukhtiyarova M, Yang R, Ray B. Analysis of the pediocin AcH gene cluster from plasmid pSMB74 and its expression in a pediocin-negative Pediococcus acidilactici strain. Appl Environ Microbiol. 1994;60:3405–3408. doi: 10.1128/aem.60.9.3405-3408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Shapira R, Eisenstein M, Montville T J. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl Environ Microbiol. 1997;63:524–531. doi: 10.1128/aem.63.2.524-531.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Ludescher R D, Montville T J. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl Environ Microbiol. 1997;63:4770–4777. doi: 10.1128/aem.63.12.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikindas M L, Garcia-Garcera M J, Driessen A J M, Ledeboer A M, Nissen-Meyer J, Nes I F, Abee T, Konings W N, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC 1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan S W, Rosenbusch J P. Folding pattern diversity of integral membrane proteins. Science. 1994;264:914–916. doi: 10.1126/science.8178151. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 10.Engelman D M, Steitz T A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 11.Fath M J, Kolter R. ABC-transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fimland G, Blingsmo O R, Sletten K, Jung G, Nes I F, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson J T, Chopko A L, van Wassenaar P D. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 16.Holck A, Axelsson L, Birkeland S-E, Aukrust T, Bloom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake LB706. J Gen Microbiol. 1992;138:2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- 17.Holck A, Axelsson L, Huhne K, Krockel L. Purification and cloning of sakacin 674, a bacteriocin from Lactobacillus sake LB674. FEMS Microbiol Lett. 1994;115:143–150. doi: 10.1111/j.1574-6968.1994.tb06629.x. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson E G, Thornton J M. A revised set of potentials for β-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser E T, Kezdy F J. Amphiphilic secondary structure: design of peptide hormones. Science. 1984;223:249–255. doi: 10.1126/science.6322295. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 23.Liu L-P, Deber C M. Anionic phospholipids modulate peptide insertion into membranes. Biochemistry. 1997;36:5476–5482. doi: 10.1021/bi970030n. [DOI] [PubMed] [Google Scholar]
- 24.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller K W, Schamber R, Chen Y, Ray B. Production of active chimeric pediocin AcH in Escherichia coli in the absence of processing and secretion genes from the Pediococcus pap operon. Appl Environ Microbiol. 1998;64:14–20. doi: 10.1128/aem.64.1.14-20.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motlagh A M, Bukhtiyarova M, Ray B. Complete nucleotide sequence of pSMB74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol. 1994;18:305–312. doi: 10.1111/j.1472-765x.1994.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 27.Ray B. Characteristics and application of pediocin(s) of Pediococcus acidilactici: pediocin PA-1/AcH. In: Bozoglu T F, Ray B, editors. Lactic acid bacteria: current advances in metabolism, genetics, and applications. New York, N.Y: Springer; 1996. pp. 155–203. [Google Scholar]
- 28.Sailer M, Helms G L, Henkel T, Niemczura W P, Stiles M E, Vederas J C. 15N- and 13C-labeled media from Anabaena sp. for universal isotopic labeling of bacteriocins: NMR resonance assignments of leucocin A from Leuconostoc gelidum and nisin A from Lactococcus lactis. Biochemistry. 1993;32:310–318. doi: 10.1021/bi00052a039. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Taylor J W, Ott J, Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985;13:8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tichaczek P S, Vogel R F, Hammes W P. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch Microbiol. 1993;160:279–283. doi: 10.1007/BF00292077. [DOI] [PubMed] [Google Scholar]
- 32.Tichaczek P S, Vogel R F, Hammes W. Cloning and sequencing sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH673. Microbiology. 1994;140:361–367. doi: 10.1099/13500872-140-2-361. [DOI] [PubMed] [Google Scholar]
- 33.Venema K, Kok J, Marugg J D, Toonen M Y, Ledeboer A M, Venema G, Chikindas M L. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC 1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 34.Wieprecht T, Dathe M, Beyermann M, Krause E, Maloy W L, MacDonald D L, Bienert M. Peptide hydrophobicity controls the activity and selectivity of magainin 2 amide in interaction with membranes. Biochemistry. 1997;36:6124–6132. doi: 10.1021/bi9619987. [DOI] [PubMed] [Google Scholar]
- 35.Yang R, Johnson M C, Ray B. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1992;58:3355–3359. doi: 10.1128/aem.58.10.3355-3359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]