Abstract
Across all walks of life, certain tRNA transcripts contain introns. Pre-tRNAs with introns require splicing to form the mature anticodon stem loop (ASL). In eukaryotes, tRNA splicing is initiated by the heterotetrameric tRNA splicing endonuclease (TSEN) complex. All TSEN subunits are essential and mutations within the complex are associated with a family of neurodevelopmental disorders known as pontocerebellar hypoplasia (PCH). Here, we report cryo-EM structures of the human TSEN•pre-tRNA complex. These structures reveal the overall architecture of the complex and the extensive tRNA binding interfaces. The structures share homology with archaeal TSENs, but contain additional features important for pre-tRNA recognition. The TSEN54 subunit functions as a pivotal scaffold for the pre-tRNA and the two endonuclease subunits. Finally, the TSEN structures enable visualization of the molecular environments of PCH-causing missense mutations, providing insight into the mechanism of pre-tRNA splicing and PCH.
Introduction
Prior to their involvement in translation, tRNAs undergo extensive processing and modification1. Although most tRNA genes are intronless, a subset of pre-tRNAs contain an intron that forms base pairing interactions with the anticodon2. These intervening sequences must be removed from the precursor prior to translation. Within the human genome, there are several isodecoder families that all contain introns; thus tRNA splicing is essential3,4. Eukaryotic tRNA intron cleavage is catalyzed by the heterotetrameric TSEN complex5,6. The TSEN complex contains two presumptive structural subunits, TSEN54 and TSEN15, along with two endonuclease subunits, TSEN34 and TSEN2, that catalyze cleavage at the 3’ and 5’-splice sites, respectively7,8. Beyond providing structural support, the TSEN54 subunit is thought to function as a molecular ruler that properly positions the 5’-splice site for cleavage by TSEN29–11. However, the overall molecular architecture of the TSEN complexes of both yeast7 and humans8 has remained elusive. Moreover, the lack of structures for any eukaryotic TSEN complex either on its own or in complex with a pre-tRNA has hindered our understanding of tRNA recognition and processing. The only available TSEN structures include an NMR structure of TSEN1512, and a crystal structure of TSEN15 bound to the C-terminal region of TSEN3413. The eukaryotic TSEN proteins are thought to have evolved from an archaeal ancestor called EndA, which forms homo-oligomers that recognize a stereotypical bulge-helix-bulge (BHB) RNA motif. How the eukaryotic TSEN complexes evolved to accommodate recognition of full pre-tRNA substrates is unknown8,14–16.
The human TSEN complex is known to associate with the polyribonucleotide 5’-hydroxyl-kinase CLP18, which also functions in mRNA 3’-end processing17. CLP1’s association with the TSEN complex is perplexing, given that its kinase activity would block RTCB mediated ligation of the exon halves. In vitro reconstitution of with the human TSEN complex has established that CLP1 is not required for intron removal13,18. In vivo, CLP1 likely plays an important regulatory role in tRNA splicing by inhibiting exon ligation and/or targeting exonucleolytic decay of the intron2,5,18. Mutations in CLP1 and all four TSEN subunits are associated with multiple subtypes of the PCH family of inherited neurodevelopmental disorders19–22. PCH subtypes are characterized by genotype and clinical features including microcephaly, intellectual disability, locomotor dysfunction and premature death23. Recent studies utilizing PCH models of CLP1 and in vitro derived human motor neurons, suggest that impaired mRNA 3’-end processing and not tRNA splicing may underscore PCH-1024,25. However, the etiology of TSEN associated PCH subtypes remains poorly understood.
Here, we report near atomic resolution cryo-EM structures of the human TSEN complex bound to intron-containing pre-tRNAs. The structures unveil the arrangement and function of all four subunits and reveal a structural core within the complex that is conserved from archaeal ancestors. The structures identify extensive eukaryotic specific interfaces within the complex that mediate recognition of the full-length pre-tRNA. Our structures also establish the TSEN54 subunit as more than a simple molecular ruler, it supports tRNA binding and serves as a scaffold for the two endonuclease subunits, TSEN2 and TSEN34. Finally, the human TSEN structures reveal the structural environment of PCH-causing missense mutations, suggesting that loss of TSEN stability may contribute to neurodevelopmental disease.
Results and Discussion
Structure of the human TSEN complex
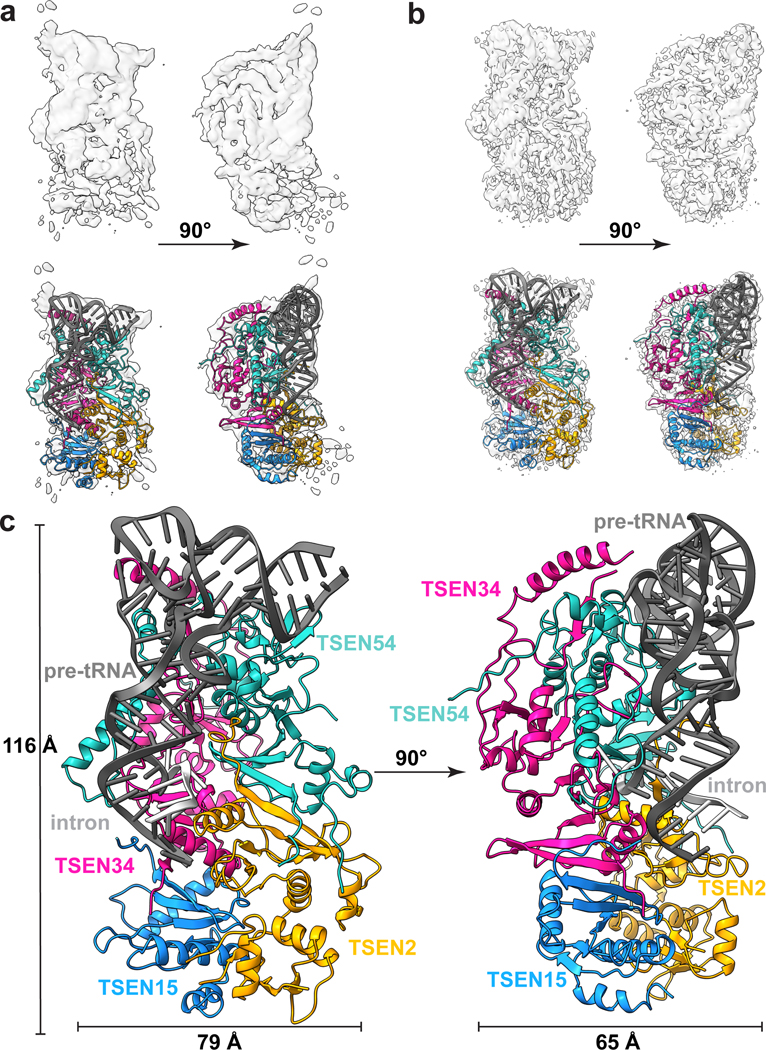
We solved two cryo-EM structures of the human TSEN complex bound to intron-containing pre-tRNA-ARGTCT-2–1 (Fig. 1). Two different approaches were used to trap the complex in a pre-cleavage state with the intron intact. First, we purified the recombinant wildtype(wt)-TSEN complex using a multi-cistronic bacterial expression system, which we previously reported is sufficient for intron cleavage18. We incubated wt-TSEN with pre-tRNA-ARG, containing a 15-nt intron wherein the 2’-hydroxyl at both the 3’ and 5’ cleavage sites was modified to fluorine to prevent catalysis26. The structure of wt-TSEN was solved to a resolution of 3.9 Å (Fig. 1a, Extended Data Table 1) and was sufficient for rigid-body docking of the tRNA and the four TSEN subunits using a combination of previously determined structures and AlphaFold models (Extended Data Fig. 1)12,13,27,28. The overall density of the pre-tRNA was well resolved, except in the area surrounding the 5’ cleavage site.
Fig. 1|. Cryo-EM structures of the human TSEN complex.
Cryo-EM reconstructions of: a. wt-TSEN•pre-tRNA and b. endoX-TSEN•pre-tRNA, with cartoon overlays below. Due to its improved resolution, the endoX reconstruction was used to build and refine the overall structure. c. Model of the human TSEN complex at 0° and 90°, with overall dimensions of 116 × 79 × 65 Å. The subunits are colored as follows: TSEN54 (teal), TSEN34 (pink), TSEN2 (orange), TSEN15 (blue). The pre-tRNA is shaded in dark grey, with the intron in light grey.
We also solved a structure of the TSEN complex in the presence of the CLP1 RNA kinase. Although the TSEN complex is sufficient for cleaving tRNA introns in vitro13,18, CLP1 is a well-established interaction partner17,22. To prevent intron cleavage, we mutated the three catalytic residues of both TSEN endonucleases to alanine (TSEN34H255A/K286A/Y247A/ TSEN2Y369A/H377A/K416A). We purified the endonuclease-deficient (endoX)-TSEN•CLP1 complex from a mammalian overexpression system using an affinity tag on CLP1, and incubated the complex with an in vitro transcribed unmodified pre-tRNA-ARGTCT-2–1. The structure of the endoX-TSEN•CLP1 complex was solved to an overall resolution of 3.3 Å (Fig. 1b, Extended Data Fig. 2 and Extended Data Table 1). Well resolved density was visible for all four TSEN subunits and the pre-tRNA (Extended Data Fig. 3), but we did not observe any well-ordered density for which we could accurately assign to CLP1. Overall, both the wt-TSEN and endoX-TSEN cryo-EM reconstructions are very similar. Neither reconstruction contains well resolved density for the intron surrounding the 5’ cleavage site, suggesting that this region is flexible. Due to its improved resolution, the endoX-TSEN•CLP1 reconstruction was used to build the atomic model of the TSEN complex (Fig. 1c, Extended Data Fig. 2).
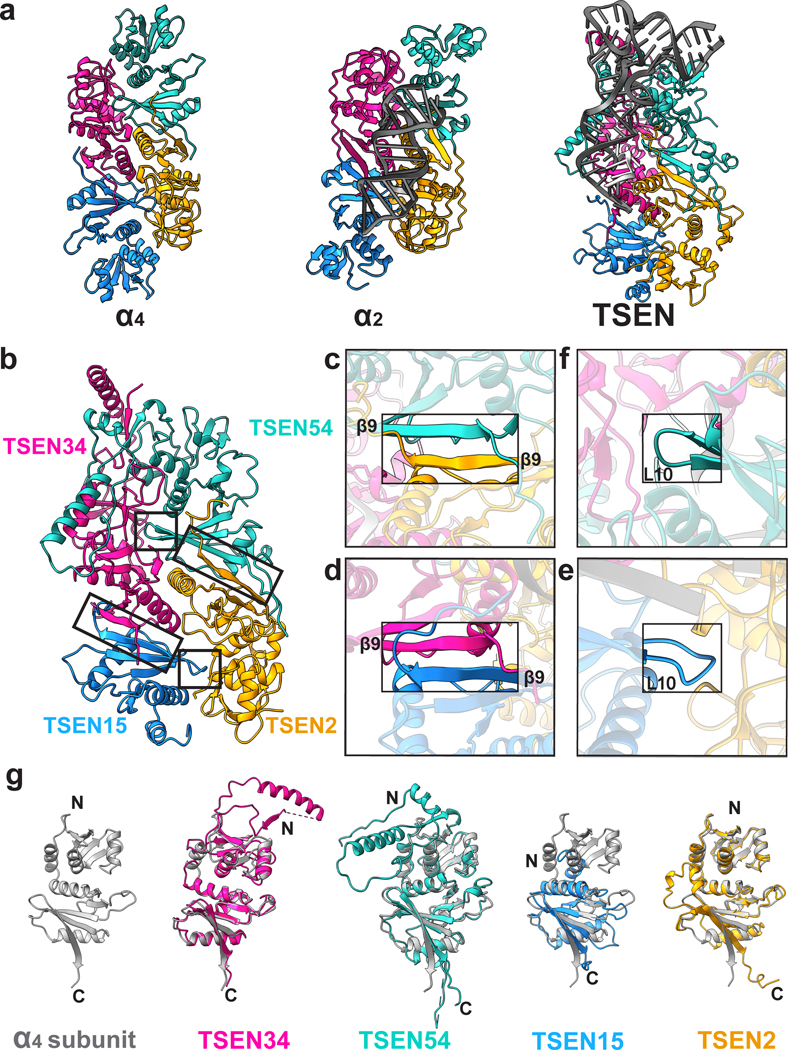
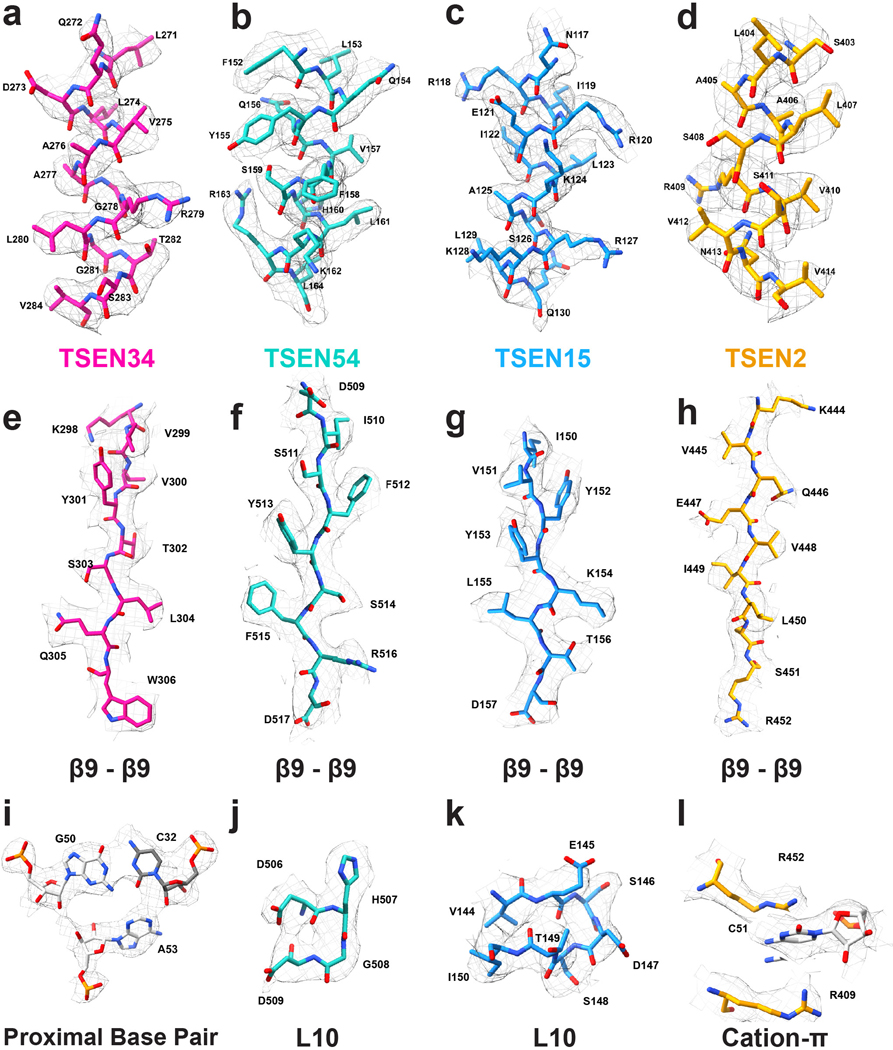
The four subunits of the TSEN complex form an interconnected, rectangular shaped structure. Overall, the core of the human TSEN complex is structurally similar to the four different families of archaeal TSEN complexes (Fig. 2 a). Previous structural studies of the archaeal enzymes established that the interfaces within the complex are mediated by two key features: a C-terminal β-sheet (β9) and loop (L10)14,16,29. Both structural features are conserved in the human TSEN complex, reinforcing the hypothesis that the eukaryotic TSEN complex evolved from a common archaeal ancestor30. Both the TSEN54-TSEN2 and TSEN34-TSEN15 interactions are mediated by anti-parallel β-β interfaces formed by the final β-strand from each subunit (Fig. 2 b–d). The same TSEN34-TSEN15 interface was also recently observed in a crystal structure of a portion of the C-terminal domain of TSEN34 bound to TSEN1513. The TSEN34-TSEN15 and TSEN54-TSEN2 dimers are held together by the L10 loop interface (Fig. 2 b,e,f). The negatively charged L10 loop in TSEN15 (residues 144–150) inserts into a positively charged cleft in the TSEN2 subunit (Fig. 2 e). Similarly, the L10 loop in TSEN54 (residues 506–509) inserts into a cleft in the TSEN34 subunit (Fig. 2 f). Beyond these conserved interfaces, the human TSEN complex also contains additional interfaces not found in the archaeal enzymes (Fig. 2 g). The N-terminus of TSEN34 and TSEN54 contain large insertions with no structural homology to the archaeal enzymes and TSEN54 forms an extensive interface with TSEN34. The TSEN34 and TSEN54 subunits also have the largest buried surface area within the complex. Additionally, while there are no interfaces between TSEN54 and TSEN15 in the structure, TSEN2 and TSEN34 form interfaces near the active sites.
Fig. 2|. The human TSEN complex retains the core architecture from the archaeal complex.
a. Structures of a homotetrameric (α4, PDBID: 1A79), homodimeric BHB bound (α2, PDBID:2GJW), and the pre-tRNA bound TSEN complex, colored to mimic the arrangement of the subunits within the TSEN complex (TSEN54 – teal, endonuclease TSEN2 - orange, endonuclease TSEN34 – pink, TSEN15 – medium blue). b. The TSEN complex retains β9-β9 interactions at the C-terminal domain of endonuclease/structural protein interfaces: c. TSEN2:TSEN54 and d. TSEN34:TSEN15. The L10-loop of the structural proteins e. TSEN54 and f. TSEN15 link the β9-β9 heterodimers together. g. A single archaeal homotetramer α subunit (PDBID: 1A79, grey) superimposed with each individual TSEN subunit.
TSEN54 mediates CLP1 binding
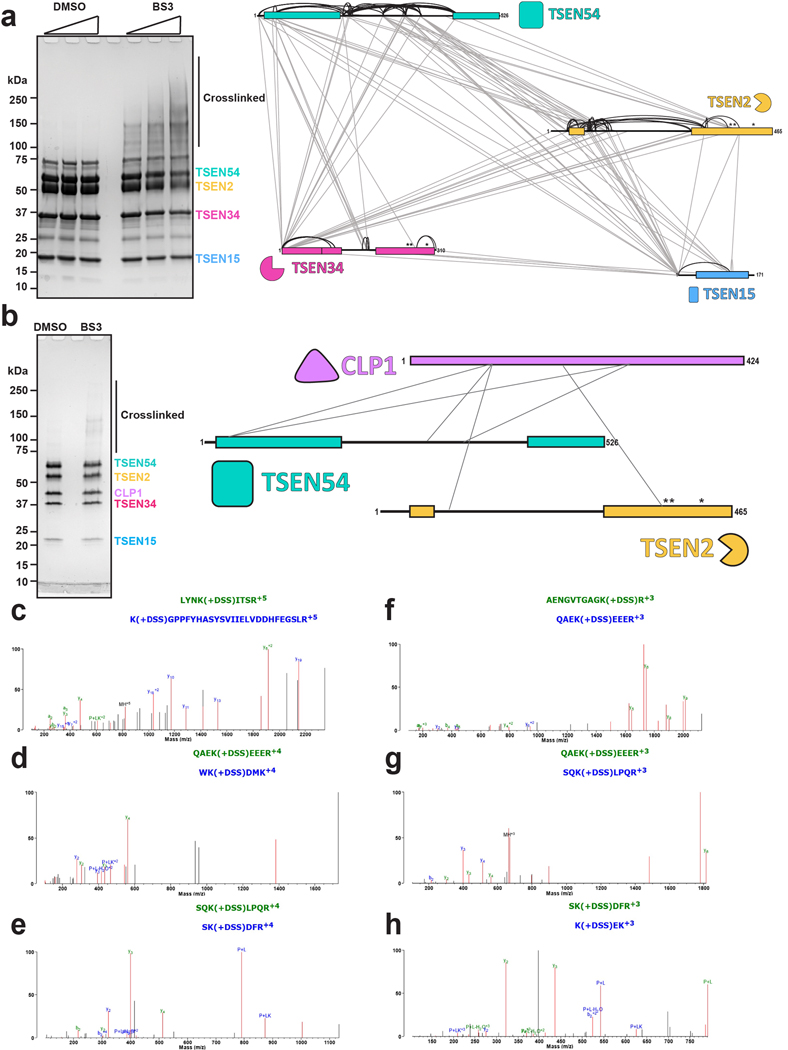
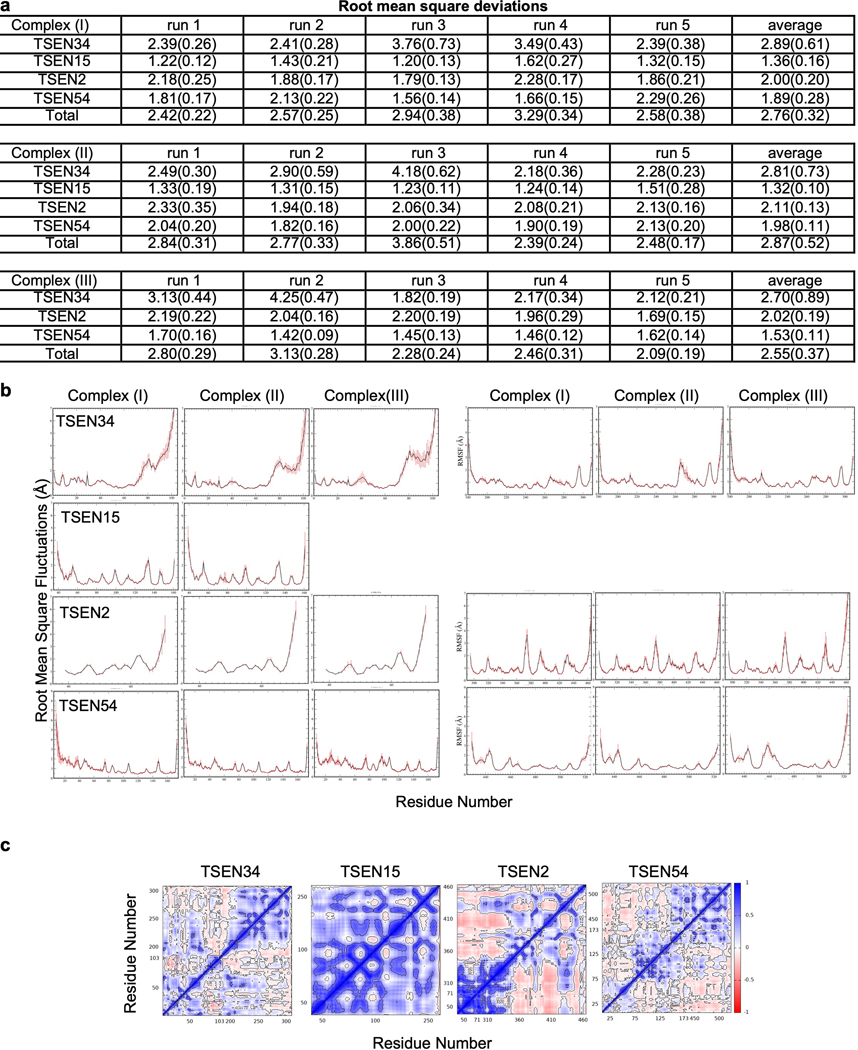
We performed cross-linking mass spectrometry analysis with a lysine reactive cross-linker on the recombinant TSEN complex to gain additional insight into the subunit interfaces (Extended Data Fig. 4). Three of the four TSEN subunits contain large, disordered regions that are not visible in the reconstructions5. Further, we did not observe well-ordered density for the CLP1 kinase in the reconstruction, suggesting that it either associates with the complex through a dynamic/disordered region, or it dissociates from the complex during cryo-EM grid preparation. We detected several crosslinks within structured regions of the TSEN core that support the cryo-EM structure such as crosslinks between the C-terminal halves of TSEN54 and TSEN2 (Extended Data Fig. 4a). However, the majority of the crosslinks that we detected are mediated across disordered regions of TSEN2, TSEN34, and TSEN54. These crosslinks provide additional insight into interactions mediated by these flexible and disordered regions (Extended Data Fig. 4a). Crosslinking was carried out in the absence of tRNA, revealing that the core complex likely does not undergo large scale conformational changes upon binding tRNA. This was supported by molecular dynamics (MD) simulations of the complex, with and without RNA bound, in which we did not observe large conformational changes (Extended Data Fig. 5). MD simulations further revealed the occasional loss of the TSEN15 subunit from the rest of the TSEN complex, likely because of its overall negative charge. The RMSD plots for the TSEN complex with and without RNA as well as the RNA bound complex without TSEN15 were stable over the trajectory used for MD simulations and across three replicates (Extended Data Fig. 5a).
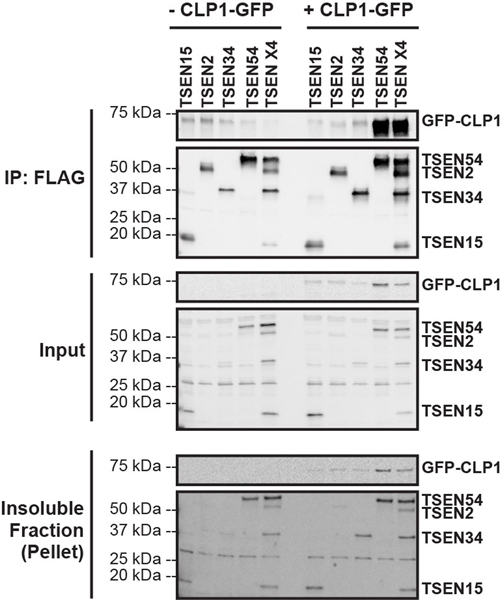
We also conducted cross-linking mass spectrometry analysis of the TSEN•CLP1 complex. We observed fewer crosslinks because the overall protein concentration was lower for the TSEN•CLP1 sample, but we detected high confidence crosslinks between CLP1 and both TSEN2 and TSEN54 (Extended Data Fig. 4b–h), suggesting that TSEN2 and TSEN54 may mediate CLP1 binding to the TSEN complex. To test this hypothesis, we carried out immunoprecipitations from cells transfected with CLP1 and either individual TSEN subunits or the full TSEN complex and found that TSEN54 was sufficient to mediate co-immunoprecipitating CLP1 (Extended Data Fig. 6).
Structure of pre-tRNA reveals similarities to mature tRNAs
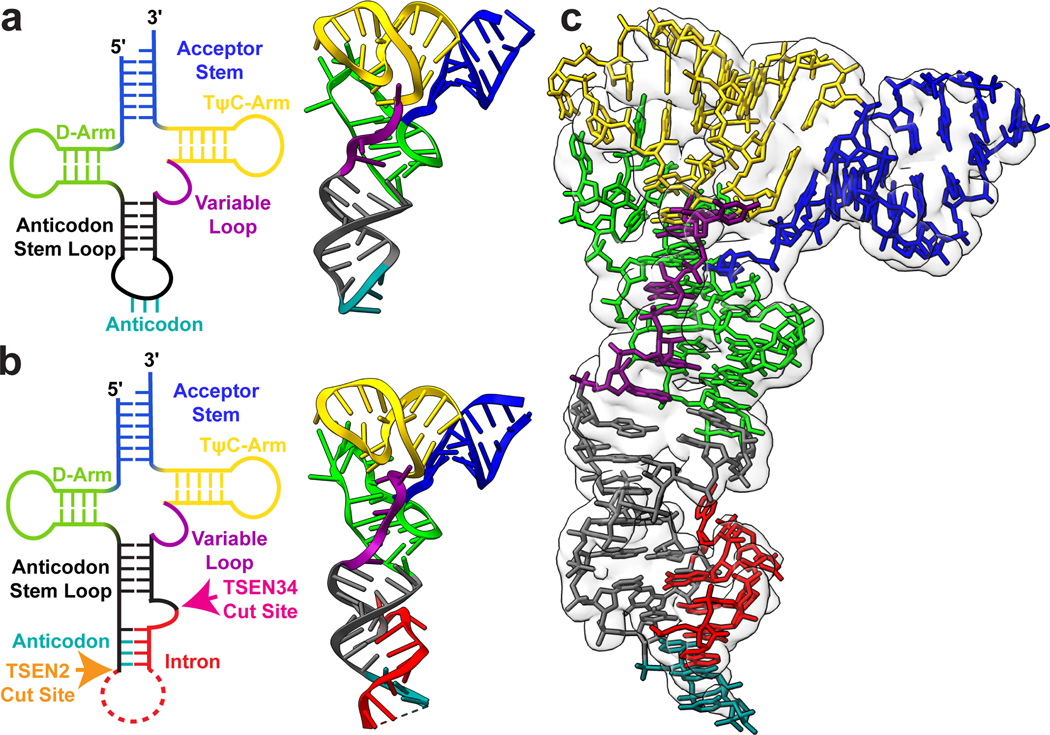
Beyond the architecture of the TSEN complex, our cryo-EM reconstruction also revealed the overall structure of an intron-containing pre-tRNA. Mature tRNAs contain several distinct structural features including, the accepter stem, D and TΨC arms, a variable loop, and ASL (Fig. 3). Chemical probing experiments suggested that the accepter stem along with the D and TΨC arms of intron-containing tRNAs is identical to mature tRNAs31. Overall, our cryo-EM structure confirms that the structure of TSEN bound pre-tRNA is very similar to the structure of mature tRNA, except for the ASL containing the intron (Fig. 3). Bending in the ASL region of the bound pre-tRNA suggests that the TSEN complex causes a slight melting of the ASL to position it for cleavage. Previous studies have established that eukaryotic introns are located between nucleotides 37 and 38 of the mature tRNA, forming a specific RNA structure known as the bulge-helix-bulge (BHB) or a more relaxed bulge-helix-loop (BHL) motif2. The anticodon-intron (A-I) helix contains the anticodon which base pairs with the intron and is flanked by single-stranded bulges/loops where cleavage occurs. Within our cryo-EM reconstructions, the density was sufficient to build a model for the A-I helix along with the bulge around the 3′-cut site (Fig. 3c). The loop encompassing the 5′-cut site is unstructured in both cryo-EM reconstructions, suggesting that this portion of the intron is highly flexible. This is not surprising given that the TSEN complex can accommodate large insertions in this loop that do not disrupt cleavage2,32.
Fig. 3 |. Structure of the pre-tRNA reveals a familiar architecture to mature tRNA.
2D and 3D structures of a. unmodified mature tRNA-PHE (PDBID: 3L0U) and b. intron containing pre-tRNA-ARG structure, with the acceptor stem (blue), D-Arm (lime green), anticodon stem loop (dark grey), anticodon (teal), intron (red), variable loop (purple) and TψC-Arm (yellow) labeled. The cut sites for TSEN2 (orange) and TSEN34 (pink) are labelled on the pre-tRNA 2D structure. c. Electron density map around the pre-tRNA, with the pre-tRNA structure docked into the density map.
pre-tRNA Recognition by the TSEN complex
How the eukaryotic TSEN complex recognizes pre-tRNAs has been a long-standing question. A crystal structure of an archaeal EndA engaging a BHB substrate established how conserved features surrounding the EndA active sites position the bulged residues for cleavage9. For EndA enzymes, the BHB motif is strictly required for recognition and cleavage, which is important because archaeal tRNA genes have introns located in various regions of the pre-tRNA33. In contrast, the human TSEN complex relies on recognition of additional tRNA features that mediate pre-tRNA recognition and it can cleave tRNA substrates with a more relaxed bulge-helix-loop (BHL) motif10,34. However, the eukaryotic complex can also cleave the universal BHB motif independent of the rest of the tRNA suggesting that recognition and cleavage of the BHB motif is a universal feature of TSEN nucleases35,36.
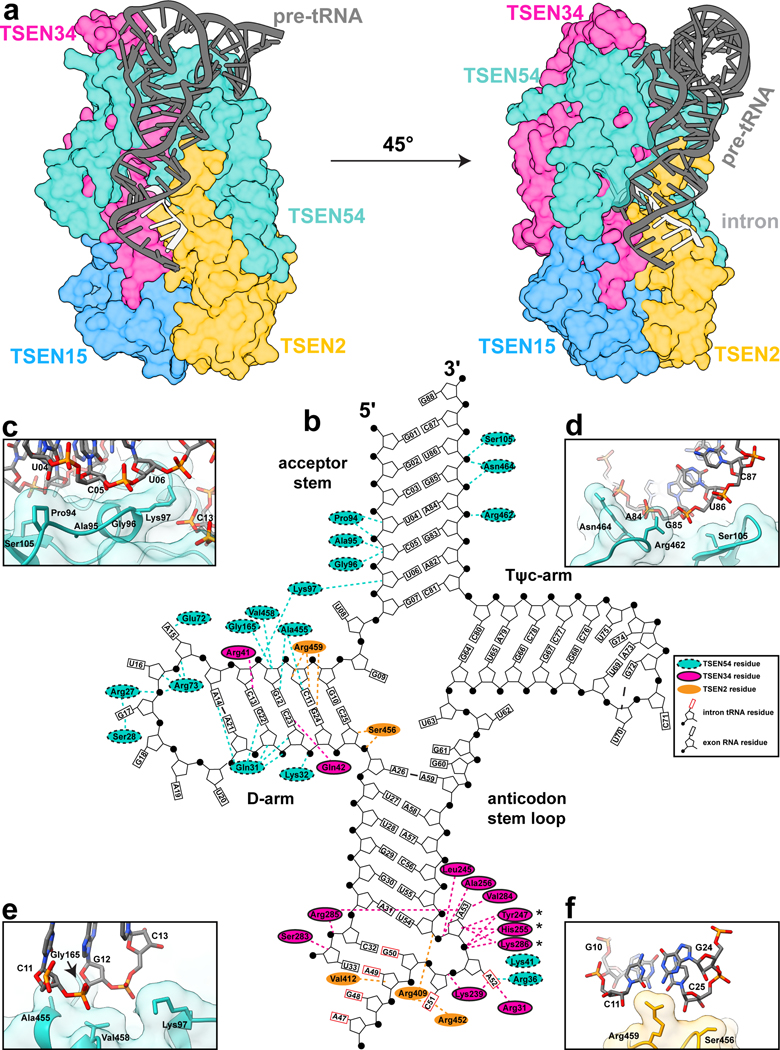
The cryo-EM structure revealed an extensive interface formed between the TSEN complex and several regions of the tRNA (Fig. 4a). The TSEN complex primarily interacts with the upside-down L-shaped tRNA along the under-side of the L, at the intersection of the acceptor-stem and stacking of the D-arm and ASL. TSEN54 was previously designated as a ‘molecular ruler’ based on studies demonstrating that insertions in specific regions of the pre-tRNA can impact the cleavage site9–11. TSEN54 contains the largest RNA interface and specifically interacts with the acceptor stem and D-arm of the tRNA, predominantly through interactions with the phosphate backbone (Fig. 4b–e). The acceptor stem is positioned along the top of the complex by TSEN54 and there are no interactions with the T-arm. Previous work revealed that while the acceptor stem is not required for intron cleavage, its removal significantly impacts the kinetic parameters of cleavage37, confirming the significance of the TSEN54-acceptor stem interface in anchoring the tRNA within the TSEN complex.
Fig. 4 |. Recognition of pre-tRNA features by the TSEN complex.
a. Surface representation of the TSEN complex with a cartoon of the pre-tRNA, showing significant interaction between TSEN54 and multiple regions of the pre-tRNA. Colors: exons (dark grey), intron (light grey), TSEN54 (teal), TSEN34 (pink), TSEN2 (orange), TSEN15 (blue). b. Cloverleaf 2D tRNA structure with selected TSEN interfaces labeled. TSEN-tRNA interfaces mediated by hydrogen bonds and hydrophobic contacts were identified with NucPlot41 and visual inspection of the cryo-EM reconstruction. Regions of the tRNA from the intron are indicated with the rectangle, representing the base, boxed in red. * indicates the TSEN34 catalytic residues which were all mutated to alanine to prevent cleavage. Interfaces between TSEN54 and c. and d. the acceptor stem e. the D-arm. f. Interface between the D-arm and TSEN2.
Our structure supports previous biochemical data suggesting a central role for TSEN54 in tRNA recognition, but also provides detailed insight into how TSEN54 engages tRNA and supports the overall architecture of the complex. Many of the TSEN54-tRNA interactions are mediated by the N-terminal region of TSEN54, which has no sequence or structural homology to the archaeal EndA proteins (Fig. 2g). In addition to TSEN54, the two endonucleases also mediate TSEN-RNA interfaces within the D-arm and the ASL (Fig. 4b,f). Similar to TSEN54, TSEN34 contains several insertions, not found in archaeal EndA, that mediate tRNA binding. These include a loop (TSEN34 residues 33–48) in the N-terminal domain that interacts with the D-arm, and a large alpha-helix positioned near the T-arm. The local resolution of this alpha-helix is poor, suggesting it is dynamic.
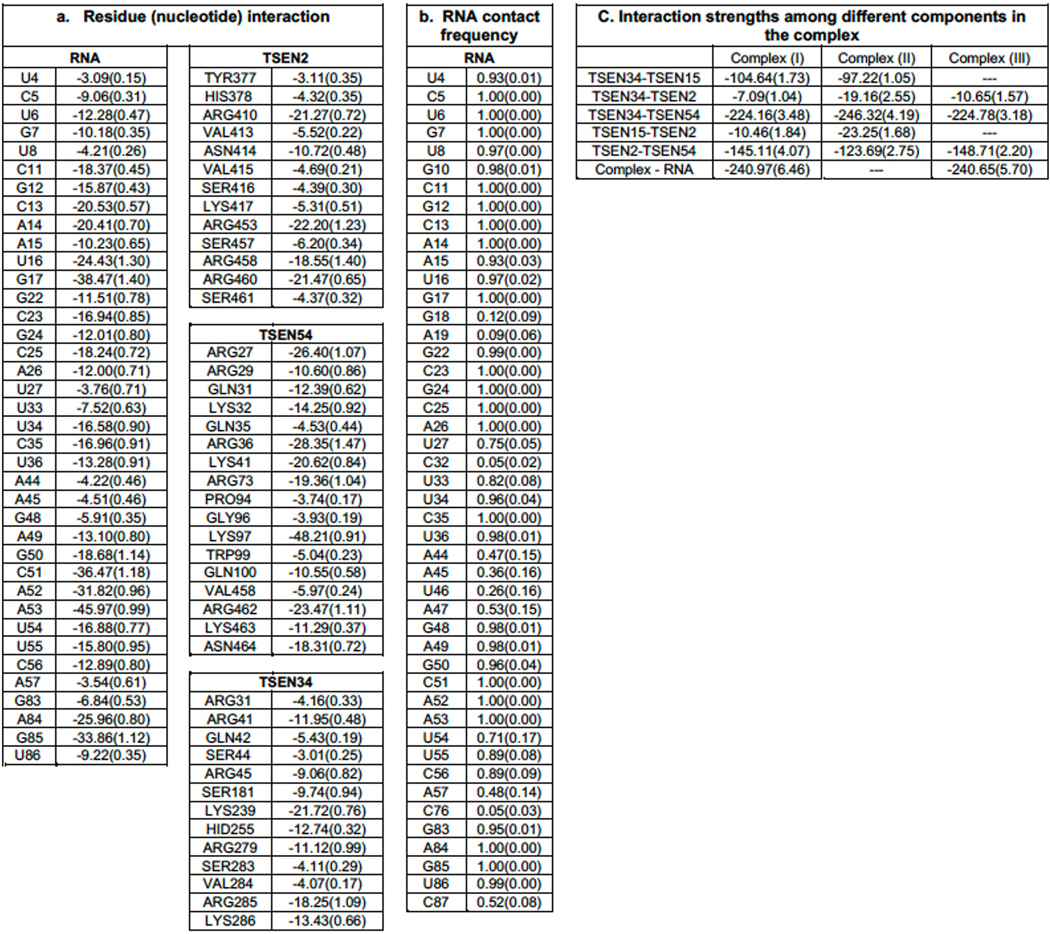
To further highlight the significance of the RNA-protein interfaces within the cryo-EM structure, we estimated the free energy of binding by the Molecular Mechanics-Generalized Born and Surface Area (MM-GBSA) approach using the configurations collected from our MD simulations. The calculated free energy of binding for residues from TSEN2, TSEN34, and TSEN54 is shown in Extended Data Table 2. From this analysis, we observed that TSEN54 provides the largest contribution to RNA binding. Collectively, our data suggest that TSEN54, TSEN34, and TSEN2 are each important for recognition of the tRNA. Although not visible in the structure, we hypothesize that TSEN15 may support the placement of the intron surrounding the 5′ splice site by providing structural support to TSEN34 and TSEN2. The overall negative charge of TSEN15 is also perplexing but could be important to promote the orientation the pre-tRNA and/or release of the cleavage products. Additional structures with ordered RNA in the 5′ splice site will be needed to fully understand if TSEN15 contributes directly or indirectly to tRNA binding.
The TSEN complex has a conserved catalytic core
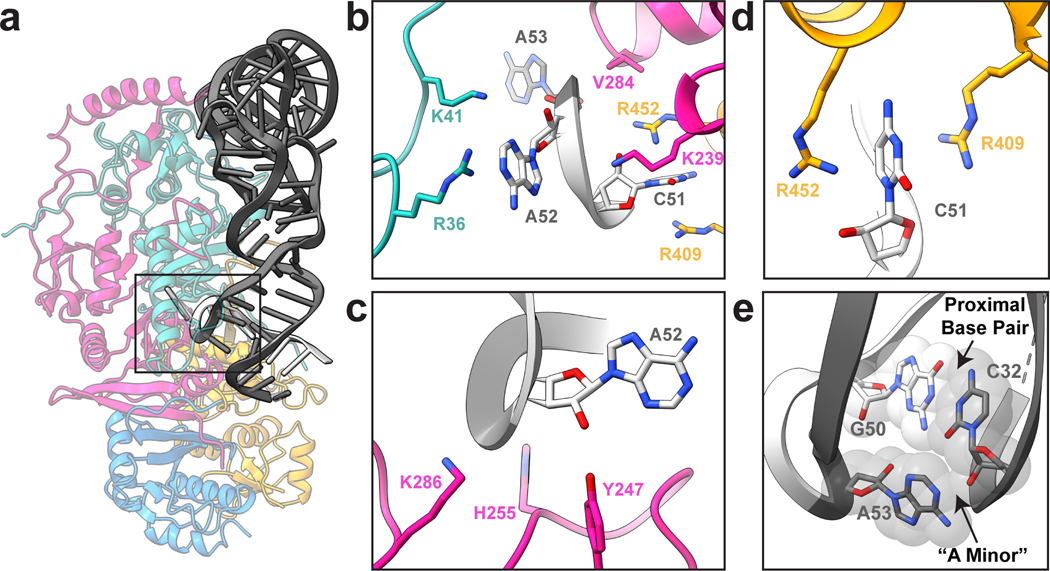
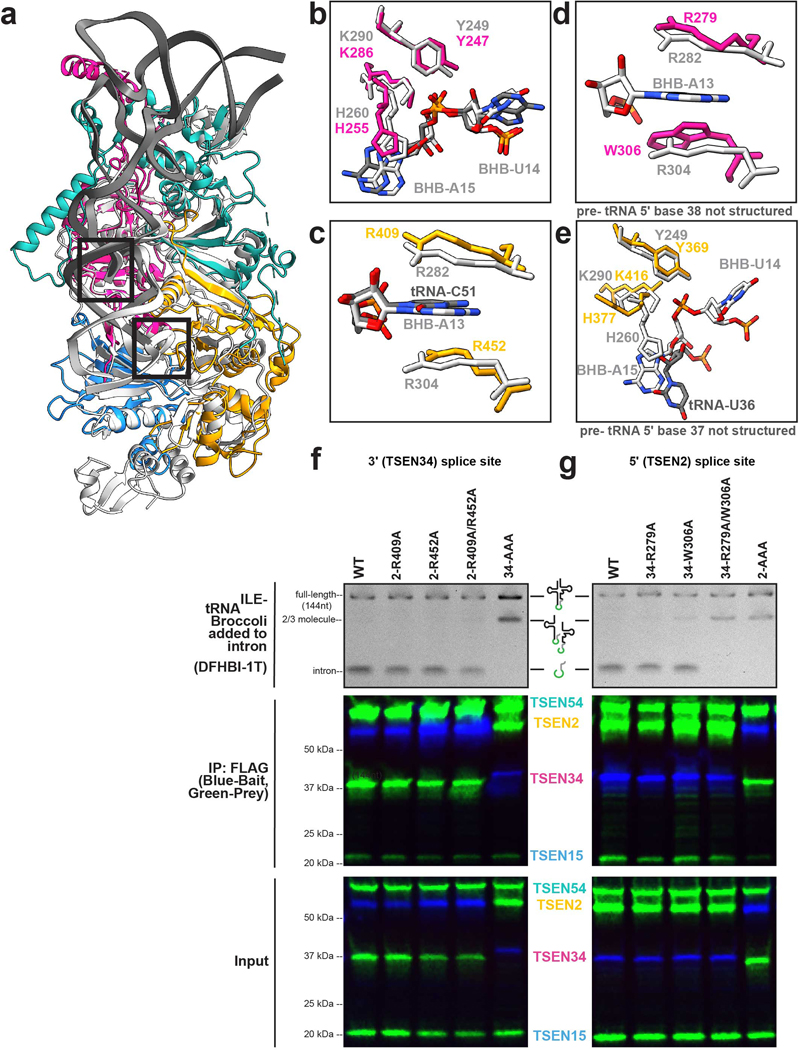
The cryo-EM structure also provided insight into cleavage at the 3′ splice site (Fig. 5a). The 3-nt bulge in the 3′ splice site is well ordered in our cryo-EM structure, in contrast to the 5′ splice site. Analogous to the archaeal EndA•BHB structure from Archaeglobus fulgidus, the 3-nt bulge forms an open knot-like conformation, presumably to orient the RNA for cleavage9. This interaction is mediated primarily by residues from TSEN34 including V284 and K239, but is further mediated by TSEN2(R409, R452) and TSEN54(R36, K41) (Fig. 5b). The close proximity of residues from TSEN54 to the 3′ splice site appears to be a feature unique to the eukaryotic TSEN complex. To prevent cleavage, we mutated the three catalytic residues from TSEN34 (H255, K286, Y247) to alanine, however in our reconstruction we observe a low occupancy of these active site residues, sufficient to roughly model side chain positions for these residues (Fig 3c). This heterogeneity is the result of purifying the TSEN complex, using CLP1 as bait, from a mammalian expression line containing the endogenous enzymes. We attempted to resolve this heterogeneity through cryo-EM data processing but were unsuccessful at generating a high-resolution map with strong density for the WT TSEN34 catalytic residues, most likely because the population of these particles is very small. The catalytic triad composed of histidine, lysine, and tyrosine residues is well conserved across tRNA splicing nucleases in archaea and eukaryoties7,18,38. Superposition of the A. fulgidus EndA•BHB structure (Extended Data Fig. 7a) reveals that these catalytic residues are in similar positions (Extended Data Fig. 7b–e).
Fig. 5 |. Conserved catalytic core at the 3’ splice site.
a. Cartoon structure of the complex with a box highlighting the 3’ splice site. b. Overview of the three nucleotide bulge in the 3’ splice site with important interactions labeled including TSEN54 (teal) (R36 and K41), TSEN34 (pink) (V284 and K239), and TSEN2 (orange) (R409 and R452). c. Modeled positions of the catalytic residues from TSEN34 (Y247, H255, and K286), which were mutated to alanine to prevent cleavage. d. TSEN2 residues R409 and R452 form a cation-π interaction with the C51 RNA base of the intron to stabilize the bulge. e. The ASL contains the proximal base pair (C32 and G50), important for splicing in vivo. The C32 base forms an A-minor like motif with A53 from the bulge.
The 3′ splice site is further supported by a cross-subunit interaction that stabilizes the first bulged nucleotide. The A. fulgidus EndA•BHB crystal structure revealed that there is cooperativity between the splice sites in archaeal nucleases that is mediated by a cross-subunit cation-π interface9. The bulged nucleotide is held in place by a pair of residues (R:R or R:Y/W) from the opposite endonuclease, forming a cation-π sandwich. Earlier studies of the yeast TSEN complex suggested that this cross-subunit cooperativity was required for cleavage at the 5′ splice site but dispensable for cleavage at the 3′ splice site39. Sequence comparisons across multiple EndA/TSEN nucleases suggest that the equivalent residues in human TSEN2 that form the cation-π interface are R409 and W45339; however, our structure revealed that R452, not W453, mediates the cation-π sandwich in the 3′ splice site (Fig. 5d, Extended Data Fig. 7c). Although we do not observe tRNA intron density in the 5′ splice site, we modeled RNA into the active site by superposition with the A. fulgidus EndA•BHB structure. We observe that the TSEN34 cation-π interface is formed by TSEN34 residues R279 and W306 (Extended Data Fig. 7d) and this superposition enabled us to roughly model the TSEN2 catalytic residues (Extended Data Fig. 7e). Both the EM-structure and modeling of the TSEN2 active site suggests that cross-subunit stabilization is a feature conserved across EndA/TSEN endonucleases.
We determined the significance of the human TSEN34 and TSEN2 cation-π interfaces in mediating tRNA cleavage within the human complex using nuclease assays. We generated single and double mutants of the TSEN2 (R409A, R452A) and TSEN34 (R279A, W306A) and isolated the TSEN complex containing the specific mutants by co-IP with an affinity tag on either TSEN2 or TSEN34 (Extended Data Fig. 7f,g). We incubated resin bound TSEN complex with a pre-tRNA containing the broccoli aptamer in the intron and then ran the quenched reaction on a denaturing gel. All of the single mutants were active, but the TSEN34 (R279A/W306A) double mutant abolished cleavage at the 5′ splice site (Extended Data Fig. 7g), supporting earlier work with the yeast complex39. In contrast, the TSEN2 (R409A/R452A) mutant retained activity (Extended Data Fig. 7f), revealing that this cation-π interface is dispensable for cleavage at the 3′ splice site39. Whereas the TSEN2 cation-π interface at the 3′ splice site is not required for pre-tRNA cleavage, its conservation suggests it plays an important role. We hypothesize that the TSEN2 cation-π interface is not essential because of the extensive contacts that can hold the pre-tRNA in the absence of the cation-π sandwich, although this interface may support cleavage of non-tRNA substrates. Given that the 5′ splice site is not well ordered in our structure, it likely has a weaker RNA interface and requires the cation-π sandwich.
The TSEN structure further revealed that the 3′ bulge is supported by the well characterized anticodon-intron interaction known as the proximal base-pair (PBP, C32-G50). Previous work has established the significance of the PBP in mediating cleavage at the 3′ splice site32,40. In Drosophila and human intronic tRNAs, this base pair is strictly a C-G, and the strength of the PBP impacts cleavage4,5,32. Analogous to the EndA•BHB structure, we observe that the PBP forms an A-minor interaction with the last nucleotide in the 3′ bulge, A53 (Fig. 5e). This A-minor motif stabilizes A53 within the 3′ splice site. Within the EndA•BHB structure, this motif is further supported by base stacking with the catalytic histidine9. Although the corresponding residue in TSEN34 (H255) is mutated in our structure, given its equivalent position to the archaeal enzymes, we assume it plays a similar role (Extended Data Fig. 7b). Overall, the structure reveals a remarkable amount of similarity between the 3′ splice sites of TSEN/EndA nucleases, supporting earlier work establishing that these enzymes employ common mechanisms to recognize the universal BHB motif and cleave the RNA by an RNAse-A like transesterification reaction35.
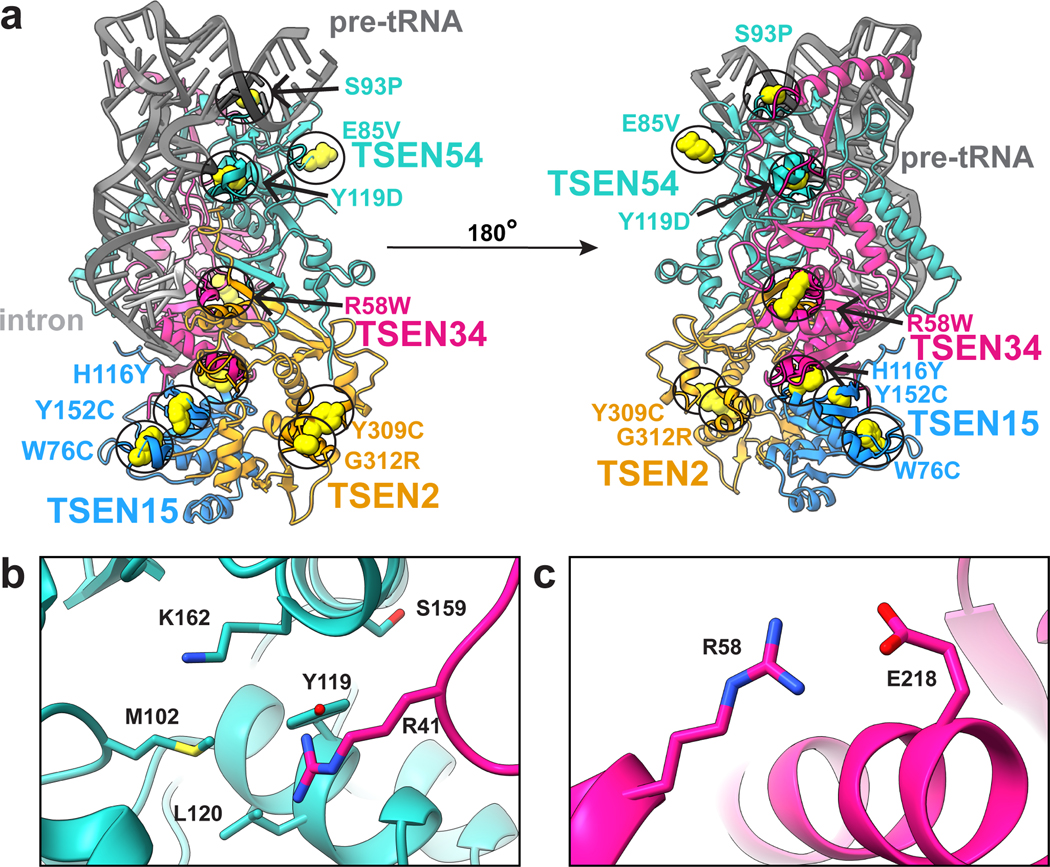
Structure reveals location of many PCH disease mutations
We mapped known PCH disease-associated mutations onto our TSEN structure, with the exception of TSEN54(A307S) which lies in a disordered region of TSEN54 not in the structure (Fig. 6a)5. Interestingly, none of the mutations appear within active site regions of TSEN34 or TSEN2, but instead are found on surface exposed interfaces that may be sites of interaction with other proteins or in regions that appear important for complex stability. For example, we predict that TSEN54-Y119 (Fig. 6b) will decrease protein stability as Y119 is buried within the core of the TSEN54 N-terminal domain. TSEN54-Y119 could also potentially interfere with tRNA binding as it lies within hydrogen bonding distance of TSEN34-R41, which associates with the D-arm of the pre-tRNA. We predict that TSEN34(R58W) (Fig. 6c) would decrease stability with the TSEN34 subunit as this would disrupt the salt bridge formed between TSEN34-R58 and TSEN34-E218. This supposition agrees with recent studies showing that a recombinant TSEN34(R58W) mutant fails to hinder complex formation or activity but impacts the thermal stability of the complex13. Analysis of the thermal stability of additional PCH mutants combined with structural mapping of the mutants suggests that loss of TSEN complex stability may be an underlying cause of PCH13.
Fig. 6 |. PCH mutations mapped onto the TSEN complex structure.
a. Previously reported PCH mutations are indicated with yellow spheres. b. TSEN54-Y119 is buried within the TSEN54 N-terminal domain and is in close proximity to TSEN34-R41 and several other TSEN54 (M102, L120, S159, K162) residues. c. TSEN34-R58 forms a salt bridge with TSEN34-E2.
Discussion
Our data provide significant insight into pre-tRNA recognition and processing by the human TSEN complex. The structure revealed an extensive tRNA-protein interface mediated by the TSEN54 subunit, which recognizes structural elements located in the acceptor stem, D-arm, and ASL of the pre-tRNA. Thus, instead of serving as a simple molecular ruler, TSEN54 works more like a set square that positions the tRNA along both sides of its L-shape. Beyond its capacity as a measuring device, TSEN54 provides structural support for the two endonuclease subunits. Our structure also revealed many similarities and differences between archaeal and eukaryotic TSEN complexes. Both share a conserved core that facilitates recognition of the universal BHB motif and cleavage of the phosphodiester backbone. Beyond the conserved core, the structure revealed how additional regions of the eukaryotic TSEN subunits mediate pre-tRNA binding. The eukaryotic TSEN subunits also contain large unstructured regions which were not resolved in the structure and that could be important for supporting recruitment of other RNA substrates or mediating interactions with regulatory factors such as the CLP1 kinase. Finally, this study provides a mechanistic framework for understanding how mutations within the human TSEN complex impact its structure and cause PCH.
Methods
Preparation of wt-TSEN
The wt-TSEN complex was purified from E.coli using a multi-cistronic expression vector18. Briefly, following IPTG overexpression cells were harvested, resuspended in lysis buffer (50 mM Tris pH 8.0, 10 % glycerol, 300 mM NaCl, 5 mM MgCl2, 0.1% Triton-X-100) with the addition of a cOmplete EDTA-free protease inhibitor tablet (Roche) , and lysed by sonication. Clarified lysate was incubated with HIS-60 (Takara) resin for 30–60 minutes and eluted with 250 mM lysis buffer supplemented with 250 mM Imidazole. The complex was further purified over a Superdex-200 (Cytivia) with the following buffer: 50 mM Tris pH 8.0, 200 mM NaCl, 5% glycerol, 5 mM MgCl2. For BS3 crosslinking, the complex was purified over a Superdex-200 (Cytivia) with the following buffer: 50 mM HEPES pH 8.0, 200 mM NaCl, 5% glycerol, 5 mM MgCl2. For cryo-EM, the purified complex was subsequently incubated with tRNA-ARG-2’F (purchased HPLC purified from Horizon) and further purified over a Superdex-200 pre-equilibrated with 20 mM HEPES pH 8.0, 100 mM NaCl, 5 mM MgCl2, and 2% glycerol. Protein-tRNA fractions were pooled and concentrated and then diluted 1:1 with 20 mM HEPES pH 8.0, 100 mM NaCl, 5 mM MgCl2 buffer.
Preparation of endoX-TSEN-CLP1 from HEK293 cells
The endoX-TSEN•CLP1 complex was expressed and purified from large scale suspension cultures of HEK293 freestyle cells following an established protocol18. Briefly, harvested cells were resuspended in HEK IP Buffer (50 mM Tris pH 8.0, 10% glycerol, 100 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 0.05% NP-40) with a cOmplete EDTA-free protease inhibitor tablet (Roche), lysed by nutation at 4 °C for 1 hour, clarified, and then incubated with anti GFP nanobody resin42. The resin was washed three times with IP buffer with the addition of 5 mM ATP. The complex was eluted from the resin by overnight TEV digestion to remove the C-terminal GFP tag from CLP1. The complex was then incubated with in vitro transcribed pre-tRNA-ARG18 followed by Superdex-200 purification. Protein-tRNA complexes were prepared as described above for wt-TSEN. For BS3 crosslinking, CLP1 contained a C-terminal FLAG-Tag and the sample was purified using ANTI-FLAG M2 Affinity Gel (Sigma), eluted with Pierce 3X FLAG peptide (Thermo) and then further purified as described above for wt-TSEN.
Protein-protein crosslinking
The TSEN•CLP1 (0.74 mg/mL) complex was crosslinked with 75 μM bis(sulfosuccinimidyl)suberate (BS3; Sigma). The wt-TSEN complex (1.6 mg/mL) was crosslinked with BS3 (75, 250, 500 μM). Crosslinking was quenched with Tris pH 8.0 to a final concentration of 30 mM on ice. Each crosslinked sample had a matching DMSO control. Samples were stored at −80 °C until processing and analysis. Sample processing and interpretation was performed as previously described43.
HEK293 cell immunoprecipitations and nuclease assays
The constructs for the TSEN•CLP1 experiments were pLexM-CLP1–TEV-GFP, pCDNA3.1-TSEN15-Flag, pCDNA3.1- TSEN2-Flag, pCAG-Strep-Flag-TSEN34, pLexM-TSEN54-Flag. For the cation-π experiments, the constructs were generated and cloned by Genscript into pCDNA3.1 vectors with a C-terminal (TSEN34) or N-terminal (TSEN2) Flag tag. The endonuclease triple mutants were cloned into pCAG-Strep-Flag-TSEN34 and pCDNA3.1- TSEN2-Flag. The non-bait TSEN proteins contained MYC tags.
Co-immunoprecipitation experiments were conducted using established methods18.
Briefly, 40mL suspension cultures of HEK293 Freestyle cells were transfected with 1 mg/mL total DNA using polyethylenimine (PEI) (1 μg/ml) and grown for approximately 72 hours. Cells were harvested by centrifugation and stored at −80 °C. 20mL of cells were then lysed on a nutator at 4 °C for ~ 30 minutes in 1.5 mL of lysis buffer containing 50 mM Tris pH 8.0 (@ RT), 100 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 10% glycerol, 0.05% NP-40, and cOmplete EDTA-free protease inhibitor (Roche). Cell lysates were cleared by centrifugation at 21,000 x g at 4 °C for 30 minutes. Cleared lysates were incubated with Anti-FLAG (Pierce™ Anti-DYKDDDDK Magnetic Agarose) resin, for approximately 1 hour, after which point the resin was washed three times with lysis buffer, using a magnetic stand to hold resin between washes. After the final wash, all lysis buffer was removed.
For the cleavage assays, reaction buffer (50 mM Tris pH 7.5 (@ RT), 50 mM KCl, 5 mM MgCl2, and 1 mM DTT with 1 unit/mL RNAsin (Promega), containing 500ng per reaction pre-tRNA-ILETAT-broccoli) was then added directly to the resin and allowed to incubate for 30 minutes at room temperature. Reactions were quenched using a 1:1 ratio of 4M UREA containing 3X RNA loading dye. The sample was then split and the RNA samples were separated on TBE-UREA gels (Novex), washed three times with water, and stained with 10 μM (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylene]-3,5-dihydro-2-methyl-3-(2,2,2-trifluoroethyl)-4H-imidazol-4-one (DFHBI-1T)(Tocris) in Tris pH 7.5 (@RT), 50 mM KCl, and 5 mM MgCl2. DFHBI-1T stained gels were imaged on a typhoon. The protein samples for both experiments were separated on SDS-PAGE gels (Bio-Rad) and transferred to PDVF (Bio-Rad). Blots were blocked using Intercept® (TBS) Blocking Buffer plus Tween-20 (Licor) and incubated overnight at 4 °C with respective primary antibodies (1:3000 anti-Flag (Sigma), 1:5000 anti-Myc (Millipore), 1:5000 anti-GFP (Roche)) in Intercept® (TBS) Blocking Buffer, washed three times with TBS-T, and incubated with secondary antibodies (1:5000 anti-mouse and anti-rabbit, Licor) in Intercept® (TBS) Blocking Buffer plus SDS and Tween-20. Blots were imaged by fluorescence on a Bio-Rad ChemiDocMP.
Cryo-EM grid preparation
Prior to freezing, grids were glow discharged (30 s, 15 mA at 0.37 mBar; Pelco Easiglow) and vitrified (3 μL sample, 3 s blot time) using an automatic plunge freezer (Leica). EndoX-TSEN•CLP1 was frozen on R1.2/1.3 Cu 300 mesh grids (Quantifoil) and TSEN (WT) was frozen on R2.4 Cu 300 mesh grids (Quantifoil).
Cryo-EM data collection
TSEN micrographs were collected using SerialEM on a Talos Arctica electron microscope at 200 keV with a Gatan K2 Summit detector and a Titan Krios at 300 keV with a K3 Bioquantum detector. Beam-induced motion and drift were corrected using MotionCor244 using Scipion3 software.
Cryo-EM image processing
Image processing was conducted using CryoSPARC45. The CTF parameters of the dose-weighted images were calculated using CTFFIND446. The complex 2D projections were initially selected using Blob Picker in CryoSPARC45 and extracted with box size of 400. Picked particles were cleaned using a series of 2D Classifications and then a subset of cleaned particles were used for training template for Topaz Extraction which uses machine-learning. The Topaz picked particles were extracted using a box size of 320, with a down sampling factor of 2. Particles from different datasets of the same sample were then combined and further processed with additional iterations and selections of 2D Classifications. Ab-initio Model Reconstruction was then used to generate initial 3D models, which were used to further separate particles at the 3D level by Heterogeneous Refinements. The cleaned particles were then re-extracted to remove the down sampling factor, prior to final refinement steps. The 3D maps for both endoX-TSEN and wt-TSEN were further refined using unsymmetrized Homogeneous Refinement, Global CTF Refinement, Non-Uniform Refinement (endoX-TSEN only) and tight-masked Local Refinements.
Model building and refinement
A combination of deposited PDB structures and alpha-fold models were used to construct the initial model of the TSEN complex and pre-tRNA. An unmodified tRNA-phe (PDB ID: 3L0U) was used as the starting point for building the pre-tRNA. The crystal structure of TSEN15-TSEN34 was used as the starting point for building TSEN15 (PDB ID: 6Z9U) and Alpha-fold models24,25 were used for TSEN34, TSEN54, and TSEN2. The starting models were individually docked into the reconstruction using UCSF Chimera47, followed by manual model building in Coot48. The structure was then refined through iterative rounds of manual model building and real-space refinement in Phenix49. The structure was validated using MolProbity50. Figures were prepared with ChimeraX51.
Molecular dynamics
Three different complexes were used in MD simulations; Complex (I) – TSEN34 (residues 1–103:181–309)/TSEN15 (residues 39–161)/TSEN2 (residues 39–71:295–464)/TSEN54 (residues 8–173:429–523) with the RNA present in the Cryo-EM structure (PDBID: 7UXA) , Complex (II) – all proteins from Complex (I) without the RNA, Complex (III) – all but TSEN15 from Complex (I). In the initial model of the RNA bound TSEN complex, only the missing segment of residues from 372 to 376 of TSEN2 was modeled in using the program, Modeller52. Protonation states of the histidine residues were selected using the web-based program, Molprobity50. All missing protons were introduced by using the leap module of Amber.1853. The complexes were solvated in a box of TIP3P water with the box boundary extending to 20 Å from the nearest protein atom (resulting in a total of 205708, 196569, and 204729 atoms in the rectangular simulation box of Complex (I), Complex (II), and Complex (III), respectively). Complex (I) contained 83 Na+ counter ions for neutralizing the charges while Complex (II) and Complex (III) had 2 and 74 Na+ ions, respectively. There were additional 116 Na+ and 116 Cl- ions added to the solution of Complex (I) providing the 100 mM effective salt concentration approximating that at the physiological conditions whereas Complex (II) contained additional 112 Na+ and 112 Cl- ions and Complex (III) had 116 Na+ and 116 Cl- ions. Prior to equilibration, the solvated complexes were sequentially subjected to (i) 500 ps of dynamics of water and all ions with fixed (or frozen) proteins and RNA (if present), (ii) 5000 steps of energy minimization of all atoms (the first 2000 steps with the steepest decent method followed by the conjugate gradient method for the rest), (iii) an initial constant temperature (300 K) - constant pressure (at 1 atm with the isotropic position scaling) dynamics at fixed protein to assure a reasonable starting density (~1 ns) while keeping the protein positions under constraints with a 10 kcal/mol force constant for 50 ps, (iv) a conjugate-gradient minimization for 1000 steps, (v) step-wise heating MD at constant volume (from 0 to 300K in 3ns), and 6) constant volume simulation for 10 ns with a constraint force constant of 10 kcal/mol applied only on backbone heavy atoms. After releasing all constraining forces within the first 10 ns of the next 40 ns equilibration period, the constant-temperature (300 K) constant-volume (NVT) production runs were performed for a 1.0 μs. Sampling was increased by performing 5 independent 1.0 μs NVT molecular dynamics simulations for each of the three complexes, resulting in a total of 15 microsecond simulations. The constant temperature was maintained using the Langevin dynamics with the collision frequency of 0.5 ps-1. All trajectories were calculated using the PMEMD module of Amber.18 with 1 fs time step. Long-range coulombic interactions were handled using the PME method with the cut-off of 10 Å for the direct interactions. The amino acid parameters were selected from the FF14SB force field of Amber.18. RNA was represented by the FF99bsc0-chiOL3 force field. Root mean square deviations, root mean square fluctuations, dynamic cross correlation matrices, and various types of relevant analysis were performed using the CPPTRAJ module of Amber.18 in conjunction with some in-house programs. At the salt concentration of 100 mM with the standard parameters, the MM/GBSA module was used to estimate residue-residue interaction energies for all protein residues. A total of 5000 configurations was selected at each nanosecond from the five 1.0 μs MD trajectories for each of the complex studied.
Reporting summary
Further information on research design in available in the Research Reporting Summary.
Data availability
Cryo-EM maps for wt-TSEN and endoX-TSEN have been deposited in the EMDB under the accession codes EMD-28755 and EMD-26856 respectively. The atomic model for endoX-TSEN has been deposited in the PDB under the accession code PDBID-7UXA. Mass spectrometry data has been deposited at MassIVE with the dataset identifier of MSV000091153. Source data including cross-linking mass spectrometry data and all un-cropped western blots are provided.
Extended Data
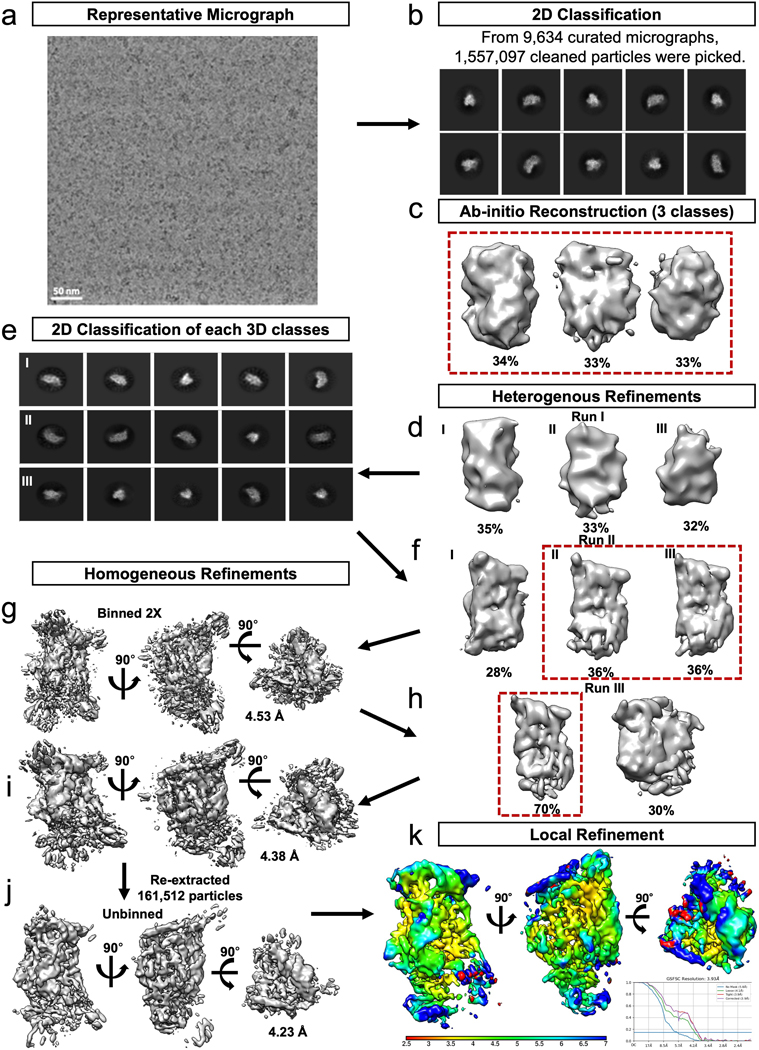
Extended Data Fig. 1 |. WT-TSEN cryo-EM processing workflow.
a. A representative micrograph, of curated 9634 micrographs. b. 1,557,097 particles were picked from the 9,634 curated micrographs, 10 representative 2D classes are shown. Particles contained in good classes were used to generate c. ab-initio reconstructions, three of which were selected and further refined using d. heterogenous refinement. The particles contained in the best three classes were further filtered using e. 2D classification and f. heterogeneous refinement prior to two classes being further refined using g. homogenous refinement, with 2x binned particles. h. A final round of heterogenous refinement resulted in one 3D class which was i. refined to an estimated 4.38 Å using homogenous refinement. j. Refinement continued with another round of homogeneous refinement resulting in a resolution of 4. Å following the unbinning of the final 161,512 particles with an estimated resolution of 4.2 Å. k. A local refinement resulted in a final map with an estimated resolution of 3.9 Å.
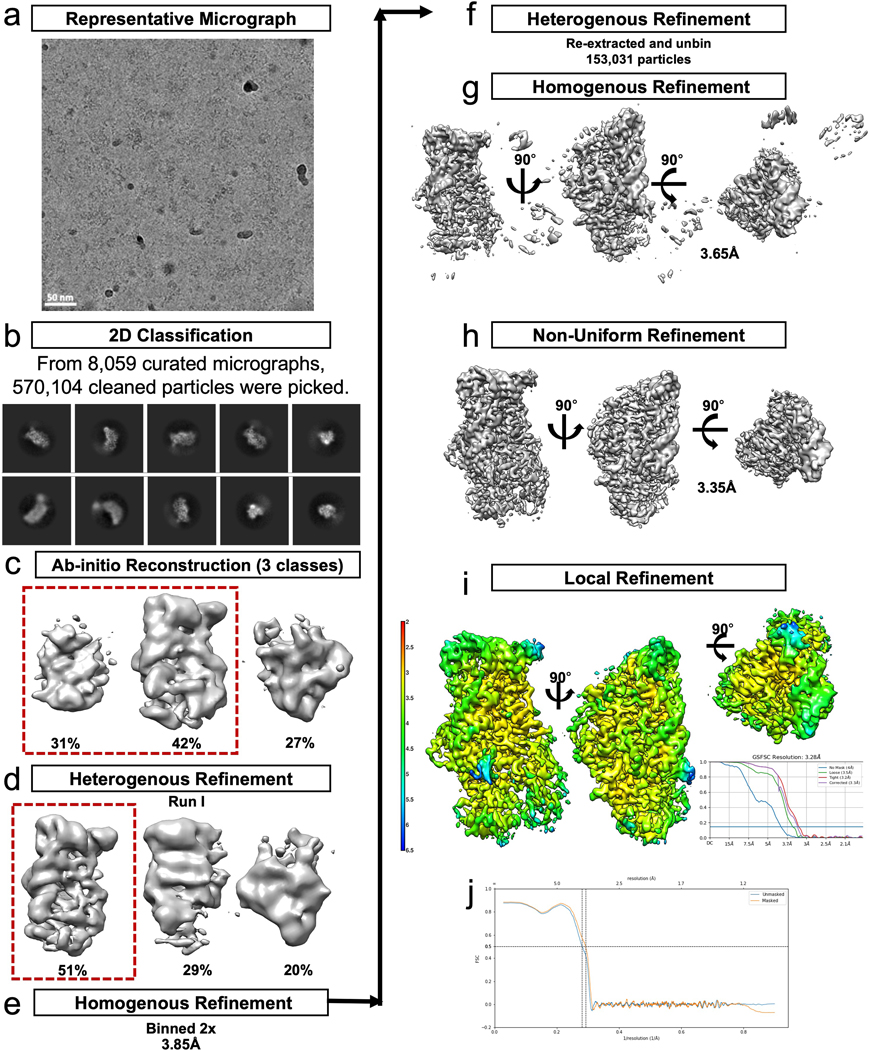
Extended Data Fig. 2. EndoX-TSEN structure cryo-EM processing workflow.
a. A representative micrograph of 8095 curated micrographs. b. 570,104 particles were picked from the 8059 curated micrographs and used for 2D classification. Particles from the good classes were used to generate c. ab-initio reconstructions, two of which were selected and further refined using d. heterogenous refinement. e. The particles contained in the best class were binned and refined with homogenous refinement. f. The particles were then reextracted and unbinned and used for another round of g. homogenous refinement followed by h. non-uniform refinement and i. local refinement resulting in a final map with an estimated resolution of 3.28 Å. j. FSC of the model to map
Extended Data Fig. 3. Example density for the TSEN complex.
Example densities for residues along helices from a. TSEN34, b. TSEN54, c. TSEN15, d. TSEN2, the β9-β9 interfaces for β-sheets for e. TSEN34, f. TSEN54, g. TSEN15, h. TSEN2 and density for the i. proximal base pair of the tRNA as well as the L10 loops for j. TSEN54, k. TSEN15, and the l. cation-π
Extended Data Fig. 4. BS3 crosslinking of TSEN complexes.
SDS-Page gels showing the DMSO control and crosslinked samples for a. wt-TSEN complex and b. TSEN Complex and CLP1. c-h. MSMS spectra of CLP1 peptides crosslinked to other members of the complex. Panels c (m/z 822.4292) and d (466.4795 m/z) show MSMS spectra of peptides arising from CLP1 and TSEN2. Panels e (m/z 412.2311), f (m/z 739.0369), g (m/z 671.3516), and h (m/z 398.5568) show MSMS spectra arising from crosslinks from CLP1 and TSEN54. Fragment ions that arise from green colored peptides are shown in green, fragments arising from blue colored peptides are shown in blue, and unassigned ions shown in black.
Extended Data Fig. 5. Molecular dynamics simulations account for stable complexes during dynamics.
a. Root mean square deviations (RMSD) of individual proteins and the complex averaged over each run and averaged over the five runs. TSEN34 has the largest deviations while TSEN15 shows smallest deviations irrespective of the RNA binding. Standard deviations are shown in parenthesis. The reference (Complex I) was the starting Cryo-EM configuration. Complex (II) is without tRNA and Complex (III) is without TSEN15. b. Root mean square fluctuations of individual residues averaged calculated during the microsecond dynamics and averaged over all runs. Standard deviations are shown as error bars. c. Representative dynamic cross correlation matrices of the protein components from Complex (I).
Extended Data Fig. 6. TSEN54 is the anchor that mediates the interaction of CLP1 with the TSEN complex.
Overexpression (in HEK cells) and immunoprecipitations of the individual TSEN proteins (lanes 1–4 and lanes 7–10) or full TSEN complex (lanes 5 and 11) in the absence (lanes 1–5) and presence (lanes 7–11) of CLP1 reveals strong association between CLP1 and TSEN54 (lane 10) as well as the full TSEN complex (lane 11). The experiments were conducted as previously reported1 using CLP1-TEV-GFP.
Extended Data Fig. 7 |. Structural comparison of the TSEN and EndA active site residues.
a. Overlay of TSEN + tRNA (colored as before) and EndA + BHB (grey, PDBID:2GJW) showing overall similar RNA binding. b. Overlay of the active site residues for the 3’ splice site and the c. cation-π residues from TSEN2 shown. d. Overlay of the cation-π residues from TSEN34 and the e. TSEN2 active site residues for the 5’ splice site. f. tRNA cleavage assays of a broccoli RNA -aptamer containing pre-tRNA-ILE using samples immunoprecipitated via the mutants of the 3’ splice site (pulled down by a FLAG tag on TSEN2, except for the 34 mutant) or g. 5’ splice site (pulled down by a FLAG tag on TSEN34, except for TSEN2 active site mutant). TSEN proteins that weren’t the pull-down target had a MYC-tag. The data shown is one representative experiment of three biological replicates.
Extended Data Table. 1. Energy contributions and contact frequencies from molecular dynamic simulations.
a. MMGBSA energy calculated from 5000 configurations collected from 5 MD runs. The standard errors are given in parenthesis. Only energies with magnitudes greater than 3 kcals/mol are shown. b. Contact frequencies of nucleotides with the TSEN complex. The frequencies were calculated from 5000 configurations and if any heavy atom of a nucleotide is within 3.2 A to any heavy atoms of the protein complex, then the nucleotide is selected to be in contact with the complex. Only 35 out of 80 nucleotides are in contact 75% or more of the time of the simulation. c. interaction energies among various components in each system, calculated using MMGBSA.
|
Supplementary Material
Table 1.
Cryo-EM data collection, refinement, and validation statistics
| CLP1-TSEN(AAA)-tRNA-Arg (EMDB-26856) (7UXA) | TSEN(WT)-2’F-tRNA-Arg (EMDB-28755) | ||
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 45000x | 81000x | 81000x |
| Voltage (kV) | 200 | 300 | 300 |
| Electron exposure (e–/Å2) | 54 | 60 | 60 |
| Defocus range (μm) | −0.8 to −1.8 | −1.2 to 2.2 | −1.2 to 2.2 |
| Pixel size (Å) | 0.93 | 0.53 | 0.53 |
| Symmetry imposed | C1 | C1 | |
| Initial particle images (no.) | 570,104 | 1,557,097 | |
| Final particle images (no.) | 153,031 | 161,512 | |
| Map resolution (Å) | 3.32 | 3.93 | |
| FSC threshold | 0.143 | 0.143 | |
| Map resolution range (Å) | 2.8–50.8 | 3.24–60.53 | |
| Refinement | |||
| Initial model used (PDB code) | 6Z9U, 3L0U | ||
| Model resolution (Å) | 3.25 | ||
| FSC threshold | 0.143 | ||
| Model resolution range (Å) | 3.2–3.4 | ||
| Map sharpening B factor (Å2) | 121.9 | ||
| Model composition | |||
| Non-hydrogen atoms | 8120 | ||
| Protein residues | 820 | ||
| Nucleotides | 78 | ||
| Ligands | 2 (Mg) | ||
| B factors (Å2) | |||
| Protein | 76.24 | ||
| Nucleotide | 81.90 | ||
| Ligand | 22.87 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 (0) | ||
| Bond angles (°) | 0.832 (0) | ||
| Validation | |||
| MolProbity score | 1.77 | ||
| Clashscore | 7.47 | ||
| Poor rotamers (%) | 0.88 | ||
| Ramachandran plot | |||
| Favored (%) | 94.78 | ||
| Allowed (%) | 5.22 | ||
| Disallowed (%) | 0.00 | ||
Acknowledgements
We thank Drs. Percy Tumbale and Joseph Rodriguez for their critical reading of this manuscript. We would like to thank Dr. Rick Huang and Allison Zeher for help with cryo-EM data collection. This work utilized the Krios at the NCI/NICE Cryo-EM Facility. This work was supported by the US National Institute of Health Intramural Research Program; US National Institute of Environmental Health Sciences (ZIA ES103247 to R.E.S., 1ZIC ES102488 to J.G.W., 1ZIC ES103206 to L.J.D., 1ZIC ES102487 to R.M.P., 1ZI ES043010 to L.P., and 1ZIC ES103326 to M.J.B.). This work was also supported by the US National Institute of Health Extramural Research Program; US National Institute of General Medical Sciences (R35-GM136435, to A.G.M. and 1K99-GM143534 to C.K.H).
Footnotes
Competing interests
The authors declare no conflict of interest.
References
- 1.Hopper AK & Nostramo RT tRNA Processing and Subcellular Trafficking Proteins Multitask in Pathways for Other RNAs. Front Genet 10, 96 (2019). 10.3389/fgene.2019.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt CA & Matera AG tRNA introns: Presence, processing, and purpose. Wiley Interdiscip Rev RNA 11, e1583 (2020). 10.1002/wrna.1583 [DOI] [PubMed] [Google Scholar]
- 3.Gogakos T. et al. Characterizing Expression and Processing of Precursor and Mature Human tRNAs by Hydro-tRNAseq and PAR-CLIP. Cell Rep 20, 1463–1475 (2017). 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan PP & Lowe TM GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44, D184–189 (2016). 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayne CK, Lewis TA & Stanley RE Recent insights into the structure, function, and regulation of the eukaryotic transfer RNA splicing endonuclease complex. Wiley Interdiscip Rev RNA, e1717 (2022). 10.1002/wrna.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekulovski S. & Trowitzsch S. Transfer RNA processing - from a structural and disease perspective. Biol Chem 403, 749–763 (2022). 10.1515/hsz-2021-0406 [DOI] [PubMed] [Google Scholar]
- 7.Trotta CR et al. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858 (1997). 10.1016/s0092-8674(00)80270-6 [DOI] [PubMed] [Google Scholar]
- 8.Paushkin SV, Patel M, Furia BS, Peltz SW & Trotta CR Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell 117, 311–321 (2004). 10.1016/s0092-8674(04)00342-3 [DOI] [PubMed] [Google Scholar]
- 9.Xue S, Calvin K. & Li H. RNA recognition and cleavage by a splicing endonuclease. Science 312, 906–910 (2006). 10.1126/science.1126629 [DOI] [PubMed] [Google Scholar]
- 10.Reyes VM & Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55, 719–730 (1988). 10.1016/0092-8674(88)90230-9 [DOI] [PubMed] [Google Scholar]
- 11.Greer CL, Soll D. & Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol 7, 76–84 (1987). 10.1128/mcb.7.1.76-84.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song J. & Markley JL Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold. J Mol Biol 366, 155–164 (2007). 10.1016/j.jmb.2006.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekulovski S. et al. Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia. Nat Commun 12, 5610 (2021). 10.1038/s41467-021-25870-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata A. Recent Insights Into the Structure, Function, and Evolution of the RNA-Splicing Endonucleases. Front Genet 10, 103 (2019). 10.3389/fgene.2019.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihisa T. Handling tRNA introns, archaeal way and eukaryotic way. Front Genet 5, 213 (2014). 10.3389/fgene.2014.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvin K. & Li H. RNA-splicing endonuclease structure and function. Cell Mol Life Sci 65, 1176–1185 (2008). 10.1007/s00018-008-7393-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weitzer S, Hanada T, Penninger JM & Martinez J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley Interdiscip Rev RNA 6, 47–63 (2015). 10.1002/wrna.1255 [DOI] [PubMed] [Google Scholar]
- 18.Hayne CK, Schmidt CA, Haque MI, Matera AG & Stanley RE Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage. Nucleic Acids Res 48, 7609–7622 (2020). 10.1093/nar/gkaa438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacheva IH et al. TSEN54 Gene-Related Pontocerebellar Hypoplasia Type 2 Could Mimic Dyskinetic Cerebral Palsy with Severe Psychomotor Retardation. Front Pediatr 6, 1 (2018). 10.3389/fped.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffer AE et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157, 651–663 (2014). 10.1016/j.cell.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian Y. et al. A familial lateonset hereditary ataxia mimicking pontocerebellar hypoplasia caused by a novel TSEN54 mutation. Mol Med Rep 10, 1423–1425 (2014). 10.3892/mmr.2014.2342 [DOI] [PubMed] [Google Scholar]
- 22.Karaca E. et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157, 636–650 (2014). 10.1016/j.cell.2014.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laugwitz L. et al. Pontocerebellar hypoplasia type 11: Does the genetic defect determine timing of cerebellar pathology? Eur J Med Genet 63, 103938 (2020). 10.1016/j.ejmg.2020.103938 [DOI] [PubMed] [Google Scholar]
- 24.Monaghan CE, Adamson SI, Kapur M, Chuang JH & Ackerman SL The Clp1 R140H mutation alters tRNA metabolism and mRNA 3’ processing in mouse models of pontocerebellar hypoplasia. Proc Natl Acad Sci U S A 118 (2021). 10.1073/pnas.2110730118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaForce GR et al. Suppression of premature transcription termination leads to reduced mRNA isoform diversity and neurodegeneration. Neuron 110, 1340–1357.e1347 (2022). 10.1016/j.neuron.2022.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forconi M. et al. 2’-Fluoro substituents can mimic native 2’-hydroxyls within structured RNA. Chem Biol 18, 949–954 (2011). 10.1016/j.chembiol.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tunyasuvunakool K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). 10.1038/s41586-021-03828-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Trotta CR & Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280, 279–284 (1998). 10.1126/science.280.5361.279 [DOI] [PubMed] [Google Scholar]
- 30.Abelson J, Trotta CR & Li H. tRNA splicing. J Biol Chem 273, 12685–12688 (1998). 10.1074/jbc.273.21.12685 [DOI] [PubMed] [Google Scholar]
- 31.Swerdlow H. & Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem 259, 5197–5207 (1984). [PubMed] [Google Scholar]
- 32.Schmidt CA, Giusto JD, Bao A, Hopper AK & Matera AG Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47, 6452–6465 (2019). 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tocchini-Valentini GD, Fruscoloni P. & Tocchini-Valentini GP Evolution of introns in the archaeal world. Proc Natl Acad Sci U S A 108, 4782–4787 (2011). 10.1073/pnas.1100862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattoccia E, Baldi IM, Gandini-Attardi D, Ciafre S. & Tocchini-Valentini GP Site selection by the tRNA splicing endonuclease of Xenopus laevis. Cell 55, 731–738 (1988). 10.1016/0092-8674(88)90231-0 [DOI] [PubMed] [Google Scholar]
- 35.Fabbri S. et al. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science (New York, N.Y.) 280, 284–286 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Fruscoloni P, Baldi MI & Tocchini-Valentini GP Cleavage of non-tRNA substrates by eukaryal tRNA splicing endonucleases. EMBO Rep 2, 217–221 (2001). 10.1093/embo-reports/kve040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Nicola Negri E. et al. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89, 859–866 (1997). 10.1016/s0092-8674(00)80271-8 [DOI] [PubMed] [Google Scholar]
- 38.Hayne CK, Lewis TA & Stanley RE Recent insights into the structure, function, and regulation of the eukaryotic transfer RNA splicing endonuclease complex. Wiley Interdiscip Rev RNA 13, e1717 (2022). 10.1002/wrna.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trotta CR, Paushkin SV, Patel M, Li H. & Peltz SW Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441, 375–377 (2006). 10.1038/nature04741 [DOI] [PubMed] [Google Scholar]
- 40.Baldi MI, Mattoccia E, Bufardeci E, Fabbri S. & Tocchini-Valentini GP Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 255, 1404–1408 (1992). 10.1126/science.1542788 [DOI] [PubMed] [Google Scholar]
- 41.Luscombe NM, Laskowski RA & Thornton JM NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res 25, 4940–4945 (1997). 10.1093/nar/25.24.4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references:
- 42.Schellenberg MJ, Petrovich RM, Malone CC & Williams RS Selectable high-yield recombinant protein production in human cells using a GFP/YFP nanobody affinity support. Protein Sci 27, 1083–1092 (2018). 10.1002/pro.3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillon MC et al. Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex. Nature structural & molecular biology 26, 830–839 (2019). 10.1038/s41594-019-0289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 46.Rohou A. & Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG & Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams CJ et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315 (2018). 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webb B. & Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics 54, 5 6 1–5 6 37 (2016). 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Case DA, B.-S. IY, Brozell SR, Cerutti DS, Cheatham TE III, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, R Harris N. Homeyer Y. Huang, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, Merz KM, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith J, Salomon- Ferrer R, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, York DM and Kollman AMBER PA 2018. University of California , San Francisco. (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps for wt-TSEN and endoX-TSEN have been deposited in the EMDB under the accession codes EMD-28755 and EMD-26856 respectively. The atomic model for endoX-TSEN has been deposited in the PDB under the accession code PDBID-7UXA. Mass spectrometry data has been deposited at MassIVE with the dataset identifier of MSV000091153. Source data including cross-linking mass spectrometry data and all un-cropped western blots are provided.