Abstract
The gal genes from the chromosome of Lactobacillus casei 64H were cloned by complementation of the galK2 mutation of Escherichia coli HB101. The pUC19 derivative pKBL1 in one complementation-positive clone contained a 5.8-kb DNA HindIII fragment. Detailed studies with other E. coli K-12 strains indicated that plasmid pKBL1 contains the genes coding for a galactokinase (GalK), a galactose 1-phosphate-uridyltransferase (GalT), and a UDP-galactose 4-epimerase (GalE). In vitro assays demonstrated that the three enzymatic activities are expressed from pKBL1. Sequence analysis revealed that pKBL1 contained two additional genes, one coding for a repressor protein of the LacI-GalR-family and the other coding for an aldose 1-epimerase (mutarotase). The gene order of the L. casei gal operon is galKETRM. Because parts of the gene for the mutarotase as well as the promoter region upstream of galK were not cloned on pKBL1, the regions flanking the HindIII fragment of pKBL1 were amplified by inverse PCR. Northern blot analysis showed that the gal genes constitute an operon that is transcribed from two promoters. The galKp promoter is inducible by galactose in the medium, while galEp constitutes a semiconstitutive promoter located in galK.
Lactobacilli are frequently involved in food fermentation processes, many of which are applied on an industrial scale for the production of such diverse products as sausages, sauerkraut, olives, pickled vegetables, cheese, or yoghurt. Although Lactobacillus casei may not always be the dominant organism, it influences the final texture and organoleptic properties of the product (e.g., by the production of exopolysaccharides or efficient acidification of the substrate).
Recent developments in the application of molecular biology to lactic acid bacteria have shown that it could be feasible to engineer metabolic pathways to either enhance specific metabolic fluxes or to divert metabolites for the production of different or new end products. However, this engineering requires detailed knowledge of metabolism and regulation within the targeted organism. We have chosen to investigate the complex of galactose and lactose metabolism. Lactose is initially the only carbohydrate available in milk fermentations, while galactose would, e.g., be derived from the fermentation of lactose by streptococci, which can expell the galactose moiety of lactose by using the galactose-lactose antiport mode of the lactose transporter (36).
Galactose metabolism via the Leloir pathway is a ubiquitous trait in bacterial cells. It can be used as a catabolic pathway for the degradation of galactose as an energy and carbon source while it links as an anabolic pathway the metabolism of carbohydrates, e.g., to the synthesis of lipopolysaccharides, of cell wall components, and of exopolysaccharides, for which galactosides are frequently required as building blocks. The Leloir pathway represents in the absence of an external galactose source the only means to provide this carbohydrate by biological interconversion from glucose to galactose (14).
Galactose can enter the cells by several types of transport systems. In the gram-negative bacterium Escherichia coli K-12, at least seven different systems have been characterized (18, 37), all of which release unmodified galactose into the cytoplasm. Galactose is also released as a hydrolysis product of internalized galactosides, e.g., from lactose, melibiose, or raffinose. The cleavage of β-galactosides generates β-d-galactose, which is converted into α-d-galactose by an aldose 1-epimerase (mutarotase [GalM]) prior to phosphorylation (5). The free α-d-galactose moiety can be activated by an ATP-dependent galactokinase (GalK) to initiate further metabolism via the Leloir pathway. Galactose 1-phosphate, the product of this reaction, is subsequently transferred to UDP-glucose in exchange with glucose 1-phosphate by galactose 1-phosphate-uridyltransferase (GalT). The resulting UDP-galactose is a substrate for the reaction catalyzed by UDP-galactose 4-epimerase (GalE), resulting in UDP-glucose. Recently, gal genes have been cloned from several lactic acid bacteria. They have been found to either constitute operons of their own, as is the case for the galKTM operon of Lactobacillus helveticus (32), or they are linked to genes of lactose metabolism like those in Streptococcus thermophilus (35). Galactose metabolism in L. casei 64H is mediated by two alternative pathways: the Leloir pathway and the tagatose 1,6-bisphosphate pathway (4, 11). The latter pathway catabolizes galactose that has been transported via a galactose-specific phosphotransferase system (Gal-PTS) which releases galactose 6-P into the cytoplasm. Galactose 6-phosphate results also from the metabolism of lactose, which in L. casei 64H is exclusively transported via the Lac-PTS (8, 9).
We report here the results of our investigations of the Leloir pathway from L. casei 64H. The gal genes of L. casei 64H are the first ones to be described that are derived from a facultative heterofermentative lactobacillus that has two pathways for the degradation of galactose.
MATERIALS AND METHODS
Bacterial strains and media.
L. casei subsp. casei 64H (15) was maintained at 4°C in calcium carbonate-fortified litmus milk containing 1% glucose (12). Experimental cultures were grown in lactobacillus carrying medium (LCM) supplemented with 0.5% carbohydrate (13). Plating of lactobacilli was performed on LCM media solidified with 14 g of agar per liter. E. coli strains are listed in Table 1. They were grown in LB0 or plated on LB0 solidified with 12 g of agar per liter. LB0 contains 10 g of Bacto tryptone, 10 g of yeast extract, and 5 g of NaCl per liter (26). Selection for E. coli cells transformed with the appropriate plasmids was performed by using 100 μg of ampicillin per ml or 25 μg of chloramphenicol per ml. Where required, 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml was used for blue-white selection in subcloning procedures. Marker tests with E. coli were performed on MacConkey agar-base indicator plates supplied with 1% of the carbohydrate to be tested (27). For T7 overexpression, cells were grown in phosphate-buffered minimal medium (47).
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype or phenotype | Reference |
|---|---|---|
| DH5α | ΔlacU169(φ80 dlacZΔM15) Gal+ | 17 |
| HB101 | galK2 mutant | 6 |
| JWL184-1 | galT6 galP63 | 25 |
| JWL191 | galT6 galP63 ptsI191 | 24 |
| S165 | Δ(galETK) | 42 |
| JM1100 | galK2 mgl-50 galP64 ptsG23 ptsM8 fruA10 | 18 |
| BL21(λDE3) | F−hsdS (rB− mB−) gal [λDE3(BamHI) int::lacUV5P T7 gene 1.0] | 45 |
DNA isolation and manipulation.
Chromosomal DNA was isolated from L. casei by the method of Neumann et al. (33). The precipitated DNA was resuspended in Tris-EDTA-RNase (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 20 μg of RNase H per ml) and incubated for several hours at 37°C before an additional extraction by phenol-chloroform and a final ethanol precipitation. Other DNA manipulations and plasmid preparations were performed according to standard procedures as described by Sambrook et al. (40). Restriction enzyme digests and ligations were performed as recommended by the suppliers. The sources for enzymes and other materials used for molecular biological procedures were Boehringer Mannheim (Mannheim, Germany), GIBCO BRL (Gaithersburg, Md.), New England Biolabs (Beverly, Mass.), Pharmacia Biotech Europe (Freiburg, Germany), and Qiagen (Hilden, Germany).
Construction of pKBL1 and subclones.
Chromosomal DNA of L. casei 64H was digested with HindIII, EcoRI, BamHI, PstI, or XbaI. The digested DNA was size fractionated on 0.7% agarose gels, and fragments of 2 to 20 kb were isolated with the Qiaquick Gel Extraction Kit from Qiagen. Fragments were ligated to dephosphorylated pUC19 vector (53).
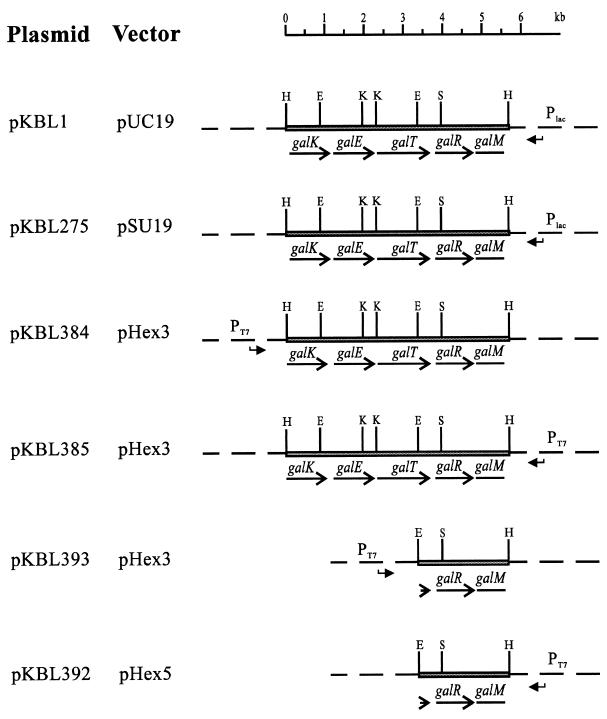
For T7 overexpression, the HindIII insert of pKBL1 was cloned into pHEX3 (20) in the direction with the T7p promoter (pKBL384) or against the T7p promoter (pKBL385). For overexpression of GalR, the EcoRI-HindIII fragment of pKBL1 containing the complete galR gene was cloned into pHEX3 in the direction of T7p, giving pKBL393, and in pHEX5 against T7p, giving pKBL392. For sequencing, subclones were constructed with EcoRI and the KpnI restriction sites on pKBL1. Additional subclones were obtained by creating deletion clones as described by S. Henikoff (19). Graphical representations of relevant plasmids are given in Fig. 1.
FIG. 1.
Schematic representation of relevant plasmids. Bent arrows indicate the direction of promoters present on the vectors. The restriction sites indicated are H, HindIII; E, EcoRI; K, KpnI; S, SalI.
Cloning of the 5′ and 3′ regions of the gal genes.
Cloning of the 5′ region was achieved by inverse PCR (49). Approximately 50 μg of chromosomal DNA from L. casei 64H was digested with KpnI. After inactivation of the restriction endonuclease by extraction with phenol, the fragments were diluted to approximately 0.05 μg of DNA per μl and ligated overnight. The ligation mixture was inactivated by phenol treatment, precipitated with ethanol, and resuspended in 50 μl of H2O. One microliter of the ligation mixture was used as a template in a 10-μl PCR mixture by the method of Saiki et al. (39) in an Air Thermo Cycler from Idaho Technologies (Idaho Falls, Idaho) according to the protocol supplied with the cycler. A product of approximately 600 bp from a reaction with primers PgalK1 (priming site nucleotides [nt] 550 to 530 [nucleotide numbers refer throughout the paper to GenBank accession no. AF005933]) and PgalK2 (priming site nt 2170 to 2191) was cloned into vector pUC19. After the sequence flanking the KpnI site was determined, the fragment was amplified from chromosomal DNA by PCR with the primers PgalPr1 (priming site nt 1 to 20) and PgalK1. The cloning of the 3′ region was performed similar to the cloning of the 5′ region with primers PgalR1 and PgalR2 (priming at sites 5926 to 5945 and 2767 to 2748). The product of approximately 300 bp was cloned into pUC19.
Sequencing and analysis.
DNA sequencing was performed by the dideoxynucleotide chain termination method of Sanger et al. (41) with the T7 sequencing kit from Pharmacia according to the instructions supplied by the manufacturer. The sequences obtained by this method were analyzed by the BLAST family of programs (2) or the DNAsis for Windows DNA and protein analysis system (Hitachi Software Engineering America, South San Francisco, Calif.).
T7 expression.
Expression and labelling of gene products were performed as described by Tabor and Richardson (46) according to the following protocol. Cells of strain BL21(λDE3) harboring plasmid pKBL384, pKBL385, pKBL393, or pKBL392 were grown overnight at 37°C in LB0 with chloramphenicol. The overnight cultures were diluted in 20 ml of LB0 to 108 cells per ml and grown to a density of 5 × 108 cells per ml. Cells were washed in minimal medium, resuspended in 20 ml of minimal medium with glucose (0.2%) containing Difco Met assay medium (0.25%), and divided into two 8-ml cultures. The cultures were incubated at 37°C for 60 min and induced with isopropyl-β-d-thiogalactoside (1 mM). After incubation for 45 min, rifampin was added to one of the two cultures to 0.4 mg per ml, and the cultures were incubated for an additional 20 min. Half a milliliter of cells of each culture was labeled with 10 μCi of [35S]methionine at 37°C for 10 min. After centrifugation, the cell pellet was resuspended in 0.1 ml of cracking buffer (60 mM Tris-HCl [pH 6.8], 1% 2-mercaptoethanol, 1% sodium dodecyl sulfate [SDS], 10% glycerol, 0.01% bromophenol blue). Samples of 20 μl were treated for 5 min in a boiling water bath before they were separated by SDS-polyacrylamide gel electrophoresis.
RNA hybridization.
For preparation of RNA, cells from an overnight culture of L. casei 64H were diluted into 200 ml of LCM supplemented with 20 mM dl-threonine and 0.5% galactose, lactose, or glucose to a density of 5 × 107 cells per ml (optical density at 600 nm of 0.05, Shimadzu UV-1202, Shimadzu Europe, Duisburg, Germany) and incubated at 30°C until a density of 4 × 108 cells per ml was reached. Cells were harvested and resuspended in 10 ml of SET buffer (0.45% sucrose, 8 mM EDTA, 15 mM Tris-HCl [pH 8]). After incubation with 10 mg of lysozyme per ml at room temperature for 30 min, protoplasts were harvested by centrifugation at 5,000 × g for 10 min, and the RNA was isolated according to the method of van Rooijen and deVos (50). Fractionation of RNA and blotting on nylon membranes were performed according to the method of Pellé and Murphy (34) with 1% agarose gels. Hybridization was performed with the DIG chemiluminescent detection kit of Boehringer Mannheim. Deviating from the kit’s instructions, we used a hybridization buffer containing 7% SDS, 50% formamide, 5× SSC buffer (0.75 M NaCl, 75 mM sodium citrate [pH 7.0]), 2% blocking reagent, 50 mM sodium phosphate buffer (pH 7.0), and 0.1% N-lauroylsarcosine.
Primer extension experiments.
For primer extension, the method of S. J. Triezenberg (48) was used. The labeled oligonucleotides were purified with MicroSpin Sephacryl HR resins from Pharmacia.
Enzyme assays.
For measuring the enzymatic activities, extracts were prepared from E. coli K-12 strains carrying pKBL1 or its pSU19 derivative (3), pKBL275, with the complete HindIII insert from pKBL1. For some tests, extracts were prepared from E. coli DH5α or L. casei 64H which had been induced with 0.2 or 0.5% galactose, respectively, for 3 h. Cells were harvested in mid-logarithmic phase and broken by shaking with zirconia beads in a Retsch mill MM2 (Retsch, Haan, Germany). Extracts were centrifuged at 15,000 × g for 5 min. Protein concentrations were determined by the bicinchoninic acid method of Smith et al. (44). Galactokinase activity was determined according to the method of Sherman (43) and a protocol described by Lengeler et al. (23). Probes were taken at 30, 60, and 90 s after the start of the reaction.
Galactose 1-phosphate-uridyltransferase activity was determined by the method of Kuruhashi and Anderson (22) as described in Methoden der enzymatischen Analyse by Isselbacher (21). Concentrations of phosphoglucomutase and of glucose-6-phosphate dehydrogenase had to be increased by a factor of 10 to give detectable enzyme activities. UDP-galactose 4-epimerase activity was determined by the method of Wilson and Hogness (52).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been assigned GenBank accession no. AF005933.
RESULTS
Cloning the gal genes.
In order to isolate the Leloir pathway genes of L. casei 64H, the galK2 mutation of E. coli HB101 was complemented with shotgun clones carrying fragments from chromosomal DNA of L. casei 64H in pUC19. One colony of approximately 5,000 transformants carrying insert sizes larger than 2 kb showed a Gal+ phenotype when tested on MacConkey galactose plates. This clone contained a 5.8-kb insert of HindIII-digested DNA and was designated pKBL1.
Complementation analysis with pKBL1.
In order to identify in vivo further functions encoded by pKBL1, several E. coli Gal− mutants were tested for complementation with either pKBL1 or the reference plasmid pNP5 carrying galKTM′ from L. helveticus (32). pKBL1 conferred a Gal+ phenotype to strains JWL184-1 (galT6 galP63) and S165 (ΔgalETK), indicating that the insert codes for the enzymes of the Leloir pathway. Complementation was also possible in the ptsI background of JWL191 (ptsI191 galT6 galP63), indicating that the Gal+ phenotype was not caused by the enzymes of the tagatose 1,6-bisphosphate pathway, which depend on a functional PTS. Interestingly the GalK− mutant JM1100, which supposedly lacks the known galactose transport systems (14), is also complemented by pKBL1 and, to a lesser extent, by pNP5. Because pNP5 and pKBL1 do not carry any transport genes in addition to the Leloir pathway genes (reference 32 and see below), strain JM1100 must contain an as yet unidentified galactose transport system.
Determination of enzymatic activities encoded by pKBL1.
The phenotypic characterization of pKBL1 is supported by the in vitro analysis of enzymatic activities. GalK, GalE, and GalT activities are expressed from pKBL275, the pSU19 derivative of pKBL1 (Table 2). Comparison of the activity levels of enzymes is problematic if they are expressed from different plasmids or from the chromosome and in different organisms. Host-specific factors such as codon usage or ribosome binding site specificity can further affect the results. However, the expression of active Leloir pathway enzymes is unequivocally shown by the data given.
TABLE 2.
GalK, GalT, and GalE activities encoded by pKBL275 in the Δgal mutant S165 of E. coli K-12 and in control strains
| Strain/plasmid | Activity (nmol/mg/min) of:
|
||
|---|---|---|---|
| GalK | GalT | GalE | |
| L. casei 64H (Gal+) | 83 | 115 | 900 |
| E. coli DH5α (Gal+) | 24 | 57 | 1,400 |
| S165/pSU19 | 0 | 0 | 0 |
| S165/pNP5 (galKTM′) | 45 | 448 | NDa |
| S165/pKBL275b (galKETRM′) | 66 | 44 | 900 |
ND not determined.
pKBL275 is a pSU19 derivative carrying the insert of pKBL1.
Cloning of the regions flanking the insert from pKBL1.
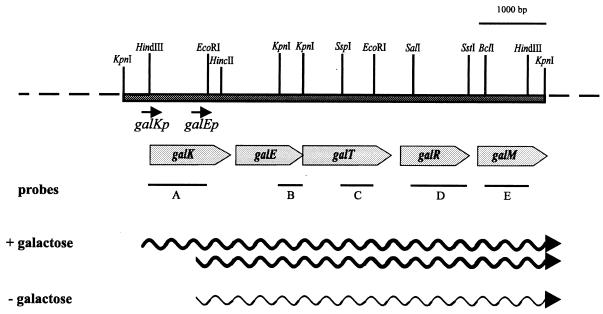
Initial sequencing data indicated the presence of an incomplete open reading frame (galM′) at the 3′ end of the cloned HindIII fragment. They also indicated that the promoter region upstream of galK was not cloned into pKBL1. The chromosomal regions flanking the HindIII fragment were amplified by inverse PCR and cloned into pUC19 as described under Materials and Methods. Using a 0.8-kbp HindIII-EcoRI fragment (Fig. 2) as a probe for the 5′ end, a KpnI fragment of 2.6 kbp was detected, indicating the presence of a KpnI site approximately 0.6 kbp upstream of the HindIII site. A 0.6-kbp product was obtained after inverted PCR (49) and cloned into pUC18. Sequencing the ends of this fragment revealed known sequences up to the HindIII and KpnI sites respectively. A PCR with primers PgalK1 and PgalPr1 with uncut chromosomal DNA as a template gave a product of 0.6 kbp, thus proving the continuity at the level of chromosomal DNA.
FIG. 2.
Arrangement of the gal operon of L. casei 64H. Shaded grey arrows indicate the location and length of the genes. Black lines show fragments that were used as probes in the Northern hybridization studies. The sizes of the detected mRNAs are schematically shown as wavy lines. Restriction sites used for the construction of subclones are given.
A similar procedure was used in order to clone the 3′ end of the galM gene. Southern hybridization indicated that there should be an extension of 3 kbp beyond the 3′ HindIII site of the gal genes when EcoRI-digested chromosomal DNA was used for the inverse PCR technique. A PCR product of the expected size of 3 kbp was obtained, but various attempts to clone it into different vectors were unsuccessful. However, a smaller fragment comprising some 180 nt up to a KpnI site could be added by this technique to the 3′ end of galM. Based on comparison with known sequences, there are still about 50 codons missing in the galM reading frame and possibly the whole gal gene cluster (see also below). The total established DNA sequence of the cloned region is 6,168 bp. It contains the genes galKETR and the incomplete gene, galM′.
T7 overexpression of the gal gene products.
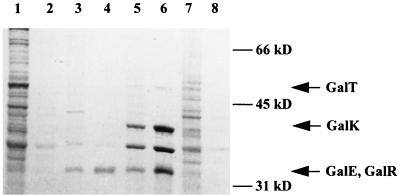
The enzymatic assays indicated that the galK, galE, and galT genes are translated into active proteins, but the expression of galR cannot easily be tested. In order to demonstrate its synthesis, a labelling experiment with the T7 expression system of Tabor and Richardson (46) was performed with either the complete HindIII fragment of pKBL1 or with the 2.3-kbp EcoRI-HindIII fragment carrying galR (Fig. 2). There appear three additional bands upon induction of the system when clone pKBL384 is used (Fig. 3). A prominent band of 36 kDa also present in the controls is probably a protein encoded by the vector or the host strain. The insert-dependent bands with apparent molecular masses of 52, 42, and 34 kDa correspond very well to the DNA-derived molecular masses of the encoded proteins. GalE and GalR probably run as a double band with similar masses, although it cannot be excluded that one product was not labeled in the experiment. The presence and expression of GalR are clearly demonstrated in lanes 3 and 4 of Fig. 3, since pKBL393 codes only for GalR and parts of GalT and GalM. The 34-kDa band in these lanes should therefore represent GalR and not GalE.
FIG. 3.
T7 overexpression of the gal genes of L. casei 64H. Probes were prepared from BL21(λDE3) containing plasmids with different fragments of the gal operon of L. casei 64H. Lanes: 1 and 2, pKBL392; 3 and 4, pKBL393; 5 and 6, pKBL384; 7 and 8, pKBL285. Probes in lanes 2, 4, 6, and 8 were prepared from cultures containing rifampin.
Analysis of gal gene expression and primer extension analysis.
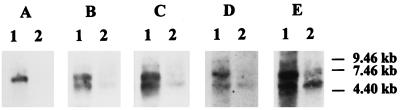
The hypothesis that the gal genes are organized as an operon was tested by Northern hybridization with probes against various parts of the HindIII insert from pKBL1. Two sizes of mRNAs could be detected (Fig. 4). (i) A signal at 6.3 kb appeared with all probes in RNA from galactose-induced cells. It was not present when RNA from glucose-grown cells was tested. This signal corresponds to a polycistronic mRNA, expressing galKETRM. (ii) A shorter mRNA of 5.6 kb was present in both samples. It could not be detected with a probe against galK, suggesting that this messenger carries galETRM. The signal was weaker in RNA from glucose-grown cells than in RNA from galactose-grown cells, indicating a semiconstitutive expression. Differential expression of the genes as indicated by the Northern hybridization experiment would require the activity of two promoters, galKp and galEp. Both transcription start sites were analyzed by primer extension and mapped to nt 334 for galKp and to nt 1208 for galEp (data not shown). As expected, the reactions for the galKp extension were only positive with RNA isolated from induced cells, while the reaction for galEp was positive with RNA from induced and uninduced cells. Potential −10 and −35 regions could be located at nt 1188 to 1193 and 1162 to 1167, respectively. The galEp promoter shows only a low degree of similarity with other known promoters from lactobacilli (10, 31). This could be due to the fact that galEp is a semiconstitutive promoter.
FIG. 4.
Northern hybridization. A, B, C, D, and E show hybridization experiments with the probes indicated in Fig. 2. Lanes: 1, RNA prepared from L. casei 64H induced with galactose; 2, RNA prepared from L. casei 64H grown with glucose.
GalK, GalT, and GalE activities during growth on different carbohydrates.
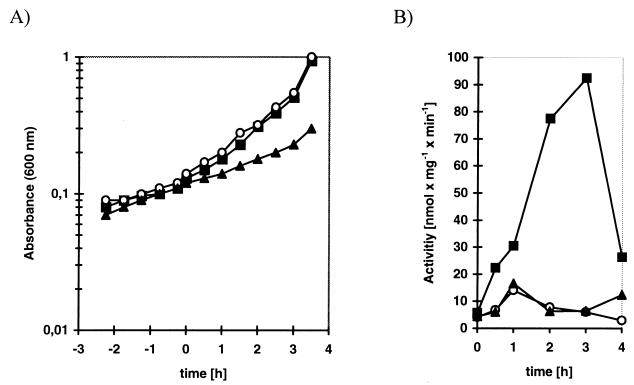
The activities of GalK, GalT, and GalE of L. casei 64H were measured during growth on different carbohydrates and during growth in LCM in the absence of carbohydrates (Table 3). All activities are induced about 10-fold during growth with galactose, and GalK activity is slightly repressed in the presence of glucose. As expected, growth on lactose has no inducing effect, since lactose transport and metabolism in L. casei 64H are exclusively mediated by a lactose PTS and the tagatose 1,6-bisphosphate pathway. In order to study the induction by galactose in more detail, we measured the activity of the galactokinase after growth in LCM without additional carbohydrate or with galactose or lactose. The GalK activity reached a maximum after approximately 3 h and then reproducibly dropped off sharply (Fig. 5).
TABLE 3.
GalK, GalE, and GalT activities in L. casei 64Ha
| Carbohydrate | Activity, [nmol/mg/min] (relative activity) of:
|
||
|---|---|---|---|
| GalK | GalE | GalT | |
| None | 17.5 (1.0) | 230 (1.0) | 7.3 (1.0) |
| Galactose | 150.0 (8.5) | 2,500 (10.9) | 71.3 (9.8) |
| Lactose | 18.9 (1.1) | 167 (0.7) | 6.3 (0.9) |
| Glucose | 7.6 (0.4) | 250 (1.1) | 4.7 (0.6) |
Extracts were prepared as described in Materials and Methods. Cells were grown in LCM alone or in LCM with galactose, lactose, or glucose for 3 h prior to harvesting.
FIG. 5.
Induction of GalK. (A) Growth of L. casei 64H cultures. At time zero, carbohydrates were added to the cultures. (B) Specific activity of GalK at different points after induction. The curve is representative for five independent experiments, which all showed the drastic drop of activity for the 4-h value. ▴, LCM; ▪, LCM plus galactose; ○, LCM plus lactose.
DISCUSSION
We report the cloning, sequencing, and analysis of the major part of the gal operon of L. casei 64H. Analysis of the DNA sequence supports this hypothesis, because the deduced amino acid sequences of the five open reading frames show significant similarities to the known enzymes of the Leloir pathway from different organisms. GalK (386 amino acids, molecular mass of 42.3 kDa) shows 57% identical amino acids to GalK of Streptococcus mutans and is also highly similar to other GalK proteins. GalE (331 amino acids, molecular mass of 36.3 kDa) shows high similarity to different GalE proteins (e.g., 68% identical amino acids to Streptococcus mutans and 41% identical amino acids to E. coli). GalT (486 amino acids, molecular mass of 54.1 kDa) exhibits similarities to GalT of several gram-positive bacteria (55% identical residues compared to GalT of L. helveticus and S. mutans) but not to those of E. coli or other gram-negative bacteria. At present, GalT of this type seems therefore to be limited to gram-positive bacteria. However, the complementation studies and enzyme tests clearly show that this type of transferase is also active in E. coli. The proposed start codon of galT is a TTG and overlaps the stop codon TGA of galE. The use of the TTG start codon is supported by the low expression of galT in the T7 overexpression experiment. TTG as a start codon, as it is suggested here, is seldom used in E. coli and could therefore be the reason for the poor expression of galT in this organism. However, TTG as a start codon has been observed in L. casei (1, 16).
GalR (331 amino acids, molecular mass of 36.5 kDa) is a member of the LacI/GalR family of repressor proteins, because it shows up to 41% identical amino acids with repressor proteins of this type. It resembles, e.g., GalR and GalS, the repressors of the gal regulon of E. coli (7, 28, 38, 51). Because of its similarity to other repressors and because of its location within the gal gene cluster, we propose that GalR could be the repressor of the L. casei 64H gal operon, although the experiments presented provide no direct proof for this hypothesis. Galactose seems the most likely inducer, as indicated by the induction studies. Our Northern blot analysis clearly shows that galR is transcribed on policistronic messengers which carry galKETRM or galETRM. The operon may therefore be autoregulated. Although gal operons are frequently regulated by repressor proteins of the LacI/GalR type, autoregulation has not been reported before for any of these operons. Repressors of the LacI/GalR type bind to palindromic operator sequences. Two nearly perfect inverted repeats are found in front of the operon. They are the palindromic sequences TTTTAGTAAAA centered around −15 and −115 which could serve as operators.
Downstream of galR, there is a fifth partial open reading frame of 290 codons (32.1 kDa). This reading frame extends beyond the KpnI site at the end of the sequenced region. Its derived amino acid sequence shows similarities to different aldose 1-epimerases (mutarotases). Three different start codons are possible for this open reading frame, which are all in frame with the proposed reading frame starting at nt 5299. The use of this start site is most probable, since there are no potential ribosome binding sites upstream of the alternative start sites. Also, the predicted additional N-terminal sequence of the longer proteins does not exhibit any similarity to other known sequences, while the protein translated from the proposed start site shares significant similarity to the so far identified GalM proteins. Like the enzymes of L. helveticus (32), E. coli (5, 29), Haemophilus influenzae (30), and S. thermophilus (35), the protein does not contain a leader peptide, indicating that it is localized within the cell. This gene is not completely cloned on pKBL1. Judging from amino acid sequence alignment, there are still about 50 codons missing to complete the reading frame of galM. It is likely that the addition of the missing codons of galM would also complete the gal operon, because the deduced length of a polycistronic messenger transcribing galKETRM corresponds to the length of the messenger detected by the Northern blot experiments.
The derived gene order for the gal region in L. casei 64H is galKETRM and represents a new arrangement of gal genes. As has been shown by the induction studies, the operon is clearly inducible by galactose in the medium. A slight repression of GalK and GalT activities is observed with glucose. In contrast to this, GalE is not repressed by glucose. This difference could result from the transcription of the operon by two different mRNAs.
The gal genes of L. casei 64H are the first ones to be cloned from a lactobacillus which can metabolize galactose by two distinct pathways. The gal genes of L. casei 64H resemble those previously cloned from other organisms. However, a feature which distinguishes the gal operon of L. casei 64H from other known gal operons is that it could be autoregulated. Autoregulation may be a requirement for the coordination of the activity of the two metabolic pathways. It will be interesting to study the regulation of the tagatose 1,6-bisphosphate pathway too, to find out whether there is any higher control mechanism in order to coordinate the two pathways.
ACKNOWLEDGMENTS
The constant advice and generous support of Joseph W. Lengeler during this work are gratefully acknowledged. We thank Uli Siebers for excellent technical assistance.
This research was supported by grant SFB171 TP C17 from Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Alpert C-A, Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the BglG family of transcriptional antiterminators. J Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bartolome B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 4.Bettenbrock K. Molekulare Untersuchung eines PTS-abhängigen und eines Leloir-Abbauweges für d-Galaktose bei Lactobacillus casei 64H. Ph.D. thesis. Osnabrück, Germany: Universität Osnabrück; 1997. [Google Scholar]
- 5.Bouffard G, Rudd K, Adhya S L. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J Mol Biol. 1994;244:269–278. doi: 10.1006/jmbi.1994.1728. [DOI] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Buttin G. Mechanismes regulateur dans la biosynthese des enzymes du metabolisme du galactose chez Escherichia coli K-12 II. Le determinisme genetique de la regulation. J Mol Biol. 1963;7:183–205. doi: 10.1016/s0022-2836(63)80045-5. [DOI] [PubMed] [Google Scholar]
- 8.Chassy B M, Alpert C-A. Molecular characterization of the plasmid-encoded lactose-PTS of Lactobacillus casei. FEMS Microbiol Rev. 1989;63:157–166. doi: 10.1016/0168-6445(89)90020-x. [DOI] [PubMed] [Google Scholar]
- 9.Chassy B M, Gibson E M, Giuffrida A. Evidence for plasmid-associated lactose metabolism in Lactobacillus casei subsp. casei. Curr Microbiol. 1978;1:141–144. doi: 10.1007/BF02601666. [DOI] [PubMed] [Google Scholar]
- 10.Chassy B M, Murphy C M. Lactococcus and lactobacillus. In: Sonenschein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 65–82. [Google Scholar]
- 11.Chassy B M, Thompson J. Regulation and characterization of the galactose-phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J Bacteriol. 1983;154:1204–1214. doi: 10.1128/jb.154.3.1204-1214.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chassy B M, Thompson J. Regulation of lactose-phosphoenolypruvate-dependent phosphotransferase system and β-d-phosphogalactoside galactohydrolase activities in Lactobacillus casei. J Bacteriol. 1983;154:1195–1203. doi: 10.1128/jb.154.3.1195-1203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efthymiou C, Hansen P A. An antigenic analysis of Lactobacillus acidophilus. J Infect Dis. 1962;110:258–267. doi: 10.1093/infdis/110.3.258. [DOI] [PubMed] [Google Scholar]
- 14.Frey P A. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 15.Gasser F, Mandel M. Deoxyribonucleic acid base composition of the genus Lactobacillus. J Bacteriol. 1968;96:580–588. doi: 10.1128/jb.96.3.580-588.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosalbes M J, Monedero V, Alpert C-A, Pérez-Martinez G. Establishing a model to study regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol Lett. 1997;148:83–89. doi: 10.1111/j.1574-6968.1997.tb10271.x. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Henderson P J F, Giddens R A, Jones-Mortimer M C. Transport of galactose, glucose and their analogues by Escherichia coli K12. Biochem J. 1977;162:309–320. doi: 10.1042/bj1620309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S. Unidirectional digestion with exonucleaseIII in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- 20.Heuel H, Turgut S, Schmid K, Lengeler J W. Substrate recognition domains as revealed by active hybrids between the d-arabinitol and ribitol transporters from Klebsiella pneumoniae. J Bacteriol. 1997;179:6014–6019. doi: 10.1128/jb.179.19.6014-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isselbacher K J. Uridyl-Transferase. In: Bergmeyer H U, editor. Methoden der enzymatischen Analyse. 3rd ed. Weinheim, Germany: Verlag Chemie; 1974. pp. 830–833. [Google Scholar]
- 22.Kuruhashi K, Anderson E P. Galactose-1-phosphate uridyl transferase, its purification and application. Biochim Biophys Acta. 1958;29:498–502. doi: 10.1016/0006-3002(58)90004-0. [DOI] [PubMed] [Google Scholar]
- 23.Lengeler J, Herman K O, Unsöld H J, Boos W. The regulation of the β-methylgalactoside transport system and of the galactose binding protein of Escherichia coli K12. Eur J Biochem. 1971;19:457–470. doi: 10.1111/j.1432-1033.1971.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 24.Lengeler J W. Characterization of mutants of Escherichia coli K12, selected by resistance to streptozotocin. Mol Gen Genet. 1980;79:49–54. doi: 10.1007/BF00268445. [DOI] [PubMed] [Google Scholar]
- 25.Lengeler J W, Auburger A M, Mayer R, Pecher A. The phosphoenolpyruvate dependent carbohydrate phosphotransferase system enzymes II as chemoreceptors in chemotaxis of Escherichia coli K12. Mol Gen Genet. 1981;183:163–170. doi: 10.1007/BF00270156. [DOI] [PubMed] [Google Scholar]
- 26.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 27.MacConkey A. Lactose fermenting bacteria in faeces. J Hyg. 1905;8:333–379. doi: 10.1017/s002217240000259x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majumdar A, Adhya S. Demonstration of two operator elements in gal: in vitro repressor binding studies. Proc Natl Acad Sci USA. 1984;81:6100–6104. doi: 10.1073/pnas.81.19.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maskell D. Protein sequence from downstream of Escherichia coli galK is homologous with galM from other organisms. Mol Microbiol. 1992;6:2211. doi: 10.1111/j.1365-2958.1992.tb01396.x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 30.Maskell D J, Szabo M J, Deadman M E, Moxon E R. The gal locus from Haemophilus influenzae: cloning, sequencing and the use of gal mutants to study lipopolysaccharide. Mol Microbiol. 1992;6:3051–3063. doi: 10.1111/j.1365-2958.1992.tb01763.x. [DOI] [PubMed] [Google Scholar]
- 31.Matern H T, Klein J R, Henrich B, Plapp R. Determination and comparison of Lactobacillus delbrueckii ssp. lactis DSM7290 promoter sequences. FEMS Microbiol Lett. 1994;122:121–128. doi: 10.1111/j.1574-6968.1994.tb07154.x. [DOI] [PubMed] [Google Scholar]
- 32.Mollet B, Pilloud N. Galactose utilization in Lactobacillus helveticus: isolation and characterization of the galactokinase (galK) and galactose-1-phosphate uridyl transferase (galT) genes. J Bacteriol. 1991;173:4464–4473. doi: 10.1128/jb.173.14.4464-4473.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann B, Pospiech A, Schairer H U. Rapid isolation of genomic DNA from gram negative bacteria. Trends Genet. 1992;8:332–333. doi: 10.1016/0168-9525(92)90269-a. [DOI] [PubMed] [Google Scholar]
- 34.Pellé R, Murphy N B. Northern hybridization: rapid and simple electrophoretic conditions. Nucleic Acids Res. 1993;21:2783–2785. doi: 10.1093/nar/21.11.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDP-glucose 4-epimerase. J Bacteriol. 1990;172:4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–148. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 37.Rotman B, Ganesan A K, Guzman R. Transport systems for galactose and galactosides in Escherichia coli. II. Substrate and inducer specificities. J Mol Biol. 1968;36:247–260. doi: 10.1016/0022-2836(68)90379-3. [DOI] [PubMed] [Google Scholar]
- 38.Saedler H, Gullon A, Fiethen L, Starlinger P. Negative control of the galactose operon in E. coli. Mol Gen Genet. 1968;102:79–88. doi: 10.1007/BF00341872. [DOI] [PubMed] [Google Scholar]
- 39.Saiki R, Scharf S, Faloona F, Mullis K B, Horn G T, Ehrlich H A, Arnheim M. Enzymatic amplification of β-globin genomic sequences and restriction sites of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro J A, Adhya S L. The galactose operon of E. coli K-12. II. A deletion analysis of operon structure and polarity. Genetics. 1969;62:249–264. doi: 10.1093/genetics/62.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman J R. Rapid enzyme assay technique utilizing radioactive substrate, ion-exchange paper and scintillation counting. Anal Biochem. 1963;5:548. doi: 10.1016/0003-2697(63)90075-7. [DOI] [PubMed] [Google Scholar]
- 44.Smith P K, Krohn R I, Hermahnson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto M K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 45.Studier F W, Moffat B A. Use of the bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 46.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system of controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka S, Lerner S A, Lin E C C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol. 1967;93:642–648. doi: 10.1128/jb.93.2.642-648.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triezenberg S J. Primer extension. Unit 4.8. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1990. [Google Scholar]
- 49.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;19:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 51.Weickert M J, Adhya S. Isorepressor of the gal regulon in Escherichia coli. J Mol Biol. 1992;226:69–83. doi: 10.1016/0022-2836(92)90125-4. [DOI] [PubMed] [Google Scholar]
- 52.Wilson D B, Hogness D S. Galactokinase and uridine diphosphogalactose 4-epimerase from Escherichia coli. Methods Enzymol. 1966;VIII:229–240. [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]