Abstract
Lettuce (Lactuca sativa L.) is a leafy vegetable crop with ongoing breeding efforts related to quality, resilience, and innovative production systems. To breed resilient and resistant lettuce in the future, valuable genetic variation found in close relatives could be further exploited. Lactuca virosa (2x = 2n = 18), a wild relative assigned to the tertiary lettuce gene pool, has a much larger genome (3.7 Gbp) than Lactuca sativa (2.5 Gbp). It has been used in interspecific crosses and is a donor to modern crisphead lettuce cultivars. Here, we present a de novo reference assembly of L. virosa with high continuity and complete gene space. This assembly facilitated comparisons to the genome of L. sativa and to that of the wild species L. saligna, a representative of the secondary lettuce gene pool. To assess the diversity in gene content, we classified the genes of the 3 Lactuca species as core, accessory, and unique. In addition, we identified 3 interspecific chromosomal inversions compared to L. sativa, which each may cause recombination suppression and thus hamper future introgression breeding. Using 3-way comparisons in both reference-based and reference-free manners, we show that the proliferation of long-terminal repeat elements has driven the genome expansion of L. virosa. Further, we performed a genome-wide comparison of immune genes, nucleotide-binding leucine-rich repeat, and receptor-like kinases among Lactuca spp. and indicated the evolutionary patterns and mechanisms behind their expansions. These genome analyses greatly facilitate the understanding of genetic variation in L. virosa, which is beneficial for the breeding of improved lettuce varieties.
Keywords: lettuce, genome assembly, comparative genomics, transposable elements (TEs), immune genes, Plant Genetics and Genomics
Introduction
Lettuce (Lactuca sativa L.) is a crop with an economic value of ∼3 billion USD per year (Food and Agriculture Organization of the United Nations 2019). To develop better lettuce cultivars, breeders often search for novel genetic variations in lettuce wild relatives. Lactuca virosa (biennial) is a donor for resistance to different pests and pathogens and a representative species in the lettuce gene pool (Maisonneuve et al. 1991, 2018; Maisonneuve 2003; Parra et al. 2016). The exploitation of L. virosa for lettuce breeding has had both challenges and successes. For example, despite reproductive barriers for direct intercrossing with lettuce, breeders and scientists were able to execute interspecific hybridization bridged by L. serriola to introduce traits like robust root architecture and resistance to currant-lettuce aphid, downy mildew, and viruses (Thompson and Ryder 1961; Eenink et al. 1982; Maisonneuve et al. 1995). Such interspecific crosses are part of the breeding pedigrees of the well-known cultivars Vanguard and Salinas, representing modern crisphead lettuce cultivars (Mikel 2007, 2013). Novel introgressions of desired genes and traits from L. virosa into cultivated lettuce could be realized through an improved understanding of its genomic content.
A reference genome and derived molecular markers are essential for breeders to select traits accurately and trace introgressions in cultivated lettuce from L. virosa. For example, genome-wide association studies (GWAS) have been performed to identify SNP variants (Mikel 2013) that are associated with interesting traits in lettuce (Walley et al. 2017; Sthapit Kandel et al. 2020; Simko et al. 2022) using the assembled lettuce (L. sativa) reference genome (Reyes-Chin-Wo et al. 2017), which can be used to develop markers for lettuce breeding to accelerate selection in offspring (Simko 2013). In addition to GWAS, a reference genome of L. virosa will also facilitate genomic analyses for various biological questions. A whole-genome screening can search genetic determinants, for instance, that trigger a resistance response. Genome rearrangements can be detected between L. virosa and other Lactuca species via comparative genomics.
The creation of a reference genome for L. virosa is challenging, because, even though it is a diploid species (2n = 2x = 18), it has a considerably larger genome (3.7 Gbp) than L. sativa (2.5 Gbp) and L. saligna (2.3 Gbp) (Doležalová et al. 2002). This is likely due to transposable elements (TEs) (Wendel et al. 2016). To date, there is only a single available genome assembly of L. virosa (CGN04683) (Wei et al. 2021), which is a short-read based and highly fragmented assembly (3,694,810 scaffolds; N50 = 4,910 bp) with relatively high completeness (BUSCO (Benchmarking Universal Single-Copy Orthologue) = 92.7%). Long-read sequencing could significantly improve the accuracy and continuity of a L. virosa genome assembly.
Here, we present a near chromosome-level de novo assembly of L. virosa (CGN04683) using a combination of long-read and short-read sequencing plus Bionano and Dovetail scaffolding. We contextualize the L. virosa genome within the lettuce gene pool together with the L. sativa and L. saligna (Xiong et al. 2023) genomes. First, we show shared and specific homology groups across the 3 species. Based on homologs, we show interspecific collinearity with an emphasis on inversions in different chromosomes. Next, we demonstrate that the proliferation of long-terminal repeat (LTR) superfamilies underlies the genome expansion of L. virosa. Finally, we describe a well-classified inventory of the 2 important resistance-related gene types encoding nucleotide-binding (NB) leucine-rich repeat (NLR) receptors and receptor-like kinases (RLK).
Materials and methods
DNA and RNA sequencing
L. virosa accession CGN04683, is also known as IVT280 and is resistant to Nasonovia ribisnigri (currant-lettuce aphid) (Eenink et al. 1982). Single seed descent of accession “IVT280” (seeds obtained from a breeding company) was grown for whole-genome sequencing. The seeds were stratified at 4°C for 3 days to improve germination. Subsequently, seedlings were grown in a growth chamber at 18–21°C and a relative humidity of 75–78%. After 8 weeks, plants were transplanted to larger pots containing potting soil and grown under greenhouse conditions. Tissue sampling was performed when plants were close to bolting, and DNA was extracted using the same protocol described in Xiong et al. (2023). DNA material was used to prepare libraries with the SMRTbell Template Prep Kit 1.0 and SMRTbell Damage Repair Kit. For library construction, we used the Procedure & Checklist –Preparing >30 kb SMRTbell Libraries Using the Megaruptor Shearing and BluePippin Size-Selection System. Then, we produced a 20-fold coverage of long-read data generated by PacBio Sequel technology using 20 SMRT cells. For the Illumina data, the Illumina TruSeq DNA Sample Preparation kit was used. Then, mechanical DNA shearing using Covaris E210, Illumina TruSeq DNA Sample Preparation Guide. Flowcell cluster generation was done using an Illumina cBot device, sequencing was done using an Illumina HiSeq2000 platform. We used an insertion size of 500 bp and a read length of 125 bp to obtain a 69-fold coverage of paired-end (PE) reads. An optical mapping library of 130 × coverage was produced by Bionano mapping for hybrid scaffolding. For Bionano, DLE-1 (Direct Label Enzyme) labeling enzyme was used at a density of 17.54/100 kbp. The Bionano Genomics Direct Label and Stain (DLS) Kit was used and 30206-Bionano-Prep-Direct-Label-and-Stain-DLS-Protocol_rev D was used for library construction. A Hi-C library produced by Dovetail Genomics provided 10,492 × physical coverage of the genome (10 kbp–10 Mbp pairs) for in vitro proximity ligation (Supplementary Table 1). Finally, 10X sequencing was performed as well with the DNA material (150 bp read length). For this, we used the Chromium Genome Library, Gel Bead & Multiplex kit. Libraries were subsequently constructed using the Chromium Genome Reagent Kits User Guide, using a 10X Genomics Chromium controller. As additional evidence for gene prediction, RNA was isolated from pooled samples of leaf, root, and flower tissues (pooled from different floral stages) using a Direct Zol RNA Miniprep Plus kit (Zymo Research) followed by treatment with DNAse. RNA was purified by ethanol precipitation. The concentration and purity of RNA samples were measured with a Nanodrop 2000c spectrophotometer and a Qubit 4.0 fluorometer using an RNA Broad Range assay (Thermo Fisher Scientific). PE sequencing (2 × 125 bp) was performed on an Illumina HiSeq2500 platform (Supplementary Table 1). All library preparation, construction, and sequencing were performed in-house at the genomics facility of Wageningen University and Research Business Unit Bioscience.
Genome assembly and annotation process
Genome size estimation
After trimming, PE Illumina reads of L. virosa were used for genome size estimation (∼1,590 million reads; ∼183 Gb). Jellyfish v2.3.0 was used with a k-mer size of 21 to count k-mer frequencies (maximum 1 million count) (Marçais and Kingsford 2011). The Jellyfish output was used by GenomeScope (v1.0) to estimate haploid genome length, percentage of repetitive DNA, and heterozygosity of the L. virosa genome (Ranallo-Benavidez et al. 2020).
Genome assembly
PacBio reads were assembled using Canu (v1.3) and then polished by Pilon (v1.20) using Illumina data (Walker et al. 2014; Koren et al. 2017). Then, we performed hybrid scaffolding for the assembly using the Bionano optical mapping data with the Bionano solve software. Mis-joins in assembled contigs were corrected using the HiRise pipeline with the Hi-C data (Putnam et al. 2016). Since the resulting assembly of Hi-C scaffolding was only 75.2% BUSCO complete, the publicly available—but highly fragmented—assembly for L. virosa (Wei et al. 2021) was used to augment the completeness of our assembly. The newly generated PE Illumina reads were trimmed before use with Trimmomatic v0.39 ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 (Bolger et al. 2014), and the barcodes of the 10X mate pairs were stripped with Longranger v2.2.2 basic. Before combining the assemblies, we first polished our assembly for a second time with the PE Illumina reads and the 10X mate pair reads (treated as single-end reads) using Pilon v1.24 –changes –diploid –fix all (Walker et al. 2014). Mapping of sequencing reads for combining these 2 assemblies was performed with bwa-mem2 v2.2.1 (Vasimuddin et al. 2019). Next, we combined our assembly with all sequences >1 kb in the Wei et al. (2021) assembly by concatenating the 2 genome assemblies. We then aligned all PE Illumina and 10X data to the combined genome. The coverage of this data was used to get the best haplotype representation of the complete genome with purge_haplotigs v1.1.1 (cutoffs were 10, 85, 180) (Roach et al. 2018). Since the number of sequences in the resulting assembly increased from 29 to 54,814, we applied several filtering steps to reduce the number of small, uninformative sequences. We filtered out possible mitochondrial and plastid sequences by blasting (BlastN) all sequences to the mitochondrial and plastid NCBI databases (dd. 2021 August 3). We filtered out non-Viridiplantae sequences as identified by a Blastn search against the NCBI database. Then, we polished the newly added sequences using the same method we used to polish our original genome assembly before (with Pilon v1.24). Based on coverage of PE Illumina and 10X data, we used purge_haplotigs to check whether any duplications were introduced, but since this was not the case, we did not apply purge_haplotigs a second time. For scaffolding the newly added sequences, we mapped the original PacBio data to the genome with minimap2 v2.21-cxmap-pb (Li 2018). Scaffolding was done with LRScaf v1.12-misl 3-t mm (Qin et al. 2019). To keep only potential gene coding sequences, we mapped the RNA-seq data with STAR v2.7.7a (Dobin et al. 2013) and removed all sequences lacking a single alignment. Finally, we also removed all sequences smaller than 5 kb.
Assessment of genome completeness
Genome and proteome (annotation) completeness were assessed using BUSCO v5.2.0 with the “eudicots_odb10” dataset (Manni et al. 2021). K-mer completeness was assessed with KAT v2.4.1 with a k-mer value of 31 (Mapleson et al. 2016).
Repeat annotation
To annotate the repetitive elements in the L. virosa genome, a custom library was created by combining different sources: a de novo library of TEs created by RepeatModeler (v2.0.1) with -LTRStruct parameter, a de novo library of miniature inverted-repeat transposable elements (MITEs) searched by MITE-hunter, and a specific database for the genus Lactuca extracted from a combined database of Dfam (20170127) and Repbase (20170127) (Han and Wessler 2010; Bao et al. 2015; Hubley et al. 2016; Flynn et al. 2020). Then, RepeatMasker (v4.0.7) was used to soft mask the L. virosa genome assembly (Smit et al. 2019). The same pipeline was also applied to create a TE library and mask the genome assembly of L. saligna version 4 (PRJEB35809) and L. sativa version 7 (GCF_002870075.2), which were used in reference-based repeatome comparison. The 3 generated TE libraries were used for a reference-free approach to TE classification (see Individual and comparative clustering analysis of repetitive elements below). The RepeatMasker outputs were further processed to summarize the different categories of repeat elements. Moreover, the LTR elements were extracted from the cross_match output of RepeatMasker and compared to the genome to determine their relationship to the genetic regions using bedmap in BEDOPS toolkit (v2.4.40) (Neph et al. 2012).
Gene prediction
Protein-encoding genes in the nuclear assembly were annotated using MAKER2, which combines de novo gene prediction and homology prediction (Holt and Yandell 2011). rRNA reads were filtered out from the RNA-seq data by SortMeRNA version 4.3.4 (Kopylova et al. 2012) using all databases to remove noncoding rRNA. Subsequently, HISAT2 (v2.2.1) was applied to map the remaining RNA-seq reads to the final genome assembly, which includes nuclear sequences, the mitochondrion assembly of CGN013357 (MZ159960.1), and plastid assembly of TKI-404/CGN04683 (CNP0000335 on Chinese National GeneBank (CNGB)) (Kim et al. 2015; Fertet et al. 2021; Wei et al. 2021). The alignment to the nuclear sequences was used as input to BRAKER (version 2) and Stringtie (v2.1.6) to conduct de novo gene prediction and transcriptome assembly, respectively, both with default settings (Pertea et al. 2015; Hoff et al. 2016). The protein alignment was done by BLAST in MAKER2 during the integration with protein databases of A. thaliana (Araport11), L. sativa, Helianthus annuus (HA412.v1), and Uniprot (SwissProt set only: release-2019_10). The predicted transcripts were then filtered using the following criteria: eAED >0.9 (computed by MAKER2), protein length <50, identical isoforms, and missing start and stop codon.
Functional annotation
Potential biological function of proteins was inferred using 3 criteria: (1) best-hit matches in SwissProt, TrEMBL using DIAMOND version 2.0.14 at E-value cutoff of 1e-5 (Buchfink et al. 2015); (2) protein domains/structure identified by InterProscan 5.53–87.0 against the Pfam, Coils, Gene3D, PANTHER, SUPERFAMILY, ModiDBLite, and TIGRFAM databases (Zdobnov and Apweiler 2001; El-Gebali et al. 2019); and (3) orthology searches for pathway information were conducted by Kofamscan (Aramaki et al. 2020) using a customized HMM database of KEGG orthologs (Kanehisa 2000) with an E-value cutoff of 1e-5.
Homology analysis
Gene space analysis
To enable a comparison between L. virosa, L. saligna, and L. sativa, we used PanTools v3.4.0 (Jonkheer et al. 2022) to calculate homologous relationships in a predicted panproteome of these 3 species. We used the longest isoform for each gene. Based on an optimal distribution of BUSCO genes, we decided to use “pantools group -rn 2” for homology grouping. Subsequent gene classification of the homology groups was also done with PanTools. The number of shared groups was visualized with ComplexUpset (Krassowski 2020). Functional enrichment analyses were performed and visualized for the unique sets of genes with ClusterProfiler v3.18.1 (Yu et al. 2012).
Synteny detection
MCScanX (Wang et al. 2012) was utilized to detect syntenic blocks (default settings) among the 3 Lactuca species using the calculated homology groups from PanTools v3.4.0. The interspecific collinearities were visualized using SynVisio (Bandi and Gutwin 2020). MCScanX was run a second time to detect the tandem arrayed genes using DIAMOND (version 2.0.14) on proteomes for each species.
Individual and comparative clustering analysis of repetitive elements
RepeatExplorer2 on a Galaxy server was used (https://repeatexplorer-elixir.cerit-sc.cz/) to conduct individual and comparative clustering of Illumina PE reads (all trimmed to a length of 120 bp) for 3 Lactuca species (L. sativa, L. saligna, and L. virosa) (Novák et al. 2020). Resequencing data of these 3 Lactuca species were retrieved from the European Nucleotide Archive (ENA) database (PRJEB36060). Trimmed FASTQ reads were converted to FASTA format and interlaced before the clustering analysis. In addition, a 4-letter prefix identity code was added to each sample dataset (i.e. Lsat for L. sativa, Lsal for L. saligna, and Lvir for L. virosa). After a preliminary round, each set of reads was randomly subsampled with the same proportion to maximize the repeat detection and annotation accuracy. For individual analysis, reads representing 20% of the genome size were separately clustered for each Lactuca species (i.e. genome proportion = 0.2X, L. sativa = 4,166,668 reads, L. saligna = 3,833,334 reads, and L. virosa = 6,166,668 reads). For comparative analysis, a mixed dataset of reads equal to 0.07 × depth for all species were clustered at once (i.e. genome proportion = 0.07 ×, L. sativa = 1,307,006 reads, L. saligna = 1,420,966 reads, and L. virosa = 2,103,018 reads). For both analyses, the reads were clustered based on the default settings (90% similarity, 55% coverage), and clusters containing more than 0.01% reads were classified at a supercluster level.
After clustering, repeat reads were annotated based on a similarity search to REXdb (protein domain in retrotransposons, Viridiplantae version 3) (Neumann et al. 2019) using BLAST on the Galaxy server. Additionally, the custom libraries previously created by reference-based searches were utilized as an additional custom library to further annotate the repeat clusters (see previous section: Repeat annotation). After annotation, clusters from plastid and mitochondrial origins were identified and excluded for downstream analysis. Next, we quantified different TE categories based on clusters and their connections to superclusters. To characterize the interspecific difference, the clusters resulting from the comparative analysis were sorted via hierarchical clustering (ward.D2) using transformed read number [log2(count + 1)] in each cluster for every species.
Analysis of immune gene repertoire
NLRs were searched for in the predicted proteomes of L. virosa and L. sativa, and retrieved from the L. saligna genome (Xiong et al. 2023). HMMER v3.3.2 (Finn et al. 2011) was used to search Hidden Markov Models (HMMs) profiles obtained from Pfam or the UC Davis database for structural domains of NLR proteins (E-value cutoff = 1e-10): PF00931.23 and NBS_712.hmm (https://niblrrs.ucdavis.edu/At_RGenes/HMM_Model/HMM_Model_NBS_Ath.html) for the NB domain; PF01582.20 and PF13676.6 for TIR (TOLL/interleukin-1 receptor); PF05659.11 and PF18052.1 for CC (coiled-coil); and 8 HMMs for the LRR (leucine-rich repeat) domain (PF00560.33, PF07723.13, PF07725.13, PF12799.7, PF13306.6, PF13516.6, PF13855.6, PF14580.6). Furthermore, NB and LRR domains identified by InterProScan (see Functional annotation), and CC motifs predicted by Paircoil2 (P scores <0.025) were combined with the HMMER output (Zdobnov and Apweiler 2001; McDonnell et al. 2006). The identified NLRs were classified as TNL [Toll-interleukin-1 receptor-like NB site Leucine-rich repeat (NBS-LRR)] or CNL [Coiled Coil (CC), Resistance to powdery mildew8 (RPW8), or potato R protein domain (Rx_N) NBS-LRR] based on the presence of either the TIR or CC domain, respectively. To further solve the unclassified NLRs (TNL or CNL), a phylogenetic tree for amino-acid (aa) sequences with NB domains was constructed. First, aa sequences were aligned using HmmerAlign (Finn et al. 2011). The alignment was then trimmed by trimAl using -automated1 mode and retained 727 residues for phylogenetic construction (Capella-Gutiérrez et al. 2009). A maximum-likelihood (ML) tree was inferred by IQTREE version 1.6.12 (-m PMB + F + R10) with 1,000 ultrafasta bootstrap (UFBoot) replicates (Nguyen et al. 2015). The phylogenetic tree was visualized and annotated using iTOL v6 (Letunic and Bork 2021).
An Inventory of RLKs was also performed for L. virosa and L. sativa. First HMMER (v3.3.2) was used to search the Pkinase domain (PF00069; E-value cutoff = 1e-10). Then, proteins containing Pkinase were examined for the existence of extracellular domains using HMMER (E-value cutoff = 1e-3) and transmembrane regions using TMHMM (v2.0) and SCAMPI (v2) (Krogh et al. 2001; Peters et al. 2016).
Results and discussion
Genome assembly and annotation
We created a complete and structurally informative genome assembly for L. virosa with a total length of 3.45 Gbp (Table 1). Based on a k-mer analysis of Illumina data, we estimated the genome size to be 3.3 Gbp with 73% repeat content and 0.169% heterozygosity rate (Supplementary Fig. 1). This predicted size was lower than the previously measured C-value (3.7 Gbp) (Doležalová et al. 2002), which might be caused by the large repeat content of L. virosa (Ranallo-Benavidez et al. 2020). The long-read assembly was based on PacBio and Illumina data and scaffolded using Bionano and Hi-C data. The longest 12 scaffolds out of the 29 scaffolds comprised 99.8% of the total length (3.3 Gbp) of this first assembly, yet not all chromosomes were reconstructed in full. Therefore, we completed the assembly through additional polishing and leveraging the fragmented, short-read-based genome assembly of the same L. virosa accession (Wei et al. 2021) which we combined in a nonredundant way (Supplementary Data 1 and Supplementary Figs. 2b and 2d). The final combined assembly consisted of 5,855 contigs spanning a total of 3.45 Gbp with an N90 score of 116,478,781 and an L90 score of 10 (Supplementary Fig. 2c and Table 2). The BUSCO completeness score was 96.2% (the duplication score was 4.5%; Supplementary Table 3 and Fig. 2d).
Table 1.
Summary of assemblies for Lactuca spp. in this paper.
| Characteristic | L. sativa | L. saligna | L. virosa |
|---|---|---|---|
| Accession ID | GCF_002870075.2 | PRJEB35809 | PRJEB50301 |
| Source | RefSeq (NCBI) | ENA | This study |
| Assembly size (Gb) | 2.39 | 2.17 | 3.45 |
| # seq | 8,325 | 10 | 5,855 |
| N50 scaffold | 257.9 Mb | 238.6 Mb | 316.9 Mb |
| L50 scaffold | 4 | 4 | 5 |
| Genome complete BUSCO | 97.8% (2,273) | 92.4% (2,147) | 96.2% (2,236) |
| # protein-coding genes | 36,136 | 42,908 | 39,887 |
| # transcripts | 46,867 | 45,476 | 42,791 |
| Proteome complete BUSCO | 98.5% (2,291) | 88.8% (2,065) | 90.2% (2,096) |
Based on both expression and orthology evidence, 39,887 protein-coding genes with a total of 42,791 transcripts were annotated. We mapped RNA-seq data from root, leaf, and flower tissue to the genome assembly to support de novo gene prediction. Next, we aligned protein sequences of model plant species to the genome and used MAKER for merging all gene predictions. We filtered the predicted genes to only retain annotations that were in accordance with the provided evidence. The BUSCO score on the resulting proteome was 90.2%, indicating a high level of completeness. Furthermore, we were able to predict functional domains in 93% (37,106) of the genes for various databases (Supplementary Table 4 and Data 2a and 2b). This structural and functional annotation is vital for the biological interpretation of L. virosa data.
Homology grouping of 3 representative Lactuca spp.
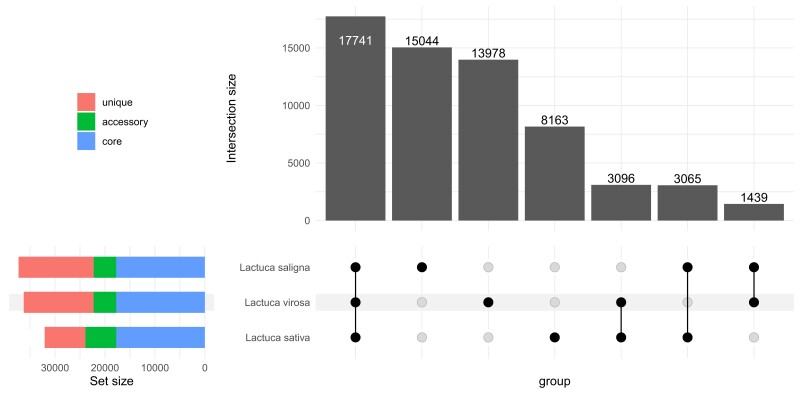
Even though the genome size of L. virosa is substantially larger than L. sativa and L. saligna, the number of genes annotated across species was similar (Table 1). A comparison of L. virosa with L. saligna and L. sativa showed that about half of the homology groups are shared across Lactuca (Fig. 1; Supplementary Data 2c). These 17,741 homology groups in Lactuca contained 19,270 L. virosa genes, meaning that about half of the L. virosa genes are part of the core Lactuca genome. This is comparable to what was found in other interspecies comparisons. For example, in rice ∼62% of core genes were reported between 2 species (Zhao et al. 2018), and in Raphanus, ∼50% of core genes were reported among 11 accessions belonging to 2 species (Zhang et al. 2021). Both L. virosa and L. saligna share fewer homologous genes with each other than with L. sativa. This stresses the importance of wild species in breeding as they contain a large pool of novel genes. The large, unique genomes of both L. virosa and L. saligna indicate that these wild species are rich sources of genetic diversity that thus far has been unexploited for lettuce breeding. We performed a functional annotation for the proteomes of the 3 species with InterProScan to perform functional enrichment for the unique content of L. virosa (15,048 genes; Supplementary Data 2d). The InterProScan domain enrichment found disease resistance proteins to be among the set of significantly enriched domains (Supplementary Fig. 3). Therefore, the genome of L. virosa is a resource for potential novel genes needed for resilience breeding in lettuce.
Fig. 1.
Overview of homology grouping for L. sativa, L. saligna, and L. virosa in an upset plot. The numbers on bars are groups of homologous genes. In total, there are 62,526 homology groups.
Furthermore, it will be relevant to sequence and produce high-quality assemblies of other wild relatives of lettuce, such as L. georgica, L. serriola, and L. aculeata, to obtain an overview of the entire Lactuca gene space (Wei et al. 2021; Guo et al. 2023). Using high-quality genetic resources will enable the construction of a comprehensive pangenome that covers the variation in the genus Lactuca.
Synteny detection between 3 Lactuca spp. via comparative genomics
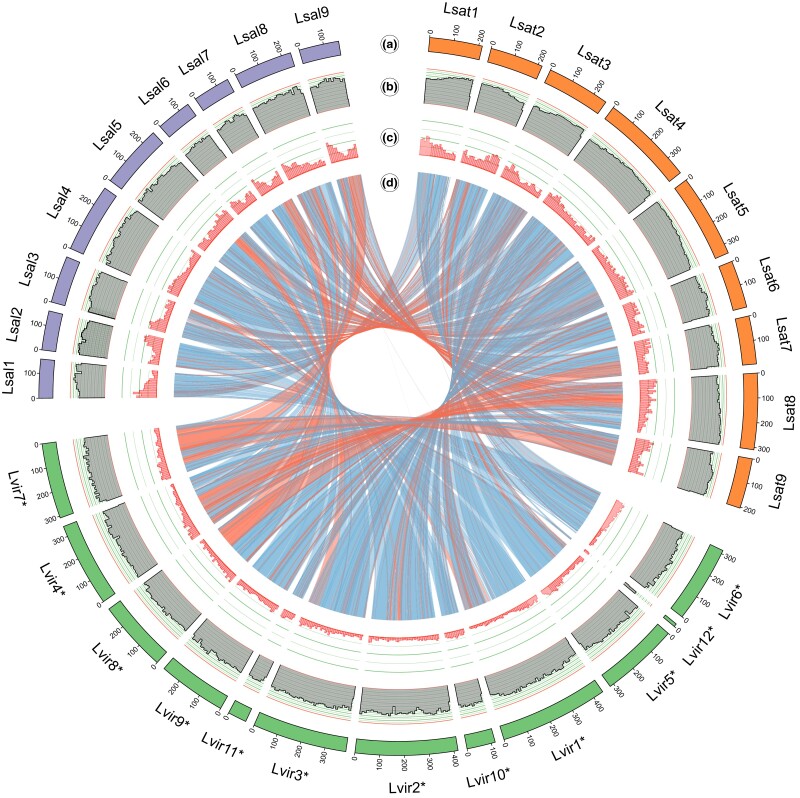
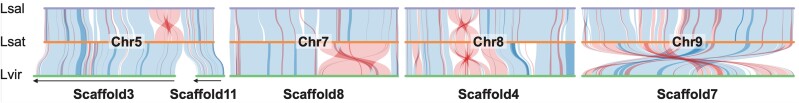
By synteny detection of homologous pairs, we identified major chromosomal inversions between the 3 Lactuca genomes. Overall, there was whole-genome collinearity (synteny) among Lactuca species (Fig. 2). Based on the collinearity, we determined the major 12 scaffolds that comprised 96% (3.30 Gbp) of the total genome assembly (Supplementary Table 5). Compared to the L. sativa genome, 3 species-specific inversions were identified on different chromosomes (Fig. 3). Two of the 3 inversions that were previously described between L. saligna and L. sativa were validated and further characterized: one is specific to L. saligna on Chr5 and one is specific to L. sativa on Chr8 (Xiong et al. 2023). Furthermore, synteny also revealed a large inversion specific to L. virosa on tentative Chr7 (Scaffold8) (Fig. 3; Supplementary Table 5). These inversions could hamper genetic mapping of interesting traits and further introgression. The syntenic patterns between L. virosa Chr9 (scaffold7) and the other 2 species showed complicated inverted and translocated regions, which might be due to a reversed joining indicated by the mapping of Hi-C data (Supplementary Fig. 4).
Fig. 2.
Circos plot of L. virosa genome compared with the L. sativa and L. saligna genomes. a) For each of the 3 genomes (Lsat: L. sativa; Lvir: L. virosa; Lsal: L. saligna), only sequences larger than 1 Mbp are shown. For L. sativa and L. saligna, the number of sequences correspond to their chromosomes. Since L. virosa is near-chromosome level, its sequence numbers were indicated with an asterisk (*). The sequences were sorted based on their collinearity to other Lactuca species, and some of the sequences for L. virosa are inverted to match the orientation in previously published genomes (d); the sequence coordinates (in Mbp) show this. b) The repeat density for each sequence is calculated per 10 Mbp and shown here as a fraction. Since the genome assembly for L. virosa has more N bases, repeats are more difficult to find than in the other 2 genomes. The scale ranges between 0 and 1. c) The gene density for each sequence is calculated per 10 Mbp and shown here as a fraction. As the 3 genomes contain approximately the same number of genes but their genome sizes differ, L. virosa has a lower overall gene density. The scale goes from 0 to 0.2. d) Synteny between the 3 genomes. Inversions are shown in red as opposed to noninverted syntenic blocks, which are shown in blue.
Fig. 3.
Synteny discloses species-specific inversions across 3 Lactuca species. Through genomic comparison, major interspecific inversions (red) were identified among the reference genomes of 3 Lactuca species. Here, the synteny in 4 sets of scaffolds/chromosomes reveals species-specific inversions: L. saligna (Lsal: purple), L. sativa (Lsat: orange), and L. virosa (Lvir: green). The chromosome numbers are labeled in the middle. Black arrows at the bottom indicate reversed scaffolds in the L. virosa assembly. Supported by Supplementary Table 5.
Comparative repeatomics between 3 Lactuca spp. via reference-based and reference-free approaches
In the 3 reference assemblies, we annotated repeat elements and classified them into TEs and other repeats (Supplementary Data 3a). The genomes of all 3 Lactuca species contained a major proportion of TEs, in agreement with previous studies (Supplementary Table 6) (Simko et al. 2022). Unsurprisingly, the TE content of the L. virosa (60%) assembly is proportionally lower than that of both L. sativa (74%) and L. saligna (77%), which is likely caused by an incomplete search due to the high N content in the L. virosa assembly. After excluding the N content of each genome, the percentage of TEs for all Lactuca genomes exceeded 80% (Supplementary Fig. 5; Supplementary Table 6). Moreover, almost all identified LTRs (99%) were located in the intergenic regions (Supplementary Fig. 6). To conclude, this reference-based repeat annotation showed that TEs are the most abundant components of Lactuca spp. genomes. However, genome incompleteness and N content of the reference genome assemblies hamper a precise estimation of TEs.
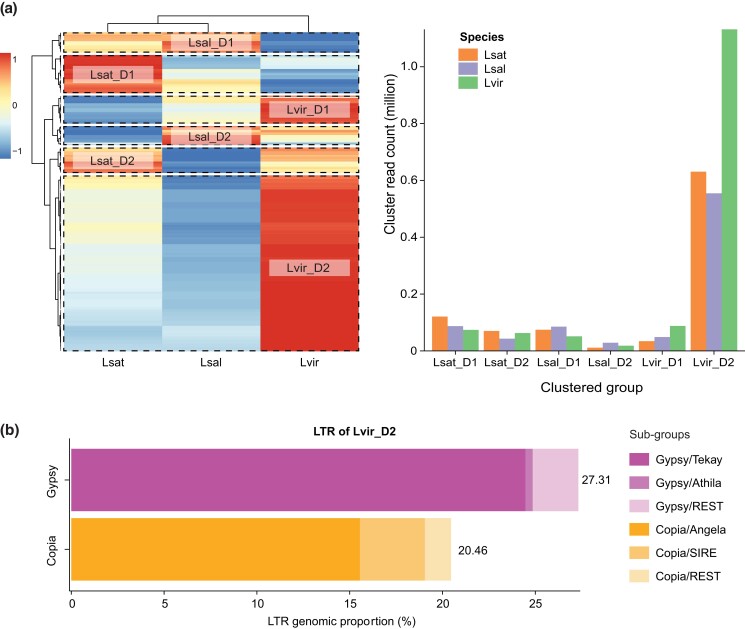
In addition to reference-based repeat annotation, we also classified repeat components and estimated their composition for 3 Lactuca spp. using a reference-free approach. First, the same depth of reads (0.2×) were sampled and clustered for each species for repeat classification (Supplementary Tables 7 and 8). L. virosa had the highest percentage of repeated reads assembled as clusters (82%). The genomic proportion of repeated sequences annotated as TEs was more than 60% for all species, with L. virosa having the highest amount of LTRs (68.34%). Another comparative analysis (read depth = 0.07×) indicated that L. virosa carries a higher percentage of repeats compared to the other 2 species (Supplementary Data 3b and Table 8). For example, cluster 10 was annotated as LTR/Gypsy and mainly composed of L. virosa reads (Supplementary Fig. 7). Moreover, in-depth cluster analysis showed that LTR proliferation in L. virosa drove its genome expansion (Supplementary Data 3c and 3d). The heatmap of hierarchical clustering shows 6 groups that were either dominated (D) by one of the 3 Lactuca species: L. sativa (Lsat), L. saligna (Lsal), or L. virosa (Lvir) (Fig. 4a: left). The bar plot in Fig. 4a (right) further decomposes the read sources for each group. The Lvir_D2 group is the largest and dominated by L. virosa reads. This group mainly consisted of LTR subfamilies Gypsy (27.31%) and Copia (20.46%) (Supplementary Table 9). Additionally, the subgroups Tekay and Angela were the primary elements for the Gypsy and Copia clusters within the Lvir_D2 group (Fig. 4b; Supplementary Table 9).
Fig. 4.
Proliferation of long-terminal repeats (LTR) drives the expansion of the L. virosa genome. Read clusters assembled by RepeatExplorer2 using a mix of resequencing data (coverage = 0.07×) from 3 Lactuca species references to detect the major difference of repeat elements. a) Heatmap shows the scaled read-count of individual cluster (row) for each species (column). Clusters and species were sorted by hierarchical clustering. Six groups (squared by dash lines) were dominated by reads either from L. sativa (Lsat), L. saligna (Lsal), or L. virosa (Lvir) and suffixed with a D (dominant). Bar plot shows the size (y-axis) of 6 clustered groups (x-axis) for each species: L. sativa (orange), L. saligna (purple), and L. virosa (green). b) A stacked bar chart shows the composition of subgroups for the 2 major LTR superfamilies: Gypsy (gradient purple) and Copia (gradient yellow). Supported by Supplementary Table 9.
L. virosa is estimated to have a significantly larger genome (3.7 Gbp) than L. sativa (2.5 Gbp) and L. saligna (2.3 Gbp) (Doležalová et al. 2002). TEs have been shown to drive plant genome expansion (Wendel et al. 2016); for example, within the genus of rice (Ma and Bennetzen 2004; Piegu et al. 2006; Ammiraju et al. 2007). Based on our combined findings, we conclude that the subgroups of transposon LTR, Tekay in Gypsy, and Angela in Copia drove the genome expansion of L. virosa.
Comparison of NLR and RLK genes between 3 Lactuca spp.
Besides the difference within TEs, there is also sizable variation in the number of genes as shown by the homology grouping (accessory/unique genes) among these 3 Lactuca species (Supplementary Fig. 3), which might convey resilience to important traits like resistance against various pathogens or pests. In our previous study, an extensive search of resistance genes was performed for lettuce and its wild relative L. saligna (Xiong et al. 2023). Using the new L. virosa assembly, we identified and classified immunity-related genes encoding NLR and RLK proteins for L. virosa and compared them to L. sativa and L. saligna.
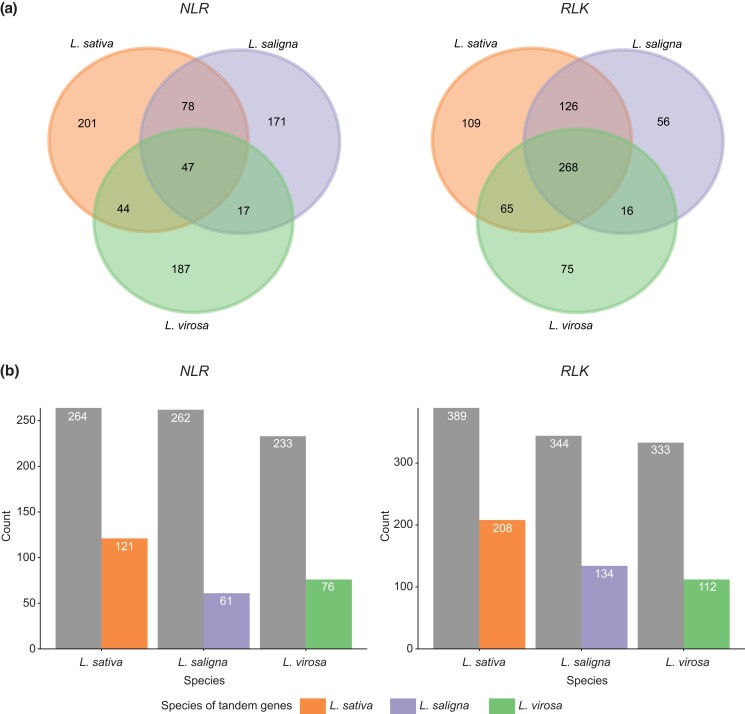
The L. sativa genome was found to have the highest number of NLRs (385), followed by L. saligna (323), and L. virosa (309) (Table 2; Supplementary Table 10). In association with the homology grouping, a Venn diagram showed that the NLRs identified in 3 Lactuca spp. are highly diverged, where more than 50% of NLRs in each species belong to specific homology groups (Fig. 5a: left; Supplementary Data 5a). This observation is in line with our enrichment study of homologs specific to L. virosa, where InterProScan domains were significantly enriched with terms related to NLR proteins (Supplementary Fig. 3). Furthermore, NLR proteins were classified into TNL and CNL types based on the N-terminal domain (TIR or CC domain, respectively) and curated by the phylogeny of a NB domain alignment (Supplementary Fig. 8; Supplementary Data 4a and 4b). The difference between L. sativa, L. saligna, and L. virosa was mainly contributed to TNL genes (227 vs 184 and 180), and the difference between L. saligna and L. virosa can be explained by the CNL type (139 vs 162). Due to the unequal completeness of the proteomes, we applied the ratio of complete BUSCOs for proteomes as a benchmark to anticipate whether NLR genes expand or contract between the 3 Lactuca spp.: L. sativa (2,291), L. saligna (2,065), and L. virosa (2,096). The ratio of BUSCOs (1.10 : 1.00 : 1.02) reflects the NLR ratio across species (1.25 : 1.05 : 1.00), where L. sativa showed a slight inflation. For different NLR types, the number of CNLs was similar in the examined species L. sativa, L. saligna, and L. virosa (1.14 : 1.00 : 1.06); however, the ratio of TNL numbers highly deviated from the BUSCO ratio (1.41 : 1.14 : 1.00; Supplementary Table 10). Such comparison suggests an expansion of NLRs in L. sativa, which is possibly caused by tandem duplication events as in most studied angiosperms (Wu et al. 2021). This hypothesis is supported by a whole-genome search of tandem duplicates (TDs) clusters between 3 Lactuca spp. genomes (Supplementary Data 5b). The number of TDs encoding NLRs in L. sativa (121) was approximately 2-times larger than that in L. saligna (61) and L. virosa (76), which principally explains the number difference among the 3 species (Fig. 5b: left). In addition to tandem duplication, transposon activities (e.g. LTRs) could also greatly elevate the number of NLRs by retroduplication as reported in the chili genome (Kim et al. 2017). The retroduplicated NLRs could partially explain the lineage-specific homologs among Lactuca species (Fig. 5a: left).
Table 2.
Identification and classification of candidate immunity-related genes for Lactuca spp.
| Immune genes | Species | |||
|---|---|---|---|---|
| Family | Classification | L. sativa | L. saligna | L. virosa |
| NLR | CNLa | 158 | 139 | 148 |
| TNL | 227 | 184 | 161 | |
| Total | 385 | 323 | 309 | |
| RLKb | Rcc1-RK | 5 | 5 | 2 |
| WAK | 61 | 48 | 36 | |
| G-LecRK | 132 | 79 | 70 | |
| L-LecRK | 31 | 29 | 21 | |
| C-LecRK | 1 | 1 | 1 | |
| CRK | 41 | 35 | 38 | |
| Malectin-RK | 55 | 55 | 32 | |
| LysM-RK | 12 | 12 | 11 | |
| LRR-RK | 258 | 213 | 233 | |
| PERK | 1 | 1 | 1 | |
| Total | 597 | 478 | 445 | |
RPW8 and Rx_N type of CNL included in this study.
RLK classification based on the extracellular domain (Supplementary Tables 11 and 12).
Fig. 5.
Homology and tandem duplication relationships of immune genes across Lactuca species. a) Venn diagrams of homology groups for NB leucine-rich repeats (NLRs) and RLKs in Lactuca spp. Homology grouping was done by PanTools. b) Bar plots show the count of tandem (colors) and nontandem (gray) NLRs and RLKs in 3 Lactuca species. Tandem-arrayed genes were identified by MCScanX. Supported by Supplementary Data 5.
We next identified RLK proteins by searching for the extracellular, transmembrane, and intracellular domains. Then, resulting RLKs were classified into 9 types based on their extracellular and kinase domains (Supplementary Tables 11 and 12 and Data 4c and 4d). Like NLRs, we found more genes encoding RLK proteins in the L. sativa (597) genome assembly than in L. saligna (478) or L. virosa (445; Table 2). Sequence similarity shows that RLKs were much more conserved in Lactuca spp. compared to NLRs, where 70% of RLKs in each Lactuca species were homologous to another RLK from at least one sister species (Fig. 5a: right). Compared to the BUSCO completeness, the RLK ratio (1.25: 1.00: 1.00) showed an increase of RLKs in L. sativa, suggesting a possible expansion of the RLK family. The majority of expansions in L. sativa were due to G-LecRK, followed by Malectin-RK and WAK, while other types of RLKs were either similar in all species or slightly inflated in L. sativa. The extra G-LecRK and WAK copies might confer specific immunity in L. sativa. For example, G-LecRK and WAK can both mediate resistance to Phytophthora spp. (oomycete) in tobacco and melon plants (Wang et al. 2020; Pi et al. 2022). On the contrary, the expansion of Malectin-RK might benefit pathogen invasion in L. sativa, like the increased susceptibility to Hyaloperonospora arabidopsidis (oomycete) observed in Arabidopsis (Hok et al. 2011). Similar to NLRs, RLKs also commonly expand via tandem duplications. For example, a G-LecRK expansion was reported in soybean (Rodgers-Melnick et al. 2012; Liu et al. 2018). The number of tandem arrayed RLKs in L. sativa was 1.5 and 1.9 times that of the RLKs in L. saligna and L. virosa, respectively, which constitutes more than 60% of the difference between L. sativa and other 2 species (Fig. 5b: right; Supplementary Data 5b). Especially for G-LecRK, the number of tandem genes appeared to more than doubled in L. sativa (Supplementary Data 5b).
Conclusions
Here, we present a near chromosome-level genome assembly for L. virosa (accession CGN04683) that has a high level of completeness. As a representative of the tertiary lettuce gene pool, this L. virosa genome assembly enables comparisons with L. sativa of the primary gene pool and L. saligna of the secondary gene pool. For gene content, L. virosa harbors a large number of genes absent from L. saligna and L. sativa and may thus constitute an important source of novel genes for lettuce breeding. Based on synteny, a 3-way genome comparison uncovered species-specific major inversions. These inversions should be considered as likely barriers to gene introgression in future breeding. In addition, we demonstrated that genome expansion in L. virosa is driven by the proliferation of LTR elements. An assembly-based comparison of NLR and RLK genes between Lactuca spp. found more immune system-related genes in the L. sativa genome than in those of the L. virosa and L. saligna genomes. These findings may contribute to future research on gene expression and regulation in L. virosa. Using this novel genome assembly, researchers can subsequently study the genetic variation in L. virosa populations to fully release its potential for lettuce breeding.
Supplementary Material
Acknowledgments
We thank Jiri Macas and Floris Breman for their help with RepeatExplorer2 analysis. We also thank Elizabeth Marie Georgian (UC Davis) for writing support.
Contributor Information
Wei Xiong, Biosystematics Group, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Dirk-Jan M van Workum, Bioinformatics Group, Wageningen University & Research, P.O. Box 633, Wageningen, 6700 AP, The Netherlands.
Lidija Berke, Biosystematics Group, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Linda V Bakker, Bioscience, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Elio Schijlen, Bioscience, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Frank F M Becker, Biosystematics Group, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands; Laboratory of Genetics, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Henri van de Geest, Bioscience, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Sander Peters, Bioscience, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Richard Michelmore, The Genome Center, Genome & Biomedical Sciences Facility, University of California, Davis, 451 East Health Sciences Drive, Davis, CA 95616-8816, USA.
Rob van Treuren, Centre for Genetic Resources, the Netherlands (CGN), Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Marieke Jeuken, Plant Breeding Group, Wageningen University & Research, P.O. Box 386, Wageningen, 6700 AJ, The Netherlands.
Sandra Smit, Bioinformatics Group, Wageningen University & Research, P.O. Box 633, Wageningen, 6700 AP, The Netherlands.
M Eric Schranz, Biosystematics Group, Wageningen University & Research, P.O. Box 16, Wageningen, 6700 AA, The Netherlands.
Data availability
The genome assembly of L. virosa, is available under the BioProject PRJEB50301 (and available under CAKMRJ010000000.1 from ENA). All raw sequencing reads have been deposited in the ENA database under BioProject PRJEB56289. This includes the Illumina, PacBio, 10X, Bionano, and Hi-C whole-genome sequences as well as RNA sequencing data for genome annotation. Supplementary data are available at https://doi.org/10.4121/21900588.
Supplemental material available at G3 online.
Funding
This research was supported by a grant from the International Lettuce Genomics Consortium (ILGC) funded by the Top Consortium for Knowledge and Innovation Horticultural and Starting Materials (grant number 1406-039). W.X. is financially supported by a fellowship from the China Scholarship Council (CSC). D.-J.M.W. is financially supported by the LettuceKnow consortium (https://lettuceknow.nl/), which is a public-private partnership funded by NWO (Nederlandse organisatie voor Wetenschappelijk Onderzoek) (P17-19) and 7 involved plant breeding companies.
Author contributions
W.X.: Software, Formal analysis, Investigation, Data Curation, Writing—Original Draft, Writing—Review & Editing, Visualization. D.-J.M.W.: Software, Formal analysis, Data Curation, Writing—Original Draft, Writing—Review & Editing, Visualization. L.B.: Methodology, Software, Investigation, Formal analysis, Writing—Review & Editing. L.V.B.: Software, Formal analysis. E.S.: Resources. F.F.M.B.: Methodology. H.G.: Software, Formal analysis. S.P.: Conceptualization, Methodology, Writing—Review & Editing. R.M.: Resources, Writing—Review & Editing. R.T.: Conceptualization, Resources, Writing—Review & Editing. M.J.: Conceptualization, Methodology, Investigation, Resources, Writing—Review & Editing. S.S.: Software, Supervision, Writing—Review & Editing. M.E.S.: Conceptualization, Methodology, Supervision, Writing—Review & Editing, Project administration, Funding acquisition.
Literature cited
- Ammiraju JSS, Zuccolo A, Yu Y, Song X, Piegu B, Chevalier F, Walling JG, Ma J, Talag J, Brar DS, et al. 2007. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 52(2):342–351. doi: 10.1111/j.1365-313X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 36(7):2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi V, Gutwin C. 2020. Interactive exploration of genomic conservation. In Proceedings of the 46th Graphics Interface Conference on Proceedings of Graphics Interface 2020 (GI’20), Waterloo.
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6(1):11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležalová I, Lebeda A, Janeček J, Číhalíková J, Křístková E, Vránová O. 2002. Variation in chromosome numbers and nuclear DNA contents in genetic resources of Lactuca L. species (Asteraceae). Genet Resour Crop Evol. 49(4):385–397. doi: 10.1023/A:1020610129424. [DOI] [Google Scholar]
- Eenink AH, Groenwold R, Dieleman FL. 1982. Resistance of lettuce (Lactuca) to the leaf aphid Nasonovia ribis nigri. 1. Transfer of resistance from L. virosa to L. sativa by interspecific crosses and selection of resistant breeding lines. Euphytica. 31(2):291–299. doi: 10.1007/BF00021643. [DOI] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2019. The pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertet A, Graindorge S, Koechler S, de Boer G-J, Guilloteau-Fonteny E, Gualberto JM. 2021. Sequence of the mitochondrial genome of Lactuca virosa suggests an unexpected role in Lactuca sativa's Evolution. Front Plant Sci. 12:1565. doi: 10.3389/fpls.2021.697136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. Repeatmodeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT . 2019. Food and Agriculture Organization of the United Nations. Rome (Italy): FAOSTAT. [Google Scholar]
- Guo Z, Li B, Du J, Shen F, Zhao Y, Deng Y, Kuang Z, Tao Y, Wan M, Lu X, et al. 2023. LettuceGDB: the community database for lettuce genetics and omics. Plant Commun. 4:100425. doi: 10.1016/j.xplc.2022.100425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wessler SR. 2010. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38(22):e199–e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 32(5):767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hok S, Danchin EGJ, Allasia V, Panabiéres F, Attard A, 2011. An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 34(11):1944–1957. doi: 10.1111/j.1365-3040.2011.02390.x. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 12(1):491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AFA, Wheeler TJ. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44(D1):D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkheer EM, van Workum D-JM, Sheikhizadeh Anari S, Brankovics B, de Haan JR, Berke L, van der Lee TAJ, de Ridder D, Smit S. 2022. Pantools v3: functional annotation, classification and phylogenomics. Bioinformatics. 38(18):4403–4405. doi: 10.1093/bioinformatics/btac506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park J, Yeom SI, Kim YM, Seo E, Kim K-T, Kim M-S, Lee JM, Cheong K, Shin H-S, et al. 2017. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18(1):210. doi: 10.1186/s13059-017-1341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassowski M. 2020. ComplexUpset.
- Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PL, Huang Y, Shi PH, Yu M, Xie JB, Xie L. 2018. Duplication and diversification of lectin receptor-like kinases (LecRLK) genes in soybean. Sci Rep. 8(1):5861. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 101(34):12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve B. 2003. Lactuca virosa, a source of disease resistance genes for lettuce breeding: results and difficulties for gene introgression. In: Eucarpia leafy vegetables, pp. 61–67.
- Maisonneuve B, Chovelon V, Lot H. 1991. Inheritance of resistance to beet western yellows virus in Lactuca virosa L. HortScience. 26(12):1543–1545. doi: 10.21273/HORTSCI.26.12.1543. [DOI] [Google Scholar]
- Maisonneuve B, Chupeau MC, Bellec Y, Chupeau Y. 1995. Sexual and somatic hybridization in the genus Lactuca. Euphytica. 85(1–3):281–285. doi: 10.1007/BF00023957. [DOI] [Google Scholar]
- Maisonneuve B, Pitrat M, Gognalons P, Moury B. 2018. Growth stage-dependent resistance to the potyviruses lettuce Italian necrotic virus and lettuce mosaic virus displayed by Lactuca sativa introgression lines carrying the Mo3 locus from L. virosa. Plant Pathol. 67(9):2013–2018. doi: 10.1111/ppa.12909. [DOI] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ. 2016. KAT: a k-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 33:574–576. doi: 10.1093/bioinformatics/btw663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell AV, Jiang T, Keating AE, Berger B. 2006. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 22(3):356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- Mikel MA. 2007. Genealogy of contemporary north American lettuce. HortScience. 42(3):489–493. doi: 10.21273/HORTSCI.42.3.489. [DOI] [Google Scholar]
- Mikel MA. 2013. Genetic composition of contemporary proprietary U.S. lettuce (Lactuca sativa L.) cultivars. Genet Resour Crop Evol. 60(1):89–96. doi: 10.1007/s10722-012-9818-6. [DOI] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics. 28(14):1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Novák P, Hoštáková N, MacAs J. 2019. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob DNA. 10(1):1–17. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J. 2020. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat Protoc. 15(11):3745–3776. doi: 10.1038/s41596-020-0400-y. [DOI] [PubMed] [Google Scholar]
- Parra L, Maisonneuve B, Lebeda A, Schut J, Christopoulou M, Jeuken M, McHale L, Truco M-J, Crute I, Michelmore R. 2016. Rationalization of genes for resistance to Bremia lactucae in lettuce. Euphytica. 210(3):309–326. doi: 10.1007/s10681-016-1687-1. [DOI] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Tsirigos KD, Shu N, Elofsson A. 2016. Improved topology prediction using the terminal hydrophobic helices rule. Bioinformatics. 32(8):1158–1162. doi: 10.1093/bioinformatics/btv709. [DOI] [PubMed] [Google Scholar]
- Pi L, Yin Z, Duan W, Wang N, Zhang Y, Wang J, Dou D. 2022. A G-type lectin receptor-like kinase regulates the perception of oomycete apoplastic expansin-like proteins. J Integr Plant Biol. 64(1):183–201. doi: 10.1111/jipb.13194. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16(10):1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26(3):342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Wu S, Li A, Zhao F, Feng H, Ding L, Ruan J. 2019. LRScaf: improving draft genomes using long noisy reads. BMC Genomics. 20(1):955. doi: 10.1186/s12864-019-6337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. 2020. Genomescope 2.0 and smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 11(1):1–10. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song Chi, Xia L, Froenicke L, Lavelle DO, Truco M-J, et al. 2017. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun. 8(1):14953. doi: 10.1038/ncomms14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. 2018. Purge haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Mane SP, Dharmawardhana P, Slavov GT, Crasta OR, Strauss SH, Brunner AM, DiFazio SP. 2012. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22(1):95–105. doi: 10.1101/gr.125146.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I. 2013. Marker-assisted selection for disease resistance in Lettuce. In: Translational Genomics for Crop Breeding, Volume I: Biotic Stress. Wiley. p. 267–289. [Google Scholar]
- Simko I, Peng H, Sthapit Kandel J, Zhao R. 2022. Genome-wide association mapping reveals genomic regions frequently associated with lettuce field resistance to downy mildew. Theor Appl Genet. 135(6):2009–2024. doi: 10.1007/s00122-022-04090-3. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2019. 2013–2015. RepeatMasker Open-4.0.
- Sthapit Kandel J, Peng H, Hayes RJ, Mou B, Simko I. 2020. Genome-wide association mapping reveals loci for shelf life and developmental rate of lettuce. Theor Appl Genet. 133(6):1947–1966. doi: 10.1007/s00122-020-03568-2. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Ryder EJ. 1961. Description and pedigrees of nine varieties of lettuce. Technical bulletin (United States. Department of Agriculture). 1244:1–19. [Google Scholar]
- Vasimuddin M, Misra S, Li H, Aluru S. 2019. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In: Proceedings of the IEEE 33rd International Parallel and Distributed Processing Symposium, IPDPS 2019. Rio de Janeiro (Brazil): Institute of Electrical and Electronics Engineers Inc. p. 314–324.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK. et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley PG, Hough G, Moore JD, Carder J, Elliott M, Mead A, Jones J, Teakle G, Barker G, Buchanan-Wollaston V, et al. 2017. Towards new sources of resistance to the currant-lettuce aphid (Nasonovia ribisnigri). Mol Breed. 37(1):4. doi: 10.1007/s11032-016-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H, et al. 2012. MCScanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49–e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xu X, Zhao G, He Y, Hou C, Kong W, Zhang J, Liu S, Xu Y, Xu Z. 2020. Genetic mapping and candidate gene analysis for melon resistance to Phytophthora capsici. Sci Rep. 10(1):20456. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, van Treuren R, Liu X, Zhang Z, Chen J, Liu Y, Dong S, Sun P, Yang T, Lan T, et al. 2021. Whole-genome resequencing of 445 Lactuca accessions reveals the domestication history of cultivated lettuce. Nat Genet. 53(5):752–760. doi: 10.1038/s41588-021-00831-0. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Jackson SA, Meyers BC, Wing RA. 2016. Evolution of plant genome architecture. Genome Biol. 17(1):1–14. doi: 10.1186/s13059-016-0908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Y, Xue J-Y, Van de Peer Y. 2021. Evolution of NLR resistance genes in Magnoliids: dramatic expansions of CNLs and multiple losses of TNLs. Front Plant Sci. 12:2998. doi: 10.3389/fpls.2021.777157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Berke L, Michelmore R, van Workum DM, Becker FFM, Schijlen E, Bakker LV, Peters S, van Treuren R, Jeuken M, et al. 2023. The genome of Lactuca saligna, a wild relative of lettuce, provides insight into non-host resistance to the downy mildew Bremia lactucae. Plant J. 115:108–126. doi: 10.1111/tpj.16212. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. Clusterprofiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. 2001. Interproscan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu T, Wang J, Wang P, Qiu Y, Zhao W, Pang S, Li X, Wang H, Song J, et al. 2021. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol Plant. 14(12):2032–2055. doi: 10.1016/j.molp.2021.08.005. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, et al. 2018. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet. 50(2):278–284. doi: 10.1038/s41588-018-0041-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly of L. virosa, is available under the BioProject PRJEB50301 (and available under CAKMRJ010000000.1 from ENA). All raw sequencing reads have been deposited in the ENA database under BioProject PRJEB56289. This includes the Illumina, PacBio, 10X, Bionano, and Hi-C whole-genome sequences as well as RNA sequencing data for genome annotation. Supplementary data are available at https://doi.org/10.4121/21900588.
Supplemental material available at G3 online.