Abstract
Uncontrolled inflammation is responsible for acute and chronic diseases in the lung. Regulating expression of pro-inflammatory genes in pulmonary tissue using small interfering RNA (siRNA) is a promising approach to combatting respiratory diseases. However, siRNA therapeutics are generally hindered at the cellular level by endosomal entrapment of delivered cargo, and at the organismal level by inefficient localization in pulmonary tissue. Here we report efficient anti-inflammatory activity in vitro and in vivo using polyplexes of siRNA and an engineered cationic polymer (PONI-Guan). PONI-Guan/siRNA polyplexes efficiently deliver siRNA cargo to the cytosol for highly efficient gene knockdown. Significantly, these polyplexes exhibit inherent targeting to inflamed lung tissue following intravenous administration in vivo. This strategy achieved effective (>70%) knockdown of gene expression in vitro and efficient (>80%) silencing of TNF-α expression in LPS-challenged mice using a low (0.28 mg/kg) siRNA dosage.
Keywords: Anti-inflammatory, siRNA, polymer, polyplex, cytosolic delivery, lung inflammation
INTRODUCTION
Inflammation underlies the pathogenesis of many autoimmune1,2 and chronic diseases3 in the lung including emphysema4 and chronic obstructive pulmonary disease (COPD).5 Acute inflammation in response to pulmonary insult is responsible for many particularly severe conditions6 including acute respiratory distress syndrome (ARDS),7 and contributes to the high mortality of diseases such as COVID-19.8 Corticosteroids administered through inhalation, in tandem with bronchodilators to relax airway muscles, generally serve as the first-line therapy for attenuating pulmonary inflammation,9 but side effects associated with long-term use are a concern.10,11 In recent years antibody therapeutics have emerged as a promising alternative for the treatment of pulmonary inflammation12,13 through inhibition of cytokines14 and chemokines15 or their receptors.16 However, limited distribution to the lungs17 coupled with a high degree of immunogenicity18 and the risk of cytokine release syndrome19 have prevented these methods from reaching clinical translation.
Gene knockdown using small interfering RNA (siRNA) is a promising approach for combatting inflammatory disease through controlled modulation of essential inflammatory pathways.20 , 21 Localization of siRNA in the cytosol is a prerequisite for activity,22 and carrier vehicles23,24 have been developed to promote cellular uptake of anti-inflammatory siRNA.25 Therapeutic delivery of siRNA for treatment of lung inflammation faces challenges related to inefficient delivery on a cellular level due to endosomal entrapment.26 Current siRNA delivery systems generally enter cells through inefficient endosomal uptake pathways, severely curtailing efficiency of cytosolic entry (<10%) and promoting endo/lysosomal degradation of cargo.27
Therapeutic delivery to the lungs in vivo faces challenges due to poor agent localization in pulmonary tissue following intravenous administration.28 In recent years, lipid-based nanoparticle formulations have been used to circumvent these obstacles, but they require a high dosage to achieve effective knockdown in pulmonary tissue. Aerosolization of siRNA-loaded carriers29,30 provides a direct means of delivering RNA cargo to pulmonary tissue,31 however obstacles related to formulation32,33 and inefficient penetration through mucosa must be overcome.34 Intravenous administration provides a straightforward approach to accessing therapeutic sites of action, but generally results in limited localization to organs other than those of the reticuloendothelial system (RES) such as the liver, spleen, and kidneys.35 These challenges collectively increase the dosage requirement for effective treatment, leading to more off-target effects and cytotoxicity in the treatment of lung inflammation.36
Polymeric delivery systems have the potential to address delivery challenges at both cellular and systemic levels, providing an effective therapeutic approach for treatment of lung inflammation. Previous work by our group showed that self-assembly between oligo(glutamate)-tagged proteins and guanidiniumfunctionalized poly(oxanorbornene)imide (PONI-Guan) polymers resulted in discrete supramolecular complexes (~200 nm) with cationic surface charges.37 These complexes delivered protein cargo directly to the cytosol with high efficiency (>90%) through a membrane fusion-like uptake process, evidenced by diffuse cytosolic/nuclear fluorescence of fluorescent protein cargo and negligible overlap with endo/lysosomal markers.38
On a systemic level, PONI-Guan/protein complexes demonstrated inherent tropism for inflamed lung tissue following intravenous administration in murine models of ARDS.39 These studies utilized lipopolysaccharide (LPS) administered through retro-orbital injection to induce inflammation. Delivery using PONI-Guan polymers resulted in substantial accumulation of polymer/protein complexes in inflamed lungs, quantified through radiolabeling. The lungs serve as a major reservoir of neutrophils and other phagocytic immune cells during the innate immune response,40 potentially driving ‘homing’ to the lungs through recognition by neutrophils and macrophages.39
We hypothesized that self-assembly of nucleic acids could proceed in a similar fashion to that observed using anionic oligoglutamate peptide tags. PONI-Guan polymers self-assembled with siRNA through electrostatic interactions to generate discrete (~170 nm) polyplexes with cationic surface charges. The observed physiochemical similarities by size and zeta potential with PONI-Guan/protein complexes suggested that these polyplexes would interact similarly with the cell membrane, promoting cytosolic uptake of siRNA cargo. Systemically, we hypothesized that siRNA behavior would be similar to that observed with proteins, facilitating localization in pulmonary tissue following intravenous administration. Together, these attributes would overcome two major challenges faced by current siRNA delivery strategies.41,42
Here we describe a multi-scale approach for combatting lung inflammation through knockdown of tumor necrosis factor α (TNF-α) in pulmonary cells using systemically administered PONI-Guan/siRNA polyplexes (Figure 1). TNF-α is a potent pro-inflammatory cytokine, heavily implicated in the pathogenesis of severe inflammatory diseases such as pneumonitis.43,44 Targeting TNF-α using siRNA provides an approach to mitigate inflammation and combat inflammatory disease by controlling cytokine expression at an organ-specific level.45 , 46 PONI-Guan homopolymers formed discrete nanoscale polyplexes with siRNA through electrostatic complementarity and delivered cargo to the cytosol in vitro with efficient (>70%) knockdown of a GFP reporter in macrophagess (RAW 264.7) cells. Intravenous administration of PONI-Guan/siRNA polyplexes in LPS-challenged mice resulted in a 3-fold increase in pulmonary localization as compared to naïve mice, with concomitantly effective serum TNF-α knockdown (>80%). Taken together, polymer-siRNA polyplexes immediately provide a powerful tool for silencing of pro-inflammatory genes with clear potential for clinical translation.
Figure 1.
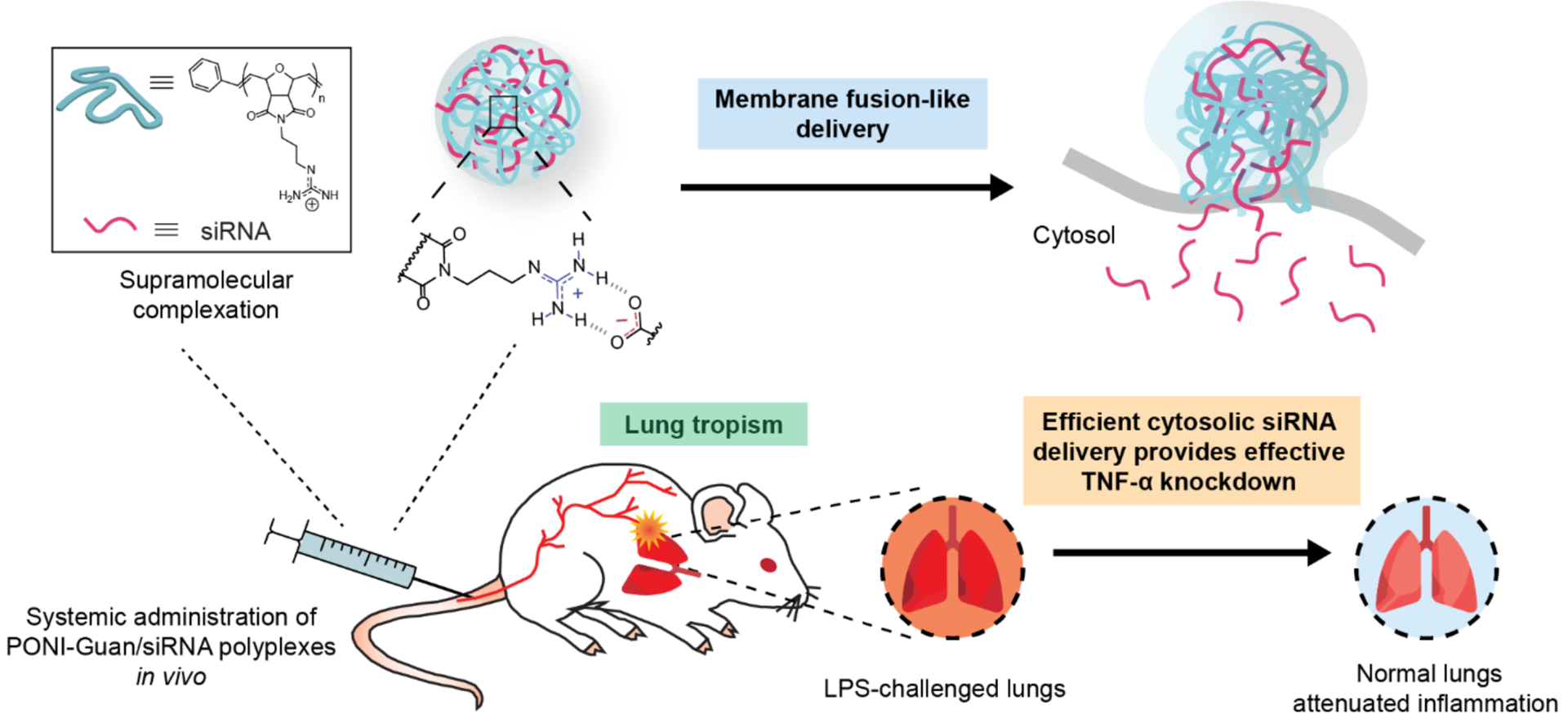
Schematic of study design. Polyplexes form between PONI-Guan polymers and siRNA through self-assembly of the two components, with competent polyplexes releasing siRNA through membrane fusion-like delivery. Systemically administered polyplexes distribute to lung tissue, with preferential localization to inflamed lungs.
RESULTS AND DISCUSSION
Characterization of PONI-Guan/siRNA polyplexes.
PONI-Guan homopolymer (~60 kDa MW) was synthesized, characterized and used for experiments, as it provided the highest efficacy according to our previous reports37 Briefly, PONI-Guan homopolymer was synthesized using ring opening metathesis polymerization (ROMP) using third generation Grubbs’ catalyst, yielding polymers with low polydispersity (Figure S1). Moreover, ROMP provides high control over molecular weights and avoids variation across different batches as shown in GPC characterization (Figure S2). Polyplexes were formed through simple co-incubation with siRNA. Details are available in Methods. Complexation between PONI-Guan and siRNA at varied guanidinium/phosphate (G/P) ratios was confirmed by gel mobility shift assay with complete complexation of siRNA observed (Figure 2a) with sufficient polymer. The sizes and zeta potentials of the PONI-Guan/siRNA polyplexes were measured in triplicate using dynamic light scattering (DLS) (Figure 2b, 2c, and S2). Polyplexes displayed an average diameter of 170 nm with low PDI (less than 0.1). Zeta potential analysis revealed that average charge increased with G/P ratio due to increased charge contribution by cationic PONI-Guan polymers. Transmission electron microscopy (TEM) of PONI-Guan/siRNA polyplexes confirmed spherical morphology with low dispersity (Figure 2d). The encapsulation efficiency of siRNA was verified by Ribogreen assay (Figure 2e). The siRNA encapsulation efficiency in polyplex was ≥97% at mostG/P ratios. Further, functional encapsulation of siRNA in polyplex was verified using nuclease degradation assay. Efficient protection of siRNA against nuclease degradation is a prerequisite for intracellular delivery. Polyplexes were prepared and incubated with RNase A to simulate physiological conditions. The encapsulation of siRNA by PONI-polymers efficiently protected siRNA from enzymatic degradation by RNase A, while free siRNA was fully degraded (Figure 2f). Similarly, polyplexes efficiently protected siRNA against degradation in serum and whole blood (Figure S4). Naked siRNA was hardly detected after being incubated with 10% serum and human whole blood for 1 h. However, the band of siRNA in polyplex could be detected even after being incubated for 12 h, suggesting the stability of siRNA in the polyplex was obviously improved, and indicating that our delivery platform protects the siRNA payload from degradation by plasma proteins.
Figure 2.
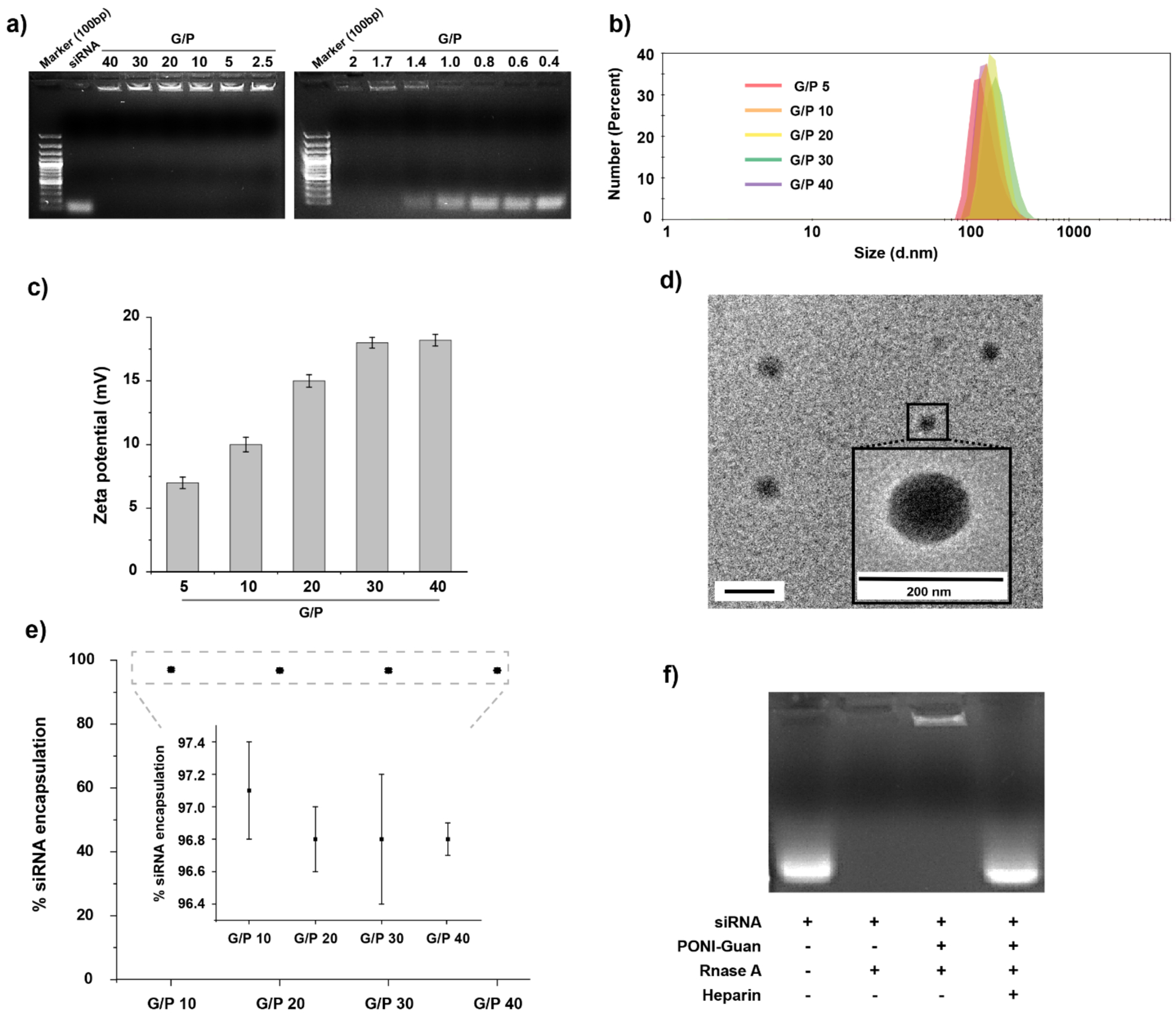
Characterization of PONI-Guan/siRNA polyplexes. a) Agarose gel electrophoresis of PONI-Guan/siRNA polyplexes at varied G/P ratios. b) Hydrodynamic diameters by number and c) zeta potential of PONI-Guan/siRNA polyplexes at varied guanidinium-phophate (G/P) ratios. Polyplex sizes by intensity and volume are available in Supporting information (Figure S2). d) Representative transmission light microscopy (TEM) micrographs of polyplexes at G/P 30. e) encapsulation efficiency of siRNA in polyplexes at varied G/P ratios. f) Nuclease protection of PONI-Guan/siRNA polyplexes at G/P 30 incubated with RNase A.
Evaluation of siRNA delivery using PONI-Guan/siRNA polyplexes.
Delivery into the cytosol is necessary to provide siRNA with access to intracellular machinery for gene silencing. We evaluated the uptake of siRNA mediated by PONI-Guan polymer in macrophages (RAW 264.7 cells) as a proxy cell line for neutrophils in vitro,47 considering the potential tropism of PONI-Guan/protein nanocomposites for phagocytes in inflamed lungs.39 Macrophages play key roles as primary mediators of inflammation and immune cell recruitment in innate immunity,48 making them prime targets for immunomodulation. Polyplexes were prepared with fluorophore-tagged, scrambled-sequence siRNA (Cy3-siRNA) and incubated with RAW 264.7 cells in serum-containing (10% fetal bovine serum, FBS) culture media to simulate physiologically relevant conditions. After incubation, cells were washed with phosphatebuffered saline (PBS) and cellular uptake was visualized by confocal laser scanning microscopy (CLSM). Diffuse fluorescence signals were observed in cell cytosols after incubation with PONI-Guan/Cy3-siRNA polyplexes and cellular internalization of the Cy3-siRNA was confirmed using a Z-stack image (Figure 3a). Cytosolic siRNA delivery was observed in the majority of cells after 6 h and negligible overlap with endo/lysosomes was found after being exposed to Cy3-siRNA polyplexes for 6 h, as visualized by Lysotracker Deep Red counterstaining. We also compared the delivery of Cy3-siRNA with a commercially available transfection reagent (RNAiMAX). Significantly, the PONI-Guan/Cy-3-siRNA signal was evenly distributed within the cytosol while with RNAiMAX had obviously punctate fluorescence, with less overall signal (Figure 3b). Previous studies of siRNA delivery demonstrated when dye labeled siRNA is internalized primarily by endocytosis, it shows a punctate fluorescence pattern.49,50 The diffuse fluorescence observed with the PONI-Guan delivery system is consistent with a non-endocytic entry route, however an endocytitic pathway followed by extraordinarily rapid endosomal escape cannot be fully excluded.
Figure 3.
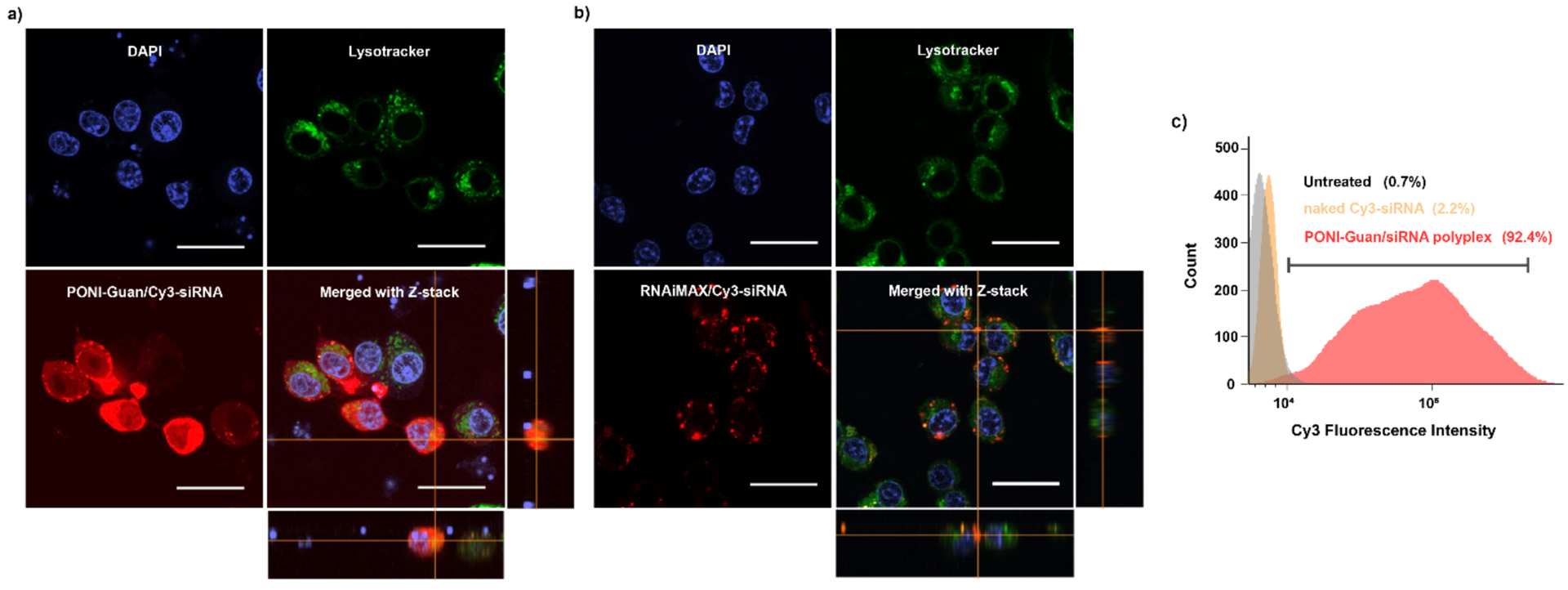
Cytosolic delivery of fluorophore-labeled siRNA by PONI-Guan/siRNA polyplexes. a-b) Representative confocal laser scanning microscopy (CLSM) images of Cy3-labeled siRNA delivery (Red, 25 nM siRNA) using PONI-Guan/siRNA polyplexes at G/P 30 or a commercially available reagent (RNAiMAX) in RAW 264.7 macrophage cells. Endo/lysosomes were stained with Lysotracker Deep Red (Cyan). Diffuse fluorescence and lack of co-localization with Lysotracker indicates cytosolic access of siRNA. Scale bars: 50 μm. c) Flow cytometric histogram profiles of fluorescence intensity of RAW 264.7 cells after 24 h incubation with naked Cy3-siRNA or PONI-Guan/Cy3-siRNA polyplexes at G/P 30 (25 nM siRNA).
Imaging flow cytometry provides a powerful tool for quantifying delivery efficiency using large data sets.51 This technique was utilized to quantitatively determine macrophage cellular uptake of Cy3-siRNA (Figure 3c). PONI-Guan/Cy3-siRNA was prepared as described and incubated with RAW 264.7 cells for 24 h in serum-containing media. After incubation with PONI-Guan/Cy3-siRNA, the percentage of Cy3 positive macrophage cells was measured, which was significantly higher (92.4%) than that of incubation with naked siRNA (2.2%) or non-treated control cells (0.7%). To quantify delivery efficiency, a robust approach was applied to the data as a series of gates, as shown in supporting information (Figure S4). Fluorescence was observed to be diffuse throughout the cytosols and efficient cytosolic delivery was revealed, consistent with the results obtained by CLSM. These studies demonstrate that PONI-Guan/siRNA polyplexes can effectively transport siRNA into the cytosol.
Reporter gene silencing and evaluation of uptake mechanism.
Having demonstrated efficient cytosolic delivery of siRNA, we next investigated gene knockdown efficacy in macrophages. Stably transfected eGFP-expressing murine macrophages (RAW 264.7:eGFP) were incubated with PONI-Guan/siRNA polyplex targeting eGFP (PONI/si_GFP) at varied G/P ratios, or scrambled siRNA (PONI/si_scramble) as a control. After 48 hours of incubation, the eGFP expression profile was monitored by CLSM and flow cytometry. Analysis by flow cytometry revealed effective knockdown (>70%) of eGFP expression at G/P 30 (50 nM siRNA), with eGFP expression reduced in an siRNA concentration-dependent manner (Figure 4a and Figure S6). This optimized ratio was used for subsequent studies. Incubation with scrambled siRNA or naked siRNA had no effect on eGFP expression. Delivery efficiency was also compared with a commercially available transfection reagent for siRNA (RNAiMAX). PONI-Guan/si_GFP polyplexes exhibited gene silencing efficiency superior to the commercial transfection reagent by ~25%.
Figure 4.
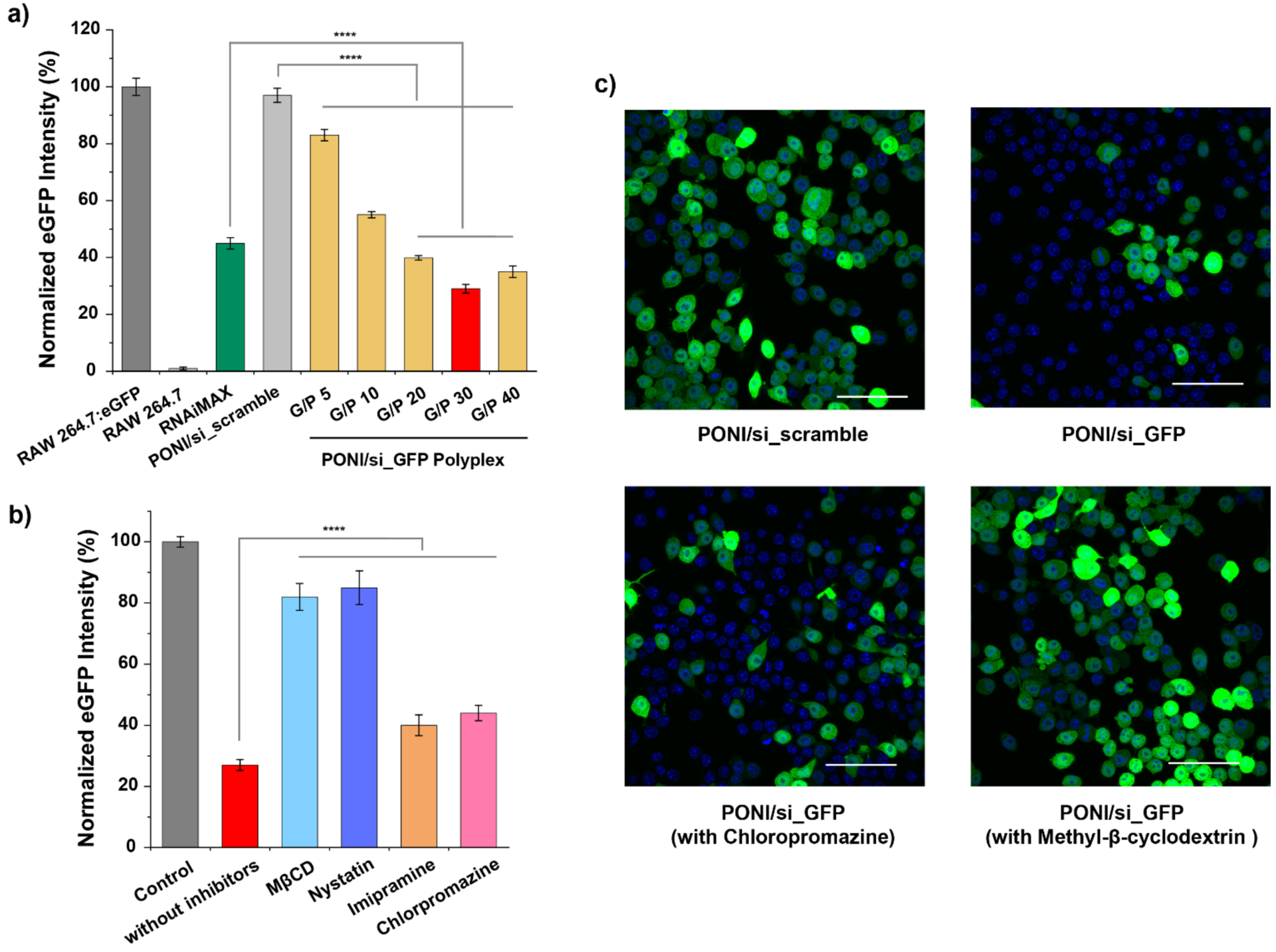
Fluorescent reporter gene silencing and evaluation of uptake mechanism. a) Quantification of eGFP knockdown in RAW 264.7:eGFP cells following treatment with PONI-Guan/si_GFP (50 nM of siRNA) polyplexes as function of G/P ratio, as quantified by cytometry. Flow cytometry histogram profiles of eGFP intensity with PONI-Guan/si_GFP polyplexes is available in Supporting information (Figure S5). b) Quantification of eGFP knockdown in RAW 264.7:eGFP cells following pretreatment with small molecule inhibitors of canonicalendocytosis or cholesterol depletion agents. Fluorescence intensity was measured by flow cytometry and normalized. Error bars represent standard deviation (SD) of three experimental replicates (Data are presented as mean ± SD, one-way Anova and Tukey multiple comparisons, ****p < 0.001) MβCD = methyl-β-cyclodextrin. c) Representative CLSM images of RAW 264.7:eGFP cells after treatment with PONI-Guan/scramble-siRNA polyplexes (top left), PONI-Guan/GFP-siRNA polyplexes (top right), PONI-Guan/GFP-siRNA with chlorpromazine pretreatment (bottom left), and with PONI/GFP-siRNA with methyl-β-cyclodextrin pretreatment (bottom right). Cell nuclei stained with DAPI (blue). Deliveries for b) and c) were performed with polyplexes formulated at G/P 30 ratio with 50 nM of siRNA. Scale bar: 50 μm.
Previous studies of PONI-Guan/protein delivery demonstrated diffuse cytosolic fluorescence of GFP and internalization via non-endocytic uptake37,38 Diffusion of Cy3-siRNA throughout the cytosol after delivery thus suggested that PONI-Guan/siRNA polyplexes followed a similar delivery mechanism. To investigate this possibility, macrophage cells were pretreated with established small molecule inhibitors of uptake mechanisms prior to delivery. RAW 264.7:eGFP cells were pretreated with chlorpromazine and imipramine to target clathrin-mediated endocytosis and micropinocytosis, respectively as canonical endocytic pathways.. Nystatin52 and methyl-β-cyclodextrin (MβCD)53 both deplete cholesterol from the cell membrane, affecting membrane structure and dynamics. The membrane fusion-like process hypothesized for the PON-Guan deliver system relies on choleserol-mediated membrane flexibility,.54,55 Knockdown efficiency was evaluated by flow cytometry (Figure 4b). Consistent with our previous reports that cholesterol provided membrane fluidity important for promoting cytosolic delivery of protein through vehicle fusion with cell membranes,37, 56 pretreatment with membrane cholesterol depletion agents reduced knockdown efficiency by >75%, while endocytosis-related inhibitors had minimal effect on knockdown efficacy (<10%). Comparable results were obtained from analyses via CLSM (Figure 4c). Taken together, these results collectively support our hypothesis that PONI-Guan polyplexes deliver siRNA cargo into cells through a membrane fusion-type mechanism, accessing a highly efficient means of entry for biologic cargo. However, nystatin and MβCD have also been reported to inhibit caveolar uptake mechanisms57, so the possibility of caveolar uptake followed by an unusually fast escape cannot be completely ruled out.
TNF-α gene silencing in vitro.
Pulmonary inflammation in response to infection or injury is a leading cause of mortality for multiple diseases.43 Since this inflammation is strongly modulated by tumor necrosis factor α (TNF-α),44 a potent pro-inflammatory cytokine implicated in systemic inflammation as well as pathogenesis of lung-specific inflammatory diseases, we investigated the delivery of functional anti-TNF-α siRNA (si_TNF-α) as a model anti-inflammatory payload. PONI-Guan/si_TNF-α (50nM of siRNA) polyplexes were incubated with RAW 264.7 cells for 24 h. To stimulate inflammation, cells were then exposed to bacterial lipopolysaccharide (LPS, 1 μg/mL) for 3 h. After incubation, supernatant was collected and TNF-α levels were measured by ELISA, which displayed effective (>65%) decreases in TNF-α secretion (Figure 5a). Degradation of target mRNA was measured by qRT-PCR and revealed significant (80%) inhibition when compared with controls (Figure 5b). The western blot experiments confirmed that the protein levels of TNF-α were also reduced in PONI-Guan/si_TNF-α polyplexes treated RAW 264.7 cells (Figure 5c).
Figure 5.
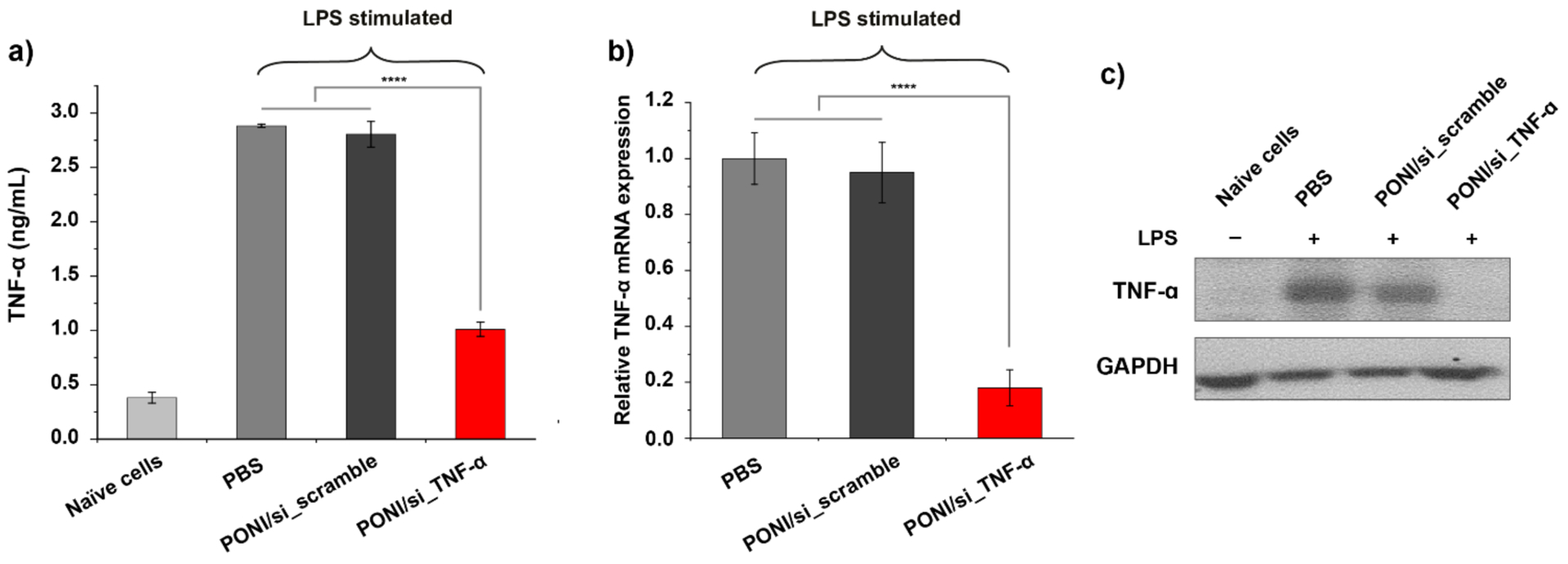
TNF-α gene silencing with PONI-Guan/si_TNF-α polyplexes. a) in vitro delivery of PONI-Guan/si_TNF-α polyplexes decreased TNF-α production in LPS-stimulated RAW 264.7 macrophages. Cells were incubated with PONI-Guan/si_scramble or PONI-Guan/si_TNF-α polyplexes for 24 h, followed by 3 h LPS stimulation (1 μg/ml). Supernatant TNF-α levels were measured by ELISA. Naïve cells indicate RAW 264.7 cells that received no lipopolysaccharide (LPS) treatment. b) Relative TNF-α mRNA expression was verified using quantitative RT-PCR (qRT-PCR). Error bars represent standard deviation (SD) of three experimental replicates (Data are presented as mean ± SD, one-way Anova and Tukey multiple comparisons, ****p < 0.001). c) Western blot analysis of TNF-α in RAW 264.7 cells that were untreated or treated with the indicated siRNA for 48 h. GAPDH was included as a loading control.
We also evaluated the toxicity of the PONI-Guan/siRNA polyplexes delivery platform followed by cell viability assay, immune response tests, and hemolysis assay. Alamar blue assay and cytokine immune response tests demonstrated that PONI-Guan/siRNA polyplexes have negligible toxicity and low immune response at relevant working conditions (Figure S7a–S7c). Hemolysis assays were also performed with human red blood cells to evaluate the biocompatibility of PONI-Guan/siRNA polyplexes (Figure S7d). Similar to Alamar blue assay, PONI-Guan polymers did not show hemolytic activity inferring excellent biocompatibility with the blood compartment.
in vivo polyplex biodistribution in murine models of inflammation.
Motivated by the efficient gene knockdown observed in vitro, we sought to investigate the polyplex-mediated knockdown efficiency of TNF-α in vivo using a model of lung inflammation. A murine model of indirect pulmonary insult model was established through intraperitoneal (I.P.) injection of LPS in BALB/c mice. An initial biodistribution (BD) profile at 24 h after intravenous (I.V.) administration with naked Cy5.5-siRNA (0.28 mg/kg) revealed highest accumulation in the kidney (Figure S8). BD of Cy5.5-labeled polymer alone showed notably enhanced accumulation in the lungs, followed by the liver and the spleen. To investigate the pharmacokinetic differences between polyplexes, siRNA, and polymer alone, BD studies with PONI-Guan/siRNA polyplexes (at G/P 30 with 0.28 mg/kg of siRNA) were performed in parallel using either Cy5.5-labeled polymer or Cy5.5-labeled scrambled siRNA both naïve BALB/c mice and those challenged with 5 mg/kg of LPS. Polyplexes were I.V. administered through the tail vein after induction of lung inflammation, and after 24 h major organs were harvested and imaged using an in vivo imaging system (IVIS). BD results indicated that the majority of polyplexes accumulated in the liver and spleen, with significant accumulation in the lungs as well (Figure 6a). To evaluate % of the injected dose accumulated in the lungs, total radiant efficiency given as (p/s/cm2/sr)/(μW/cm2) from all major organs was first calculated and then the radiant efficiency from the lungs was divided by the total radiant efficiency. Notably, LPS stimulation enhanced lung tropism, eliciting a 2.5-fold increase in the injected polyplex concentration in pulmonary tissue as compared to naïve mice (60% in inflamed lungs versus 25% in naïve mice) (Figure 6b–6c). Imaging using polyplexes with Cy5.5-siRNA likewise showed enhanced accumulation in the lungs of LPS-challenged mice (Figure S9); no changes in body weight or animal health were noted relative to a PBS control (data not shown). Macrophages and neutrophils are key modulators of innate immunity, and crucial players in the pulmonary inflammatory response.47 The results obtained were consistent with our previous reports of lung tropism with PONI-Guan/protein nanocomposites,39 suggesting tropism of PONI-Guan/siRNA polyplexes to inflamed lungs through interaction with these pro-inflammatory phagocytes. Coupled with the effective cytosolic delivery of siRNA to macrophages observed in vitro, these studies demonstrate the potential for therapeutic immunomodulation in a systemic inflammation model.
Figure 6.
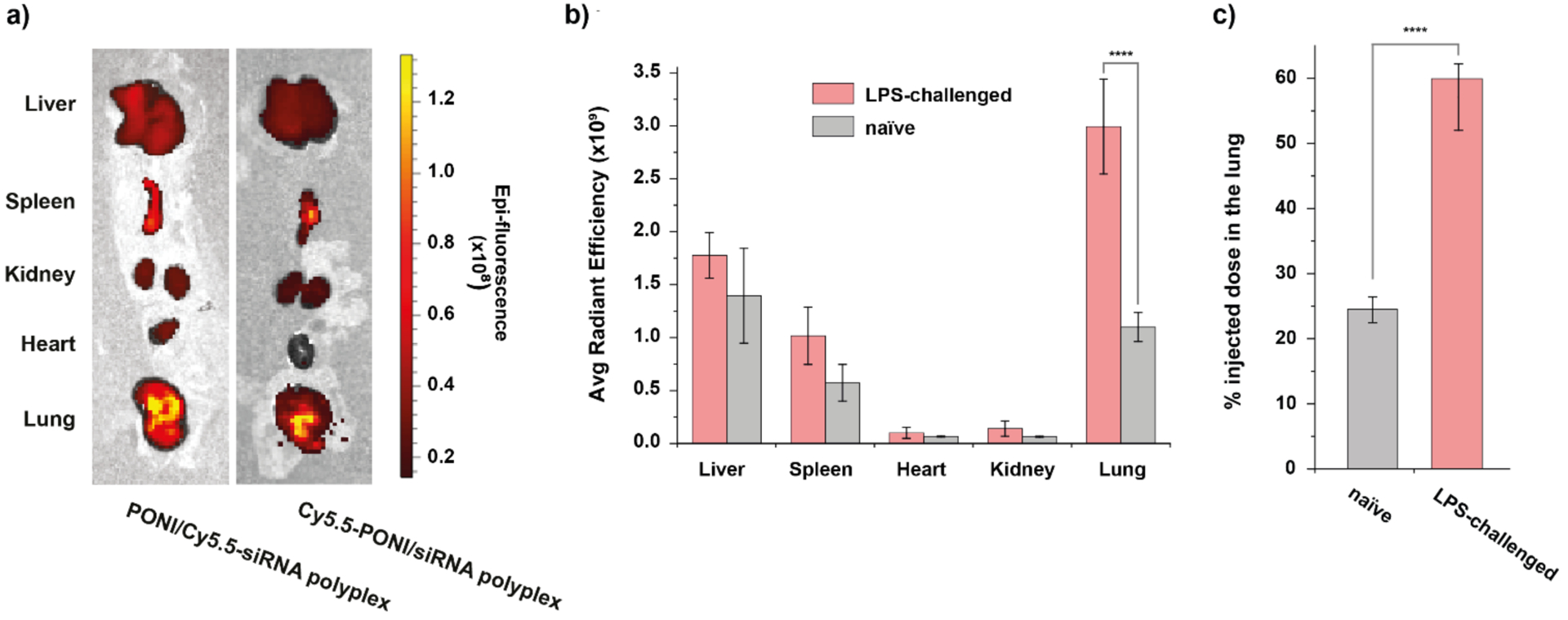
in vivo biodistribution of PONI-Guan/siRNA polyplexes in LPS-challenged BALB/c mice. a) Representative in vivo imaging system (IVIS) organ images of polyplexes labeled with Cy5.5-PONI-Guan (left) or Cy5.5-siRNA (right), showing colocalization and lung accumulation following systemic administration in LPS-challenged mice. b) Tissue-level biodistribution of polyplexes (Cy5.5-PONI-Guan) reveals preferential lung tropism, enhanced by LPS challenge (n=3). Radiant efficiency given as (p/s/cm2/sr)/(μW/cm2). c) % of injected dose in the lung 24 h after systemic administration. Error bars represent the mean ± the standard error of the mean (SEM) (n=3, one-way Anova and Tukey multiple comparisons, ****p < 0.001)
in vivo therapeutic efficacy of PONI-Guan/siRNA polyplexes.
We finally investigated the ability of polyplex-mediated siRNA delivery to regulate TNF-α levels and modulate immune responses in an LPS-challenged BALB/c mouse model of inflammation. LPS challenge (5 mg/kg) and polyplex administration were performed as above. After sacrifice, blood and organs were collected and harvested for a TNF-α ELISA assay, qRT-PCR, and tissue analysis. Mice treated with PONI/si_TNF-α polyplexes (0.28 mg/kg siRNA) displayed ~80% reduction in serum TNF-α secretion compared to those treated with PBS or PONI/si_scramble (Figure 7a). Treatment with 0.14 mg/kg siRNA also exhibited efficient knockdown of ~40%. Both dosages utilized are notably lower than are required for contemporary delivery platforms.58,59 TNF-α mRNA expression was also evaluated using RT-PCR. Significant downregulation (70–80%) was observed in pulmonary tissue as compared with PONI/si_scramble treatment, validating localization of siRNA to the lungs (Figure 7b). There was no significant difference in serum TNF-α production or mRNA levels between naïve mice, indicating that polyplexes are biocompatible and nonimmunogenic, and that attenuation of LPS-induced inflammation in vivo resulted only from siRNA-mediated knockdown.
Figure 7.
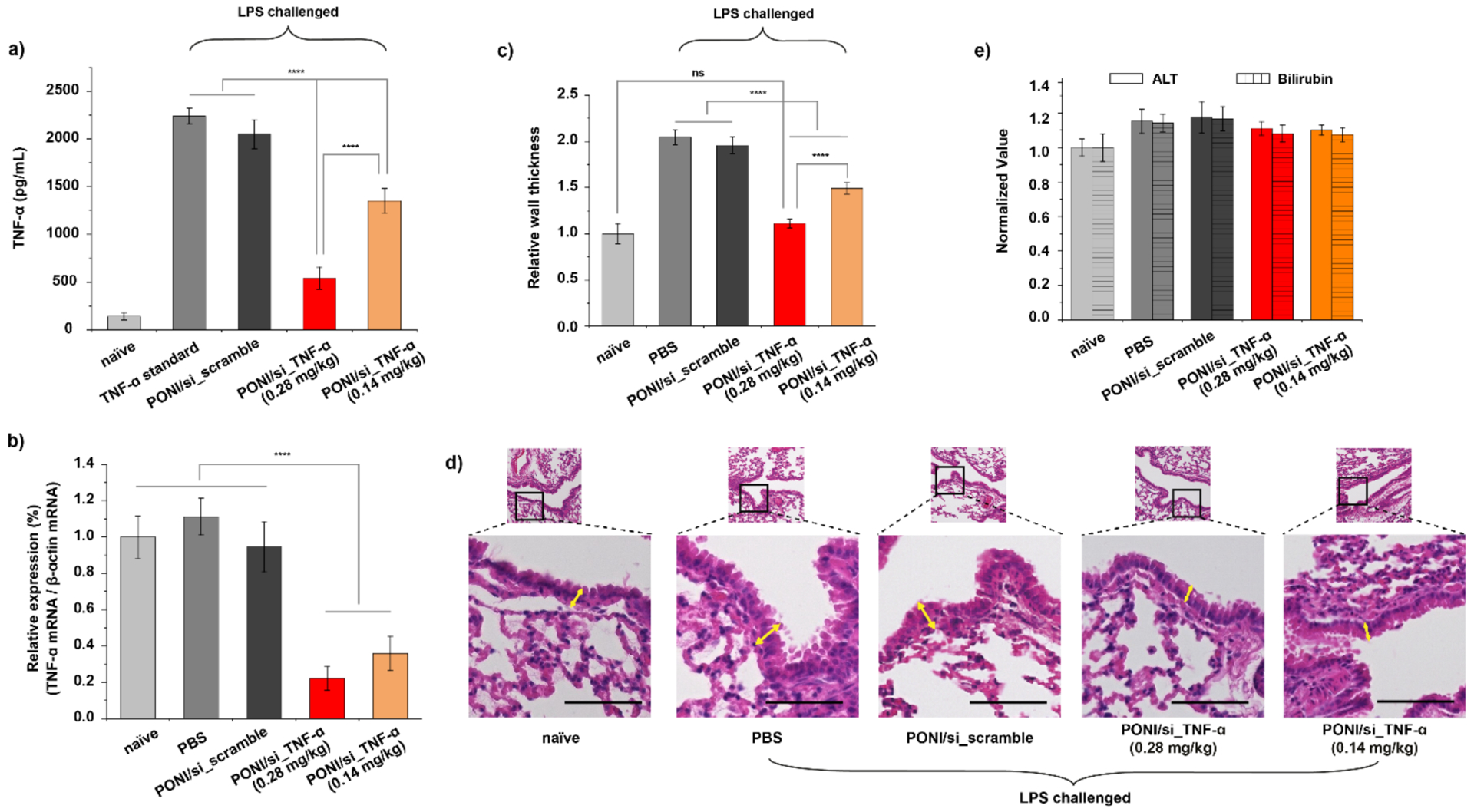
Therapeutic efficacy of PONI-Guan/siRNA polyplex-mediated gene knockdown. a) I.V. delivery of PONI/si_TNF-α polyplexes decreased serum TNF-α production. BALB/c mice were injected with LPS (5 mg/mouse, i.p.), followed by systemic administration of polyplexes at an siRNA dose of 0.14 or 0.28 mg/kg, 5 h later. Serum TNF-α was measured by ELISA 24 h after polyplexes dosing (n=3). b) In vivo delivery of PONI/si_TNF-α decreased TNF-α mRNA levels. BALB/c mice were systematically injected with the polyplexes at a siRNA dose of 0.14 or 0.28 mg/kg, and the lungs were harvested after 24 h. c) Relative bronchial wall thickness analysis 24 h after systemic administration of polyplexes at an siRNA dose of 0.14 or 0.28 mg/kg. Error bars represent the mean ± the standard error of the mean (SEM) (n=3, one-way Anova and Tukey multiple comparisons, ****p < 0.001). d) Representative hematoxylin and eosin-stained lung histology sections of mice. The bottom depicts the zoomed-in images of the top with axis through the bronchi. Yellow arrows indicate bronchial wall thickness. Mice were challenged with LPS intraperitonially and then injected intravenously with PBS, PONI/si_scramble, or PONI/si_TNF-α treatments (scale bar, 100 μm). e) in vivo determination of hepatic function using ALT and Bilirubin analysis of the blood plasma. No statistically significant changes in any parameters were observed for treatment groups (siRNA dose of 0.14 or 0.28 mg/kg) compared to naïve mice.
Leukocyte recruitment and peribronchial wall thickening are two major hallmarks of pulmonary inflammation.60,61 We evaluated tissue sections with hematoxylin and eosin (H&E) staining to observe the efficacy of polyplex-mediated siRNA delivery against lung inflammation, revealing minimal leukocyte recruitment and negligible peribronchial thickening in mice treated with PONI/si_TNF-α polyplexes (Figure 7c and 7d). In contrast, significant peribronchial thickening (a 2-fold increase) was observed in LPS-challenged mice with PBS or PONI/si_scramble treatment.
Nanoparticles are widely used vehicles for siRNA delivery but can cause significant liver damage or fibrosis during clearance.62,63 We examined serum alanine aminotransferase (ALT) and bilirubin levels in blood after sacrifice to determine the effects of PONI-Guan/siRNA polyplexes on liver function (Figure 7e). Clinical chemistry parameters indicated that mice treated with polyplexes had similar results to those that experienced PBS treatment, indicating no notable liver toxicity. Taken together, these results suggest that systemic delivery of PONI-Guan/siRNA polyplexes targeting TNF-α effectively and safely attenuates pulmonary inflammation induced by LPS challenge.
CONCLUSIONS
In summary, PONI-Guan/siRNA polyplexes provide an effective platform for immunomodulation in vitro and in vivo. Efficient cytosolic delivery of siRNA in combination with natural homing to inflamed lungs provide a potent RNAi-based therapeutic approach to anti-inflammatory immunomodulation. This method notably requires a low dose of siRNA (0.14 – 0.28 mg/kg) to achieve efficient (>70%) knockdown. This platform overcomes two key barriers to in vitro and in vivo siRNA delivery through 1) efficient delivery into the cytosol and 2) dosing at significantly lower levels than required contemporary clinical studies, reducing the risk of off-target effects. Moreover, the modular properties of the PONI system allow for future modification to target other organs and parts of the immune system. Further studies will explore targeting of other organs and tissues, as well as determine cellular tropism of these vehicles. In summary, this polymer platform provides a means of greatly enhancing the potential of therapeutic siRNA in the treatment of pulmonary autoimmune disorders and other inflammatory conditions.
EXPERIMENTAL METHODS
General.
All reagents or chemicals used were purchased from Fisher Scientific or Sigma-Aldrich unless otherwise stated. All siRNA (si_scramble, sense strand, 5’-UUCUCCGAACGUGUCACGU-3’; si_GFP, sense strand, 5’-AUGAUAUAGACGUUGUGGC-3’; si_TNF-α, sense strand, 5’-GCCGAUGGGUUGUACCUUG-3’; Cy3-labeled scramble) were purchased from Sigma-Aldrich. Confocal microscopy images were obtained on a Nikon A1 Sepectral Detector Confocal Microscope using a 40X or 60X objective and flow cytometry was performed on a Amnis ImageStream Mark II Imaging Flow Cytometry by counting 1,000 events.
Cell culture.
RAW 264.7 cells, A549 cells, and HT1299 cells were purchased from American Type Culture Ceollection (ATCC, Manassas, VA). eGFP-expressing RAW 264.7 (RAW 264.7:eGFP) cells were a generous provided from Prof. Michelle Farkas (University of Massachusetts Amherst). GFP is used as a reporter for iNOS in RAW 264.7:eGFP cells64, but no inflammatory responses are observed following treatments with the agents. All cells were cultured at 37°C under a humidified atmosphere of 5% CO2. High-glucose Dulbecco’s modified Eagle’s medium (DMEM, 4.5 g/L glucose) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic (100 U/mL penicillin and 100 μg/mL streptomycin) was used for cell culture.
Preparation and Charaterization of PONI/siRNA polyplex.
PONI backbone C3 guanidinium monomer was prepared as previously described65 and PONI-Guan Homopolymer (M.W. 60 kDa) was synthesized by ring-opening metathesis polymerization using third generation Grubbs’ catalyst according to our previous report.37 Briefly, solutions of guanidium functionalized monomer and Grubbs’ catalyst were dissolved in dichloromethane (DCM) and subjected to multiple freeze-thaw cycles for degassing. After warming the solutions to room temperature, the degassed monomer solution was added to degassed catalyst solutions and allowed to stir for 30 min under nitrogen. The polymerization was terminated by the addition of excess ethyl vinyl ether. The reaction mixture was stirred further for another 30 min. The resultant polymer were precipitated from excess hexane or anhydrous diethyl ether, filtered, washed, and dried under vacuum to yield a light-yellow powder. The reaction yields varied from 90 to 98%. The resultant polymer wascharacterized by 1H NMR and gel permeation chromatography (GPC) to assess chemical compositions and molecular weight distributions, respectively. For removal of the Boc functionalities, the polymer was dissolved in DCM with addition of trifluoroacetic acid (TFA) at 1:1 ratio. The reaction was allowed to stir for 4 hours following which excess TFA was removed by azeotropic distillation with methanol. Afterwards, the resultant polymer was re-dissolved in minimal DCM and precipitated in anhydrous diethyl ether, filtered, washed, and dried. Polymers were then dissolved in water and transferred to Biotech CE dialysis tubing membranes with a MWCO of 10000 g/mol and dialyzed against Milli-Q water (2−3 days). The polymers were then lyophilized to yield a light white powder. Polyplexes were formed through simple co-incubation. Briefly, polymer stocks were prepared in Milli-Q water, and added to sterile Eppendorf tubes followed by siRNA at varied G/P ratios. Mixed well by pipetting and then incubated at ambient temperature for 10 min, and added complete media (10% FBS, 1% antibiotic) to PONI/siRNA polyplex. To optimize the conditions for siRNA encapsulation, 10 nM siRNA was mixed with PONI-Guan at different G/P ratios for 10 min and then subjected to electrophoresis on 1% agarose gel. To evaluate the protection of siRNA against RNase A digestion, PONI/siRNA polyplexes were incubated with RNase A (35 mU) at 37 °C for 1 h and then the enzymatic reaction was stopped by adding 50 mM sodium dodecyl sulfate (SDS) to denature RNase A at 60 °C for 5 min. To the above reaction mixture was added 50 mg/ml heparin solution, followed by another 10 min incubation to displace siRNA from PONI/siRNA polyplexes, and then subjected to electrophoresis on agarose gel. For serum stability study, polyplexes were incubated with 10% FBS at 37°C for 12 h or human whole blood (BioIVT Elevating Science) for 24 h. At a predetermined time, samples were taken out. Then, nucleases in FBS were denatured by adding 50 mM SDS solution, followed by an additional adding 50 mg/ml heparin to dissociate siRNA from polyplexes before executing an agarose gel retardation assay. Size and zeta potential were measured by dynamic light scattering (DLS, Nano series, Malvern Instruments Inc). The morphology of PONI/siRNA polyplexes was analyzed and photographed via a FEI Tecnai-T12 electron microscope. The transmission electron microscopy (TEM) sample was prepared by dipping a 300-mech copper grid coated with carbon film in the solution and drying at room temperature for two days.
Determination of the extent of siRNA loading efficiency using the RiboGreen assay.
The extent of siRNA loading in the prepared polyplexes was measured using the Quant-iT Ribogreen RNA Assay Kit by following the manufacturer’s instructions. Total siRNA content is defined as the amount of encapsulated and non-encapsulated/free siRNA in the polyplexes. The difference between free siRNA and the total siRNA was used to calculate the amount of siRNA encapsulated within the polyplexes. The fluorescence were measured at an excitation wavelength of 485 nm and an emission wavelength of 528 nm. The % of encapsulation efficiency was calculated using the following formula.
Cytoxoticity of PONI-Guan/siRNA polyplexes.
Cells were seeded (3×103 cells/well) in a 96-well plate 24 h prior to the experiment. At the day of experiment, cells were washed by PBS and treated with varied concentration of PONI-Guan polymers or PONI-Guan/siRNA polyplexes followed by an incubation of additional 48 h with complete media (10% FBS, 1% antibiotic). The cell viability was measured using AlamarBlue assay (Invitrogen, CA) by following the manufactures’ instruction.
Analysis of inflammatory cytokines.
Enzyme-linked immunosorbent assay (ELISA, BD Bioscience) was used to detect the secretion of TNF-α or IL-6 in cells according to the manufacturer’s instructions. Briefly, supernatatnt of cells with varied concentration of PONI-Guan/siRNA polyplexes was collected and then subjected to ELISA assay. Cytokine status was inspected in regards with the recombinant biotinylated murine TNF-α or IL-6 standards supplied with the kits.
Homolysis assay to evaluate biocompatibility.
Human whole blood (pooled, mixed gender) was purchased (BioIVT Elevating Science) and precessed as soon as received. Red blood cells were colloced through centrifugation at 5000 rpm for 5 min followed by washing 4 times with PBS and then diluted in PBS to a final concentration of approximately 5% (v/v). Vaied concentration of PONI-Guan polymers were serially diluted using PBS and incubated in 96-well plates (200 μL/well). The blood cell suspension (20 μL/well) was added to each well and the plates were incubated at 37°C for 1 h while shaking at r.p.m. PBS and 1% of Triton X-100 were used as negative and positive controls, respectively. After incubation, the mixture was centrifuged at 3000 r.p.m. for 7 min and 120 μL of supernatant was transferred to a new 96-well plate. The absorvance was recorded at 560 nm in each well, and hemolysis was calculated using the following formula.
Confocal laser scanning microscope (CLSM) for fluorescently labeled Cy3-siRNA delivery.
Confocal laser scanning microscopy (CLSM) imaging was performed using a Nikon A1 Sepectral Detector Confocal Microscope. For imaging of the cellular uptake of PONI/siRNA polyplex, cells (1.5 ×105 cells) were seeded in a 35mm glass bottom culture dish (MatTek, MA) a day before the experiment. On the day of treatment, cells were washed with phosphate buffer saline (PBS) followed by replacement with complete media (10% FBS and 1% antibiotic) containing PONI/Cy3-siRNA (25nM), followed by another 6 h of incubation at 37 °C. After removing medium, the cells were washed with PBS, stained with Lysotracker Deep Red (Invitrogen), and visualized for fluorescence imaging using the confocal microscope. Images were processed and analyzed using the NIS-Elements Advanced Research software (Nikon).
Imaging flow cytometry for uptake mechanism knockdown evaluation with endocytic inhibiitors.
RAW 264.7:eGFP cells were seeded for 24 h prior to treatments. On the day of the experiment, cells were washed with PBS and incubated with the PONI/si_GFP polyplexes in complete media (10% FBS and 1% antibiotic) containing a dissolved quantity of small molecule, chlorpromazine (1.5 μg/mL), imipramine (3 μg/mL), nystatin (50 μg/mL), or methyl-β-cyclodextrin (MβCD, 7.5 μg/mL) for 4 h at 37 °C, 5% CO2. For the comparison of commercial transfection reagent, RNAiMAX and the same amount of siRNA were mixed according to manufacturer’s instructions. Cells were cultured for additional 48 h before CSLM imaging studies and flow cytometry analysis. The cells were trypsinized, harvested, and resuspended in PBS for flow cytometry analysis on Amnis ImageStream Mark II Imaging Flow Cytometry (Luminex). At least 1000 events were analyzed for each sample according to the standard instrumentation protocol from Amnis. Briefly, after incubation with PONI-Guan/Cy3-siRNA, single cells were isolated and gated using a scatterplot of Aspect Ratio versus Area of the Brightfield area. Then, fluorescence positive cells were gated, and the internalization wizard provided by the Amnis IDEAS software was utilized as a preliminary gate during the screening process. The isolated population from the gating stage was manually confirmed by the user and provided separation between cytosolic delivery and surface-bound cells.
In vitro TNF-α knockdown.
RAW 264.7 (2.0 × 105 cells/well) were cultured in a 48-well plate for 24 h prior to the experiment. At the day of experiment, cells were washed with PBS and treated with media containing PONI/siRNA (50 nM) for 24 h, followed by 3 h LPS stimulation (1 μg/mL in PBS). At the end of incubation, cultured media was collected for TNF-α level measurement by ELISA (R&D System, MN, USA) using a mouse TNF-α ELISA kit according to the manufacturer’s protocol.
Animal care and in vivo biodistribution.
All animal experiments were conducted under Protocol Number: 2492 in accordance with the guidelines of Institutional Animal Care and Use Committee (IACUC) at University of Massachusets Amherst. Female BALB/c mice at least 5 weeks of age used for biodistribution and therapeutic studies were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were allowed to rest for at least one week in the animal facilities before any procedure was performed. To induce lung inflammation in mice, 5mg/kg of LPS in PBS was injected intraperitoneally into mice. 6 h after LPS injection, Cy5.5-siRNA, Cy5.5-PONI-Guan, Cy5.5-PONI/siRNA polyplex, or PONI/Cy5.5-siRNA polyplex in PBS at a siRNA dose of 0.28 mg/kg were administered intravenously. After 24 hours, mice were sacrificed and organs were collected for ex vivo imaging using an IVIS system.
Inflammation treatment studies and in vivo gene silencing.
To treat lung inflammation, female BALB/c mice were first intraperitoneally challenged with LPS (5 mg/kg) in PBS. 6 h after the challenge, Cy5.5-siRNA, Cy5.5-PONI-Guan, Cy5.5-PONI/siRNA polyplex, or PONI/Cy5.5-siRNA polyplex in PBS at a siRNA dose of 0.14 or 0.28 mg/kg were administered intravenously. After 24 hours, mice were sacrificed and blood was collected for the TNF-α knockdown study and centrifuged in serum separator tubes at 5000 r.p.m. for 10 min, and then supernatant serum was carefully collected for TNF-α analysis ELISA (R&D Systems, MN, USA) using a mouse TNF-α ELISA kit according to the manufacturer’s protocol. Blood was also collected for the liver function assays. Serum total bilirubin and alanine aminotrasferase (ALT) were measured using commercial kits (Teco Diagnostics, Anaheim, CA) according to the manufacturer’s protocols. The lungs were collected for histology analysis after 24 hours and fixed in 4% paraformaldegyde (Sigma-Aldrich) overnight at 4°C. The lungs were dehydrated in ethanol, cleared in xylenes, embedded in paraffin, and sectioned at 7 μm. Hematoxylin and eosin staining was performed as previously described.66 Images were obtained using a Nikon Eclipse Ti2 inverted microscope with a DS-Ri2 color camera and analyzed using the NIS-Elements software and Image J software.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR).
RAW 264.7 cells (2×105 cells/well) were cultured in a 6-well plate for 24 h prior the experiment. At the day of experiment, cells were washed by PBS and treated with PBS, PONI/si_scramble, PONI/si_TNF-α polyplexes at the indicated siRNA concentration (dose of 50 nM at G/P 30) for 24 h, followed by 3 h LPS stimulation (1 μg/mL). At the end of incubation, cells were washed with PBS and then harvested with TRIZol reagent. For in vivo samples, lungs were harvested once mice were sacrificed. The lungs were washed with PBS and homogenized with Trizol reagent. RNA extraction was performed using the Pure Link RNA Mini kit (Ambion) following the manufacturer’s protocol. Superscript IV reverse transcriptase (Invitrogen) was used for conversion of approximately 100 ng of RNA to cDNA, along with RNaseOut, also following manufacturer’s instructions. RT-PCR was performed on cDNA as prepared above using a CFX Connect Real Time System with iTag Universal SYBR Green Supermix (Biorad). All primers were purchased from Integrated DNA Technologies (Caralville, Iowa). The following sequences were used: TNF-α Forward (5’-CCT GTA GCC CAC GTC GTA A-3’), Reverse (5’-GGG AGT CAA GGT ACA ACC C-3’); β-actin Forward (5′-GAT CAG CAA GCA GGA GTA CGA-3′), Reverse (5′-AAA ACG CAG CTC AGT AAC AGT C-3′). At least three biological replicates were performed for each control group and three technical replicates were used for each biological replicate. All TNF-α mRNA measurements was normalized to β-actin.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to thank Dr. J. Chambers and the Light Microscopy Core Facility at UMass Amherst as well as Dr. A. S. Burnside and the Flow Cytometry Core Facility for establishing confocal microscopy and flow cytometric assays respectively.
FUNDING SOURCES
This work is supported by NIH R01 EB022641 and NIH R21 CA267260.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no conflict of interests.
REFERENCES
- (1).Bo M; Jasemi S; Uras G; Erre GL; Passiu G; Sechi LA Role of Infections in the Pathogenesis of Rheumatoid Arthritis: Focus on Mycobacteria. Microorganisms 2020, 8 (10), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Demoruelle MK; Wilson TM; Deane KD Lung Inflammation in the Pathogenesis of Rheumatoid Arthritis. Immunol. Rev 2020, 294 (1), 124–132. [DOI] [PubMed] [Google Scholar]
- (3).Racanelli AC; Kikkers SA; Choi AMK; Cloonan SM Autophagy and Inflammation in Chronic Respiratory Disease. Autophagy 2018, 14 (2), 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sharafkhaneh A; Hanania NA; Kim V Pathogenesis of Emphysema: From the Bench to the Bedside. Proc. Am. Thorac. Soc 2008, 5 (4), 475–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Matera MG; Cazzola M; Page C Prospects for COPD Treatment. Curr. Opin. Pharmacol 2021, 56, 74–84. [DOI] [PubMed] [Google Scholar]
- (6).Aslan A; Aslan C; Zolbanin NM; Jafari R Acute Respiratory Distress Syndrome in COVID-19: Possible Mechanisms and Therapeutic Management. Pneumonia (Nathan Qld.) 2021, 13 (1), 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lupu L; Palmer A; Huber-Lang M Inflammation, Thrombosis, and Destruction: The Three-Headed Cerberus of Trauma- and SARS-CoV-2-Induced ARDS. Front. Immunol 2020, 11, 2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wu C; Chen X; Cai Y; Xia J; Zhou X; Xu S; Huang H; Zhang L; Zhou X; Du C; Zhang Y; Song J; Wang S; Chao Y; Yang Z; Xu J; Zhou X; Chen D; Xiong W; Xu L; Zhou F; Jiang J; Bai C; Zheng J; Song Y Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med 2020, 180 (7), 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kainthola A; Haritwal T; Tiwari M; Gupta N; Parvez S; Tiwari M; Prakash H; Agrawala PK Immunological Aspect of Radiation-Induced Pneumonitis, Current Treatment Strategies, and Future Prospects. Front. Immunol 2017, 8, 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mukker JK; Singh RSP; Derendorf H Ciclesonide: A Pro-Soft Drug Approach for Mitigation of Side Effects of Inhaled Corticosteroids. J. Pharm. Sci 2016, 105 (9), 2509–2514. [DOI] [PubMed] [Google Scholar]
- (11).Rajapaksa AE; Do LAH; Suryawijaya Ong D; Sourial M; Veysey D; Beare R; Hughes W; Yang W; Bischof RJ; McDonnell A; Eu P; Yeo LY; Licciardi PV; Mulholland EK Pulmonary Deposition of Radionucleotide-Labeled Palivizumab: Proof-of-Concept Study. Front. Pharmacol 2020, 11, 1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Patil HP; Freches D; Karmani L; Duncan GA; Ucakar B; Suk JS; Hanes J; Gallez B; Vanbever R Fate of PEGylated Antibody Fragments Following Delivery to the Lungs: Influence of Delivery Site, PEG Size and Lung Inflammation. J. Control. Release 2018, 272, 62–71. [DOI] [PubMed] [Google Scholar]
- (13).Nixon J; Newbold P; Mustelin T; Anderson GP; Kolbeck R Monoclonal Antibody Therapy for the Treatment of Asthma and Chronic Obstructive Pulmonary Disease with Eosinophilic Inflammation. Pharmacol. Ther 2017, 169, 57–77. [DOI] [PubMed] [Google Scholar]
- (14).Shibabaw T Inflammatory Cytokine: IL-17A Signaling Pathway in Patients Present with COVID-19 and Current Treatment Strategy. J. Inflamm. Res 2020, 13, 673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kirsten A; Watz H; Pedersen F; Holz O; Smith R; Bruin G; Koehne-Voss S; Magnussen H; Waltz DA The Anti-IL-17A Antibody Secukinumab Does Not Attenuate Ozone-Induced Airway Neutrophilia in Healthy Volunteers. Eur. Respir. J 2013, 41 (1), 239–241 [DOI] [PubMed] [Google Scholar]
- (16).Matera MG; Page C; Rogliani P; Calzetta L; Cazzola M Therapeutic Monoclonal Antibodies for the Treatment of Chronic Obstructive Pulmonary Disease. Drugs 2016, 76 (13), 1257–1270. [DOI] [PubMed] [Google Scholar]
- (17).Parray HA; Shukla S; Perween R; Khatri R; Shrivastava T; Singh V; Murugavelu P; Ahmed S; Samal S; Sharma C; Sinha S; Luthra K; Kumar R Inhalation Monoclonal Antibody Therapy: A New Way to Treat and Manage Respiratory Infections. Appl. Microbiol. Biotechnol 2021, 105 (16–17), 6315–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hall AP; Tepper JS; Boyle MH; Cary MG; Flandre TG; Piaia A; Tarnow I; Macri NP; Freke MC; Nikula KJ; Paul GR; Cauvin A; Gregori M; Haworth R; Naylor S; Price M; Robinson IN; Allen A; Gelzleichter T; Hohlbaum AM; Manetz S; Wolfreys A; Colman K; Fleurance R; Jones D; Mukaratirwa S BSTP Review of 12 Case Studies Discussing the Challenges, Pathology, Immunogenicity, and Mechanisms of Inhaled Biologics. Toxicol. Pathol 2021, 49 (2), 235–260. [DOI] [PubMed] [Google Scholar]
- (19).Guilleminault L; Azzopardi N; Arnoult C; Sobilo J; Hervé V; Montharu J; Guillon A; Andres C; Herault O; Le Pape A; Diot P; Lemarié E; Paintaud G; Gouilleux-Gruart V; Heuzé-Vourc’H N Fate of Inhaled Monoclonal Antibodies after the Deposition of Aerosolized Particles in the Respiratory System. J. Control. Release 2014, 196, 344–354. [DOI] [PubMed] [Google Scholar]
- (20).Kelly C; Yadav AB; Lawlor C; Nolan K; O’Dwyer J; Greene CM; McElvaney NG; Sivadas N; Ramsey JM; Cryan SA Therapeutic Aerosol Bioengineering of SiRNA for the Treatment of Inflammatory Lung Disease by TNFα Gene Silencing in Macrophages. Mol. Pharm 2014, 11, 4270–4279. [DOI] [PubMed] [Google Scholar]
- (21).Jiang Y; Hardie J; Liu Y; Ray M; Luo X; Das R; Landis RF; Farkas ME; Rotello VM Nanocapsule-Mediated Cytosolic SiRNA Delivery for Anti-Inflammatory Treatment. J. Control. Release 2018, 283, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang H; Zhang S; Lv J; Cheng Y Design of Polymers for SiRNA Delivery: Recent Progress and Challenges. View 2021, 2 (3), 20200026 [Google Scholar]
- (23).Lokras A; Thakur A; Wadhwa A; Thanki K; Franzyk H; Foged C Optimizing the Intracellular Delivery of Therapeutic Anti-Inflammatory TNF-α SiRNA to Activated Macrophages Using Lipidoid-Polymer Hybrid Nanoparticles. Front. Bioeng. Biotechnol 2021, 8, 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Xu X; Yang W; Liang Q; Shi Y; Zhang W; Wang X; Meng F; Zhong Z; Yin L Efficient and Targeted Drug/SiRNA Co-Delivery Mediated by Reversibly Crosslinked Polymersomes toward Anti-Inflammatory Treatment of Ulcerative Colitis (UC). Nano Res 2019, 12 (3), 659–667. [Google Scholar]
- (25).Gupta A; Andresen JL; Manan RS; Langer R Nucleic Acid Delivery for Therapeutic Applications. Adv. Drug Deliv. Rev 2021, 178, 113834. [DOI] [PubMed] [Google Scholar]
- (26).Du Rietz H; Hedlund H; Wilhelmson S; Nordenfelt P; Wittrup A Imaging Small Molecule-Induced Endosomal Escape of SiRNA. Nat. Commun 2020, 11 (1), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yoo YJ; Lee CH; Park SH; Lim YT Nanoparticle-Based Delivery Strategies of Multifaceted Immunomodulatory RNA for Cancer Immunotherapy. J. Control. Release 2022, 343, 564–583. [DOI] [PubMed] [Google Scholar]
- (28).McCaskill J; Singhania R; Burgess M; Allavena R; Wu S; Blumenthal A; McMillan NA Efficient Biodistribution and Gene Silencing in the Lung Epithelium via Intravenous Liposomal Delivery of SiRNA. Mol. Ther. Nucleic Acids 2013, 2 (6), e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Merckx P; De Backer L; Van Hoecke L; Guagliardo R; Echaide M; Baatsen P; Olmeda B; Saelens X; Pérez-Gil J; De Smedt SC; Raemdonck K Surfactant Protein B (SP-B) Enhances the Cellular SiRNA Delivery of Proteolipid Coated Nanogels for Inhalation Therapy. Acta Biomater 2018, 78, 236–246. [DOI] [PubMed] [Google Scholar]
- (30).Xu PY; Kankala RK; Pan YJ; Yuan H; Wang S. Bin; Chen AZ Overcoming Multidrug Resistance through Inhalable SiRNA Nanoparticles-Decorated Porous Microparticles Based on Supercritical Fluid Technology. Int. J. Nanomedicine 2018, 13, 4685–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zoulikha M; Xiao Q; Boafo GF; Sallam MA; Chen Z; He W Pulmonary Delivery of SiRNA against Acute Lung Injury/Acute Respiratory Distress Syndrome. Acta Pharm. Sin. B 2022, 12 (2), 600–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yeo LY; Friend JR; McIntosh MP; Meeusen EN; Morton DA Ultrasonic Nebulization Platforms for Pulmonary Drug Delivery. Expert Opin. Drug Deliv 2010, 7 (6), 663–679. [DOI] [PubMed] [Google Scholar]
- (33).Mayor A; Thibert B; Huille S; Respaud R; Audat H; Heuzé-Vourc’h N Inhaled Antibodies: Formulations Require Specific Development to Overcome Instability Due to Nebulization. Drug Deliv. Transl. Res 2021, 11 (4), 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rudokas M; Najlah M; Alhnan MA; Elhissi A Liposome Delivery Systems for Inhalation: A Critical Review Highlighting Formulation Issues and Anticancer Applications. Med. Princ. Pract 2016, 25 Suppl 2 (Suppl 2), 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Whitehead KA; Langer R; Anderson DG Knocking down Barriers: Advances in SiRNA Delivery. Nat. Rev. Drug Discov 2009, 8 (2), 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wittrup A; Ai A; Liu X; Hamar P; Trifonova R; Charisse K; Manoharan M; Kirchhausen T; Lieberman J Visualizing Lipid-Formulated SiRNA Release from Endosomes and Target Gene Knockdown. Nat. Biotechnol 2015, 33 (8), 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lee YW; Luther DC; Goswami R; Jeon T; Clark V; Elia J; Gopalakrishnan S; Rotello VM Direct Cytosolic Delivery of Proteins through Coengineering of Proteins and Polymeric Delivery Vehicles. J. Am. Chem. Soc 2020, 142 (9), 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Luther DC; Jeon T; Goswami R; Nagaraj H; Kim D; Lee YW; Rotello VM Protein Delivery: If Your GFP (or Other Small Protein) Is in the Cytosol, It Will Also Be in the Nucleus. Bioconjug. Chem 2021, 32 (5), 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Myerson JW; Patel PN; Rubey KM; Zamora ME; Zaleski MH; Habibi N; Walsh LR; Lee YW; Luther DC; Ferguson LT; Marcos-Contreras OA; Glassman PM; Mazaleuskaya LL; Johnston I; Hood ED; Shuvaeva T; Wu J; Zhang HY; Gregory JV; Kiseleva RY; Nong J; Grosser T; Greineder CF; Mitragotri S; Worthen GS; Rotello VM; Lahann J; Muzykantov VR; Brenner JS Supramolecular Arrangement of Protein in Nanoparticle Structures Predicts Nanoparticle Tropism for Neutrophils in Acute Lung Inflammation. Nat. Nanotechnol 2021, 17 (1), 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Aulakh GK Neutrophils in the Lung: “The First Responders.” Cell Tissue Res 2018, 371 (3), 577–588. [DOI] [PubMed] [Google Scholar]
- (41).Hu B; Zhong L; Weng Y; Peng L; Huang Y; Zhao Y; Liang XJ Therapeutic SiRNA: State of the Art. Signal Transduct. Target. Ther 2020, 5 (1), 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).de Fougerolles A; Vornlocher HP; Maraganore J; Lieberman J Interfering with Disease: A Progress Report on SiRNA-Based Therapeutics. Nat. Rev. Drug Discov 2007, 6 (6), 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Batah SS; Fabro AT Pulmonary Pathology of ARDS in COVID-19: A Pathological Review for Clinicians. Respir. Med 2021, 176, 106239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Mukhopadhyay S; Hoidal JR; Mukherjee TK Role of TNFalpha in Pulmonary Pathophysiology. Respir. Res 2006, 7 (1), 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).McCarthy J; O’Neill MJ; Bourre L; Walsh D; Quinlan A; Hurley G; Ogier J; Shanahan F; Melgar S; Darcy R; O’Driscoll CM Gene Silencing of TNF-Alpha in a Murine Model of Acute Colitis Using a Modified Cyclodextrin Delivery System. J. Control. Release 2013, 168 (1), 28–34. [DOI] [PubMed] [Google Scholar]
- (46).Ding L; Tang S; Wyatt TA; Knoell DL; Oupický D Pulmonary SiRNA Delivery for Lung Disease: Review of Recent Progress and Challenges. J. Control. Release 2021, 330, 977–991. [DOI] [PubMed] [Google Scholar]
- (47).Pelletier MGH; Szymczak K; Barbeau AM; Prata GN; O’Fallon KS; Gaines P Characterization of Neutrophils and Macrophages from Ex Vivo Cultured Murine Bone Marrow for Morphologic Maturation and Functional Responses by Imaging Flow Cytometry. Methods 2017, 112, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Abdulkhaleq LA; Assi MA; Abdullah R; Zamri-Saad M; Taufiq-Yap YH; Hezmee MNM The Crucial Roles of Inflammatory Mediators in Inflammation: A Review. Vet. world 2018, 11 (5), 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sahay G; Querbes W; Alabi C; Eltoukhy A; Sarkar S; Zurenko C; Karagiannis E; Love K; Chen D; Zoncu R; Buganim Y; Schroeder A; Langer R; Anderson DG Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Nat. Biotechnol. 2013 317 2013, 31 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Gomes-Da-Silva LC; Fonseca NA; Moura V; Pedroso De Lima MC; Simões S; Moreira JN Lipid-Based Nanoparticles for SiRNA Delivery in Cancer Therapy: Paradigms and Challenges. Acc. Chem. Res 2012, 45 (7), 1163–1171 [DOI] [PubMed] [Google Scholar]
- (51).Rane AS; Rutkauskaite J; deMello A; Stavrakis S High-Throughput Multi-Parametric Imaging Flow Cytometry. Chem 2017, 3 (4), 588–602. [Google Scholar]
- (52).Lu JJ; Langer R; Chen J A Novel Mechanism Is Involved in Cationic Lipid-Mediated Functional SiRNA Delivery. Mol. Pharm 2009, 6 (3), 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Mandal P; Noutsi P; Chaieb S Cholesterol Depletion from a Ceramide/Cholesterol Mixed Monolayer: A Brewster Angle Microscope Study. Sci. Reports 2016, 6 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Mout R; Ray M; Yesilbag Tonga G; Lee YW; Tay T; Sasaki K; Rotello VM Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 2017, 11 (3), 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Jiang Y; Tang R; Duncan B; Jiang Z; Yan B; Mout R; Rotello VM Direct Cytosolic Delivery of SiRNA Using Nanoparticle-Stabilized Nanocapsules. Angew. Chem. Int. Ed. Engl 2015, 54 (2), 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Luther DC; Lee YW; Nagaraj H; Clark V; Jeon T; Goswami R; Gopalakrishnan S; Fedeli S; Jerome W; Elia JL; Rotello VM Cytosolic Protein Delivery Using Modular Biotin-Streptavidin Assembly of Nanocomposites. ACS Nano 2021, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Li Z; Zhang Y; Zhu D; Li S; Yu X; Zhao Y; Ouyang X; Xie Z; Li L Transporting Carriers for Intracellular Targeting Delivery via Non-Endocytic Uptake Pathways. Drug Deliv 2017, 24 (sup1), 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Osborn MF; Khvorova A Improving SiRNA Delivery in vivo Through Lipid Conjugation. Nucleic Acid Ther 2018, 28 (3), 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Shim MS; Kwon YJ Efficient and Targeted Delivery of SiRNA in Vivo. FEBS J 2010, 277 (23), 4814– 4827. [DOI] [PubMed] [Google Scholar]
- (60).Ren Y; Su X; Kong L; Li M; Zhao X; Yu N; Kang J Therapeutic Effects of Histone Deacetylase Inhibitors in a Murine Asthma Model. Inflamm. Res 2016, 65 (12), 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Marini T; Hobbs SK; Chaturvedi A; Kaproth-Joslin K Beyond Bronchitis: A Review of the Congenital and Acquired Abnormalities of the Bronchus. Insights Imaging 2017, 8 (1), 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Whitehead KA; Dorkin JR; Vegas AJ; Chang PH; Veiseh O; Matthews J; Fenton OS; Zhang Y; Olejnik KT; Yesilyurt V; Chen D; Barros S; Klebanov B; Novobrantseva T; Langer R; Anderson DG Degradable Lipid Nanoparticles with Predictable in vivo SiRNA Delivery Activity. Nat. Commun 2014, 5 (1), 1– 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Yao Y; Zang Y; Qu J; Tang M; Zhang T The Toxicity Of Metallic Nanoparticles On Liver: The Subcellular Damages, Mechanisms, And Outcomes. Int. J. Nanomedicine 2019, 14, 8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Mas-Rosario JA, Medor JD, Jeffway MI, Martinez-Montes JM, Farkas ME “Macrophage-Based iNos Reporter Reveals Polarization and Reprogramming in the Context of Breast Cancer.” BioRxiv 20 22, 08, 504212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Peveler WJ; Landis RF; Yazdani M; Day JW; Modi R; Carmalt CJ; Rosenberg WM; Rotello VM A Rapid and Robust Diagnostic for Liver Fibrosis Using a Multichannel Polymer Sensor Array. Adv. Mater 2018, 30 (28), 1800634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Archambault D; Cheong A; Iverson E; Tremblay KD; Mager J Protein Phosphatase 1 Regulatory Subunit 35 Is Required for Ciliogenesis, Notochord Morphogenesis, and Cell-Cycle Progression during Murine Development. Dev. Biol 2020, 465 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


