SUMMARY
Clostridioides difficile produces toxins that damage the colonic epithelium, causing colitis. Variation in disease severity is poorly understood and has been attributed to host factors and virulence differences between C. difficile strains. We test 23 epidemic ST1 C. difficile clinical isolates for their virulence in mice. All isolates encode a complete Tcd pathogenicity locus and achieve similar colonization densities. However, disease severity varies from lethal to avirulent infections. Genomic analysis of avirulent isolates reveals a 69-bp deletion in the cdtR gene, which encodes a response regulator for binary toxin expression. Deleting the 69-bp sequence in virulent R20291 strain renders it avirulent in mice with reduced toxin gene transcription. Our study demonstrates that a natural deletion within cdtR attenuates virulence in the epidemic ST1 C. difficile isolates without reducing colonization and persistence. Distinguishing strains on the basis of cdtR may enhance the specificity of diagnostic tests for C. difficile colitis.
In brief
Dong et al. show that clinical ST1 C. difficile isolates with identical pathogenicity loci have variable virulence in mice. A natural deletion within the regulatory gene (cdtR) for binary toxin attenuates virulence in epidemic ST1 C. difficile isolates without reducing colonization and persistence.
Graphical Abstract
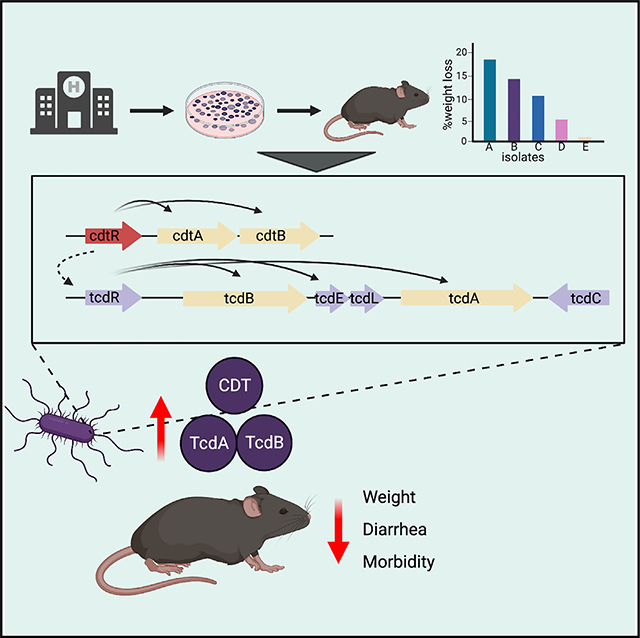
INTRODUCTION
Clostridioides difficile is a gram-positive, spore-forming anaerobic bacterium and the leading cause of nosocomial infections in the United States.1–3 Infections are acquired by oral ingestion of C. difficile spores, which are prevalent in the environment and can survive for extended periods of time on contaminated surfaces. Upon ingestion, C. difficile spores germinate, produce toxins, and cause colitis and, in severe cases, can result in mortality. The major virulence factors of C. difficile are toxins A (tcdA) and B (tcdB), which are encoded in the pathogenicity locus (PaLoc).4,5 These toxins glycosylate and thereby inactivate host GTPases, triggering the death of intestinal epithelial cells and leading to gut inflammation.6
The C. difficile species is comprised of hundreds of strain types across more than 6 phylogenetic clades. PCR- and sequencing-based approaches, including PCR-ribotyping (RT) and multilocus sequencing typing (MLST or ST), have been used to characterize C. difficile strain types. Recently, whole-genome sequencing (WGS) has greatly contributed to our understanding of C. difficile diversity, evolution, and epidemiology.7 Almost two decades ago, the BI/NAP1/027 strain, characterized as ST1 by MLST, emerged as a cause of severe nosocomial outbreaks and increased C. difficile infection (CDI) incidence in North America and Europe. Since then, the prevalence of ST1 has declined, but it remains among the most frequently isolated strains in hospital- and community-acquired CDI cases in the United States.2,8–11 The ST1 C. difficile strain encodes an additional CDT toxin (encoded by cdtA and cdtB and also referred to as binary toxin), which is an ADP-ribosyltransferase that modifies actin and disrupts cellular cytoskeleton organization.12 The ST1 strains have higher minimal inhibitory concentrations (MICs) to several antibiotics, most notably fluroquinolones, and produce higher amounts of TcdA and TcdB compared to non-ST1 strains.13,14 The relative virulence of the ST1 strain is controversial, however, with some studies demonstrating clinical disease severities similar to other strains.15–17 Host factors can impact the severity of CDI, including underlying diseases, immune competence, and microbiome composition.18,19 Whether genetic variants of ST1 explain diverse disease manifestations is unknown.
To determine intra-strain type virulence diversity, we used an antibiotic-treated mouse model of CDI to test a panel of PaLoc- and CdtLoc-encoding ST1 C. difficile clinical isolates to quantify disease severity.20 Clinical C. difficile isolates with identical PaLocs caused a range of disease severities, with two isolates causing no detectable disease in antibiotic-treated wild-type, germ-free mice or MyD88-deficient mice. We identified a 69-bp deletion in the cdtR gene of these two avirulent isolates that encodes a LytTR family response regulator that regulates CDT expression. The 69-bp deletion in the cdtR leads to reduced CDT toxin and PaLoc gene expression, resulting in loss of virulence and confirming previous studies implicating CdtR as regulator of CDT and Tcd toxin expression.21 Overall, our study describes virulence diversity within a single strain type and demonstrates the critical role of CdtR for ST1 C. difficile virulence.
RESULTS
Clinical ST1 C. difficile isolates demonstrate variable severities in mice
We focus on a group of 23 C. difficile isolates belonging to ribotype 027 epidemic strains (here referred to as ST1) isolated from patients with diarrhea during a molecular surveillance program at Memorial Sloan Kettering Cancer Center 2013–2017.22 Whole-genome Illumina sequencing of these isolates allows us to compare them to public collections. We plotted a uniform manifold approximation and projection (UMAP) analysis of the presence or the absence of unique coding sequences (annotated proteins or unannotated protein clusters) across the top 10 STs of C. difficile strains in Patric (date: February 10, 2021).23 Different STs cluster individually, and our ST1 isolates overlap with other ST1 C. difficile included in the analysis, confirming their strain type (Figure 1A). These 23 isolates demonstrate high genome-wide similarity by average nucleotide identity (ANI) scores above 99.8% and encode identical PaLoc sequences (Figures S1A and S1B). To study if close-related C. difficile isolates may have variable virulence, mice treated with antibiotics (metronidazole, neomycin, vancomycin in drinking water with clindamycin intraperitoneal injection) were orally infected with each of these isolates at a dose of 200 spores, and C. difficile pathogenicity was monitored throughout a 7-day time course (Figure 1B). Mice infected with different ST1 isolates displayed a spectrum of disease severity, including variable weight loss and mortality (Figures 1C and S1C). The widely used ST1 lab strain R20291 was included in parallel for virulence comparison. Within our ST1 collection, 5 isolates resulted in mortality in mice. A few isolates caused more severe weight loss than R20291, including ST1-49, ST1-11, and ST1-12, while most ST1 isolates caused moderate and non-lethal infections. Two isolates, ST1-75 and ST1-35, demonstrated no impact on mouse body weights. No apparent colonization deficiency was observed in any of these isolates (Figure S1D). The variable pathogenicity induced by a group of ST1 isolates with identical PaLocs suggested additional regulatory mechanisms of C. difficile virulence. Therefore, we sought to examine other genomic factors that are responsible for attenuated virulence of C. difficile isolates ST1-75 and ST1-35.
Figure 1. Clinical ST1 C. difficile isolates demonstrated variable virulence in mice treated with antibiotics.
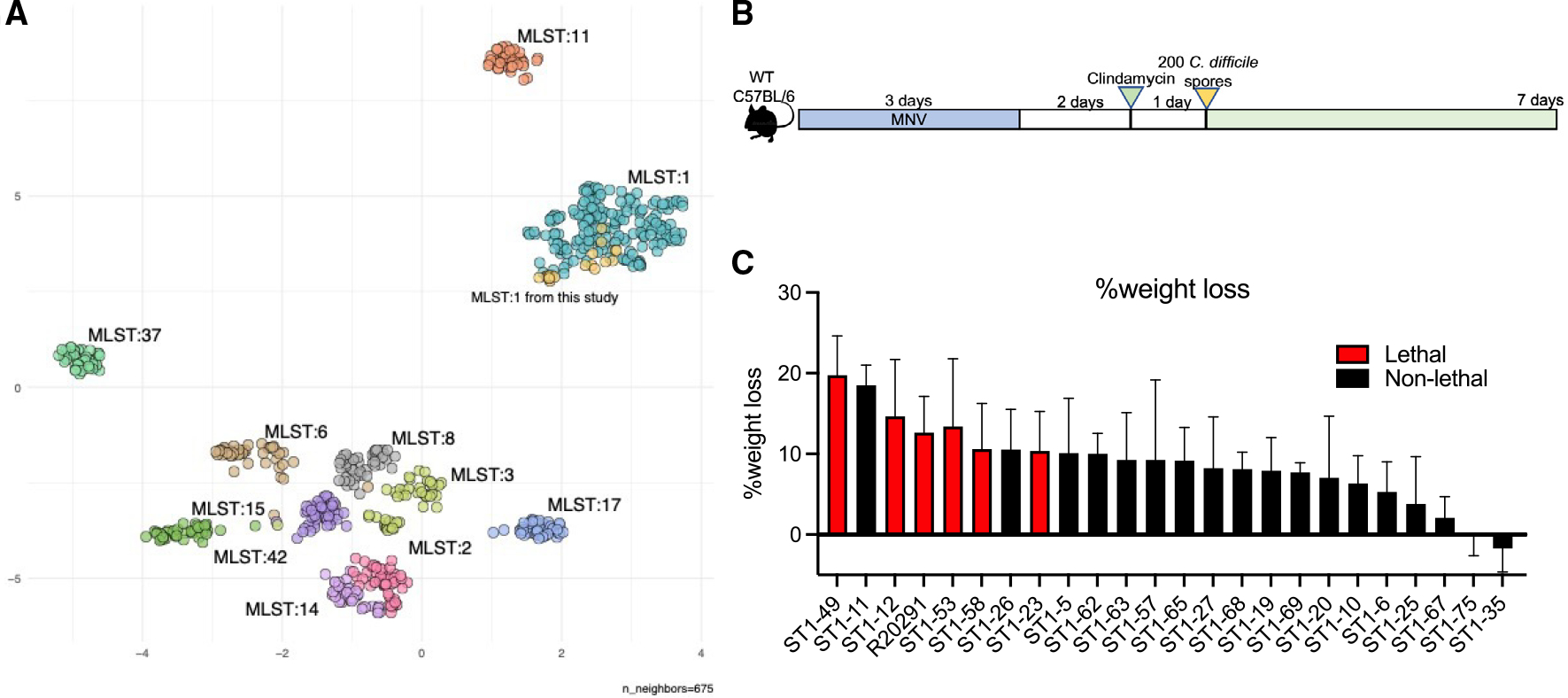
(A) Plot of the UMAP analysis of the presence or absence of unique coding sequences (annotated proteins or unannotated protein clusters) across the top 10 STsof C. difficile strains in Patric.
(B) Mouse experiment schematic: wild-type C57BL/6 mice were treated with metronidazole, vancomycin, and neomycin (MNV; 0.25 g/L for each) in drinking water for 3 days, followed by one intraperitoneal injection of clindamycin (200 μg/mouse) 2 days after antibiotic recess. Then, mice were inoculated with 200 C. difficile spores via oral gavage. Daily body weight and acute disease scores were monitored for 7 days post-infection.
(C) Maximum percentage of weight loss to baseline was calculated using the lowest weights within 7 days post-infection divided by day 0 weights. n (number of mice per strain-infected group) = 5–8 except for ST1-62 and ST1-68, which have 2 mice per group. Results represent means ± SD.
Two ST1 C. difficile isolates demonstrate avirulent phenotype
Among the C. difficile isolates that we examined using antibiotic-treated mice, two isolates, ST1-75 and ST1-35, caught our attention due to their strikingly attenuated phenotypes (Figure 1C). Almost no weight loss was observed throughout the 7-day time course, and low acute disease scores were displayed in mice infected with ST1-75 or ST1-35 compared with mice infected with R20291 (Figures 2A, 2B, 2D, and 2E). This avirulent phenotype was not due to colonization deficiency of ST1-75 or ST1-35, as the fecal colony-forming units (CFUs) recovered from the mice infected with these two isolates were comparable to R20291-infected mice on both early and late days post-infection (Figures 2C and 2F). Fecal levels of TcdA and TcdB were also measured, and similar levels were seen in the feces at day +1 post-infection from mice infected with ST1-75 and ST1-35 compared with R20291 (Figures 2G and 2H).
Figure 2. Two isolated clinical strains of C. difficile have no virulence in mice treated with antibiotics.
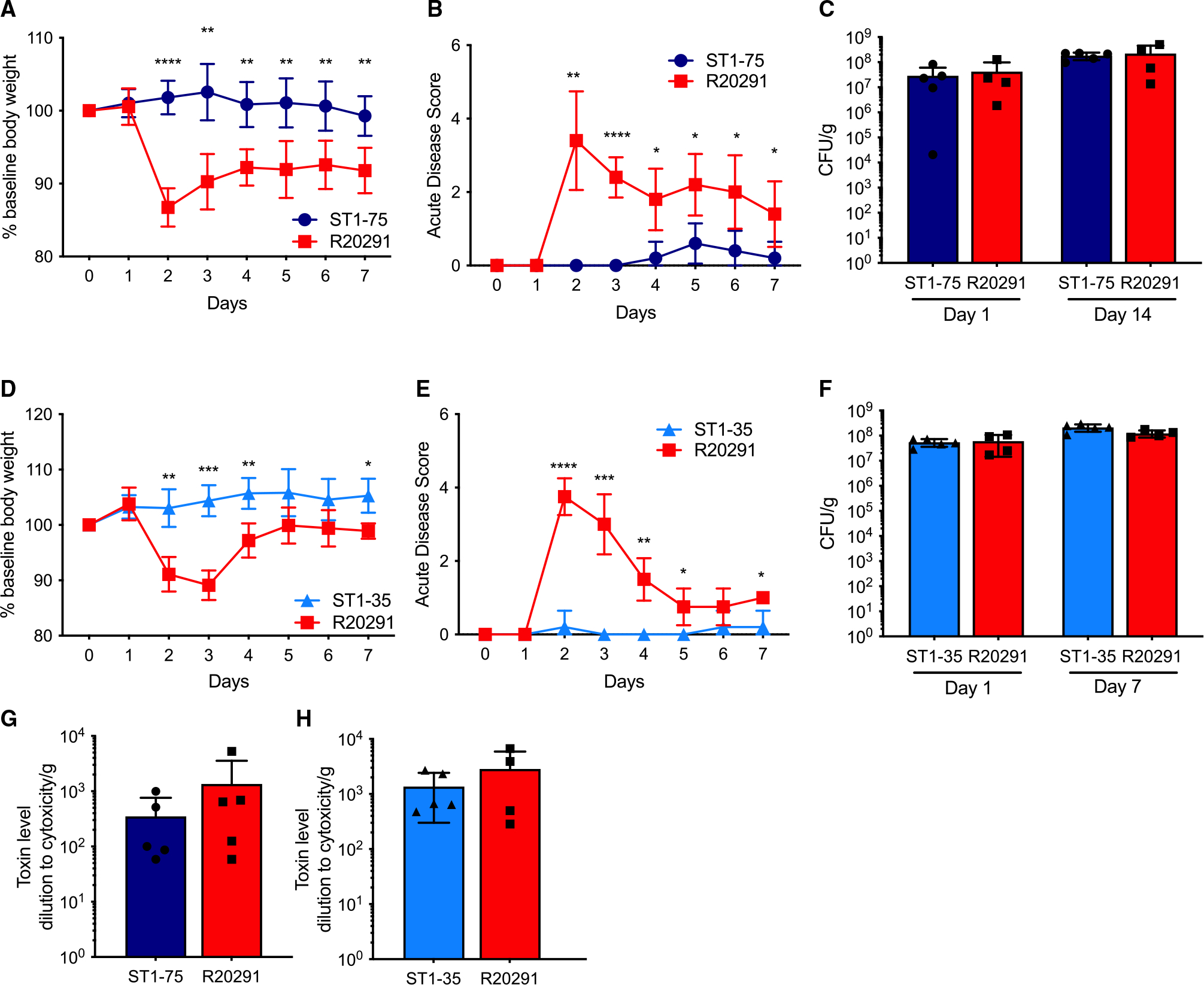
Wild-type C57BL/6 mice (n = 3–5 per group) were treated with MNV (0.25 g/L for each) in drinking water for 3 days, followed by one intraperitoneal injection of clindamycin (200 μg/mouse) 2 days after antibiotic recess. Then, mice were inoculated with 200 C. difficile spores via oral gavage. Daily body weight and acute disease scores were monitored for 7 days post-infection.
(A and D) Percentage of weight loss to baseline of mice infected with indicated strains.
(B and E) Acute disease scores comprising weight loss, body temperature drop, diarrhea, and morbidity of mice infected with indicated strains.
(C and F) Fecal colony-forming units measured by plating on selective agar on indicated days.
(G and H) Fecal Tcd toxins measured by CHO cell rounding assay 1 day post-infection. Results represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further investigate this avirulent phenotype, we inoculated ST1-75 into MyD88−/− mice, which lack the adaptor protein for Toll-like receptor signaling.24 MyD88−/− mice fail to recruit neutrophils to the colonic tissue during early stages of CDI and display markedly increased susceptibility to C. difficile-induced colitis.25 Here, MyD88−/− mice were treated with antibiotics and infected with either ST1-75 or R20291. Mice infected with R20291 quickly succumbed to infection 2 days after spore inoculation, whereas all MyD88−/− mice infected with ST1-75 survived the experiment with minimal weight loss or disease scores (Figures 3A and 3B). Consistent with our results with wild-type mice, no deficiencies of colonization or Tcd toxin production were observed day +1 post-infection of MyD88−/− mice (Figures 3C and 3D). These data suggest that the attenuation of the avirulent strain is independent of MyD88-mediated host innate immunity.
Figure 3. Avirulent C. difficile strain demonstrates no virulence in innate immune-deficient mice and germ-free mice.
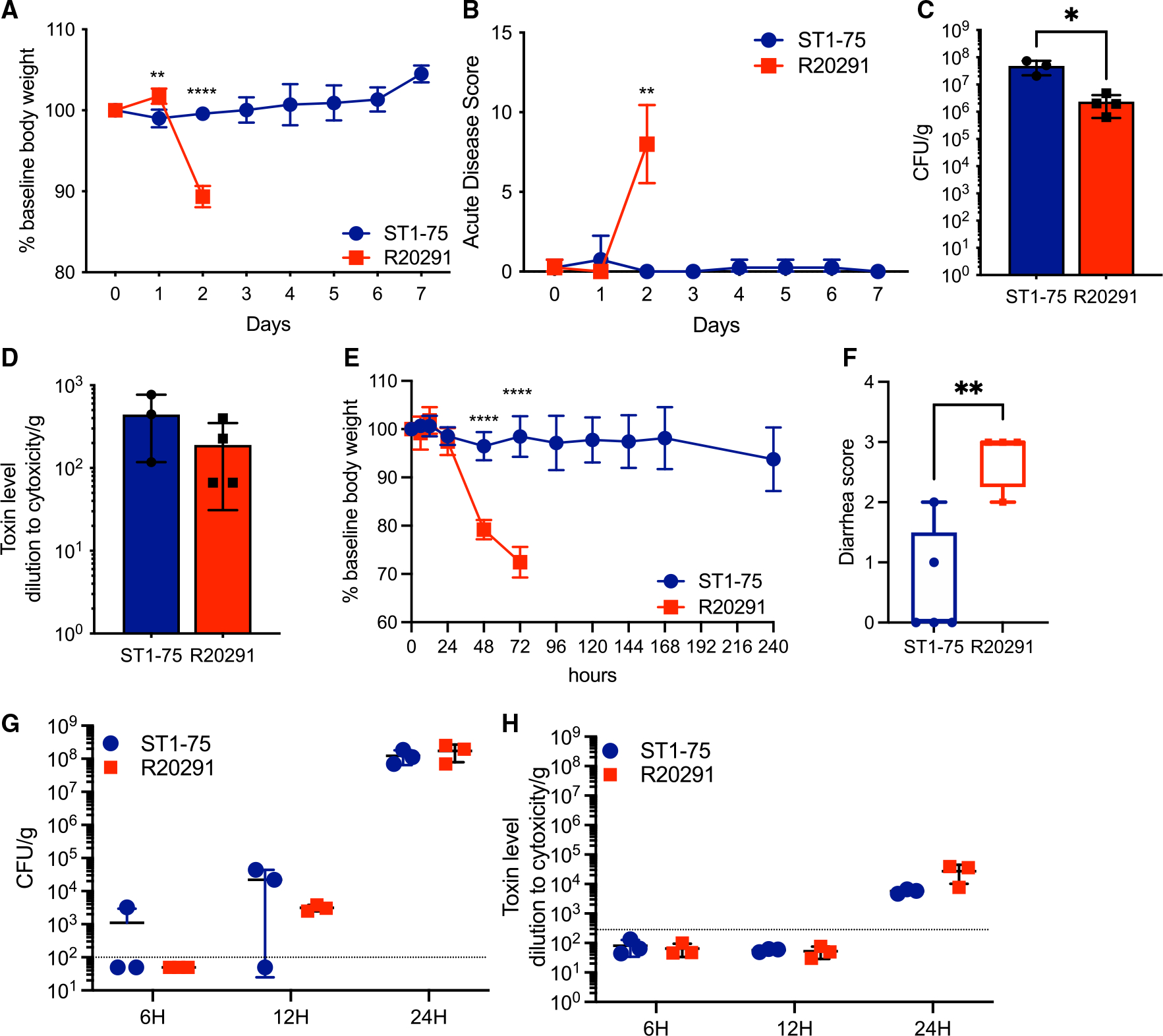
(A–D) MyD88−/− mice (n = 4 per group) were treated with MNV and clindamycin before being orally administered with 200 spores of C. difficile strains. Daily body weight and acute disease scores were monitored for 7 days post-infection.
(A) Percentage of weight loss to baseline of mice infected with indicated strains.
(B) Acute disease scores comprising weight loss, body temperature drop, diarrhea, and morbidity of mice infected with indicated strains.
(C) Fecal colony-forming units measured by plating on selective agar 1 day post-infection.
(D) Fecal Tcd toxins measured by CHO cell rounding assay 1 day post-infection.
(E–H) Germ-free mice (n = 3 to 5) orally administered with 200 spores of indicated C. difficile strains. Daily body weight and acute disease scores were monitored for 10 days post-infection.
(E) Percentage of weight loss to baseline of mice infected with indicated strains.
(F) Diarrhea scores of mice infected with indicated strains 2 days post-infection. Results represent min to max showing all points.
(G) Fecal colony-forming units measured by plating on selective agar at 6, 12, and 24 h post-infection.
(H) Fecal Tcd toxins measured by CHO cell rounding assay at 6, 12, and 24 h post-infection. Results represent means ± SD. *p < 0.05, **p < 0.01, ****p < 0.0001.
Germ-free mice are highly susceptible to CDI because they lack microbiome-mediated colonization resistance against C. difficile.26,27 To investigate whether the gut microbiome renders ST1-75 avirulent, germ-free mice were infected with ST1-75 or R20291. Similarly, we observed no mortality or weight loss in the mice with ST1-75 infection, whereas mice infected with R20291 quickly lost weight and died (or had >20% weight loss) (Figure 3E). Milder diarrhea was observed in mice with ST1-75 compared with R20291 (Figure 3F). We observed no differences in colonization or fecal Tcd toxins between ST1-75 and R20291 up to 24 h post-infection (Figures 3G and 3H). Similar attenuation was also seen in ST1-35-infected germ-free mice (Figure S2). In contrast, isolates that demonstrated relatively mild pathogenicity in antibiotic-treated mice, such as ST1-25 and ST1-67 (Figure 1C), led to severe weight loss and diarrhea in germ-free mice (Figure S2), reaffirming the protective role of the gut microbiome during CDI. However, the attenuation of ST1-75 and ST1-35 in mice is independent of the gut microbiome.
Prophages identified in avirulent strains do not impact ST1 C. difficile virulence
We next sought to determine the genetic factors that may abrogate C. difficile virulence in ST1-75 and ST1-35. Fully circularized genomes of 14 ST1 isolates were successfully obtained using Nanopore and Illumina hybrid assembly, and pangenomic analysis was conducted on these 14 genomes and R20291 using the anvi’o pangenomics workflow.28 A group of gene clusters that are unique to ST1-75 and ST1-35, which are enriched for phage-related genes, stood out (Figure S3A). We then applied PHASTER, a tool for phage identification in bacterial genomes, to discover two prophages in the genomes of ST1-75 and ST1-35. One prophage resides on a 41-kb plasmid in ST1-75 and ST1-35 with 4–5 copies per cell and here is named phiCD75-2. Blasting phiCD75-2 found high similarities to reported C. difficile phages phiCD38-2 (99.8% identity) and phiCDHS1 (94.7% identity).29–32 In addition, a 54-kb segment was found as inserted into the chromosomal DNA of ST1-75 and ST1-35 around position 1.29 Mbp and here is named phiCD75-3. phiCD75-3 does not show high similarity to any described C. difficile phages to date.
Lysogenic bacteriophages have been identified in many C. difficile genomes and play an important role in shaping C. difficile evolution. However, their roles in C. difficile biology, especially virulence, are not well characterized.33,34 To investigate the potential role of these two prophages on C. difficile virulence, we induced lytic phage particles of phiCD75-2 and phiCD75-3 from ST1-75 culture and infected R20291 to generate R20291 lysogens harboring these prophages. We were able to generate R20291 derivatives carrying phiCD75-2, phiCD75-3, or both prophages in their genomes (Figure S3B). WGS of lysogenic R20291 strains confirmed that phiCD75-3 was inserted in situ as in ST1-75 at 1.29 Mbp. Antibiotic-treated mice infected with R20291 lysogenic strains (Figure S3B) followed the curve of virulent infection, as 10% weight loss and 4–6 disease scores were seen during the peak of symptomatic infection (Figures S3C and S4D). The seemingly faster recovery in the lysogens was not a reproducible finding. Similar levels of colonization and Tcd toxin production were also observed (Figures S3E–S3H). Here, we discover two prophages in avirulent strains ST1-75 and ST1-35 that are not present in R20291 or other ST1 strains from our collection. However, these two prophages do not appear to impact the virulence of R20291 in antibiotic-treated mice.
Mutations in the cdtR gene eliminate ST1 C. difficile virulence in mice
Lysogenic R20291 strains with either or both prophages did not recapitulate the avirulent phenotype of ST1-75 or ST1-35. A closer look at the chromosomal genomes of ST1-75 and ST1-35 led us discover a common mutation in their cdtR gene, which was reported previously as a transcriptional regulator for binary toxin (CDT) genes, cdtA and cdtB.35 A 69-bp deletion was found in the cdtR gene of ST1-75 and ST1-35, leading to an in-frame deletion of 23 amino acids (Figure 4A). To investigate if there is a possible loss of function of CdtR resulting from the deletion, we accessed the transcriptional level of cdtB in mouse cecum following infection of ST1-75 or R20291. More than a 2-log reduction of cdtB transcripts was observed in the ST1-75 group (Figure 4B), suggesting an important role of these 69 bp for a fully functional cdtR gene. Next, we applied a CRISPR-mediated genome editing approach to generate CdtR mutants using the parental R20291 strain to study the contribution of CdtR to C. difficile virulence (Figures S4A and 4A). In accordance with a previous report,21 knocking out cdtR either by deleting the whole gene (CdtRKO8.1 and CdtRKO10.3) or by introducing a proximal premature stop codon (CdtRstop4.2 and CdtRstop8) led to a loss of pathogenicity in antibiotic-treated mice (Figures S4B and S4C), confirming a critical role of CdtR for C. difficile virulence. Moreover, deleting the exact same 69-bp region, as in ST1-75/35, in the cdtR of R20291 (CdtRmut6.1 and CdtRmut8.1) again eliminated the virulence of C. difficile (Figures 4C and 4D). Thus, loss of the 69 bp in cdtR explains the avirulence phenotype of ST1-75/35. On the other hand, colonization of these CdtR mutants, assessed by CFUs at day+1 post-infection, was comparable to that of R20291 (Figures S4D and S4G), suggesting that CdtR is not required for colonization. Interestingly, while the fecal levels of Tcd toxins of CdtR mutants were comparable to R20291 in the early phase (day+1 post-infection), a significantly reduced level at a later time point (7 days post-infection) was observed upon infection of CdtR mutants (Figures S4E, S4F, 4E and S4H), supporting a role of CdtR in regulating Tcd toxin production. Further, infecting germ-free mice with CdtRmut6.1 results in no weight loss or diarrhea, perfectly recapitulating ST1-75 and ST1-35 phenotypes in germ-free mice (Figures 4F, 4G, and S4I). Collectively, CRISPR-edited CdtR mutant strains mimic phenotypes of ST1-75 and ST1-35 in mice, demonstrating that the cdtR gene is necessary for in vivo virulence. Furthermore, the 69-bp region in cdtR, which is deleted in ST1-75/35, is necessary for proper CdtR function through mechanisms yet to be determined.
Figure 4. Binary toxin regulator cdtR contributes to C. difficile virulence in mice.
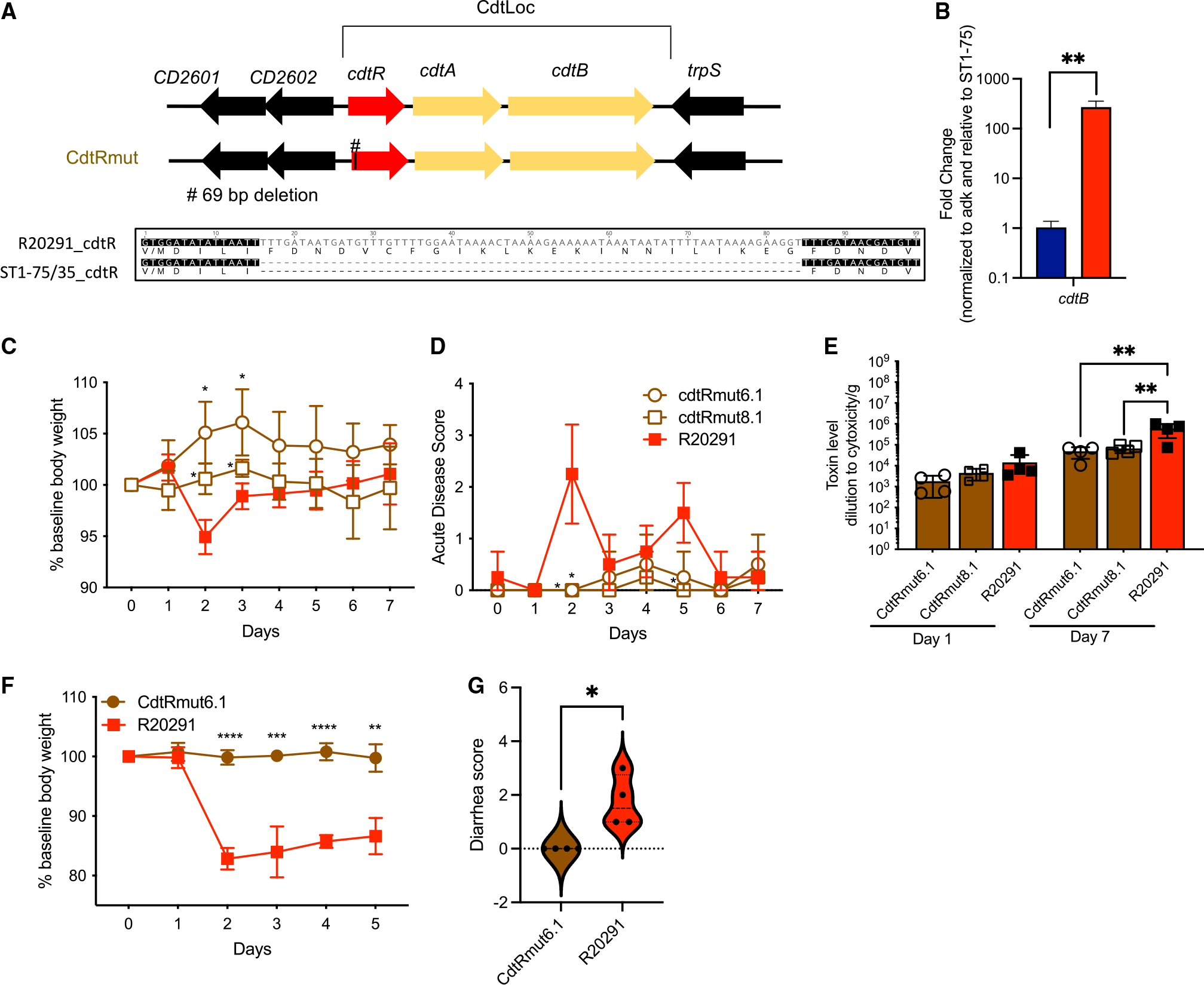
(A) Deletion identified in ST1-35/75 and schematic of cdtR mutants generated using R20291 C. difficile strain.
(B) Germ-free mice (n = 3 per group) orally administered with 200 spores of indicated C. difficile strains. Binary toxin gene cdtB transcripts were measured by RT-qPCR on cecal contents harvested at 24 h post-infection with 2 technical replicates per sample. Transcripts were normalized to the adk, and fold change is relative to ST1-75 condition.
(C–E) Wild-type C57BL/6 mice (n = 3 to 5 per group) were treated with MNV and clindamycin as previously described. Then, mice were inoculated with 200 C. difficile spores via oral gavage. Daily body weight and acute disease scores were monitored for 7 days post-infection.
(C) Percentage of weight loss to baseline of mice infected with indicated strains. Both CdtRmut6.1 and CdtRmut8.1 have significant differences on days 2 and 3 compared with R20291.
(D) Acute disease scores comprising weight loss, body temperature drop, diarrhea, and morbidity of mice infected with indicated strains. CdtRmut6.1 has a significant difference on day 2 compared with R20291. CdtRmut8.1 has a significant difference on days 2 and 5 compared with R20291.
(E) Fecal Tcd toxins measured by CHO cell rounding assay on indicated days.
(F and G) Germ-free mice (n = 4) orally administered with 200 spores of indicated C. difficile strains. Daily body weight and were monitored for 5 days post-infection.
(F) Percentage of weight loss to baseline of mice infected with indicated strains.
(G) Diarrhea scores of mice infected with indicated strains 3 days post-infection (violin plot showing all points). Results represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Mutation in cdtR reduce Tcd toxin transcription in vivo
CdtR mutants produce significantly reduced fecal Tcd toxins at a later time point (7 days post-infection) (Figures 4E and S4E), a phenotype that was confirmed in ST1-75 and ST1-35 (Figures S5A and S5B). To examine whether the 69-bp deletion in cdtR impacts Tcd toxin production, we harvested cecal contents from germ-free mice infected with ST1-75 or R20291. In contrast to fecal toxin concentrations, we observed a significantly reduced cecal Tcd toxin concentration in mice infected with ST1-75 compared with R20291 (Figure 5A). This was not due to a slightly lower CFU of cecal ST1-75 in germ-free mice (Figures S5C and S5D). We further validated the reduced toxin production in cecal content by RT-qPCR, and we observed a 50-fold reduction of the tcdA and tcdB transcripts in the cecum of mice infected with ST1-75 (Figure 5B). Additionally, transcripts of other PaLoc genes including tcdE and tcdR were also reduced in the cecum of mice infected with ST1-75 (Figure 5B). TcdE is a putative holin that mediates toxin secretion.36–38 Reduced tcdE likely further impacts the amount of toxins that may reach the intestinal epithelium. TcdR is a positive regulator of the PaLoc39,40 and is likely the common target of CdtR, which results in the observed downregulation of many PaLoc genes. Interestingly, cdtR transcripts were comparable between ST1-75 and R20291, suggesting that the 69-bp deletion does not impact the transcript’s stability. These results were further confirmed with the CdtRmut6.1 strain, though to a lesser extent (Figure S5E). Overexpressing wild-type CdtR (but not CdtR with a 69-bp deletion) restored and enhanced the transcription of both PaLoc genes and binary toxin (Figures S5F and S5G). Collectively, we demonstrate that a natural mutation found in cdtR of two ST1 clinical isolates results in reduced binary toxin production, reduced Tcd toxin production (and likely secretion) in cecum of infected mice, and attenuated C. difficile virulence, independent of host innate immunity, colonization burden, microbiome constitution, or any noticeable impact of incidentally discovered prophages within these strains. This difference of toxin production in cecum at 24 h post-infection is, however, not reflected in feces in parallel but could be reflected at later time points, likely due to cumulative differences over time.
Figure 5. CdtR regulates Tcd toxins transcription in vivo.
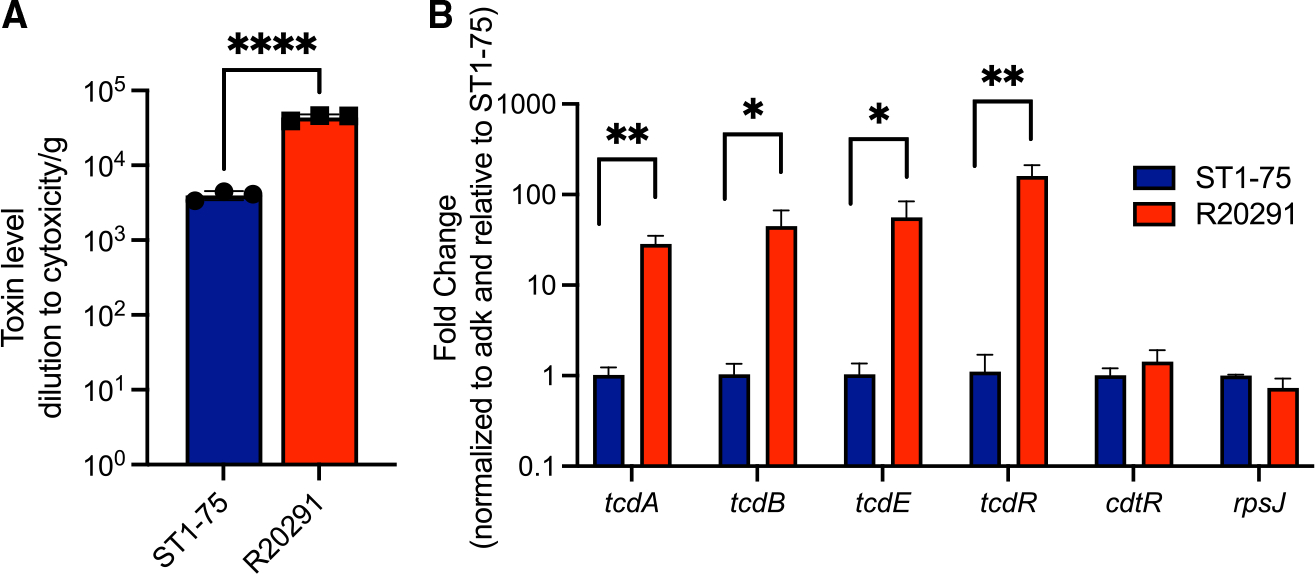
Germ-free mice (n = 4) were orally administered with 200 spores of indicated C. difficile strains and cecal contents were harvested at 24 h post-infection.
(A) Cecal Tcd toxins measured by CHO cell rounding assay.
(B) Indicated gene transcripts were measured by RT-qPCR with 2 technical replicates per sample. Transcripts were all normalized to the adk, and fold change is relative to ST1-75 for each of the genes. Results represent means ± SD. *p < 0.05, **p < 0.01, ****p < 0.0001.
cdtR is versatile and more prevalent than cdtA and cdtB
Our data support a regulatory role of CdtR outside CdtLoc, so we hypothesize that CdtR may have evolved to impact virulence beyond regulating CDT binary toxins. To test this possibility, we surveyed the presence of cdtR, cdtA, and cdtB in two major C. difficile clinical collections.23,41 As expected, the majority of clade 2 strains, including the epidemic ST1/RT027 strains, contain the CdtLoc with the presence of all three genes. Other subgroups of C. difficile strains, including MLST5 and MLST11, were also reported to encode CDT (Figure 6).42,43 Unexpectedly, many strain types of C. difficile that were reported as CDT negative also encode cdtR, such as MLST2 and MLST8 from clade 1 (Figure 6). The higher prevalence of cdtR than cdtA and cdtB supports the possibility that CdtR functions beyond regulating CDT. Additional work is needed to evaluate the functions of CdtR in these CDT-negative strains. Alternatively, CdtR may also lose its function in CDT-positive strains. Two such cases are ST11 strains, in which cdtR has lost a premature stop codon resulting in a pseudogene,42 as well as here with the mutation in the cdtR of ST1-75/35. To evaluate the prevalence of cdtR mutations that may lead to a loss of function, we aligned all cdtR genes in MLST1 strains from the two described collections and found that several strains had similar truncations at the proximal end that may have lost CdtR function, yet we did not find, within almost 500 strains, the exact deletion we identified in ST1-75/35 (Figure S6A).
Figure 6. Binary toxin regulator cdtR is prevalent in clinical C. difficile isolates.
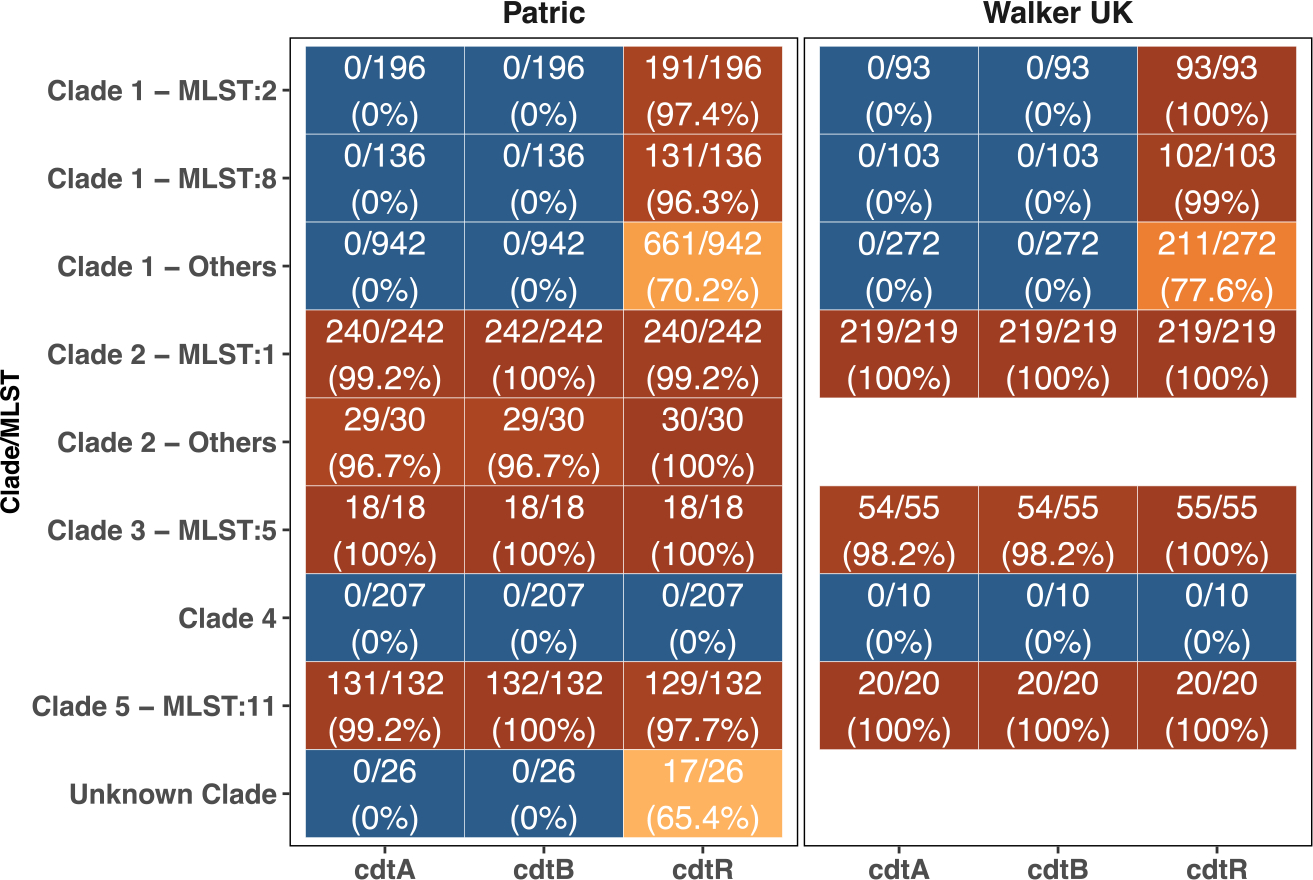
Binary toxins cdtA, cdtB, and cdtR from R20291 were used as query to BLAST against the assembled contigs. Hits with at least 85% identity and 85% coverage of the query were considered a valid match. Numbers of match in total and percentages are presented.
The high genetic and phenotypic similarity between ST1-75 and ST-35 led us to investigate their potential clonality. We performed a core-genome SNP analysis across all ST1 isolates from our collection with R20291 as the reference. ST1-75 and ST1-35 shared all SNPs when compared to the genome of R20291 (Figure S6B). Additional clinical evidence supporting that ST1-35 and ST1-75 are clonal is that these strains were isolated from two patients who were hospitalized in the same hospital room within 2 weeks of each other (Figure S6C).
DISCUSSION
Mouse models are valuable tools to study how C. difficile strain variations may result in variable disease severities, thanks to the advantages of their identical genetic, immune background and controlled microbiome compositions. Here, we focused on a group of clinical C. difficile isolates belonging to the RT027/MLST1, with high genomic similarity, that all encode PaLoc and CdtLoc. We found that these similar C. difficile isolates caused variable disease severities in mice and that a very specific mutation in the cdtR gene rendered two clinical isolates, ST1-75 and ST1-35, avirulent. Avirulence was solely dependent on the cdtR mutation, as we obtained similar observations using MyD88−/− mice and germ-free mice, which was also further validated with CRISPR-edited cdtR mutants. Lower transcripts of binary toxin gene cdtB, toxin A tcdA, and toxin B tcdB, together with other PaLoc genes including regulator tcdR and putative holin tcdE, were observed in germ-free mouse cecum infected with the CdtR mutants. Our data support a critical role of CdtR in regulating C. difficile toxin production and secretion, which are essential to ST1 virulence. However, all the other ST1 isolates in this study encoded an intact CdtLoc with wild-type cdtR, whose variations in virulence are likely attributable to alternative mechanisms.
The presence of a binary toxin locus has been associated with epidemic strains and hypervirulence of C. difficile.44,45 CDT belongs to the family of ADP-ribosylating toxins that consist of two components: CDTa (cdtA), the enzymatic active ADP-ribosyltransferase that modifies cellular actin, and CDTb (cdtB), the binding component that facilitates CDTa translocation. However, despite knowing their enzymatic activities, experimental evidence is very limited to support critical roles of CDT in C. difficile virulence.46 CDTb was reported to induce Toll-like receptor 2 (TLR2)-dependent pathogenic inflammation, which suppresses a protective eosinophilic response and enhances virulence of RT027 strains; however, C. difficile lacking CDTb still causes acute disease in mice.47 On the other hand, CdtR, as the transcriptional regulator of cdtA and cdtB,6,35,48 has been previously linked to Tcd toxin production,21 suggesting a role as a major virulence regulator. Here, we demonstrated a critical role of CdtR as a determinant of C. difficile virulence within ST1 strains. A natural 69-bp deletion in cdtR that was found in two clinical isolates can reverse the virulence of a wild-type strain by downregulating the expression of PaLoc genes and binary toxin genes. Additionally, higher prevalence of cdtR over cdtA or cdtB was found while surveying CdtLoc on clinical isolates from public databases. This suggests that CdtR may have evolved to function beyond regulating cdtA and cdtB. Systematically examining the target genes of CdtR may give us insights on its additional functions, which may also help unveil the mechanisms by which CdtR regulates the PaLoc genes.
ST1-75 and ST1-35 are avirulent in susceptible mouse models despite producing toxins, albeit at reduced levels. This is intriguing because it is well appreciated that toxin expression is necessary for C. difficile virulence.46,49 However, our data indicate that toxin production is not sufficient for causing CDI. The amount of toxin being produced and released likely impacts the development of disease. The patients from whom we isolated ST1-75 or ST1-35 had an overall mild clinical assessment, and their symptoms may be attributable to causes other than CDI. Current CDI diagnoses largely depend on the detection of the TcdB gene or toxin B positivity in feces and may lead to overdiagnosis of CDI. We, together with other reports, suggest the importance of quantifying toxins to evaluate CDI cases.50–52 Incorporating adjunctive biomarkers, such as interleukin-1β (IL-1β), better distinguishes CDI from asymptomatic carriage and non-CDI diarrhea.53 Here, CdtR regulates both toxin production and secretion and is essential for C. difficile virulence in mice, suggesting that it may serve as an adjunctive biomarker for CDI diagnosis.
Apart from characterizing CdtR, we also identified two prophages in ST1-75 and ST1-35. Prophages have been identified in many C. difficile genomes and play important roles in shaping C. difficile evolution.33 While prophages are highly prevalent in C. difficile, little is known about how prophages impact C. difficile biology. A couple of pioneering studies have shown that prophages can affect C. difficile gene expression, impacting toxin production.29,54,55 In this study, we identified two prophages in ST1-75/35 and named them phiCD75-2 and phiCD75-3. By making R20291 lysogenic strains harboring either or both prophages, we observed minimal impacts on C. difficile virulence by both of the prophages in antibiotic-treated mice. PhiCD38-2 was shown to increase PaLoc gene expression and toxin production in some RT027 isolates but not in all of them, suggesting that the genetic background influences the impact of a newly acquired prophage.29 This may explain why phiCD75-2 (a phiCD38-2 derivative) did not increase toxin production in ST1-75 isolates. Certain phages also impact phase variation of the cell surface protein and biofilm formation and carry genes involved in quorum sensing, inferring their roles in bacterial fitness.30,56,57 It would be very intriguing to investigate how phiCD75-2 and phiCD75-3 may impact C. difficile fitness, including gene expression, antibiotic resistance, and interspecies competition.
In summary, we demonstrate that ST1 C. difficile clinical isolates with identical PaLoc display variable virulence in vivo. Among them, two clonal clinical isolates, ST1-75 and ST1-35, were avirulent in mice due to a 69-bp deletion mutation in their cdtR genes. These data suggest that specific cdtR genetic variants within the same strain type may predict disease occurrence and severity. Routine detection of these variants may enhance the specificity of nucleic acid amplification tests (NAATs) for CDI diagnosis. Our data also corroborate recent clinical observations that toxin detection is unreliable as the sole criterion to distinguish between CDI and colonization.
Limitations of the study
It is acknowledged that the mouse model of CDI does not mimic all aspects of the human infection. The wide range of disease severities we observed in mice does not perfectly mirror the clinical data. Thus, phenotypes in mice cannot be used to infer CDI severity in humans. However, the identical background of mice led us to discover essential factors for C. difficile virulence that were masked by the complex host factors within clinical data. We discovered an important role of CdtR in regulating Tcd and CDT toxins in clinical ST1, a prevalent C. difficile strain type. This critical role of CdtR for virulence may not be generalizable to all other strain types. For example, ST11 has a CdtR pseudogene, and ST37 has no CDT genes, while both still cause CDI in patients. Better understanding the roles of CdtR in other strain types will further help dissect the contributions of Tcd and CDT toxins in clinical CDI.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Qiwen Dong (qiwendong0721@gmail.com)
Materials availability
All unique reagents and plasmids generated in this study are available from the lead contact Qiwen Dong (qiwendong0721@gmail.com) with a completed materials transfer agreement.
Data and code availability
Whole-genome sequence data were uploaded to National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject: PRJNA885086 and PRJNA595724.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Bacterial strains and growth conditions
C. difficile isolates were grown on brain heart infusion (BHI) agar plates supplemented with yeast extract and L-cysteine (BHIS) or in BHIS broth at 37°C in an anaerobic chamber (Coylabs). Antibiotics may be supplemented as described in the detailed methods. B. subtilis and E. coli were routinely grown on Luria-Bertani (LB) agar or in LB broth at 37°C aerobically. Transformed B. subtilis and E. coli with plasmids were selected with media containing 15 μg/mL chloramphenicol or 100 μg/mL ampicillin as needed.
Cell line
Chinese hamster ovary cells (CHO/dhFr-, ATCC#CRL-9096) were grown in complete media (alpha-modified MEM supplemented with 10% FBS, 2% HEPES, 0.4% L-Glutamine, 0.007% β-mercaptoethanol, 1% Penicilin/Streptimycin/Gentamycin) at 37°C with 5% CO2 and split every 2–3 days for subculture.
Mice
Wild-type female C57BL/6 mice, aged 6 to 8 weeks, were purchased from the Jackson Laboratories. Male and female MyD88−/− mice were maintained in augmentin (0.48 g/L and 0.07 mg/L of amoxicillin and clavulanate respectively) in the drinking water in specific-pathogen-free (SPF) facility at the University of Chicago. Male and female germ-free C57Bl/6J mice were bred and maintained in plastic gnotobiotic isolators within the University of Chicago Gnotobiotic Core Facility and fed ad libitum autoclaved standard chow diet (LabDiets 5K67) before transferring to BSL2 room for infection. Mice housed in the BSL2 animal room are fed irradiated feed and provided with acidified water. All mouse experiments were performed in compliance with University of Chicago’s institutional guidelines and were approved by its Institutional Animal Care and Use Committee.
METHOD DETAILS
C. difficile clinical isolate collection and classification
Toxigenic C. difficile-positive stool specimens were collected at Memorial Sloan Kettering Cancer Center between 2013 and 2017. C. difficile isolates were recovered by plating onto BHIS agar, supplemented with antibiotics D-cycloserine and cefoxitin (BHI and yeast extract were from BD Biosciences, and the other components were from Sigma-Aldrich) in an anaerobic chamber (Coylabs). Individual colonies that were able to grow in the presence of these antibiotics and that had the characteristic phenotype of C. difficile were selected, isolated, and subjected to whole-genome sequencing and MLST classification.75
C. difficile spore preparation and numeration
C. difficile sporulation and preparation was processed as described previously76 with minor modifications. Briefly, single colonies of C. difficile isolates were inoculated in deoxygenated BHIS broth and incubated anaerobically for 40–50 days. C. difficile cells were harvested by centrifugation and five washes with ice-cold water. The cells were then suspended in 20% (w/v) HistoDenz (Sigma, St. Louis, MO) and layered onto a 50% (w/v) HistoDenz solution before centrifugating at 15,000 × g for 15 min to separate spores from vegetative cells. The purified spores pelleted at the bottom were then collected and washed for four times with ice-cold water to remove traces of HistoDenz, and finally resuspended in sterile water. Prepared spores were heated to 60°C for 20 min to kill vegetative cells, diluted and plated on both BHIS agar and BHIS agar containing 0.1% (w/v) taurocholic acid (BHIS-TA) for numeration. Spore stocks for mouse infection were verified to have less than 1 vegetative cell per 200 spores (as the infection dose).
Virulence assessment of clinical isolates in mice
SPF mice were treated with antibiotic cocktail containing metronidazole, neomycin and vancomycin (MNV) in drinking water (0.25 g/L for each antibiotic) for 3 days, 2 days after removing MNV, the mice were received one dose of clindamycin (200 μg/mouse) via intraperitoneal injection. Mice were then the next day infected with 200 C difficile spores via oral gavage. Germ-free mice were infected with 200 C difficile spores via oral gavage without antibiotic treatments.
Following infection, mice were monitored and scored for disease severity by four parameters77: weight loss (>95% of initial weight = 0, 95%–90% initial weight = 1, 90%–80% initial weight = 2, <80% = 3), surface body temperature (>95% of initial temp = 0, 95%–90% initial temp = 1, 90%–85% initial temp = 2, <85% = 3), diarrhea severity (formed pellets = 0, loose pellets = 1, liquid discharge = 2, no pellets/caked to fur = 3), morbidity (score of 1 for each symptoms with max score of 3; ruffled fur, hunched back, lethargy, ocular discharge).
Quantification of fecal colony forming units
Fecal pellets or cecal content from C. difficile infected mice were harvested and resuspended in deoxygenated phosphate-buffed saline (PBS), diluted and plated on BHI agar supplemented with yeast extract, taurocholic acid, L-cysteine, D-cycloserine and cefoxitin (CC-BHIS-TA) at 37°C anaerobically for overnight.78
Cell-based assay to quantify fecal and cecal Tcd toxin
The presence of C. difficile Tcd toxins was determined using a cell-based cytotoxicity assay as previously described with minor modifications.78 Briefly, Chinese hamster ovary cells (CHO/dhFr-, ATCC#CRL-9096) were incubated in a 96-well plate overnight at 37°C. 10-fold dilutions of supernatant from resuspended fecal or cecal content were added to CHO/dhFr-cells, incubated overnight at 37°C. Cell rounding and death was scored the next day. The presence of C. difficile Tcd toxins was confirmed by neutralization by antitoxin antisera (Techlab, Blacksburg, VA). The data are expressed as the log10 reciprocal value of the last dilution where cell rounding was observed.
DNA extraction, RNA extraction and reverse transcription
Fecal DNA was extracted using DNeasy PowerSoil Pro Kit (Qiagen), and RNA was isolated from cecal contents or bacterial culture using RNeasy PowerMicrobiome Kit (Qiagen) according to the manufacturer’s instructions, respectively. Complementary DNA was generated using the QuantiTect reverse transcriptase kit (Qiagen) according to the manufacturer’s instructions.
Quantitative polymerase chain reaction (qPCR)
Quantitative PCR was performed on genomic DNA or complementary DNA using primers (listed in Table S1) with PowerTrack SYBR Green Master Mix (Thermo Fisher). Reactions were run on a QuantStudio 6 pro (Thermo Fisher). Relative abundance was normalized by ΔΔCt.
Generation of C. difficile cdtR mutants using CRISPR
CRISPR editing on C. difficile strains R20291 was performed as described in.61 The primers were listed in Table S1.79–83 Briefly, donor regions for homology were generated by separately amplifying regions ~500 bp upstream and ~500 bp downstream of the target of interest. The resulting regions were cloned into pCE677 between NotI and XhoI sites by Gibson Assembly. Geneious Prime (v11) was used to design sgRNAs targeting each deleted target. sgRNA fragments were then amplified by PCR from pCE677, using an upstream primer that introduces the altered guide and inserted at the MscI and MluI sites of the pCE677-derivative with the appropriate homology region. Regions of plasmids constructed using PCR were verified by Sanger sequencing. Plasmids were then passaged through NEBturbo E. coli strain before transformation into Bacillus subtilis strain BS49. The CRISPR-Cas9 deletion plasmids which harbor the oriT (Tn916) origin of transfer, were then introduced into C. difficile strains by conjugation.84 C. difficile colonies were then screened for proper mutations in the genomes by PCR and correct clones were further validated by whole-genome sequencing.
Whole-genome sequencing and assembly
DNA was extracted using the QIAamp PowerFecal Pro DNA kit (Qiagen). Libraries were prepared using 100 ng of genomic DNA using the QIAseq FX DNA library kit (Qiagen). Briefly, DNA was fragmented enzymatically into smaller fragments and desired insert size was achieved by adjusting fragmentation conditions. Fragmented DNA was end repaired and ‘A’s’ were added to the 3′ ends to stage inserts for ligation. During ligation step, Illumina compatible Unique Dual Index (UDI) adapters were added to the inserts and prepared library was PCR amplified. Amplified libraries were cleaned up, and QC was performed using Tapestation 4200 (Agilent Technologies). Libraries were sequenced on an Illumina NextSeq 500 or MiSeq platform to generate 2 × 150 or 2 × 250 bp reads respectively. Illumina reads were assembled into contigs using SPAdes63 and genes were called and annotated using Prokka (v1.14.6).85
Samples for Nanopore and Illumina hybrid assemblies were extracted using the NEB Monarch Genomic DNA Purification Kit. DNA was QC’ed using genomic Tapestation 4200. Nanopore libraries were prepared using the Ligation Sequencing Kit (SQK-LSK109), the Native Barcoding Expansions 1–12 (EXP-NBD104) and 13–24 (EXP-NBD114), and the NebNext Companion Module for Oxford Nanopore Technologies (E7180S). The shearing steps and first ethanol wash were eliminated to ensure high concentrations of long fragments. Using R9.4.1 flow cells, libraries were run on a MinION for 72 h at 180V. The Nanopore and Illumina hybrid assemblies were completed using Unicycler (v0.4.8)64 either with the untrimmed or trimmed Illumina reads. The assemblies with less number of circularized contigs were used for genome analysis.
Binary toxin genes prevalence analysis
C. difficile isolates (N = 827) from BioProject: PRJEB4556 were downloaded from NCBI, and assembled into contigs using SPAdes.63 A collection of 2143 C difficile genomes from Patric (date: Feb. 10 2021)23 were also downloaded. MLST was determined on those contigs by mlst.86 ST type with less than 3 isolates were removed. Binary toxin cdtA, cdtB and cdtR from R20291 (NCBI: NC_013316) were used as query to BLAST65 against the assembled contigs, and hits with at least 85% identity and 85% coverage of the query are considered a valid match.
UMAP (uniform manifold approximation and projection) analysis
A subset of isolate contigs of 199 ST1, 50 ST2, 50 ST3, 49 ST6, 50 ST8, 50 ST11, 42 ST14, 50 ST15, 50 ST17, 50 ST37 and 50 ST42, totaling 690 isolates were selected from the above Patric collection. They were all sequenced by short read technology, and they are the top 10 abundant ST groups except ST1 in the Patric collection. Genes were called and annotated from their contigs using Prokka (v1.14.6).85 By combining the 23 isolates from this study, we constructed a matrix of 731 isolates by 8025 annotated genes and hypothetical protein clusters. Specifically, hypothetical protein clusters were formed by clustering hypothetical proteins at 50% identity using cd-hit.87,88 Any protein sequences that were at least 50% similar fall into an artificially cluster. UMAP analysis was performed on the basis of the presence/absence of the genes/hypothetical protein clusters by setting the n_neighbors parameter to 675.
QUANTIFICATION AND STATISTICAL ANALYSIS
Results represent means ± SD. Statistical significance was determined by t test and one-way ANOVA test. Multiple comparisons were corrected with False Discovery Rate with desired FDR at 0.05. Statistical analyses were performed using Prism GraphPad software v9.3.1 (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001)
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| B. subtilis BS49 | Wilson and Bott58 | N/A |
| C.difficile R_cdtRKO10.3 | This manuscript | BioSample: SAMN31149530 |
| C.difficile R_cdtRKO8.1 | This manuscript | BioSample:SAMN31149531 |
| C.difficile R_cdtRmut6.1 | This manuscript | BioSample:SAMN31149534 |
| C.difficile R_cdtRmut8.1 | This manuscript | BioSample:SAMN31149535 |
| C.difficile R_cdtRstop4.2 | This manuscript | BioSample:SAMN31149532 |
| C.difficile R_cdtRstop8 | This manuscript | BioSample:SAMN31149533 |
| C.difficile R_phi75-2 | This manuscript | BioSample:SAMN31149536 |
| C.difficile R_phi75-2/3 | This manuscript | BioSample:SAMN31149537 |
| C.difficile R_phi75-3 | This manuscript | BioSample:SAMN31149538 |
| C.difficile R20291 | Human isolate | NCBI:NC_013316 |
| C.difficile ST1-10 | Human isolate | BioSample:SAMN13566327 |
| C.difficile ST1-11 | Human isolate | BioSample:SAMN13566371 |
| C.difficile ST1-12 | Human isolate | BioSample:SAMN13566370 |
| C.difficile ST1-19 | Human isolate | BioSample:SAMN13566326 |
| C.difficile ST1-20 | Human isolate | BioSample:SAMN13566335 |
| C.difficile ST1-23 | Human isolate | BioSample:SAMN13566336 |
| C.difficile ST1-25 | Human isolate | BioSample:SAMN13566328 |
| C.difficile ST1-26 | Human isolate | BioSample:SAMN13566333 |
| C.difficile ST1-27 | Human isolate | BioSample:SAMN13566330 |
| C.difficile ST1-35 | Human isolate | BioSample:SAMN13566365 |
| C.difficile ST1-49 | Human isolate | BioSample:SAMN13566378 |
| C.difficile ST1-5 | Human isolate | BioSample:SAMN13566334 |
| C.difficile ST1-53 | Human isolate | BioSample:SAMN13566406 |
| C.difficile ST1-57 | Human isolate | BioSample:SAMN13566317 |
| C.difficile ST1-58 | Human isolate | BioSample:SAMN13566423 |
| C.difficile ST1-6 | Human isolate | BioSample:SAMN13566324 |
| C.difficile ST1-62 | Human isolate | BioSample:SAMN13566360 |
| C.difficile ST1-63 | Human isolate | BioSample:SAMN13566320 |
| C.difficile ST1-65 | Human isolate | BioSample:SAMN13566318 |
| C.difficile ST1-67 | Human isolate | BioSample:SAMN13566325 |
| C.difficile ST1-68 | Human isolate | BioSample:SAMN13566403 |
| C.difficile ST1-69 | Human isolate | BioSample:SAMN13566435 |
| C.difficile ST1-75 | Human isolate | BioSample:SAMN13566366 |
| C.difficile VPI10463 | ATCC | ATCC #43255 |
| E.coli NEB5-alpha | NEB | NEB #C2987 |
| E.coli NEBturbo | NEB | NEB #C2984 |
| E.coli HB101(RP4) | Maikova et al.59 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Metronidazole | Sigma-Aldrich | Cat# M3761 |
| Neomycin Sulfate | Fisher scientific | Cat# BP2669 |
| Vancomycin Hydrochloride | Hospira | UoS NDC # 00409-1319-01 |
| Clindamycin hydrochloride | Sigma-Aldrich | Cat# C5296 |
| Mitomycin C | Novus Biologicals | Cat# NB3258 |
| Histodenz | Sigma-Aldrich | Cat# D2158 |
| Amoxicillin/Potassium Clavulanate | NorthStar Rx | NDC# 16714029301 |
| Chloramphenicol | Sigma-Aldrich | Cat# C1919 |
| Thiamphenicol | Sigma-Aldrich | Cat# T0261 |
| Taurocholic acid sodium salt hydrate | Sigma-Aldrich | Cat# T4009 |
| Xylose | Sigma-Aldrich | Cat# X3877 |
| Cefoxitin sodium salt | Sigma-Aldrich | Cat# C4786 |
| D-Cycloserine | Sigma-Aldrich | Cat# C6880 |
| L-Cysteine | Sigma-Aldrich | Cat# C7352 |
| Minimum Essential Medium Eagle | Sigma-Aldrich | Cat# M8042 |
| HEPES | Gibco | Cat# 845-1344 |
| PEN/STREP | Gibco | Cat# 15140-122 |
| L-Glutamine | Gibco | Cat# 810-1051 |
| 2-Mercaptoethanol | Applied Biosystems | Cat# AB1340 |
| Gentamycin Sulfate | Gemini | Cat# 400-108 |
|
| ||
| Critical commercial assays | ||
|
| ||
| QuantiTect Reverse Transcription Kit | Qiagen | Cat# 205311 |
| RNeasy PowerMicrobiome Kit | Qiagen | Cat# 26000 |
| QiAamp PowerFecal pro DNA Kit | Qiagen | Cat# 51804 |
| DNeasy PowerSoil Pro Kit | Qiagen | Cat# 47016 |
| Gibson Assembly® Cloning Kit | NEB | Cat# E5510S |
| QIAquick PCR Purification Kit | Qiagen | Cat# 28104 |
| C. DIFFICILE TOXIN/ANTITOXIN KIT | TechLab | Cat# T5000 |
| PowerTrack SYBR Green Master Mix | Thermo Fisher | Cat# A46109 |
| QIAseq FX DNA library kit | Qiagen | Cat# 180479 |
| NEB Monarch Genomic DNA Purification Kit | NEB | Cat# T3010S |
| Ligation Sequencing Kit | Oxford Nanopore | SQK-LSK109 |
| Native Barcoding Expansions 1-12 and 13-24 | Oxford Nanopore | EXP-NBD104 and EXP-NBD114 |
| NebNext Companion Module for Oxford Nanopore Technologies | NEB | Cat# E7180S |
| Illumina MiSeq Reagent kit v2 | Illumina | MS-102-2001 |
|
| ||
| Deposited data | ||
|
| ||
| Whole-genome sequencing assembly | This manuscript | BioProject: PRJNA885086 and PRJNA595724 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Hamster: CHO/dhFr- | ATCC | ATCC# CRL-9096 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX: 000664 |
| Mouse: inhouse Germ-free | University of Chicago | N/A |
| Mouse: MyD88~/~ (bred to C57BL/6J) | Adachi et al.60 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 | IDT | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pCE677 | Kaus et al.61 | N/A |
| Plasmid:pRPF144 | Fagan and Fairweather62 | Addgene# 106372 |
| Plasmid:pRPF144-WTcdtR | This manuscript | N/A |
| Plasmid:pRPF144-MutcdtR | This manuscript | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| SPAdes | Prjibelski et al.63 | N/A |
| GraphPad Prism v. 9 | GraphPad Software | N/A |
| Geneious Prime v.11 | Geneious by Dotmatics | N/A |
| PATRIC web resources | Wattam et al.23 | N/A |
| Unicycler v0.4.8 | Wick et al.64 | N/A |
| BLAST | Camacho et al.65 | N/A |
| mlst | Jolley and Maiden66 | N/A |
| FastANI (v 1.32) | Jain et al.67 | N/A |
| seqtk | https://github.com/lh3/seqtk 68 | N/A |
| snippy | Seemann. T, Snippy: rapid haploid variant calling, Githubhttps://github.com/tseemann/snippy 69 | N/A |
| gubbins | Croucher et al.70 | N/A |
| Anvi'o | Eren et al.28 | N/A |
| PHASTER | Arndt et al.71 | N/A |
| R | R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URLhttps://www.R-project.org/. | N/A |
| R package: tidyverse | Wickham et al.72 | N/A |
| R package: Biostrings | Pages et al.73 R package version 2.64.1,<https://bioconductor.org/packages/Biostrings>. |
N/A |
| R package: genoPlotR | Guy et al.74 | N/A |
Highlights.
Clinical ST1 C. difficile isolates with identical PaLoc can differ remarkably in virulence
Two avirulent C. difficile isolates remain avirulent in immune-deficient and germ-free mice
Avirulent C. difficile isolates contain a deletion in the regulatory gene for binary toxin
CdtR regulates Tcd and CDT toxins, thus broadly impacting the virulence of C. difficile
ACKNOWLEDGMENTS
We thank Dr. Craig D. Ellermeier for generously providing us the CRISPR editing system of C. difficile and Dr. Olga Soutourina for providing C. difficile R20291 and E. coli HB101(RP4) strains. We thank Melanie Spedale and the animal facilities of the University of Chicago for their help with mouse experiments. We thank Dr. Nhu Nguyen, Dr. Zhenrun Zhang, and the rest of the Pamer lab members for helpful discussions. This work was supported by National Institutes of Health R01 AI095706 and by the Duchossois Family Institute of the University of Chicago. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The graphical abstract and Figure S3B were created with BioRender.com.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112861.
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, et al. (2015). Burden of Clostridium difficile Infection in the United States. N. Engl. J. Med. 372, 825–834. 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, et al. (2020). Trends in U.S. Burden of Clostridioides difficile Infection and Outcomes. N. Engl. J. Med. 382, 1320–1330. 10.1056/NEJMoa1910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abt MC, McKenney PT, and Pamer EG (2016). Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 14, 609–620. 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupnik M, Wilcox MH, and Gerding DN (2009). Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536. 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 5.Cohen SH, Tang YJ, and Silva J (2000). Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 181, 659–663. 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 6.Kordus SL, Thomas AK, and Lacy DB (2022). Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat. Rev. Microbiol. 20, 285–298. 10.1038/s41579-021-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight DR, Elliott B, Chang BJ, Perkins TT, and Riley TV (2015). Diversity and Evolution in the Genome of Clostridium difficile. Clin. Microbiol. Rev. 28, 721–741. 10.1128/CMR.00127-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao K, Micic D, Natarajan M, Winters S, Kiel MJ, Walk ST, Santhosh K, Mogle JA, Galecki AT, LeBar W, et al. (2015). Clostridium difficile ribotype 027: relationship to age, detectability of toxins A or B in stool with rapid testing, severe infection, and mortality. Clin. Infect. Dis. 61, 233–241. 10.1093/cid/civ254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See I, Mu Y, Cohen J, Beldavs ZG, Winston LG, Dumyati G, Holzbauer S, Dunn J, Farley MM, Lyons C, et al. (2014). NAP1 Strain Type Predicts Outcomes From Clostridium difficile Infection. Clin. Infect. Dis. 58, 1394–1400. 10.1093/cid/ciu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giancola SE, Williams RJ, and Gentry CA (2018). Prevalence of the Clostridium difficile BI/NAP1/027 strain across the United States Veterans Health Administration. Clin. Microbiol. Infect. 24, 877–881. 10.1016/j.cmi.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 11. C. difficile Infections | A.R. & Patient Safety Portal. https://arpsp.cdc.gov/profile/eip/cdi. [Google Scholar]
- 12.Gerding DN, Johnson S, Rupnik M, and Aktories K (2014). Clostridium difficile binary toxin CDT. Gut Microb. 5, 15–27. 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gargis AS, Karlsson M, Paulick AL, Anderson KF, Adamczyk M, Vlachos N, Kent AG, McAllister G, McKay SL, Halpin AL, et al. (2022). Reference Susceptibility Testing and Genomic Surveillance of Clostridioides Difficile, United States, 2012–17 (Clinical Infectious Diseases), ciac817.. 10.1093/cid/ciac817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J,Frost E, and McDonald LC (2005). Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366, 1079–1084. 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 15.Cloud J, Noddin L, Pressman A, Hu M, and Kelly C (2009). Clostridium difficile strain NAP-1 is not associated with severe disease in a nonepidemic setting. Clin. Gastroenterol. Hepatol. 7, 868–873.e2. 10.1016/j.cgh.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, Almassalha LM, Ewing SA, Ring C, Galecki AT, et al. (2012). Clostridium difficile ribotype does not predict severe infection. Clin. Infect. Dis. 55, 1661–1668. 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer KA, Johnston JEW, Wenzler E, Goff DA, Cook CH, Balada-Llasat J-M, Pancholi P, and Mangino JE (2017). Impact of the NAP-1 strain on disease severity, mortality, and recurrence of healthcare-associated Clostridium difficile infection. Anaerobe 48, 1–6. 10.1016/j.anaerobe.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Revolinski SL, and Munoz-Price LS (2019). Clostridium difficile inImmunocompromised Hosts: A Review of Epidemiology, Risk Factors, Treatment, and Prevention. Clin. Infect. Dis. 68, 2144–2153. 10.1093/cid/ciy845. [DOI] [PubMed] [Google Scholar]
- 19.Seekatz AM, and Young VB (2014). Clostridium difficile and the microbiota. J. Clin. Invest. 124, 4182–4189. 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BB, Carter RA, Ling L, Leiner I, Taur Y, Kamboj M, Dubberke ER, Xavier J, and Pamer EG (2017). Pathogenicity Locus, Core Genome, and Accessory Gene Contributions to Clostridium difficile Virulence. mBio 8, e00885–17. 10.1128/mBio.00885-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyon SA, Hutton ML, Rood JI, Cheung JK, and Lyras D (2016). CdtR Regulates TcdA and TcdB Production in Clostridium difficile. PLoS Pathog. 12, e1005758. 10.1371/journal.ppat.1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miles-Jay A, Young VB, Pamer EG, Savidge TC, Kamboj M,Garey KW, and Snitkin ES (2021). A multisite genomic epidemiology study of Clostridioides difficile infections in the USA supports differential roles of healthcare versus community spread for two common strains. Microb. Genom. 7, 000590. 10.1099/mgen.0.000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, et al. (2017). Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 45, D535–D542. 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebeis SL, Bommarius B, Parkos CA, Sherman MA, and Kalman D(2007). TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J. Immunol. 179, 566–577. 10.4049/jimmunol.179.1.566. [DOI] [PubMed] [Google Scholar]
- 25.Jarchum I, Liu M, Shi C, Equinda M, and Pamer EG (2012). Criticalrole for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect. Immun. 80, 2989–2996. 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves AE, Koenigsknecht MJ, Bergin IL, and Young VB (2012). Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect. Immun. 80, 3786–3794. 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, et al. (2021). Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 6, 3–6. 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekulovic O, Meessen-Pinard M, and Fortier L-C (2011). Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 193, 2726–2734. 10.1128/JB.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekulovic O, Ospina Bedoya M, Fivian-Hughes AS, Fairweather NF, and Fortier L-C (2015). The Clostridium difficile cell wall protein CwpV confers phase-variable phage resistance. Mol. Microbiol. 98, 329–342. 10.1111/mmi.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepinski P, Douce GR, and Clokie MRJ (2016). Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob. Agents Chemother. 60, 968–981. 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan J, Ramachandran A, Thanki AM, Vukusic FBI, Barylski J, and Clokie MRJ (2018). Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 8, 5091. 10.1038/s41598-018-23418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fortier L-C (2018). Bacteriophages Contribute to Shaping Clostridioides (Clostridium) difficile Species. Front. Microbiol. 9, 2033. 10.3389/fmicb.2018.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heuler J, Fortier L-C, and Sun X (2021). Clostridioides difficile phage biology and application. FEMS Microbiol. Rev. 45, fuab012, fuab012. 10.1093/femsre/fuab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter GP, Lyras D, Allen DL, Mackin KE, Howarth PM, O’Connor JR, and Rood JI (2007). Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J. Bacteriol. 189, 7290–7301. 10.1128/JB.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govind R, and Dupuy B (2012). Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 8, e1002727. 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan KS, Wee BY, and Song KP (2001). Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J. Med. Microbiol. 50, 613–619. 10.1099/0022-1317-50-7-613. [DOI] [PubMed] [Google Scholar]
- 38.Govind R, Fitzwater L, and Nichols R (2015). Observations on the Role of TcdE Isoforms in Clostridium difficile Toxin Secretion. J. Bacteriol. 197, 2600–2609. 10.1128/JB.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupuy B, Raffestin S, Matamouros S, Mani N, Popoff MR, and Sonenshein AL (2006). Regulation of toxin and bacteriocin gene expression in Clostridium by interchangeable RNA polymerase sigma factors. Mol. Microbiol. 60, 1044–1057. 10.1111/j.1365-2958.2006.05159.x. [DOI] [PubMed] [Google Scholar]
- 40.Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, and Dupuy B (2002). Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J. Bacteriol. 184, 5971–5978. 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O’Connor L, Ip CLC,Golubchik T,Batty EM,Finney JM,et al.(2013).Diversesources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 369, 1195–1205. 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bilverstone TW, Minton NP, and Kuehne SA (2019). Phosphorylation and functionality of CdtR in Clostridium difficile. Anaerobe 58, 103–109. 10.1016/j.anaerobe.2019.102074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R, Feng Y, Wang X, Yang J, Zhang X, Lu X, and Zong Z (2017). Whole genome sequences of three Clade 3 Clostridium difficile strains carrying binary toxin genes in China. Sci. Rep. 7, 43555. 10.1038/srep43555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbut F, Decré D, Lalande V, Burghoffer B, Noussair L, Gigandon A, Espinasse F, Raskine L, Robert J, Mangeol A, et al. (2005). Clinical features of Clostridium difficile-associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J. Med. Microbiol. 54, 181–185. 10.1099/jmm.0.45804-0. [DOI] [PubMed] [Google Scholar]
- 45.Young MK, Leslie JL, Madden GR, Lyerly DM, Carman RJ, Lyerly MW, Stewart DB, Abhyankar MM, and Petri WA (2022). Binary Toxin Expression by Clostridioides difficile Is Associated With Worse Disease. Open Forum Infect. Dis. 9, ofac001. 10.1093/ofid/ofac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, et al. (2015). Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. mBio 6, e00551. 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D, et al. (2016). The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat. Microbiol. 1, 16108. 10.1038/nmicrobiol.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metcalf DS, and Weese JS (2011). Binary toxin locus analysis in Clostridium difficile. J. Med. Microbiol. 60, 1137–1145. 10.1099/jmm.0.028498-0. [DOI] [PubMed] [Google Scholar]
- 49.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, andMinton NP (2010). The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713. 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 50.Alonso CD, Pollock NR, Garey KW, Gonzales-Luna AJ, Williams DN, Daugherty K, Cuddemi C, Villafuerte-Gálvez J, White NC, Chen X, et al. (2022). Higher In Vivo Fecal Concentrations of Clostridioides difficile Toxins A and B in Patients With North American Pulsed-Field Gel Electrophoresis Type 1/Ribotype 027 Strain Infection. Clin. Infect. Dis. 75, 2019–2022. 10.1093/cid/ciac406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alonso CD, Kelly CP, Garey KW, Gonzales-Luna AJ, Williams D, Daugherty K, Cuddemi C, Villafuerte-Gálvez J, White NC, Chen X, et al. (2022). Ultrasensitive and Quantitative Toxin Measurement Correlates With Baseline Severity, Severe Outcomes, and Recurrence Among Hospitalized Patients With Clostridioides difficile Infection. Clin. Infect. Dis. 74, 2142–2149. 10.1093/cid/ciab826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guh AY, Hatfield KM, Winston LG, Martin B, Johnston H, Brousseau G, Farley MM, Wilson L, Perlmutter R, Phipps EC, et al. (2019). Toxin Enzyme Immunoassays Detect Clostridioides difficile Infection With Greater Severity and Higher Recurrence Rates. Clin. Infect. Dis. 69, 1667–1674. 10.1093/cid/ciz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gálvez JAV, Banz A, Chen X, Williams D, Xu H, Cuddemi CA, Cui AX, Perrotta M, Alhassan E, Riou B, et al. (2018). Stool interleukin-1b differentiates Clostridioides difficile infection from asymptomatic carriage and non-C.difficile infection diarrhea. Clin. Infect. Dis. ciac624. 10.1093/cid/ciac624. [DOI] [Google Scholar]
- 54.Govind R, Vediyappan G, Rolfe RD, Dupuy B, and Fralick JA (2009). Bacteriophage-Mediated Toxin Gene Regulation in Clostridium difficile. J. Virol. 83, 12037–12045. 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goh S, Chang BJ, and Riley TV (2005). Effect of phage infection ontoxin production by Clostridium difficile. J. Med. Microbiol. 54, 129–135. 10.1099/jmm.0.45821-0. [DOI] [PubMed] [Google Scholar]
- 56.Hargreaves KR, Kropinski AM, and Clokie MRJ (2014). What does the talking?: quorum sensing signalling genes discovered in a bacteriophage genome. PLoS One 9, e85131. 10.1371/journal.pone.0085131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nale JY, Chutia M, Carr P, Hickenbotham PT, and Clokie MRJ (2016). Get in Early’’; Biofilm and Wax Moth (Galleria mellonella) Models Reveal New Insights into the Therapeutic Potential of Clostridium difficile Bacteriophages. Front. Microbiol. 7, 1383. 10.3389/fmicb.2016.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson GA, and Bott KF (1968). Nutritional Factors Influencing the Development of Competence in the Bacillus subtilis Transformation System. J. Bacteriol. 95, 1439–1449. 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maikova A, Kreis V, Boutserin A, Severinov K, and Soutourina O (2019). Using an Endogenous CRISPR-Cas System for Genome Editing in the Human Pathogen Clostridium difficile. Appl. Environ. Microbiol. 85, e01416–19. 10.1128/AEM.01416-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, and Akira S (1998). Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9, 143–150. 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 61.Kaus GM, Snyder LF, Muh U, Flores MJ, Popham DL, and Ellermeier CD (2020). Lysozyme Resistance in Clostridioides difficile Is Dependent on Two Peptidoglycan Deacetylases. J. Bacteriol. 202, e004211–20. 10.1128/JB.00421-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fagan RP, and Fairweather NF (2011). Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286, 27483–27493. 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prjibelski A, Antipov D, Meleshko D, Lapidus A, and Korobeynikov A (2020). Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatics 70, e102. 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 64.Wick RR, Judd LM, Gorrie CL, and Holt KE (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jolley KA, and Maiden MCJ (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinf. 11, 595. 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, and Aluru S (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H (2023). lh3/seqtk. https://github.com/lh3/seqtk. [Google Scholar]
- 69.Seemann T (2023). Snippy. https://github.com/tseemann/snippy. [Google Scholar]
- 70.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, and Harris SR (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15. 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, and Wishart DS (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. (2019). Welcome to the Tidyverse. J. Open Source Softw. 4, 1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 73. Biostrings: Efficient Manipulation of Biological Strings Version 2.58.0 from Bioconductor. https://rdrr.io/bioc/Biostrings/. [Google Scholar]
- 74.Guy L, Kultima JR, and Andersson SGE (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335. 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJM, Jolley KA, et al. (2010). Multilocus Sequence Typing of Clostridium difficile. J. Clin. Microbiol. 48, 770–778. 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sorg JA, and Sonenshein AL (2008). Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512. 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, Ling L, Leiner I, and Pamer EG (2015). Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe 18, 27–37. 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keith JW, Dong Q, Sorbara MT, Becattini S, Sia JK, Gjonbalaj M, Seok R, Leiner IM, Littmann ER, and Pamer EG (2020). Impact of Antibiotic-Resistant Bacteria on Immune Activation and Clostridioides difficile Infection in the Mouse Intestine. Infect. Immun. 88, e00362–19. 10.1128/IAI.00362-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards AN, Krall EG, and McBride SM (2020). Strain-Dependent RstA Regulation of Clostridioides difficile Toxin Production and Sporulation. J. Bacteriol. 202, e00586–19. 10.1128/JB.00586-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angione SL, Sarma AA, Novikov A, Seward L, Fieber JH, Mermel LA, and Tripathi A (2014). A novel subtyping assay for detection of Clostridium difficile virulence genes. J. Mol. Diagn. 16, 244–252. 10.1016/j.jmoldx.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Babakhani F, Bouillaut L, Sears P, Sims C, Gomez A, and Sonenshein AL (2013). Fidaxomicin inhibits toxin production in Clostridium difficile. J. Antimicrob. Chemother. 68, 515–522. 10.1093/jac/dks450. [DOI] [PubMed] [Google Scholar]
- 82.Wroblewski D, Hannett GE, Bopp DJ, Dumyati GK, Halse TA, Dumas NB, and Musser KA (2009). Rapid molecular characterization of Clostridium difficile and assessment of populations of C. difficile in stool specimens. J. Clin. Microbiol. 47, 2142–2148. 10.1128/JCM.02498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Metcalf D, Sharif S, and Weese JS (2010). Evaluation of candidate reference genes in Clostridium difficile for gene expression normalization. Anaerobe 16, 439–443. 10.1016/j.anaerobe.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 84.McAllister KN, Bouillaut L, Kahn JN, Self WT, and Sorg JA (2017). Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci. Rep. 7, 14672. 10.1038/s41598-017-15236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seemann T (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 86.Seemann T (2023). mlst. https://github.com/tseemann/mlst. [Google Scholar]
- 87.Li W, and Godzik A (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 88.Fu L, Niu B, Zhu Z, Wu S, W, L., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequence data were uploaded to National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject: PRJNA885086 and PRJNA595724.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


