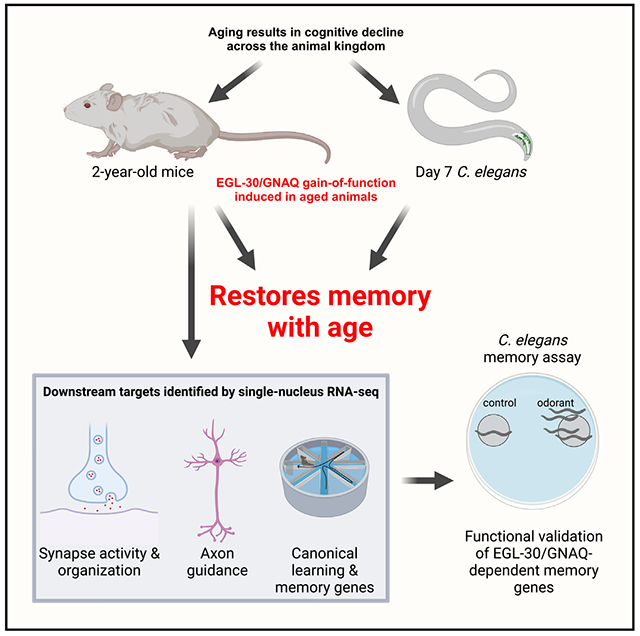
SUMMARY
Loss of cognitive function with age is devastating. EGL-30/GNAQ and Gαq signaling pathways are highly conserved between C. elegans and mammals, and murine Gnaq is enriched in hippocampal neurons and declines with age. We found that activation of EGL-30 in aged worms triples memory span, and GNAQ gain of function significantly improved memory in aged mice: GNAQ(gf) in hippocampal neurons of 24-month-old mice (equivalent to 70- to 80-year-old humans) rescued age-related impairments in well-being and memory. Single-nucleus RNA sequencing revealed increased expression of genes regulating synaptic function, axon guidance, and memory in GNAQ-treated mice, and worm orthologs of these genes were required for long-term memory extension in worms. These experiments demonstrate that C. elegans is a powerful model to identify mammalian regulators of memory, leading to the identification of a pathway that improves memory in extremely old mice. To our knowledge, this is the oldest age at which an intervention has improved age-related cognitive decline.
In brief
Stevenson et al. find that activation of GNAQ and Gαq signaling extends long-term memory in 2-year-old mice. GNAQ gain of function upregulates genes involved in plasticity, and worm orthologs were required for C. elegans memory. Invertebrates are a powerful model for identifying pathways that can improve memory in extremely old mammals.
Graphical Abstract
INTRODUCTION
One debilitating feature of aging is loss of cognitive abilities. Like humans, C. elegans and mice show learning and memory decline with age.1 The cAMP element binding protein (CREB) transcription factor, whose expression and function decline with age,2 activates the plasticity mechanisms necessary for long-term memory consolidation in vertebrates and invertebrates.3–6 Activation of CREB through rejuvenation methods,2,7,8 such as exercise9–11 and parabiosis,2 or CREB overexpression,12 improves synaptic plasticity and memory with age.
EGL-30/Gαq is a guanine nucleotide-binding protein G(q) subunit α (Gαq) that regulates pre-synaptic transmission and other neuronal functions in C. elegans.13–17 Its constitutive activation (EGL-30 [Q205L and V180M]) extends memory through activation of CREB.18 Like EGL-30(Q205L), the constitutively active form of the highly conserved mammalian GNAQ(Q209L) leads to reduced GTPase activity and continued activation of downstream Gαq signaling.19 Therefore, we wondered if Gnaq(gf). like egl-30(gf), could improve memory and slow cognitive decline in aged mammals and, if so, whether the downstream molecular mechanisms are similarly well conserved across species. We found that the analogous activating mutation in GNAQ rejuvenates memory in very old mice through well-conserved neuronal molecular mechanisms.
RESULTS
Activated EGL-30 extends memory in C. elegans until memory can no longer be measured
We previously found that a gain-of-function mutation (Q205L) in the C. elegans Gαq subunit EGL-30 increased memory in both young and aging (day 5) worms, through the activation of the CREB transcription factor.18 However, how late in life EGL-30(gf) induction could rescue memory was unknown. By day 4 of adulthood, wild-type worms have no ability to form long-term associative memories, but can still learn, perform chemotaxis, and move.4 We used heat shock induction to express EGL-30(gf) only in AWC sensory neurons18 on days 6, 7, and 8 (Figures 1A–1C, S1B, and S1C); in day 6 and day 7 worms, 24-h long-term associative memory (LTAM) is restored (worms are trained with odorant [butanone] and food, and subsequent chemotaxis to the odorant assesses this association; Figure S1A).3,4,18 By day 8, the worms have difficulty moving, preventing memory assessment (Figures 1D and 1E). That is, while wild-type worms experience the decline in LTAM well before short-term memory loss,4 induced AWC::egl-30(gf) worms retain memory function until they can no longer move well, at least tripling “memory span” (Figure 1F). In addition to sensing butanone, the AWC neuron senses benzaldehyde.20 Like LTAM, late-in-life expression of activated EGL-30 in the AWC neuron significantly improves chemotaxis toward benzaldehyde with age (Figure 1G) with no effect on maximum velocity decline (Figure 1H) or lifespan (Figure 1I). That is, EGL-30(gf) does not affect other aspects of worm health or behavior, but specifically rescues memory—previously the function that is most sensitive to aging—beyond the point when the animals have lost their ability to move.
Figure 1. Late-in-life induction of egl-30(gf) extends memory in old worms.
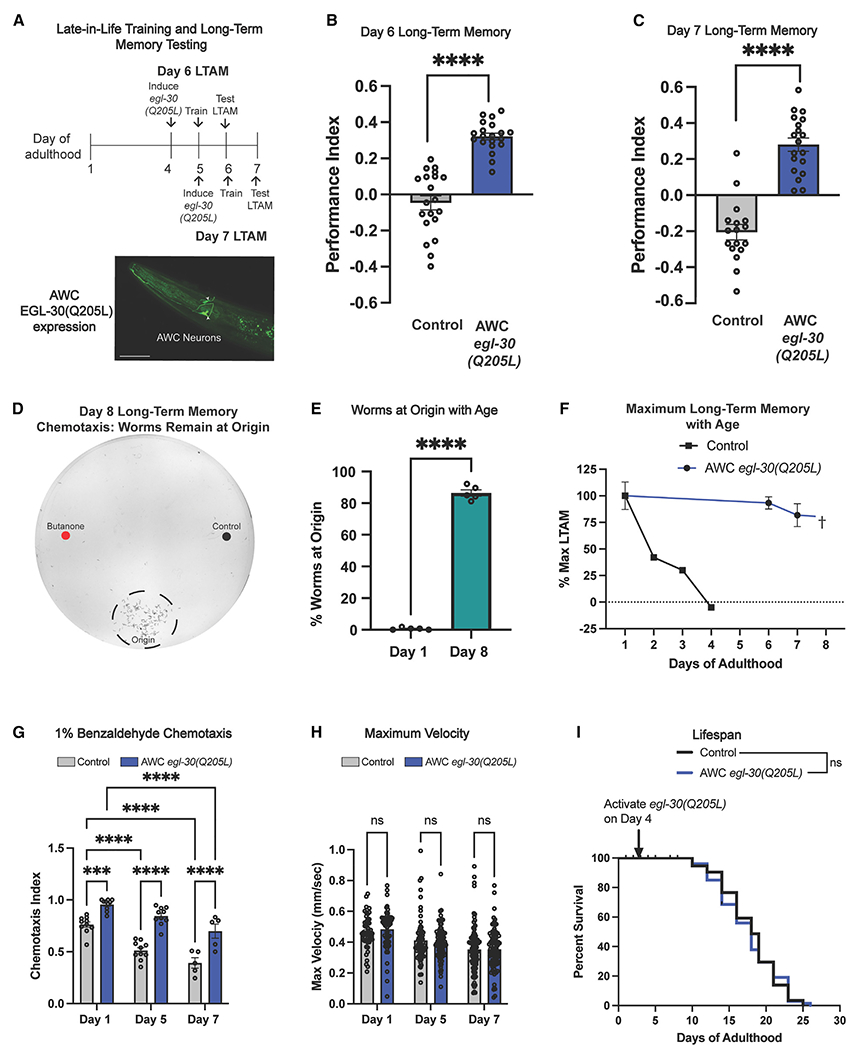
(A) Schematic of experiments. An FLP-inducible GFP-tagged egl-30(Q205L) expressed in AWC neurons was permanently activated after heat shock and long-term associative memory (LTAM) tested 24 h later. Scale bar, 50 μm.
(B and C) Twenty-four-hour LTAM was extended on (B) day 6 and (C) day 7. For memory assays, each dot represents an individual assay plate.
(D and E) Representative day 8 chemotaxis plate and quantification. By day 8, worms lose the ability to move to the odorant spots and instead remain on the origin.
(F) Late-in-life induction of egl-30(gf) in the AWC extends memory until worms can no longer move (day 7), well beyond wild-type loss of LTAM (day 4).
(G) AWC egl-30(Q205L) results in a higher preference for benzaldehyde (AWC-sensed odorant).
(H and I) AWC egl-30(Q205L) does not affect (H) maximum velocity with age or (I) lifespan. For maximum velocity, each dot represents an individual worm. Mean ± SEM. Two-way ANOVA (B, C, G, H) with Bonferroni correction or unpaired Student’s t test (E) were performed. Kaplan-Meier analysis with log-rank (Mantel-Cox) method was used to compare lifespans (I). ***p < 0.001; ****p < 0.0001; ns, not significant.
Gnaq is expressed in the mouse hippocampus and decreases with age
EGL-30 is highly conserved, sharing 82% amino acid identity with mammalian Gnaq/GNAQ (Figure 2A). Changes in G-protein-coupled receptor (GPCR) expression or activity contribute to the pathology of Alzheimer’s disease,21 and Gnaq was identified as one of 64 genes downregulated across several brain regions in aging humans.22 Given the role of the hippocampus in learning and memory, and evidence that Gnaq is enriched in the hippocampus and declines with age, we focused on this region. Gnaq is enriched in excitatory neurons in the dentate gyrus (DG), CA1, and CA3 hippocampal subregions of a publicly available single-cell RNA-sequencing dataset of the adult mouse hippocampus23 (Figure 2B). We measured levels of Gnaq in young (3-month) and old (24-month) mouse hippocampi (Figures 2C and 2D); Gnaq levels decrease significantly with age in all hippocampal subfields, including the DG, CA1, and CA3 (Figures 2D and 2E).
Figure 2. GNAQ declines with age, and the constitutively active mutation is required for extended memory.
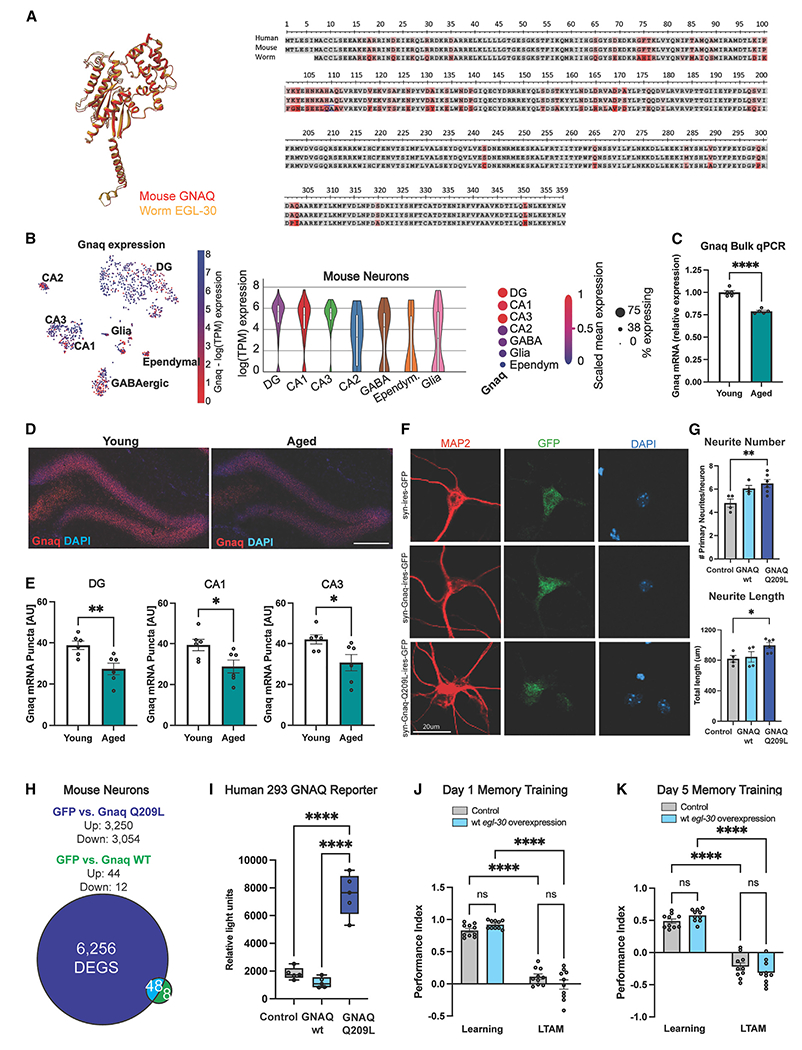
(A) EGL-30 and mammalian GNAQ share 82% amino acid identity.
(B) Gnaq is enriched in CA1, CA3, and dentate gyrus (DG) in snRNA sequencing data of the mouse brain.23
(C) Gnaq mRNA expression decreases in the hippocampi of 24-month (aged) vs. 3-month (young) mice.
(D) Representative images of Gnaq RNAscope in the dentate gyrus. Scale bar, 150 μm.
(E) Hippocampal Gnaq declines with age comparing 3-month (young) and 24-month-old (aged) mice.
(F) Neurite morphology was assessed using MAP2 immunostaining (red). Infected neurons were labeled with eGFP. Nuclei were labeled with DAPI. Scale bar, 20 μm.
(G) GNAQ(Q209L) treatment in primary mouse neurons increased neurite number and length.
(H) GNAQ(Q209L) transfection of primary neurons upregulates more genes than does wild-type (WT) GNAQ.
(I) In a GNAQ reporter human cell line, only GNAQ(Q209L) increases luminescence.
(J and K) egl-30 overexpression does not improve memory in young (J) day 1 or (K) day 5 aged worms. Learning, 0 h post-training; LTAM, 24 h post-training. Each dot represents an individual mouse, an individual well (in vitro experiments), or an individual chemotaxis assay plate (worm memory). Mean ± SEM. One-way ANOVA (G, I) and two-way ANOVA (J, K) with Bonferroni correction or unpaired Student’s t test (C, D) was performed. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant.
Overexpressing Gnaq/egl-30 does not impact neurons or increase memory
Before embarking on tests of Gnaq in aged mice in vivo, we determined whether the gain-of-function mutation was necessary for neuronal effects or if simply overexpressing the wild-type form of Gnaq/egl-30 was sufficient. The analogous constitutively activating mutation to EGL-30(Q205L) in mammalian Gnaq/GNAQ is Q209L.19 We infected mature primary mouse neurons with lentiviral constructs encoding mouse wild-type Gnaq, activated Gnaq(Q209L), or GFP control under the control of the neuron-specific Synapsin 1 promoter. While overexpression of wild-type GNAQ in primary mouse neurons did not elicit morphological changes, Gnaq(Q209L) increased neuronal complexity, resulting in longer primary neurites (Figures 2F and 2G). These results demonstrate that this hyperactivating mutation—rather than simple overexpression—induces plasticity in a mammalian system, making it of interest to test in vivo. Concordantly, overexpression of wild-type mouse GNAQ in primary mouse neurons induces fewer changes in differential gene expression compared with activated GNAQ(Q209L) (Figures 2H and S2A). Interestingly, GNAQ(Q209L) expression also increased immediate-early gene (IEG) expression (Figure S2B). In cells expressing a luminescent reporter for GNAQ pathway activity, wild-type human GNAQ does not activate the pathway to the same degree as expression of the gain-of-function form of human GNAQ(Q209L) (Figure 2I).
Like the mammalian GNAQ, overexpression of wild-type EGL-30 in C. elegans causes no increase in 24-h LTAM on day 1 or 5 of adulthood (Figures 2J and 2K), unlike expression of the gain-of-function mutation EGL-30(Q205L) (Figures 1B and 1C). Therefore, the gain-of-function mutation in EGL-30 or GNAQ is necessary to observe improvements in memory in C. elegans and to induce expression changes in the pathway in both mouse and human cells.
Expression of activated GNAQ increases IEG expression in old mice
To test EGL-30/GNAQ(gf)’s memory function in aged worms in mice, we injected viral vectors expressing either GFP control or GNAQ(Q209L) under the neuron-specific Synapsin 1 promoter into the hippocampi of 24-month-old mice (Figures 3A and 3B). The mice recovered for 5 weeks and then were tested in behavioral assays, and, at ~26 months of age, tissue was collected for molecular analyses (Figure 3C).
Figure 3. Gnaq(Q209L) increases immediate early gene expression and improves memory in aged mice (A and B).
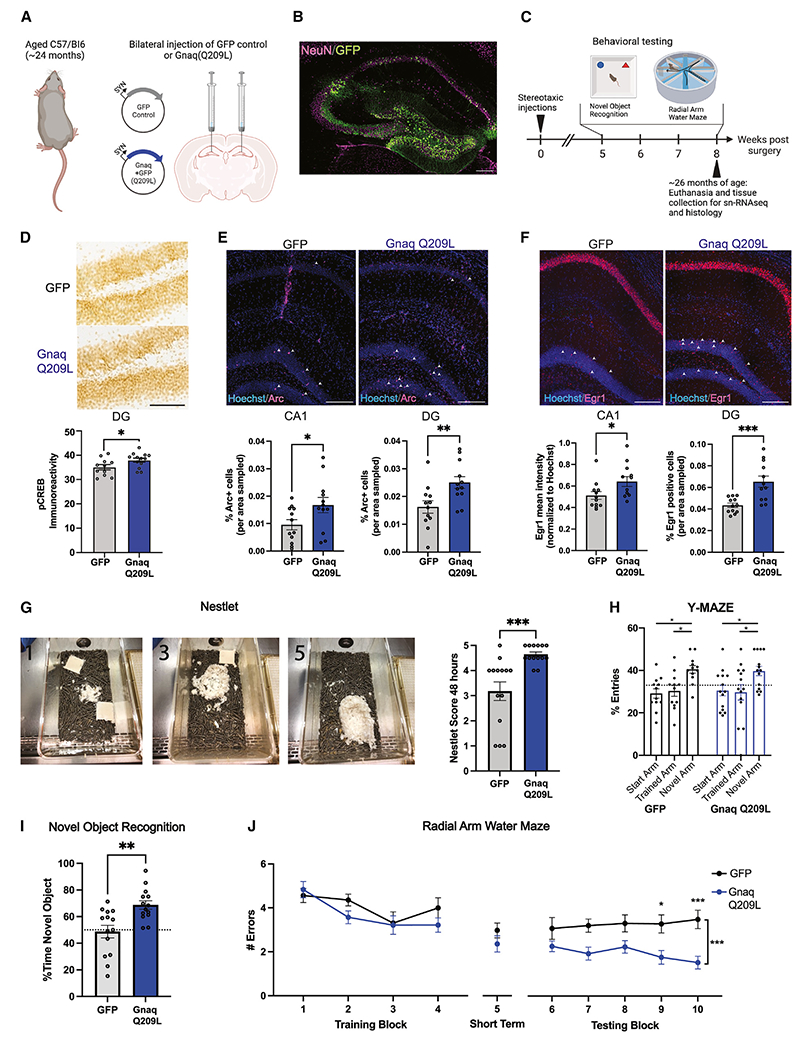
(A) Twenty-four-month-old mice were injected with a viral vector expressing either GFP or GNAQ(Q209L) in adult hippocampal neurons. (B) Representative image of viral expression (GFP) in hippocampus. Scale bar, 200 μm.
(C) After recovery, aged mice were tested in novel object recognition and radial arm water maze tasks and then euthanized at ~26 months of age for histology and single-nucleus RNA sequencing.
(D) Expression of phosphorylated Creb (pCreb) protein in the dentate gyrus (DG) of the hippocampi of aged mice following neuronal Gnaq(Q209L) was increased relative to GFP control expression. Representative image; scale bar, 100 μm.
(E and F) Expression of (E) Arc and (F) Egr1 was increased in the CA1 and DG regions in Gnaq(Q209L) mice relative to GFP control. Representative images; scale bars, 200 μm.
(G) Representative nests, where 1 shows a poor, 3 an average, and 5 a perfect nest. GNAQ(Q209L) mice built significantly better nests than GFP controls, indicating better well-being.
(H) GNAQ(Q209L) mice showed no differences from GFP controls in the Y-maze test (short-term memory).
(I) GNAQ(Q209L) mice had improved 24-h long-term memory on the novel object recognition task.
(J) GNAQ(Q209L) mice had improved long-term memory (blocks 6–10) compared with GFP controls. Each dot represents one mouse. Mean ± SEM. Unpaired Student’s t test (D–G, I) or ANOVAs followed by post hoc corrections (H, J) were used. *p < 0.05, **p < 0.01, ***p < 0.001.
Because egl-30(gf) activates CREB and gene transcription in C. elegans,18 we first tested whether there were differences in the levels of pCREB and IEG expression in aged GNAQ(Q209L)-treated mice. Arc and Egr1 are IEGs downstream of CREB that regulate DNA methylation, neuronal activity, synaptic plasticity, and memory formation.24–26 All were elevated in GNAQ-treated hippocampi (Figures 3D–3F), suggesting that GNAQ(Q209L) treatment may activate the CREB pathway.
Expression of activated GNAQ increases long-term memory in old mice
We next tested the aged mice in behavioral assays. Improved nest building is a metric of better well-being27 and declines with age28; GNAQ(Q209L)-treated mice displayed nestlet behavior reminiscent of younger animals (Figure 3G). While there were no differences in short-term working memory, as measured through a Y-maze assay (Figure 3H), GNAQ(Q209L)-treated mice showed significant improvements in metrics of long-term memory. Specifically, GNAQ(Q209L)-treated mice spent more time with the novel object when tested 24 h later, indicating better long-term memory (Figure 3I). Perhaps most strikingly, while they showed no differences from controls in the training blocks (learning) or during the short-term memory testing (block 5) of the radial arm water maze (RAWM), GNAQ(Q209L)-treated mice had significantly better performance in long-term memory (the 6–10 testing block) than did GFP control mice when tested the following day (Figure 3J); that is, like egl-30(gf) worms, aged mice with increased GNAQ activity showed improvements in long-term memory. Thus, Gnaq(Q205L) expression in old mice has a significant rejuvenating effect, rescuing long-term memory function and increasing quality of life of aged animals.
GNAQ(Q209L)-treated mice have significant changes in transcription
To determine the mechanisms that GNAQ(Q209L)-treated mice use to improve their long-term memory, we performed single-nucleus RNA sequencing (snSeq) of hippocampal cells of control and GNAQ(Q209L)-treated mice. All major hippocampal cell types were represented in our snSeq data (Figure 4A); activated GNAQ(Q209L) treatment changes gene expression (Table S1) but does not affect cell type (Figures 4B and S3A). The CA1 pyramidal and DG granule neurons–the cells targeted by our intervention–were the major sites of differential gene expression (Figure 4A; examples in Figure 4C). In addition, there were many upregulated genes in the oligodendrocyte cluster, indicating at least some cell non-autonomous effects of GNAQ(Q209L) treatment (Table S1).
Figure 4. Gnaq treatment upregulates synaptic plasticity genes and increases synapse number and neurite complexity.
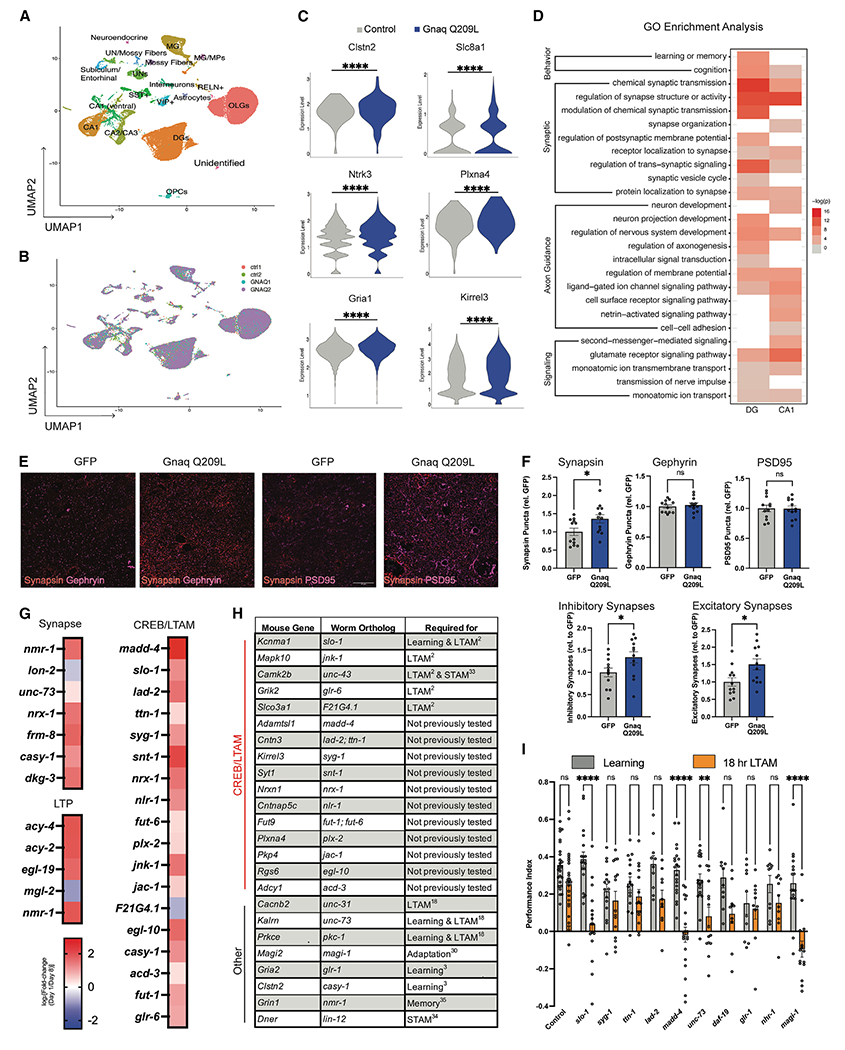
(A) Hippocampal cell type clusters following single-nucleus RNA sequencing of hippocampi after GNAQ(Q209L) treatment.
(B) No new cell types were induced by GNAQ(Q209L) treatment.
(C and D) Differential expression analysis of significantly upregulated genes in CA1 and DG. (C) All violin plots shown have met the minimum cutoff of logFC > 0.1 and padj < 0.05 (Wilcox test). (D) Gene ontology analysis revealed enrichment in CA1 and DG for genes involved in learning and memory, synaptic plasticity, axon guidance, and signaling.
(E) Representative images of pre-synaptic (Synapsin) and post-synaptic (PSD95, Gephyrin) labeling in CA1 to quantify synapses in GNAQ(Q209L) mice. Scale bar, 20 μm.
(F) GNAQ(Q209L) increases inhibitory and excitatory synapses (co-localization of Synapsin with post-synaptic markers), with Synapsin expression also elevated in CA1.
(G) Orthologs of CREB/LTAM genes, “regulation of synapse structure/activity,” and long-term potentiation (LTP) genes generally decline with age.
(H) Many homologs of Gnaq-induced genes following single-nucleus RNA sequencing identified here overlapped with our CREB-dependent long-term associative memory list and are required for worm memory.
(I) Reduction of worm orthologs (madd-4, slo-1, unc-73, magi-1) in the egl-30 background impairs 18-h long-term associative memory extension. Each dot represents an individual mouse (synapse experiments) or one plate (worm experiments). Mean ± SEM. Unpaired Student’s t test (F) and a two-way ANOVA with Bonferroni correction (I) was performed. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant.
Synapse formation is induced by GNAQ activation
Because our snSeq analysis indicated an enrichment of synaptic-related changes (Figures 4A–4D), we tested whether GNAQ activation affects synapse number in vivo in aged mice. We labeled hippocampal sections from GFP control and GNAQ-treated mice with synapse markers (Synapsin, Gephyrin, and PSD95) and found that activated GNAQ(Q209L)-treated mice have significantly more inhibitory and excitatory synapses in CA1 than controls (Figures 4E and 4F). The increase in synapse number appeared to be driven by an increase in pre-synaptic densities (Synapsin) (Figure 4F). Together, our data suggest that activated GNAQ(Q209L) restores synaptic density, even at an advanced age.
GNAQ activation upregulates synaptic plasticity, axon guidance, and learning and memory genes
Genes that function in synapse organization and structure were upregulated in hippocampal DG and CA1 excitatory neurons (Figure 4D), including genes necessary for learning and memory in mammals and C. elegans, such as Clstn2/casy-1, Kalrn/unc-73, and Magi2/magi-1.3,4,18,29–31 These genes decline in aging C. elegans neurons (Figure 4G) (Y.W. and C.T.M., unpublished data). Other genes known to be required for hippocampal neuron function are also upregulated in the brains of activated GNAQ(Q209L)-treated mice, including neurotransmitter/glutamate receptor activity (Gria1/glr-1, Grik2, Grm1, Grm7/mgl-2), potassium channels (Kcnh7/unc-103, Kcnd2/shl-1), calcium channels (Cacan1c/egl-19, Cacnb2/ccb-1), synapse development and function (synaptotagmin Syt1/snt-1, neuregulin Ngr3, neurexin Nrxn1/nrx-1, Kalrn/unc-73, Stxbp5/tom-1, synaptoporin Synpr/sph-1, Gephyrin Gphn/moc-1, and transmembrane proteins such as SynDIG1), cytoskeletal structure and function (Mid1/madd-2, Carmil1/crml-1, Mast4/kin-4, Arhgef4), GABA B receptor (Gabrb1, 2/gab-1), GPCR activity (Adgrb2), CREB activity regulators (Camk2b/unc-43, Mapk10/jnk-1), solute carrier transporters (Slc2a13/hmit-1.3, Slco3a1/F21G4.1, Slc4a4/abts-1), and genes required for learning and memory (Tafa2) (Figure 4H). Surprisingly, early development function genes, including axon guidance genes (Dcc/unc-40, Unc5c/unc-5, Slit1, Robo1,2, Dscam, Epha3,6, Sema5a, 6d), are also upregulated by activated GNAQ.
While many of the GNAQ(Q209L)-upregulated genes are known to be important for neuronal function in mammals, we previously found that many of their C. elegans homologs are required for LTAM (e.g., Slco3a1/F21G4.1, Kcnma1/slo-1, Mapk10/jnk-1, Camk2b/unc-43, Kalrn/unc-73, Cacnb2/unc-31, and Prkce/pkc-1)3,18 (Figure 4H). Furthermore, Cacnb2/unc-31, which is involved in neuropeptide release, is also required for LTAM in the egl-30 background.18 Other genes from our list also overlap with CREB-dependent LTAM genes, including Adamtsl1/madd-4, Cntn3/lad-2 and ttn-1, Kirrel3/syg-1, Syt1/snt-1, Nrxn1/nrx-1, Cntnap5c/nlr-1, Fut9/fut-6, Plxna4/plx-2, Pkp4/jac-1, Rgs6/elg-10, Adcy1/acd-3, Fut9/fut-1, and Grik2/glr-6 (Figure 4H)3. Many of these genes also decline with age in worms by day 8 (Figure 4G), an age when worms no longer have long-term memory (Figure 1F). Others are homologs of genes that are already known to function in C. elegans learning or short-term associative memory (e.g., Magi2/magi-1, Gria2/glr-1, Grin1 glutamate receptor/nmr-1, Dner Notch/lin-12, Camk2b/unc-43, and Clstn2/casy-1; Figure 4H).4,32–34
Upregulated GNAQ(Q209L) gene orthologs are required for long-term memory
To test whether the gene expression changes we observed in mice are required for memory, we returned to C. elegans, as cognitively testing multiple gene candidates in aged mice would be prohibitively slow. Almost every gene we found to be changed in activated GNAQ(Q209L)-treated hippocampus cells has a well-conserved worm ortholog (Figure 4H; Table S1). In fact, a significant subset of the orthologs was already identified as downstream CREB targets in LTAM,3 and of those, we previously found several to be required for memory in wild-type worms3 (Figure 4H).
To test whether activated GNAQ(Q209L)-induced genes (Figures 4G and 4H) are required for GNAQ/EGL-30 pathway-regulated memory, we used RNAi to knock down specific genes in egl-30(gf) adults, trained the animals for food-butanone association on day 2, and then tested long-term memory 18 h later (Figure 4I). We focused on genes involved in synapse activity and organization, that decline with age in worms, that overlapped with our previously generated CREB/LTAM, or are known to be involved in other forms of memory in C. elegans (e.g., Kcmna/slo-1, Kirrel3/syg-1, Cntn2/ttn-1/lad-2, Adamtsl1/madd-4, Kalrn/unc-73, Gria2/glr-1, Magi2/magi-1). Several of these genes are required for extended egl-30(gf) memory, including slo-1/Kcnma1, the α subunit of a calcium-activated BK channel; madd-4/Adamtsl1, which is involved in axon guidance and synapse organization; unc-73/Kalrn, a known downstream target of Gαq signaling,18 and magi-1/Magi2, a membrane-associated guanylate kinase homolog (MAGUK) family member involved in habituation and long-term memory.3,29 Thus, we have used C. elegans to identify a pathway that rejuvenates memory in aged mice, and the orthologs of gene expression targets altered in the hippocampal cells of these activated GNAQ(Q209L)-treated mice are also required for long-term memory function in the orthologous pathway in C. elegans.
DISCUSSION
Here, we found that a constitutively active form of the murine Gαq protein GNAQ, when expressed in hippocampal neurons of 2-year-old mice, significantly enhances long-term memory. These results highlight how well conserved the molecular and genetic pathways required for memory are from worms to mammals and suggest that C. elegans is a powerful model for the study of memory maintenance with age.
The ability to add new synaptic connections is limited in adulthood and only worsens with age; however, activated GNAQ(Q209L) treatment rescues this ability. Our analyses suggested that GNAQ(gf) treatment rejuvenates memory function of aged mice through the coordinated expression of genes that increase synaptic structure and function, axon guidance, and neuronal activity. We also found that GNAQ(Q209L) increases the expression of phosphorylated CREB and IEGs, like rejuvenation via heterochronic parabiosis, young blood plasma,2 or aerobic exercise11 in aged mice. This suggests these rejuvenation methods may converge on similar pathways, all mediated in part through CREB signaling in aged neurons. CREB activation is required for the extended memory effects of C. elegans EGL-30(gf). Gnaq(gf) intervention targeted adult neurons in the aged hippocampus and improved function without requiring any obvious markers of stem cell activation, according to our snSeq data. Neurogenesis declines dramatically with age; the neurogenesis capability of 9-month-old mice is less than 5% that of 3-month-old mice.35 Thus, it is unlikely that neurogenesis drives the memory improvement of 2-year-old GNAQ(Q209L)-treated mice. Instead, our results suggest that the memory improvements are the result of changes to existing neurons. Furthermore, we find no activation of microglia or neuroinflammatory responses in GNAQ(Q209L)-treated mice, suggesting that the treatment is not toxic. Excitingly, the degree of memory improvement is on the scale of that seen in aged mice with parabiosis and exercise interventions,2,11 which may function in parallel to the GNAQ pathway.
GNAQ(Q209L) treatment upregulates genes that play central roles in consolidation of memory, including synapse organization and activity and long-term potentiation. For example, genes that are directly involved in synaptogenesis and spinogenesis, such as Nrxn1/nrx-1, a critical regulator of synaptic function, synaptogenesis, and memory in mice36–38 and C. elegans,39 were upregulated in activated GNAQ(Q209L)-treated mice. CAMKIIB/UNC-43 interacts directly with the cytoskeleton and regulates spinogenesis and maintenance in adulthood via interactions with F-actin in rodents40 and is important for memory in C. elegans.3,32 CAMKIIB’s structural role at the synapse is important for Schaffer collateral-CA1 synaptic plasticity and hippocampal learning.41 We also found that genes that regulate synapse activity, many of which have direct involvement in long-term potentiation, were upregulated; for example, Grin1/nmr-1 encodes GluN1, an essential component of the NMDA receptor, and alternative splicing of Grin1 exon 5 controls the magnitude of long-term potentiation in CA1 and thus the ability to learn and remember.42 Gria1 encodes for the GluA1 AMPA receptor subunit, and reduction of GluA1 in CA2/CA3 impairs short-term memory.43
Axon guidance genes, including Semaphorins and Netrin, Ephrin, and Slit receptors, were also upregulated in activated GNAQ-treated mice. Although axon guidance has been studied in the context of development, they can guide synaptic plasticity in adulthood. Netrin-1 infusion can recover long-term potentiation and memory impairments that result from amyloid-β treatment in mice,44 and ephrin receptors are involved in long-term potentiation, promote neurotransmitter release, and regulate spine morphology and synapse formation.45,46 However, the roles of these proteins in aged animals have not been previously examined.
Although neurons are the primary site of effects, we also observed an enrichment in genes involved in signal transduction and synaptic transmission in oligodendrocytes. Myelination is inhibited in aged mice, and loss of myelin impairs spatial memory in young mice.47 Activation of existing oligodendrocytes can also enhance myelin thickness and conduction speed, leading to better hippocampal memory.48
Testing in aged animals allows the discovery of beneficial mechanisms and acute treatments that might be deleterious with chronic administration or in younger animals. The pathways that C. elegans neurons use to function are highly conserved with molecular pathways in mouse neurons, which allowed us to use worms for rapid discovery and testing of possible mechanisms that can rejuvenate aged mammalian neurons. Distinct from the negative effects induced by chronic Gq activation of ventral hippocampal networks early in life,49 late-life, temporally controlled acute activation of Gαq/GNAQ may offer a mechanism to slow or even reverse memory decline in aged individuals.
To our knowledge, GNAQ(Q209L) treatment in 2-year-old mice is the latest age at which a rejuvenation method has improved memory. Mice between 18 and 24 months are considered old, but cognition declines still more in older animals. For example, 22-month-old mice were more impaired on the Morris water maze than 18-month-old mice.50 Here, we treated mice with activated GNAQ when they were 24 months of age, and they were tested for learning and memory ability when they were 25–26 months of age. This is a remarkably old age to employ rejuvenation methods and test cognition in mice, as most previous experiments begin interventions and cognitive testing between 18 and 20 months of age, including treatments that continue during memory training (although it is possible that other interventions could also improve memory in 2-year-old mice). For example, exercise9 and treatment with parabiosis or plasma factors from young or exercised mice2,11 improve memory when interventions begin around 18 months. The effects of young cerebral spinal fluid on aged mouse memory were tested around 20 months of age.51 Similarly, transplantation of young microbiota improved long-term memory in 19- to 20-month-old mice.52 One month of environmental enrichment improves memory in mice as old as 21 months of age.53 Treatment with ISRIB, a drug-like small molecule that inhibits the integrated stress response, improves memory in 19-month-old mice,54 but later ages were not tested. Therefore, the fact that activated GNAQ(Q209L) treatment significantly improves cognition in mice more than 2 years of age is unprecedented, as far as we know.
Activated GNAQ upregulates synaptic plasticity mechanisms that improve memory. Moreover, GNAQ(Q209L)-treated mice display signs of improved well-being and increased synapse formation. GNAQ(Q209L) intervention late in life can rejuvenate post-mitotic neurons in the aged hippocampus, producing an environment that supports plasticity and in turn memory consolidation, with no evidence of neuroinflammation or microglia activation. The fact that memory in such aged mice—the equivalent of 70- to 80-year-old humans— could be rescued by activated GNAQ treatment suggests that aged neurons may have more potential to remodel than has been previously recognized.
Possible limitations
Because our primary goal was to treat cognitive aging, our analysis was limited to testing Gαq/GNAQ activation in aged mice; it does not rule out the possibility that Gαq/GNAQ could improve memory in young mice, as we previously found in young, day 1 adult worms.18 Furthermore, this effect seems to be specific to Gαq, as we previously found that Gαs gain of function impairs short-term memory in C. elegans.32 Although we did not test Gαi signaling, this pathway is inhibitory, and is unlikely to produce a similar effect on memory. However, it is possible that activation of GNA11, which is highly homologous to EGL-30 and has 90% amino acid identity with GNAQ, could improve memory.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Dr. Coleen Murphy (ctmurphy@princeton.edu).
Materials availability
Plasmids or strains generated for this study are available upon request.
Data and code availability
With the exception of single-nucleus RNA-sequencing and mouse neuron sequencing data, all other data are available in the main text or the supplementary materials. Sequencing data are available at BioProject #PRJNA972702.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
C. elegans maintenance
Strains were maintained at 20°C for the duration of experiments. Maintenance plates were made with high growth medium (HGM: 3 g/L NaCl, 20 g/L Bacto-peptone, 30 g/L Bacto-agar in distilled water, with 4 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1M CaCl2, 1 mL/L 1M MgSO4, and 25 mL/L 1M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving). All assays were performed on plates made with standard nematode growth medium (NGM: 3 g/L NaCl, 2.5 g/L Bacto-peptone, 17 g/L Bacto-agar in distilled water, with 1 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1M CaCl2, 1 mL/L 1M MgSO4, and 25 mL/L 1M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving57. All experiments that did not involve RNAi treatment were seeded with OP50 E. Coli (BactoBeads) for ad libitium feeding. Hypochlorite synchronization was used to develop-mentally synchronize experimental worms, where gravid hermaphrodites were exposed to an alkaline-bleach solution (e.g. 6 mL sodium hypochlorite, 2.5 mL KOH, 41.5 mL distilled water) to collect eggs, followed by repeated washes with M9 buffer (6 g/L Na2HPO4, 3 g/L KH2PO4, 5 g/L NaCl and 1 mL/L 1M MgSO4 in distilled water).57
C. elegans strains
C. elegans strain N2 var. Bristol: wild-type/CGC/N2
C. elegans strain NM1380: egl-30(js126)/CGC/NM1380
C. elegans strain CQ429: pHSP16-48::FLPase;Podr-1::FRT::egl-30(Q205L); Pmyo-2::mCherry/Arey et al., 2018/CQ429
C. elegans strain: CQ701: wtEx74 Topo-Prab-3::GFP-egl-30(WT)::rab3 UTR; Pmyo-2::mCherry/this paper/CQ701
C. elegans strain CQ601: egl-30(Q205L);vls69[pCFJ90(Pmyo-2::mCherry+Punc-119::sid-1)]/this paper/CQ601
Construction of C. elegans transgenic lines
Extrachromosomal transgenic arrays were generated for experiments testing the effects of wild-type EGL-30 overexpression. Line CQ701: wtEx74 Prab-3:GFP-egl-30(WT)::rab-3 UTR; Pmyo-2::mCherry was generated by injecting wild-type worms with 25 ng/μl of Prab-3::GFP-egl-30(WT)::rab-3 UTR and 1 ng/μl Pmyo-2::mcherry. Prab-3::GFP-egl-30(WT)::rab-3 UTR was made by Gibson cloning of 1215bp of the rab-3 promoter upstream of gfp N-terminally fused to the egl-30 cDNA, followed by the rab-3 3’UTR into a Topo backbone.
Animal models
The C57BL/6 mouse line was used for all experiments (The Jackson Laboratory and National Institutes of Aging). All other studies were performed with male mice. The numbers of mice used to result in statistically significant differences was calculated using standard power calculations with α = 0.05 and a power of 0.8. We used an online tool (http://www.stat.uiowa.edu/~rlenth/Power/index.html) to calculate power and sample size based on experience with the respective tests, variability of the assays, and inter-individual differences within groups. Animals utilized for each individual experiment were from the same vendor and aged together. All animals from Jackson Laboratories were acquired at two months of age and aged in-house. Animals were moved to a new location for behavioral assessment at the Villeda Lab Behavioral Suite. Mice were housed under specific pathogen-free conditions under a 12-hour light-dark cycle, and all animal handling and use was in accordance with institutional guidelines approved by the University of California San Francisco IACUC.
METHOD DETAILS
C. elegans experiments
FUdR treatment
For aging experiments, worms were transferred at the L4 larval stage to HGM plates supplemented with 500 μl/L 0.1M FUdR (5-Fluoro-2’-deoxyuridine; Sigma Aldrich, Cat. # F0503) for a final concentration of 0.05M FUdR at the L4 larval stage. Worms were moved off FUdR plates, back to HGM plates, 24 hours prior to behavioral assays.
RNAi treatment
For adult only-RNAi experiments, worms were transferred at the L4 larval stage to HGM plates supplemented with 1 mL/L 1M IPTG (isopropyl β-d-1-thiogalacto-pyrano-side) and 1 mL/L 100 mg/mL carbenicillin. Plates were seeded with HT115 E. coli for ad libitium feeding and induced with 200 μl of 0.1 M IPTG. RNAi treatment was performed using standard RNAi feeding methods. Worms were maintained on HGM RNAi plates until Day 2 of adulthood. All bacterial clones, including the control vector (pL4440), were sequenced prior to use.
Heat-shock inducible EGL-30 strain
For experiments using CQ429: pHSP16-48::FLPase;Podr-1::FRT::egl-30(Q205L); Pmyo-2::mCherry18, transgenic and non-transgenic siblings were heat-shocked at 34°C for 1 hour to permanently activate the FLP-inducible GFP-tagged gain-of-function EGL-30 in the AWC of transgenic worms (Figure S1). The heat-shock always occurred 24 hours prior to behavioral assays, to allow worms to recover. The heat shocked, non-transgenic siblings served as the controls for all experiments. To verify GFP-tagged gain-of-function EGL-30 was expressed in the AWC, a subset of worms were imaged using a Nikon AXR confocal microscope with a 60x oil objective (Figure 1A).
Recombinant DNA
Plasmid: pL4440 RNAi control/Addgene/Fire Lab
Plasmid: pL4440-slo-1 RNAi/Ahringer
Plasmid: pL4440-syg-1 RNAi/Ahringer
Plasmid: pL4440-ttn-1 RNAi/Ahringer
Plasmid: pL4440-lad-2 RNAi/Ahringer
Plasmid: pL4440-madd-4 RNAi/Ahringer
Plasmid: pL4440-unc-73 RNAi/Ahringer
Plasmid: pL4440-daf-19 RNAi/Ahringer
Plamid: pL4440-nhr-1 RNAi/Ahringer
Plasmid: pL4440-glr-1 RNAi/Ahringer
Plasmid: pL4440-magi-1 RNAi/Ahringer
Positive olfactory associative memory assay
Worms completed short-term associative memory assays and were trained and tested as described previously.3,4,18 For the short-term memory training, when worms were at the appropriate age of adulthood, they were washed from HGM or HGM+RNAi plates with M9 buffer, allowed to settle by gravity and washed a total of 3x. Next worms completed a 1-hour starvation in M9 buffer followed by one, 1 hour CS-US pairing, where worms were transferred to NGM plates seeded with OP50 E. Coli with 18 μl of 10% 2-butanone (Acros Organics; Cat. # 332828-25ML) in ethanol streaked on the lid at 20°C. After conditioning, trained worms were tested for chemotaxis to 10% butanone vs. ethanol control either immediately after training (0 hr/learning) or held on seeded NGM plates at 20°C with fresh OP50 E. Coli for 1-24 hours. Short-term memory (1 hour) and long-term memory (18 or 24 hours) were tested by chemotaxis to 10% butanone vs. ethanol control. This is compared with the naïve chemotaxis index to butanone, completed on a subset of worms that did not complete the memory training. Standard chemotaxis assay conditions were used.20 If fewer than 20 worms left origin, the plate was excluded from analysis. All memory assay experiments included five technical replicates and three biological replicates.
For each plate, chemotaxis indices were calculated as follows: (#wormsButanone – # wormsEthanol)/(#wormstotal – #wormsorigin). Performance indices were calculated to account for naïve butanone chemotaxis: Chemotaxis IndexTrained – Chemotaxis IndexNaïve. For extrachromosomal transgenic strains, chemotaxis indices were manually counted for fluorescent transgenic and non-fluorescent wild-type siblings at individual timepoints on the chemotaxis plates.
Chemotaxis assay
Chemotaxis towards 1% benzaldehyde (Millipore Sigma, Cat. # B1334-100G) in ethanol was accessed with age in synchronized Day 1, Day 5, and Day 7 pHSP16-48::FLPase;Podr-1::FRT::egl-30(Q205L); Pmyo-2::mCherry transgenic worms or their wild-type siblings using standard chemotaxis assay conditions.20 Assays included five technical replicates and three biological replicates.
Maximum velocity measurement
Maximum velocity was assessed for pHSP16-48::FLPase;Podr-1::FRT::egl-30(Q205L); Pmyo-2::mCherry transgenic worms or their wild-type siblings with age (Day 1, Day 5, Day 7). Worms were picked off HGM plates onto unseeded 60 mm NGM plates for the analysis, avoiding the carryover of any bacteria to the assay plate. Movements were recorded for 30 sec at a rate of 25 frames per second using a Nikon DS-Fi3 camera mounted on a Nikon Smz645 Stereo microscope and recorded with Nikon Elements software. Locomotion velocity was recorded as mm/sec. Recorded images were analyzed by ImageJ and wrMTrck (plugin for ImageJ). Only worms that were in the field of view for at least 15 sec were included in the analysis. Data was imported into an Excel spreadsheet and the peak locomotion velocity observed was used for maximum velocity. n > 47 worms per day/group.
Lifespan analysis
Lifespan analyses were completed for pHSP16-48::FLPase;Podr-1::FRT::egl-30(Q205L); Pmyo-2::mCherry transgenic worms or their wild-type siblings to test of late-in-life egl-30 gain-of-function only in the AWC affected lifespan. At the L4 larval stage worms picked for the assay. Every other day, worms were transferred to freshly seeded NGM plates. The first day of adulthood was defined at T=0. To permanently activate egl-30 in the AWC late-in-life, all worms were heat-shocked at 34°C for 1 hour on Day 4 of adulthood. Kaplin-Meier analysis with log-rank (Mantel-Cox) method was used to compare lifespans between transgenic and non-transgenic siblings. Worms that “escaped” or “bagged” were censored on the day of the event. n = 96 per strain, three biological replicates were performed.
Human cell line GNAQ luciferase report assay
The GloResponse NFAT-RE-luc2P HEK293 Cell Line (Promega, Cat. # E8510) was used to compare luciferase activity between control GFP, GNAQ wild-type, and GNAQ(Q209L) expressing cells. Cells were grown in at 37°C with 5% CO2 in DMEM, high glucose medium (Gibco 11965118; Thermo Fisher Cat. # 11-965-118) with 10% FBS (Thermo Fisher, Cat. # 16140089) and Penicillin-Streptomycin-Glutamine (Thermo Fisher Cat. # 10378016). Cells were plated in white-bottom, 96-well plates (Falcon 353296, Fisher Scientific Cat. # 09-771-26) at a concentration of 2x104 and transfected the following day with the pcDNA3.1-mGreenLantern control (Addgene #161912), pcDNA3.1- human GNAQ WT, and pcDNA-3.1-human GNAQ (Q209L) following the Lipofectamine 3000 Transfection Reagent (Thermo Fisher, Cat. # L3000008) protocol. Forty-eight hours later, the luciferase activity was determined using the Bright-Glo Luciferase Assay System (Promega, Cat. # E2620) reagents, which were prepared following manufacturer instructions and added 8 min prior to reading the plate on a Biotek Synergy Mx plate reader. Luminescence (relative light units) was recorded and averaged within conditions, and three biological replicates were completed.
Mouse experiments
RNAscope for Gnaq expression in young and aged mice
RNAscope was used to compare Gnaq expression in young (3 mo. old) and aged (24 mo. old) mice. In situ hybridization experiments and fluorescent staining for Gnaq was performed using the RNAScope Multiplex Fluorescent Reagent Kit V2 and the RNA-Protein Co-Detection Ancillary Kit (ACD Bio, Cat. # 323100 and 323180). RNAscope processing followed the manufacturer’s instructions. Cryo-preserved 40 μm thick coronal sections that included the hippocampus were washed in PBS and pre-treated in 200 μl of hydrogen peroxide provided by the RNAscope reagent kit. Sections were treated for 10 min with RNAScope Protease III, incubated for 2 hours with Gnaq RNAScope probe (Cat. # 1067091-C1) in a HybEZ oven set at 40°C. After amplification steps, mouse Gnaq transcripts were detected using TSA Plus Cy3 reagents (Akoya Biosciences, Cat. # SKU NEL744001KT). Sections were placed on slides, coverslipped with Prolong Diamond (Molecular Probes, Thermo Fisher, Cat. # P35970) and stored at 4°C. Slides were imaged within 2 weeks using a Zeiss LSM 800 confocal microscope with 20x objective. For quantifications, stacks of 1 μm thick were acquired from the dentate gyrus, CA1, and CA3 subfields of the hippocampus. The number of puncta per ROI were counted as previously describe58 and averaged from 3 hippocampal sections per animal; n = 6 mice/group.
Primary neuron cultures (for sequencing and neurite analysis)
E17 mouse embryo (C57B1/6J) brains were used to dissociate primary mouse hippocampal neuron suspensions using the papain dissociation system (Worthington, Cat. # LK003153). Neurons were seeded on poly-L-lysine (Sigma Aldrich, Cat. # P6282) coated plates [0.1% (wt/vol)] and grown in a humidified chamber at 37°C with 5% CO2. Neurobasal medium (Thermo Fisher, Cat. # 21103049) that was supplemented with B-27 serum free supplement (Thermo Fisher, Cat. # 17504044), GlutaMAX (Thermo Fisher, Cat. #35050061) and penicillin-streptomycin (Thermo Fisher, Cat. #15140122). Half-media changes were completed every 4-5 days. Neurons were plated on 12 mm glass coverslips (Carolina Biological Supplies Cat. #633009) in 24-well plates (100,000 cells/well). For immunocytochemistry, cells were fixed for 10 min with 4% paraformaldehyde. Cells were then washed and stained with MAP2 antibody (Sigma-Aldrich, Cat. # M1406) and AlexaFluor 555 conjugated secondary antibody (Thermo Fisher Cat. # A31570). For the neurite analysis, 5 randomly selected image stacks were acquired (10 slices spaced 1.5 μm apart) for each coverslip using a Zeiss LSM900 confocal microscope with a 20x objective. Neurite length and number was assessed using the NeuronJ ImageJ plug. 20-25 neurons were traced and averaged for each coverslip.
Bulk RNA-sequencing analysis
Total RNA was isolated from primary neuron cultures using Trizol extraction combined with columns from PureLink RNA Mini kit (ThermoFisher, Cat# 12183025) following the manufacturer’s instructions. To quantify mRNA expression levels, equal amounts of cDNA were synthesized using the HighCapacity cDNA Reverse Transcription kit (ThermoFisher, Cat# 4374966) and mixed with the KAPA SYBR Fast mix (Roche, Cat# KK4601), and Gnaq-specific primers.
(GCCTACACAACAAGACGTGC, GACCTTTGGCCCCCTACATC). GAPDH mRNA was amplified as an internal control. Quantitative RT-PCR was carried out in a CFX384 Real Time System (Bio-Rad). RNA-sequencing libraries were generated using and sequenced using Genewiz RNA-seq services (New Jersey). Alignment of RNA-sequencing reads to the mouse mm10 transcriptome was performed using STAR v2.7.3a59 following ENCODE standard options, read counts were generated using RSEM v1.3.1,56 and differential expression analysis was performed in R v4.0.2 using the DESeq2 v1.28.1.55 Detailed pipeline v2.1.2 and options available on https://github.com/emc2cube/Bioinformatics/.
Viral vectors
The lentiviral murine Gnaq overexpression construct was generated in a two-step cloning process. First, the Gnaq coding sequence (CDS) and part of the 5’ and 3’ untranslated regions (UTRs) was amplified from adult mouse hippocampal cDNA and cloned into the pENTR-D-TOPO vector (Thermo Fischer, Cat # K240020). After sequence validation, the CDS was further PCR amplified and restriction enzyme sites (NheI and BamHI) were incorporated into the forward and reverse primers. Gnaq was then ligated into a Synapsin promoter-based IRES eGFP lentiviral plasmid using traditional restriction enzyme-based cloning strategy. The Q209L mutation was generated using the QuikChange Lighting site-directed mutagenesis kit (Agilent). A Synapsin-GFP construct based on the same lentiviral plasmid was used as a control.60 All plasmids were validated by Sanger sequencing prior to virus production. Lentiviral particles were generated as previously described.61 Briefly, HEK293T cells were lipotransfected with 4:3:1 mg of lentiviral vector: psPax2:pCMV-VSVG: (psPAX2 was a gift from Didier Trono (Addgene plasmid #12260); pCMV-VSV-G was a gift from Bob Weinberg (Addgene plasmid # 8454, RRID:Addgene_8454). After 48 hours lentivirus-containing media was centrifuged for 5 min at 1000g and the supernatant filtered through a 45μm filter to remove cellular debris. Media underwent ultracentrifugation (24000 RPM for 1.5 hours) to concentrate virus. Viral pellets were gently resuspended in PBS. Lentiviral titers were between 1-3 x 10^9 viral particles per mL. Primary neuron cell cultures were infected at an ROI of 1 on 5DIV and analyzed on 11DIV. For in vivo applications, viral solutions were diluted to 1.0 x 10^8 viral particles/mL prior to stereotaxic delivery.
Stereotaxic injections
Sterotaxic injections followed previously described protocols.58,61 Mice were placed in stereotaxic frames and anesthetized with 2% isoflurane (Patterson Veterinary; 2L per min oxygen flow rate). Fur near the incision was trimmed and ophthalmic eye ointment was applied to prevent desiccation for the cornea during surgery. Viral solutions (Gnaq(Q209L) or GFP control) were injected bilaterally into hippocampal CA1 and dentate gyrus using the following coordinates: (from bregma) anterior: −2 mm and lateral: 1.5 mm and (from skull surface) height: −1.7 mm and −2.1 mm. A volume of 2 μL of viral solution was injected into each hemisphere over 10 min (injection speed of 0.1 μL per min) using a 5 μL 30s-guage Hamilton syringe. The needle was maintained in place for 8 min, slowly retracted half-way and kept in position for 2 min, and then removed, to prevent backflow along the injection tract. The incision was closed with a silk suture. Mice then received a subcutaneous injection of saline, enrofloxacin (Bayer) antibiotic, carprofen (Patterson) and buprenorphine (Butler Schein) for pain. Mice were housed alone and monitored during recovery. 14-16 mice were injected for each group.
Mouse behavior
Nestlet.
The nestlet assay was completed to assess well-being following protocols described previously.27 Mice were provided with pressed cotton nestlets and given 48 hours to build nests. After 48 hours, a nestlet score (1-5) was given based on the quality of nests. A score of 1 = no nest built and a score of 5 = an enclosed nest (Figure 3G). The scores from two separate nesting experiments performed at a one-week interval were averaged for each mouse.
Novel object recognition.
The novel object recognition task was performed following the protocol previously described.58,61 During habituation (day 1) mice could freely explore an empty, 40 cm x 40 cm square arena. During training (day 2), two identical objects were placed into the arena, and mice could explore the objects for 5 min. For testing (day 3), one object was replaced with a novel object, and mice could explore the objects for 5 min. Time spent exploring each object was quantified using Smart Video Tracking Software (Panlab; Harvard Apparatus). To control for any inherent object preference, two different sets of objects were used for experiments. To control for any potential object-independent location preference, the location of the novel object, relative to the trained object, was varied. Objects were selected based on their ability to capture mouse interest, regardless of genetic background or age. The time spent with the novel object was calculated by: ((Time with novel object)/(Time with trained object + Time with novel object)) * 100. In this object preference index, 100% indicates full preference for the novel object and 0% indicates full preference for the trained object. A score of 50% would mean the mouse spend equal times exploring both objects. If a mouse did not explore either object during training, they were excluded from the analysis.
Radial arm water maze.
The radial arm water maze (RAWM) to assess spatial learning and memory was completed following previously described protocols.58 For this task, the goal arm containing the platform remained in the same location for training and testing phases, but the starting arm was changed during each trial. Spatial cues were posted on all four walls in the RAWM area. Entry into the incorrect arm was scored as an error, and errors were averaged over training blocks (three consecutive trials). During the training phase (day 1), mice trained for 15 trials, with trials alternating between a visible and hidden platform for blocks 1-4 and then switching to only a hidden platform in block 5. During the testing phase (day 2) mice were tested for 15 trials with a hidden platform for blocks 6-10. Scoring was performed blind to treatment condition.
Tissue collection
Following the completion of behavioral assays, all mice were anesthetized with 87.5 mg/kg ketamine and 12.5mg/kg xylazine, transcardially perfused with ice-cold phos-phate buffered saline and brains dissected. For one hemisphere, hippocampi were subdissected, flash frozen, and stored at −80°C for snRNAseq analysis. The second hemisphere was post-fixed in 4% paraformaldehyde, pH 7.4, at 4°C for 48 hours before cryprotection with 30% sucrose. Brains were sectioned coronally at 40 μm with a freeze-stage microtome (Leica Camera, Inc.) and stored at −20°C in cryo-protective media until immunofluorescence, immunohistochemistry, or RNA-scope.
Immunofluorescence and microscopy
Immunofluorescence methods followed those described previously.2,58 One hemisphere of each mouse brain was sectioned at 40 μm though the entire hippocampus and stored in cryoprotective media at −20°C until use. Sections for each mouse were stored in tubes, with each tube containing sections representative of the entire hippocampus. From each tube, 3-4 sections containing the dorsal hippocampus was selected for each immunofluorescence experiment. Sections were first washed 3x in TBST (Millipore Sigma, Cat. # 91414) for 5 min each, and then permeabilized with 0.1% Triton X-100 (Millipore Sigma, Cat. # 11332481001) for 30 min at room temperature. Sections were then washed 3x, for 5 min each in TBST and blocked in donkey serum (Southern Biotech, Cat. # 0030-01) for 1 hour at room temperature. Sections incubated shaking, overnight at 4°C in primary antibody solutions in TBST with 3% donkey serum. The dilutions were as follows: synapsin: 1:1000 (Anti-Synapsin I antibody; Abcam, Cat. # ab64581), PSD95: 1:500 (Anti-PSD95 antibody; Sigma-Aldrich, Cat. # P246), gephyrin: 1:200 (Gephryin antibody; Synaptic Systems, Cat. # 147 318), Arc: 1:500 (Arc (C-7); Santa Cruz Biotech, Cat. # sc-17839), and Egr1:1:500 (Egr1 Antibody; Thermo Scientific, Cat. # MA5-15009). The next day, sections were washed 3x for 5 min each in TBST and then incubated in secondary antibody solution in TBST with 3% donkey serum, shaking for 1 hour at room temperature. All secondary antibodies were used at a 1:500 dilution and included the following: Anti-Mouse IgG (H+L), highly cross-adsorbed, CF 647 antibody (Sigma-Aldrich, Cat. # SAB4600176), Anti-Rabbit IgG (H+L), highly cross-adsorbed, CF 555 antibody (Sigma-Aldrich, Cat. # SAB4600061), Alexa Fluor 647-AffiniPure Donkey Anti-Guinea Pig IgG (H+L) (Jackson ImmunoResearch, Cat. #706-605-148). All samples were then washed 3x in TBST and stained with Hoechst (1:1000 in TBST; Molecular Probes Hoechst 33342, Thermo Fisher, Cat. # H3570) for 10 min at room temperature. Samples were lastly washed 3x in TBST and mounted using Prolong Gold Medium (Molecular Probes, Thermo Fisher, Cat. #P36934). For each analysis, n = 12 mice per group.
For the synaptic proteins (protocol modified from62), slides were imaged using a Nikon A1R Confocal Microscope equipped with a 60x oil objective. Z-stacks through the entire section, spaced at 0.33 μm apart, were taken from the same region of DG and CA1 from all hippocampal sections for each mouse on 3-4 different sections. Using FIJI, Z-stacks were analyzed by max intensity projections for clusters of 3 consecutive stacks, to avoid false positives. A macro was written to identify colocalized synaptic puncta with the following processing parameters: despeckle, subtract background (rolling ball, radius of 10 pixels), thresholding with triangle method, and watershed to separate overlapping particles. Colocalized and non-colocalized points were then counted using the co-localization plugin followed by analyze particles. An average for synapsin, gephyrin, PSD95, and excitatory and inhibitory synapses was calculated for each mouse by averaging values across sections.
To image immediate early genes Arc and Egr1, a Spinning Disk CSU-W1 Nikon Confocal Microscope (Princeton University Microscopy Core) with a 20x objective was used, and z-stacks spaced 3 μm apart were taken through the entire section and stitched together to include the entire hippocampus. After max intensity projection, FIJI was used for analysis. The ROI (CA1 or DG) was outlined in each section. To identify the percent positive cells per ROI the following parameters were used: Gaussian filtering (sigma of 2), thresholding using the triangle method, and counting particles greater than 15 pixels. This value was then divided by the ROI area sampled. Mean signal (normalized to Hoechst) was measured when there were not punctate particles to count. For all analyses, an average was calculated for each mouse across all sections for CA1 and DG.
Neuronal nuclei isolation
Neuronal nuclei were isolated similar to previously described methods.58,63 One hemisphere of the hippocampus dissected, flash frozen, and stored at −80°C until use. Samples were kept on ice during the experiment (n = 4 GFP hippocampi and 4 GNAQ hippocampi). The hippocampi were dounce homogenized using 1 mL Dounce tissue grinders (Wheaton, DWK Life Sciences, Cat. # 357538) in 300 L of NP40 lysis buffer (10 mM Tris-HCl (Sigma-Aldrich, Cat. # T2194; pH 7.4), 10 mM NaCl (Sigma-Aldrich, Cat. # 59222C; 5M), 3 mM MgCl2 (Sigma-Aldrich, Cat. # M1028; 1M), 0.1% Nonidet P40 Substitute (Sigma-Aldrich, Cat. # 74385), 1 mM DTT (Sigma-Aldrich, Cat. # 646563), 1 U/μL RNase inhibitor (Sigma Protector RNase inhibitor; Sigma-Aldrich, Cat. # 3335402001), Nuclease-free water) with 15 loose stokes followed by 15 tight strokes. Samples were incubated for 7 min on ice, and pipetted twice after 2 min and 5 min, 3x at each point, with a P200 pipette. Suspensions were passed through a 40 μm filter into a 15 mL conical tube, and then transferred to a 2-mL low bind microcentrifuge tube. Samples were centrifuged at 500 rcf for 5 min at 4°C. The supernatant was removed and 1 mL of wash buffer (PBS + 1% BSA (Miltenyi Biotec, Cat. # 130-091-376) + 1 U/uL of RNAase inhibitor) was added with no mixing for 5 min on ice. The pellet was then resuspended by pipette mixing and samples were centrifuged again at 500 rcf for 5 min at 4°C. The supernatant was removed and the cells were resuspended in 1 mL of the same wash buffer. Next, 10 μL of Hoechst stain (1:10,000 dilution; Molecular Probes Hoechst 33342, Thermo Fisher, Cat. # H3570) was added to each sample, prior to filtering into the FACS tube and incubating on ice for 5 min on the way to the FACS core.
FACS, snRNA-seq library preparation, and sequencing
The Princeton Flow Cytometry Resource Facility sorted Hoechst+ and GFP+ nuclei using a 70μm nozzle at 70psi on a BD Biosciences FACSAria Fusion sorter into a 1.5 mL low bind microcentrifuge tube containing 500 μL collection buffer (500 μL of 1% BSA + 1.5 U/μL RNase Inhibitor). Samples and sort collection tubes were kept at 4°C during sorting.
After FACS, samples were centrifuged at 500 rcf for 5 min at 4°C. The supernatant was removed and nuclei were resuspended in 20 μL of collection buffer. For each sample, 2 μL of nuclei were diluted 1:5 in PBS + Trypan Blue (Gibco Trypan Blue Solution, Fisher Scientific, Cat. # 15-250-061) and counted on a hemocytometer (Millicell Disposable Hemocytometer, Millipore Sigma, Cat. # MDH-2N1-50PK). The number of nuclei/μL was estimated for each sample and provided to the Princeton Genomics Core. Single nuclei suspension samples were loaded to the 10X Genomics Chromium X system using the Single Cell 3’ v3.1 Reagent Kits (10X Genomics Inc., CA) to generate and amplify cDNA. The amplified cDNA samples were purified with Ampure XP magnetic beads (Beckman Coulter, CA), quantified by Qubit fluorometer (Invitrogen, CA), and examined on Bioanalyzer with High Sensitivity DNA chips (Agilent, CA) for size distribution. Illumina sequencing libraries were generated from the amplified cDNA samples using the Illumina Tagment DNA Enzyme and Buffer kit (Illumina, CA). These libraries were examined by Qubit and Bioanalyzer, then pooled at equal molar amount and sequenced on Illumina NovaSeq 6000 S Prime flowcells as 28+94 nt pair-end reads following the standard protocol. Raw sequencing reads were filtered by Illumina NovaSeq Control Software and only the Pass-Filter (PF) reads were used for further analysis.
snRNA-seq analysis
Raw data handling.
After sequencing, output fastq files were mapped to an mm10 reference genome that was edited to include eGFP and filtered using Cell Ranger version 7.1.0. The pooled samples consisted of GFP control samples: four samples, pooled in groups of two, and GNAQ samples: four samples, pooled in groups of two. For the GFP control samples, Cell Ranger estimated 12,771 cells or 8,852 cells for the two pooled samples, with ~4500 median UMIs per cell. In the GNAQ samples, Cell Ranger estimated 10,860 and 9,832 cells in each of the two pooled samples, with ~4000 median UMIs per cell.
Quality control.
Ambient RNA contamination was detected and removed using the SoupX package in R. Full Cell Ranger out files were input and ambient mRNA expression profile was estimated from empty droplets, the contamination fraction of each cell was calculated, and then the UMIs for all cells were corrected based on the contamination fraction. The global contamination fraction of all samples was estimated to be 0.03% which falls between the normal range of 0.02-0.05%. Note: SoupX removes ambient RNA contamination, it does not remove any cells from the dataset.
Additional quality control was done in R using the Seurat package. Violin Plots of the nFeatures/cell and the mtRNA/cell were generated, and cells were removed based on Seurat’s suggested cutoffs. For our samples, cells with <200 nFeatures (potentially damaged cells) or >5000 nFeatures (potential doublets) were filtered out. Cells with mitochondrial contamination >0.25% were also removed from the dataset.
Following quality control, our final cell numbers for GFP control samples were 12,227 and 8,183. Our two GNAQ samples had final cell numbers of 9,325 and 10,231 cells. Approximately 500-600 cells were filtered out of each condition after all quality control.
Normalization, integration, and clustering.
Prior to normalization, all datasets were merged into a single Seurat object. The data was normalized using SCTransform to fit the gene expression counts along a negative binomial distribution. 3000 integration features were selected from the dataset and used to find integration anchors. Integration was then performed on the SCTransform normalized counts.
Next, principal component analysis was performed, and an elbow plot of the principal components was generated to help choose the number of components needed to accurately represent the data. We chose 40 principal components to perform clustering. Clustering was performed using the Louvain algorithm for network clustering at a resolution of 0.5 which resulted in 31 clusters.
Cell type Annotation.
Finally, clusters were annotated using known hippocampal markers for different cell types (Allen Mouse Brain Atlas (http://mouse.brain-map.org/), HippoSeq (https://hipposeq.janelia.org/),64–66). The markers are in Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
For worm experiments, data are expressed at mean ± SEM. Statistical analysis was performed with Prism 8.0 or 9.0 software (GraphPad Software). Means between two groups were compared with two-tailed, unpaired Student’s t-test. Comparisons of means from multiple groups with each other were analyzed with one-way or two-way ANOVAs followed by a Bonferroni post hoc tests for multiple comparisons, as indicated in the figure legends. When worms were tested across days, separate cohorts of worms were analyzed at each timepoint. All worm experiments included five technical replicates and three biological replicates, with the exception Figure 4I, where two biological replicates were completed for non-significant effects. All replicates are graphed in the paper, but statistical results were the same when each biological replicate was analyzed individually. Additional statistical details are reported in figure legends.
All mouse experiments were randomized and blinded by an independent researcher before behavioral testing. Researchers remained blinded throughout histological, biochemical and behavioral assessments. Groups were un-blinded at the end of each experiment upon statistical analysis. Data are expressed as mean ± SEM. The distribution of data in each set of experiments was tested for normality using D’Agostino-Pearson omnibus test or Shapiro-Wilk test. Statistical analysis was performed with Prism 8.0 or 9.0 software (GraphPad Software). Means between two groups were compared with two-tailed, unpaired Student’s t test. Comparisons of means from multiple groups with each other were analyzed with one-way ANOVA followed by the appropriate post hoc test, as indicated in the figure legends. Trial by group interactions were analyzed using repeated measures ANOVA and with Śídák’s correction for multiple comparisons. Additional statistic details are indicated in the respective figure legends. All data generated or analyzed in this study are included in this article.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| MAP2 antibody | Sigma-Aldrich | Cat. # M1406; RRID:AB_477171 |
| AlexaFluor 555 conjugated secondary antibody | Thermo Fisher | Cat. # A31570; RRID:AB_2536180 |
| Anti-Synapsin I antibody | Abcam | Cat. # ab64581; RRID:AB_1281135 |
| Anti-PSD95 antibody | Sigma-Aldrich | Cat. # P246; RRID:AB_260911 |
| Gephryin antibody | Synaptic Systems | Cat. # 147 318; RRID:AB_2661777 |
| Arc (C-7) antibody | Santa Cruz Biotech | Cat. # sc-17839; RRID:AB_626696 |
| Egr1 Antibody | Thermo Scientific | Cat. # MA5-15009; RRID:AB_10982091 |
| Anti-Mouse IgG (H+L), highly cross-adsorbed, CF 647 antibody | Sigma-Aldrich | Cat. # SAB4600176; RRID:AB_3064834 |
| Anti-Rabbit IgG (H+L), highly cross-adsorbed, CF 555 antibody | Sigma-Aldrich | Cat. # SAB4600061; RRID:AB_3064836 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch | Cat. # 706-605-148; RRID:AB_2340476 |
| Bacterial and virus strains | ||
| OP50 E. Coli (BactoBeads) | Sigma-Aldrich | Cat. #S29021 |
| HT115 E coli | Caenorhabditis Genetics Center | HT115 |
| Chemicals, peptides, and recombinant proteins | ||
| 2-butanone | Acros Organics | Cat. # 332828-25ML |
| Benzaldehyde | Millipore Sigma | Cat. # B1334-100G |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher | Cat. # L3000008 |
| Sigma Protector RNase inhibitor | Sigma-Aldrich | Cat. # 3335402001 |
| Molecular Probes Hoechst 33342 | Thermo Fisher | Cat. # H3570 |
| Ampure XP magnetic beads | Beckman Coulter | Cat. # A63882 |
| Critical commercial assays | ||
| Bright-Glo Luciferase Assay System | Promega | Cat. # E2620 |
| RNAScope Multiplex Fluorescent Reagent Kit V2 | ACD Bio | Cat. # 323100 |
| RNA-Protein Co-Detection Ancillary Kit | ACD Bio | Cat. # 323180 |
| PureLink RNA Mini kit | Thermo Fisher | Cat. # 12183025 |
| HighCapacity cDNA Reverse Transcription kit | Thermo Fisher | Cat. # 4374966 |
| KAPA SYBR Fast mix | Roche | Cat. # KK4601 |
| Papain dissociation system | Worthington | Cat. # LK003153 |
| 10X Genomics Chromium X system using the Single Cell 3’ v3.1 Reagent Kits | 10X Genomics | Cat. # 1000 |
| Illumina Tagment DNA Enzyme and Buffer kit | Illumina | Cat. # 20034198 |
| Deposited data | ||
| RNA-seq data | This paper | BioProject #PRJNA972702. |
| Experimental models: Cell lines | ||
| GloResponse NFAT-RE-luc2P HEK293 Cell Line | Promega | E8510 |
| Experimental models: Organisms/strains | ||
| C. elegans strain: N2 var. Bristol: wild-type | Caenorhabditis Genetics Center | N2 |
| C. elegans strain: NM1380:elg-30(js126) | Caenorhabditis Genetics Center | NM1380 |
| C. elegans strain: CQ429: pHSP16-48::FLPase;Podr-1::FRT::egl-30(Q205L); Pmyo-2::mCherry | Arey et al.18 | CQ429 |
| C. elegans strain: CQ701: wtEx74 Topo-Prab-3:: GFP-egl-30(WT)::rab3 UTR; Pmyo-2::mCherry/this paper | This paper | CQ701 |
| C. elegans strain: CQ601:egl-30(Q205L);vls69[pCFJ90(Pmyo-2::mCherry+Punc-119::sid-1)] | This paper | CQ601 |
| Mouse: C57BL/6 | The Jackson Laboratory/NIA | RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Gnaq RNAScope probe | ACD Bio | Cat. # 1067091-C1 |
| Gnaq-specific primer: GCCTACACAACAAGACGTGC | This paper | N/A |
| Gnaq-specific primer: GACCTTTGGCCCCCTACATC | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1-mGreenLantern control | Addgene | Cat. # 161912 |
| psPax2:pCMV-VSVG | Addgene | Plasmid # 12260 |
| pCMV-VSV-G | Addgene | Plasmid # 8454; RRID:Addgene_8454 |
| pENTR-D-TOPO vector | Thermo Fisher | Cat # K240020 |
| Plasmid pL4440 RNAi | Addgene/Fire Lab | L4440/control |
| Plasmid: pL4440-slo-1 RNAi | Ahringer | slo-1 |
| Plasmid: pL4440-syg-1 RNAi | Ahringer | syg-1 |
| Plasmid: pL4440-ttn-1 RNAi | Ahringer | ttn-1 |
| Plasmid: pL4440-lad-2 RNAi | Ahringer | lad-2 |
| Plasmid: pL4440-madd-4 RNAi | Ahringer | madd-4 |
| Plasmid: pL4440-unc-73 RNAi | Ahringer | unc-73 |
| Plasmid: pL4440-glr-1 RNAi | Ahringer | glr-1 |
| Plasmid: pL4440-magi-1 RNAi | Ahringer | magi-1 |
| Plasmid: pL4440-daf-19 RNAi | Ahringer | daf-19 |
| Plasmid: PL4440-nhr-1 RNAi | Ahringer | nhr-1 |
| Software and algorithms | ||
| GraphPad Prism version 8.0 or 9.0 | GraphPad Software | https://www.graphpad.com |
| FIJI or ImageJ | ImageJ | https://imagej.net/software/fiji/ |
| DESeq2 v1.28 | Love et al.55 | https://github.com/emc2cube/Bioinformatics/ |
| RSEM v1.3.1 | Li and Dewey56 | https://github.com/deweylab/RSEM |
| Cell Ranger version 7.1.0. | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| R software for statistical computing v4.0.2 | R Core Team, 2022 | https://www.r-project.org/ |
Highlights.
EGL-30/GNAQ and Gαq signaling is highly conserved between worms and mammals
GNAQ gain of function in the hippocampus extends memory in 2-year-old mice
GNAQ gain of function also upregulates synaptic function, axon guidance, and memory genes
Worm orthologs of these genes are required for extended long-term memory
ACKNOWLEDGMENTS
We thank the Princeton FACS Core and Christina DeCoste for her assistance, the Princeton Confocal Microscopy Core, the Princeton Genomics Core and Jennifer Miller and Jean Arly Vomar for their assistance, the C. elegans Genetics Center, and members of the Murphy lab for suggestions on the manuscript. We acknowledge the UCSF Parnassus Flow Core (RRID:SCR_018206), UCSF Center for Advanced Technology, and Stanford Sherlock cluster. We also thank the Murphy and Villeda labs for feedback on the manuscript. Illustrations were created using BioRender.com. Funding for this work was provided by the Simons Collaboration on Plasticity in the Aging Brain (SCPAB) to C.T.M. and to S.A.V., the Mikheev Fellowship and an NIH F32 award to M.E.S. (AG079490), an NIH F32 award (AG081038) and a Hillblom postdoctoral fellowship to G.B., an NSF pre-doctoral award to J.S.A., an NIA R01 (AG077816) to S.A.V., and an NIH Director’s Pioneer Award to C.T.M. (DP1AG077430).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113151.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research.
REFERENCES
- 1.Arey RN, and Murphy CT (2017). Conserved regulators of cognitive aging: From worms to humans. Behav. Brain Res 322, 299–310. 10.1016/j.bbr.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, Smith LK, Bieri G, Lin K, Berdnik D, et al. (2014). Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med 20, 659–663. 10.1038/nm.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakhina V, Arey RN, Kaletsky R, Kauffman A, Stein G, Keyes W, Xu D, and Murphy CT (2015). Genome-wide Functional Analysis of CREB/Long-Term Memory-Dependent Transcription Reveals Distinct Basal and Memory Gene Expression Programs. Neuron 85, 330–345. 10.1016/j.neuron.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, and Murphy CT (2010). Insulin Signaling and Dietary Restriction Differentially Influence the Decline of Learning and Memory with Age. PLoS Biol. 8, e1000372. 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin JC, and Tully T (1996). CREB and the formation of long-term memory. Curr. Opin. Neurobiol 6, 264–268. 10.1016/s0959-4388(96)80082-1. [DOI] [PubMed] [Google Scholar]
- 6.Silva AJ, Kogan JH, Frankland PW, and Kida S (1998). CREB and memory. Annu. Rev. Neurosci 21, 127–148. 10.1146/an-nurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 7.Bieri G, Schroer AB, and Villeda SA (2023). Blood-to-brain communication in aging and rejuvenation. Nat. Neurosci 26, 379–393. 10.1038/s41593-022-01238-8. [DOI] [PubMed] [Google Scholar]
- 8.Fan X, Wheatley EG, and Villeda SA (2017). Mechanisms of Hippocampal Aging and the Potential for Rejuvenation. Annu. Rev. Neurosci 40, 251–272. 10.1146/annurev-neuro-072116-031357. [DOI] [PubMed] [Google Scholar]
- 9.van Praag H, Shubert T, Zhao C, and Gage FH (2005). Exercise enhances learning and hippocampal neurogenesis in aged mice. J. Neurosci 25, 8680–8685. 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer AF, Hahn S, Cohen NJ, Banich MT, McAuley E, Harrison CR, Chason J, Vakil E, Bardell L, Boileau RA, and Colcombe A (1999). Ageing, fitness and neurocognitive function. Nature 400, 418–419. 10.1038/22682. [DOI] [PubMed] [Google Scholar]
- 11.Horowitz AM, Fan X, Bieri G, Smith LK, Sanchez-Diaz CI, Schroer AB, Gontier G, Casaletto KB, Kramer JH, Williams KE, and Villeda SA (2020). Blood factors transfer beneficial effects of exercise on neurogenesis and cognition to the aged brain. Science 360, 167–173. 10.1126/science.aaw2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X-W, Curlik DM, Oh MM, Yin JC, and Disterhoft JF (2017). CREB overexpression in dorsal CA1 ameliorates long-term memory deficits in aged rats. Elife 6, e19358. 10.7554/eLife.19358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brundage L, Avery L, Katz A, Kim U-J, Mendel JE, Sternberg PW, and Simon MI (1996). Mutations in a C. elegans Gqα Gene Disrupt Movement, Egg Laying, and Viability. Neuron 16, 999–1009. 10.1016/S0896-6273(00)80123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lackner MR, Nurrish SJ, and Kaplan JM (1999). Facilitation of Synaptic Transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG Binding to UNC-13 Is Required to Stimulate Acetylcholine Release. Neuron 24, 335–346. 10.1016/S0896-6273(00)80848-X. [DOI] [PubMed] [Google Scholar]
- 15.Miller KG, Emerson MD, and Rand JB (1999). Goα and Diacylglycerol Kinase Negatively Regulate the Gqα Pathway in C. elegans. Neuron 24, 323–333. 10.1016/S0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ch’ng Q, Sieburth D, and Kaplan JM (2008). Profiling Synaptic Proteins Identifies Regulators of Insulin Secretion and Lifespan. PLoS Genet. 4, e1000283. 10.1371/journal.pgen.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi T, Kunitomo H, Tomioka M, Ohno H, Okochi Y, Mori I, and Iino Y (2010). Reversal of Salt Preference Is Directed by the Insulin/PI3K and Gq/PKC Signaling in Caenorhabditis elegans. Genetics 186, 13091319. 10.1534/genetics.110.119768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arey RN, Stein GM, Kaletsky R, Kauffman A, and Murphy CT (2018). Activation of Gαq Signaling Enhances Memory Consolidation and Slows Cognitive Decline. Neuron 98, 562–574.e5. 10.1016/j.neuron.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian NX, Winitz S, and Johnson GL (1993). Epitope-tagged Gq alpha subunits: expression of GTPase-deficient alpha subunits persistently stimulates phosphatidylinositol-specific phospholipase C but not mitogen-activated protein kinase activity regulated by the M1 muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA 90, 4077–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bargmann CI, Hartwieg E, and Horvitz HR (1993). Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527. 10.1016/0092-8674(93)80053-H. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Todd N, and Thathiah A (2017). The role of GPCRs in neurodegenerative diseases: avenues for therapeutic intervention. Curr. Opin. Pharmacol 32, 96–110. 10.1016/j.coph.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Peng S, Zeng L, Haure-Mirande J-V, Wang M, Huffman DM, Haroutunian V, Ehrlich ME, Zhang B, and Tu Z (2021). Transcriptomic Changes Highly Similar to Alzheimer’s Disease Are Observed in a Subpopulation of Individuals During Normal Brain Aging. Front. Aging Neurosci 13, 711524. 10.3389/fnagi.2021.711524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberini CM (1999). Genes to remember. J. Exp. Biol 202, 2887–2891. 10.1242/jeb.202.21.2887. [DOI] [PubMed] [Google Scholar]
- 25.Alberini CM, and Kandel ER (2014). The Regulation of Transcription in Memory Consolidation. Cold Spring Harb. Perspect. Biol 7, a021741. 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarome TJ, and Lubin FD (2014). Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol. Learn. Mem 115, 116–127. 10.1016/j.nlm.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deacon RMJ (2006). Assessing nest building in mice. Nat. Protoc 1, 1117–1119. 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- 28.Nolte ED, Nolte KA, and Yan SS (2019). Anxiety and task performance changes in an aging mouse model. Biochem. Biophys. Res. Commun 514, 246–251. 10.1016/j.bbrc.2019.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danielson E, Zhang N, Metallo J, Kaleka K, Shin SM, Gerges N, and Lee SH (2012). S-SCAM/MAGI-2 is an essential synaptic scaffolding molecule for the GluA2-containing maintenance pool of AMPA receptors. J. Neurosci 32, 6967–6980. 10.1523/JNEUROSCI.0025-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cissé M, Duplan E, Lorivel T, Dunys J, Bauer C, Meckler X, Gerakis Y, Lauritzen I, and Checler F (2017). The transcription factor XBP1s restores hippocampal synaptic plasticity and memory by control of the Kalirin-7 pathway in Alzheimer model. Mol. Psychiatry 22, 1562–1575. 10.1038/mp.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranneva SV, Pavlov KS, Gromova AV, Amstislavskaya TG, and Lipina TV (2017). Features of emotional and social behavioral phenotypes of calsyntenin2 knockout mice. Behav. Brain Res 332, 343–354. 10.1016/j.bbr.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 32.Stein GM, and Murphy CT (2014). C. elegans positive olfactory associative memory is a molecularly conserved behavioral paradigm. Neurobiol. Learn. Mem 115, 86–94. 10.1016/j.nlm.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Zhang Y, Kaletsky R, Toraason E, Zhang W, Dong M-Q, and Murphy CT (2023). Signaling from the C. elegans Hypodermis Non-autonomously Facilitates Short-Term Associative Memory. Preprint at bioRxiv. 10.1101/2023.02.16.528821. [DOI] [Google Scholar]
- 34.Kano T, Brockie PJ, Sassa T, Fujimoto H, Kawahara Y, Iino Y, Mellem JE, Madsen DM, Hosono R, and Maricq AV (2008). Memory in Caenorhabditis elegans is Mediated By NMDA-Type Ionotropic Glutamate Receptors. Curr. Biol 18,1010–1015. 10.1016/j.cub.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang T-T, Lo C-P, Tsai P-S, Wu S-Y, Wang T-F, Chen Y-W, Jiang-Shieh Y-F, and Kuo Y-M (2015). Aging and Exercise Affect Hippocampal Neurogenesis via Different Mechanisms. PLoS One 10, e0132152. 10.1371/journal.pone.0132152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsetsenis T, Boucard AA, Araç D, Brunger AT, and Südhof TC (2014). Direct Visualization of Trans-Synaptic Neurexin–Neuroligin Interactions during Synapse Formation. J. Neurosci 34, 15083–15096. 10.1523/JNEUROSCI.0348-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalva MB, McClelland AC, and Kayser MS (2007). Cell adhesion molecules: signalling functions at the synapse. Nat. Rev. Neurosci 8, 206–220. 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding X, Liu S, Tian M, Zhang W, Zhu T, Li D, Wu J, Deng H, Jia Y, Xie W, et al. (2017). Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. Nat. Neurosci 20, 690–699. 10.1038/nn.4536. [DOI] [PubMed] [Google Scholar]
- 39.Philbrook A, Ramachandran S, Lambert CM, Oliver D, Florman J, Alkema MJ, Lemons M, and Francis MM (2018). Neurexin directs partner-specific synaptic connectivity in C. elegans. Elife 7, e35692. 10.7554/eLife.35692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto K-I, Narayanan R, Lee SH, Murata K, and Hayashi Y (2007). The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc. Natl. Acad. Sci. USA 104,64186423. 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borgesius NZ, van Woerden GM, Buitendijk GHS, Keijzer N, Jaarsma D, Hoogenraad CC, and Elgersma Y (2011). βCaMKII Plays a Nonenzymatic Role in Hippocampal Synaptic Plasticity and Learning by Targeting αCaMKII to Synapses. J. Neurosci 31, 10141–10148. 10.1523/JNEUROSCI.5105-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sengar AS, Li H, Zhang W, Leung C, Ramani AK, Saw NM, Wang Y, Tu Y, Ross PJ, Scherer SW, et al. (2019). Control of Long-Term Synaptic Potentiation and Learning by Alternative Splicing of the NMDA Receptor Subunit GluN1. Cell Rep. 29, 4285–4294.e5. 10.1016/j.celrep.2019.11.087. [DOI] [PubMed] [Google Scholar]
- 43.Kilonzo K, Strahnen D, Prex V, Gems J, van der Veen B, Kapanaiah SKT, Murthy BKB, Schulz S, Sprengel R, Bannerman D, and Kätzel D (2022). Distinct contributions of GluA1-containing AMPA receptors of different hippocampal subfields to salience processing, memory and impulse control. Transl. Psychiatry 12, 102–113. 10.1038/s41398-022-01863-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shabani M, Haghani M, Tazangi PE, Bayat M, Shid Moosavi SM, and Ranjbar H (2017). Netrin-1 improves the amyloid-β-mediated suppression of memory and synaptic plasticity. Brain Res. Bull 131, 107–116. 10.1016/j.brainresbull.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Klein R (2009). Bidirectional modulation of synaptic functions by Eph/ephrin signaling. Nat. Neurosci 12, 15–20. 10.1038/nn.2231. [DOI] [PubMed] [Google Scholar]
- 46.Lamprecht R, and LeDoux J (2004). Structural plasticity and memory. Nat. Rev. Neurosci 5, 45–54. 10.1038/nrn1301. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Ren S-Y, Chen J-F, Liu K, Li R-X, Li Z-F, Hu B, Niu JQ, Xiao L, Chan JR, and Mei F (2020). Myelin degeneration and diminished myelin renewal contribute to age-related deficits in memory. Nat. Neurosci 23, 481–486. 10.1038/s41593-020-0588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffries MA, Urbanek K, Torres L, Wendell SG, Rubio ME, and Fyffe-Maricich SL (2016). ERK1/2 Activation in Preexisting Oligodendrocytes of Adult Mice Drives New Myelin Synthesis and Enhanced CNS Function. J. Neurosci 36, 9186–9200. 10.1523/JNEURO-SCI.1444-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suthard RL, Jellinger AL, Surets M, Shpokayte M, Pyo AY, Buzharsky MD, Senne RA, Dorst K, Leblanc H, and Ramirez S (2023). Chronic Gq activation of ventral hippocampal neurons and astrocytes differentially affects memory and behavior. Neurobiol. Aging 125, 9–31. 10.1016/j.neurobiolaging.2023.01.007. [DOI] [PubMed] [Google Scholar]
- 50.Yanai S, and Endo S (2021). Functional Aging in Male C57BL/6J Mice Across the Life-Span: A Systematic Behavioral Analysis of Motor, Emotional, and Memory Function to Define an Aging Phenotype. Front. Aging Neurosci 13, 697621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iram T, Kern F, Kaur A, Myneni S, Morningstar AR, Shin H, Garcia MA, Yerra L, Palovics R, Yang AC, et al. (2022). Young CSF restores oligodendrogenesis and memory in aged mice via Fgf17. Nature 605, 509–515. 10.1038/s41586-022-04722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boehme M, Guzzetta KE, Bastiaanssen TFS, van de Wouw M, Moloney GM, Gual-Grau A, Spichak S, Olavarría-Ramírez L, Fitzgerald P, Morillas E, et al. (2021). Microbiota from young mice counteracts selective age-associated behavioral deficits. Nat. Aging 1, 666–676. 10.1038/s43587-021-00093-9. [DOI] [PubMed] [Google Scholar]
- 53.Harburger LL, Lambert TJ, and Frick KM (2007). Age-dependent effects of environmental enrichment on spatial reference memory in male mice. Behav. Brain Res 185, 43–48. 10.1016/j.bbr.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krukowski K, Nolan A, Frias ES, Boone M, Ureta G, Grue K, Paladini M-S, Elizarraras E, Delgado L, Bernales S, et al. (2020). Small molecule cognitive enhancer reverses age-related memory decline in mice. Elife 9, e62048. 10.7554/eLife.62048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, and Anders S (2014). Moderated estimation off old change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner S (1974). THE GENETICS OF CAENORHABDITIS ELEGANS. Genetics 77, 71–94. 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pratt KJB, Shea JM, Remesal-Gomez L, Bieri G, Smith LK, Couthouis J, Chen CP, Roy IJ, Gontier G, and Villeda SA (2022). Loss of neuronal Tet2 enhances hippocampal-dependent cognitive function. Cell Rep. 41, 111612. 10.1016/j.celrep.2022.111612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, et al. (2003). Lenti-virus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin K, Bieri G, Gontier G, Muller S, Smith LK, Snethlage CE, White CW, Maybury-Lewis SY, and Villeda SA (2021). MHC class I H2-Kb negatively regulates neural progenitor cell proliferation by inhibiting FGFR signaling. PLoS Biol. 19, e3001311. 10.1371/journal.pbio.3001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ippolito DM, and Eroglu C (2010). Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. J. Vis. Exp 2270, 2270. 10.3791/2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, et al. (2016). Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc 11, 499–524. 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C, et al. (2019). Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci 22, 1696–1708. 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- 65.Zalocusky KA, Najm R, Taubes AL, Hao Y, Yoon SY, Koutsodendris N, Nelson MR, Rao A, Bennett DA, Bant J, et al. (2021). Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer’s disease. Nat. Neurosci 24, 786–798. 10.1038/s41593-021-00851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cembrowski MS, Wang L, Sugino K, Shields BC, and Spruston N (2016). Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. Elife 5, e14997. 10.7554/eLife.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
With the exception of single-nucleus RNA-sequencing and mouse neuron sequencing data, all other data are available in the main text or the supplementary materials. Sequencing data are available at BioProject #PRJNA972702.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


