SUMMARY
Reports that mouse sperm gain small RNAs from the epididymosomes secreted by epididymal epithelial cells and that these “foreign” small RNAs act as an epigenetic information carrier mediating the transmission of acquired paternal traits have drawn great attention because the findings suggest that heritable information can flow from soma to germline, thus invalidating the long-standing Weismann’s barrier theory on heritable information flow. Using small RNA sequencing (sRNA-seq), Northern blots, sRNA in situ hybridization and immunofluorescence, we detected substantial changes in the small RNA profile in murine caput epididymal sperm (sperm in the head of the epididymis) and further determined that the changes resulted from sperm exchanging small RNAs, mainly tsRNAs and rsRNAs, with cytoplasmic droplets rather than the epididymosomes. Moreover, the murine sperm-borne small RNAs were mainly derived from the nuclear small RNAs in late spermatids. Thus, caution is needed regarding sperm gaining “foreign” small RNAs as an underlying mechanism of epigenetic inheritance.
Keywords: sperm RNA, small RNA, epididymis, sperm maturation, epigenetic inheritance, epididymosomes
Graphical Abstract
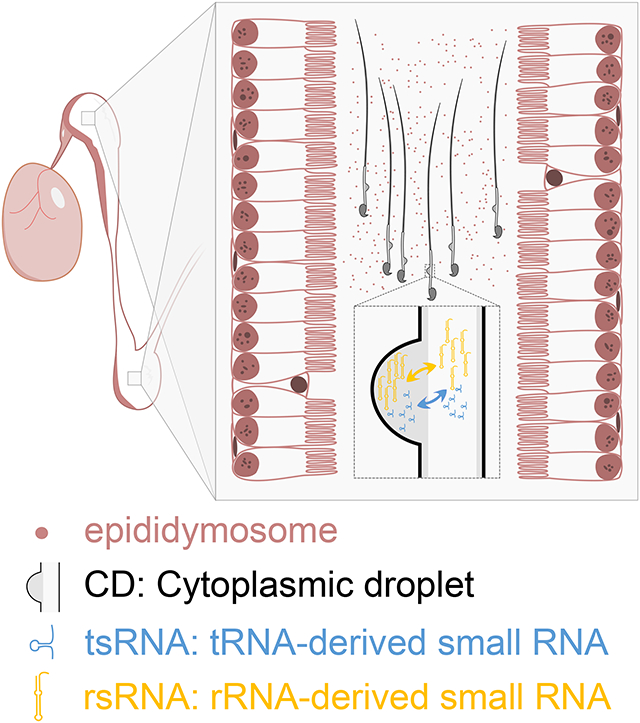
In brief
Wang et al. report that small RNA exchanges occur between sperm and their associated cytoplasmic droplets (CDs) during sperm maturation inside the epididymis. Their finding challenges the notion that sperm gain “foreign” small RNAs from the exosomes released by epididymal epithelial cells.
INTRODUCTION
As the male gametes, spermatozoa (sperm) function to deliver the paternal genome into the oocytes during fertilization. In addition to DNA, sperm also carry RNAs, both large and small, and these sperm-borne RNAs are also delivered into the oocytes during fertilization1. Although the exact physiological roles of the sperm-borne RNAs remain to be determined, emerging data have shown that these RNAs are critical for early embryonic development and might participate in the transmission of acquired paternal traits to offspring2–7. For example, Drosha- or Dicer-deficient sperm fail to support early embryonic development, and this defect can be partially rescued by supplementing total sperm small RNAs in the zygotes8. Microinjection of sperm total or small RNAs including microRNAs (miRNAs) or tRNA-derived small RNAs (tsRNAs) into the wildtype zygotes can recapitulate the paternal phenotypes caused by Kit paramutation, stress, and over/undernutrition in offspring9–15. These data strongly support the role of sperm-borne small RNAs as an epigenetic information carrier to mediate epigenetic inheritance of acquired paternal traits16,17. However, an outstanding question remains: where do sperm-borne RNAs come from? One obvious source of sperm-borne small RNAs is their precursor cells, i.e., spermatogenic cells including spermatogonia, spermatocytes, and spermatids. Small RNAs may be synthesized and loaded into sperm chromatin/head and other compartments during late spermiogenesis. Alternatively, sperm might be able to gain small RNAs from their microenvironments during their transit through the male reproductive tracts, including the efferent ducts, epididymis, and vas deference. Of interest, earlier reports have shown that sperm gain a large number of small RNAs, especially tsRNAs, from epididymosomes, the exosomes produced by caput epididymal epithelial cells, and that these epididymal small RNAs appear to be capable of inducing paternal phenotypes in offspring when injected into the wildtype zygotes18,19. This claim was based on the sRNA-seq data, showing that the small RNA compositions in testicular, caput, and cauda epididymal sperm change drastically, and that numerous novel tsRNAs and miRNAs show up when sperm migrate from the caput to cauda epididymis. Since some of these newly gained small RNAs are also present in the epididymosomes, it has been proposed that sperm gain small RNAs from the epididymis18,19. Further supporting this notion, caput sperm appear to be able to take up small RNAs from the purified epididymosomes in vitro18. These findings are both astonishing and significant because it implicates that heritable information can flow from soma (the epididymis) to the germline (sperm) and induce paternal phenotypes in offspring, thus invalidating the long-standing Weismann’s barrier theory on inheritance, which states that heritable information can only flow from germline to soma20,21. Given the extraordinary significance of such a claim, it is critical to re-confirm that sperm indeed exchange small RNAs with the epididymosomes or epididymal epithelial cells during their transit through the epididymis.
Since one mouse sperm contains ~0.1 pg of RNA, which is ~1/100 of a somatic cell22, any contamination, even in trace amount, would potentially bias the small RNA transcriptome. We, therefore, first optimized the somatic cell lysis steps in the protocol used for the purification of sperm collected from the testis, caput, and cauda epididymis18,19, so that ultrapure sperm (less than one somatic cell per 1,000 sperm) could be obtained for sRNA-seq. Our sRNA-seq data confirmed drastic changes in small RNA composition in the caput epididymal sperm characterized by the significantly increased proportional abundance of tsRNAs compared to testicular and cauda epididymal sperm. However, the up- or down-regulated small RNAs in the caput epididymal sperm were mostly from the cytoplasmic droplets (CDs) rather than the epididymosomes. Moreover, the sperm-borne small RNA appeared to be mainly derived from the nuclear small RNAs in spermatids.
RESULTS
Dynamic changes in small RNA compositions during sperm transit through the epididymis
Consistent with previous data22, we found that one mouse cauda epididymal sperm contained ~0.13 pg RNA, whereas one mouse epididymal epithelial cell and one NIH-3T3 cell had ~16.7 pg and ~37.8 pg RNA, respectively (Fig. S1A). Therefore, trace amounts of somatic cell contamination could potentially bias the sperm RNA transcriptome. We first used the method reported in the study showing that sperm gain small RNAs from the epididymosomes18,19, to remove somatic cells in sperm suspensions prepared from the testis and caput and cauda epididymis (Figs. S1B–E). Comparable purity was obtained for caput (98.7%), cauda (99.6%) and testicular sperm (96.0%) when sperm suspensions were treated by the somatic cell lysis buffer (SCLB) for 15 min (Table S1 and Figs. S1F–K). To further improve purity, we increased the SDS concentration from 0.01% to 0.05% and shortened the treatment duration to 5 min for caput and cauda epididymal sperm suspensions, and obtained even higher purity, with only 1–2 contaminating somatic cells per 1,000 caput and cauda epididymal sperm (⩾99.8%) (Table S1 and Figs. S1H–K). To purify testicular sperm, the SDS concentration was increased to 0.025%, and the purity was improved to ~98.5% (Table S1 and Figs. S1F–G). Using these ultrapure sperm samples (Fig. 1A), we isolated small RNAs and constructed libraries for sRNA-seq.
Figure 1. sRNA-seq analyses of small RNA compositions in testicular, caput and cauda epididymal sperm, see alsoFigures S1-S3, andTables S1-S2.
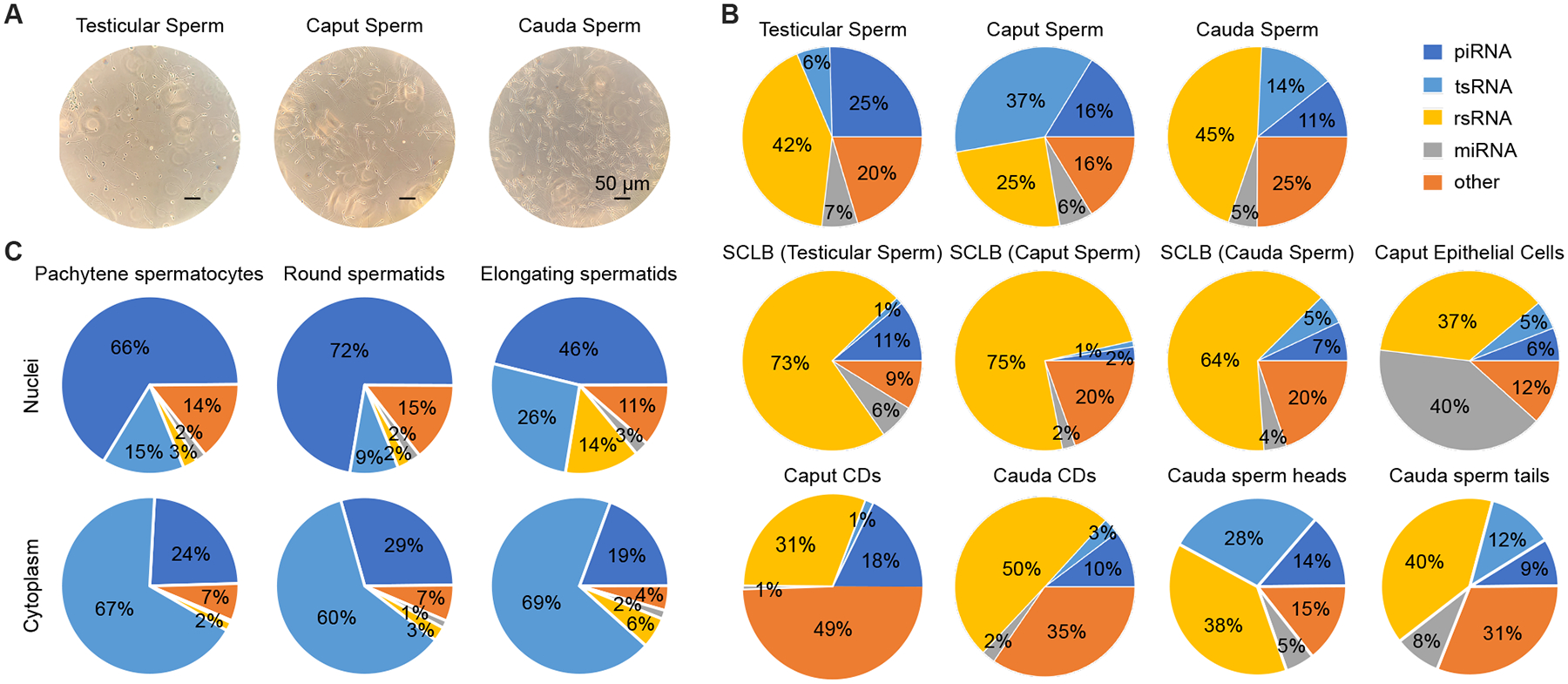
(A) Representative phase-contrast images of sperm after purification with the optimized SCLB method. Scale bar = 50 μm.
(B) Proportional distribution patterns of small RNAs in eleven samples, including purified three types of sperm (testicular, caput, and cauda epididymal), supernatants from the three sperm purification procedures, caput epididymal epithelial cells, CDs purified from caput and cauda epididymal sperm, heads and tails of cauda epididymal sperm. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(C) Proportional distribution patterns of small RNAs in the nuclei and cytoplasm of pachytene spermatocytes, round and elongating spermatids. Two biological replicates were sequenced, with each collected and pooled from two to three mice.
To determine the source of major contaminants in each of the three types of sperm, the supernatants after treatment with corresponding SCLB were also subjected to Srna-seq. Similarly, small RNAs from highly enriched caput epididymal epithelial cells (purity>90%) (Figs. S1L–P) and CDs (purity >98%) (Figs. S1Q–U) of cauda epididymal sperm were also sequenced. In the past, ribosomal RNAs (rRNAs) were often removed before library constructions, thus preventing the identification of rRNA-derived small RNAs (rsRNAs). In the present study, we directly isolated small RNAs from sperm samples for library constructions. SPORTS1.023 was used to determine the abundance and size of four major small RNA species, including miRNAs, piRNAs (PIWI-interacting RNAs), tsRNAs and rsRNAs (Fig. 1B, Figs. S2A–H and Table S2). The proportional distribution pattern of small RNAs was different in the caput epididymal sperm, characterized by increased proportions of tsRNAs and reduced relative abundance of rsRNAs, as compared to testicular and cauda epididymal sperm (Fig. 1B and Table S2). While rsRNAs represented the dominant small RNA species in both the testicular and cauda epididymal sperm, tsRNAs were the major small RNAs in the caput epididymal sperm (Fig. 1B and Table S2). Of interest, the SCLB supernatants from the three sperm preparations displayed similar small RNA proportional distribution patterns, with rsRNAs being the predominant small RNA species, followed by piRNAs and tsRNAs (Fig. 1B and Table S2). The small RNA compositions were more similar between the SCLB supernatants and CDs, but far more different from those in epididymal epithelial cells (Fig. 1B and Table S2), suggesting that the SCLB treatment mainly removed the CDs and that epididymal epithelial cells are not the major source of contamination.
By downloading the sRNA-seq datasets from the previous study reporting that sperm gain small RNAs from the epididymis18,19, we compared the small RNA compositions in testicular, caput and cauda epididymal sperm between the two independent studies (Fig. S2). The small RNA compositions in the caput and cauda epididymal sperm were similar between the two studies (Figs. S2B, C, J and K), and the major difference existed in the testicular sperm (Figs. S2A and I), probably reflecting the different purity of testicular sperm used for sRNA-seq in their (~96.0%) and our (~98.5%) studies. Of interest, while rsRNAs were dominant in both cauda epididymal sperm heads and tails, tsRNAs appeared to be much more enriched in sperm heads than in sperm tails (Fig. 1B). CDs, as a special transient organelle uniquely present in testicular and epididymal sperm, are initially located near the connecting piece/neck of testicular sperm and then migrate gradually toward the junction of the mid to principal piece of sperm flagella24. The membrane covering the CDs is continuous with sperm plasm membrane; therefore, small RNAs in the CDs, in theory, can move back and forth between CDs and sperm, especially the tails and the perinuclear theca of sperm heads. Together with previous studies showing that abundant small RNAs are present in sperm heads and membrane-free sperm nuclei25,26, our data strongly hint that sperm-borne small RNAs, at least those in sperm heads, may have been preloaded during late spermiogenesis, the last phase of spermatogenesis through which round spermatids differentiate into elongating and elongated spermatids. To test this hypothesis, we purified pachytene spermatocytes, round and elongating spermatids, and prepared the nuclei and cytoplasm for sRNA-seq (Fig. 1C). While piRNAs were dominant in the nuclei of the three types of spermatogenic cells, tsRNAs were the most abundant small RNA species in the cytoplasm (Fig. 1C). From pachytene spermatocytes to round and elongating spermatids, the proportions of both tsRNAs and rsRNAs increased, and the small RNA compositions in the nuclei of elongating spermatids started to resemble those of testicular and epididymal sperm. Of great interest, the small RNA compositions in the cytoplasm of elongating spermatids were relatively stable, with tsRNAs and piRNAs being the most dominant small RNA species (Fig. 1C). The small RNA profiles in both cauda epididymal sperm heads and tails showed greater correlation with the nuclei than with the cytoplasm of the three types of spermatogenic cells, especially the elongating spermatids (Fig. S3A), suggesting that sperm-borne small RNAs are mainly derived from the nuclear small RNAs of late spermatids during spermiogenesis. To ensure the reproducibility of our data, we repeated all of our sequencing experiments using samples collected and prepared independently, and similar results were obtained (Fig. S3B).
Among the 21 tsRNAs detected in mouse spermatozoa, four (tsRNA-Asp, tsRNA-Glu, tsRNA-Gly, tsRNA-His) were proportionally enriched in the caput sperm compared to the testicular and cauda epididymal sperm (Fig. 2A and Fig. S4A). Five tsRNAs (tsRNA-Ala, tsRNA-Asn, tsRNA-Ile, tsRNA-Leu and tsRNA-Trp) were enriched in the cauda epididymal sperm (Fig. S4B), whereas the remaining 12 tsRNAs were dominant in testicular sperm (Fig. 2A and Fig. S4C). Using SPORTS1.023, we further mapped the rsRNA reads to six major ribosomal RNA types, including 4.5S, 5S, 5.8S, 18S, 28S, and 45S, thus allowing us to further identify the rsRNA subtypes. Interestingly, the distribution patterns of various rsRNA subtypes were similar among testicular, caput and cauda epididymal sperm (Figs. S4D–F). The rsRNAs identified in sperm were derived from specific regions within various ribosomal RNAs, suggesting that the biogenesis of these rsRNAs is strictly regulated and that these rsRNAs are not intermediates or random degradation products.
Figure 2. Expression and localization of tsRNAs and rsRNAs in testicular, caput and cauda epididymal sperm, see alsoFigure S4.
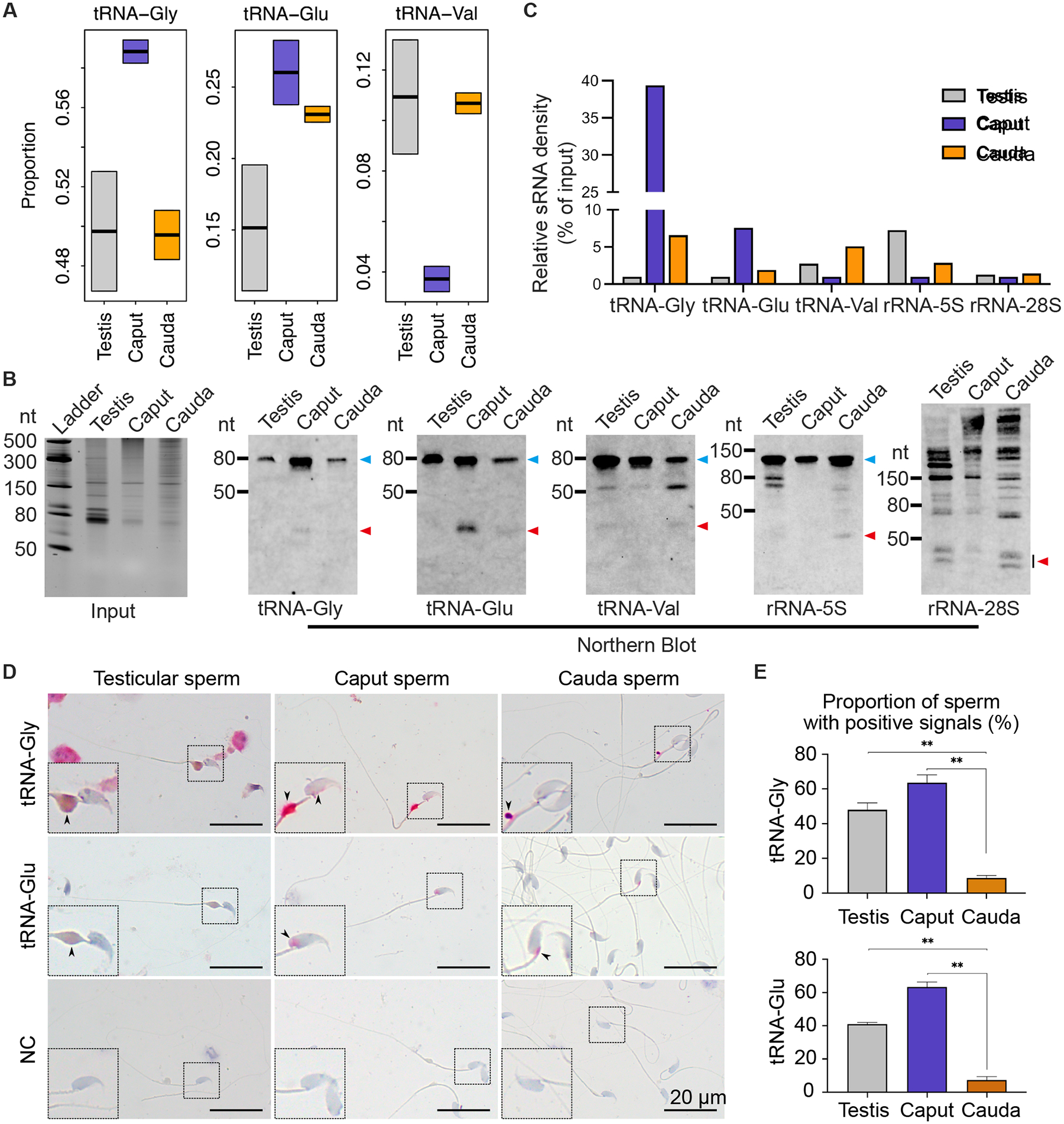
(A) Relative abundance of tRNA-Gly, tRNA-Glu, and tRNA-Val in testicular, caput, and cauda sperm as determined by sRNA-seq. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(B) Representative Northern blots showing expression levels of three tRNAs/tsRNAs and two rRNAs/rsRNAs in testicular, caput, and cauda epididymal sperm. Equal amounts of RNA were loaded, as shown in the input panel to the very left. Blue and red arrowheads point to intact and cleaved tRNAs and rRNAs, respectively. Two biological replicates were conducted, with each collected and pooled from five mice.
(C) Quantitative analyses of Northern blot results from two biological replicates, with each collected and pooled from five mice.
(D) sRNA ISH localization of tRNAs/tsRNAs in testicular, caput, and cauda epididymal sperm. Pink color represents specific hybridization signals. Negative control (NC) yielded no labeling. Insets are digitally enlarged framed areas. Scale bar = 20 μm. Three biological replicates were conducted.
(E) Quantitative analyses of proportions of sperm with specific sRNA ISH signals. At least 300 sperm were counted for each sample. Data are presented as mean ± SEM, n=3. **, p<0.01, Student’s t-test.
To further validate the sRNA-seq data, Northern blots were performed to quantify three tRNAs (tRNA-Glu, tRNA-Gly and tRNA-Val) and their corresponding tsRNAs in testicular, caput and cauda epididymal sperm (Figs. 2B–C). Northern blots showed that levels of both intact tRNA-Glu and tRNA-Gly and their tsRNAs increased when sperm transited from the testis to caput epididymis and decreased from the caput to cauda epididymis. In contrast, tRNA-Val and its tsRNAs showed the opposite trend (Figs. 2B–C). In testicular and cauda epididymal sperm, tRNA-Glu and tRNA-Gly remained largely intact (Figs. 2B–C). Of interest, levels of both the intact tRNA-Gly and its tsRNAs increased in the caput sperm, suggesting that tRNA-Gly is likely gained from other sources rather than active degradation. Both 5S and 28S rsRNAs showed lower levels of cleavage products (i.e., rsRNAs) in the caput epididymis, but higher levels of rsRNAs in testicular and cauda sperm (Figs. 2B–C). To determine the localization of the tsRNAs, small RNA in situ hybridization (sRNA-ISH) was carried out. sRNA-ISH results showed that tRNA-Gly and its tsRNAs were mainly localized to the CDs of testicular, caput, and cauda sperm; lower levels of tRNA-Gly and its tsRNAs were also present in the head of caput sperm but absent in the head of testicular or cauda sperm (Figs. 2D–E). This may explain the increased abundance of both intact tRNA-Gly and its tsRNAs in the caput epididymal sperm, as observed in Northern blots (Figs. 2B–C). The tRNA-Glu and its tsRNAs were mainly localized to the CDs of testicular sperm, but confined to the perinuclear theca in caput and cauda epididymal sperm (Figs. 2D–E). Together, our data show that caput sperm display a different small RNA profile compared to testicular and cauda epididymal sperm, characterized by significant enrichment of tsRNAs, and sperm-borne small RNAs are likely derived from the nuclear small RNAs in late spermatids. The cytoplasm of elongating spermatids contains abundant tsRNAs, whereas the CDs are enriched with rsRNAs.
Sperm gain and lose small RNAs during their transit through the epididymis
The small RNA compositions in caput epididymal sperm differ from those in testicular and cauda epididymal sperm, suggesting dynamic changes in sperm-borne small RNA abundance when sperm transit from the testis to the caput and then to the cauda epididymis. By comparing the differentially expressed small RNAs in testicular and caput epididymal sperm, we found 2,635 significantly up-regulated and 4,075 significantly down-regulated small RNAs, suggesting that when the sperm migrated from the testis to the caput epididymis, 2,635 small RNAs were gained and 4,075 small RNAs got lost (Fig. 3A and Table S3). Similarly, the caput epididymal sperm gained 3,266 and lost 910 small RNAs when reaching the cauda epididymis (Fig. 3A and Table S3). Surprisingly, when testicular sperm were compared with cauda epididymal sperm, only 509 and 829 small RNAs were found to be up- and down-regulated, respectively (Fig. 3A and Table S3). These numbers strongly suggest that the small RNAs gained between the testis and caput epididymis get lost when sperm further move from the caput to cauda epididymis, whereas those lost between the testis and caput epididymis are regained from the caput to cauda epididymis, thus leading to a small net change in sperm small RNA profiles between the testis and cauda epididymis. Indeed, 657 of the 2,635 small RNAs that sperm gained during their migration from the testis to caput epididymis were found among those 910 small RNAs lost when caput epididymal sperm reached the cauda epididymis (Fig. 3B and Table S4), whereas 1,741 of the 4,075 small RNAs lost between the testis and caput epididymis were regained when sperm further move from the caput to cauda epididymis (Fig. 3C and Table S4). 83% (545 out of 657) of the small RNAs that were first gained and later lost were tsRNAs, whereas 99% (1717 out of 1741) of the small RNAs that were lost first and later regained were rsRNAs. Overall, tsRNAs and rsRNAs represent the two major small RNA species accounting for the dynamic changes in small RNA profiles during sperm transit through the epididymis.
Figure 3. Dynamic changes in small RNA composition during sperm epididymal maturation, see alsoTables S3 andS4.
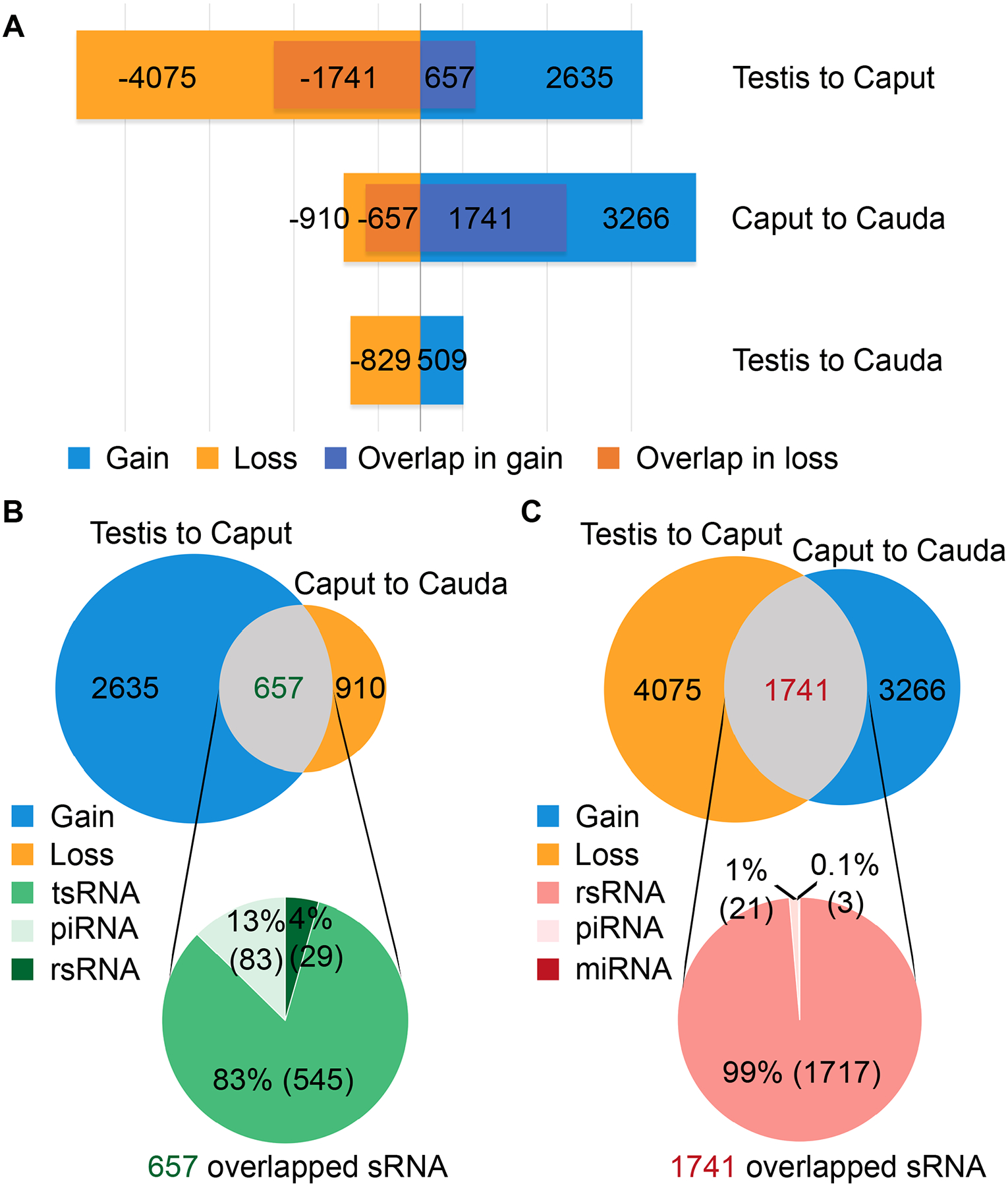
(A) The number of significantly up-regulated (gained) and down-regulated (lost) small RNAs when sperm transit from the testis to the caput and then to cauda epididymis. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(B) The number of overlapping small RNAs between up-regulated small RNAs during testis to caput transition and down-regulated small RNAs during caput to cauda migration. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(C) The number of overlapping small RNAs between down-regulated small RNAs during testis to caput transition and up-regulated small RNAs during caput to cauda migration. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
Cytoplasmic droplets, rather than epididymosomes or epididymal epithelial cells, are the source of the small RNAs gained and lost during sperm transit through the epididymis
Our data suggest that sperm gain and lose small RNAs during their epididymal maturation. Where do those “gained” small RNAs come from? Where do those “lost” small RNAs go? A previous study concluded that epididymosomes are the source of small RNAs, based on the detection of those upregulated tsRNAs in the epididymosomes, and the transfer of small RNAs from the epididymosomes to sperm after incubation of caput sperm with purified epididymosomes in vitro18. While this notion sounds possible, it does not consider that both testicular and epididymal sperm carry CDs, an important organelle uniquely and transiently present in testicular and epididymal sperm27,28. CDs are present on almost all of the testicular sperm, and CDs are gradually lost while sperm transit through the epididymis, with ~70% and ~30% of the caput and cauda epididymal sperm having CDs, respectively27. CDs are completely lost during ejaculation. Consequently, ejaculated sperm do not have CDs, and the ejaculated sperm with retained CDs are mostly functionally defective27. Under physiological conditions, both testicular and epididymal sperm have their CDs attached. However, the SCLB treatment not only lysed the contaminating somatic cells but also removed the CDs (Fig. 1A and Figs. S1F–U). Therefore, it is highly likely that those gained and lost small RNAs are from CDs rather than the epididymal epithelial cells or epididymosomes. To test this hypothesis, we purposely included the purified epididymal epithelial cells and purified CDs in our sequencing analyses. Principal component analyses (PCA) revealed distinct small RNA expression signatures among the three types of sperm and their corresponding supernatants after SCLB treatment (Fig. 4A). Of interest, caput epididymal sperm, caput epididymal epithelial cells and cauda CDs were clustered closely, suggesting similar expression profiles among these three samples (Fig. 4A). Hierarchical clustering analyses on the four major small RNA species indicate that the caput epididymal epithelial cells and cauda CDs contained a large number of tsRNAs and rsRNAs, which were shared by the caput epididymal sperm (Fig. S5), which may explain why the small RNA profiles of the caput sperm appear to be similar to those of the epididymal epithelial cells and CDs. To further determine the source of those “gained” or “lost” small RNAs, we examined the small RNA transcriptomes of CDs, epididymosomes, caput, and cauda epididymides to find their presence. Notably, the vast majority of the small RNAs that were gained or lost during sperm transit through the epididymis were found in CDs, whereas only a small fraction was found in epididymosomes and epididymis (Fig. 4B). We then analyzed correlations among the small RNA transcriptomes of the testicular, caput, and cauda epididymal sperm and those of purified CDs, caput and cauda epididymis, as well as purified epididymosomes (Fig. 4C). Clearly, the sperm small RNA profiles are more closely correlated with those of CDs (R>0.73) than those of epididymosomes (R<0.43) or epididymis (R<0.5) (Fig. 4C). To test whether small RNAs in CDs result from interactions and/or fusion with the epididymosomes, we performed co-immunofluorescent analyses to detect KRT19 (an epididymal epithelial marker) and LDHC (a marker for spermatids and sperm) using the testis, caput and cauda epididymis, as well as cell suspensions from the testis, caput and cauda epididymis. Although it is practically impossible to visualize the epididymosomes using conventional immunofluorescent staining due to their nano-size (30–150nm, as compared to the size of CDs, which is ~1–5μm), an exchange of KRT19 signals would be expected if fusion between CDs and epididymosomes occurs. In the caput and cauda epididymis, KRT19 was detected in the epididymal epithelial cells, whereas LDHC was found in the caput and cauda sperm in the lumen (Fig. 4D). However, no KRT19 signals were observed in sperm, and no LDHC signals were detected in the epididymal epithelial cells. In the testis, LDHC was abundant in spermatids and spermatozoa, whereas KRT19 was absent (Fig. 4D). In the sperm smears prepared using the testis, caput and cauda epididymis, we further confirmed that LDHC was predominately detected in sperm tails and CDs, but no KRT19 signals were observed in either sperm or their CDs (Fig. 4D). These results suggest no interactions between the epididymosomes and CDs; thus, the likelihood that small RNAs are transferred from the epididymosomes to CDs is remote. Our data from immunofluorescent staining of the epididymal epithelial marker, sRNA-seq-based small RNA profiling, Northern blot and sRNA ISH analyses (Figs. 1–2), all support that the changing small RNA compositions in the caput epididymal sperm result from small RNA exchanges between epididymal sperm and their CDs rather than the epididymosomes.
Figure 4. Small RNAs up- or down-regulated in caput epididymal sperm are likely derived from CDs rather than epididymosomes, see alsoFigure S5.
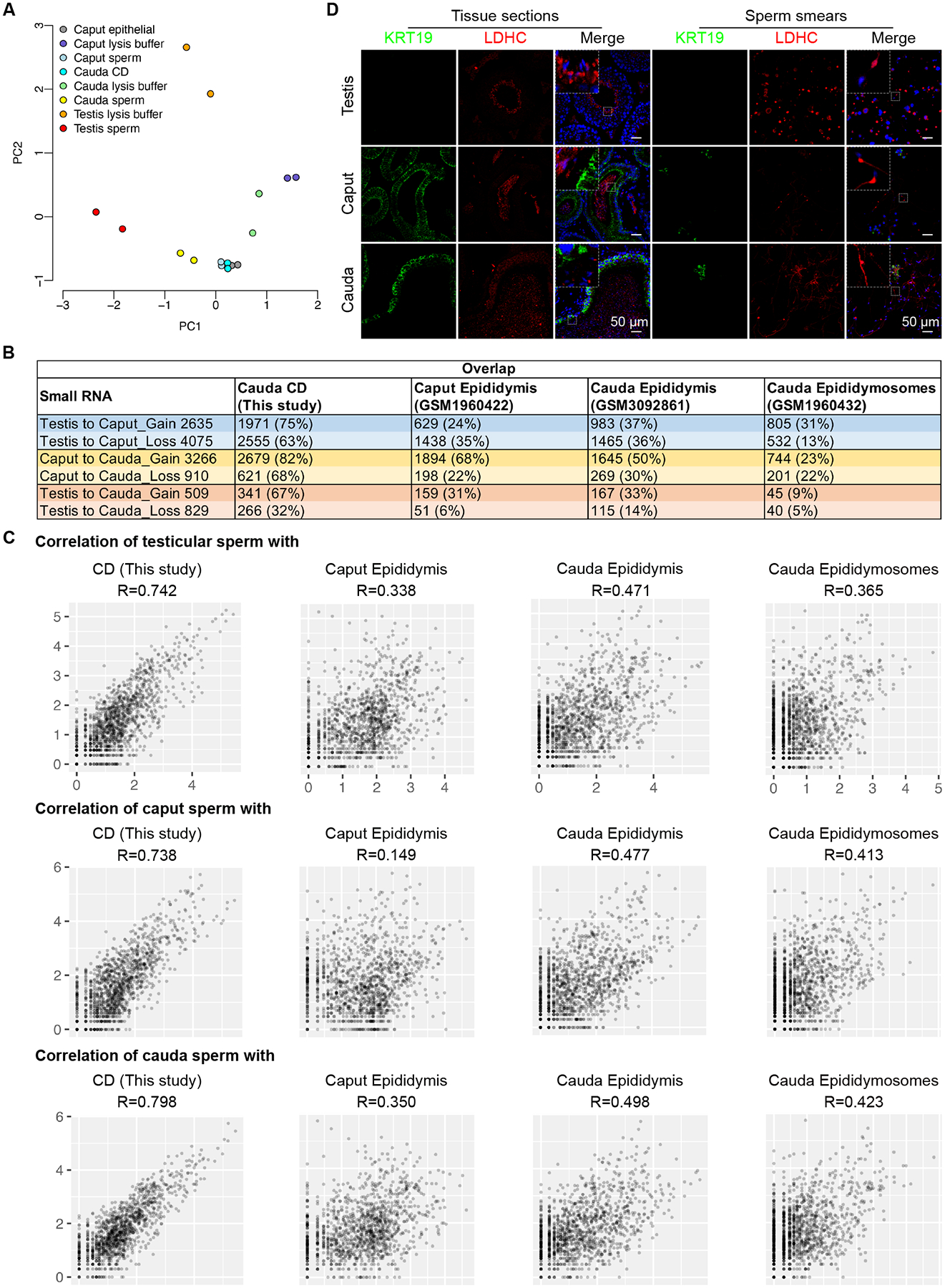
(A) Principal components analysis (PCA) of the small RNA transcriptomes of the eight samples. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(B) Summary of overlapped small RNAs between the gained or lost during sperm transit through the epididymis and small RNAs identified in CDs, caput epididymis, cauda epididymis and epididymosomes. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(C) Scattered plots showing correlations between the small transcriptomes of testicular sperm (top panels), the caput (middle panels) or cauda (bottom panels) epididymal sperm and CD, caput epididymis, cauda epididymis, or epididymosomes. Correlation is in the log10 expression. Two biological replicates were sequenced, with each collected and pooled from at least four mice.
(D) Representative immunofluorescent images of KRT19 and LDHC in testis, caput and cauda epididymis cross-sections and sperm smears. Scale bar = 50 μm. Three biological replicates were conducted.
DISCUSSION
The report that sperm deliver RNAs into the oocytes during fertilization has inspired many studies aiming to determine the exact sperm-borne RNA profiles and their physiological roles5,26,29–34. Most of the published data were from lab animals, especially rodents. Since there are no convenient methods for collecting ejaculated sperm from rats and mice, epididymal sperm are often used to isolate DNA, RNA and proteins for molecular analyses. However, epididymal sperm are not equivalent to ejaculated sperm because epididymal sperm represent immature sperm, which must go through the epididymis to gain fertilizing capability35,36. Both morphological and biochemical changes occur when sperm migrate from the testis to the caput and then to cauda epididymis, e.g., the CDs move downward from the neck region to the junction of the mid to principal piece; the lipid and protein compositions on the sperm plasm membrane change; sperm chromatin becomes more compacted through the formation of disulfate begrudges between protamines associated with epigenetic remodeling36–38. To obtain pure sperm from the testis and epididymis, methods have been developed to remove the common contaminants, including somatic cells and spermatogenic cells18,26. The methods are mostly based on short and gentle treatment of sperm suspensions with low concentrations of detergents since sperm are more resistant to detergent-induced membrane lysis. While the methods allow for obtaining highly pure sperm, the CDs, which are normally attached to the epididymal sperm, are also removed. In other words, the sperm purification protocol commonly used to yield sperm of high purity generates artificial (CD-less), rather than natural (CD-bearing) epididymal sperm, as these purified epididymal sperm no longer have their CDs attached. Consequently, when changes in small RNA compositions are detected among these artificially generated, CD-less sperm, one would naturally come to the conclusion that sperm gain or lose small RNAs from their environment. But the fact is that both testicular and epididymal sperm have CDs, which contain abundant small RNAs and proteins28,39. Small RNAs can be acquired from and discharged to their own CDs. Our data show that this is indeed the case as the vast majority of the small RNAs up- or down-regulated among testicular, caput, and cauda epididymal sperm are mostly present in CDs.
One major difference between the present and our earlier sperm small RNA profiling studies26,40,41 lies in the discovery that rsRNAs, rather than tsRNAs, are the most abundant small RNA species in both testicular and cauda epididymal sperm. This is because ribosome RNA depletion was performed before the construction of RNA libraries on the belief that rRNAs and their degradation products, due to their extremely high abundance, would make the identification of low-abundance small RNAs difficult. Recent studies have revealed that rsRNAs are not simply degradation products of rRNAs, and their biogenesis is strictly regulated, suggesting that rsRNAs may play specific roles, like other small RNA species42–44.
Why do caput sperm display a small RNA profile so different from that in testicular or cauda epididymal sperm? Given that small RNAs can move freely between the sperm and their CDs, the drastic changes in the small RNA composition in caput epididymal sperm may simply reflect the changes in small RNAs in CDs. This appears to be the case, as our sequencing data reveal that tsRNAs are the most abundant in both the cytoplasm and nuclei of elongating spermatids, whereas rsRNAs are the most enriched in the CDs of caput and cauda sperm. Since tsRNAs and rsRNAs are derived from tRNAs and rRNAs, respectively, and both play an essential role in translation, we reasoned that the different small RNA profiles among testicular, caput, and cauda sperm might reflect the translational status. In other words, CDs may persist after the cytoplasm is shed off upon spermiation (i.e., sperm release from the seminiferous epithelium) to maintain the continuous translation of proteins critical for sperm maturation in the epididymis. If the CDs indeed serve as a translational apparatus during sperm transit from the testis to caput epididymis, the enrichment of tsRNAs and rsRNAs in caput sperm may reflect the increased cleavage of tRNAs and rRNAs, which is known to occur during the translational shutdown. This interesting hypothesis is worth testing in future studies.
Where do sperm-borne small RNAs come from? Are they from CDs or preloaded during spermiogenesis? The answer appears to be “both”. To understand small RNA trafficking between sperm and their CDs, it would be ideal to compare the small RNA profiles of testicular, caput and cauda sperm with those of their corresponding CDs. However, it is almost impossible to purify CDs from testicular sperm because the lysis and centrifugation steps for enriching the testicular sperm would have removed their CDs. To test whether sperm-borne small RNA payloads are purposely packed into sperm during spermiogenesis, we purified pachytene spermatocytes, round and elongating spermatids, and then nuclei and cytoplasm were purified for small RNA sequencing. Since CDs are derived from the cytoplasm of elongating and elongated spermatids, we could use the small RNA profiles in the cytoplasm of elongating spermatids to estimate the small RNA profiles in the CDs of testicular sperm. tsRNAs were the most abundant small RNAs in the cytoplasm of elongating spermatids, whereas rsRNAs were the most predominant ones in the CDs of caput and cauda sperm. The abundant tsRNAs and rsRNAs in CDs appear to serve as a reservoir of small RNAs, which could migrate into the sperm tail or maybe even the perinuclear theca of the sperm head. Since the absolute number of non-redundant rsRNAs is much greater than that of tsRNAs, the proportional changes in tsRNAs in the CDs of testicular, caput and cauda sperm may not be perfectly complementary to those in sperm. As CDs are located in the tails of maturing sperm, the small RNAs trafficking between CDs and sperm most likely end up in sperm tails, thereby contributing to the drastic changes in tsRNA and rsRNA abundance in the caput epididymal sperm. Taken together, our data suggest that sperm-borne small RNAs are mainly derived from the nuclear small RNAs in spermatids and sperm small RNA contents can be modified by CD small RNAs during sperm epididymal transit.
What roles do the sperm-borne small RNAs play? Evidence supporting their roles in early embryonic development continues to accumulate, but the more inspiring findings are the possibility of an epigenetic role in transmitting acquired paternal traits. Earlier studies have demonstrated that the sperm small RNAs, including tsRNAs, miRNAs, and rsRNAs, can induce the paternal phenotypes induced by a special diet (high fat or low protein), stress, or kit paramutations in offspring when injected into the wildtype oocytes9–15 although the exact molecular mechanisms remain unknown. If sperm-borne small RNAs indeed mediate epigenetic inheritance, then determining the source of these sperm-borne small RNAs would be of paramount importance as it is critical to understand the underlying mechanism. The notion that sperm gain small RNAs from the epididymosomes during their epididymal transit has gained tremendous attention over the last several years because it represents a pathway allowing the heritable information to flow from soma to the germline. If this pathway indeed exists, then the long-standing Weismann’s barrier theory on heritable information flow would be invalidated. However, our data reported here suggest that the increased proportions of tsRNAs in the caput epididymal sperm are mostly derived from their CDs, not from the epididymosomes. Due to the high abundance of rsRNAs and tsRNAs in both epididymal epithelial cells and CDs, many small RNAs that shuffle between sperm and their CDs are also present in epididymal epithelial cells, but the number is much smaller in epithelial cells than in CDs. Although the possibility that these small RNAs might come from the epididymal epithelial cells or epididymosomes cannot be completely excluded, the major source of the small RNAs that display drastic changes is indeed the CDs. In the physiological context, shuffling small RNAs between sperm and their CDs is more likely than trafficking small RNAs between sperm and epididymosomes. In future studies, one should look into whether testicular and cauda epididymal sperm can transmit the paternal epigenetic phenotypes to offspring with comparable efficiency, as it would tell whether gaining small RNAs from the epididymis is a prerequisite for the successful transmission of acquired traits from the father to his offspring.
In summary, our data strongly support that the sperm-borne small RNA profile is largely set up during late spermiogenesis, and this small RNA profile undergoes a transient change characterized by a significant increase in tsRNA abundance when sperm reach the caput epididymis, but the original profile is largely restored when sperm get to the cauda epididymis. The CDs, rather than the epididymosomes, serve as the source of increased tsRNAs in caput epididymal sperm. Therefore, the notion that sperm gain small RNAs from the epididymis might need to be revisited, and Weissmann’s barrier theory remains valid.
Limitations of the Study
It is noteworthy that the quantitative analyses of small RNAs in the present study were based on proportional changes of all small RNAs detected in sperm and other samples. As millions of small RNA reads can be detected in sperm, small changes in the absolute abundance of a particular small RNA may not be physiologically significant. Therefore, we focused on the proportional changes of four major small RNA species (miRNAs, piRNAs, tsRNAs and rsRNAs). Consequently, only correlations among various samples were analyzed based on their total small RNA profiles. Absolute quantification of individual sRNAs can be achieved by using the same amount of sperm supplemented with an equal amount of spike-ins during library construction. However, it would not change the interpretation of our data and the conclusion drawn. Another technical limitation lies in the conventional sRNA library construction method used in the present study, which requires adaptor ligation to the 3’ and 5’ ends of the small RNAs before reverse transcription. The RNA modifications on the 3’ end (e.g., 2’,3’-cyclic phosphate or 3’-phosphate) and in the middle (e.g., N1-methyladenosine (m1A)) may hinder the ligation and reverse transcription, respectively. Therefore, new methods that use AlkB enzyme and/or T4 Polynucleotide Kinase to reduce RNA modifications45,46 and/or group II intron/non-LTR reverse transcriptases (e.g., TGIRT III (InGex), BoMoC47, or Induro (NEB)) with high-processive and template switching/jumping activities to bypass the RNA modifications and to eliminate the ligation steps may help obtain more comprehensive sRNA profiles.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Wei Yan (wei.yan@lundquist.org).
Materials availability
Unique materials generated in this study are available from the lead contact without restriction.
Data and code availability
The RNA-seq data have been deposited into the National Center for Biotechnology Information Sequence Read Achieve database (accession no. PRJNA815339). Any additional information relating to the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Laboratory mice
Adult (8–12 weeks of age) FVB male mice used in this study were housed in a temperature and humidity-controlled, specific pathogen-free facility under a light-dark cycle (12:12 light-dark) with food and water ad libitum. Animal use protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nevada, Reno, and is in accordance with the “Guide for the Care and Use of Experimental Animals” established by the National Institutes of Health (1996, revised 2011).
METHOD DETAILS
Collection of spermatozoa
Testicular, caput and cauda epididymal sperm were collected as described18. Briefly, the epididymides were dissected and the fat pads and overlying connective tissue were removed (Fig. S1B). Caput and cauda epididymis were carefully dissected and placed in a human tubal fluid (HTF) medium (FUJIFILM Irvine Scientific, Cat#90125) at 37°C. To collect caput sperm, multiple small incisions were made at the proximal end of the caput, followed by piercing the remaining tissue with a 26G needle to release the caput luminal contents. To collect cauda sperm, the cauda epididymis was gently squeezed with a tweezer to release the luminal contents. Sperm-containing medium was incubated at 37°C for 30 min, then sperm were collected and transferred to a new tube. For testicular spermatozoa collection, testes were finely minced in a 35 mm Petri dish containing 1ml 150 mM NaCl and the contents were dispersed by vigorous pipetting and then subjected to the downstream purification procedures.
Purification of spermatozoa
Four to six male mice were used in each preparation of sperm suspensions from the testis, caput and cauda epididymis. The caput and cauda epididymal sperm suspensions were first filtered through a 40 μm cell strainer (Olympus) to remove tissue blocks and then gently washed with 1× PBS. After centrifugation at 1000× g for 5 min, the sperm suspension was split into three aliquots: the pellet from the first aliquot was resuspended in the SCLB (0.01% SDS, 0.005% Triton-X) followed by incubation on ice for 10 min as described18. The pellet from the second aliquot was resuspended with the same SCLB (0.01% SDS, 0.005% Triton-X) followed by incubation on ice for 15 min. The pellet from the third aliquot was resuspended in a different SCLB (0.05% SDS and 0.005% Triton-X) followed by incubation on ice for 3–5 min. After treatment, sperm were pelleted by centrifugation at 3000× g for 5 min. The supernatants were collected and the sperm pellets were washed with 1× PBS. The purity of the sperm samples was determined by microscopic examination.
For testicular sperm purification, the sperm-containing medium was first filtered through a 40 μm cell strainer. Next, the collected cell suspension was loaded onto 10 ml of 52% isotonic Percoll (Sigma-Aldrich, Cat.# P1644) then centrifuged at 12000× g for 15 min at 10°C. The pellet was resuspended in 5 ml of 150 mM NaCl spun at 600× g at 4°C for 10 min, and then divided four equal fractions to pellet sperm by centrifugation: one was washed with 75 mM NaCl solution 3 times followed by treatment with SCLB (0.01% SDS, 0.005% Triton-X); the other three groups were directly subjected to treatment with SCLB at three different concentrations (0.01%, 0.0125% and 0.025% SDS) by incubation on ice for 10 min, 5 min and 5 min, respectively. After SCLB treatment, sperm were pelleted by centrifugation at 3000× g 5 min. After collecting the supernatants, the sperm pellets were washed with 1× PBS. The purity of the sperm samples was determined by microscopic examination.
Preparation of highly pure epididymal epithelial cells
The caput epididymis was cut into small pieces (2 mm × 2 mm) and washed in HBSS to remove spermatozoa. Isolation of high enriched epithelial cells was performed as described54. In brief, the caput epididymal cell suspension was cultured in DMEM for 4 hours. The non-epithelial cells, such as blood cells and fibroblasts became attached to the culture dish, whereas the epithelial cells and a few sperm remained in suspension. After centrifugation at 800× g for 5 min, the pellet was resuspended in SCLB followed by incubation on ice for 10 min. After centrifugation at 3000× g for 5 min to remove the sperm, the supernatants were collected for RNA extraction for RNA-seq.
CDs purification
Adult FVB male mice were euthanized and the cauda epididymides were dissected and transferred into 2.5 ml of HTF. The epididymides were further dissected into smaller pieces (5 mm × 5 mm) followed by incubation in a humidity incubator at 37°C for about 30 min. The spermatozoa-containing supernatants (2 ml) were collected and subjected to CDs purification using a discontinuous sucrose density gradient centrifugation as described28.
Purification of nuclei and cytoplasm
Pachytene spermatocytes, round and elongating spermatids were purified from adult testes using the STA-PUT method as described55. The purified spermatogenic cells were lysed in 1ml of the nucleus lysis buffer (Sigma-Aldrich, Cat.# NUC101-1KT, 1:10 dilution in 10mM Tris-HCl (pH 7.4), 10mM NaCl, 3mM MgCl2, and 1× PBS) on ice for 1–3 minutes, and stopped by adding 6 ml 4% BSA-PBS. The lysate was centrifuged at 500× g for 5 min at 4 °C. The pellets containing the nuclei were washed with 1× PBS followed by storage at −80°C, and the supernatants containing cytoplasmic contents were stored at −80°C.
Isolation of sperm heads and tails
Sperm were washed in 1× PBS and resuspended in 5 ml NIM consisting of 121.6 mM KCl, 7.8 mM NaH2PO4, 1.4 mM KH2PO4, 10 mM EDTA disodium salt, and 0.01% PVA. The pH was adjusted to 7.2 using 1 M KOH. Sperm suspensions were sonicated on ice using a sonicator (Misonix XL-2000) with the power setting at 2 for 5s until >95% of sperm heads and tails were dissociated. Sonicated sperm heads and tails were loaded onto 62% (w/v) sucrose diluted in 1× PBS followed by centrifugation using the SW28Ti rotor (Beckman, Cat.# 342207) at 5,000 rpm (~4,500 × g) and 4°C for 10 min. After centrifugation, the heads were pelleted at the bottom of the tube and the tails were located in the interface. The sperm heads and tails were collected into separate tubes, followed by washing with 1× PBS. The pellets were collected and stored at −80°C for later use.
Northern blot
Northern blot was conducted as described56,57 with modifications. In brief, azide-labeled DNA oligos were ordered from IDT and covalently bonded to IRDye 680RD DBCO Infrared Dye (Li-Cor, Cat.# 929-50005) by incubating them at room temperature in 1× PBS in the dark. 15% Novex™ TBE-Urea Gels (Thermo Fisher Scientific, Cat.# EC6885BOX) was prerun at 300 V for 30 min. 150 ng of sperm total RNA was mixed with an equal volume of 2× Formamide Loading Dye (Thermo Fisher Scientific, Cat.# AM1560) followed by denaturing at 95°C for 3 min, and immediately chilling on ice. The denatured RNA samples were separated by the polyacrylamide gel electrophoresis at 180 V for 1h in 1× TBE buffer, and transferred onto BrightStar™-Plus Positively Charged Nylon Membrane (Thermo Fisher Scientific, Cat.# AM10100) using Novex™ Semi-Dry Blotter (Thermo Fisher Scientific, Cat.# SD1000) in the 0.5×TBE buffer at 250 mA for 60 min. The membrane was briefly dried on a papertowel and crosslinked twice at 120 mJ/cm2 using Spectrolinker™ XL-1500 UV crosslinker (Spectronics Corporation). The crosslinked membrane was pre-hybridized in 10 ml ExpressHyb hybridization solution (Clontech, Cat.# 636832) at 42°C for 30 min, followed by incubation in the IRDye 680RD-labeled probe diluted in ExpressHyb hybridization solution (1pmol/ml) at 42°C overnight. After washing with 2× SSC (Saline-sodium citrate) buffer (Sigma-Aldrich, Cat.# S6639) containing 0.1% SDS (Sigma-Aldrich, Cat.# 71736) and a second wash with 1× SSC-0.1% SDS wash buffer for 10 min at room temperature, the membrane was visualized by ChemiDoc MP Imaging System (Bio-Rad, Cat.# 12003154) at IRDye 680RD wavelength (700 nm). The band intensity was quantified using Fiji software. Probes used in this study are listed in Table S5.
Small RNA in situ Hybridization (sRNA-ISH)
sRNA-ISH was performed using the miRNAscope™ HD reagent kit (ACD biotechne, Cat.# QG324500) according to the manufacturer’s instructions. Briefly, sperm were fixed in 4% paraformaldehyde (Thermo Fisher Scientific, Cat.# J19943-K2) for 15 min at room temperature, then spread onto Superfrost Plus slides (Thermo Fisher Scientific, Cat.# 22-037-246), dried at room temperature, and stored at 4°C until further use. The slides were incubated at 4°C for 20 min in the −20°C prechilled 100% Acetone (Fisher Scientific, Cat.# A949-1), then proceeded to two changes of 100% ethanol (Fisher Scientific, Cat.# HC-800-1GAL) for 2 min each time with agitation, followed by post-fixation for 16 h in 10% Normal buffered Formalin (Sigma-Aldrich, Cat.# HT501128-4L) at room temperature. After quenching the endogenous peroxidase with RNAscope® Hydrogen Peroxide for 10 min, the slides were treated with 1× RNAscope® Target Retrieval Solution (ACD biotechne, Cat.# 322000) in a Hamilton Beach steamer preheated to at least 99°C for 15 min, followed by a 30-min incubation in Protease III at 40°C in an ACD HybEZ™ II Hybridization System (ACD biotechne, Cat.# 321710-R). Then the slides were hybridized with probes from ACD biotechne at 40°C for 2 h in the ACD HybEZ™ II Hybridization System, followed by signal amplification, red signal detection, counterstain with 50% hematoxylin (Sigma-Aldrich, Cat.# MHS32-1L), and bluing at 0.02% Ammonium Hydroxide (Sigma-Aldrich, Cat.# 221228-500ML-PCA). The slides were dried in a 60°C dry oven for 15 min, then dipped in two changes of 100% xylene (Fisher Scientific, Cat.# HC-700-1GAL) for 2 min before mounting with Vectamount (Vector Lab, Cat.# H-5000). Images were taken under 100× Ph2 magnification.
Immunofluorescence
Immunofluorescent staining of sperm and CDs was performed as described previously28. Briefly, sperm and the purified CD were fixed in 4% paraformaldehyde for 15 min (Thermo Fisher Scientific, Cat.# J19943-K2), then spread onto Superfrost Plus slides (Thermo Fisher Scientific, Cat.# 22-037-246), air-dried, and washed with 1× PBS 3 times (5 min/wash). Heat-induced antigen retrieval was performed in 0.01 M citric acid buffer with high power for 4 min once, and three times with low power for 4 min. Slides were cooled down to room temperature and washed with 1× PBS 3 times (5 min/wash). Following permeabilization with 0.5% Triton X-100 (Sigma-Aldrich, Cat.# T8787) in PBS for 5 min at room temperature, the slides were washed with 1× PBS 3 times (5 min/wash), and blocked with 1% BSA at room temperature for 30 min. Then the slides were incubated with primary antibodies at 4°C overnight. The goat anti-LDHC antibody (Thermo Fisher Scientific, Cat.# PA5-18779) and the rabbit anti-Cytokeratin 19 (KRT19) antibody (Abcam, Cat.# ab52625) were used at dilutions of 1:500. After overnight primary antibody incubation, the slides were moved to room temperature for 30 min before 3 washes with 1× PBS (10 min/wash). Slides were then incubated with the Donkey anti-Goat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 594 (for LDHC, 1: 500 in PBS, Thermo Fisher Scientific, Cat.# A32758) and Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 (for Cytokeratin 19, 1:1000 in PBS, Thermo Fisher Scientific, Cat.# A32790) at room temperature for 1 h with gentle shaking, followed by 3 times 1x PBS washes (10 min/wash). Finally, the slides were mounted and counterstained in Antifade Mounting Medium with DAPI (Vector Lab, Cat.# H-1800). Images were taken using a confocal microscope.
Total RNA extraction
Sperm pooled from four mice were subjected to RNA extraction using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Cat#AM1560). The Qubit RNA High Sensitivity Assay Kit (Invitrogen, Cat#Q32855) was used to quantify the extracted RNA on a Qubit 2.0 Fluorometer (Invitrogen).
RNA library construction
Small RNA libraries were constructed using NEBNext® Small RNA Library Prep Set for Illumina® (Multiplex Compatible) (NEB, Cat#E7330L) according to the manufacturer’s instructions, as described previously58. Duplicate samples were used with each collected and pooled from four mice. The libraries were sequenced using HiSeq 2500 system for single-end 50 bp sequencing.
QUANTITATION AND STATISTICAL ANALYSIS
RNA-Seq data analysis
The small RNA-seq data were parsed and annotated using the SPORTS tool with one mismatch tolerance23. Briefly, SPORTS was applied to generate trimmed sequencing reads by removing adapter sequences. The trimmed sequencing reads with lengths out of the defined range were excluded from further analyses. The remaining trimmed reads were next aligned against miRBase, rRNA sequences, GtRNAdb, piRBase, and piRNABank, respectively48–51. Both tsRNAs and rsRNAs were grouped into individual subcategories according to their parent RNAs, i.e., tRNA and rRNAs. We used the edgeR tool to prioritize the small RNA species that were differentially expressed between groups52. The TMM algorithm was applied to perform the sequencing read count normalization and effective library size estimation53. The correlation in RNA expression between individual samples was plotted by the GGally package within the R platform. The hierarchical clustering and heatmaps were generated by the gplots package within the R platform.
Statistical analyses
Statistical analyses were performed using the SPSS program (IBM, SPSS) and shown as mean ± standard error of the mean (SEM). Statistical differences between the two groups were assessed by the Student’s t-test. *, **, *** and ns represent p< 0.05, p< 0.01, p< 0.001 and no significance, respectively.
Supplementary Material
Table S1. The average ratio of somatic cell contaminations in sperm after SCLB treatment, related to STAR Methods and Figure 1.
The sheet shows ratios of somatic cell contaminations in testicular, caput, and cauda sperm after different SCLB treatments.
Table S2. Small RNA-seq datasets, related to Figure 1.
Sheets show raw read counts for all eight libraries (sperm from testis, caput and cauda epididymis; SCLB in testicular, caput and cauda sperm; caput epithelial cells and cauda CDs).
Table S3. Gain and loss of small RNA during sperm epididymal transit, related to Figure 3.
Sheets show significantly up-regulated (gained) and down-regulated (lost) small RNAs when sperm transit from the testis to the caput and then to cauda epididymis.
Table S4. Overlapping small RNA that sperm first gained then lost and those first lost then regained, related to Figure 3.
Sheets show the overlapping small RNAs between up-regulated small RNAs during testis to caput transition and down-regulated small RNAs during caput to cauda migration, as well as between down-regulated small RNAs during testis to caput transition and up-regulated small RNAs during caput to cauda migration.
Table S5. Probes used in this study, related to STAR Methods.
Figure 5. Schematics showing exchanges of tsRNAs and rsRNAs between testicular/epididymal sperm and their CDs during sperm epididymal maturation.
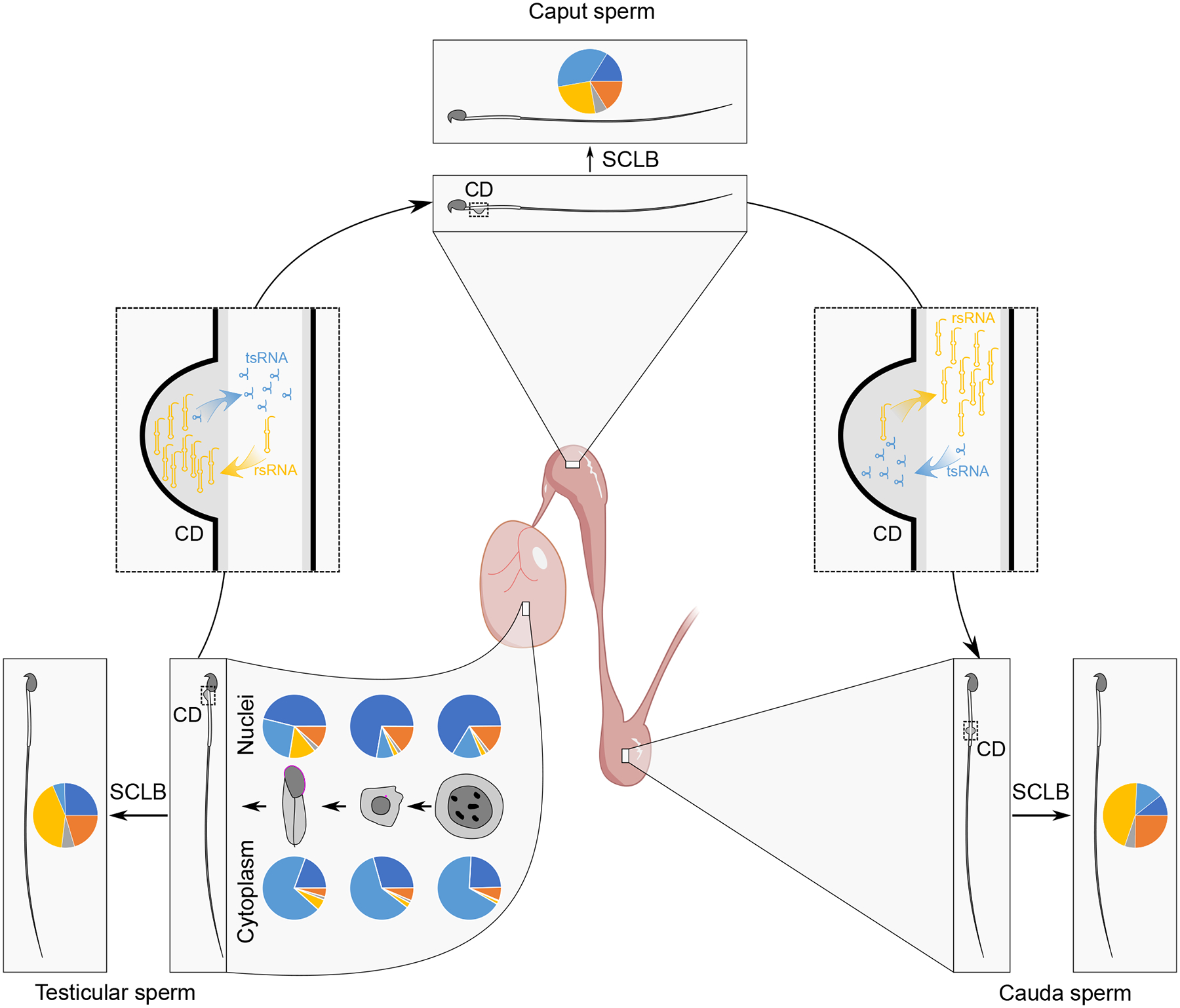
From pachytene spermatocytes to round and then elongating spermatids, the small RNA proportional distribution patterns are very different between the nucleus and cytoplasm, and the nuclear small RNA profile is gradually shaped into that of the testicular sperm during murine spermiogenesis. Once testicular sperm reach the caput epididymis, numerous tsRNAs shift from CDs to the sperm flagellum and neck regions, whereas sperm rsRNAs translocate into CDs. The opposite movements of tsRNAs and rsRNAs occur between sperm and CDs when sperm further migrate from the caput to cauda epididymis, leading to similar small RNA compositions between testicular and cauda epididymal sperm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-LDHC | Thermo Fisher Scientific | Cat.# PA5-18779 |
| Rabbit anti-Cytokeratin 19 | Abcam | Cat.# ab52625 |
| Donkey anti-Goat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 594 | Thermo Fisher Scientific | Cat.# A32758 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 | Thermo Fisher Scientific | Cat.# A32790 |
| Chemicals, peptides, and recombinant proteins | ||
| Percoll | Sigma-Aldrich | Cat.# P1644 |
| IRDye 680RD DBCO Infrared Dye | Li-Cor | Cat.# 929-50005 |
| DAPI | Vector Lab | Cat.# H-1800 |
| Human tubal fluid medium | FUJIFILM Irvine Scientific | Cat.# 90125 |
| 15% Novex™ TBE-Urea Gels | Thermo Fisher Scientific | Cat.# EC6885BOX |
| 2× Formamide Loading Dye | Thermo Fisher Scientific | Cat.# AM1560 |
| Advanced Cell Strainers, 40μm | Olympus | Cat.# 25-375 |
| 4% paraformaldehyde | Thermo Fisher Scientific | Cat.# J19943-K2 |
| 100% Acetone | Fisher Scientific | Cat.# A949-1 |
| 100% ethanol | Fisher Scientific | Cat.# HC-800-1GAL |
| 10% Normal buffered Formalin | Sigma-Aldrich | Cat.# HT501128-4L |
| BrightStar™-Plus Positively Charged Nylon Membrane | Thermo Fisher Scientific | Cat.# AM10100 |
| Novex™ Semi-Dry Blotter | Thermo Fisher Scientific | Cat.# SD1000 |
| ExpressHyb hybridization solution | Clontech | Cat.# 636832 |
| 2× SSC (Saline-sodium citrate) buffer | Sigma-Aldrich | Cat.# S6639 |
| SDS | Sigma-Aldrich | Cat.# 71736 |
| 1× RNAscope® Target Retrieval Solution | ACD biotechne | Cat.# 322000 |
| ACD HybEZ™ II Hybridization System | ACD biotechne | Cat.# 321710-R |
| 50% hematoxylin | Sigma-Aldrich | Cat.# MHS32-1L |
| Ammonium Hydroxide | Sigma-Aldrich | Cat.# 221228-500ML-PCA |
| 100% xylene | Fisher Scientific | Cat.# HC-700-1GAL |
| Vectamount | Vector Lab | Cat.# H-5000 |
| Triton X-100 | Sigma-Aldrich | Cat.# T8787 |
| Critical commercial assays | ||
| mirVana miRNA isolation kit | ThermoFisher | Cat.# AM1560 |
| Nuclei Isolation Kit | Sigma-Aldrich | Cat.# NUC101-1KT |
| Qubit RNA high sensitivity assay kit | Invitrogen | Cat.# Q32855 |
| NEBNext® small RNA library prep set | Illumina | Cat.# E7330L |
| miRNAscope™ HD reagent kit | ACD biotechne | Cat.# QG324500 |
| Deposited data | ||
| RNA-seq data | This paper | PRJNA815339 |
| Experimental models: Cell lines | ||
| NIH/3T3 cells | ATCC | CRL-1658 |
| Experimental models: Organisms/strains | ||
| Mouse: FVB | Charles River Laboratories | Strain Code: 207 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| SPORTS | Shi et al.23 | https://github.com/junchaoshi/sports1.1 |
| miRBase | Kozomara and Griffiths-Jones.48 | http://www.mirbase.org/ |
| GtRNAdb | Chan and Lowe.49 | http://gtrnadb.ucsc.edu/ |
| piRBase | Zhang et al.50 | http://www.regulatoryrna.org/database/piRNA/ |
| piRNABank | Lakshmi and Agrawal.51 | http://pirnabank.ibab.ac.in/ |
| edgeR | Robinson et al.52 | http://bioconductor.org/ |
| TMM algorithm | Robinson and Oshlack.53 | N/A |
| SPSS | IBM | https://www.ibm.com/analvtics/spss-statistics-software |
Highlights.
Sperm-borne small RNAs are mainly derived from late spermatids in the testis
The small RNA profile changes when sperm transit through the epididymis
Sperm exchange small RNAs with cytoplasmic droplets (CDs) rather than epididymosomes
ACKNOWLEDGMENTS
This work was supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) (HD071736, HD0085506, HD098593, HD099924 and P30GM110767 to WY), National Center for Advancing Translational Science (NCATS) UCLA CTSI (UL1TR001881-01 to ZW and WY), and John Templeton Foundation (PID: 61174 to WY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, and Krawetz SA (2004). Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429, 154. 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Meng N, Wang Y, Zhou T, Li M, Wang S, Chen S, Zheng H, Kong S, Wang H, and Yan W (2022). Ablation of the miR-465 Cluster Causes a Skewed Sex Ratio in Mice. Front Endocrinol (Lausanne) 13, 893854. 10.3389/fendo.2022.893854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santiago J, Silva JV, Howl J, Santos MAS, and Fardilha M (2021). All you need to know about sperm RNAs. Hum Reprod Update 28, 67–91. 10.1093/humupd/dmab034. [DOI] [PubMed] [Google Scholar]
- 4.Godia M, Swanson G, and Krawetz SA (2018). A history of why fathers’ RNA matters. Biol Reprod 99, 147–159. 10.1093/biolre/ioy007. [DOI] [PubMed] [Google Scholar]
- 5.Le Blevec E, Muronova J, Ray PF, and Arnoult C (2020). Paternal epigenetics: Mammalian sperm provide much more than DNA at fertilization. Mol Cell Endocrinol 518, 110964. 10.1016/j.mce.2020.110964. [DOI] [PubMed] [Google Scholar]
- 6.Sharma U (2019). Paternal Contributions to Offspring Health: Role of Sperm Small RNAs in Intergenerational Transmission of Epigenetic Information. Front Cell Dev Biol 7, 215. 10.3389/fcell.2019.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jodar M (2019). Sperm and seminal plasma RNAs: what roles do they play beyond fertilization? Reproduction 158, R113–R123. 10.1530/REP-18-0639. [DOI] [PubMed] [Google Scholar]
- 8.Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, and Yan W (2016). Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143, 635–647. 10.1242/dev.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan S, Oliver D, Schuster A, Zheng H, and Yan W (2015). Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice. Sci Rep 5, 9266. 10.1038/srep09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, and Rassoulzadegan M (2009). The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development 136, 3647–3655. 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 11.Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, and Rassoulzadegan M (2008). RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell 14, 962–969. 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, and Cuzin F (2006). RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474. 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 13.Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, and Mansuy IM (2014). Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17, 667–669. 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodgers AB, Morgan CP, Leu NA, and Bale TL (2015). Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A 112, 13699–13704. 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400. 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 16.Yan W (2014). Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol Cell Endocrinol 398, 24–30. 10.1016/j.mce.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, Yan W, and Duan E (2016). Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 17, 733–743. 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, and Rando OJ (2018). Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev. Cell 46, 481–494.e486. 10.1016/j.devcel.2018.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S AE (1893). Das Keimplasma Eine Theorie der Vererbung. Nature 47, 265–266. 10.1038/047265a0. [DOI] [Google Scholar]
- 21.Surani MA (2016). Breaking the germ line-soma barrier. Nat Rev Mol Cell Biol 17, 136. 10.1038/nrm.2016.12. [DOI] [PubMed] [Google Scholar]
- 22.Cappallo-Obermann H, and Spiess AN (2016). Comment on “Absence of sperm RNA elements correlates with idiopathic male infertility”. Sci Transl Med 8, 353tc351. 10.1126/scitranslmed.aaf2396. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Ko EA, Sanders KM, Chen Q, and Zhou T (2018). SPORTS1.0: A Tool for Annotating and Profiling Non-coding RNAs Optimized for rRNA- and tRNA-derived Small RNAs. Genomics Proteomics Bioinformatics 16, 144–151. 10.1016/j.gpb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Yuan S-Q, Zheng Z-H, and Yan W (2013). The cytoplasmic droplet may be indicative of sperm motility and normal spermiogenesis. Asian journal of andrology 15, 799–805. 10.1038/aja.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan W, Morozumi K, Zhang J, Ro S, Park C, and Yanagimachi R (2008). Birth of Mice after Intracytoplasmic Injection of Single Purified Sperm Nuclei and Detection of Messenger RNAs and MicroRNAs in the Sperm Nuclei1. Biology of Reproduction 78, 896–902. 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- 26.Schuster A, Tang C, Xie Y, Ortogero N, Yuan S, and Yan W (2016). SpermBase: A Database for Sperm-Borne RNA Contents. Biol Reprod 95, 99. 10.1095/biolreprod.116.142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Yuan SQ, Zheng ZH, and Yan W (2013). The cytoplasmic droplet may be indicative of sperm motility and normal spermiogenesis. Asian J Androl 15, 799–805. 10.1038/aja.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan S, Zheng H, Zheng Z, and Yan W (2013). Proteomic analyses reveal a role of cytoplasmic droplets as an energy source during epididymal sperm maturation. PLoS One 8, e77466. 10.1371/journal.pone.0077466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi E, Stermer A, Nolan T, Li H, Hall S, Boekelheide K, Sigman M, and Hwang K (2021). Highly conserved sperm function-related transcripts across three species: human, rat and mouse. Reprod Toxicol 104, 44–51. 10.1016/j.reprotox.2021.06.012. [DOI] [PubMed] [Google Scholar]
- 30.El Fekih S, Nguyen MH, Perrin A, Beauvillard D, Morel F, Saad A, Ben Ali H, and De Braekeleer M (2017). Sperm RNA preparation for transcriptomic analysis: Review of the techniques and personal experience. Andrologia 49. 10.1111/and.12767. [DOI] [PubMed] [Google Scholar]
- 31.Ostermeier GC, Dix DJ, Miller D, Khatri P, and Krawetz SA (2002). Spermatozoal RNA profiles of normal fertile men. Lancet 360, 772–777. 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- 32.Sellem E, Jammes H, and Schibler L (2021). Sperm-borne sncRNAs: potential biomarkers for semen fertility? Reprod Fertil Dev 34, 160–173. 10.1071/RD21276. [DOI] [PubMed] [Google Scholar]
- 33.Tarozzi N, Nadalini M, Coticchio G, Zaca C, Lagalla C, and Borini A (2021). The paternal toolbox for embryo development and health. Mol Hum Reprod 27. 10.1093/molehr/gaab042. [DOI] [PubMed] [Google Scholar]
- 34.Boerke A, Dieleman SJ, and Gadella BM (2007). A possible role for sperm RNA in early embryo development. Theriogenology 68 Suppl 1, S147–155. 10.1016/j.theriogenology.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 35.Zhou W, De Iuliis GN, Dun MD, and Nixon B (2018). Characteristics of the Epididymal Luminal Environment Responsible for Sperm Maturation and Storage. Front Endocrinol (Lausanne) 9, 59. 10.3389/fendo.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dacheux JL, and Dacheux F (2014). New insights into epididymal function in relation to sperm maturation. Reproduction 147, R27–42. 10.1530/REP-13-0420. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan R, and Mieusset R (2016). The human epididymis: its function in sperm maturation. Hum Reprod Update 22, 574–587. 10.1093/humupd/dmw015. [DOI] [PubMed] [Google Scholar]
- 38.Hermo L, Oliveira RL, Smith CE, Au CE, and Bergeron JJM (2019). Dark side of the epididymis: tails of sperm maturation. Andrology 7, 566–580. 10.1111/andr.12641. [DOI] [PubMed] [Google Scholar]
- 39.Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ, Tang Y, Zhang X, Qin W, Zhang Y, et al. (2020). m(6)A-dependent biogenesis of circular RNAs in male germ cells. Cell Res 30, 211–228. 10.1038/s41422-020-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song R, Hennig GW, Wu Q, Jose C, Zheng H, and Yan W (2011). Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci U S A 108, 13159–13164. 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T, Xie Y, Tang C, Wang Y, Yuan S, Zheng H, and Yan W (2021). Dnmt2-null sperm block maternal transmission of a paramutant phenotypedagger. Biol Reprod 105, 603–612. 10.1093/biolre/ioab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert M, Benmoussa A, and Provost P (2019). Small Non-Coding RNAs Derived From Eukaryotic Ribosomal RNA. Noncoding RNA 5. 10.3390/ncrna5010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. (2017). A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62. 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu S, et al. (2021). PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol 23, 424–436. 10.1038/s41556-021-00652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12, 835–837. 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, and Lowe TM (2015). ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12, 879–884. 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Upton HE, Ferguson L, Temoche-Diaz MM, Liu XM, Pimentel SC, Ingolia NT, Schekman R, and Collins K (2021). Low-bias ncRNA libraries using ordered two-template relay: Serial template jumping by a modified retroelement reverse transcriptase. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2107900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozomara A, and Griffiths-Jones S (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42, D68–73. 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan PP, and Lowe TM (2016). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–189. 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang P, Si X, Skogerbo G, Wang J, Cui D, Li Y, Sun X, Liu L, Sun B, Chen R, et al. (2014). piRBase: a web resource assisting piRNA functional study. Database (Oxford) 2014, bau110. 10.1093/database/bau110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sai Lakshmi S, and Agrawal S (2008). piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 36, D173–177. 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kierszenbaum AL, Lea O, Petrusz P, French FS, and Tres LL (1981). Isolation, culture, and immunocytochemical characterization of epididymal epithelial cells from pubertal and adult rats. Proc Natl Acad Sci U S A 78, 1675–1679. 10.1073/pnas.78.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo M, Luo C, Wang Z, Chen S, Morris D, Ruan F, Chen Z, Yang L, Wei X, Wu C, et al. (2022). Uncoupling transcription and translation through miRNA-dependent poly(A) length control in haploid male germ cells. Development 149. 10.1242/dev.199573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fields C, Sheng P, Miller B, Wei T, and Xie M (2019). Northern Blot with IR Fluorescent Probes: Strategies for Probe Preparation. Bio Protoc 9. 10.21769/BioProtoc.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, McSwiggin H, Newkirk SJ, Wang Y, Oliver D, Tang C, Lee S, Wang S, Yuan S, Zheng H, et al. (2019). Insertion of a chimeric retrotransposon sequence in mouse Axin1 locus causes metastable kinky tail phenotype. Mob DNA 10, 17. 10.1186/s13100-019-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Xie Y, Wang Y, Morris D, Wang S, Oliver D, Yuan S, Zayac K, Bloomquist S, Zheng H, and Yan W (2020). X-linked miR-506 family miRNAs promote FMRP expression in mouse spermatogonia. EMBO Rep. 21, e49024. 10.15252/embr.201949024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The average ratio of somatic cell contaminations in sperm after SCLB treatment, related to STAR Methods and Figure 1.
The sheet shows ratios of somatic cell contaminations in testicular, caput, and cauda sperm after different SCLB treatments.
Table S2. Small RNA-seq datasets, related to Figure 1.
Sheets show raw read counts for all eight libraries (sperm from testis, caput and cauda epididymis; SCLB in testicular, caput and cauda sperm; caput epithelial cells and cauda CDs).
Table S3. Gain and loss of small RNA during sperm epididymal transit, related to Figure 3.
Sheets show significantly up-regulated (gained) and down-regulated (lost) small RNAs when sperm transit from the testis to the caput and then to cauda epididymis.
Table S4. Overlapping small RNA that sperm first gained then lost and those first lost then regained, related to Figure 3.
Sheets show the overlapping small RNAs between up-regulated small RNAs during testis to caput transition and down-regulated small RNAs during caput to cauda migration, as well as between down-regulated small RNAs during testis to caput transition and up-regulated small RNAs during caput to cauda migration.
Table S5. Probes used in this study, related to STAR Methods.
Data Availability Statement
The RNA-seq data have been deposited into the National Center for Biotechnology Information Sequence Read Achieve database (accession no. PRJNA815339). Any additional information relating to the data reported in this paper is available from the lead contact upon request. This paper does not report original code.


