Abstract
Optogenetic techniques have been developed to allow control over the activity of selected cells within a highly heterogeneous tissue, using a combination of genetic engineering and light. Optogenetics employs natural and engineered photoreceptors, mostly of microbial origin, to be genetically introduced into the cells of interest. As a result, cells that are naturally light-insensitive can be made photosensitive and addressable by illumination and precisely controllable in time and space. The selectivity of expression and subcellular targeting in the host is enabled by applying control elements such as promoters, enhancers and specific targeting sequences to the employed photoreceptor-encoding DNA. This powerful approach allows precise characterization and manipulation of cellular functions and has motivated the development of advanced optical methods for patterned photostimulation. Optogenetics has revolutionized neuroscience during the past 15 years and is primed to have a similar impact in other fields, including cardiology, cell biology and plant sciences. In this Primer, we describe the principles of optogenetics, review the most commonly used optogenetic tools, illumination approaches and scientific applications and discuss the possibilities and limitations associated with optogenetic manipulations across a wide variety of optical techniques, cells, circuits and organisms.
Light-dependent processes are abundant in nature, occurring in diverse organisms from bacteria and algae to plants and animals, and are used for energy capture and storage, to regulate developmental processes and to mediate orientation1–3. Although the photoreceptors involved in light-sensing have been studied for decades, the use of such proteins for actuation of naturally light-insensitive cells began only in 2002 with the expression of the Drosophila rhodopsin and its associated signalling proteins in neurons4. The discovery of channelrhodopsin (ChR), identified in the same year in the green alga Chlamydomonas, in conjunction with the almost universal cellular availability of the chromophore all-trans retinal (vitamin A) in most cells and organisms, accelerated the progress of this new technology. Almost in parallel with the initial application of channelrhodopsin 2 (ChR2) in isolated neurons in 2005 (REF.5) and brain slices in 2006 (REF.6), ChRs were rapidly adapted for use in living model organisms, including chicken embryos7 and Caenorhabditis elegans in 2005 (REF.8), Drosophila in 2006 (REF.9), freely moving mice in 2007 (REF.10), zebrafish in 2008 (REF.11) and even non-human primates in 2009 (REF.12). The first experiments that pointed towards potential therapeutic applications were performed in 2006, pioneered by the expression of ChR in inner retinal cells to restore vision in blind mice13. Optogenetics is based on sensory photoreceptor sequences from microalgae, fungi or bacteria. The combination of photoreceptor-encoding DNA with control elements such as promoters and targeting sequences, typically derived from genes expressed selectively in the target tissue, allows the protein specificity not only in the choice of target cell population but also in the subcellular compartments to be manipulated. The DNA constructs are incorporated into target cell populations, tissues or living organisms using vectors such as plasmids, viral vectors or bacteria using established transformation technologies (FIG. 1).
Fig. 1 |. Principles of optogenetics.

DNA encoding a sensory photoreceptor derived from a microorganism, plant or animal (orange) is cloned under regulation of control elements that allow targeting of specific host cells (blue), packed into a vector such as a viral vector or bacteria and injected into the tissue, organ or organism of interest. Targeted cell (orange) now expresses light-sensitive protein and can be controlled with light in various ways, depending on the specific photoreceptor expressed.
The robust function and revolutionary utility of ChR2 in neuroscience resulted in the description and application of many photoreceptor subtypes, engineered or retrieved from genomic or cDNA databases, progress in protein expression and targeting, microelectrode and optrode technology and, finally, the combination of optogenetic actuators with optical fluorescent reporter systems and high-resolution subcellular imaging, accelerating the interdisciplinary growth of optogenetic technology with unprecedented pace. The need to control neuronal activity with increased spatial resolution has, in turn, motivated the development of advanced optical methods for patterned photostimulation. Digital mirror devices or liquid crystal spatial light modulators coupled to single-photon or two-photon excitation have enabled single and multi-target excitation in vitro and in vivo with single-spike precision and cellular resolution in head-restrained and freely moving animals14. Optogenetics has developed as a basic science methodology for dissecting biological functions; whereas it was initially adopted by neuroscientists to study brain function and dysfunction, optogenetics has expanded into new research fields such as cardiology, microbiology, immunology, parasitology and plant science. These developments are culminating in highly anticipated clinical applications, as envisioned in the early days of optogenetics, including multiple clinical trials currently in progress for selected human disorders. A crude timeline of key breakthroughs in optogenetic technology is displayed in FIG. 2.
Fig. 2 |. The optogenetic actuator toolbox.

a | Key advances in development of optogenetic tools. Not all available tools are highlighted. Major developments are shown above the arrow, and first applications of channelrhodopsins (ChRs) to model organisms including humans are shown below. b | Tools for optogenetic manipulation of membrane voltage and local ion concentrations (top), second messenger, G-protein signalling and kinase signalling (middle) and the light-controlled interaction of photoreceptors with tethered partner proteins for subcellular application (bottom). Light–oxygen–voltage (LOV) domain-based dimerizers expose an ‘aged’ signalling peptide after light-triggered unfolding of the Jɑ-helix251. Cryptochrome 2 (Cry2) and phytochrome B (PhyB) interact with CiBN or PIF domains after blue or red light absorption, respectively252,253. c | Commonly used optogenetic tools for excitation or inhibition of neuronal activity including cation-conducting ChRs eTsChR254, Cheriff203, CoChR30, CrChR2TC (REF.255), ChroME77 and derivatives, SSFO/Soul120,256, ChRmine, bReaChES257 and f-Chrimson115, chloride and potassium-conducting ChRs (for example, GtACR1, GtACR2 (REF.217) and HcKCR1 (REF.22)), inward directed proton pumps (for example, NsXeR258) and outward-directed proton, sodium and chloride pumps (for example, Arch3.0 (REF.169), eKR2 (REF.259), eNpHR3.0 (REF.260)), all plotted according to their peak excitation wavelength and temporal kinetics. d | Soluble enzyme bPAC38 and rhodopsin–guanyl cyclase CaRhGC67 produce cAMP and cGMP following illumination, whereas non-bleaching opsins mOPN4 (REF.51), eOPN3 (REF.57), PPO56 and JellyOP50 activate different G-protein pathways. e | Genetically encoded sensors with diverse excitation spectra (x axis) can be used to monitor changes in Ca2+ voltage and pH, such as GCaMP and R-CaMP156 and FRGeco261 for Ca2+ (REF.262), ASAP3 (REF.263), Voltron264, VARNAM265, Quasar203 and Archon266 for voltage, and pHluorin267 and pHmScarlet for pH. In experiments combining sensors and actuators, both need to be chosen carefully to minimize optical crosstalk. ec, extracellular; GPCR, G-protein-coupled receptor; ic, intracellular; optoGPCR, hybrid between structurally related opsin and GPCR; RTK, receptor tyrosine kinase.
With the growth of optogenetic technology came an abundance of tools with diverse functional properties. This Primer is focused predominantly on rhodopsin-based optogenetic tools, which are the most widely used within the growing optogenetic toolbox. Although the differences between tools can be subtle, their spectral sensitivity, kinetic properties and ion selectivity can have a major influence on the outcome of an optogenetic experiment. Understanding these features and careful design are therefore crucial for the success and interpretability of optogenetic experiments. As the technology matures and gains popularity across multiple fields of biology, this Primer aims to provide experimentalists with the most relevant knowledge needed to design, perform and interpret optogenetic experiments.
Experimentation
Optogenetic experiments are based on the combination of several fundamental components: a genetically encoded actuator that, after reconstitution with an organic molecule serving as a chromophore, responds to light and can be used to influence the function of the tool-expressing cell or tissue in a light-dependent manner; a light source providing light at the appropriate wavelength and intensity; and a light delivery system, which allows for illumination of targeted cells for temporally precise activation of the optogenetic actuator. Together, these components allow the experimenter to modulate the biological system and interrogate its function.
Selecting the correct actuator
When designing an optogenetic experiment, the first considerations should be the cellular parameter to be modulated and the available optogenetic actuators for such an endeavour. An enormous number of light-switchable tools have been developed for controlling ion fluxes and membrane voltage, G-protein signalling, regulation of second messengers such as Ca2+, cAMP, cGMP, IP3 and receptor tyrosine kinases (RTKs), organelle repositioning, transcription and translation (FIG. 2). Most actuators rely on photoreceptors or light-sensing modules of natural origin, although photoswitchable synthetic organic compounds have also been employed15. The use of photoswitchable synthetic organic compounds is also known as chemo-optogenetics or photopharmacology, and the interested reader might consult related reviews15,16. Many light-modulated actuators have been described that do not rely on opsin proteins. Whereas this Primer is focused on the opsin-based toolbox, the reader might find more information about non-opsin-based optogenetic tools in several excellent recent reviews17,18.
Light-activated ion channels.
Until recently, the most widely applied optogenetic photoreceptor was ChR2 from the alga Chlamydomonas reinhardtii (known as CrChR2 or simply C2) and its variant ChR2-H134R8,19. Presently, almost 900 ChR sequences have been identified, including many with properties superior to those of the original prototypes20 (FIG. 2). ChRs may be subdivided into cation or anion-conducting channels, termed CCRs and ACRs, respectively. CCRs typically conduct multiple types of cations with high preference for protons. Na+ selectivity varies widely among different CCRs21 and divalent cations are only poorly conducted under most physiological host conditions. Whereas there are no Ca2+-selective CCRs available to date, continuous metagenomic screening recently revealed a new class of potassium-selective channels (KCRs)22. ACRs are selective for numerous anions, similar to most human anion channels23.
In host cells, Na+ and H+-conducting CCRs can be used as depolarizing actuators, whereas the action of ACRs depends on the chloride reversal potential in the targeted cells or subcellular compartment (Box 1). ACRs may clamp the voltage to near the resting potential and inhibit action potential firing by shunting inhibition (Box 1). However, in cardiac cells, immature neurons and presynaptic terminals, chloride gradients are less pronounced and ACRs may depolarize the cell membrane24,25. In plants, the chloride gradient is always directed outward, and ACR activation will generally lead to membrane depolarization. Thus far, KCRs have been applied under highly controlled in vitro conditions, but — once established for in vivo experiments — hold major promise for optogenetic inhibition in all variants of cells and host model systems.
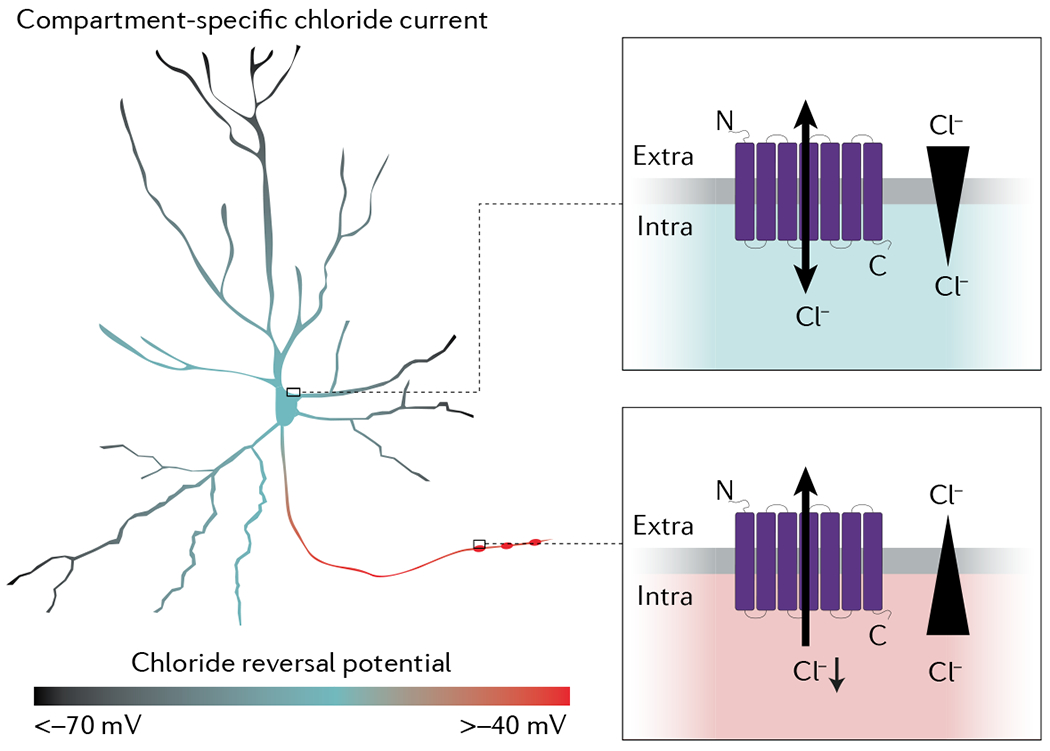
Box 1 |. Compartment-specific functions of microbial rhodopsins.
Anion-conducting channels (ACRs) (such as GtACR2, iC++ and iChloc) can be used to control chloride conductance in a light-dependent manner. However, the effect on neuronal excitability depends on the chloride reversal potential, which can differ between subcellular compartments, as well as on the membrane potential. The figure (left) shows a neuron coloured according to the typical chloride reversal potential. In the somatic compartment (top inset), shunting inhibition occurs if the reversal potential of a channel is close to the resting membrane potential of the cell. In this case, channel activation does not lead to a substantial current but, rather, to a conductance that will oppose any change of the membrane potential from the channel reversal potential, referred to as shunting conductance. The input resistance of the neuron is reduced, causing a smaller amplitude of subsequent excitatory potentials. If the channel reversal potential lies between the resting potential and the action potential threshold, however, the effects of shunting inhibition are more complex. In this case, ACR activation will lead to a depolarization, but excitatory potential amplitudes will still be reduced. Consequently, ACR activation will still lead to a reduced spike rate. Upon channel closing, the input resistance then increases while the membrane potential is still depolarized, leading to a transiently increased excitability. Furthermore, if ACRs are activated over extended periods of excitatory drive, Cl− can accumulate in the cell, and the depolarizing phase of shunting inhibition will become more accentuated, leading to activity-dependent effects of shunting inhibition. At the presynaptic terminal (bottom), ACR activation leads to depolarization, and potentially even action potential initiation, especially at light onset when the pool of activatable voltage-gated sodium channels is large. ACR-based optogenetic manipulations should thus take the unique features of compartment-specific physiology into account.
Our current molecular understanding of ChRs mostly relies on CrChR2, which has been extensively studied and modified with respect to kinetics, ion selectivity, inactivation and absorption wavelength26,27, revealing principles that have been successfully transferred to other CCRs (see Supplementary Fig. 1). Recently discovered ChRs such as ChRmine and KCRs belonging to a new family of cation-conducting ChRs hold great promise, but understanding of their molecular mechanism is only beginning to emerge28,29. The maximal colour sensitivity of known ChRs so far spans from 445 nm for TsChR to 610 nm for the ChrimsonSA mutant and Ruby-ACR30–32 (FIG. 2). Such distinct colour sensitivity may allow the combination of different ChRs within the same experiment for activation and inhibition of the same or different cells. However, all rhodopsins absorb blue or UVA light to a certain extent due to transition to higher excited state levels. This has to be taken into consideration when combining multiple rhodopsins in single or multiple cell populations (see Supplementary Fig. 2). For bidirectional voltage modulation, for example, the more potent actuator should be selected to absorb at the shorter wavelength (FIG. 2), thereby allowing for lower light powers used in the blue range, which will, in turn, minimize the undesired activation of the red-shifted actuator. Another consideration is the reversal potential of the conducted ion. In nature, as well as in neuronal experiments, ACRs operate closer to the reversal potential than Na+ or H+-conducting depolarizing CCRs. Although it is possible to co-express two opsins using two separate viral vectors, this approach inevitably leads to incomplete co-expression. To overcome this drawback, several constructs have been engineered that allow tandem expression of two opsins from the same vector. The most prominent examples are eNPAC, which co-expresses eNpHR3.0 and ChR2(H134R) initially linked by a 2A self-cleaving peptide33, and BiPOLES34, which combines the red-shifted CCR Chrimson with the blue-shifted GtACR2 in a single targeting-optimized fusion construct. Owing to the stoichiometric membrane expression, equal photocurrents near the cellular resting potential and comparable light sensitivities of both channel modules, BiPOLES outperforms previous bi-cistronic combinations of ChR2 with different ion pumps35,36 and guarantees subcellular co-localization and selective red-light excitation for multicolour applications. A combination of optogenetics and chemogenetics has been exemplified by direct fusion of slow cycling step function rhodopsins (SFOs) with a luciferase that produces light upon peripheral injection of its small-molecule substrate. These luminopsins allow direct light stimulation by optical fibres, while at the same time providing chemogenetic access in awake and anaesthetized animals in vivo37,38.
Light-driven pumps.
The first application of optogenetics for neuronal silencing was achieved with the chloride pump halorhodopsin39. However, since the discovery of ACRs, the interest in optogenetic silencing of animal cells by light-driven pumps has decreased in animal cells because pumps require higher expression levels and higher light intensities for sufficient ion turnover (see Supplementary Fig. 3). By contrast, in plants — which naturally hyperpolarize their membranes and drive secondary transporters via H+ pumps — light-driven H+ pumps are valuable tools. The advantage of light-driven pumps is their high ion specificity and robust electric response that depends less on the ionic composition of the surrounding buffers and the membrane voltage. Light-driven chloride pumps such as NpHR40 or Jaws41 allow reliable — although often weak — neuronal inhibition in synaptic terminals, where the action of ACRs is difficult to predict owing to variable and elevated intracellular chloride concentrations42. Pumps may be successfully used in small compartments such as neuronal vesicles, lysosomes43, mitochondria or thylakoids, where the action of ion channels is poorly defined owing to the lack of free ions44. In the plasma membrane, the use of light-driven ion pumps requires caution because both proton and chloride pumps can drive non-physiological ion concentrations in neurons and trigger off-target effects, including a transient increase of the chloride reversal potential, leading to excitatory actions of the inhibitory neurotransmitter GABA and alkalization of presynaptic terminals, leading to increased spontaneous neurotransmission42,45.
Optogenetic control of biochemical signalling pathways.
Animal rhodopsins are G-protein-coupled receptors (GPCRs) and animal vision is the most studied G-protein signalling pathway. A pioneering study demonstrated that bovine rhodopsin expression may be used to activate G-protein signalling in Xenopus oocytes but without describing the signalling mechanism46. However, the off response of rod-rhodopsins remained uncontrollable in the absence of rhodopsin kinase and arrestin, and responses severely declined upon repetitive stimulation. The responses of Gi/o activating cone rhodopsins47–49 or Gs-specific box jellyfish opsins50 declined faster, but were still not tightly controllable. The solution was approached by revitalizing melanopsin OPN4, which can be switched on and off with blue and yellow light, albeit incompletely owing to substantial overlapping spectra of the dark state and signalling state51–53, and only the UV-sensitive Lamprey parapinopsin (PPO) with its green-absorbing signalling state offered efficient on and off switching with a dual-colour light source54–57.
GPCR signalling depends on many properties of the receptors, including substrate binding kinetics, G-protein specificity and timing of activation and receptor inactivation, which in total cannot be fully mimicked by rhodopsins. One way to more selectively mimic the activity of a specific GPCR is to engineer hybrids between structurally related opsins and GPCRs (optoGPCRs)58,59. OptoGPCRs open new, and possibly more specific, routes for the analysis of intracellular signalling pathways compared with unmodified rhodopsins, whereas the dynamics of G-protein coupling and pathway recruitment still has to be approached by testing various expression levels and light regimes. However, these optoGPCRs cannot be simply transferred to another cell type because G-protein promiscuity might activate unwanted pathways51,60. With optoGPCRs, the application of G-protein activation has enormously broadened the optogenetic actuator toolbox. These tools will be well suited for temporally defined modulation of non-excitable cells, potentially including glial cells in the brain and other non-neuronal cell types61.
RTKs are another large family of cell surface receptors that sense growth factors and hormones to regulate various cellular behaviours by target phosphorylation. Engineered light-sensitive epidermal growth factor receptor (EGFR1) and the fibroblast growth factor receptor 1 (FGFR1) have shown robust light activation of both RTKs and cellular signalling in human cancer and endothelial cells, and faithful mimicking of complex mitogenic and morphogenic cell behaviour35. The cobalamin-binding domain (CBD) and tropomyosin receptor kinase B (TRKB) have been fused to RTKs to yield light-sensitive receptors36,37. Fusions with TRKB have high specificity for the target proteins, although their application range is narrow and the constructs need to be optimized for every new application. Moreover, one drawback is that cobalamin-based light sensors or phytochrome-based light sensors generally require addition or cellular synthesis of the cofactor molecules, making their potential for in vivo applications more complex than the application of the retinal-based photoreceptors.
Second messengers.
Photoactivated cyclases (PACs) have been employed for direct control of the second messengers cAMP and cGMP. The soluble bPAC from Beggiatoa spp. is a tandem of BLUF-type light sensors (blue light sensors using FAD (flavin adenine dinucleotide)) (FIG. 2) with carboxy-terminal adenylyl cyclases. These optogenetic actuators show millisecond-range on-kinetics upon photostimulation and second-range off-kinetics in the dark (bPAC τoff = 12 s)38. Co-expression of bPAC with the small prokaryotic potassium channel SthK (PAC-K silencer) in two-component optogenetic approaches has been exploited for long-lasting neuronal hyperpolarization in cardiomyocytes (CM) as well as in fly, mouse and zebrafish neurons, providing high operational light sensitivity but low time resolution38,62–64. However, colour modification is only possible within a small range around 470 nm and occasional residual dark activity has been observed65. New spectral windows were opened by introducing rhodopsin guanylyl cyclases (RGCs), which are cyclases with amino-terminally linked rhodopsins. These rhodopsin cyclases (RhCs) are characterized by low dark activity, effective light absorption (ε > 32,000 M−1 cm−1) and the promise of flexible colour tuning66–68. RhCs show millisecond-range off-kinetics, are naturally GTP selective and are convertible into ATP cyclases by genetic engineering. Some members of the fungal Chytridiomycota may use heterodimeric RhGCs, with one blue or green sensitive rhodopsin catalyst, and a second near-infrared sensitive modulator (NeoR, λmax = 660–700 nm). These NeoRs might allow to extend the usable spectral range into the superior infrared spectral window69.
Protein abundance.
Control over the concentration of selected proteins within a cell has been a long-standing goal and has stimulated the interest of protein engineers for decades. The most obvious point of intervention is the regulation of transcription. Previously explored concepts were based on the connection of DNA-binding proteins to a photoreceptor such as Phytochrome, FKF1 or VIVID (light–oxygen–voltage (LOV) proteins), or CRY. Upon illumination, these photoreceptors bind to their signalling partner proteins PIF3, GIGANTEA/Tulips or CIB, respectively, with bound components of the transcription machinery as VP16 or VP64. In light, the transcription component is attracted to the promoter region of interest by the photoreceptor and signal–protein interaction, leading to the assembly of the transcription complex and initiation of transcription. However, the chosen GAL4 DNA-binding domains have to be incorporated into the model organism (reviewed elsewhere70,71). To address any promoter of interest, programmable DNA-binding proteins such as zinc finger DNA-binding proteins72, TALEs73 and deactivated Cas9 have been functionalized as the second generation of transcription regulators74,75. The main caveat for Cas9 application is the prolonged occupancy of Cas9 at its DNA-binding site, especially in situations where the DNA is not cleaved, which disturbs gene expression prior to the intended start of the experiment76. Inserting a LOV domain into an anti-CRISPR protein such as AcrIIA4 or AcrIIC3 (CASANOVAs) overcomes this problem and makes Cas9 binding better controllable. This approach works reliably in HEK cells, but has not been rigorously tested for non-embryonic cells such as neurons77.
Targeting strategies
Optogenetics was first applied in neuroscience, driven by the complexity of neural circuits and the demand for improved selectivity in perturbational approaches for studying neural circuits. Genetic techniques, viral vector technology and optical methods have grown rapidly around the developing optogenetic toolbox. As a result, the tools and enabling technologies for optogenetic experimentation in neuroscience, as well as the fundamental understanding of the caveats and constraints of their application, are more advanced in neuroscience than in other fields. In the following section, we review some of the major targeting approaches for expression of optogenetic tools in neural circuits.
One of the major benefits of the optogenetic paradigm is its selectivity to defined cells and circuits. In neuroscience applications, genetic targeting of optogenetic tools has advanced considerably, and has profited greatly from developments in viral vector technologies. As optogenetic tools are genetically encoded and mostly single-component actuators (requiring the introduction of only one gene to the target cell population), multiple delivery methods can be used to introduce them into the cells of interest. Targeting strategies are either based on promoter specificity directly or through a combination of a conditional transgene expression cassette that can be switched on or off using a recombinase.
Transgenic expression of optogenetic tools.
Transgenic expression is the simplest approach to implement as it requires only the maintenance of an opsin-expressing animal strain78 (FIG. 3a) or the crossing of two strains of animals. The latter involves a driver line — engineered to express a recombinase or transcription regulator such as Cre and Flpo (in rodents) or a Gal4 driver (in zebrafish) in a particular cell population — and an animal strain expressing a conditional opsin gene under the control of the relevant driver (FIG. 3b). The F1 progeny of such a cross will express the opsin gene in all cells in which the driver protein is expressed and will therefore be amenable to optogenetic manipulation simply by illuminating the targeted brain region. The approach is simple to implement, but one should consider potential caveats, including the presence of axons from neurons in other brain regions, which might be activated along with the cell bodies in the illuminated region. In mice, expression of ChR2 or eNpHR3.0 from the ROSA26 locus79 can be quite weak and not universally sufficient to drive activity in every neuron subtype. Expression of opsin genes from the TIGER locus80 showed stronger opsin expression and might, therefore, be useful for some target neuron populations. However, this approach requires generation and/or breeding of a dedicated animal strain for every targeted neuron population, and thus lacks the versatility and cost-efficiency of viral vector-based approaches. Another potential confound is unintentional targeting in some driver lines (see for instance REF.81), making the verification of driver lines advisable82.
Fig. 3 |. Cell type-specific targeting of optogenetic tools.
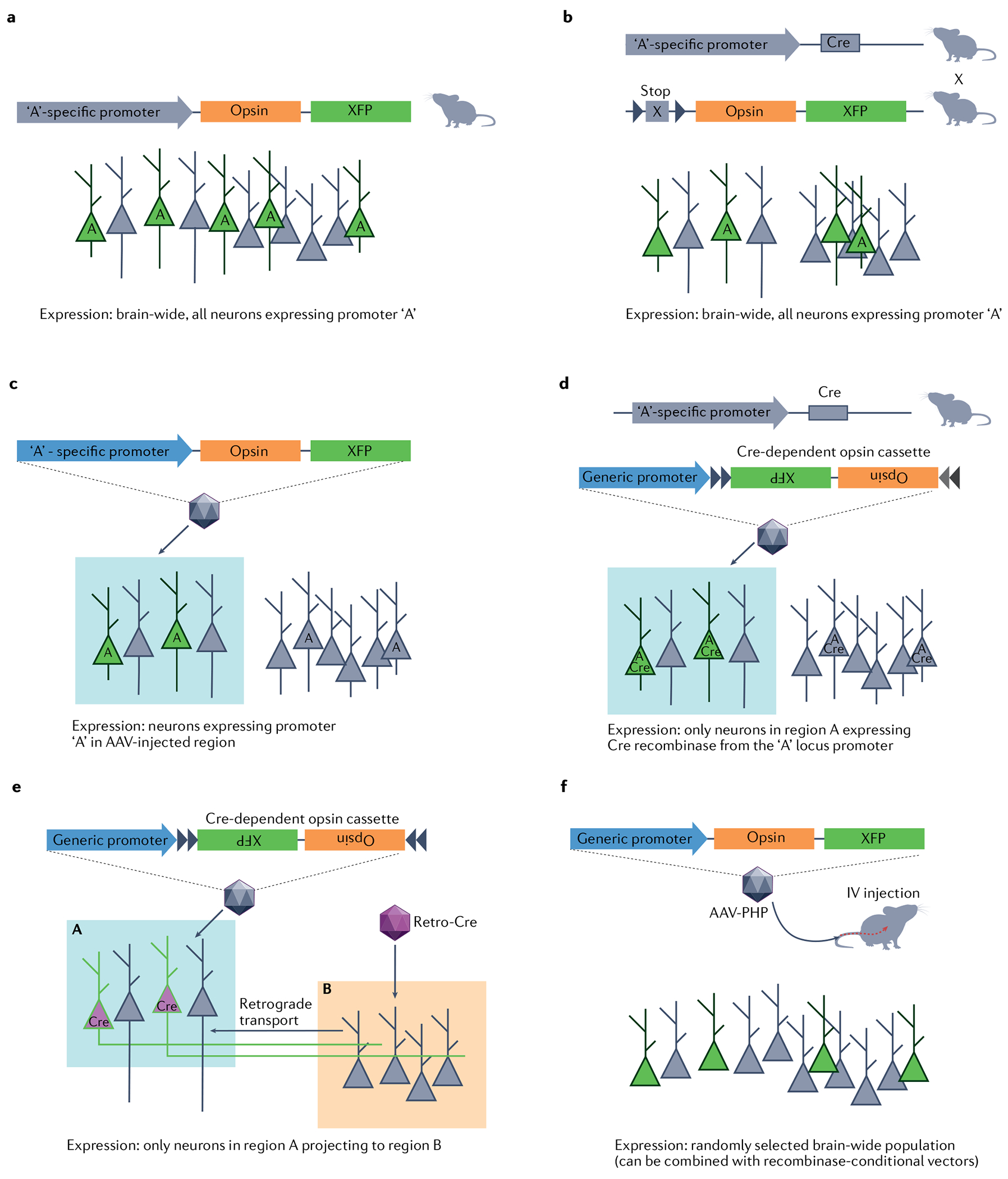
a | Transgenic mice constitutively expressing an opsin gene from their genome allow simple experiments that only require addition of light delivery apparatus. Promoter ‘A’ activity (indicated by A) will lead to transgene expression (green). b | Transgenic animal expressing a recombinase such as Cre under control of a cell type-specific promoter is crossed with a second line carrying a conditional expression cassette encoding the desired opsin. Dual transgenic offspring will then show organism-wide expression of the opsin in all cells that underwent promoter activation at any stage of development (green). Cre expression (indicated by A) is unnecessary once the conditional expression cassette was activated. c | Where a short minimal promoter sequence is available, targeted viral vector injections can be used to restrict expression spatially as well as by the gene expression profile. A viral vector containing the specific minimal promoter sequence upstream of the opsin gene will lead to expression in specific cells expressing the promoter (indicated by A), only in the region targeted with the injection (blue box). d | Approaches in parts a and b can be combined to achieve both spatial and gene expression specificity in cases where short specific promoters are not available, or where promoter activity is not specific during development. e | Projection neurons can be addressed by injection of an axon terminal-transducing, retrograde travelling viral vector encoding for the opsin or a recombinase into the target region. Recombinase-encoding viral vector is injected in a projection target (area B, red box) and travels retrogradely. A second viral injection of conditional expression cassette encoding the desired opsin into an upstream region (area A, blue box) will then lead to opsin expression only in neurons within area A that project to area B. f | Adeno-associated virus (AAV) capsids engineered for improved blood–brain barrier penetration allow brain-wide (mostly sparse) expression of an opsin through intravenous injection of the viral vector. IV, intravenous.
Viral vector targeting.
Lentiviral or adeno-associated virus (AAV) vectors can be engineered to encode optogenetic actuators and delivered either directly to the brain parenchyma or through systemic injection to target either specific brain regions or brain-wide populations, respectively. Targeting of genetically identified neuronal populations is achieved either by using the tissue tropism of the virus serotype and a cell type-specific promoter or enhancer (FIG. 3c), or by injecting the viral vector into a transgenic recombinase-expressing animal strain (FIG. 3d). Promoter-based viral vector targeting is attractive as it does not require the maintenance of a specific animal strain for every target neuron population and can also be applied in non-genetic models. However, the limited viral payload size — particularly of AAVs — prohibits the use of most native promoters. The list of minimal promoter or enhancer sequences that have been validated to specifically express in defined neuron populations is quite restricted. However, this field is rapidly expanding83,84 and is further diversified by synthetic approaches85.
Circuit-based viral vector targeting.
The most commonly used retrograde viral tracer is AAVretro86, which can be taken up by presynaptic terminals and travel in retrograde to express at the soma of long-range projecting neurons (FIG. 3e). The herpes simplex virus 1 (HSV1) and canine adenovirus 2 (CAV2) both have retrograde targeting abilities, but these are less readily available and have been shown to impair the health of targeted neurons, particularly over longer expression times of weeks to months87.
Systemic delivery of AAV-PHP capsids.
Targeting sparse brain-wide populations is beneficial for some experimental configurations. For example, structural imaging of dendritic spines in the cortex or excitation of a randomly selected sparse ensemble in a given brain region. For this purpose, AAV-PHP vectors have been engineered to cross the blood–brain barrier with high efficiency (FIG. 3f). The AAV-PHP serotypes allow targeting of diverse central and peripheral nervous system neurons88,89. The same capsids can be used with Cre-dependent AAV expression plasmids to allow sparse brain-wide expression in a genetically defined neuronal subtype. However, the efficiency of AAV-PHP serotypes in crossing of the blood–brain barrier can vary in different mouse strains90.
Electroporation.
Concentrated DNA can also be injected into the cerebral ventricles followed by in utero electroporation91–93, enabling the study of neural cell fate determination and migration or cortical layer-specific expression.
Compartment-specific functions
The effective current resulting from light-gated channel conductance can vary dramatically owing to local ion concentration gradient differences. For neuroscience applications, this is particularly crucial for use of ACRs. At the somatic and dendritic compartments, this is an advantage, as they can be used for shunting inhibition. In contrast, ACRs can exert excitatory effects in axons and presynaptic terminals, in which the intracellular chloride concentration is higher (Box 1). Ion pumping rhodopsins, on the other hand, translocate the ion over the membrane in a predetermined direction, which can be an advantage owing to the increased control of ion flux. However, the pumping-induced hyperpolarization and elevation in ion concentration can also have side effects, such as the alkalinization of presynaptic boutons94 or an artificial increase in intracellular chloride45. Similarly, the effects of G-protein-coupled animal rhodopsins on neuronal activity strongly depend on the given second messenger cascade in the local compartment. For instance, in the soma and dendrite, Gi/o signalling can activate G-protein-coupled inward rectifying potassium channels, whereas in the presynaptic compartment the Gi/o pathway mainly acts through inhibition of voltage-gated calcium channels and cAMP signalling57.
Optimizing expression and targeting
Beyond single-channel conductance, one of the main factors determining maximal photocurrent is the number of functional opsin molecules in the membrane, which in turn depends on the expression level, protein-folding efficacy, retinal binding affinity, membrane trafficking and protein turnover rate. The expression level of a transgene can be controlled via promoter strength and transgene copy number. The opsin-folding efficacy and protein stability were shown to depend on the availability of the chromophore retinal95. Although retinal availability does not seem to be a limiting factor in mammalian tissues, it needs to be routinely supplemented in the food of invertebrate model systems and some cultured cell lines. In plants, the absence of retinal can be compensated for by its synthesis via expression of a bacterial β-dioxygenase that facilitates rhodopsin expression. A common issue with unmodified opsin expression cassettes is aggregation of the synthetized protein in the endoplasmic reticulum. To overcome this limitation, trafficking motifs involved in transport of membrane proteins along the secretory pathway to the cell surface were utilized to improve plasma membrane targeting (see Supplementary Fig. 4). The most widely used trafficking motifs utilized were first described for the potassium channel Kir2.1 — these motifs enhance endoplasmic reticulum export as well as Golgi to plasma membrane trafficking96, resulting in higher plasma membrane localization and increased photocurrents in animal97 as well as plant98 cells.
Further optimization of functional expression can be achieved by adjusting the linkers between the opsin and the often co-expressed fluorophore, mutating potential ubiquitination sites and screening random mutations in the opsin coding sequence99. Beyond improved photocurrents, targeting an opsin to a selected subcellular compartment can be used to investigate the function of the chosen compartment, such as the mitochondria, synaptic vesicles, lysosomes or endoplasmic reticulum (see Supplementary Fig. 4), or to use the differential effects of ion channels discussed above. Somatic restriction has been successful in increasing the specificity of single-cell stimulation by reducing inadvertent modulation of nearby neurites77,100–104, as well as in reducing ACR-mediated axonal excitation24. Somatic restriction has the added effect of accelerating the effective photocurrent off-kinetics, owing to the elimination of photocurrents arising from distal neurites in the illuminated tissue volume, as these are low-pass filtered while travelling along the neurite to the somatic compartment.
Although targeting microbial rhodopsins to presynaptic vesicles is feasible43, enrichment of rhodopsin abundance in the axonal plasma membrane has not been achieved. Cytosolic proteins can be enriched in the axon by mRNA shuttling motifs. However, local rhodopsin translation in the axon has not been successfully applied, potentially owing to a lack of transmembrane protein synthesis in the vertebrate axon105.
Light delivery techniques
Although the vast majority of advanced light targeting approaches have been developed with the specific applications of neuronal and cardiac optogenetics in mind, these methods are generalizable and are beginning to be applied to other systems106. Optogenetics is readily applicable to light-accessible preparations such as cultured cells, tissue slices, transparent organisms, such as zebrafish larvae, or the cortical surface of the mammalian brain, allowing for extensive flexibility in light delivery. For whole circuit or brain region optogenetics, light needs to reach the target with sufficient irradiance to induce opsin activation. Ideally, light should be guided into the target structure with minimum damage to the tissue. In behaving animals, stimulation should also be conducted with minimal disruption to the measured behaviour, limiting implantable weight and tether stiffness. Whole circuit/region optogenetic stimulation is typically carried out using a multimode optical fibre, guiding the light from the source to the target (FIG. 4a). Optical fibres targeting a deep brain region can be permanently implanted by attaching a fibre-optic implant to the skull using dental cement. The dimensions of the fibre and its optical properties strongly influence the spatial profile of light reaching the brain. Most commonly, flat-cleaved optical fibres are used. However, the high radiant flux density necessary at the fibre tip to achieve sufficient irradiance within the targeted volume can lead to heat-induced changes in neuronal activity and behaviour107,108. It is therefore advisable to consider tissue heating when planning the experiment and to use opsin-free light-stimulated controls. One approach to minimizing the irradiance required in optogenetic experiments is to maximize the operational light sensitivity of the opsin used (see Supplementary Fig. 3). Another factor is wavelength, as absorption is higher for shorter wavelengths and therefore the peak temperature increase is lower for longer wavelengths at the same radiant flux density. Increased optical fibre diameter also reduces the peak light power density. However, wider fibres also cause more tissue damage and have a higher chance of illuminating blood vessels, which strongly absorb visible light and thus increase potential heating-related artefacts. This trade-off can be, at least partially, mitigated by the use of tapered optical fibres (FIG. 4b), which can be used to flexibly illuminate a large brain volume109.
Fig. 4 |. Optical approaches for optogenetic stimulation.
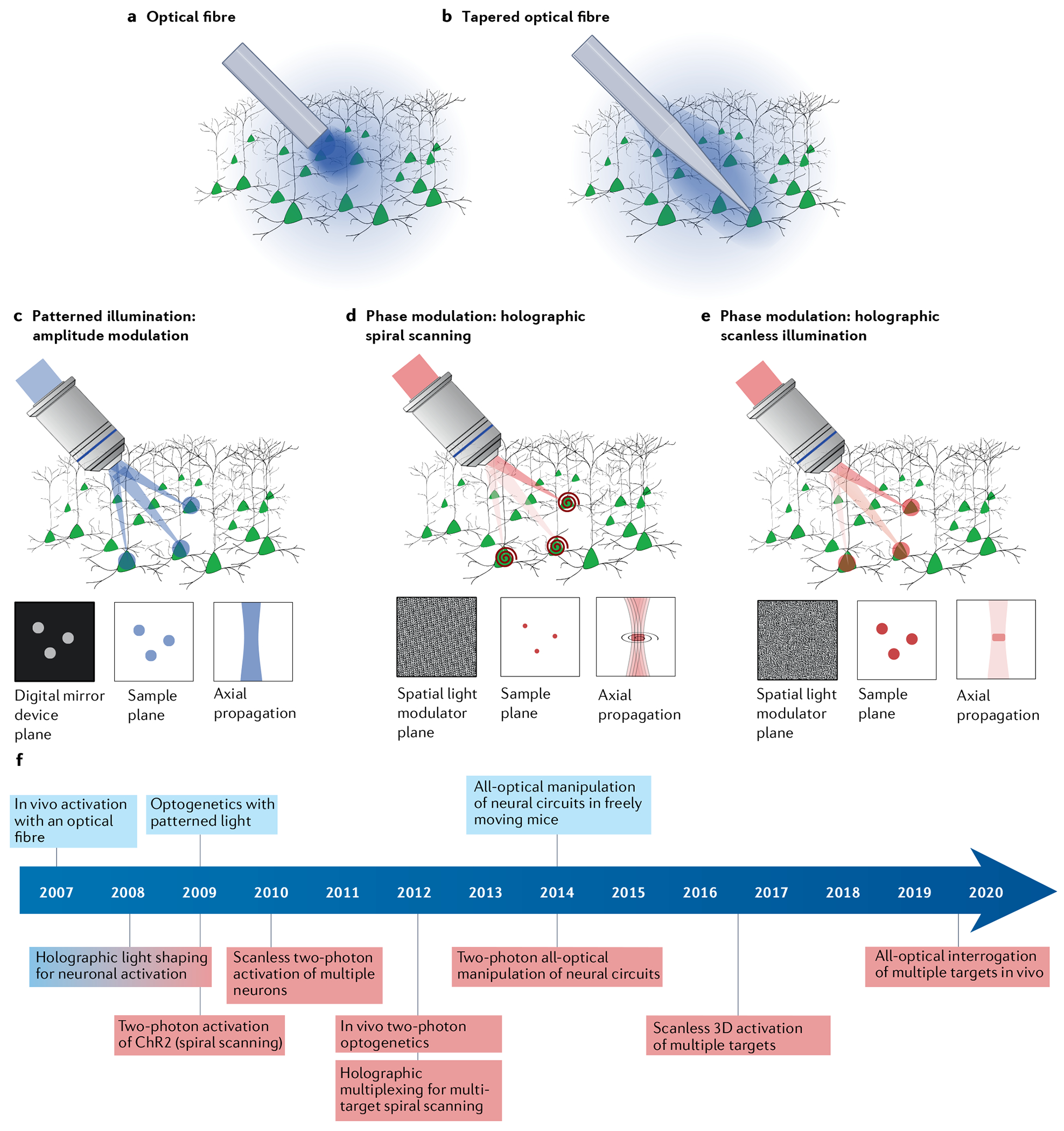
a–c | Single-photon wide-field illumination (blue) of all genetically targeted opsin-expressing neurons using excitation through optical fibres: illumination using a flat-cleaved optical fibre causes high peak light power density at the fibre–tissue interface (part a); a tapered fibre increases the optical fibre–tissue interface resulting in a reduced peak light power density (part b); and single-photon multi-target patterned illumination by spatially shaping the intensity of the excitation beam by means of a digital micromirror device, placed in a plane conjugated to the sample plane (part c). Light distribution at the digital mirror device plane and at the sample plane only differ by a spatial scaling factor corresponding to the magnification of the optical system. Axial resolution is proportional to the square of the lateral spot dimensions. d,e | Two-photon multi-target illumination by holographic light shaping: a spatial light modulator placed at a plane conjugated with the objective back aperture generates a 3D distribution of holographic spots, which are scanned with a spiral trajectory to cover the cell surface — axial extension of the generated spot is optimized to illuminate upper and lower cell membranes (part d); and a spatial light modulator is used to generate multiple extended spots with a size large enough to cover the whole cell soma — temporal focusing is used to maintain micrometre axial resolution independently of lateral spot size (part e). f | Timeline indicating critical optical developments that have enabled new optogenetic experiments throughout the past 15 years. Single-photon and two-photon milestones coloured blue and red, respectively. Holographic light shaping for neuronal activation was developed simultaneously for single-photon and two-photon activation, indicated by red–blue gradient for the milestone in part f. ChR2, channelrhodopsin 2.
In these conventional optogenetic experiments, visible light is mostly delivered non-specifically to large tissue regions and genetic targeting strategies are used to express the optogenetic actuator in specific cell types. This approach has enabled tissue function to be mapped with unprecedented anatomical and cell type specificity. However, wide-field illumination synchronously activates or silences entire populations of all opsin-expressing cells, which does not replicate the physiological case: adjacent cells belonging to genetically defined classes have been observed to exhibit divergent activity patterns. To investigate complex population activity patterns, whole-region optogenetics is insufficient. Digital mirror devices coupled to single-photon excitation have enabled single-target and multi-target excitation in head-restrained and freely moving animals and found in situ applications in control of excitation waves underlying cardiac arrythmias110–113. However, the use of visible light has limited these approaches to superficial brain layers or low scattering samples. Recent developments in opsin engineering, optical microscopy and multiphoton laser source development have given rise to circuit optogenetics114, which allows modulation of neuronal activity deep in scattering tissue with single-spike precision and single-cell resolution (FIG. 4c–e). Specifically, combining variants with enhanced kinetics30,77,115,116, higher conductance117–119 or shifted absorption peaks30,119,120 with optimized targeting and expression strategies77,101–103 enables neuronal control with single-cell, single-spike precision at millisecond temporal resolution and the generation of action potential trains with high (50–100 Hz) spiking rates116,121. In parallel, advanced optical techniques based on two-photon excitation (BOX 2) have been developed to precisely guide light through tissue. The small single-channel conductance of commonly used optogenetic actuators such as ChR2 (40–90 fS)122, and the limited number of channels or pumps recruited within a conventional two-photon focal volume, mean that it is generally necessary to use spiral scanning or parallel light shaping using computer-generated holography or the generalized phase contrast method (see Supplementary Fig. 5) combined with temporal focusing14 (see Supplementary Fig. 6) to increase the portion of excited membrane123,124 and to sufficiently depolarize a neuron to firing threshold or, effectively, silence it. Holographic light multiplexing with spiral scanning125 or ad hoc spatio-temporal shaping approaches (see Supplementary Fig. 7) have been used to generate patterned illumination at multiple axially distinct planes117,126,127. Multiplexing divides the available laser power between targets and, thus, requires powerful lasers. Owing to the higher peak photon density, amplified low repetition rate (200 kHz–10 MHz) fibre lasers enable higher rates of two-photon absorption than titanium:sapphire oscillators (at the same average power), and can therefore be used to reduce the necessary power to generate physiological signals128. Additionally, these sources deliver tens of watts of power, facilitating the simultaneous photostimulation of hundreds of cells throughout cubic millimetre volumes. The combination of these technologies has recently led to the first demonstrations of multi-target neural circuit manipulation77,118,129.
Box 2 |. Single-photon versus two-photon excitation, mechanism and focal volume.
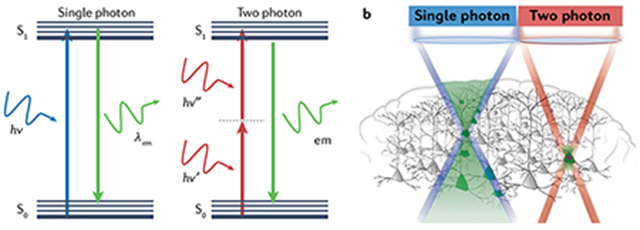
In single-photon excitation, the absorption of a photon by a chromophore induces a molecular transition from the ground state (S0) to the excited electronic state (S1), whereas in two-photon excitation the same transition can be induced by the quasi-simultaneous absorption of two photons. As two-photon cross-sections are typically much smaller than those for single-photon absorption, significantly higher photon fluxes are generally required to generate similar excitation rates, requiring more complex and expensive components such as ultra-fast lasers. There are two main implications of two-photon absorption in microscopy. First, as the probability of excitation is a quadratic function of the instantaneous photon density, targets at the focal plane are much more likely excited than out of focus targets, whereas in single-photon excitation all targets throughout the light path can be excited. Second, the use of photons of lower energy, and therefore of longer wavelengths (deep red and infrared), can penetrate more deeply (~700 μm) into scattering tissue.
The ability to control neuronal activity with single-cell precision and millisecond temporal resolution allows to functionally probe neuronal networks beyond the resolution of synchronous modulation of entire networks or genetically defined network components. For instance, using temporally precise single-cell excitation in the visual cortex and olfactory bulb, the minimal number of co-activated cortical neurons necessary for visual perception130 and the dependence of olfactory perceptual detection on both the number of activated neurons and their relative spiking latency were characterized131. The requirement of high numerical aperture objectives has limited two-photon optogenetics to circuits in superficial (≤500 μm) cortical areas of mouse brain, transparent zebrafish larvae132 or in vitro applications. Micro-endoscopes are small optical probes that can be inserted into living tissues, and represent a promising solution to extend optical brain manipulation to deeper brain structures both in combination with holographic spiral scanning133 or using multi-temporally focused light-shaping approaches134. Three-photon optogenetics, which relies on longer wavelengths and exhibits a cubic dependence of excitation efficiency on excitation power, could potentially be used to stimulate neuronal circuits in deeper brain regions (600 μm–1 mm) with single-cell resolution. However, to date, three-photon photostimulation has only been demonstrated in vitro135.
Results
Output analysis
When designing optogenetic experiments, care should be taken to verify the impact of the optogenetic manipulation on the targeted cells. This can be achieved in numerous ways, including electrophysiological recordings in vitro or in vivo, optical recordings with genetically encoded sensors, immediate early gene labelling and non-invasive imaging modalities. Below we outline the major techniques used in such experiments, and the considerations that should be taken into account when designing and performing such experiments.
Electrophysiological recordings.
To interpret the results of optogenetic manipulations, it is often necessary to determine the extent of optogenetic tool expression and its physiological effects on the targeted neurons. In the case of light-gated ion channels or pumps, recording the electrophysiological changes induced by the optogenetic manipulation is the most direct way to characterize light-mediated effects (FIG. 5a–c). As these effects can vary greatly between cell types, brain regions and even viral serotypes136, it is crucial to validate the optogenetic effector in every new experimental system before proceeding to behavioural or other functional read-outs. To describe effects on the level of spike rates and timing, whole-cell recordings are often not necessary. Instead, extracellular recordings are often used (FIG. 5d–f), given their higher throughput and minimal crosstalk with light delivery (see REF.137 for discussion of light-induced electrical artefacts). However, higher frequency spiking activity does not necessarily indicate increased synaptic transmission from the stimulated neurons136. Synaptic depression and depletion of neurotransmitter release can lead to erroneous interpretation and should be taken into account when performing optogenetic excitation experiments, particularly with neuromodulatory and neuropeptide-releasing neuronal populations.
Fig. 5 |. Expected results in optogenetic experiments.
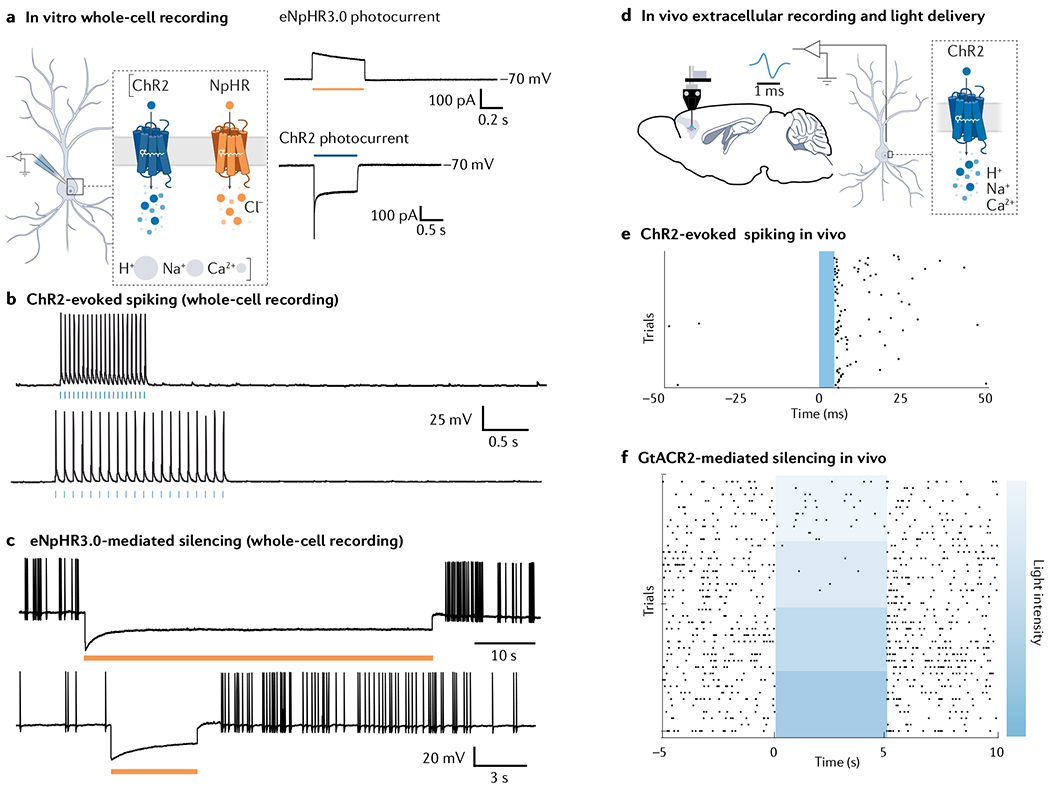
a | Expression of optogenetic actuators such as channelrhodopsin 2 (ChR2) or NpHR in neurons leads to emergence of light-driven photocurrents, which can be recorded using the whole-cell patch clamp technique (left). Cells expressing chloride-conducting NpHR will show an outward current (top right, voltage clamp recording with cell resting at −70 mV) whereas cells expressing cation-conducting ChR2 will show an inward photocurrent (bottom right, voltage clamp recording with cell resting at −70 mV). b | Whole-cell current-clamp recordings in a neuron expressing excitatory ChR2, showing action potentials evoked by brief light pulses (blue bars). c | Hyperpolarization and silencing of spontaneously occurring action potentials in a neuron expressing eNpHR3.0. d | Extracellular recordings, coupled with local light delivery, used to reveal activity of neurons in vivo, using the awake behaving optrode configuration268. e | Raster plot showing action potentials (black dots) occurring rapidly after a 5-ms blue light pulse delivered into the target brain region. f | Raster plot showing activity of neurons expressing inhibitory anion-conducting GtACR2, showing increased inhibition of action potential firing with increasing light intensity. Part f is reprinted from REF.24, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Optical recordings.
Fluorescent reporters are another common method for monitoring the effects of optogenetic manipulations. These techniques enable recording from the same cells over several recording sessions and the concurrent recording of high numbers of cells. However, given that optogenetics itself relies on light delivery, fluorescent reporters can be efficiently integrated only if a spectral or light power separation can be achieved to minimize the crosstalk between the recording and manipulation modalities (see Supplementary Fig. 2). Additionally, when combining red-shifted Ca2+ indicators with optogenetic actuators, extra care must be taken, as these can show blue light-activated photo-switching behaviour that can resemble Ca2+ activity in amplitude and kinetics138.
Whereas genetically encoded calcium sensors continue to be the state of the art in terms of optical activity read-out, voltage indicators are gradually reaching a level of maturity that could allow for wider adoption by the field139. Novel fluorescent sensors for neurotransmitters, neuromodulators and other small molecules are continuously developed140–142. Another approach to read out gross neuronal firing rate changes is to characterize the expression of immediate early genes, for instance via immunohistochemistry on the protein level143 or on the mRNA level using quantitative PCR, in situ hybridization or single-cell RNA sequencing144. Immediate early gene expression can be used to determine the relationship between the modulation of specific neuronal populations and global brain activity145. However, the temporal precision of this approach is limited to the average neuronal activity over minutes to hours and, unless combined with targeted recombination approaches146, only a single manipulation can be characterized per animal.
Although ChR variants with peak single-photon excitation wavelengths spanning the visible region of the electromagnetic spectrum have been engineered26, performing crosstalk-free, multicolour two-photon experiments is not trivial. Ideally, spectrally orthogonal ChRs and activity reporters would be chosen, but, unfortunately, the two-photon action spectra of commonly used opsins are extremely broad26 (see Supplementary Fig. 8). As previously introduced, opsins with red-shifted action spectra exhibit persistent activation in the blue range, which coincides with wavelengths used for two-photon imaging of commonly used activity reporters (920–980 nm). One approach to reduce crosstalk is to use opsins with fast kinetics. Although this approach does not eliminate sub-threshold network perturbation, the (relatively) fast repolarization of neurons expressing ChRs with fast off-kinetics means they are unlikely to fire action potentials owing to excitation by the imaging laser during scanning. Successful employment of this method requires careful titration of imaging parameters, including the imaging power, frame rate and field of view. This is an interim approach until high-efficiency blue-shifted opsins, red-shifted activity indicators and amplified lasers in the appropriate spectral range become more widely available.
Alternative recording modalities.
Electrophysiological and optical recording modalities both suffer from potential interactions with the light required to excite optogenetic actuators. The haemodynamic response is an alternative physiological response to neural activity that can be exploited to report the impact of optogenetic modulation. For superficial brain areas such as the cortex, the haemodynamic response can be measured via intrinsic imaging147,148, whereas functional MRI149 can be utilized to record brain-wide haemodynamics. Although the non-invasive nature and the ability to measure the haemodynamic response throughout the entire brain are major advantages, the main drawbacks are that the temporal resolution of this approach is fundamentally limited by the specificity and kinetics of the haemodynamic response itself and the limited spatial resolution of neurovascular coupling150. Heating should also be taken into account here as it can directly impact the haemodynamic response151. Functional ultrasound imaging is a rapidly developing technology that could be used to perform brain-wide detection of neural activity triggered by localized optogenetic stimulation. Although this method still relies on changes in neurovascular blood volume changes, it can be performed at a fraction of the cost of functional MRI recordings and is rapidly advancing to allow better spatio-temporal resolution and portability152.
Linking neural to behavioural read-outs
The exquisite spatial and temporal control of genetically defined cells with optogenetics are attractive features for experiments aiming at establishing causal links between neural activity and behaviour. The growing understanding of neuronal coding has also led to nuanced insight of the limits of interpretability of such experiments. However, when appropriately designed and controlled, optogenetic experiments can provide important information on how neural circuits drive behaviour.
Choosing the locus of intervention may be instructed by previous literature, lesion experiments and behavioural pharmacology. For example, we know that silencing the motor cortex with compounds such as muscimol or baclofen causes motor impairment whereas optogenetic stimulation elicits muscle contraction153. Although gain-of-function experiments may be a starting point, cell type-specific optogenetic inhibition of genetically defined neurons in the motor cortex would provide a more accurate picture, better dissociating the physiological motor response from an artificial perturbation154. Another way to determine the brain region and cell types of interest is the use of activity markers such as the immediate early genes Fos or Arc. Finally, technological advances in wide-field optical monitoring of intracellular calcium may allow to visualize the activity of large cortical areas150,155 and selectively silence defined cortical regions transcranially156. Alternatively, high-density electrical recordings157,158 can elucidate the activity of many neurons in deeper structures. This allows the experimenter to identify circuits with activity patterns that may be relevant to the behaviour to be studied.
Next, observational experiments should be implemented to characterize the functional properties of the cell population to be modulated (FIG. 6). This may be achieved using electrophysiology in vivo — for example, by tetrode recordings of photo-tagged neurons159 or genetically encoded calcium sensor imaging160. The choice of the optogenetic intervention should ideally be instructed by these observational investigations and match the dynamic range of the activity observed. Additional selectivity can be achieved by aiming at axon terminals rather than cell bodies. Effectors aiming at hyperpolarizing terminals or creating shunting inhibition may not always be efficient or, at times, even perturb para-membranous ion concentrations such that the effect is difficult to predict42. With the advent of Gi/o-coupled effectors56,57, presynaptic inhibition is more straightforward, but it remains good practice to validate the efficacy of inhibition, as well as its spatial selectivity, particularly with the highly light-sensitive effectors. It is particularly important to take into account the firing frequency of the cells under investigation as presynaptic inhibition is potentially less efficacious at higher firing rates.
Fig. 6 |. Establishing links of causality with optogenetics.
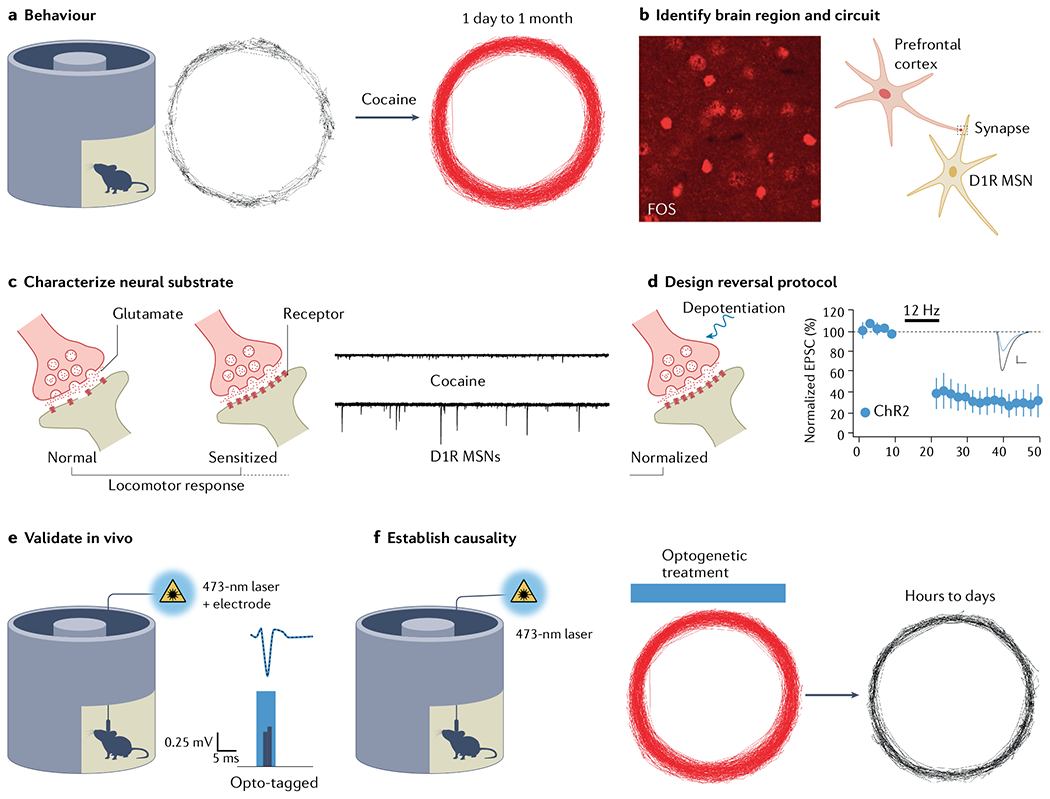
Experimental road map based on identifying the neural correlate of behavioural sensitization to cocaine269. a | When injected, cocaine elicits a locomotor response quantified in a cyclotron. Response is enhanced upon second injection of same dose. b | Fos is an immediate early gene highlighting the neurons particularly active, which provided the entry point to identifying the medial prefrontal cortex to nucleus accumbens projection as the behavioural relevant circuit. c | Slice electrophysiology enables observation of selective potentiation of glutamate transmission onto dopamine type 1 receptor medium spiny neurons (D1R MSNs)270. d | Depotentiation protocol (long-term depression (LTD) at 12 Hz) validated in slices restores standard transmission. e | In vivo validation involves opto-tagging, where spontaneously occurring spikes (grey, dashed trace) are compared with optogenetically evoked spikes (blue trace). Waveform and latency are important parameters. f | LTD protocol is eventually applied in vivo to reverse sensitization. ChR2, channelrhodopsin 2; EPSC, excitatory postsynaptic current. Part d adapted with permission from REF.269, AAAS.
There are two distinct approaches for optogenetic manipulations, one with an acute effect and the other with long-lasting effects. Acute manipulations require behavioural observations in real time. Ideally, a small set of optogenetic trials should be randomly interleaved with control trials. This assesses the acute effects on optogenetic trials along with longer-lasting changes to the subsequent control trials. Such laser on–off protocols can be used to control for adaptive behavioural changes throughout a given session. However, often the particular structure of the behavioural paradigm does not allow for hundreds of trials. The timing of the optogenetic stimulation or inhibition should therefore occur in a behaviourally defined window, and be only as long as is strictly necessary.
Long-term observation is appropriate when optogenetic interventions exploit synaptic plasticity mechanisms. For example, synaptic potentiation typically is achieved by high stimulation frequency, whereas depression requires sustained low-frequency stimulation. Optogenetic synaptic plasticity protocols are particularly suited to study learned and adaptive behaviour. The goal of long-term observation experiments is to induce synaptic plasticity at identified synapses and observe the effect on behaviour at a later time point when optogenetic stimulation is no longer active. For example, low-frequency optogenetic stimulation can restore baseline transmission in cortico-accumbal synapses that have been potentiated by cocaine exposure161. Similarly, daily optogenetic stimulation of orbitofrontal to dorsal striatum axons for 10 min triggered long-term changes in synaptic strength and inhibited compulsion162.
Applications
The vast majority of applications of optogenetics have involved neuroscience and brain research. Many of the general principles and approaches of optogenetics can be extended to other organs, particularly to those with excitable cells — such as skeletal muscle, heart, retina and gut — as well as to microorganisms and plants. These newer applications often present unique challenges and opportunities. Below, we illustrate some of these aspects with three select examples from visual, cardiac and plant applications.
Vision restoration
Retinal degenerative diseases, such as retinitis pigmentosa and age-related macular degeneration, result in the loss of rod and cone photoreceptor cells, leading to partial or complete blindness163,164. Rendering inner retinal neurons responsive to light is one of the most obvious medical applications for optogenetics (FIG. 7A). The first proof-of-concept study involved the ubiquitous expression of ChR2 in retinal ganglion cells in retinal degenerated mice13. The approach since then has been reported by numerous studies using different optogenetic tools, retinal cell targeting strategies and animal models (see Supplementary Table 1).
Fig. 7 |. Optogenetic application for vision restoration, cardiac research and plant modification.

A | Strategies for optogenetic restoration of vision following photoreceptor degeneration. Visual processing pathways in normal retina, illustrating the rod/cone, ON/OFF pathways and antagonistic centre–surround receptive fields of retinal ganglion cells (ON cells, including rod bipolar cells and AII amacrine cells (AII), shown in grey tones; OFF cells shown in black; ON and OFF regions receptive field of retinal ganglion cells indicated + and −, respectively) (part Aa). Ubiquitous expression of a depolarizing optogenetic tool (green) in all retinal ganglion cells to convert them into ON cells (part Ab). Targeting a depolarizing optogenetic tool in ON bipolar cells to produce ON and OFF response in retinal ganglion cells and, possibly, the centre–surround receptive fields (part Ac). B | Optogenetics in cardiac research. Cell-specific targeting used for sympathetic (red) and parasympathetic (blue) nervous control of the heart using tyrosine hydroxylase (TH) and choline acetyltransferase (ChAT) promoters; cardiomyocytes (CM) from upper or lower chambers of the heart (atria (A) or ventricles (V)) can be selectively light-sensitized; and specific targeting of the fast conduction system (CS), cardiac fibroblasts (FB), vascular cells (VC) or macrophages (M) is also of interest (part Ba). Rhythm control can include optical pacemaking by short pulses (top trace), heart rate modulation by low-level constant (middle trace) or pulsed light by activating sympathetic nervous system (increase) or parasympathetic nervous system (decrease), and arrhythmias can be terminated to restore normal rhythm through a single long pulse (bottom trace), series of pulses and/or spatially patterned light (part Bb). Cardiotoxicity testing, a required component in drug development, enabled by high-throughput screening (HTS) optogenetic platforms, which can integrate patient-derived induced pluripotent stem cell-derived CM (iPSC-CM) for personalized therapy (part Bc). C | Optogenetic approaches in plants. Carbon dioxide entering through stomata with loss of water and oxygen (part Ca); and (Cb–Cg) expression of rhodopsins to control plant cell behaviour (scale bars: 15 μm): absorbance spectra of anion channelrhodopsins GtACR1 (black) in relation to endogenous relevant plant photoreceptors (part Cb); optical fibre illumination of a leaf from an Arabidopsis plant mounted in a microscope set-up (part Cc) for simultaneous optical stimulation and electric recordings of guard cells embedded in the leaf epidermis (part Cd); representative membrane voltage recording from wild-type tobacco (red) and tobacco with stable GtACR1-expressing guard cell (black) in response to a 525 nm light pulse (10 s) of 0.57 mW mm−2 in presence of background red light (630 nm, 0.018 mW mm−2) to elicit stomatal opening (part Ce); and closure of stomatal aperture only induced in GtACR1-expressing cells in presence of green light (green bar in part Cf; green light spot in part Cg). BC, bipolar cells; GC, guanyl cyclase; RBC, rod bipolar cells. Part A adapted with permission from REF.245, Annual Reviews. Part Cc, image courtesy of S. Scherzer and A. Reyer. Parts Cd, Cf and Cg adapted with permission from REF.215, AAAS.
Multiple clinical trials using ChRs for treating retinitis pigmentosa-related blindness have been initiated since 2015, with encouraging results (see Supplementary Table 2). Recently, the first published case study reported the partial restoration of vision (in the form of perceiving, locating and counting objects) in a blind patient with retinitis pigmentosa165. Positive preliminary results have also been reported in other clinical trials (see Supplementary Table 2). However, further efforts will be required to improve the outcome of optogenetic vision restoration, including the development of effective optogenetic tools and treatment strategies, and the improvement of gene delivery efficiency.
Optogenetic tools.
ChRs have been the more commonly used optogenetic tools for vision restoration in animal models and the ones used so far in clinical trials. Two main issues should be considered when choosing an optogenetic tool for vision restoration. The first is the tool’s expression efficiency and long-term safety in mammalian neurons; problems with the expression of an optogenetic tool are difficult to correct and, usually, result in cell toxicity in the long term. The second issue is the low operational light sensitivity of ChR-expressing retinal neurons, in general, caused by the small unitary conductance and substantial inactivation. The requirement of high light intensity to activate the ChR-expressing retinal neurons constrains this application and also raises concerns regarding tissue photochemical damage, especially for short-wavelength sensitive ChRs. One solution to mitigate the potential photochemical damage is to use red-shifted ChRs, such as Chrimson, as the threshold of light intensity causing tissue photochemical damage is shifted to higher light intensities for longer wavelengths166–168. Another solution is to improve the light sensitivity of a ChR-expressing cell by slowing its closing kinetics or off-rate with molecular engineering169 (see Supplementary Figs 1 and 2) combined with genome mining for more potent ChRs30. This strategy has been recently used to further optimize the more effective ChR variant CoChR. Functional vision is restored with improved CoChR mutants under ambient light conditions in a blind mouse model170. A third solution is to use GPCRs, including animal opsins (for example, rhodopsin and cone opsins)171–173 or engineering of optoGPCR chimeras174, taking advantage of their high light sensitivity due to intracellular signal amplification. Further studies will need to evaluate the most effective optogenetic tools or develop better ones for this application.
Gene delivery.
AAV vectors are the current choice for transgene delivery in the retina both in animal studies and in clinical trials175. Intravitreal injection is a preferred route of viral vector administration owing to its safe operation and ability to achieve widespread delivery to the retina. However, in non-human primates and in humans, virus transduction was mainly conferred to a narrow region surrounding the fovea or parafoveal region176,177, owing to the barrier of a thick limiting membrane in the retinal surface of primates22, which is one of the major factors limiting the outcome of AAV-mediated optogenetic therapy. Further development of more efficient gene delivery vehicles or techniques is required.
Retinal cell targeting.
Most animal studies and clinical trials have employed ubiquitous promoters to express depolarizing ChRs in retinal ganglion cells. However, unlike the normal visual processing features in the retina including the segregation of ON and OFF signal pathways and the presence of antagonistic centre–surround receptive fields (FIG. 7Aa), this treatment strategy converts all retinal ganglion cells into ON cells (FIG. 7Ab). Although useful vision could still be generated as demonstrated in animal studies and reported from clinical trials, it is commonly believed that restoration of vision to mimic the intrinsic visual processing features in the retina would result in a better outcome. To this end, one strategy is to target an optogenetic tool to distal retinal neurons. Targeting a depolarizing ChR to ON bipolar cells using the mGluR6 promoter has been the most commonly employed strategy (FIG. 7Ac). Owing to the unique rod pathway in the mammalian retina, this could lead to ON and OFF responses at the level of retinal ganglion cells178–181, and possibly centre–surround receptive fields. Targeting surviving cone photoreceptors with a hyperpolarizing optogenetic tool, such as eNpHR, has also been reported182. As a limitation for this strategy, the distal retinal neurons are more susceptible to severe retinal deterioration or remodelling than retinal ganglion cells after the death of photoreceptors183. Multiple treatment strategies will need to be developed for treating patients with different retinal degenerative conditions.
Cardiac research
The key benefits for clinical translation are sought in more versatile optogenetic pacing or suppression of wave propagation during arrhythmias, compared with currently used cardiac devices such as pacemakers and cardioverter/defibrillators184–191 (FIG. 7B). Strategies for rhythm control enabled by optogenetic actuators aim to lower the energy needed to power cardiac devices and extend battery life by delivering longer lower-energy light pulses — electrical pulse duration is limited owing to electrochemical toxicity via Faraday effects. Optogenetic actuators also eliminate discomfort and pain during classic cardioversion/defibrillation for better quality of life by using cell-specific genetic targeting to engage the fast conduction system188,192 or to specifically target myocytes and avoid unintended contractions of thoracic skeletal muscle, diaphragm and vocal cords such as pain-inducing electrical defibrillation193. Computational modelling of the action of optogenetic tools in the heart helps explore strategies for control of arrhythmias, both with excitatory/depolarizing opsins and with inhibitory/hyperpolarizing opsins112,187,188,194,195. Longer-term in vivo clinical applications face the challenges of genetic modification of the hard to access cardiac muscle, potential immune responses and realizing embedded miniaturized light control devices that are reliable and safe191. Light penetration in the haemoglobin-rich heart muscle requires operation in the near-infrared and opsins excitable within that range, along with stabilization techniques to counter mechanical contractions. The atria are thinner (human atria are <5 mm) and present an easier target, along with more accessible autonomic nerves, such as the vagus nerve196.
AAV9 is the most efficient AAV serotype for targeting the ventricular myocytes in vivo when using a ubiquitous or a specific promoter, such as Myh6 (REF.197). The heart atria can be targeted optogenetically using the NPPA promoter and local viral gene delivery190. Cre–lox transgenic mouse models with suitable promoters have been used to transform the fast conduction system cells (Cx40)192, sympathetic neurons (tyrosine hydroxylase (TH))198 and parasympathetic neurons (choline acetyltransferase (ChAT))196,199 (FIG. 7Ba). To translate the approaches from rodents to larger animals, more work is needed in finding minimally invasive ways of transgene delivery to the heart and in minimizing immune responses. Previous clinical trials on gene therapy for cardiac disorders found that a large portion of the patients had antibodies against the viral vectors used, thus reducing the efficacy of the therapy200. Most of the published studies have used ChR2-H134R as an excitatory opsin. In general, more efficient and fast inhibitory opsins are desirable for arrhythmia control applications. There may also be a niche for step function-like depolarizing opsins that have fast recovery from inactivation as clamping tools in arrhythmia management. Bidirectional closed-loop control could make an all-optical approach named optical clamp at the whole organ level a reality. However, this will require spectral compatibility to accommodate not only for an excitatory and an inhibitory opsin but also for the optical read-out of a voltage indicator.
Overall, clinical applications of optogenetics in the heart face many challenges compared with the more accessible, immune-privileged applications to the eye that have seen translational advances. Considering the potential impact for control of arrhythmias, efforts should continue to improve the genetic targeting by more specific promoters, safer viral vectors, longer-wavelength opsins for better penetration and miniaturized distributed light sources. Basic science experiments with optogenetic tools provide invaluable insights for improvement of current cardiac devices and may yield new strategies for arrhythmia control111,113,191,201,202. These new strategies take advantage of the ability to produce complex space–time control patterns by light (unlike discrete signals from electrode arrays) to steer waves of excitation towards non-arrhythmic behaviour at very low energy. Optogenetics-empowered high-throughput systems can more immediately improve cardiotoxicity testing and drug development. All-optical cardiac electrophysiology, which combines optogenetic actuators and optical/optogenetic sensors203–205, offers immediate adoption and translation (FIG. 7Bc). Cardiotoxicity testing is crucial in the development of any new pharmaceutical, and high-throughput optogenetic methods with patient-derived cells represent impactful technology for personalized medicine206,207. Optogenetic techniques using hyperpolarizing opsins such as ArchT have been used to dynamically alter the action potential characteristics of induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) towards a more mature phenotype to better predict drug responses208. The maturity of tissue-engineered constructs of such patient-derived iPSC-CMs can be improved through chronic optogenetic pacing207 towards new regenerative solutions for the heart (see Supplementary Fig. 9).
Plants
A large set of photoreceptors that control phototropism, diurnal rhythms and photomorphogenesis play fundamental roles in plant growth and development. Blue light-absorbing phototropins and cryptochromes or red/far-red light-absorbing phytochromes are found in almost all plant tissues (FIG. 7Ca). Therefore, when using optogenetics tools in plants, the light regime used needs to be considered. The light required for plant growth will activate optogenetic tools when light of the entire visible spectrum is used; this can be avoided by combining a blue light-regulated transcriptional repressor with a red light-triggered switch209, allowing plants to grow in ambient white light. The use of flavoprotein-based optogenetic tools in plants has been described in detail recently210,211. Based on the LOV domain, a synthetic light-gated K+ channel with considerable dark activity, called BLINK1, was recently expressed in Arabidopsis guard cells for control of stomatal behaviour212. The mechanism of BLINK1 light activation that clamps the membrane potential to EK and facilitates stomatal opening and closing in the same way remains to be clarified. A rather simple but valuable technique to avoid non-specific activation of rhodopsin-based optogenetic tools is to grow plants exclusively in red light98,213. Both chlorophyll a and chlorophyll b absorb red light (FIG. 7Cb), and tobacco plants exclusively grown in red light are hardly distinguishable from those grown in white light98. Green light is the least absorbed wavelength by endogenous plant photoreceptors; therefore, green light allows for optogenetic manipulation with only minimal crosstalk98 (FIG. 7Cb), especially with GtACR1.
Rhodopsin-based plant optogenetic approaches are still in their infancy compared with their long-standing use in animals. The combination of ubiquitous rhodopsin expression with global or local light-emitting diodes (LEDs) or laser light applications have been used in plants98,213–215. However, cell type-specific expression with global green light exposure certainly bears great potential, when combined with red light growth conditions. Use of the LeLAT52 pollen-specific promoter216 allows plants to be grown in white light under green-house conditions for optogenetics-inspired research on pollen tubes98. For local rhodopsin stimulation at the single-cell level, optical fibres or laser light pulses have been successfully applied98,215. The fluorescence recovery after photobleaching module of conventional laser scanning microscopes allows local optogenetic stimulation of plant cells when using rhodopsins, such as GtACR1, with activation kinetics in the lower millisecond range98.
To perform plant optogenetics with rhodopsins, retinal can be added externally213 (see Supplementary Fig. 10a–c) or plants can be empowered to produce retinal by expressing a β-dioxygenase from a marine bacterium targeted to the chloroplasts to synthesize retinal from carotenoids efficiently98 (see Supplementary Fig. 10d–f). In contrast to animal cells, the plant cell extracellular medium is low in ions and mostly moderately acidic, which may result in different electrical responses in plant and animal cells using the same rhodopsin (see Supplementary Figure 11). Activation of ACRs in the soma of neurons leads to membrane hyperpolarization217, whereas depolarization occurs in plant cells98 due to the outward-directed anion gradient. When expressed in leaves or pollen tubes, activation of GtACR1 by green light (530 nm) resulted in membrane depolarization by about 60–100 mV within milliseconds98. Local GtACR1 activation on one side of the dome of apically growing pollen tubes has been used to demonstrate the involvement of an anion efflux in polar growth98 (see Supplementary Fig. 10e,f), supporting earlier studies on the role of anion transport in polar growth218,219. In guard cells, native anion channel activity can be mimicked when GtACR1 is triggered by a series of light pulses (FIG. 7Cc–Cg), demonstrating that anion channel-driven depolarization is sufficient to close stomata215. Although plants do not have neuronal-like networks, voltage changes in the form of depolarization waves are transmitted between leaves or even between different organs220–222. The role of these long-range electrical signals can now be investigated with the help of GtACR1. Through GtACR1-induced anion efflux, depolarizations of any shape and intensity can be optogenetically generated to mimic the voltage changes observed in plants such as variation potentials, system potentials or action potentials223–225.
A wide range of processes in plants are induced by changes in cytoplasmic Ca2+ and H+ levels226,227. For both ions, there is a strong inward gradient, in contrast to animal cells where there are minimal differences in intracellular and extracellular pH (see Supplementary Fig. 11). The slow cycling ChR2 variant XXL with high proton conductance228 is excellent to impose light-induced pH changes, and has already been used to feed the P-type plasma membrane H+ pump with substrate and study its voltage dependence213 (see Supplementary Fig. 10b,c). The resting potential of plants is negative with respect to EK (−120 to −180 mV) due to the voltage-dependent activity of P-type plasma membrane H+ pumps. The latter hyperpolarize the membrane and acidify the cell wall space229. This voltage deflection is used by the plant to open hyperpolarization active Shaker-type K+ channels230 and electrophoretically move K+ ions into the cell231. The combined driving proton motive force of the electrical gradient and that of the H+ is used by solute transporters using protons as co-substrate. The plant optogenetics toolbox therefore needs to be complemented by light-driven H+ pumps such as Arch3. Of great potential for the study of Ca2+ signalling is the ChR2 variant XXM, with increased Ca2+ conductivity and medium open-state lifetime232. Combined with electrophysiology and Ca2+ imaging, the molecular mechanisms for long-distance Ca2+ signalling could be resolved. Ca2+ signatures can represent either single events or rhythmically recurring signals. Whether and how different Ca2+ signatures control specific processes in plants is still largely unexplored. In the future, Ca2+-permeable ChRs could be used to elicit defined Ca2+ signatures.
Reproducibility and data deposition
Reproducibility of optogenetic tools
Reproducibility in optogenetics experiments depends on the consistency of the tools used, the organism/cell type, genetic transformation procedures and light delivery. Adherence to minimum reporting standards for all relevant parameters of an experiment can help increase reproducibility.
Optogenetic actuators are used in a diverse range of organisms, tissues and cell types. Because of differences in codon usage between the original host and the organisms in which the tools are applied, it is common to codon-optimize the coding sequence to facilitate translation in these heterologous systems. New codon-optimized sequences should be tested for expression, membrane targeting and function before applying these novel constructs in optogenetic experiments. The sequences of codon-optimized constructs should be appropriately reported in publications to allow reproduction of findings in other laboratories. However, even with codon optimization or adding trafficking motifs, the intracellular aggregation of many optogenetic actuators can still pose a problem for their applicability, particularly for translational applications. A thorough evaluation in targeted organism/cell type is needed because the intracellular aggregation not only reduces expression efficacy but also affects cell health or causes cell death.
Viral vectors are a popular gene delivery system for optogenetic tools. The quality of viral vectors, purity and viral titre can profoundly affect the transduction efficiency and experiment outcome. The quality of viral vectors produced by different laboratories, centralized viral vector cores and companies can vary widely. Variation can even occur from batch to batch produced at the same facility. Therefore, even when produced by centralized viral vector cores, service centres and major laboratories, viral vector preparations can vary in quality and efficiency. To minimize the variation, standardized purification and titration methods should be used. Each batch needs to be verified before scaling up experiments in order to obtain reproducible results.
Reproducibility of opsin expression
Evaluation of the viral titre is needed to optimize viral vector spread and expression level, and to minimize overexpression-mediated off-target effects. Many opsin viral vectors were designed to co-express a fluorophore. Standard histological methods can be used to visualize the strength and spatial extent of viral vector expression. Characterizing viral expression for every experimental animal can increase interpretability by correlating the variability in behavioural effect to the variability in expression area and, for instance, optical fibre placement. Even when an experiment is planned based on published work, the experimental design should be validated in each new experiment owing to the potential variability of viral vector batches, optical hardware and mouse strain. When presenting results obtained using viral vectors, the source of the viral vector, its purification and titration methods, and the duration of expression should be reported.
Viral vector expression can impact cell health or change the electrophysiological properties of the targeted neurons. It is therefore necessary to include a control group injected with a titre-matched virus that expresses a control transgene. Researchers often use a virus encoding the same fluorophore that is co-expressed with the opsin. This control group can be used to evaluate direct effects of the virus injection surgery and potential phototoxic or heating effects due to the light delivery paradigm. Strong opsin expression has been reported to affect cell physiology233. It is therefore advisable to include an opsin-expressing group where no light is applied. Where the experiment allows for multiple repeats of the same manipulation, light and no light conditions can be tested in the same group, which presents a within-animal control.
Transgenic animals for optogenetics research should be genotyped continuously to confirm suitability for the experiments. For in vivo optogenetics with viral delivery, even when using the same tools in the same organism type, variations in responses may be due to variations in the immune response of the subjects (animals or humans) to the viral capsid, or the cargo (opsin and/or fluorescent reporter). To obtain reproducible data with viral delivery, testing for neutralizing antibodies can be implemented234. Appropriate control groups, immunohistochemistry and histology should be done routinely in animal experiments to demonstrate consistency of the optogenetic transformation.
Reproducibility of light delivery
Activation of optogenetic tools depends on the photon irradiance or photon exposure in case of short flashes and the spectral profile of the delivered light. The spectral profile should be reported by listing the light source, all filters and optical components used in the experiments. Insufficient irradiance may lead to failure to engage the optogenetic tools and, therefore, failure to reproduce the phenotypic changes; excessive irradiance may lead to adverse thermal effects and photoreceptor bleaching that also affect reproducibility. For single-photon excitation, the spatial pattern of the delivered light is variable and highly depends on the positioning of the light source and the tissue properties. Although total power is trivial to report, the normalized values of irradiance are influenced by the uncertainties of area estimation and the non-uniform spatial profile of light delivery. At a minimum, effort should be made to measure and report irradiance at the tissue point of entry. Whenever possible, light–tissue interactions can be simulated235 to yield relevant estimates of irradiance at points of interest.
Under optimal conditions, two-photon optogenetics is capable of stimulating individual neurons within a circuit with single-spike and single-cell resolution. Irrespective of the light sculpting method used (spiral scanning or parallel illumination), one must keep in mind that the effective spatio-temporal resolution of optogenetic stimulation depends on several factors, including the functional expression level of the opsin, the targeting specificity and the photon density required for sufficient actuation. Once a reliable and reproducible experimental preparation has been established, and the average incident powers required identified, the physiological resolution should be measured experimentally rather than drawing any conclusions about the confinement of actuation based on the optical resolution of the light targeting method.
Data and metadata sharing
The data type and format from optogenetics experiments can be extremely diverse. Outputs may include spectra, ion channel recordings, functional recordings of responses by different measurement technologies, images of altered responses and behavioural analysis, among others. For each subfield where optogenetics is deployed, minimum standards of reporting and guidance of data sharing will help determine best practices. In general, specifics of the instruments used, the acquisition and the analysis software need to be included. GitHub, figshare and other general repositories for data and analysis tools can be used to increase reproducibility.
Limitations and optimizations
Tissue heating and photodamage
Single-photon optogenetics.
Optogenetic experiments based on illumination with visible light excitation (450–630 nm) typically use optical fibres coupled to lasers or high-power LEDs for large (approximately cubic millimetre) field illumination, relatively long (0.5–60 s) exposure times and excitation powers in the order of milliwatts (0.5–20 mW). Under these conditions, the main cause for concern with respect to photodamage is heating due to light absorption. This has been investigated both theoretically, using Monte Carlo with finite-difference time-domain simulations107 or the finite element method236, and experimentally using thermocouples107,236, infrared cameras237 or electrophysiological recordings238. Depending on the precise stimulation protocol used, these experimental and theoretical studies report a wavelength and power density-dependent temperature increase between 0.3 and 6 K throughout the volume of illuminated tissue107,237. Temperature variations in the order of only 2 K can affect ion channel kinetics and conductance239, synaptic transmission240 and neuronal firing rate107, and lead to a bias in turning behaviour across various brain regions238. Importantly, changes in temperature can induce physiological changes in the absence of detectable changes in behaviour241. It is extremely important to carefully design optogenetic experiments to minimize photon exposure and absorption, for instance by using short illumination duty cycles237 and opsins with long open-state lifetimes and red-shifted absorption peaks30,77,242. Simulations107,237,243,244 can be used to guide experimental design, but, as the effects of heating vary between cell types and brain regions, opsin-negative controls should always be performed.
Multiphoton optogenetics.
Generating sufficient rates of multiphoton excitation requires the use of pulsed lasers with high peak energies, but as typical optogenetic stimulation protocols irradiate cells on millisecond timescales, the temperature rises induced by single-cell multiphoton photostimulation are in the order of 10−1 K (REF.244). Much larger temperature rises are induced during multi-target excitation due to the diffusion of heat from each target into the surrounding tissue. The resultant temperature increase occurs over hundreds of milliseconds and can approach or even exceed the 2 K threshold for thermal damage244. This effect can be mitigated by ensuring that the separation between adjacent targets is larger than the thermal diffusion length.
The risk of non-linear photodamage increases with peak fluence and could be a dominant source of photodamage in the case of spiral scanning, which typically requires higher photon density than parallel illumination. Non-linear photodamage can be reduced by increasing the repetition rate of the pulsed laser source, although this will increase photo-induced temperature rises244. In all-optical experiments that combine two-photon optogenetics with two-photon imaging, the possibility of thermal or non-linear damage induced by the imaging laser should also be considered243.
Interpreting optogenetic experiments
Light delivery schemes based on single-photon excitation are not generally capable of recapitulating physiological activity patterns. In most optogenetic gain and loss-of-function experiments, a set of cells is activated or silenced, and the effects of this manipulation are subsequently characterized by functional or behavioural read-outs to probe causal dependencies. Light delivery via an optic fibre can be precisely controlled in terms of output power and temporal pattern to influence neuronal functions such as the spike rate and spike pattern, and may be restricted to specific short behavioural epochs. However, such optogenetic manipulations typically lead to highly synchronous activity patterns, and might drive the circuits to states that are outside their physiological activity range, potentially confounding any causal inference regarding the natural functions of the circuit245. One major current effort aimed at overcoming these constraints is the development of tools for evoking naturalistic network activity patterns. Such manipulations would enable causal inference of the effects of an activity pattern on a given behaviour.
Non-physiological activity patterns can occur at the single-cell level as well as at the broader circuit scale226,227. On the single-cell level, ion pump-mediated hyperpolarization, for instance, can lead to rebound excitation upon inhibition release246 or to supraphysiological ion concentrations45,94. High-frequency light pulse trains or constant illumination of an excitatory pyramidal neuron expressing a CCR can, for instance, lead to depolarization block, effectively reducing rather than increasing its firing rate247. Whether such rebound excitation or depolarization block occurs, and to what extent, is hard to predict, as it depends on many experiment-specific parameters which can greatly vary between laboratories. Although axonal stimulation can be used to effectively isolate the activity of an anatomically defined projection pathway, optogenetic stimulation of axons can cause antidromic activation of both neuronal cell bodies as well as collaterals to other brain regions, leading to reduced specificity which should be taken into account.
At the circuit level, particularly when a large portion of cells express ion translocation-based optogenetic tools such as ion channels or ion pumps, the simultaneous activation of these tools can lead to transient but significant changes in the ion composition of the local extracellular space, thereby indirectly affecting nearby non-opsin-expressing neurons248. Electrophysiological characterization of the optogenetic manipulation can be performed to quantify the extent of such unintended effects, allowing the optimization of light power and illumination paradigms. Optogenetic tools that modulate biochemical activity within the cells or ones that act on slower timescales, or only induce sub-threshold depolarization, are less prone to the caveats imposed by highly synchronous neuronal activation249. Finally, optogenetic firing rate modulation experiments are mostly designed to acutely alter the firing rates of targeted cells, which can have different effects from chronic manipulations. Brain circuits regulate their overall activity to achieve a homeostatic equilibrium, such that when the firing rate of a circuit is transiently increased or decreased it can acutely affect the independent functions of downstream circuits and lead to markedly different results compared with chronic manipulations249. Acute effects are normally more severe, and could lead to overestimation of the roles of targeted regions in a given behaviour. Although chronic manipulations such as lesions do not suffer from this limitation, plastic changes during lesion recovery can also lead to an underestimation of the necessity of a given input to a local circuit. In summary, a sound experimental strategy should balance the use of acute and powerful optogenetic approaches with chronic experiments, pharmacological manipulations or lesions, and use caution in claims of causality based purely on manipulations that might suffer from any of the above-mentioned artefacts.
Outlook
Refinement of the optogenetic toolbox
We anticipate a further optimization of existing tools in terms of kinetics, ion or substrate selectivity, and widening of the spectral range from UV to the near-infrared to enhance the use of optogenetics. Additional light-activated enzymes allowing for optogenetic control beyond cell excitability are still to be discovered. Efforts are currently directed at optogenetic control of translation, transcription, nucleotide modification and epigenetics, as well as protein degradation. We are also expecting better tools for the control of cellular mechanics, development and differentiation.
Enhancement of basic research
Optogenetics will further advance the investigation of neural circuits. This will not only establish links of causality between neural activity and behaviour but, eventually, generate sufficient knowledge for a theory of the brain to emerge. Empirically observed neural activity in optogenetic experiments taking into account the activity of individual neurons may eventually allow deriving the neural code, which, when integrated into a solid theoretical framework, will bring the neurosciences on a par with other fields of natural sciences.
Optogenetics may also drive basic knowledge in other fields of life science, from cardiac physiology to plant physiology. For plant optogenetics, which is still in its infancy, there is great potential through the recently introduced in planta retinal synthesis, which now allows access for many rhodopsin-based manipulations. Implementing optogenetic approaches in any system of excitable cells will allow for the investigation of so far intangible questions. This may apply, for example, to the control of muscle contraction in the heart as well as the insulin secretion in the pancreas.
Open routes of translation
Beyond advancing basic science, optogenetics also has translational potential, either by inspiring novel protocols of existing therapies or as a therapy in humans. Several possible optogenetically inspired medical interventions and therapies are already outlined in this Primer. Optogenetics can be used in vitro to analyse cellular processes in single cells, cultured tissue or brain slices. Optogenetics can also simulate clinically relevant scenarios in animal models of brain diseases, including optogenetics-informed electrical stimulation protocols or closed-feedback control schemes. Possible indications are epilepsy, Parkinson disease or addiction. Some of these interventions may eventually be emulated in humans, for example with refined deep brain stimulation protocols or pharmacology (see Supplementary Fig. 12).
With the recent proof of principle of optogenetic vision restoration in humans with retinitis pigmentosa, longer-term gene therapy options remain open for optogenetics, although several challenges need to be addressed. Optimization of gene delivery vectors that are safe and produce long-lasting expression and optimization of light delivery to the desired organ are essential. Novel methods for light delivery deep into the tissue, overcoming the limited optical penetration depth and thereby minimizing the use of optical fibres, are also needed. One non-conventional solution is to introduce in situ sources of biological light, such as luminopsins. Triggered by a chemical process such as simple substrate delivery, these luminopsins do not require device implantation and can be tuned to control inhibitory or excitatory actuators. Further remote trigger methods involve energy-conversion schemes via mechano-luminescent nanoparticles. The energy could be provided by intermittent focused ultrasound, thus recharging light-emitting materials that can deliver short opsin-engaging pulses. The mechanosensitive TRAAK K+ channel, for example, could be specifically activated by ultrasound with submillisecond kinetics250, providing a new, orthogonal dimension for external non-invasive manipulation of neural circuits.
Clinical applications of optogenetics to the heart face many challenges compared with the more accessible, immune-privileged applications to the eye that have seen translational advances. Considering the potential impact for control of arrhythmias, efforts should continue to improve the genetic targeting by more specific promoters, safer viral vectors, longer-wavelength opsins for better tissue penetration and miniaturized distributed light sources. Basic science experiments with optogenetic tools provide invaluable insights for improvement of current cardiac devices and may yield new strategies for arrhythmia control111,191. In the meantime, optogenetics-empowered high-throughput systems can more immediately improve cardiotoxicity testing and drug development. Similarly, in vitro assays for drug development and personalized medicine may use humanized optogenetic tools, patient-derived cells and engineered tissues coupled with highly parallel all-optical electrophysiology techniques to yield a low-cost, faster and more efficient drug development pipeline.
Likewise, scalable optogenetic control of living plants, as discussed here, or microorganisms can be leveraged to address problems related to energy, food, biotechnology and climate challenges. As these do not involve deployment in the human body, they can be implemented on a shorter timescale, with less technical and regulatory obstacles.
Supplementary Material
Acknowledgements
This work was supported by the following funding sources: V.E. is supported by the IHU FOReSIGHT grant (Grant P-ALLOP3-IHU-000), the Axa Research Fund, the European Research Council (ERC) (ERC-2019-AdG; no. 885090) and the Agence National pour la Recherche (ANR-17-CE16-0021). E.E. is supported, in part, by the grants National Institutes of Health (NIH) R01 HL144157, NIH R21EB026152, National Science Foundation (NSF) 1705645 and NSF 1830941. J.V. is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC-2049 – 390688087). R.H. is supported by the German Research Foundation (DFG) (Koselleck award HE1640/42-1). P.H. is supported by the DFG (SFB1078, SPP1926, UniSysCat and Neurocure) and the ERC (Stardust 767092). P.H. is Hertie Professor for Neuroscience and supported by the Hertie Foundation. C.L. is supported by the ERC (ERC-2020-AdG, F-addict) and the Swiss National Science Foundations (no. 310030_189188 and CRSII5_186266). M.M. has received funding from the European Union’s Horizon 2020 research and innovation programme under Marie Skłodowska-Curie grant agreement no. 844492. Z.-H.P. is supported by the Ligon Research Center of Vision at Kresge Eye Institute, Dryer Foundation, Herrick Foundation and Research to Prevent Blindness to Department of Ophthalmology, Visual and Anatomical Sciences at Wayne State University School of Medicine. O.Y. is supported by the Joseph and Wolf Lebovic Charitable Foundation Chair for Research in Neuroscience, the ERC (grant no. 819496), the EU Horizon 2020 programme (H2020-ICT-2018-20 DEEPER 101016787) and the Israel Science Foundation (grant no. 3131/20).
Glossary
- Microelectrode
An electrode with a micrometre-sized tip used to record single-neuron activity.
- Optrode
An electrode coupled to an optical fibre used to record and manipulate neural activity in cells expressing an optogenetic actuator.
- Optogenetic actuators
Light-sensitive proteins that transiently modify cellular properties during illumination.
- Bidirectional voltage modulation
Changing the voltage in the depolarizing (excitatory) or hyperpolarizing (inhibitory) direction.
- Immediate early genes
Genes that are rapidly induced by elevated neural activity such as Fos.
- Optical clamp
A technique using light and real-time feedback to keep membrane electrical parameters, such as voltage or action potential shape, at a set desired value.
- Antidromic activation
Retrograde propagation of an action potential from the axon to the neuronal soma.
Footnotes
Competing interests
Z.-H.P. is a co-inventor on patents related to optogenetic vision restoration and is also a co-founder and scientific advisor of Ray Therapeutics. The other authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s43586-022-00136-4.
References
- 1.Famintzin A Die Wirkung der Lichtes auf die Bewegung der Chlamidomonas pulvisculus Ehr., Euglena viridis Ehr. und Orcillatoria insignis Tw. in Melanges Biologiques tires du Bulletin de I’Ácademie Imperial des Sciences De St. Petersbourg 73–93 (1866).
- 2.Kuhne WF Zur Photochemie der Netzhaut [German] (Carl Winter’s Universitatsbuchhandlung, 1877). [Google Scholar]
- 3.Darwin C The Power of Movements in Plants (Appleton, 1881). [Google Scholar]
- 4.Zemelman BV, Lee GA, Ng M & Miesenböck G Selective photostimulation of genetically chARGed neurons. Neuron 33, 15–22 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Boyden ES, Zhang F, Bamberg E, Nagel G & Deisseroth K Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Ishizuka T, Kakuda M, Araki R & Yawo H Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res 54, 85–94 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Li X et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopin and green algae channelrhodopsin. Proc. Natl Acad. Sci. USA 102, 17816–17821 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagel G et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol 15, 2279–2284 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Schroll C et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr. Biol 16, 1741–1747 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K & De Lecea L Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450, 420–424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglass AD, Kraves S, Deisseroth K, Schier AF & Engert F Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr. Biol 18, 1133–1137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron 62, 191–198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi AD et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron 50, 23–33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronzitti E et al. Recent advances in patterned photostimulation for optogenetics. J. Opt 19, 113001 (2017). [Google Scholar]
- 15.Hull K, Morstein J & Trauner D In vivo photopharmacology. Chem. Rev 118, 10710–10747 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Velema WA, Szymanski W & Feringa BL Photopharmacology: beyond proof of principle. J. Am. Chem. Soc 136, 2178–2191 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Kolar K, Knobloch C, Stork H, ŽnidariČ M & Weber W OptoBase: a web platform for molecular optogenetics. ACS Synth. Biol 7, 1825–1828 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Manoilov KY, Verkhusha VV & Shcherbakova DM A guide to the optogenetic regulation of endogenous molecules. Nat. Methods 18, 1027–1037 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Nagel G et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl Acad. Sci. USA 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozenberg A et al. Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses. Curr. Biol 30, 4910–4920.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Vierock J, Grimm C, Nitzan N & Hegemann P Molecular determinants of proton selectivity and gating in the red-light activated channelrhodopsin Chrimson. Sci. Rep 7, 9928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govorunova EG et al. Kalium channelrhodopsins are natural light-gated potassium channels that mediate optogenetic inhibition. Nat. Neurosci 10.1038/s41593-022-01094-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govorunova EG et al. The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep 7, 43358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahn M et al. High-efficiency optogenetic silencing with soma-targeted anion-conducting channelrhodopsins. Nat. Commun 9, 4125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Ari Y Excitatory actions of GABA during development: the nature of the nurture. Nat. Rev. Neurosci 3, 728–739 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Schneider F, Grimm C & Hegemann P Biophysics of channelrhodopsin. Annu. Rev. Biophys 44, 167–186 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Kuhne J et al. Unifying photocycle model for light adaptation and temporal evolution of cation conductance in channelrhodopsin-2. Proc. Natl Acad. Sci. USA 116, 9380–9389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sineshchekov OA, Govorunova EG, Li H & Spudich JL Bacteriorhodopsin-like channelrhodopsins: alternative mechanism for control of cation conductance. Proc. Natl Acad. Sci. USA 114, E9512–E9519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishi KE et al. Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689.e23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klapoetke NC et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda K et al. Crystal structure of the red light-activated channelrhodopsin Chrimson. Nat. Commun 9, 3949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govorunova EG et al. RubyACRs, non-algal anion channelrhodopsins with highly red-shifted absorption. Proc. Natl Acad. Sci. USA 117, 22833–22840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gradinaru V et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vierock J et al. BiPOLES is an optogenetic tool developed for bidirectional dual-color control of neurons. Nat. Commun 12, 4527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grusch M et al. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 33, 1713–1726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kainrath S, Stadler M, Reichhart E, Distel M & Janovjak H Green-light-induced inactivation of receptor signaling using cobalamin-binding domains. Angew. Chem. Int. Ed 56, 4608–4611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang K-Y et al. Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat. Commun 5, 4057 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Stierl M et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem 286, 1181–1188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–U634 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Parker D Neuronal network analyses: premises, promises and uncertainties. Philos. Trans. R. Soc. B Biol. Sci 365, 2315–2328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuong AS et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci 17, 1123–1129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiegert JS, Mahn M, Prigge M, Printz Y & Yizhar O Silencing neurons: tools, applications, and experimental constraints. Neuron 95, 504–529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rost BR et al. Optogenetic acidification of synaptic vesicles and lysosomes. Nat. Neurosci 18, 1845–1852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Junge W Protons, the thylakoid membrane, and the chloroplast ATP synthase. Ann. N. Y. Acad. Sci 574, 268–286 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Raimondo JV, Kay L, Ellender TJ & Akerman CJ Optogenetic silencing strategies differ in their effects on inhibitory synaptic transmission. Nat. Neurosci 15, 1102–1104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khorana HG, Knox BE, Nasi E, Swanson R & Thompson DA Expression of a bovine rhodopsin gene in Xenopus oocytes — demonstration of light-dependent ionic currents. Proc. Natl Acad. Sci. USA 85, 7917–7921 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karunarathne WA, Giri L, Patel AK, Venkatesh KV & Gautam N Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc. Natl Acad. Sci 110, E1575–E1583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karunarathne WA, Giri L, Kalyanaraman V & Gautam N Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc. Natl Acad. Sci. USA 110, E1565–E1574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masseck OA et al. Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron 81, 1263–1273 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Bailes HJ, Zhuang L-Y & Lucas RJ Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS ONE 7, e30774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spoida K et al. Melanopsin variants as intrinsic optogenetic on and off switches for transient versus sustained activation of G protein pathways. Curr. Biol 26, 1206–1212 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Lin B, Koizumi A, Tanaka N, Panda S & Masland RH Restoration of visual function in retinal degeneration mice by ectopic expression of melanopsin. Proc. Natl Acad. Sci. USA 105, 16009–16014 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsunematsu T, Tanaka KF, Yamanaka A & Koizumi A Ectopic expression of melanopsin in orexin/hypocretin neurons enables control of wakefulness of mice in vivo by blue light. Neurosci. Res 75, 23–28 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Koyanagi M et al. in Optogenetics 141–151 (Springer, 2021). [Google Scholar]
- 55.Eickelbeck D et al. Lamprey parapinopsin (“UVLamP”): a bistable UV-sensitive optogenetic switch for ultrafast control of GPCR pathways. ChemBioChem 21, 612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Copits BA et al. A photoswitchable GPCR-based opsin for presynaptic inhibition. Neuron 109, 1791–1809. e1711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahn M et al. Efficient optogenetic silencing of neurotransmitter release with a mosquito rhodopsin. Neuron 109, 1621–1635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J-M et al. Light-driven activation of β2-adrenergic receptor signaling by a chimeric rhodopsin containing the β2-adrenergic receptor cytoplasmic loops. Biochemistry 44, 2284–2292 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Airan RD, Thompson KR, Fenno LE, Bernstein H & Deisseroth K Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Osorno T et al. Light control of G protein signaling pathways by a novel photopigment. PLoS ONE 13, e0205015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradley SJ & Challiss RJ G protein-coupled receptor signalling in astrocytes in health and disease: a focus on metabotropic glutamate receptors. Biochem. Pharmacol 84, 249–259 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Bernal Sierra YA et al. Potassium channel-based optogenetic silencing. Nat. Commun 9, 4611 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Marco RJ, Groneberg AH, Yeh CM, Castillo Ramirez LA & Ryu S Optogenetic elevation of endogenous glucocorticoid level in larval zebrafish. Front. Neural Circuits 7, 82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jansen V et al. Controlling fertilization and cAMP signaling in sperm flagella by optogenetics. eLife 4, e05161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang SX et al. Hypothalamic dopamine neurons motivate mating through persistent cAMP signalling. Nature 597, 245–249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheib U et al. The rhodopsin–guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Sci. Signal 8, rs8 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Scheib U et al. Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 Å structure of the adenylyl cyclase domain. Nat. Commun 9, 2046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao S et al. Optogenetic manipulation of cGMP in cells and animals by the tightly light-regulated guanylyl-cyclase opsin CyclOp. Nat. Commun 6, 8046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Broser M et al. NeoR, a near-infrared absorbing rhodopsin. Nat. Commun 11, 5682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rost BR, Schneider-Warme F, Schmitz D & Hegemann P Optogenetic tools for subcellular applications in neuroscience. Neuron 96, 572–603 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Polesskaya O et al. Optogenetic regulation of transcription. BMC Neurosci. 19, 3–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polstein LR & Gersbach CA Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J. Am. Chem. Soc 134, 16480–16483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konermann S et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nihongaki Y, Kawano F, Nakajima T & Sato M Photoactivatable CRISPR–Cas9 for optogenetic genome editing. Nat. Biotechnol 33, 755–760 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Nihongaki Y et al. CRISPR–Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 14, 963–966 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Raper AT, Stephenson AA & Suo Z Functional insights revealed by the kinetic mechanism of CRISPR/Cas9. J. Am. Chem. Soc 140, 2971–2984 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Mardinly AR et al. Precise multimodal optical control of neural ensemble activity. Nat. Neurosci 21, 881–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arenkiel BR et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54, 205–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madisen L et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci 15, 793–802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lammel S et al. Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron 85, 429–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song AJ & Palmiter RD Detecting and avoiding problems when using the Cre–lox system. Trends Genet. 34, 333–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graybuck LT et al. Enhancer viruses and a transgenic platform for combinatorial cell subclass-specific labeling. Preprint at bioRxiv 10.1101/525014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nair RR, Blankvoort S, Lagartos MJ & Kentros C Enhancer-driven gene expression (EDGE) enables the generation of viral vectors specific to neuronal subtypes. iScience 23, 100888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jüttner J et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci 22, 1345–1356 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Tervo DGR et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang S et al. Long-range and local circuits for top-down modulation of visual cortex processing. Science 345, 660–665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Challis RC et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc 14, 379–414 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Mathiesen SN, Lock JL, Schoderboeck L, Abraham WC & Hughes SM CNS transduction benefits of AAV-PHP.eB over AAV9 are dependent on administration route and mouse strain. Mol. Ther. Methods Clin. Dev 19, 447–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.LoTurco J, Manent J-B & Sidiqi F New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb. Cortex 19, i120–i125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dal Maschio M et al. High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat. Commun 3, 960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosin JM & Kurrasch DM In utero electroporation induces cell death and alters embryonic microglia morphology and expression signatures in the developing hypothalamus. J. Neuroinflamm 15, 181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mahn M, Prigge M, Ron S, Levy R & Yizhar O Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat. Neurosci 19, 554–556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ullrich S, Gueta R & Nagel G Degradation of channelopsin-2 in the absence of retinal and degradation resistance in certain mutants. Biol. Chem 394, 271–280 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Hofherr A, Fakler B & Klöcker N Selective Golgi export of Kir2.1 controls the stoichiometry of functional Kir2.x channel heteromers. J. Cell Sci 118, 1935–1943 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Mattis J et al. An analysis of new and existing opsins for scientific application. Eur. Biophys. J. Biophys. Lett 40, 68–69 (2011). [Google Scholar]
- 98.Zhou Y et al. Optogenetic control of plant growth by a microbial rhodopsin. Nat. Plants 7, 144–151 (2021). [DOI] [PubMed] [Google Scholar]
- 99.Adam Y et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu C, Ivanova E, Zhang Y & Pan Z-H rAAV-mediated subcellular targeting of optogenetic tools in retinal ganglion cells in vivo. PLoS ONE 8, e66332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baker CA, Elyada YM, Parra A & Bolton MM Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. eLife 5, e14193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shemesh OA et al. Temporally precise single-cell-resolution optogenetics. Nat. Neurosci 20, 1796–1806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forli A et al. Two-photon bidirectional control and imaging of neuronal excitability with high spatial resolution in vivo. Cell Rep. 22, 3087–3098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Forli A, Pisoni M, Printz Y, Yizhar O & Fellin T Optogenetic strategies for high-efficiency all-optical interrogation using blue-light-sensitive opsins. eLife 10, e63359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Spaulding EL & Burgess RW Accumulating evidence for axonal translation in neuronal homeostasis. Front. Neurosci 11, 312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson HE & Toettcher JE Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol 52, 42–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stujenske JM, Spellman T & Gordon JA Modeling the spatiotemporal dynamics of light and heat propagation for in vivo optogenetics. Cell Rep. 12, 525–534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rungta RL, Osmanski B-F, Boido D, Tanter M & Charpak S Light controls cerebral blood flow in naive animals. Nat. Commun 8, 14191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pisanello M et al. Tailoring light delivery for optogenetics by modal demultiplexing in tapered optical fibers. Sci. Rep 8, 4467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arrenberg AB, Stainier DY, Baier H & Huisken J Optogenetic control of cardiac function. Science 330, 971–974 (2010). [DOI] [PubMed] [Google Scholar]
- 111.Burton RA et al. Optical control of excitation waves in cardiac tissue. Nat. Photonics 9, 813–816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Majumder R et al. Optogenetics enables real-time spatiotemporal control over spiral wave dynamics in an excitable cardiac system. eLife 7, e41076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scardigli M et al. Real-time optical manipulation of cardiac conduction in intact hearts. J. Physiol 596, 3841–3858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen I-W, Papagiakoumou E & Emiliani V Towards circuit optogenetics. Curr. Opin. Neurobiol 50, 179–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mager T et al. High frequency neural spiking and auditory signaling by ultrafast red-shifted optogenetics. Nat. Commun 9, 1750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ronzitti E et al. Submillisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of chronos. J. Neurosci 37, 10679–10689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pégard NC et al. Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT). Nat. Commun 8, 1228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marshel JH et al. Cortical layer-specific critical dynamics triggering perception. Science 365, eaaw5202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin J, Knutsen P, Muller A, Kleinfeld D & Tsien R ReaChR: a red-shifted variant of channelrhodopsin enables neuronal activation through the intact skull. Nat. Neurosci 16, 1499–1508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yizhar O et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen I-W et al. In vivo submillisecond two-photon optogenetics with temporally focused patterned light. J. Neurosci 39, 3484–3497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feldbauer K et al. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl Acad. Sci. USA 106, 12317–12322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Papagiakoumou E et al. Scanless two-photon excitation of channelrhodopsin-2. Nat. Methods 7, 848–854 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Andrasfalvy BK, Zemelman BV, Tang J & Vaziri A Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc. Natl Acad. Sci. USA 107, 11981–11986 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Packer AM, Russell LE, Dalgleish HW & Häusser M Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods 12, 140–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hernandez O et al. Three-dimensional spatiotemporal focusing of holographic patterns. Nat. Commun 7, 11928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Accanto N et al. Multiplexed temporally focused light shaping for high-resolution multi-cell targeting. Optica 5, 1478–1491 (2018). [Google Scholar]
- 128.Chaigneau E et al. Two-photon holographic stimulation of ReaChR. Front. Cell. Neurosci 10, 234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Robinson NT et al. Targeted activation of hippocampal place cells drives memory-guided spatial behavior. Cell 183, 1586–1599.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dalgleish HW et al. How many neurons are sufficient for perception of cortical activity? eLife 9, e58889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gill JV et al. Precise holographic manipulation of olfactory circuits reveals coding features determining perceptual detection. Neuron 108, 382–393.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dal Maschio M, Donovan JC, Helmbrecht TO & Baier H Linking neurons to network function and behavior by two-photon holographic optogenetics and volumetric imaging. Neuron 94, 774–789.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 133.Jennings JH et al. Interacting neural ensembles in orbitofrontal cortex for social and feeding behaviour. Nature 565, 645–649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Accanto N et al. Multiplexed temporally focused light shaping through a gradient index lens for precise in-depth optogenetic photostimulation. Sci. Rep 9, 7603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rowlands CJ et al. Wide-field three-photon excitation in biological samples. Light Sci. Appl 6, e16255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jackman SL, Beneduce BM, Drew IR & Regehr WG Achieving high-frequency optical control of synaptic transmission. J. Neurosci 34, 7704–7714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cardin JA et al. Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of channelrhodopsin-2. Nat. Protoc 5, 247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu J et al. Improved orange and red Ca2+ indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci 4, 963–972 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Platisa J & Pieribone VA Genetically encoded fluorescent voltage indicators: are we there yet? Curr. Opin. Neurobiol 50, 146–153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Urakubo H, Yagishita S, Kasai H, Kubota Y & Ishii S The critical balance between dopamine D2 receptor and RGS for the sensitive detection of a transient decay in dopamine signal. PLoS Comput. Biol 17, e1009364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang H, Jing M & Li Y Lighting up the brain: genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Curr. Opin. Neurobiol 50, 171–178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leopold AV, Shcherbakova DM & Verkhusha VV Fluorescent biosensors for neurotransmission and neuromodulation: engineering and applications. Front. Cell. Neurosci 13, 474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gradinaru V, Mogri M, Thompson KR, Henderson JM & Deisseroth K Optical deconstruction of parkinsonian neural circuitry. Science 324, 354–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu YE, Pan L, Zuo Y, Li X & Hong W Detecting activated cell populations using single-cell RNA-seq. Neuron 96, 313–329 (2017). [DOI] [PubMed] [Google Scholar]
- 145.Franceschini A, Costantini I, Pavone FS & Silvestri L Dissecting neuronal activation on a brain-wide scale with immediate early genes. Front. Neurosci 14, 1111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guenthner CJ, Miyamichi K, Yang HH, Heller HC & Luo L Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Grinvald A, Lieke E, Frostig RD, Gilbert CD & Wiesel TN Functional architecture of cortex revealed by optical imaging of intrinsic signals. Nature 324, 361–364 (1986). [DOI] [PubMed] [Google Scholar]
- 148.Juavinett AL, Nauhaus I, Garrett ME, Zhuang J & Callaway EM Automated identification of mouse visual areas with intrinsic signal imaging. Nat. Protoc 12, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee JH et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature 465, 788–792 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Uludağ K & Blinder P Linking brain vascular physiology to hemodynamic response in ultra-high field MRI. Neuroimage 168, 279–295 (2018). [DOI] [PubMed] [Google Scholar]
- 151.Albers F, Wachsmuth L, Schache D, Lambers H & Faber C Functional MRI readouts from bold and diffusion measurements differentially respond to optogenetic activation and tissue heating. Front. Neurosci 13, 1104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Edelman BJ & Macé E Functional ultrasound brain imaging: bridging networks, neurons and behavior. Curr. Opin. Biomed. Eng 18, 100286 (2021). [Google Scholar]
- 153.Brecht M et al. Organization of rat vibrissa motor cortex and adjacent areas according to cytoarchitectonics, microstimulation, and intracellular stimulation of identified cells. J. Comp. Neurol 479, 360–373 (2004). [DOI] [PubMed] [Google Scholar]
- 154.Heindorf M, Arber S & Keller GB Mouse motor cortex coordinates the behavioral response to unpredicted sensory feedback. Neuron 99, 1040–1054.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dana H et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019). [DOI] [PubMed] [Google Scholar]
- 156.Guo ZV et al. Flow of cortical activity underlying a tactile decision in mice. Neuron 81, 179–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jun JJ et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Steinmetz NA et al. Neuropixels 2.0: a miniaturized high-density probe for stable, long-term brain recordings. Science 372, eabf4588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kvitsiani D et al. Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature 498, 363–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ferreira-Pinto MJ et al. Functional diversity for body actions in the mesencephalic locomotor region. Cell 184, 4564–4578.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pascoli V et al. Contrasting forms of cocaine-evoked plasticity control components of relapse. Nature 509, 459–464 (2014). [DOI] [PubMed] [Google Scholar]
- 162.Burguière E, Monteiro P, Feng G & Graybiel AM Optogenetic stimulation of lateral orbitofronto-striatal pathway suppresses compulsive behaviors. Science 340, 1243–1246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hartong DT, Berson EL & Dryja TP Retinitis pigmentosa. Lancet 368, 1795–1809 (2006). [DOI] [PubMed] [Google Scholar]
- 164.de Jong EK, Geerlings MJ & den Hollander AI Age-related macular degeneration. Genet. Genomics Eye Dis 10.1016/B978-0-12-816222-4.00010-1 (2020). [DOI] [Google Scholar]
- 165.Sahel J-A et al. Partial recovery of visual function in a blind patient after optogenetic therapy. Nat. Med 27, 1223–1229 (2021). [DOI] [PubMed] [Google Scholar]
- 166.Tomita H et al. Restoration of the majority of the visual spectrum by using modified Volvox channelrhodopsin-1. Mol. Ther 22, 1434–1440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sengupta A et al. Red-shifted channelrhodopsin stimulation restores light responses in blind mice, macaque retina, and human retina. EMBO Mol. Med 8, 1248–1264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gauvain G et al. Optogenetic therapy: high spatiotemporal resolution and pattern discrimination compatible with vision restoration in non-human primates. Commun. Biol 4, 125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mattis J et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods 9, 159–172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ganjawala TH, Lu Q, Fenner MD, Abrams GW & Pan Z-H Improved CoChR variants restore visual acuity and contrast sensitivity in a mouse model of blindness under ambient light conditions. Mol. Ther 27, 1195–1205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cehajic-Kapetanovic J et al. Restoration of vision with ectopic expression of human rod opsin. Curr. Biol 25, 2111–2122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gaub BM, Berry MH, Holt AE, Isacoff EY & Flannery JG Optogenetic vision restoration using rhodopsin for enhanced sensitivity. Mol. Ther 23, 1562–1571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Berry MH et al. Restoration of high-sensitivity and adapting vision with a cone opsin. Nat. Commun 10, 1221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.van Wyk M, Pielecka-Fortuna J, Löwel S & Kleinlogel S Restoring the ON switch in blind retinas: opto-mGluR6, a next-generation, cell-tailored optogenetic tool. PLoS Biol. 13, e1002143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Surace EM & Auricchio A Versatility of AAV vectors for retinal gene transfer. Vis. Res 48, 353–359 (2008). [DOI] [PubMed] [Google Scholar]
- 176.Ivanova E, Hwang G-S, Pan Z-H & Troilo D Evaluation of AAV-mediated expression of Chop2-GFP in the marmoset retina. Investig. Ophthalmol. Vis. Sci 51, 5288–5296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yin L et al. Intravitreal injection of AAV2 transduces macaque inner retina. Investig. Ophthalmol. Vis. Sci 52, 2775–2783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lagali PS et al. Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration. Nat. Neurosci 11, 667–675 (2008). [DOI] [PubMed] [Google Scholar]
- 179.Cronin T et al. Efficient transduction and optogenetic stimulation of retinal bipolar cells by a synthetic adeno-associated virus capsid and promoter. EMBO Mol. Med 6, 1175–1190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Macé E et al. Targeting channelrhodopsin-2 to ON-bipolar cells with vitreally administered AAV restores ON and OFF visual responses in blind mice. Mol. Ther 23, 7–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lu Q, Ganjawala TH, Krstevski A, Abrams GW & Pan Z-H Comparison of AAV-mediated optogenetic vision restoration between retinal ganglion cell expression and ON bipolar cell targeting. Mol. Ther. Methods Clin. Dev 18, 15–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Busskamp V et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science 329, 413–417 (2010). [DOI] [PubMed] [Google Scholar]
- 183.Marc RE, Jones BW, Watt CB & Strettoi E Neural remodeling in retinal degeneration. Prog. Retin. Eye Res 22, 607–655 (2003). [DOI] [PubMed] [Google Scholar]
- 184.Bruegmann T, Beiert T, Vogt CC, Schrickel JW & Sasse P Optogenetic termination of atrial fibrillation in mice. Cardiovas. Res 114, 713–723 (2018). [DOI] [PubMed] [Google Scholar]
- 185.Bruegmann T et al. Optogenetic control of heart muscle in vitro and in vivo. Nat. Methods 7, 897–900 (2010). [DOI] [PubMed] [Google Scholar]
- 186.Nussinovitch U & Gepstein L Optogenetics for in vivo cardiac pacing and resynchronization therapies. Nat. Biotechnol 33, 750–754 (2015). [DOI] [PubMed] [Google Scholar]
- 187.Bruegmann T et al. Optogenetic defibrillation terminates ventricular arrhythmia in mouse hearts and human simulations. J. Clin. Invest 126, 3894–3904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Boyle PM, Williams JC, Ambrosi CM, Entcheva E & Trayanova NA A comprehensive multiscale framework for simulating optogenetics in the heart. Nat. Commun 4, 2370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Quiñonez Uribe RA, Luther S, Diaz-Maue L & Richter C Energy-reduced arrhythmia termination using global photostimulation in optogenetic murine hearts. Front. Physiol 9, 1651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nyns EC et al. An automated hybrid bioelectronic system for autogenous restoration of sinus rhythm in atrial fibrillation. Sci. Transl. Med 11, eaau6447 (2019). [DOI] [PubMed] [Google Scholar]
- 191.Entcheva E & Kay MW Cardiac optogenetics: a decade of enlightenment. Nat. Rev. Cardiol 18, 349–367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zaglia T et al. Optogenetic determination of the myocardial requirements for extrasystoles by cell type-specific targeting of channelrhodopsin-2. Proc. Natl Acad. Sci. USA 112, E4495–E4504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ambrosi CM & Entcheva E Optogenetics’ promise: pacing and cardioversion by light? Future Cardiol. 10, 1–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Williams JC et al. Computational optogenetics: empirically-derived voltage-and light-sensitive channelrhodopsin-2 model. PLoS Comput. Biol 9, e1003220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Karathanos TV, Boyle PM & Trayanova NA Optogenetics-enabled dynamic modulation of action potential duration in atrial tissue: feasibility of a novel therapeutic approach. Europace 16, iv69–iv76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rajendran PS et al. Identification of peripheral neural circuits that regulate heart rate using optogenetic and viral vector strategies. Nat. Commun 10, 1944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ambrosi CM, Sadananda G, Han JL & Entcheva E Adeno-associated virus mediated gene delivery: implications for scalable in vitro and in vivo cardiac optogenetic models. Front. Physiol 10, 168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wengrowski AM et al. Optogenetic release of norepinephrine from cardiac sympathetic neurons alters mechanical and electrical function. Cardiovasc. Res 105, 143–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Moreno A et al. Sudden heart rate reduction upon optogenetic release of acetylcholine from cardiac parasympathetic neurons in perfused hearts. Front. Physiol 10, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Greenberg B et al. Prevalence of AAV1 neutralizing antibodies and consequences for a clinical trial of gene transfer for advanced heart failure. Gene Ther. 23, 313–319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Crocini C et al. Optogenetics design of mechanistically-based stimulation patterns for cardiac defibrillation. Sci. Rep 6, 35628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hussaini S et al. Drift and termination of spiral waves in optogenetically modified cardiac tissue at sub-threshold illumination. eLife 10, e59954 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hochbaum DR et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Klimas A et al. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat. Commun 7, 11542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Klimas A, Ortiz G, Boggess SC, Miller EW & Entcheva E Multimodal on-axis platform for all-optical electrophysiology with near-infrared probes in human stem-cell-derived cardiomyocytes. Prog. Biophys. Mol. Biol 154, 62–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dempsey GT et al. Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging. J. Pharmacol. Toxicol. Meth 81, 240–250 (2016). [DOI] [PubMed] [Google Scholar]
- 207.Dwenger M et al. Chronic optical pacing conditioning of h-iPSC engineered cardiac tissues. J. Tissue Eng 10, 2041731419841748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Quach B, Krogh-Madsen T, Entcheva E & Christini DJ Light-activated dynamic clamp using iPSC-derived cardiomyocytes. Biophys. J 115, 2206–2217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ochoa-Fernandez R et al. Optogenetic control of gene expression in plants in the presence of ambient white light. Nat. Methods 17, 717–725 (2020). [DOI] [PubMed] [Google Scholar]
- 210.Christie JM & Zurbriggen MD Optogenetics in plants. New Phytol. 229, 3108–3115 (2021). [DOI] [PubMed] [Google Scholar]
- 211.Omelina ES et al. Optogenetic and chemical induction systems for regulation of transgene expression in plants: use in basic and applied research. Int. J. Mol. Sci 23, 1737 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Papanatsiou M et al. Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 363, 1456–1459 (2019). [DOI] [PubMed] [Google Scholar]
- 213.Reyer A et al. Channelrhodopsin-mediated optogenetics highlights a central role of depolarization-dependent plant proton pumps. Proc. Natl Acad. Sci. USA 117, 20920–20925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhou Y et al. Extending the anion channelrhodopsin-based toolbox for plant optogenetics. Membranes 11, 287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang S et al. Optogenetic control of the guard cell membrane potential and stomatal movement by the light-gated anion channel Gt ACR1. Sci. Adv 7, eabg4619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Twell D, Yamaguchi J & McCORMICK S Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109, 705–713 (1990). [DOI] [PubMed] [Google Scholar]
- 217.Govorunova EG, Sineshchekov OA, Janz R, Liu X & Spudich JL NEUROSCIENCE. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gutermuth T et al. Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 25, 4525–4543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gutermuth T et al. Tip-localized Ca2+-permeable channels control pollen tube growth via kinasedependent R-and S-type anion channel regulation. New Phytol. 218, 1089–1105 (2018). [DOI] [PubMed] [Google Scholar]
- 220.Gilroy S et al. ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 171, 1606–1615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Choi W-G et al. Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant. J 90, 698–707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fromm J & Lautner S Electrical signals and their physiological significance in plants. Plant Cell Environ. 30, 249–257 (2007). [DOI] [PubMed] [Google Scholar]
- 223.Vodeneev V, Katicheva L & Sukhov V Electrical signals in higher plants: mechanisms of generation and propagation. Biophysics 61, 505–512 (2016). [Google Scholar]
- 224.Szechynska-Hebda M, Lewandowska M & Karpiński S Electrical signaling, photosynthesis and systemic acquired acclimation. Front. Physiol 8, 684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hedrich R & Neher E Venus flytrap: how an excitable, carnivorous plant works. Trends Plant. Sci 23, 220–234 (2018). [DOI] [PubMed] [Google Scholar]
- 226.Kravitz A & Bonci A Optogenetics, physiology, and emotions. Front. Behav. Neurosci 7, 169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Häusser M Optogenetics: the age of light. Nat. Methods 11, 1012–1014 (2014). [DOI] [PubMed] [Google Scholar]
- 228.Dawydow A et al. Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl Acad. Sci. USA 111, 13972–13977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Inoue S.-i & Kinoshita T Blue light regulation of stomatal opening and the plasma membrane H+-ATPase. Plant Physiol. 174, 531–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pilot G et al. Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J. Biol. Chem 276, 3215–3221 (2001). [DOI] [PubMed] [Google Scholar]
- 231.Catterall WA, Wisedchaisri G & Zheng N The chemical basis for electrical signaling. Nat. Chem. Biol 13, 455–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Duan X, Nagel G & Gao S Mutated channelrhodopsins with increased sodium and calcium permeability. Appl. Sci 9, 664 (2019). [Google Scholar]
- 233.Miyashita T, Shao YR, Chung J, Pourzia O & Feldman D Long-term channelrhodopsin-2 (ChR2) expression can induce abnormal axonal morphology and targeting in cerebral cortex. Front. Neural Circuits 7, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rapti K et al. Neutralizing antibodies against AAV serotypes 1, 2, 6, and 9 in sera of commonly used animal models. Mol. Ther 20, 73–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Al-Juboori SI et al. Light scattering properties vary across different regions of the adult mouse brain. PLoS ONE 8, e67626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shin Y et al. Characterization of fiber-optic light delivery and light-induced temperature changes in a rodent brain for precise optogenetic neuromodulation. Biomed. Opt. Express 7, 4450–4471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Arias-Gil G, Ohl FW, Takagaki K & Lippert MT Measurement, modeling, and prediction of temperature rise due to optogenetic brain stimulation. Neurophotonics 3, 045007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Owen SF, Liu MH & Kreitzer AC Thermal constraints on in vivo optogenetic manipulations. Nat. Neurosci 22, 1061–1065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yang F & Zheng J High temperature sensitivity is intrinsic to voltage-gated potassium channels. eLife 3, e03255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sabatini BL & Regehr WG Timing of neurotransmission at fast synapses in the mammalian brain. Nature 384, 170–172 (1996). [DOI] [PubMed] [Google Scholar]
- 241.Moser EI & Andersen P Conserved spatial learning in cooled rats in spite of slowing of dentate field potentials. J. Neurosci 14, 4458–4466 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chen R et al. Deep brain optogenetics without intracranial surgery. Nat. Biotechnol 39, 161–164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Podgorski K & Ranganathan G Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol 116, 1012–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Picot A et al. Temperature rise under two-photon optogenetic brain stimulation. Cell Rep. 24, 1243–1253.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 245.Pan Z-H, Lu Q, Bi A, Dizhoor AM & Abrams GW Optogenetic approaches to restoring vision. Annu. Rev. Vis. Sci 1, 185–210 (2015). [DOI] [PubMed] [Google Scholar]
- 246.Chuong A, Miri M & Acker L Non-invasive optogenetic neural silencing. Nat. Neurosci 17, 1123–1129 (2014).24997763 [Google Scholar]
- 247.Herman AM, Huang L, Murphey DK, Garcia I & Arenkiel BR Cell type-specific and time-dependent light exposure contribute to silencing in neurons expressing channelrhodopsin-2. eLife 3, e01481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ferenczi EA et al. Optogenetic approaches addressing extracellular modulation of neural excitability. Sci. Rep 6, 23947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Otchy TM et al. Acute off-target effects of neural circuit manipulations. Nature 528, 358–363 (2015). [DOI] [PubMed] [Google Scholar]
- 250.Sorum B, Rietmeijer RA, Gopakumar K, Adesnik H & Brohawn SG Ultrasound activates mechanosensitive TRAAK K+ channels through the lipid membrane. Proc. Natl Acad. Sci. USA 118, e2006980118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Strickland D et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kennedy MJ et al. Rapid blue-light–mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Levskaya A, Weiner OD, Lim WA & Voigt CA Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Farhi SL et al. Wide-area all-optical neurophysiology in acute brain slices. J. Neurosci 39, 4889–4908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Berndt A et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl Acad. Sci. USA 108, 7595–7600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Gong X et al. An ultra-sensitive step-function opsin for minimally invasive optogenetic stimulation in mice and macaques. Neuron 107, 38–51.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rajasethupathy P et al. Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Shevchenko V et al. Inward H+ pump xenorhodopsin: mechanism and alternative optogenetic approach. Sci. Adv 3, e1603187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Grimm C, Silapetere A, Vogt A, Sierra YAB & Hegemann P Electrical properties, substrate specificity and optogenetic potential of the engineered light-driven sodium pump eKR2. Sci. Rep 8, 9316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gradinaru V, Thompson KR & Deisseroth K eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 36, 129–139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Subach OM et al. FRCaMP, a red fluorescent genetically encoded calcium indicator based on calmodulin from Schizosaccharomyces pombe fungus. Int. J. Mol. Sci 22, 111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dalangin R et al. Far-red fluorescent genetically encoded calcium ion indicators. Preprint at bioRxiv 10.1101/2020.11.12.380089 (2020). [DOI] [Google Scholar]
- 263.Villette V et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Abdelfattah AS et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019). [DOI] [PubMed] [Google Scholar]
- 265.Kannan M et al. Fast, in vivo voltage imaging using a red fluorescent indicator. Nat. Methods 15, 1108–1116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Piatkevich KD et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol 14, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Miesenböck G, De Angelis DA & Rothman JE Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 (1998). [DOI] [PubMed] [Google Scholar]
- 268.Anikeeva P et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat. Neurosci. 15, 163–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Creed M, Pascoli VJ & Lüscher C Refining deep brain stimulation to emulate optogenetic treatment of synaptic pathology. Science 347, 659–664 (2015). [DOI] [PubMed] [Google Scholar]
- 270.Kuhn J, Gruendler TO, Klosterkötter J & Bartsch C Stimulating the addictive brain. Front. Hum. Neurosci 6, 220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


