Abstract
Active biological containment systems consist of two components, a killing element designed to induce cell death and a control element which modulates the expression of the killing function. We constructed a mini-Tn5 transposon bearing a fusion of the Plac promoter to the gef killing gene and a fusion of the Pm promoter to the lacI gene plus the positive regulator of the Pm promoter, the xylS gene. This mini-Tn5 transposon was transferred to the chromosome of Pseudomonas putida CMC4, and in culture this strain survived in the presence of 3-methylbenzoate (an XylS effector) and committed suicide in the absence of this aromatic compound. The rate of killing escape was on the order of 10−8 per cell and per generation. This contained strain and an uncontained control strain were used in outdoor tests performed in the spring-summer and autumn-winter periods to determine their survival in planted and unplanted soils with and without 3-methylbenzoate. In unplanted soils the numbers of both the contained strain and the uncontained strain per gram of soil tended to decrease, but the numbers of the contained strain decreased faster in soils without 3-methylbenzoate. The decrease in the number of CFU per gram of soil was faster in the spring-summer period than in the autumn-winter period. In planted soils survival in the rhizosphere and survival in bulk soil were studied. In the rhizosphere the uncontained control strain tended to become established at levels on the order of 105 to 106 CFU/g of soil regardless of the presence of 3-methylbenzoate. In the bulk soil the numbers of bacterial cells were 2 to 3 orders of magnitude lower. In planted soils the contained strain tended to disappear, but this tendency was more pronounced in the absence of 3-methylbenzoate and occurred faster in the summer assay than in the winter assay. We found no evidence of dispersal of the test strains outside the experimental plots.
Genetic engineering tools make it possible to clone and manipulate almost any given piece of DNA and to transfer this material to a wide range of organisms. This procedure has many applications in medicine, including expression in heterologous hosts of a number of genes that code for proteins and hormones of medical and pharmaceutical interest and biosynthesis of new antibiotics and other chemicals with added value (4, 11, 14). In recent years a number of tools have been specifically designed to develop microorganisms as biopesticides and as enhancers of plant growth which have potential uses in agriculture (7). In the area of bioremediation, recombinant microorganisms with improved activities for amelioration of pollution have also been constructed (17, 19). Although genetically engineered microorganisms (GEMs) can be used for bioproduction of added-value pharmaceuticals and other chemicals in physically contained sites, GEMs that are used to biologically treat polluted sites or stimulate plant growth and plant protection will eventually be used in open environments. Although the benefits of cleaning a polluted site by biological means are obvious, the long-term behavior of GEMs released in open environments is an unexplored area. The consequences of the persistence of GEMs at sites and their effects on recolonization of the sites by indigenous microbes are also unknown. These issues raise serious concerns (15).
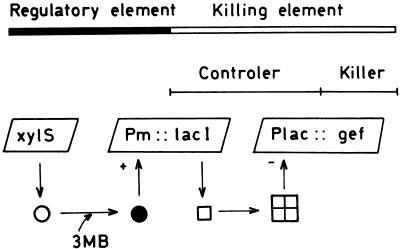
One way to decrease the persistence of GEMs in the environment is to provide them with active biological containment (ABC) systems. ABC systems are based on control of the expression of a lethal function (e.g., a porinlike protein or a nuclease) via sensory systems that recognize physical or chemical signals in the surrounding environment (15). Contreras et al. (5) designed the first circuit for biological containment of bacteria that degrade pollutants. This system was based on the well-characterized regulatory circuit for expression of the meta-cleavage pathway of the Pseudomonas putida TOL plasmid pWW0, the xylS gene and its cognate Pm promoter (21), and the gef gene of Escherichia coli, which encodes a membrane protein that collapses the cell membrane potential by generating pores (15). The model shown in Fig. 1 predicts that in the presence of XylS effectors (i.e., a wide range of alkyl-substituted, chloro-substituted, and other halosubstituted benzoates) (22), expression of the porinlike Gef protein is prevented and the strain survives and removes the pollutant. When the target compound is exhausted (or the microbe is spread to a nonpolluted site), the lack of induction of Pm results in the absence of the LacI protein in the host cell, which leads to expression of the lethal protein and cell death.
FIG. 1.
ABC model system. This model consists of the Pm promoter, which drives transcription of the meta-cleavage pathway of the TOL plasmid, and the xylS gene, which encodes the sensor protein (○) that interacts with alkylbenzoates and stimulates transcription from Pm (•). In the containment system the lacI gene coding for the LacI repressor protein (□) was cloned downstream from Pm. The lethal element consists of the Plac promoter fused to the gef gene of E. coli, a member of the hok gene family (15), which encodes a pore-forming protein. The system performs as described in the text. 3MB, 3-methylbenzoate.
This model system was shown to be functional in E. coli (5) and in Pseudomonas putida (12, 26), but it exhibited a relatively high rate of killing escape, which was found to be associated with the fact that the regulatory element of the ABC system was plasmid borne (5, 12, 25). In this study we describe transfer of the elements of the ABC system to the chromosome of P. putida KT2440 (9). The resulting new biologically contained strain was shown to function under laboratory conditions in accordance with the model proposed in Fig. 1, and the frequency of killing escape was less than 1 in 108 cells per cell and per generation. The strain with the ABC system was introduced into planted and unplanted pots with and without 3-methylbenzoate. Inoculated pots were kept under environmental conditions during the spring-summer and autumn-winter periods, and the survival of the contained strain and the survival of an uncontained control strain under these conditions were determined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study and their relevant characteristics are shown in Table 1. Bacterial strains were grown with shaking at 30°C. E. coli strains were grown on Luria-Bertani (LB) medium at 37°C (27). P. putida strains were grown in modified M9 minimal medium (1) supplemented with 28 mM glucose, 5 mM benzoate, 5 to 15 mM 3-methylbenzoate, or 10 mM p-hydroxyphenylacetic acid as a carbon source. When appropriate, antibiotics were used at the following final concentrations: chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; rifampin, 20 μg/ml; and tetracycline, 15 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| P. putida strains | ||
| CMC4 | (pWW0), Kmr 3MB+ mini-Tn5/ Km::(xylS Pm::lacI Plac::gef) | This study |
| CMC5 | Kmr, pCC102, mini-Tn5/ Tc::Pm::′luxAB | This study |
| EEZ32 | (pWW0), pCC102, Kmr Tcr 3MB+ | 26 |
| KT2440 | Prototroph, Cmr | 8 |
| UWC1 | Rifr derivative of KT2440 | 23 |
| E. coli strains | ||
| CC118λpir | Rifr, host for pSM1350 | 9 |
| HB101 | Smr, host for pRK600 | 13 |
| Plasmids | ||
| pCC102 | Kmr, xylS2 Pm::lacI | 5 |
| pRK600 | Cmr, ColE1 oriV, RK2mob+tra+ | 13 |
| pSM1350 | Kmr, mini-Tn5, xylS Pm::lacI, Plac::gef | This study |
| pWW0 | IncP9, mob+ tra+ 3MB+ | 21 |
Kmr, Apr, Cmr, and Rifr, resistance to kanamycin, ampicillin, chloramphenicol, and rifampin, respectively; 3MB+, ability to grow on 3-methylbenzoate.
Matings.
Triparental matings were used to mobilize nonautotransmissible plasmids into P. putida strains. Equal numbers (about 108 cells) of the recipient strain P. putida 2440 (benzoate positive, Cmr), the donor strain E. coli CC118λpir bearing the suicide pSM1350 plasmid, and the helper strain E. coli HB101 (pRK600) were mixed and deposited on a nitrocellulose filter placed on the surface of an LB agar plate containing 5 mM 3-methylbenzoate (6). Appropriate controls containing unmixed cells were included. P. putida transconjugants were selected on M9 minimal medium plates supplemented with kanamycin, chloramphenicol, 10 mM benzoate, and 5 mM 3-methylbenzoate. (Note that P. putida 2440 cannot use 3-methylbenzoate as a C source because it does not bear the TOL plasmid pWW0; in this case the aromatic carboxylic acid was used as a gratuitous inducer of the XylS protein to prevent cell killing.) Biparental matings were used to transfer the TOL plasmid pWW0 between P. putida strains under the conditions described previously with the controls described previously (23).
Transmission electron microscopy.
P. putida cells were harvested by centrifugation (4,000 × g, 5 min) and then immediately fixed with 2% (vol/vol) glutaraldehyde–1% (vol/vol) formaldehyde in cacodylate buffer, postfixed with osmium tetroxide in the presence of 2% (wt/vol) potassium ferrocyanide, and embedded in Eponate 12. Thin sections were poststained with uranyl acetate and lead citrate and examined with a Zeiss transmission electron microscope at an accelerating voltage of 75 kV.
Light emission measurements.
Soil leachates obtained from rhizosphere and bulk soils were prepared as follows: 10 g of soil was suspended in 90 ml of phosphate buffer, and after shaking for 1 h the soil was decanted and the liquid suspension was used for assays. To 1 ml of each soil leachate suspension we added 100 μl of a culture of P. putida CMC5 (Pm:′luxAB xylS) containing about 108 CFU/ml and then incubated the resulting culture for 1 h at 30°C to allow induction of Pm. To determine luciferase activity, the turbidity of the culture at 660 nm was adjusted to 0.1, and to 1 ml of this P. putida CMC5 cell suspension we added 0.1 ml of 0.01% (vol/vol) n-decylaldehyde and then recorded the time course of light emission immediately thereafter for 1 min with an LKB model 1250 luminometer (16). Activity was expressed as the peak height in relative light units per turbidity unit.
Seed coating and soil inoculation.
Cells in the mid-log phase in 1 liter of culture medium were harvested by centrifugation, washed twice in 50 mM phosphate–100 mM NaCl, and resuspended in the same buffer to a concentration of about 108 CFU/ml. About 300 seeds of corn (Zea mays) or broad bean (Vicia faba) were soaked in 200 to 400 ml of the two cell suspensions used for 30 min with gentle shaking at 30°C. The number of bacteria attached per seed was estimated as follows. Two seeds coated with P. putida were air dried and then transferred to a test tube containing 1 to 5 ml of 1× M9 minimal medium without a C source. The preparations were vortexed for 2 min, and then serial dilutions were spread onto plates. Each seed was coated with about 106 CFU (corn seeds) or 107 CFU (broad bean seeds). Two seeds per pot were sown at a depth of 2 cm in pots that were 40 cm in diameter and contained 40 kg of soil. Twelve pots per treatment were used; one pot was used per sample, and each pot was analyzed in triplicate.
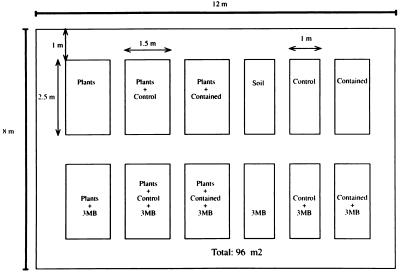
When bacteria were introduced into nonplanted soils, the pots used were 10 cm in diameter and contained about 1 kg of soil. Ten milliliters of bacterial inoculum was added to each pot, and the soil was mixed thoroughly to obtain a density of about 106 CFU/g of soil. The pots were grouped in subsets as shown in Fig. 2.
FIG. 2.
Distribution of subareas in the field release assay. In the spring-summer assay corn (Z. mays) plants were used, and in the autumn-winter assay the species used was broad bean (V. faba). Bacteria were introduced in the form of a biofilm on each seed or were homogeneously mixed with soil in pots without seeds. 3MB, 3-methylbenzoate.
Environmental release field design.
The Spanish Ministry of Environment provided the necessary permits to carry out outdoor assays with P. putida EEZ32 (26) and CMC4 (this study). On the advice of the authorities, the soil was kept in pots, which were placed in different subareas in a controlled, fenced-in, 96-m2 site located within the 2,000-m2 experimental area at the Estación Experimental del Zaidín of the Consejo Superior de Investigaciones Científicas, Granada, Spain (Fig. 2). The granulometric composition of this soil was as follows: sand, 43%; silt, 41%; and clay, 16%. The pH of the soil was 7.8, and its CaCO3 content was 6% (wt/wt). The experimental subareas in the agricultural experimental field site consisted of groups of pots filled with this soil. One group of pots contained seeds coated with the control bacteria or the contained bacteria, and other series of pots contained soil inoculated with the GEMs but no seeds. Some pots were supplemented with 0.01% (wt/wt) 3-methylbenzoate. To minimize edge effects and provide material to assess the dispersal of the GEMs, groups of pots were separated from each other by 1 m and the entire release area was completely surrounded by a buffer zone where GEMs were not introduced (Fig. 2). Z. mays (corn) seeds were used in the field trials performed during the spring and summer. The assay period lasted for 112 days from May to September 1996. During this period the daytime temperatures ranged from 25 to 45°C and the nighttime temperatures ranged from 10 to 20°C. V. faba (broad bean) seeds were used in the field trials performed during the autumn and winter. This assay was performed from December 1995 to March 1996. The temperature ranged from −3 to 13°C during the night and from 5 to 27°C during the day.
Monitoring bacteria in soil and the rhizosphere.
After germination, the first samples were obtained from individual plants after the appearance of the first true leaf (11 days after sowing for corn and 21 days after sowing for broad bean), and samples were obtained at subsequent times. Whole plants were gently removed from the soil, and the bacteria in the soil attached to the roots (rhizosphere soil) and in the rest of the soil (bulk soil) were counted.
Regardless of the type of soil sample, 10 g of soil was placed in a 250-ml Erlenmeyer flask containing 90 ml of M9 minimal medium without a C source and shaken for 30 min on a Heidolph bench shaker at 200 strokes per min. The soil suspensions were then serially diluted 10-fold, and 0.1-ml aliquots were spread in triplicate onto selective medium. The contained and control strain counts were determined on minimal medium supplemented with 5 mM 3-methylbenzoate as the sole C source and the appropriate antibiotics. No indigenous bacteria grew on the plates.
RESULTS
Construction of P. putida CMC4, a strain with the ABC system on the host chromosome.
A mini-Tn5-Km transposon bearing the Plac::gef fusion was transferred to the chromosome of P. putida 2440 via triparental mating as described in Materials and Methods. Twenty Kmr transconjugants, which appeared at a frequency of about 2 × 10−8 transconjugant per recipient cell, were selected for further study. As expected, all transconjugant cells grew on minimal medium containing glucose as the sole C source only if 3-methylbenzoate was also present, which confirmed the functionality of the ABC system. When the clones were grown with glucose as the C source and with 3-methylbenzoate in the absence of kanamycin for about 50 generations, we always found that 100% of the cells retained the Kmr marker and died when they were spread onto plates without 3-methylbenzoate. This suggested that the mini-Tn5 element bearing the ABC system was stably maintained in the host.
Ramos-González et al. (23) have shown that the TOL plasmid is able to mobilize the host chromosome and that the rate of mobilization is influenced by the physical location of the marker. For outdoor assays we were interested in selecting a clone bearing the ABC system in a region with a low mobilization rate. To find such a clone, we first transferred the TOL plasmid to each of the original transconjugants with the ABC system and then carried out mobilization studies. The donor strains used were the clones with the ABC system and the TOL plasmid (Rifs Kmr 3MB+), and P. putida UWC1 (Rifr Kms 3MB−) was the recipient. We obtained Rifr 3MB+ transconjugants of P. putida UWC1 cells at a frequency of 10−1 to 10−2 transconjugant per recipient, whereas the frequency of derivatives that had received the Kmr marker varied between 10−6 and less than 10−8 transconjugant per recipient, in agreement with previous findings (23). The clones whose Kmr markers were mobilized at a rate equal to or less than 10−8 were kept, and fluctuation tests (26) were performed to determine the rate of mutant escape from cell killing. It was found that the rate of mutation was around 10−8 per cell and per generation. This was at least 2 orders of magnitude lower than the rate of mutation reported previously for strains in which the killing element had been introduced on plasmids (5, 12, 13). One of these clones was chosen for further study and was designated P. putida CMC4.
The control strain used for the P. putida CMC4 study was a strain previously generated by workers in our group, P. putida EEZ32 (26). This strain is also a derivative of P. putida 2440 and carries the control element of the ABC system on low-copy-number plasmid pCC102, but it lacks the killing element.
To further confirm that P. putida CMC4 commits suicide when it is transferred to a medium without 3-methylbenzoate, we carried out a series of assays in which the contained strain P. putida CMC4 and the control strain P. putida EEZ32 were grown in M9 minimal medium containing glucose as the sole C source and 15 mM 3-methylbenzoate as a gratuitous inducer. Cells in the exponential phase were harvested by filtration and washed with 50 mM phosphate buffer, and then the cells were resuspended at a high density (about 108 CFU/ml) in minimal medium containing glucose but not 3-methylbenzoate. The sample was divided into two halves. To one of these halves we added 5 mM 3-methylbenzoate, and the other half was maintained without modification. All of the samples were then incubated at 30°C with shaking.
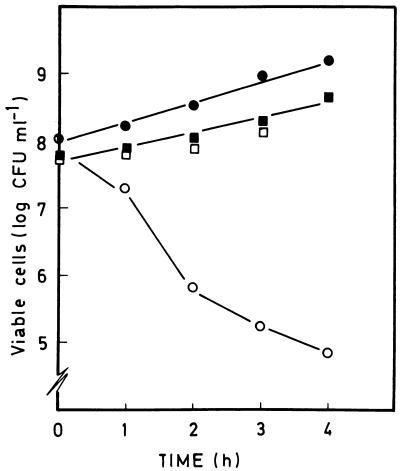
The number of CFU of both strains per milliliter increased with time in medium containing 3-methylbenzoate; this was also the case for the control strain in the absence of the aromatic carboxylic acid (Fig. 3). However, the number of cells of the contained strain P. putida CMC4 decreased with time in the absence of 3-methylbenzoate (Fig. 3).
FIG. 3.
Effect of removal of 3-methylbenzoate from the culture medium of P. putida EEZ32 and the contained strain P. putida CMC4. Cells of P. putida EEZ32 (▪ and □) and P. putida CMC4 (• and ○) growing exponentially on M9 minimal medium containing glucose and 15 mM 3-methylbenzoate were filtered, thoroughly washed with 50 mM phosphate buffer, and resuspended in M9 minimal medium containing glucose but not 3-methylbenzoate (□ and ○) or in M9 minimal medium containing glucose and 15 mM 3-methylbenzoate (▪ and •). At the times indicated the numbers of viable cells (CFU per milliliter) were determined in triplicate in LB medium supplemented with 5 mM 3-methylbenzoate and appropriate antibiotics.
The decrease in the number of P. putida CMC4 cells after transfer to 3-methylbenzoate-free medium represented cell death. As determined by transmission electron microscopy, P. putida CMC4 growing in the presence of 3-methylbenzoate exhibited typical P. putida morphology, whereas after 1.5 h in the culture medium without 3-methylbenzoate the P. putida CMC4 cells appeared to be deformed and lysed. These changes were not observed when the control strain was transferred to culture medium without 3-methylbenzoate (data not shown). The above results suggested that P. putida CMC4 and EEZ32 were appropriate strains for large-scale assays, such as field release studies.
Autumn-winter outdoor assay.
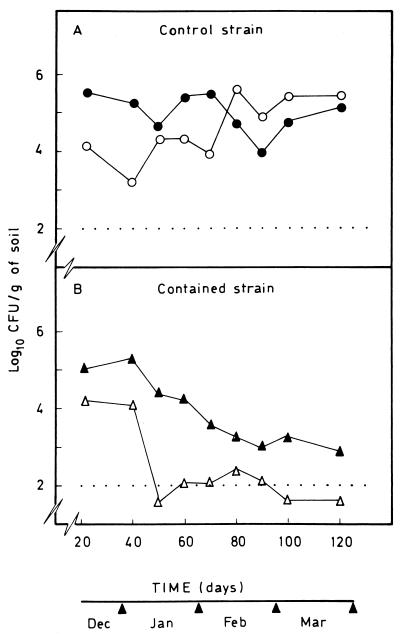
We examined the behavior of the two strains in pots in which broad beans were planted. After germination, the first sample was obtained when the first leaf appeared, 21 days after sowing. The results are presented in Fig. 4. Both in the absence and in the presence of 3-methylbenzoate the control strain tended to become established at a concentration of about 105 CFU per g of rhizosphere soil. During the assay the number of organisms ranged from 103 to 106 CFU/g of rhizosphere soil. Control strain P. putida EEZ32 became established in pots containing 3-methylbenzoate faster than it became established in the absence of this aromatic compound, but at the end of the study the total counts were similar to the total counts obtained in the absence of 3-methylbenzoate (Fig. 4A). The number of CFU of the contained strain per gram of rhizosphere soil tended to decrease with time, and this phenomenon was more pronounced in the absence of 3-methylbenzoate, so that after 100 days the concentration of the strain was below our detection limit. In the presence of 3-methylbenzoate the number of CFU per gram of rhizosphere soil was 1 to 2 orders of magnitude higher in any given sample, and at the end of the study we found almost 103 CFU/g of soil. Confirmation that the CFU in the pots into which the contained strain was introduced indeed represented contained strain cells was obtained from the fact that none of the bacteria selected on plates containing 3-methylbenzoate was able to grow on minimal medium supplemented with glucose, whereas all of the control bacteria survived in this medium.
FIG. 4.
Survival of P. putida CMC4 and EEZ32 under outdoor conditions in pots. Broad bean seeds were coated with P. putida EEZ32 (A) or P. putida CMC4 (B) and sown in pots in which the soil was not supplemented (○ and ▵) or was supplemented (• and ▴) with 3-methylbenzoate. The numbers of CFU per gram of rhizosphere soil were determined at the times indicated. Data are the averages of values from three independent counts, and the standard deviations were less than 10%.
At present, no method is available for repetitive extraction and reliable quantification of the amount of 3-methylbenzoate remaining in soil. For this reason we used an indirect method to detect the presence of 3-methylbenzoate (see Materials and Methods), which was based on light emission by P. putida CMC5 bearing xylS and a Pm::′luxAB fusion. This construct emits light when cells are exposed to a concentration of 3-methylbenzoate higher than 10 μM (16). We found that throughout the assay, leaching of the soil that had been supplemented with 3-methylbenzoate induced specific light emission in this strain (100 to 200 relative light units), whereas leaching of soils without 3-methylbenzoate induced no or very low levels of light emission (about 5 relative light units) (data not shown). This suggests that part of the 3-methylbenzoate initially added to the soil was still present and probably bioavailable.
For bulk soil (i.e., soil not attached to the root systems in pots), we observed that during the assay the number of control strain organisms ranged from about 103 to 3 × 104 CFU/g of soil regardless of the presence of 3-methylbenzoate (Table 2). For the contained strain P. putida CMC4, in the presence of 3-methylbenzoate the number of organisms ranged from about 102 to 5 × 103 CFU/g of soil (Table 2). In the absence of 3-methylbenzoate P. putida CMC4 was detected during the first 40 days of the assay; thereafter the number of CFU per gram of bulk soil was always below our detection limit. This suggests that colonization of the bulk soil by the contained strain was limited.
TABLE 2.
Colonization of bulk soil by P. putida CMC4 and EEZ32 in the autumn-winter outdoor assaya
| Strain | Presence of 3-methylbenzoate | CFU/g of soil after:
|
||
|---|---|---|---|---|
| 40 days | 60 days | 100 days | ||
| Control strain EEZ32 | + | 104 | 3 × 104 | 103 |
| − | 103 | 103 | 3 × 103 | |
| Contained strain CMC4 | + | 5 × 103 | 102 | 6 × 102 |
| − | 103 | <102 | <102 | |
The conditions were the conditions described in the legend to Fig. 4 except that the bacteria in the bulk soil were counted at the times indicated. The values are the averages of values for duplicate samples counted in triplicate, and the standard deviations were on the order of 10% of the values given.
The contained and control strains were also introduced into unplanted pots, and the number of CFU of both strains per gram of soil tended to decrease. The decreases in the control strain were similar in soils with and without 3-methylbenzoate, and this strain became established at levels below our detection limit (i.e., 100 CFU/g of soil) about 40 days after the start of the assay (data not shown). For the contained strain P. putida CMC4 we found that in the absence of 3-methylbenzoate the number of CFU per gram of soil after day 15 had decreased to just above our detection limit; thereafter the number of CFU per gram of soil fell below our detection limit (data not shown). In the presence of 3-methylbenzoate the number of contained cells also tended to decrease, but at a slightly slower rate than in the absence of 3-methylbenzoate, so that the number of CFU per gram of soil had fallen below our detection limit at day 35 after the start of the assay (data not shown).
In all of the assays described above we were unable to detect mutants of the contained strain that escaped killing and became established in the rhizosphere or bulk soil in the planted pots or in unplanted soils.
The population of indigenous soil microbes was estimated by determining the number of CFU of p-hydroxyphenylacetic acid utilizers per gram of soil. These organisms were known to be members of one of the predominant populations in the soil used (26). The numbers of these microbes, regardless of the presence of broad beans and of 3-methylbenzoate, were on the order of 106 CFU/g of rhizosphere soil and were 1 order of magnitude lower in the bulk soil.
Spring-summer outdoor assay.
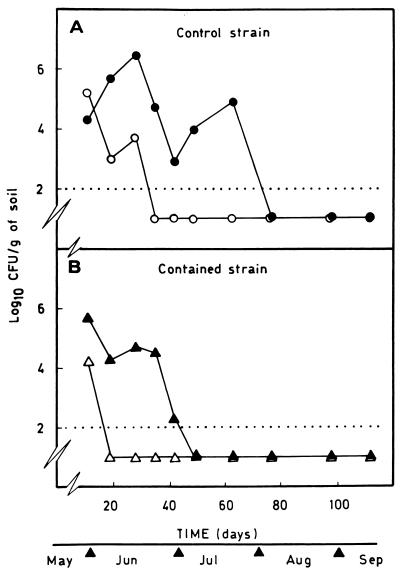
In the spring-summer outdoor assay corn seeds were coated with the control or contained strain, and the survival of the bacteria was investigated. In soil without 3-methylbenzoate, the contained strain P. putida CMC4 became unrecoverable from the rhizosphere and bulk soils 19 and 11 days, respectively, after the appearance of the first true leaves of corn plants (Fig. 5 and Table 3). In contrast, the control strain was recovered at a level of about 103 CFU/g of rhizosphere soil for about 28 days (Fig. 5); however, it was recovered from bulk soil only during the first 11 days after seed germination (Table 3). In soils containing 3-methylbenzoate the control strain was detected at levels of 103 to 106 CFU/g of rhizosphere soil during the first 70 days, whereas the contained strain was recovered at a level higher than 100 CFU/g of soil only during the first 40 days of the assay (Fig. 5). The contained and control strains became established poorly (if at all) in the bulk soil, and their levels never were greater than 103 to 104 CFU per g of soil (Table 3). The control strain could be recovered from bulk soil up to 28 days after sowing but not thereafter (Table 3), whereas the contained strain was recovered from bulk soil only up to day 11.
FIG. 5.
Survival of P. putida CMC4 and EEZ32 under outdoor conditions in pots. The conditions were the same as those described in the legend to Fig. 4 except that corn seeds were used.
TABLE 3.
Colonization of bulk soil by P. putida CMC4 and EEZ32 in the spring-summer outdoor assaya
| Strain | Presence of 3-methylbenzoate | CFU/g of soil after:
|
|||
|---|---|---|---|---|---|
| 11 days | 19 days | 28 days | 98 days | ||
| Control strain EEZ32 | + | 2 × 104 | 2 × 103 | 103 | <102 |
| − | 6 × 102 | <102 | <102 | <102 | |
| Contained strain CMC4 | + | 5 × 103 | <102 | <102 | <102 |
| − | 2 × 102 | <102 | <102 | <102 | |
The conditions were the conditions described in the legend to Fig. 5 except that bacteria were counted in the bulk soil at the times indicated. The values are the averages of values for duplicate samples counted in triplicate, and the standard deviations were on the order of 1 to 10% of the values given.
In unplanted soils the numbers of CFU of both the contained strain P. putida CMC4 and the control strain P. putida EEZ32 decreased rapidly regardless of the presence of 3-methylbenzoate, although bacteria disappeared faster in the absence of 3-methylbenzoate than in the presence of 3-methylbenzoate. After 3 days the contained strain was undetectable, and the control strain disappeared after 1 week. This rapid decrease was most likely due to the relatively high temperatures during the first few days of the release (up to 40°C), which is consistent with previous findings that suggested that the survival of P. putida 2440 derivatives in soil is notably influenced by soil temperature (18). From these results we concluded that the introduced P. putida strains were particularly sensitive to high temperatures during the summer.
Regardless of the strain introduced and the presence of 3-methylbenzoate, the sizes of the p-hydroxyphenylacetic acid-degrading population were around 105 to 106 CFU per g of rhizosphere soil and around 105 CFU per g of bulk soil throughout the assay.
Lack of dispersal of GEMs during the field releases.
During the outdoor trials reported in this study, five locations in the experimental field into which no GEMs had been introduced were used to monitor undesired dispersion of GEMs. Samples were taken every 14 days. Control or contained bacteria were never detected outside the pots. At the conclusion of the assay soil from the site was checked every fortnight for 6 months for the presence of the control or contained strain. Neither strain was detected in these tests.
DISCUSSION
Xenobiotic compounds contain structures or substituents rarely found in natural products. These compounds are usually not metabolized by microbes, and as a consequence they accumulate in the biosphere and contribute to the burden of pollution (17). Many microbes are able to evolve new catabolic activities against some of these recalcitrant compounds (3, 20, 28, 29), but most xenobiotic compounds remain in the biosphere unattacked (2). New catabolic pathways have been constructed for some of these pollutants, and in recent years it has become possible with molecular biology techniques to engineer bacteria that are able to degrade toxic compounds (17, 19). In the future recombinant microbes bearing genetically engineered catabolic pathways will probably be used in open environments, particularly in sites polluted with toxic chemicals (15). These GEMs should survive and perform for as long as the pollutant is present, but it is desirable that they should commit suicide once the compound is consumed (5, 26) to reduce concerns regarding unanticipated consequences. This behavior can be achieved by providing recombinant microbes with ABC systems.
In all genetic systems mutations appear at a certain frequency, and in the case of containment systems mutations that lead to killing escape have been reported (5, 12, 13). To decrease the rate of killing escape when cell death was induced, the control and killing elements of the ABC system were incorporated into the host chromosome of P. putida CMC4, where they were stably maintained even in the absence of selective pressure. When 3-methylbenzoate was exhausted, the killing genes were induced and the cells died (Fig. 3). Interestingly, the rate of killing escape under laboratory conditions was around 10−8 mutants per cell and per generation.
The effectively contained strain P. putida CMC4 and the control strain P. putida EEZ32 were released under controlled conditions. Survival of P. putida EEZ32 and the contained strain P. putida CMC4 was better in the rhizospheres of plants than in bulk soil and unplanted soils. In the rhizospheres of corn or broad bean plants the control strain became established at similar levels regardless of the presence of the target compound. In contrast, the contained strain tended to die, although survival in the presence of the aromatic compound was better than survival in its absence (Fig. 4 and 5). It should nonetheless be noted that even in the presence of 3-methylbenzoate the contained strain survived poorly compared to the control. This may have been due to the reduced availability of the 3-methylbenzoate added to the soil. The ability to colonize the soil rhizosphere is a characteristic of P. putida 2440, which can become established around the root systems of many herbaceous plants (21, 24). Therefore, the marked decrease in the number of CFU of the contained strain per gram of rhizosphere soil can be attributed unequivocally to the containment system that this strain bears. However, under outdoor conditions a decrease in the number of CFU of the contained bacteria per gram of soil required long periods of time (weeks), whereas in the laboratory the counts decreased within hours (25). This difference may reflect the following two facts: (i) when the contained cells were introduced into the soil, they were full of LacI protein, and the killing gene was not expressed until degradation of the LacI protein occurred; and (ii) functioning of the Gef protein requires cells in an active metabolic state (25), as reported previously for cell lysis mediated by other porins (8). Once added to the soil, the cells were probably metabolically less active.
The contained strain colonized bulk soil less efficiently than the uncontained control strain. This reflects the situation described above for unplanted soils, and it follows that dispersal of the contained strain may be severely limited in a number of environments.
To monitor the possible effects of the release of P. putida CMC4 and EEZ32 on the natural bacterial populations, we selected p-hydroxyphenylacetic acid utilizers as the target population to monitor because the numbers of these organisms in the soil remained stable at around 105 to 106 CFU per g of soil throughout the year (26). The presence of the control strain or the contained strain had no significant effect on survival of this natural population.
Our results show that an ABC system based on killing genes can be developed in the laboratory and that such a system can work under outdoor conditions.
ACKNOWLEDGMENTS
This work was supported in part by GX-Biosystems and by grant BIO4-CT97-2313 from the Commission of the European Communities.
REFERENCES
- 1.Abril M A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander M. Nonbiodegradable and other recalcitrant molecules. Biotechnol Bioeng. 1973;15:611–647. [Google Scholar]
- 3.Cain R B. Biodegradation of detergents. Curr Opin Biotechnol. 1994;5:266–274. [Google Scholar]
- 4.Carter B L A, Irani M, Mackay V L, Scale R L, Sledziewski A V, Smith R A. Expression and secretion of foreign genes in yeast. In: Glover D M, editor. DNA cloning. III. Oxford, United Kingdom: IRL Press; 1987. pp. 141–162. [Google Scholar]
- 5.Contreras A, Molin S, Ramos J L. Conditional-suicide containment system for bacteria which mineralize aromatics. Appl Environ Microbiol. 1991;57:1504–1508. doi: 10.1128/aem.57.5.1504-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived mini-transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 7.Dowling D N, O’Gara F. Metabolites of Pseudomonas involved in the control of plant disease. Trends Biotechnol. 1994;12:133–141. [Google Scholar]
- 8.Eko F O, Szostak M P, Wanner G, Lubitz W. Production of Vibrio cholerae ghosts (VCG) by expression of a cloned phage lysis gene: potential for vaccine development. Vaccine. 1994;12:1231–1237. doi: 10.1016/0264-410x(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 9.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood D A, Chater K F, Bibb M J. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology. 1995;28:65–102. doi: 10.1016/b978-0-7506-9095-9.50009-5. [DOI] [PubMed] [Google Scholar]
- 12.Jensen L B, Ramos J L, Kaneva Z, Molin S. A substrate-dependent biological containment system for Pseudomonas putida based on the Escherichia coli gef gene. Appl Environ Microbiol. 1993;59:3713–3717. doi: 10.1128/aem.59.11.3713-3717.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knudsen S M, Karlström O H. Development of efficient suicide mechanisms for biological containment of bacteria. Appl Environ Microbiol. 1991;57:85–92. doi: 10.1128/aem.57.1.85-92.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marston F A O. The purification of eukaryotic polypeptides expressed in Escherichia coli. In: Glover D M, editor. DNA cloning. III. Oxford, United Kingdom: IRL Press; 1987. pp. 59–88. [Google Scholar]
- 15.Molin S, Boe L, Jensen L B, Kristensen C S, Givskov M, Ramos J L, Bej A K. Suicidal genetic elements and their use in biological containment of bacteria. Annu Rev Microbiol. 1993;47:139–166. doi: 10.1146/annurev.mi.47.100193.001035. [DOI] [PubMed] [Google Scholar]
- 16.Ramos, C., L. Molina, S. Molin, and J. L. Ramos. Colonization of the rhizosphere of corn (Zea mays) by a bioluminescent Pseudomonas putida strain and its competitiveness advantages on soil isolates. Submitted for publication.
- 17.Ramos J L, Díaz E, Dowling D, de Lorenzo V, Molin S, O’Gara F, Ramos C, Timmis K N. The behavior of bacteria designed for biodegradation. Bio/Technology. 1994;12:1349–1356. doi: 10.1038/nbt1294-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos J L, Duque E, Ramos-González M I. Survival in soils of an herbicide-resistant Pseudomonas putida strain bearing a recombinant TOL plasmid. Appl Environ Microbiol. 1991;57:260–266. doi: 10.1128/aem.57.1.260-266.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos J L, Haïdour A, Delgado A, Duque E, Fandila M-D, Gil M, Piñar G. Potential of toluene-degrading systems for the construction of hybrid pathways for nitrotoluene metabolism. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 53–68. [Google Scholar]
- 20.Ramos J L, Haïdour A, Duque E, Piñar G, Calvo V, Oliva J M. Metabolism of nitrate esters by a consortium of two bacteria. Nat Biotechnol. 1996;14:320–322. doi: 10.1038/nbt0396-320. [DOI] [PubMed] [Google Scholar]
- 21.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 22.Ramos J L, Stolz A, Reineke W, Timmis K N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci USA. 1986;83:8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos-González M I, Ramos-Díaz M A, Ramos J L. Chromosomal gene capture mediated by the Pseudomonas putida TOL catabolic plasmid. J Bacteriol. 1994;176:4635–4641. doi: 10.1128/jb.176.15.4635-4641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Herva, J. J., D. Reniero, M. A. Ramos-Díaz, L. Molina, C. Ramos, E. Galli, and J. L. Ramos. LPS and OprL mutants of Pseudomonas putida exhibit increased sensitivity to drugs and limited capacity to colonize soils. Submitted for publication.
- 25.Ronchel M C. Construcción de cepas de Pseudomonas putida portadoras de sistemas condicionales de contención biológica para la eliminación de contaminantes ambientales. Ph.D. thesis. Granada, Spain: University of Granada; 1997. [Google Scholar]
- 26.Ronchel M C, Ramos C, Jensen L B, Molin S, Ramos J L. Construction and behavior of biologically contained bacteria for environmental applications in bioremediation. Appl Environ Microbiol. 1995;61:2990–2994. doi: 10.1128/aem.61.8.2990-2994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Soberón-Chávez G, Campos J, Haïdour A, Ramos J L, Ortigoza J. Selection and preliminary characterization of a Pseudomonas aeruginosa strain mineralizing selected isomers in a branched-chain dodecylbenzenesulphonate mixture. World J Microbiol Biotechnol. 1996;12:367–372. doi: 10.1007/BF00340213. [DOI] [PubMed] [Google Scholar]
- 29.White G F, Snape J R. Microbial cleavage of nitrate esters: defusing the environment. J Gen Microbiol. 1993;139:1947–1957. doi: 10.1099/00221287-139-9-1947. [DOI] [PubMed] [Google Scholar]