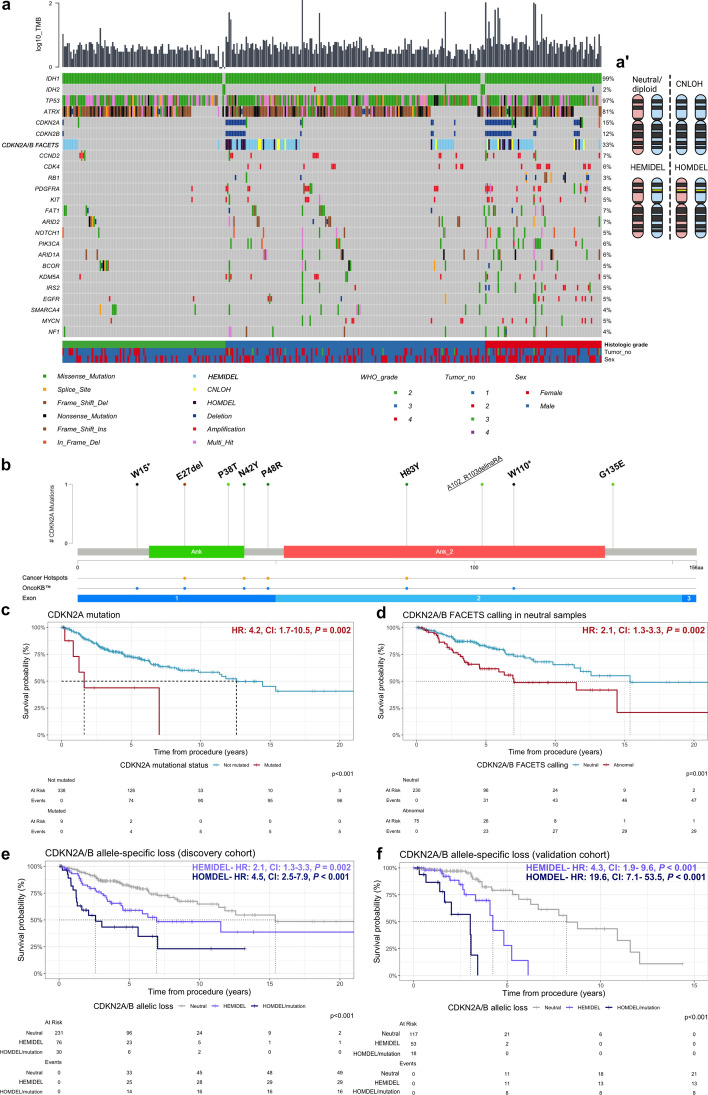
IDH-mutant astrocytomas (IDHA) are initially slow-growing tumors that almost inevitably progress [4]. Homozygous deletion (HOMDEL) of CDKN2A and/or CDKN2B (CDKN2A/B), now a WHO grade 4 defining criterion for IDHA, is associated with rapid progression and poor overall survival (OS) [1, 9]. We examined the prognostic significance of additional CDKN2A/B inactivating events, including nonsynonymous mutations and allele-specific copy number alterations (ASCNA) by analyzing 347 prospectively tumor-matched normal sequenced IDHA (347 patients, supplemental table 1) using FACETS (Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing), a SNP-based algorithm to assess ASCNA across genomic targets [8], with validation using The Cancer Genome Atlas (TCGA, 188 tumors/patients, supplemental methods).
CDKN2A/B alterations were found in up to 15% of IDHA (Fig. 1a, b): 12% exhibited CDKN2A/B loss (n = 42/347) and were associated with higher histologic grade (supplemental table 2). CDKN2A/B loss, PDGFRA gain, and CDK4 gain associated with shorter OS by multivariable Cox proportional hazards modelling (supplemental Fig. 1e). Nonsynonymous mutations in CDKN2A were uncommon (n = 9, 2.6%), but were mostly classified as oncogenic or likely oncogenic by OncoKB™ (n = 6, Fig. 1b, supplemental table 3) [2]. CDKN2A-mutant tumors were higher grade and had shortened OS (median OS: 1.6 years, 95% CI: 0.8 years–not reached [NR]) versus non-mutant tumors (median OS: 12.6 years, 95% CI: 11.4 years–NR, P < 0.001, Fig. 1c), approximating the OS of tumors with CDKN2A/B copy loss (median survival: 3.0 years, CI: 1.4 years–NR) even after excluding hypermutant tumors [10].
Fig. 1.
a Oncoplot depicting frequently altered genes and log10 tumor mutational burden (TMB) in 347 IDHA. a' CDKN2A/B allele-specific copy number states at 9p21.3. Yellow band = CDKN2A/B deletion, red and blue shades discern parental alleles. b Lollipop plots display variants in copy neutral CDKN2A/B (underlined: hypermutant). c Nonsynonymous CDKN2A mutations shorten OS. d ASCNA in CDKN2A/B copy neutral samples conferred worse OS than FACETS neutral. e CDKN2A/B HEMIDEL (n = 76) confers intermediate OS between patients with CDKN2A/B HOMDEL/mutation and CDKN2A/B neutral/without mutation. f TCGA validation cohort; HR—hazard ratios, CI—confidence intervals, P values within graphs refer to univariable Cox-proportional hazards regression models, log-rank test
FACETS detected hemizygous deletion (HEMIDEL) in 23% of IDHA (n = 81/347), HOMDEL in 6% (n = 21), and copy-neutral loss of heterozygosity (CNLOH) in 3% (n = 11), mostly in high histologic grade tumors (supplemental table 4). The remaining 67% (n = 234) were neutral (supplemental Fig. 2a). Of 305 copy neutral tumors, FACETS detected ASCNA in 75 (25%) demonstrating shorter OS (median 7.0 years, 95% CI: 5.9 years–NR) vs. FACETS neutral (15.4 years, 95% CI: 11.8 years–NR, P = 0.0014, Fig. 1d). HEMIDEL IDHA had shorter OS than neutral (median survival: 6.9 years (95% CI: 4.5 years–NR) vs. 15.4 years (95% CI: 11.8 years–NR, P < 0.001), but longer OS than HOMDEL/mutant cases (median survival: 2.6 years, 95% CI: 1.3 years–NR, P = 0.018, Fig. 1e). CNLOH of CDKN2A/B portended poor prognosis (median survival: 2.3 years, 95% CI: 1.6 years–NR, P < 0.001, supplemental Fig. 2b). We verified intermediate OS of CDKN2A/B HEMIDEL between CDKN2A/B HOMDEL/mutant and neutral tumors (Fig. 1f) in the TCGA cohort (supplemental methods). Gains of CDK4 and/or CCND2 worsened OS in CDKN2A/B HEMIDEL tumors (median survival: 3.2 years (95% CI: 1.4 years–NR) versus 11.5 years (95% CI: 4.5 years-NR, P = 0.011, supplemental Fig. 2c, d).
Matched tumor-normal NGS with FACETS analysis detects a range of prognostically relevant alterations in CDKN2A/B and other genes. FACETS showed good concordance with SNP-microarray in a subset of cases (supplemental Fig. 3). Kocakavuk et al. recently described shortened OS across multiple cohorts of IDH-mutant, 1p/19q intact gliomas with CDKN2A HEMIDEL using copy number profiles from NGS/methylation data [3]. These findings emphasize the clinical relevance of detection of CDKN2A/B HEMIDEL in IDHA. [6, 7]. As a molecularly-targeted therapeutic has shown promise in IDH-mutant grade 2 gliomas [5], extending the utility of NGS with bioinformatic tools like FACETS may help to both improve patient management and guide optimal treatment in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
This work was funded by P30-CA008748.
Data availability
The datasets analyzed in the current study are available from the corresponding author on reasonable request.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Appay R, Dehais C, Maurage C-A, Alentorn A, Carpentier C, Colin C, Ducray F, Escande F, Idbaih A, Kamoun A, et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019;21:1519–1528. doi: 10.1093/neuonc/noz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017. 2017 doi: 10.1200/po.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kocakavuk E, Johnson KC, Sabedot TS, Reinhardt HC, Noushmehr H, Verhaak RGW. Hemizygous CDKN2A deletion confers worse survival outcomes in IDHmut-noncodel gliomas. Neuro Oncol. 2023 doi: 10.1093/neuonc/noad095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandonnet E, Delattre JY, Tanguy ML, Swanson KR, Carpentier AF, Duffau H, Cornu P, Van Effenterre R, Alvord EC, Jr, Capelle L. Continuous growth of mean tumor diameter in a subset of grade II gliomas. Ann Neurol. 2003;53:524–528. doi: 10.1002/ana.10528. [DOI] [PubMed] [Google Scholar]
- 5.Mellinghoff IK, van den Bent MJ, Blumenthal DT, Touat M, Peters KB, Clarke J, Mendez J, Yust-Katz S, Welsh L, Mason WP, et al. Vorasidenib in IDH1- or IDH2-mutant low-grade glioma. New Engl J Med. 2023 doi: 10.1056/NEJMoa2304194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry A, Anderl K, Borell TJ, Kimmel DW, Wang CH, O'Fallon JR, Feuerstein BG, Scheithauer BW, Jenkins RB. Detection of p16, RB, CDK4, and p53 gene deletion and amplification by fluorescence in situ hybridization in 96 gliomas. Am J Clin Pathol. 1999;112:801–809. doi: 10.1093/ajcp/112.6.801. [DOI] [PubMed] [Google Scholar]
- 7.Reis GF, Pekmezci M, Hansen HM, Rice T, Marshall RE, Molinaro AM, Phillips JJ, Vogel H, Wiencke JK, Wrensch MR, et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol. 2015;74:442–452. doi: 10.1097/nen.0000000000000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131–e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirahata M, Ono T, Stichel D, Schrimpf D, Reuss DE, Sahm F, Koelsche C, Wefers A, Reinhardt A, Huang K, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136:153–166. doi: 10.1007/s00401-018-1849-4. [DOI] [PubMed] [Google Scholar]
- 10.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed in the current study are available from the corresponding author on reasonable request.