Abstract
The Bacillus subtilis strain VTT E-68013 was chosen for purification and characterization of its excreted phytase. Purified enzyme had maximal phytase activity at pH 7 and 55°C. Isolated enzyme required calcium for its activity and/or stability and was readily inhibited by EDTA. The enzyme proved to be highly specific since, of the substrates tested, only phytate, ADP, and ATP were hydrolyzed (100, 75, and 50% of the relative activity, respectively). The phytase gene (phyC) was cloned from the B. subtilis VTT E-68013 genomic library. The deduced amino acid sequence (383 residues) showed no homology to the sequences of other phytases nor to those of any known phosphatases. PhyC did not have the conserved RHGXRXP sequence found in the active site of known phytases, and therefore PhyC appears not to be a member of the phytase subfamily of histidine acid phosphatases but a novel enzyme having phytase activity. Due to its pH profile and optimum, it could be an interesting candidate for feed applications.
Cereals, legumes, and oilseed crops are grown in over 90% of the world’s harvested area. These crops serve as a major source of nutrients for humans and animals. An important constituent in these crops is phytic acid (myo-inositol hexaphosphate). The salt form, phytate, is the major storage form of phosphorus and accounts for more than 80% of the total phosphorus in cereals and legumes (27). Phytases are enzymes capable of hydrolyzing phytic acid to less-phosphorylated myo-inositol derivates. Monogastric animals, such as pig, poultry and fish, are not able to metabolize phytic acid, and therefore inorganic phosphate is added to their diets to satisfy the phosphorus requirement. This consequently contributes to phosphorus pollution problems in areas of intensive livestock production (1, 19, 20). Phytic acid also acts as an antinutritional agent in monogastric animals by chelating various metal ions needed by the animal, such as calcium, copper, and zinc (5, 13, 14). Therefore, the enzymatic hydrolysis of phytic acid into less-phosphorylated myo-inositol derivatives in the intestine of monogastric animals is desirable. Many attempts to enzymatically hydrolyze phytic acid have been made to improve the nutritional value of feed and to decrease the amount of phosphorus excreted by animals (12, 24, 32). There have been reports of partially purified microbial phytase preparations from a variety of microbial species (4, 6, 7, 8, 10, 30, 33), the best characterized being those from Aspergillus ficuum (34) and Aspergillus niger (3). There are two previous reports on partial purification of phytase from Bacillus subtilis (26, 31). Genes encoding fungal phytases from Aspergillus niger (3, 25, 36), Aspergillus fumigatus (22), Aspergillus terrus (16), Myceliophthora thermophila (16), Aspergillus nidulans (23), and Talaromyces thermophila (23) have been cloned and sequenced. The only bacterial phytase cloned so far is the Escherichia coli gene appA, which encodes periplasmic phosphoanhydride phosphohydrolase (2). However, due to the kinetic parameters, this enzyme should be designated a phytase (8).
In the present study, we screened several food-grade bacterial strains belonging to the genus Bacillus for extracellular phytase production. Phytase from the strain showing the highest phytase production was purified and partially characterized, and the gene was cloned, sequenced, and recombinantly produced. Therefore, we report here the first cloned, sequenced, and recombinantly produced food-grade bacterial phytase.
MATERIALS AND METHODS
Chemicals and bacterial strains.
Phytic acid, dodecasodium salt, was purchased from Sigma Chemical Co., St. Louis, Mo. Wheat bran was purchased locally (Melia Ltd., Raisio, Finland). All other chemicals were of the analytical grade commercially available. The following strains were obtained from the culture collection of the Technical Research Centre of Finland (VTT): Bacillus amyloliquefaciens VTT E-71014, VTT E-71015, VTT E-80124, and VTT E-90408; Bacillus coagulans VTT E-82150; Bacillus licheniformis VTT E-80117, VTT E-80118, VTT E-80119, and VTT E-83175; Bacillus stearothermophilus VTT E-81128, VTT E-81129, VTT E-84208, and VTT E-88318; and B. subtilis VTT E-68012, VTT E-68013, VTT E-70009, VTT E-83176, VTT E-83177, VTT E-83178, VTT E-84207, and VTT E-85178. E. coli XL-1 Blue MRF′ and SOLR′ (Stratagene, San Diego, Calif.) were used as a host for DNA manipulations and gene expressions. E. coli RV308 expression host was obtained from Kristiina Takkinen, Technical Research Centre of Finland. A. niger phytase Natuphos was obtained from Gist-brocades, Delft, The Netherlands.
Screening of Bacillus strains for phytase production.
Strains were tested for phytase production in Luria broth, in Luria broth supplemented with phytate, and in wheat bran extract medium described by Powar and Jagannathan (26). Samples were withdrawn from the culture media at different time points, cleared by centrifugation, and passed through a PD-10 gel filtration column (Pharmacia Inc., Uppsala, Sweden). These crude enzyme preparations were assayed for phytase activity as initially described by Shimizu (31).
Purification of native phytase.
All purification steps were carried out at 0 to 4°C unless otherwise stated. Bacteria grown on wheat bran extract were collected by centrifugation at 7,000 × g for 30 min. CaCl2 was added to a final concentration of 1 mM in the collected supernatant. The enzyme was precipitated by adding 3 volumes of cold (−20°C) ethanol with constant stirring. Stirring was continued for 45 min, and the precipitation was continued overnight. The precipitate was collected by centrifugation at 1,800 × g for 20 min. The collected precipitate was washed once with cold (−20°C) ethanol and once with cold (−20°C) acetone. Excess acetone was evaporated under nitrogen gas flow. The drying was completed by lyophilization. Dried precipitate was dissolved in 100 mM Tris-HCl (pH 7.5) supplemented with 1 mM CaCl2, and then ammonium sulfate was added slowly with constant stirring to give 65% saturation. The solution was incubated overnight and centrifuged at 9,000 × g for 60 min, and the supernatant was collected. Ammonium sulfate was added to the supernatant to give 85% saturation. The solution was incubated overnight. Precipitate was collected by centrifugation at 9,000 × g for 60 min. The pellet was dissolved in 100 mM Tris-HCl (pH 7.5) supplemented with 1 mM CaCl2. Aliquots of enzyme preparation were stored at −20°C. For the enzyme assays in defined buffers, an aliquot of the enzyme preparation was thawed and passed through a PD-10 gel filtration column (Pharmacia) into an appropriate buffer. For the enzyme assays in wheat bran buffer systems, aliquots of enzyme preparation were passed through a PD-10 gel filtration column (Pharmacia) into a 100 mM Tris-HCl (pH 7.5) buffer supplemented with 1 mM CaCl2. The molecular weight was determined by using 8 to 25% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Pharmacia). The isoelectric point was determined with the same system by using PhastGel IEF 3-9 isoelectric focusing gels and the Pharmacia IEF calibration kit as the standard. Other protein samples were separated by SDS–12.5% PAGE as described by Laemmli (11) and stained with Coomassie brilliant blue. For sequencing purposes, final purification of the PhyC protein was performed by reversed-phase high-performance liquid chromatography (RP-HPLC) on a 0.21- by 10-cm (TSK Tosoh Corporation, Tokyo, Japan) TMS-250 (C1) column by elution with a linear gradient of acetonitrile (3 to 100% in 60 min) in 0.1% trifluoroacetic acid. Chromatography was performed at a flow rate of 200 μl/min, and the protein was detected by UV absorbance at 214 nm. The collected protein fraction was dried in a vacuum centrifuge and dissolved in 40 μl of 6 M guanidine-HCl–2 mM EDTA–0.5 M Tris-Cl (pH 7.5).
Production of B. subtilis VTT E-68013 phytase in defined media.
B. subtilis VTT E-68013 colonies were grown on M9 minimal medium, M9 minimal medium supplemented with 2 mM phytate, and phytase screening medium (2% d-glucose, 0.4% sodium phytate, 0.2% CaCl2, 0.5% NH4NO3, 0.05% KCl, 0.05% MgSO4 · 7H2O, 0.001% FeSO4 · 7H2O, 0.001% MnSO4 · H2O per liter adjusted to pH 7). The culture broth was clarified by centrifugation, proteins were precipitated by adding 3 volumes of cold ethanol (−20°C), and precipitate was dissolved in 100 mM Tris-HCl (pH 7.5) supplemented with 1 mM CaCl2 and assayed for phytase activity.
Alkylation, enzymatic digestion, and peptide separation.
Dithiothreitol (5 μmol) was added, and reduction was performed for 20 min at room temperature; this was followed by addition of 1 μl of 4-vinylpyridine (Sigma). Alkylation was performed at room temperature for 15 min, followed by addition of 5 μl of dithiothreitol (1 μmol/μl). The alkylated protein (about 15 μg) was separated from the remaining reagents by C1 RP-HPLC as described above, dried in a vacuum centrifuge, and dissolved in 50 μl of 0.1 M Tris-Cl (pH 9.2); addition of 0.2 μg of Lysylendopeptidase C (LysC; Wako GmbH, Neuss, Germany) followed. Digestion was performed overnight at 37°C. Generated peptides were separated by narrow-bore RP-HPLC on a 1.0- by 15-cm Vydac C8 column (300Å, 5 μm; LC-Packings, Amsterdam, The Netherlands). Elution was performed with a linear gradient of acetonitrile (0 to 40% in 120 min) in 0.1% trifluoroacetic acid. Peptides were monitored at 214 nm and automatically collected with a SMART system (Pharmacia Biotech, Uppsala, Sweden).
Mass spectrometry, protein N-terminal sequencing, and internal peptide sequencing.
The collected peptides were subjected to MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) mass spectrometry in the delayed extraction mode with a BIFLEX mass spectrometer (Bruker-Franzen Analytik, Bremen, Germany) by using a 337-nm nitrogen laser. A thin-layer matrix preparation with saturated α-cyano-4-hydroxycinnamic acid in acetone was used. One-half microliter of matrix was deposited on a stainless-steel target plate and allowed to dry, after which 0.5 μl of sample was added on top of the matrix spot. External calibration was performed with insulin (human; Sigma) and cytochrome c (horse heart; Sigma). Protein N-terminal sequencing and internal peptide sequencing were performed with a Procise 494A sequencer (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.).
Phytase activity assays.
Enzyme assays were preformed as described by Shimizu (31). One unit of enzyme activity was defined as the amount of enzyme hydrolyzing 1 μmol of Pi per min under assay conditions. The specific activity was expressed in units of enzyme activity per milligram of protein. Enzyme activity assays were performed in defined buffers and in a wheat bran buffer system as described below. All enzyme assays were run in duplicate. Defined buffers used in enzyme activity assays were as follows: 100 mM glycine (pH 3.0), 100 mM succinate (pH 5.0), 100 mM Tris-maleate (pH 5.0, 6.0, 7.0, and 8.0), 100 mM Tris-HCl (pH 7.5, 8.0, and 9.0). All buffers were supplemented with 2 mM sodium phytate and 1 mM CaCl2. Enzyme assays were performed in these buffers at five different temperatures (37, 45, 55, 65, and 75°C). Six-hundred-microliter aliquots of buffer were preincubated at the relevant temperature for 5 min, and the enzyme reactions were started by adding 150 μl of enzyme preparation. Since enzyme addition tends to affect the pH of the reaction mixture, the true pH of each assay mixture was measured at the beginning and at the end of the 30-min incubation. After 30 min of incubation, reactions were stopped with 750 μl of 5% trichloroacetic acid and the released inorganic orthophosphate was measured as described previously (31). The protein concentration of each enzyme preparation was measured with the Bio-Rad protein assay (Bio-Rad Life Science Group, Hercules, Calif.), and the specific activity of enzyme at the different pH and temperature levels was calculated.
Wheat bran extract used in the enzyme activity assay was prepared by dissolving 50 g of wheat bran in 500 ml of distilled water, followed by autoclaving at 121°C for 60 min. The extract was filtered through a cheesecloth, and the volume was adjusted to 500 ml with distilled water and clarified by centrifugation. The supernatant was adjusted to five different pH levels by HCl or NaOH additions (pH 3.0, 5.5, 7.0, 8.0, or 9.0), diluted 1:10 in distilled water, and supplemented with 2 mM sodium phytate and 1 mM CaCl2. Six hundred microliters of the wheat bran buffer described above was preincubated at the desired reaction temperature (37, 55, and 75°C), and the enzyme reactions were then run as described above.
Substrate specificity.
Substrate specificity of the PhyC was determined by using the standard activity assay in 100 mM Tris-HCl (pH 7.5) supplemented with 1 mM CaCl2 and 2 mM tested substrate. Besides phytic acid, β-glycerophosphate, d-glucose 6-phosphate, p-nitrophenylphosphate, ATP, ADP, AMP, fructose 1,6-diphosphate, 3-phosphoglyceric acid, bis-(p-nitrophenyl)phosphate, and α,β-methyleneadenosine-5′-disphosphate were tested as substrates.
General DNA techniques.
All PCRs were performed by using a PTC-255 DNA Engine (MJ Research Inc., Watertown, Mass.) and Taq polymerase (Perkin-Elmer, Roche Molecular Systems Inc., Branchburg, N.J.). On the basis of N-terminal and internal PhyC peptide sequences, several degenerate PCR primers were designed. PCR was performed with these primers by using B. subtilis VTT E-68013 DNA as a template at different annealing temperatures and at different magnesium concentrations. The following PCR protocol was chosen: premelting at 94°C for 4 min, followed by 30 cycles of melting at 92°C for 60 s, annealing at 50°C for 60 s, and extension at 72°C for 120 s in 2.5 mM magnesium. The largest PCR fragment was cloned into pCR 2.1 (Invitrogen, San Diego, Calif.) vector and sequenced. Southern blotting was performed as described by Sambrook et al. (29) by using the largest PCR fragment, labelled with digoxigenin (PCR DIG probe synthesis kit; Boehringer-Mannheim, Mannheim, Germany), as the hybridization probe. The nucleotide sequence of the phyC gene was determined with the ABI Prism Dye Terminator Cycle Sequencing kit with an ABI 377 DNA sequencer. Nucleotide and amino acid sequence homology searches were performed on National Center for Biotechnology Information (NCBI) databases by Blast searches.
B. subtilis VTT E-68013 genomic library construction.
Partially EcoRI-digested genomic B. subtilis VTT E-68013 DNA was cloned into Lambda ZapII and packaged into lambda particles by using a Lambda ZapII/EcoRI/CIAP GigaPack Gold III cloning kit (Stratagene) as described in the recommendations provided by the manufacturer. Genomic B. subtilis VTT E-68013 library was screened with an EasyToHyb hybridization kit (Boehringer-Mannheim) as described in the recommendations provided by the manufacturer by using the largest PCR fragment, labelled with digoxigenin, as the hybridization probe. Positive lambda clones were cored and excised with ExAssist helper phage (Stratagene) to obtain phagemids. The phagemids obtained were transformed into SOLR′ E. coli host cells (Stratagene), and plasmid DNA was purified with the Qiagen (Santa Clara, Calif.) plasmid kit and used in analysis of insert DNA and DNA sequencing.
Construction of clones overexpressing recombinant PhyC-His6 fusion protein.
The phyC gene fragment encoding mature enzyme was amplified by PCR with insertion of SphI and BglII sites at the 5′ and 3′ ends, respectively. Primers used were pBsf (5′ CTCGCATGCTGTCCGATCCTTATCATTTTTACCG 3′) and pBsr (5′ GGCAGATCTTTTTCCGCTTCTGTCGGTCAGTTC 3′). The amplified PCR fragment was purified with the QIAquick DNA purification kit (Qiagen) and cloned into SphI/BglII-cut pQE-70 expression vector harboring C-terminal His6 tag (Qiagen) to generate plasmid pBsm. Another forward primer, pBssf (5′ CGTTCAATTGAGGAGGAAGTAAAATGAATC 3′), with insertion of an MfeI site (compatible with EcoRI), was designed to amplify the phyC gene fragment with its natural signal sequence and ribosomal binding site. The reverse primer used in this amplification was the pBsr primer. The amplified PCR fragment was purified with the QIAquick DNA purification kit (Qiagen) and cloned into EcoRI/BglII-cut pQE-70 expression vector harboring C-terminal His6 tag to generate plasmid pBss. Primer phytac(+) (5′ CGCGGATCCATGGCCCTGTCCGATCCTTATCATTTTACC 3′), with insertion of BamHI and NcoI sites, and primer phytac(−) (5′ GCTAGTCTAGATTTTCCGCTTCTGTCGGTCAG 3′), with insertion of an XbaI site, was designed to subclone the mature phyC gene fragment into BamHI/XbaI-cut pUC19. The amplified PCR fragment was cut with XbaI and partially cut with NcoI due to the internal NcoI site in the phyC gene. Partially cut fragments were separated on agarose, and the NcoI/XbaI phyC fragment was cut from the gel and purified with the QIAquick DNA purification kit (Qiagen). Purified phyC NcoI/XbaI fragment was cloned into an NcoI/XbaI-cut pKKtac E. coli expression vector harboring C-terminal His6 tag to generate pKKtacBs. The pBsm and pBss plasmids were transformed into XL-1 Blue MRF′ as described by Hanahan (9). Plasmid pKKtacBs was transformed into the CaCl2-competent E. coli RV308 expression host.
The pBsm and pBss transformants were grown in LB broth containing 100 μg of ampicillin per ml, induced, and purified as described in QIAexpressionist (Qiagen) with growth and induction times varied and temperature as well as the amount of isopropyl-β-d-thiogalactopyranoside (IPTG) used as an inducer to optimize expression. For pKKtacBs expression in RV308, an overnight culture was diluted 1:50 into fresh LB broth supplemented with 100 μg of ampicillin per ml and grown at 37°C and 200 rpm until the A600 was 1.0. IPTG was then added to 1 mM, and the culture was shifted to 30°C since the production level of the recombinant PhyC was found to be higher at 30 than at 37°C. After 1 h of induction, CaCl2 was added to 1 mM to stabilize the enzyme produced. For production analysis, samples were withdrawn at various times after induction, cells were pelleted, and recombinant proteins from culture supernatant were purified and assayed for phytase activity. Purification was performed in the following manner: the sample was applied to a Ni-nitrilotriacetic acid matrix (Qiagen) and washed first with 50 mM Tris-HCl–300 mM NaCl (pH 8.0) supplemented with 1 mM CaCl2 and then with 50 mM Tris-HCl–300 mM NaCl (pH 8.0) supplemented with 1 mM CaCl2 and 20 mM imidazole. Recombinant protein was eluted with the same buffer except that the imidazole concentration used was 500 mM.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper has been deposited in the GenBank nucleotide sequence database under accession no. AF029053.
RESULTS
Screening of Bacillus strains for phytase production.
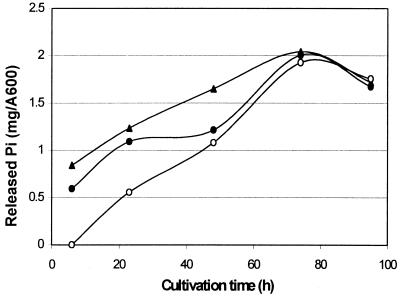
Twenty-one strains from the genus Bacillus were tested for extracellular phytase production in Luria broth, in Luria broth supplemented with phytate, and in wheat bran extract medium. None of the strains produced phytase activity in the Luria broth, whether or not it was supplemented with phytate (data not shown). However, in the wheat bran medium, two B. amyloliquefaciens strains and one B. subtilis strain produced significant amounts of phytase activity. The amount of inorganic phosphate released per cell density (A600) during the cultivation of these three strains is shown in Fig. 1. The B. subtilis strain VTT E-68013 showed the highest phytase activity production and was therefore chosen for phytase enzyme production.
FIG. 1.
Phytase activities of B. subtilis VTT E-68013 (▴), B. amyloliquefaciens VTT E-71015 (•), and B. amyloliquefaciens VTT E-90408 (○) during cultivation in wheat bran extract. The phytase activities are expressed as the amount of released inorganic phosphate per cell density (A600). Enzyme assays were run in duplicate, and the standard error in all assays was below 0.04.
Induction studies.
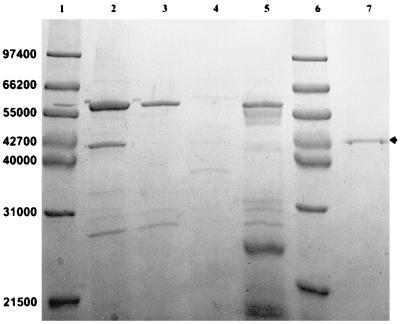
Induction studies were carried out to exclude the possibility that the phytase was readily expressed in phytate-containing media other than wheat bran medium but became instantly and irreversibly inactivated or proteolytically cleaved when secreted to media other than wheat bran. Strain VTT E-68013 was cultivated in a wheat bran extract, Luria broth, Luria broth supplemented with 10 mM phytate, and Luria broth supplemented with 2% bovine serum albumin in order to protect the produced phytase from possible proteolysis. After different time points, samples were withdrawn and assayed for phytase activity. After 50 h of cultivation, when phytase activity was at its highest in wheat bran extract but still no activity was detected in Luria broth, samples of cleared culture media were subjected to SDS-PAGE. At this point of cultivation, cells from each cultivation were examined under the microscope and observed to be undergoing sporulation. No phytase band was detected for any Luria cultivation (Fig. 2), verifying that these media did not support phytase enzyme production even in an inactive form. It was clear that phytate did not induce phytase production but instead proved to repress protein expression of B. subtilis VTT E-68013 since the major ca. 58,000 band (most likely amylase) and other bands detected in other cultivation media were barely detectable although cell densities (A600s) in each cultivation were about the same.
FIG. 2.
SDS-PAGE separation of culture supernatant samples of secreted protein of B. subtilis VTT E-68013 in different culture media after 50 h of cultivation. Lanes: 1 and 6, molecular weight markers; 2, wheat bran; 3, Luria broth; 4, Luria broth supplemented with 10 mM phytate; 5, Luria broth supplemented with 2% bovine serum albumin; 7, purified recombinant phytase (phytase band indicated by arrow).
To verify that the phytase enzyme was not proteolytically cleaved by excreted proteases in Luria broth, purified phytase was incubated with Luria broth spent medium. There was no drop in phytase activity after 1 h of incubation with Luria broth spent medium at 37°C, indicating that phytase was not cleaved by proteases excreted by B. subtilis VTT E-68013.
B. subtilis VTT E-68013 was also grown on defined media. Minimal medium containing inorganic phosphate as well as phytate did not induce phytase production, but defined medium in which phytate was the sole source of phosphate (phytase screening medium; see Materials and Methods) induced phytase production.
Production and purification of native phytase.
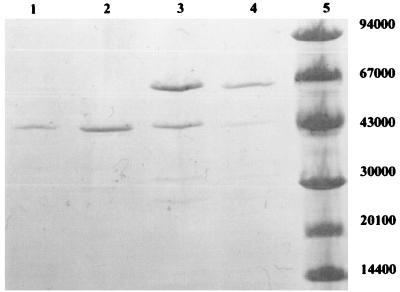
Phytase proved to be very sensitive to commonly used chromatographic purification methods such as ion exchange and gel filtration. The enzyme required CaCl2 in all purification steps to maintain activity and lost activity if EDTA was used in buffers. A combination of purification by ethanol and ammonium sulfate precipitation proved to be the best purification method and was therefore used to purify protein for enzyme characterization. The purification of phytase is described in Table 1. Redissolved pellet from 85% ammonium sulfate precipitate with high phytase protein purity, but not optimum specific activity, was used in all enzyme characterization experiments described and, after further purification (described in Materials and Methods), was used in N-terminal and internal peptide sequencing. Phytase purification was monitored by SDS-PAGE as shown in Fig. 3.
TABLE 1.
Purification of phytase from B. subtilis VTT E-68013
| Enzyme sample | Sp act (U/mg)a | Recovery (%) | Purification factor (fold) |
|---|---|---|---|
| Culture supernatant | 8 | 100 | 1 |
| Redissolved ethanol precipitate | 15 | 93 | 1.9 |
| Supernatant from 65% ammonium sulfate | 88 | 56 | 11.2 |
| Redissolved pellet from 85% ammonium sulfate | 29 | 22 | 3.7 |
One unit is defined as the amount of enzyme required to liberate 1 μmol of Pi per min under assay conditions. The specific activity is expressed in units of enzyme activity per milligram of protein in an activity assay.
FIG. 3.
Purity monitoring of the PhyC by SDS-PAGE. Lanes: 1, 85% saturation ammonium sulfate precipitate; 2, 65% saturation ammonium sulfate supernatant; 3, ethanol precipitate; 4, culture supernatant; 5, molecular weight markers.
Chemical and physical characteristics and substrate specificity of the purified phytase.
The molecular mass of the mature PhyC was 43 kDa as determined by SDS-PAGE (Fig. 3). The determined molecular mass was 5 kDa more than that of the phytase purified from B. subtilis (natto) N-77 described by Shimizu (31). The isoelectric point of PhyC was 6.5 as determined by isoelectric focusing (data not shown). PhyC proved to be highly specific for phytate, hydrolyzing in addition to phytate only ATP and ADP (50 and 75% of the activity with phytate, respectively) of the substrates tested (see Materials and Methods).
Effect of pH and temperature on the phytase activity.
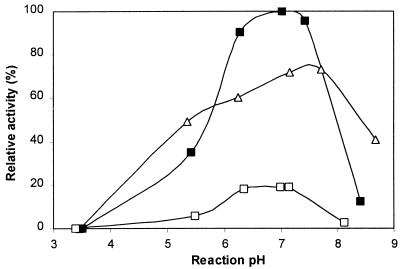
The activity of native PhyC was determined at different pHs and different temperatures as described in Materials and Methods. During the reaction, the changes in pH proved to be insignificant whether the reaction was performed in defined buffer or in wheat bran extract. The final pHs were plotted, and these were within 0.3 pH unit of the initial pH. Figure 4 shows the effect of pH on phytase activity in defined buffers at different temperatures (for clarity, only data for temperatures of 37, 55, and 75°C are shown). The optimum temperature proved to be 55°C. Irrespective of the reaction temperature, PhyC showed the highest phytase activity at neutral pH.
FIG. 4.
Effect of pH on phytase activity on defined buffers at three different temperatures (37°C [▵], 55°C [▪], and 75°C [○]). Phytase activities are expressed as relative activity. Enzyme assays were run in duplicate, and the standard error in all assays was below 0.04.
We also determined the PhyC pH activity profiles in a wheat bran buffer system because it is likely to provide an environment somewhat closer to that encountered in feed applications. The optimum pH and temperature as well as the pH profiles as a whole in the wheat bran extract buffer system proved to be very similar to those determined in defined buffers.
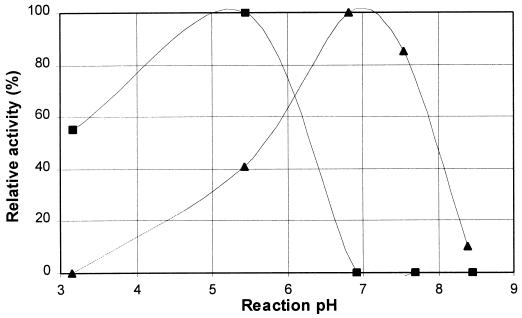
To compare PhyC to commercially available fungal phytase used in feed applications, the pH activity profile of Natuphos (an A. niger phytase) was also determined. Figure 5 shows the pH activity profiles of PhyC and Natuphos in a wheat bran buffer system at 55°C (optimum temperature of Natuphos as well). Figure 5 clearly shows that PhyC is functional at neutral pH whereas A. niger phytase is completely inactive.
FIG. 5.
The pH activity profiles of Natuphos (▪), an A. niger phytase, and PhyC (▴) at 55°C in a wheat bran buffer system. Phytase activities are expressed as relative activities. Enzyme assays were run in duplicate, and the standard error in all assays was below 0.04.
N-terminal and internal peptide sequencing and degenerate primer design.
The sequence of 25 amino acid residues was obtained from protein N-terminal sequencing. A total of nineteen RP-HPLC-purified internal peptides from alkylated, LysC-digested PhyC was sequenced. The molecular weights of the peptides were measured with a mass spectrometer and compared with calculated molecular weights. LysC digestion was also performed on nonalkylated PhyC; this was followed by RP-HPLC purification of peptides. There was no difference between RP-HPLC results for alkylated and nonalkylated LysC-digested PhyC, indicating the absence of sulfur bridges. Fourteen sequenced internal peptides including the N-terminal peptide showed no overlap with one another and gave a total of 227 amino acid residues. On the basis of these peptide sequences, degenerate primers for PCR were designed. All sequenced peptides and the degenerate primers designed are shown in Table 2.
TABLE 2.
N-terminal and internal peptide sequences of PhyC and degenerate primers for PCR designed thereof
| Mol wt of peptide
|
Amino acid sequenceb | Degenerate primer
|
||
|---|---|---|---|---|
| Determineda | Calculated | Nucleotide sequencec | Designation | |
| LSDPYHFTVNAAAETEPVDTAGDAA | TCIGATCCITATCATTTTACIGT | 6465 | ||
| LSDPYHFTVNAAAETEPVDTAGDAADDPAILD | ||||
| 932 | 932.1 | YYAMVTGK | TTTICCIGTIACCATIGC | 6544 |
| 1,271.4 | 1,271.3 | EGEFEQYELK | TTCATA(T/C)TGTTCAAATTCICC | 6472 |
| 1,050.3 | 1,050.2 | MLHSYNTGK | TTICCIGT(A/G)TTATAIGAATGIA(A/G)CAT | 6473 |
| 798.9 | 798.9 | IVPWER | ||
| 2,951.2 | 2,948.4 | IVPWERIADQIGFRPLANEQVDPRK | TGATCIGC(G/A)ATIC(G/T)TTCCCA | 6470 |
| NGTLQSMTDPDHPIATAINEVYGFTLWHSQ | GC(G/A)AT(C/A)GGATGATC(C/A)GGATC | 6471 | ||
| YVADFRITDGPETDGTSDDDGII | TCIGATTCIGGICCATCIGT | 6468 | ||
| 775.7 | 775.8 | LTDRSGK | TTTICCI(G/C)(T/A)IC(G/T)ATCIGT | 6543 |
| 1,317.9 | 1,317.4 | VDIAAASNRSEGK | CTTCIGAIC(G/T)(G/A)TTIGAIGCIGC | 6469 |
| 2,167.4 | 2,167.4 | IADQIGFRPLANEQVDPRK | ||
| 720.7 | 720.8 | ANQNFK | TTTAAA(G/A)TT(C/T)TG(G/A)TTIGC | 6541 |
| 619.6 | 619.7 | VRAFK | ||
| LNNVDIRYDFP | AG(C/A)GGAAAATCATAIC(C/T)(G/A)ATATC | 6467 | ||
| 1,779.4 | 1,778 | LNNVDIRYDFPLNGK | ||
| 1,236.3 | 1,236.4 | NTIEIYAIDGK | CCATC(G/A)ATIGCATA(G/A)ATTTC | 6474 |
| 1,137.4 | 1,137.3 | SGLVVYSLDGK | TTICCATCIA(G/T)I(G/C)(T/A)ATAIAC | 6542 |
| FSAEPDGGSNGTVIDRADGRHL | CCATCIGCIC(G/T)ATC(G/A)ATIAC | 6475 | ||
Molecular weight of peptide determined by mass spectrometer.
Sequences of LysC-digested purified peptides, the first one being the N-terminal peptide. The region from which the degenerate primer was designed is underlined.
Designed degenerate primers, the first one being the forward primer and the others being reverse primers. I, inosine. All primer sequences are written in 5′→3′ direction.
Molecular cloning and nucleotide sequence of the gene encoding PhyC.
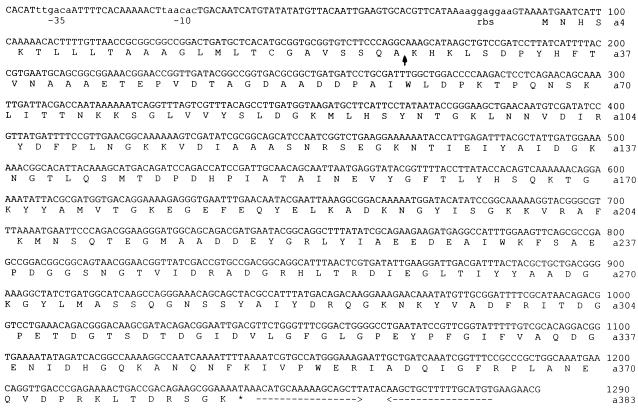
PCR was performed with designed degenerate primers by using genomic B. subtilis VTT E-68013 DNA as the template. Under PCR conditions described in Materials and Methods, nine reverse primers amplified a single fragment with the forward primer 6465. Primers 6465 and 6470 amplified the largest PCR fragment, which was cloned to a pCR 2.1 vector and sequenced. This resulted in determination of the partial phytase gene sequence of 989 bp. This partial gene fragment was translated into an amino acid sequence, revealing an open reading frame of 330 amino acid residues, and corresponded to the peptide sequences obtained from N-terminal and internal peptide sequencing of purified PhyC. A total of 14 peptides were found in the translated amino acid sequence. Southern hybridization revealed two fragments of 6 and 2.4 kbp, respectively. The genomic B. subtilis VTT E-68013 library was screened, and positive clones carrying 6 and 2.4 kbp inserts were obtained. Sequences from these clones were determined by using both vector-specific and gene-specific primers. The sequence of the phyC gene, the deduced amino acid sequence, putative −35 and −10 sequences, a ribosomal binding site, and a transcription terminator are shown in Fig. 6. The −35 sequence was the consensus sequence for Bacillus expression. However, the −10 sequence was not similar to anything listed (17), suggesting a specific sigma factor. The stop codon (TAA) is followed by a sequence of dyad symmetry (18-nucleotide perfect repeat) which could form a stem-loop structure and therefore be a transcription terminator. The putative ribosomal binding site is 9 nucleotides in length, contains a canonical GGAGG consensus sequence, and is optimally spaced from the start codon (37).
FIG. 6.
Nucleotide sequence and deduced amino acid sequence of the phyC gene. The putative −35 and −10 sequences are indicated (sequence in lowercase letters) as well as a putative ribosome binding site (rbs; sequence in lowercase letters). A possible transcription terminator downstream of phyC gene is indicated by horizontal arrows under the sequence. The possible signal peptide cleavage site is indicated by a vertical arrow.
Deduced amino acid sequence of PhyC.
The primary amino acid sequence deduced from the nucleotide sequence of the phyC gene revealed a fragment of 383 amino acid residues after putative ribosomal binding. The putative signal peptide cleavage site according to Nielsen et al. (21) is located between residues 26 and 27 (SQA-KH). The N-terminal sequence analysis of the purified protein would indicate that the first 29 amino acids are cleaved, but it is possible that the first 26 amino acids are a true signal peptide since the first 26 amino acids closely match the consensus of secreted proteins. Residues 27 to 29 might be analogous to a propeptide because of the positively charged residues (KHK). However, three amino acids would be very short for a propeptide (18). The molecular masses of PhyC preprotein and mature PhyC as deduced from the amino acid sequence were ca. 41.9 kDa and ca. 39 kDa (i.e., without the first 29 residues), respectively. The deduced amino acid sequence was compared to the NCBI protein database by Blast search. The only sequence with homology to PhyC was a hypothetical open reading frame (73% amino acid identity and 86% amino acid similarity to PhyC) from sequence analysis of the B. subtilis chromosome region between the odhAB and sspC loci cloned in a yeast artificial chromosome (38). This hypothetical open reading frame is identical to a hypothetical partial open reading frame (orf181) from the B. subtilis cgeAB gene cluster region (28).
Overexpression and purification of recombinant PhyC-His6 fusion proteins.
The phyC gene fragment encoding mature PhyC (clone pBsm) as well as a fragment encoding mature PhyC with its own signal peptide (clone pBss) was cloned into an overexpression vector, pQE-70, as a C-terminal His6 tag fusion protein, under the control of T5 promoter as described in Materials and Methods. The pBsm clone overexpressed a fusion protein which had the same molecular mass as native, mature protein as determined by SDS-PAGE (data not shown). No active form was obtained from this construct under the expression conditions tested. It appeared that the fusion protein encoded by the pBsm construct was toxic to E. coli, since the growth rate of the expression strain XL-1 Blue MRF′ harboring plasmid pBsm after induction was significantly lower than that of the strain carrying the vector alone. This is possibly due to the ATPase and ADPase activities of PhyC enzyme. Furthermore, more than 90% of the expressed fusion protein was found in the insoluble cytoplasmic fraction resulting from the formation of inclusion bodies. Likewise, no active enzyme was obtained with the pBss construct that was designed to direct the fusion protein to the periplasmic space by using the natural signal peptide of PhyC. A fusion protein encoded by pBss construct having a molecular mass about 3 kDa larger than that of the nonrecombinant protein as determined by SDS-PAGE was also found in the insoluble cytoplasmic fraction, indicating that the PhyC signal peptide was not able to direct fusion protein to the periplasmic space of the E. coli host strain (data not shown). However, the pKKtacBs construct, in which the phyC gene fragment encoding the mature enzyme was cloned downstream of a pectate lyase (pelB) signal sequence from Erwinia carotovora, overexpressed a fusion protein in an active form to the culture medium of E. coli RV308 host. The highest phytase activity was obtained after 20 h of induction at 30°C. The fusion protein was purified from the culture medium as described in Materials and Methods. It had the same molecular mass as that of the native, mature protein as determined by SDS-PAGE (Fig. 2), and it also showed the same pH and temperature optima and the same substrate specificity.
DISCUSSION
The chemical, physical, and enzymatic characteristics of the purified PhyC as well as the requirement for calcium and the inhibition by EDTA suggest that this phytase is similar to the phytase from B. subtilis (natto) N-77 described by Shimizu (31) and that from B. subtilis described by Powar and Jagannathan (26). It could be assumed that the phytase gene is present in the genome of B. subtilis since DNA sequences homologous (67% on a DNA level) to phyC from other B. subtilis strains have been reported (28, 38). B. subtilis VTT E-68013 is not likely very close to B. subtilis 168 since another gene cloned from the same genomic B. subtilis VTT E-68013 library proved to be only 80% identical to the corresponding gene from B. subtilis 168 on a DNA level (15).
The fact that commonly used defined and complex media containing inorganic phosphate in the presence or absence of phytate did not induce the production of PhyC from B. subtilis VTT E-68013 suggests that synthesis of PhyC is not upregulated only by phytate itself. However, phytate as a sole source of phosphate induced PhyC production. This finding suggests that PhyC production is induced only when inorganic phosphate is a limiting factor.
All cloned and sequenced microbial phytases have significant homology to each other, and their active sites show remarkable homology to the active site residues of the members of a particular class of acid phosphatases (histidine acid phosphatases), therefore forming the phytase subfamily of histidine acid phosphatases (16, 23, 35). The deduced amino acid sequence of PhyC did not have homology to the sequences of any phytases nor to those of any phosphatases listed in the databases. Most of all, PhyC did not have the RHGXRXP sequence which is the most conserved sequence in the active site of cloned phytases (35); thus, PhyC is not a member of the phytase subfamily of histidine acid phosphatases but is a novel enzyme having a phytase activity.
The inhibition of PhyC by EDTA and the requirement for calcium indicate the presence of a metal, most likely calcium, in the enzyme. We are currently working on identification of this metal, something that is important to know if the PhyC enzyme is to be used in animal feed applications.
ACKNOWLEDGMENTS
We are indebted to Walter Callen and Keith Kretz for sequencing the phyC gene and for the excellent sequence analysis and, especially to Keith, for fruitful discussion. We are grateful to Kristiina Takkinen for providing the pKKtac expression vector and the E. coli RV308 expression host strain. Many thanks to Osmo Siikanen for excellent technical assistance. We also thank Pekka Hilden and Andrei Miasnikov for critically reading the manuscript and Andrew Morgan for correcting the English manuscript.
REFERENCES
- 1.Common F H. Biological availability of phosphorus for pigs. Nature. 1989;143:370–380. [Google Scholar]
- 2.Dassa J, Marck C, Boquet P L. The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2.5 acid phosphatase and glucose-1-phosphatase. J Bacteriol. 1990;172:5497–5500. doi: 10.1128/jb.172.9.5497-5500.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich K C, Montalbano B G, Mullaney E J, Dischinger H C, Jr, Ullah A H. Identification and cloning of a second phytase gene (phyB) from Aspergillus niger (ficuum) Biochem Biophys Res Commun. 1993;195:53–57. doi: 10.1006/bbrc.1993.2008. [DOI] [PubMed] [Google Scholar]
- 4.Ghareib M. Biosynthesis, purification and some properties of extracellular phytase from Aspergillus carneus. Acta Microbiol Hung. 1990;37:159–164. [PubMed] [Google Scholar]
- 5.Graf E. Calsium binding to phytic acid. J Agric Food Chem. 1983;31:851–855. [Google Scholar]
- 6.Greaves M P, Anderson G, Webley D M. The hydrolysis of inositol phosphates by Aerobacter aerogenes. Biochim Biophys Acta. 1967;132:412–418. doi: 10.1016/0005-2744(67)90160-x. [DOI] [PubMed] [Google Scholar]
- 7.Greiner R, Haller E, Konietzny U, Jany K D. Purification and characterization of a phytase from Klebsiella terrigena. Arch Biochem Biophys. 1997;341:201–206. doi: 10.1006/abbi.1997.9942. [DOI] [PubMed] [Google Scholar]
- 8.Greiner R, Konietzny U, Jany K D. Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys. 1993;303:107–113. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Irving G C, Cosgrove D J. Inositol phosphate phosphatase of microbial origin. Observations on the nature of the active centre of a bacterial (Pseudomanas sp.) phytase. Aust J Biol Sci. 1971;24:559–564. doi: 10.1071/bi9710559. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Lambrechts C, Boze H, Moulin G, Galzv P. Utilization of phytate by some yeast. Biotechnol Lett. 1992;14:61–66. [Google Scholar]
- 13.Lee D, Schroeder J, Gordon D T. Enhancement of Cu bioavailability in the rat by phytic acid. J Nutr. 1988;118:712–717. doi: 10.1093/jn/118.6.712. [DOI] [PubMed] [Google Scholar]
- 14.Lei X, Pao K, Elwyn R M, Ullrey D E, Yokoyama M T. Supplemental microbial phytase improves bioavailability of dietary zinc to weanling pigs. J Nutr. 1993;123:1117–1123. doi: 10.1093/jn/123.6.1117. [DOI] [PubMed] [Google Scholar]
- 15.Miasnikov, A. Unpublished data.
- 16.Mitchell D B, Vogel K, Weimann B J, Pasamontes L, van Loon A P. The phytase subfamily of histidine acid phosphatases: isolation of two genes for two novel phytases from the fungi Aspergillus terrus and Mycoliophthora thermophila. Microbiology. 1997;143:245–252. doi: 10.1099/00221287-143-1-245. [DOI] [PubMed] [Google Scholar]
- 17.Moran C P., Jr . RNA polymerases and transcription factors. In: Sonenshein A, Hoch J, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 18.Nagarajan V. Protein secretion. In: Sonenshein A, Hoch J, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 713–726. [Google Scholar]
- 19.Nasi M. Microbial phytase supplementation for improving availability of plants phosphorus in the diets of growing pigs. J Agric Sci. 1990;62:435–442. [Google Scholar]
- 20.Nayini N R, Markakis P. Effects of inositol phosphates on mineral utilization. Fed Proc. 1983;45:819–826. [Google Scholar]
- 21.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Pasamontes L, Haiker M, Wyss M, Tessier M, Loon A P. Gene cloning, purification and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl Environ Microbiol. 1997;63:1696–1700. doi: 10.1128/aem.63.5.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasamontes, L., M. Haiker, M. Henriquez-Huecas, D. B. Mitchell, C. Broger, and A. P. van Loon. Cloning of the phytases of Aspergillus nidulans and Talaromyces thermophilus and their evolutionary relation to other histidine acid phosphatases. Unpublished data.
- 24.Pen J, Verwoerd T C, Hoekema A. Phytase-containing transgenic seeds as novel feed additive for improved phosphorus utilization. Bio/Technology. 1993;11:811–814. [Google Scholar]
- 25.Piddington C S, Houston C S, Paloheimo M, Cantrell M, Miettinen-Oinonen A, Nevalainen H, Rambosek J. The cloning and sequencing of the genes encoding phytase (phy) and pH 2.5-optimum acid phosphatase (aph) from Aspergillus niger var. awamori. Gene. 1993;133:55–62. doi: 10.1016/0378-1119(93)90224-q. [DOI] [PubMed] [Google Scholar]
- 26.Powar V K, Jagannathan V. Purification and properties of phytate-specific phosphatase from Bacillus subtilis. J Bacteriol. 1982;151:1102–1108. doi: 10.1128/jb.151.3.1102-1108.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy N R, Pierson M D, Sathe S K, Salunkhe D K. Phytates in cereals and legumes. Boca Raton, Fla: CRC Press, Inc.; 1989. [Google Scholar]
- 28.Roels S, Losick R. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J Bacteriol. 1995;177:6263–6275. doi: 10.1128/jb.177.21.6263-6275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Shah V, Parekh L J. Phytase from Klebsiella sp. no. PG-2: purification and properties. Indian J Biochem Biophys. 1990;27:98–102. [PubMed] [Google Scholar]
- 31.Shimizu M. Purification and characterization of phytase from Bacillus subtilis (natto) N-77. Biosci Biotechnol Biochem. 1992;56:1266–1269. [Google Scholar]
- 32.Simons P C, Versteegh H A J. Improvement of phosphorus availability by microbial phytase in broilers and pigs. Br J Nutr. 1990;64:525–540. doi: 10.1079/bjn19900052. [DOI] [PubMed] [Google Scholar]
- 33.Sutardi •, Buckle K A. Characterization of extra- and intracellular phytases from Rhizopus oligoporus used in tempeh production. Int J Food Microbiol. 1988;6:67–79. doi: 10.1016/0168-1605(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 34.Ullah A H, Gibson D M. Extracellular phytase (E.C.3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization. Prep Biochem. 1987;17:63–91. doi: 10.1080/00327488708062477. [DOI] [PubMed] [Google Scholar]
- 35.Ullah A H, Wodzinski R J. Phytase. Adv Appl Microbiol. 1996;42:263–302. doi: 10.1016/s0065-2164(08)70375-7. [DOI] [PubMed] [Google Scholar]
- 36.van Harttingsveldt W, et al. Cloning, characterization and over expression of the phytase-encoding gene (phyA) of Aspergillus niger. Gene. 1993;127:87–94. doi: 10.1016/0378-1119(93)90620-i. [DOI] [PubMed] [Google Scholar]
- 37.Vellanoweth R L. Translation and its regulation. In: Sonenshein A, Hoch J, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 699–711. [Google Scholar]
- 38.Wambutt R, Wedler H, Lapidus A, Sorokin A, Ehrlich D. Sequence analysis of the Bacillus subtilis chromosome region between the odhAB and sspC loci in a yeast artificial chromosome. Accession no. AF015775. 1997. Unpublished data. [Google Scholar]