Abstract
Background
Long non-coding RNAs (lncRNAs)-related immune genes (lrRIGs) play a crucial role in the development and progression of lung adenocarcinoma (LUAD). However, reliable prognostic signatures based on lrRIGs have not yet been identified.
Methods
We screened lrRIGs associated with the prognosis of LUAD using The Cancer Genome Atlas (TCGA) database and then established a novel prognostic nine-gene signature composed of CD79A, INHA, SHC3, LIFR, TNFRSF11A, GPI, F2RL1, SEMA7A and WFDC2 through bioinformatic approaches. A risk score derived from this gene signature was used to divide LUAD patients into the low- and high-risk groups. The latter was confirmed to have markedly worse overall survival (O.S.). A nomogram was developed using the risk score and other independent prognostic elements, demonstrating excellent performance in predicting the O.S. rate of LUAD patients.
Results
We observed that the infiltration of diverse immune cell subtypes and response to immunotherapy and chemotherapy significantly differed between the low- and high-risk groups.
Conclusions
Overall, stratification based on this gene signature could be used to guide better therapeutic management and improve outcomes for LUAD patients.
Subject terms: Prognostic markers, Cancer genomics
Background
Lung cancer is the leading cause of cancer-related death worldwide [1, 2]. Lung adenocarcinoma (LUAD) is the primary histological subtype of lung cancer for men and women. Although treatment techniques have greatly improved over the past few decades, the 5-year survival rate for LUAD is only ~10% due to a lack of early detection and a tendency to metastasize early [3, 4]. Currently, known clinicopathological risk factors cannot effectively distinguish these LUAD patients with a high risk of disease progression. Therefore, identifying novel prognostic biomarkers would be one of the promising ways to develop new diagnostic and therapeutic strategies for LUAD.
Increasing evidence has shown that immune system disorders may be a significant cause of cancer development, and immunotherapy has become a promising cancer treatment strategy [5, 6]. In recent years, the breakthrough of immunomodulatory therapies targeting the programmed death 1 (PI-1)/PD-1 ligand (PD-L1) signaling has shown considerable success in multiple cancers by promoting anti-tumor immune function [7]. Moreover, a recent study observed that the blockade of PD-1/PD-L1 signaling promoted cytotoxic T lymphocytes’ activity, inhibiting tumor growth and increasing the survival rate in the mouse metastasis model [8]. Thus, immune-oncology has attracted extensive attention, and immune-related genes (IRGs) and immune infiltrating cells are considered to be determinants of the development and progression of various tumors [9, 10].
Long non-coding RNAs (lncRNAs), which are non-coding transcripts with a length longer than 200 nucleotides [11], can influence the tumor microenvironment by regulating immune gene expression and participating in inflammation [12, 13]. For example, the lncRNA NRON could maintain a resting state of T cells by sequestering the phosphorylated nuclear factor of activated T cells (NFAT) in the cytoplasm [14]. Lnc-chop enhances the immunosuppressive function of myeloid-derived suppressor cells in the tumor environment by activating C/EBPβ and increasing the expression of cyclooxygenase-2, NO synthase 2, arginase-1, and NADPH oxidase2 [15]. The NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death [16]. Besides, in 2019, Wang et al. reported that lncRNA UCA1 increased PD-L1 expression by inhibiting miR-214 and miR-193a, thus contributing to the immune escape of gastric cancer cells [17]. However, only a few immune genes associated with lncRNAs have been found to play an essential role in tumorigenesis and malignant transformation so far [11, 13, 18, 19]. The overall biological role and underlying mechanism of the lncRNAs-related immune genes (lrRIGs) in LUAD are still unclear. Whether lrRIGs could serve as a predictor for LUAD prognosis remains unknown.
Herein, we extensively analyzed the relationship of lncRNAs with 2483 immune-related genes based on the transcript and clinical data obtained from the TCGA and GEO databases. We explored the expression variations of 329 differentially expressed lrRIGs in LUAD and investigated the potency as biomarkers. Then, a lrRIGs prognostic signature composed of CD79A, INHA, SHC3, LIFR, TNFRSF11A, GPI, F2RL1, SEMA7A, and WFDC2 was established and validated using external databases and clinical samples. Moreover, we showed the correlation between the prognostic risk model and tumor-infiltrating immune cells and therapeutic response. Independent prognostic factors were further explored and combined into a predictive nomogram for survival prediction of LUAD patients. Additionally, in vitro studies revealed that the prognostic signature gene GPI affected the prognosis of LUAD patients by activating the mTORC1 signaling pathway.
Methods
Data collection
Transcriptome and clinical data were downloaded from the TCGA (https://portal.gdc.cancer.gov/) and GEO (http://www.ncbi.nlm.nih.gov/geo/). The immune-related genes were obtained from the ImmPort website (available online: https://www.immport.org/shared/home, accessed on July 2020) [20]. The Ensembl database was used to screen lncRNAs. The correlation between lncRNAs and immune-related genes was calculated to obtain lrRIGs based on the TCGA_LUAD database. Correlation coefficients >0.4 and P < 0.001 were used as the threshold.
Bioinformatic analysis
Differentially expressed lrRIGs (DE-lrRIGs) were screened using the R package “limma”. Screening condition: false-discovery rate [FDR] < 0.05, |logFC | > 1. Volcano plots and DE-lrRIGs heatmap analyses were done using ggplot2 and pheatmap package, respectively. The R survival package was used to assess relationships between DE-lrRIGs and overall survival (O.S.) and to plot survival curves. ROC maps were created using the survival ROC package. Univariate and multivariate independent prognostic analyses by the survival package were used to determine the prognostic values of specific gene signatures. The least absolute shrinkage and selection operator (LASSO) Cox regression method was adopted to construct multivariable models with DE-lrRIGs using the R package “glmnet”.
Estimation of tumor-infiltrating immune cells and biomarkers for immunotherapy based on the risk signature
The fraction of 22 immune cell types in LUAD samples was estimated using the CIBERSORT algorithm based on the TCGA_LUAD database. Samples with a CIBERPORT output value of P < 0.05 were considered to meet the conditions for further analysis. The difference of immune cells in the proportion between the high- and low-risk groups was examined by the Wilcoxon rank sum test. The R package “limma” was used to analyze the correlation between the signature of the lncRNAs-related immune genes and key immune checkpoints, tumor mutation load (TMB), and HLA family members.
Prediction of chemotherapeutic response based on the lrRIGs signature
Seven commonly used chemotherapy drugs (cisplatin, docetaxel, doxorubicin, gemcitabine, etoposide, paclitaxel and cytarabine) and two targeted drugs (axitinib and gefitinib) were selected for the chemotherapeutic response prediction through the ridge regression using the “pRRophetic” R package based on the Genomics of Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/). The half-maximal inhibitory concentration (IC50) predicted for each TCGA_LUAD patient was used to assess differential chemotherapeutic response [21].
Gene set enrichment analysis and development of nomogram
The signaling pathways and biological processes of differentially expressed gene enrichment between the high- and low-risk subgroups were investigated using Gene Set Enrichment Analysis (GSEA) (http://software.broadinstitute.org/gsea). Stage, recurrence, and risk scores were used to develop a nomogram by the “survival” and the “rms” package for R. Calibration curves were plotted to assess the consistency between actual and predicted survival. The concordance index (C-index) was calculated to evaluate the performance of the model predicting prognosis.
Clinical lung adenocarcinoma and adjacent no-tumor lung tissues
Thirty matched LUAD and adjacent normal lung tissues were obtained from the First Affiliated Hospital of Henan University and Puyang Hospital of traditional Chinese medicine, China. All patients gave informed consent before sample collection. This study was approved by the Ethics Committee of Medical School of Henan University, China (HUSOM-2018-282). All methods in this study were carried out following the approved guidelines. Clinicopathological qualities of the LUAD patients are provided in Supplementary Table S1.
Cell culture and stable transfection of shRNA
Detailed information on cell culture and plasmid construction are described in the Supplementary Materials and Methods.
Colony formation assay, wound-healing assay, and in vivo lung metastasis and subcutaneous tumor model
Cell proliferation and migration were assessed by colony formation and wound-healing assay. Details of the relevant contents have been described previously [22]. Five-week-old specific pathogen-free (SPF) female BALB/C nude mice and C57BL/6J were obtained from the Weitong Lihua Animal Co. (Beijing, China) and housed in a SPF-grade facility on our campus. A549 cells steadily infected with lentiviruses carrying shGPI or control shRNA were collected for pulmonary metastasis analysis. A single-cell suspension containing 2 × 106 cells in 200 μL of PBS was injected into the tail veins of BALB/C nude mice (n = 5). All mice were euthanized, and lung tissues were collected seven weeks after injection. In the CMT167 subcutaneous tumor model, CMT167/shNC or CMT167/sh3mGPI (2.5 × 105) cells were subcutaneously injected into the right flank of C57BL/6 J mice. Seven days later, when the tumors reached about 60 mm3, the mice were randomly grouped using a random number table and identified by ear tags. Subsequently, they were intravenously administered either a vehicle or an anti-CTLA-4 antibody (cat. no. BE0131; Bio X Cell, West Lebanon, NH; 10 mg/kg, every 3 days). Mice were sacrificed when the tumor length reached 1.5 cm. The experimental protocol was approved by the Animal Care and Research Committee of Henan University.
Western blot
Tissue proteins were extracted using RIPA lysis buffer containing 500 mM NaCl, 1 mM EDTA, 1% NP-40, 50 mM Tris pH 8.0, and 1×cocktail of protease inhibitors (Roche, Lewes, UK). The protein concentration was examined using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Equal amounts of proteins per sample were isolated by SDS-PAGE and then blotted onto polyvinylidene difluoride (PVDF) membrane and blocked with 5% nonfat milk for 1 h at room temperature. Blots were probed with the indicated primary antibodies overnight at 4 °C and followed by incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody. Protein bands were visualized using the enhanced chemiluminescence (ECL) detection kit (GE Healthcare Biosciences). The detailed information on antibodies used in this study was provided in Supplementary Table S2.
Statistical analysis
All statistical analyses were executed using SPSS21.0 (SPSS Inc., Chicago, IL, USA) and R software (version 3.6.0). Pearson’s chi-square test and the Wilcoxon test were used to examine the significance level of correlation amongst variables and compare the data from different groups, respectively. The length of time from the date of diagnosis to death from any cause was the O.S. time. The continuous data are expressed as the mean ± standard deviation (SD). The association between risk score and O.S. were assessed by univariate and multivariate Cox regression analyses. Kaplan–Meier curves were drawn, and the significant difference in O.S. between groups was examined using the long-rank test. Unless otherwise specified, all statistical tests were two-tailed, and P < 0.05 was considered statistically significant.
Results
Development of a prognostic model based on lrRIGs in LUAD
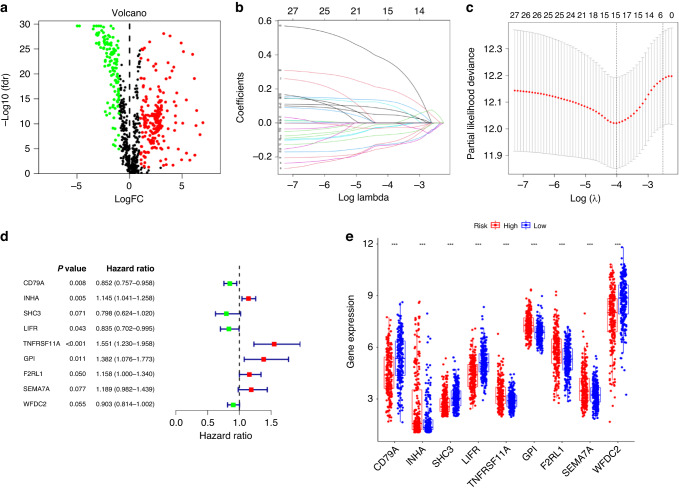
We downloaded the transcriptome profiling data of LUAD, including 54 normal and 497 tumor samples, from the TCGA database. Then, the data were annotated according to gene transfer format (GTF) files from Ensemble, and a co-expression analysis was conducted between lncRNAs and 2483 immune-related genes (Supplementary Table S3) by correlation analysis using the cor. test in R with |correlation coefficient | >0.4 and P < 0.01. A total of 889 lrRIGs were identified (Supplementary Table S4), and 329 were distinguished as differentially expressed lrRIGs (Supplementary Table S5), among which 123 were downregulated and 206 were upregulated (Fig. 1a). To further explore the potential predictive value of the 329 lrRIGs in LUAD, a univariate Cox regression analysis was conducted. At Hazard Ratio (HR) ≠ 1, P < 0.01, 27 genes were identified as prognostic genes for O.S. in patients with LUAD from the TCGA database (Supplementary Table S6). Next, LASSO regression with tenfold cross-validation was performed to obtain the optimal lambda value from the minimum partial likelihood deviance, which was related to 15 of the 27 prognostic genes significantly associated with O.S. (Fig. 1b, c). Finally, we used the TCGA_LUAD database as a training set (n = 468) and the GSE31210 database as an external validation set (n = 226). Multivariate Cox regression analysis observed that nine genes constructed a significant prognostic signature for LUAD: CD79A, INHA, SHC3, LIFR, TNFRSF11A, GPI, F2RL1, SEMA7A and WFDC2 (Supplementary Table S7 and Fig. 1d, e). The prognostic gene signatures were shown as risk Score=sum [gene expression × coefficient].
Fig. 1. Identifying prognostic risk genes and analyzing their expression based on differentially expressed lncRNA-related immune genes (DE-lrRIGs) using the TCGA_LUAD database.
a Volcano plot representing DE-lrRIGs in the TCGA_LUAD database. The red dots in the plot represent upregulated genes, green dots represent downregulated genes with statistical significance, and black dots represent no differentially expressed genes. b LASSO coefficient profiles of the 27 DE-lrRIGs with non-zero coefficients were determined by the optimal lambda in the TCGA_LUAD database. c LASSO regression with the screening of optimal parameters (lambda) obtained fifteen prognostic genes. d Multivariate Cox regression of 15 prognostic genes from LASSO regression analysis. e Differential expression analysis of 9 prognostic signature genes in the high- and low-risk groups.
Evaluation of the performance of lrRIGs prognostic signature
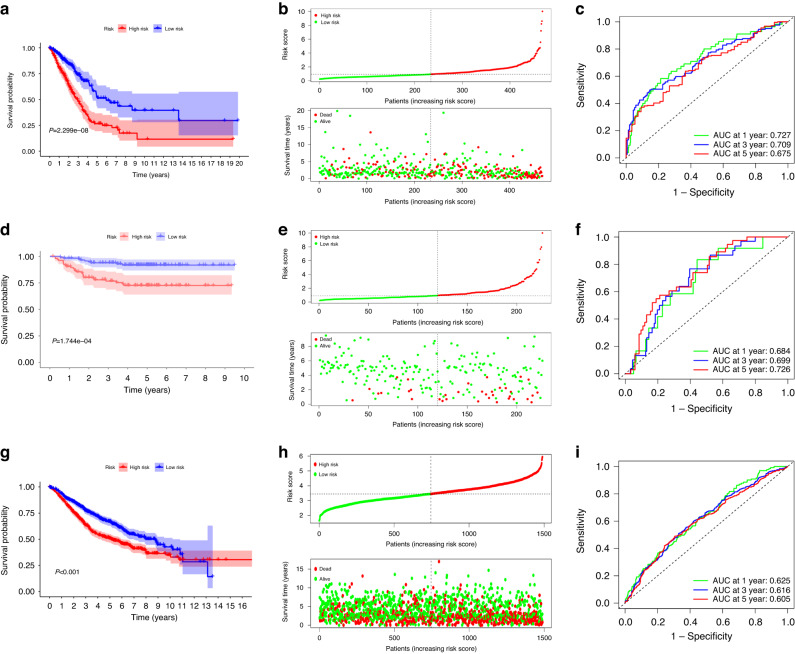
The risk score of each patient in the TCGA_LUAD dataset was calculated according to lrRIGs prognostic signature using the “survminer” R software package. Patients were divided into the high- and low-risk groups by the median of risk scores. Kaplan–Meier analysis demonstrated that the O.S. of the high-risk group was worse than that of the low-risk group (Fig. 2a; P < 0.05). The distribution of risk scores and survival status were plotted in Fig. 2b, showing poorer survival in high-risk patients. Then, ROC curve analysis was performed to evaluate the predictive ability of the lrRIGs signature. The AUC for lrRIGs risk signature was 0.727, 0.709, and 0.675 at 1-, 3- and 5- year for O.S., respectively (Fig. 2c). Last, validation of the lrRIGs risk signature was conducted using the GSE31210 database and an integrated dataset consisting of GSE11969, GSE13213, GSE41271, GSE42127, GSE50081, GSE68465 and GSE72094, which included 1489 LUAD patients with survival data. We carried out risk scoring and risk grouping for LUAD patients in the manner mentioned above. Consistently, survival analysis observed markedly longer O.S. in the low-risk group than in the high-risk group (Fig. 2d, e, g, h; P < 0.05). Moreover, the analyses of ROC curves proved that this nine-gene signature had a robust predictive ability in the above two databases (Fig. 2f, i).
Fig. 2. Prognostic performance of the lrRIGs signature.
a, d, g Kaplan–Meier curve shows a better O.S. in the low-risk group than in the high-risk group in the TCGA (a), GSE31210 (d) and the integrated dataset (g). b, e, h Patients’ risk score and survival status distribution based on lrRIGs signature in the TCGA (b), GSE31210 (e) and the integrated database (h). c, f, i Receiver operating characteristic (ROC) curves showed the performance of the signature in predicting 1/3/5-year O.S. in the TCGA (c), GSE31210 (f) and the integrated database (i).
Validation of the expression level and the prognostic value of prognostic signature genes
We performed external validation of these nine prognostic signature genes. The GSE31210 and GSE75037 database analyses demonstrated that the mRNA expression levels of INHA, TNFRSF11A, GPI, F2RL1, WFDC2, CD79A and SEMA7A in LUAD tissues were significantly higher than in normal lung tissues (Supplementary Fig. S1A, B; all P < 0.05). Inversely, the expression levels of SHC3 and LIFR in tumor tissues were lower. Moreover, the protein expression levels of these nine signature genes were further confirmed in 30 matched clinical LUAD tissues (T) and adjacent non-tumor tissues (N) by western blot analysis. (Supplementary Figs. S1C, D and S2). Additionally, we further analyzed the expression characteristics of prognostic signature genes and their correlation with lung adenocarcinoma based on the immunohistochemistry data from Human Protein Atlas (HPA) database (https://www.proteinatlas.org/). We selected the GPI gene for subsequent research work because GPI has the highest HR value among the nine risk genes. The results demonstrated that the expression level of GPI was relatively low in normal lung tissues and adjacent tissues, while a high proportion of non-small cell lung cancer (NSCLC) tissues and LUAD tissues displayed high (NSCLC, 5/12; LUAD, 1/6) and moderate (NSCLC, 6/12; LUAD, 4/6) GPI staining, which is mainly located in the cytoplasm and membrane (Supplementary Fig. S3). Subsequently, the predictive effects of these signature genes for the O.S. and recurrence-free survival (RFS) were evaluated by Kaplan–Meier survival analysis and the log-rank test using the Kaplan–Meier plotter, respectively. The results showed that high expression of INHA, GPI, F2RL1, CD79A, and SEMA7A, and low expression of SHC3 and LIFR, were associated with shortened O.S. and RFS in LUAD (Supplementary Figs. S1E-M and S4).
Performance comparison of the lrRIGs signature with other reported gene signatures
To further evaluate the prediction performance of the lrRIGs signature, four published gene signatures obtained from Sun’s [23], Cao’s [24], Zhang’s [25], and Li’s [26] were selected for comparison. According to the corresponding genes in these four risk models, the risk score of each patient was calculated using the same method in the training cohort. Then the analyses of Kaplan–Meier survival and the time-dependent ROC were performed. We observed that the prognosis difference for the high- and low-risk groups was significant across all four risk models. However, the ROC analysis showed that the AUC of the lrRIGs signature for 1-, 3-, and 5-year O.S. were 0.727, 0.709, and 0.675, respectively. These AUCs were significantly larger than those of Sun’s, Cao’s, Zhang’s and Li’s gene signatures (Fig. 2c and Supplementary Fig. S5A–D). Additionally, Supplementary Fig. S5E showed that our risk model based on lrRIGs had the highest C-index with 0.68. Moreover, the RMS time curve of all five prognostic models further demonstrated that this 9-gene signature had the largest slope (Supplementary Fig. S5F), indicating superior estimation of LUAD survival with lrRIGs prognostic signature.
Prognostic risk score was associated with clinical outcome
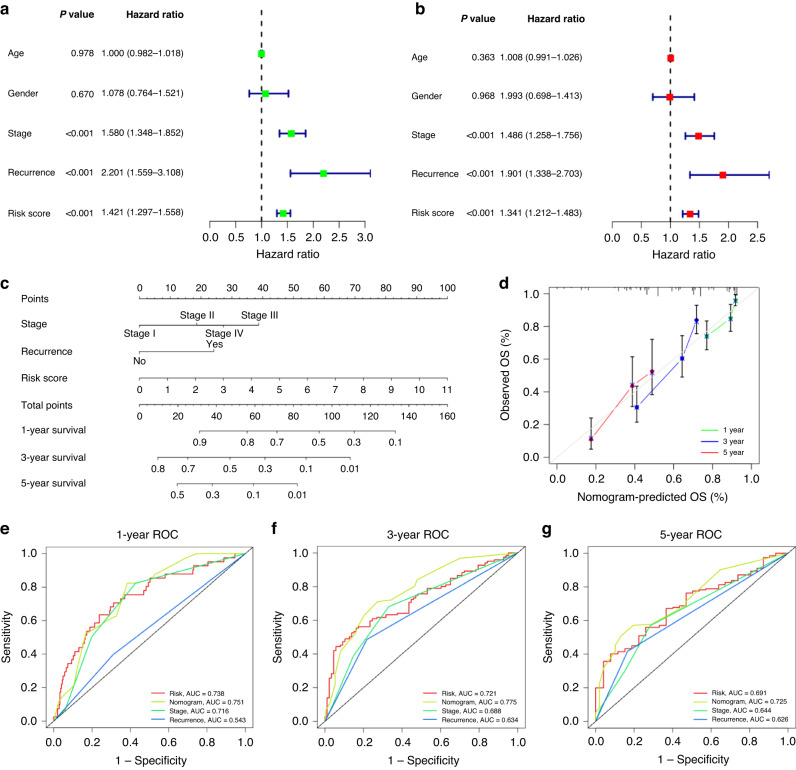
We utilized univariable and multivariable Cox regression to analyze the relationship between clinical parameters, risk scores, and O.S. (Fig. 3a, b). The results revealed that the risk score is an independent risk factor with a hazard ratio (HR) of 1.341. Similar results were indicated by stage (HR = 1.486) and recurrence (HR = 1.901). Age and gender showed no significance.
Fig. 3. Association between indicated variables and prognosis of patients with LUAD.
a, b Univariate Cox regression analyses (a) and multivariate Cox regression analyses regarding the indicated variables and O.S. of LUAD patients (b) in the TCGA_LUAD database. c Construction of nomogram using indicated variables to predict individuals’ probability of survival. d Calibration plots of a nomogram to predict 1-, 3-, and 5-year O.S. of patients to actual survival. e–g The time-dependent ROC curves of the nomograms compared with indicated variables in 1-, 3-, and 5-year O.S. AUC, area under curve.
To elevate the accuracy and reliability of the predictive model in LUAD, we then integrated stage, recurrence, and risk scores to build a nomogram model. It showed that risk score had the greatest impact on the prediction of survival rate (Fig. 3c). The calibration plots of the nomogram for predicting 1-, 3- and 5-year O.S. of LUAD revealed the excellent concordance between actual observation and our nomogram prediction (Fig. 3d), and the nomogram model’s C-index was 0.742 (95% CI = 0.701–0.784, P = 2.93e-30). Moreover, the AUC value of the nomogram for predicting 1-, 3- and 5-year O.S. was larger than that of the stage, risk score, and recurrence, indicating our nomogram served as an essential factor in predicting clinical outcomes of LUAD patients (Fig. 3e–g).
Association of the risk model with tumor-infiltrating immune cells
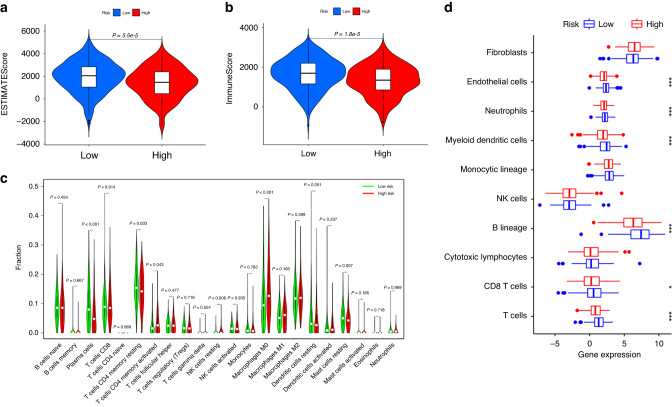
To investigate the relationship between immune-cell features and the lrRIGs risk model, we first calculated the ESTIMATE score and the immune score of each LUAD sample from the TCGA_LUAD database based on the ESTIMATE algorithm. The results showed that the ESTIMATE score (1927.23 vs. 1404.48, P < 0.05) and immune score (1642.43 vs. 1323.45, P < 0.05) markedly increased in the low-risk group compared with those in the high-risk group (Fig. 4a, b). Next, we analyzed the relative proportion of 22 tumor-infiltrating immune cells in each LUAD sample using the Cell type identification by estimating relative subsets of RNA transcripts (CIBERSORT) method. We observed significant differences in immune cell infiltration between the high- and low-risk groups. In particular, the high-risk group was characterized by a relatively high proportion of T cells CD4 memory activated, NK cells resting, and macrophages M0, while the low-risk group displayed a relatively high proportion of plasma cells, T4 cells CD4 memory resting, and mast cells resting (Fig. 4c; P < 0.05). Subsequently, we further evaluated the abundance of eight immune-related cells and two stromal cells using the microenvironment cell population count (MCP-counter) algorithm. Compared with the high-risk group, the abundance of endothelial cells, neutrophils, myeloid dendritic cells, B lineage, CD8 T cells, and T cells in the low-risk group increased significantly (Fig. 4d; P < 0.05).
Fig. 4. Comparison of immune microenvironments between high- and low-risk groups defined by the 9-gene signature.
a, b Analysis of the ESTIMATE algorithm showed significantly high ESTIMATE score (a) and immune score (b) in the low-risk group than in the high-risk group. c Estimation of 22 immune cell infiltration using the CIBERSORT method. d Evaluation of the abundance of indicated cells using the MCP-counter algorithm. ESTIMATE, Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data. *P < 0.05; ***P < 0.001.
Association of the risk model with immunotherapies
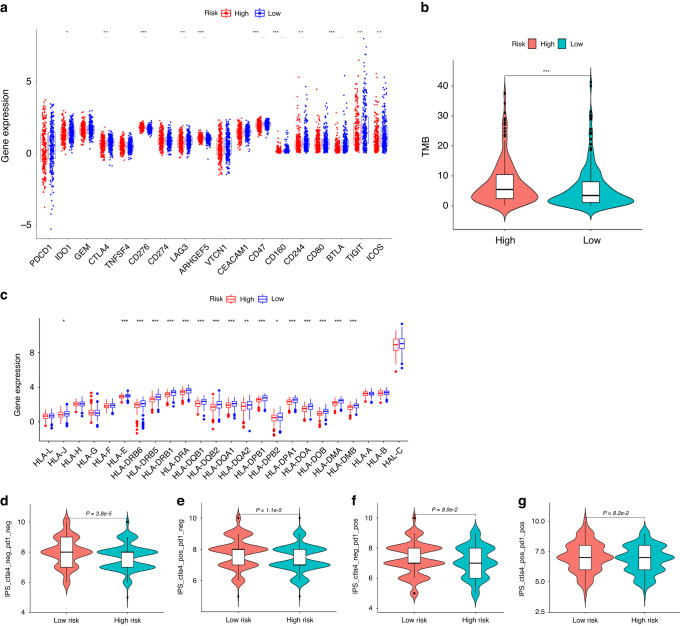
Some patients with LUAD have obtained great clinical benefits through immunotherapy, especially immune checkpoint inhibitors. However, selecting patients who respond to immunotherapy remains challenging. At present, several biomarkers have shown potential in predicting immunotherapy response, including the expression levels of immune checkpoint proteins [27, 28], class I human leukocyte antigen (HLA) family members [29], and TMB [30–32]. Therefore, we first examined whether immune checkpoints are differentially expressed between the high- and low-risk groups. As shown in Fig. 5a, compared with the high-risk group, the low-risk group had markedly higher expression levels of IDO1, CTLA-4, LAG3, CD47, CD160, CD244, BTLA, TIGIT, and ICOS. Conversely, the expression levels of CD276, and ARHGEF5 were higher in the high-risk group. Next, the single nucleotide mutation data of 562 LUAD patients were downloaded from the TCGA database and processed using the maftools package. The results showed that the TMB in the high-risk group was markedly higher than that of the low-risk group (7.57 vs. 5.99, P = 4.9e-4) (Fig. 5b).
Fig. 5. Analysis of immune checkpoints, HLA and mismatch repair genes expression and TMB.
a Analysis of differential expression of immune checkpoints in the high- and low-risk groups. b The high-risk group exhibited markedly higher TMB than the low-risk group. c Analysis of the profile of HLA member expression levels in the high- and low-risk groups. d–g Immunophenoscore (IPS) difference of LUAD with different status of CTLA-4 or PD-1 between the high- and low-risk groups. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, we further investigated the expression of class I human leukocyte antigen (HLA) family members because HLA is responsible for neoantigen presentation and cytolytic T cell activity by presenting intracellular peptides on the cell surface. The lack of HLA may impair the ability of cells to present new antigens and lead to immune tolerance [33]. The results demonstrated that the expression of various HLA family members differed significantly between the two risk groups (Fig. 5c; P < 0.05). In fact, most of the HLA family members, including HLA-J, HLA-E, HLA-DRB6, HLA-DRB5, HLA-DRB1, HLA-DRA, HLA-DQB1, HLA-DQB2, HLA-DQA1, HLA-DQA2, HLA-DPB1, HLA-DPB2, HLA-DPA1, HLA-DOA, HLA-DOB, HLA-DMA, and HLA-DMB, showed decreased expression in the high-risk group relative to that in the low-risk group. Additionally, the cancer immunome atlas (TCIA) is a database that provides comprehensive immunogenomic analyses based on the TCGA (https://tcia.at/). Herein, we used the TCIA database to evaluate the immunotherapy response of LUAD patients with different risk scores through the immunophenoscore (IPS). The results revealed that the total IPS and IPS for CTLA-4 blocker in the low-risk group were significantly higher than that in the high-risk group (Fig. 5d, e), which strongly predicted that LUAD patients with lower risk scores would have better immunotherapy response, especially for CTLA-4 blockers. In comparison, the IPS for PD1 pus CTLA-4 blocker and PD1 blocker did not differ significantly between risk groups (Fig. 5f, g).
Finally, we attempted to validate the above results of the bioinformatics analysis by silencing key genes that are positively correlated with the risk score in the risk model. Given GPI has the most significant effect on the O.S. and RFS of LUAD (Supplementary Figs. S1I and S4E). We silenced the mGPI gene in CMT167 cells using shRNA and obtained a stable cell line. We next investigated whether GPI silencing could affect the anti-tumor effect produced by immune checkpoint blockade. In vivo experimental results showed that CMT167 tumors could respond to anti-CTLA-4 antibody monotherapy. Interestingly, mGPI knockdown-mediated by sh3mGPI in CMT167 cells rendered the cell line more sensitive to anti-CTLA-4 antibodies [tumor inhibition rate (%) = 93.3%] in the CMT167/sh3mGPI subcutaneous tumor model than control CMT167/shNC cells [tumor inhibition rate (%) = 80.7%] (n = 5/group). Compared with the anti-CTLA-4 therapy, combined mGPI knockdown with anti-CTLA-4 treatment markedly reduced tumor weight and repressed tumor growth (Supplementary Fig. S6). No discernible adverse events were observed during the experiment. These data indicated that the expression of risk genes was associated with the efficacy of immune checkpoint blockade.
Association of the risk model with chemotherapeutic responses in LUAD
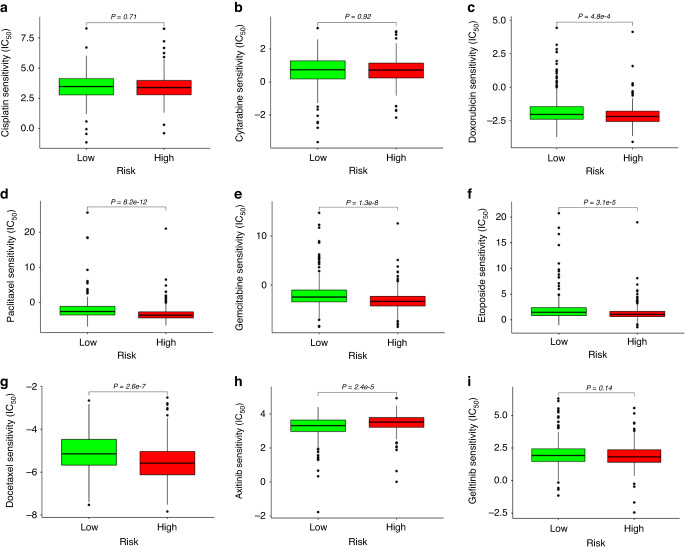
Given that chemotherapy is one of the critical means of tumor treatment in the clinic, we explored the relationship between the risk score and clinical response to chemotherapeutic drugs by calculating the IC50 using the R package pRRophetic [21, 34]. Based on the Cancer Genome Project (CGP) database, we screened seven chemotherapy drugs (cisplatin, docetaxel, doxorubicin, gemcitabine, etoposide, paclitaxel, and cytarabine) and two targeted drugs (axitinib and gefitinib), which had been used in the clinical treatment of lung adenocarcinoma. We observed that the drug response differed in the high-risk and low-risk groups. In the chemotherapy drug for lung cancer, doxorubicin, paclitaxel, gemcitabine, etoposide, and docetaxel had a high drug response in the high-risk group. For the targeted drug of lung cancer, axitinib had a favorable response in the low-risk group (Fig. 6). According to multivariable Cox analysis, TNFRSF11A and GPI in prognostic risk genes had a high-risk coefficient (Supplementary Table S7), indicating that their expression significantly impacts the risk score of LUAD patients. Therefore, we chose to silence the TNFRSF11A or GPI gene in H1299 and A549 cells using shRNA to explore whether the expression of these genes affects the sensitivity of tumor cells to commonly used chemotherapy drugs, such as doxorubicin (sc-280681A, SANTA) and docetaxel (HY-B0011, MCE). Tumor cells were treated with various concentrations of doxorubicin (0.1, 1, 2.5, 5, 10 and 20 μM) or docetaxel (0.1, 1, 10, 20, 50 and 100 μM) for 24 h, and the inhibitory concentration 50 (IC50) value was calculated in GraphPad Prism 6 program. In vitro experiments demonstrated that GPI knockdown in A549 and H1299 cells by sh1GPI made both cell lines less sensitive to doxorubicin (IC50 = 4.548 μM, 8.549 μM, respectively) and docetaxel (IC50 = 24.85 μM, 30.81 μM, respectively) than control A549/shNC (IC50 = 1.531 μM, 4.605 μM, respectively) and H1299/shNC (IC50 = 2.252 μM, 8.042 μM, respectively). However, silencing the TNFRSF11A gene in A549 and H1299 cells did not significantly affect the sensitivity of these two cell lines to doxorubicin and docetaxel (Supplementary Fig. S7). These results suggested that the prognostic risk gene GPI plays a potential role in the chemosensitivity of LUAD cells.
Fig. 6. Predicted patients' response in TCGA database to targeted therapies and standard chemotherapies in different risk groups.
IC50 was calculated for cisplatin (a), cytarabine (b), doxorubicin (c), paclitaxel (d), gemcitabine (e), etoposide (f), docetaxel (g), axitinib (h), and gefitinib (i) in the high- and low-risk groups by coupling baseline gene expression levels of patients and drug sensitivity data in cancer cell lines.
Prognostic signature gene GPI affects the prognosis of LUAD patients, involving the activation of mTORC1 signaling pathway
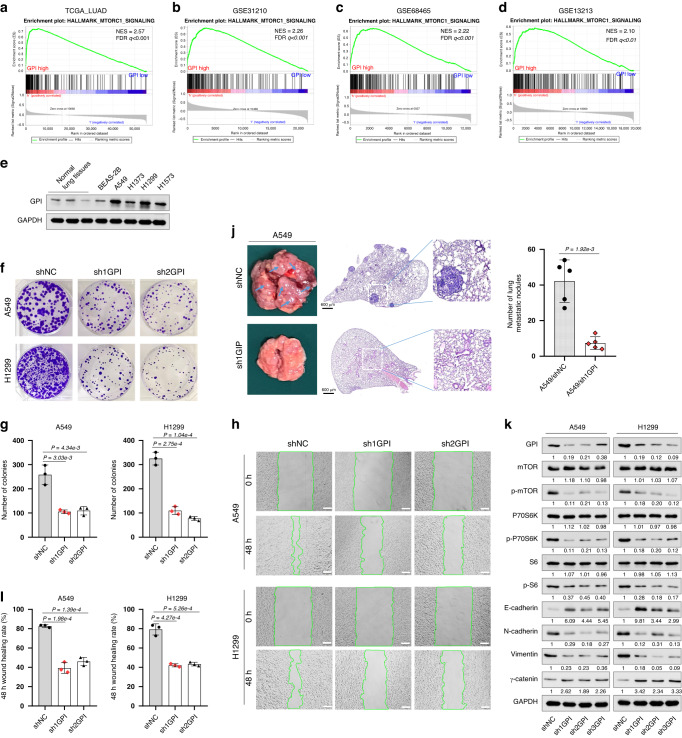
To gain more insights into the potential mechanisms of the prognostic risk genes in LUAD, GSEA was performed by comparing the high and low expression of these signature genes based on the TCGA_LUAD, GSE31210, GES68465, and GSE13213 databases. GPI is one of the nine prognostic risk genes; it has the most significant effect on the O.S. and RFS of LUAD. Previous studies reported that GPI was related to poor metastasis in LUAD, colorectal cancer, renal cell carcinoma, breast cancer, and endometrial cancer [35–38]. However, the effects of GPI on malignant biological behaviors and downstream signaling transduction in LUAD were unclear. Therefore, GPI was selected for further study. The results showed that several vital regulatory genes involved in mTORC1 signaling were consistently enriched in cells with high GPI expression in the TCGA_LUAD, GSE31210, GES68465, and GSE13213 databases, indicating that GPI expression was positively associated with the mTORC1 signaling pathway in LUAD patients (Supplementary Table S8 and Fig. 7a–d).
Fig. 7. To investigate the possible mechanism of prognostic signature gene GPI affecting the prognosis of LUAD patients.
a–d GSEA analysis was performed using TCGA_LUAD, GSE31210, GSE68465, and GSE13213 databases to compare the high and low expression of GPI, respectively. e The protein expression level of GPI in normal lung tissues and indicated cell lines was analyzed by western blot. f, g The effect of GPI knockdown on the proliferation of A549 and H1299 cells was assessed by colony formation assay. h, i The effect of GPI knockdown on the migration of A549 and H1299 cells was examined by wound-healing assay (scale bar, 100 μm). j GPI knockdown inhibited tumor cells’ lung metastasis in vivo. Each group was assigned five mice, and the metastatic tumor lesion in each mouse lung was analyzed by Haematoxylin and eosin staining (H&E staining). Appearance and representative images of H&E-stained sections of the lung tissues were shown. The nodule numbers of A549/shNC and A549/sh1GPI groups were 42.2 ± 11.9 and 7.4 ± 3.5. k Western blot analysis of indicated protein expression level. GSEA, Gene Set Enrichment Analysis. The experiments were conducted in triplicate, ensuring independent replication.
Afterward, we performed Western blot analysis to examine the expression level of GPI in A549, H1299, H1373, H1573, and BEAS-2B cell lines. Compared with normal lung tissues and the BEAS-2B cell line, GPI was markedly overexpressed in A549 and H1299 cells (Fig. 7e). Endogenous GPI silencing was conducted by lentivirus transfection of shRNA targeting GPI in A549 and H1299 cells. We observed that compared with the control group, GPI knockdown significantly inhibited the proliferation and migration of both cell lines as determined by colony formation assay (Fig. 7f, g) and wound-healing assay (Fig. 7h, i), respectively. Furthermore, in vivo lung metastasis assay showed that mice with A549/sh1GPI developed fewer pulmonary metastasis nodules than the mock control (Fig. 7j). Additionally, Western blot analysis showed that knockdown of GPI led to a decrease of p-mTOR, p-P70S6K, and p-S6, which are critical downstream molecules of the mTORC1 signaling pathway (Fig. 7k). Previous studies reported that the activation of this pathway is closely related to the malignant transformation of a variety of tumors and the proliferation and metastasis of tumor cells [39–41]. Therefore, we further examined the expression of the key proteins associated with epithelial-mesenchymal transition (EMT). We observed that GPI silencing in A549 and H1299 cells markedly decreased the expression of N-cadherin and Vimentin proteins and increased the levels of E-cadherin and γ-catenin (Fig. 7k). These results suggested that the regulation of GPI on multiple biological traits of lung adenocarcinoma cells could involve the activation of the mTORC1 signaling pathway.
Discussion
Multiple lines of evidence indicate that immune-related biomarkers are associated with the prognosis of various tumor types [42, 43]. However, biomarkers that can be used directly to determine the prognosis of patients and the efficacy of tumor immunotherapy remain to be investigated. Additionally, the regulation of immune-related genes is often closely related to the corresponding lncRNAs. The current study represents the first systemic analysis of 889 lrRIGs expression levels in LUAD tissues. Then, according to the multi-step selection, a prognostic risk model markedly associated with the O.S., immunotherapeutic response, chemotherapeutic response, and tumor microenvironment of LUAD patients was developed based on 9 differentially expressed lrRIGs, which potentially serves as an indicator for evaluating the effectiveness of immunotherapy and chemotherapy in LUAD.
In recent years, numerous studies have shown that lncRNAs play indispensable roles in the initiation and progression of various cancer, such as breast cancer [44, 45], prostate cancer [46], gastric cancer [47], and lung cancer [48, 49] by shaping the tumor immune microenvironment and regulating immune-related genes. In 2019, Hu et al. observed a decrease of MHC-I and β-2M expression in patients with higher lncRNA LINK-A expression. Mechanistically, LINK-A degrades TAP1/2, TPSN, and CALR proteins of the peptide-loading complex (PLC), thus affecting the loading and editing of MHC-I and antigen presentation [50]; In addition, Zhou et al. found that LINC00471 upregulates the expression of PD-L1 by sponging miR-195-5p in pancreatic cancer, suppresses the function of enhanced CD8 + T cells and promotes the development of cancer [51]. However, hitherto, little is known about the immune-related genes regulated by lncRNAs and their role in tumors. In the present study, we comprehensively evaluated immune genes related to lncRNAs using bioinformatics analysis based on the TCGA_LUAD database. 27 genes, such as FURIN, ADRB2, INHA, WFDC2, etc., were markedly associated with O.S. in patients with LUAD. According to the correlation analysis and hypergeometric testing, a nine-gene prognostic signature including CD79A, INHA, SHC3, LIFR, TNFRSF11A, GPI, F2RL1, SEMA7A and WFDC2 was developed. To our knowledge, this is the first established prognostic gene signature associated with differentially expressed lrRIGs in LUAD. This nine-gene signature could provide a new method for evaluating LUAD patients, guiding prognosis prediction and the choice of immunotherapy and chemotherapy.
Previous studies have reported some model genes’ biological function and expression patterns. For example, CD79A, a subunit of CD79, is mainly expressed on the surface of B cell and plays a crucial role in the transduction of BCR-recognized antigen signals into the cytoplasm. It was confirmed that overexpressed CD79A contributed to the malignant transformation of B-cell neoplasms, plasma cell tumors, and chronic lymphocytic leukemia [52, 53]. LIFR (receptor of leukemia inhibitory factor (LIF)) is commonly overexpressed in many solid cancers and recent studies have implicated that LIF/LIFR activates JAK2/STATs, MAPK, AKT, mTOR and other oncogenic signaling pathways, which play key roles in tumor progression, metastasis, stemness and therapy resistance. Additionally, LIF/LIFR signaling also plays a role in modulating multiple immune cell types present in the tumor microenvironment [54]. GPI (glucose-6-phosphate isomerase) is a housekeeping cytoplasmic enzyme which is frequently upregulated in various cancer types [55]. Besides its role as a glycolytic enzyme, mammalian GPI can function as a tumor-secreted cytokine and an angiogenic factor that stimulates endothelial cell motility [56]. SEMA7A (semaphorin 7A), also known as CD108, is an immune semaphorin that modulates diverse immunoinflammatory processes, including cytokine production, inflammatory infiltration, and immune cell interactions. Additionally, SEMA7A also regulates the migration, invasion, lymph formation, and angiogenesis of multiple types of tumor cells by interaction of SEMA7A with PLXNC1 and integrins [57]. However, the relationship between these nine genes and LUAD progression remains to be clarified, as few studies have reported the study of these genes. In this study, CD79A, INHA, TNFRSF11A, GPI, F2RL1, SEMA7A, and WFDC2 were all found to be upregulated in LUAD tissues and overexpressed INHA, GPI, F2RL1, CD79A, and SEMA7A were significantly correlated with a short O.S. and RFS. Importantly, by Western blotting analysis, we verified the overexpression patterns of the above seven genes in 30 pairs of LUAD tissues and adjacent normal lung tissues. Moreover, we further investigated the expression characteristic of the prognostic signature gene GPI and its correlation with LUAD using immunohistochemistry data from HPA dataset.
The mammalian target of rapamycin (mTOR) is a crucial signaling node that integrates environmental cues to regulate cell metabolism, proliferation and survival, and is often deregulated in human cancer. Activation of the mTOR signaling is involved in some of the cancer hallmarks described by Hanahan and Weinberg [58]. It is known that mTOR encompasses two functionally distinct complexes, mTOR complex 1 (mTORC1) and 2 (mTORC2). Recent studies suggested that aberrant mTORC1 pathway activation contributes to tumor growth, angiogenesis, and metastasis [59]. In this study, we found that the high expression of GPI was closely related to the activation of the mTORC1 pathway through GSEA analysis. GPI knockdown caused reduced phosphorylation of mTOR, P70S6K, and S6, increased expression of E-cadherin and decreased N-cadherin in A549 and H1299 cells. We speculated that the mTORC1 signaling pathway activation might be a critical process in the progression of GPI-overexpression tumors. Of course, this study has several limitations. Our research is mainly based on the public database and limited clinical tissue specimens of LUAD. Additional studies are needed using large-scale, prospective, and multicenter clinical trials to verify the robustness and reproducibility of this nine-gene signature.
Supplementary information
Cell Culture and Stable Transfection of shRNA
Table S1. The clinicopathologic characteristics of 30 matched clinical LUAD samples employed for Western blot assay of protein expression of candidate genes.
Table S2. The antibodies used in this study.
Table S3. Immune-related genes matrix based on TCGA_LUAD database.
Table S4. A total of 889 lncRNAs-related immune genes (lrRIGs) were identified.
Table S5. 329 differentially expressed lncRNAs-related immune genes were identified.
Table S6. 27 lncRNAs-related immune genes were significantly associated with the OS of LUAD patients through univariate Cox regression analysis.
Table S7. Nine genes were screened to construct a significant prognostic signature by multivariate Cox regression analysis.
Table S8. Gene Set Enrichment Analysis (GSEA) was performed by comparing the high expression and low expression of the GPI gene based on the TCGA_LUAD, GSE31210, GSE68465 and GSE13213 databases.
Acknowledgements
We sincerely acknowledge the contributions from the TCGA, GEO, innateDB, GDSC and GSEA databases. This work was supported by the National Natural Science Foundation of China (Nos. 81372147, 81803575, 31902287), Henan University support grant CX3070A0780502, the Key Science and Technology Research and Development and Promotion Special Project of Henan Province (No. 232102311205), and the Key R&D and promotion projects of Kaifeng (No. 2203008).
Author contributions
FL conceived and designed this article. ZY, JZ, TY, WT, and XZ participated in the experimental data collection; SJ provided technical assistance; FL drafted the manuscript; ZR revised the study draft. All authors contributed to the article and approved the submitted version.
Data availability
Relevant research data have been presented in the text. All data will be provided upon request if necessary.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Ethics Committee of Medical School of Henan University, China (HUSOM-2018-282). Informed consent was obtained from all subjects involved in the study.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhengyan Yang, Jianling Zhu.
Contributor Information
Zhiguang Ren, Email: Renzhiguang66@outlook.com.
Feng Lu, Email: lufeng@henu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02379-8.
References
- 1.Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103:463–73. doi: 10.1016/j.mcna.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.Epi-15-0578. [DOI] [PubMed] [Google Scholar]
- 3.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 4.Mazza F, Ferrari E, Maineri P, Dozin B, Ratto GB. Pleural lavage cytology predicts recurrence and survival, even in early non-small cell lung cancer. Surg Today. 2015;45:322–8. doi: 10.1007/s00595-014-0915-3. [DOI] [PubMed] [Google Scholar]
- 5.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–71. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–84. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lussier DM, O'Neill L, Nieves LM, McAfee MS, Holechek SA, Collins AW, et al. Enhanced T-cell immunity to osteosarcoma through antibody blockade of PD-1/PD-L1 interactions. J Immunother. 2015;38:96–106. doi: 10.1097/cji.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnjatic S, Bronte V, Brunet LR, Butler MO, Disis ML, Galon J, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer. 2017;5:44. doi: 10.1186/s40425-017-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Ma X, Ma Z, Ma Y, Wang J, Cao B. Research progress of biomarkers for immune checkpoint inhibitors on digestive system cancers. Front Immunol. 2022;13:810539. doi: 10.3389/fimmu.2022.810539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375–82. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–72. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci USA. 2011;108:11381–6. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Wang T, Li Y, Zhang Y, Yang R. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J Immunol. 2018;200:2603–14. doi: 10.4049/jimmunol.1701721. [DOI] [PubMed] [Google Scholar]
- 16.Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19:1112–25. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 17.Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18:115. doi: 10.1186/s12943-019-1032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non-coding RNAs regulation of macrophage polarization in cancer. Mol Cancer. 2021;20:24. doi: 10.1186/s12943-021-01313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pi YN, Qi WC, Xia BR, Lou G, Jin WL. Long non-coding RNAs in the tumor immune microenvironment: biological properties and therapeutic potential. Front Immunol. 2021;12:697083. doi: 10.3389/fimmu.2021.697083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun. 2020;11:1000. doi: 10.1038/s41467-020-14802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15:R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang HJ, Chang WJ, Jia CY, Qiao L, Zhou J, Chen Q, et al. Destrin contributes to lung adenocarcinoma progression by activating Wnt/β-catenin signaling pathway. Mol Cancer Res. 2020;18:1789–802. doi: 10.1158/1541-7786.Mcr-20-0187. [DOI] [PubMed] [Google Scholar]
- 23.Sun S, Guo W, Wang Z, Wang X, Zhang G, Zhang H, et al. Development and validation of an immune-related prognostic signature in lung adenocarcinoma. Cancer Med. 2020;9:5960–75. doi: 10.1002/cam4.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y, Lu X, Li Y, Fu J, Li H, Li X, et al. Identification of a six-gene metabolic signature predicting overall survival for patients with lung adenocarcinoma. PeerJ. 2020;8:e10320. doi: 10.7717/peerj.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Zhang Z, Yu Z. Identification of a novel glycolysis-related gene signature for predicting metastasis and survival in patients with lung adenocarcinoma. J Transl Med. 2019;17:423. doi: 10.1186/s12967-019-02173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Xuan Y, Gao B, Sun X, Miao S, Lu T, et al. Identification of an eight-gene prognostic signature for lung adenocarcinoma. Cancer Manag Res. 2018;10:3383–92. doi: 10.2147/cmar.S173941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruf B, Heinrich B, Greten TF. Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol. 2021;18:112–27. doi: 10.1038/s41423-020-00572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9:eaah3560. doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi E, Yamazaki K, Torigoe T, Cho Y, Miyamoto M, Oizumi S, et al. HLA class I antigen expression is associated with a favorable prognosis in early stage non-small cell lung cancer. Cancer SCI. 2007;98:1424–30. doi: 10.1111/j.1349-7006.2007.00558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–1. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–71. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J, Deng X, Sun R, Luo M, Liang M, Gu B, et al. GPI is a prognostic biomarker and correlates with immune infiltrates in lung adenocarcinoma. Front Oncol. 2021;11:752642. doi: 10.3389/fonc.2021.752642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Veen M, Matas-Rico E, van de Wetering K, Leyton-Puig D, Kedziora KM, De Lorenzi V, et al. Negative regulation of urokinase receptor activity by a GPI-specific phospholipase C in breast cancer cells. eLife 2017;6. 10.7554/eLife.23649. [DOI] [PMC free article] [PubMed]
- 37.Wu ST, Liu B, Ai ZZ, Hong ZC, You PT, Wu HZ, et al. Esculetin inhibits cancer cell glycolysis by binding tumor PGK2, GPD2, and GPI. Front Pharmacol. 2020;11:379. doi: 10.3389/fphar.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 2016;529:532–6. doi: 10.1038/nature16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol. 2003;3:371–7. doi: 10.1016/s1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 40.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Shen S, Wang G, Zhang R, Zhao Y, Yu H, Wei Y, et al. Development and validation of an immune gene-set based prognostic signature in ovarian cancer. EBioMedicine. 2019;40:318–26. doi: 10.1016/j.ebiom.2018.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P(3) to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–51. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sang LJ, Ju HQ, Liu GP, Tian T, Ma GL, Lu YX, et al. LncRNA CamK-A regulates Ca(2+)-signaling-mediated tumor microenvironment remodeling. Mol Cell. 2018;72:601. doi: 10.1016/j.molcel.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 46.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, et al. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–9. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Jia F, Bai P, Liang Y, Sun R, Yuan F, et al. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of gastric cancer. Tumour Biol. 2016;37:299–303. doi: 10.1007/s13277-015-3750-2. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Wang Y, Song G, Zhang X, Gao S, Liu H. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019;450:14–21. doi: 10.1016/j.canlet.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 49.Teng C, Huang G, Luo Y, Pan Y, Wang H, Liao X, et al. Differential long noncoding RNAs expression in cancer-associated fibroblasts of non-small-cell lung cancer. Pharmacogenomics. 2019;20:143–53. doi: 10.2217/pgs-2018-0102. [DOI] [PubMed] [Google Scholar]
- 50.Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20:835–51. doi: 10.1038/s41590-019-0400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol. 2019;234:23176–89. doi: 10.1002/jcp.28884. [DOI] [PubMed] [Google Scholar]
- 52.Mason DY, Cordell JL, Brown MH, Borst J, Jones M, Pulford K, et al. CD79a: a novel marker for B-cell neoplasms in routinely processed tissue samples. Blood. 1995;86:1453–9. doi: 10.1182/blood.V86.4.1453.bloodjournal8641453. [DOI] [PubMed] [Google Scholar]
- 53.Zomas AP, Matutes E, Morilla R, Owusu-Ankomah K, Seon BK, Catovsky D. Expression of the immunoglobulin-associated protein B29 in B cell disorders with the monoclonal antibody SN8 (CD79b) Leukemia. 1996;10:1966–70. [PubMed] [Google Scholar]
- 54.Viswanadhapalli S, Dileep KV, Zhang KYJ, Nair HB, Vadlamudi RK. Targeting LIF/LIFR signaling in cancer. Genes Dis. 2022;9:973–80. doi: 10.1016/j.gendis.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pusapati RV, Daemen A, Wilson C, Sandoval W, Gao M, Haley B, et al. mTORC1-dependent metabolic reprogramming underlies escape from glycolysis addiction in cancer cells. Cancer Cell. 2016;29:548–62. doi: 10.1016/j.ccell.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 56.Liotta LA, Mandler R, Murano G, Katz DA, Gordon RK, Chiang PK, et al. Tumor cell autocrine motility factor. Proc Natl Acad Sci USA. 1986;83:3302–6. doi: 10.1073/pnas.83.10.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song Y, Wang L, Zhang L, Huang D. The involvement of semaphorin 7A in tumorigenic and immunoinflammatory regulation. J Cell Physiol. 2021;236:6235–48. doi: 10.1002/jcp.30340. [DOI] [PubMed] [Google Scholar]
- 58.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–76. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell Culture and Stable Transfection of shRNA
Table S1. The clinicopathologic characteristics of 30 matched clinical LUAD samples employed for Western blot assay of protein expression of candidate genes.
Table S2. The antibodies used in this study.
Table S3. Immune-related genes matrix based on TCGA_LUAD database.
Table S4. A total of 889 lncRNAs-related immune genes (lrRIGs) were identified.
Table S5. 329 differentially expressed lncRNAs-related immune genes were identified.
Table S6. 27 lncRNAs-related immune genes were significantly associated with the OS of LUAD patients through univariate Cox regression analysis.
Table S7. Nine genes were screened to construct a significant prognostic signature by multivariate Cox regression analysis.
Table S8. Gene Set Enrichment Analysis (GSEA) was performed by comparing the high expression and low expression of the GPI gene based on the TCGA_LUAD, GSE31210, GSE68465 and GSE13213 databases.
Data Availability Statement
Relevant research data have been presented in the text. All data will be provided upon request if necessary.